Abstract
Early diagnosis and monitoring of SARS-CoV-2 virus is essential to control COVID-19 outbreak. In this study, we propose a promising surface enhanced Raman scattering (SERS)-based COVID-19 biosensor for ultrasensitive detection of SARS-CoV-2 virus in untreated saliva. The SERS-immune substrate was fabricated by a novel oil/water/oil (O/W/O) three-phase liquid-liquid interfaces self-assembly method, forming two layers of dense and uniform gold nanoparticle films to ensure the reproducibility and sensitivity of SERS immunoassay. The detection was performed by an immunoreaction between the SARS-CoV-2 spike antibody modified SERS-immune substrate, spike antigen protein and Raman reporter-labeled immuno-Ag nanoparticles. This SERS-based biosensor was able to detect the SARS-CoV-2 spike protein at concentrations of 0.77 fg mL−1 in phosphate-buffered saline and 6.07 fg mL−1 in untreated saliva. The designed SERS-based biosensor exhibited excellent specificity and sensitivity for SARS-CoV-2 virus without any sample pretreatment, providing a potential choice for the early diagnosis of COVID-19.
Keywords: COVID-19, SARS-CoV-2, SERS, Biosensor, Early diagnosis
Graphical abstract
1. Introduction
Rapid spread of the COVID-19 pandemic, which was caused by a novel coronavirus named severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2), has created alarming situation all over the world (Yüce et al., 2021; Ji et al., 2020). There is an urgent requirement to develop a sensitive, accurate, rapid and low-cost diagnostic tool to early screening infected individuals so that proper isolation and treatment can be facilitated.
Currently, COVID-19 diagnostic tests can be divided into two broad categories. The first category includes the molecular diagnostic test for the identification of SARS-CoV-2 viral RNA using reverse transcriptase real-time polymerase chain reaction (RT-PCR) and nucleic acid hybridization strategies (Pan et al., 2020; Li et al., 2020). Although RT-qPCR method has become the gold standard for SARS-CoV-2 virus detection, it is time-intensive, requires highly qualified personnel, and the risk of infection should be strictly controlled during the operation. The second category includes serological and immunological tests that primarily focus on the detection of antibodies (Roda et al., 2021; Funari et al., 2020; Liu et al., 2020). While these tests are rapid and require minimal equipment, it is not suitable for screening of early and asymptomatic cases, as most patients have antibody response at about 10 days after onset of symptoms (Jadhav et al., 2021; Cui and Zhou, 2020; Younes et al., 2020). Hence, highly sensitive immunological diagnostic methods that directly detect viral antigens are necessary for early and accurate diagnosis of COVID-19.
The whole SARS-CoV-2 virus and its structural proteins, including the spike (S) protein, small envelope (E) protein, and also several accessory proteins, can theoretically be used as antigens for COVID-19 diagnosis (Wu et al., 2020). Among them, S protein may be one of the most valuable antigen biomarkers for diagnosis of COVID-19 (Liu et al., 2021). Kim's group has prepared a lateral flow-based COVID-19 diagnostic technology to detect viral proteins (Kim et al., 2021). It can be a point-of-care disease diagnostic tool because it is portable, inexpensive, and without requiring power. Newly, several nanoscale integrated architectures based on optical and electronic systems have been introduced to detect SARS-CoV-2 spike protein with much higher sensitivity, such as terahertz plasmonic biosensor (Ahmadivand et al., 2021) and gated graphene-enhanced field-effect transistor (FET)-based biosensor (Seo et al., 2020). Generally, most of these devices have reasonably sensitive detection of SARS-CoV-2 virus and possess the advantages of low cost, simplicity, and are more easily miniaturized and mass fabricated. However, there is still a continuous demand for biosensors that are fast, ultrasensitive and accurate in detecting antigens.
Surface enhanced Raman scattering (SERS) is known as an ultrasensitive molecular spectroscopy technique that has no interference from water, making it a distinct advantage in the identification of bio-samples (Wang et al., 2017, Wang et al., 2013; Granger et al., 2016; Porter et al., 2008; Laing et al., 2016). Compared with lateral flow immunoassay, photothermal method and electrical biosensor (Khanmohammadi et al., 2020; Karimian et al., 2020), SERS-based immunoassay technique does not require sample pretreatment, has high sensitivity for detecting trace amounts of bioparticles, and even be capable of single-molecule detection in special cases, providing an efficient method for the SARS-CoV-2 virus detection (Jadhav et al., 2021).
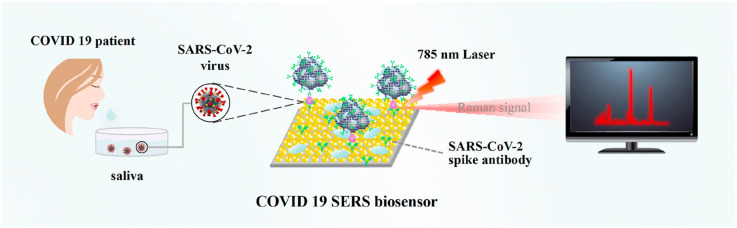
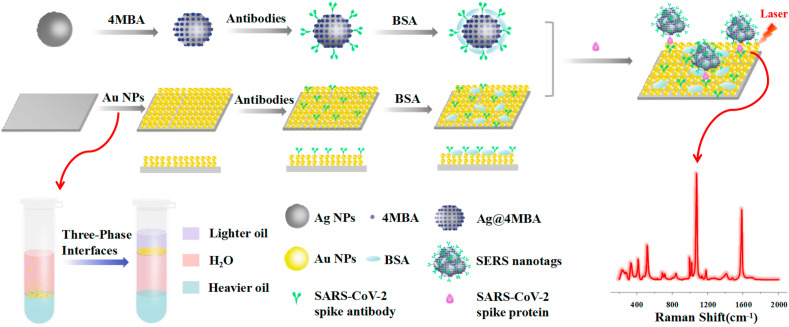
In this work, we developed an ultrasensitive and specific SERS-based biosensor for detecting SARS-CoV-2 virus in untreated saliva (COVID-19 SERS sensor). In such an immunoassay structure (Scheme 1 ), two layers of gold nanoparticles were immobilized onto the silicon wafer by a novel oil/water/oil three-phase liquid-liquid interfaces self-assembly process (Lin et al., 2020) to form a SERS-active substrate. Through this three-phase self-assembly approach, the nanoparticle monolayer possesses excellent uniformity and reproducibility, guaranteeing high sensitivity, stability and repeatability of SERS immunoassay. Afterwards, the specific SARS-CoV-2 spike antibodies were attached on the SERS-active substrate to capture the spike protein, and Raman reporter-labeled immuno-Ag nanoparticles were used as the SERS nanotags to recognize the species and concentration of the antigen. With target antigen presented in the sample, a sandwich immunoassay structure would be formed between the SERS-immune substrate, target antigen and SERS nanotags. Finally, the target antigen was read out qualitatively and quantitatively via the strong SERS signals of Raman reporter. This novel SERS-based SARS-CoV-2 diagnostic tool offers a new opportunity for direct and sensitive screening of symptomatic as well as asymptomatic individuals of COVID-19 at early stage.
Scheme 1.
Schematic illustration of the SERS-based immunoassay.
2. Material and methods
2.1. Materials and chemicals
Silver nitrate (AgNO3), sodium citrate, cetyltrimethylammonium bromide (CTAB), ascorbic acid (AA), dichloromethane, n-hexane, polyvinylpyrrolidone (PVP) and 4-mercaptobenzoic acid (4-MBA) were all bought from Aladdin Reagent Co., Ltd. Gold(III) chloride trihydrate (HAuCl4·3H2O), sodium borohydride, 11-Mercaptoundecanoic acid (MUA), bovine serum albumin (BSA), polyvinyl pyrrolidone (PVP) and cetyltrimethylammonium chloride (CTAC) were all obtained from Sigma Aldrich. Phosphate-buffered saline (PBS), 1 × TBST/Tween 20 buffer were prepared in-house. SARS-CoV-2 spike antigen protein, human immunodeficiency virus type 1 p24 antigen (HIV-1 p24), Epstein-Barr virus glycoprotein 350 (EBV GP350), carcinoembryonic antigen (CEA), alpha fetoprotein (AFP), SARS-CoV-2 spike antibody were purchased from Beijing Yiqiao Shenzhou Science and Technology Co., Ltd. The clinical saliva samples, clinical serum samples and whole blood were obtained from The First Hospital of Jilin University. The deionized water with the resistivity of 18.2 MΩ was used in all experiments.
2.2. Preparation of the SERS nanotags
The SERS nanotags were prepared by a process that consisted of the following three steps. First, the silver nanoparticles (Ag NPs) were fabricated according to Qin's method (Qin et al., 2010). Briefly, AgNO3 solution (2.4 mL, 0.1 M) was added into 240 mL aqueous solution including ascorbic acid (0.6 μM) and trisodium citrate (3 μM) under slight stirring. The reaction was executed at 30 °C for 15 min until the color of the solution stopped changing, then aged at 100 °C for 2 h. Second, the Ag@4MBA was obtained through adding 4MBA (200 μL, 0.1 mM) solution into 1 mL Ag NPs under stirring at 30 °C for 12 h, followed by a centrifugation at 8500 rpm for 15 min. Finally, the SERS nanotags were prepared by the following process: 30 μL of 10 μg mL−1 SARS-CoV-2 spike antibody was added into 1 mL of 4MBA-labeled Ag NPs and incubated at 4 °C for 6 h. Then the mixture was purified by centrifugation at 6500 rpm for 15 min and blocked by 50 μL of 5% BSA solution. After being centrifuged at 6500 rpm for 15 min, the precipitates were re-dispersed in 1 mL PBS solution and stored at 4 °C for future use.
2.3. Preparation of the SERS-immune substrate
The SERS-immune substrate was prepared according to the following three steps. First, the gold nanoparticles (Au NPs) were prepared using the typical seed-mediated growth method (Zheng et al., 2014). Briefly, HAuCl4 solution (6 mL, 0.5 mM) was added into a mixture of CTAC solution (6 mL, 0.1 M), AA solution (390 μL, 0.01 M) and 30 μL 10 nm seeds, then the reaction was executed at 27 °C for 10 min and centrifugation at 8500 rpm for 10 min. Second, the prepared Au NPs were self-assembly on the hydrophilically treated silicon wafer by oil/water/oil three-phase liquid-liquid interfaces self-assembly method (Lin et al., 2020). CTAC-coated Au NPs were converted to PVP-coated Au NPs through two rounds of the centrifugation and finally transferred into ethanol. Then, 1 mL dichloromethane (heavier oil) and 1.8 mL water were added into 120 μL as-prepared Au NPs in a 5 mL centrifuge tube, respectively. The mixture was vigorously shaken for 30 s, then 400 μL n-hexane (lighter oil) was slowly added along the centrifuge tube wall. After the container had slightly tilted and rotated, a monolayer film with a metallic screen was gradually formed onto the n-hexane/water interface, then the n-hexane was slowly removed using pipette and the hydrophilic silicon wafer was slowly immersed in the intermediate water layer along the centrifuge tube wall to take the monolayer nanoparticles out. Finally, the SERS-immune substrate was prepared by the following process: SERS-active substrate was immersed into 20 μM MUA solution for 2 h, then 15 μL of 10 μg mL−1 SARS-CoV-2 spike antibody was dropped onto the SERS-active substrate and incubated at 4 °C for 6 h. The nonspecific bindings were prevented by pipetting 5% BSA onto the SERS-immune substrate and incubated at 4 °C for 2 h. After being rinsed with TBST and deionized water successively, the SERS-immune substrate was stored at 4 °C for further use.
2.4. Sandwich immunoassay
The SARS-CoV-2 spike protein standard solution was prepared by dissolving 100 μg SARS-CoV-2 spike protein powder into 400 μL sterile water. PBS solutions of SARS-CoV-2 spike protein with gradient concentrations were prepared by diluting SARS-CoV-2 spike protein standard solution with different proportions. Saliva samples were prepared by adding 20 μL of different concentrations of SARS-CoV-2 protein solutions to 180 μL of saliva. In this way, saliva samples of graded concentrations were obtained. Serum and blood samples were prepared in the same way as saliva samples.
The sandwich immunoassay was carried out according to the typical ELISA method. Typically, 10 μL target antigen with different concentrations was pipetted onto the SERS-immune substrate and incubated at 37 °C for 2 h. After the immune recognition, the SERS-immune substrate was rinsed with TBST and deionized water successively. Subsequently, the recognition sites were covered with the as-prepared SERS nanotags and incubated at 4 °C for 2 h. After that, the SERS-immune substrate was rinsed by TBST and deionized water and then stocked at 4 °C.
2.5. Instruments
Transmission electron microscopy (TEM) images and energy dispersive X-ray spectroscopy (EDS) were obtained by transmission electron microscope (JEOL, JEM-2100). Scanning electron microscopy (SEM) images were characterized by field emission scanning electron microscopy (JEOL, JSM-7500). UV–vis spectra were performed employing spectrophotometer (UV-3600, Shimadzu, Japan). The Raman measurements were recorded by a Raman spectrometer (HORIBA, France) using a 785 nm excitation laser, the acquisition time was 5s and the accumulation was 2 times, the spectra were only scanned once and baseline correction was performed to obtain the final spectra with the background subtracted. Each sample was measured five times and each spectrum was the average of five measurements.
3. Results and discussion
3.1. Preparation and characterization of the SERS nanotags
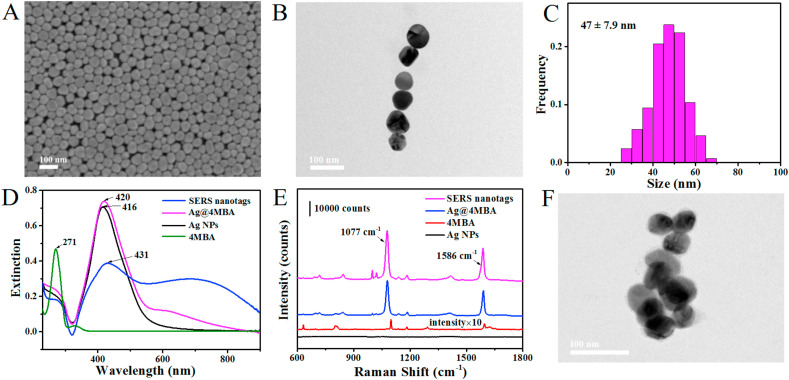
The silver nanoparticles (Ag NPs) were synthesized through the chemical reduction method (Qin et al., 2010). SEM image and TEM image of the as-prepared Ag NPs suggested that the nanoparticles exhibited good monodispersity (Fig. 1 A and B), which could be employed to fabricate sensitive SERS nanotags. The size histogram distribution revealed that the average size of the Ag NPs is about 47.0 ± 7.9 nm (Fig. 1C). Studies show that the localized Surface Plasmon Resonance (LSPR) band of nanoparticles is related to numerous parameters such as diameter distribution, morphology, and dielectric property of surrounding medium (Song et al., 2016; Reyes et al., 2017). Therefore, the sequential surface modification of Ag NPs was monitored by UV–vis spectrum (Fig. 1D). The 4-MBA molecules had an absorption maximum at 271 nm which is consistent with the previous literature (Liang et al., 2012). The as-prepared Ag NPs had a strong extinction maximum at 416 nm and then slightly red-shifted to 420 nm after labeling with Raman reporter via the Ag–S bond (Song et al., 2016), which was caused by the variation in the refractive index around the nanoparticles (Li et al., 2019). After conjugating with anti-SARS-CoV-2 antibodies, the LSPR band significantly red-shifted from 420 nm to 431 nm, indicating that the SARS-CoV-2 spike antibodies were successfully anchored on the Ag NPs@4MBA (Yang et al., 2019). Besides, a new extinction peak appeared at around 670 nm, which was caused by the plasmon resonance of Ag NPs aggregates (Zheng et al., 2018; Shu et al., 2014). The successful binding of antibodies and Raman reporter on Ag NPs was also evidenced by zeta-potential (Supplementary Fig. S1). The surface charge of Ag NPs was changed from −4.21 mV to −9.76 mV after conjugating with Raman reporter, and then decreased to −14.90 mV after incubating with SARS-CoV-2 spike antibodies. In addition, SERS detection was further carried out to characterize the manufacturing process of SERS nanotags (Fig. 1E). The red line was the normal Raman spectrum of the 4-MBA molecules and the intensity was magnified 10 times. SERS spectrum would induce a slight shift in the Raman peak position which is consistent with the literature (Lin et al., 2020). Compared with bare Ag NPs, two dominated Raman peaks were observed in the spectra of Ag@4MBA and SERS nanotags at 1077 cm−1 and 1586 cm−1, which were considered as the characteristic peaks of 4MBA and attributed to the in-plane ring breathing mode coupled with ν(C–S) and aromatic ring ν(C–C) vibrations mode, respectively (Zhou et al., 2018). Notably, the Raman signal of SERS nanotags was stronger than that of Ag@4MBA, owing to the slight aggregation of Ag NPs after antibodies incubation. The same conclusion was also obtained in the TEM image of SERS nanotags (Fig. 1F). Compare with the Ag NPs, the as-prepared SERS nanotags present slight aggregation which could further serve more hot spots to strengthen SERS response.
Fig. 1.
Preparation and characterization of the SERS nanotags. (A) SEM image of the Ag NPs. (B) TEM image of the Ag NPs. (C) Dimensional distribution map of the Ag NPs. (D) UV–vis extinction spectra of 4-MBA, Ag NPs, Ag@4MBA and SERS nanotags. (E) Normal Raman spectrum of 4-MBA and SERS spectra of Ag NPs, Ag@4MBA and SERS nanotags. (F) TEM image of the SERS nanotags.
3.2. Preparation and optimization of the SERS-immune substrate
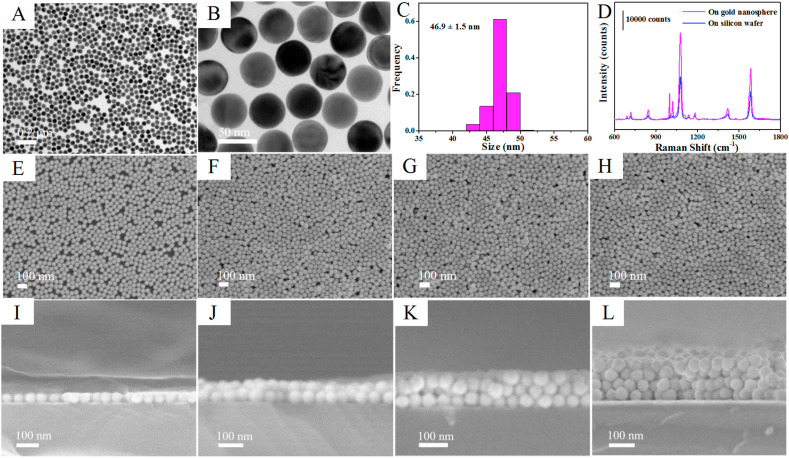
The gold nanoparticles (Au NPs) were synthesized through the seed growth method (Zheng et al., 2014). TEM images with low and high magnification indicate that the Au NPs possess typical spherical profile and homogeneity dimensional (Fig. 2 A and B). The average diameter of Au NPs is about 46.9 ± 1.5 nm (Fig. 2C). The role of gold nanospheres for the SERS response was investigated by dropping SERS nanotags directly onto the gold nanospheres and bare silicon wafer (Fig. 2D). A significant enhancement was obtained when dropping SERS nanotags on gold nanospheres compared with that on silicon wafer. This was owing to the electromagnetic field coupling between the gold and silver nanoparticles could further strengthen the SERS response, affording the ability to ultrasensitive detection of low-concentration target antigen (Jiang et al., 2018).
Fig. 2.
Preparation and characterization of the SERS-active substrate. (A, B) TEM images of the Au NPs with low and high magnification. (C) Dimensional distribution map of the Au NPs. (D) SERS spectra of the SERS nanotags on gold nanosphere and on silicon wafer. (E–H) SEM images of SERS-active substrate assembled by 1–4 layers Au NPs. (I–L) Cross-sectional SEM images of SERS-active substrate assembled by 1–4 layers Au NPs. (For interpretation of the references to color in this figure legend, the reader is referred to the Web version of this article.)
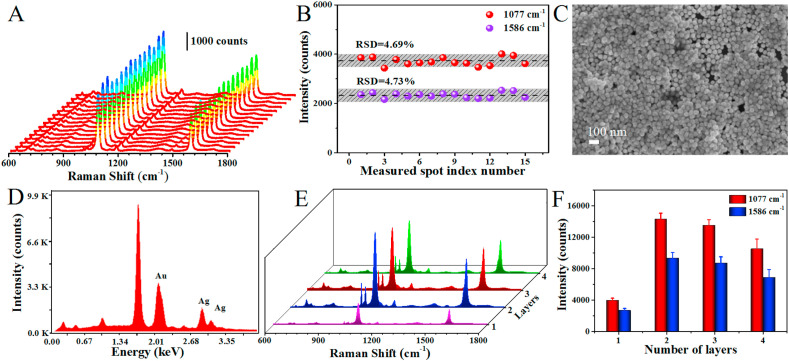
The reproducibility of SERS-active substrate is crucial for final SERS immunoassay. However, the SERS-active substrate prepared by conventional electrostatic interaction always requires a time-consuming process and has poor uniformity due to the random adsorption between nanoparticles and substrate (Pei et al., 2013), leading to inaccurate detection results. In order to enhance the reproducibility of detection, a novel oil/water/oil (O/W/O) three-phase liquid-liquid interfaces self-assembly method (Lin et al., 2020), was applied to construct the SERS-active substrate. As shown in Scheme 1, the system was initially composed of heavier oil phase and aqueous phase, and the Au NPs monolayer distributed at the oil-water (O/W) interface was sparse and inhomogeneous. After adding lighter oil phase, the system comprised an “upper O/W interface” and a “lower O/W interface” around the intermediate aqueous layer. When the interfacial tension of the upper O/W interface is greater than that of the lower O/W interface, the Au NPs could be transferred from the lower O/W interface to the upper O/W interface and compressed into a dense and uniform monolayer driven by Marangoni effect (Shim et al., 2014; Wang et al., 2019). Subsequently, this nanoparticle monolayer could be transferred onto hydrophilic silicon wafer after removing the lighter oil phase, and the SERS-active substrate with multilayers Au NPs could be successfully fabricated by repeating this step constantly. The SEM images of the substrates deposited with 1–4 layers of Au NPs were given in Fig. 2E–H, which clearly indicate that the Au NPs films were densely and uniformly packed without obvious aggregates and large void spaces, and the coverage rate was nearly 100%. The cross-sectional SEM images (Fig. 2I-L) clearly show that the number of assembled layers of Au NPs and almost no clusters and aggregates. The thicknesses of 1–4 layers of Au NPs were approximately 46 nm, 80 nm, 100 nm and 140 nm, respectively (Supplementary Fig. S2). Then, the reproducibility of the SERS-active substrate before the SERS nanotags adsorbed was investigated (Fig. 3 A and B). The SERS-active substrate was immersed into 0.1 mM 4MBA ethanol solution for 3 h and the SERS spectra were collected from picking 15 spots randomly on the substrate. As shown in Fig. 3B, there were almost no significant differences in SERS intensities of spot-to-spot distributions at 1077 cm−1 and 1586 cm−1, which clearly indicates that the SERS-active substrate possess excellent uniformity and reproducibility over a large area. The relative standard deviation (RSD) was as low as 4.69% at 1077 cm−1 and 4.73% at 1586 cm−1, respectively. The Raman mapping from 400 measurement points over a 1 mm1mm area was also performed to investigate the reproducibility of the SERS-active substrate (Supplementary Fig. S3). The step size was 50 μm and the RSD of the spot-to-spot intensity at 1077 cm−1 was calculated to be 7.58%, which indicates that the SERS-active substrate has good uniformity. The uniform nanoparticle monolayers were able to significantly improve the reproducibility of Raman signals and overcome the limitations of conventional electrostatic interaction methods (Lin et al., 2019).
Fig. 3.
(A, B) SERS spectra and intensities of 4-MBA from randomly fifteen measured sites on SERS-active substrate. (C) SEM image of the as-prepared sandwich immunoassay structure. (D) EDS spectrum of the as-prepared sandwich immunoassay structure. (E, F) SERS spectra of the sandwich immunoassay structure with 1–4 layers of Au NPs and the corresponding intensities at 1077 cm−1 and 1586 cm−1.
The SERS activity of the substrates decorated with nanoparticles is closely related to the number of assembled layers. Therefore, the SERS enhancement effect of the immune substrate assembled by 1–4 layers of Au NPs was investigated. After the SARS-CoV-2 spike antibodies were immobilized on the SERS-active substrate, the SERS immunoassay was carried out by sequentially incubating the antigen protein and SERS nanotags on the SERS-immune substrate to form a sandwich immunoassay structure through the antigen-antibody binding. As can be seen in Fig. 3C, a large amount of SERS nanotags were anchored on the SERS-immune substrate after immunoreaction, demonstrating the successful construction of sandwich immunoassay structure. This result could be further confirmed by EDS mapping (Jiang et al., 2018). Strong silver peaks were observed in EDS spectrum except gold peak after the immunoreaction (Fig. 3D). The SERS spectra of the sandwich immunoassay structure with 1–4 layers of Au NPs were shown in Fig. 3E and F. The SERS intensity of the Raman reporter increased with increasing the thickness of the Au NPs films until two layers and then decreased. This phenomenon was due to the formation of the interlayer plasmonic coupling and the limited penetration depth of light (Gao et al., 2019). As the layers of Au NPs increases, interlayer plasmonic coupling would generate between the nanoparticle layers, which could induce strong localized electromagnetic field and achieve more intense SERS response. On the other hand, with increase of the thickness, the substrates would suffer more light scattering loss and the Au NPs were more likely to aggregate, thus reducing the chance of hot spots occurring and weakening the SERS response of the substrates. Therefore, two layers of Au NPs was the optimal condition for obtaining the strongest SERS signal, and this SERS-immune substrate with two layers of Au NPs was further employed for SARS-CoV-2 virus detection.
3.3. Measurement and analysis of clinical saliva samples
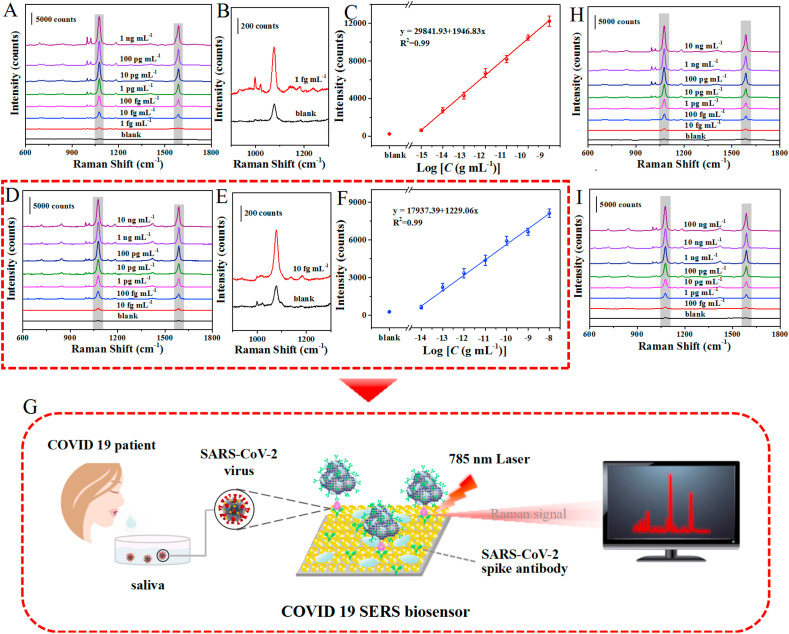
As a proof-of-concept study, the potential application of the as-proposed SERS-based immunoassay platform for quantitative analysis of SARS-CoV-2 spike protein was investigated by incubating the target antigen with different concentrations on the SERS-immune substrate. The peak intensity at 1077 cm−1 was used to monitor indirectly the corresponding concentration of spike protein. Control experiment was also performed without adding any antigens for SERS immunoassay. As expected, the SERS intensity of the sandwich immunoassay structure continuously increased with the increasing of the target antigen concentration within the range from 1 fg mL−1 to 1 ng mL−1 (Fig. 4 A–C). Meanwhile, a good linearity relationship was established between the SERS intensity at 1077 cm−1 and the logarithm of target antigen concentration. The linear calibration curve was fitted as y = 29841.93 + 1946.83x (R2 = 0.99). Error bars in the plot represent the standard deviations from five measurements of different spots for each concentration. The limit of detection (LOD) of SARS-CoV-2 spike protein was 0.77 fg mL−1 in phosphate-buffered saline. The calculation method refers to the definition of LOD by the International Union of Pure and Applied Chemistry (IUPAC) (Long and Winefordner, 1983), as detailed in the Supporting Information.
Fig. 4.
SERS immunoassay of SARS-CoV-2 spike protein. (A) SERS immunoassay for SARS-CoV-2 spike protein at different concentrations ranging from 1 fg mL−1 to 1 ng mL−1 in PBS. (B) Raman spectrum of sandwich immunoassay structure with SARS-CoV-2 spike protein at 1 fg mL−1 in PBS and the blank signal. (C) Corresponding calibration curve of the peak intensity at 1077 cm−1. (D) Average SERS spectra of the sandwich immunoassay structure with SARS-CoV-2 spike protein at different concentrations from 10 fg mL−1 to 10 ng mL−1 in saliva. (E) Raman spectrum of sandwich immunoassay structure with SARS-CoV-2 spike protein at 10 fg mL−1 in saliva and the blank signal. (F) The calibration plots of the peak intensity at 1077 cm−1. Each error bar indicates the standard deviation of five different measurements. (G) Schematic illustration of the SERS-based immunoassay in untreated saliva. (H) SERS immunoassay for SARS-CoV-2 spike protein at different concentrations from 10 fg mL−1 to 10 ng mL−1 in serum. (I) Raman spectra of sandwich immunoassay structure at different SARS-CoV-2 spike protein concentrations from 100 fg mL−1 to 100 ng mL−1 in blood.
In order to evaluate the practical application prospect of the SERS-based immunoassay platform for early diagnosis of COVID-19, we further carried out the SERS immunoassay of the target antigen with varying concentrations in unprocessed saliva to simulate the real immunodetection environment (Fig. 4D–F), and the schematic illustration of SERS-based immunoassay in untreated saliva was shown in Fig. 4G. Similarly, the intensity of Raman peak at 1077 cm−1 was positively correlated to the logarithm of the SARS-CoV-2 spike protein concentration at the range of 10 fg mL−1 to 10 ng mL−1. The linear equation was y = 17937.39 + 1229.06x (R2 = 0.99) and the LOD could reach to 6.07 fg mL−1. In addition, for demonstrating that our SERS-based biosensor was almost undisturbed by complex biological environment, the SERS immunoassay for the SARS-CoV-2 spike protein was also performed in serum and blood (Fig. 4H and I), and the LOD was calculated to be 7.60 fg mL−1 and 0.10 pg mL−1, respectively. The calibration curve of the peak intensity at 1077 cm−1 in serum and blood are presented in Figs. S4 and S5 (Supporting Information). These results indicate that the proposed SERS-based biosensor has good potential for practical immunoassay both in untreated saliva and blood.
3.4. Specificity and reproducibility of the SERS immunoassay
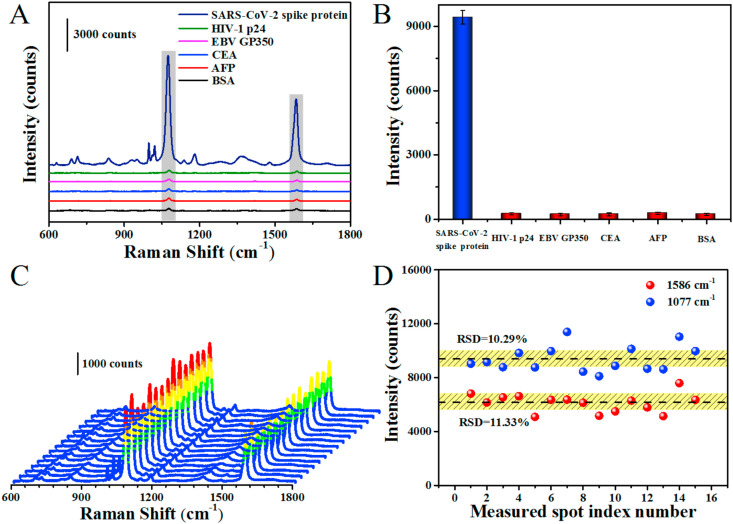
We further investigate the specificity of the SERS-based immunoassay platform. The target protein and other interfering proteins, including HIV-1 p24, EBV GP350, CEA, AFP and BSA were incubated on the SERS-immune substrate, respectively. The Raman intensity at 1077 cm−1 was chosen as a reference to evaluate the SERS performance of SERS-immune substrate incubated with different antigens. As shown in Fig. 5 A and B, remarkable SERS response was only generated from the immunoassay of target protein at 0.1 ng mL−1. Whereas, the neglected SERS signals were observed from other interfering proteins even at the concentration of 1 μg mL−1, owing to the failure of sandwich immunoassay structure construction. These results clearly indicate that the high specificity and selectivity of the proposed SERS-based biosensor for target antigen. Additionally, the reproducibility of the SERS-based biosensor was also explored by randomly recording 15 Raman spectra from the sandwich immunoassay structure (Fig. 5C and D). The RSD was calculated to be 10.29% at 1077 cm−1 and 11.33% at 1586 cm−1, respectively. It should be noted that the reproducibility of the SERS immunoassay platform was a little less than that of the SERS-active substrate mentioned above with the RSD of 4.69% at 1077 cm−1 and 4.73% at 1586 cm−1. This phenomenon can be explained by the nonuniform distribution or aggregation of SERS nanotags after multiple steps, including reagents adding, continuous purification and separation processes, etc., (Jiang et al., 2014; Lee et al., 2011), but this had little significant impact on the final SERS immunoassay. Moreover, we also collected the Raman spectra of the sandwich immunoassay structure for 15 consecutive days to investigate its stability (Supplementary Fig. S6), the intensity of Raman peak at 1077 cm−1 hardly changed significantly, indicating the good stability of the proposed SERS-based biosensor. Therefore, we believe that the as-prepared SERS-based immunoassay platform possess good specificity, reproducibility and stability for practical application.
Fig. 5.
Specificity and reproducibility of the SERS immunoassay. (A, B) The SERS spectra from the sandwich specificity immunoassay of SARS-CoV-2 spike protein, HIV-1 p24, EBV GP350, CEA, AFP and BSA and corresponding peak intensities at 1077 cm−1. The concentration of target antigen is 0.1 ng mL−1 while the other proteins are 1 μg mL−1. (C) Reproducibility of SERS spectra of sandwich immunoassay structure. (D) Relative standard deviation (RSD) of the peak intensity at 1077 cm−1 and 1586 cm−1, respectively. The spectra were collected from different points of sandwich immunoassay structure.
To date, many promising technologies and techniques have been developed for the diagnosis of SARS-CoV-2 virus. Our proposed SERS-based biosensor exhibited an ultrasensitive detection of SARS-CoV-2 spike protein with LOD of 6.07 fg mL−1 in untreated saliva, which is a very satisfactory result, comparing with the detection results of the other recently reported biosensors (Supplementary Table S1). Although the SERS-based biosensor seems to have good performance, but problems related to non-specific response outcoming from the biorecognition element were still the main limitation that need to be solved, and the SERS nanotags and SERS-immune substrate should be robust to ensure that the retrieved signal is accurate. Furthermore, the SERS-based biosensor could not be reused after the SERS immunoassay because the antigen and SERS nanotags could not be removed from SERS-immune substrate alone after Raman detection.
4. Conclusions
In conclusion, ultrasensitive and specific laboratory diagnostic methods are critical for controlling the rapidly evolving SARS-CoV-2 associated COVID-19 pandemic. We reported a SERS-based COVID-19 biosensor for early and ultrasensitive detection of SARS-CoV-2 virus. The SERS-immune substrate was fabricated by a novel oil/water/oil (O/W/O) three-phase liquid-liquid interfaces self-assembly method, by which two layers of dense and uniform gold nanoparticle films were immobilized onto the silicon wafer, guaranteeing the excellent reproducibility and sensitivity of the SERS immunoassay. Meanwhile, SARS-CoV-2 spike antibodies were conjugated to the SERS-active substrate to ensure the simultaneous capture and detection of spike protein, and Raman reporter-labeled immuno-Ag nanoparticles were employed as the SERS nanotags to qualitatively and quantitatively read out the SARS-CoV-2 virus. Based on the peculiar design, the SERS immunoassay with high sensitivity, specificity, as well as reproducibility was performed successfully. The SARS-CoV-2 spike protein could be detected at ultra-low concentrations of 0.77 fg mL−1 in phosphate-buffered saline and 6.07 fg mL−1 in untreated saliva. Moreover, the SERS-based biosensor successfully detected SARS-CoV-2 spike protein in serum and blood with the LOD of 7.60 fg mL−1 and 0.10 pg mL−1, respectively, demonstrating the anti-interference capability of the designed SERS immunoassay platform. It is expected that both in untreated saliva and blood, our proposed SERS-based biosensor might hold promising potential for sensitive screening of symptomatic and asymptomatic individuals of COVID-19 during early infection.
CRediT authorship contribution statement
Meiling Zhang: Conceptualization, Methodology, Writing – original draft. Xiaodan Li: Methodology. Jialin Pan: Methodology, Writing – original draft. Youlin Zhang: Methodology. Ling Zhang: Methodology, Writing – original draft. Chenguang Wang: Methodology, Writing – original draft. Xu Yan: Methodology, Writing – original draft. Xiaomin Liu: Conceptualization, Methodology, Writing – original draft, Funding acquisition, Supervision. Geyu Lu: Funding acquisition, Supervision.
Declaration of competing interest
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Acknowledgements
This work is supported by the National Nature Science Foundation of China (Nos. 61875191, U2003127, 61722305, 61833006, 11874355 and 21902057) and Foundation of Jilin Province Department of Finance (2018SCZWSZX-033).
Footnotes
Supplementary data to this article can be found online at https://doi.org/10.1016/j.bios.2021.113421.
Appendix A. Supplementary data
The following is the Supplementary data to this article:
References
- Ahmadivand A., Gerislioglu B., Ramezani Z., Kaushik A., Manickam P., Ghoreishi S.A. Biosens. Bioelectron. 2021;117:112971. doi: 10.1016/j.bios.2021.112971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cui F., Zhou H.S. Biosens. Bioelectron. 2020;165:112349. doi: 10.1016/j.bios.2020.112349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Funari R., Chu K.Y., Shen A.Q. Biosens. Bioelectron. 2020;169:112578. doi: 10.1016/j.bios.2020.112578. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gao M., Lin X., Li Z., Wang X., Qiao Y., Zhao H., Zhang J., Wang L. Nanotechnology. 2019;30(34):345604. doi: 10.1088/1361-6528/ab1ff2. [DOI] [PubMed] [Google Scholar]
- Granger J.H., Schlotter N.E., Crawford A.C., Porter M.D. Chem. Soc. Rev. 2016;45(14):3865–3882. doi: 10.1039/c5cs00828j. [DOI] [PubMed] [Google Scholar]
- Jadhav S.A., Biji P., Panthalingal M.K., Krishna C.M., Rajkumar S., Joshi D.S., Sundaram N. Med. Hypotheses. 2021;146:110356. doi: 10.1016/j.mehy.2020.110356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ji T., Liu Z., Wang G., Guo X., khan S.A., Lai C., Chen H., Huang S., Xia S., Chen B., Jia H., Chen Y., Zhou Q. Biosens. Bioelectron. 2020;166:112455. doi: 10.1016/j.bios.2020.112455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiang T., Wang X., Zhou J., Jin H. Sensor. Actuator. B Chem. 2018;258:105–114. [Google Scholar]
- Jiang T., Zhang L., Zhou J. Analyst. 2014;139:5893–5900. doi: 10.1039/c4an01293c. [DOI] [PubMed] [Google Scholar]
- Karimian N., Hashemi P., Khanmohammadi A., Afkhami A., Bagheri H. Anal. Bioanal. Chem. Res. 2020;7(3):281–301. [Google Scholar]
- Khanmohammadi A., Ghazizadeh A.J., Hashemi P., Afkhami A., Arduini F., Bagheri H. J. Iran. Chem. Soc. 2020;17:2429–2447. [Google Scholar]
- Kim H.Y., Lee J.H., Kim M.J., Park S.C., Choi M., Lee W., Ku K.B., Kim B.T., Park E.C., Kim H.G., Kim S.I. Biosens. Bioelectron. 2021;175:112868. doi: 10.1016/j.bios.2020.112868. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laing S., Gracie K., Faulds K. Chem. Soc. Rev. 2016;45:1901–1918. doi: 10.1039/c5cs00644a. [DOI] [PubMed] [Google Scholar]
- Lee M., Lee S., Lee J., Lim H., Seong G.H., Lee E.K., Chang S.I., Oh C.H., Choo J. Biosens. Bioelectron. 2011;26:2135–2141. doi: 10.1016/j.bios.2010.09.021. [DOI] [PubMed] [Google Scholar]
- Liang Y., Thorne J.E., Parkinson B.A. Langmuir. 2012;28(30):11072–11077. doi: 10.1021/la301237p. [DOI] [PubMed] [Google Scholar]
- Li D., Jiang L., Piper J.A., Maksymov I.S., Greentree A.D., Wang E., Wang Y. ACS Sens. 2019;4:2507–2514. doi: 10.1021/acssensors.9b01211. [DOI] [PubMed] [Google Scholar]
- Li Y., Yao L., Li J., Chen L., Song Y., Cai Z., Yang C. J. Med. Virol. 2020;92(7):903–908. doi: 10.1002/jmv.25786. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin S., Li X., Fang G., Zhao H., Wang L., Dong B. ACS Appl. Mater. Interfaces. 2020;12:56350–56360. doi: 10.1021/acsami.0c17471. [DOI] [PubMed] [Google Scholar]
- Lin S., Lin X., Shang Y., Han S., Hasi W., Wang L. J. Phys. Chem. C. 2019;123(40):24714–24722. [Google Scholar]
- Lin X., Fang G., Liu Y., He Y., Wang L., Dong B. J. Phys. Chem. Lett. 2020;11:3573–3581. doi: 10.1021/acs.jpclett.0c01116. [DOI] [PubMed] [Google Scholar]
- Liu D., Ju C., Han C., Shi R., Chen X., Duan D., Yan J., Yan X. Biosens. Bioelectron. 2021;173:112817. doi: 10.1016/j.bios.2020.112817. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu H., Dai E., Xiao R., Zhou Z., Zhang M., Bai Z., Shao Y., Qi K., Tu J., Wang C., Wang S. Sensor. Actuator. B Chem. 2020;329:129196. doi: 10.1016/j.snb.2020.129196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Long G.L., Winefordner J.D. Anal. Chem. 1983;55(7):712A–724A. [Google Scholar]
- Pan Y., Zhang D., Yang P., Poon L.L., Wang Q. Lancet Infect. Dis. 2020;20(4):411–412. doi: 10.1016/S1473-3099(20)30113-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pei Y., Wang Z., Zong S., Cui Y. J. Mat. Chem. B. 2013;1(32):3992–3998. doi: 10.1039/c3tb00519d. [DOI] [PubMed] [Google Scholar]
- Porter M.D., Lipert R.J., Siperko L.M., Wang G., Narayanan R. Chem. Soc. Rev. 2008;37(5):1001–1011. doi: 10.1039/b708461g. [DOI] [PubMed] [Google Scholar]
- Qin Y., Ji X., Jing J., Liu H., Wu H., Yang W. Colloid. Surface. Physicochem. Eng. Aspect. 2010;372(1–3):172–176. [Google Scholar]
- Reyes M., Piotrowski M., Ang S.K., Chan J., He S., Chu J.J.H., Kah J.C.Y. Anal. Chem. 2017;89:5373–5381. doi: 10.1021/acs.analchem.7b00066. [DOI] [PubMed] [Google Scholar]
- Roda A., Cavalera S., Di Nardo F., Calabria D., Rosati S., Simoni P., Colitti B., Baggiani C., Roda M., Anfossi L. Biosens. Bioelectron. 2021;172:112765. doi: 10.1016/j.bios.2020.112765. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seo G., Lee G., Kim M.J., Baek S.H., Choi M., Ku K.B., Lee C.S., Jun S., Park D., Kim H.G., Kim S.J., Lee J.O., Kim B.T., Park E.C., Kim S.I. ACS Nano. 2020;14(4):5135–5142. doi: 10.1021/acsnano.0c02823. [DOI] [PubMed] [Google Scholar]
- Shim J., Yun J.M., Yun T., Kim P., Lee K.E., Lee W.J., Ryoo R., Pine D.J., Yi G.R., Kim S.O. Nano Lett. 2014;14(3):1388–1393. doi: 10.1021/nl404446f. [DOI] [PubMed] [Google Scholar]
- Shu L., Zhou J., Yuan X., Petti L., Chen J., Jia Z., Mormile P. Talanta. 2014;123:161–168. doi: 10.1016/j.talanta.2014.02.015. [DOI] [PubMed] [Google Scholar]
- Song C., Yang Y., Yang B., Min L., Wang L. J. Mat. Chem. B. 2016;4:1811–1817. doi: 10.1039/c5tb02780b. [DOI] [PubMed] [Google Scholar]
- Wang J., Teng C., Jiang Y., Zhu Y., Jiang L. Adv. Mater. 2019;31(10):1806742. doi: 10.1002/adma.201806742. [DOI] [PubMed] [Google Scholar]
- Wang Y., Yan B., Chen L. Chem. Rev. 2013;113(3):1391–1428. doi: 10.1021/cr300120g. [DOI] [PubMed] [Google Scholar]
- Wang Z., Zong S., Wu L., Zhu D., Cui Y. Chem. Rev. 2017;117(12):7910–7963. doi: 10.1021/acs.chemrev.7b00027. [DOI] [PubMed] [Google Scholar]
- Wu A., Peng Y., Huang B., Ding X., Wang X., Niu P., Meng J., Zhu Z., Zhang Z., Wang J., Sheng J., Quan L., Xia Z., Tan W., Cheng G., Jiang T. Cell Host Microbe. 2020;27(3):325–328. doi: 10.1016/j.chom.2020.02.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang Y., Zhu J., Zhao J., Weng G.J., Li J.J., Zhao J.W. ACS Appl. Mater. Interfaces. 2019;11(3):3617–3626. doi: 10.1021/acsami.8b21238. [DOI] [PubMed] [Google Scholar]
- Younes N., Al-Sadeq D.W., Al-Jighefee H., Younes S., Al-Jamal O., Daas H.I., Yassine H.M., Nasrallah G.K. Viruses. 2020;12(6):582. doi: 10.3390/v12060582. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yüce M., Filiztekin E., Ozkaya K.G. Biosens. Bioelectron. 2021;172:112752. doi: 10.1016/j.bios.2020.112752. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zheng Y., Zhong X., Li Z., Xia Y. Part. Part. Syst. Char. 2014;31(2):266–273. [Google Scholar]
- Zheng Z., Wu L., Li L., Zong S., Wang Z., Cui Y. Talanta. 2018;188:507–515. doi: 10.1016/j.talanta.2018.06.013. [DOI] [PubMed] [Google Scholar]
- Zhou L., Liu Y., Wang F., Jia Z., Zhou J., Jiang T., Petti L., Chen Y., Xiong Q., Wang X. Talanta. 2018;188:238–244. doi: 10.1016/j.talanta.2018.05.070. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.