Abstract
Recently, nine Caenorhabditis elegans genes, grouped into two pathways/clusters, were found to be implicated in healthspan in C. elegans and their homologues in humans, based on literature curation, WormBase data mining and bioinformatics analyses. Here, we further validated these genes experimentally in C. elegans. We downregulated the nine genes via RNA interference (RNAi), and their effects on physical function (locomotion in a swim assay) and on physiological function (survival after heat stress) were analysed in aged nematodes. Swim performance was negatively affected by the downregulation of acox-1.1, pept-1, pak-2, gsk-3 and C25G6.3 in worms with advanced age (twelfth day of adulthood) and heat stress resistance was decreased by RNAi targeting of acox-1.1, daf-22, cat-4, pig-1, pak-2, gsk-3 and C25G6.3 in moderately (seventh day of adulthood) or advanced aged nematodes. Only one gene, sad-1, could not be linked to a health-related function in C. elegans with the bioassays we selected. Thus, most of the healthspan genes could be re-confirmed by health measurements in old worms.
Keywords: C. elegans, healthspan, ageing, locomotion, stress resistance, RNAi
1. Introduction
Human life expectancy has increased in recent decades worldwide. For example, in Germany, life expectancy at birth was 66.69 years in 1950. By 2020, this had increased by 22% to 81.41 years [1]. However, the age at onset of morbidity and ageing-associated diseases has not changed as much [2]. Thus, people spend more time in poor health: in the UK, the time in poor health increased from 15.4/18.1 to 16.1/19.1 years for males/females between 2000/2002 and 2012/2014 (https://www.gov.uk/government/publications/health-profile-for-england/chapter-1-life-expectancy-and-healthy-life-expectancy). Furthermore, in a global study, male and female healthy life expectancy at birth was 54.8 and 58.7 years, respectively, in 1990 and 59 and 63.2 years, respectively, in 2010 [3]. This corresponds to an increase of healthspan by 4.2 and 4.5 years, respectively, whereas the life expectancy during the same time period increased by 4.7 and 5.1 years, respectively. To increase our understanding of the genetic basis of health and ageing, the model organism Caenorhabditis elegans is frequently used. This nematode is about 1 mm-long (measured at the first day of adulthood [4]) and characterized by easy handling, cheap maintenance, a simple body plan, a short generation time and easy storage. While key genetic players for an extended lifespan are known in C. elegans, few researchers have examined whether extended lifespan is accompanied by extended healthspan, with conflicting results [5,6].
Recently, we calculated ‘healthspan pathway maps' and their overlap in humans and C. elegans [7]. To compile the maps, we first defined healthspan as the time spent in the absence of disease and dysfunction [8]. Based on literature, gene/protein interaction and annotation data, we could then infer two healthspan pathway maps (human and worm), and their overlap comprised nine human genes, arranged in two interconnected sets. These were ACOX1, SCP2, GCH1, SLC15A1 and MELK, PAK4, BRSK2, GSK3B, CDKN2B and their C. elegans homologues were acox-1.1, daf-22, cat-4, pept-1 and pig-1, pak-2, sad-1, gsk-3, C25G6.3. Here, we test the two overlaps of the healthspan pathway maps by health-related bioassays.
To validate that the nine genes play a role in the maintenance of health in C. elegans, health-relevant phenotypes were measured in aged nematodes after downregulation via RNA interference (RNAi) [9]. According to Fuellen et al. [8], the most important features of health that decline during ageing can be categorized as belonging to physiological, physical, cognitive or reproductive functions. Here, we test the hypothesis that downregulation of each of the nine genes will lead to changes in stress resistance (physiological function) and locomotion (physical function) in aged worms. Furthermore, it is assumed that the genes involved in stress resistance are similarly involved in lifespan determination. We recently found a very high correlation between lifespan and stress resistance in C. elegans [10]. These findings corroborated previous studies with similar results [11].
2. Material and methods
(a) . Maintenance and treatment of Caenorhabditis elegans
The wild-type N2 Bristol C. elegans strain was obtained from the Caenorhabditis Genetics Center (Minneapolis, MN, USA) and maintained at 22°C on NGM agar plates according to Brenner [12]. Sodium hypochlorite (3%) treatment, inspired by T. Stiernagle [13], was performed to synchronize the population. Nematodes were fed with Escherichia coli HT115 bacteria strains containing an empty vector (EV) (Ahringer Library, Source BioScience, Nottingham, UK). From the fourth larval stage (L4) on, nematodes were distributed to NGM plates containing 100 µM floxuridine (FUdR), 1 mM isopropyl β-d-1-thiogalactopyranoside (IPTG) and 2 mg ml−1 carbenicillin as well as the appropriate RNAi feeding strain to downregulate the target genes. The acox-1.1 and pig-1 feeding strains were constructed by restriction-based plasmid cloning. The target sequences from genomic C. elegans DNA were amplified using the following primer pairs including restriction sites: acox-1.1 forward (GCAGTCCTCGAGGCACAGATTTATGGGGGAGA) and reverse (GGCGGCAGATCTTGATCTTTGCATGTGGTGGT) primer, as well as pig-1 forward (GCAGTCCTCGAGAATGTCGGTGAGGAGCAGTG) and reverse (GGCGGCAGATCTTGTTGGTTTTCTCGACGGTA) primer, and ligated into a L4440 plasmid. The plasmid was first transformed into E. coli DH5α cells and finally into E. coli HT115 cells for feeding C. elegans. The other feeding strains were obtained from the Ahringer library [14], and all strains were sequenced to verify the correct plasmid insert sequences.
Prior to use, fresh feeding bacteria were treated with 1 mM IPTG for 2 h, subsequently washed with 3 g l−1 NaCl and adjusted to a density of OD595 = 9. Nematodes fed with EV-feeding bacteria served as a control. Animals were kept in the dark until the third (A3), seventh (A7) and twelfth (A12) day of adulthood. The efficiencies of the RNAi treatments were tested via RT-qPCR (see electronic supplementary material, data).
(b) . Heat stress assay
At the chosen ages (A3, A7, A12), a heat stress assay was performed by incubating the worms for 3 h at 37°C. From the next day on, animals were counted blinded as dead or alive and lost animals were noted. For statistical evaluations, the log-rank test with subsequent Bonferroni correction provided by OASIS 2 [15] was used. Significance is deemed to be reached if the p-value is less than 0.05.
(c) . Swim assay
In addition, a swim assay was carried out at ages A3, A7 and A12 by transferring the animals into wells (0.5 mm deep and 10 mm diameter) on object slides, which were filled with 42 µl of M9 buffer. The wells were covered with a coverslip, and the movement of the nematodes was filmed for 60 s. Videos were recorded with a Müller Optronic MicroAnalytics HD camera and software (MHDC-500 microscope camera) with a 9.5 times magnification. Every other frame of each MP4 video was converted into a jpg file, which was edited with Adobe Photoshop to obtain binary and inverted pictures. The pictures were analysed with the program CeleST (v. 3.1, [16]), which tracks the movement of nematodes and calculates, among others, the parameters: ‘wave initiation rate’, ‘body wavenumber’, ‘brush stroke’ and ‘activity index’. An unpaired t-test with subsequent Bonferroni correction was performed to evaluate significance, i.e. if the corrected p-value is below 0.05.
3. Results
(a) . Downregulation of acox-1.1, daf-22, cat-4, pig-1, pak-2, gsk-3 and C25G6.3 leads to a decreased thermal tolerance in aged nematodes
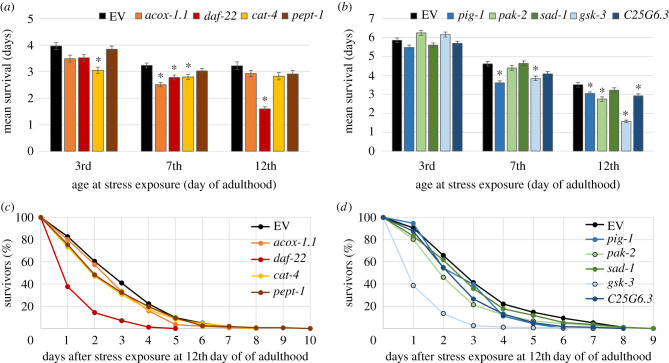
Stress resistance is a key feature for the maintenance of healthspan and declines with age in C. elegans as well as in humans [17–19]. Therefore, heat stress resistance was selected to assess the health status of the nematodes. The experiments were conducted separately for the first (acox-1.1, daf-22, cat-4 and pept-1) and the second (pig-1, pak-2, sad-1, gsk-3 and C25G6.3) overlap. Following the heat stress, we observed a significantly lower thermal tolerance in A12-old animals treated with RNAi for daf-22, pig-1, pak-2 and gsk-3 or C25G6.3 (figure 1a–d). While untreated animals survived for 3.22 (figure 1c) to 3.5 (figure 1d) days after application of heat stress, the decreased mean survival of treated animals ranged from 1.57 days (gsk-3) to 3.04 days (pig-1) (table 1). Interestingly, nematodes with downregulation in pept-1 and sad-1 displayed no significant differences in their mean survival at any tested age after heat stress, compared to the control group. However, a slight but significant decrease of median and minimum survival was triggered by the downregulation of pept-1 at A7 and A12, respectively (table 1). Therefore, these two genes may not be strongly involved in thermal tolerance. The role of acox-1.1 and cat-4 appears to depend on age. Nematodes deficient in cat-4 displayed a remarkably decreased mean survival in young (−30%) and A7 old (−15%) adults, whereas acox-1.1 deficient animals only revealed a decreased survival at the age A7 (−29%) compared to the untreated group. It should be noted that all nine treatments led to a decreased survival at A12, although not all reached the significance level (table 1).
Figure 1.
Survival after heat stress. Nematodes at the L4 stage were treated with nine different RNAi strains or the EV strain. On the third, seventh and twelfth day of adulthood, they were exposed to heat stress at 37°C for 3 h, prior to monitoring their survival. The mean survival ± s.e.m. is plotted (a,b) and differences compared to control were considered significant at p < 0.05 (*) by using a two-sided log-rank test and subsequent Bonferroni correction. In addition, survival curves are shown for nematodes which were heat shocked on the twelfth day of adulthood (c,d). The number of tested individuals is collected in table 1.
Table 1.
Heat stress survival characteristics during RNAi treatments. Differences compared to control were considered significant at p < 0.05 (*). p-value determination was realized with log-rank test and subsequent Bonferroni correction for the mean survival and Fisher's Exact test for specific time points. n = number of worms; min./med./max. = minimum/median/maximum; s.e.m. = standard error of the mean.
| days until deaths of population reached |
||||||||
|---|---|---|---|---|---|---|---|---|
| age at heat stress exposure | RNAi treatment | n | 25% (min. survival) | 50% (med. survival) | 90% (max. survival) | mean survival (days) | s.e.m. | change relative to control [%] |
| third day of adulthood | EV | 157 | 2.38 | 3.48 | 5.55 | 3.96 | 0.13 | |
| acox-1.1 | 151 | 1.90* | 2.87* | 5.17 | 3.49 | 0.13 | −13.47 | |
| daf-22 | 152 | 1.93 | 3.21 | 4.86 | 3.52 | 0.12 | −12.50 | |
| cat-4 | 152 | 1.43* | 2.41* | 4.65 | 3.05* | 0.12 | −29.84 | |
| pept-1 | 151 | 2.28 | 3.38 | 5.22 | 3.84 | 0.12 | −3.13 | |
| EV | 244 | 4.05 | 4.77 | 8.28 | 5.85 | 0.13 | ||
| pig-1 | 249 | 3.43* | 5.05 | 7.48* | 5.47 | 0.12 | −6.95 | |
| pak-2 | 240 | 4.26* | 5.21* | 8.57 | 6.24 | 0.13 | +6.25 | |
| sad-1 | 243 | 3.85 | 4.69 | 7.59 | 5.59 | 0.12 | −4.65 | |
| gsk-3 | 244 | 3.89 | 5.32* | 8.46 | 6.15 | 0.13 | +4.88 | |
| C25G6.3 | 241 | 3.74* | 4.74 | 7.74 | 5.69 | 0.11 | −2.81 | |
| seventh day of adulthood | EV | 158 | 1.81 | 2.73 | 4.34 | 3.23 | 0.09 | |
| acox-1.1 | 153 | 1.26* | 1.88* | 3.47* | 2.51* | 0.08 | −28.69 | |
| daf-22 | 156 | 1.43* | 2.13* | 3.97 | 2.78* | 0.09 | −16.19 | |
| cat-4 | 158 | 1.35* | 2.11* | 3.95 | 2.80* | 0.10 | −15.36 | |
| pept-1 | 156 | 1.65 | 2.41* | 3.98 | 3.03 | 0.09 | −6.60 | |
| EV | 228 | 2.60 | 4.05 | 6.67 | 4.61 | 0.12 | ||
| pig-1 | 263 | 1.87* | 2.90* | 5.47* | 3.61* | 0.10 | −27.70 | |
| pak-2 | 255 | 2.23 | 3.73 | 6.61 | 4.39 | 0.13 | −5.01 | |
| sad-1 | 253 | 2.54 | 4.16 | 6.71 | 4.63 | 0.12 | +0.43 | |
| gsk-3 | 246 | 1.77* | 2.84* | 6.22 | 3.84* | 0.12 | −20.05 | |
| C25G6.3 | 238 | 2.04* | 3.16* | 6.41 | 4.08 | 0.12 | −12.99 | |
| twelfth day of adulthood | EV | 144 | 1.34 | 2.54 | 4.98 | 3.22 | 0.14 | |
| acox-1.1 | 150 | 1.22 | 2.31 | 4.48 | 2.93 | 0.12 | −9.90 | |
| daf-22 | 154 | 0.40* | 0.80* | 2.60* | 1.60* | 0.08 | −101.25 | |
| cat-4 | 157 | 0.93* | 1.89 | 4.81 | 2.83 | 0.14 | −13.78 | |
| pept-1 | 159 | 1.02* | 1.93 | 4.95 | 2.91 | 0.14 | −10.65 | |
| EV | 193 | 1.63 | 2.65 | 5.89 | 3.50 | 0.13 | ||
| pig-1 | 224 | 1.49* | 2.30 | 4.17* | 3.04* | 0.08 | −15.13 | |
| pak-2 | 178 | 1.16* | 1.89* | 4.42 | 2.75* | 0.12 | −27.27 | |
| sad-1 | 186 | 1.39 | 2.46 | 5.28 | 3.21 | 0.13 | −9.03 | |
| gsk-3 | 155 | 0.41* | 0.82* | 2.32* | 1.57* | 0.07 | −122.93 | |
| C25G6.3 | 169 | 1.39* | 2.18* | 4.39* | 2.91* | 0.11 | −20.27 | |
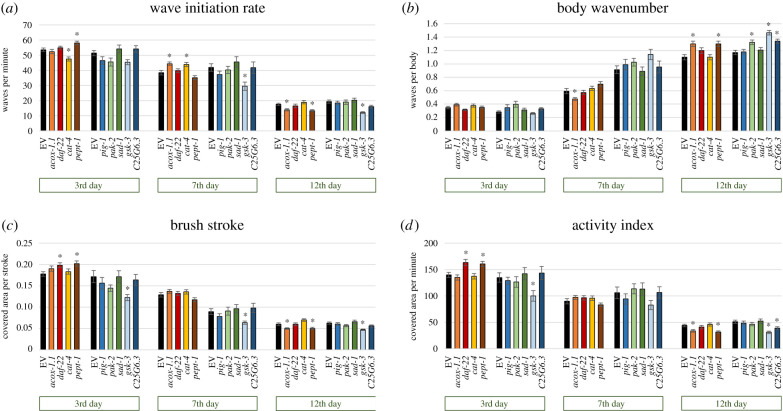
(b) . ACOX-1.1, PEPT-1, C25G6.3 and GSK-3 are important to maintain locomotor fitness in old nematodes
The reduction of physical function during ageing is a serious health deterioration in humans and represents also a clear sign of ageing in nematodes [16]. Therefore, the movement of RNAi-treated nematodes was evaluated by analysing four age-dependent swim parameters: the wave initiation rate (figure 2a) indicates the speed of movement by determining the number of body thrashes per minute; the body wavenumber (figure 2b) reflects the waviness of the body at each time point by counting the waves per body; the brush stroke (figure 2c) indicates the flexibility of the body by summarizing the number of pixels that are covered by the body during one stroke; and the activity index (figure 2d) is an indicator for the vigorousness of bending over time measured by the number of pixels that are covered by the body per minute.
Figure 2.
Swim performance of C. elegans after RNAi treatment. Nematodes at the L4 stage were treated with nine different RNAi strains or the EV strain. Wave initiation rate (a), body wavenumber (b), brush stroke (c) and activity index (d) were determined on the third, seventh and twelfth days of adulthood. The mean values ± s.e.m. are plotted and differences compared to control were considered significant at p < 0.05 (*) by using a two-sided t-test with subsequent Bonferroni correction. Each bar represents n ≥ 42 nematodes from two independent trials (concrete details are presented in the electronic supplementary material, raw data).
As shown in Restif et al. [16] and in figure 2, only the body wavenumber increases with age, whereas the other parameters decrease. Thus, the RNAi treatments targeting acox-1.1, pept-1 and gsk-3 appear to accelerate ageing given the decreased wave initiation rate, brush stroke and activity index (figure 2a,c,d) as well as the increased body wavenumber (figure 2b) in A12-old worms compared to the respective EV-control. The strongest changes were observed after gsk-3 RNAi treatment at A12, with a reduction of up to −60% in the wave initiation rate and activity index. At A3 and A7, the downregulation of gsk-3 resulted in similar, but not always significant, effects, whereas pept-1 and acox-1.1 targeting RNAi led to contrasting phenotypes at A3 or A7, respectively. C25G6.3-deficient nematodes worsened the body wavenumber and activity index at A12. Interestingly, the downregulation of pak-2 solely led to an increase of waves per body at A12 (figure 2b) and cat-4 targeted RNAi led to contrasting changes of the wave initiation rate at A3 and A7 (figure 2a). In addition, worms treated with daf-22 RNAi showed locomotion improvements at A3 (figure 2c,d). RNAi treatments targeting pig-1 and sad-1 did not provoke any significant changes in swimming behaviour.
4. Discussion
Several proteins encoded by the genes in our study are involved in lipid metabolism, which is known to affect life- as well as healthspan in model organisms and humans [20]. ACOX-1.1 belongs to the class of acyl-coenzyme A oxidases that are important in lipid metabolism; thus, the knockout of acox-1.1 via mutation leads to increased intestinal fat deposits [21]. DAF-22 exhibits propanoyl-CoA C-acyltransferase activity. This enzyme participates in a signalling pathway including transcription factors called peroxisome proliferator-activated receptors, which (among others) regulate lipid metabolism [22]. The peptide transporter PEPT-1 interacts with the sodium–proton exchanger NHX-2 to regulate intracellular pH, which is important for the influx of free fatty acids. Furthermore, the absence of PEPT-1 in a C. elegans mutant strain leads to a twofold increase in total body fat [23]. The last member of the first overlap, CAT-4, is involved in dopamine biosynthesis, as shown with the aid of mutant strains [24], and its expression was decreased in skn-1(RNAi) worms, thus it is a target of the stress-related transcription factor SKN-1 [25].
PIG-1, PAK-2, SAD-1 and GSK-3 all exhibit serine/threonine kinase signalling activity. Furthermore, SAD-1 and PIG-1 are involved in neuronal development as shown in mutant analyses [26,27], and PIG-1 and GSK-3 in apoptotic processes, proven via RNAi and mutant strains [28–30]. In addition, GSK-3 also plays a role in Wnt signalling [31]. Finally, gene expression studies suggest a role of pak-2 during ageing [32] and C25G6.3 during pathogenic stress [33]. Of note, analyses based on RNAi can differ from results obtained with mutant strains, due to unspecific effects elicited by the RNAi vector L4440 [34], or off-target effects of the dsRNA [35]. Furthermore, in mutant analyses, the knockout of the target usually impacts the complete larval development phase, whereas RNAi treatments are often started in older larvae or even adults.
Despite their health-related evidence, the effect of downregulating the nine target genes on health parameters in aged worms was not studied previously. Interestingly, all RNAi treatment effects on swim performance (acox-1.1, pept-1, pak-2, gsk-3 and C25G6.3) at A12 and stress resistance (acox-1.1, daf-22, cat-4, pig-1, pak-2, gsk-3 and C25G6.3) at A7 and A12 were deleterious. Remarkably, such effects were mostly absent or negligible in young worms or are even contrary in locomotion parameters of young or middle-aged worms. Thus, it is suggested that the corresponding gene products are beneficial to locomotion and/or stress resistance in ageing C. elegans. The reasons for the age dependence of the observed effects are not fully understood so far. However, it is assumed that ageing-related treatments show stronger phenotypic expressions in older individuals that feature at least first signs of ageing. Furthermore, in older individuals, the respective treatment had much more time to produce any effect compared to young worms (see also [10]).
The downregulation of sad-1 did not significantly affect the performance of the worms, which might be related to poor RNAi efficiency. In conclusion, eight of the nine genes implicated in healthspan were shown to be relevant for physical and/or physiological function, in line with Möller et al. [7].
Acknowledgements
We thank the Caenorhabditis Genetics Centre, which is funded by the National Institutes of Health National Centre for Research Resources, for the supply of the Caenorhabditis elegans strains and the Braeckman Lab (Ghent University, Belgium) for providing the RNAi strains. Furthermore, we thank Thea Böttcher and Shumon Chakrabarti for their technical support in the laboratory, and Christian E.W. Steinberg for enabling this project.
Data accessibility
The datasets supporting this article have been uploaded as part of the electronic supplementary material.
Authors' contributions
J.D. carried out the laboratory work and statistical analysis, participated in data analysis and interpretation and drafted the manuscript. G.F. participated in the design of the study. S.M. participated in the design of the study. W.L. conceived the study. C.S.-L. participated in the design of the study and in data analysis. N.S. participated in the design of the study and in data analysis and coordinated the study. All authors critically revised the manuscript, gave final approval for its publication and agree to be held accountable for the work performed therein.
Competing interests
We declare we have no competing interests
Funding
This work was supported by the European Union's Horizon 2020 research and innovation programme (grant agreement No 633589; Project ‘Ageing with Elegans’). This publication reflects only the authors' views and the Commission is not responsible for any use that may be made of the information it contains.
References
- 1.United Nations, Department of Economic and Social Affairs, Population Division. 2019. Total population (both sexes combined) by single age, region, subregion and country, annually for 1950–2100 (File INT/3-1). World Population Prospects 2019; Online Edition. Rev. 1.
- 2.Crimmins EM. 2015. Lifespan and healthspan: past, present, and promise. Gerontologist 55, 901-911. ( 10.1093/geront/gnv130) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Salomon JA, Wang H, Freeman MK, Vos T, Flaxman AD, Lopez AD, Murray CJ. 2012. Healthy life expectancy for 187 countries, 1990–2010: a systematic analysis for the Global Burden Disease Study 2010. Lancet 380, 2144-2162. ( 10.1016/S0140-6736(12)61690-0) [DOI] [PubMed] [Google Scholar]
- 4.Altun Z, Hall D. 2009. Introduction. In WormAtlas. ( 10.3908/wormatlas.1.1) [DOI]
- 5.Bansal A, Zhu LJ, Yen K, Tissenbaum HA. 2015. Uncoupling lifespan and healthspan in Caenorhabditis elegans longevity mutants. Proc. Natl Acad. Sci. USA 112, E277-E286. ( 10.1073/pnas.1412192112) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Hahm JH, Kim S, DiLoreto R, Shi C, Lee SJ, Murphy CT, Nam HG. 2015. C. elegans maximum velocity correlates with healthspan and is maintained in worms with an insulin receptor mutation. Nat. Commun. 6, 8919. ( 10.1038/ncomms9919) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Möller S, et al. 2020. Healthspan pathway maps in C. elegans and humans highlight transcription, proliferation/biosynthesis and lipids. Aging (Albany NY) 12, 12 534-12 581. ( 10.18632/aging.103514) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Fuellen G, et al. 2019. Health and aging: unifying concepts, scores, biomarkers and pathways. Aging Dis. 10, 883-900. ( 10.14336/AD.2018.1030) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Luyten W, et al. 2016. Ageing with elegans: a research proposal to map healthspan pathways. Biogerontology 17, 771-782. ( 10.1007/s10522-016-9644-x) [DOI] [PubMed] [Google Scholar]
- 10.Saul N, Möller S, Cirulli F, Berry A, Luyten W, Fuellen G. 2021. Health and longevity studies in C. elegans: the ‘healthy worm database’ reveals strengths, weaknesses and gaps of test compound-based studies. Biogerontology 22, 215-236. ( 10.1007/s10522-021-09913-2) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Lithgow GJ, Walker GA. 2002. Stress resistance as a determinate of C. elegans lifespan. Mech. Ageing Dev. 123, 765-771. ( 10.1016/S0047-6374(01)00422-5) [DOI] [PubMed] [Google Scholar]
- 12.Brenner S. 1974. The genetics of Caenorhabditis elegans. Genetics 77, 71-94. ( 10.1093/genetics/77.1.71) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Stiernagle T. 1999. Maintenance of C. elegans. In C. elegans: a practical approach (ed. Hope IA), pp. 51-67. Oxford, UK: Oxford University Press. [Google Scholar]
- 14.Kamath RS, Ahringer J. 2003. Genome-wide RNAi screening in Caenorhabditis elegans. Methods 30, 313-321. ( 10.1016/S1046-2023(03)00050-1) [DOI] [PubMed] [Google Scholar]
- 15.Han SK, Lee D, Lee H, Kim D, Son HG, Yang JS, Lee SV, Kim S. 2016. OASIS 2: online application for survival analysis 2 with features for the analysis of maximal lifespan and healthspan in aging research. Oncotarget 7, 56 147-56 152. ( 10.18632/oncotarget.11269) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Restif C, Ibáñez-Ventoso C, Vora MM, Guo S, Metaxas D, Driscoll M. 2014. CeleST: computer vision software for quantitative analysis of C. elegans swim behavior reveals novel features of locomotion. PLoS Comput. Biol. 10, e1003702. ( 10.1371/journal.pcbi.1003702) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Everman ER, Morgan TJ. 2018. Antagonistic pleiotropy and mutation accumulation contribute to age-related decline in stress response. Evolution 72, 303-317. ( 10.1111/evo.13408) [DOI] [PubMed] [Google Scholar]
- 18.Kourtis N, Tavernarakis N. 2011. Cellular stress response pathways and ageing: intricate molecular relationships. EMBO J. 30, 2520-2531. ( 10.1038/emboj.2011.162) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Chen K, Shen W, Zhang Z, Xiong F, Ouyang Q, Luo C. 2020. Age-dependent decline in stress response capacity revealed by proteins dynamics analysis. Sci. Rep. 10, 1-13. ( 10.1038/s41598-020-72167-4) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Johnson AA, Stolzing A. 2019. The role of lipid metabolism in aging, lifespan regulation, and age-related disease. Aging Cell 18, e13048. ( 10.1111/acel.13048) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Zhang J, Bakheet R, Parhar RS, Huang CH, Hussain MM, Pan X, Siddiqui SS, Hashmi S. 2011. Regulation of fat storage and reproduction by Krüppel-like transcription factor KLF3 and fat-associated genes in Caenorhabditis elegans. J. Mol. Biol. 411, 537-553. ( 10.1016/j.jmb.2011.06.011) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Dunning KR, Anastasi MR, Zhang VJ, Russell DL, Robker RL. 2014. Regulation of fatty acid oxidation in mouse cumulus-oocyte complexes during maturation and modulation by PPAR agonists. PLoS ONE 9, e87327. ( 10.1371/journal.pone.0087327) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Spanier B, Lasch K, Marsch S, Benner J, Liao W, Hu H, Kienberger H, Eisenreich W, Daniel H. 2009. How the intestinal peptide transporter PEPT-1 contributes to an obesity phenotype in Caenorhabditits elegans. PLoS ONE 4, e6279. ( 10.1371/journal.pone.0006279) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Sawin ER, Ranganathan R, Horvitz HR. 2000. C. elegans locomotory rate is modulated by the environment through a dopaminergic pathway and by experience through a serotonergic pathway. Neuron 26, 619-631. ( 10.1016/S0896-6273(00)81199-X) [DOI] [PubMed] [Google Scholar]
- 25.Oliveira RP, Porter Abate J, Dilks K, Landis J, Ashraf J, Murphy CT, Blackwell TK. 2009. Condition-adapted stress and longevity gene regulation by Caenorhabditis elegans SKN-1/Nrf. Aging Cell 8, 524-541. ( 10.1111/j.1474-9726.2009.00501.x) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Hung W, Hwang C, Po MD, Zhen M. 2007. Neuronal polarity is regulated by a direct interaction between a scaffolding protein, Neurabin, and a presynaptic SAD-1 kinase in Caenorhabditis elegans. Development 134, 237-249. ( 10.1242/dev.02725) [DOI] [PubMed] [Google Scholar]
- 27.Cordes S, Frank CA, Garriga G. 2006. The C. elegans MELK ortholog PIG-1 regulates cell size asymmetry and daughter cell fate in asymmetric neuroblast divisions. Development 133, 2747-2756. ( 10.1242/dev.02447) [DOI] [PubMed] [Google Scholar]
- 28.Hirose T, Horvitz HR. 2013. An Sp1 transcription factor coordinates caspase-dependent and -independent apoptotic pathways. Nature 500, 354-358. ( 10.1038/nature12329) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Pinto SM, Hengartner MO. 2012. Cleaning up the mess: cell corpse clearance in Caenorhabditis elegans. Curr. Opin Cell Biol. 24, 881-888. ( 10.1016/j.ceb.2012.11.002) [DOI] [PubMed] [Google Scholar]
- 30.Cabello J, Neukomm LJ, Günesdogan U, Burkart K, Charette SJ, Lochnit G, Hengartner MO, Schnabel R. 2010. The Wnt pathway controls cell death engulfment, spindle orientation, and migration through CED-10/Rac. PLoS Biol. 8, e1000297. ( 10.1371/journal.pbio.1000297) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Thorpe CJ, Schlesinger A, Bowerman B. 2000. Wnt signalling in Caenorhabditis elegans: regulating repressors and polarizing the cytoskeleton. Trends Cell Biol. 10, 10-17. ( 10.1016/S0962-8924(99)01672-4) [DOI] [PubMed] [Google Scholar]
- 32.Mansfeld J, et al. 2015. Branched-chain amino acid catabolism is a conserved regulator of physiological ageing. Nat. Commun. 6, 10043. ( 10.1038/ncomms10043) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Engelmann I, Griffon A, Tichit L, Montañana-Sanchis F, Wang G, Reinke V, Waterston RH, Hillier LW, Ewbank JJ. 2011. A comprehensive analysis of gene expression changes provoked by bacterial and fungal infection in C. elegans. PLoS ONE 6, e19055. ( 10.1371/journal.pone.0019055) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.De-Souza EA, et al. 2019. RNA interference may result in unexpected phenotypes in Caenorhabditis elegans. Nucleic Acids Res. 47, 3957-3969. ( 10.1093/nar/gkz154) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Qiu S, Adema CM, Lane T. 2005. A computational study of off-target effects of RNA interference. Nucleic Acids Res. 33, 1834-1847. ( 10.1093/nar/gki324) [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
The datasets supporting this article have been uploaded as part of the electronic supplementary material.