Graphical abstract
Keywords: High-dimensional mediation analysis, Mediation effect, High-throughput genomics studies, Composite null hypothesis testing, Bayesian model, Penalization regression
Abstract
Mediation analysis investigates the intermediate mechanism through which an exposure exerts its influence on the outcome of interest. Mediation analysis is becoming increasingly popular in high-throughput genomics studies where a common goal is to identify molecular-level traits, such as gene expression or methylation, which actively mediate the genetic or environmental effects on the outcome. Mediation analysis in genomics studies is particularly challenging, however, thanks to the large number of potential mediators measured in these studies as well as the composite null nature of the mediation effect hypothesis. Indeed, while the standard univariate and multivariate mediation methods have been well-established for analyzing one or multiple mediators, they are not well-suited for genomics studies with a large number of mediators and often yield conservative p-values and limited power. Consequently, over the past few years many new high-dimensional mediation methods have been developed for analyzing the large number of potential mediators collected in high-throughput genomics studies. In this work, we present a thorough review of these important recent methodological advances in high-dimensional mediation analysis. Specifically, we describe in detail more than ten high-dimensional mediation methods, focusing on their motivations, basic modeling ideas, specific modeling assumptions, practical successes, methodological limitations, as well as future directions. We hope our review will serve as a useful guidance for statisticians and computational biologists who develop methods of high-dimensional mediation analysis as well as for analysts who apply mediation methods to high-throughput genomics studies.
1. Introduction
Advances of various high-throughput biological technologies have revolutionized the field of genomics. In particular, both array-based and sequencing-based techniques have enabled genomics studies to be performed at the genome-wide scale [1], [2], [3], [4], [5], [6], [7], [8], [9], [10], [11], providing unprecedented insights into many fundamental biological questions that are previously impossible to address [7], [12], [13], [14], [15], [16], [17], [18]. These genomics studies produce various molecular-level traits by measuring gene expression profiles and characterizing different covalent modifications of DNA and histone proteins. The molecular-level traits, including both expression and methylation, have been revealed to mediate the effects of DNA, environments and/or behaviors on many diseases and traits [5], [19], [20], [21], [22], [23], [24], [25], [26], [27], [28], [29], [30], [31], and hold the key to understanding the genetic and environmental foundations of disease susceptibility and phenotypic variation.
As one example, genome-wide association studies (GWASs) have recently identified hundreds of thousands of genetic loci associated with complex diseases and traits, but most of the discovered genetic variants are located outside protein-coding regions and are of unknown functions [7], [13], [14], [32]. It has been hypothesized that genetic associations with diseases in many cases might be mediated at the epigenomic level through molecular-level traits [33], [34]. Evidence that supports the mediating role of molecular-level traits includes differential gene expression analyses which have detected plentiful associations between expressions and disease status [35], [36], [37], and expression quantitative trait loci (eQTL) mapping studies, as well as allelic specific expression analyses which have detected associations between expressions of specific transcripts with individual genetic alleles [38], [39]. Evidence also includes other genomics studies which have identified differentially methylated GpG sites or regions with respect to disease statuses [35], [40] and have linked differences in methylation to specific genetic alleles [41]. Therefore, mounting evidence suggests that the molecular-level traits such as expression and methylation can mediate the genetic effects on disease susceptibility.
As another example, many epidemiological studies have also been performed to elaborate the environmental and socioeconomic basis of disease susceptibility. It has been well established that socioeconomic indicators (such as education, income, wealth, and occupation) and overall socioeconomic status (SES)/position (SEP) are associated with cardiovascular disease (CVD) risk [42], [43], [44], [45], [46], [47]; such that CVD incidence, prevalence and mortality are all higher in persons with lower socioeconomic status. The effects of these socioeconomic factors are also confirmed to be mediated, as least in part, through molecular-level traits including methylation, as socioeconomic status/position are associated with changes in DNA methylations [26], [48], [49], [50], [51], [52], which in turn are also related to CVD risk [53], [54]. Therefore, integrating various molecular-level traits from omics studies with data from either GWASs or epidemiological studies has become an important topic in the genomics era. Such integrative analysis can improve our understanding of the molecular basis of complex diseases and traits. Indeed, treating molecular-level traits as mediators and evaluating how they mediate the genetic or socioeconomic effects on disease susceptibility or phenotypic variation has become an important first step towards better characterization of disease etiology and phenotypic distinction [55], [56], [57], [58], [59], [60], [61], [62], [63].
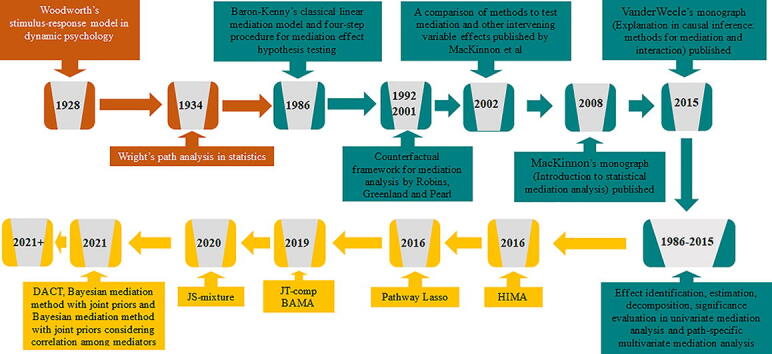
Mediation analysis is a contemporary statistical method that can be employed to elucidate the mediating role of various molecular-level traits in genomics. Conceptually, mediation analysis aims to investigate how an intermediate variable, commonly referred to as a mediator, explains the mechanism or pathway through which an exposure affects an outcome [64], [65]. The rudimentary idea of mediation analysis can be at least dated back to Woodworth’s stimulus–response model in dynamic psychology in 1928 [66] and Wright’s path analysis in statistics in 1934 [67]. Since Baron and Kenny (1986) [68] established the classical statistical formula for mediation analysis, there is a tremendous growth in both methodological development and applications of mediation methods over the past two decades (Fig. 1A), across a wide variety of research areas (Fig. 1B). Mediation analysis is now being routinely carried out in the fields of psychology [69], [70], [71], sociology [65], [72], [73], epidemiology [74], [75], [76], [77], environmental science [19], genetics [78], [79], [80], [81], [82], [83], [84], and appears in a substantial proportion, sometimes more than a third, of research articles published in many disciplines [69], [85]. Various mediation analyses performed thus far have helped establish the foundation of many important psychological and sociological theories. Overall, mediation analysis has become an effective statistical tool for understanding the causal and mediating mechanism underlying the exposure effect on the outcome across a wide range of applications [64], [65].
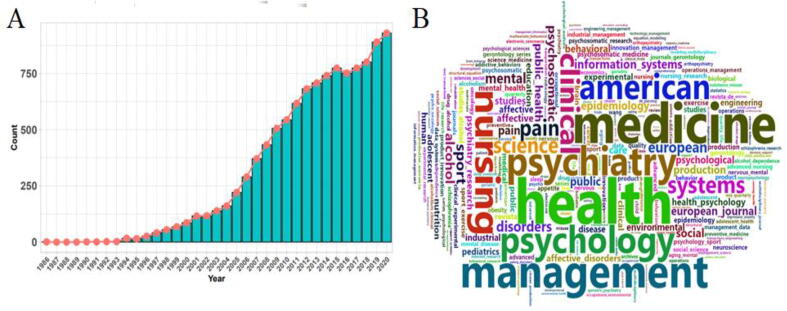
Fig. 1.
(A) Citations of the Baron-Kenny’s 1986 classical mediation analysis article in terms of PubMed retrieval, which reflects the popularity of mediation analysis in biomedical research areas. (B) Word cloud shows the key words of the names of journals that published articles citing the Baron-Kenny’s work, reflecing the diversity of biomedical research displines which employ various mediation methods.
Detailed statistical methodology for mediation analysis is generally constructed under the counterfactual framework, which is also known as the potential outcome framework or Rubin’s model [64], [65], [86], [87], [88], [89], [90], [91], [92], developed in the field of causal statistical inference. The counterfactual framework facilitates methodological establishment for mediation analysis to accommodate different outcome types that include continuous [64], [65], [69], [70], binary [76], [88], [93] and survival outcomes with censoring [74], [80], [92], [94], [95], [96], as well as to account for possible interactions between exposure and mediator [64], [88]. Under the counterfactual framework, mediation analysis makes further modeling assumptions to effectively represent the relationship among the exposure, mediator, and outcome through a directed acyclic graph that links the exposure to the outcome through the mediator (see below) [97], [98]. Mediation analysis then proceeds by decomposing the total effect of the exposure on the outcome into two parts: a direct effect, which represents the exposure on outcome effect not mediated through the mediator; and an indirect effect, which represents the exposure on outcome effect mediated through the mediator. Decomposition of the total effect allows for a mechanistic characterization of the exposure effect on the outcome, facilitating the investigation of causal mediating role of the mediator.
Methodological development for mediation analysis in the past three decades has been primarily focused on univariate mediation analysis where only one mediator is present or multivariate mediation analysis where a few mediators are present [64], [65], [69], [70]. Methods for univariate and multivariate mediation analyses have been thoroughly reviewed by multiple excellent review articles [64], [75], [85], [87], [69], [70], [71], [72], [99], [100], [101], [102], [103], [104], [105], [106], which describe at length the identification, estimation, inference, decomposition and explanation of causal effects in various application fields [65], [68], [70], [85], [86], [107], [108]. Unfortunately, it has become increasingly challenging and often infeasible to directly apply them towards the large and complex data collected in genomics due to several reasons listed as follows [78], [79], [81], [109]. First, the number of potential mediators in the form of molecular-level traits collected in high-throughput genomics studies is in general large, often in the order of thousands to hundreds of thousands, exceeding the collected sample size and thus the capacity accommodated by the univariate and multivariate mediation analyses. For example, the Illumina Infinium HumanMethylation450 BeadChip can array approximately half a million GpG sites but the sample size in methylation studies is typically restricted to be at most a few hundreds or thousands due to heavy experimental costs [78], [79], [81], [109]. Second, genome-wide genomics studies are generally interested in identifying among a large number of potential mediators the ones which exhibit non-zero mediation effects. While univariate mediation methods can perform hypothesis test to examine one potential mediator at a time [70], these methods are often overly conservative and do not fare well for large-scale multiple testing tasks [78], [79], [81], [109]. In particular, the univariate mediation methods rely on asymptotics for hypothesis testing and do not directly accommodate for the composite nature of the null hypothesis as will be detailed below. Third, the potential mediators in genomics are often correlated with each other, sometimes quite strongly. For example, methylation measurements on proximal CpG sites are generally similar to each other and genes in the same pathway also show coordinated co-expression pattern [110]. However, existing univariate and multivariate mediation methods do not explicitly model correlation among potential mediators. Altogether, the above new challenges brought by high-throughput genomics have motivated the intense recent development of high-dimensional mediation methods that aim to accommodate a large number of potentially correlated mediators [78], [79], [81], [82], [96], [109], [111], [112], [113], [114], [115].
Here, we present a thorough literature review on statistical methods that have been developed in recent years for performing high-dimensional mediation analysis in high-throughput genomics studies. A timeline of these methods is shown in Fig. 2. In the review, we begin with the classical univariate and multivariate mediation methods to setup notations and basic statistical formula for mediation analysis. These classical methods include the univariate Baron-Kenny linear mediation model and its extensions to multiple mediators. There, we will introduce the basic concepts, modeling assumptions, effect estimation and decomposition, inference, and significance test for mediation analysis. We will then review recently developed high-dimensional mediation methods for genomics studies that can model thousands of correlated mediations jointly or perform hypothesis tests that can account for the composite nature of the null hypothesis. We discuss their detailed modeling assumptions, important methodological benefits, as well as potential practical drawbacks. We finally conclude our review with future directions for high-dimensional mediation analysis.
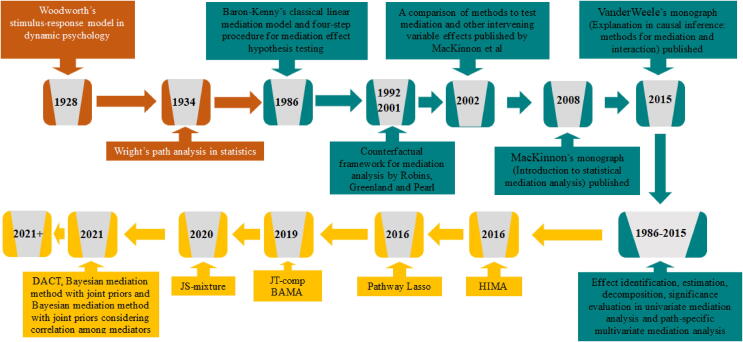
Fig. 2.
Timeline of several key mediation methods developed over the years. Orange color represents initial methods with rudimentary ideas for mediation analysis. Green color represents classical mediation methods developed for univariate and multivariate mediation analysis. Yellow color represents high-dimensional mediation methods targeted for a large number of potential mediators. HIMA: high-dimensional mediation analysis, BAMA: Bayesian mediation method, DACT: divide-aggregate composite-null test. (For interpretation of the references to color in this figure legend, the reader is referred to the web version of this article.)
2. Modeling framework for univariate mediation analysis
We first describe the classical univariate mediation analysis framework [64], [65], [68], [86], [101], [116], also known as Baron-Kenny mediation model [68]. This classical mediation model describes the relationship among a triplet that includes an exposure variable (X), a continuous mediator variable (M), and a continuous outcome variable (Y) (Fig. 3A and 3B)
Fig. 3.
Directed acyclic graph depicting the relationship among an exposure (X), a mediator (M) or multiple mediators (M = (M1, …, Mm)), and an outcome (Y) in the classical mediation analysis. (A) Relationship between the exposure and the outcome, without considering the mediator. Here, c is the total exposure effect on outcome. (B) Relationship between the exposure, the mediator, and the outcome. Here, c′ is the direct effect of exposure on outcome, a is the exposure effect on the mediator, and b is the mediator effect on the outcome. The product of a and b (i.e., ab) represents the indirect/mediation effect. (C) Relationship between the exposure and the outcome with multiple mediators M = (M1, …, Mm). Here, m is the total number of potential mediators; a = (a1, …, am) is a vector of exposure effects on the mediators; and b = (b1, …, bm) is a vector of mediator effects on the outcome. The product of a and b (i.e., aTb) represents the indirect/mediation effects.
where c is the total exposure effect on the outcome; a is the exposure effect on the mediator; c′ is the direct exposure effect on the outcome after controlling for the mediator; b is the mediator effect on the outcome; UX, UM and UXM are confounding effects from known covariates; and eX, eM and eXM are residual errors that are mutually independent of each other. For simplicity of presentation, we assume that the mediator and the outcome are standardized to have mean zero, thus ignoring the intercept terms in the above models.
These effects (i.e., a, b, c and c′) in model can be interpreted in a causal way when the mediation model is correctly specified and certain identifiability assumptions are satisfied [75], [85], [86]. The required identifiability assumptions are known as the sequential ignorability assumptions and are sometimes also referred to as the no unmeasured confounding assumptions. In particular, besides the implicit assumption of temporal ordering between the exposure, mediator and outcome, we assume that all confounders in the three equations are correctly controlled for and that no confounders affecting both the mediator and the outcome are affected by the exposure [72], [78], [101], [103], [104]. While some of these assumptions can be enforced in observational studies, some of them cannot [72], [117], [118], [119]. For example, it is sometimes feasible to control for unmeasured confounders in the exposure-mediator model and in the exposure-outcome model through randomizing the exposure. However, it is challenging to randomize the mediator to control for confounders in the mediator-outcome model. Nevertheless, mediation analysis methods can still be applied for screening purposes, identifying potential biomarkers and genetic regions for further exploration, even when the above causality conditions are not completely satisfied.
3. Extensions of the univariate mediation analysis
The univariate mediation model in has been extended to accommodate a binary mediator and/or a binary outcome [76], [88], [93]. In this case, a logistic regression model is applied in place of the corresponding linear regression to model the exposure-mediator and/or the mediator-outcome relationship. The univariate mediation model has also been extended to accommodate interactions between the exposure and the mediator by adding an interaction term to the mediator-outcome model [75], [88]. In addition, the univariate mediation model has been generalized to accommodate multiple mediators [77], [111], [120], [121], [122]. With m continuous mediators M = (M1, …, Mm) (Fig. 3C), the relationship among the exposure, mediators and outcome can be characterized by the following equations
where a = (a1, …, am) is the m-vector of the exposure on mediator effects; and b = (b1, …, bm) is the m-vector of the mediator on outcome effects. The multivariate mediation model defined in equation can easily accommodate binary mediators and/or binary outcome through logistic regressions. The model effectively treats all mediators en bloc, where the indirect effect is defined as the exposure on outcome effect mediated through at least one of the mediators while the direct effect is defined as the exposure on outcome effect acting around all mediators [64], [65].
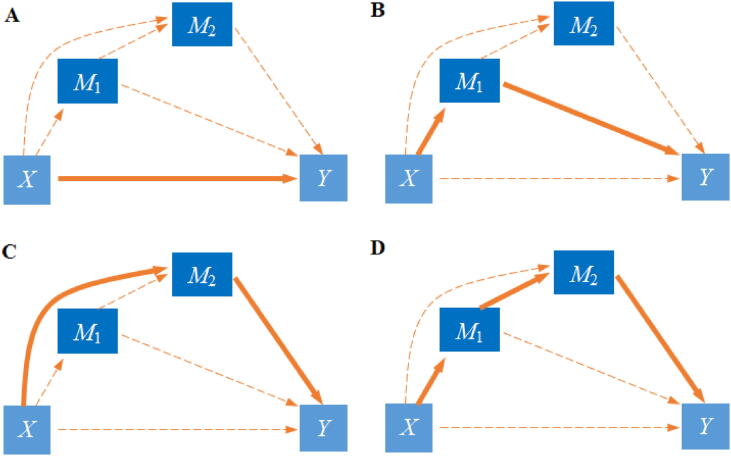
The model is not the only multivariate mediation extension. Indeed, other than treating all mediators en bloc, one can also attempt to incorporate the ordering of mediators into consideration and decompose the total exposure on outcome effect as the sum of separate effects along each possible pathway that consists of a set of ordered mediators [77], [121]. An example of the relationship among the exposure, outcome and two ordered mediators is displayed in Fig. 4. If the detailed ordering of the mediators and their relationship with respect to the exposure and outcome is known, one can perform proper decomposition of mediation effect and potentially interpret the causal mediation effect under the counterfactual framework. Certainly, the ordering of potential mediators is generally unknown in most genomics studies, making the causal interpretation of the mediation estimation challenging. In addition, exploring the exponential number of possible orderings among mediators can quickly become computationally infeasible in high-dimensional settings.
Fig. 4.
Possible relationships among the exposure (X), outcome (Y), and two ordered mediators (M1 and M2). (A) Only the direct effect from the exposure to the outcome is present; neither M1 nor M2 has a mediating role. (B) The indirect effect of the exposure is mediated by M1 only. (C) The indirect effect of the exposure is mediated by M2 only. (D) The indirect effect of the exposure is mediated by M1, followed by M2. The case where the indirect effect of the exposure is mediated by M2 followed by M1 is not displayed. In all panels, the solid line stands for the presence of a relationship while the dot line stands for the absence of a relationship. Here, we ignore the residual terms that are shown in Fig. 3.
Fig. 1. (A) Citations of the Baron-Kenny’s 1986 classical mediation analysis article in terms of PubMed retrieval, which reflects the popularity of mediation analysis in biomedical research areas. (B) Word cloud shows the key words of the names of journals that published articles citing the Baron-Kenny’s work, reflecing the diversity of biomedical research displines which employ various mediation methods.
Fig. 2. Timeline of several key mediation methods developed over the years. Orange color represents initial methods with rudimentary ideas for mediation analysis. Green color represents classical mediation methods developed for univariate and multivariate mediation analysis. Yellow color represents high-dimensional mediation methods targeted for a large number of potential mediators. HIMA: high-dimensional mediation analysis, BAMA: Bayesian mediation method, DACT: divide-aggregate composite-null test.
Fig. 3. Directed acyclic graph depicting the relationship among an exposure (X), a mediator (M) or multiple mediators (M = (M1, …, Mm)), and an outcome (Y) in the classical mediation analysis. (A) Relationship between the exposure and the outcome, without considering the mediator. Here, c is the total exposure effect on outcome. (B) Relationship between the exposure, the mediator, and the outcome. Here, c′ is the direct effect of exposure on outcome, a is the exposure effect on the mediator, and b is the mediator effect on the outcome. The product of a and b (i.e., ab) represents the indirect/mediation effect. (C) Relationship between the exposure and the outcome with multiple mediators M = (M1, …, Mm). Here, m is the total number of potential mediators; a = (a1, …, am) is a vector of exposure effects on the mediators; and b = (b1, …, bm) is a vector of mediator effects on the outcome. The product of a and b (i.e., aTb) represents the indirect/mediation effects.
4. Partition of total effect: Natural direct effect and natural indirect effect
As brought up earlier, the total effect (TE) of the exposure on the outcome can be partitioned into two parts: the natural direct effect (NDE) and the natural indirect effect (NIE); the latter is also known as the mediation effect. The partition of total effect is formally derived under the counterfactual framework [64], [65], [86], [87], [88], [89], [90], [91], [92]. Conceptually, NDE quantifies how much the outcome will change on average when the exposure changes from x0 to x1 but the mediator is fixed at the level it would be in the absence of the exposure. NIE measures how much the outcome will change on average when the exposure is controlled at the level of x1, but the mediator changes from the level it would be at the exposure level of x0 to the level it would be at the exposure level of x1. TE quantifies how much the outcome will change overall for a change of the exposure varying from x0 to x1. In the univariate mediation model with the absence of the interaction effect, NDE equals to c′ while NIE equals ab if assuming x1 = 1 and x0 = 0, which are exactly the same direct and indirect effects obtained through the classical Baron-Kenny mediation analysis [68].
One advantage of partitioning the total effect within the counterfactual framework is that the equation of TE = NDE + NIE holds regardless whether the relationship among exposure, mediator and outcome is linear or non-linear, and regardless whether there is a presence or absence of exposure-mediator interactions. The definition of NDE and NIE in non-linear mediation models for a binary mediator/outcome is also well studied in the literature [72], [75], [76], [88], [100], [103]. There, the mediation effect is no longer in a simple product form when the outcome is binary as the exposure-outcome model and the mediator-outcome model are both expressed as logistic regressions [93], [123], [124], [125]. However, an approximation exists. That is, in the mediation analysis with a binary outcome, in the absence of the interaction effect, one can adopt the approximate log-scale formula to express the log transformed NIE as log(NIE)≈ab, which is an effective approximation when the binary outcome is rare in the population [75], [76], [81].
During the partitioning of the total effect, one can also calculate two ratio quantities: the ratio of the mediation effect to the total effect, PM = θ/c = ab/(ab + c′), which quantifies the proportion of the exposure effect on the outcome mediated by the mediator; and the ratio of the mediation effect to the direct effect, PD = θ/c′=ab/c′, which quantifies the relative strength of the direct and indirect effects [101], [126]. These ratio quantities are defined on the latent variable scale when the outcome is binary [127], [128], [129], [130]. A potential drawback of the two quantities is that they can only be estimated accurately when the sample size is large (e.g., >5000 as demonstrated by simulations in [126]) and are only meaningful when ab and c (or c′) have the same sign. When ab and c (or c′) have the opposite signs, the two ratio quantities may exceed 100% or become negative, making interpretation challenging [126]. Therefore, PM and PD are only recommended to be reported in practical mediation analysis when the calculated values are reasonable [71], [85], [131].
5. Methods for testing mediation effect in univariate mediation analysis
5.1. Testing for mediation effect and the composite nature of the null hypothesis
The significance test of the mediation effect, commonly referred to as the mediation test, is of great scientific interest in many application areas. Baron and Kenny (1986) [68] initially advocated on carrying out the mediation test in a four-step procedure, where the next test step is proceeded only when the test in the previous step is significant. The four steps include: (i) test whether there is a presence of a non-zero total effect (H0: c = 0) in the exposure-outcome model; (ii) test whether the exposure is associated with the mediator (H0: a = 0) in the exposure-mediator model; (iii) test whether the mediator is associated with the outcome after controlling for the direct effect of the exposure (H0: b = 0) in the mediator-outcome model; and (iv) test whether the mediator fully mediates the exposure on outcome effect (H0: c′=0).
Because the four-step procedure requires a series of statistical tests that each has a different statistical power, its results from different steps are not always consistent with each other [69], [70]. For example, when the direct effect c′ is in the opposite sign of the indirect effect ab and both effects are comparable to each other in magnitude, it is possible that the total effect c is not significantly different from zero but the direct effect c′ is, resulting in suppression or inconsistent mediation [70], [71], [88], [132], [133]. To avoid results inconsistency, most studies have recommended on testing the mediation effect directly through the null hypothesis H0: ab = 0, based on steps (ii) and (iii) [71], [88]. The last step of testing c′ can sometimes also be beneficial as it facilitates the further interpretation of the mediation test results: complete or perfect mediation occurs when c′=0 [68] while partial mediation occurs when c′≠0 [68], [70], [88].
Direct hypothesis test on the mediation effect based on H0: ab = 0, however, turns out to be challenging [70], [78], [81], [109], [112]. In particular, such test is complicated by the composite nature of the null hypothesis, which corresponds to three sub-null scenarios
where H01 represents an absence of the exposure on mediator effect but a presence of the mediator on outcome effect; H10 represents a presence of the exposure on mediator effect but an absence of the mediator on outcome effect; and H00 represents an absence of both the exposure on mediator effect and the mediator on outcome effect. Ignoring the composite nature of the null hypothesis testing can lead to uncalibrated test statistics [70], [78], [81], [109], [112].
5.2. Sobel test
Many methods have been previously developed to evaluate the mediation effect based on H0: ab = 0 [70], [81], [126]. Among them, the Sobel test and the joint significance test (JST) are two most common ones, though both yield conservative p-values (more details in Section 5.4). The Sobel test, also known as the product method, assesses the mediation effect ab directly as a product of the exposure on mediator effect and the mediator on outcome effect [68], [134]. The Sobel test depends on the following statistic
where and are the estimates of a and b obtained in the exposure-mediator model and mediator-outcome model, respectively; and are the corresponding standard errors; is the point estimate of mediation effect ab; is the corresponding standard error obtained through the multivariate delta method [68], [123], [134]; and z is the resulting Sobel test statistic. The Sobel test assumes that the test statistics z follows a standard normal distribution and obtains a p-value from the z statistics accordingly. However, the standard normal distribution is invalid even under H00 as the multivariate delta method to arrive at equation does not hold, implying that the Sobel test is not appropriate under all null scenarios listed in (more details in Section 5.4). Other alternative formulas for computing also exist [126] and these include the second-order exact solution [68], [134], [135] as well as the Goodman estimator [136].
It assumes that the Sobel z-statistic in asymptotically follows a standard normal distribution and thus can be further converted to a P value [68], [134], [137]. Confidence intervals for the mediation effect estimate can also be constructed under the same asymptotic normality assumption in the form of (, ). Because of the composite nature of the null, however, the asymptotic normality of the Sobel z-statistic is not guaranteed even when both and respectively follow a normal distribution. Indeed, simulations have shown that the symmetric confidence intervals often generate asymmetric error rates and are generally conservative, especially when the sample size and mediation effect are small, resulting in uncalibrated type I error control and reduced power [70], [71], [123].
5.3. Joint significance test
JST, also known as the causal path method or the maximum P-value method, is another popular traditional mediation test [138]. JST proceeds by performing two separate tests: one for a in the exposure-mediator model and the other for b in the mediator-outcome model. Through the two tests, JST obtains two z-statistics ( and ) and converts them into two P values (Pa and Pb) based on an asymptotic standard normal null distribution. Afterwards, JST obtains the maximum value among the two P values, Pmax = max(Pa, Pb), to serve as the mediation effect test statistic [70]; then it deems the mediation effect to be significant at a level α if and only if Pmax is significant at the level α (i.e., Pmax < α) [70], [139], [140]. JST is essentially an intersection–union test (IUT) and a level-α test with type I error guaranteed to be at most α. Due to the composite nature of the null, however, type I error generated from JST is also conservative and equals α only under certain regularity conditions [141], [142], [143].
5.4. Conservativeness of the Sobel test and the joint significance test
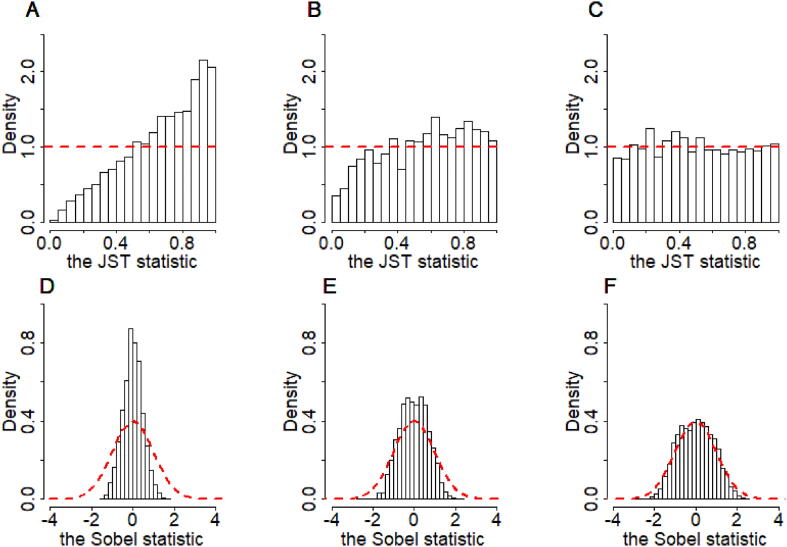
Extensive simulations have been previously conducted to assess the performance of the Sobel test and JST under different sub-null scenarios [70], [81]. For JST, it has been shown that it is extremely conservative under H00 as the true null distribution for Pmax is no longer a uniform distribution there but skewed towards to one (Fig. 5A). Indeed, Pmax follows Beta(2,1) under H00 [109]. In addition, the uniform distribution of Pmax under H01 or H10 holds only under some regularity conditions (Fig. 5B-C). For example, the uniform distribution of Pmax holds under H10 only when the null hypothesis testing of a = 0 against a ≠ 0 is dominantly more powerful as compared to the test of b = 0, which guarantees a Pa consistently smaller than Pb [81], [109], [112]. For the Sobel test, it has also been demonstrated that it is underpowered under H00 because the null distribution of the Sobel test statistic does not follow a standard normal distribution even with large sample sizes (Fig. 5D) [70], [78], [81], [126]. In addition, the Sobel test statistic follows a standard normal distribution under H01 or H10 only when the exposure on mediator effect size or the mediator on outcome effect size is far from zero, respectively (Fig. 5E-F).
Fig. 5.
Distribution of the JST statistic (top: A-C) and the Sobel test statistic (bottom: D-F) under three sub-null scenarios. Simulations were performed based on model under H00: a = 0 and b = 0 (A and D); under H10: b = 0, with a relatively weak exposure-to-mediator effect a = 0.05 (B and E); or under H10: b = 0, with a relatively strong exposure-to-mediator effect a = 0.10 (C and F). In all scenarios, we set c′=0.50 and draw the exposure X and the residuals for the exposure-mediator model and the mediator-outcome model from independent standard normal distributions. The sample size was set to 103. For each dataset consisting of simulated exposure, mediator, and outcome, we separately estimated and inferred a or b in the exposure-mediator model or in the mediator-outcome model using the ordinary least squares estimation procedure. Afterwards, we obtained , and Pa as well as , and Pb, based on which we obtained the Sobel test statistic and the JST statistic. In each simulation scenario we ran 104 replicates. The red dashed line in each panel represents the asymptotic distribution: uniform for A-C and standard normal for D-F. (For interpretation of the references to color in this figure legend, the reader is referred to the web version of this article.)
Because the asymptotic distribution for the Sobel test statistic or JST statistic is challenging to obtain accurately in an analytic form, various bootstrap sampling approaches, including parametric, nonparametric, and bias-corrected versions, have been proposed to obtain their empirical null distributions [71], [140], [144], [145], [146], [147], [148], [149], [150]. Constructing an empirical null distribution through sampling has been shown to lead to accurate confidence intervals and P value in certain settings but not all settings [81]. In addition, constructing the empirical null distribution remains difficult if one does not know the accurate proportions of the three sub-null scenarios [78], [109]. We will discuss these details in the high-dimensional mediation analysis section.
6. Mediation analysis approaches in the presence of high-dimensional mediators
The univariate and multivariate mediation methods described in the previous sections, unfortunately, are not directly applicable for performing mediation effect test in the presence of high-dimensional mediators collected from high-throughput genomics studies. In particular, when the number of mediators exceeds the sample size (i.e., m≫n), the multivariate mediation model defined in becomes unidentifiable, making it infeasible to detect active mediators through joint mediator modeling. In addition, examining one mediator at a time using the traditional univariate mediation model is not feasible either, as the P values from the univariate methods are not calibrated due to the composite nature of the null hypothesis. Because of these drawbacks of univariate and multivariate methods, many high-dimensional mediation methods have been recently developed to model high-dimensional mediators (Table 1). These recent mediation methods can be generally classified into three methodological categories. The first category of mediation methods performs dimension reduction or mediator screening on the high-dimensional mediators to extract a set of low-dimensional variables. Afterwards, they directly apply standard univariate or multivariate mediation methods to these extracted low-dimensional variables to detect active ones that are involved in mediation. The second category of mediation methods examines either one or a few mediators that are within a genomic testing unit (e.g., a gene) one at a time. However, different from standard testing approaches, these new methods explicitly account for the composite nature of the null mediation effect hypothesis to obtain calibrated test statistics. The third category of mediation methods models all potentially mediators jointly in the mediation model by specifying additional modeling assumptions on the mediator on outcome effects as well as on the exposure on mediator effects to ensure model identifiability. We describe the three categories of high-dimensional mediation methods in the following sections.
Table 1.
Summary of statistical methods for mediation analysis in the presence of multiple or high-dimensional mediators.
| First category: Mediation methods based on dimension reduction or mediator screening | |||
|---|---|---|---|
| Methods | Test Statistics | Null Distribution | References |
| correlation-based method | Pmax | permutation | [120] |
| Huang-Pan method | marginal and component-wise ME based on PCA | Monte Carlo (normal-based or bootstrapping) | [122] |
| causal inference test (CIT) | Pmax | permutation | [157] |
| direction of mediation | PCA-based | bootstrapping | [158] |
| MCP-subset | Pmax | screening followed by multiple comparison procedure | [106] |
| MCP-subset based on Westfall-Young | Pmax | screening followed by multiple comparison procedure | [106] |
| MCP-subset based on multivariate | Pmax | screening followed by multiple comparison procedure | [106] |
| HDMA | Pmax | screening followed by debiased estimation | [159] |
| gHMA# | ACAT combining gHMA-L and gHMA-NL | screening followed by multiple comparison procedure | [160] |
| global test + ScreenMin# | Pmin followed by Pmax | screening followed by multiple comparison procedure | [161] |
| Second category: Mediation methods accounting for the composite nature of the null | |||
| Methods | Test Statistics | Null Distribution | References |
| JTV-comp# | mixture of multiple-mediator based P value without estimating the proportions | composite null | [79] |
| JT-comp | mixture of single-mediator based P value without estimating the proportions | composite null | [78] |
| DACT | mixture of single-mediator based P value with estimated proportion | composite null | [109] |
| JS-mixture | mixture of single-mediator based P value with estimated proportion | composite null | [112] |
| Third category: Penalization-based mediation regression methods and Bayesian mediation methods | |||
| Methods | Prior Effects Assumptions | Optimization Procedure | References |
| pathway Lasso | penalization based method | ADMM | [162] |
| HIMA | Pmax | screening followed by minimax concave penalty estimation | [111] |
| BAMA | spike-and-slab prior | MCMC | [163] |
| BAMA with joint priors | Gaussian mixture prior and, product threshold Gaussian prior | MCMC | [164] |
| BAMA with joint priors considering correlation among mediators | the Potts prior and logistic normal prior | MCMC | [165] |
Note: we focus primarily on methodological papers and have not listed applied work that employs mediation methods similar to these listed above (e.g., Wu et al. (2018) [166], Luo et al. (2020) [96]). In addition, we only focus on methods that aim to detect active mediators and have not listed mediation methods for effect estimation and decomposition (e.g., VanderWeele and Vansteelandt (2014) [77], Daniel et al. (2015) [121], Huang and Yang (2017) [92], Steen et al. (2017) [167], Taguri et al. (2018) [168], Zhou et al. (2020) [115], and Zhao et al. (2020) [114]. ADMM: alternating direction method of multipliers; MCMC: Markov chain Monte Carlo; BAMA: Bayesian medaition analysis method; gHMA: gene based high-dimensional mediation analysis; PCA: principal component analysis; MCP: multiple comparison procedure; DACT: divide-aggregate composite-null test; HDMA: high-dimensional mediation analysis; ACAT: aggregated Cauchy association test. Above, # denotes a gene-centric mediation method.
Certainly, one common challenge for all high-dimensional mediation analysis methods is on establishing the causal interpretation of mediation effect. Mediation effects in high-dimensional mediation analysis can be causally interpreted when the required sequential ignorability assumptions hold [78], [79]. However, these sequential ignitability assumptions can be generally challenging to establish in practice as they require additional biological knowledge. For example, to investigate the role of DNA methylations in mediating the effect of smoking behavior on gene expression [79], one needs to assume a priori that smoking can lead to methylation alterations, which in turn can regulate gene expression [151], [152], [153], [154]. Such assumption may be violated as the expression of certain genes can sometimes influence methylation, thus potentially reversing the order of mediator and outcome. In addition, due to the consequence of global epigenetic remodeling mechanism [155], [156], altered DNA methylation may simply represent a passenger event rather than a mediation event. In particular, the expression of key genes may influence the outcome directly while at the same time affects DNA methylation at multiple CpG sites as a by-product. Therefore, when studying the role of gene expression in mediating the impact of DNA methylation on an outcome, it is important to distinguish passenger methylation events from mediation methylation events that likely exert a direct or indirect effect on the outcome. Distinguishing between passenger events and mediation events will also require additional domain knowledge.
6.1. High-dimensional mediation methods based on dimension reduction or mediator screening
The first category of methods attempts to apply multivariate mediation methods directly on a suitable set of potential mediators or a transformed version of mediators that have a dimensionality below the sample size (Table 1). This category primarily includes gene-centric methods such as gene high-dimensional mediation analysis (gHMA) [160], principal component analysis (PCA) based methods [122] as well as several mediator-screening based methods [111]. In brief, gHMA examines one gene at a time and perform mediation analysis on the potential mediators that reside within the examined gene together. Such gene-centric analysis limits the number of analyzed potential mediators to be those within the gene, thus making the standard multivariate mediation methods applicable. Alternatively, the PCA-based mediation method aims to transform the original set of high-dimensional mediators into a low-dimensional space and then analyzes the resulting low-dimensional components using standard multivariate mediation analysis. Finally, a group of mediation methods perform variable screening on mediators to select a subset of potentially active mediator candidates. These methods then apply multivariate mediation methods to analyze these mediator candidates. We describe all these methods in detail below.
6.1.1. Gene-centric mediation methods
The gHMA method adapts linear or non-linear kernels to characterize the relationship between multiple mediators and the outcome [160]. Specifically, the linear-version of gHMA (gHMA-L) examines one gene at a time and focuses on potential mediators that reside in the gene. For the gene of interest, gHMA tests the significance of the exposure on mediator effect (i.e., a in Equation) for each mediator using a univarate mediation model and obtain a corresponding P value; then it combines the P values aross all mediators in that gene using a new Fisher’s combination method that accounts for correlation among P values [169]. Afterwards, gHMA evaluates the significance of multiple mediator on outcome effects (i.e., b in Equation) through the likelihood ratio test, and further integrates the evidience of testing a and b through the same principle of JST. Besides the linear version gHMA-L, gHMA also provides an alternative version gHMA-NL that accommodates non-linear relationship between the mediators and the outcome using kernel principal components (KPC) [160]. The P values from the linear and non-linear versions of gHMA can be further combined through the ACAT (aggregated Cauchy association test) procedur to achieve robust power across various application scenarios while accounting for positive dependence among test statistics of gHMA-L and gHMA-NL [170], [171].
6.1.2. PCA-based mediation methods
Huang and Pan (2016) [122] proposed to first transform the potentially correlated mediators into independent ones based on PCA. Afterwards, Huang-Pan’s method performs multivariate mediation analysis on the resulting principal components using Monte-Carlo resampling. Huang-Pan’s method of testing for marginal mediation effects is equivalent to the integrative statistical framework proposed in Zhao et al (2014) [172]. A similar PCA-based dimension reduction strategy was also applied for mediation analysis in neuroimaging datasets [66]. While the dimension reduction-based mediation analysis effectively addresses the high number of potential mediators and the correlation among them, it is not always easy to interpret the results from the subsequent mediation analysis — after all, the transformed variables are a linear combination of the original mediators and may not have direct biological interpretation. Subsequently, the PCA-based mediation analysis [114] is recently extended to rely on sparse PCA [173] for dimension reduction, which improves the interpretability of the obtained principle components and subsequent mediation analysis results.
6.1.3. Screening-based mediation methods
Finally, several mediation methods perform mediator screening and include only potentially active mediators to the multivariate mediation model for final analysis (Table 1). For example, borrowing ideas in replicability analysis [174], Sampson et al. (2018) first selected mediators that had potential mediating effects based on the marginal mediator-outcome model [106]. The selected mediators were then analyzed in a multiple comparison procedure to ensure correct control of familywise error rate (FWER) or false discovery rate (FDR). Similar screening strategy is also employed by the penalization-based mediation analysis in the third category of high dimensional mediation methods discussed below.
6.2. High-dimensional mediation methods accounting for the composite nature of the null hypothesis
The second category of high-dimensional mediation methods perform hypothesis testing by examining one potential mediator at a time (or a set of potential mediators within a testing unit, one unit at a time) (Table 1). Different from standard univariate methods such as the Sobel test or JST, however, these methods borrow information across all potential mediators to infer key parameters of the three sub-null scenarios as shown in , thus allowing for the computation of calibrated P values. Calibrated P values from these methods lead to correct type I error control across all potential mediators and are essential for detecting promising mediators at the genome-wide significance threshold. Four methods belong to this category, including JT-comp [78], DACT (divide-aggregate composite-null test) [109], JS-mixture [112], and JTV-comp [79]. The first three examine potential mediators one at a time, while the last one is a gene-centric method.
6.2.1. JT-comp
JT-comp is the first method that attempts to accomodate the composite nature of the null hypothesis for testing mediation effect in high-dimensional setting [78]. JT-comp examines one mediator at a time. For each mediator in turn, JT-comp calculates two z-statistics ( and ) by performing two separate tests: one for a in the exposure-mediator model and the other for b in the mediator-outcome model. Afterwards, it derives the null distribution of the product of za and zb by carefully examining the two z-statistics under the three sub-null scenarios. Specifically, under H01, za follows a standard normal distribution while zb follows a normal distribution N(μb,1) with the mean parameter μb characterized by the mediator on outcome effect b. Under H10, zb follows a standard normal distribution while za follows a normal distribution N(μa,1) with the mean parameter μa characterized by the exposure on mediator effect a. Under H00, both za and zb asymptotically follow the standard normal distribution. Therefore, one can compute the P value for the product of za and zb under H00 directly, though computing the P value under either H01 or H10 would require knowing μa and μb which rely on the true effects a and b. JT-comp overcomes the difficulty of unknown μa and μb by making additional assumptions on how they distribute across all potential mediators. In particular, it assumes that μa follows a normal distribution N(0,τa) and μb follows another normal distribution N(0,τb). Consequently, if τa and τb are known, then becomes a product of two independent standard normal distributions under H01 and becomes a product of two independent standard normal distributions under H10. As a result, one can obtain a P value under each of the three sub-null hypotheses if τa and τb are given. If one knows further the probability of each sub-null hypothesis, then one can compute a final P value for testing H0: ab = 0 as [78]
where π01, π10, and π00 represent the probabilities of H01, H10, and H00, respectively; is the right-sided tail probability of a normal product distribution evaluated at point z, with f(x) = K0(x)/π (-∞<x<∞) being the probability density function of the normal product distribution and K0(x) being the modified Bessel function of the second kind with order 0. Note that, computing the P value based on requires estimating the two variance parameters (τa and τb) and the three probability parameters (π01, π10, and π00). Estimating these parameters individually through, for example mixture modeling, may not be accurate. To circumvent the difficulty of estimating each of these parameters individually, JT-comp makes further modeling assumptions. In particular, it assumes that τa and τb are relatively small and that the mediation signals are sparse [78], so that the sample variance of za and zb across mediators, var(za) and var(zb), are related to the individual parameters through var(za) = 1 + π10τa and var(zb) = 1 + π01τb. With these additional assumptions, JT-comp can now approximately compute the final P value for each mediator through
Extensive simulations have shown that the JT-comp testing procedure described above is more powerful than JST and maintains calibrated type I error control when the JT-comp approximation assumptions hold [78]. A drawback of JT-comp, however, is that it may fail to control for type I error well when the sample size is large (e.g., n > 500) and var(za) or var(zb) is>1.5 [78], [109]. A large sample size makes the two assumptions of JT-comp — small effect sizes of a and b characterized by small τa and τb as well as the sparse mediation effects — harder to satisfy in practice.
6.2.2. DACt
DACT is another mediation method that attempts to borrow information across mediators to produce calibrated P values [109]. Different from JT-comp, DACT relies on a modified test statistic and directly estimates the proportions of the three sub-null hypotheses across whole genome mediators. Like JT-comp, DACT examines one potential mediator at a time and performs the same two tests explained before: one for a in the mediator-exposure model and the other for b in the outcome-mediator model. Afterwards, DACT calculates two P values, Pa and Pb, from these two tests based on asymptotic normality. Intuitively, Under H01, if the mediator-to-outcome effect is non-zero (i.e., b ≠ 0), then one only needs to test H01: a = 0 and uses Pa for assessing the significance of meditation effect. Under H10, if the exposure-to-mediator effect is non-zero (i.e., a ≠ 0), then one only needs to evaluate test H10: b = 0 and uses Pb for evaluating meditation effect. Under H00, the maximum P value of the two, Pmax, follows Beta(2,1); thus follows a uniform distribution [109]. Taking these together, DACT generates the P value for testing H0: ab = 0 as a weighted summation of P values under the three sub-null hypotheses
where the weights are given as
with πa0 and πb0 being the probabilities of H01 and H10, respectively. The equation makes an implicit assumption that the effects of a and b are independent of each other, such that the probability of a = 0 is not influenced by b, and vice versa. Such independence is guaranteed by the sequential ignorability assumptions described before. DACT estimates πa0 and πb0 using novel methods that have been well-established in prior FDR work [111], [122], [175], [176], [177], [178], [179], [180], [181], [182], such as Efron’s approach [183] using either the central matching method [177] or the empirical characteristic function and Fourier analysis [175]. DACT has been shown to be comparable to or more powerful than JT-comp across various simulation scenarios [109] and, in contrast to JT-comp, is not sensitive to sample size.
6.2.3. JS-mixture
JS-mixture is also a recent mediation method that is designed to produce calibrated P values and bares some conceptual similarity with JT-comp and DACT. JS-mixture aims to directly construct the null distribution for the JST statistic, Pmax, to correct for its conservative type I error control [112]. Specifically, JS-mixture applies JST to examine one potential mediator at a time and obtains the maximum P value for the jth mediator as Pmax, j (j = 1, …, m); then it estimates the proportions of the three sub-null hypotheses and constructs a null distribution for Pmax, j through
where π01, π10 and π00 are the proportions of the three sub-null scenarios; u is a given cut-off value for significance evaluation; p01 is the probability/power of rejecting b = 0 under H01, j; and p10 is the probability/power of rejecting a = 0 under H10, j. Both p01 and p10 can be estimated via the Grenander method [184]. When sample size is large, equation can be further approximated by
where F1 is the standard uniform distribution U(0,1); and F2 is a right skewed distribution for the maximum of two independent random variables drawn from the standard uniform distribution, with a cumulative distribution function Pr(Pmax ≤ u) = u2. It is easy to see that JS-mixture given in becomes very similar to DACT shown in under the case of large sample sizes, with the only difference being that JS-mixture controls FWER or FDR based on a newly estimated significance rule while DACT directly controls FWER or FDR based on the final weighted P values. The additional estimation of power functions p01 and p10 in JS-mixture might lead to higher statistical power compared to DACT which instead fixes them to be one. The proportion parameters required in JS-mixture can be also estimated with the same methods as used in DACT [111], [122], [175], [176], [177], [178], [179], [180], [181], [182].
6.2.4. JTV-comp
Unlike the three methods discussed above, JTV-comp [79] performs the gene-centric mediation analysis by examining one gene at a time and analyizng the mutiple mediators located within a gene together. JTV-comp accounts for the composite nature of the null hypothesis by making additional assumptions. Specifically, JTV-comp treats the mediator effects as random effects and assumes the exposure on mediator effect (i.e., a) and mediator on outcome effect (i.e., b) for the kth mediator in a given gene follow arbitrary dsitributions with mean zero and a variance being either κa and κb, respectively [79]. That is
Because κa = 0 implies a = 0 and κb = 0 implies b = 0, testing a = 0 and b = 0 can be translated into testing κa and κb, respectively. By this way, JTV-comp therefore translates testing H0: a = 0 or b = 0 into two variance component tests: a multivariate variance component test on κa and a univariate variance component test on κb. JTV-comp performs the variance component tests based on score statistics [185], [186], [187], [188], [189] within the framework of kernel machine learning [79], [187], [189]. Alternatively, JTV-comp can also perform the variance component test on κa = 0 based on the inverse regression framework [161]. With the variance component tests, JTV-comp obtains two P values on testing κa and κb and converts them into two z-statistics (z′a and z′b) using the probit function. With the transformed statistics z′a and z′b, JTV-comp proceeds with the same procedure of JT-comp to perform hypothesis tests [79]. Because JTV-comp relies on the same procedure of JT-comp to account for the composite nature of the null, it has similar advantages and limitations of JT-comp as mentioned in the previous subsection.
6.3. High-dimensional mediation methods jointly modeling exposure on mediator effects and mediator on outcome effects
The last category of high-dimensional mediation methods directly models all mediators jointly. To account for the large number of mediators, these methods either penalize mediation effects or specify particular priors on them to make the mediation model identifiable. Two types of methods belong to this category: penalization-based regression methods such as HIMA (high-dimensional mediation analysis) [190] and pathway Lasso [162]; and Bayesian methods such as BAMA (Bayesian variable selection mediation method) [163] and its extensions (Table 1). Both types of methods can also rely on an initial screening procedure in the mediator-outcome model to reduce the number of mediators to a reasonable size before analyzing them collectively.
6.3.1. Penalization-based high-dimensional mediation methods
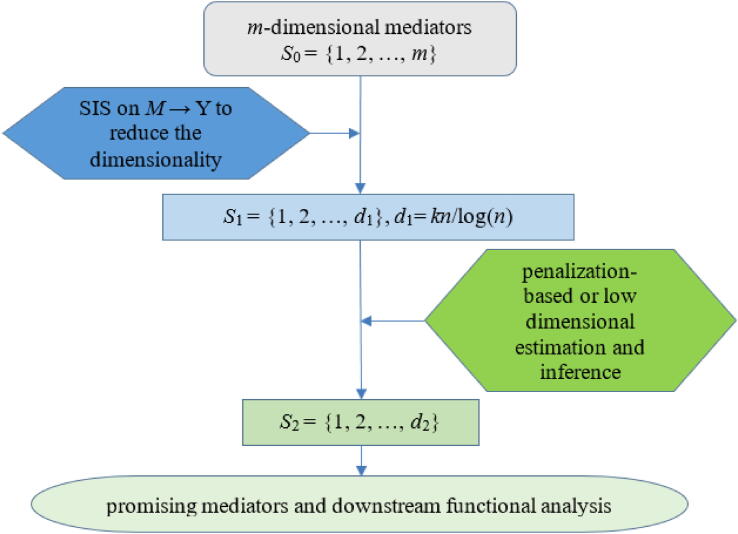
HIMA is one of the first penalization-based regression methods for high-dimensional mediation analysis [111] and includes the following three steps (Fig. 6). First, HIMA applies the sure independence screening (SIS) [191], [192] in the mediator-outcome model to reduce the dimensionality of mediators from ultra-high to high. This first step of HIMA can reduce the number of mediators from m to d1 = [kn/log(n)], where k is assigned by the user. Second, HIMA applies the multivariate mediator-outcome model to model the selected candidate mediators together and relies on the minimax concave penalty (MCP) [190] to shrink the mediator on outcome effects towards zero
where λ > 0 is the tuning parameter that controls the shrinkage and δ > 0 determines the concavity of MCP. By optimizing , HIMA can select mediators that have non-zero mediator on outcome effects. In particular, the P values of testing for b for mediators with non-zero effects can be calculated as
where is the MCP estimate of bs and is the corresponding standard error obtained based on the oracle property of MCP [190]. Besides b, HIMA also examines the mediator on outcome effects a in the exposure-mediator model and obtains the corresponding P values, Pas (s = 1, …, S2), based on the linear regression. Finally, HIMA evaluates the significance of the mediation effect using Pmax, s = max(, Pas) and adjusts for multiple comparisons through Bonferroni correction. HIMA has been recently extended to survival outcomes [96] and to yield unbiased mediator on outcome effects with debiased Lasso [159].
Fig. 6.
Overall workflow for HIMA with screening and penalization-based selection on mediators. HIMA includes the three main processes: (i) applying variable selection techniques for preliminary screening to reduce the high-dimensionality of mediators to tractable level (from m to d1); (ii) conducting penalization-based variable selection and estimation for the remaining mediators to reduce dimensionality further (from d1 to d2); (iii) performing the joint significance test for mediation effects. In the screening procedure, k is a tuning parameter determined by users.
The pathway Lasso is another penalization-based mediation method [162], which imposes a convex Lasso-type penalty on the mediation effects. In particular, the pathway Lasso attempts to minimize the following penalized likelihood function
where , λ1 and λ2 are tunning parameters. In the pathway Lasso, the first penalty aims to shrink the mediation effect as the product of aj and bj, while the second term penalizes individual aj and bj. The pathway Lasso applies the alternating direction method of multipliers (ADMM) for parameter estimation [193]. By optimizing , pathway Lasso can select active mediators with non-zero ajbj.
6.3.2. Bayesian high-dimensional mediation methods
BAMA is a Bayesian mediation method for selecting active mediators [163]. BAMA specifies a Bayesian sparse linear mixed model (BSLMM) prior [194], also known as the two-component spike-and-slab prior [195], on both the exposure on mediator effects and the mediator on outcome effects. The BSLMM prior assumes that each effect for the jth mediator, aj or bj, follows a mixture of two normal distributrions, one with a large variance and the other with a small variance
where κa1 > κa0 and κb1 > κb0, and πa (or πb) denote the probability that the effect belongs to the normal distribution with a larger variance. BAMA applies a Markov chain Monte Carlo (MCMC) sampling algorithm to obtain posterior samples [163] and uses the posterior inclusion probability (PIP) to select active mediators with both a and b belonging to the large normal components.
Song et al (2020) [164] extended the separate priors on aj and bj in BAMA towards joint modeling of both sets of effects. In particular, the authors depended on the four-component Gaussian mixture model (GMM) developed in genome-wide association studies [196] to decompose the exposure on mediator and mediator on outcome effects into four components
where δ0 is a point mass at zero; is the prior variance for the exposure on mediator effect; is the prior variance for the mediator on outcome effect; ρ is the correlation between the two sets of effects; and π00, π10, π01, and π11 are the probabilities of the four components, respectively. The components in capture all possible relationship among the exposure, mediator and outcome: the first three components directly correspond to the three sub-null hypotheses while the last component representing the alternative. The GMM-based mediation method has been shown to enjoy excellent and robust performance for mediator selection and mediation effect estimation [164]. A potential drawback of the GMM prior, however, is that it does not directly impose sparsity on the mediation effects for mediator selection. Therefore, Song et al (2020) [164] provided a second method, based on a product threshold Gaussian (PTG) prior, to directly sparsity on the mediation effects. Song et al (2020) [164] relied on a latent variable-based MCMC algorithm for parameter estimation in both these two models.
Song et al. (2020) [165] further extended the above Bayesian approaches towards direct modeling of the correlation structure among active mediators. Intuitively, if the active mediators are correlated with each other, then accounting for such correlation would improve power to detect them. Song et al (2020) [165] developed two methods to account for such correlation among active mediators under the GMM-based Bayesian joint mediation model. The first is to use the Potts distribution [197], which is a generalization of the Ising distribution and accounts for the complex dependency structures among multiple groups. The second is based on joint modeling of the mediator-specific mixing probabilities via a logistic normal distribution [198] similar to that used in Zeng et al (2018) [196], where the group probabilities reflect the underlying correlation structure. Therefore, this newly developed joint mediatin method allows for identifying correlated active mediators that could be missed by other methods [165].
7. Conclusions
We have presented a systematacial review of statistical methods for mediation analysis, with a special emphasis on recent methods developed for high-dimensional mediators commonly encountered in high-throughput genomics studies. In spite of current successes of these newly developed high-dimensional mediation methods, many challenges remain. For example, accurately estimating the proportion parameters of different sub-null hypotheses is critical for generating calibrated P values from both DACT [109] and JS-mixture [112], but accurate estimation of these parameters may be hard to achieve. As another example, the empirical null distribution of JS-mixture is currently constructed in a nonparametric manner [112], and it remains important to explore whether parametric mixture distributions such as Beta mixture can help improve power further. For Bayesian mediation methods [163], [164], [165], the current sampling-based algorithms are not computationally efficent. Future algorithmic development is needed to adapt them for truly genome-wide mediation studies with large sample sizes. In addition, various extensions of these high-dimensional mediation methods towards modeling non-linear relationship between mediators and outcomes, accomodating missing data [199], [200] and exposure-mediator interaction, performing sensitivity analysis under model misspecifications [72], [117], [119], accounting for multilevel or longitudinal outcomes [201], [202], [203], could all yield fruitful results. Finally, a comprehensive comparison among methods for high-dimensional mediation analysis is warrented for evaluating their relative performance and understanding their practical advantages and drawbacks in high-throughput genomics applications.
CRediT authorship contribution statement
Ping Zeng: Conceptualization, Methodology, Writing - original draft, Writing - review & editing, Project administration. Zhonghe Shao: Writing - original draft, Visualization. Xiang Zhou: Supervision.
Declaration of Competing Interest
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Acknowledgements
We would like to thank the editor and reviewers for their helpful and constructive comments which improved our manuscript substantially. The research of Ping Zeng was supported in part by the Youth Foundation of Humanity and Social Science funded by Ministry of Education of China (18YJC910002), the Natural Science Foundation of Jiangsu Province of China (BK20181472), the China Postdoctoral Science Foundation (2018 M630607 and 2019 T120465), the QingLan Research Project of Jiangsu Province for Outstanding Young Teachers, the Six-Talent Peaks Project in Jiangsu Province of China (WSN-087), the Training Project for Youth Teams of Science and Technology Innovation at Xuzhou Medical University (TD202008), the Postdoctoral Science Foundation of Xuzhou Medical University, the National Natural Science Foundation of China (81402765), and the Statistical Science Research Project from National Bureau of Statistics of China (2014LY112). The research of Xiang Zhou was supported by the University of Michigan.
Contributor Information
Ping Zeng, Email: zpstat@xzhmu.edu.cn.
Zhonghe Shao, Email: 301911011600@stu.xzhmu.edu.cn.
Xiang Zhou, Email: xzhousph@umich.edu.
References
- 1.GTEx Consortium., Genetic effects on gene expression across human tissues. Nature, 2017. 550(7675): p. 204-213. [DOI] [PMC free article] [PubMed]
- 2.Roadmap Epigenomics Consortium Integrative analysis of 111 reference human epigenomes. Nature. 2015;518(7539):317–330. doi: 10.1038/nature14248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Schizophrenia Consortium Biological insights from 108 schizophrenia-associated genetic loci. Nature. 2014;511(7510):421–427. doi: 10.1038/nature13595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.The 1000 Genomes Project Consortium, A global reference for human genetic variation. Nature, 2015. 526(7571): p. 68-74. [DOI] [PMC free article] [PubMed]
- 5.Liu Y., Aryee M., Padyukov L., Fallin M., Hesselberg E., Runarsson A. Epigenome-wide association data implicate DNA methylation as an intermediary of genetic risk in rheumatoid arthritis. Nat Biotechnol. 2013;31:142–147. doi: 10.1038/nbt.2487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Wood A.R., Esko T., Yang J., Vedantam S., Pers T.H., Gustafsson S. Defining the role of common variation in the genomic and biological architecture of adult human height. Nat Genet. 2014;46(11):1173–1186. doi: 10.1038/ng.3097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Tam V., Patel N., Turcotte M., Bossé Y., Paré G., Meyre D. Benefits and limitations of genome-wide association studies. Nat Rev Genet. 2019;20(8):467–484. doi: 10.1038/s41576-019-0127-1. [DOI] [PubMed] [Google Scholar]
- 8.McMahon A., Malangone C., Suveges D., Sollis E., Cunningham F., Riat H.S. The NHGRI-EBI GWAS Catalog of published genome-wide association studies, targeted arrays and summary statistics 2019. Nucleic Acids Res. 2019;47(D1):D1005–D1012. doi: 10.1093/nar/gky1120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Bush W.S., Moore J.H. Genome-wide association studies. PLoS Comput Biol. 2012;8(12) doi: 10.1371/journal.pcbi.1002822. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Kim-Hellmuth, S., F. Aguet, M. Oliva, M. Muñoz-Aguirre, S. Kasela, V. Wucher, et al., Cell type–specific genetic regulation of gene expression across human tissues. Science, 2020. 369(6509): p. eaaz8528. [DOI] [PMC free article] [PubMed]
- 11.The GTEx Consortium The GTEx Consortium atlas of genetic regulatory effects across human tissues. Science. 2020;369(6509):1318–1330. doi: 10.1126/science.aaz1776. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Edwards Stacey L., Beesley J., French Juliet D., Dunning Alison M. Beyond GWASs: Illuminating the Dark Road from Association to Function. Am J Human Genet. 2013;93(5):779–797. doi: 10.1016/j.ajhg.2013.10.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Gallagher M.D., Chen-Plotkin A.S. The Post-GWAS Era: From Association to Function. Am J Human Genet. 2018;102(5):717–730. doi: 10.1016/j.ajhg.2018.04.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Visscher P.M., Wray N.R., Zhang Q., Sklar P., McCarthy M.I., Brown M.A. 10 Years of GWAS Discovery: Biology, Function, and Translation. Am J Human Genet. 2017;101(1):5–22. doi: 10.1016/j.ajhg.2017.06.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Boyle E.A., Li Y.I., Pritchard J.K. An Expanded View of Complex Traits: From Polygenic to Omnigenic. Cell. 2017;169(7):1177–1186. doi: 10.1016/j.cell.2017.05.038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Wray N.R., Wijmenga C., Sullivan P.F., Yang J., Visscher P.M. Common Disease Is More Complex Than Implied by the Core Gene Omnigenic Model. Cell. 2018;173(7):1573–1580. doi: 10.1016/j.cell.2018.05.051. [DOI] [PubMed] [Google Scholar]
- 17.Gibson G. Rare and common variants: twenty arguments. Nat Rev Genet. 2012;13(2):135–145. doi: 10.1038/nrg3118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Price A.L., Spencer C.C.A., Donnelly P. Progress and promise in understanding the genetic basis of common diseases. Proceedings of the Royal Society B: Biological Sciences. 2015;282(1821) doi: 10.1098/rspb.2015.1684. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Aung M.T., Song Y., Ferguson K.K., Cantonwine D.E., Zeng L., McElrath T.F. Application of an analytical framework for multivariate mediation analysis of environmental data. Nat Commun. 2020;11(1):5624. doi: 10.1038/s41467-020-19335-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Vanderweele T.J., Vansteelandt S., Robins J.M. Effect decomposition in the presence of an exposure-induced mediator-outcome confounder. Epidemiology. 2014;25(2):300–306. doi: 10.1097/EDE.0000000000000034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Boya P., Reggiori F., Codogno P. Emerging regulation and functions of autophagy. Nat Cell Biol. 2013;15(7):713–720. doi: 10.1038/ncb2788. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Albert J.M., Nelson S. Generalized Causal Mediation Analysis. Biometrics. 2011;67(3):1028–1038. doi: 10.1111/j.1541-0420.2010.01547.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Avin, C., I. Shpitser and J. Pearl, Identifiability of path-specific effects, in Proceedings of the 19th international joint conference on Artificial intelligence. 2005, Morgan Kaufmann Publishers Inc.: Edinburgh, Scotland. p. 357–363.
- 24.Taguri M., Chiba Y. A principal stratification approach for evaluating natural direct and indirect effects in the presence of treatment-induced intermediate confounding. Stat Med. 2015;34(1):131–144. doi: 10.1002/sim.6329. [DOI] [PubMed] [Google Scholar]
- 25.MacKinnon D. Contrasts in multiple mediator models. Contrasts In Multiple Mediator Models. 2000:141–160. [Google Scholar]
- 26.Needham B.L., Smith J.A., Zhao W., Wang X., Mukherjee B., Kardia S.L.R. Life course socioeconomic status and DNA methylation in genes related to stress reactivity and inflammation: The multi-ethnic study of atherosclerosis. Epigenetics. 2015;10(10):958–969. doi: 10.1080/15592294.2015.1085139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Petronis A. Epigenetics as a unifying principle in the aetiology of complex traits and diseases. Nature. 2010;465(7299):721–727. doi: 10.1038/nature09230. [DOI] [PubMed] [Google Scholar]
- 28.MacKinnon, D. Contrasts in multiple mediator models. In J. S. Rose, L. Chassin, C. C. Presson, & S. J. Sherman (Eds.), Multivariate applications in substance use research: New methods for new questions (p. 141–160). Lawrence Erlbaum Associates Publishers. 2000.
- 29.Lange T., Rasmussen M., Thygesen L.C. Assessing natural direct and indirect effects through multiple pathways. Am J Epidemiol. 2014;179(4):513–518. doi: 10.1093/aje/kwt270. [DOI] [PubMed] [Google Scholar]
- 30.Stone C.A., Sobel M.E. The robustness of estimates of total indirect effects in covariance structure models estimated by maximum. Psychometrika. 1990;55(2):337–352. [Google Scholar]
- 31.Tchetgen Tchetgen E.J., Vanderweele T.J. Identification of natural direct effects when a confounder of the mediator is directly affected by exposure. Epidemiology. 2014;25(2):282–291. doi: 10.1097/EDE.0000000000000054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Hindorff L.A., Sethupathy P., Junkins H.A., Ramos E.M., Mehta J.P., Collins F.S. Potential etiologic and functional implications of genome-wide association loci for human diseases and traits. PNAS. 2009;106(23):9362–9367. doi: 10.1073/pnas.0903103106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Yuan Z., Zhu H., Zeng P., Yang S., Sun S., Yang C. Testing and controlling for horizontal pleiotropy with the probabilistic Mendelian randomization in transcriptome-wide association studies. Nat Commun. 2020;11(1):3861. doi: 10.1038/s41467-020-17668-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Zeng P., Zhou X. Non-parametric genetic prediction of complex traits with latent Dirichlet process regression models. Nat Commun. 2017;8(1):456. doi: 10.1038/s41467-017-00470-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Sun S., Zhu J., Mozaffari S., Ober C., Chen M., Zhou X. Heritability estimation and differential analysis of count data with generalized linear mixed models in genomic sequencing studies. Bioinformatics. 2019;35(3):487–496. doi: 10.1093/bioinformatics/bty644. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Sun S., Zhu J., Zhou X. Statistical analysis of spatial expression patterns for spatially resolved transcriptomic studies. Nat Methods. 2020;17(2):193–200. doi: 10.1038/s41592-019-0701-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Sun S., Hood M., Scott L., Peng Q., Mukherjee S., Tung J. Differential expression analysis for RNAseq using Poisson mixed models. Nucleic Acids Res. 2017;45(11) doi: 10.1093/nar/gkx204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Shang L., Smith J.A., Zhao W., Kho M., Turner S.T., Mosley T.H. Genetic Architecture of Gene Expression in European and African Americans: An eQTL Mapping Study in GENOA. Am J Human Genet. 2020;106(4):496–512. doi: 10.1016/j.ajhg.2020.03.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Tung J., Zhou X., Alberts S.C., Stephens M., Gilad Y. The genetic architecture of gene expression levels in wild baboons. Elife. 2015;4 doi: 10.7554/eLife.04729. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Lea A.J., Tung J., Zhou X. A Flexible, Efficient Binomial Mixed Model for Identifying Differential DNA Methylation in Bisulfite Sequencing Data. PLoS Genet. 2015;11(11) doi: 10.1371/journal.pgen.1005650. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Fan Y., Vilgalys T.P., Sun S., Peng Q., Tung J., Zhou X. IMAGE: high-powered detection of genetic effects on DNA methylation using integrated methylation QTL mapping and allele-specific analysis. Genome Biol. 2019;20(1):220. doi: 10.1186/s13059-019-1813-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Stringhini S., Zaninotto P., Kumari M., Kivimäki M., Lassale C., Batty G.D. Socio-economic trajectories and cardiovascular disease mortality in older people: the English Longitudinal Study of Ageing. Int J Epidemiol. 2017;47(1):36–46. doi: 10.1093/ije/dyx106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Kaplan G.A., Keil J.E. Socioeconomic factors and cardiovascular disease: a review of the literature. Circulation. 1993;88(4):1973–1998. doi: 10.1161/01.cir.88.4.1973. [DOI] [PubMed] [Google Scholar]
- 44.Kivimäki M., Lawlor D.A., Smith G.D., Kouvonen A., Virtanen M., Elovainio M. Socioeconomic Position, Co-Occurrence of Behavior-Related Risk Factors, and Coronary Heart Disease: the Finnish Public Sector Study. Am J Public Health. 2007;97(5):874–879. doi: 10.2105/AJPH.2005.078691. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Kilander L., Berglund L., Boberg M., Vessby B., Lithell H. Education, lifestyle factors and mortality from cardiovascular disease and cancer. A 25-year follow-up of Swedish 50-year-old men. Int J Epidemiol. 2001;30(5):1119–1126. doi: 10.1093/ije/30.5.1119. [DOI] [PubMed] [Google Scholar]
- 46.Frankel S., Smith G.D., Gunnell D. Childhood Socioeconomic Position and Adult Cardiovascular Mortality: The Boyd Orr Cohort. Am J Epidemiol. 1999;150(10):1081–1084. doi: 10.1093/oxfordjournals.aje.a009932. [DOI] [PubMed] [Google Scholar]
- 47.Barker D.J.P., Osmond C. Infant mortality, childhood nutrition, and ischaemic heart disease in England and Wales. The Lancet. 1986;327(8489):1077–1081. doi: 10.1016/s0140-6736(86)91340-1. [DOI] [PubMed] [Google Scholar]
- 48.Tehranifar P., Wu H.-C., Fan X., Flom J.D., Ferris J.S., Cho Y.H. Early life socioeconomic factors and genomic DNA methylation in mid-life. Epigenetics. 2013;8(1):23–27. doi: 10.4161/epi.22989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Stringhini S., Polidoro S., Sacerdote C., Kelly R.S., van Veldhoven K., Agnoli C. Life-course socioeconomic status and DNA methylation of genes regulating inflammation. Int J Epidemiol. 2015;44(4):1320–1330. doi: 10.1093/ije/dyv060. [DOI] [PubMed] [Google Scholar]
- 50.McGuinness D., McGlynn L.M., Johnson P.C., MacIntyre A., Batty G.D., Burns H. Socio-economic status is associated with epigenetic differences in the pSoBid cohort. Int J Epidemiol. 2012;41(1):151–160. doi: 10.1093/ije/dyr215. [DOI] [PubMed] [Google Scholar]
- 51.Borghol N., Suderman M., McArdle W., Racine A., Hallett M., Pembrey M. Associations with early-life socio-economic position in adult DNA methylation. Int J Epidemiol. 2011;41(1):62–74. doi: 10.1093/ije/dyr147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Martin E.M., Fry R.C. Environmental influences on the epigenome: exposure-associated DNA methylation in human populations. Annu Rev Public Health. 2018;39:309–333. doi: 10.1146/annurev-publhealth-040617-014629. [DOI] [PubMed] [Google Scholar]
- 53.Hang C.T., Yang J., Han P., Cheng H.-L., Shang C., Ashley E. Chromatin regulation by Brg1 underlies heart muscle development and disease. Nature. 2010;466(7302):62–67. doi: 10.1038/nature09130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Huan T., Joehanes R., Song C., Peng F., Guo Y., Mendelson M. Genome-wide identification of DNA methylation QTLs in whole blood highlights pathways for cardiovascular disease. Nat Commun. 2019;10(1):4267. doi: 10.1038/s41467-019-12228-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Tobi, E.W., R.C. Slieker, R. Luijk, K.F. Dekkers, A.D. Stein, K.M. Xu, et al., DNA methylation as a mediator of the association between prenatal adversity and risk factors for metabolic disease in adulthood. Science Advances, 2018. 4(1): p. eaao4364. [DOI] [PMC free article] [PubMed]
- 56.Huang J.Y., Gavin A.R., Richardson T.S., Rowhani-Rahbar A., Siscovick D.S., Hochner H. Accounting for Life-Course Exposures in Epigenetic Biomarker Association Studies: Early Life Socioeconomic Position, Candidate Gene DNA Methylation, and Adult Cardiometabolic Risk. Am J Epidemiol. 2016;184(7):520–531. doi: 10.1093/aje/kww014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Schiele M.A., Domschke K. Epigenetics at the crossroads between genes, environment and resilience in anxiety disorders. Genes, Brain and Behavior. 2018;17(3) doi: 10.1111/gbb.12423. [DOI] [PubMed] [Google Scholar]
- 58.Gottschalk M.G., Domschke K., Schiele M.A. Epigenetics Underlying Susceptibility and Resilience Relating to Daily Life Stress, Work Stress, and Socioeconomic Status. Front Psychiatry. 2020;11:163. doi: 10.3389/fpsyt.2020.00163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Bush N.R., Lane R.D., McLaughlin K.A. Mechanisms Underlying the Association Between Early-Life Adversity and Physical Health: Charting a Course for the Future. Psychosom Med. 2016;78(9):1114–1119. doi: 10.1097/PSY.0000000000000421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Juarez P.D., Hood D.B., Song M.-A., Ramesh A. Use of an Exposome Approach to Understand the Effects of Exposures From the Natural, Built, and Social Environments on Cardio-Vascular Disease Onset, Progression, and Outcomes. Front Public Health. 2020;8:379–380. doi: 10.3389/fpubh.2020.00379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Hao G., Youssef N.A., Davis C.L., Su S. The role of DNA methylation in the association between childhood adversity and cardiometabolic disease. Int J Cardiol. 2018;255:168–174. doi: 10.1016/j.ijcard.2017.12.063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.McLaughlin K.A., Lane R.D., Bush N.R. Introduction to the special issue of psychosomatic medicine: Mechanisms linking early-life adversity to physical health. Psychosom Med. 2016;78(9):976. doi: 10.1097/PSY.0000000000000420. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Loucks E.B., Huang Y.-T., Agha G., Chu S., Eaton C.B., Gilman S.E. Epigenetic Mediators Between Childhood Socioeconomic Disadvantage and Mid-Life Body Mass Index: The New England Family Study. Psychosom Med. 2016;78(9):1053–1065. doi: 10.1097/PSY.0000000000000411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.VanderWeele, T., Explanation in causal inference: methods for mediation and interaction. 2015: Oxford University Press.
- 65.MacKinnon, D.P., Introduction to statistical mediation analysis. 2008: Routledge.
- 66.Chén O.Y., Crainiceanu C., Ogburn E.L., Caffo B.S., Wager T.D., Lindquist M.A. High-dimensional multivariate mediation with application to neuroimaging data. Biostatistics. 2018;19(2):121–136. doi: 10.1093/biostatistics/kxx027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Wright S. The Method of Path Coefficients. Ann Math Stat. 1934;5(3):161–215. [Google Scholar]
- 68.Baron R.M., Kenny D.A. The moderator-mediator variable distinction in social psychological research: Conceptual, strategic, and statistical considerations. J Pers Soc Psychol. 1986;51(6):1173–1182. doi: 10.1037//0022-3514.51.6.1173. [DOI] [PubMed] [Google Scholar]
- 69.Rucker D.D., Preacher K.J., Tormala Z.L., Petty R.E. Mediation analysis in social psychology: Current practices and new recommendations. Soc Pers Psychol Compass. 2011;5(6):359–371. [Google Scholar]
- 70.MacKinnon D.P., Lockwood C.M., Hoffman J.M., West S.G., Sheets V. A comparison of methods to test mediation and other intervening variable effects. Psychol Methods. 2002;7(1):83–104. doi: 10.1037/1082-989x.7.1.83. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Shrout P.E., Bolger N. Mediation in experimental and nonexperimental studies: new procedures and recommendations. Psychol Methods. 2002;7(4):422–445. [PubMed] [Google Scholar]
- 72.Imai K., Keele L., Yamamoto T. Identification, inference and sensitivity analysis for causal mediation effects. Statistical Science. 2010;25(1):51–71. [Google Scholar]
- 73.Huang Y.-T. Joint significance tests for mediation effects of socioeconomic adversity on adiposity via epigenetics. The Annals of Applied Statistics. 2018;12(3):1535–1557. [Google Scholar]
- 74.VanderWeele T.J. Causal mediation analysis with survival data. Epidemiology. 2011;22(4):582. doi: 10.1097/EDE.0b013e31821db37e. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.VanderWeele T.J. Mediation analysis: a practitioner's guide. Annu Rev Public Health. 2016;37:17–32. doi: 10.1146/annurev-publhealth-032315-021402. [DOI] [PubMed] [Google Scholar]
- 76.Vanderweele T.J., Vansteelandt S. Odds ratios for mediation analysis for a dichotomous outcome. Am J Epidemiol. 2010;172(12):1339–1348. doi: 10.1093/aje/kwq332. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.VanderWeele T.J., Vansteelandt S. Mediation Analysis with Multiple Mediators. Epidemiologic Methods. 2013;2(1):95–115. doi: 10.1515/em-2012-0010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Huang Y.-T. Genome-wide analyses of sparse mediation effects under composite null hypotheses. Ann Appl Statist. 2019;13(1):60–84. [Google Scholar]
- 79.Huang Y.-T. Variance component tests of multivariate mediation effects under composite null hypotheses. Biometrics. 2019;75(4):1191–1204. doi: 10.1111/biom.13073. [DOI] [PubMed] [Google Scholar]
- 80.Huang Y.-T., Cai T. Mediation analysis for survival data using semiparametric probit models. Biometrics. 2016;72(2):563–574. doi: 10.1111/biom.12445. [DOI] [PubMed] [Google Scholar]
- 81.Barfield R., Shen J., Just A.C., Vokonas P.S., Schwartz J., Baccarelli A.A. Testing for the indirect effect under the null for genome-wide mediation analyses. Genet Epidemiol. 2017;41(8):824–833. doi: 10.1002/gepi.22084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Schaid D.J., Sinnwell J.P. Penalized models for analysis of multiple mediators. Genet Epidemiol. 2020;44(5):408–424. doi: 10.1002/gepi.22296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Shan N., Wang Z., Hou L. Identification of trans-eQTLs using mediation analysis with multiple mediators. BMC Bioinf. 2019;20(3):126. doi: 10.1186/s12859-019-2651-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Yang, F., J. Wang, T.G. Consortium, B.L. Pierce and L.S. Chen, Identifying cis-mediators for trans-eQTLs across many human tissues using genomic mediation analysis. Genome Res; 2017. [DOI] [PMC free article] [PubMed]
- 85.MacKinnon D.P., Fairchild A.J., Fritz M.S. Mediation analysis. Annu Rev Psychol. 2007;58:593–614. doi: 10.1146/annurev.psych.58.110405.085542. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Robins J.M., Greenland S. Identifiability and exchangeability for direct and indirect effects. Epidemiology. 1992:143–155. doi: 10.1097/00001648-199203000-00013. [DOI] [PubMed] [Google Scholar]
- 87.Pearl J. Causal inference in statistics: An overview. Statistics Surveys. 2009;3:96–146. [Google Scholar]
- 88.Valeri L., VanderWeele T.J. Mediation analysis allowing for exposure–mediator interactions and causal interpretation: Theoretical assumptions and implementation with SAS and SPSS macros. Psychol Methods. 2013;18(2):137. doi: 10.1037/a0031034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Ito Z., Takakura K., Suka M., Kanai T., Saito R., Fujioka S. Prognostic impact of carbohydrate sulfotransferase 15 in patients with pancreatic ductal adenocarcinoma. Oncol Lett. 2017;13(6):4799–4805. doi: 10.3892/ol.2017.6071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Bakulski K.M., Dou J., Lin N., London S.J., Colacino J.A. DNA methylation signature of smoking in lung cancer is enriched for exposure signatures in newborn and adult blood. Sci Rep. 2019;9(1):4576. doi: 10.1038/s41598-019-40963-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Lee K.W.K., Pausova Z. Cigarette smoking and DNA methylation. Front Genet. 2013;4:132. doi: 10.3389/fgene.2013.00132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Huang Y.-T., Yang H.-I. Causal Mediation Analysis of Survival Outcome with Multiple Mediators. Epidemiology. 2017;28(3):370–378. doi: 10.1097/EDE.0000000000000651. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Gaynor S.M., Schwartz J., Lin X. Mediation analysis for common binary outcomes. Stat Med. 2019;38(4):512–529. doi: 10.1002/sim.7945. [DOI] [PubMed] [Google Scholar]
- 94.Lange T., Hansen J. Direct and Indirect Effects in a Survival Context. Epidemiology. 2011;22:575–581. doi: 10.1097/EDE.0b013e31821c680c. [DOI] [PubMed] [Google Scholar]
- 95.Wang L., Zhang Z. Estimating and Testing Mediation Effects with Censored Data. Struct Eq Model Multidiscipl J. 2011;18(1):18–34. [Google Scholar]
- 96.Luo C., Fa B., Yan Y., Wang Y., Zhou Y., Zhang Y. High-dimensional mediation analysis in survival models. PLoS Comput Biol. 2020;16(4) doi: 10.1371/journal.pcbi.1007768. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Joffe M.M., Small D., Hsu C.-Y. Defining and Estimating Intervention Effects for Groups that will Develop an Auxiliary Outcome. Statistical Science. 2007;22(1):74–97. [Google Scholar]
- 98.Pearl, J., Direct and Indirect Effects, in Proceedings of the 17th Conference in Uncertainty in Artificial Intelligence. 2001, Morgan Kaufmann Publishers Inc. p. 411–420.
- 99.Mackinnon D.P., Fairchild A.J. Current Directions in Mediation Analysis. Current directions in psychological science. 2009;18(1):16–19. doi: 10.1111/j.1467-8721.2009.01598.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.VanderWeele T., Vansteelandt S. Conceptual issues concerning mediation, interventions and composition. Statistics and its. Interface. 2009;4 [Google Scholar]
- 101.Imai K., Keele L., Tingley D. A general approach to causal mediation analysis. Psychol Methods. 2010;15(4):309. doi: 10.1037/a0020761. [DOI] [PubMed] [Google Scholar]
- 102.Ten Have T.R., Joffe M.M. A review of causal estimation of effects in mediation analyses. Stat Methods Med Res. 2012;21(1):77–107. doi: 10.1177/0962280210391076. [DOI] [PubMed] [Google Scholar]
- 103.Pearl J. The causal mediation formula—a guide to the assessment of pathways and mechanisms. Prev Sci. 2012;13(4):426–436. doi: 10.1007/s11121-011-0270-1. [DOI] [PubMed] [Google Scholar]
- 104.Pearl J. Interpretation and identification of causal mediation. Psychol Methods. 2014;19(4):459–481. doi: 10.1037/a0036434. [DOI] [PubMed] [Google Scholar]
- 105.Preacher K.J. Advances in mediation analysis: A survey and synthesis of new developments. Annu Rev Psychol. 2015;66:825–852. doi: 10.1146/annurev-psych-010814-015258. [DOI] [PubMed] [Google Scholar]
- 106.Sampson J.N., Boca S.M., Moore S.C., Heller R. FWER and FDR control when testing multiple mediators. Bioinformatics. 2018;34(14):2418–2424. doi: 10.1093/bioinformatics/bty064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.MacKinnon D.P. Analysis of mediating variables in prevention and intervention research. NIDA Res Monogr. 1994;139:127–153. [PubMed] [Google Scholar]
- 108.Fairchild A.J., MacKinnon D.P. A general model for testing mediation and moderation effects. Prevent Sci Off J Soc Prevent Res. 2009;10(2):87–99. doi: 10.1007/s11121-008-0109-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Liu Z., Shen J., Barfield R., Schwartz J., Baccarelli A.A., Lin X. Large-Scale Hypothesis Testing for Causal Mediation Effects with Applications in Genome-wide Epigenetic Studies. J Am Stat Assoc. 2021:1–39. doi: 10.1080/01621459.2021.1914634. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Shang L., Smith J.A., Zhou X. Leveraging gene co-expression patterns to infer trait-relevant tissues in genome-wide association studies. PLoS Genet. 2020;16(4) doi: 10.1371/journal.pgen.1008734. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Zhang H., Zheng Y., Zhang Z., Gao T., Joyce B., Yoon G. Estimating and testing high-dimensional mediation effects in epigenetic studies. Bioinformatics. 2016;32(20):3150–3154. doi: 10.1093/bioinformatics/btw351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Dai J.Y., Stanford J.L., LeBlanc M. A Multiple-Testing Procedure for High-Dimensional Mediation Hypotheses. J Am Stat Assoc. 2020:1–16. doi: 10.1080/01621459.2020.1765785. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Luo X., Schwartz J., Baccarelli A., Liu Z. Testing cell-type-specific mediation effects in genome-wide epigenetic studies. Briefings Bioinf. 2020 doi: 10.1093/bib/bbaa131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Zhao Y., Lindquist M.A., Caffo B.S. Sparse principal component based high-dimensional mediation analysis. Comput Stat Data Anal. 2020;142 doi: 10.1016/j.csda.2019.106835. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Zhou R.R., Wang L., Zhao S.D. Estimation and inference for the indirect effect in high-dimensional linear mediation models. Biometrika. 2020;107(3):573–589. doi: 10.1093/biomet/asaa016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Hicks, R., Tingley, D. Causal mediation analysis. Stata J; 2011. 11(4): p. 605.
- 117.Albert J.M., Wang W. Sensitivity analyses for parametric causal mediation effect estimation. Biostatistics. 2015;16(2):339–351. doi: 10.1093/biostatistics/kxu048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.VanderWeele T.J., Chiba Y. Sensitivity analysis for direct and indirect effects in the presence of exposure-induced mediator-outcome confounders. Epidemiology, biostatistics and public health. 2014;11(2) doi: 10.2427/9027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Ding P., Vanderweele T.J. Sharp sensitivity bounds for mediation under unmeasured mediator-outcome confounding. Biometrika. 2016;103(2):483–490. doi: 10.1093/biomet/asw012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Boca S.M., Sinha R., Cross A.J., Moore S.C., Sampson J.N. Testing multiple biological mediators simultaneously. Bioinformatics. 2014;30(2):214–220. doi: 10.1093/bioinformatics/btt633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Daniel R., De Stavola B., Cousens S., Vansteelandt S. Causal mediation analysis with multiple mediators. Biometrics. 2015;71(1):1–14. doi: 10.1111/biom.12248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Huang Y.-T., Pan W.-C. Hypothesis test of mediation effect in causal mediation model with high-dimensional continuous mediators. Biometrics. 2016;72(2):402–413. doi: 10.1111/biom.12421. [DOI] [PubMed] [Google Scholar]
- 123.Mackinnon D.P., Dwyer J.H. Estimating Mediated Effects in Prevention Studies. Evaluation Review. 1993;17(2):144–158. [Google Scholar]
- 124.Winship C., Mare R.D. Structural Equations and Path Analysis for Discrete Data. Am J Sociol. 1983;89(1):54–110. [Google Scholar]
- 125.Buis M.L. Direct and indirect effects in a logit model. Stata J. 2010;10(1):11–29. [PMC free article] [PubMed] [Google Scholar]
- 126.Mackinnon D.P., Warsi G., Dwyer J.H. A Simulation Study of Mediated Effect Measures. Multivar Behav Res. 1995;30(1):41–45. doi: 10.1207/s15327906mbr3001_3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127.Ditlevsen S., Christensen U., Lynch J., Damsgaard M., Keiding N. The mediation proportion: a structural equation approach for estimating the proportion of exposure effect on outcome explained by an intermediate variable. Epidemiology. 2005;16:114–120. doi: 10.1097/01.ede.0000147107.76079.07. [DOI] [PubMed] [Google Scholar]
- 128.Freedman L.S., Graubard B.I., Schatzkin A. Statistical validation of intermediate endpoints for chronic diseases. Stat Med. 1992;11(2):167–178. doi: 10.1002/sim.4780110204. [DOI] [PubMed] [Google Scholar]
- 129.MacKinnon D.P., Lockwood C.M., Brown C.H., Wang W., Hoffman J.M. The intermediate endpoint effect in logistic and probit regression. Clinical trials. 2007;4(5):499–513. doi: 10.1177/1740774507083434. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130.Wang Y., Taylor J. A Measure of the Proportion of Treatment Effect Explained by a Surrogate Marker. Biometrics. 2003;58:803–812. doi: 10.1111/j.0006-341x.2002.00803.x. [DOI] [PubMed] [Google Scholar]
- 131.Preacher K., Kelley K. Effect Size Measures for Mediation Models: Quantitative Strategies for Communicating Indirect Effects. Psychol Methods. 2011;16:93–115. doi: 10.1037/a0022658. [DOI] [PubMed] [Google Scholar]
- 132.Fairchild A.J., McDaniel H.L. Best (but oft-forgotten) practices: mediation analysis. Am J Clin Nutrit. 2017;105(6):1259–1271. doi: 10.3945/ajcn.117.152546. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133.MacKinnon, D.P., Fairchild A.J. Fritz, M.S. Mediation analysis, in Annual Rev Psychol; 2007. p. 593-614. [DOI] [PMC free article] [PubMed]
- 134.Sobel M.E. Asymptotic Confidence Intervals for Indirect Effects in Structural Equation Models. Sociol Methodol. 1982;13:290–312. [Google Scholar]
- 135.Aroian L.A. The Probability Function of the Product of Two Normally Distributed Variables. Ann Math Stat. 1947;18(2):265–271. [Google Scholar]
- 136.Goodman L.A. On the Exact Variance of Products. J Am Stat Assoc. 1960;55(292):708–713. [Google Scholar]
- 137.Tofighi D., MacKinnon D.P. RMediation: An R package for mediation analysis confidence interval. Behavior Research Methods. 2011;43(3):692–700. doi: 10.3758/s13428-011-0076-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 138.!!! INVALID CITATION !!!
- 139.Fritz M.S., Mackinnon D.P. Required sample size to detect the mediated effect. Psychol Sci. 2007;18(3):233–239. doi: 10.1111/j.1467-9280.2007.01882.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 140.Hayes A.F., Scharkow M. The relative trustworthiness of inferential tests of the indirect effect in statistical mediation analysis: does method really matter? Psychol Sci. 2013;24(10):1918–1927. doi: 10.1177/0956797613480187. [DOI] [PubMed] [Google Scholar]
- 141.Berger R.L. Multiparameter Hypothesis Testing and Acceptance Sampling. Technometrics. 1982;24(4):295–300. [Google Scholar]
- 142.Berger R.L., Hsu J.C. Bioequivalence trials, intersection-union tests and equivalence confidence sets. Statistical Science. 1996;11(4):283–319. [Google Scholar]
- 143.Berger, R.L., Likelihood Ratio Tests and Intersection-Union Tests, in Advances in Statistical Decision Theory and Applications, S. Panchapakesan and N. Balakrishnan, Editors. 1997, Birkhäuser Boston: Boston, MA. p. 225-237.
- 144.Preacher K.J., Hayes A.F. Asymptotic and resampling strategies for assessing and comparing indirect effects in multiple mediator models. Behavior Research Methods. 2008;40(3):879–891. doi: 10.3758/brm.40.3.879. [DOI] [PubMed] [Google Scholar]
- 145.Preacher K.J., Hayes A.F. SPSS and SAS procedures for estimating indirect effects in simple mediation models. Behavior Research Methods, Instruments, & Computers. 2004;36(4):717–731. doi: 10.3758/bf03206553. [DOI] [PubMed] [Google Scholar]
- 146.Burgess S., Thompson D.J., Rees J.M.B., Day F.R., Perry J.R., Ong K.K. Dissecting Causal Pathways Using Mendelian Randomization with Summarized Genetic Data: Application to Age at Menarche and Risk of Breast Cancer. Genetics. 2017;207(2):481–487. doi: 10.1534/genetics.117.300191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 147.Bollen. Kenneth A and R. Stine, Direct and Indirect Effects: Classical and Bootstrap Estimates of Variability. Sociological Methodology, 1990. 20: p. 115-140.
- 148.Fritz M.S., Taylor A.B., Mackinnon D.P. Explanation of Two Anomalous Results in Statistical Mediation Analysis. Multivar Behav Res. 2012;47(1):61–87. doi: 10.1080/00273171.2012.640596. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 149.Mackinnon D.P., Lockwood C.M., Williams J. Confidence Limits for the Indirect Effect: Distribution of the Product and Resampling Methods. Multivar Behav Res. 2004;39(1):99–109. doi: 10.1207/s15327906mbr3901_4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 150.Williams J., Mackinnon D.P. Resampling and Distribution of the Product Methods for Testing Indirect Effects in Complex Models. Struct Eq Model Multidiscipl J. 2008;15(1):23–51. doi: 10.1080/10705510701758166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 151.Glinsky G.V. Integration of HapMap-Based SNP Pattern Analysis and Gene Expression Profiling Reveals Common SNP Profiles for Cancer Therapy Outcome Predictor Genes*. Cell Cycle. 2006;5(22):2613–2625. doi: 10.4161/cc.5.22.3498. [DOI] [PubMed] [Google Scholar]
- 152.Fabiani E., Leone G., Giachelia M., D'Alo F., Greco M., Criscuolo M. Analysis of genome-wide methylation and gene expression induced by 5-aza-2'-deoxycytidine identifies BCL2L10 as a frequent methylation target in acute myeloid leukemia. Leukemia Lymphoma. 2010;51(12):2275–2284. doi: 10.3109/10428194.2010.528093. [DOI] [PubMed] [Google Scholar]
- 153.Wang W., Baladandayuthapani V., Morris J.S., Broom B.M., Manyam G., Do K.-A. iBAG: integrative Bayesian analysis of high-dimensional multiplatform genomics data. Bioinformatics. 2013;29(2):149–159. doi: 10.1093/bioinformatics/bts655. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 154.de Tayrac M., Lê S., Aubry M., Mosser J., Husson F. Simultaneous analysis of distinct Omics data sets with integration of biological knowledge: Multiple Factor Analysis approach. BMC Genomics. 2009;10(1):32. doi: 10.1186/1471-2164-10-32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 155.Zhou W., Dinh H.Q., Ramjan Z., Weisenberger D.J., Nicolet C.M., Shen H. DNA methylation loss in late-replicating domains is linked to mitotic cell division. Nat Genet. 2018;50(4):591–602. doi: 10.1038/s41588-018-0073-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 156.Meir Z., Mukamel Z., Chomsky E., Lifshitz A., Tanay A. Single-cell analysis of clonal maintenance of transcriptional and epigenetic states in cancer cells. Nat Genet. 2020;52(7):709–718. doi: 10.1038/s41588-020-0645-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 157.Millstein J., Chen G.K., Breton C.V. cit: hypothesis testing software for mediation analysis in genomic applications. Bioinformatics. 2016;32(15):2364–2365. doi: 10.1093/bioinformatics/btw135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 158.Djordjilović, V., J. Hemerik and M. Thoresen, On optimal two-stage testing of multiple mediators. arXiv preprint arXiv:2007.02844, 2020. [DOI] [PMC free article] [PubMed]
- 159.Gao Y., Yang H., Fang R., Zhang Y., Goode E.L., Cui Y. Testing Mediation Effects in High-Dimensional Epigenetic Studies. Front Genet. 2019;10:1195. doi: 10.3389/fgene.2019.01195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 160.Fang R., Yang H., Gao Y., Cao H., Goode E.L., Cui Y. Gene-based mediation analysis in epigenetic studies. Briefings Bioinf. 2020 doi: 10.1093/bib/bbaa113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 161.Djordjilović V., Page C.M., Gran J.M., Nøst T.H., Sandanger T.M., Veierød M.B. Global test for high-dimensional mediation: Testing groups of potential mediators. Stat Med. 2019;38(18):3346–3360. doi: 10.1002/sim.8199. [DOI] [PubMed] [Google Scholar]
- 162.Zhao, Y. and X. Luo, Pathway Lasso: Estimate and Select Sparse Mediation Pathways with High Dimensional Mediators. 2016.
- 163.Song Y., Zhou X., Zhang M., Zhao W., Liu Y., Kardia S.L.R. Bayesian shrinkage estimation of high dimensional causal mediation effects in omics studies. Biometrics. 2020;76(3):700–710. doi: 10.1111/biom.13189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 164.Song, Y., X. Zhou, J. Kang, M. Aung, M. Zhang, W. Zhao, et al., Bayesian Sparse Mediation Analysis with Targeted Penalization of Natural Indirect Effects. 2020. [DOI] [PMC free article] [PubMed]
- 165.Song, Y., Zhou, X. Kang, J. Aung, M. Zhang, M. Zhao, W. et al., Bayesian Hierarchical Models for High-Dimensional Mediation Analysis with Coordinated Selection of Correlated Mediators; 2020. [DOI] [PMC free article] [PubMed]
- 166.Wu D., Yang H., Winham S.J., Natanzon Y., Koestler D.C., Luo T. Mediation analysis of alcohol consumption, DNA methylation, and epithelial ovarian cancer. J Hum Genet. 2018;63(3):339–348. doi: 10.1038/s10038-017-0385-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 167.Steen J., Loeys T., Moerkerke B., Vansteelandt S. Flexible Mediation Analysis With Multiple Mediators. Am J Epidemiol. 2017;186(2):184–193. doi: 10.1093/aje/kwx051. [DOI] [PubMed] [Google Scholar]
- 168.Taguri M., Featherstone J., Cheng J. Causal mediation analysis with multiple causally non-ordered mediators. Stat Methods Med Res. 2018;27(1) doi: 10.1177/0962280215615899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 169.Yang J.J., Li J., Williams L.K., Buu A. An efficient genome-wide association test for multivariate phenotypes based on the Fisher combination function. BMC Bioinf. 2016;17:19–29. doi: 10.1186/s12859-015-0868-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 170.Liu Y., Xie J. Cauchy Combination Test: A Powerful Test With Analytic p-Value Calculation Under Arbitrary Dependency Structures. J Am Stat Assoc. 2020;115(529):393–402. doi: 10.1080/01621459.2018.1554485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 171.Liu Y., Chen S., Li Z., Morrison A.C., Boerwinkle E., Lin X. ACAT: A Fast and Powerful p Value Combination Method for Rare-Variant Analysis in Sequencing Studies. Am J Human Genet. 2019;104(3):410–421. doi: 10.1016/j.ajhg.2019.01.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 172.Zhao S.D., Cai T.T., Li H. More powerful genetic association testing via a new statistical framework for integrative genomics. Biometrics. 2014;70(4):881–890. doi: 10.1111/biom.12206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 173.Zou H., Hastie T., Tibshirani R. Sparse Principal Component Analysis. J Computat Graph Statist. 2006;15(2):265–286. [Google Scholar]
- 174.Bogomolov M., Heller R. Assessing replicability of findings across two studies of multiple features. Biometrika. 2018;105(3):505–516. [Google Scholar]
- 175.Jin J., Cai T.T. Estimating the Null and the Proportion of Nonnull Effects in Large-Scale Multiple Comparisons. J Am Stat Assoc. 2007;102(478):495–506. [Google Scholar]
- 176.Jiang H., Doerge R.W. Estimating the proportion of true null hypotheses for multiple comparisons. Cancer Inf. 2008;6:25–32. [PMC free article] [PubMed] [Google Scholar]
- 177.Efron B. Size, power and false discovery rates. Ann Statist. 2007;35(4):1351–1377. [Google Scholar]
- 178.Storey J. The positive false discovery rate: a Bayesian interpretation and the q-value. Ann Statist. 2003;31:2013–2035. [Google Scholar]
- 179.Efron B., Zhang N.R. False discovery rates and copy number variation. Biometrika. 2011;98(2):251–271. [Google Scholar]
- 180.Storey J., Tibshirani R. Statistical significance for genomewide studies. PNAS. 2003;100:9440–9445. doi: 10.1073/pnas.1530509100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 181.Storey J. A direct approach to false discovery rates. J Roy Statist Soc: Ser B (Statistical Methodology) 2002;64:479–498. [Google Scholar]
- 182.Efron B., Tibshirani R. Empirical bayes methods and false discovery rates for microarrays. Genet Epidemiol. 2002;23(1):70–86. doi: 10.1002/gepi.1124. [DOI] [PubMed] [Google Scholar]
- 183.Efron B. Large-Scale Simultaneous Hypothesis Testing. J Am Stat Assoc. 2004;99(465):96–104. [Google Scholar]
- 184.Langaas M., Lindqvist B.H., Ferkingstad E. Estimating the proportion of true null hypotheses, with application to DNA microarray data. J Roy Statist Soc: Series B (Statistical Methodology) 2005;67(4):555–572. [Google Scholar]
- 185.Kwee L.C., Liu D., Lin X., Ghosh D., Epstein M.P. A Powerful and Flexible Multilocus Association Test for Quantitative Traits. Am J Hum Genet. 2008;82(2):386–397. doi: 10.1016/j.ajhg.2007.10.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 186.Wu M.C., Kraft P., Epstein M.P., Taylor D.M., Chanock S.J., Hunter D.J. Powerful SNP-Set Analysis for Case-Control Genome-wide Association Studies. Am J Human Genet. 2010;86(6):929–942. doi: 10.1016/j.ajhg.2010.05.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 187.Wu Michael C., Lee S., Cai T., Li Y., Boehnke M., Lin X. Rare-Variant Association Testing for Sequencing Data with the Sequence Kernel Association Test. Am J Human Genet. 2011;89(1):82–93. doi: 10.1016/j.ajhg.2011.05.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 188.Lin X. Variance component testing in generalised linear models with random effects. Biometrika. 1997;84(2):309–326. [Google Scholar]
- 189.Goeman J.J., Van De Geer S.A., Van Houwelingen H.C. Testing against a high dimensional alternative. J Roy Statist Soc Ser B (Statistical Methodology) 2006;68(3):477–493. [Google Scholar]
- 190.Zhang C.-H. Nearly unbiased variable selection under minimax concave penalty. Ann Statist. 2010;38(2):894–942. [Google Scholar]
- 191.Fan J., Lv J. Sure independence screening for ultrahigh dimensional feature space. J Roy Statist Soc: Ser B (Statistical Methodology) 2008;70(5):849–911. doi: 10.1111/j.1467-9868.2008.00674.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 192.Fan J., Lv J. Statistics Reference Online; Wiley StatsRef: 2018. Sure Independence Screening; pp. 1–8. [Google Scholar]
- 193.Boyd S., Parikh N., Chu E., Peleato B., Eckstein J. Distributed Optimization and Statistical Learning via the Alternating Direction Method of Multipliers. Foundations and Trends®. Machine Learning. 2011;3(1):1–122. [Google Scholar]
- 194.Zhou X., Carbonetto P., Stephens M. Polygenic modeling with Bayesian sparse linear mixed models. PLoS Genet. 2013;9(2) doi: 10.1371/journal.pgen.1003264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 195.Ročková V., George E.I. The Spike-and-Slab LASSO. J Am Stat Assoc. 2016 [Google Scholar]
- 196.Zeng P., Hao X., Zhou X. Pleiotropic mapping and annotation selection in genome-wide association studies with penalized Gaussian mixture models. Bioinformatics. 2018;34(16):2797–2807. doi: 10.1093/bioinformatics/bty204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 197.Potts R.B. Some generalized order-disorder transformations. Math Proc Cambridge Philos Soc. 1952;48(1):106–109. [Google Scholar]
- 198.Aitchison J., Shen S.M. Logistic-Normal Distributions: Some Properties and Uses. Biometrika. 1980;67(2):261–272. [Google Scholar]
- 199.Li W., Zhou X.-H. Identifiability and estimation of causal mediation effects with missing data. Stat Med. 2017;36(25):3948–3965. doi: 10.1002/sim.7413. [DOI] [PubMed] [Google Scholar]
- 200.Zheng C., Zhou X.-H. Causal mediation analysis in the multilevel intervention and multicomponent mediator case. J Roy Statist Soc: Series B (Statistical Methodology) 2015;77(3):581–615. [Google Scholar]
- 201.Bind M.A., VanderWeele T.J., Schwartz J.D., Coull B.A. Quantile causal mediation analysis allowing longitudinal data. Stat Med. 2017;36(26):4182–4195. doi: 10.1002/sim.7423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 202.Bind M.A.C., Vanderweele T.J., Coull B.A., Schwartz J.D. Causal mediation analysis for longitudinal data with exogenous exposure. Biostatistics. 2016;17(1):122–134. doi: 10.1093/biostatistics/kxv029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 203.Qin H., Niu T., Zhao J. Identifying Multi-Omics Causers and Causal Pathways for Complex Traits. Front Genet. 2019;10(110) doi: 10.3389/fgene.2019.00110. [DOI] [PMC free article] [PubMed] [Google Scholar]