Abstract
Herpesviruses, known as large DNA viruses, have a wide host range. In addition to human beings, cattle, and horses, even carp can be hosts for herpesvirus infection. Herpesviruses are pathogens possessing elaborate mechanisms that regulate host cell components for its replication, assembly and generating mature virus particles that can infect humans and most animals, usually causing multiple and lifelong infections. In addition, several human diseases, such as genital or mouth herpes, mononucleosis, and Burkitt lymphoma, are usually associated with herpesvirus infection. Blocking the steps of viral infection, such as entry, replication and assembly, may be an effective way for many different herpes viruses and their related diseases. Therefore, we aim to describe antiviral agents that are able to prevent herpesvirus entry, replication and assembly in host cells. We summarize antiviral strategies, including certain small molecular drugs, RNA interference and CRISPR/Cas9 system-based antiviral approaches, which represent promising approaches.
Keywords: Herpesvirus, DNA virus, Antiviral drug, RNAi, CRISPR/Cas9
1. Introduction
Herpesviruses consist of enveloped double-stranded DNA and primarily affect the skin, mucous membranes, and nerve tissues of hosts, often resulting in severe morbidity and mortality.1 A wide range of hosts can be infected by herpesviruses, including mammals, primates, amphibians, poultry, and carp. To date, more than 200 distinct herpesvirus species have been identified.2 This old viral lineage is diverse, with multiple naming schemes: 1) according to the hosts of certain isolated viruses, such as avian herpesvirus, monkey herpesvirus, and others; 2) depending on the diseases resulting from herpesvirus infection, such as herpes simplex virus and frog kidney adenocarcinoma herpes virus; 3) by the person(s) who discovered it, like the Epstein-Barr virus, Lucke herpes virus, Marek herpes virus, and others. Nevertheless, on the basis of their distinct biological and genetic structure, herpesviruses are divided into three groups of alpha (α), beta (β) and gamma (γ). Especially, herpes simplex virus (HSV) type 1 and 2, varicella zoster virus (VZV), cytomegalovirus (CMV) and Epstein–Barr virus (EBV) which are members of the Alphaherpesvirinae, Betaherpesvirinae, Gammaherpesvirinae subfamily, caused the most health concerns, thus have received extensive attention.
2. Herpesvirus infection
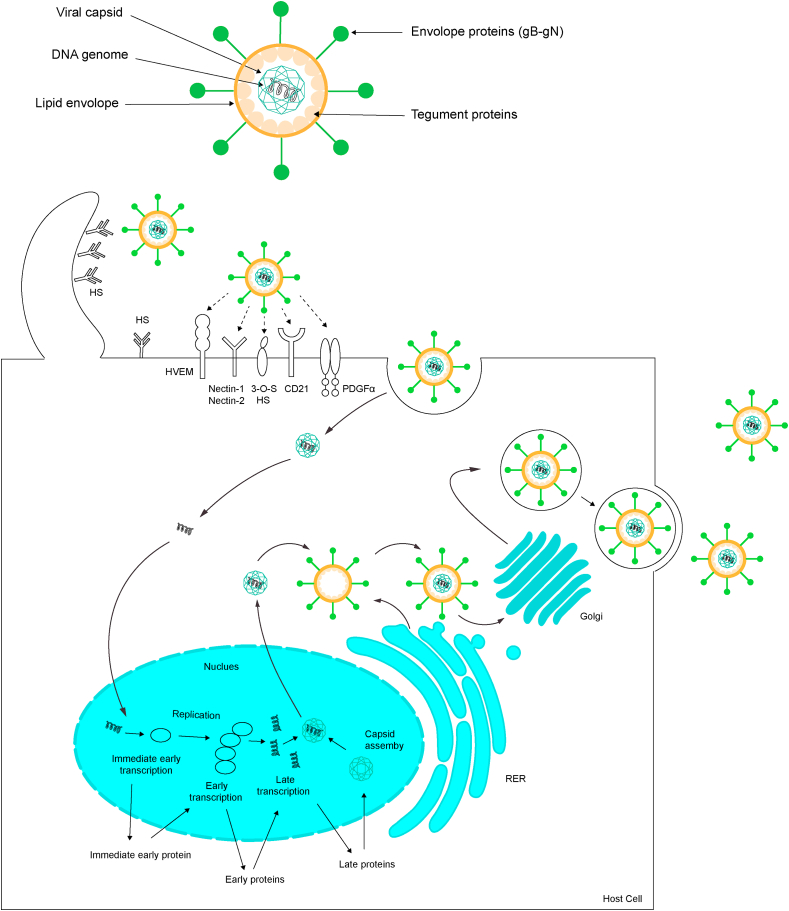
Herpesvirus infection is a complex process (Fig. 1) that includes virus attachment to the host cell, virus-host membrane fusion and internalization of the virus particles, viral genome replication, capsid assembly and virion release. The infection begins when a large number of virions are attached to heparan sulfate (HS), located on host cell surfaces by gB and/or gC, promoting gD or gE binding to relevant receptors, it usually in the form of proteoglycans, known as heparan sulfate proteoglycans (HSPG). Interaction between gB and heparan sulfate (HS) is required for HSV-1 surfing. The binding of these viral envelope proteins and host membrane receptors induces membrane fusion, resulting in internalization of the envelope and capsid into the host cytoplasm. Interestingly, several reports have found complete virions in the host cytoplasm, indicating that herpesvirus may infect the host by inducing cell endocytosis. Moreover, herpesvirus can enter the body in other ways. For instance, Nicola et al. found that HSV infection occurs partly through PH-dependent endocytosis in certain types of host cells.3 In addition, many herpesviruses have also been reported to enter cells through a phagocytosis-like mechanism, such as HSV-1, HHV-8 and CMV.4, 5, [6]
Fig. 1.
Model of the herpesvirus life cycle. All members of the Herpesviridae share a common structure, a mature virion composed of these four parts: genomic DNA, capsid, tegument, and envelope with glycoproteins. The innermost layer is viral DNA which located in an icosahedral protein cage called the capsid surrounded by a tegument proteins layer. The outermost layer is the envelope containing multiple glycoproteins. Infection is initiated when viral envelope glycoproteins bind to the specific types of cell surface receptors include HVEM, nectin-1/2, PDGFα, CD21 and 3-O-S HS etc. Post-binding events: viral particles enter into cells mainly through direct fusion with cell membrane or via an endocytic pathway. Next, viral capsid is dismantled and viral DNA enters host cell nucleus through nuclear pore. Within the nucleus, viral IE and E genes will be transcribed mediating the replication of viral genomic DNA. After 24h infection, transcript of the late (L) gene occurs and the produce capsid proteins so that a new nucleocapsid will assemble in host cell nucleus and guiding viral DNA to enter the capsid. After these immature particles pass through the nuclear membrane, they acquire an envelope. A secondary envelopment occurs in host cytoplasm to help virus forming mature virions, finally the virions leave the host cell via exocytosis.
After passing through the host membrane, capsid containing viral genomic DNA is transferred by the host cell movement protein along the microtubules to the host nucleus. Subsequently, the viral genomic DNA comes out of the capsid via a portal and enters host cell nucleus through the nuclear pore to initiate lytic-cycle infection. Immediate early (IE) proteins are first produced to promote the expression of early proteins (E proteins). Some of early proteins are critical for inducing viral DNA replication in replication compartments (RCs). Next, late gene of capsid proteins are transcribed, and the synthesized viral genomes are packaged into nucleocapsids.7 By budding, the incomplete virions leave the host nucleus to the cytoplasm through inner and outer nuclear layers and acquire most of tegument proteins in there. Then, those incomplete virions are wrapped by secondary envelopment on the Golgi membrane and/or endosome membrane, which also provides a transport vesicle. These specialized transitional vesicles take all virus particles to the cell plasma membrane and release them via exocytosis.8 All herpesviruses can establish lifelong latency in the nuclei of cells, and then distribute to its daughter cells. During the latent period, there is almost no obvious viral replication or viral gene expression as a strategy to successfully elude the immune system, thereby causing chronic infection throughout the whole life of their hosts. This brief description about viral replication applies to most, herpesviruses.
3. Antiviral strategies for treating herpesviruses
As a common contagious virus, although most herpes viruses infections are asymptomatic, some herpesvirus usually cause serious disease. Drugs against herpesviruses infections in the clinic are mainly of two types: nucleoside analogs such as acyclovir targeting DNA polymerase/thymidine kinase and helicase–primase inhibitors such as amenamevir (ASP2151), with both types having the ability to reduce or stop the replication of the viral genome. However, these therapeutic applications often induce drug resistance, making it necessary to gain a deeper understanding of the viral replication mechanism for use in exploring more suitable antiviral targets.
With the continuous development of biotechnology, new approaches to suppress herpesvirus at the gene level have been developed; for instance, RNA interference (RNAi) be could represented as a powerful tool to silence gene expression at the mRNA level and has demonstrated promise in combatting several herpesviruses, such as HSV-1, HHV-6B and CyHV-39-10-.11 At the DNA level, zinc finger nucleases (ZFNs), transcription activator-like effector nucleases (TALENs) and clustered regularly interspersed short palindromic repeat (CRISPR)-associated protein 9 (Cas9) make editing viral DNA genomes feasible.12 The specific editing of viral DNA may lead to better effects during the acute and latent periods with reduced virus-induced toxicity and a strong host immune response. Taken together, both gene-specific siRNA and DNA can in theory repress viral attachment to host cells, membrane fusion and internalization of viral particles, DNA replication, virion assembly and release. In this section, we mainly discuss antiviral approaches that impact on multiple aspects of virus such as entry, replication and assembly and mechanisms by which these treatments have enhanced efficacy, reduced viral resistance to treatment and potential for treatment failure.
3.1. Antiviral strategies for herpesvirus infection at the protein, DNA and RNA levels
3.1.1. Suppressing herpesvirus attachments and invasion by targeting viral proteins
Attachment to host cells is an important step to successfully establish herpesvirus infection, and one strategy is to directly block viral attachment factors such as an ideal and classic antiviral approach involves directly blocking viral attachment factors such as HSV gB and gC at the protein level. Bushra et al. showed that retrocyclin 2 can directly bind to the HSV-2 gB2 with high affinity and thereby prevent HSV-2 from entering target cells.13 A mandelic acid condensation polymer named SAMMA is also effective against HSV-2 and targets gB2, reducing both viral attachment and invasion.14 Agents that prevent the virus from entering or spreading by forming a stable complex with gB, include PRO 2000, polystyrene sulfonate, cellulose sulfate, and polymethylene hydroquinone sulfonate. Among these, PRO 2000 has been tested in clinical phase I trials and has displayed good safety and tolerability.15 In addition, when using anti-gC, it can target the cell-binding domain of gC protein and neutralize the virus infectivity in human immortalized keratinocytes (HaCaT).16 Additionally, gD, gH and gL also play a key role during the related processes. Subash et al. isolated a series of RNA aptamers that can inhibit viral entry by specifically binding to the HSV-1 gD protein.17 A 45-nt-long DNA aptamer also showed high affinity for HSV-1 gD, with an impact on virus entry and replication in vivo and in vitro.18 Thus, these studies have demonstrated that it is feasible to inhibit viral entry by targeting HSV-1 entry glycoproteins.
Varicella zoster virus (VZV) is different from other α-herpesviruses, the gE is critical for binding to the specific entry receptors of host cells since it does not have a gD. During VZV infection, gE is associated with gI targeting of specific regions in the host cell. However, there are few current reports showing VZV inhibition by blocking gE.
Similar to VZV, the human cytomegalovirus (HCMV) has no gD, the gB and gH/L complex play an important role in invasion. During the initial interaction, gB can bind not only cell surface heparan sulfate (HS) but also interact with epidermal growth factor receptor (EGFR), platelet-derived growth factor receptor-α (PDGFRα), BST/tetherin and various integrins to promote HCMV entry, but the potential of these targets are still unclear and needs to be studied further.
The Epstein-Barr virus (EBV) gp350/220 first binds to B-cell receptor CD21. Studies have shown that soluble recombinant CD21 can inhibit EBV infection in vitro, indicating that soluble CD21 may have potential as an effective agent for treating human EBV infection.19 In addition, EphA2 is essential for virus entry, as it triggers fusion upon virus attachment to epithelial cells.20 However, few works have been published investigating whether the inhibitor of EphA2 can suppress EBV infection.
3.1.2. Suppressing herpesvirus genome replication at the protein level
The results from genetic analysis show substantial similarity between HSV-1-and 2-induced diseases. Acyclovir (ACV) and related nucleoside analogs have been used to treat infection and suppress HSV transmission. These drugs can be phosphorylated by TK and form triphosphates through host enzymes, which compete with deoxyguanosine triphosphate to inhibit HSV-encoded DNA polymerase and selectively inhibit viral DNA replication with low host cell toxicity.21 Notably, inhibitors targeting viral DNA polymerase can suppress only viruses at the outbreak stage with rapid replication but do not inhibit viruses in the latent phase. Thus, drug-resistant HSV can occur during clinical therapy, especially in immunocompromised patients. Genotypic identification of these drug-resistant strains revealed that the mutations are located either in the UL23 gene, which encodes the activating/phosphorylating TK enzyme, or in the UL30 gene, which encodes the viral DNA polymerase.22 However, these aforementioned inhibitors are not suitable for TK-deficient HSV or VZV strains, as acyclic nucleoside analogs require phosphorylation by the virus-encoded TK. Interrupting the interaction of cofactors within herpesvirus DNA replication may also suppress herpesvirus infection; for example, the small molecule BP5 is capable of depressing HSV-1 replication by interfering with the UL30/UL42 complex formation in vitro.23
Similar to the effects on suppressing HSV, suppressing viral DNA polymerase is effective against CMV, with inhibitors such as foscarnet, cidofovir and ganciclovir. Targeting CMV terminase is also an attractive strategy, the CMV terminase consists of the products of UL89 and UL56 gene. Letermovir is the new CMV terminase inhibitor that has been approved by the FDA for prevention of CMV in allogeneic hematopoietic stem cell transplantation (allo-HSCT) recipients.24 However, there is appearance of a benzimidazole ribonucleoside-resistant CMV strain with mutated UL89.25 By high-throughput screening, Arianna et al. obtained five compounds that can specifically inhibit the UL54/UL44 interaction and then hamper the replication of CMV.26 The successful inhibition of these functional complexes provides potential new anti-CMV agents; that is, agents may be designed to inhibit not only the target protein itself but also the formation of functional complexes.
In fact, most DNA polymerase inhibitors, such as penciclovir and foscarnet, are able to suppress genome replication of all three herpesvirus subfamilies. Similarly, protein kinase enzyme inhibitors cyclopropavir and maribavir (1263W94) are also effective antiviral drugs against EBV. However, ACV can block EBV replication in lytically infected cells but does not have the same effect in latently infected cells. In contrast to nucleoside analogs, glycyrrhizic acid (GL) has been confirmed to impact on the early step of the EBV replication cycle.27
3.1.3. Inhibition of herpesvirus replication at the DNA and RNA levels
Virus-host interactions are dynamic and complex processes, and interfering with virus receptor genes to block virus binding at the DNA level is an alternative method. With the development of specific genome editing technologies, such as ZFNs, TALENs and the CRISPR/Cas9 system, targeting certain genes to inhibit viral infections appears feasible. However, ZFNs and TALENs have not been widely applied as ideal methods in the treatment of herpesvirus since the cost and complexity of designing these custom-built endonucleases are limiting factors. CRISPR-Cas, found in bacteria and archaea, is an adaptive (acquired) system that can target foreign sequences and then silence or modify these genetic elements in hosts, furthermore, in this system, multiple single guide RNA (sgRNAs) can be used simultaneously for the specific editing of different genes.28 As a double stranded DNA, the herpesvirus genome is easily edited by CRISPR/Cas9. To identify the most effective target sites for limiting HSV productive and latent infections, Ferdy et al. used CRISPR/Cas9 to disrupt a series of essential viral genes, including seven involved in DNA replication: UL30, UL42, UL9, UL5, UL8, UL52, and UL29; two genes encoding tegument proteins, namely, UL36 and UL37; the terminase UL15, which is related to DNA packaging; and UL27, which encodes capsid-associated gB and UL54, a multifunctional regulator of gene expression. The results suggest that the CRISPR/Cas9 system is able to impair HSV-1 replication in Vero cells by targeting all the aforementioned genes.29 Additionally, Xu et al. demonstrated that the CRISPR/Cas9-induced UL7 mutation, encoding a tegument protein, reduced HSV-1 replication capacity.30 Editing ICP0, ICP4 or ICP27 by CRISPR/Cas9 can also effectively reduce HSV-1 infection.31 Similar to other herpesviruses, the treatment of HCMV infection by CRISPR/Cas9 is feasible. Ferdy et al. designed sgRNAs targeting seven essential genes: viral DNA polymerase UL54, the polymerase accessory factor UL44, the single-stranded DNA-binding protein UL57, the primase UL70, the DNA helicase UL105, the major capsid protein UL86, and UL84, and almost completely inhibited viral replication by one or more sgRNAs. In his study, modification of the nonessential genes US6, US7, and US11 did not impact HCMV replication, because of the longer replication cycle of HCMV, resulting in the virus having sufficient time to repair the double-strand breaks (DSBs) of the viral genome.32 Indeed, a study has indicated that ±75% of DSBs can be repaired within 24 h in MRC5 cells.33 In addition, the selection pressure from CRISPR/Cas9 may drive viral evolution to produce mutants. Therefore, to minimize the escape of the virus to the greatest extent possible, targeting the viral genome using a single sgRNA was insufficient; at least two sgRNAs need to be used simultaneously to target multiple genes or different loci of one gene. The incurability of EBV infection is mainly due to viral latency in B lymphocytes. In vitro studies have shown that the CRISPR/Cas9 system can successfully eliminate latent EBV in cells. For example, Wang et al. reported that utilizing CRISPR/Cas9 technology generated a small deletion in EBV nuclear antigen 1 (EBNA1), EBV nuclear antigen 3C (EBNA3C) and latent membrane protein-1 genes (LMP1), which are essential for the regulation of latent genome replication and host cell transformation, resulting in a dramatic decrease in viral load in a Burkitt lymphoma cell line.34 Thereafter, Ferdy et al. designed sgRNAs targeting EBV EBNA1 and EBV origin of replication (OriP), and the results indicated that CRISPR/Cas9 could effectively reduce the EBV viral load in Akata-BX1 cells.29 Subsequently, Yuen et al. confirmed that the CRISPR/Cas9 system targeting 3 different genomic elements, namely EBNA1, OriP and W repeats can inhibit EBV DNA accumulation in nasopharyngeal carcinoma cells.35
At the RNA level, synthetic siRNA is a potential treatment strategy for viral genome replication. For HSV-1, Fang et al. knocked down the immediate-early (IE) gene ICP4, a major regulatory gene involved in lytic infection, and viral inhibition was observed in HCE cells, human trabecular meshwork cells (HTMs) and Vero cells.36 To test whether RNAi technology can be used as a method to prevent HSV-2 infection, Guo et al. showed that UL54-specific siRNA can significantly reduce the HSV-2 virus titer between 12 and 24 h.37 In vitro studies have shown that siRNAs can effectively inhibit HCMV infection by targeting the viral DNA polymerase enzyme pUL54, kinase pUL97, and immediate-early genes UL123 and UL122. Because treatment with siUL54B, siUL97A and siUL122B led to a 52.9%, 49.2% and 58.3% reduction in the number of infected cells, respectively.38 EBNA1, the EBV latency replication factor, is uniquely expressed in all EBV-positive proliferating cells. Currently, many reports have shown that inhibition of EBNA1 mRNA can cause loss of EBV episomal maintenance.39,40 In addition, Epstein-Barr virus latent membrane protein 1 (EBV-LMP1) is also useful for affecting EBV infection, Yu et al. constructed a vector encoding a short hairpin RNA (shRNA) against LMP1, an integral membrane protein gene, and which induced apoptosis in EBV-positive lymphoma cells.41
4. Conclusion and future directions
Currently, clinical therapy against herpesviruses does not cure the infection, it only reduces virus replication. Here, we summarize various antiviral strategies in herpesvirus treatment (Supplementary Table 1).
Although CRISPR/Cas9 and similar systems seem to be very promising for eradicating herpesviruses at the latent stage, it is possible that CRISPR/Cas9-resistant variants could arise due to survival selective pressure, which may limit the application of specific DNA-editing technologies. Several bottlenecks limit the use of siRNA, low transfection efficiency and original siRNA instability make totally silencing the target genes difficult. High levels of agent-facilitated transfection may lead to a host interferon overresponse and off-target side-effects. In addition to traditional siRNA, modified antisense oligonucleotides (AODs) are able to effectively suppress these important viral genes with suitable stability and low toxicity. For example, fomivirsen, an antisense thiodeoxynucleotide molecular drug targeting CMV immediate-early mRNA, has been approved by the FDA.42 In addition, recent studies have shown that DNA tetrahedrons and carbon nanotubes, which are recently developed nanophase biomaterials, have the ability to deliver exogenous biomolecules, such as siRNA, to target cells with enhanced stability.43,44
In theory, directly suppressing viral functional proteins is the efficient strategy since the polypeptide sequence is relatively conservative, compared to that of viral DNA or RNA, which are diverse among strains. Several novel methods for screening new small molecule inhibitors targeting known target proteins have been developed, such as virtual screening (VS), high-throughput screening (HTS), and high-content screening (HCS). By calculating the electrostatic field, hydrophobic field, and hydrogen bond distribution of the binding sites in silico, VS is used for screening a large number of conditional compounds for subsequent detection in a short time.
Early studies have shown that several chemical inhibitors are able to induce the degradation of target proteins. For example, canertinib (CI-1033) is an effective TK inhibitor that can also promote ErbB-2 (HER2/neu) degradation. Fulvestrant can inhibit the function of Era (estrogen receptor alpha) and induce its degradation45 by selectively degrading functional viral proteins, rather than attenuating their activity. Currently, proteolysis targeting chimera (PROTAC) technology offers an opportunity to degrade intracellular proteins by coupling small molecule inhibitors and E3 ubiquitin ligases. Compared with other antiviral strategies, a PROTAC has many advantages, including cell membrane permeability and high stability, and can be repeatedly used in cells. Similar to PROTAC technology, hydrophobic tagging of the HaloTag protein has also been employed to degrade target proteins.46 However, the current application of these technologies has been mainly focused on cancer research.
The discovery of new, effective targets associated with herpesvirus progression is a matter of urgency. The heparanase (HPSE), a host enzyme, is upregulated through NF-kB and translocated to the cell surface upon HSV-1 infection for the removal of cell surface heparan sulfate (HS) to facilitate viral release, it may be a potent target for inhibiting viral release.47,48 In order to better understand the infection mechanism of the herpesvirus and develop more antiviral drug targets, more attention to host proteins is necessary. To improve screening efficiency, sensitive in vitro and in vivo evaluation systems are required; for instance, Rui et al. developed a cell culture-based green fluorescent protein reporter system for determining EBV drug sensitivity.49 Employing known effective small molecules to identify targets by immunoprecipitation, surface plasmon resonance and other technologies are also important tools.
In this review, we highlight the current methodologies and strategies including small molecular drugs, RNA interference and CRISPR/Cas9 system-based antiviral approaches that impact on multiple aspects of virus such as entry, replication and assembly and hope to inform present and future drug development against herpes viruses.
Funding
This work was supported by grants from the National Natural Science Foundation of China (No. 81804013 to Z.W. and No. 31502030 to Y.Z.), the TCM Clinical Research Center for Bone Diseases of Jilin Province (No. 20180623048TC to X.L.) and the Jilin Province Science and Technology Development Project (No. 2020122218JC).
Author contributions
H.D., Z.W., D.Z., X.L., and Y.Z. wrote the paper. All authors have read and agreed to the published version of the manuscript.
Declaration of competing interest
The authors declare no conflict of interest.
Footnotes
Supplementary data to this article can be found online at https://doi.org/10.1016/j.jve.2021.100047.
Contributor Information
Xiangyang Leng, Email: leng_xiangyang@163.com.
Yicheng Zhao, Email: yichengzhao@live.cn.
Appendix A. Supplementary data
The following is the Supplementary data to this article:
References
- 1.Thomas C., Mettenleiter B.E., Thomas Müller, Kyoung‐Jin Yoon, Jens Peter Teifke. herpesviruses. Diseases of Swine. 2019 doi: 10.1002/97811193.50927.ch35. [DOI] [Google Scholar]
- 2.Wibbelt G. Discovery of herpesviruses in bats. J Gen Virol. 2007;88:2651–2655. doi: 10.1099/vir.0.83045-0. [DOI] [PubMed] [Google Scholar]
- 3.Nicola A.V., Hou J., Major E.O., Straus S.E. Herpes simplex virus type 1 enters human epidermal keratinocytes, but not neurons, via a pH-dependent endocytic pathway. J Virol. 2005;79:7609–7616. doi: 10.1128/JVI.79.12.7609.7616.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bandyopadhyay C., Valiya-Veettil M., Dutta D., Chakraborty S., Chandran B. CIB1 synergizes with EphrinA2 to regulate Kaposi's sarcoma-associated herpesvirus macropinocytic entry in human microvascular dermal endothelial cells. PLoS Pathog. 2014;10 doi: 10.1371/journal.ppat.1003941. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Clement C. A novel role for phagocytosis-like uptake in herpes simplex virus entry. J Cell Biol. 2006;174:1009–1021. doi: 10.1083/jcb.200509155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Tiwari V., Shukla D. Nonprofessional phagocytosis can facilitate herpesvirus entry into ocular cells. Clin Dev Immunol. 2012:651691. doi: 10.1155/2012/651691. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Kobiler O., Brodersen P., Taylor M.P., Ludmir E.B., Enquist L.W. Herpesvirus replication compartments originate with single incoming viral genomes. mBio. 2011;2 doi: 10.1128/mBio.00278-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Owen D.J., Crump C.M., Graham S.C. Tegument assembly and secondary envelopment of alphaherpesviruses. Viruses. 2015;7:5084–5114. doi: 10.3390/v7092861. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Gotesman M., Soliman H., Besch R., El-Matbouli M. In vitro inhibition of Cyprinid herpesvirus-3 replication by RNAi. J Virol Methods. 2014;206:63–66. doi: 10.1016/j.jviromet.2014.05.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Paavilainen H. Inhibition of clinical pathogenic herpes simplex virus 1 strains with enzymatically created siRNA pools. J Med Virol. 2016;88:2196–2205. doi: 10.1002/jmv.24578. [DOI] [PubMed] [Google Scholar]
- 11.Yoon J.S. Inhibition of herpesvirus-6B RNA replication by short interference RNAs. J Biochem Mol Biol. 2004;37:383–385. doi: 10.5483/bmbrep.2004.37.3.383. [DOI] [PubMed] [Google Scholar]
- 12.Saayman S., Ali S.A., Morris K.V., Weinberg M.S. The therapeutic application of CRISPR/Cas9 technologies for HIV. Expet Opin Biol Ther. 2015;15:819–830. doi: 10.1517/14712598.2015.1036736. 10.1517/14712598.2015.1036736. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Yasin B. Theta defensins protect cells from infection by herpes simplex virus by inhibiting viral adhesion and entry. J Virol. 2004;78:5147–5156. doi: 10.1128/jvi.78.10.5147.5156.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Herold B.C. Mandelic acid condensation polymer: novel candidate microbicide for prevention of human immunodeficiency virus and herpes simplex virus entry. J Virol. 2002;76:11236–11244. doi: 10.1128/jvi.76.22.11236.11244.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Cheshenko N. Candidate topical microbicides bind herpes simplex virus glycoprotein B and prevent viral entry and cell-to-cell spread. Antimicrob Agents Chemother. 2004;48:2025–2036. doi: 10.1128/AAC.48.6.2025-2036.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Adamiak B. Human antibodies to herpes simplex virus type 1 glycoprotein C are neutralizing and target the heparan sulfate-binding domain. Virology. 2010;400:197–206. doi: 10.1016/j.virol.2010.01.032. [DOI] [PubMed] [Google Scholar]
- 17.Gopinath S.C.B., Hayashi K., Kumar P.K.R. Aptamer that binds to the gD protein of herpes simplex virus 1 and efficiently inhibits viral entry. J Virol. 2012;86:6732–6744. doi: 10.1128/Jvi.00377-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Yadavalli T. Targeting herpes simplex virus-1 gD by a DNA aptamer can be an effective new strategy to curb viral infection. Mol Ther Nucleic Acids. 2017;9:365–378. doi: 10.1016/j.omtn.2017.10.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Nemerow G.R., Mullen J.J., 3rd, Dickson P.W., Cooper N.R. Soluble recombinant CR2 (CD21) inhibits Epstein-Barr virus infection. J Virol. 1990;64:1348–1352. doi: 10.1128/jvi.64.3.1348-1352.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Zhang H. Ephrin receptor A2 is an epithelial cell receptor for Epstein-Barr virus entry (vol 3, pg 1, 2018) Nat Microbiol. 2018;3:1075. doi: 10.1038/s41564-018-0155-1. 1075. [DOI] [PubMed] [Google Scholar]
- 21.De Clercq E. Antivirals and antiviral strategies. Nat Rev Microbiol. 2004;2:704–720. doi: 10.1038/nrmicro975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Piret J., Boivin G. Resistance of herpes simplex viruses to nucleoside analogues: mechanisms, prevalence, and management. Antimicrob Agents Chemother. 2011;55:459–472. doi: 10.1128/AAC.00615-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Pilger B.D., Cui C., Coen D.M. Identification of a small molecule that inhibits herpes simplex virus DNA Polymerase subunit interactions and viral replication. Chem Biol. 2004;11:647–654. doi: 10.1016/j.chembiol.2004.01.018. [DOI] [PubMed] [Google Scholar]
- 24.El Helou G., Razonable R.R. Letermovir for the prevention of cytomegalovirus infection and disease in transplant recipients: an evidence-based review. Infect Drug Resist. 2019;12:1481–1491. doi: 10.2147/IDR.S180908. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Buerger I. A Novel nonnucleoside inhibitor specifically targets cytomegalovirus DNA maturation via the UL89 and UL56 gene products. J Virol. 2001;75:9077–9086. doi: 10.1128/jvi.75.19.9077-9086.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Loregian A., Coen D.M. Selective anti-cytomegalovirus compounds discovered by screening for inhibitors of subunit interactions of the viral polymerase. Chem Biol. 2006;13:191–200. doi: 10.1016/j.chembiol.2005.12.002. [DOI] [PubMed] [Google Scholar]
- 27.Bentz G.L. Using glycyrrhizic acid to target sumoylation processes during Epstein-Barr virus latency. PloS One. 2019;14 doi: 10.1371/journal.pone.0217578. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Koonin E.V., Makarova K.S., Zhang F. Diversity, classification and evolution of CRISPR-Cas systems. Curr Opin Microbiol. 2017;37:67–78. doi: 10.1016/j.mib.2017.05.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Nelson J.A. CRISPR/Cas9-mediated genome editing of herpesviruses limits productive and latent infections. PLoS Pathog. 2016;12 doi: 10.1371/journal.ppat.1005701. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Xu X. The mutated tegument protein UL7 attenuates the virulence of herpes simplex virus 1 by reducing the modulation of alpha-4 gene transcription. Virol J. 2016;13:152. doi: 10.1186/s12985-016-0600-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Roehm P.C. Inhibition of HSV-1 replication by gene editing strategy. Sci Rep. 2016;6:23146. doi: 10.1038/srep23146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Rajcani J., Durmanova V. Mechanisms of replication of alpha- and betaherpesviruses and their pathogenesis. Bratisl Lek Listy. 2001;102:505–514. [PubMed] [Google Scholar]
- 33.Kuhne C., Tjornhammar M.L., Pongor S., Banks L., Simoncsits A. Repair of a minimal DNA double-strand break by NHEJ requires DNA-PKcs and is controlled by the ATM/ATR checkpoint. Nucleic Acids Res. 2003;31:7227–7237. doi: 10.1093/nar/gkg937. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Wang J., Quake S.R. RNA-guided endonuclease provides a therapeutic strategy to cure latent herpesviridae infection. Proc Natl Acad Sci U S A. 2014;111:13157–13162. doi: 10.1073/pnas.1410785111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Yuen K.S. Suppression of Epstein-Barr virus DNA load in latently infected nasopharyngeal carcinoma cells by CRISPR/Cas9. Virus Res. 2018;244:296–303. doi: 10.1016/j.virusres.2017.04.019. 10.1016/j.virusres.2017.04.019. [DOI] [PubMed] [Google Scholar]
- 36.Duan F., Ni S., Nie Y., Huang Q., Wu K. Small interfering RNA targeting for infected-cell polypeptide 4 inhibits herpes simplex virus type 1 replication in retinal pigment epithelial cells. Clin Exp Ophthalmol. 2012;40:195–204. doi: 10.1111/j.1442-9071.2011.02668.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Qing G. Research of UL54-specific siRNA on herpes simplex virus type II replication. Int J Dermatol. 2011;50:362–366. doi: 10.1111/j.1365-4632.2010.\04732.x. [DOI] [PubMed] [Google Scholar]
- 38.Hamilton S.T., Milbradt J., Marschall M., Rawlinson W.D. Human cytomegalovirus replication is strictly inhibited by siRNAs targeting UL54, UL97 or UL122/123 gene transcripts. PloS One. 2014;9 doi: 10.1371/journal.pone.0097231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Noh K.W., Park J., Kang M.S. Targeted disruption of EBNA1 in EBV-infected cells attenuated cell growth. BMB Rep. 2016;49:226–231. doi: 10.5483/bmbrep.2016.49.4.260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Messick T.E. Structure-based design of small-molecule inhibitors of EBNA1 DNA binding blocks Epstein-Barr virus latent infection and tumor growth. Sci Transl Med. 2019;11 doi: 10.1126/scitranslmed.aau5612. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Mei Y.P. SiRNA targeting LMP1-induced apoptosis in EBV-positive lymphoma cells is associated with inhibition of telomerase activity and expression. Canc Lett. 2006;232:189–198. doi: 10.1016/j.canlet.2005.02.010. [DOI] [PubMed] [Google Scholar]
- 42.Lalevee S., Feil R. Long noncoding RNAs in human disease: emerging mechanisms and therapeutic strategies. Epigenomics. 2015;7:877–879. doi: 10.2217/epi.15.55. [DOI] [PubMed] [Google Scholar]
- 43.Zhang H. DNA nanostructures coordinate gene silencing in mature plants. Proc Natl Acad Sci U S A. 2019;116:7543–7548. doi: 10.1073/pnas.1818290116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Hu F. Carbon nanotube-based DNA vaccine against koi herpesvirus given by intramuscular injection. Fish Shellfish Immunol. 2019 doi: 10.1016/j.fsi.2019.11.035. [DOI] [PubMed] [Google Scholar]
- 45.Citri A. Drug-induced ubiquitylation and degradation of ErbB receptor tyrosine kinases: implications for cancer therapy. EMBO J. 2002;21:2407–2417. doi: 10.1093/emboj/21.10.2407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Neklesa T.K. Small-molecule hydrophobic tagging-induced degradation of HaloTag fusion proteins. Nat Chem Biol. 2011;7:538–543. doi: 10.1038/nchembio.597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Agelidis A.M., Hadigal S.R., Jaishankar D., Shukla D. Viral activation of heparanase drives pathogenesis of herpes simplex virus-1. Cell Rep. 2017;20:439–450. doi: 10.1016/j.celrep.2017.06.041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Hadigal S.R. Heparanase is a host enzyme required for herpes simplex virus-1 release from cells. Nat Commun. 2015;6 doi: 10.1038/ncomms7985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Lin R. Development of a robust, higher throughput green fluorescent protein (GFP)-based Epstein-Barr Virus (EBV) micro-neutralization assay. J Virol Methods. 2017;247:15–21. doi: 10.1016/j.jviromet.2017.04.012. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.