Abstract
Egg-laying performance is one of the most important economic traits in the poultry industry. Commercial layers can lay one egg almost every day during their peak-laying period. However, many Chinese indigenous chicken breeds show a relatively low egg-laying rate, even during their peak-laying period. To understand what makes the difference in egg production, we compared the hypothalamus transcriptome profiles of Lushi blue-shelled-egg chickens (LBS), a Chinese indigenous breed with low egg-laying rate and Rhode Island Red chickens (RIR), a commercial layer with relatively high egg-laying rate using RNA-seq. A total of 753 differentially expressed genes (DEGs) were obtained. Of these DEGs, 38 genes were enriched in 2 Gene Ontology (GO) terms, namely reproduction term and the reproductive process term, and 6 KEGG pathways, namely Wnt signaling pathway, Oocyte meiosis, GnRH signaling pathway, Thyroid hormone signaling pathway, Thyroid hormone synthesis and MAPK signaling pathway, which have been long known to be involved in egg production regulation. To further determine the core genes from the 38 DEGs, protein-protein interaction (PPI) network, co-expression network and transcriptional regulatory network analyses were carried out. After integrated analysis and experimental validation, 4 core genes including RAC1, MRE11A, MAP7 and SOX5 were identified as the potential core genes that are responsible for the laying-rate difference between the 2 breeds. These findings paved the way for future investigating the mechanism of egg-laying regulation and enriched the chicken reproductive regulation theory.
Key words: laying hen, hypothalamus, reproductive regulation, RNA-Seq, egg-laying trait
INTRODUCTION
Egg-laying performance is an important economical trait, which reflects the productive capacity of layers. Egg numbers or egg laying rate as the selected indicator have been used in layers breeding programs and helped make considerable progress with genetic selection in commercial egg-layer breeds over recent decades (Johnson et al., 2015). Some commercial layers with a high laying rate can lay one egg almost every day during the peak-laying period, which almost reaches their physiological limits (Crawford, 1990). However, the peak-laying rates of many native chicken breeds in China are only between 70% and 80%. Moreover, the peak-laying period is relatively shorter in the indigenous hens compared to that of commercial layers. The low egg production performance has been a bottleneck that constrains the development of the indigenous chicken industry (Li and Tian, 2018). Therefore, what genes are responsible for the difference in egg production between native chickens and high-yielding layers, and how to accelerate the genetic progress of egg-laying traits have always been the principal issues in indigenous chicken breeding programs.
With the development of next-generation sequencing and high-density single nucleotide polymorphisms (SNP) genotyping techniques, a number of studies based on genome-wide genetic variance have identified several quantitative trait loci (QTLs) and SNPs associated with reproductive traits of chickens. A genome-wide association study (GWAS) performed on both White Leghorn and dwarf brown-egg layers and revealed 8 SNPs that were associated with egg production and egg quality traits (Liu et al., 2011). Wolc et al. (2014) mapped several QTLs on chromosomes 2, 12 and 17 of a brown egg layer line, explaining > 5% of the genetic variance for albumen height, egg shell color and egg production. A SNP for accumulative egg number from 21 to 40 wk was identified on F2 hens produced by reciprocal crosses between White Leghorn and Dongxiang Blue-shelled chickens, which created phenotypic differences of 6.86 eggs between 2 homozygous genotypes (Yuan et al., 2015). However, reproductive traits are polygenic traits with low to moderate heritability, and these QTLs and SNPs identified by GWAS or other methods account for only some of the genetic variation for chicken reproduction performance and often show breed or population specificity, which makes it difficult to clarify the molecular mechanism and to estimate the level of genetic improvement in each generation (Biscarini et al., 2010).
Transcriptome sequencing (RNA-seq) data analysis showed growing importance in understanding how altered expression of genetic variants contributes to complex traits such as growth, meat quality (Piorkowska et al., 2018), fat deposition/ metabolism (Li et al., 2018) and reproduction (Sun et al., 2016). For instance, the gene expression profiles of chicken follicles from different developmental stages provided novel insights into the molecular mechanisms of follicular physiology (Zhu et al., 2015). A whole-transcriptome analysis between atrophic ovaries and normal ovaries from equivalent-aged egg-laying hens using RNA sequencing revealed that protein-coding genes, miRNAs, and lncRNA transcripts are candidate regulators of ovary development in broody chickens (Liu et al., 2018). Yin et al. (2019) performed the transcriptome analysis of ovary and 3 segments of oviduct of chicken using RNA-seq and obtained functional genes involved in egg formation by combining QTL, transcriptome and proteome data. Wang and Ma (2019) analyzed the expression profiles of hypothalamus and pituitary in high- and low-yielding laying Chinese Dagu Chickens, and identified increased expression of genes involved in amino acid metabolism, glycosaminoglycan biosynthesis, and estrogen negative feedback systems in low-yielding laying hens. All of the above studies were carried out with tissues collected from different physiological stages of animals within the same population sharing similar genetic backgrounds.
Increasing studies showed that transcriptome analysis of the specific tissues obtained from animals with different genetic backgrounds could also provide valuable information about genes or pathways regulating the target traits. Kong et al. (2017) compared the transcriptome profiles of breast muscles between the 2 lines, BPR (Barred Plymouth Rock), an unselected foundational slow growth and lower efficient line and PeM (Pedigree male), a modern inbred male line highly selected for growth and feed efficiency, and identified crucial genes which were response for muscle growth. Zhang et al. (2017a) analyzed the breast muscle transcriptomes of Gushi chicken, a typical dual-purpose Chinese indigenous chicken breed and Arbor Acres (AA) broiler, a commercial meat-type breed and discovered significant signal transduction pathways and genes related to muscle formation and fat metabolism. Similarly, Monson et al. (2019) performed comparative studies on transcriptomes of thymus of 2 genetic lines exposed to acute heat stress and/or immune simulation with lipopolysaccharide fund that a large number of significant genes both at homeostasis and in response to treatment. A genome-wide analysis of mRNAs and lncRNAs from Bashang long-tail chickens and Hy-Line brown layers ovarian follicles suggested that some differentially expressed genes were involved in ovarian follicular development through oocyte meiosis, progesterone-mediated oocyte maturation, and cell cycle (Peng et al., 2019).
The Rhode Island Red (RIR) chicken is a commercial layer breed with a peak laying rate between 90% and 95%, whereas the Lushi blue-shelled-egg (LBS) chicken is a Chinese local breed with relatively low egg performance, with the peak laying rate being between 70% and 75%. The hypothalamus is the center regulating reproductive activity, which alone or works in conjunction with the pituitary to affect follicular development, ovulation and spawning. Hypothalamus tissue should be the top priority for exploring the possible key genes that cause differences in egg production using RNA-seq (Brady, et al., 2020; Mishra, et al., 2020; Wu, et al., 2020). Herein, we compared and analyzed the expression profiles of the hypothalamus in RIR hens and LBS hens during the peak laying period. Our study aimed to identify the pathways and core genes regulating the peak egg production rate by the integrated analysis of gene networks, which probably cause high egg-laying performance in RIR hens and low egg-laying performance in LBS hens. Our results provide novel information on the regulatory mechanisms of egg-laying performance in chickens.
MATERIALS AND METHODS
Ethics Statement
Animal experiments were approved by the Institutional Animal Care and Use Committee (IACUC) of Henan Agricultural University Zhengzhou, P.R. China (Permit Number: 11-0085) and performed in accordance with the protocol outlined in the “Guide for Care and Use of Laboratory Animals”.
Experimental Animals and Samples Preparation
The RIR and LBS hens used in this study were raised in the Animal Center of Henan Agricultural University under the same husbandry conditions with a standard commercial hen food and ad libitum access to water during the whole experimental stage. Each of the birds was housed separately in a single cage. Hens entering egg-laying stage (at 20 wk of age) were exposed to a 16 h / 8 h light / dark regime. The egg numbers of individuals from 25 to 30 wk were recorded. The RIR and LBS birds produced 39.19 ± 2.06 and 30.38 ± 4.16 eggs during this period, respectively. The difference of the average numbers of eggs between RIR hens and LBS hens reached a significant level (P < 0.05). In each breed, 3 birds who displayed the similar egg number to the group average at the age of 30 wk old were randomly selected and slaughtered. The hypothalamic samples were isolated, snap-frozen in nitrogen immediately, and stored at -80°C until use.
The Gushi (GS) hens, a Chinese indigenous breed with a similar egg production to LBS hens, were obtained from Gushi Chicken Breeding Farm at Henan Sangao Agriculture and Animal Husbandry Co. LTD, Henan Province, China. The hens were raised in individual cages with normal feeding procedure. The egg number of all hens was recorded daily. Six hypothalamic samples were collected from peak-laying hens (30 wk old) with high egg-laying rate (ELR) (ELR from 25 to 30 wk were more than 90%, GS30w-H) and low ELR (ELR were from 20% to 30%, GS30wL), respectively. The birds were euthanized and the samples were processed as mentioned above.
RNA Preparation
Total RNA was extracted from each hypothalamus tissue sample using the Trizol Reagent (Invitrogen, Carlsbad, CA) according to the manufacturer's instructions. The concentration and purity of total RNA were assessed by the NanoDrop 2000 spectrophotometer (Thermo Scientific, Waltham, MA). The integrity was estimated using the RNA Nano 6000 Assay Kit of the Agilent Bioanalyzer 2100 system (Agilent Technologies, Palo Alto, California), and the RNA Integrity Number (RIN) value of each sample ≥ 7.0 was used to construct the sequencing libraries.
Library Construction and Sequencing
Six hypothalamus RNA samples from 3 LBS hens and 3 RIR hens were used to construct RNA-seq libraries. A total amount of 4 μg RNA per sample was used as input material for the RNA sample preparations. Firstly, the RNA samples were treated by using the Ribo-Zero rRNA Removal Kit (Epicentre, Madison, WI) to remove the ribosomal RNA. Then, sequencing libraries were generated using Illumina TruSeqTM RNA Sample Preparation Kit (Illumina, San Diego, CA) following the manufacturer's instructions. In brief, mRNA was cleaved into short fragments by adding Fragmentation buffer. Then, the short fragments were used as templates to synthesize the first-strand cDNA using random hexamer primers. The first-strand cDNA was used to form double-stranded cDNA with DNA polymerase I and RNase H (Invitrogen, Carlsbad, CA). The cDNA fragments of desired length were purified using an AMPure XP system (Beckman Coulter, Brea, CA), end repaired and linked with sequencing adapters. The purified cDNA fragments were used as templates to perform PCR with Phusion High-Fidelity DNA polymerase, Universal PCR primers and Index (X) Primer. Finally, the purified PCR products were assessed on the Agilent Bioanalyzer 2100 system, and subsequently used to construct the sequencing libraries. The 6 libraries were sequenced on an Illumina HiSeqTM 2500 plateform (Illumina, San Diego, CA) followed by Illumina's RNA-seq instructions. And 150 bp paired-end reads were generated for further analysis. All data from this article have been deposited in NCBI-SRA database under the accession numbers of SRR8573648, SRR8573649, SRR8573646, SRR8573647, SRR8573650 and SRR8573651.
RNA-Seq Data Analysis
FASTQ sequencing data were primarily processed by using the FASTX-Toolkit (version: 0.0.13) (http://hannonlab.cshl.edu/fastx_toolkit/index.html). Clean reads were obtained by removing reads containing adapter, poly-N, and low-quality reads from the raw data. The clean reads were mapped to the chicken genome sequence downloaded from the website: (ftp://ftp.ncbi.nlm.nih.gov/genomes/all /GCF_000002315.4_ Gallus_gallus-5. 0_genomic.gff.gz) with TopHat2 (version: 2.0.14) (Kim et al., 2013). Transcript assembly and abundance estimation was performed using Cufflinks (Lei et al., 2007). The gene expression level was calculated by HTSeq (version: 0.6.1) (Anders et al., 2015). And each gene expression level was normalized with FPKM (fragments per kilobase of exon model per million mapped reads). The differential expression analysis of the 2 groups was performed using the DESeq. 2 R package (1.16.1) based on the read count data. To identify differentially expressed genes (DEGs), Cuffdiff was used to calculate the P-value and the false discovery ratio (FDR). If |log2 Foldchange| was > 1 and FDR was < 0.05, the expression levels of the genes were considered significantly different.
Function Annotation of DEGs
Gene Ontology (GO) and Kyoto Encyclopedia of Genes and Genomes (KEGG) pathway analysis of the DEGs was performed with the software DAVID (Dennis et al., 2003). GO enrichment analysis was used to analyze the main function of DEGs. The DEGs were annotated from 3 definitions including biological process, molecular function and cellular component. The P-value was computed for the GO terms. The P-value < 0.05 was defined as a significantly enriched GO terms associated with the DEGs. Pathway enrichment analysis was used to identify the significant pathways involving the DEGs by pathway annotations using KEGG. The ClusterProfiler R package was used to test the statistical enrichment of DEGs in KEGG pathways. The P-value < 0.05 was defined as a significantly enriched KEGG pathway.
Protein Interaction Network Analysis
The protein-protein interaction (PPI) network of DEGs that were predicted to be associated with reproductive regulation by GO and KEGG pathway analysis, was built up using the STRING protein interaction database (http://string-db.org/) by calculating the combined score (threshold: score > 0.4). In the PPI network, each node represents a protein and each edge represents an interaction between these 2 proteins. Visual editing of differential PPI network data files was performed by Cytoscape software version 3.5.1 (http://www.cytoscape.org/).
Co-expression-Network Analysis
The co-expression-network was built according to the normalized signal intensity of DEGs that were selected from significant GO terms and pathways. Pearson's correlation coefficient was calculated for each pair of genes, and the significant correlated pairs (FDR < 0.05) were used for establishing the network (Prieto et al., 2008). Within the co-expression-network, for locating the key regulatory genes, k-core and | Dif _ k-core | were calculated according to the method described by Barabasi (Barabási and Oltvai, 2004). The value of | Dif _ k-core | reflects the relative importance of genes which cause the phenotypic changes. The greater the value of | Dif _ k-core |, the bigger the possibility of the gene functioning in the phenotype. The DEGs were considered as key regulatory genes to causing the phenotypic changes, if the | Dif _ k-core | was > 10 (Chen et al., 2016).
Transcription Regulatory Network Analysis
TFBSTools provides a toolkit for efficiently investigating transcription factor binding sites genome-wide (Tan and Lenhard, 2016). To investigate the effects of transcription factors on reproductive regulation, the DEGs were introduced into the TFBSTools database. The interactional relationships between the transcription factor (TF) and the targeted DEGs were identified and the transcription factor-target gene regulatory network was established by Cytoscape software (Smoot et al., 2010). The TFs highly connected with target genes are considered as the key TFs affecting phenotypic changes (Chen et al., 2016).
RT-qPCR Verification
To confirm the repeatability and accuracy of the RNA-Seq data, ten DEGs were randomly selected for real time fluorescent quantitative PCR (RT-qPCR) validation. The hypothalamic tissue samples were obtained from 6 LGS hens and 6 RIR hens, and the number of eggs in each group was the same or close to those used for RNA sequencing. After extracting total RNA, the first-strand cDNA was synthesized using the PrimeScript RT Reagent kit with gDNA.
The RT-qPCR was conducted in a 10μL reaction volume containing 1μL of first-strand cDNA (diluted to 500 ng/μL), 5μL of 2×SYBR Premix Ex TaqTM II (Tli RNaseH Plus) (TaKaRa), 0.5μL of each forward and reverse primer (10μM), and 3μL deionized water. The RT-qPCR was performed on a LightCycler 96 Real Time PCR System (Roche Applied Science, Indianapolis, IN). The optimized cycling conditions were as follows: one cycle of preincubation at 95℃ for 3 min and 40 cycles of amplification (95℃ for 10 s, 61℃ for 30 s, and 72℃ for 30 s). Each sample was tested in triplicate. The specificities of RT-qPCR products were verified by agarose gel electrophoresis followed by melting curve analysis. The relative expression level of each mRNA normalized to beta-actin was calculated by the 2−ΔΔCT method (Schmittgen and Livak, 2008). All primers were designed by using the Primer-BLAST tool (https://www.ncbi.nlm.nih.gov/tools/primer-blast/). The primer sequences are listed in Supplementary Table S1.
RESULTS
Identification of Expressed Transcripts in the Hypothalamus Transcriptome
In this study, a total of 6 cDNA libraries were constructed from the hypothalamus of LBS laying hens (L-ln30-1, L-ln30-2, L-ln30-3) and RIR laying hens (R-ln30-1, R-ln30-2, R-ln30-3), and the raw reads from the 6 libraries ranged from 75.71 to 103.64 million (Supplementary Table S2). The raw reads were filtered and mapped to the NCBI GCF_000002315.4 Gallus_gallus-5.0 version of the chicken genome assembly with a unique mapping rate from 0.850 to 0.864 (Supplementary Table S2). The mapped reads were distributed on all chromosomes but less on micro-chromosomes 30, 31 and 32 (Supplementary Figure S1a). Gene structure of mapped reads showed that the read content was highest on exons (Supplementary Figure S1b).
Identification of DEGs Between the Two Groups of Birds
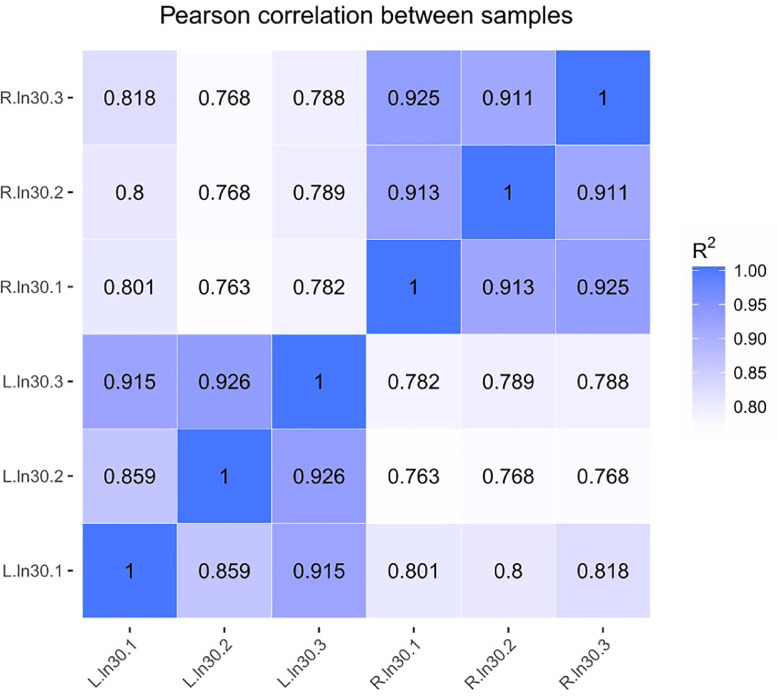
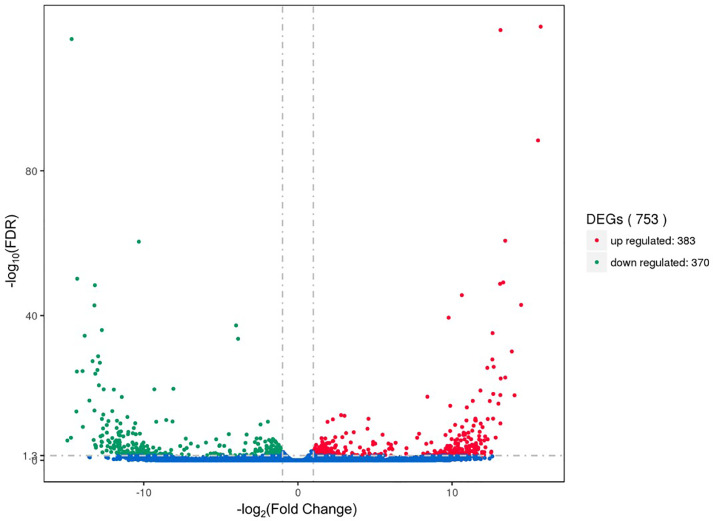
To investigate the different hypothalamus properties of Chinese indigenous hens and high-yielding layers at peak laying periods from a genetic perspective, the expression levels of the genes were quantified by the FPKM method. The Pearson correlation coefficient of gene expression levels between samples was more than 0.859, which indicated the selected experimental samples could be regarded as effective biological replicates (Figure 1). A total of 14,190 genes were identified in the 6 hypothalamus cDNA libraries, among which 5,295 genes were detected as novel genes. Among all the genes, a total of 753 DEGs were identified (|log2 Foldchange| > 1 and FDR < 0.05), including 383 upregulated and 370 downregulated genes in hypothalamus tissues of RIR hens compared to that of LBS hens (Figure 2). The FPKM hierarchical cluster analysis showed that the expression of DEGs in LBS samples was accurately distinguished from that in the RIR samples and the data were repeatable and credible (Supplementary Figure S2).
Figure 1.
Correlation analysis between samples. R-ln30-1∼R-ln30-3 are hypothalamic samples of high-yielding RIR hens from 1 to 3, and L-ln30-1∼L-ln30-3 are hypothalamic samples of low-yielding LBS hens from 1 to 3. The abscissa and ordinate are log10 (FPKM + 1) of the samples compared with each other.
Figure 2.
Volcano plot of differentially expressed genes. The abscissa represents the fold change of gene expression in hypothalamic samples of high-yielding RIR hens compared with that of low-yielding LBS hens; The ordinate represents the statistical significance of the difference in the amount of gene expression; The red dot indicates the significantly upregulated genes (Fold Change > 2, FDR < 0.05), and the green dot indicates the significantly downregulated genes (Fold Change > 2, FDR < 0.05).
GO and KEGG Pathway Analyses of DEGs
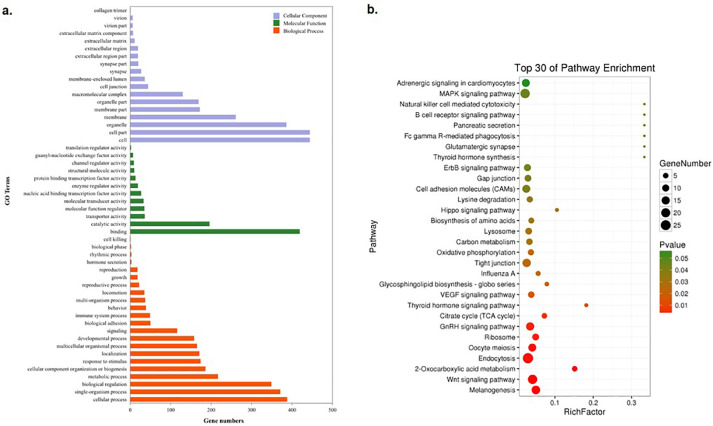
To elucidate the function of DEGs, GO enrichment analysis was used to annotate DEGs and to study their distribution. A total of 623 DEGs were enriched in 52 GO terms (Figure 3A) that were classified into 3 categories: biological process, molecular function and cellular component. The most abundant DEGs terms were in the cellular process term of biological processes, binding term of molecular functions and cell term of cellular components, in which there were 388, 419 and 444 DEGs involved, respectively.
Figure 3.
GO and pathway enrichment analysis of differentially expressed genes (DEGs) identified in hypothalamus of RIR vs. LBS. (A). Gene ontology classification of DEGs. Orange indicates biological processes; purple indicates cellular components; green indicates molecular function. (B). Top 30 of KEGG pathway enrichment classifications of DEGs. The horizontal axis represents the Rich factor, and the vertical axis represents the name of the pathway. The point size indicates the number of DEGs enriched in the pathway, and the point color corresponds to a different P-value range.
Here, we mainly focused on the differences in egg production between the 2 varieties during peak laying periods. Out of the 52 GO terms, 2 terms involved in reproductive regulation, namely reproduction (12 DEGs) and the reproductive process (15 DEGs). And a total of 15 DEGs were involved in the 2 GO terms which may relate with the reproductive regulation action (Supplementary Table S3).
To gain insight into the regulation network of DEGs produced by comparing the hypothalamic transcriptome of the 2 varieties during peak laying periods, pathway enrichment analysis was performed. The DEGs were enriched in 134 KEGG pathways, and the top 30 of pathways were showed in Figure 3B. Among the 30 pathways, 6 played pivotal roles in reproductive regulation, and which included the Wnt signaling pathway, Oocyte meiosis, GnRH signaling pathway, thyroid hormone synthesis, thyroid hormone signaling pathway and MAPK signaling pathway. There were 24 DEGs involved in the 6 pathways that were speculated to be related to regulation of egg production (Supplementary Table S3).
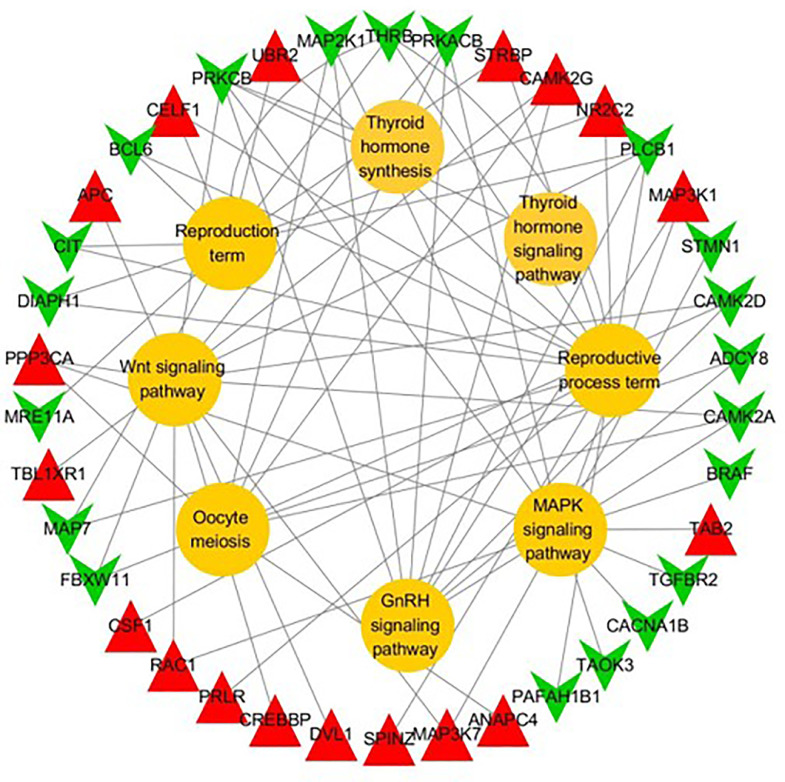
Based on the above enriched 2 GO terms and 6 pathways, a total of 38 DEGs related to egg-laying regulation were identified (Table 1, Figure 4).
Table 1.
The 38 DEGs involved in egg-laying regulation.
| Genes | Description | log2FC | FDR |
|---|---|---|---|
| BRAF | serine/threonine protein kinase | -13.171 | 4.15504E-49 |
| PLCB1 | 1-phosphatidylinositol 4,5-bisphosphate phosphodiesterase beta-1 | -12.307 | 3.09113E-06 |
| TGFBR2 | TGF-beta receptor type-2 | -10.897 | 0.01600927 |
| TAOK3 | serine/threonine-protein kinase TAO3 | -10.757 | 0.037503691 |
| CAMK2D | calcium/calmodulin-dependent protein kinase type II subunit delta | -10.553 | 5.74164E-11 |
| DIAPH1 | protein diaphanous homolog 1 | -10.430 | 0.002104531 |
| CACNA1B | voltage-dependent N-type calcium channel subunit alpha-1B | -10.050 | 0.010224589 |
| ADCY8 | adenylate cyclase type 8 | -10.013 | 0.015088767 |
| MRE11A | double-strand break repair protein MRE11 | -7.375 | 0.001188869 |
| BCL6 | B-cell lymphoma 6 protein homolog | -5.073 | 6.80152E-05 |
| FBXW11 | F-box/WD repeat-containing protein 11 | -5.028 | 0.024085914 |
| CAMK2A | calcium/calmodulin-dependent protein kinase type II subunit alpha | -1.753 | 0.000738379 |
| PAFAH1B1 | lissencephaly-1 homolog | -1.744 | 0.00894066 |
| MAP2K1 | dual specificity mitogen-activated protein kinase kinase 1 | -1.726 | 8.97582E-06 |
| CIT | citron Rho-interacting kinase | -1.489 | 5.8911E-06 |
| PRKACB | cAMP-dependent protein kinase catalytic subunit beta | -1.163 | 0.028770997 |
| STMN1 | stathmin | -1.122 | 0.000284834 |
| PRKCB | protein kinase C beta type | -1.097 | 0.006704126 |
| THRB | thyroid hormone receptor beta | 1.142 | 0.018880847 |
| TBL1XR1 | F-box-like/WD repeat-containing protein TBL1XR1 | 1.165 | 0.012134297 |
| NR2C2 | nuclear receptor subfamily 2 group C member 2 | 1.325 | 0.037436713 |
| RAC1 | ras-related C3 botulinum toxin substrate 1 | 1.380 | 0.014360939 |
| TAB2 | TGF-beta-activated kinase 1 and MAP3K7-binding protein 2 | 1.415 | 0.007693178 |
| APC2 | adenomatous polyposis coli protein 2 | 1.919 | 0.000654916 |
| SPINZ | spindlin-Z | 2.060 | 0.001629958 |
| ANAPC4 | anaphase-promoting complex subunit 4 | 2.596 | 0.002890362 |
| MAP3K7 | mitogen-activated protein kinase kinase kinase 7 | 6.044 | 0.049967263 |
| MAP3K1 | mitogen-activated protein kinase kinase kinase 1 | 9.261 | 0.006215996 |
| PRLR | prolactin receptor | 9.971 | 1.33865E-06 |
| CAMK2G | calcium/calmodulin-dependent protein kinase type II subunit gamma | 10.318 | 0.041858546 |
| CREBBP | CREB-binding protein | 10.472 | 1.20896E-07 |
| UBR2 | E3 ubiquitin-protein ligase UBR2 | 10.869 | 0.009675688 |
| CELF1 | CUGBP Elav-like family member 1 | 10.961 | 2.41486E-15 |
| DVL1 | segment polarity protein dishevelled homolog DVL-1 | 11.544 | 1.04803E-11 |
| STRBP | spermatid perinuclear RNA-binding protein | 11.758 | 1.7915E-06 |
| CSF1 | colony-stimulating factor 1 | 11.980 | 0.04534749 |
| PPP3CA | serine/threonine-protein phosphatase 2B catalytic subunit alpha | 12.320 | 3.84468E-12 |
Figure 4.
The functional network of differentially expressed genes (DEGs) associated with reproductive regulation. The red triangles indicate downregulated DEGs, and the green triangles indicate upregulated DEGs. The yellow circles represent the GO terms or pathways. If there is a link between DEG and GO term/pathway, the DEG is involved in the GO term/pathway.
Analysis of PPI Network for DEGs
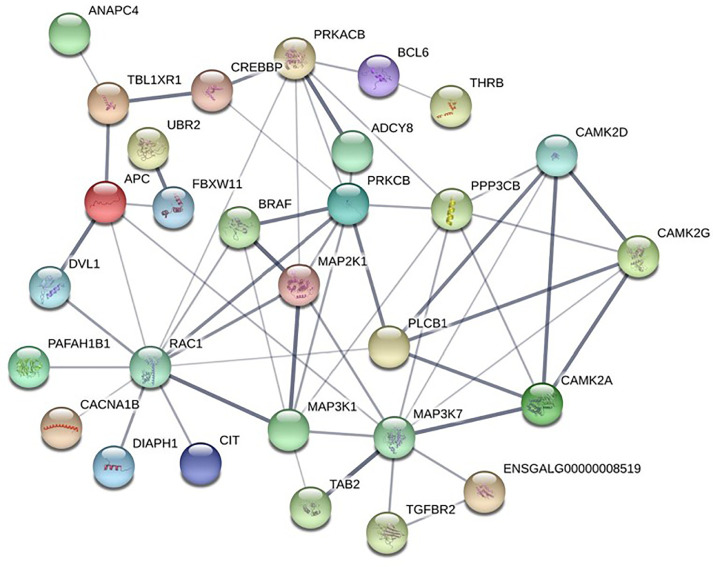
To further explore the potential key genes regulating egg-laying performance from the above 38 DEGs, the PPI network was constructed (Figure 5). RAC1 (Degree=12) and MAP3K7 (Degree=10) showed higher degrees of connection degrees than other DEGs in the PPI network and were considered core genes regulating egg-laying performance.
Figure 5.
Map of protein-protein interaction networks in hypothalamus. Nodes represent proteins. Edges represent protein-protein associations. The relationship between the 2 proteins is expressed through the thickness of the line; the thicker the line, the closer the relationship.
Co-expression Analysis for DEGs
A co-expression network based on the 753 DEGs in the hypothalamic tissues between the LBS and RIR hens was constructed. A total of 397 network nodes and 2,281 connections were formed in the LBS group (Supplementary Figure S3a), while 394 network nodes and 2,009 connections were formed in the RIR group (Supplementary Figure S3b). The k-core and | Dif _ k-core | were calculated to identify the key genes regulating reproductive performance (Supplementary Table S4 and Supplementary Table S5). A total of 15 DEGs with | Dif _ k-core | > 10 between the LBS and RIR groups were identified (Supplementary Table S6). Among the 15 DEGs, MRE11A and MAP7 were also listed among the 38 DEGs related to egg-laying regulation, which were identified by GO and KEGG pathway analyses of DEGs. Therefore, MRE11A and MAP7 were speculated to be 2 of the core genes for regulating egg-laying performance.
Transcription Regulatory Network Analysis
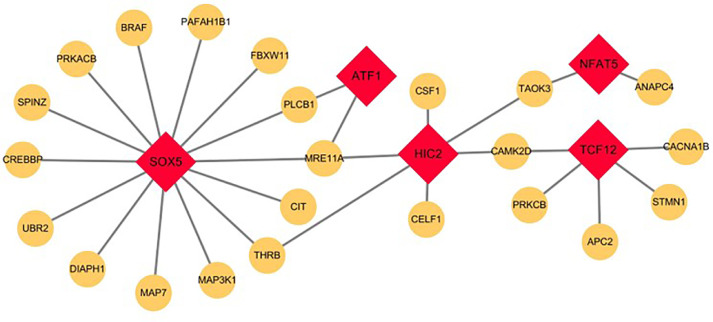
Transcription factors play crucial roles in the regulation of gene expression. To explore the effects of TFs on egg-laying regulation, a transcription regulatory network was constructed. The 753 DEGs were first imported into the TFBS Tools database. Six DEGs were detected as transcription factors, and 302 DEGs were predicted to be their target genes (Supplementary Table S7). Among the 302 DEGs, 23 DEGs were listed among the 38 DEGs that were related to egg-laying regulation by GO and KEGG analysis. There were 5 TFs targeted to the 23 DEGs. A transcription regulatory network of the 5 TFs and the 23 DEGs was constructed (Figure 6). The transcription factor SOX5 had the most relationships with 14 target genes. Therefore, the transcription factor SOX5 was considered a key transcription factor in the egg-laying regulation process.
Figure 6.
Transcription regulatory network of DEGs. Construction of transcription regulatory network centered around the DEGs that related to reproductive regulation. Red squares represent transcription factors, yellow circles represent target DEGs.
RT-qPCR Validation of Gene Expression Profile
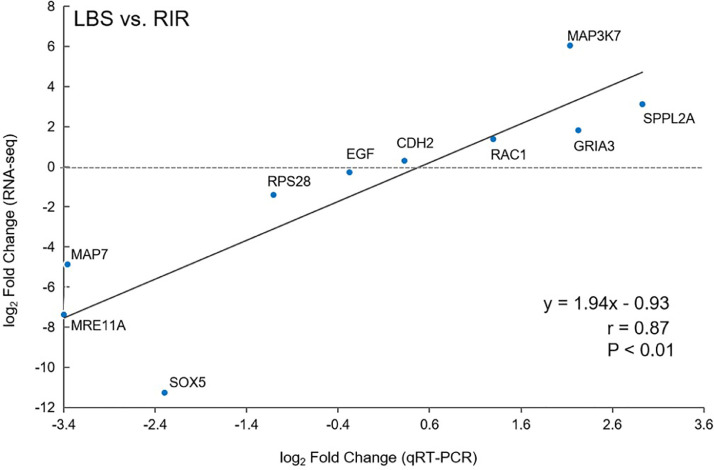
To validate the RNA-Seq transcriptome data, genes were selected as follows: significantly upregulated genes RAC1, MAP3K7, SPPL2A, GRIA3; significantly downregulated genes RPS28, MRE11A, MAP7, SOX5; and nonsignificantly different genes EGF and CDH2. The expression levels of the selected genes were confirmed by RT-qPCR, which consistent with the results obtained by RNA-Seq (Figure 7). The log2 Fold Change of gene expression calculated from RNA-Seq and RT-qPCR were significantly related in LBS vs. RIR. Pearson correlation coefficient is equal to 0.87 with P < 0.01. This correlation showed the results of the RNA-Seq analysis to be reliable.
Figure 7.
RT-qPCR validation of gene expression profile. Plot of gene expression log2 Fold Change (LBS vs. RIR) determined by the RT-qPCR (horizontal axis) and RNA-Seq (vertical axis) for 10 selected genes (Supplementary Table S1). Correlation between RT-qPCR and RNA-Seq was calculated by Pearson product moment correlation. Each blue dot represents one tested gene, and the plot presents linear regression line, P value and correlation coefficient (r).
Validation of Key Genes in GS Hens With High and Low Egg-Laying Rate
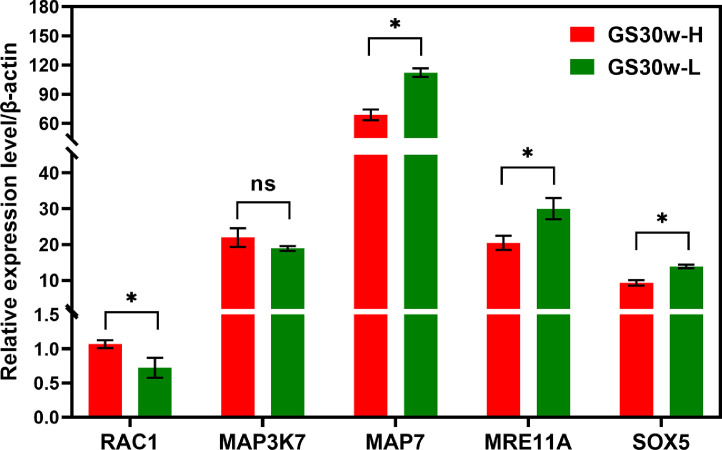
To confirm the 5 key genes identified above that were related with ELR, the gene expression levels were quantified in hypothalamus tissues of GS hens with high egg-laying rate and low egg-laying rate. The expression patterns of 5 genes between high-yielding group and low-yielding group were consistent with that between RIR and LBS hens. And among the 5 genes, the expression levels of the 4 genes including RAC1, MAP7, MRE11A and SOX5 were significant differences between high-yielding group and low-yielding group (P < 0.05) (Figure 8).
Figure 8.
Identification the expression of the 5 key genes in GS30w-H and GS30w-L. GS30w-H, laying hens with higher egg-laying rate from 25 to 30 wk were more than 90% were collected at the age of 30 wk old; GS30w-L, laying hens with lower egg-laying rate from 20% to 30% were collected at the age of 30 wk old.
DISCUSSION
Egg-laying performance is one of the most important economic traits in the poultry industry (Johnson et al., 2015). Compared with high-yield commercial laying hens, the meat and egg dual-purpose type local chicken has a low egg production performance. The hypothalamus is a key component of the HPG system, and it plays a key role in regulating hormone synthesis and release and in chicken reproductive performance. In this study, we selected 2 breeds (RIR vs. LBS) with significant differences in egg-laying rates during peak periods. To investigate the different genetic mechanisms of hypothalamus involvement in egg production between high-yielding layers and local chickens, a comparative transcriptome analysis of hypothalamus in the 2 chicken breeds was performed. Therefore, the present study provides more new information regarding regulation of egg-laying in chicken based on the previous working in the field.
Here, RNA-seq analysis revealed 753 DEGs including 383 upregulated and 370 downregulated DEGs in hypothalamus of RIR layers vs. LBS chickens. Fifteen DEGs were enriched in reproduction terms and reproductive process terms. Additionally, 6 key significant pathways, including Wnt signaling pathway, GnRH signaling pathway, MAPK signaling pathway, oocytes meiosis, thyroid hormone signaling pathway and thyroid hormone synthesis pathway, that probably play pivotal roles in egg-laying regulation, were identified by KEGG analysis. In the 6 pathways, the Wnt signaling pathway regulates several key developmental regions including the midbrain, central nervous system, kidney, limbs, and the reproductive system (Heikkilä et al., 2001). The GnRH signaling pathway is the central to regulate and maintain normal fertility (Bliss et al., 2010). In addition, GnRH also induces activation of MAPK pathway, which further mediates transcriptional activation of glycoprotein hormones alpha chain (CGA) (Maurer et al., 1999). In our study, MAP3K1 and CGA were enriched in the GnRH signaling pathway and MAPK signaling pathway, and significantly upregulated in hypothalamus of RIR high-yielding layers. MAP3K1 is an important mitogen-activated protein kinase in the MAPK signaling pathway, which is responsible for the transmission of GnRH-induced correlation signals to Jun N-terminal kinases (JNKs), which in turn phosphorylates transcription factor c-JUN to regulate the expression of CGA (Geh, 2011). The CGA is required to enable the biosynthesis of luteinizing hormone, and follicle-stimulating hormone (Das and Kumar, 2018). Therefore, the differential expression of MAP3K1 and CGA between the 2 breeds may be one of the reasons for the difference in egg-laying rate during peak-laying period. In addition, during the process of meiosis in oocytes, many reproductive-related signaling pathways are activated, such as the oocyte meiotic pathway (Kajiura-Kobayashi et al., 2000; Schmitt and Nebreda, 2002), thyroid hormone signaling pathway and thyroid hormone synthesis pathway (Baxter et al., 2004). Finally, there were 38 DEGs enriched in the 2 GO terms and 6 pathways as previously mentioned. This indicates that the 38 DEGs could be considered as underlying the difference in egg production performance seen between the 2 chicken breeds.
To gain further insight into the possible causative genes, a PPI network analysis of the 38 DEGs was carried out. Two genes, RAC1 and MAP3K7, that showed a higher degree of connection than other DEGs in the PPI network were identified. RAC1 is a small signaling G protein, which is a member of the RAC subfamily of RHO family of GTPases, and can regulate a wide variety of functions, including cell migration, cycle progression, and survival (Li et al., 2011). Previous research reported that RAC1 modulates the formation of primordial follicles via STAT3, Jagged1, GDF-9, and BMP-15 (Zhao et al., 2016), and disturbs follicular development by negatively affecting expression and activity of RAC1 (Maurya et al., 2014). Besides, RAC1 gene deletion disrupts mammary gland development and causes long-term tissue malfunction (Akhtar et al., 2016). In our study, RAC1 was located in the center of the PPI network (the network contains 35 genes involved in reproduction regulation). Compared with the GS30w-L, RAC1 was significantly upregulated in GS30w-H. It is therefore speculated that RAC1 may play an important role in egg-laying performance through regulating the development of ovarian follicle. MAP3K7, also known as TGF1aor TAK1, is a member of the serine / threonine protein kinase family and participates as a mediator in multiple signaling pathways, including the IL1 signaling pathway (Ninomiya-Tsuji et al., 1999), TGFβ signaling pathway, MAPK pathway, and Wnt pathway(Shim et al., 2005). Recently, it was reported that candidate gene MAP3K7 was identified for egg weight traits through a 600K Affymetrix chip of an egg-laying hen population (Farahani et al., 2020). In our study, MAP3K7 was located in the center of the PPI network related with reproduction, and its expression level was significantly higher in the hypothalamus of RIR than that in LBS, and but no significant difference was found between GS3w-H and GS3w-L. Therefore, it was suggested that MAP3K7 may not related with egg laying performance in GS chicken.
To investigate the possible causative genes in another direction, a co-expression-network analysis was conducted. Two genes, MRE11A and MAP7, which were | Dif_Kcore | > 10, and common to the 38 DEGs related to reproductive regulation identified by GO and KEGG pathway analyses, were identified. The MRE11A plays key roles in the regulation of disease (Regal et al., 2013),but few reports have described its role in the regulation of reproduction. In our study, MRE11A was more highly expressed in hypothalamus of LBS chicken than that in RIR chicken, and also highly expressed in the hypothalamus of GS3w-L than that in GS3w-H. The MAP7 is a ubiquitous MAP that organizes the microtubule cytoskeleton in mitosis and neuronal branching (Chaudhary et al., 2019). It is essential for cell polarization and differentiation (Fabrejonca et al., 2010). Previous research has shown that MAP7 is related to the process of generating sperm in mice (Komada et al., 2000). Métivier (2019) demonstrated that MAP7 promotes binding of kinesin-1 to microtubules to enable centrosome separation in brain neuroblasts (NBs) and asymmetric transport in oocytes. It was proved that loss of MAP7 in oocytes results in impaired plus-ended motility (Sung et al., 2008) and defects in nuclear positioning in muscle cells (Metzgeret al., 2012). Other research showed that the MAP7 gene was related to the development of the testes in broiler chickens (Zhang et al., 2017b). In this study, MAP7 was downregulated in hypothalamus of RIR chicken in comparison with LBS chicken, and also downregulated in GS30w-H in comparison with GS30w-L. It indicated that MAP7 might involve in the egg-laying performance. However, further work needed to be done to validate the function.
In addition, TFs have a significant role in phenotypic performance. Transcriptional regulatory network analysis identified 5 TFs that could interact with 23 DEGs which were related to reproductive regulation. Of the 5 TFs, SOX5 showed the most extensive relationships with target genes and was considered one of the core genes underlying the phenotypic differences in egg production performance between the 2 breeds. It was well known that SOX5 was one member of the SOX (SRY-related HMG-box) transcription factor family, and involved in regulating the embryonic development and cell fate determination (Kurtsdotter et al., 2017). It was also reported that SOX5 was related with chondrogenic differentiation of human adipose-derived stem cells (Xu et al., 2012), melanocyte development (Claus et al., 2008) and testis development (Mikella et al., 2015). A study showed that SOX5 was related to the classical Pea-comb phenotype in chickens (Wright et al., 2009), its expression was associated with testis development and sperm motility in chickens (Xu et al., 2018). It must be mentioned that although 5 genes were identified as associated with reproductive regulation, further in-depth studies are necessary to validate their functions in reproductive regulation in chickens.
The HPG axis regulates reproduction in all vertebrates. Classical theory suggests that gonadotropin-releasing hormone (GnRH) and other signals secreted by the hypothalamus regulate the synthesis and release of pituitary luteinizing hormone (LH) and follicle stimulating hormone (FSH), to further control gonadal development and sex hormone production (Thompson and Kaiser, 2014). While estrogen, progesterone, and other hormones produced by the developing gonads can act on the pituitary or hypothalamus through the endocrine system and regulate the synthesis and release of gonadotropin and neuropeptide hormones through negative feedback (Johnson, 1993). In our data, the expression of GnRH in the hypothalamus of RIR high-yielding layers did not change. Meanwhile, ESR1 and ESR2 expression was nonsignificantly downregulated and ESRRG was significantly downregulated in the hypothalamus. These results are similar to the hypothalamus expression profile results observed in high- and low-yielding laying Chinese Dagu Chickens (Wang and Ma, 2019). Combined with our analysis results of these 2 breeds and other RNA-seq results in hypothalamus and pituitary of one breed (Wang and Ma, 2019), we also speculate that the GnRH-FSH/LH pathway may not be the direct pathway of determining egg laying performance in chickens.
In summary, we report for the first time the transcriptomic profiles of the hypothalamus from 2 chicken breeds that show different egg-laying performances using RNA-Seq. Comprehensive analysis suggests that RAC1, MRE11A, MAP7 and SOX5 are the potential core genes that are responsible for egg-laying performance in LBS hens and RIR layers. Our study paved the way for future investigation into the mechanism of egg-laying regulation and enrich the chicken reproductive regulation theory.
ACKNOWLEDGMENTS
The work was supported by the Key Project of NSFC-Henan Province Joint Fund (U1704233); the Lifting Engineering Project of Henan Province for Young Talent (2018HYTP015); and the Open Projects of Key Laboratory of Chicken Genetics and Breeding, Ministry of Agriculture (CGB-201701).
DISCLOSURES
The authors declare no conflict of interest.
Footnotes
Supplementary material associated with this article can be found in the online version at doi:10.1016/j.psj.2021.101110.
Appendix. Supplementary materials
REFERENCES
- Akhtar N., Li W.P., Mironov A., Streuli C.H. Rac1 controls both the secretory function of the mammary gland and its remodeling for successive gestations. Dev. Cell. 2016;38:522–535. doi: 10.1016/j.devcel.2016.08.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anders S., Pyl P.T., Huber W. HTSeq–a Python framework to work with high-throughput sequencing data. Bioinformatics. 2015;31:166–169. doi: 10.1093/bioinformatics/btu638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barabási A.L., Oltvai Z.N. Network biology: understanding the cell's functional organization. Nat. Rev. Genet. 2004;5:101–113. doi: 10.1038/nrg1272. [DOI] [PubMed] [Google Scholar]
- Baxter J.D., Webb P., Grover G., Scanlan T.S. Selective activation of thyroid hormone signaling pathways by GC-1: a new approach to controlling cholesterol and body weight. Trends. Endocrinol. Metab. 2004;15:0–157. doi: 10.1016/j.tem.2004.03.008. [DOI] [PubMed] [Google Scholar]
- Biscarini F., Bovenhuis H., Ellen E.D., Addo S., van Arendonk J.A.M. Estimation of heritability and breeding values for early egg production in laying hens from pooled data. Poult. Sci. 2010;89:1842–1849. doi: 10.3382/ps.2010-00730. [DOI] [PubMed] [Google Scholar]
- Bliss S.P., Navratil A.M., Xie J., Roberson M.S. GnRH signaling, the gonadotrope and endocrine control of fertility. Front. Neuroendocrinol. 2010;31:322–340. doi: 10.1016/j.yfrne.2010.04.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brady K., Liu H.C., Hicks J.A., Long J.A., Porter T.E. Transcriptome analysis of the hypothalamus and pituitary of turkey hens with low and high egg production. BMC Genomics. 2020;21:647. doi: 10.1186/s12864-020-07075-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chaudhary A.R., Lu H., Krementsova E.B., Bookwalter C.S., Trybus K.M., Hendricks A.G. MAP7 regulates organelle transport by recruiting kinesin-1 to microtubules. J. Biol. Chem. 2019;294:10160–10171. doi: 10.1074/jbc.RA119.008052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen J.Q., Yu. L., Zhang S.W., Chen X. Network analysis-based approach for exploring the potential diagnostic biomarkers of acute myocardial infarction. Front. Physiol. 2016;7:615. doi: 10.3389/fphys.2016.00615. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Claus S.C., Petra L., Simone H., Michael W. The transcription factor Sox5 modulates Sox10 function during melanocyte development. Nucleic. Acids. Res. 2008;36:5427–5440. doi: 10.1093/nar/gkn527. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crawford, R. D. 1990. Poultry Breeding and Genetics. R.D. Crawford, ed. Elsevier, Amsterdam, the Netherlands, New York, NY.
- Das N., Kumar T.R. Molecular regulation of follicle-stimulating hormone synthesis, secretion and action. J. Mol. Endocrinol. 2018;60 doi: 10.1530/JME-17-0308. R131–R155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dennis G., Sherman B.T., Hosack D.A., Yang J., Gao W., Lane H.C., Lempicki R.A. DAVID: database for annotation, visualization, and integrated discovery. Genome. Biol. 2003;4:R60. [PubMed] [Google Scholar]
- Fabrejonca N., Allaman J., Radlgruber G., Meda P., Kiss J.Z., French L.E., Masson D. The distribution of murine 115-kDa epithelial microtubule-associated protein (E-MAP-115) during embryogenesis and in adult organs suggests a role in epithelial polarization and differentiation. Differentiation. 2010;63:169–180. doi: 10.1111/j.1432-0436.1998.00169.x. [DOI] [PubMed] [Google Scholar]
- Farahani A.H.Khaltabadi., Mohammadi H., Moradi M.H., Ghasemi H.A. Identification of potential genomic regions for egg weight by a haplotype-based genome-wide association study using Bayesian methods. Br. Poult. Sci. 2020;61:251–257. doi: 10.1080/00071668.2020.1724879. [DOI] [PubMed] [Google Scholar]
- Geh E.N. PhD Diss. University of Cincinnati, OH; 2011. The Molecular Regulation of MAP3K1 in Eyelid Development. [Google Scholar]
- Heikkilä M., Peltoketo H., Vainio S. Wnts and the female reproductive system. J. Exp. Zool. 2001;290:616–623. doi: 10.1002/jez.1112. [DOI] [PubMed] [Google Scholar]
- Johnson A.L. Regulation of follicle differentiation by gonadotropins and growth factors. Poult. Sci. 1993;72:867–873. doi: 10.3382/ps.0720867. [DOI] [PubMed] [Google Scholar]
- Johnson P.A., Stephens C.S., Giles J.R. The domestic chicken: Causes and consequences of an egg a day. Poult. Sci. 2015;94:816–820. doi: 10.3382/ps/peu083. [DOI] [PubMed] [Google Scholar]
- Kajiura-Kobayashi H., Yoshida N., Sagata N., Yamashita M., Nagahama Y. The Mos/MAPK pathway is involved in metaphase II arrest as a cytostatic factor but is neither necessary nor sufficient for initiating oocyte maturation in goldfish. Dev. Genes. Evol. 2000;210:416–425. doi: 10.1007/s004270000083. [DOI] [PubMed] [Google Scholar]
- Kim D., Pertea G., Trapnell C., Pimentel H., Kelley R., Salzberg S.L. TopHat2: accurate alignment of transcriptomes in the presence of insertions, deletions and gene fusions. Genome Biol. 2013;14:R36. doi: 10.1186/gb-2013-14-4-r36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Komada M., Mclean D.J., Griswold M.D., Russell L.D., Soriano P. E-MAP-115, encoding a microtubule-associated protein, is a retinoic acid-inducible gene required for spermatogenesis. Genes. Dev. 2000;14:1332–1342. [PMC free article] [PubMed] [Google Scholar]
- Kong B.W., Hudson N., Seo D., Lee S., Khatri B., Lassiter K., Cook D., Piekarski A., Dridi S., Anthony N., Bottje W. RNA sequencing for global gene expression associated with muscle growth in a single male modern broiler line compared to a foundational Barred Plymouth Rock chicken line. BMC Genomics. 2017;18:82. doi: 10.1186/s12864-016-3471-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kurtsdotter I., Topcic D., Karlen A., Singla B., Hagey D.W., Bergsland M., Siesjo P., Nister M., Carlson J.W., Lefebvre V., Persson O., Holmberg J., Muhr J. SOX5/6/21 prevent oncogene-driven transformation of brain stem cells. Cancer Res. 2017;77:4985–4997. doi: 10.1158/0008-5472.CAN-17-0704. [DOI] [PubMed] [Google Scholar]
- Li A., Ma Y.F., Yu X.Z., Mort R.L., Lindsay C., Stevenson D., Strathdee D., Insall R.H., Chernoff J., Snapper S.B., Jackson I.J., Larue L., Sansom O.J., Machesky L.M. Rac1 drives melanoblast organization during mouse development by orchestrating pseudopod-driven motility and cell-cycle progression. Dev. Cell. 2011;21:722–734. doi: 10.1016/j.devcel.2011.07.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li B.J, Qiao L.Y., An L.X., Wang W.W., Liu J.H., Ren Y.S., Pan Y.Y., Jing J.J., Liu W.Z. Transcriptome analysis of adipose tissues from two fat-tailed sheep breeds reveals key genes involved in fat deposition. BMC Genomics. 2018;19:338. doi: 10.1186/s12864-018-4747-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li H., Tian J.D. Research progress in genetic control of reproductive performance in chicken by high-throughout sequencing technology. In: Liu X.J., editor. Pages 89–99 in Application of Genetics and Genomics in Poultry Science. 2018. IntechOpen, Rijeka, Croatia. [Google Scholar]
- Liu L., Xiao Q., Gilbert E.R., Cui Z.F., Zhao X.L., Zhao X.L., Wang Y., Yin H.D., Li D.Y., Zhang H.H., Zhu Q. Whole-transcriptome analysis of atrophic ovaries in broody chickens reveals regulatory pathways associated with proliferation and apoptosis. Sci. Rep. 2018;8 doi: 10.1038/s41598-018-25103-6. 7231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu W.B, Li D.F., Liu J.F., Chen S.R., Qu L.J., Zheng J.X., Yun G.Y., Yang N. A genome-wide SNP scan reveals novel loci for egg production and quality traits in white leghorn and brown-egg dwarf layers. Plos One. 2011;6:e28600. doi: 10.1371/journal.pone.0028600. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maurer R.A., Kim K.E., Schoderbek W.E., Roberson M.S., Glenn D.J. Regulation of glycoprotein hormone alpha-subunit gene expression. Recent. Prog. Horm. Res. 1999;54:455–484. [PubMed] [Google Scholar]
- Maurya V.K, Sangappa C., Kumar V., Mahfooz S., Singh A., Rajender S., Jha R.K. Expression and activity of Rac1 is negatively affected in the dehydroepiandrosterone induced polycystic ovary of mouse. J. Ovarian. Res. 2014;7:32. doi: 10.1186/1757-2215-7-32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Métivier M., Monroy B.Y., Gallaud E., Caous R., Pascal A., Richard-Parpaillon L., Guichet A., Ori-McKenney K.M., Giet R. Dual control of Kinesin-1 recruitment to microtubules by Ensconsin in Drosophila neuroblasts and oocytes. Development. 2019;146 doi: 10.1242/dev.171579. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Metzger T., Gache V., Xu M.U., Cadot B., Folker E.S., Richardson B., Gomes E.R., Baylies M.K. MAP and kinesin-dependent nuclear positioning is required for skeletal muscle function. Nature. 2012;484:120–124. doi: 10.1038/nature10914. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mikella D., Roumaud P., Martin L.J. Expressions of Sox9, Sox5, and Sox13 transcription factors in mice testis during postnatal development. Mol. Cell. Biochem. 2015;407:209–221. doi: 10.1007/s11010-015-2470-7. [DOI] [PubMed] [Google Scholar]
- Mishra S.K., Chen B, Zhu Q., Xu Z.X., Ning C.Y., Yin H.D., Wang Y., Zhao X.L., Fan X.L., Yang M.Y., Yang D.Y., Ni Q.Y., Zhang M.W., Li D.Y. Transcriptome analysis reveals differentially expressed genes associated with high rates of egg production in chicken hypothalamic-pituitary-ovarian axis. Sci.Rep. 2020;10:5976. doi: 10.1038/s41598-020-62886-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Monson M.S., Goor A.V., Persia M.E., Rothschild M.F., Schmidt C.J., Lamont S.J. Genetic lines respond uniquely within the chicken thymic transcriptome to acute heat stress and low dose lipopolysaccharide. Sci. Rep. 2019;9:13649. doi: 10.1038/s41598-019-50051-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ninomiya-Tsuji J., Kishimoto K., Hiyama A., Inoue J., Cao Z., Matsumoto K. The kinase TAK1 can activate the NIK-I kappaB as well as the MAP kinase cascade in the IL-1 signalling pathway. Nature. 1999;398:252–256. doi: 10.1038/18465. [DOI] [PubMed] [Google Scholar]
- Peng Y.D., Chang C., Wang Y.Q., Hu L.L., Zhao Z.Y., Zhao L.Y., Geng L.Y., Liu Z.Z., Gong Y.F., Li J.S., Li X.L., Zhang C.S. Genome-wide differential expression of long noncoding RNAs and mRNAs in ovarian follicles of two different chicken breeds. Genomics. 2019;111:1395–1403. doi: 10.1016/j.ygeno.2018.09.012. [DOI] [PubMed] [Google Scholar]
- Piorkowska K., Żukowski K., Ropkamolik K., Tyra M., Gurgul A. A comprehensive transcriptome analysis of skeletal muscles in two Polish pig breeds differing in fat and meat quality traits. Genet. Mol. Biol. 2018;41:125–136. doi: 10.1590/1678-4685-GMB-2016-0101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prieto C., Risueño A., Fontanillo C., Rivas J.D.L. Human gene coexpression landscape: confident network derived from tissue transcriptomic profiles. Plos One. 2008;3:e3911. doi: 10.1371/journal.pone.0003911. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Regal J.A., Festerling T.A., Buis J.M., Ferguson D.O. Disease-associated MRE11 mutants impact ATM/ATR DNA damage signaling by distinct mechanisms. Hum. Mol. Genet. 2013;22:5146–5159. doi: 10.1093/hmg/ddt368. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmitt A., Nebreda A.R. Signalling pathways in oocyte meiotic maturation. J. Cell. Sci. 2002;115:2457–2459. doi: 10.1242/jcs.115.12.2457. [DOI] [PubMed] [Google Scholar]
- Schmittgen T.D., Livak K.J. Analyzing real-time PCR data by the comparative CT method. Nat. Protoc. 2008;3:1101–1108. doi: 10.1038/nprot.2008.73. [DOI] [PubMed] [Google Scholar]
- Shim J.H., Xiao C.C., Paschal A.E., Bailey S.T., Rao P., Hayden M.S., Lee K.Y., Bussey C., Steckel M., Tanaka N., Yamada G., Akira S., Matsumoto K., Ghosh S. TAK1, but not TAB1 or TAB2, plays an essential role in multiple signaling pathways in vivo. Genes. Dev. 2005;19:2668–2681. doi: 10.1101/gad.1360605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smoot M.E., Ono K., Ruscheinski J., Wang P.L., Ideker T. Cytoscape 2.8: new features for data integration and network visualization. Bioinformatics. 2010;27:431–432. doi: 10.1093/bioinformatics/btq675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun L.M, Bai M., Xiang L.J., Zhang G.S., Ma W., Jiang H.Z. Comparative transcriptome profiling of longissimus muscle tissues from Qianhua Mutton Merino and Small Tail Han sheep. Sci. Rep. 2016;6:33586. doi: 10.1038/srep33586. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sung H.H., Telley I.A., Papadaki P., Ephrussi A., Surrey T., Rørth P. Drosophila ensconsin promotes productive recruitment of Kinesin-1 to microtubules. Dev. Cell. 2008;15:866–876. doi: 10.1016/j.devcel.2008.10.006. [DOI] [PubMed] [Google Scholar]
- Tan G., Lenhard B. TFBSTools: an R/Bioconductor package for transcription factor binding site analysis. Bioinformatics. 2016;32:1555–1556. doi: 10.1093/bioinformatics/btw024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thompson I.R., Kaiser U.B. GnRH pulse frequency-dependent differential regulation of LH and FSH gene expression. Mol. Cell. Endocrinol. 2014;385:28–35. doi: 10.1016/j.mce.2013.09.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang C.Q., Ma W. Hypothalamic and pituitary transcriptome profiling using RNA-sequencing in high-yielding and low-yielding laying hens. Sci. Rep. 2019;9:10285. doi: 10.1038/s41598-019-46807-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wolc A., Arango J., Jankowski T., Dunn I., Settar P., Fulton J.E., Sullivan N.P.O., Preisinger R., Fernando R.L., Garrick D.J., Dekkers J.C.M. Genome-wide association study for egg production and quality in layer chickens. J. Anim. Breed. Genet. 2014;131:173–182. doi: 10.1111/jbg.12086. [DOI] [PubMed] [Google Scholar]
- Wright D., Boije H., Meadows J.R.S., Bed'Hom B., Gourichon D., Vieaud A., Tixier-Boichard M., Rubin C.J., Imsland F., Hallböök F., Andersson L. Copy number variation in intron 1 of SOX5 causes the pea-comb phenotype in chickens. Plos. Genet. 2009;5 doi: 10.1371/journal.pgen.1000512. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu Y.P., Zhao X.Y., Chen L., Wang J.H., Duan Y.Q., Li H.Y., Lu L.Z. Transcriptomic Analyses of the Hypothalamic-Pituitary-Gonadal Axis Identify Candidate Genes Related to Egg Production in Xinjiang Yili Geese. Animals. 2020;10:90. doi: 10.3390/ani10010090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu H., Sun Y.Y., Shi L., Liu Y.F., Bai H., Li Y.L., .Huang Z.Y, Ye J.H., Jia Y.X., Wang L., Chen J.L. The Effect of SOX5 Protein in Spermatogenesis and Sperm Motility Regulation and Its Expression Localization in the Testis of Roosters. Acta Veterinaria Et Zootechnica Sinica. 2018;49:718–724. [Google Scholar]
- Xu J., Kang Y., Liao W.M., Yu L. MiR-194 regulates chondrogenic differentiation of human adipose-derived stem cells by targeting Sox5. PLoS One. 2012;7:e31861. doi: 10.1371/journal.pone.0031861. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yin Z.T., Lian L., Zhu F., Zhang Z.H., Hincke M., Yang N., Hou Z.C. The transcriptome landscapes of ovary and three oviduct segments during chicken (Gallus gallus) egg formation. Genomics. 2019;112:243–251. doi: 10.1016/j.ygeno.2019.02.003. [DOI] [PubMed] [Google Scholar]
- Yuan J.W., Sun C.J., Dou T.C., Yi G.Q., Qu L.J., Qu L., Wang K.H., Yang N. Identification of promising mutants associated with egg production traits revealed by genome-wide association study. PLoS One. 2015;10 doi: 10.1371/journal.pone.0140615. e0140615. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang H., Na W., Zhang H.L., Wang N., Du Z.Q., Wang S.Z., Wang Z.P., Zha Z.W., Li H. TCF21 is related to testis growth and development in broiler chickens. Genet. Sel. Evol. 2017;49:25. doi: 10.1186/s12711-017-0299-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Y.H., Li D.H., Han R.L., Wang Y.B., Li G.X., Liu X.J., Tian Y.D., Kang X.T., Li Z.J. Transcriptome analysis of the pectoral muscles of local chickens and commercial broilers using Ribo-Zero ribonucleic acid sequencing. PLoS One. 2017;12 doi: 10.1371/journal.pone.0184115. e0184115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao L.H, Du X.H., Huang K., Zhang T., Teng Z., Niu W.B., Wang C., Xia G.L. Rac1 modulates the formation of primordial follicles by facilitating STAT3-directed Jagged1, GDF9 and BMP15 transcription in mice. Sci. Rep. 2016;6:23972. doi: 10.1038/srep23972. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu G., Mao Y., Zhou W., Jiang Y. Transcriptome and Promoter DNA Methylation pattern of steroidogenic genes in chicken follicles throughout the ovulation cycle. PLoS One. 2015;10 doi: 10.1371/journal.pone.0146028. e0146028. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.