Abstract
Since a substantial difference in the prevalence of genetic causes of rod-cone dystrophy (RCD) was found among different populations, we conducted a systematic review of the genetic findings associated with RCD in Arab countries. Of the 816 articles retrieved from PubMed, 31 studies conducted on 407 participants from 11 countries were reviewed. Next-generation sequencing (NGS) was the most commonly used technique (68%). Autosomal recessive pattern was the most common pattern of inheritance (97%) and half of the known genes associated with RCD (32/63) were identified. In the Kingdom of Saudi Arabia, in addition to RP1 (20%) and TULP1 (20%), gene defects in EYS (8%) and CRB1 (7%) were also prevalently mutated. In North Africa, the main gene defects were in MERTK (18%) and RLBP1 (18%). Considering all countries, RP1 and TULP1 remained the most prevalently mutated. Variants in TULP1, RP1, EYS, MERTK, and RLBP1 were the most prevalent, possibly because of founder effects. On the other hand, only ten Individuals were found to have dominant or X-linked RCD. This is the first time a catalog of RCD genetic variations has been established in subjects from the Arabi countries. Although the last decade has seen significant interest, expertise, and an increase in RCD scientific publication, much work needs to be conducted.
Subject terms: Diagnostic markers, Risk factors
Introduction
Rod-cone dystrophy (RCD), also known as retinitis pigmentosa (RP), is a heterogeneous group of inherited disorders affecting primary rod photoreceptors in the majority of cases with secondary cone degeneration [1, 2]. The dysfunction in rod photoreceptors causes night blindness followed by progressive visual field constriction, abnormal color vision, and eventually loss of central vision due to cone photoreceptor involvement [1, 2].
The worldwide prevalence of RCD is around 1:4000 individuals [2]. RCD is transmitted as a Mendelian disorder, where the phenotype is usually caused by variant(s) in a single gene and can be inherited in an autosomal recessive (ar) (50–60%), autosomal dominant (ad) (30–40%), or X-linked (xl) pattern (5–15%) with also rare cases of mitochondrial transmission [2, 3]. RCD is exceptionally heterogeneous [4]. At the genotype level, variants in different genes may cause the same phenotype and numerous disease-causing variants are reported in each gene [4]. At the phenotype level, different variants in the same gene may cause different clinical consequences and the same variant may produce different phenotypes even among siblings [4]. Until today, more than 89 genes were associated with three modes of inheritance of RCD (https://sph.uth.edu/retnet/; last accessed on 14 February 2020). Of note, variants in six genes that are NR2E3, NRL, RHO, RP1, RPE65, and SAG are simultaneously found to cause both dominant and recessive forms (https://sph.uth.edu/retnet/).
Particular gene defects were reported to be responsible for significant proportions of the disease; most of these reports focused on European and North American populations. For instance, RHO variants are the most prevalent cause of adRCD in those populations (29–40%) [3, 5]. Besides, RP1 is mutated in 5–10% of adRCD cases [6, 7]. Variants in USH2A that can also cause Usher syndrome are one of the most frequent causes of autosomal recessive rod-cone dystrophy (arRCD) (10–23%) [8–10]. EYS variants account also for about 10–20% of arRCD cases [11, 12]. For instance, these gene defects are the most common cause of RCD in the Japanese population [13, 14]. As for the least frequently inherited xlRCD, RPGR variants are responsible for more than 70% of all cases [15]. Genetic studies showed a substantial difference in the genetic causes of RCD when cohorts of different populations were compared [8–10]. This might be true for Arab countries having large family size, older parental age (for both genders), high consanguinity rate (40–68%), and first-cousin marriages [16]. Therefore, we conducted a systematic analysis of the entire genetic findings of RCD in Arab countries. The aims of this analysis were to identify the prevalence of causative genes per country and overall (all Arab countries), and to detect the most prevalent variants with possible founder effect in the Arab countries.
Methods
The current review was conducted in adherence to the Preferred Reporting Items for Systematic Review and Meta-Analyses guidelines [17]. The protocol was registered in the International prospective register of systematic reviews (PROSPERO) as “The genetics of RCD in Arab countries: a systematic review (CRD42018086992)”.
Inclusion and exclusion criteria
All studies focusing on the genetics of RCD were included if they met the following criteria: (1) Any report including RCD phenotype that was diagnosed according to standard ophthalmic examinations even if it was among other cases of retinal dystrophies; (2) original articles written in English and having a full text.
Studies were excluded if (1) not reporting genetic variants. (2) Reporting individuals that belong to non-Arab Middle Eastern countries (i.e. Iran, Turkey) or those where a country is not specified (Middle East region, Africa). (3) Not in English or (4) not on humans. (5) Not dealing with RCD or RP (i.e. Stargardt, cone-rod dystrophy, Leber congenital amaurosis (LCA), Usher syndrome, Bardet–Biedl Syndrome, and other syndromic dystrophies). (6) No full text was available.
Search strategy and information source
LJ and SES performed independently the literature search. PubMed database was systematically screened using six combinations of MeSH terms (Supplementary Table 3). LJ and SES also performed a manual search using free text search terms (i.e., RCD or RP and Arab countries) to retrieve other articles that had not been identified via the initial search strategy. Moreover, the reference lists of articles were carefully checked to ensure that all relevant studies had been identified. The publication date was not considered an exclusion criterion.
Data collection process and data items
LJ and SES authors independently assessed the title and abstract of each paper for language suitability and subject matter relevance. The papers thereby selected were then scrutinized for their appropriateness for inclusion. The first author, year of publication, family/sample ID, sample size, variant, status, gene, Refseq accession numbers for mRNA (NM), and country are shown in Supplementary Table 1 and were submitted to ClinVar database (accession numbers: SCV001434591–SCV001434724).
Results and discussion
The initial search retrieved 816 papers that were filtered to leave 37 human genetics papers (Fig. 1). When screened according to content, 7 more were excluded, leaving 30 articles published from 2001 till the first of February 2020 (Fig. 2A). During the first decade (2001 till 2010), only 5 articles were published and this number increased by 5× in the second decade (2010 till 2020) (Fig. 2A).
Fig. 1. Flow chart for identifying eligible articles.
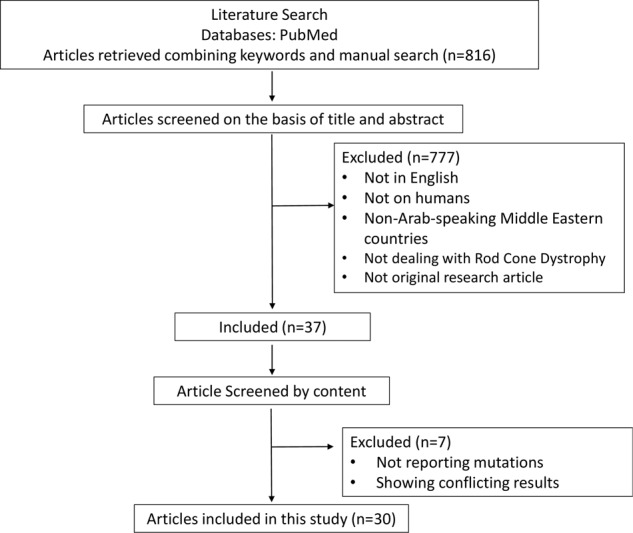
The initial search retrieved 816 papers that were filtered to leave 37 human genetics papers. When screened according to content, seven more were excluded, leaving 30 articles published from 2001 till the first of February 2020.
Fig. 2. Articles and variant identification techniques.
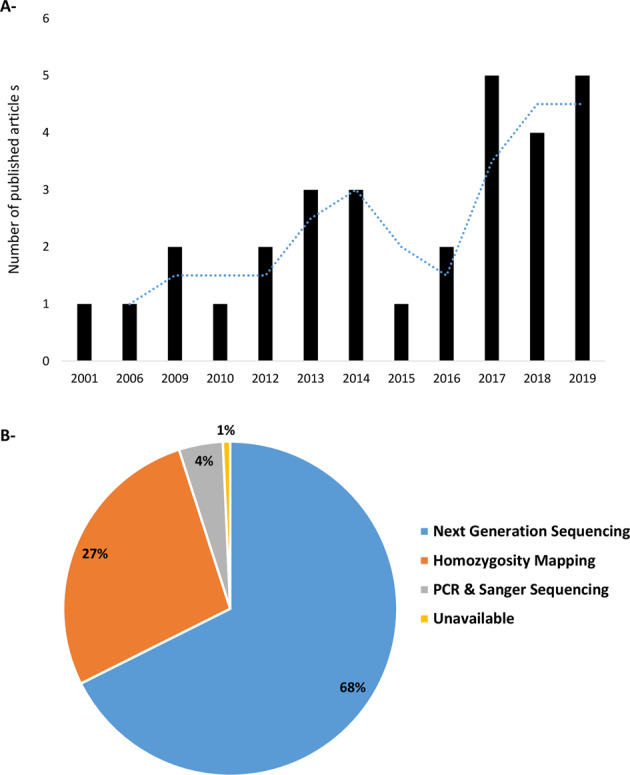
A Number of articles investigating the genetics of rod-cone dystrophy in Arab speaking countries until the first of February 2020. B Variant identification techniques used to report disease-causing variants.
When individuals were stratified according to the screening method; next-generation sequencing (NGS) was the most commonly used (68%) followed by homozygosity mapping (27%). Sanger sequencing as a first test was the less common (4%, Fig. 2B). This result is not surprising because NGS platforms have become widely available, reducing the cost of DNA sequencing [18].
It is noteworthy to mention that all studies performing NGS have validated the identified variants by Sanger sequencing. Variant co-segregation in available family members was also performed in the majority of the studies (Table 1). Bioinformatics analysis varied between studies, while many such as Audo et al. [19, 20], Gerth-Kahlert et al. [21], Patel et al. [22, 23], and Habibi et al. [24, 25] performed a detailed bioinformatics analysis (Table 1), others did not mention any bioinformatics analysis (Table 1).
Table 1.
Co-segregation analysis, in silico pathogenicity assessment and genotyping of controls in included studies.
| Authors | Year | Segregation analysis | In silico pathogenicity assessment | Genotyping of controls | ||
|---|---|---|---|---|---|---|
| Conservation across species | MAF and zygosity search in public databases (Gnomad, ExAC, 1000 Genomes,..) | Prediction tool (s) for missense | ||||
| Humbert et al. | 2006 | No | N.A | − | +, Unspecified tool | + |
| Mackay et al. | 2010 | Yes | N.A | − | − | + |
| Abu-safieh et al. | 2013 | Yes | + | − | + | + |
| Al rashed et al. | 2012 | Yes | − | − | +, Unspecified tool | − |
| Dessalces et al. | 2013 | Yes | N.A | − | − | + |
| Davidson et al. | 2013 | Yes | + | + | + | + |
| Coppieters et al. | 2014 | Yes | − | No | + | − |
| Méjécase et al. | 2017 | No | + | + | + | − |
| Gerth et al. | 2017 | Yes | − | + | +, Unspecified tool | − |
| Astuti et al. | 2018 | No | + | + | + | + |
| Albarry et al. | 2019 | Yes | − | − | +, Unspecified tool | − |
| Abu-Ameerh et al. | 2019 | Yes | + | + | + | − |
| Azab et al. | 2019 | Yes | − | + | + | − |
| Patel et al. | 2016 | Yes | + | + | + | − |
| Patel et al. | 2018 | Yes | − | + | + | − |
| Khan et al. | 2015 | No | N.A | − | + | + |
| Katsanis et. al | 2001 | Yes | + | − | − | + |
| Aldahmesh et al. | 2009 | Yes | + | − | − | + |
| Nair et al. | 2017 | Yes | N.A | + | N.A | − |
| Hmani-Aifa et al. | 2009 | Yes | + | − | − | + |
| Ksantini et al. | 2012 | Yes | − | − | − | + |
| Habibi et al. | 2017 | Yes | − | + | + | − |
| Habibi et al. | 2016 | Yes | + | + | + | − |
| Audo et al. | 2018 | Yes | + | + | + | − |
| Audo et al. | 2017 | Yes | + | + | + | − |
| Zobor et al. | 2014 | Yes | − | − | − | + |
| Hashmi et al. | 2018 | Yes | N.A | − | N.A | − |
| Jalkh et al. | 2014 | Yes | − | − | + | − |
| Khan et al. | 2019 | Yes | − | − | − | − |
| Khan AO | 2019 | Yes | − | − | − | − |
In many studies, the conservation across species and predictions tools were not applicable since the identified variants were either insertion/deletions, splice sites, or nonsense.
MAF minor allele frequency, + yes, − no, N.A not applicable.
Variant spectrum in sporadic and autosomal recessive cases of rod-cone dysrophy
Among the identified individuals, 46 individuals were excluded because they were either reported in different studies making them duplicates or from unspecified country (Supplementary Table 4); this left 407 individuals with sporadic or arRCD (Supplementary Table 1).
To study the phenotype–genotype correlations and prevalence of gene defects in arRCD, we have exclusively included the genes known for their association with arRCD in RetNet (https://sph.uth.edu/RETNET/, last accessed on February 14, 2020). Using these criteria, a total of 356 individuals carrying arRCD variants remained for further analysis. All the details of the analysed articles are shown in Supplementary Table 1.
The ar mode was the most common pattern of inheritance where out of the 63 known genes associated with RCD in RetNet, 33 (52%) were identified. The spectrum included missense (30%), nonsense (29%), deletion (24%), splice-site (6%), insertion (5%), duplication (5%), and exon deletion (2%) variants (Fig. 2).
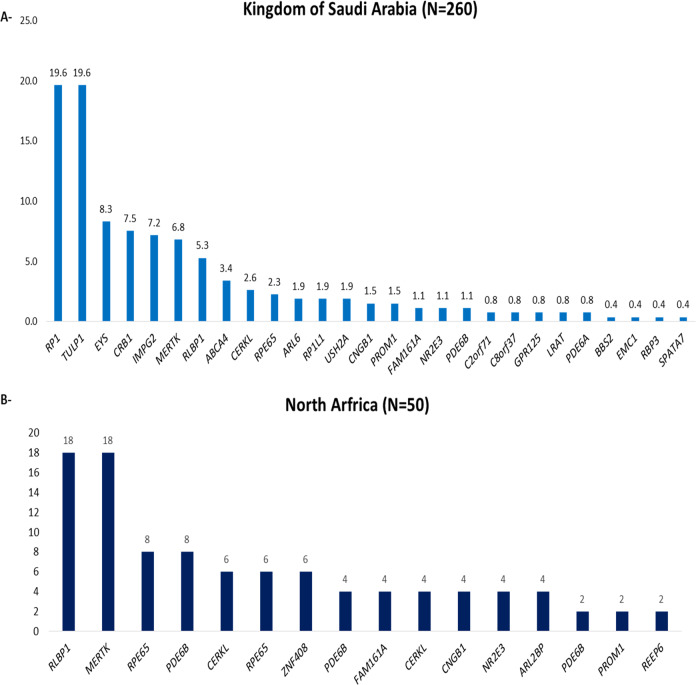
In 260 individuals from Kingdom of Saudi Arabia (KSA), we found that variants in RP1 (~20%), TULP1 (~20%), EYS (~8%), CRB1 (~7%), MERTK (~7%), and RLBP1 (~5%) account for two-thirds of the known variants (Fig. 3A).
Fig. 3. Variants spectrum in analyzed individuals from Kingdom of Saudi Arabia and North Africa.
A In individuals from Kingdom of Saudi Arabia (KSA), variants in RP1 (~20%), TULP1 (~20%), EYS (~8%), CRB1 (~7%), MERTK (~7%), and RLBP1 (~5%) account for two-thirds of the known variants. B In North Africa, the main identified genes defects were harbored in: MERTK (18%), RLBP1 (18%), RPE65 (8%), and PDE6B (8%), accounting for half of the solved cases.
In North Africa, the main identified genes defects were harbored in: MERTK (18%), RLBP1 (18%), RPE65 (8%), and PDE6B (8%), accounting for half of the solved cases (Fig. 3B). No analysis was performed on the Levantine (Palestinians, Jordanians, Lebanese, and Syrians) because the sample size was limited and thus no conclusive results can be drawn.
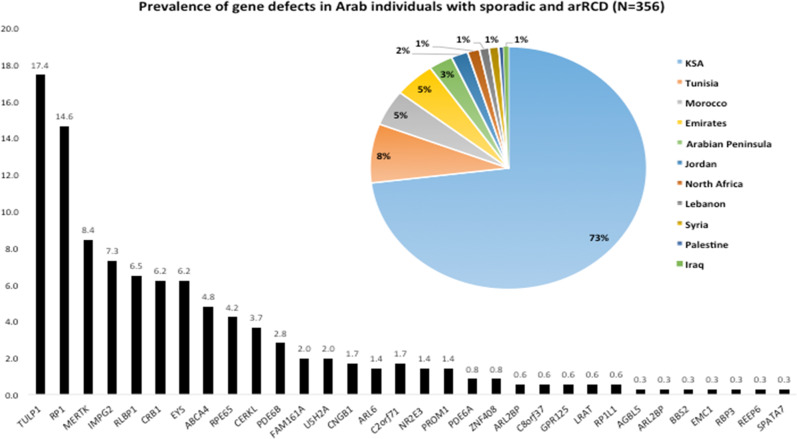
Considering all countries, we found that variants in RP1 (17%), TULP1 (14%), MERTK (8%), IMPG2 (7%), and RLBP1 (6%) were the most prevalently mutated genes accounting for half of the known genes. Variants in other genes such as EYS (6%), ABCA4 (5%), PDE6B (3%), family with sequence similarity 161, member A (FAM161A) (2%), USH2A (2%), PCARE (1.5%), and PDE6A (1%) had a minor implication (Fig. 4).
Fig. 4. A histogram showing gene prevalence in Arab individuals with sporadic and autosomal recessive rod-cone dystrophy, along with a pie-chart showing their repartition in the Arab countries.
Pie-chart shows the repartition of different individuals making part of the prevalence study in the Arab countries.
RP1 variants were initially reported for underlying adRCD [26–28]; however, since 2005, reports have shed light on their association to arRCD [29]. RP1 variants were shown to account for ~5.5% and ~1% of adRCD and arRCD cases, respectively [29, 30], a prevalence significantly lower than the one found in the arRCD Arab patients included in this review (~17%). Interestingly, Avila-Fernandez et al. reported a founder nonsense RP1 variant in the Spanish population p.(Ser542∗) responsible for 4.5% of arRCD cases [31]. Along with the current results, this suggests that RP1 variants are more prevalent in arRCD than previously thought.
In addition to RP1, variants in TULP1 were found in a significant proportion of the reported Arab individuals (~14%), while they account for 1–2% of arRCD cases in different ethnic background worldwide including Pakistanis [32]. Among the disease-causing variants in TULP1, the p.(Lys498Arg) was reported as the most abundant RCD-associated allele with a particular prevalence in German and Pakistani populations [32]. This missense change has not yet been reported in any participant from the Arab populations, instead, c.901C>T; p.(Gln301*) is the most prevalent allele reported in at least 95% of individuals with TULP1 variants [22, 23, 33, 34].
MERTK variants were also associated with 8.4% of the reported arRCD Arab individuals. These numbers are significantly higher than previous reports showing MERTK variants ranging from 1% [35] to ∼4% [36] in other populations. In contrast, MERTK variants are responsible for up to 30% in the Faroe Islands because of a founder deletion spanning exons 1–7 [37]. Comparing our results with Patel et al. [22], we find a higher prevalence (8.4% vs. 3%); the reason may be that in our meta-analysis, MERTK variants were also reported in seven different studies including individuals from North Africa, Lebanon, and Emirates (Supplementary Table 1).
In contrast with previous studies reporting USH2A as a major arRCD gene in Europeans, only 2% of Arab individuals harbored variants in USH2A (Fig. 4) in our analysis implying that it has a minor implication in Arabs [8]. Additionally, ~6.2% had variants in EYS (Fig. 4), that was reported to be the major causativ gene for RCD in French [11] and Japanese populations [13, 14]. These findings highlight differences in the genetic variations in Arab countries in comparison with other countries or populations, which can be due to the presence of distinct founder variants. This may indicate that Arabs have different RCD mutational spectra, but this needs to be validated via more and larger studies.
Studying the country of origin revealed that 73% of the participants were from KSA, 13% from North African countries (8 and 5% from Tunisia and Morocco respectively and 1.5% unspecified), 4% from Emirates, 6% equally divided between Lebanon and Arabian Peninsula (Fig. 4). The remaining 4% were from Jordan, Iraq, Syria, and Palestine (Fig. 4). The high number of publication records from KSA is not surprising because estimates suggest that Arab countries spend in the order of 10 billion dollars on research, of which half is for the Gulf States, one-third for Egypt and Levant and the remaining one-fifth for the Maghreb [38].
Sporadic and autosomal recessive rod-cone dysrophy
Genes known to be associated with autosomal recessive rod-cone dysrophy
All variants described below are located in genes identified in RetNet as associated with RCD. Most of the variants associated with arRCD were homozygous (97%), except a few that were compound heterozygous (3%). The details related to these variants are available in Supplementary Table 1, noting that we checked and adjusted the nomenclature when needed, according to Mutalyzer tool [39] (Supplementary Table 1).
ATP-binding cassette, subfamily A (ABCA4) (MIM: 601691)
In 2013, Abu-Safieh et al. investigated four Saudi Arabians cases using whole exome sequencing (WES) and reported two missense variants: c.4316G>A; p.(Gly1439Asp) and c.4793C>A; p.(Ala1598Asp) [33]. From 2016 to 2018, Patel et al. identified one deletion; c.5391_5392del; p.(Ala1798*), one duplication; c.1630_1633dup; p.(Asn545Argfs*12), and three compound heterozygous variants; [c.2815G>T];[c.1615del]; p.[(Glu939*);(Leu539Serfs*29)], [c.1140T>A];[c.5642C>G]; p.[(Asn380Lys);(Ala1881Gly)], [c.3482G>A];[c.5917del]; p.[(Arg1161His);(Val1973*)] [22, 23]. Recently, Abu-Ameerh identified a splice-site donor variant; c.5460 + 1G>A predicted to cause exon skipping in a family affected with RCD [40]. Since this variant is known to be associated with Stargardt disease [41], the fact that it is also associated with RCD expands the phenotypic spectrum for ABCA4 variants.
ATP/GTP binding protein like 5 (AGBL5) (MIM: 615900)
Recently, a frameshift duplication; c.1255dup; p.(Thr419Asnfs*32) in AGBL5 was reported to be associated with RCD in one Emirati individual [42].
ADP ribosylation factor like GTPase 2 binding protein (ARL2BP) (MIM: 615407)
Davidson et al. identified the first variant in ARL2BP in an Arab-Muslim family consisting of three affected siblings [43]. Specifically, a splice-site variant; c.101-1G>C, was identified in a large interval using homozygosity mapping followed by WES [43]. Audo et al., in 2017, reported a second splice-site variant; c.207+1G>T carried by a Moroccan family with two affected young sisters diagnosed in their teens [19].
ADP ribosylation factor like GTPase 6 (ARL6) (MIM: 608845)
Two missense variants were identified; c.266C>T; p.(Ala89Val) and c.362G>A; p.(Arg121His) in individuals from Saudi Arabia [22, 44].
Bardet–Biedl syndrome 2 (BBS2) (MIM: 606151)
Only one missense variant; c.943C>T; p.(Arg315Trp) in BBS2 was found, it was carried by a Saudi Arabian individual [22]. This genotype-RCD data should be taken with caution since BBS2 is known for its association with syndromic forms of RCD.
Ceramide kinase-like (CERKL) (MIM: 608381)
Abu-Safieh et al. investigated the DNA of Saudi Arabian using WES and reported a missense variant; c.734T>C; p.(Leu245Pro) in CERKL [33]. Two years later, p.(Leu245Pro) was also identified in another Saudi Arabian individual along with a nonsense variant c.999C>A; p.(Cys333*) [45].
Not far from Leu245Pro, Patel et al. in 2016 identified a missense variant; c.890T>C; p.(Ile297Thr), in addition to a splice-site variant; c.238+1G>A, both carried by Saudi Arabian individuals [22]. Habibi et al. investigated four Tunisian families having four affected index individuals (three males and a female, age of onset 4–25 years) and identified a splice-site deletion; c.1151 + 3_1151 + 6del [24]. This variant was first identified by WES and Sanger sequencing; additional genotyping revealed the same deletion in three seemingly unrelated families [24]. Recently, an additional variant; c.1187_1188del; p.(Gln396Argfs*20) was detected in a Jordanian individual [46].
Cyclic nucleotide gated channel beta 1 (CNGB1) (MIM: 600724)
The first variant in CNGB1; c.2957A>T; p.(Lys986Met) was identified by Abu-Safieh et al. in Saudi Arabian individuals using WES [33]. Two missense variants; c.2294G>T; p.(Arg765Leu) and c.2957A>T; p.(Asn986Ile) were identified by Patel et al. in 2016 in Saudi Arabian individuals [22]. The same year, Habibi et al. reported an additional missense variant in an affected Tunisian female (age of onset 6 years); c.2293C>T; p.(Arg765Cys) [24].
Crumbs 1 (CRB1) (MIM: 604210)
Aldahmesh et al. in 2009 identified two homozygous variants; c.3159T>G; p.(Cys1053Trp) and c.80G>C; p.(Cys27Phe) in CRB1 among two consanguineous Saudi families [44]. Four missense variants; c.80G>T; p.(Cys27Phe), c.1429G>A; p.(Gly477Arg), c.3495T>G; p.(Cys1165Trp), c.2234C>T; p.(Thr745Met) and one nonsense; c.2024G>A; p.(Trp675*) were found by Abu Safieh et al. [33]. A year later, in a large Lebanese family having two branches each with a specific retinal dystrophy disease; RCD and LCA, Jalkh et al. performed homozygosity mapping followed by Sanger sequencing [47]. This approach identified a deletion; c.1772_1775del; p.(Cys591Serfs*29) in CRB1 carried by two affected females with RCD, diagnosed at a young age (10–12 years old) [47]. Finally, a frameshift deletion; c.2330_2336del; p.(Pro777Leufs*4) and three additional missense; c.1180T>C; p.(Cys394Arg), c.2701G>T; p.(Val901Phe), and c.1463T>C; p.(Phe488Ser) were reported by Patel et al. in 2016 [25]. Thus, except for the c.1772_1775delGCAT; p.(Cys591Serfs*29) found in Lebanese, all other CRB1 variants were found in Saudi Arabian individuals.
Chromosome 8 open reading frame 37 (C8orf37) (MIM: 614477)
One missense; c.529C>T; p.(Arg177Trp) was identified in two Saudi Arabian individuals [22].
ER membrane protein complex subunit 1 (EMC1) (MIM: 616846)
One missense; c.430G>A; p.(Ala144Thr) was identified in a Saudi Arabian individual [33].
Eyes shut homolog (EYS) (MIM: 612424)
Abu-Safieh et al. reported a missense variant; c.6050G>T, p.(Gly2017Val) and a frameshift duplication c.32dup, p.(Met12Aspfs*14) [33]. Two additional frameshift variants; c.179del; p.(Leu60Trpfs*3) and c.875_888delinsTTT; p.(Glu292Valfs*17) were reported by Patel et al. [22, 23]. Hashmi et al. in 2018 performed WES to detect the underlying genetic defect in a large Saudi Arabian family with 12 affected (nine males and three females) individuals [48]. WES identified a 1-bp insertion in EYS; c.910dup; p.(Trp304Leufs*9) [48].
Family with sequence similarity 161, member A (FAM161A) (MIM: 613596)
Zobor et al. in 2014 applied homozygosity mapping in a consanguineous Palestinian family with three siblings affected (two males and one female) diagnosed at age <27 years. This approach identified several homozygous genomic regions including FAM161A [49]. Subsequent Sanger sequencing analysis revealed the presence of a nonsense variant; c.1003C>T; p.(Arg335*) in the index patient and the affected sister [49]. Co-segregation revealed that both the father and one of the unaffected brothers are heterozygous for the variant whereas the other unaffected brother does not carry the variant. Arg335* is predicted to result in a truncated polypeptide that lacks the carboxyterminus of the FAM161A genuine protein [49]. Two years later using IROme technology, Habibi et al., identified a frameshift deletion; c.678_681del; p.(Lys227Asnfs*17) in a Tunisian female index (age of onset 12 years) [24]. In the same year, Patel et al. identified a nonsense variant; c.685C>T; p.(Arg229*) associated with arRCD in three individuals [22].
G protein-coupled receptor 125 (GPR125) (MIM: 612303)
The same missense; c.2504C>G; p.(Ser835Cys), was identified twice by Abu-Safieh et al. [33] and Patel et al. [22] in two Saudi Arabian individuals.
Interphotoreceptor matrix proteoglycan 2 (IMPG2) (MIM: 607056)
Until 2016, three variants: a frameshift deletion in IMPG2; c.2346_2347del, p.(Arg782Serfs*24) and two nonsense c.2274G>A; p.(Trp758*) and c.513T>G; p.(Tyr171*) were identified among nine cases of RCD [22, 33]. In 2018, an additional deletion; c.909-802 _1154-539del; p.(Gly304Glufs*9) was reported in two different KSA patients [23]. In seven Emirati individuals with early onset RCD, all issued from first-cousin marriages, Khan et al. uncovered three additional variants; c.189dup; p.(Gln64Thrfs*9) (in five individuals), c.533 + 4_533 + 7del (in one individual) and c.3262C>T; p.(Arg1088*) (in one individual) [50].
Lecithin retinol acyltransferase (LRAT) (MIM: 604863)
Two frameshift deletions; c.233_242del; p.(Leu78Argfs*85) and c.241_242del; p.(Leu81Aspfs*40) in LRAT were subsequently identified by Patel et al. in 2016 and 2018 [22, 23].
Tyrosine kinase protooncogene (MERTK) (MIM: 604705)
Aldahmesh et al. in 2009 identified a small frameshift deletion; c.1335_1336del; p.(Ala446Serfs*28) in MERTK in a consanguineous Saudi Arabian family [44]. Mackay et al. in 2010, identified a ~9 kb deletion in the eighth exon c.(1144 + 1_1145-1)_(1296 + 1_1297-1) delin a Saudi Arabian family [51]. Haplotyping analysis revealed the deletion was heterozygous in parents and the unaffected siblings, and homozygous in the two affected ones [51]. Screening of 100 Saudi Arabian probands with arRCD identified a second family with the same deletion. All affected members of the second family were homozygous for this deletion [51]. In 2012, Ksantini et al. used homozygosity mapping followed by Sanger sequencing in a consanguineous Moroccan family [52]. This approach showed that the three young (7–18 years) affected patients (two females and one male) had a missense variant; c.2323C>T; p.(Arg775*) [52]. Segregation analysis showed that unaffected brothers, sister, parents, and paternal grandfather were heterozygous for this variant [52]. A recurrent frameshift deletion; c.2214del; p.(Cys738Trpfs*32) in addition to two nonsense and two splice-site variants; c.325A>T; p.(Lys109*), c.2262C>G; p.(Tyr754*), c.1604 + 2T>G and c.2189 + 1G>T were also identified by Patel et al. in Saudi Arabians using NGS [22]. Audo et al. reported different classes of variants including frameshift; c.1301_1302del; p.(Glu434Alafs*40), splice-site; c.2079 + 2T>G, c.1604 + 2T>G, missense; c.2219C>T; p.(Ala740Val), and nonsense; c.1951C>T; p.(Arg651*) variants in individuals from different Arab countries [20]. Noting that variants c.2079 + 2T>G and c.1951C>T; p.(Arg651*) were identified as compound heterozygous [20].
Nuclear receptor subfamily 2, group E, member 3 in (NR2E3) (MIM: 604485)
A frameshift deletion in NR2E3; c.951del; p.(Thr318Argfs*6) was reported by Abu Safieh et al. in 2013 using WES in two affected individuals from Saudi Arabia [33]. In 2016, Habibi et al. identified a missense variant; c.932G>A; p.(Arg311Gln) in a Tunisian family having a male index individual (age of onset 4 years) [24]. The genotype–phenotype associations identified for NR2E3 should be taken with caution since this gene is known for its association with Enhanced S-cone dystrophy.
Photoreceptor cilium actin regulator (PCARE) (MIM: 613425)
Using autozygome guided sequencing, Abu-Safieh et al. reported a frameshift deletion; c.1525del; p.(Thr509Leufs*32) carried by a Saudi Arabian individual [33]. Gerth-Kahlert et al. in 2017 investigated the DNA of a Lebanese/Armenian individuals with RCD and identified a compound heterozygous variant [c.802C>T]; [c.2756_2768del]; p.[(Gln268*);(Lys919Thrfs*2)] [21]. Two additional frameshift deletion were recently identified; c.1377del; p.(Phe459Leufs*39) and c.2967del; p.(Val990Trpfs*45) in Iraqi and Emirati individuals respectively [23].
Phosphodiesterase 6A (PDE6A) (MIM: 180071)
A missense variant; c.304C>A; p.(Arg102Ser), was identified in Saudi patients by Abu Safieh et al. in 2013 [33]. Nair et al. in 2017 identified a 2-bp deletion; c.1358_1359del, resulting in a frameshift and premature termination p.(Ile453Serfs*8) in an 8-year-old male Emirati patient with arRCD [53].
Phosphodiesterase 6B (PDE6B) (MIM: 180072)
In an extended consanguineous Tunisian family where RCD and USH segregate each in a specific family branch, linkage analysis followed by Sanger sequencing were conducted by Hmani-Aifa et al. in 2009 [54]. This approach revealed a variant; c.2419T>A; p.(Trp807Arg) [54]. In a Moroccan family with arRCD, Coppieters et al. in 2014 identified an insertion-deletion variant; c.121_125del; p.(Pro41Glufs*123) using homozygosity mapping followed by WES [55]. Habibi et al., in 2016 investigated a Tunisian family having a male index individual (age of onset 4 years) [24]. Using IROme technology, the authors identified a missense variant; c.1010A>G; p.(His337Arg) [24]. In Saudi Arabia, a splice-site; c.992 + 1G>A and a nonsense variant; c.810C>A; p.(Cys270*) were reported [22, 33].
Prominin 1 (PROM1) (MIM: 604365)
Abu-Safieh et al. reported a nonsense variant in PROM1; c.1530C>G, p.(Tyr510*) in a Saudi Arabian individual [33]. Three years later, three additional variants; c.2130 + 2del; c.604C>G; p.(Arg202Gly) and c.1354dup; p.(Tyr452Leufs*13) were reported by Patel et al. and Habibi et al. [22, 24].
Retinol-binding protein 3 (RBP3) (MIM: 180290)
One nonsense variant;c.1162C>T; p.(Arg338*) was reported using autozygome guided sequencing [33].
Receptor expression-enhancing protein 6 (REEP6) (MIM: 609346)
Mejecase et al. identified in 2018 a homozygous nonsense variant; c.267G>A; p.(Trp89*) in REEP6 using WES in a 60-year-old North African woman [56].
Retinaldehyde-binding protein 1 (RLBP1) (MIM: 180090)
In 2001 Katsanis et al. performed genetic analyses of Saudi Arabian consanguineous kindred having two running clinical diagnosis, both retinitis punctate albecens (RPA) and fundus albipunctatus (FA) using Sanger sequencing [57]. RPA is considered an RCD type that typically features early childhood-onset night blindness, white dots and a progression towards rod-cone degeneration [58] whereas FA is classified as a congenital stationary night blindness with rare cases of macular atrophy, associated with RDH5 variants [59]. The investigation of all known genes associated with flecked retinal dystrophies revealed a missense variant; the c.452G>A; p.(Arg151Gln) [57]. This variant was consistent with FA during the first 3 decades of life and an RPA-like phenotype at later ages. The authors performed co-segregation, analyzed 112 unrelated Saudi controls and did not find this variant in homozygous state [57]. Humbert et al. in 2006, identified a homozygous 7.36-kb deletion that includes the last three exons of RLBP1 (the c.(?_526)_(*418_?)del) in a 24-year-old Moroccan male with RPA, born to first-cousin parents [60]. Long-distance PCR and cloning of genomic DNA were performed to characterize the deletion. Segregation analysis in his unaffected brother showed an absence of the deletion [60]. Dessalces et al. also identified the same deletion in eight Moroccan patients with RPA [58]. On the other hand in KSA, two missense variants; c.446C>T; p.(Ser149Phe) and c.452G>A; p.(Arg151Gln) and one frameshift deletion; c.286_297del; p.(Phe96_Phe99del) were reported [22, 33].
Retinitis pigmentosa (RP1) (MIM: 603937)
Until today, all reported RP1 variants were found in Saudi Arabian and all were homozygous. In 2009 Aldahmesh et al. identified two variants; c.662del; p.(Ala221Glyfs*43) and c.606C>A; p.(Asp202Glu) [44]. Al Rashed et al., in 2012 analyzed a sporadic case and nineteen individuals from four arRCD families [61]. This study identified four variants consisting of two nonsense variants; c.4552A>T; p.(Lys1518*) and c.3396G>A; p.(Trp1132*), a single base deletion; c.3428del; p.(Asn1143Ilefs*25) and a frameshift variant; c.3677_3678dup; p.(Glu1227Metfs*29) [61]. Similarly, Abu-Safieh et al. identified three additional variants consisting of a frameshift deletion; c.4242_4243del; p.(His1414Glnfs*5), a nonsense; c.1012C>T; p.(Arg338*) and a missense; c.5008G>A; p.(Ala1670Thr) [33]. In 2016, Patel et al. reported two frameshift deletions c.662del; p.(Ala221Glyfs*43) and c.1719_1723del; p.Ser574Cysfs*7) [22]. Recently, in a family harboring two affected members with typical symptoms of RCD, Abdullah Albarry et al., identified an insertion variant c.3544_3545insAGAAAAGCTG; p.(Ala1182Glufs*20) [62].
Retinitis pigmentosa 1-Like 1 (RP1L1) (MIM: 608581)
Patel et al in 2016. reported the first RP1L1 variant in Arab countries, more specifically it was a nonsense variant; c.5959C>T; p.(Gln1987*) [22]. Very recently, Albarry et al. reported a large insertion of 48 nucleotides in the coding region leading to p.(Glu1318_Ala1319insGlyThrLysValIleGluGlyLeuGlnGluGluArgValGlnLeuGlu) [62].
Retinal pigment epithelium-specific protein 65 (RPE65) (MIM: 180069)
RPE65 variants were reported in three countries; KSA, Tunisia, and Emirates. In KSA, Abu-Safieh et al. and Patel et al. reported two missense and one frameshift deletion: c.310G>C; p.(Gly104Arg), c.131G>A; p.(Arg44Gln) and c.1366del; p.(Glu456Lysfs*30) respectively [22, 33]. In Tunisia, Habibi et al. in 2016, investigated two families having each a male index (age of onset 14–46 years). Using WES and IROme technologies, the authors identified two variants in compound heterozygous state; [c.271C>T];[c.515T>A]; p.[(Arg91Trp);(Val172Asp)], one missense variant; c.544C>T; p.(His182Tyr) and one splice-site variant; c.[1129-2A>G] in homozygous state [24]. Recently, two additional variants; c.993G>A; p.(Trp331*) and c.(?_-1)_(1128 + 1_1129–1)del were reported in Emirati individuals [42]. It is noteworthy to mention that variants in RPE65 are usually associated with LCA or early onset of RCD.
Spermatogenesis-associated protein 7 (SPATA7) (MIM: 609868)
In Saudi Arabia, a nonsense variant; c.288T>A; p.(Cys96*) was reported by Patel et al. in 2016 [22].
Tubby-related protein 1 (TULP1) (MIM: 602280)
Until today all the disease-causing variants in TULP1 were identified in the gulf region (specifically in 60 individuals from KSA and Arabian Peninsula). Despite the significant abundance of TULP1 variants, only three different variants have been reported; c.901C>T; p.(Gln301*), c.1256G>A; p.(Arg419Gln) and c.1495 + 1G> A [22, 23, 33, 34]. Notably, the c.901C>T; p.(Gln301*) occurred in ~95% of the TULP1 mutated individuals. Similarly to RPE65, variants in TULP1 are usually associated with LCA or early onset of RCD.
Usherin 2A (USH2A) (MIM: 608400)
Until today, seven individuals with RCD were reported to carry variants in USH2A, five out of seven of Saudi origin [22, 23]. Three different classes of variants (missense, nonsense, and splice site) were documented by Patel et al. between 2016 and 2018; comprising compound heterozygous variants: [c.2276G>T];[c.5776 + 1G>A]; p.[(Cys759Phe);p.(=)], and [c.1923T>A];[c.14294T>C]; p.[(Cys641*);(Val4765Ala)], in addition to two other homozygous variants; c.842C>A; p.(Thr281Lys), and c.4033G>C; p.(Ala1345Pro) [22, 23].
Zinc finger 408 (ZNF408) (MIM: 616454)
Habibi et al. in 2017, performed WES on DNA of a consanguineous Tunisian family where three sisters (diagnosed at age <46 years old) out of four members were affected with RCD. The authors reported c.653-1G>T as a splice-site variant shared by the affected family members [25].
Genes possibly associated with sporadic or autosomal recessive rod-cone dysrophy
In addition to the above-mentioned genes, more genotype–phenotype associations were reported in the literature. However, the implicated genes were not included in RetNet as RCD genes, and phenotypic data were not available to confirm the RCD phenotype in the majority of the included papers. These facts led to their classification as possible arRCD genes that were described in Supplementary Table 4.
Autosomal dominant and x-linked rod-cone dystrophy
Only ten Individuals were found to have adRCD or xlRCD (Supplementary Table 2). Because of the limited number, no further analysis was performed. Instead, a description of the identified variants was reported below. The information on co-segregation analysis, in silico pathogenicity assessment and genotyping of controls are found in Table 1.
Cone-rod homeobox (CRX) (MIM: 602225)
Three variants; c.425A>G; p.(Tyr142Cys), c.695del; p.(Pro232Argfs*139) and c.274G>A; p.(Ala92Thr) (possibly dominant) were found in two Saudi Arabian individuals affected with adRCD [22]. Variants in CRX, especially the c.425A>G; p.(Tyr142Cys), are known to be associated with dominant cone-rod dystrophy and with de novo LCA, however, since this gene encodes a transcription factor for several retinal genes, it might also be associated with adRCD [63].
Pre-mRNA-processing-splicing factor 8 (PRPF8) (MIM: 607300)
In an Emirati patient with adRCD, Khan et al. identified one heterozygous missense variant; c.6928A>G; p.(Arg2310Gly) to be disease-causing [42].
Small nuclear ribonucleoprotein, 200-Kd; (SNRNP200) (MIM: 601664)
Two variants in SNRNP200; c.6308A>G; p.(Asn2103Ser) and c.2593G>A; p.(Gly865Ser) (possibly heterozygous since this information and the co-segregation analysis for this variant were not mentioned) were carried by two sporadic cases of adRCD from Saudi Arabia [22].
Retinitis pigmentosa GTPase regulator (RPGR) (MIM: 312610)
Using Sanger sequencing, Haddad et al. in 2016, detected a missense variant; p.(Ala196Thr) associated with xlRCD. This variant was found in all affected males, and one unaffected male member. In addition, this variant was absent in fathers and present heterozygous in mothers of affected males [64]. All the previous observations led the authors to conclude that this variant may be pathogenic, with incomplete penetrance [64]. This variant was not included in Table 2 because of an error in the nomenclature and the absence of enough information (genomic, or cDNA position) for correction.
Table 2.
Variants with possible founder effect in Arab populations.
| Gene | Ref_Seq | DNA change | Protein change | N | Country |
|---|---|---|---|---|---|
| TULP1 | NM_003322.3 | c.901C>T | p.(Gln301*) | 62 | KSA, Arabian Peninsula |
| RP1 | NM_006269.1 | c.3428del | p.(Asn1143Ilefs*25) | 16 | KSA |
| EYS | NM_001142800.1 | c.910_911insT | p.(Trp304Leufs*8) | 12 | KSA |
| RLBP1 | NG_008116.1 (NM_000326.4) | c.(525 + 1_526 + 1)_(*418_?)del | - | 9 | Morocco |
| MERTK | NM_006343.2 | c.2214del | p.(Cys738Trpfs*32) | 8 | KSA, Emirates |
Ref_Seq reference sequence, N number of individuals, KSA Kingdom of Saudi Arabia.
Retinitis pigmentosa 2 (RP2) (MIM: 300757)
In addition to RPGR, two hemizygous muations in RP2; c.2593G>A; p.(Gly865Ser) and c.2T>C;p.? were carried by two Saudi Arabian individuals with xlRCD [22].
Variants with possible founder effect in Arab populations
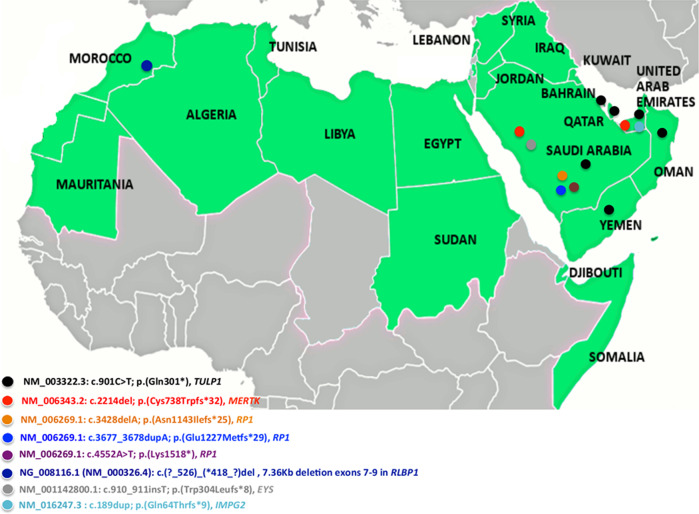
Table 2 shows the most prevalent variants in the Arab countries. One variant in TULP1, the c.901C>T; p.(Gln301*) had the utmost number of reported cases with 60 patients divided between KSA (50 individuals) and the Arabian Peninsula (10 individuals) (Table 2);[22, 23, 33, 34] this account for 16% of the total participants. Khan et al. already described p.(Gln301*) as being a founder variant in the Arabian Peninsula and supported this statement by haplotype analysis in ten children harbouring the variant in a homozygous state [34]. Other variants were also reported several times in multiple families signifying they may represent a founder effect in certain Arab populations. These include the RP1 variant; c.3428del; p.(Asn1143Ilefs*25) detected in 16 Saudi Arabians belonging to different families (Table 2) [22, 33, 61], the c.910_911insT; p.(Trp304Leufs*8) in EYS carried by 12 KSA individuals, the MERTK variant c.2214del; p.(Cys738Trpfs*32) detected in 8 individuals from KSA and Emirates [22, 42], and the RLBP1 deletion (c.(525 + 1_526 + 1)_(*418_?)dell) removing exons 7–9 and detected in nine different Moroccan individuals belonging to five families [58, 60] (Table 2). The variations with possible founder effects and associated with sporadic and arRCD were also shown in a map of the Arab countries (Fig. 5).
Fig. 5. Map of the Arab countries showing the variations with possible founder effects and associated with sporadic and autosomal recessive rod-cone dystrophy.
Almost all variations with possible founder effects were specific to one country except; (2) c.901C>T; p.(Gln301*) in TULP1 shared in the Kingdom of Saudi Arabia and the Arabian Peninsula, (2) c.2214del; p.(Cys738Trpfs*32) in MERTK shared in the Kingdom of Saudi Arabia and the United Arab Emirates.
Limitations
Three limitations should be mentioned; (1) A large proportion of the included participants were from Saudi Arabia. This fact impacted significantly our prevalence results in the Arab world and implies that this result should be taken with caution. Besides, it was impossible to estimate the gene prevalence per country for the populations with a limited sample size (Jordanians, Lebanese…). (2) Among the included papers, two of the largest studies had around 41% of unsolved cases [22, 33]. This rate indicates that additional hidden genetic defects are to be uncovered in Arab populations; an issue that can be ameliorated by implicating copy number variation analysis and investigating noncoding regions in genetic studies [65]. (3) The majority of reported RCD patients lack published phenotypic data, thus we were unable to check the exact phenotype and consequently the adequacy of genotype-RCD association. More specifically, some papers may not have made the distinction between RCD and cone-rod dystrophy as well as between early onset retinal dystrophies, mild forms of LCA and RCD. For that matter, we adopted the genotype-RCD associations known in RetNet but we also recognized its limitation due to overlapping phenotype/genotype association.
Conclusion and perspectives
In conclusion, the current work is the first step towards organizing and updating existing genetic data, cataloging the genetic variations associated with RCD in the Arab world and highlighting the different distribution of causative genes among its populations. Although, the last decade has seen significant interest, expertise, and an increase in RCD scientific publication, much work needs to be conducted. Therefore, more studies in the Arab world are needed before definitive and general conclusions about gene prevalence are drawn.
Supplementary information
Data availability
The data that support the findings of this study are openly available in PubMed repository at https://www.ncbi.nlm.nih.gov/clinvar/submitters/507726.
Compliance with ethical standards
Conflict of interest
The authors declare that they have no conflict of interest.
Footnotes
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
These authors jointly supervised: Christina Zeitz, Isabelle Audo
These authors contributed equally: Hawraa Joumaa, Zamzam Mrad
Supplementary information
The online version of this article (10.1038/s41431-020-00754-0) contains supplementary material, which is available to authorized users.
References
- 1.Hamel C. Retinitis pigmentosa. Orphanet J Rare Dis. 2006;1:40. doi: 10.1186/1750-1172-1-40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Hartong DT, Berson EL, Dryja TP. Retinitis pigmentosa. Lancet. 2006;368:1795–809. doi: 10.1016/S0140-6736(06)69740-7. [DOI] [PubMed] [Google Scholar]
- 3.Ferrari S, Di Iorio E, Barbaro V, Ponzin D, Sorrentino FS, Parmeggiani F. Retinitis pigmentosa: genes and disease mechanisms. Curr Genom. 2011;12:238–49. doi: 10.2174/138920211795860107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Daiger SP, Sullivan LS, Bowne SJ. Genes and mutations causing retinitis pigmentosa. Clin Genet. 2013;84:132–41. doi: 10.1111/cge.12203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Dryja TP, Hahn LB, Cowley GS, McGee TL, Berson EL. Mutation spectrum of the rhodopsin gene among patients with autosomal dominant retinitis pigmentosa. Proc Natl Acad Sci USA. 1991;88:9370–4. doi: 10.1073/pnas.88.20.9370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Berson EL, Grimsby JL, Adams SM, McGee TL, Sweklo E, Pierce EA, et al. Clinical features and mutations in patients with dominant retinitis pigmentosa-1 (RP1) Investig Ophthalmol Vis Sci. 2001;42:2217–24. [PubMed] [Google Scholar]
- 7.Payne A, Vithana E, Khaliq S, Hameed A, Deller J, Abu-Safieh L, et al. RP1 protein truncating mutations predominate at the RP1 adRP locus. Investig Ophthalmol Vis Sci. 2000;41:4069–73. [PubMed] [Google Scholar]
- 8.Avila-Fernandez A, Cantalapiedra D, Aller E, Vallespin E, Aguirre-Lamban J, Blanco-Kelly F, et al. Mutation analysis of 272 Spanish families affected by autosomal recessive retinitis pigmentosa using a genotyping microarray. Mol Vis. 2010;16:2550–8. [PMC free article] [PubMed] [Google Scholar]
- 9.Lenassi E, Vincent A, Li Z, Saihan Z, Coffey AJ, Steele-Stallard HB, et al. A detailed clinical and molecular survey of subjects with nonsyndromic USH2A retinopathy reveals an allelic hierarchy of disease-causing variants. Eur J Hum Genet. 2015;23:1318–27. doi: 10.1038/ejhg.2014.283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.McGee TL, Seyedahmadi BJ, Sweeney MO, Dryja TP, Berson EL. Novel mutations in the long isoform of the USH2A gene in patients with Usher syndrome type II or non-syndromic retinitis pigmentosa. J Med Genet. 2010;47:499–506. doi: 10.1136/jmg.2009.075143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Audo I, Sahel JA, Mohand-Said S, Lancelot ME, Antonio A, Moskova-Doumanova V, et al. EYS is a major gene for rod-cone dystrophies in France. Hum Mutat. 2010;31:E1406–35. doi: 10.1002/humu.21249. [DOI] [PubMed] [Google Scholar]
- 12.Barragan I, Abd El-Aziz MM, Borrego S, El-Ashry MF, O’Driscoll C, Bhattacharya SS, et al. Linkage validation of RP25 Using the 10 K genechip array and further refinement of the locus by new linked families. Ann Hum Genet. 2008;72:454–62. doi: 10.1111/j.1469-1809.2008.00448.x. [DOI] [PubMed] [Google Scholar]
- 13.Arai Y, Maeda A, Hirami Y, Ishigami C, Kosugi S, Mandai M, et al. Retinitis pigmentosa with EYS mutations is the most prevalent inherited retinal dystrophy in Japanese populations. J Ophthalmol. 2015;2015:819760. [DOI] [PMC free article] [PubMed]
- 14.Oishi M, Oishi A, Gotoh N, Ogino K, Higasa K, Iida K, et al. Comprehensive molecular diagnosis of a large cohort of Japanese retinitis pigmentosa and Usher syndrome patients by next-generation sequencing. Investig Ophthalmol Vis Sci. 2014;55:7369–75. doi: 10.1167/iovs.14-15458. [DOI] [PubMed] [Google Scholar]
- 15.Vervoort R, Lennon A, Bird AC, Tulloch B, Axton R, Miano MG, et al. Mutational hot spot within a new RPGR exon in X-linked retinitis pigmentosa. Nat Genet. 2000;25:462–6. doi: 10.1038/78182. [DOI] [PubMed] [Google Scholar]
- 16.Fortina P, Al Khaja N, Al Ali MT, Hamzeh AR, Nair P, Innocenti F, et al. Genomics into Healthcare: the 5th Pan Arab Human Genetics Conference and 2013 Golden Helix Symposium. Hum Mutat. 2014;35:637–40. doi: 10.1002/humu.22530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Liberati A, Altman DG, Tetzlaff J, Mulrow C, Gotzsche PC, Ioannidis JP, et al. The PRISMA statement for reporting systematic reviews and meta-analyses of studies that evaluate healthcare interventions: explanation and elaboration. BMJ. 2009;339:b2700. doi: 10.1136/bmj.b2700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Metzker ML. Sequencing technologies—the next generation. Nat Rev Genet. 2010;11:31–46. doi: 10.1038/nrg2626. [DOI] [PubMed] [Google Scholar]
- 19.Audo I, El Shamieh S, Mejecase C, Michiels C, Demontant V, Antonio A, et al. ARL2BP mutations account for 0.1% of autosomal recessive rod-cone dystrophies with the report of a novel splice variant. Clin Genet. 2017;92:109–11. doi: 10.1111/cge.12909. [DOI] [PubMed] [Google Scholar]
- 20.Audo I, Mohand-Said S, Boulanger-Scemama E, Zanlonghi X, Condroyer C, Demontant V, et al. MERTK mutation update in inherited retinal diseases. Hum Mutat. 2018;39:887–913. doi: 10.1002/humu.23431. [DOI] [PubMed] [Google Scholar]
- 21.Gerth-Kahlert C, Tiwari A, Hanson JVM, Batmanabane V, Traboulsi E, Pennesi ME, et al. C2orf71 mutations as a frequent cause of autosomal-recessive retinitis pigmentosa: clinical analysis and presentation of 8 novel mutations. Investig Ophthalmol Vis Sci. 2017;58:3840–50. doi: 10.1167/iovs.17-21597. [DOI] [PubMed] [Google Scholar]
- 22.Patel N, Aldahmesh MA, Alkuraya H, Anazi S, Alsharif H, Khan AO, et al. Expanding the clinical, allelic, and locus heterogeneity of retinal dystrophies. Genet Med. 2016;18:554–62. doi: 10.1038/gim.2015.127. [DOI] [PubMed] [Google Scholar]
- 23.Patel N, Alkuraya H, Alzahrani SS, Nowailaty SR, Seidahmed MZ, Alhemidan A, et al. Mutations in known disease genes account for the majority of autosomal recessive retinal dystrophies. Clin Genet. 2018;94:554–63. doi: 10.1111/cge.13426. [DOI] [PubMed] [Google Scholar]
- 24.Habibi I, Chebil A, Falfoul Y, Allaman-Pillet N, Kort F, Schorderet DF, et al. Identifying mutations in Tunisian families with retinal dystrophy. Sci Rep. 2016;6:37455. doi: 10.1038/srep37455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Habibi I, Chebil A, Kort F, Schorderet DF, El Matri L. Exome sequencing confirms ZNF408 mutations as a cause of familial retinitis pigmentosa. Ophthalmic Genet. 2017;38:494–7. doi: 10.1080/13816810.2016.1275020. [DOI] [PubMed] [Google Scholar]
- 26.Guillonneau X, Piriev NI, Danciger M, Kozak CA, Cideciyan AV, Jacobson SG, et al. A nonsense mutation in a novel gene is associated with retinitis pigmentosa in a family linked to the RP1 locus. Hum Mol Genet. 1999;8:1541–6. doi: 10.1093/hmg/8.8.1541. [DOI] [PubMed] [Google Scholar]
- 27.Pierce EA, Quinn T, Meehan T, McGee TL, Berson EL, Dryja TP. Mutations in a gene encoding a new oxygen-regulated photoreceptor protein cause dominant retinitis pigmentosa. Nat Genet. 1999;22:248–54. doi: 10.1038/10305. [DOI] [PubMed] [Google Scholar]
- 28.Sullivan LS, Heckenlively JR, Bowne SJ, Zuo J, Hide WA, Gal A, et al. Mutations in a novel retina-specific gene cause autosomal dominant retinitis pigmentosa. Nat Genet. 1999;22:255–9. doi: 10.1038/10314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Audo I, Mohand-Said S, Dhaenens CM, Germain A, Orhan E, Antonio A, et al. RP1 and autosomal dominant rod-cone dystrophy: novel mutations, a review of published variants, and genotype-phenotype correlation. Hum Mutat. 2012;33:73–80. doi: 10.1002/humu.21640. [DOI] [PubMed] [Google Scholar]
- 30.El Shamieh S, Boulanger-Scemama E, Lancelot ME, Antonio A, Demontant V, Condroyer C, et al. Targeted next generation sequencing identifies novel mutations in RP1 as a relatively common cause of autosomal recessive rod-cone dystrophy. BioMed Res Int. 2015;2015:485624. [DOI] [PMC free article] [PubMed]
- 31.Avila-Fernandez A, Corton M, Nishiguchi KM, Munoz-Sanz N, Benavides-Mori B, Blanco-Kelly F, et al. Identification of an RP1 prevalent founder mutation and related phenotype in Spanish patients with early-onset autosomal recessive retinitis. Ophthalmology. 2012;119:2616–21. doi: 10.1016/j.ophtha.2012.06.033. [DOI] [PubMed] [Google Scholar]
- 32.Ullah I, Kabir F, Iqbal M, Gottsch CB, Naeem MA, Assir MZ, et al. Pathogenic mutations in TULP1 responsible for retinitis pigmentosa identified in consanguineous familial cases. Mol Vis. 2016;22:797–815. [PMC free article] [PubMed] [Google Scholar]
- 33.Abu-Safieh L, Alrashed M, Anazi S, Alkuraya H, Khan AO, Al-Owain M, et al. Autozygome-guided exome sequencing in retinal dystrophy patients reveals pathogenetic mutations and novel candidate disease genes. Genome Res. 2013;23:236–47. doi: 10.1101/gr.144105.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Khan AO, Bergmann C, Eisenberger T, Bolz HJ. A TULP1 founder mutation, p.Gln301*, underlies a recognisable congenital rod-cone dystrophy phenotype on the Arabian Peninsula. Br J Ophthalmol. 2015;99:488–92. doi: 10.1136/bjophthalmol-2014-305836. [DOI] [PubMed] [Google Scholar]
- 35.Strick DJ, Vollrath D. Focus on molecules: MERTK. Exp Eye Res. 2010;91:786–7. doi: 10.1016/j.exer.2010.05.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Ge Z, Bowles K, Goetz K, Scholl HP, Wang F, Wang X, et al. NGS-based Molecular diagnosis of 105 eyeGENE((R)) probands with Retinitis Pigmentosa. Sci Rep. 2015;5:18287. doi: 10.1038/srep18287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Ostergaard E, Duno M, Batbayli M, Vilhelmsen K, Rosenberg T. A novel MERTK deletion is a common founder mutation in the Faroe Islands and is responsible for a high proportion of retinitis pigmentosa cases. Mol Vis. 2011;17:1485–92. [PMC free article] [PubMed] [Google Scholar]
- 38.Currie-Alder B, Arvanitis R, Hanafi S. Research in Arabic-speaking countries: funding competitions, international collaboration, and career incentives. Sci Publ Policy. 2017;45:74–82. [Google Scholar]
- 39.Wildeman M, van Ophuizen E, den Dunnen JT, Taschner PE. Improving sequence variant descriptions in mutation databases and literature using the Mutalyzer sequence variation nomenclature checker. Hum Mutat. 2008;29:6–13. doi: 10.1002/humu.20654. [DOI] [PubMed] [Google Scholar]
- 40.Abu-Ameerh M, Mohammad H, Dardas Z, Barham R, Ali D, Bijawi M, et al. Extending the spectrum of CLRN1- and ABCA4-associated inherited retinal dystrophies caused by novel and recurrent variants using exome sequencing. Mol Genet Genom Med. 2020;8:e1123. doi: 10.1002/mgg3.1123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Rivera A, White K, Stohr H, Steiner K, Hemmrich N, Grimm T, et al. A comprehensive survey of sequence variation in the ABCA4 (ABCR) gene in Stargardt disease and age-related macular degeneration. Am J Hum Genet. 2000;67:800–13. doi: 10.1086/303090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Khan AO. Phenotype-guided genetic testing of pediatric inherited retinal disease in the United Arab Emirates. Retina. 2019;40:1829–37. [DOI] [PubMed]
- 43.Davidson AE, Schwarz N, Zelinger L, Stern-Schneider G, Shoemark A, Spitzbarth B, et al. Mutations in ARL2BP, encoding ADP-ribosylation-factor-like 2 binding protein, cause autosomal-recessive retinitis pigmentosa. Am J Hum Genet. 2013;93:321–9. doi: 10.1016/j.ajhg.2013.06.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Aldahmesh MA, Safieh LA, Alkuraya H, Al-Rajhi A, Shamseldin H, Hashem M, et al. Molecular characterization of retinitis pigmentosa in Saudi Arabia. Mol Vis. 2009;15:2464–9. [PMC free article] [PubMed] [Google Scholar]
- 45.Khan AO, Abu-Safieh L. Rod-cone dystrophy with initially preserved visual acuity despite early macular involvement suggests recessive CERKL mutations. Ophthalmic Genet. 2015;36:369–72. doi: 10.3109/13816810.2014.889168. [DOI] [PubMed] [Google Scholar]
- 46.Azab B, Barham R, Ali D, Dardas Z, Rashdan L, Bijawi M, et al. Novel CERKL variant in consanguineous Jordanian pedigrees with inherited retinal dystrophies. Can J Ophthalmol. 2019;54:51–9. doi: 10.1016/j.jcjo.2018.02.018. [DOI] [PubMed] [Google Scholar]
- 47.Jalkh N, Guissart C, Chouery E, Yammine T, El Ali N, Farah HA, et al. Report of a novel mutation in CRB1 in a Lebanese family presenting retinal dystrophy. Ophthalmic Genet. 2014;35:57–62. doi: 10.3109/13816810.2013.763995. [DOI] [PubMed] [Google Scholar]
- 48.Hashmi JA, Albarry MA, Almatrafi AM, Albalawi AM, Mahmood A, Basit S. Whole exome sequencing identified a novel single base pair insertion mutation in the EYS gene in a six generation family with retinitis pigmentosa. Congenit Anom. 2018;58:10–5. doi: 10.1111/cga.12225. [DOI] [PubMed] [Google Scholar]
- 49.Zobor D, Balousha G, Baumann B, Wissinger B. Homozygosity mapping reveals new nonsense mutation in the FAM161A gene causing autosomal recessive retinitis pigmentosa in a Palestinian family. Mol Vis. 2014;20:178–82. [PMC free article] [PubMed] [Google Scholar]
- 50.Khan AO, Al Teneiji AM. Homozygous and heterozygous retinal phenotypes in families harbouring IMPG2 mutations. Ophthalmic Genet. 2019;40:247–51. doi: 10.1080/13816810.2019.1627467. [DOI] [PubMed] [Google Scholar]
- 51.Mackay DS, Henderson RH, Sergouniotis PI, Li Z, Moradi P, Holder GE, et al. Novel mutations in MERTK associated with childhood onset rod-cone dystrophy. Mol Vis. 2010;16:369–77. [PMC free article] [PubMed] [Google Scholar]
- 52.Ksantini M, Lafont E, Bocquet B, Meunier I, Hamel CP. Homozygous mutation in MERTK causes severe autosomal recessive retinitis pigmentosa. Eur J Ophthalmol. 2012;22:647–53. doi: 10.5301/ejo.5000096. [DOI] [PubMed] [Google Scholar]
- 53.Nair P, Hamzeh AR, Malik EM, Oberoi D, Al-Ali MT, Bastaki F. Novel PDE6A mutation in an Emirati patient with retinitis pigmentosa. Oman J Ophthalmol. 2017;10:228–31. doi: 10.4103/ojo.OJO_213_2016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Hmani-Aifa M, Benzina Z, Zulfiqar F, Dhouib H, Shahzadi A, Ghorbel A, et al. Identification of two new mutations in the GPR98 and the PDE6B genes segregating in a Tunisian family. Eur J Hum Genet. 2009;17:474–82. doi: 10.1038/ejhg.2008.167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Coppieters F, Van Schil K, Bauwens M, Verdin H, De Jaegher A, Syx D, et al. Identity-by-descent-guided mutation analysis and exome sequencing in consanguineous families reveals unusual clinical and molecular findings in retinal dystrophy. Genet Med. 2014;16:671–80. doi: 10.1038/gim.2014.24. [DOI] [PubMed] [Google Scholar]
- 56.Mejecase C, Mohand-Said S, El Shamieh S, Antonio A, Condroyer C, Blanchard S, et al. A novel nonsense variant in REEP6 is involved in a sporadic rod-cone dystrophy case. Clin Genet. 2018;93:707–11. doi: 10.1111/cge.13171. [DOI] [PubMed] [Google Scholar]
- 57.Katsanis N, Shroyer NF, Lewis RA, Cavender JC, Al-Rajhi AA, Jabak M, et al. Fundus albipunctatus and retinitis punctata albescens in a pedigree with an R150Q mutation in RLBP1. Clin Genet. 2001;59:424–9. doi: 10.1034/j.1399-0004.2001.590607.x. [DOI] [PubMed] [Google Scholar]
- 58.Dessalces E, Bocquet B, Bourien J, Zanlonghi X, Verdet R, Meunier I, et al. Early-onset foveal involvement in retinitis punctata albescens with mutations in RLBP1. JAMA Ophthalmol. 2013;131:1314–23. doi: 10.1001/jamaophthalmol.2013.4476. [DOI] [PubMed] [Google Scholar]
- 59.Yamamoto H, Simon A, Eriksson U, Harris E, Berson EL, Dryja TP. Mutations in the gene encoding 11-cis retinol dehydrogenase cause delayed dark adaptation and fundus albipunctatus. Nat Genet. 1999;22:188–91. doi: 10.1038/9707. [DOI] [PubMed] [Google Scholar]
- 60.Humbert G, Delettre C, Senechal A, Bazalgette C, Barakat A, Arnaud B, et al. Homozygous deletion related to Alu repeats in RLBP1 causes retinitis punctata albescens. Investig Ophthalmol Vis Sci. 2006;47:4719–24. doi: 10.1167/iovs.05-1488. [DOI] [PubMed] [Google Scholar]
- 61.Al-Rashed M, Abu Safieh L, Alkuraya H, Aldahmesh MA, Alzahrani J, Diya M, et al. RP1 and retinitis pigmentosa: report of novel mutations and insight into mutational mechanism. Br J Ophthalmol. 2012;96:1018–22. doi: 10.1136/bjophthalmol-2011-301134. [DOI] [PubMed] [Google Scholar]
- 62.Albarry MA, Hashmi JA, Alreheli AQ, Albalawi AM, Khan B, Ramzan K, et al. Novel homozygous loss-of-function mutations in RP1 and RP1L1 genes in retinitis pigmentosa patients. Ophthalmic Genet. 2019;40:507–13. doi: 10.1080/13816810.2019.1703014. [DOI] [PubMed] [Google Scholar]
- 63.Sohocki MM, Sullivan LS, Mintz-Hittner HA, Birch D, Heckenlively JR, Freund CL, et al. A range of clinical phenotypes associated with mutations in CRX, a photoreceptor transcription-factor gene. Am J Hum Genet. 1998;63:1307–15. doi: 10.1086/302101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Haddad MF, Khabour OF, Abuzaideh KA, Shihadeh W. Screening for mutations in RPGR and RP2 genes in Jordanian families with X-linked retinitis pigmentosa. Genet Mol Res. 2016;15:gmr7842. [DOI] [PubMed]
- 65.Eisenberger T, Neuhaus C, Khan AO, Decker C, Preising MN, Friedburg C, et al. Increasing the yield in targeted next-generation sequencing by implicating CNV analysis, non-coding exons and the overall variant load: the example of retinal dystrophies. PloS One. 2013;8:e78496. doi: 10.1371/journal.pone.0078496. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The data that support the findings of this study are openly available in PubMed repository at https://www.ncbi.nlm.nih.gov/clinvar/submitters/507726.