Abstract
Background
Children infected with SARS-CoV-2 are often asymptomatic or have only mild symptoms, leading to underestimation of disease prevalence in symptom-based testing strategies.
Objectives
This study sought to determine pediatric SARS-CoV-2 disease burden during local mitigation efforts by using antibody testing to compare seroprevalence estimates to cumulative PCR prevalence estimates.
Study design
In this cross-sectional study, we collected 1142 strict phase and 1196 relaxed phase remnant blood specimens from patients less than 19-years-old in southwestern Pennsylvania (SWPA). Patients were excluded if their residential zip code was outside the region of interest, if they were under 6-months-old, or they had recently received antibody-modifying treatments. Demographic, encounter, and laboratory electronic medical record information was extracted. Samples were tested for SARS-CoV-2 spike protein IgG using an EUA ELISA, and PCR results were recorded from county health department data. Seroprevalence and Clopper-Pearson exact 95% confidence intervals were calculated.
Results
The observed seroprevalence of SARS-CoV-2 spike protein antibodies in children during strictest mitigation was 0.53% (95% CI 0.19, 1.14) and 0.92% (95% CI 0.46,1.64) during moderately relaxed. Strict and relaxed phase PCR-based prevalence were significantly higher, 2.87% (95% CI 1.95, 4.08) and 3.64 (95% CI 3.01, 4.38), respectively.
Conclusions
Estimates of pediatric seroprevalence were significantly lower than cumulative PCR prevalence estimates, and less than adult seroprevalence estimates, potentially due to biological, population, or sampling differences. Biological differences in pediatric immune responses to SARS-CoV-2 may make serosurvey interpretation challenging and these differences warrant further study.
Keywords: SARS-CoV-2, Antibodies, Epidemiology, Seroprevalence
1. Background
Coronavirus disease 2019 (COVID-19), caused by Severe Acute Respiratory Syndrome Coronavirus 2 (SARS-CoV-2), has a spectrum of disease severity, from asymptomatic infection to death [1], [2], [3]. Children have the same disease spectrum as adults, but pediatric mortality is far lower at 0–0.2% [4], [5], [6], [7]. Pediatric asymptomatic or mildly symptomatic infection may allow children to act as undetected vectors [8], [9], [10]. Due to transmission risk from children to more vulnerable populations, an understanding of pediatric infection burden and how government-imposed mitigation strategies altered pediatric infection burden is vital to policy making.
Polymerase chain reaction (PCR) testing is the standard for diagnosing acute respiratory viral infections, including SARS-CoV-2 infection. Test availability limitations have led to prioritizing symptomatic individuals, but significant asymptomatic viral shedding occurs [11], [12], [13]. Symptom-based testing, and minimal symptoms in children, likely resulted in insufficient characterization of pediatric infections early in the US epidemic. Antibody testing allows for retrospectively characterizing case numbers as it does not require active viral infection for detection.
The first cases of SARS-CoV-2 in southwestern Pennsylvania (SWPA) were reported on March 11, 2020 [14]. On March 23, 2020, a statewide stay-at-home order went into effect and began “red phase” mitigation efforts whereby only life sustaining businesses were permitted to be open. This phase continued until May 15, 2020. In SWPA, the “yellow phase” mitigation strategy, when stay-at-home orders were lifted and retail stores, offices, and childcare were permitted to be open with required masking and reduced capacity, was in effect from May 15, 2020 through June 5, 2020.
To better understand the prevalence of COVID-19 in children who live in SWPA in the two phases, we tested seroprevalence in pediatric residual blood specimens.
2. Study design
2.1. Cohort
This study was approved by the Institutional Review Board at the University of Pittsburgh (IRB 20040027) for collecting all residual blood samples obtained as part of routine clinical care and for EMR data at the University of Pittsburgh Medical Center Children's Hospital of Pittsburgh (UPMC CHP). Patient information was stored de-identified in a REDCap database with a research code matched to specimens [15,16].
Published reports suggest that SARS-CoV-2 IgG antibodies are detectable 7–14 days from positive nasopharyngeal PCR; thus, blood samples collection began approximately two weeks after the peak of the first wave of cases of COVID-19 in Allegheny county [14,17,18]. PA DOH relaxed SWPA mitigation standards on May 15, 2020, thus, samples from April 27-May 19 were analyzed to reflect the red phase. Collection of the second set of samples began 6 weeks after the first day mitigation standards were relaxed as yellow phase, June 22-July 3.
All samples that arrived in the lab were screened for inclusion, and samples from patients aged less than 19 years-old whose residential zip code was within 11 SWPA counties were included. The SWPA region, defined by the PA DOH, includes Allegheny, Armstrong, Beaver, Butler, Cambria, Fayette, Greene, Indiana, Somerset, Washington, and Westmoreland counties. Additional exclusion criteria were: 1) age ≤ 6 months due to confounding maternal antibodies, 2) hospital length of stay > 30 days when the sample was collected due to lack of community exposure, or 3) receipt of treatments that alter antibody production or antibody profile (e.g. immunoglobulin, rituximab, or bortezomib) in the six months prior to collection. Total positive and PCR tests done by May 15 for red phase and June 22 for yellow phase were collected from publicly available ACHD data [14].
2.2. SARS-CoV-2 antibody testing
SARS-CoV-2 IgG testing was performed in the clinical laboratory (Euroimmun, PerkinElmer, Lübeck, Germany). This assay was validated for use in the CLIA-certified high complexity clinical laboratories at UPMC (internally validated sensitivity of 98.7% at 14 days post symptom onset, specificity of 98.9%). Positive specimens were confirmed by repeat testing with spike protein pre-treatment as previously described [19]. Positive samples with sufficient volume were also confirmed by the Beckman Coulter SARS-CoV-2 IgG (sensitivity 100%, specificity 99.6%) (Brea, CA, USA). Specimens with discrepancies between first and second tests were assessed by Siemens Centaur SARS-CoV-2 Total assay (sensitivity 100%, specificity 99.1%) (Munich, Germany). Adult sera were tested for SARS-CoV-2 IgG in the clinical laboratory (Euroimmun, PerkinElmer, Lübeck, Germany) with positive tests confirmed by repeat testing with spike protein pre-treatment as previously described [19].
2.3. Statistical analysis
A minimum sample size of 1124 for each sample set was calculated using power calculation for proportionality in a binomial distribution to be 95% confident that the true seroprevalence would be within 1% of an estimated seroprevalence of 2%. The estimated prevalence of 2% was based on seroprevalence reported in Santa Clara County, a county with similar population and presumed PCR based prevalence as Allegheny county at the time [20].
We calculated observed seroprevalence as the proportion of total specimens that confirmed reactive. PCR observed prevalence was calculated as the cumulative proportion of total tests that were positive as publicly reported by the ACHD by the end of each mitigation phase [14]. 95% Clopper-Pearson exact confidence intervals for prevalence estimates were calculated using StatXact [20,21]. Observed seroprevalence and 95% intervals within selected subgroups were calculated in the same manner. Estimated prevalence and 95% confidence intervals adjusting for test characteristics (98.7% sensitivity, 99.99% combined specificity of initial and confirmation tests) were calculated as previously described [22]. Seroprevalence in each phase was compared to the PCR-based prevalence at the same time point using exact tests.
We also examined patient characteristics in the two cohorts. Differences in age and BMI categories were tested using the Kruskal-Wallis test. Differences in the proportions of the cohorts by sex, race, and primary insurance were assessed using Pearson exact test. For county of origin, Monte-Carlo estimation was used due to the large number of cells. All of these tests were calculated using StatXact, v.11.1.0 (Cytel,Inc.).
3. Results
From April 27-May 19, 2020, 1142 unique samples were collected during the “red phase”. During these first four months of the epidemic, both adult and pediatric cases rose slowly, but community spread was evident (Fig. S1, online). For the moderately relaxed mitigation yellow phase, 1196 samples were collected six weeks to eight weeks after stay-at-home orders were lifted.
3.1. Subject characteristics in each phase
The two sets of subjects were similar with respect to the characteristics examined ( Table 1 ). Mean ages were around 11 years in both phases. The racial distribution was also similar between groups (79.1% and 77.5% identifying as Caucasian) and is similar to the general population of Allegheny County, PA [23]. As expected, the majority of subjects (56.0% and 55.8%) in each group were residents of Allegheny County, the most populous county in SWPA and site of UPMC CHP. The only statistically significant differences between the two phases were seen in sex (higher proportion of males in the yellow phase), subjects with co-morbid conditions (higher proportion without any in the yellow phase), and immunocompromised (defined in Supplemental Methods) (lower proportion in the yellow phase) patients ( Table 1 ). Co-morbidity and immunocompromised differences are likely because otherwise healthy children were less likely to seek care unless absolutely necessary during the red phase. Selected subject comorbid conditions are shown in the supplementary materials (Table S1, online).
Table 1.
Demographics of subjects with residual clinical specimens tested for SARS-CoV-2 Spike IgG antibodies in each phase of mitigation.
| Red Phase [Number (% of total)] | Yellow Phase [Number (% of total)] | P-Value | |
|---|---|---|---|
| Total | 1142 | 1196 | |
| Age | |||
| Mean | 10.8 ± 5.5 | 11.13 ± 5.12 | |
| Median | 11.6 | 11.94 | |
| Range | 0.6–18.9 | 0.87–18.99 | |
| .29◊ | |||
| 0 to 6 | 290 (25.4%) | 262 (21.9%) | |
| 6 to 12 | 294 (25.7%) | 341 (28.5%) | |
| 12 to 18 | 558 (48.9%) | 593 (49.6%) | |
| Sex | .03# | ||
| Male | 523 (45.8%) | 601 (50.3%) | |
| Female | 619 (54.2%) | 595 (49.7%) | |
| Race | 0.87, 0.74# | ||
| Caucasian | 903 (79.1,81.7%) | 927 (77.5,80.2%) | |
| African American | 176 (15.4,15.9%) | 197 (16.5,17.0%) | |
| Latinx | 5 (0.4,0.5%) | 8 (0.7,0.7%) | |
| Asian American | 21 (1.8,1.9%) | 24 (2.0,2.1%) | |
| Unable to Determine | 37 (3.2,NA%) | 40 (3.3,NA%) | |
| Primary Insurance Type | 0.63, 0.97# | ||
| Medicaid | 455 (39.8, 40.3%) | 477 (39.9, 40.2%) | |
| Private Insurance | 674 (59.0, 59.7%) | 710 (59.59.84%) | |
| Unable to Determine | 13 (1.1, NA%) | 9 (0.8,NA%) | |
| BMI (patients >2 yo, with sufficient data) | 957 | 1017 | 0.46◊ |
| <25 | 760 (79.4%) | 794 (78.1%) | |
| 25 to <30 | 93 (9.7%) | 104 (10.2%) | |
| 30 or greater | 104 (10.9%) | 119 (11.7%) | |
| County of Origin | 0.66$ | ||
| Allegheny | 640 (56.0%) | 667 (55.8%) | |
| Armstrong | 13 (1.1%) | 22 (1.8%) | |
| Beaver | 65 (5.7%) | 73 (6.1%) | |
| Butler | 84 (7.4%) | 93 (7.8%) | |
| Cambria | 34 (3.0%) | 30 (2.5%) | |
| Fayette | 41 (3.6%) | 35 (2.9%) | |
| Greene | 6 (0.5%) | 8 (0.7%) | |
| Indiana | 12 (1.1%) | 21 (1.8%) | |
| Somerset | 12 (1.1%) | 18 (1.5%) | |
| Washington | 118 (10.3%) | 108 (9.0%) | |
| Westmoreland | 117 (10.2%) | 121 (10.1%) | |
| Comorbid Conditions | .0001# | ||
| None | 399 (34.9%) | 511 (42.7%) | |
| 1 or more | 743 (65.1%) | 685 (57.3%) | |
| Immunocompetent | <0.0001# | ||
| Yes | 869 (76.1%) | 989 (82.7%) | |
| No | 273 (23.9%) | 207 (17.3%) |
yo – years old, ◊ - tested by Kruskal-Wallis, # tested by Pearson exact, $ tested by exact test, Monte-Carlo. For primary insurance and race percentages, the first percent and p-value, represents inclusion of the “unable to determine” groups and the second percent and p-value represents if that group is excluded.
3.2. Seroprevalence and antibody positive subjects
For the red phase specimens, six of 1142 patients tested positive for SARS-CoV-2 antibodies, an 0.53% (95% CI 0.19,1.14) observed prevalence (OP) ( Table 2 ), and an adjusted prevalence (AP) for test performance characteristics of 0.52% (95% CI 0.23, 1.15). For the yellow phase, eleven of 1196 patient samples tested positive, OP of 0.92% (95% CI 0.46,1.64) and an AP of 0.91% (95% CI 0.51,1.65). Due to overall low prevalence, subgroups within the cohort did not demonstrate any unique findings in either mitigation phase ( Table 2 ). A contemporaneous serosurvey conducted in adults during the strict phase of mitigation at Pittsburgh-based UPMC hospitals utilizing pre-operative blood specimens demonstrated seroreactivity of 1.35% (95% CI 0.62, 2.55) of samples from April 27-May 19 (personal communication, GH).
Table 2.
Antibody status and seroprevalence rates of subjects with residual clinical specimens tested for SARS-CoV-2 Spike IgG antibodies in each phase of mitigation broken down by subgroup.
| Red Phase |
Yellow Phase |
|||
|---|---|---|---|---|
| Number Positive/Total Subgroup | % Seropositive (95% CI) | Number Positive/Total Subgroup | % Seropositive (95% CI) | |
| Total | 6/1142 | 0.53 (0.19, 1.14) | 11/1196 | 0.92 (0.46, 1.64) |
| 0 to 6 | 2/290 | 0.69 (0.08, 2.47) | 1/262 | 0.38 (0.00, 2.11) |
| 6 to 12 | 3/294 | 1.02 (0.21, 2.95) | 4/341 | 1.17 (0.32, 2.98) |
| 12 to 18 | 1/558 | 0.18 (0.00, 0.99) | 6/593 | 1.01 (0.37, 2.19) |
| Sex | ||||
| Male | 2/523 | 0.38 (0.05, 1.37) | 6/601 | 1.00 (0.37, 2.16) |
| Female | 4/619 | 0.65 (0.18, 1.65) | 5/595 | 0.84 (0.27, 1.95) |
| Race | ||||
| Caucasian | 5/903 | 0.55 (0.18, 1.29) | 6/927 | 0.65 (0.24,1.40) |
| African American | 1/176 | 0.57 (0.01, 3.12) | 4/197 | 2.03 (0.56, 5.12) |
| Latinx | 0/5 | ND | 0/8 | ND |
| Asian American | 0/21 | ND | 0/24 | ND |
| Not Listed | 0/37 | ND | 1/40 | ND |
| Primary Insurance | ||||
| Medicaid | 1/455 | 0.22 (0.01, 1.22) | 5/477 | 1.05 (0.34, 2.43) |
| Private Insurance | 5/674 | 0.74 (0.24, 1.72) | 6/710 | 0.85 (0.31, 1.83) |
| Unable to Determine | 0/13 | ND | 0/9 | ND |
| BMI (patients >2 yo, with sufficient data) | 957 | 1017 | ||
| <25 | 4/760 | 0.53 (0.14, 1.34) | 10/794 | 1.26 (0.61, 2.30) |
| 25 to <30 | 0/93 | 0.00 | 1/104 | 0.96 (0.02, 5.24) |
| 30 or greater | 0/104 | 0.00 | 0/119 | 0.00 (0.00, 3.05) |
| County of Origin | ||||
| Allegheny | 4/640 | 0.63 (0.17, 1.59) | 5/667 | 0.75 (0.24, 1.74) |
| Other Counties in SWPA | 2/502 | 0.40 (0.04, 1.43) | 6/529 | 1.13 (0.42, 2.45) |
| Comorbid Conditions | ||||
| None | 1/399 | 0.25 (0.01, 1.39) | 4/511 | 0.78 (0.21, 1.99) |
| 1 or more | 5/743 | 0.67 (0.22, 1.56) | 7/685 | 1.02 (0.41, 2.09) |
| Immunocompromised | ||||
| No | 3/869 | 0.35 (0.07, 1.01) | 8/989 | 0.81 (0.35, 1.59) |
| Yes | 3/273 | 1.10 (0.23, 3.18) | 3/207 | 1.45 (0.30, 4.18) |
| SARS-CoV-2 PCR Positive | 1/2 | 50.0 (1.26, 98.74) | 2/5 | 40.0 (5.27, 85.34) |
| Patients with Documented Respiratory Illness (01/2020 to Sample Date) | 3/226 | 1.33 (0.27, 3.83) | 5/238 | 2.10 (0.69, 4.83) |
| Patients with Documented COVID Exposure | 2/6 | 33.3 (4.33, 77.7) | 2/11 | 18.18 (2.28, 51.78) |
yo: years old, CI: confidence interval, SWPA: southwestern Pennsylvania. ND: Confidence intervals (CI) not calculated when number in subgroup<50 with 0 observed positive cases. Upper limit of the 95% CI for these cells is >7%.
Combining the findings of all antibody positive patients, only three of the 16 patients who were antibody positive required admission (one patient presented in both phases). Of the admitted patients, one was hospitalized for surgical complications unrelated to COVID course, one was hospitalized for less than 24 h for COVID, and the last met criteria for MIS-C and was hospitalized in the ICU. Five of the 16 were PCR tested, and of those five, three were positive and two were negative prior to their antibody sample collection. Five of the 16 antibody positive patients had documented respiratory illnesses and four had documented exposure to a COVID positive person. Interestingly, patients with EMR documented positive SARS-CoV-2 PCR did not always test positive for antibodies. Three of seven PCR-positive patients were antibody positive. Of the remaining four subjects, two had blood collected within a week of positive PCR, perhaps too early to detect an antibody response, but surprisingly, two others were antibody negative more than a month after their positive PCR result.
3.3. Cumulative PCR-based prevalence was higher than seroprevalence
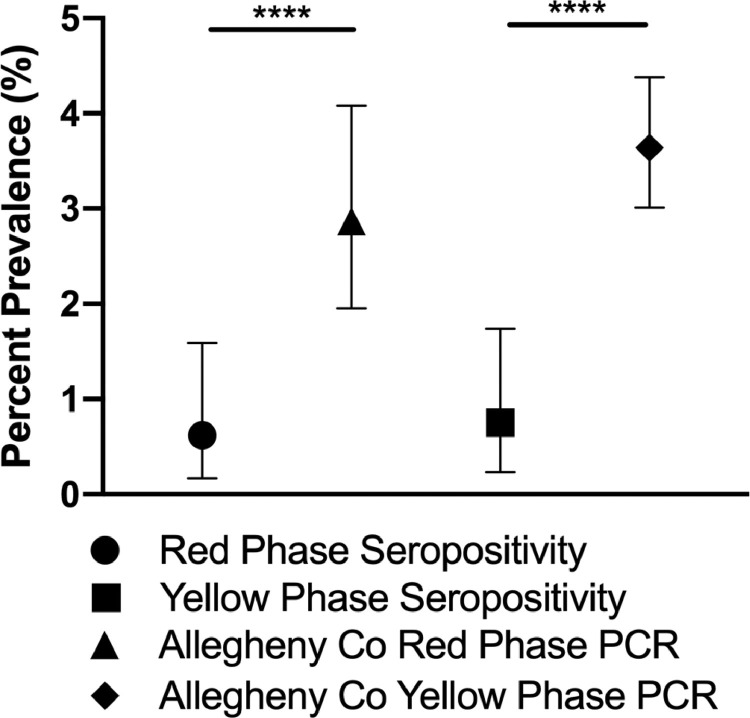
ACHD maintains publicly accessible COVID-19 data [14]. With the mitigation changes, PCR data from ACHD showed that cases in the 10–19-year-old children increased more quickly in yellow phase, 0.89 cases/day, compared to 0–9-year-old children, 0.57 cases/day (Fig. S2, online). Because seroprevalence estimates should detect the cumulative prevalence of all cases prior to the dates of collection, all positive PCR tests and total PCR tests as of the end of each mitigation phase were used to calculate a cumulative infection prevalence as of that date. PCR testing showed 30 positive PCR tests out of 1044 total tests, an OP of 2.87% (95% CI 1.95, 4.08), throughout the red phase (ending May 15), and total of 99 positive results out of 2814, an OP of 3.64% (95% CI 3.01, 4.38), by the end of the yellow mitigation ( Fig. 1 ). While these prevalence estimates were derived by two different mechanisms, the estimate of cumulative prevalence by PCR in Allegheny county was significantly higher than the estimate of seroprevalence in Allegheny county during both phases (p < 0.001). Similar to the findings of seroprevalence, there was not a significant difference in cumulative PCR prevalence between the two phases (p = 0.36).
Fig. 1.
Seroprevalence in Allegheny County compared to cumulative PCR based prevalence as of the end of the indicated mitigation phase in Allegheny County as reported by the ACHD COVID-19 Dashboard. Symbols represent prevalence estimates at different times and error bars represent Clopper Pearson 95% confidence interval. **** - p-value < 0.0001.
4. Discussion
In this study, we characterized SARS-CoV-2 seroprevalence in children by collecting two sets of specimens reflective of two different phases of mitigation strategies in order to assess pediatric infection burden. While seroprevalence almost doubled between red and yellow phase, we did not detect a significant change in the seroprevalence from the red to the yellow phase mitigation efforts likely due to insufficient power for comparison. However, we found a significant difference in the prevalence measured by PCR as compared to seroprevalence during each of the sampling periods.
The significantly higher prevalence estimates based on PCR versus serology may be due to the differences in sampling. PCR testing limitations imposed criteria that increase the pretest probability of a positive result, biasing prevalence data. While this is vital to ensure sufficient resources for continued testing, it limits the generalizability of the results due to the selection bias. Bias also exists since our convenience sampling methodology is not truly random. Despite these biases, the existence of this difference raises questions about the true prevalence in the community and how it could be best measured given the circumstances.
These data raise questions about the utility of serology to retrospectively estimate the prevalence in children, potentially due to biologic differences between pediatric and adult immune responses to SARS-CoV-2 [24,25]. Specifically, children have been shown to produce a narrower spectrum of antibodies to SARS-CoV-2 antigens than adults, and their antibodies have less neutralizing activity than adult antibodies [24]. Additionally, another study showed differing cytokine profiles, less robust T-cell responses, and decreased Fc receptor mediated phagocytosis activity when comparing children infected with SARS-CoV-2 with adults [25]. T-cells reactive to SARS-CoV-2 have been found in patients who have never been infected with SARS-CoV-2, perhaps due to exposure to endemic coronaviruses [26]. Endemic coronaviruses circulate annually and may increase children's likelihood of having T-cells cross-reactive to SARS-CoV-2 [27,28]. It is possible that CD8+ T-cell responses in children clear the infection before measurable antibodies are produced, a potential problem for pediatric SARS-CoV-2 serosurveillance studies.
To our knowledge at the time of writing, no pediatric specific serosurveys have been published, but several serosurveys have included pediatric subsets. The largest of these that utilized specimens from a similar time frame as the current study, organized by the CDC, completed a convenience serosurvey of 16,025 specimens at several national sites of which 7.5% (1205 samples/10 sites) were children. They demonstrated generally lower seroprevalence in children compared to adults at most sites, in concordance with our findings [29]. Two other serosurveys of children and adults suggest comparatively low seroprevalence in children as well [20,30]. The New Vaccine Surveillance Network (NSVN), which surveils pediatric acute respiratory illnesses at seven sites across the US, showed that only 0.1% of ill patients were COVID19 positive by PCR around the time of our sampling [31] supporting the idea that the prevalence in sick children is lower than in adults likely due to disease severity in children. While school closures may initially provide protection from infection in children, exposure of children to household members working outside the home should lead to similar seroprevalence between pediatric and adult populations in the absence of biological differences in responses to infection [8]. Biologically, if children do not reliably mount detectable antibody responses after infection, as seen with the two PCR positive patients without detectable antibodies greater than one month after infection, this could account for the discrepancy between adult and pediatric seroprevalence.
The timing of sampling could also confound serosurvey results, as antibodies may have waned or not yet be measurable at the time of sampling, a factor that will need to be considered when trying to utilize serology to retrospectively characterize this epidemic. Data suggest that milder infection leads to longer time to seroconversion and lower antibody titers in adults [32,33]. These factors may have contributed to the two subjects with positive PCR for SARS-CoV-2 but absent antibodies upon sampling more than a month afterwards. The implications of the biologic differences between the immune response of children and adults is important as vaccines are tested in children.
We recognize some limitations in this study. First, this study was done with sampling medically indicated blood specimens and not random community sampling. As a result, there was a difference in the proportions of previously healthy patients and immunocompetent patients seen in the two phases. PCR data were collected from ACHD rather than sampled directly from patients with longitudinal follow-up, a limitation necessitated by the need to conserve reagents for clinically indicated testing. While sufficient for independent prevalence estimates, samples sizes were insufficient to compare seroprevalence between the two phases. Additionally, a substantial proportion of serologic samples came from patients outside Allegheny County while the PCR data was exclusively from that county, and the adult samples did not have residence identified. Lastly, the overall low disease burden in our community during sampling decreased our ability to draw conclusions about factors that influence the likelihood of seropositivity.
In summary, we demonstrated the seroprevalence of SARS-CoV-2 in children early in the US epidemic at two separate times. Seroprevalence estimates were lower than the PCR prevalence estimates in children and lower than seroprevalence estimates in adults, potentially due to biological, population, and sampling differences. The biologic differences may make pediatric SARS-CoV-2 serosurveys more challenging to interpret. Primarily, this study affirms the importance of studying the immune response to SARS-CoV-2 in children, especially as these biologic differences could have implications not only for testing, but also for vaccine development and efficacy.
Declaration of Competing Interest
None of the authors have any conflicts of interest that relate to the work conducted for this manuscript.
Acknowledgements
The authors would like to acknowledge the UPMC lab personnel, who were instrumental in ensuring the convenience specimens were retained and helped with entering specimens into our database, Jill Lewandowski, Jayne Zona, and Jayne Rasmussen.
This work was supported by the University of Pittsburgh Clinical and Translational Science Institute COVID-19 Pilot Study Program and the David Scaife Family (DSF) Charitable Foundation, Pediatric Infectious Disease Society and St. Jude Children's Research Hospital Fellowship Program in Basic and Translational Research [MCF], Pediatric Scientist Development Program (PSDP) sponsored by the Association of Medical School Pediatric Department Chairs (AMSPDC) funded by Eunice Kennedy Shriver National Institute of Child Health and Human Development (K12HD000850–34[GJR]).
Footnotes
Supplementary material associated with this article can be found, in the online version, at doi:10.1016/j.jcvp.2021.100026.
Appendix. Supplementary materials
References
- 1.Guan W.J., Ni Z.Y., Hu Y., Liang W.H., Ou C.Q., He J.X. Clinical characteristics of coronavirus disease 2019 in China. N. Eng. J. Med. 2020;382:1708–1720. doi: 10.1056/NEJMoa2002032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Goyal P., Choi J.J., Pinheiro L.C., Schenck E.J., Chen R., Jabri A. Clinical characteristics of COVID-19 in New York City. N Engl J Med. 2020;382:2372–2374. doi: 10.1056/NEJMc2010419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Richardson S., Hirsch J.S., Narasimhan M., Crawford J.M., McGinn T., Davidson K.W. Presenting characteristics, comorbidities, and outcomes among 5700 patients hospitalized with COVID-19 in the New York City Area. JAMA. 2020;323:2052–2059. doi: 10.1001/jama.2020.6775. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Dong Y., Mo X., Hu Y., Qi X., Jiang F., Jiang Z. Epidemiology of COVID-19 among children in China. Pediatrics. 2020:145. doi: 10.1542/peds.2020-0702. [DOI] [PubMed] [Google Scholar]
- 5.Lu X., Zhang L., Du H., Zhang J., Li Y.Y., Qu J. SARS-CoV-2 infection in children. N. Eng. J. Med. 2020;382:1663–1665. doi: 10.1056/NEJMc2005073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Team CC-R Coronavirus disease 2019 in children - United States, February 12-April 2, 2020. MMWR Morb. Mortal. Wkly. Rep. 2020;69:422–426. doi: 10.15585/mmwr.mm6914e4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Pediatrics AAo, Association CsH . In: Children and COVID-19: State Data Report. Sisk B., Harris M., editors. 2020. Children and COVID-19: state data report.https://services.aap.org/en/pages/2019-novel-coronavirus-covid-19-infections/children-and-covid-19-state-level-data-report/ [Google Scholar]
- 8.Posfay-Barbe K.M., Wagner N., Gauthey M., Moussaoui D., Loevy N., Diana A. COVID-19 in children and the dynamics of infection in families. Pediatrics. 2020:146. doi: 10.1542/peds.2020-1576. [DOI] [PubMed] [Google Scholar]
- 9.Park Y.J., Choe Y.J., Park O., Park S.Y., Kim Y.M., Kim J. Contact tracing during coronavirus disease outbreak, South Korea, 2020. Emerg. Infect. Dis. 2020;26:2465–2468. doi: 10.3201/eid2610.201315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Grijalva C.G., Rolfes M.A., Zhu Y., McLean H.Q., Hanson K.E., Belongia E.A. Transmission of SARS-COV-2 infections in households - Tennessee and Wisconsin, April-September 2020. MMWR Morb. Mortal. Wkly. Rep. 2020;69:1631–1634. doi: 10.15585/mmwr.mm6944e1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Sola A.M., David A.P., Rosbe K.W., Baba A., Ramirez-Avila L., Chan D.K. Prevalence of SARS-CoV-2 infection in children without symptoms of coronavirus disease 2019. JAMA Pediatr. 2021;175:198–201. doi: 10.1001/jamapediatrics.2020.4095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Arons M.M., Hatfield K.M., Reddy S.C., Kimball A., James A., Jacobs J.R. Presymptomatic SARS-CoV-2 infections and transmission in a skilled nursing facility. N. Eng. J. Med. 2020;382:2081–2090. doi: 10.1056/NEJMoa2008457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Gudbjartsson D.F., Helgason A., Jonsson H., Magnusson O.T., Melsted P., Norddahl G.L. Spread of SARS-CoV-2 in the Icelandic population. N. Eng. J. Med. 2020;382:2302–2315. doi: 10.1056/NEJMoa2006100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Department ACH . Alleghency County Health Department; 2020. Allegheny County Health Department COVID-19 Dashboard. [Google Scholar]
- 15.Harris P.A., Taylor R., Thielke R., Payne J., Gonzalez N., Conde J.G. Research electronic data capture (REDCap)–a metadata-driven methodology and workflow process for providing translational research informatics support. J. Biomed. Inform. 2009;42:377–381. doi: 10.1016/j.jbi.2008.08.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Harris P.A., Taylor R., Minor B.L., Elliott V., Fernandez M., O'Neal L. The REDCap consortium: building an international community of software platform partners. J. Biomed. Inform. 2019;95 doi: 10.1016/j.jbi.2019.103208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Wolfel R., Corman V.M., Guggemos W., Seilmaier M., Zange S., Muller M.A. Virological assessment of hospitalized patients with COVID-2019. Nature. 2020;581:465–469. doi: 10.1038/s41586-020-2196-x. [DOI] [PubMed] [Google Scholar]
- 18.Dispinseri S., Secchi M., Pirillo M.F., Tolazzi M., Borghi M., Brigatti C. Neutralizing antibody responses to SARS-CoV-2 in symptomatic COVID-19 is persistent and critical for survival. Nat. Commun. 2021;12:2670. doi: 10.1038/s41467-021-22958-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Wheeler S.E., Shurin G.V., Keetch C., Mitchell G., Kattel G., McBreen J. Evaluation of SARS-CoV-2 prototype serologic test in hospitalized patients. Clin. Biochem. 2020;86:8–14. doi: 10.1016/j.clinbiochem.2020.08.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Bendavid E., Mulaney B., Sood N., Shah S., Bromley-Dulfano R., Lai C. COVID-19 antibody seroprevalence in Santa Clara County, California. Int. J. Epidemiol. 2021;50:410–419. doi: 10.1093/ije/dyab010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Clopper C.J., Pearson E.S. The use of confidence or fiducial limits illustrated in the case of the binomial. Biometrika. 1934;26:404–413. [Google Scholar]
- 22.Diggle P.J. Estimating prevalence using an imperfect test. Epidemiol. Res. Int. 2011;2011:1–5. [Google Scholar]
- 23.Bureau USC . 2019. QuickFacts County Search. Census.gov: United States Census Bureau. [Google Scholar]
- 24.Weisberg S.P., Connors T.J., Zhu Y., Baldwin M.R., Lin W.H., Wontakal S. Distinct antibody responses to SARS-CoV-2 in children and adults across the COVID-19 clinical spectrum. Nat. Immunol. 2021;22:25–31. doi: 10.1038/s41590-020-00826-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Pierce C.A., Preston-Hurlburt P., Dai Y., Aschner C.B., Cheshenko N., Galen B. Immune responses to SARS-CoV-2 infection in hospitalized pediatric and adult patients. Sci. Transl. Med. 2020:12. doi: 10.1126/scitranslmed.abd5487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Grifoni A., Weiskopf D., Ramirez S.I., Mateus J., Dan J.M., Moderbacher C.R. Targets of T Cell Responses to SARS-CoV-2 coronavirus in humans with COVID-19 disease and unexposed individuals. Cell. 2020;181 doi: 10.1016/j.cell.2020.05.015. 1489-501 e15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.National Respiratory and Enteric Virus Surveillance System . Centers for Disease Control and Prevention; 2020. National Trends for Common Human Coronaviruses.https://www.cdc.gov/surveillance/nrevss/coronavirus/natl-trends.htmlCDC.gov [Google Scholar]
- 28.Killerby M.E., Biggs H.M., Haynes A., Dahl R.M., Mustaquim D., Gerber S.I. Human coronavirus circulation in the United States 2014-2017. J. Clin. Virol. 2018;101:52–56. doi: 10.1016/j.jcv.2018.01.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Havers F.P., Reed C., Lim T., Montgomery J.M., Klena J.D., Hall A.J. Seroprevalence of antibodies to SARS-CoV-2 in 10 sites in the United States, March 23-May 12, 2020. JAMA Intern. Med. 2020;180:1576–1586. doi: 10.1001/jamainternmed.2020.4130. [DOI] [PubMed] [Google Scholar]
- 30.Bryan A., Pepper G., Wener M.H., Fink S.L., Morishima C., Chaudhary A. Performance characteristics of the Abbott architect SARS-CoV-2 IgG assay and seroprevalence in Boise, Idaho. J. Clin. Microbiol. 2020:58. doi: 10.1128/JCM.00941-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Rha B., Lively J.Y., Englund J.A., Staat M.A., Weinberg G.A., Selvarangan R. Severe acute respiratory syndrome coronavirus 2 infections in children: multicenter surveillance, United States, January-March 2020. J. Pediatric Infect. Dis. Soc. 2020;9:609–612. doi: 10.1093/jpids/piaa075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Bahar B., Jacquot C., Mo Y.D., DeBiasi R.L., Campos J., Delaney M. Kinetics of viral clearance and antibody production across age groups in children with severe acute respiratory syndrome coronavirus 2 infection. J. Pediatr. 2020;227:31–37. doi: 10.1016/j.jpeds.2020.08.078. el. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Rijkers G., Murk J.L., Wintermans B., van Looy B., van den Berge M., Veenemans J. Differences in antibody kinetics and functionality between severe and mild severe acute respiratory syndrome coronavirus 2 infections. J. Infect. Dis. 2020;222:1265–1269. doi: 10.1093/infdis/jiaa463. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.