Abstract
Resistance to antimalarial drugs, and in particular to the artemisinin derivatives and their partner drugs, threatens recent progress toward regional malaria elimination and eventual global malaria eradication. Population-level studies utilizing whole-genome sequencing approaches have facilitated the identification of regions of the parasite genome associated with both clinical and in vitro drug-resistance phenotypes. However, the biological relevance of genes identified in these analyses and the establishment of a causal relationship between genotype and phenotype requires functional characterization. Here we examined data from population genomic and transcriptomic studies in the context of data generated from recent functional studies, using a new population genetic approach designed to identify potential favored mutations within the region of a selective sweep (iSAFE). We identified several genes functioning in pathways now known to be associated with artemisinin resistance that were supported in early population genomic studies, as well as potential new drug targets/pathways for further validation and consideration for treatment of artemisinin-resistant Plasmodium falciparum. In addition, we establish the utility of iSAFE in identifying positively-selected mutations in population genomic studies, potentially accelerating the time to functional validation of candidate genes.
Keywords: P. falciparum, Drug resistance, iSAFE, Functional genomics
Graphical abstract
Highlights
-
•
Artemisinin resistance mechanisms identified in recent functional studies are supported in previous population-level studies.
-
•
iSAFE can identify possible favored mutations within a selective sweep and accelerate time to functional validation.
-
•
The role of exported proteins and host cell remodeling in artemisinin resistance warrants further investigation.
1. Introduction
Although malaria incidence has declined over the last decade, progress toward malaria elimination has stalled over the last 5 years as incidence has plateaued (World Health Organization, 2019). In addition, these gains are threatened by the continued evolution of resistance in both the parasite and mosquito vector in response to interventions. For example, the parasite continues to evolve resistance to antimalarial drugs, with Plasmodium falciparum having developed resistance to most antimalarials used to treat clinical infection, including former first-line antimalarials such as chloroquine, the antifolates, and most recently the artemisinin derivatives and their partner drugs used in artemisinin-based combination therapies (ACTs) [Reviewed in (Menard and Dondorp, 2017)]. Indeed, in some malaria endemic areas, such as the eastern Greater Mekong Subregion, P. falciparum is resistant to multiple approved ACTs, severely limiting treatment options (World Health Organization, 2019).
The advent of technologies allowing whole-genome sequencing of P. falciparum at epidemiological scales has facilitated the use of population-level approaches to identify regions of the parasite genome associated with drug-resistance phenotypes. These studies have been successful in identifying molecular markers of resistance and the genetic architecture of resistant parasites and have provided insights into the underlying mechanisms of resistance (Agrawal et al., 2017; Amato et al., 2017; Ariey et al., 2014; Borrmann et al., 2013; Cerqueira et al., 2017; Cheeseman et al., 2012, 2015; Demas et al., 2018; Miotto et al., 2013, 2015; Mu et al., 2010; Mukherjee et al., 2017; Park et al., 2012; Takala-Harrison et al., 2013, 2015; Van Tyne et al., 2011; Wang et al., 2016; Wendler et al., 2014; Witkowski et al., 2010; Yuan et al., 2011; Zhu et al., 2018). However, these studies are limited by the fact that observed associations are not necessarily causal and that, in the region of a selective sweep, several polymorphisms in multiple genes may be associated with the phenotype of interest, making it difficult to pinpoint the gene driving the observed associations. For example, our initial genome-wide association study of delayed parasite clearance following artemisinin treatment identified a region of chromosome 13 (containing 12 genes) that was associated with parasite clearance half-life (Takala-Harrison et al., 2013). In a similar timeframe, other population-genomic studies also identified a region of chromosome 13 that appeared to be under recent positive selection and highly differentiated in artemisinin resistant parasite populations (Cheeseman et al., 2012; Miotto et al., 2013). In these studies, it was not clear which gene(s) contained the causal mutation until the following year, when results from in vitro selection experiments indicated that mutations within the kelch13 gene were responsible for artemisinin resistance (Ariey et al., 2014). Since these initial studies, several subsequent population-level genomic and transcriptomic studies have identified multiple genomic regions and differentially expressed genes associated with resistance to the artemisinin derivatives or their partner drugs (Agrawal et al., 2017; Amato et al., 2017; Borrmann et al., 2013; Cerqueira et al., 2017; Cheeseman et al., 2015; Demas et al., 2018; Miotto et al., 2013, 2015; Mok et al., 2015; Mukherjee et al., 2017; Takala-Harrison et al., 2015; Wang et al., 2016; Wendler et al., 2014; Zhu et al., 2018); however, as in the case of kelch13, the function and biological relevance of these genes has not always been clear.
The development of higher-throughput experimental genetic approaches has increased the capacity for functional characterization of gene products as they relate to various phenotypes of interest (Birnbaum et al., 2020; Cowell et al., 2018; Demas et al., 2018; Gnädig et al., 2020; Li et al., 2019; Oberstaller et al., 2021; Zhang et al., 2020), including drug resistance. Results from such genome-scale screens provide an opportunity to re-interpret previous findings of population-level genomic and transcriptomic studies for which the functional relevance of identified genes is not understood. In this study, we aimed to integrate findings from previous population-level studies with new functional data to better understand drug resistance and compensatory mechanisms utilized by multidrug-resistant malaria parasites. In addition, we utilized a new population-genetic approach (i.e., Integrated Selection of Allele Favored by Evolution (iSAFE)), designed to identify favored mutation(s) within the genomic region of a positive selective sweep (Akbari et al., 2018), to identify potentially novel functional targets that may inform rational strategies to identify new drugs to counter resistance.
2. Methods
2.1. Literature review
We mined the literature for genes/genomic regions identified in multiple population-genomic or transcriptomic studies of resistance to artemisinin derivatives or ACTs. We curated gene IDs from 22 published population-genomic or transcriptomic studies and 8 published functional screens encompassing 28 total publications (Table 1). Studies were published between the years 2010 and 2020 and were conducted in malaria-endemic areas of Asia, Africa, South America, and Oceania. Genes in genomic regions either significantly associated with a resistance phenotype or identified based on estimated signals of positive selection were compiled, and any outdated gene IDs were corrected using PlasmoDB release 48 (https://plasmodb.org) (Aurrecoechea et al., 2009). Each gene was given an evidence score equal to the number of independent genomic, transcriptomic or functional studies in which it was identified (Supplementary Table 1). Genes containing more than one SNP associated with a phenotype (e.g., kelch13) were only counted once per publication.
Table 1.
Publications providing source-data for this meta-analysis by evidence type.
2.2. Down-selection of genes for iSAFE analysis
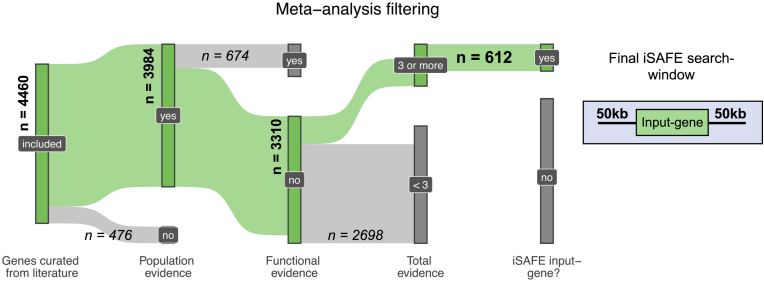
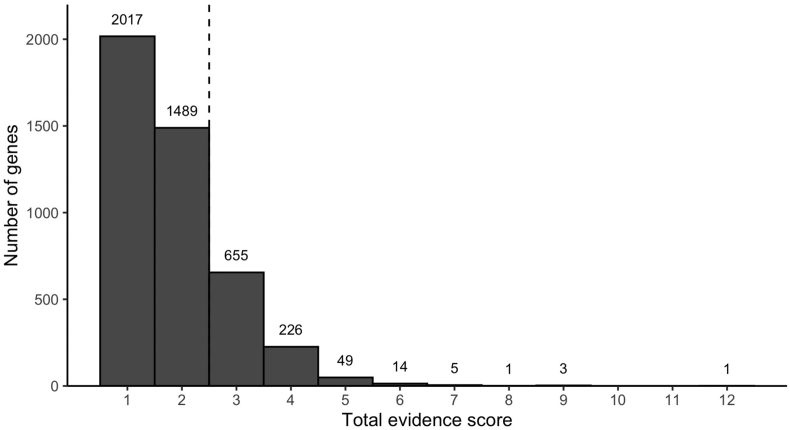
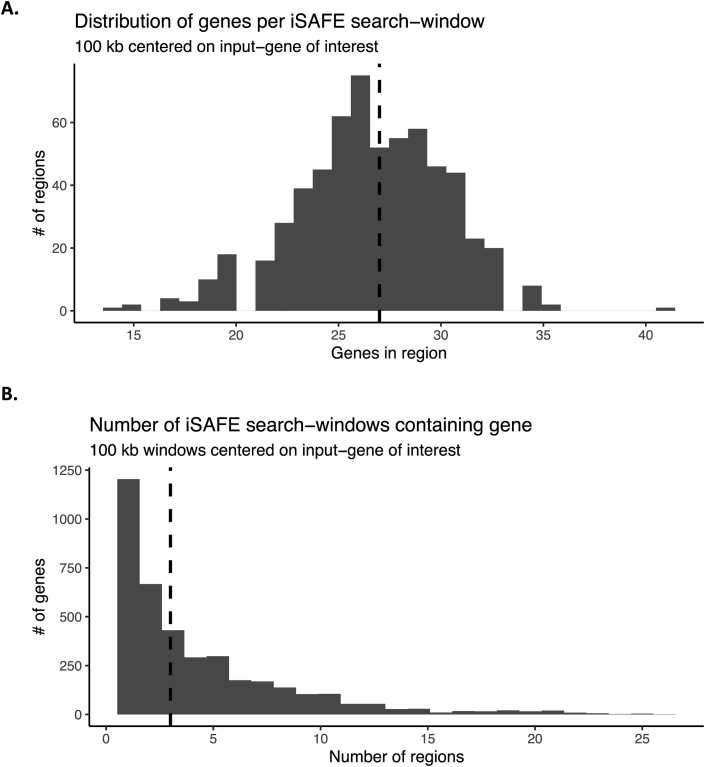
We aimed to 1) estimate the degree to which population-level associations or selection signals can be explained by new functional evidence and 2) identify potentially novel functions contributing to resistance. To accomplish the latter goal, we first sought to identify genes with population-genomic or transcriptomic evidence, but not functional evidence, for further analysis to identify favored mutations driving associations with resistance. The down-selection strategy is illustrated in Fig. 1. From among genes identified in the literature search, we first excluded genes that were not identified in at least one population-genomics study. We then excluded genes that were identified in one or more functional screens (to identify potential new functional targets). Finally, based on the inflection point of a plot of the number of citations per gene (Fig. 2), we excluded all genes observed in fewer than three independent studies.
Fig. 1.
Down-selection strategy for genes to include in iSAFE analysis. The green flows and associated bolded counts indicate genes continuing through filtering at each step, while grey flows and italicized counts indicate genes filtered out at each step. 612 genes met our criteria for iSAFE analysis. Final iSAFE search-windows were 100 kb centered around each of the 612 input-genes, containing on average 27 genes per window. (For interpretation of the references to color in this figure legend, the reader is referred to the Web version of this article.)
Fig. 2.
Distribution of total evidence-scores across all genes. Each publication contributes one point to the total evidence score. Genes implicated by < 3 publications (dashed line) were not considered as input genes for the iSAFE analysis.
2.3. Integrated Selection of Allele Favored by evolution (iSAFE) analysis
The remaining genes were analyzed using Integrated Selection of Allele Favored by Evolution (iSAFE), a new approach that uses population genetic signals to identify the favored mutation within the broad genomic region contained within a recent positive selective sweep (Akbari et al., 2018). The iSAFE score is based on the distribution of each derived mutation in sampled haplotypes and the relative frequency of those haplotypes, where the favored mutation is expected to be associated with a small number of distinct haplotypes that are at high frequency in the population. First, a SAFE score is calculated for each derived mutation, which is a function of the fraction of haplotypes containing the mutation, the total number of derived mutations in those haplotypes, and the relative frequency of each haplotype (Ronen et al., 2015). The iSAFE score for each mutation then combines the SAFE score across several consecutive, small (low-recombination) genomic windows. iSAFE exploits slight differences in signal expected between the selected mutation and those closely linked to it, which lie on the shoulders of the sweep. iSAFE becomes less accurate when favored mutations are at or near fixation (derived allele frequency > 0.9). To account for these types of mutations, samples chosen randomly from a non-target population can be used as a control, to modulate the derived allele frequency of the favored mutation.
Using iSAFE, we analyzed publicly available whole-genome sequencing data from 61 P. falciparum isolates (Supplementary Table 2), including 34 isolates that harbor the Kelch13 C580Y mutation and belong to the same genetic subpopulation (ie., “cases”) and 27 isolates that lack Kelch13 mutations and belong to the pre-artemisinin resistance Southeast Asian genetic subpopulation (i.e., “controls”) (Dwivedi et al., 2017; Miotto et al., 2013; Takala-Harrison et al., 2015). Variant-calling was performed using Haplotype Caller to generate genomic variant call format (GVCF) files for each sample and joint single nucleotide polymorphism (SNP) calling was performed using GATK (v4.1) (McKenna et al., 2010). Variant calls were filtered for quality based on the following criteria: (QUAL < 50 || DP < 12 || FS > 14.5 || MQ < 20.0).
We examined 100 kb regions centered around the 612 genes of interest to identify genes containing putative favored mutations within that genomic region. The 612 windows included an average of 27 genes/window, and combined, covered 2727 unique genes across the genome (Fig. 1, Supplementary Figure 1). Several iSAFE parameters were modified from defaults (designed for human genomes) to be appropriate for the much-smaller P. falciparum genome: minimum region-size was decreased to 20 bp or 20 polymorphic sites, with a sliding window of 10 polymorphic sites and a step-parameter of 5 polymorphic sites. SNPs with an iSAFE score >0.5 and iSAFE rank above 5 were considered possible favored mutations (Supplementary Table 3; Supplementary Figure 2). Variant annotation information, including predicted phenotypic consequences of each candidate mutation was assessed using SNPeff (v.4.3) (Supplementary Table 4). Genes of interest were grouped into three categories based on whether they contained possible favored mutations: 1) genes linked to known artemisinin resistance determinants or genetic background genes as identified by Miotto and colleagues (Miotto et al., 2015) (e.g., Kelch13, PfCRT, etc.); 2) genes predicted to contain a putative favored mutation, and 3) genes not containing a putative favored mutation and not “hitchhiking” with known artemisinin resistance determinants. Genes linked to known artemisinin resistance determinants were excluded from further analysis. Genes predicted to contain a putative favored mutation underwent Gene Ontology (GO) enrichment analysis to identify significantly enriched functions representing potential new functional targets. Genomic regions centered on genes that were neither hitchhiking with known resistance determinants nor harboring possible favored mutations themselves were examined for linked genes predicted to contain a favored mutation. These linked genes with favored mutations also underwent a GO analysis to identify enriched functions and were cross-referenced with genes having functional evidence to determine the extent to which population-level signals are corroborated by data from recent functional screens.
2.4. Gene ontology (GO) enrichment analysis
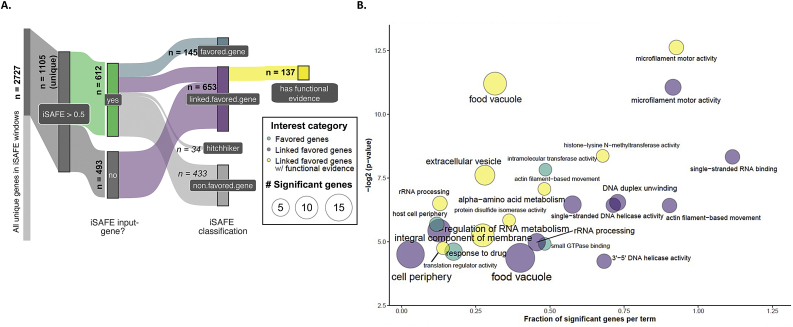
All GOenrichment analyses were performed testing GOterms mapped to genes in the category of interest against a background of GOterms mapped to all other genes in the analysis. The GOterm database was created from the latest curated P. falciparum ontology available at the time of analysis, downloaded from GeneDB (accessed May 2, 2019) (Logan-Klumpler et al., 2012). GO enrichment analyses were performed in R using the topGO package from bioconductor (v. 2.40.0) (Alexa and Rahnenfuhrer, 2020). Genes selected for iSAFE analysis containing favored mutations (n = 145; Fig. 3A, Supplementary Table 6) were compared to a background of all genes with population genomic evidence but not functional evidence (n = 3310 genes; Fig. 3A, Supplementary Table 1). Genes driving the signal of nearby iSAFE input genes of interest (n = 653; Fig. 3A, Supplementary Table 6) were compared to a background of all genes with population genomic evidence identified in the literature search, including those with functional evidence (n = 3984; Fig. 3A, Supplementary Table 1). GO terms with a weighted Fisher/elim hybrid p-value ≤ 0.05 were further examined as significantly enriched biological processes, molecular functions, or cellular compartments.
Fig. 3.
Functional enrichment of genes containing favored mutations. A. iSAFE classification for functional enrichment analyses. A total of 2727 unique genes were covered in iSAFE search-windows centered on the 612 input-genes satisfying our criteria. All genes with an iSAFE score >0.5 were classified by favored mutation: 145 iSAFE input-genes contained one or more favored mutations (“favored.gene”, blue). 653 genes contained favored mutations linked to input genes (“favored.linked.genes”, purple), and 137 of those had functional support from at least one screen (yellow). Genes linked to favored mutations in a known determinant of artemisinin resistance were discarded (“hitchhikers”, n = 34; see Methods). Counts are provided in bold for each total iSAFE classification category-size. Genes filtered at each step are indicated in italics. Flows are colored for final categories tested for functional enrichment against a database of all other genes included in the analysis. B. Functional enrichment of favored mutations. Colors are as in 3A. Circle-size indicates the number of significant genes mapped to a GO term in the given category of interest. The x-axis indicates proportion of significant genes vs. background-genes mapped to that term in the analysis. Positions are approximate. . (For interpretation of the references to color in this figure legend, the reader is referred to the Web version of this article.)
3. Results
3.1. Genomic regions identified in multiple independent studies of artemisinin resistance or ACTs
A total of 4460 genes were identified within the 28 publications reviewed for this analysis (Table 1, Fig. 1). Of these 4460 genes, 78.6% were only observed in one or two independent studies, while 0.5% were observed in ≥5 independent studies (Fig. 1), including genes encoding Kelch13 (PF3D7_1343700), PfCRT (PF3D7_0709000), prodrug activation and resistance esterase (PF3D7_0709700), autophagy-related protein 7 (PF3D7_1126100), heat shock protein 90 (PF3D7_0708400), and K13-interacting candidates 1 and 5 (KIC1, KIC5), among others (Supplementary Table 1). As indicated in Fig. 1, 612 genes were down-selected for inclusion in the iSAFE analysis, including genes with genomic evidence score ≥1, functional score = 0, and total evidence score ≥3 (Supplementary Table 1).
3.2. iSAFE analysis
To verify that iSAFE was working as expected with our parameters and genomic data set (representative of a pre- and post-Kelch13 C580Y selective sweep), we performed iSAFE on the 100 kb region centered around the gene encoding Kelch13. The top favored mutation identified by iSAFE encodes C580Y, indicating that iSAFE is detecting known mutations favored by selection (Supplementary Table 5). We next applied iSAFE to examine 100 kb genomic regions centered on the 612 genes of interest (n = 2727 unique genes) and identified 34 genes that were linked to known artemisinin-resistance determinants or genetic background genes (Supplementary Table 6). 145 of the 612 genes of interest were predicted to contain favored mutations that were likely driving the selection signal (Supplementary Tables 3-6) and represent potential new functional targets that require validation. Genomic windows comprising the 433 genes that were neither “hitchhiking” with known resistance determinants nor harboring favored mutations themselves contained 653 genes that were previously identified in at least one population genomics study and were predicted to contain a putative favored mutation. Of these 653 genes, 137 have been identified in recent genome-scale functional screens (Supplementary Table 7), representing a significant enrichment of genes with functional evidence among genes with putative favored mutations in genomic regions identified in population-level studies (Fisher's exact test, p-value < 0.001).
3.3. Genes identified in functional screens that are supported by population-level studies
A GO enrichment analysis was performed on the 653 genes containing favored mutations that were linked to genes identified in population-level studies. Significantly enriched GO terms (hybrid Fisher/elim p-value<0.05; Supplementary Table 8) are shown in Fig. 3B (purple), with genes corresponding to those terms shown in Supplementary Table 9. Significantly enriched molecular functions or biological processes include microfilament motor activity (GO:0000146) and actin filament-based movement (GO:0030048), single-stranded RNA binding (GO:0003727), rRNA processing (GO:0006364) and regulation of RNA metabolic process (GO:0051252), DNA helicase activity (GO:0017116, GO:0032508, GO:0043138), and alpha-amino acid metabolic process (GO:1901605). Significantly enriched cellular compartments include the cell periphery, food vacuole, nucleus, and nuclear periphery.
Of these 653 genes, 137 have been identified in recent functional screens of antimalarial drug resistance (Supplementary Table 7), including 40 conserved proteins of unknown function. Among these 137 genes, several have been shown to encode products involved in processes implicated in artemisinin resistance, such as proteasomal degradation, endocytosis, vesicular trafficking, or stress response (Fig. 3B, yellow; Table 2; Supplementary Tables 10-11). Notable examples include amino acid transporter AAT1 (PF3D7_0629500), clathrin heavy chain (PF3D7_1219100), DER-1 like protein (PF3D7_1032500), Eps15-like protein (PF3D7_1025000), actin II (PF3D7_1412500), myosins F and J (PF3D7_1329100, PF3D7_1229800), ubiquitin conjugating enzyme E2 (PF3D7_0812600), K13 interacting protein KIC6 (PF3D7_0609700), and other proteins shown to interact with K13 (Gnädig et al., 2020). In addition, proteins involved in erythrocyte invasion, such as apical membrane antigen 1 (AMA1), merozoite surface protein 1 (MSP1), and rhoptry neck protein 2 (RON2) were also among those proteins having both functional and population-level evidence. Interestingly, a number of genes encoding exported proteins were among the 137 genes with functional evidence, including genes encoding members of the PfEMP1, PHIST, RIFIN, and CLAG protein families (Table 2).
Table 2.
Selected genes in functional categories of interest implicated in artemisinin resistance by multiple population-level studies.
| Gene_ID | Total score | Annotation | Functional Categories |
|---|---|---|---|
| PF3D7_1118200 | 5 | Heat shock protein 90, putative | Stress response |
| PF3D7_1329100 | 5 | Myosin F, putative | Vesicular trafficking |
| PF3D7_1412500 | 4 | Actin II | Vesicular trafficking |
| PF3D7_0629500 | 4 | Amino acid transporter AAT1 | Hemoglobin digestion/digestive vacuole |
| PF3D7_1133400 | 4 | Apical membrane antigen 1 | Exported protein, invasion |
| PF3D7_0220800 | 4 | Cytoadherence linked asexual protein 2 | Exported protein, host-cell remodeling |
| PF3D7_0629200 | 4 | DnaJ protein, putative | Exported protein, host-cell remodeling |
| PF3D7_0823800 | 4 | DnaJ protein, putative | Exported protein, host-cell remodeling |
| PF3D7_0930300 | 4 | Merozoite surface protein 1 | Exported protein, invasion |
| PF3D7_0902700 | 4 | Plasmodium exported protein (PHISTb) | Exported protein, host-cell remodeling |
| PF3D7_1016800 | 4 | Plasmodium exported protein (PHISTc) | Exported protein, host-cell remodeling |
| PF3D7_0609700 | 4 | Protein KIC6 | K13-mediated endocytosis |
| PF3D7_0822600 | 4 | Protein transport protein SEC23 | Vesicular transport; protein export |
| PF3D7_1452000 | 4 | Rhoptry neck protein 2 | Exported protein, invasion |
| PF3D7_0702400 | 4 | Small, exported membrane protein 1 | Exported protein |
| PF3D7_0319700 | 3 | ABC transporter I family member 1, putative | Stress response |
| PF3D7_0935800 | 3 | Cytoadherence linked asexual protein 9 | Exported protein, host-cell remodeling |
| PF3D7_0711700 | 3 | Erythrocyte membrane protein 1, PfEMP1 | Exported protein, host-cell remodeling |
| PF3D7_1229800 | 3 | Myosin J, putative | Vesicular trafficking |
| PF3D7_0608800 | 3 | Ornithine aminotransferase | Stress response |
| PF3D7_1119900 | 3 | Protein transport protein SEC16, putative | Vesicular trafficking |
| PF3D7_0935900 | 3 | Ring-exported protein 1 | Exported protein, host-cell remodeling |
| PF3D7_1145600 | 3 | TMEM65 domain-containing protein, putative | Stress response |
| PF3D7_1414700 | 3 | Ubiquitin carboxyl-terminal hydrolase, putative | Protein degradation |
| PF3D7_1025000 | 2 | Eps15-like protein | K13 interactome/protein degradation |
| PF3D7_0533100 | 2 | Erythrocyte membrane protein 1 (PfEMP1) | Exported protein, host-cell remodeling |
| PF3D7_0712900 | 2 | Erythrocyte membrane protein 1, PfEMP1 | Exported protein, host-cell remodeling |
| PF3D7_1476600 | 2 | Plasmodium exported protein, unknown function | Exported protein |
| PF3D7_1472600 | 2 | protein disulfide-isomerase | Oxidative stress (redox)/protein degradation |
| PF3D7_1340700 | 2 | ras-related protein Rab-11B | Vesicular trafficking |
| PF3D7_0712500 | 2 | rifin, pseudogene | Exported protein, host-cell remodeling |
| PF3D7_1352500 | 2 | thioredoxin-related protein, putative | Oxidative stress |
| PF3D7_0812600 | 2 | ubiquitin-conjugating enzyme E2, putative | Protein degradation |
3.4. Potential new targets from population-genomic studies
145 genes evaluated in the iSAFE analysis contained favored mutations (Supplementary Table 6). A GO enrichment analysis indicated significant enrichment (hybrid Fisher/elim p-value<0.05; Supplementary Table 11) in gene products involved in intramolecular transport activity (GO:0016866), small GTPase binding (GO:0031267), and response to drug (GO:0042493), as well as in gene products found in the host cell periphery (Fig. 3, turquoise). Ten genes were associated with these GO terms (Table 3; Supplementary Table 12), including (among others) the gene encoding phosphatidylinositol-4-phosphate 5-kinase, PIP5K (PF3D7_0110600), serine/threonine kinase, FIKK4.2 (PF3D7_0424700), histone acetyltransferase, MYST (PF3D7_1118600), and calcium-dependent protein kinase 5, CDPK5 (PF3D7_1337800).
Table 3.
Genes mapped to categories functionally enriched in favored linked genes.
| GO_ID | Term | Gene_ID | Gene_Name | Product |
|---|---|---|---|---|
| GO:0031267 | small GTPase binding | PF3D7_0110600 | PIP5K | phosphatidylinositol-4-phosphate 5-kinase |
| GO:0044538 | host cell periphery | PF3D7_0424700 | FIKK4.2 | serine/threonine protein kinase FIKK family |
| GO:0042493 | response to drug | PF3D7_0615100 | ENR | enoyl-acyl carrier reductase |
| GO:0044538 | host cell periphery | PF3D7_0701900 | Plasmodium exported protein unknown function | |
| GO:0031267 | small GTPase binding | PF3D7_0711500 | regulator of chromosome condensation putative | |
| GO:0042493 | response to drug | PF3D7_0810800 | PPPK-DHPS | dihydropteroate synthase |
| GO:0016866 | intramolecular transferase activity | PF3D7_1017400 | PMM | haloacid dehalogenase-like hydrolase putative |
| GO:0042493 | response to drug | PF3D7_1118600 | MYST | histone acetyltransferase MYST |
| GO:0016866 | intramolecular transferase activity | PF3D7_1130000 | PAGM | phosphoacetylglucosamine mutase putative |
| GO:0042493 | response to drug | PF3D7_1337800 | CDPK5 | calcium-dependent protein kinase 5 |
4. Discussion
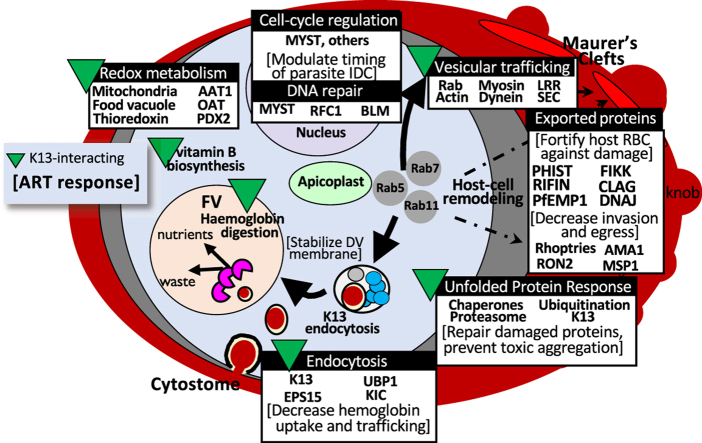
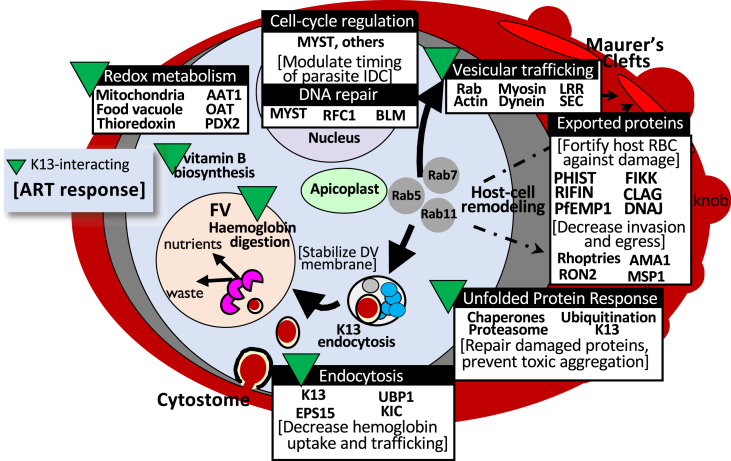
Population-genomic and transcriptomic studies have identified genomic regions and differentially expressed genes associated with resistance phenotypes; however, the function and biological relevance of these genes has not always been clear. Recent advances in both targeted and forward-genetic approaches are providing insight into the function of many Plasmodium genes as it relates to phenotypes of public health interest, including drug resistance. In this study, we conducted a meta-analysis of population-level studies of resistance to artemisinin derivatives or ACTs, identified genes or genomic regions identified in multiple studies, and used a new population-genomic tool, iSAFE, to identify genes likely to contain favored mutations driving a selective sweep or that are linked to genes with possible favored mutations. We found that multiple prongs of the increasingly complex network of interactions implicated in artemisinin resistance, including those related to proteasomal degradation, endocytosis, vesicular trafficking and stress response are well-supported in the data obtained from previous population-level studies (Fig. 4). Our results suggested a role for exported proteins in artemisinin resistance and identified potential new drug targets/pathways to consider for treatment of artemisinin-resistant P. falciparum. The large number of uncharacterized gene products with both population-level and functional evidence emphasizes the need for further genetic screens to understand the role of these gene products in established or yet-undiscovered mechanisms contributing to parasite artemisinin resistance.
Fig. 4.
Summary of processes functionally implicated in artemisinin resistance and supported in earlier population-level data.
Artemisinin resistance was first reported in western Cambodia in 2006–2007 (Dondorp et al., 2009; Noedl et al., 2008), and primarily manifested as delayed clearance of parasitemia following treatment with artemisinin derivatives or ACTs. Although Kelch13 was identified as a major determinant of this phenotype in 2014 (Ariey et al., 2014), the molecular mechanisms underlying this clinical phenotype have been more difficult to resolve. Artemisinin derivatives are thought to be activated by reduced iron that results from parasite hemoglobin digestion (Klonis et al., 2011), resulting in reactive carbon-centered radicals and oxygen species that cause extensive cellular damage and lethal proteotoxic stress. Parasite artemisinin resistance/tolerance has been hypothesized to result from decreased endocytosis and hemoglobin digestion, as well as increased cellular repair in surviving parasites (reviewed in (Sutherland et al., 2020; Xie et al., 2020)). The complete picture of all Kelch13 functions has not yet been elucidated; however, Kelch13 and its interacting partners have been localized to parasite cytostomes where they are hypothesized to play a role in hemoglobin uptake from the host cytosol (Birnbaum et al., 2020; Yang et al., 2019). Kelch13 abundance is reduced in mutant parasites (Gnädig et al., 2020), leading to decreased hemoglobin uptake, disrupted hemoglobin catabolism, and reduced artemisinin activation. Several proteins involved in endocytosis and vesicular trafficking, including proteins shown to interact with Kelch13 (Birnbaum et al., 2020; Gnädig et al., 2020), have been identified in this meta-analysis, offering additional support for this resistance mechanism based on earlier population-level studies. Likewise, gene products involved in oxidative stress and protein damage responses (e.g., thioredoxin-related protein, PF3D7_1352500; Der1-like protein, PF3D7_1032500) are well-supported in previous population genomic studies, based on our analysis. Using iSAFE, we have also identified genes with favored mutations that are functionally-related to these established processes, that are well-supported by population-level studies, but have not been characterized in functional studies for their specific contributions to artemisinin resistance, including genes encoding phosphatidylinositol-4-phosphate 5-kinase, PIP5K (PF3D7_0110600), the histone acetyltransferase, PfMYST (PF3D7_1118600), and calcium-dependent protein kinase 5, CDPK5 (PF3D7_1337800) (Table 3, Fig. 4).
Phosphatidylinositol-4-phosphate 5-kinase, PIP5K (PF3D7_0110600) is thought to be a bifunctional molecule with a PIP5K domain at its C-terminus and a neuronal calcium sensor (NCS)-like domain (i.e., calcium trigger that causes protein conformational changes in response to calcium) at its N-terminus (Leber et al., 2009). In eukaryotes, PIP5K plays a role in membrane trafficking, cytoskeletal reorganization and cell signaling by regulating the levels of PtdIns(4,5)P2, in conjunction with phosphatidylinositol 4-kinases (PI4Ks) (Martin, 2012). Based on experimental data from other eukaryotes (Kakiuchi et al., 2007; Liu et al., 2018; Voisinne et al., 2016), PI4Ks on both chromosomes 4 (PF3D7_0419900) and 5 (PF3D7_0509800) are predicted to be functional partners of PIP5K (Szklarczyk et al., 2019), a mutation in the former having been identified by Cerqueira et al. as increasing in prevalence along the same timeline as the Kelch13 C580Y protein (Cerqueira et al., 2017; Van Tyne et al., 2011). Other members of the inositol phosphate metabolism pathway, namely PI3K (PF3D7_0515300), have also been functionally implicated in artemisinin resistance (Mbengue et al., 2015), adding further support for the prospect that this pathway may be useful to target to counter artemisinin resistance.
PfMYST (PF3D7_1118600) is a histone acetyltransferase (HAT) that has been demonstrated to play a role in regulation of gene expression, cell cycle control, and DNA repair (Miao et al., 2010). PfMYST is the only HAT of its kind in P. falciparum. It is expressed in both long and short forms and localizes to both the nucleus and the cytoplasm (Miao et al., 2010). Efforts to knock out pfmyst have not been successful, suggesting that it is an essential gene; however, overexpression of PfMYST led to changes in cell cycle progression. Specifically, parasites with overexpression of full-length PfMYST displayed a shorter intraerythrocytic developmental cycle (IDC), while parasites expressing truncated PfMYST had a more prolonged IDC compared to control parasites, with IDC changes occurring primarily in the schizont stage (Miao et al., 2010). Changes in the duration of the parasite IDC are also a hallmark of artemisinin-resistant parasites, although it is the ring stage that has been shown to be prolonged, likely as a result of reduced hemoglobin catabolism. PfMYST was also demonstrated to participate in var gene activation, as well as DNA repair, and to modulate sensitivity to genotoxic agents. HAT inhibitors have been shown in previous studies to have parasiticidal effects (Andrews et al., 2000; Cui et al., 2007; Darkin-Rattray et al., 1996; Sen et al., 2020), lending credence to the possibility of histone acetylation enzymes as antimalarial drug targets.
Multiple studies have implicated DNA damage-repair mechanisms in K13-associated drug-resistance, supporting that DNA repair functions are utilized by the parasite to cope with cellular damage induced by activated artemisinins (for example (Gibbons et al., 2018; Sheriff et al., 2021; Suthram et al., 2020; Xiong et al., 2020),). An enhanced capacity for DNA-repair may be an underlying advantage common to resistance-phenotypes more generally that may also hasten their spread (Xiong et al., 2020). Several DNA damage-response genes met our criteria for iSAFE analysis, with two recently characterized double-strand break repair proteins identified as harboring possible favored mutations (DNA replication-factor C subunit 1, RFC1; the RecQ helicase BLM) (Sheriff et al., 2021; Suthram et al., 2020). A small molecule-inhibitor of PfBLM was also found to act synergistically with both artemisinin and chloroquine against multi drug-resistant parasites, suggesting the utility of targeting these pathways for promising co-therapies (Suthram et al., 2020).
Calcium-dependent protein kinase 5, CDPK5 (PF3D7_1337800) is an enzyme that has been shown to regulate parasite egress from the host cell (Dvorin et al., 2010). The CDPKs are unique to plants and some protists, including apicomplexans. Blomqvist et al. recently identified 50 proteins with significantly reduced phosphorylation using a conditional CDPK5 knockdown (Blomqvist et al., 2020). An enrichment analysis of these 50 proteins based on gene ontology showed significant enrichment for proteins with transport and transmembrane transport activity, as well as proteins located in vesicles and the cell periphery, consistent with artemisinin resistance mechanisms involving endocytosis. Existing compounds, including kinase inhibitors within the Tres Cantos antimalarial compound set (TCAMS) (Gamo et al., 2010), have been predicted to target CDPK5, among other kinases, and these compounds could be explored for activity on multidrug-resistant parasites.
As observed in some prior studies, exported proteins, such as those encoded by the var, phist, rifin, and clag multigene families, were enriched among gene products with functional and/or population genomic evidence (Mok et al., 2015; Rocamora et al., 2018; Siddiqui et al., 2020; Zhu et al., 2018). This observation suggests a potential role for host-cell remodeling in artemisinin resistance. Previous studies have indicated that in populations with less antimalarial immunity, “pitting” (i.e., splenic removal of parasites from infected erythrocytes) is a major mechanism for rapid parasite clearance following treatment with artemisinin derivatives (Ndour et al., 2015; Wojnarski et al., 2019). However, erythrocytes infected by artemisinin resistant parasites have been shown to have increased deformability and reduced splenic pitting, allowing them to remain in circulation (Wojnarski et al., 2019). One unexplored hypothesis is that exported proteins could contribute to the altered deformability and reduced splenic clearance of artemisinin-resistant parasites. Indeed, the var genes identified in this meta-analysis as potential contributors to artemisinin resistance encode shorter group B or C PfEMP1 proteins that bind CD36, are not associated with severe disease, and contain only four main domains, potentially impacting cytoadherence of infected host red blood cells (Rask et al., 2010). In addition, PHIST proteins, along with other effectors (e.g., RESA and KAHRP proteins) have been shown to modulate the cytoskeleton of infected host cells to increase rigidity and reduce splenic clearance [Reviewed in (de Koning-Ward et al., 2016; Matthews et al., 2019; Spillman et al., 2015),]. Further examination of the role of exported proteins and host cell remodeling in reduced artemisinin susceptibility will be important to fully understand the parasite's multipronged approach to artemisinin resistance.
In further support of a possible role for host-cell remodeling in artemisinin resistance, our analysis also identified the gene encoding FIKK4.2 as potentially containing favored mutations in artemisinin resistant parasites. FIKK4.2 (PF3D7_0424700) is an exported kinase belonging to the FIKK multigene family, which is comprised of 20 genes on 11 chromosomes (Davies et al., 2020; Nunes et al., 2007). The FIKK kinases are specific to apicomplexans and are thought to play a role in host cell remodeling in response to intracellular, and possibly extracellular, changes in infected erythrocytes (Nunes et al., 2007). FIKK4.2 is unique in that it contains a large central repetitive region and, of the human malaria parasite species, is specific to P. falciparum (Kats et al., 2014). FIKK4.2 localizes to a compartment of the infected erythrocyte distinct from the Maurer's clefts or J-dots, and disruption of the gene encoding FIKK4.2 leads to dramatically altered mechanical properties and cytoadherence of infected erythrocytes (Kats et al., 2014). Specifically, erythrocytes infected with FIKK4.2 knockout parasites were less rigid and less adhesive than erythrocytes harboring parasites with functional FIKK4.2 (Kats et al., 2014), a phenotype also observed in cells infected with artemisinin-resistant parasites (Ndour et al., 2015; Wojnarski et al., 2019). Further studies would be required to determine whether mutations in FIKK4.2 contribute to changes in deformability observed in artemisinin resistant parasites.
In addition to providing further evidence in support of recently identified mechanisms of artemisinin resistance, our meta-analysis of population genomic and transcriptomic studies identified potential new or related targets that may be leveraged to counter artemisinin-resistant parasites. These new targets will require functional characterization to validate their contribution to reduced artemisinin susceptibility, as it is possible that the selection signals identified could be driven by other selective pressures, including partner drugs or other pressures not related to drug treatment. In addition, because our iSAFE analysis compared parasites from the predominant mutant Kelch13 lineage circulating in the eastern Greater Mekong (Agrawal et al., 2017; Amato et al., 2017) to parasites from the pre-artemisinin resistant genetic subpopulation (Miotto et al., 2013), it is possible that our analysis may not have captured the contribution of gene products involved in non-kelch-mediated mechanisms of resistance. Likewise, our analysis focused on biallelic SNPs, and therefore would not have detected signals from gene copy number variants, which are also commonly observed in drug-resistant parasites (Cowell et al., 2018) and which are more difficult to detect because they can arise on different genetic backgrounds (i.e., a soft sweep).
5. Conclusions
In conclusion, we have implemented a novel approach integrating findings from previous population genomic and transcriptomic studies with new data acquired in genome-wide functional screens. By looking at the union of results from these different types of studies, we can more easily differentiate determinants of drug resistance from determinants of successful in vitro culture, as most population-level studies utilize sequencing data generated directly from clinical isolates without culture adaptation. Our analysis has corroborated the contribution of altered processes related to proteasomal degradation, endocytosis, vesicular trafficking and stress responses in artemisinin resistance, and has shown the utility of iSAFE to narrow the list of possible favored mutations within the region of a selective sweep, potentially accelerating time to functional validation. In addition, our analysis emphasizes the importance of future studies of the role of exported proteins and host-cell remodeling in artemisinin resistance.
Declaration of competing interest
The authors have nothing to declare.
Acknowledgments
This work was funded by the National Institutes of Health [Grant numbers: R01AI101713 (ST-H), R01AI125579 (ST-H), F32AI112271 (JO), R01AI117017 (JHA), R01AI30171 (JHA) and U19AI110820 (Project Leader: JCS, PI: Fraser)]. CDB was supported as part of the University of Maryland Baltimore Science Training for Advancing Biomedical Research Postbaccalaureate (STAR-PREP) Program funded by an award from the National Institutes of Health [Grant number: 2R25GM113262 (PI: Greg Carey)].
Footnotes
Supplementary data to this article can be found online at https://doi.org/10.1016/j.ijpddr.2021.05.006.
Contributor Information
Jenna Oberstaller, Email: jobersta@usf.edu.
Linda Zoungrana, Email: lindainesz@usf.edu.
Carl D. Bannerman, Email: bacarl1@umbc.edu.
Samira Jahangiri, Email: jahangiri@usf.edu.
Ankit Dwivedi, Email: ADwivedi@som.umaryland.edu.
Joana C. Silva, Email: jcsilva@som.umaryland.edu.
John H. Adams, Email: ja2@usf.edu.
Shannon Takala-Harrison, Email: stakala@som.umaryland.edu.
Appendix A. Supplementary data
The following are the Supplementary data to this article:
figs1.
figs2.
References
- Agrawal S., Moser K.A., Morton L., Cummings M.P., Parihar A., Dwivedi A., Shetty A.C., Drabek E.F., Jacob C.G., Henrich P.P., Parobek C.M., Jongsakul K., Huy R., Spring M.D., Lanteri C.A., Chaorattanakawee S., Lon C., Fukuda M.M., Saunders D.L., Fidock D.A., Lin J.T., Juliano J.J., Plowe C.V., Silva J.C., Takala-Harrison S. Association of a novel mutation in the Plasmodium falciparum chloroquine resistance transporter with decreased piperaquine sensitivity. J. Infect. Dis. 2017;216:468–476. doi: 10.1093/infdis/jix334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Akbari A., Vitti J.J., Iranmehr A., Bakhtiari M., Sabeti P.C., Mirarab S., Bafna V. Identifying the favored mutation in a positive selective sweep. Nat. Methods. 2018;15:279–282. doi: 10.1038/nmeth.4606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alexa A., Rahnenfuhrer J. 2020. topGO: Enrichment Analysis for Gene Ontology, R Package. version 2.40.0 ed. [Google Scholar]
- Amato R., Lim P., Miotto O., Amaratunga C., Dek D., Pearson R.D., Almagro-Garcia J., Neal A.T., Sreng S., Suon S., Drury E., Jyothi D., Stalker J., Kwiatkowski D.P., Fairhurst R.M. Genetic markers associated with dihydroartemisinin-piperaquine failure in Plasmodium falciparum malaria in Cambodia: a genotype-phenotype association study. Lancet Infect. Dis. 2017;17:164–173. doi: 10.1016/S1473-3099(16)30409-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Andrews K.T., Walduck A., Kelso M.J., Fairlie D.P., Saul A., Parsons P.G. Anti-malarial effect of histone deacetylation inhibitors and mammalian tumour cytodifferentiating agents. Int. J. Parasitol. 2000;30:761–768. doi: 10.1016/s0020-7519(00)00043-6. [DOI] [PubMed] [Google Scholar]
- Ariey F., Witkowski B., Amaratunga C., Beghain J., Langlois A.C., Khim N., Kim S., Duru V., Bouchier C., Ma L., Lim P., Leang R., Duong S., Sreng S., Suon S., Chuor C.M., Bout D.M., Ménard S., Rogers W.O., Genton B., Fandeur T., Miotto O., Ringwald P., Le Bras J., Berry A., Barale J.C., Fairhurst R.M., Benoit-Vical F., Mercereau-Puijalon O., Ménard D. A molecular marker of artemisinin-resistant Plasmodium falciparum malaria. Nature. 2014;505:50–55. doi: 10.1038/nature12876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aurrecoechea C., Brestelli J., Brunk B.P., Dommer J., Fischer S., Gajria B., Gao X., Gingle A., Grant G., Harb O.S., Heiges M., Innamorato F., Iodice J., Kissinger J.C., Kraemer E., Li W., Miller J.A., Nayak V., Pennington C., Pinney D.F., Roos D.S., Ross C., Stoeckert C.J., Jr., Treatman C., Wang H. PlasmoDB: a functional genomic database for malaria parasites. Nucleic Acids Res. 2009;37:D539–D543. doi: 10.1093/nar/gkn814. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Birnbaum J., Scharf S., Schmidt S., Jonscher E., Hoeijmakers W.A.M., Flemming S., Toenhake C.G., Schmitt M., Sabitzki R., Bergmann B., Fröhlke U., Mesén-Ramírez P., Blancke Soares A., Herrmann H., Bártfai R., Spielmann T. A Kelch13-defined endocytosis pathway mediates artemisinin resistance in malaria parasites. Science. 2020;367:51. doi: 10.1126/science.aax4735. [DOI] [PubMed] [Google Scholar]
- Blomqvist K., Helmel M., Wang C., Absalon S., Labunska T., Rudlaff R.M., Adapa S., Jiang R., Steen H., Dvorin J.D. Influence of Plasmodium falciparum calcium-dependent protein kinase 5 (PfCDPK5) on the late schizont stage phosphoproteome. mSphere. 2020;5 doi: 10.1128/mSphere.00921-19. e00921-919. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Borrmann S., Straimer J., Mwai L., Abdi A., Rippert A., Okombo J., Muriithi S., Sasi P., Kortok M.M., Lowe B., Campino S., Assefa S., Auburn S., Manske M., Maslen G., Peshu N., Kwiatkowski D.P., Marsh K., Nzila A., Clark T.G. Genome-wide screen identifies new candidate genes associated with artemisinin susceptibility in Plasmodium falciparum in Kenya. Sci. Rep. 2013;3:3318. doi: 10.1038/srep03318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cerqueira G.C., Cheeseman I.H., Schaffner S.F., Nair S., McDew-White M., Phyo A.P., Ashley E.A., Melnikov A., Rogov P., Birren B.W., Nosten F., Anderson T.J.C., Neafsey D.E. Longitudinal genomic surveillance of Plasmodium falciparum malaria parasites reveals complex genomic architecture of emerging artemisinin resistance. Genome Biol. 2017;18:78. doi: 10.1186/s13059-017-1204-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheeseman I.H., McDew-White M., Phyo A.P., Sriprawat K., Nosten F., Anderson T.J. Pooled sequencing and rare variant association tests for identifying the determinants of emerging drug resistance in malaria parasites. Mol. Biol. Evol. 2015;32:1080–1090. doi: 10.1093/molbev/msu397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheeseman I.H., Miller B.A., Nair S., Nkhoma S., Tan A., Tan J.C., Al S.S., Phyo A.P., Moo C.L., Lwin K.M., McGready R., Ashley E., Imwong M., Stepniewska K., Yi P., Dondorp A.M., Mayxay M., Newton P.N., White N.J., Nosten F., Ferdig M.T., Anderson T.J. A major genome region underlying artemisinin resistance in malaria. Science. 2012;336:79–82. doi: 10.1126/science.1215966. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cowell A.N., Istvan E.S., Lukens A.K., Gomez-Lorenzo M.G., Vanaerschot M., Sakata-Kato T., Flannery E.L., Magistrado P., Owen E., Abraham M., LaMonte G., Painter H.J., Williams R.M., Franco V., Linares M., Arriaga I., Bopp S., Corey V.C., Gnadig N.F., Coburn-Flynn O., Reimer C., Gupta P., Murithi J.M., Moura P.A., Fuchs O., Sasaki E., Kim S.W., Teng C.H., Wang L.T., Akidil A., Adjalley S., Willis P.A., Siegel D., Tanaseichuk O., Zhong Y., Zhou Y., Llinas M., Ottilie S., Gamo F.J., Lee M.C.S., Goldberg D.E., Fidock D.A., Wirth D.F., Winzeler E.A. Mapping the malaria parasite druggable genome by using in vitro evolution and chemogenomics. Science. 2018;359:191–199. doi: 10.1126/science.aan4472. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cui L., Miao J., Cui L. Cytotoxic effect of curcumin on malaria parasite Plasmodium falciparum: inhibition of histone acetylation and generation of reactive oxygen species. Antimicrob. Agents Chemother. 2007;51:488–494. doi: 10.1128/AAC.01238-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Darkin-Rattray S.J., Gurnett A.M., Myers R.W., Dulski P.M., Crumley T.M., Allocco J.J., Cannova C., Meinke P.T., Colletti S.L., Bednarek M.A., Singh S.B., Goetz M.A., Dombrowski A.W., Polishook J.D., Schmatz D.M. Apicidin: a novel antiprotozoal agent that inhibits parasite histone deacetylase. Proc. Natl. Acad. Sci. U.S.A. 1996;93:13143–13147. doi: 10.1073/pnas.93.23.13143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davies H., Belda H., Broncel M., Ye X., Bisson C., Introini V., Dorin-Semblat D., Semblat J.-P., Tibúrcio M., Gamain B., Kaforou M., Treeck M. An exported kinase family mediates species-specific erythrocyte remodelling and virulence in human malaria. Nature Microbiology. 2020;5:848–863. doi: 10.1038/s41564-020-0702-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Koning-Ward T.F., Dixon M.W.A., Tilley L., Gilson P.R. Plasmodium species: master renovators of their host cells. Nat. Rev. Microbiol. 2016;14:494–507. doi: 10.1038/nrmicro.2016.79. [DOI] [PubMed] [Google Scholar]
- Demas A.R., Sharma A.I., Wong W., Early A.M., Redmond S., Bopp S., Neafsey D.E., Volkman S.K., Hartl D.L., Wirth D.F. Mutations in Plasmodium falciparum actin-binding protein coronin confer reduced artemisinin susceptibility. Proc. Natl. Acad. Sci. U. S. A. 2018;115:12799–12804. doi: 10.1073/pnas.1812317115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dondorp A.M., Nosten F., Yi P., Das D., Phyo A.P., Tarning J., Lwin K.M., Ariey F., Hanpithakpong W., Lee S.J., Ringwald P., Silamut K., Imwong M., Chotivanich K., Lim P., Herdman T., An S.S., Yeung S., Singhasivanon P., Day N.P., Lindegardh N., Socheat D., White N.J. Artemisinin resistance in Plasmodium falciparum malaria. N. Engl. J. Med. 2009;361:455–467. doi: 10.1056/NEJMoa0808859. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dvorin J.D., Martyn D.C., Patel S.D., Grimley J.S., Collins C.R., Hopp C.S., Bright A.T., Westenberger S., Winzeler E., Blackman M.J., Baker D.A., Wandless T.J., Duraisingh M.T. A plant-like kinase in <em>Plasmodium falciparum</em> regulates parasite egress from erythrocytes. Science. 2010;328:910–912. doi: 10.1126/science.1188191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dwivedi A., Reynes C., Kuehn A., Roche D.B., Khim N., Hebrard M., Milanesi S., Rivals E., Frutos R., Menard D., Mamoun C.B., Colinge J., Cornillot E. Functional analysis of Plasmodium falciparum subpopulations associated with artemisinin resistance in Cambodia. Malar. J. 2017;16:493. doi: 10.1186/s12936-017-2140-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gamo F.J., Sanz L.M., Vidal J., de Cozar C., Alvarez E., Lavandera J.L., Vanderwall D.E., Green D.V., Kumar V., Hasan S., Brown J.R., Peishoff C.E., Cardon L.R., Garcia-Bustos J.F. Thousands of chemical starting points for antimalarial lead identification. Nature. 2010;465:305–310. doi: 10.1038/nature09107. [DOI] [PubMed] [Google Scholar]
- Gibbons J., Button-Simons K.A., Adapa S.R., Li S., Pietsch M., Zhang M., Liao X., Adams J.H., Ferdig M.T., Jiang R.H.Y. Altered expression of K13 disrupts DNA replication and repair in Plasmodium falciparum. BMC Genom. 2018;19:849. doi: 10.1186/s12864-018-5207-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gnädig N.F., Stokes B.H., Edwards R.L., Kalantarov G.F., Heimsch K.C., Kuderjavy M., Crane A., Lee M.C.S., Straimer J., Becker K., Trakht I.N., Odom John A.R., Mok S., Fidock D.A. Insights into the intracellular localization, protein associations and artemisinin resistance properties of Plasmodium falciparum K13. PLoS Pathog. 2020;16 doi: 10.1371/journal.ppat.1008482. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kakiuchi K., Yamauchi Y., Taoka M., Iwago M., Fujita T., Ito T., Song S.Y., Sakai A., Isobe T., Ichimura T. Proteomic analysis of in vivo 14-3-3 interactions in the yeast Saccharomyces cerevisiae. Biochemistry. 2007;46:7781–7792. doi: 10.1021/bi700501t. [DOI] [PubMed] [Google Scholar]
- Kats L.M., Fernandez K.M., Glenister F.K., Herrmann S., Buckingham D.W., Siddiqui G., Sharma L., Bamert R., Lucet I., Guillotte M., Mercereau-Puijalon O., Cooke B.M. An exported kinase (FIKK4.2) that mediates virulence-associated changes in Plasmodium falciparum-infected red blood cells. Int. J. Parasitol. 2014;44:319–328. doi: 10.1016/j.ijpara.2014.01.003. [DOI] [PubMed] [Google Scholar]
- Klonis N., Crespo-Ortiz M.P., Bottova I., Abu-Bakar N., Kenny S., Rosenthal P.J., Tilley L. Artemisinin activity against <em>Plasmodium falciparum</em> requires hemoglobin uptake and digestion. Proc. Natl. Acad. Sci. Unit. States Am. 2011;108:11405–11410. doi: 10.1073/pnas.1104063108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leber W., Skippen A., Fivelman Q.L., Bowyer P.W., Cockcroft S., Baker D.A. A unique phosphatidylinositol 4-phosphate 5-kinase is activated by ADP-ribosylation factor in Plasmodium falciparum. Int. J. Parasitol. 2009;39:645–653. doi: 10.1016/j.ijpara.2008.11.015. [DOI] [PubMed] [Google Scholar]
- Li X., Kumar S., McDew-White M., Haile M., Cheeseman I.H., Emrich S., Button-Simons K., Nosten F., Kappe S.H.I., Ferdig M.T., Anderson T.J.C., Vaughan A.M. Genetic mapping of fitness determinants across the malaria parasite Plasmodium falciparum life cycle. PLoS Genet. 2019;15 doi: 10.1371/journal.pgen.1008453. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu X., Salokas K., Tamene F., Jiu Y., Weldatsadik R.G., Öhman T., Varjosalo M. An AP-MS- and BioID-compatible MAC-tag enables comprehensive mapping of protein interactions and subcellular localizations. Nat. Commun. 2018;9:1188. doi: 10.1038/s41467-018-03523-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Logan-Klumpler F.J., De Silva N., Boehme U., Rogers M.B., Velarde G., McQuillan J.A., Carver T., Aslett M., Olsen C., Subramanian S., Phan I., Farris C., Mitra S., Ramasamy G., Wang H., Tivey A., Jackson A., Houston R., Parkhill J., Holden M., Harb O.S., Brunk B.P., Myler P.J., Roos D., Carrington M., Smith D.F., Hertz-Fowler C., Berriman M. GeneDB--an annotation database for pathogens. Nucleic Acids Res. 2012;40:D98–D108. doi: 10.1093/nar/gkr1032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martin T.F.J. Role of PI(4,5)P(2) in vesicle exocytosis and membrane fusion. Subcell. Biochem. 2012;59:111–130. doi: 10.1007/978-94-007-3015-1_4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matthews K.M., Pitman E.L., de Koning-Ward T.F. Illuminating how malaria parasites export proteins into host erythrocytes. Cell Microbiol. 2019;21 doi: 10.1111/cmi.13009. [DOI] [PubMed] [Google Scholar]
- Mbengue A., Bhattacharjee S., Pandharkar T., Liu H., Estiu G., Stahelin R.V., Rizk S.S., Njimoh D.L., Ryan Y., Chotivanich K., Nguon C., Ghorbal M., Lopez-Rubio J.-J., Pfrender M., Emrich S., Mohandas N., Dondorp A.M., Wiest O., Haldar K. A molecular mechanism of artemisinin resistance in Plasmodium falciparum malaria. Nature. 2015;520:683–687. doi: 10.1038/nature14412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McKenna A., Hanna M., Banks E., Sivachenko A., Cibulskis K., Kernytsky A., Garimella K., Altshuler D., Gabriel S., Daly M., DePristo M.A. The Genome Analysis Toolkit: a MapReduce framework for analyzing next-generation DNA sequencing data. Genome Res. 2010;20:1297–1303. doi: 10.1101/gr.107524.110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Menard D., Dondorp A. Antimalarial drug resistance: a threat to malaria elimination. Cold Spring Harb Perspect Med. 2017;7 doi: 10.1101/cshperspect.a025619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miao J., Fan Q., Cui L., Li X., Wang H., Ning G., Reese J.C., Cui L. The MYST family histone acetyltransferase regulates gene expression and cell cycle in malaria parasite Plasmodium falciparum. Mol. Microbiol. 2010;78:883–902. doi: 10.1111/j.1365-2958.2010.07371.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miotto O., Almagro-Garcia J., Manske M., MacInnis B., Campino S., Rockett K.A., Amaratunga C., Lim P., Suon S., Sreng S., Anderson J.M., Duong S., Nguon C., Chuor C.M., Saunders D., Se Y., Lon C., Fukuda M.M., Amenga-Etego L., Hodgson A.V., Asoala V., Imwong M., Takala-Harrison S., Nosten F., Su X.Z., Ringwald P., Ariey F., Dolecek C., Hien T.T., Boni M.F., Thai C.Q., Amambua-Ngwa A., Conway D.J., Djimde A.A., Doumbo O.K., Zongo I., Ouedraogo J.B., Alcock D., Drury E., Auburn S., Koch O., Sanders M., Hubbart C., Maslen G., Ruano-Rubio V., Jyothi D., Miles A., O'Brien J., Gamble C., Oyola S.O., Rayner J.C., Newbold C.I., Berriman M., Spencer C.C., McVean G., Day N.P., White N.J., Bethell D., Dondorp A.M., Plowe C.V., Fairhurst R.M., Kwiatkowski D.P. Multiple populations of artemisinin-resistant Plasmodium falciparum in Cambodia. Nat. Genet. 2013;45:648–655. doi: 10.1038/ng.2624. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miotto O., Amato R., Ashley E.A., MacInnis B., Almagro-Garcia J., Amaratunga C., Lim P., Mead D., Oyola S.O., Dhorda M., Imwong M., Woodrow C., Manske M., Stalker J., Drury E., Campino S., Amenga-Etego L., Thanh T.N., Tran H.T., Ringwald P., Bethell D., Nosten F., Phyo A.P., Pukrittayakamee S., Chotivanich K., Chuor C.M., Nguon C., Suon S., Sreng S., Newton P.N., Mayxay M., Khanthavong M., Hongvanthong B., Htut Y., Han K.T., Kyaw M.P., Faiz M.A., Fanello C.I., Onyamboko M., Mokuolu O.A., Jacob C.G., Takala-Harrison S., Plowe C.V., Day N.P., Dondorp A.M., Spencer C.C., McVean G., Fairhurst R.M., White N.J., Kwiatkowski D.P. Genetic architecture of artemisinin-resistant Plasmodium falciparum. Nat. Genet. 2015;47:226–234. doi: 10.1038/ng.3189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mok S., Ashley E.A., Ferreira P.E., Zhu L., Lin Z., Yeo T., Chotivanich K., Imwong M., Pukrittayakamee S., Dhorda M., Nguon C., Lim P., Amaratunga C., Suon S., Hien T.T., Htut Y., Faiz M.A., Onyamboko M.A., Mayxay M., Newton P.N., Tripura R., Woodrow C.J., Miotto O., Kwiatkowski D.P., Nosten F., Day N.P., Preiser P.R., White N.J., Dondorp A.M., Fairhurst R.M., Bozdech Z. Drug resistance. Population transcriptomics of human malaria parasites reveals the mechanism of artemisinin resistance. Science. 2015;347:431–435. doi: 10.1126/science.1260403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mu J., Myers R.A., Jiang H., Liu S., Ricklefs S., Waisberg M., Chotivanich K., Wilairatana P., Krudsood S., White N.J., Udomsangpetch R., Cui L., Ho M., Ou F., Li H., Song J., Li G., Wang X., Seila S., Sokunthea S., Socheat D., Sturdevant D.E., Porcella S.F., Fairhurst R.M., Wellems T.E., Awadalla P., Su X.-z. Plasmodium falciparum genome-wide scans for positive selection, recombination hot spots and resistance to antimalarial drugs. Nat. Genet. 2010;42:268–271. doi: 10.1038/ng.528. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mukherjee A., Bopp S., Magistrado P., Wong W., Daniels R., Demas A., Schaffner S., Amaratunga C., Lim P., Dhorda M., Miotto O., Woodrow C., Ashley E.A., Dondorp A.M., White N.J., Wirth D., Fairhurst R., Volkman S.K. Artemisinin resistance without pfkelch13 mutations in Plasmodium falciparum isolates from Cambodia. Malar. J. 2017;16:195. doi: 10.1186/s12936-017-1845-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ndour P.A., Lopera-Mesa T.M., Diakité S.A.S., Chiang S., Mouri O., Roussel C., Jauréguiberry S., Biligui S., Kendjo E., Claessens A., Ciceron L., Mazier D., Thellier M., Diakité M., Fairhurst R.M., Buffet P.A. Plasmodium falciparum clearance is rapid and pitting independent in immune Malian children treated with artesunate for malaria. J. Infect. Dis. 2015;211:290–297. doi: 10.1093/infdis/jiu427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Noedl H., Se Y., Schaecher K., Smith B.L., Socheat D., Fukuda M.M. Evidence of artemisinin-resistant malaria in western Cambodia. N. Engl. J. Med. 2008;359:2619–2620. doi: 10.1056/NEJMc0805011. [DOI] [PubMed] [Google Scholar]
- Nunes M.C., Goldring J.P.D., Doerig C., Scherf A. A novel protein kinase family in Plasmodium falciparum is differentially transcribed and secreted to various cellular compartments of the host cell. Mol. Microbiol. 2007;63:391–403. doi: 10.1111/j.1365-2958.2006.05521.x. [DOI] [PubMed] [Google Scholar]
- Oberstaller J., Otto T.D., Rayner J.C., Adams J.H. Essential genes of the parasitic apicomplexa. Trends Parasitol. 2021 doi: 10.1016/j.pt.2020.11.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park D.J., Lukens A.K., Neafsey D.E., Schaffner S.F., Chang H.H., Valim C., Ribacke U., Van Tyne D., Galinsky K., Galligan M., Becker J.S., Ndiaye D., Mboup S., Wiegand R.C., Hartl D.L., Sabeti P.C., Wirth D.F., Volkman S.K. Sequence-based association and selection scans identify drug resistance loci in the Plasmodium falciparum malaria parasite. Proc. Natl. Acad. Sci. U. S. A. 2012;109:13052–13057. doi: 10.1073/pnas.1210585109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pradhan A., Siwo G.H., Singh N., Martens B., Balu B., Button-Simons K.A., Tan A., Zhang M., Udenze K.O., Jiang R.H., Ferdig M.T., Adams J.H., Kyle D.E. Chemogenomic profiling of Plasmodium falciparum as a tool to aid antimalarial drug discovery. Sci. Rep. 2015;5:15930. doi: 10.1038/srep15930. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rask T.S., Hansen D.A., Theander T.G., Gorm Pedersen A., Lavstsen T. Plasmodium falciparum erythrocyte membrane protein 1 diversity in seven genomes--divide and conquer. PLoS Comput. Biol. 2010;6 doi: 10.1371/journal.pcbi.1000933. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rocamora F., Zhu L., Liong K.Y., Dondorp A., Miotto O., Mok S., Bozdech Z. Oxidative stress and protein damage responses mediate artemisinin resistance in malaria parasites. PLoS Pathog. 2018;14 doi: 10.1371/journal.ppat.1006930. e1006930. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ronen R., Tesler G., Akbari A., Zakov S., Rosenberg N.A., Bafna V. Predicting carriers of ongoing selective sweeps without knowledge of the favored allele. PLoS Genet. 2015;11 doi: 10.1371/journal.pgen.1005527. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sen U., Nayak A., Khurana J., Sharma D., Gupta A. Inhibition of PfMYST histone acetyltransferase activity blocks Plasmodium falciparum growth and survival. Antimicrob. Agents Chemother. 2020;65 doi: 10.1128/AAC.00953-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sheriff O., Yaw A., Lai S.K., loo h.l., Sze S.K., Preiser P.R. Plasmodium falciparum replication factor C subunit 1 is involved in genotoxic stress response. Cell Microbiol. 2021;23 doi: 10.1111/cmi.13277. [DOI] [PubMed] [Google Scholar]
- Siddiqui G., Proellochs N.I., Cooke B.M. Identification of essential exported Plasmodium falciparum protein kinases in malaria-infected red blood cells. Br. J. Haematol. 2020;188:774–783. doi: 10.1111/bjh.16219. [DOI] [PubMed] [Google Scholar]
- Spillman N.J., Beck J.R., Goldberg D.E. Protein export into malaria parasite–infected erythrocytes: mechanisms and functional consequences. Annu. Rev. Biochem. 2015;84:813–841. doi: 10.1146/annurev-biochem-060614-034157. [DOI] [PubMed] [Google Scholar]
- Sutherland C.J., Henrici R.C., Artavanis-Tsakonas K. Artemisinin susceptibility in the malaria parasite Plasmodium falciparum: propellers, adaptor proteins and the need for cellular healing. FEMS (Fed. Eur. Microbiol. Soc.) Microbiol. Rev. 2020 doi: 10.1093/femsre/fuaa056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suthram N., Padhi S., Jha P., Bhattacharyya S., Bulusu G., Roy A., Bhattacharyya M.K. Elucidation of DNA repair function of PfBlm and potentiation of artemisinin action by a small-molecule inhibitor of RecQ helicase. mSphere. 2020;5 doi: 10.1128/mSphere.00956-20. e00956-920. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Szklarczyk D., Gable A.L., Lyon D., Junge A., Wyder S., Huerta-Cepas J., Simonovic M., Doncheva N.T., Morris J.H., Bork P., Jensen L.J., Mering C.V. STRING v11: protein-protein association networks with increased coverage, supporting functional discovery in genome-wide experimental datasets. Nucleic Acids Res. 2019;47:D607–d613. doi: 10.1093/nar/gky1131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takala-Harrison S., Clark T.G., Jacob C.G., Cummings M.P., Miotto O., Dondorp A.M., Fukuda M.M., Nosten F., Noedl H., Imwong M., Bethell D., Se Y., Lon C., Tyner S.D., Saunders D.L., Socheat D., Ariey F., Phyo A.P., Starzengruber P., Fuehrer H.P., Swoboda P., Stepniewska K., Flegg J., Arze C., Cerqueira G.C., Silva J.C., Ricklefs S.M., Porcella S.F., Stephens R.M., Adams M., Kenefic L.J., Campino S., Auburn S., MacInnis B., Kwiatkowski D.P., Su X.Z., White N.J., Ringwald P., Plowe C.V. Genetic loci associated with delayed clearance of Plasmodium falciparum following artemisinin treatment in Southeast Asia. Proc. Natl. Acad. Sci. U. S. A. 2013;110:240–245. doi: 10.1073/pnas.1211205110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takala-Harrison S., Jacob C.G., Arze C., Cummings M.P., Silva J.C., Dondorp A.M., Fukuda M.M., Hien T.T., Mayxay M., Noedl H., Nosten F., Kyaw M.P., Nhien N.T., Imwong M., Bethell D., Se Y., Lon C., Tyner S.D., Saunders D.L., Ariey F., Mercereau-Puijalon O., Menard D., Newton P.N., Khanthavong M., Hongvanthong B., Starzengruber P., Fuehrer H.P., Swoboda P., Khan W.A., Phyo A.P., Nyunt M.M., Nyunt M.H., Brown T.S., Adams M., Pepin C.S., Bailey J., Tan J.C., Ferdig M.T., Clark T.G., Miotto O., MacInnis B., Kwiatkowski D.P., White N.J., Ringwald P., Plowe C.V. Independent emergence of artemisinin resistance mutations among Plasmodium falciparum in Southeast Asia. J. Infect. Dis. 2015;211:670–679. doi: 10.1093/infdis/jiu491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van Tyne D., Park D.J., Schaffner S.F., Neafsey D.E., Angelino E., Cortese J.F., Barnes K.G., Rosen D.M., Lukens A.K., Daniels R.F., Milner D.A., Jr., Johnson C.A., Shlyakhter I., Grossman S.R., Becker J.S., Yamins D., Karlsson E.K., Ndiaye D., Sarr O., Mboup S., Happi C., Furlotte N.A., Eskin E., Kang H.M., Hartl D.L., Birren B.W., Wiegand R.C., Lander E.S., Wirth D.F., Volkman S.K., Sabeti P.C. Identification and functional validation of the novel antimalarial resistance locus PF10_0355 in Plasmodium falciparum. PLoS Genet. 2011;7 doi: 10.1371/journal.pgen.1001383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Voisinne G., García-Blesa A., Chaoui K., Fiore F., Bergot E., Girard L., Malissen M., Burlet-Schiltz O., Gonzalez de Peredo A., Malissen B., Roncagalli R. Co-recruitment analysis of the CBL and CBLB signalosomes in primary T cells identifies CD5 as a key regulator of TCR-induced ubiquitylation. Mol. Syst. Biol. 2016;12:876. doi: 10.15252/msb.20166837. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Z., Cabrera M., Yang J., Yuan L., Gupta B., Liang X., Kemirembe K., Shrestha S., Brashear A., Li X., Porcella S.F., Miao J., Yang Z., Su X.Z., Cui L. Genome-wide association analysis identifies genetic loci associated with resistance to multiple antimalarials in Plasmodium falciparum from China-Myanmar border. Sci. Rep. 2016;6:33891. doi: 10.1038/srep33891. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wendler J.P., Okombo J., Amato R., Miotto O., Kiara S.M., Mwai L., Pole L., O'Brien J., Manske M., Alcock D., Drury E., Sanders M., Oyola S.O., Malangone C., Jyothi D., Miles A., Rockett K.A., MacInnis B.L., Marsh K., Bejon P., Nzila A., Kwiatkowski D.P. A genome wide association study of Plasmodium falciparum susceptibility to 22 antimalarial drugs in Kenya. PloS One. 2014;9 doi: 10.1371/journal.pone.0096486. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Witkowski B., Duru V., Khim N., Ross L.S., Saintpierre B., Beghain J., Chy S., Kim S., Ke S., Kloeung N., Eam R., Khean C., Ken M., Loch K., Bouillon A., Domergue A., Ma L., Bouchier C., Leang R., Huy R., Nuel G., Barale J.C., Legrand E., Ringwald P., Fidock D.A., Mercereau-Puijalon O., Ariey F., Menard D. A surrogate marker of piperaquine-resistant Plasmodium falciparum malaria: a phenotype-genotype association study. Lancet Infect. Dis. 2017;17:174–183. doi: 10.1016/S1473-3099(16)30415-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Witkowski B., Lelièvre J., Barragán M.J., Laurent V., Su X.Z., Berry A., Benoit-Vical F. Increased tolerance to artemisinin in Plasmodium falciparum is mediated by a quiescence mechanism. Antimicrob. Agents Chemother. 2010;54:1872–1877. doi: 10.1128/AAC.01636-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wojnarski M., Mouri O., Chambrion C., Roussel C., Chartrel N., Smith B., Smith P., Thellier M., Buffet P., Ndour P.A. Plasmodium falciparum clearance is pitting-dependent with artemisinin-based drugs but pitting-independent with atovaquone-proguanil or mefloquine. J. Infect. Dis. 2019;220:535–539. doi: 10.1093/infdis/jiz115. [DOI] [PubMed] [Google Scholar]
- World Health Organization . World Health Organization; Geneva: 2019. World Malaria Report 2019. [Google Scholar]
- Xie S.C., Ralph S.A., Tilley L. K13, the cytostome, and artemisinin resistance. Trends Parasitol. 2020;36:533–544. doi: 10.1016/j.pt.2020.03.006. [DOI] [PubMed] [Google Scholar]
- Xiong A., Prakash P., Gao X., Chew M., Tay I.J.J., Woodrow C.J., Engelward B.P., Han J., Preiser P.R. K13-Mediated reduced susceptibility to artemisinin in Plasmodium falciparum is overlaid on a trait of enhanced DNA damage repair. Cell Rep. 2020;32 doi: 10.1016/j.celrep.2020.107996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang T., Yeoh L.M., Tutor M.V., Dixon M.W., McMillan P.J., Xie S.C., Bridgford J.L., Gillett D.L., Duffy M.F., Ralph S.A., McConville M.J., Tilley L., Cobbold S.A. Decreased K13 abundance reduces hemoglobin catabolism and proteotoxic stress, underpinning artemisinin resistance. Cell Rep. 2019;29:2917–2928. doi: 10.1016/j.celrep.2019.10.095. e2915. [DOI] [PubMed] [Google Scholar]
- Yuan J., Cheng K.C., Johnson R.L., Huang R., Pattaradilokrat S., Liu A., Guha R., Fidock D.A., Inglese J., Wellems T.E., Austin C.P., Su X.Z. Chemical genomic profiling for antimalarial therapies, response signatures, and molecular targets. Science. 2011;333:724–729. doi: 10.1126/science.1205216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang M., Wang C., Oberstaller J., Thomas P., Otto T.D., Casandra D., Boyapalle S., Adapa S.R., Xu S., Button-Simons K., Mayho M., Rayner J.C., Ferdig M.T., Jiang R.H.Y., Adams J.H. The endosymbiotic origins of the apicoplast link fever-survival and artemisinin-resistance in the malaria parasite. bioRxiv. 2020;2012:419788. doi: 10.1038/s41467-021-24814-1. 2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu L., Tripathi J., Rocamora F.M., Miotto O., van der Pluijm R., Voss T.S., Mok S., Kwiatkowski D.P., Nosten F., Day N.P.J., White N.J., Dondorp A.M., Bozdech Z., Phyo A.P., Ashley E.A., Smithuis F., Lin K., Tun K.M., Faiz M.A., Mayxay M., Dhorda M., Thuy-Nhien N.T., Newton P.N., Pukrittayakamee S., Hlaing T.M., Hien T.T., Htut Y., Tracking Resistance to Artemisinin Collaboration, I., The origins of malaria artemisinin resistance defined by a genetic and transcriptomic background. Nat. Commun. 2018;9:5158. doi: 10.1038/s41467-018-07588-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.