Summary
Mucus-secreting goblet cells are the dominant cell type in pulmonary diseases e.g. asthma and cystic fibrosis (CF), leading to pathologic mucus metaplasia and airway obstruction. Cytokines including IL-13 are the major players in the transdifferentiation of club cells into goblet cells. Unexpectedly, we have uncovered a previously undescribed pathway promoting mucous metaplasia that involves VEGFa and its receptor KDR. Single cell RNA sequencing analysis coupled with genetic mouse modeling demonstrates that loss of epithelial VEGFa, KDR or MEK/ERK kinase, promotes excessive club-to-goblet transdifferentiation during development and regeneration. Sox9 is required for goblet cell differentiation following Kdr inhibition in both mouse and human club cells. Significantly, airway mucous metaplasia in asthmatic and CF patients is also associated with reduced KDR signaling and increased SOX9 expression. Together, these findings reveal an unexpected role for VEGFa/KDR signaling in the defense against mucous metaplasia, offering a potential therapeutic target for this common airway pathology.
eTOC blurb:
Jiang et al. demonstrate that the VEGFa/KDR pathway protects the airway epithelium against mucous metaplasia during regeneration. Suppressed VEGF signaling leads to increased levels of SOX9 which promotes goblet cell differentiation. These findings introduce critical players in airway mucous metaplasia that is commonly seen in asthma and cystic fibrosis.
Introduction
In the conducting airways of the lung, controlled mucus production and effective clearance are dependent on a balanced composition of mucus producing and multiciliated cells in the luminal epithelium. The differentiation of mucus cells occurs towards the end of gestation and, following a “burst” of production during the early neonatal period, the population is maintained as a minor proportion of the total in the normal airways. However, in pulmonary diseases, including allergic asthma, chronic obstructive pulmonary disease (COPD) and cystic fibrosis (CF), the numbers of mucous cells significantly expand, resulting in mucous metaplasia which leads to airway obstruction, infection and even mortality (Boucher, 2019; Curran and Cohn, 2010; Fahy and Dickey, 2010).
Past research into mucous metaplasia has focused on immune cells, revealing the importance of Th1 and Th2 inflammatory cytokines such as IL-1, IL-4 and IL-13 in driving the appearance of goblet cells (Gour and Wills-Karp, 2015; Li et al., 2019; Whitsett, 2018; Wills-Karp et al., 1998; Zhou-Suckow et al., 2017). Investigation of asthmatic animal models indicates that group 2 innate lymphoid cells (ILC2s) secret IL-5 and IL-13 which promote goblet cell hyperplasia following allergen challenge (Klein Wolterink et al., 2012; Kuperman et al., 2002). Moreover, transgenic overexpression of IL-5, IL-13 or its downstream player SAM pointed domain ETS factor (SPDEF) leads to extensive mucous metaplasia in mouse airways (Lee et al., 1997; Park et al., 2007; Rajavelu et al., 2015; Schmid-Grendelmeier et al., 2002; Zhu et al., 1999). Interestingly, recent research implicates the critical roles of non-hematopoietic cells/tissues in mucous metaplasia (Branchfield et al., 2016; Sui et al., 2018). For example, rare neuroendocrine cells can modulate mucous cell differentiation through neuropeptides and neurotransmitters, including CGRP and GABA (Barrios et al., 2017; Sui et al., 2018). That being said, the molecular mechanism driving the differentiation of airway progenitor cells towards mucous cell lineage remains largely unknown.
Both ciliated cells and club cells were initially thought to generate mucous cells depending on the models used (Chen et al., 2009; Evans et al., 2004; Reader et al., 2003; Tyner et al., 2006). However, lineage-tracing experiment suggested that ciliated cells do not transdifferentiate into mucous cells in the airways in an asthmatic mouse model (Pardo-Saganta et al., 2013). Instead, a subset of club cells express mucin proteins following ovalbumin (OVA) challenge (Evans et al., 2004). Electron microscopy analysis further revealed that the metaplastic mucous cells contain many ultrastructural characteristics of club cells (Hayashi et al., 2004), suggesting that club cells are the progenitors for the metaplastic mucous cells. Consistently, the newly formed mucous cells are lineage-labeled in the airways of Scgb1a1-rtTA; Otet7-CMVminCre (Otet-Cre); R26lacZ mice following OVA challenge (Chen et al., 2009). These studies support the hypothesis that club cells serve as a cell of origin for mucous cells in the intrapulmonary airways. However, club cells are heterogeneous and contain multiple subpopulations including variant club cells and newly identified MHC class I marker H2-K1high progenitor cells (Guha et al., 2017; Hong et al., 2001; Kathiriya et al., 2020; Kim et al., 2005; Reynolds et al., 2000; Yuan et al., 2019). Therefore, it remains an open question which subpopulation(s) generate mucous/goblet cells.
In this study, our single cell RNA sequencing analysis surprisingly revealed that VEGFR2 (also known as FLK1 or KDR) is expressed in club cell subpopulations. Loss of KDR, its ligand VEGFa, or downstream MEK/ERK causes excessive differentiation of club cells into mucous cells (mainly goblet cells) during development and regeneration following Naphthalene (NAPH) challenge. We further show that deletion of the transcription factor Sox9 attenuates mucous metaplasia caused by KDR loss. KDR inhibition also promotes goblet cell differentiation of human club cells that form in air-liquid interface (ALI) culture, and this is accompanied by increased levels of SOX9. Consistently, airway mucous metaplasia is associated with reduced KDR signaling and increased SOX9 expression in patients with asthma and CF. Together, these findings identify an unexpected role for VEGFa/KDR signaling in the defense against mucous metaplasia.
Results
Single cell analysis identified Kdr expression in club cell subpopulations.
To gain initial insights into club cell heterogeneity under normal and repair conditions, we performed single-cell RNA sequencing of EPCAM+ intrapulmonary cells of the adult mouse before and after treatment with a single dose of NAPH (Figure 1A and S1A). Previous studies have shown that NAPH selectively ablates the majority of club cells, and that the residual, NAPH resistant club cells can fully regenerate the epithelium (Guha et al., 2017; Hong et al., 2001; Kotton and Morrisey, 2014; Rawlins et al., 2009; Reynolds et al., 2000; Van Winkle et al., 1995; Volckaert et al., 2011). Cells were isolated 14 days after treatment when the epithelial cells are proliferating and differentiating extensively (Van Winkle et al., 1995). A total of 3298 and 3685 cells were analyzed from control and NAPH-injured mice, respectively, and cell types assigned based on the expression of specific lineage markers (Figure 1B, 1C and S1B). The major cell types included club, ciliated, neuroendocrine and alveolar type 2 (AT2) cells (Figure 1B and S1B). A minor population of immune cells also expressed EPCAM in both control and NAPH-injured lungs, which is consistent with previous findings (Figure 1B and 1C) (Gautier et al., 2012). Notably, NAPH challenge also led to the presence of a minor population of basal cells (Trp63+) (Figure 1B and S1B).
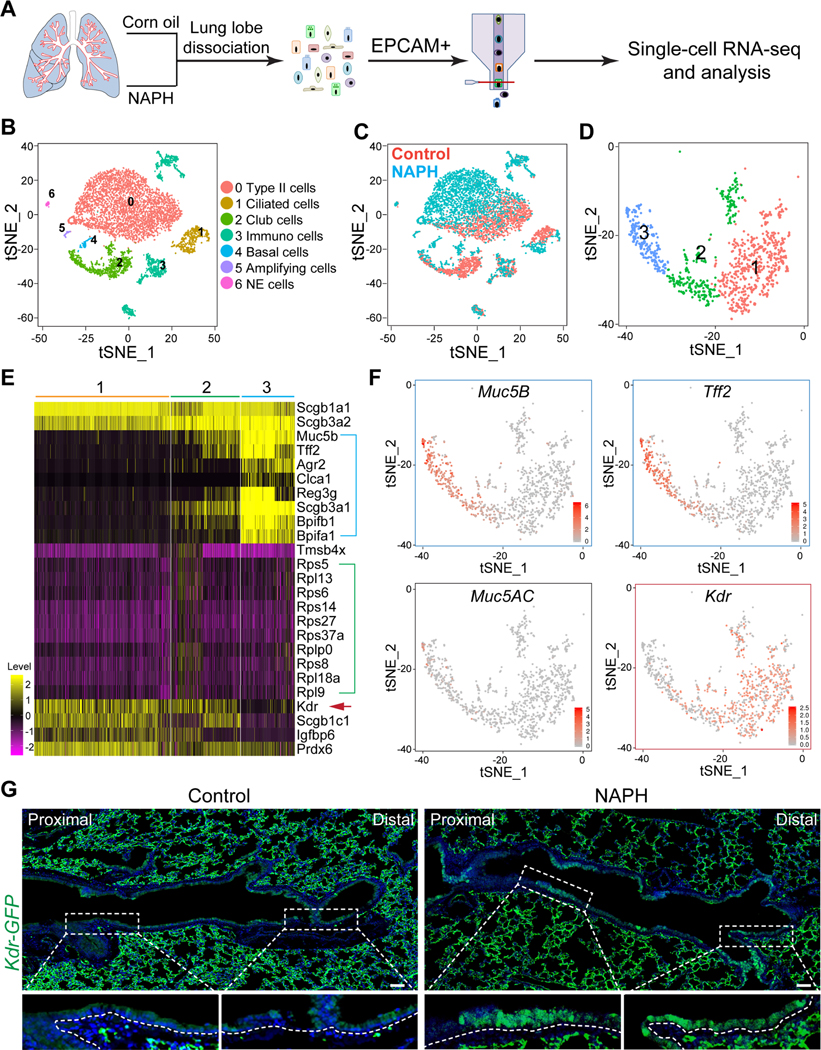
Figure 1: Kdr is expressed in subpopulations of intrapulmonary airway club cells.
(A) Schematics for single cell RNA sequencing of airway EPCAM+ cells isolated from mice treated with corn oil or Naphthalene (NAPH). (B and C) t-Distributed Stochastic Neighbor Embedding (t-SNE) plot indicative of cell population changes upon NAPH treatment. (D) Three subpopulations of club cells are present in the airways. (E) Differential gene expression in the three subpopulations of club cells. Note distinct gene expression enriched in Sub2 (green bracket) and Sub3 (blue bracket). Also note that Kdr is enriched in Sub1 and 2 (red arrow). (F) Enrichment of Kdr transcripts in Sub1/2 in contrast to expression of mucous cell-related genes Muc5B, Tff2, and Muc5AC in Sub3. (G) Increased expression of Kdr-GFP in the regenerated airway epithelium 14 days following NAPH challenge. Scale bar: 50 μm. See also Figure S1 and Table S1.
Cluster analysis revealed that club cells (Scgb1a1+, Scgb3a2+) were divided into three major subpopulations, Sub1/2/3 (Figure 1D and S1C). Gene enrichment analysis suggests that Sub1 and 2 share similar gene expression, with relatively higher expression of Igfbp6 and Prdx6 in Sub1 (Figure 1E and Table S1). By contrast, Sub3 expresses transcripts for the mucous cell markers Muc5B and Tff2 (Figure 1F), although the proteins are undetectable in the cytoplasm (data not shown). Surprisingly, Sub1/2 are characterized by the expression of transcripts of Kdr, which has previously only been found in mesoderm-derived cells (e.g., endothelial cells) and a rare population of hepatocytes (Figure 1E, 1F and S1D) (Ema et al., 2006; Goldman et al., 2013; Holmes et al., 2007). We used a Kdr-GFP knock-in mouse line to localize cells transcribing the gene under normal and repair conditions. At steady-state, GFP expression was low or undetected. However, there was clear expression in the airways during repair and this was confirmed by quantitative PCR (Figure 1G and S3B).
Transient Kdr expression prevents mucous metaplasia in the developing airways
Kdr-Cre activity was barely detected in the airway epithelium at E16.5, but became prominent at P0 (Figure S2A). Consistently, Kdr-GFP expression was limited to club cells and the levels increased from E16.5 to P0 (Figure S2B–E), correlating with the timing of club cell maturation (Reynolds et al., 2008). While Kdr-driven GFP expression in the airways progressively declined from P5 to P60, the levels of expression in the blood vessels remained unaltered (Figure S2B and S2C). Notably, the dynamic expression of Kdr in the epithelium is inversely correlated with the transient “burst” of mucous cell appearance that normally occurs in the distal trachea and intrapulmonary airways during the early postnatal period (Roy et al., 2011). When Kdr expression peaked at around P0, the number of mucous cells was minimal but increased from P0 to P10 (Figure S2B and S2C), suggesting that Kdr suppresses the differentiation of club cells towards mucous cell lineage. To test whether Kdr is indeed required for suppressing mucous cell differentiation, we generated Shh-Cre; Kdrloxp/loxp mutants in which Kdr is conditionally deleted in the lung epithelium as early as E9.0 (Harris-Johnson et al., 2009). Significantly, loss of Kdr led to an increased number of mucous (goblet) cells (Alcian blue+ Muc5AC+ Clca3+) in the airways at P10 (24.3±4.6% vs 41.4%±4.2%,p<0.05) (Figure S2F–H). However, the number of mucous cells reverted back to normal at P60 in both controls and mutants (Figure S2F–H), suggesting that Kdr is transiently required for suppressing mucous cell fate during the early neonatal stage.
Transient Kdr expression is required to block mucous metaplasia during regeneration
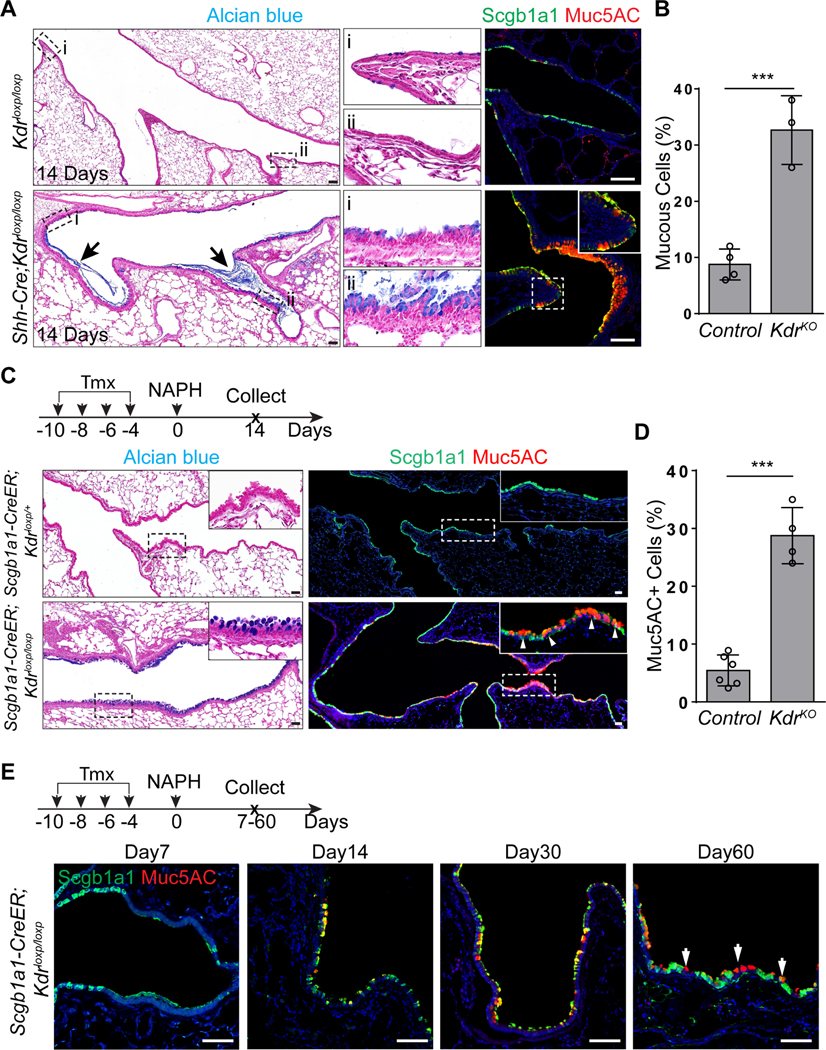
A characteristic feature of injury-repair mechanisms is the redeployment of development signaling pathways (Barker et al., 2010; Lynch et al., 2018; Mou et al., 2016; Vaughan et al., 2015; Zacharias et al., 2018). Following NAPH challenge the levels of Kdr-driven GFP transiently increased from day7 and peaked at approximately day14–21 in the distal trachea, main bronchi and intrapulmonary airways (Figure S3A, S3B and data not shown). Kdr-GFP expression reverted back to normal when the repair was accomplished (Figure S3A and S3B). To test whether re-expression of Kdr is critical for airway epithelial regeneration, we subjected Shh-Cre;Kdrloxp/loxp mutants to NAPH challenge. As described above, the airways of these mice appeared normal. However, when the airway epithelium lacking Kdr was examined 14 days after NAPH injury there was evidence for severe mucous metaplasia and the airways were filled with mucus (8.8%±2.6% vs 32.8%±6.1%,p<0.001) (Figure 2A and 2B). This result suggests that Kdr is essential for inhibiting mucous differentiation of the regenerated epithelium. Consistently, loss of Kdr in club cells led to similar extensive mucous metaplasia in the regenerated airways of Scgb1a1-CreER;Kdrloxp/loxp mutants following NAPH treatments (5.6%±2.6% vs 29.3%±4.8% p<0.001) (Figure 2C and 2D). Increased numbers of mucous cells could be detected 60 days after injury (Figure 2E). Furthermore, treatment with the selective Kdr inhibitor Semaxanib (SU5416) (Fong et al., 1999) also induced similar mucous metaplasia following NAPH challenge (4.2%±1.7% vs 24.4%±2.6%,p<0.001) (Figure S3C and S3D). Together these results confirm that transient re-expression of Kdr is required for normal airway epithelial regeneration and that loss of Kdr during the process of restoration leads to mucous metaplasia.
Figure 2: Kdr signaling is required for airway epithelial regeneration.
(A and B) Extensive mucous metaplasia in the regenerated airways of Shh-Cre;Kdrloxp/loxp mutants following NAPH challenge (n=3). (C and D) Loss of Kdr in Scgb1a1-CreER;Kdrloxp/loxp mutants promotes mucous metaplasia of regenerated club cells following NAPH challenge (n=4). (E) Mucous cells remain in the airways 60 days following NAPH challenge. Note that mucous cells co-express Muc5AC and Scgb1a1 at day 14 and day 30, but lose Scgb1a1 expression at day 60. Data represent mean ± s.e.m. ***p<0.001; statistical analysis by unpaired two-tailed Student’s t-test. Scale bar: 50 μm. See also Figure S2 and S3.
The Kdr ligand VEGFa is required for suppressing mucous metaplasia during airway regeneration
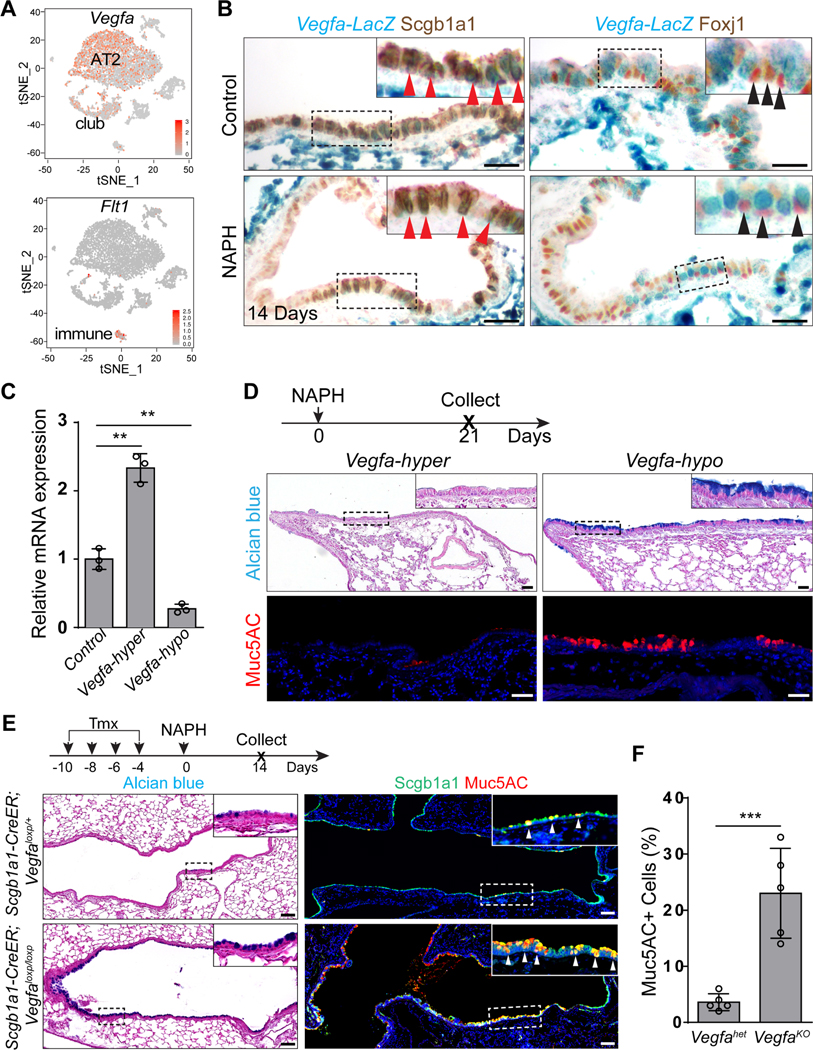
KDR has two major ligands, VEGFa and VEGFc (Hamada et al., 2000; Mustonen and Alitalo, 1995). While VEGFa/KDR signaling is critical for vascular development (Carmeliet et al., 1996; Shalaby et al., 1995), and VEGFc/KDR plays an essential role in the formation of lymphatic vessels (Makinen et al., 2001), no evidence has been described for function in airway epithelial tissues. Single-cell RNA sequencing data showed that in the adult lung epithelium VEGFa transcripts were enriched in club cells and AT2 cells, whereas VEGFc was not detected in any populations present in t-SNE plot (Figure 3A and data not shown). In addition to KDR, VEGFa also binds VEGF receptor 1 (also known as FLT1 (Ferrara et al., 2003)) which was enriched in a subpopulation of immune cells (Figure 3A). We confirmed that VEGFa expression was limited to club cells (Scgb1a1+) in both control and NAPH-injured airway epithelium as evidenced by a Vegfa-lacZ reporter mouse line (Figure 3B) (Miquerol et al., 1999). Notably, mucous metaplasia occurred in the airways of Vegfa hypomorphic mutants (Vegf-hypo) (Damert et al., 2002), in which Vegfa levels were dramatically reduced (Figure 3C and 3D). Conversely, increased expression of Vegfa protected the regenerated airway epithelium from mucous metaplasia in Vegfa hypermorphic mutants (Vegf-hyper) (Miquerol et al., 1999) (Figure 3C and 3D). To further test whether club cell-derived VEGFa is required for airway regeneration, we deleted Vegfa using Scgb1a1-CreER and subjected the mutants to NAPH challenge. Vegfa deletion resulted in extensive mucous differentiation of the regenerated epithelium in Scgb1a1-CreER;Vegfaloxp/loxp mutants (3.7%±1.5% vs 23.3%±7.7% ,p<0.001) (Figure 3E and 3F). Notably, mucous metaplasia was less severe in the terminal airways, presumably due to compensatory VEGFa secretion by the neighboring alveolar epithelial cells (Figure 3A and 3E) (Vila Ellis et al., 2020). Together these results support a model in which club cell-produced VEGFa activates KDR to block mucous cell differentiation of the airway epithelium during regeneration.
Figure 3: Club cell-derived Vegfa is required for airway epithelial regeneration.
(A) Vegfa transcripts are expressed in club cells. Note Vegfa is also enriched in AT2 cells. Also note that Flt1 is not detected in the epithelium but expressed in immune cells. (B) Vegfa-lacZ is expressed in club cells but not ciliated cells of the airway epithelium as shown by X-gal staining. (C) The transcript levels of Vegfa are increased and decreased in the isolated EPCAM+ cells of Vegf-hyper and Vegf-hypo mice, respectively (n=3). (D) Mucous metaplasia occurs in the airways of Vegf-hypo mice following NAPH challenge. (E and F) Loss of Vegfa promotes mucous metaplasia of regenerated club cells following NAPH challenge (n=5). Data represent mean ± s.e.m. **p<0.01, ***p<0.001; statistical analysis by unpaired two-tailed Student’s t-test. Scale bar: 100 μm.
Kdr suppresses mucous metaplasia of the regenerated epithelium via MEK/ERK signaling
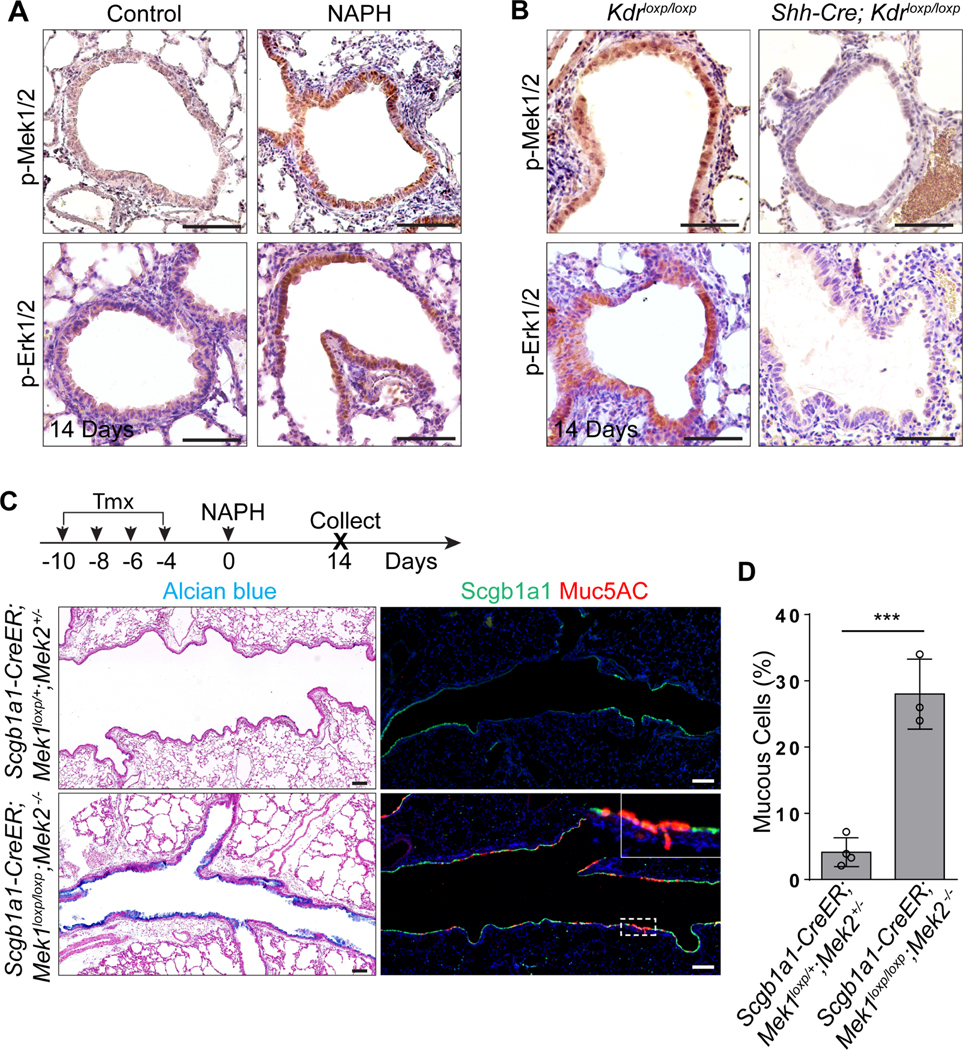
VEGFa/KDR signaling activates multiple downstream pathways, including the p38 MAPK, JNK and MEK/ERK pathways (Carmeliet and Jain, 2011; Koch and Claesson-Welsh, 2012). We found that p-MEK1/2 and p-ERK1/2 but not p-p38 or p-JNK were prominently expressed in the airway epithelium during regeneration (Figure 4A, S4A and S4B). Consistently, p-MEK1/2 and p-ERK1/2 were lost upon Kdr deletion in Shh-Cre;Kdrloxp/loxp mutants (Figure 4B), suggesting that VEGFa/KDR activates the MEK/ERK pathway during airway regeneration. In line with this finding, treatment with the MEK1/2 inhibitor PD98059 or the ERK inhibitor FR180204 but not the p38 MAPK inhibitor SB203580 or JNK inhibitor SP600125 led to severe mucous metaplasia in the airways of wildtype mice following NAPH challenge (Figure S4C and S4D). PD98059 has also been shown to block the NF-κB pathway in addition to MEK1/2 (Di Paola et al., 2010). We therefore genetically deleted Mek1/2 with the Scgb1a1-CreER mouse line and observed mucous metaplasia in the regenerated airway epithelium following NAPH challenge (4.1%±2.2% vs 27.8%±5.3%,p<0.001) (Figure 4C and 4D). Together these data support the conclusion that KDR activates MEK/ERK signaling to block mucous cell differentiation during airway regeneration.
Figure 4: Inhibition of Mek/Erk signaling promotes mucous metaplasia of the regenerated airway epithelium.
(A) Increased expression of p-Mek1/2 and p-Erk1/2 in the airway epithelium following NAPH challenge. (B) Kdr deletion reduces the expression of p-Mek1/2 and p-Erk1/2 in Shh-Cre;Kdrloxp/loxp mutants following NAPH challenge. (C and D) Genetic inhibition of Mek signaling promotes airway mucous metaplasia in Scgb1a1-CreER;Mek1loxp/loxp;Mek2−/− mutants following NAPH treatment (n=3). Data represent mean ± s.e.m. ***p<0.001; statistical analysis by unpaired two-tailed Student’s t-test. Scale bar:100 μm. See also Figure S4.
Sox9 is critical for mucous metaplasia induced by Kdr loss during airway epithelial regeneration
Sox9 is critical for goblet cell differentiation in the intestine (Bastide et al., 2007), raising the possibility that the transcription factor also plays a role in the adult lung, in addition to its well-studied role in distal epithelial progenitors during lung development (Chang et al., 2013; Rockich et al., 2013). Our single cell RNA sequencing data revealed that Sox9 was expressed in the Sub3 population of club cells in the adult lung (Figure S5A and S5B). Lineage labeling with a knockin Sox9-CreER mouse line confirmed that SOX9 was expressed in a minor population of club cells in the proximal airways. Moreover, lineage labeled cells also included a minor population of ciliated cells in the distal airways (Figure 5A and S5C). These lineage-labeled cells remained solitary throughout a chase period of three months, suggesting that they are quiescent (Figure S5D). Interestingly, SOX9 expression was seen in the abundant mucous cells present in the regenerated airways of both Shh-Cre;Kdrloxp/loxp (0.3%±0.2% vs 23.1%±3.9%,p<0.001) and Scgb1a1-CreER; Kdrloxp/loxp (0.2%±0.1% vs 9.8%±2.5%,p<0.001) mutants following NAPH challenge (Figure 5B–E). SOX9 was also enriched in the nuclei of mucous cells following challenge with PD98059 (0.1%±0.2% vs 5.1%±1.6%,p<0.001) (Figure S5E and S5F). Notably, a subpopulation of Sox9+ cells did not express Mu5AC, suggesting that these cells are club cells that are going to undergo mucous differentiation (Figure 5B–E, S5E and S5F). To test whether activation of SOX9 is critical for mucous metaplasia, we generated Scgb1a1-CreER; Kdrloxp/loxp; Sox9loxp/loxp mutants. Loss of Sox9 blocked mucous cell differentiation following NAPH challenge (28.6%±8.1% vs 9.5%±3.9%,p<0.01) (Figure 5F and 5G). We further found that Sox9 deletion reduced the expression of Spdef and Foxa3 in Scgb1a1-CreER;Kdrloxp/loxp;Sox9loxp/loxp mutants (Figure S5G and S5H). Thus, these findings suggest that SOX9 expression is critical for mucous cell differentiation upon Kdr deficiency.
Figure 5: Sox9 mediates mucous metaplasia upon Kdr loss.
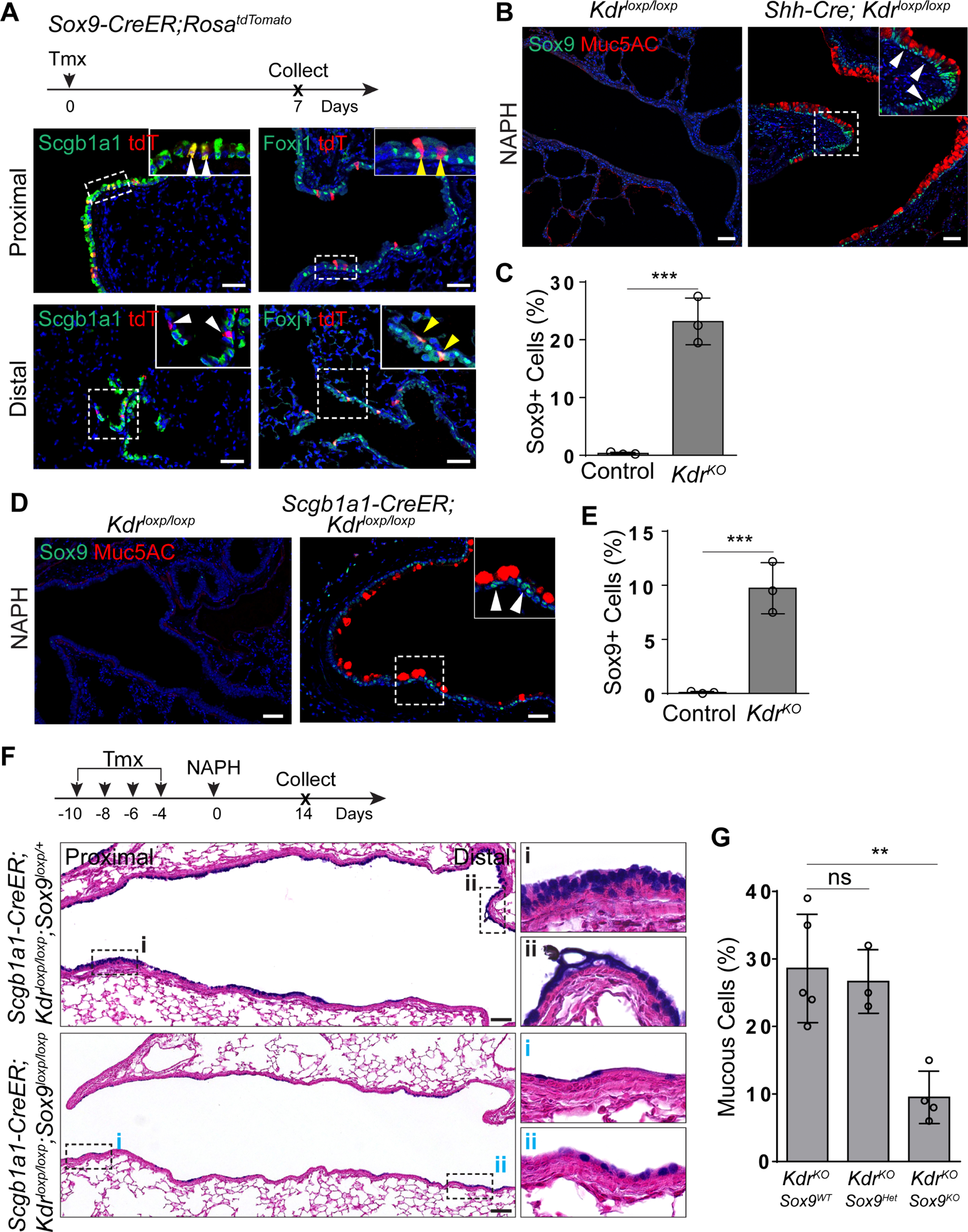
(A) Sox9 lineage-labeled cells include a limited number of club cells in the proximal airways and ciliated cells in the distal airways. (B and C) Kdr deletion leads to Sox9 expression in the metaplastic mucous cells of Shh-Cre;Kdrloxp/loxp mutants following NAPH treatment (n=3). (D and E) Sox9 is expressed in the airway mucous cells of Scgb1a1-CreER;Kdrloxp/loxp mutants following NAPH challenge (n=3). (F and G) Sox9 deletion attenuates mucous metaplasia of the regenerated airway epithelium in Scgb1a1-CreER;Kdrloxp/loxp;Sox9loxp/loxp mutants (n=4). Data represent mean ± s.e.m. ns: no significant; **p<0.01; ***p<0.001; statistical analysis by unpaired two-tailed Student’s t-test. Scale bar: 50 μm. See also Figure S5.
Reduced VEGFa/Kdr levels are associated with mucous metaplasia in influenza and asthmatic mouse models
Infection of the lung by viruses, including influenza and SARS-Cov-2, leads to severely disrupted airway epithelium and excessive mucous secretion (Fang et al., 2020; Rane et al., 2019; Wang et al., 2020). We used H1N1 PR8 influenza virus to address whether KDR is involved in these pathological changes. Excessive mucus production in the dysplastic regions of the injured lungs was observed after viral infection (Figure S6A). Notably, mucous cells were present in the regions where Kdr-GFP was not expressed. By contrast, the adjacent epithelial cells expressing Kdr-GFP contained minimal mucous cells (Figure S6B). Consistently, the transcript levels of Vegfa and Kdr were reduced in the metaplastic mucous epithelium (H score of Vegfa: 99.4±9.6 vs 70.1±5.7, p<0.05; H score of Kdr: 90.4±10.4 vs 21.5±6.6, p<0.001) (Figure S6C and S6D). Meanwhile, SOX9 was also enriched in the nuclei of mucous cells upon H1N1 infection (0.2%±0.2% vs 7.3%±1.1%,p<0.001) (Figure S6E and S6F).
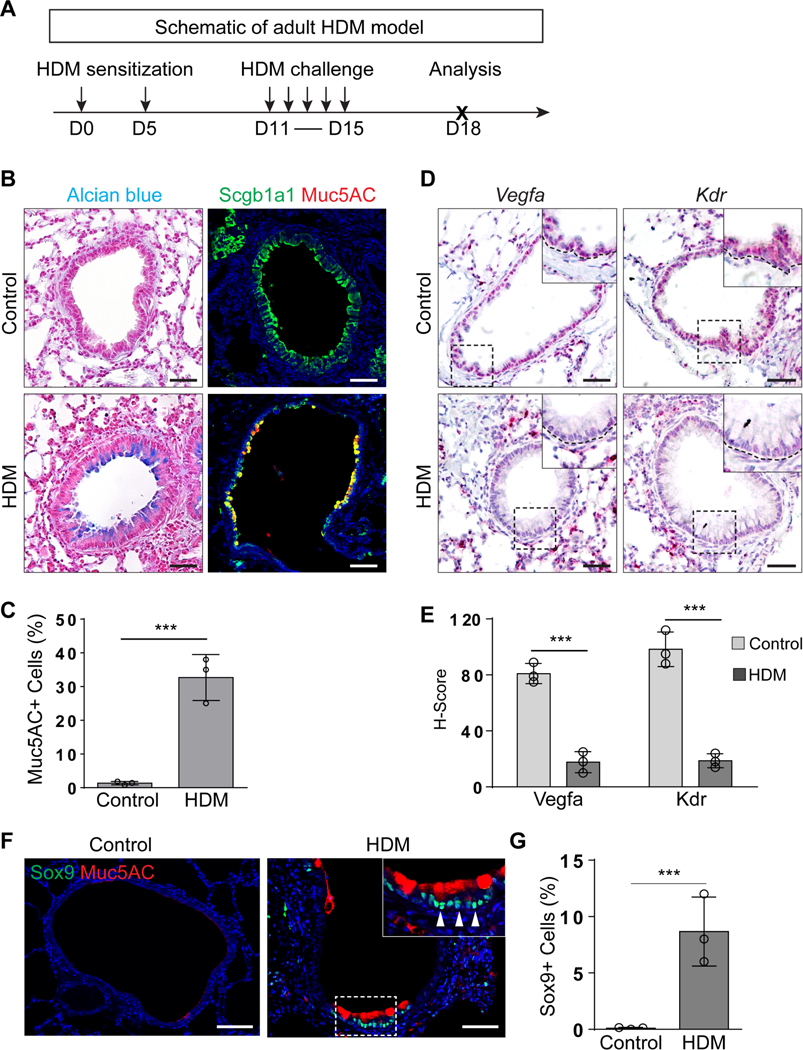
Mucous metaplasia is one of the most common phenotypes in the airways of patients with allergic Th2 dependent asthma (Busse et al., 1999; Tanizaki et al., 1993). To test whether abnormal VEGFa/KDR levels are associated with asthmatic mucous metaplasia, we employed an established murine allergic asthma model in which house dust mite (HDM) antigen is used to sensitize and re-challenge the mice (Figure 6A) (Wang et al., 2019). HDM treatment led to mucous metaplasia in the airways (1.2%±0.6% vs 32.4%±6.7%,p<0.001) (Figure 6B and 6C). Notably, mucous metaplasia was associated with the reduced transcript levels of Vegfa and Kdr (H score of Vegfa: 80.9±7.3 vs 17.3±7.3, p<0.001; H score of Kdr: 98.1±12.6 vs 18.3±4.9, p<0.001) in contrast to the unchanged levels in the blood vessels and the accompanying increase in SOX9 levels (0.1%±0.04% vs 8.6%±3.1%,p<0.001) (Figure 6D–G). Together these data support a model in which reduced Vegfa/Kdr signaling is associated with mucous metaplasia during both regeneration and asthmatic pathogenesis.
Figure 6: Asthmatic mucous metaplasia is associated with reduced Vegfa and Kdr transcripts.
(A) Schematic of the house dust mite (HDM)-induced adult mouse asthma model. (B and C) Mucous metaplasia occurs in the airway epithelium of asthmatic mice (n=3). (D and E) RNAscope analysis indicates the reduced transcripts of Vegfa and Kdr in the airway epithelium of asthmatic mice (n=3). (F and G) Sox9 is expressed in the airway mucous cells of HDM-induced asthmatic mice (n=3). Data represent mean ± s.e.m. ***p<0.001; statistical analysis by unpaired two-tailed Student’s t-test.Scale bar:100 μm. See also Figure S6.
Kdr signaling suppression is associated with mucous metaplasia of human airways
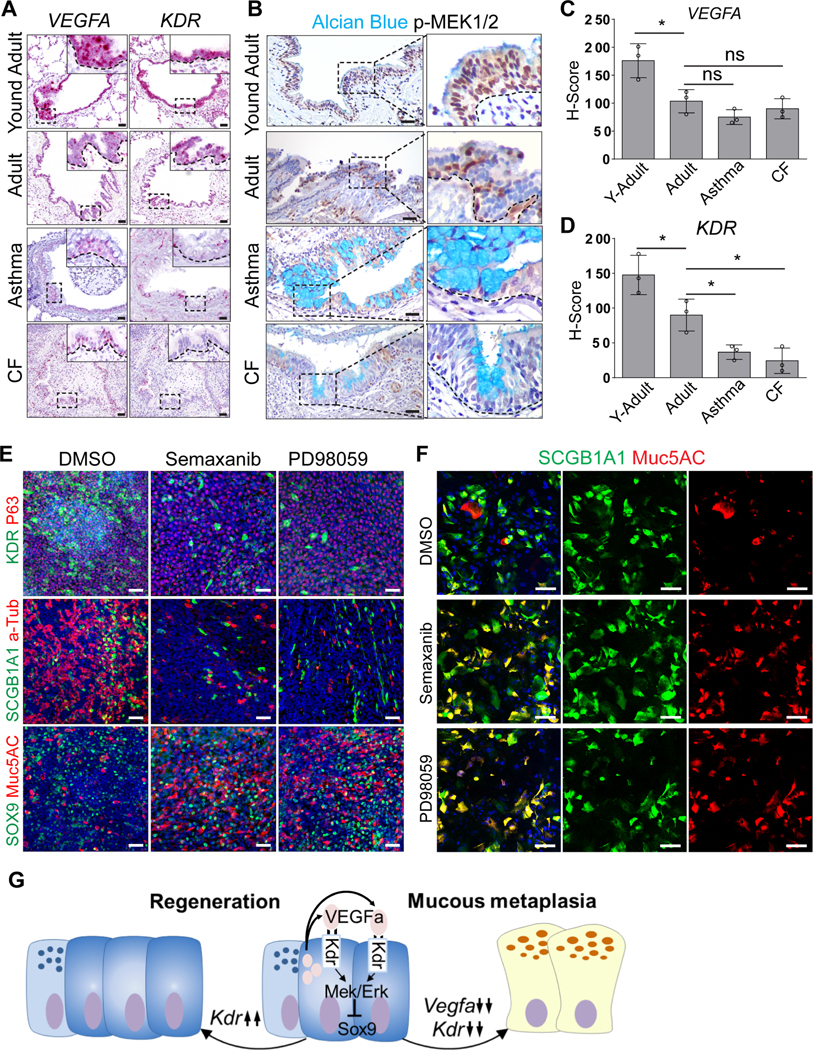
We next asked whether VEGF/KDR signaling activities are associated with mucous metaplasia in the human lung related to pulmonary diseases (asthma and CF). RNAScope analysis revealed relatively high levels of VEGFA and KDR transcripts in the conducting airway epithelium of young adolescents compared to adults (H score of VEGFA: 176.1±30.5 vs 103.3±20.8, p<0.05; H score of KDR: 147.7±28.3 vs 90.2±22.9, p<0.05) (Figure 7A, 7C and 7D). Positive staining of p-MEK1/2 and p-ERK1/2 was also observed in the epithelium of healthy controls (Figure 7B and S7A). However, KDR transcripts were reduced, although not the levels of VEGFA, in the airways of asthma (H score of KDR: 90.2±22.9 vs 36.7±10.4, p<0.05) and cystic fibrosis patients (H score of KDR: 90.2±22.9 vs 24.3±18.3, p<0.05). Consistently, the epithelium exhibited decreased p-MEK1/2 and p-ERK1/2 staining (Figure 7A–D and S7A). These results are consistent with our findings in mouse models that suppressed VEGF/KDR signaling contributes to mucous metaplasia of the airway epithelium.
Figure 7: Suppressed VEGFA-KDR signaling promotes mucous metaplasia of human airway epithelial cells.
(A) VEGFA and KDR transcripts are expressed in the airway epithelium of young adults and reduced in adults (n=3). Note further reduced levels of transcripts in asthma (n=3) and cystic fibrosis (CF) patients (n=3). (B) Reduced p-MEK1/2 expression in the airway epithelium of asthma (n=3) and CF patients (n=3). (C and D) Reduced transcript levels of VEGFA (C) and KDR (D) in asthma and CF lung samples as measured by H-score. Data represent mean ± s.e.m. ns: no significant; *p<0.05; statistical analysis by unpaired two-tailed Student’s t-test. (E) Inhibition of KDR (Semaxanib) and MEK (PD98059) promotes mucous differentiation of human small airway epithelial cells. Note the decreased numbers of club and ciliated cells following inhibitor treatments. (F) Inhibition of KDR or MEK promotes mucous metaplasia of club cells. (G) Transiently increased Vegfa-Kdr signaling is critical for airway epithelial regeneration. Reduced Vegfa-Kdr-Mek signaling promotes mucous cell differentiation of club cells through Sox9. Scale bar: 50 μm. See also Figure S7.
To further test whether VEGF/KDR signaling blocks the differentiation of the human airway epithelium towards mucous cells, we cultured human small airway epithelial cells (HSAECs) in an air-liquid interface (ALI) culture system (Whitcutt et al., 1988). Under these conditions KDR was present in a subpopulation of club but not basal or ciliated cells, similar to the mouse airway epithelium (Figure S7B and data not shown). Treatment with the KDR inhibitor Semaxanib or the MEK inhibitor PD98059 from the start of the culture reduced the numbers of club and ciliated cells while promoting mucous cells concomitant with increased SOX9 expression (Figure 7E, S7C and S7D). Previous studies have shown that basal cells give rise to mucous cells in murine airways (Hong et al., 2004; Rock et al., 2011). We therefore used FACS to exclude basal cells (p75+) and reseeded the differentiated cells into ALI culture (Figure S7E). Remarkably, a significant number of club cells differentiated into mucous cells upon treatment with Semaxinib or PD98059 (Figure 7F and S7F), confirming that VEGF/KDR signaling plays a conserved role in suppressing mucous cell differentiation of club cells.
Discussion
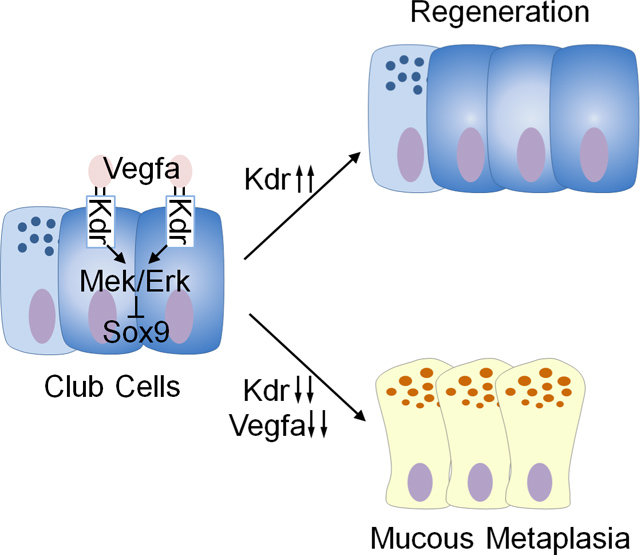
Previous studies have demonstrated the important role for inflammatory cytokines in the pathogenesis of airway mucous metaplasia (Gour and Wills-Karp, 2015; Klein Wolterink et al., 2012; Kuperman et al., 2002; Lee et al., 1997; Li et al., 2019; Schmid-Grendelmeier et al., 2002; Whitsett, 2018; Wills-Karp et al., 1998; Zhou-Suckow et al., 2017; Zhu et al., 1999). Here, we describe the unexpected yet essential function for VEGF/KDR signaling in achieving normal airway epithelial regeneration and mucous cell differentiation. Our single-cell RNA analysis identified that club cell subpopulations transiently increase KDR expression during airway regeneration. Genetic or pharmacological inhibition of VEGFa/KDR or its downstream MEK/ERK signaling disrupts normal epithelial differentiation program, leading to accumulated mucous cells. Further genetic analysis suggests that SOX9 is an important mediator of mucous metaplasia following KDR loss. ALI culture confirmed that KDR inhibition also causes mucous differentiation of human club cells, accompanied by increased SOX9 protein levels. In asthmatic and CF patients airway mucous metaplasia is associated with decreased KDR and increased SOX9 protein levels (Figure 7G).
Club cells constitute a heterogeneous population with stem/progenitor cell function, contributing to the regeneration of the epithelium of the airways and alveoli following injury (Guha et al., 2017; Hong et al., 2001; Kathiriya et al., 2020; Kim et al., 2005; Rawlins et al., 2009; Reynolds et al., 2000; Van Winkle et al., 1995; Yuan et al., 2019). More recently, single cell RNA sequencing has revealed that MHC class I marker H2-K1high progenitor cells are critical for alveolar repair (Kathiriya et al., 2020). Our single cell analysis identified three subpopulations of club cells, which are associated with the distinct expression of KDR and mucin-related genes. Numerous studies demonstrate that KDR is critical for vasculature and hematopoiesis development (Carmeliet et al., 1996; Olsson et al., 2006; Shalaby et al., 1995; Yeh et al., 2003). Here, we uncovered a critical role for KDR in the regulation of epithelial differentiation. Our in vivo and in vitro studies support the hypothesis that VEGF/KDR is a guardian against mucous cell differentiation of the regenerated airway epithelium. Interestingly, reduced levels of VEGF and KDR have been found in the bronchoalveolar lavage fluid and lungs of smokers and patients with COPD which is characterized by small airway mucous metaplasia (Kim et al., 2008; Koyama et al., 2002; Marwick et al., 2006). We found that VEGFa and KDR levels are also reduced in asthmatic and CF patients, suggesting a common role for suppressed VEGF/KDR signaling in the pathogenesis of mucous metaplasia in these pulmonary diseases.
In the developing lung the Notch signaling pathway participates in the regulation of goblet cell differentiation (Guseh et al., 2009; Lafkas et al., 2015; Tsao et al., 2011). Transgenic overexpression of the active intracellular domain of the mouse Notch1 receptor (NotchIC) using Sftpc-Cre results in increased numbers of mucous cells in the airways (Guseh et al., 2009). However, Notch signaling was also shown to prevent mucous metaplasia of the airway epithelium during postnatal development. Conditional deletion of the essential Notch pathway components protein O-fucosyltransferase1 (Pofut1) or RbpjK with Tgfb3-Cre which is activated postnatally causes goblet cell metaplasia concomitant with decreased club cell numbers. In the same study lineage tracing also suggests mucous cells are derived from club cell subpopulations (Tsao et al., 2011). In addition, inhibition of Notch signaling with an antibody against Jag1/2 attenuates IL-13-induced mucous metaplasia (Lafkas et al., 2015). We also examined the potential involvement of Notch1 signaling by evaluating the Notch1 intracellular domain (NICD1) and Jag1/2 expression in Kdr deletion mutants. No apparent changes were observed in the metaplastic airway epithelium during regeneration (unpublished data), suggesting that Notch1 signaling unlikely mediates Kdr function in the regulation of mucous cell differentiation.
Previous studies have shown that HDM-induced mucous metaplasia involves the activation of other signaling molecules such as Toll-like receptor 4 (TLR4) and EGF (Hammad et al., 2009; Jacquet and Robinson, 2020; Le Cras et al., 2011; Yasuda et al., 2020). Of note is that both EGF and TLR4 can directly or indirectly regulate MEK/ERK activities (Aomatsu et al., 2008; Athari et al., 2017; Avraham and Yarden, 2011; Le Cras et al., 2011). It is possible that reduced KDR also impacts the expression of Egf and/or TLR4 which contribute to the reduced MEK/ERK signaling. Our genetic study further indicates that Sox9 acts downstream of VEGF/KDR signaling. Deletion of Sox9 attenuates mucous metaplasia of the regenerated airway epithelium following KDR loss. Sox9 has been shown to play essential roles in the development of multiple organs (Bastide et al., 2007; Chang et al., 2013; Rockich et al., 2013). In the developing lung, Sox9 is enriched in the distal tips of the branching epithelium. Sox9 deletion impacts both lung branching and alveolar differentiation (Chang et al., 2013; Rockich et al., 2013), leading to small lungs with dilated airway branches and premature differentiation of alveolar cells (Chang et al., 2013). Further study demonstrates that FGF/Kras regulates Sox9 during lung branching morphogenesis (Chang et al., 2013). The role of Sox9 in adult lung remains largely unexplored. In the tracheal submucosal gland, Sox9 is required for the proliferation and migration of myoepithelial cells to the airways during regeneration (Tata et al., 2018). Our lineage labeling with a knockin Sox9-CreER allele suggests that the Sox9 promoter is active in a limited number of club cells in the upper airways. However, during metaplastic changes, extensive mucous cells express Sox9 following Kdr deletion, suggesting that Sox9 is required for the mucous differentiation of club cells during regeneration. Consistently, Sox9 deletion attenuated mucous metaplasia caused by Kdr deletion. Our findings are consistent with the role played by Sox9 in the development of secretory cell lineages in the intestine where Sox9 inactivation results in the loss of Paneth cells and decreased numbers of goblet cells (Bastide et al., 2007).
Taken together our findings now introduce new and unexpected players into the field of lung pathogenesis, namely Kdr, Vegf and Sox9. In the future, it will be important to understand the extent to which these factors are independent of and/or integral to cytokine signaling pathways at the cellular and tissue level. Our findings also raise the exciting possibility that new druggable pathways can be identified to inhibit mucus metaplasia in respiratory diseases.
Limitations of study
This study revealed inverse correlation of VEGF/Kdr signaling activities and mucous metaplasia. We further used the MEK1/2 inhibitor PD98059 and genetic ablation of MEK1/2 with the Scgb1a1-CreER mouse line to demonstrate that MEK1/2 act downstream of VEGF/Kdr signaling to guard against mucous metaplasia of the regenerated airway epithelium following NAPH challenge. However, we do not know whether MEK1 or MEK2 is equally important for blocking mucous differentiation since both our pharmaceutical and genetic approaches target MEK1 and 2 simultaneously. In addition, it remains unknown whether MAPK signaling is also involved in blocking premature mucous differentiation of airway progenitors during development.
STAR Methods
Resource availability
Lead contact
Further information and requests for reagents may be directed to and will be fulfilled by the lead contact, Dr. Jianwen Que (jq2240@cumc.columbia.edu)
Materials availability
This study did not generate new unique reagents.
Data and code availability
The single cell RNA-sequencing datasets of airway epithelium isolated from NAPH- or vehicle-treated mice has been deposited to the Expression Omnibus: GSE171571.
Experimental model and subject details
Mice
Shh-Cre (Harfe et al., 2004), Scgb1a1-CreER (Rawlins et al., 2009), Sox9-CreER (Soeda et al., 2010), Mek1loxp/loxp (Bissonauth et al., 2006),Mek2−/− (Belanger et al., 2003) mice were previously described. Kdr-GFP, Kdr-Cre, Kdrloxp/loxp, Sox9loxp/loxp, R26tdTomato, R26LacZ mice were purchased from the Jackson Laboratory. Vegfa-hyper, Vegfa-hypo and Vegfaloxp/loxp mice were kindly provided by Dr. Hoon-Ki Sung of The Hospital for Sick Children. To lineage trace cells derived from Kdr+ progenitors, Kdr-Cre mice were crossed with RosalacZ mice to generate Kdr-Cre;RosalacZ compounds. To lineage trace cells derived from Sox9+ epithelium, Tamoxifen (Tmx, T5648, Sigma-Aldrich) was injected intraperitoneally into Sox9-CreER;RosatdTomato mice (200 mg/kg body weight). Mice were sacrificed at defined time points as indicated in the text. Shh-Cre;Kdrloxp/loxp mice were generated to conditionally delete Kdr in the foregut epithelium. To delete genes in club cells, Scgb1a1-CreER;Kdrloxp/loxp, Scgb1a1-CreER;Vegfaloxp/loxp or Scgb1a1-CreER; Mek1loxp/loxp;Mek2−/− mice were administrated with four doses of Tmx prior to naphthalene (NAPH) injection. In addition, mice were exposed to NAPH and then treated with 10 mg/kg PD98059 (Mek inhibitor, S1177, Selleckchem), 20 mg/kg Semaxanib (Kdr inhibitor, S2845, Selleckchem), 20 mg/kg SP600125 (JNK inhibitor, S1460, Selleckchem), 10 mg/kg SB203580 (p38 inhibitor, S1076, Selleckchem) or 50 mg/kg FR180204 (Erk inhibitor, S7524, Selleckchem) at day 8–14 following NAPH challenge. Lungs were collected for analysis at various time points as indicated in the text. All mice were maintained in animal facilities of Columbia University or Zhejiang University according to institutional guidelines. All mouse experiments were conducted in accordance with procedures approved by the Institutional Animal Care and Use Committee. All relevant ethical regulations were followed to the text.
Method details
NAPH treatment to induce airway epithelial injury
NAPH solution (25 mg/ml) was prepared immediately before use by dissolving NAPH (84679, Sigma-Aldrich) in corn oil. A single dose of NAPH was administrated intraperitoneally into mice at least 8 weeks of age (275 mg/kg body weight). NAPH was injected at least 4 days after the final dose of Tmx to allow mice to recover. Approximately 90% of club cells are ablated three days after NAPH administration as measured by immunostaining or quantitative PCR of Scgb1a1. Control mice were injected with the same amount of corn oil.
H1N1 PR8 influenza viral infection
Mice at 8–12 weeks of age were anesthetized with isoflurane and infected intranasally with 250 plaque forming units (pfu) of A/Puerto Rico/8/1934 H1N1 (PR8) virus in 40 μl DMEM medium. Control mice were administrated intranasally with equal volume of DMEM medium. Successful infection was confirmed by weight loss and lung sections.
Mouse model of Asthma
House dust mite (HDM)-induced asthma model was performed as previously described with a minor modification (Wang et al., 2019). Briefly, 8-week-old mice were sensitized intra-nasally with 5 μg of HDM in PBS (XPB70D3A25, Greer Laboratories) on day 0 and day 5. Mice were then intra-nasally challenged with 25 μg HDM for five consecutive days (day 11–15) and were sacrificed on day 18 for analysis.
Lung epithelium isolation and Fluorescence-Activated Cell Sorting (FACS)
Mice were perfused with PBS from right ventricles to remove blood in the lungs. Trachea was infused with 1.5ml digested solution containing 450 U/ml Collagenase Type I (17100–017, Gibco), 4 U/ml Elastase (LS002279, Worthington Biochemical Corporation), 5 U/ml Dispase II (354235, BD Biosciences) and 0.5 U/ml Dnase I (10104159001, Roche). Lungs were removed from the chest and further incubated in the digested solution for 45 mins at room temperature with gentle shaking. After digestion lung tissues were cut into small pieces and washed with DMEM buffer (10% FBS and 1% Penicillin-Streptomycin in DMEM medium). Single-cell suspension was obtained after passing through a 40 μm cell strainer and centrifuged at 1200rpm for 5 minutes. Cell pellet was resuspended in a red blood cell (RBC) lysis buffer for 1 minute to remove RBCs. Cells were centrifuged and resuspended in 100 μl sorting buffer (5% FBS, 0.2 mM EDTA, and 1% PS in PBS buffer). Cell suspension was incubated with an APC-conjugated EPCAM primary antibody for one hour at 4°C and washed for two times with cell sorting buffer. Dead cells were excluded by LIVE/DEAD Fixable Violet Dead Cell Stain Kit (L34963, Thermo Scientific). Cells were sorted into the sorting buffer. Analysis was performed using FlowJo software.
Air-liquid interface (ALI) culture of human small airway epithelial cells
Primary human small airway epithelial cells (PCS-301–010) were purchased from ATCC (Walkersville, MD). Cells were cultured in PneumaCult-Ex Plus (PnC-Ex-PLUS) media (05008, StemCell Technologies) for expansion. Once cells reached 80–90% confluency, they were digested and reseeded in 6.5 mm Transwells (353095, Corning) coated with 0.3 mg/mL Collagen type IV (234154, Sigma-Aldrich) with a density of 50,000 cells/well. Cells were firstly expanded in the PneumaCult-Ex PLUS media to reach confluency. Medium was then removed and replaced with the PneumaCult-ALI medium (05001, StemCell Technologies) to induce differentiation in the presence/absence of various inhibitors. Medium was changed every two days, and the transwell surface was also washed with PBS every two days. All cells were grown for four weeks airlifted (37°C, 5% CO2) and physiological and characterization tests were subsequently performed. Additionally, the airway epithelial cells were initially induced to differentiate with ALI culture, and then FACS sorting was performed to exclude basal cells using the basal cell surface marker p75. The remaining cells were then re-plated in the transwells for further ALI culture as described above.
Tissue preparation, histology and immunostaining
Lungs were inflated and fixed in 4% paraformaldehyde (PFA) in PBS overnight at 4°C and processed as previously described (Jiang et al., 2017). In brief, lungs were washed with PBS after fixation and dehydrated with ethanol prior to embedding in paraffin. 6μm sections were then prepared from the paraffin blocks. To prepare cryosections, fixed lung tissues were placed in 30% sucrose and embedded in OCT. 10μm sections were cut from the embedded lung tissues. Primary antibodies used for immunostaining analysis include rabbit anti-p63 (1:200, sc-8343, Santa Cruz Biotechnology); mouse anti-p63 (1:500, CM163, Biocare); chicken anti-GFP (1:1000, GFP-1020, Aves Labs); rabbit anti-Sox9 (1:1000, AB5535, Millipore); goat anti-tdTomato (1:1000, orb182397, Biorbyt); mouse anti-Foxj1 (1:100, 14–9965-82, eBioscience); rabbit anti-pMek1/2 (1:200, 2338S, Cell Signaling Technology); rabbit anti-pErk1/2 (1:200, 4370S, Cell Signaling Technology); rabbit anti-pP38 (1:200, 9215S, Cell Signaling Technology); rabbit anti-pJNK (1:200, 9251S, Cell Signaling Technology); rabbit anti-Kdr (1:200, 9698S, Cell Signaling Technology); mouse anti-Muc5AC (1:500, MS-145-P0, Lab Vision); rabbit anti-Clca3 (1:2000, ab46512, Abcam); rabbit anti-Scgb1a1 (1:500, 07–623, Millipore); rat anti-human SCGB1A1 (1:1000, MAB4218, R&D Systems); mouse anti-acetylated-tubulin (1:2000, T7451, Sigma). Biotinylated or fluorescent secondary antibodies were used for detection and visualization. Immunohistochemistry images were obtained using Nikon SMZ1500 Inverted microscope (Nikon). Confocal images were obtained with a Zeiss LSM T-PMT confocal laser-scanning microscope (Carl Zeiss).
RNAscope in situ hybridization
RNAscope in situ hybridizations was performed using the RNAscope 2.5 HD Assay-RED kit (322360, Advanced Cell Diagnostics), according to the manufacture instructions. The following probes were used in these analyses: mm-Kdr (414811, ACD), mm-Vegfa (405131, ACD), hs-KDR (312121, ACD) and hs-VEGFA (423161, ACD). Sections were baked for one hour at 60°C before use. After deparaffinization and hydration, sections were air-dried, treated with a hydrogen peroxidase solution and heated in a target retrieval solution for 20 min at 95–100°C. Sections were then incubated in proteinase solution at 40°C for 30 min. Target probes were hybridized for two hours at 40°C, followed by a series of amplification steps using Amp1–6 in the kit. ISH signals were detected with a Fast Red substrate reaction for 10 min at room temperature. Sections were counterstained with hematoxylin. After washing in PBS, sections were completely dried on a hot plate set at 60°C and cover-slipped with Mounting medium. Positive or negative controls were performed by hybridizing with PPIB (Cat. No. 313901, ACD) or DapB (Cat. No. 310043, ACD), respectively. Sections were examined by Nikon SMZ1500 Inverted microscope (Nikon). At least five random fields (20x magnified images) were captured for each section. Semi-quantitative histological scoring (H-score) of target genes was analyzed using ACD semi-quantitative scoring system, calculated as follows: H-score = (0 × % score0+ cells) + (1 × % score1+ Cells) + (2 × % score2+ Cells) + (3 × % score3+ Cells) +(4 × % score4+ Cells) (Jolly et al., 2019).
Alcian blue and X-gal staining
Alcian blue and X-gal staining were performed as previously described (Hou et al., 2019). Briefly, for Alcian blue staining, sections were treated with 3% acetic acid solution for three minutes, then stained in Alcian blue solution (A3157, Sigma) for five minutes and counterstained with Nuclear Fast Red (N8002, Sigma). For X-gal staining, whole lungs were fixed in 4% paraformaldehyde for 30 minutes at room temperature, followed by X-gal (R0404, Thermo Scientific) staining overnight at 37°C. X-gal–stained samples were then dehydrated with isopropanol and embedded in paraffin for sectioning.
Reverse transcription and quantitative Real-Time PCR
Tissues and Cells were lysed with TRIzol reagent (15596026, Thermo Scientific), and RNA was purified using the RNeasy Mini Kit (74104, QIAGEN). RNA reverse transcription was performed using the Super-Script III First-Strand SuperMix (18080400, Invitrogen) according to the manufacturer’s instructions. cDNA was quantified by real-time PCR using the iTaq Universal SYBR Green Supermix (1725122, Bio-Rad) and the StepOnePlus Real-Time PCR Detection System (Applied Biosystems). The transcript levels of genes were normalized to Actin expression. All real-time quantitative PCR experiments were performed at least triplicate. PCR primers were designed using the Lasergene Core Suite (DNASTAR Inc.), and the sequences of primers are listed in the Key Resources Table.
KEY RESOURCES TABLE
| REAGENT OR RESOURCE | SOURCE | IDENTIFIER |
|---|---|---|
| Antibodies | ||
| Rabbit monoclonal anti-P63 | Santa Cruz | Cat#sc-8343; RRID: AB_653763 |
| Mouse monoclonal anti-P63 | Biocare Medical | Cat# CM163; RRID: AB_10583039 |
| Chicken polyclonal anti-GFP | Aves Labs | Cat#GFP-1020; RRID: AB_2307323 |
| Rabbit polyclonal anti-Sox9 | Millipore | Cat#AB5535; RRID: AB_2239761 |
| Goat polyclonal anti-tdTomato | Biorbyt | Cat#orb182397; RRID: AB_2687917 |
| Mouse monoclonal anti-Foxj1 | eBioscience | Cat#14-9965-82; RRID: AB_1548835 |
| Rabbit polyclonal anti-pMek1/2 | Cell Signaling Technology | Cat#2338S; RRID: AB_490903 |
| Rabbit polyclonal anti-pErk1/2 | Cell Signaling Technology | Cat#4370S; RRID: AB_2315112 |
| Rabbit polyclonal anti-pP38 | Cell Signaling Technology | Cat#9215S; RRID: AB_331762 |
| Rabbit polyclonal anti-pJNK | Cell Signaling Technology | Cat#9251S; RRID: AB_331659 |
| Rabbit polyclonal anti-Kdr | Cell Signaling Technology | Cat#9698S; RRID: AB_11178792 |
| Mouse monoclonal anti-Muc5AC | Lab Vision | Cat#MS-145-P0; RRID: AB_62735 |
| Rabbit polyclonal anti-Clca3 | Abcam | Cat#ab46512; RRID: AB_152837 |
| Rabbit polyclonal anti-Scgb1a1 | Millipore | Cat#07-623; RRID: AB_310759 |
| Rat monoclonal anti-human SCGB1A1 | R&D Systems | Cat#MAB4218; RRID: AB_394324 |
| Mouse monoclonal anti-Acetylated tubulin | Sigma-Aldrich | Cat#T7451; RRID: AB_ 609894 |
| Rat monoclonal APC anti-mouse Epcam | Biolegend | Cat#118213; RRID: AB_1134105 |
| Mouse monoclonal APC anti-human p75 | Biolegend | Cat#345107; RRID: AB_ 10639737 |
| Chemicals, Peptides, and Recombinant Proteins | ||
| PneumaCult-Ex Plus Medium | StemCell Technologies | Cat#05008 |
| PneumaCult-ALI Medium | StemCell Technologies | Cat#05001 |
| Collagen Type IV | Sigma-Aldrich | Cat#234154 |
| Collagenase Type I | Gibco | Cat#17100-017 |
| Elastase | Worthington Biochemical Corporation | Cat#LS002279 |
| Dispase II | BD Biosciences | Cat#354235 |
| Dnase I | Roche | Cat#10104159001 |
| Naphthalene | Sigma-Aldrich | Cat#84679 |
| HDM | Greer Laboratories | Cat#XPB70D3A25 |
| Tamoxifen | Sigma-Aldrich | Cat#T5648 |
| LIVE/DEAD Fixable Violet Dead Cell Stain Kit | Thermo Fisher Scientific | Cat#L34963 |
| Semaxanib | Selleck | Cat#S2845 |
| PD98059 | Selleck | Cat#S1177 |
| FR180204 | Selleck | Cat#S7524 |
| SB203580 | Selleck | Cat#S1076 |
| SP600125 | Selleck | Cat#S1460 |
| RNAscope 2.5 HD Assay-RED Kit | Advanced Cell Diagnostics | Cat#322360 |
| mm-Kdr probe | Advanced Cell Diagnostics | Cat#414811 |
| mm-Vegfa probe | Advanced Cell Diagnostics | Cat#405131 |
| hs-KDR probe | Advanced Cell Diagnostics | Cat#312121 |
| hs-Vegfa probe | Advanced Cell Diagnostics | Cat#423161 |
| PPIB probe | Advanced Cell Diagnostics | Cat#313901 |
| DapB probe | Advanced Cell Diagnostics | Cat#310043 |
| Deposited data | ||
| Single cell RNA sequencing data of Normal and NAPH treated lung epithelium | This paper | GEO: GSE# |
| Experimental Models: Cell Lines | ||
| Primary Human Small Airway Epithelial Cells | ATCC | PCS-301-010 |
| Experimental Models: Organisms/Strains | ||
| Shh-Cre | (Harfe et al., 2004) | N/A |
| Scgb1a1-CreER | (Rawlins et al., 2009) | N/A |
| Sox9-CreER | (Soeda et al., 2010) | N/A |
| Mek1loxp/loxp | (Bissonauth et al., 2006) | N/A |
| Mek2−/− | (Belanger et al., 2003) | N/A |
| Kdr-GFP | Jackson Laboratory | 017006 |
| Kdr-Cre | Jackson Laboratory | 018976 |
| Kdrloxp/loxp | Jackson Laboratory | 018977 |
| Sox9loxp/loxp | Jackson Laboratory | 013106 |
| R26tdTomato | Jackson Laboratory | 007914 |
| R26lacZ | Jackson Laboratory | 003474 |
| Vegf-hyper | (Miquerol et al., 1999) | N/A |
| Vegf-hypo | (Damert et al., 2002) | N/A |
| Vegfloxp/loxp | Kindly provided by Dr. Hoon-Ki Sung | N/A |
| Oligonucleotides | ||
| Primer: Mouse Kdr Forward: CGAGACCATTGAAGTGACTTGCC | This paper | N/A |
| Primer: Mouse Kdr Reverse: TTCCTCACCCTGCGGATAGTCA | This paper | N/A |
| Primer: Mouse Scgb1a1 Forward: GGTTATGTGGCATCCCTGAAGC | This paper | N/A |
| Primer: Mouse Scgb1a1 Reverse: GCTTACACAGAGGACTTGTTAGG | This paper | N/A |
| Primer: Mouse Vegfa Forward: CTGCTGTAACGATGAAGCCCTG | This paper | N/A |
| Primer: Mouse Vegfa Reverse: GCTGTAGGAAGCTCATCTCTCC | This paper | N/A |
| Primer: Human Kdr Forward: GGAACCTCACTATCCGCAGAGT | This paper | N/A |
| Primer: Human Kdr Reverse: CCAAGTTCGTCTTTTCCTGGGC | This paper | N/A |
| Primer: Human p63 Forward: CAGGAAGACAGAGTGTGCTGGT | This paper | N/A |
| Primer: Human p63 Reverse: AATTGGACGGCGGTTCATCCCT | This paper | N/A |
| Primer: Human Scgb1a1 Forward: TCATGGACACACCCTCCAGTTATGAG | This paper | N/A |
| Primer: Human Scgb1a1 Reverse: TGAGCTTAATGATGCTTTCTCTGGGC | This paper | N/A |
| Primer: Human Foxj1 Forward: GGCATAAGCGCAAACAGCCG | This paper | N/A |
| Primer: Human Foxj1 Reverse: TCGAAGATGGCCTCCCAGTCAAA | This paper | N/A |
| Primer: Human Sox9 Forward: AGGAAGCTCGCGGACCAGTAC | This paper | N/A |
| Primer: Human Sox9 Reverse: GGTGGTCCTTCTTGTGCTGCAC | This paper | N/A |
| Primer: Human Muc5AC Forward: GCACCAACGACAGGAAGGATGAG | This paper | N/A |
| Primer: Human Muc5AC Reverse: CACGTTCCAGAGGCCGGACAT | This paper | N/A |
| Software and Algorithms | ||
| Prism 6 | GraphPad Software | https://www.graphpad.com/scientific-software/prism/ |
| R | Bioconductor | https://www.bioconductor.org/ |
Quantification and statistical analysis
For cell counting in the slides at least five random fields (20x magnified images) were captured for each section. Statistical analysis was done using unpaired two-tailed student’s t-test. Results were presented as mean ± s.e.m.; p values < 0.05 were considered statistically significant. *P < 0.05; **P < 0.01; ***P < 0.005. All statistical analyses were performed using GraphPad Software Prism 6.
Single-cell RNA sequencing analysis
EPCAM+ lung cells from control or NAPH-treated mice were purified by FACS and then processed following the 10x Genomics protocol. Cell Ranger pipeline was used to process raw sequencing data, and Cell Ranger R package and Seurat v2.0 pipeline were used to perform downstream scRNA-seq analysis. Differentially expressed genes in the heatmap were identified with the expression levels > 1.5 fold differences between the club cell subpopulations.
Data availability statement
The authors declare that the main data supporting the findings of this study are available within the paper and its supplementary information. Any additional data are available from the corresponding authors upon request.
Supplementary Material
Highlights.
VEGFa/KDR suppresses mucous differentiation of club cells during regeneration.
VEGFa/KDR signaling executes its epithelial function through MEK/ERK kinases.
Sox9 mediates mucous metaplasia of the airway epithelium upon VEGFa/KDR inhibition.
VEGFa/KDR/MEK blockage is linked to mucous metaplasia in asthma and cystic fibrosis.
Acknowledgements
We thank Dr. Brigid Hogan at Duke University and Dr. Wellington Cardoso at Columbia Center for Human Development for critical reading of the manuscript. We thank the Single Cell Analysis Core of Columbia University for single-cell RNA sequencing and data analysis. We also thank Dr. Hongxu Ding at the University of California (Santa Cruz) for assisting single-cell data analysis. Human biospecimens were provided in part by the LungMAP Consortium supported by the Molecular Atlas of Lung Development Program, Human Tissue Core funded by U01HL122700 / U01HL148861 (G.H. Deutsch, T.J. Mariani, G.S. Pryhuber). We gratefully acknowledge the tissue donors. This work is partly supported by R01HL152293, R01HL132996, R01DK113144, R01DK100342 (to J.Q), the NIH Support Grant S10RR027050 for Flow Cytometry analysis and the Genetically Modified Mouse Model Shared Resource of the NIH/NCI Cancer Center Support Grant (P30CA013696) at Columbia University, R01HL132991 (to X.A) and Young Investigator Research Program (588020-D01907, to M.J).
Footnotes
Declaration of Interests
A patent application (CU21016, “SUPPRESSION OF KDR, VEGF OR THE DOWNSTREAM MEK/ERK KINASE PATHWAY”) related to this work has been filed by Columbia Technology Ventures. M.J. and J.Q. are listed as co-inventors on this application.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Aomatsu K, Kato T, Fujita H, Hato F, Oshitani N, Kamata N, Tamura T, Arakawa T, and Kitagawa S. (2008). Toll-like receptor agonists stimulate human neutrophil migration via activation of mitogen-activated protein kinases. Immunology 123, 171–180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Athari SS, Athari SM, Beyzay F, Movassaghi M, Mortaz E, and Taghavi M. (2017). Critical role of Toll-like receptors in pathophysiology of allergic asthma. Eur J Pharmacol 808, 21–27. [DOI] [PubMed] [Google Scholar]
- Avraham R, and Yarden Y. (2011). Feedback regulation of EGFR signalling: decision making by early and delayed loops. Nat Rev Mol Cell Biol 12, 104–117. [DOI] [PubMed] [Google Scholar]
- Barker N, Huch M, Kujala P, van de Wetering M, Snippert HJ, van Es JH, Sato T, Stange DE, Begthel H, van den Born M, et al. (2010). Lgr5(+ve) stem cells drive self-renewal in the stomach and build long-lived gastric units in vitro. Cell Stem Cell 6, 25–36. [DOI] [PubMed] [Google Scholar]
- Barrios J, Patel KR, Aven L, Achey R, Minns MS, Lee Y, Trinkaus-Randall VE, and Ai X. (2017). Early life allergen-induced mucus overproduction requires augmented neural stimulation of pulmonary neuroendocrine cell secretion. FASEB J 31, 4117–4128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bastide P, Darido C, Pannequin J, Kist R, Robine S, Marty-Double C, Bibeau F, Scherer G, Joubert D, Hollande F, et al. (2007). Sox9 regulates cell proliferation and is required for Paneth cell differentiation in the intestinal epithelium. J Cell Biol 178, 635–648. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Belanger LF, Roy S, Tremblay M, Brott B, Steff AM, Mourad W, Hugo P, Erikson R, and Charron J. (2003). Mek2 is dispensable for mouse growth and development. Mol Cell Biol 23, 4778–4787. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bissonauth V, Roy S, Gravel M, Guillemette S, and Charron J. (2006). Requirement for Map2k1 (Mek1) in extra-embryonic ectoderm during placentogenesis. Development 133, 3429–3440. [DOI] [PubMed] [Google Scholar]
- Boucher RC (2019). Muco-Obstructive Lung Diseases. N Engl J Med 380, 1941–1953. [DOI] [PubMed] [Google Scholar]
- Branchfield K, Nantie L, Verheyden JM, Sui PF, Wienhold MD, and Sun X. (2016). Pulmonary neuroendocrine cells function as airway sensors to control lung immune response. Science 351, 707–710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Busse W, Elias J, Sheppard D, and Banks-Schlegel S. (1999). Airway remodeling and repair. Am J Respir Crit Care Med 160, 1035–1042. [DOI] [PubMed] [Google Scholar]
- Carmeliet P, Ferreira V, Breier G, Pollefeyt S, Kieckens L, Gertsenstein M, Fahrig M, Vandenhoeck A, Harpal K, Eberhardt C, et al. (1996). Abnormal blood vessel development and lethality in embryos lacking a single VEGF allele. Nature 380, 435–439. [DOI] [PubMed] [Google Scholar]
- Carmeliet P, and Jain RK (2011). Molecular mechanisms and clinical applications of angiogenesis. Nature 473, 298–307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chang DR, Martinez Alanis D, Miller RK, Ji H, Akiyama H, McCrea PD, and Chen J. (2013). Lung epithelial branching program antagonizes alveolar differentiation. Proc Natl Acad Sci U S A 110, 18042–18051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen G, Korfhagen TR, Xu Y, Kitzmiller J, Wert SE, Maeda Y, Gregorieff A, Clevers H, and Whitsett JA (2009). SPDEF is required for mouse pulmonary goblet cell differentiation and regulates a network of genes associated with mucus production. J Clin Invest 119, 2914–2924. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Curran DR, and Cohn L. (2010). Advances in mucous cell metaplasia: a plug for mucus as a therapeutic focus in chronic airway disease. Am J Respir Cell Mol Biol 42, 268–275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Damert A, Miquerol L, Gertsenstein M, Risau W, and Nagy A. (2002). Insufficient VEGFA activity in yolk sac endoderm compromises haematopoietic and endothelial differentiation. Development 129, 1881–1892. [DOI] [PubMed] [Google Scholar]
- Di Paola R, Galuppo M, Mazzon E, Paterniti I, Bramanti P, and Cuzzocrea S. (2010). PD98059, a specific MAP kinase inhibitor, attenuates multiple organ dysfunction syndrome/failure (MODS) induced by zymosan in mice. Pharmacol Res 61, 175–187. [DOI] [PubMed] [Google Scholar]
- Ema M, Takahashi S, and Rossant J. (2006). Deletion of the selection cassette, but not cis-acting elements, in targeted Flk1-lacZ allele reveals Flk1 expression in multipotent mesodermal progenitors. Blood 107, 111–117. [DOI] [PubMed] [Google Scholar]
- Evans CM, Williams OW, Tuvim MJ, Nigam R, Mixides GP, Blackburn MR, DeMayo FJ, Burns AR, Smith C, Reynolds SD, et al. (2004). Mucin is produced by clara cells in the proximal airways of antigen-challenged mice. Am J Respir Cell Mol Biol 31, 382–394. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fahy JV, and Dickey BF (2010). Airway mucus function and dysfunction. N Engl J Med 363, 2233–2247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fang Y, Liu H, Huang H, Li H, Saqi A, Qiang L, and Que J. (2020). Distinct stem/progenitor cells proliferate to regenerate the trachea, intrapulmonary airways and alveoli in COVID-19 patients. Cell Res 30, 705–707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ferrara N, Gerber HP, and LeCouter J. (2003). The biology of VEGF and its receptors. Nat Med 9, 669–676. [DOI] [PubMed] [Google Scholar]
- Fong TA, Shawver LK, Sun L, Tang C, App H, Powell TJ, Kim YH, Schreck R, Wang X, Risau W, et al. (1999). SU5416 is a potent and selective inhibitor of the vascular endothelial growth factor receptor (Flk-1/KDR) that inhibits tyrosine kinase catalysis, tumor vascularization, and growth of multiple tumor types. Cancer Res 59, 99–106. [PubMed] [Google Scholar]
- Gautier EL, Shay T, Miller J, Greter M, Jakubzick C, Ivanov S, Helft J, Chow A, Elpek KG, Gordonov S, et al. (2012). Gene-expression profiles and transcriptional regulatory pathways that underlie the identity and diversity of mouse tissue macrophages. Nat Immunol 13, 1118–1128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goldman O, Han S, Sourisseau M, Dziedzic N, Hamou W, Corneo B, D’Souza S, Sato T, Kotton DN, Bissig KD, et al. (2013). KDR identifies a conserved human and murine hepatic progenitor and instructs early liver development. Cell Stem Cell 12, 748–760. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gour N, and Wills-Karp M. (2015). IL-4 and IL-13 signaling in allergic airway disease. Cytokine 75, 68–78. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guha A, Deshpande A, Jain A, Sebastiani P, and Cardoso WV (2017). Uroplakin 3a(+) Cells Are a Distinctive Population of Epithelial Progenitors that Contribute to Airway Maintenance and Post-injury Repair. Cell Rep 19, 246–254. [DOI] [PubMed] [Google Scholar]
- Guseh JS, Bores SA, Stanger BZ, Zhou Q, Anderson WJ, Melton DA, and Rajagopal J. (2009). Notch signaling promotes airway mucous metaplasia and inhibits alveolar development. Development 136, 1751–1759. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hamada K, Oike Y, Takakura N, Ito Y, Jussila L, Dumont DJ, Alitalo K, and Suda T. (2000). VEGF-C signaling pathways through VEGFR-2 and VEGFR-3 in vasculoangiogenesis and hematopoiesis. Blood 96, 3793–3800. [PubMed] [Google Scholar]
- Hammad H, Chieppa M, Perros F, Willart MA, Germain RN, and Lambrecht BN (2009). House dust mite allergen induces asthma via Toll-like receptor 4 triggering of airway structural cells. Nat Med 15, 410–416. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harfe BD, Scherz PJ, Nissim S, Tian H, McMahon AP, and Tabin CJ (2004). Evidence for an expansion-based temporal Shh gradient in specifying vertebrate digit identities. Cell 118, 517–528. [DOI] [PubMed] [Google Scholar]
- Harris-Johnson KS, Domyan ET, Vezina CM, and Sun X. (2009). beta-Catenin promotes respiratory progenitor identity in mouse foregut. Proc Natl Acad Sci U S A 106, 16287–16292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hayashi T, Ishii A, Nakai S, and Hasegawa K. (2004). Ultrastructure of goblet-cell metaplasia from Clara cell in the allergic asthmatic airway inflammation in a mouse model of asthma in vivo. Virchows Arch 444, 66–73. [DOI] [PubMed] [Google Scholar]
- Holmes K, Roberts OL, Thomas AM, and Cross MJ (2007). Vascular endothelial growth factor receptor-2: structure, function, intracellular signalling and therapeutic inhibition. Cell Signal 19, 2003–2012. [DOI] [PubMed] [Google Scholar]
- Hong KU, Reynolds SD, Giangreco A, Hurley CM, and Stripp BR (2001). Clara cell secretory protein-expressing cells of the airway neuroepithelial body microenvironment include a label-retaining subset and are critical for epithelial renewal after progenitor cell depletion. Am J Respir Cell Mol Biol 24, 671–681. [DOI] [PubMed] [Google Scholar]
- Hong KU, Reynolds SD, Watkins S, Fuchs E, and Stripp BR (2004). Basal cells are a multipotent progenitor capable of renewing the bronchial epithelium. Am J Pathol 164, 577–588. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hou Z, Wu Q, Sun X, Chen H, Li Y, Zhang Y, Mori M, Yang Y, Que J, and Jiang M. (2019). Wnt/Fgf crosstalk is required for the specification of basal cells in the mouse trachea. Development 146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jacquet A, and Robinson C. (2020). Proteolytic, lipidergic and polysaccharide molecular recognition shape innate responses to house dust mite allergens. Allergy 75, 33–53. [DOI] [PubMed] [Google Scholar]
- Jiang M, Li H, Zhang Y, Yang Y, Lu R, Liu K, Lin S, Lan X, Wang H, Wu H, et al. (2017). Transitional basal cells at the squamous-columnar junction generate Barrett’s oesophagus. Nature 550, 529–533. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jolly S, Lang V, Koelzer VH, Sala Frigerio C, Magno L, Salinas PC, Whiting P, and Palomer E. (2019). Single-Cell Quantification of mRNA Expression in The Human Brain. Sci Rep 9, 12353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kathiriya JJ, Brumwell AN, Jackson JR, Tang X, and Chapman HA. (2020). Distinct Airway Epithelial Stem Cells Hide among Club Cells but Mobilize to Promote Alveolar Regeneration. Cell Stem Cell 26, 346–358 e344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim CF, Jackson EL, Woolfenden AE, Lawrence S, Babar I, Vogel S, Crowley D, Bronson RT, and Jacks T. (2005). Identification of bronchioalveolar stem cells in normal lung and lung cancer. Cell 121, 823–835. [DOI] [PubMed] [Google Scholar]
- Kim V, Kelemen SE, Abuel-Haija M, Gaughan JP, Sharafkaneh A, Evans CM, Dickey BF, Solomides CC, Rogers TJ, and Criner GJ (2008). Small airway mucous metaplasia and inflammation in chronic obstructive pulmonary disease. COPD 5, 329–338. [DOI] [PubMed] [Google Scholar]
- Klein Wolterink RG, Kleinjan A, van Nimwegen M, Bergen I, de Bruijn M, Levani Y, and Hendriks RW (2012). Pulmonary innate lymphoid cells are major producers of IL-5 and IL-13 in murine models of allergic asthma. Eur J Immunol 42, 1106–1116. [DOI] [PubMed] [Google Scholar]
- Koch S, and Claesson-Welsh L. (2012). Signal transduction by vascular endothelial growth factor receptors. Cold Spring Harb Perspect Med 2, a006502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kotton DN, and Morrisey EE (2014). Lung regeneration: mechanisms, applications and emerging stem cell populations. Nat Med 20, 822–832. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koyama S, Sato E, Haniuda M, Numanami H, Nagai S, and Izumi T. (2002). Decreased level of vascular endothelial growth factor in bronchoalveolar lavage fluid of normal smokers and patients with pulmonary fibrosis. Am J Respir Crit Care Med 166, 382–385. [DOI] [PubMed] [Google Scholar]
- Kuperman DA, Huang X, Koth LL, Chang GH, Dolganov GM, Zhu Z, Elias JA, Sheppard D, and Erle DJ (2002). Direct effects of interleukin-13 on epithelial cells cause airway hyperreactivity and mucus overproduction in asthma. Nat Med 8, 885–889. [DOI] [PubMed] [Google Scholar]
- Lafkas D, Shelton A, Chiu C, de Leon Boenig G, Chen Y, Stawicki SS, Siltanen C, Reichelt M, Zhou M, Wu X, et al. (2015). Therapeutic antibodies reveal Notch control of transdifferentiation in the adult lung. Nature 528, 127–131. [DOI] [PubMed] [Google Scholar]
- Le Cras TD, Acciani TH, Mushaben EM, Kramer EL, Pastura PA, Hardie WD, Korfhagen TR, Sivaprasad U, Ericksen M, Gibson AM, et al. (2011). Epithelial EGF receptor signaling mediates airway hyperreactivity and remodeling in a mouse model of chronic asthma. Am J Physiol Lung Cell Mol Physiol 300, L414–421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee JJ, McGarry MP, Farmer SC, Denzler KL, Larson KA, Carrigan PE, Brenneise IE, Horton MA, Haczku A, Gelfand EW, et al. (1997). Interleukin-5 expression in the lung epithelium of transgenic mice leads to pulmonary changes pathognomonic of asthma. J Exp Med 185, 2143–2156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li E, Knight JM, Wu Y, Luong A, Rodriguez A, Kheradmand F, and Corry DB (2019). Airway mycosis in allergic airway disease. Adv Immunol 142, 85–140. [DOI] [PubMed] [Google Scholar]
- Lynch TJ, Anderson PJ, Rotti PG, Tyler SR, Crooke AK, Choi SH, Montoro DT, Silverman CL, Shahin W, Zhao R, et al. (2018). Submucosal Gland Myoepithelial Cells Are Reserve Stem Cells That Can Regenerate Mouse Tracheal Epithelium. Cell Stem Cell 22, 653–667 e655. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Makinen T, Veikkola T, Mustjoki S, Karpanen T, Catimel B, Nice EC, Wise L, Mercer A, Kowalski H, Kerjaschki D, et al. (2001). Isolated lymphatic endothelial cells transduce growth, survival and migratory signals via the VEGF-C/D receptor VEGFR-3. EMBO J 20, 4762–4773. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marwick JA, Stevenson CS, Giddings J, MacNee W, Butler K, Rahman I, and Kirkham PA (2006). Cigarette smoke disrupts VEGF165-VEGFR-2 receptor signaling complex in rat lungs and patients with COPD: morphological impact of VEGFR-2 inhibition. Am J Physiol Lung Cell Mol Physiol 290, L897–908. [DOI] [PubMed] [Google Scholar]
- Miquerol L, Gertsenstein M, Harpal K, Rossant J, and Nagy A. (1999). Multiple developmental roles of VEGF suggested by a LacZ-tagged allele. Dev Biol 212, 307–322. [DOI] [PubMed] [Google Scholar]
- Mou H, Vinarsky V, Tata PR, Brazauskas K, Choi SH, Crooke AK, Zhang B, Solomon GM, Turner B, Bihler H, et al. (2016). Dual SMAD Signaling Inhibition Enables Long-Term Expansion of Diverse Epithelial Basal Cells. Cell Stem Cell 19, 217–231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mustonen T, and Alitalo K. (1995). Endothelial receptor tyrosine kinases involved in angiogenesis. J Cell Biol 129, 895–898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Olsson AK, Dimberg A, Kreuger J, and Claesson-Welsh L. (2006). VEGF receptor signalling - in control of vascular function. Nat Rev Mol Cell Biol 7, 359–371. [DOI] [PubMed] [Google Scholar]
- Pardo-Saganta A, Law BM, Gonzalez-Celeiro M, Vinarsky V, and Rajagopal J. (2013). Ciliated cells of pseudostratified airway epithelium do not become mucous cells after ovalbumin challenge. Am J Respir Cell Mol Biol 48, 364–373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park KS, Korfhagen TR, Bruno MD, Kitzmiller JA, Wan H, Wert SE, Khurana Hershey GK, Chen G, and Whitsett JA (2007). SPDEF regulates goblet cell hyperplasia in the airway epithelium. J Clin Invest 117, 978–988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rajavelu P, Chen G, Xu Y, Kitzmiller JA, Korfhagen TR, and Whitsett JA (2015). Airway epithelial SPDEF integrates goblet cell differentiation and pulmonary Th2 inflammation. J Clin Invest 125, 2021–2031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rane CK, Jackson SR, Pastore CF, Zhao G, Weiner AI, Patel NN, Herbert DR, Cohen NA, and Vaughan AE (2019). Development of solitary chemosensory cells in the distal lung after severe influenza injury. Am J Physiol Lung Cell Mol Physiol 316, L1141–L1149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rawlins EL, Okubo T, Xue Y, Brass DM, Auten RL, Hasegawa H, Wang F, and Hogan BL (2009). The role of Scgb1a1+ Clara cells in the long-term maintenance and repair of lung airway, but not alveolar, epithelium. Cell Stem Cell 4, 525–534. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reader JR, Tepper JS, Schelegle ES, Aldrich MC, Putney LF, Pfeiffer JW, and Hyde DM (2003). Pathogenesis of mucous cell metaplasia in a murine asthma model. Am J Pathol 162, 2069–2078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reynolds SD, Giangreco A, Power JH, and Stripp BR (2000). Neuroepithelial bodies of pulmonary airways serve as a reservoir of progenitor cells capable of epithelial regeneration. Am J Pathol 156, 269–278. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reynolds SD, Zemke AC, Giangreco A, Brockway BL, Teisanu RM, Drake JA, Mariani T, Di PY, Taketo MM, and Stripp BR (2008). Conditional stabilization of beta-catenin expands the pool of lung stem cells. Stem Cells 26, 1337–1346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rock JR, Gao X, Xue Y, Randell SH, Kong YY, and Hogan BL (2011). Notch-dependent differentiation of adult airway basal stem cells. Cell Stem Cell 8, 639–648. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rockich BE, Hrycaj SM, Shih HP, Nagy MS, Ferguson MA, Kopp JL, Sander M, Wellik DM, and Spence JR (2013). Sox9 plays multiple roles in the lung epithelium during branching morphogenesis. Proc Natl Acad Sci U S A 110, E4456–4464. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roy MG, Rahmani M, Hernandez JR, Alexander SN, Ehre C, Ho SB, and Evans CM (2011). Mucin production during prenatal and postnatal murine lung development. Am J Respir Cell Mol Biol 44, 755–760. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmid-Grendelmeier P, Altznauer F, Fischer B, Bizer C, Straumann A, Menz G, Blaser K, Wuthrich B, and Simon HU (2002). Eosinophils express functional IL-13 in eosinophilic inflammatory diseases. J Immunol 169, 1021–1027. [DOI] [PubMed] [Google Scholar]
- Shalaby F, Rossant J, Yamaguchi TP, Gertsenstein M, Wu XF, Breitman ML, and Schuh AC (1995). Failure of blood-island formation and vasculogenesis in Flk-1-deficient mice. Nature 376, 62–66. [DOI] [PubMed] [Google Scholar]
- Soeda T, Deng JM, de Crombrugghe B, Behringer RR, Nakamura T, and Akiyama H. (2010). Sox9-expressing precursors are the cellular origin of the cruciate ligament of the knee joint and the limb tendons. Genesis 48, 635–644. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sui P, Wiesner DL, Xu J, Zhang Y, Lee J, Van Dyken S, Lashua A, Yu C, Klein BS, Locksley RM, et al. (2018). Pulmonary neuroendocrine cells amplify allergic asthma responses. Science 360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tanizaki Y, Kitani H, Okazaki M, Mifune T, Mitsunobu F, and Kimura I. (1993). Mucus hypersecretion and eosinophils in bronchoalveolar lavage fluid in adult patients with bronchial asthma. J Asthma 30, 257–262. [DOI] [PubMed] [Google Scholar]
- Tata A, Kobayashi Y, Chow RD, Tran J, Desai A, Massri AJ, McCord TJ, Gunn MD, and Tata PR (2018). Myoepithelial Cells of Submucosal Glands Can Function as Reserve Stem Cells to Regenerate Airways after Injury. Cell Stem Cell 22, 668–683 e666. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsao PN, Wei SC, Wu MF, Huang MT, Lin HY, Lee MC, Lin KM, Wang IJ, Kaartinen V, Yang LT, and Cardoso WV (2011). Notch signaling prevents mucous metaplasia in mouse conducting airways during postnatal development. Development 138, 3533–3543. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tyner JW, Kim EY, Ide K, Pelletier MR, Roswit WT, Morton JD, Battaile JT, Patel AC, Patterson GA, Castro M, et al. (2006). Blocking airway mucous cell metaplasia by inhibiting EGFR antiapoptosis and IL-13 transdifferentiation signals. J Clin Invest 116, 309–321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van Winkle LS, Buckpitt AR, Nishio SJ, Isaac JM, and Plopper CG (1995). Cellular response in naphthalene-induced Clara cell injury and bronchiolar epithelial repair in mice. Am J Physiol 269, L800–818. [DOI] [PubMed] [Google Scholar]
- Vaughan AE, Brumwell AN, Xi Y, Gotts JE, Brownfield DG, Treutlein B, Tan K, Tan V, Liu FC, Looney MR, et al. (2015). Lineage-negative progenitors mobilize to regenerate lung epithelium after major injury. Nature 517, 621–625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vila Ellis L, Cain MP, Hutchison V, Flodby P, Crandall ED, Borok Z, Zhou B, Ostrin EJ, Wythe JD, and Chen J. (2020). Epithelial Vegfa Specifies a Distinct Endothelial Population in the Mouse Lung. Dev Cell 52, 617–630 e616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Volckaert T, Dill E, Campbell A, Tiozzo C, Majka S, Bellusci S, and De Langhe SP (2011). Parabronchial smooth muscle constitutes an airway epithelial stem cell niche in the mouse lung after injury. J Clin Invest 121, 4409–4419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang C, Xie J, Zhao L, Fei X, Zhang H, Tan Y, Nie X, Zhou L, Liu Z, Ren Y, et al. (2020). Alveolar macrophage dysfunction and cytokine storm in the pathogenesis of two severe COVID-19 patients. EBioMedicine 57, 102833. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang W, Cohen JA, Wallrapp A, Trieu KG, Barrios J, Shao F, Krishnamoorthy N, Kuchroo VK, Jones MR, Fine A, et al. (2019). Age-Related Dopaminergic Innervation Augments T Helper 2-Type Allergic Inflammation in the Postnatal Lung. Immunity 51, 1102–1118 e1107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Whitcutt MJ, Adler KB, and Wu R. (1988). A biphasic chamber system for maintaining polarity of differentiation of cultured respiratory tract epithelial cells. In Vitro Cell Dev Biol 24, 420–428. [DOI] [PubMed] [Google Scholar]
- Whitsett JA (2018). Airway Epithelial Differentiation and Mucociliary Clearance. Ann Am Thorac Soc 15, S143–S148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wills-Karp M, Luyimbazi J, Xu X, Schofield B, Neben TY, Karp CL, and Donaldson DD (1998). Interleukin-13: central mediator of allergic asthma. Science 282, 2258–2261. [DOI] [PubMed] [Google Scholar]
- Yasuda Y, Nagano T, Kobayashi K, and Nishimura Y. (2020). Group 2 Innate Lymphoid Cells and the House Dust Mite-Induced Asthma Mouse Model. Cells 9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yeh ET, Zhang S, Wu HD, Korbling M, Willerson JT, and Estrov Z. (2003). Transdifferentiation of human peripheral blood CD34+-enriched cell population into cardiomyocytes, endothelial cells, and smooth muscle cells in vivo. Circulation 108, 2070–2073. [DOI] [PubMed] [Google Scholar]
- Yuan T, Volckaert T, Redente EF, Hopkins S, Klinkhammer K, Wasnick R, Chao CM, Yuan J, Zhang JS, Yao C, et al. (2019). FGF10-FGFR2B Signaling Generates Basal Cells and Drives Alveolar Epithelial Regeneration by Bronchial Epithelial Stem Cells after Lung Injury. Stem Cell Reports 12, 1041–1055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zacharias WJ, Frank DB, Zepp JA, Morley MP, Alkhaleel FA, Kong J, Zhou S, Cantu E, and Morrisey EE (2018). Regeneration of the lung alveolus by an evolutionarily conserved epithelial progenitor. Nature 555, 251–255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou-Suckow Z, Duerr J, Hagner M, Agrawal R, and Mall MA (2017). Airway mucus, inflammation and remodeling: emerging links in the pathogenesis of chronic lung diseases. Cell Tissue Res 367, 537–550. [DOI] [PubMed] [Google Scholar]
- Zhu Z, Homer RJ, Wang Z, Chen Q, Geba GP, Wang J, Zhang Y, and Elias JA (1999). Pulmonary expression of interleukin-13 causes inflammation, mucus hypersecretion, subepithelial fibrosis, physiologic abnormalities, and eotaxin production. J Clin Invest 103, 779–788. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The single cell RNA-sequencing datasets of airway epithelium isolated from NAPH- or vehicle-treated mice has been deposited to the Expression Omnibus: GSE171571.
The authors declare that the main data supporting the findings of this study are available within the paper and its supplementary information. Any additional data are available from the corresponding authors upon request.