Abstract
Traditional Chinese medicine (TCM) has exerted positive effects in controlling the COVID-19 pandemic. HuaShi XuanFei Formula (HSXFF) was developed to treat patients with mild and general COVID-19 in Zhejiang Province, China. The present study seeks to explore its potentially active compounds and pharmacological mechanisms against COVID-19 based on network pharmacology, molecular docking, and molecular dynamics (MD) simulation. All components of HSXFF were harvested from the pharmacology database of the TCMSP system. COVID-19-related targets were retrieved from using OMIM and GeneCards databases. The herb-compound-targets network was constructed by Cytoscape. The target protein–protein interaction (PPI) network, Gene Ontology (GO), and Kyoto Encyclopedia of Genes and Genomes (KEGG) were performed to discover the potential key target genes and mechanism. The main active compounds of HSXFF were docked with 3C-like (3CL) protease hydrolase and angiotensin-converting enzyme 2 (ACE2). The MD simulation confirmed the binding stability of docking results. The herbs-targets network mainly contained 52 compounds and 70 corresponding targets, including key targets such as RELA, TNF, TP53, IL6, MAPK1, CXCL8, IL-1β, and MAPK14. The GO and KEGG indicated that HSXFF may be mainly acting on the IL-17 signaling pathway, TNF signaling pathway, NF-κB signaling pathway, etc. The molecular docking results indicated that isovitexin and procyanidin B1 showed the highest affinity with 3CL and ACE2, respectively, which were confirmed by MD simulation. These findings suggested HSXFF exerted therapeutic effects involving “multi-compounds and multi-targets.” It might be working through directly inhibiting the virus, improving immune function, and reducing the inflammatory in response to anti-COVID-19. In summary, the present study would provide a valuable direction for further research of HSXFF.
Graphic abstract
Keywords: COVID-19, HuaShi XuanFei formula, SARS-CoV-2, Molecular docking, Network pharmacology
Introduction
Coronavirus disease 2019 (COVID-19) is caused by severe acute respiratory syndrome coronavirus-2 (SARS-CoV-2) [1]. Its global outbreak is attributed to the susceptibility of the population toward virus infection, acting as a continuous and serious threat to public health [2]. According to the World Health Organization, 223 countries or regions have reported 105, 805,951 confirmed cases of COVID-19 by February 8, 2021, including 2,312,278 deaths [3]. However, vaccines have been developed too late for prevention, and there has been no drug to cure SARS-CoV-2 infection.
As the country of the initial COVID-19 outbreak, China has successfully controlled it with multiple active approaches. Traditional Chinese herbal medicine has made great contributions to the prevention and control of the epidemic [4]. According to the traditional Chinese medicine (TCM) theory, therapeutic principles and formulas may differ for different syndromes, regional characteristics, and populations [5]. HuaShi XuanFei Formula (HSXFF) is a cipher prescription to treat COVID-19 at Tongde Hospital of Zhejiang Province (Zhejiang Academy of Traditional Chinese Medicine), which is consists of 6 Chinese herbal medicines: Tetrastigmae Radix (Sanyeqing, SYQ), Schizonepetae Herba (Jingjie, JJ), Saposhnikoviae Radix (Fangfeng, FF), Forsythiae Fructus (Lianqiao, LQ), Lonicerae Japonicae Flos (Jinyinhua, JYH) and Peucedani Radix (Qianhu, QH). This prescription can remove toxins and resolve dampness, ventilate the lung and remove pathogenesis. SYQ was the sovereign drug in this prescription, which is used to treat throat swelling and pain, pneumonia, and fever. It can exert numerous bioactivities, including antivirus, anti-inflammatory, immunoregulatory, enhancing immune function, antibacterial, and antioxidant, etc. [6]. Flavonoids of SYQ, such as kaempferol, quercetin, procyanidin dimmer, and epicatechin had an antiviral effect on H1N1 influenza virus [7]. JYH, another effective ingredient of HSXFF, could lower the release of nitric oxide, IL-6, and TNF-α in macrophages, which is used in multiple Chinese patent medicines [8]. In the clinic, HSXFF can effectively relieve the symptoms of early-stage COVID-19, such as fever, cough, and fatigue [9], but its active compounds and putative mechanisms remain unclear.
Currently, network pharmacology is considered to be a promising method to elucidate herbal formulas [10], screen potential bioactive compounds, discover targets, and explore the mechanisms of TCM formulation [11]. Hence, this novel approach will help to understand the relationship among components in TCM, targets, and diseases. Molecular docking is a significant pathway for drug design, which refers to the process that a small molecule compound is spatially docked into a target receptor and has its binding energy scored [12]. Based on the structures of SARS-CoV-2, 3C-like (3CL) protease hydrolase has been identified as a potential target against COVID-19, because it can inhibit the replication of CoV [13]. Meanwhile, angiotensin-converting enzyme 2 (ACE2) has a great affinity with the spike protein of SARS-CoV-2[14]. Therefore, ACE2 and 3CL were considered as receptors in molecular docking.
Network pharmacology is a key method that integrates computer science and medicine to evaluate the molecular mechanisms of TCM. Several studies have shown that network pharmacology, molecular docking, and molecular dynamic simulation were effective to explore the underlying mechanisms and active compounds of TCM formula [15, 16]. In this study, we aim to discover the active compounds and explore the potential mechanisms of HSXFF to cure COVID-19 by integrating network pharmacology and molecular docking.
Materials and methods
Screening of the potentially active compounds and the related targets in HSXFF
All compounds of HSXFF were authenticated by herb names or chemical composition names in the Traditional Chinese Medicine Systems Pharmacology database and analysis platform (TCMSP, https://tcmspw.com/index.php) [17]. The compounds of HSXFF were screened based on two key parameters of absorption, distribution, metabolism, and excretion (ADME) assessment categories, namely oral bioavailability (OB) and drug-likeness (DL). The potential active compounds in HSXFF were screened with the criterion of OB ≥ 30% and DL ≥ 0.18 [18]. In addition, compound-related targets obtained from the TCMSP database were verified using the UniProt protein database (https://www.uniprot.org/) and converted into their corresponding gene names [19].
Collection of COVID-19 related targets
The targets related to the COVID-19 pathogenesis were acquired by retrieving GeneCards (https://www.genecards.org/), [20] OMIM (http://www.ncbi.nlm.nih.gov/omim) [21], PharmGkb (https://www.pharmgkb.org/) [22], TTD (http://db.idrblab.net/ttd/) [23], and DrugBank database (https://www.drugbank.ca/) [24]. All of the collected targets were merged and had the duplicates deleted to find the unique items to construct the COVID-19 -related target set. The targets of active components in HSXFF and the COVID-19 related targets were mapped by the Venn Diagram data package. The intersection of the two was reputed to find the potential targets of HSXFF in treating COVID-19.
Construction of the herbs-compounds-target network of HSXFF in treating COVID-19
Cytoscape is a universal open-source software for large-scale integrated development of molecular interaction networks [25]. The herb-compounds-target network of HSXFF in the treating COVID-19 was constructed by the Cytoscape software (version 3.8.0), with “herb” being set as a round rectangle, “compounds” being set as an octagon, and “target” being set as a diamond. The cytoHubba plug-in (version 0.1) was employed to calculate and rank the degree of all nodes. The node size in the core network reflects the degree value.
Construction of the protein–protein interaction (PPI) network
String database (http://string-db.org/) covers the majority of known human protein–protein interaction information [26]. The HSXFF and COVID-19 related target sets were imported into the String tool to obtain the PPI network. The organism parameter was set as “Homo sapiens,” and the interaction confidence score was set as high confidence (0.400). The PPI network from String was then imported into CytoHubba plug-in in Cytoscape to investigate the critical subnetworks. Genes were filtered according to their primary score files calculated by CytoHubba, and only those with scores of betweenness centrality (BC), closeness centrality (CC), and degree centrality (DC) higher than the median value were filtered out. This method was applied to construct the critical subnetwork without checking the first-stage nodes.
Gene ontology (GO) and kyoto encyclopedia of genes and genomes (KEGG) pathway enrichment analysis
The “cluster profile” package in R software version 4.0.2 was used to process data and visualize the enrichment results of GO enrichment analysis and KEGG pathway analysis [27]. GO enrichment was conducted on the aspects of molecular function (MF), biological process (BP), and cellular components (CC). Besides, the statistical significance threshold of enrichment analysis was p ≤ 0.05.
Molecular docking prediction
The most significant targets for anti-COVID-19 are SARS-CoV-2 3CL protease hydrolase from 2019-nCoV coronavirus and ACE2 in host cells [28, 29]. Hence, molecular docking simulation between the compounds of HSXFF and the target proteins was performed. The three-dimensional (3D) protein structures of the 3CL hydrolase (PDB ID: 6lu7) and ACE2 (PDB ID: 1r42) were downloaded from the RCSB PDB database (https://www.rcsb.org/) and used as receptors, while the compounds of HSXFF were used as molecular ligands. The mol2 structures of the compounds were downloaded from the TCMSP (https://tcmspw.com/index.php), minimized energy, and transformed into PDB format using ChemBio 3D Ultra 12.0 software. AutoDock Tools 1.5.6 software was used to perform the dehydration, hydrogenation, as well as calculating the charges of the receptor protein [30]. The coordinates and box sizes for molecular docking were finalized according to the key amino acid residues Thr24, Thr26, and Asn119 in SARS-CoV-2 3CL protein [31] and Asp38, Gln42, Gln325, and Glu329 in ACE2 protein [32]. Autodock Vina 1.1.2 was used to dock molecular ligands with SARS-CoV-2 3CL and ACE2 protein, respectively. To achieve higher computational accuracy, 20 exhaustiveness parameters were generated for each receptor. The conformation with the highest affinity was selected as the final docking conformation and visualized in Pymol 2.3. Meanwhile, ritonavir and remdesivir, two potential antiviral drugs for COVID-19, were taken as a positive control [13].
Molecular dynamic (MD) simulation study
The molecular dynamics simulation study was performed using the AMBER18 software package to evaluate the stability and interaction of 3CL and ACE2 receptors with Procyanidin B1 and isovitexin. FF14Sb force field was used to process the protein system, and the GAFF force field was used to process the ligands. ANTECHAMBER module was used to calculate the atomic charge of AM1-BCC. Hydrogen atoms and sodium ions were added to protein with the tleap utility. Solvation of each complex was performed using the TIP3P water model. To start the MD simulation, protein–ligand complexes must be pre-treated as the following: the system was minimized for 10,000 steps in a constant number of atoms. The temperature was set to 300 K and, then a production run for 100 ns in NPT ensemble was performed. The protein–ligand complexes prepared in tleap were analyzed by a CPPTRAJ module [33]. Furthermore, the binding free energy of ligands and proteins was calculated using the MMPBSA.py module.
Results
Identification of potentially active compounds and related targets in HSXFF
In total, 75 potential compounds in HSXFF were retrieved from the TCMSP database with the criteria of OB ≥ 30% and DL ≥ 0.18, and 59 of them were kept after removing components that did not meet the threshold values. These active compounds were originated from SYQ (5 compounds), JJ (4 compounds), FF (11 compounds), LQ (3 compounds), JYH (11 compounds), QH (13 compounds), and 12 common compounds of 6 herbs in HSXFF (Table S1). Meanwhile, 225 target genes of these 59 compounds were acquired after removing the duplicates, 188 in SYQ, 201 in JJ, 85 in FF, 44 in LQ, 220 in JYH, and 196 in QH. The gene names were obtained with the species of “Homo Sapiens” through the UniProt protein database (Table S2).
Prediction results of HSXFF targets in treating COVID-19
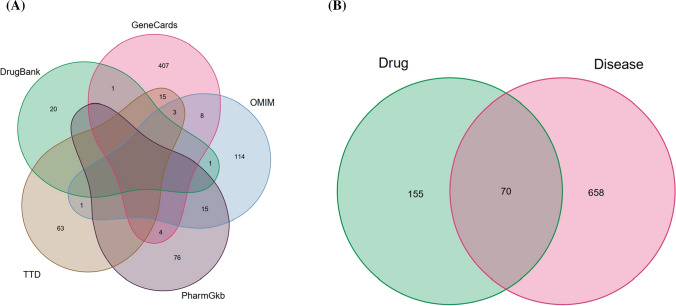
In this study, a total of 728 targets related to COVID-19 were obtained through the keywords of “COVID-19” and “Novel coronavirus pneumonia” in the GeneCards, OMIM, PharmGkb, TTD, and DrugBank databases. Then, the compound targets were mapped to COVID-19-related targets. As a result, 70 targets related to herbal compounds were identified (Fig. 1). These targets were considered as potential targets of HSXFF against COVID-19.
Fig. 1.
a Identification of the COVID-19-related genes by taking a union of all the results from 5 databases. b Venn diagram of drug targets and disease proteins
Herbs-compounds-target genes network analysis
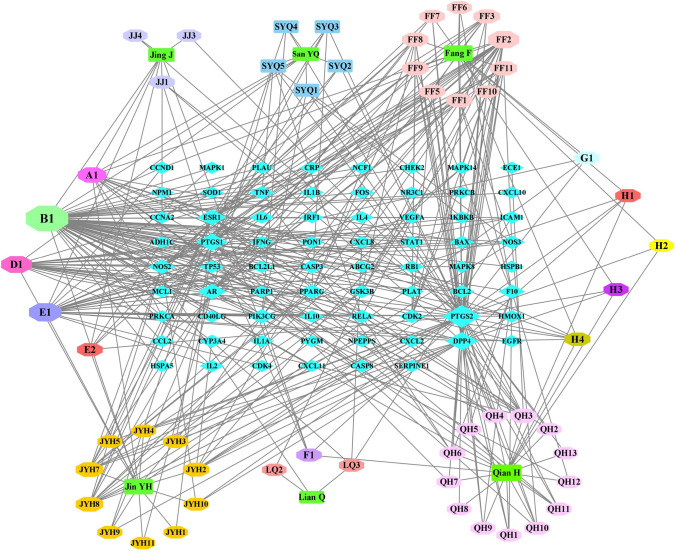
After removing the 7 compounds without targets, the herb-compound-target network was developed, involving 130 nodes (including 6 herbs, 52 compounds, and 70 genes) and 396 edges (Fig. 2). Data analysis showed that the compound nodes had a median DC value of 3, a median CC of 0.3816, and a median BC of 0.0012. Specifically, the three centralities of 46 compounds were higher than the median, and the top five compounds were B1 (MOL000098, quercetin), E1 (MOL000006, luteolin), FF2 (MOL000173, wogonin), D1 (MOL000422, kaempferol), and A1 (MOL000358, β-sitosterol), with 54°, 27°, 21°, 20°, and 14°, respectively. These results suggested that these compounds play a crucial role in HSXFF against COVID-19. Among the 70 targeted genes, 14 had DC, BC, and CC values twice higher than the median, which were PTGS2, DPP4, PTGS1, AR, ESR1, PIK3CG, NOS2, PPARG, NOS3, CDK2, RELA, CASP3, TNF, and BCL2. Therefore, they could serve as the key targets for HSXFF in treating COVID-19.
Fig. 2.
Herbs-Compounds-Target genes network of HSXFF. The round rectangle nodes represent all the herbs of HSXFF, which are surrounded by their particular compounds. The octagon nodes represent the compounds of HSXFF. The diamond nodes, arranged into a rectangular matrix, represent the relative gene targets of HSXFF
Meridian information of herbs in HSXFF
The meridian information of the herbs was used to construct the herb-meridian network. As shown in Fig. 3, organ affinity of the HSXFF herbs is the highest for the lung (5), followed by the liver (3), heart (3), spleen (1), kidney (1), small intestine (1), bladder (1), and stomach (1). Thus, HSXFF may have specific and strong effects in treating lung diseases.
Fig. 3.

The herb-meridian network of HSXFF. The triangle and circular nodes represent the herbs and meridians, respectively. The node size is proportional to the node degree. The lung meridian with the largest degree is 5, followed by the heart, liver, spleen, kidney, small intestine, bladder, stomach meridians in the network, with degree values of 3, 3, 1, 1, 1, 1, and 1, respectively
PPI network analysis
PPI network was established based on the String database. Then, Cytoscape software was adopted to perform network, visualization and topological analysis. This network consisted of 70 nodes and 994 edges. Based on the median DC, CC, and BC values, the top 28 target genes were listed in Fig. 4. Among them, RELA, TNF, TP53, IL6, MAPK1, CXCL8, IL-1β, and MAPK14 were considered to play a relatively important role in the PPI network.
Fig. 4.
a PPI network of HSXFF putative and COVID-19 related target genes. b PPI by Cytoscape: The node color was from pale yellow to red, and the corresponding degree gradually larger. c Barplot showing the significant genes in PPI network
The targets based on the PPI network analysis were compared with those from the HCT network analysis. Twelve of them were found to play a major role in both networks, namely PTGS2, NOS2, PIK3CG, PPARG, RELA, TNF, TP53, IL6, MAPK1, CXCL8, IL1B, and MAPK14. These targets were considered to be the core targets of anti-COVID-19 active component, which also played a key role in the gene regulatory network.
GO and KEGG enrichment analysis
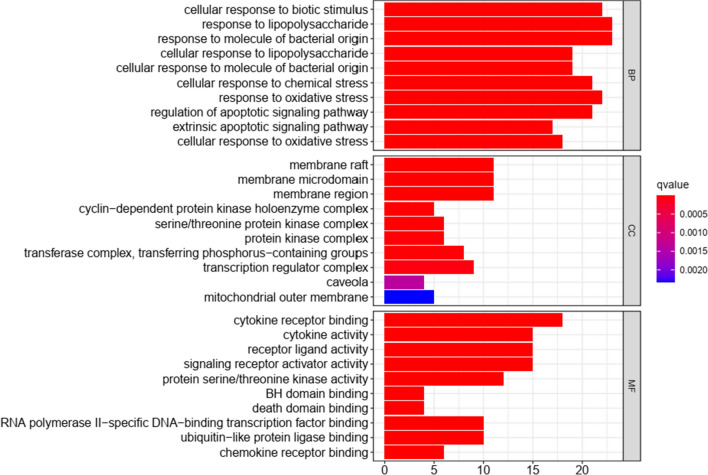
GO annotation analysis included three parts, namely BP, CC, and MF. A total of 1842 GO terms were enriched, and the top 10 terms are shown in Fig. 5. Biological processes mainly include biotic stimulus response, lipopolysaccharide response, bacterial origin response, oxidative stress response, and regulation of apoptotic signaling pathway. The top three cellular component were membrane raft, membrane microdomain, and membrane region. The top three molecular function were cytokine receptor binding, cytokine activity, and receptor-ligand activity.
Fig. 5.
Gene ontology terms of 70 potential targets. The top 10 GO functional terms were selected (p ≤ 0.05). Note: The color of terms turned from blue to red. The redder the bar was, the smaller the adjusted p value was. Abbreviations: BP: biological processes; CC: Cellular Component; MF: molecular function
KEGG enrichment was performed to cluster major pathways associated with HSXFF. A total of 156 KEGG pathways were significantly enriched (p < 0.05), and the top 20 are listed in Fig. 6. Based on the analysis of KEGG pathway data (Table 1), the core pharmacological mechanisms include the IL-17 signaling pathway, TNF signaling pathway, NF-κB signaling pathway, MAPK signaling pathway, and Toll-like receptor signaling pathway.
Fig. 6.
KEGG pathway enrichment of 70 potential targets. The top 20 pathways were identified. Color represented p value and size of the spot represented count of genes
Table 1.
The data of the top twenty KEGG pathway
| ID | Description | p value | p.adjust | q-value | Count |
|---|---|---|---|---|---|
| hsa04933 | AGE-RAGE signaling pathway in diabetic complications | 1.44E−28 | 2.95E−26 | 7.11E−27 | 22 |
| hsa04657 | IL-17 signaling pathway | 7.84E−24 | 8.04E−22 | 1.94E−22 | 19 |
| hsa05167 | Kaposi sarcoma-associated herpesvirus infection | 6.58E−22 | 4.49E−20 | 1.08E−20 | 22 |
| hsa05161 | Hepatitis B | 1.41E−20 | 7.20E−19 | 1.74E−19 | 20 |
| hsa05163 | Human cytomegalovirus infection | 2.02E−20 | 7.20E−19 | 1.74E−19 | 22 |
| hsa05162 | Measles | 2.11E−20 | 7.20E−19 | 1.74E−19 | 19 |
| hsa05142 | Chagas disease | 8.61E−20 | 2.52E−18 | 6.09E−19 | 17 |
| hsa04668 | TNF signaling pathway | 4.60E−19 | 1.18E−17 | 2.85E−18 | 17 |
| hsa05164 | Influenza A | 1.20E−18 | 2.74E−17 | 6.62E−18 | 19 |
| hsa05145 | Toxoplasmosis | 1.62E−17 | 3.32E−16 | 8.02E−17 | 16 |
| hsa05418 | Fluid shear stress and atherosclerosis | 2.07E−17 | 3.86E−16 | 9.31E−17 | 17 |
| hsa05169 | Epstein–Barr virus infection | 2.93E−17 | 5.01E−16 | 1.21E−16 | 19 |
| hsa05133 | Pertussis | 5.13E−17 | 8.09E−16 | 1.95E−16 | 14 |
| hsa05140 | Leishmaniasis | 6.24E−17 | 9.14E−16 | 2.21E−16 | 14 |
| hsa05160 | Hepatitis C | 1.70E−16 | 2.33E−15 | 5.62E−16 | 17 |
| hsa05212 | Pancreatic cancer | 2.12E−15 | 2.71E−14 | 6.54E−15 | 13 |
| hsa05166 | Human T cell leukemia virus 1 infection | 2.64E−15 | 3.19E−14 | 7.69E−15 | 18 |
| hsa04620 | Toll-like receptor signaling pathway | 5.17E−15 | 5.30E−14 | 1.28E−14 | 14 |
| hsa04625 | C-type lectin receptor signaling pathway | 5.17E−15 | 5.30E−14 | 1.28E−14 | 14 |
| hsa04660 | T cell receptor signaling pathway | 5.17E−15 | 5.30E−14 | 1.28E−14 | 14 |
Docking results
The 59 compounds of HSXFF were docking with 3CL and ACE2. Compared with the positive control drugs (ritonavir and remdesivir), 16 compounds showed strong affinity with 3CL, and 12 compounds showed strong affinity with ACE2. The binding energies are shown in Table 2 (3CL protein) and Table 3 (ACE2 protein). Isovitexin was found as the most stable active compound in binding 3CL, while β-sitosterol showed the best affinity with ACE2. Furthermore, procyanidin B1, isovitexin, β-carotene, chrysoeriol, nodakenin, and β-sitosterol all had strong affinities with both 3CL and ACE2.
Table 2.
Binding energies of 16 compounds and positive drug to 3CL potential targets
| Mol ID | Molecule Name | 3CL(kal/mol) |
|---|---|---|
| MOL000004 | Procyanidin B1 | − 7.3 |
| MOL000073 | Epicatechin | − 6.9 |
| MOL002322 | Isovitexin | − 8.2 |
| MOL002881 | Diosmetin | − 6.5 |
| MOL003315 | 3β-Acetyl-20,25-epoxydammarane-24alpha-ol | − 6.5 |
| MOL002773 | β-carotene | − 6.6 |
| MOL002914 | Eriodyctiol | − 6.9 |
| MOL003044 | Chrysoeriol | − 6.6 |
| MOL004792 | Nodakenin | − 6.7 |
| MOL007154 | Tanshinone IIA | − 6.9 |
| MOL013100 | (2S)-2-(1-hydroxy-1-methyl-ethyl)-9-[(2S,3R,4R,5S,6R)-3,4,5-trihydroxy-6-methylol-tetrahydropyran-2-yl]oxy-2,3-dihydrofuro[3,2-g]chromen-7-one | − 7.3 |
| MOL000358 | β-sitosterol | − 7.0 |
| MOL000098 | Quercetin | − 6.5 |
| MOL000422 | Kaempferol | − 6.7 |
| MOL000006 | Luteolin | − 6.7 |
| MOL000449 | Stigmasterol | − 6.7 |
| Positive drug | Ritonavir | − 6.5 |
| Positive drug | Remdesivir | − 6.5 |
Table 3.
Binding energies of 22 compounds and positive drug to ACE2 targets
| Mol ID | Molecule Name | ACE2(kal/mol) |
|---|---|---|
| MOL000004 | Procyanidin B1 | − 9.1 |
| MOL000096 | Catechin | − 8.2 |
| MOL001368 | 3-O-p-coumaroylquinic acid | − 8.3 |
| MOL002322 | Isovitexin | − 8.2 |
| MOL011730 | 11-hydroxy-sec-o-β-d-glucosylhamaudol_qt | − 8.2 |
| MOL011740 | Divaricatol | − 8.2 |
| MOL011747 | Ledebouriellol | − 8.3 |
| MOL003315 | 3β-Acetyl-20,25-epoxydammarane-24alpha-ol | − 8.9 |
| MOL002773 | β-carotene | − 8.7 |
| MOL003044 | Chrysoeriol | − 8.6 |
| MOL004792 | Nodakenin | − 8.6 |
| MOL000358 | β-sitosterol | − 9.5 |
| Positive drug | Ritonavir | − 8.2 |
| Positive drug | Remdesivir | − 8.7 |
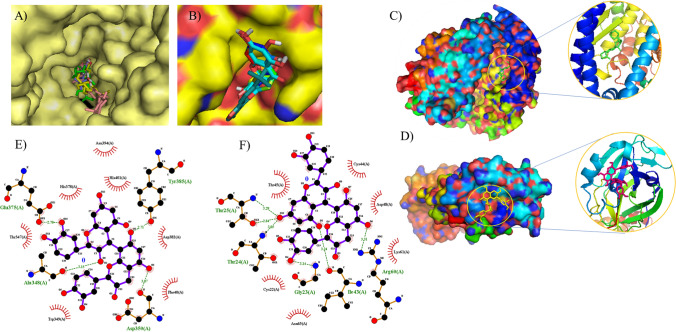
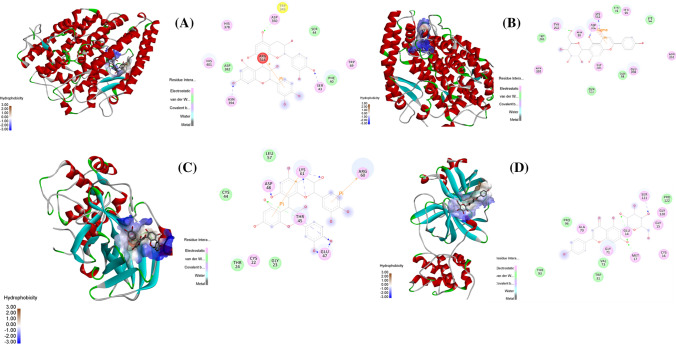
The docking patterns of candidate compounds with 3CL and ACE2 protein are shown in Fig. 7a, b. Procyanidin B1 and isovitexin showed compact binding patterns with the active pockets of two targets (Fig. 7c, d). The binding residues were predicted to explore the drug targets. Procyanidin B1 formed four hydrogen bonds with the amino acid residues Tyr385, Glu375, Ala348, and Asp350, creating a stable complex with ACE2 (Fig. 7e). Isovitexin formed six hydrogen bonds with Thr25, Thr24, Gly23, Ile43, and Arg60, creating a stable complex with 3CL protein (Fig. 7f).
Fig. 7.
The docking complex of active compounds with ACE2 and 3CL protein. a Active compounds in active pocket site of ACE2 protein. b Active compounds in an active pocket site of 3CL protein. c ACE2 protein-procyanidin B1. d 3CL protein-isovitexin. e Procyanidin B1 formed four hydrogen bonds with the amino acid residues of ACE2 protein. f Isovitexin formed six hydrogen bonds with the amino acid residues of 3CL protein
Molecular dynamics simulations results
Based on the interactions with the binding pockets and binding energy calculations (Fig. 7, Tables 2, 3), procyanidin B1 and isovitexin-docked complexes were selected to run MD simulations. To study the stability, root-mean-square deviation (RMSD) was calculated for the initial structures within 100 ns (ns), and each trajectory were found with an average RMSD of 2.0–3.5 Å for ACE2-procyanidin B1 complex (Fig. 8a), 1.5–2.5 Å for ACE2-isovitexin complex (Fig. 8b), 2.0–3.5 Å for 3CL-procyanidin B1 complex (Fig. 8c), and 1.5–2.5 Å for 3CL- isovitexin complex (Fig. 8d), respectively.
Fig. 8.
Molecular dynamics simulation. a RMSD of ACE2-procyanidin B1; b RMSD of ACE2-isovitexin; c RMSD of 3CL-procyanidin B1; d RMSD of 3CL-isovitexin
The binding free energy (ΔGbind) reflects the affinity of a ligand to its target. ΔGbind of ACE2-procyanidin B1, ACE2-isovitexin, 3CL-procyanidin B1, and 3CL-isovitexin complexes were calculated using MM-GBSA implemented in Ambertools 18. The results showed the ΔGbind of the above complexes were − 26.1 ± 0.5 kcal/mol, − 26.0 ± 0.28 kcal/mol, − 24.5 ± 2.2 kcal/mol, and − 26.4 ± 1.1 kcal/mol, respectively.
The analysis of protein–ligand interactions also plays a key role in identifying the target site. The stability of protein–ligand complex is predominantly determined by the hydrogen bonds, which are the major stabilizers of the docked complexes. In the MD simulations, the H-bonds were observed between hydroxy (OH) of procyanidin B1 with ASP350, ASN394, and SER43 residues in ACE2, and a benzene ring formed pi-( +) (cationic conjugated system) with ARG514 (Fig. 9a). The complex of ACE2-isovitexin formed H-bond between hydroxy (OH) of the pyranose sugar, carbonyl (C=O), and OH groups of isovitexin with TYR202, LYS562, and ASP206, respectively, and ASP206 also formed pi–pi interaction with a cyclic ring of isovitexin (Fig. 9b). 3CL-procyanidin B1 complex formed 4 H-bonds, namely those between LYS61 and an ether group (C–O–C), GLU47 and 2 OH groups, ASP48 and an OH group, THR45 and 2 OH groups. ARG60 and LYS61 also formed pi–pi interaction with two benzene rings of procyanidin B1(Fig. 9c). The 3CL-isovitexin complex formed 3 H-bonds, including those between MET17 and an OH group, GLU14 and 2 OH groups, SER121 and an OH group of the pyranose sugar (Fig. 9d). Furthermore, all complexes showed water-mediated interaction.
Fig. 9.
Amino acid interactions of procyanidin B1 and isovitexin in binding cavity of 3CL and ACE2 of SARS-CoV-2. a ACE2-procyanidin B1; b ACE2-isovitexin; c 3CL-procyanidin- B1; d 3CL-isovitexin
In summary, these results proved the validity of the docking results. No effect from the temperature and pressure on the structural conformation was detected.
Discussion
As COVID-19 continues to expand rapidly, the development of effective drugs has become increasingly important. The prescription of HSXFF has been applied in 9 designated COVID-19 hospitals in Zhejiang Province China. It has been proved as an effective treatment for mild and general COVID-19by relieving respiratory symptoms, and ameliorating GI tract symptoms caused by antiviral drugs. In the present study, a compounds-targets network of HSXFF with 52 compounds and 59 targets was constructed. PPI network and critical network analyses found 8 hub targets out of 59 genes. Go and KEGG analysis implicated that HSXFF can regulate the inflammatory response, oxidative stress, and virus defense. Particularly, the 3CL protease of SARS-CoV-2 and ACE2 were investigated in this study, and performed molecular docking to verify the interaction between these two targets and the active compounds of HSXFF.
The MD study was carried out to validate the results of molecular docking, and evaluate the stability and interaction of ACE2 and 3CL with ligands. After 100 ns dynamics simulation, the binding sites of small molecules did not change significantly, indicating that the molecular docking results were reliable and the binding of small molecules remained relatively stable. The results revealed that procyanidin B1 and isovitexin could efficiently activate the biological pathway without changing the conformation of the active site in 3CL and ACE2. On the other hand, the number of hydrogen bonds reflects the stability of protein–ligand complexes. In the MD study, the complexes formed by 3CL, ACE2, procyanidin B1, and isovitexin contained 3–6 hydrogen bonds. This result indicated that the strong interactions of the critical residues (most especially Asp350, SER121, CYS145, THR45, and MET17) with procyanidin B1 and isovitexin will greatly impair the dimerization and substrate binding of SARS-CoV-2 3CL and ACE2.
ACE2 is widely distributed in the human body, and it is one of the necessary receptors of SARS-CoV-2 and key substance leading to lung injury [29]. 3CL plays an important role in cleaving viral polyproteins into functional proteins, making it one of the most attractive drug targets. The inhibition of ACE2 and 3CL is considered an effective strategy to block virus replication [34]. In the present study, several key compounds, such as isovitexin, β-sitosterol, procyanidin B1, tanshinone IIA, quercetin, kaempferol, luteolin, and chrysoeriol, were screened using molecular docking. All these active compounds showed superior or similar affinity compared with ritonavir and remdesivir, two currently recommended drugs for clinical therapies [35]. The herbs-compounds-target genes network also showed similar results that quercetin, luteolin, kaempferol, β-sitosterol, chrysoeriol, tanshinone IIA, and wogonin had higher degree values.
Notably, isovitexin, procyanidin B1, quercetin, luteolin, kaempferol, chrysoeriol, and wogonin are all flavonoids, who participate in anti-inflammatory and antivirus activities [36]. It had been verified that isovitexin inhibits viral replication on mammalian H1N1, avian influenza H5N1, and avian H6N1. It also acts against the cytopathic effect induced by influenza virus infection [37, 38]. One study has suggested that procyanidin exerted inhibitory activity on influenza virus by inhibiting the formation of the Atg5-Atg12/Atg16 Heterotrimer and Beclin1/bcl2 Heterodimer [39]. Quercetin was reported to show virucidal activity, which could prevent or cure murine coronavirus, herpes simplex, influenza virus (H1N1/H3N2/H9N2) infection, and respiratory syncytium [40–43]. Quercetin could also reduce the production of reactive oxygen species (ROS) to mitigate the pro-inflammatory response [44]. Luteolin and chrysoeriol were revealed to show antioxidant, anti-inflammatory, antiviral, and antimicrobial activities[45]. Wogonin had been confirmed to exhibit anti-inflammatory, antitumor, antiviral, neuroprotective, and anxiolytic effects by modulating PI3K-Akt, p53, NF-κB, MAPK pathways [46]. Tanshinone IIA could attenuate LPS-induced neuroinflammation by inhibiting TLR4/NF-κB/MAPKs signaling pathway [47]. Sodium tanshinone IIA sulfonate had been reported to show anti-PRRSV activity by suppressing Atg5, Atg7, and Beclin1 genes in vitro, all of which were related to autophagy [48]. In a word, a large number of studies have suggested the efficacy of HSXFF in preventing COVID-19 with antiviral, antimicrobial, anti-inflammatory, and antioxidant activities.
PPI network was constructed based on the key and core genes. Network analysis showed that RELA, the core target gene, had the largest degree. The other top 7 genes, namely TNF, TP53, IL6, MAPK1, CXCL8, IL-1β, and MAPK14, had high degree values (degree > 14, 2 times higher than the median). RELA is a member of the NF-κB family and acts as an effectual transcriptional activator that activates many inflammatory responses under exposure to pathogens and inflammatory cytokines [49]. The pathophysiological process of SARS-COV-2 infection is a strong inflammatory response. COVID-19 is the result of the spike protein of SARS-COV-2 binding to ACE2, triggering the production of pro-inflammatory cytokines and chemokines [50]. The functions of MAPK1 and MAPK 14 were demonstrated to regulate oxidative stress, inflammatory, immune response, apoptosis, and cell proliferation responses [51]. In clinics, IL-6 content is higher in plasma of COVID-19 patients, and to decrease it is an effective treatment to reduce mortality of patients [52, 53]. Therefore, the anti-COVID-19 effect of the active compounds in HSXFF might be mainly regulated by RELA, TNF, TP53, IL-6, MAPK1, CXCL8, IL-1β, and MAPK14 genes.
GO analysis found that hub target genes were predominantly associated with the biological response processes to biotic stimulus, lipopolysaccharide, the molecules of bacterial origins, oxidative stress, and regulation of apoptotic signaling pathway. Moreover, KEGG enrichment analysis showed that the key targets were mainly concentrated in IL-17, TNF, and Toll-like receptor signaling pathways. Besides, RELA, TNF, TP53, IL-6, MAPK1, CXCL8, IL-1β, and MAPK14 were the hub genes according to the results of PPI network analysis. Therefore, the treatment of HSXFF on COVID-19 might act through IL-17, TNF, NF-κB/MAPK, and Toll-like receptor signaling pathways.
During viral infection, IL-17 enhanced the pro-inflammatory response, which might be crucial to regulate the immunoreaction of SARS-COV-2 infection [54, 55]. IL6, IL-1β, and TNF are involved in TNF, NF-κB, and MAPK signaling pathways, and these pathways also mediated inflammatory responses in infectious diseases [56–58]. Recently, one study showed that several p38 MAPK inhibitors applied in the clinic to cure COVID-19 can inhibit severe inflammation induced by SARS-COV-2 [59]. The candidate compounds of this study, namely isovitexin, chrysoeriol, and tanshinone IIA, have been reported to exert anti-inflammatory and antioxidant activities on lipopolysaccharide-induced inflammatory models by suppressing the MAPK and NF-κB pathways [46, 60, 61]. TLR3, TLR7, and TLR8, subfamily members of Toll-like receptors can also identify RNA [62, 63]. Quercetin, a candidate compound of this study, can inhibit the activation of TLR7-mediated signaling pathways [64].
In conclusion, the prevention and treatment of COVID-19 by HSXFF may be realized by inhibiting the virus directly, improving immune function, enhancing the antioxidative effects, and reducing the inflammatory responses by acting on the several aforementioned signaling pathways. This study provides a holistic view of the potential pharmacological mechanisms for HSXFF, establishing the foundations for further study on the optimization of experimental designs for more reliable results.
Abbreviations
- 3CL
3C-like
- 3D
Three-dimensional
- ACE2
Angiotensin-converting enzyme 2
- ADME
Absorption, distribution, metabolism, and excretion
- BC
Betweenness centrality
- BP
Biological process
- CC
Cellular components
- CC
Closeness centrality
- COVID-19
Coronavirus disease 2019
- DC
Degree centrality
- DL
Drug-likeness
- FF
Fangfeng
- GO
Gene ontology
- HSXFF
HuaShi XuanFei Formula
- JJ
Jingjie
- JYH
Jinyinhua
- KEGG
Kyoto encyclopedia of genes and genomes
- LQ
Lianqiao
- MD
Molecular dynamic
- MF
Molecular function
- NCP
Novel coronavirus pneumonia
- OB
Oral bioavailability
- PPI
Protein–protein interaction
- QH
Qianhu
- RMSD
Root-mean-square deviation
- ROS
Reactive oxygen species
- SARS-CoV-2
Severe acute respiratory syndrome coronavirus-2
- SYQ
Shanyeqing
- TCM
Traditional Chinese Medicine
- TCMSP
Traditional Chinese Medicine Systems Pharmacology Database and Analysis Platform
Authors' contributions
JW, WG designed the study and drafted the manuscript; WG performed the data collection; LY, SH and XF analyzed the data; XP revised the manuscript. All authors read and approved the final manuscript.
Funding
Ningbo City Science and Technology Innovation 2025 Major Research Project (2019B10008), Ningbo Natural Science Foundation Project (2019A610370, 2017A610263), Zhejiang Province Public Welfare Technology Project (LGN18B020001, LGF20H280009), Natural Science Foundation of Zhejiang Province (LQ19C020003), Ningbo Public Welfare Science and Technology Project (2019C50064), the scientific research projects of Zhejiang Pharmaceutical College (2019010, XY2020003), the Science and Technology Program of Fuzhou (2018-N-9).
Availability of data and material
Publicly available databases were analyzed in our study. The active ingredients and putative target genes of HSXFF from TCMSP can be found in http://tcmspw.com/tcmsp.php. COVID-19-related target genes were from GeneCards (https://www.genecards.org/) and OMIM (http://www.ncbi.nlm.nih.gov/omim) databases. Complementary data are supplied as supplementary file.
Declarations
Conflict of interest
The authors declare that they have no conflict of interest.
Footnotes
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Juan Wang and Wen Ge have contributed equally to this work.
References
- 1.Lipsitch M, Swerdlow DL, Finelli L. Defining the epidemiology of Covid-19-studies needed. N Engl J Med. 2020;382(13):1194–1196. doi: 10.1056/NEJMp2002125. [DOI] [PubMed] [Google Scholar]
- 2.Nkengasong J. China’s response to a novel coronavirus stands in stark contrast to the 2002 SARS outbreak response. Nat Med. 2020;26(3):310–311. doi: 10.1038/s41591-020-0816-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.World Health Organization. Novel Coronavirus (COVID-19) Situation dashboard. [cited 2021 February 8]. https://covid19.who.int/.
- 4.Novel Coronavirus Pneumonia Emergency Response Epidemiology Team The epidemiological characteristics of an outbreak of 2019 novel coronavirus diseases (COVID-19) in China. Chin J Epidemiol. 2020;41(2):145–151. doi: 10.1097/MD.0000000000024-315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Li YX, Li J, Zhong Y, Tian YP, Zhang YG, Jin RJ, Guo Y, Clarke M. Clinical practice guidelines and experts’ consensuses of traditional Chinese herbal medicine for novel coronavirus (COVID-19): protocol of a systematic review. Int J Med Sci. 2020;17(16):2511–2530. doi: 10.1186/s13643-020-01432-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Ji T, Ji WW, Wang J, Chen HJ, Peng X, Cheng KJ, Qiu D, Yang WJ. A comprehensive review on traditional uses, chemical compositions, pharmacology properties and toxicology of Tetrastigma hemsleyanum. J Ethnopharmacol. 2021;264:113247–113263. doi: 10.1016/j.jep.2020.113247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Ding FJ, Liu JT, Du RK. Qualitative and quantitative analysis for the chemical constituents of tetrastigma hemsleyanum diels et gilg using ultra-high performance liquid chromatography/hybrid quadrupole-orbitrap mass spectrometry and preliminary screening for anti-influenza virus components. Evid Based Compl Alt Med. 2019 doi: 10.1155/2019/9414926. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Kiyohara H, Ichino C, Kawamura Y, Nagai T, Sato N, Yamada H. Patchouli alcohol: in vitro direct anti-influenza virus sesquiterpene in Pogostemon cablin Benth. J Nat Med. 2012;66(1):55–61. doi: 10.1007/s11418-011-0550-x. [DOI] [PubMed] [Google Scholar]
- 9.Chen LL, Ge GB, Rong Y. Application and research progress of traditional Chinese medicine in prevention and treatment of corona virus disease 2019. Acad J Shanghai Univ Trad Chin Med. 2020;34(3):1–8. [Google Scholar]
- 10.Zhang GB, Li QY, Chen QL, Su SB. Network pharmacology: a new approach for Chinese herbal medicine research. Evid Based Complement Alternat Med. 2013;2013:1–9. doi: 10.1016/S1875-5364(21)60001-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Zhang R, Zhu X, Bai H, Ning K. Network pharmacology databases for traditional Chinese medicine: review and assessment. Front Pharmacol. 2019;10:123. doi: 10.3389/fphar.2019.00123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Saikia S, Bordoloi M. Molecular docking: challenges, advances and its use in drug discovery perspective. Curr Drug Targets. 2019;20(5):501–521. doi: 10.2174/1389450119666181022153016. [DOI] [PubMed] [Google Scholar]
- 13.Tao QY, Du JD, Li XT, Zeng JY, Tan B, Xu JH, Lin WJ, Chen XL. Network pharmacology and molecular docking analysis on molecular targets and mechanisms of Huashi Baidu formula in the treatment of COVID-19. Drug Dev Ind Pharm. 2020;468(8):1–9. doi: 10.1080/03639045.2020.1788070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Shen M, Liu C, Xu R, Ruan ZJ, Zhao SY, Zhang HD, et al. Predicting the animal susceptibility and therapeutic drugs to SARS-CoV-2 based on spike glycoprotein combined with ACE2. Front Genet. 2020;11:575012. doi: 10.3389/fgene.2020.575012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Chen X, Yin YH, Zhang MY, Li JY, Li R, Qu YQ. Investigating the mechanism of ShuFeng JieDu capsule for the treatment of novel coronavirus pneumonia (COVID-19) based on network pharmacology. Int J Med Sci. 2020;17(16):2511–2530. doi: 10.7150/ijms.46378. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Zhuang ZJ, Wen JM, Zhang L, Zhang MJ, Zhong XY, Chen HQ, Luo CJ. Can network pharmacology identify the anti-virus and anti-inflammatory activities of Shuanghuanglian oral liquid used in Chinese medicine for respiratory tract infection? Eur J Integr Med. 2020;37:101139. doi: 10.1016/j.eujim.2020.101139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Ru JL, Li P, Wang JN, Zhou W, Li BH, Huang C, et al. TCMSP: a database of systems pharmacology for drug discovery from herbal medicines. J Cheminform. 2014;6(1):13. doi: 10.1186/1758-2946-6-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Ning K, Zhao X, Poetsch A, Chen WH, Yang JL. Computational molecular networks and network pharmacology. Biomed Res Int. 2017;2017:7573904. doi: 10.1155/2017/7573-904. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.The UniProt Consortium UniProt: the universal protein knowledgebase in 2021. Nucl Acids Res. 2021;45(D1):D158–D169. doi: 10.1093/nar/gkaa1100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Rebhan M, Chalifa-Caspi V, Prilusky J, Lancet D. GeneCards: integrating information about genes, proteins and diseases. TIG. 1997;13(4):163. doi: 10.1016/s0168-9525(97)01103-7. [DOI] [PubMed] [Google Scholar]
- 21.Amberger JS, Bocchini CA, Schiettecatte F, Scott AF, Hamosh A. OMIM.org: online mendelian inheritance in man (OMIM®), an online catalog of human genes and genetic disorders. Nucl Acids Res. 2015;43(1):789–798. doi: 10.1093/nar/gku12-05. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Barbarino JM, Carrillo MW, Altman RB, Klein TE. PharmGKB: a worldwide resource for pharmacogenomic information. Wiley Interdisc Rev Syst Biol Med. 2018;10(4):e1417. doi: 10.1002/wsbm.1417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Chen X, Ji ZL, Chen YZ. TTD: therapeutic target database. Nucl Acids Res. 2002;30(1):412–415. doi: 10.1093/nar/30.1.412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Wishart DS, Feunang YD, Guo AC, Lo EJ, Marcu A, Grant JR, et al. DrugBank 5.0: a major update to the DrugBank database for 2018. Nucl Acids Res. 2020;46(D1):D1074–d1082. doi: 10.1093/nar/gkx1037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Shannon P, Markiel A, Ozier O, Baliga NS, Wang JT, Ramage D, Amin N, Schwikowski B, Ideker T. Cytoscape: a software environment for integrated models of biomolecular interaction networks. Genome Res. 2003;13(11):2498–2504. doi: 10.1101/gr.1239303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Szklarczyk D, Gable AL, Lyon D, Junge A, Wyder S, Huerta-Cepas J, et al. STRING v11: protein-protein association networks with increased coverage, supporting functional discovery in genome-wide experimental datasets. Nucl Acids Res. 2019;47(D1):D607–d613. doi: 10.1093/nar/gky1131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Chen L, Zhang YH, Wang S, Zhang YH, Huang T, Cai YD. Prediction and analysis of essential genes using the enrichments of gene ontology and KEGG pathways. PLoS ONE. 2017;12(9):e0184129. doi: 10.1371/journal.pone.0184129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Zhang L, Lin D, Sun X, Curth U, Drosten C, Sauerhering L, Becker S, Rox K, Hilgenfeld R. Crystal structure of SARS-CoV-2 main protease provides a basis for design of improved α-ketoamide inhibitors. Science. 2020;368(6489):409–412. doi: 10.1126/science.abb3405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Batlle D, Wysocki J, Satchell K. Soluble angiotensin- converting enzyme 2: a potential approach for coronavirus infection therapy? Clin Sci Lond. 2020;134(5):543–545. doi: 10.1042/CS20200163. [DOI] [PubMed] [Google Scholar]
- 30.Morris GM, Huey R, Lindstrom W, Sanner MF, Belew RK, Goodsell DS, Olson AJ. AutoDock4 and AutoDockTools4: automated docking with selective receptor flexibility. J Comput Chem. 2009;30(16):2785–2791. doi: 10.1002/jcc.21256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Liu X, Wang XJ. Potential inhibitors for 2019-nCoV coronavirus M protease from clinically approved medicines. J Genet Genomics. 2020;47(2):119–121. doi: 10.1016/j.jgg.2020.02.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Xu XT, Chen P, Wang JF, Zhou H, Li X, Zhong W, Hao P. Evolution of the novel coronavirus from the ongoing Wuhan outbreak and modeling of its spike protein for risk of human transmission. Sci China Life Sci. 2020;63(3):457–460. doi: 10.1007/s11427-020-1637-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Roe DR, Cheatham TE. PTRAJ and CPPTRAJ: software for processing and analysis of molecular dynamics trajectory data. J Chem Theory Comput. 2013;9(7):3084–3095. doi: 10.1021/ct400341p. [DOI] [PubMed] [Google Scholar]
- 34.Li Q, Kang C. Progress in developing inhibitors of SARS-CoV-2 3C-like protease. Microorganisms. 2020;8(8):1250. doi: 10.3390/molecules26040797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Lu H. Drug treatment options for the 2019-new coronavirus (2019-nCoV) Biosci Trends. 2020;14(1):69–71. doi: 10.5582/bst.2020.01020. [DOI] [PubMed] [Google Scholar]
- 36.Jain AS, Sushma P, Dharmashekar C, Beelagi MS, Prasad SK, Shivamallu C. In silico evaluation of flavonoids as effective antiviral agents on the spike glycoprotein of SARS-CoV-2. Saudi J Biol Sci. 2021;28(1):1040–1051. doi: 10.1016/j.sjbs.2020.11.049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Lo CW, Pi CC, Chen YT, Chen HW. Vigna radiata (L.) R. Wilczek extract inhibits influenza a virus by targeting viral attachment, penetration, assembly, and release. Front Pharmacol. 2020;11:584973. doi: 10.3389/fphar.2020.584973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Howaida AA, Nagat AG, Amal H. Antiviral activity of Aloe hijazensis against some haemagglutinating viruses infection and its phytoconstituents. Arch Pharm Res. 2012;35(8):1347–1354. doi: 10.1007/s12272-012-0804-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Dai JP, Wang GF, Li WZ, Zhang L, Yang JC, Zhao XF, et al. High-throughput screening for anti-influenza A virus drugs and study of the mechanism of procyanidin on influenza A virus-induced autophagy. J Biomol Screen. 2012;175(5):605–617. doi: 10.1177/1087057111435236. [DOI] [PubMed] [Google Scholar]
- 40.Chiow KH, Phoon MC, Putti T, Tan BKH, Chow VT. Evaluation of antiviral activities of Houttuynia cordata Thunb. extract, quercetin, quercetrin and cinanserin on murine coronavirus and dengue virus infection. Asian Pac J Trop Med. 2016;9(1):1–7. doi: 10.1016/j.apjtm.2015.12.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Zhang XX, Wu QF, Yan YL, Zhang FL. Inhibitory effects and related molecular mechanisms of total flavonoids in Mosla chinensis maxim against H1N1 influenza virus. Inflamm Res. 2018;67(2):179–189. doi: 10.1007/s00011-017-1109-4. [DOI] [PubMed] [Google Scholar]
- 42.Wu WJ, Li RC, Li XL, He J, Jiang SB, Liu SW, Yang J. Quercetin as an antiviral agent inhibits influenza a virus (IAV) entry. Viruses. 2015;8(1):6. doi: 10.3390/v8010006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Evers DL, Chao CF, Wang X, Zhang ZG, Huong SM, Huang ES. Human cytomegalovirus- inhibitory flavonoids: studies on antiviral activity and mechanism of action. Antiviral Res. 2005;68(3):124–134. doi: 10.1016/j.antiviral.2005.08.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Lugt TVD, Weseler AR, Vrolijk MF, Opperhuizen A, Bast A. Dietary advanced glycation endproducts decrease glucocorticoid sensitivity in vitro. Nutrients. 2020;12(2):441. doi: 10.3390/nu12020441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Barreca D, Mandalari G, Calderaro A, Smeriglio A, Trombetta D, Felice MR, Gattuso G. Citrus flavones: an update on sources, biological functions, and health promoting properties. Plants (Basel) 2020;93(3):288. doi: 10.3390/plants9030288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Chu Y, Lv XW, Zhang LF, Fu XL, Song SW, Su AR. Wogonin inhibits in vitro herpes simplex virus type 1 and 2 infection by modulating cellular NF-κB and MAPK pathways. BMC Microbiol. 2020;20(1):227. doi: 10.1186/s12866-020-01916-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Jin H, Peng XQ, He YY, Ruganzu JB, Yang WN. Tanshinone IIA suppresses lipopolysaccharide-induced neuroinflammatory responses through NF-κB/MAPKs signaling pathways in human U87 astrocytoma cells. Brain Res Bull. 2020;164:136–145. doi: 10.1016/j.brainresbull.2020.08.019. [DOI] [PubMed] [Google Scholar]
- 48.Sun N, Sun P, Yao M, Khan A, Sun YG, Fan KH, Yin W, Li HQ. Autophagy involved in antiviral activity of sodium tanshinone IIA sulfonate against porcine reproductive and respiratory syndrome virus infection in vitro. Antivir Ther. 2019;241(1):27–33. doi: 10.3851/IMP3268. [DOI] [PubMed] [Google Scholar]
- 49.Ngo KA, Kishimoto K, Davis-Turak J, Pimplaskar A, Cheng Z, Spreafico R, Chen EY, Hoffmann A. Dissecting the regulatory strategies of NF-kappaB rela target genes in the inflammatory response reveals differential transactivation logics. Cell Rep. 2020;30(8):2758–2775. doi: 10.1016/j.celrep.2020.01.108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Tay MZ, Poh CM, Rénia L, MacAry PA, Ng LFP. The trinity of COVID-19: immunity, inflammation and intervention. Nat Rev Immunol. 2020;20(6):363–374. doi: 10.1038/s41577-020-0311-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Li Y, Meng TT, Hao N, Tao HQ, Zou SP, Li MM, et al. Immune regulation mechanism of Astragaloside IV on RAW264.7 cells through activating the NF-κB /MAPK signaling pathway. Int Immunopharmacol. 2017;49:38–49. doi: 10.1016/j.intimp.2017.05.017. [DOI] [PubMed] [Google Scholar]
- 52.Rodríguez Y, Novelli L, Rojas M, Santis MD, Acosta-Ampudia Y, Monsalve DM, et al. Autoinflammatory and autoimmune conditions at the crossroad of COVID-19. J Autoimmun. 2020;114:102506. doi: 10.1016/j.jaut.2020.102506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Xu XL, Han MF, Li TT, Sun W, Wang DS, Fu BQ, et al. Effective treatment of severe COVID-19 patients with tocilizumab. Proc Natl Acad Sci USA. 2020;117(20):10970–10975. doi: 10.1073/pnas.2005615117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Ryzhakov G, Lai CC, Blazek K, To KW, Tracy H, Irina U. IL-17 boosts proinflammatory outcome of antiviral response in human cells. J Immunol. 2011;187(10):5357–5362. doi: 10.4049/jimmunol.1100917. [DOI] [PubMed] [Google Scholar]
- 55.Xu Z, Shi L, Wang Y, Zhang JY, Huang L, Zhang C, et al. Pathological findings of COVID-19 associated with acute respiratory distress syndrome. Lancet Resp Med. 2020;8(4):420–422. doi: 10.1016/S2213-2600(20)30076-X. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Kalliolias GD, Ivashkiv LB. TNF biology, pathogenic mechanisms and emerging therapeutic strategies. Nat Rev Rheumatol. 2016;12(1):49–62. doi: 10.1038/nrrheum.2015.169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Wang SF, Tseng SP, Yen CH, Yang JY, Tsao CH, Shen CW, Chen KH, Liu FT, Liu WT, Chen YMA, Huang JC. Antibody-dependent SARS coronavirus infection is mediated by antibodies against spike proteins. Biochem Bioph Res Co. 2014;451(2):208–214. doi: 10.1016/j.bbrc.2014.07.090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Bode JG, Ehlting C, Häussinger D. The macrophage response towards LPS and its control through the p38 (MAPK)-STAT3 axis. Cell Signal. 2012;24(6):1185–1194. doi: 10.1016/j.cellsig.2012.01.018. [DOI] [PubMed] [Google Scholar]
- 59.Grimes JM, Grimes KV. p38 MAPK inhibition: a promising therapeutic approach for COVID-19. J Mol Cell Cardiol. 2020;144:63–65. doi: 10.1016/j.yjmcc.2020.05.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Lv HM, Yu ZX, Zheng YW, Wang LD, Qin XF, Cheng GH, Ci XX. Isovitexin exerts anti-inflammatory and anti-oxidant activities on lipopolysaccharide-induced acute lung injury by inhibiting MAPK and NF-κB and activating HO-1/Nrf2 pathways. Int J Biol Sci. 2016;121(1):72–86. doi: 10.7150/ijbs.13188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Xie C, Kang J, Li Z, Schauss AG, Badger TM, Nagarajan S, Wu T, Wu XL. The açaí flavonoid velutin is a potent anti- inflammatory agent: blockade of LPS-mediated TNF-α and IL-6 production through inhibiting NF-κB activation and MAPK pathway. J Nutr Biochem. 2012;239(9):1184–1191. doi: 10.1016/j.jnutbio.2011.06.013. [DOI] [PubMed] [Google Scholar]
- 62.Kawai T, Akira S. The role of pattern-recognition receptors in innate immunity: update on toll-like receptors. Nat Immunol. 2010;11(5):373–384. doi: 10.1038/ni.1863. [DOI] [PubMed] [Google Scholar]
- 63.Heil F, Hemmi H, Hochrein H, Ampenberger F, Kirschning C, Akira S, Lipford G, Wagner H. Species-specific recognition of single-stranded RNA via toll-like receptor 7 and 8. Science. 2004;303(5663):1526–1529. doi: 10.1126/science.1093620. [DOI] [PubMed] [Google Scholar]
- 64.Yasui M, Matsushima M, Omura A, Mori K, Ogasawara N, Kodera Y, Shiga M, Ito K, Kojima S. The suppressive effect of quercetin on toll-like receptor 7-mediated activation in alveolar macrophages. Pharmacology. 2015;96(5–6):201–209. doi: 10.1159/000438993. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
Publicly available databases were analyzed in our study. The active ingredients and putative target genes of HSXFF from TCMSP can be found in http://tcmspw.com/tcmsp.php. COVID-19-related target genes were from GeneCards (https://www.genecards.org/) and OMIM (http://www.ncbi.nlm.nih.gov/omim) databases. Complementary data are supplied as supplementary file.