Abstract
Effective treatments targeting disease etiology are urgently needed for Alzheimer’s disease (AD). Although candidate AD genes have been identified and altering their levels may serve as therapeutic strategies, the consequence of such alterations remain largely unknown. Herein, we analyzed CRISPR knockout/RNAi knockdown screen data for over 700 cell lines and evaluated cellular dependencies of 104 AD-associated genes previously identified by genome-wide association studies (GWAS) and gene expression network studies. Multiple genes showed widespread cell dependencies across tissue lineages, suggesting their inhibition may yield off-target effects. Meanwhile, several genes including SPI1, MEF2C, GAB2, ABCC11, ATCG1 were identified as genes of interest since their genetic knockouts specifically affected high-expressing cells whose tissue lineages are relevant to cell types found in AD. Overall, analyses of genetic screen data identified AD-associated genes whose knockout or knockdown selectively affected cell lines of relevant tissue lineages, prioritizing targets for potential AD treatments.
Subject terms: Cancer genomics, Drug screening, Target identification, Neurological disorders
Introduction
Globally, an estimated 43.8 million individuals were affected by dementia in 2016, a more than two-fold increase from 20.2 million in 1990, and is expected to continue rising1. Alzheimer’s disease (AD) is the leading cause of dementia, and only supportive treatments are currently available for AD. AD medications consist of cholinesterase inhibitors (donepezil, galantamine, and rivastigmine), NMDA receptor uncompetitive antagonist memantine, or a combination of both (donepezil)2. These medications only address symptoms and are helpful for a limited timespan in improving patients’ cognitive abilities. They are not disease-modifying therapies and do not alter disease progression. Other novel therapies for both early-onset AD (EOAD) and late-onset AD (LOAD) have focused on APP, PSEN1, and PSEN2, identified through rare mutations in familial EOAD cases, that cause amyloid-β deposition. Testing has been carried out through the administration of anti-amyloid-β exogenous monoclonal antibodies, but this approach has not yielded promising results3–5. One such antibody is aducanumab which was recently approved for expedited review by the FDA but is yet to be validated and demonstrates inconclusive clinical efficacy6,7. Several gamma secretase inhibitors have also been tested but failed in clinical studies and were ultimately discontinued5.
Recent research has discovered new genetic risk loci for LOAD by genome-wide association studies (GWAS) or transcriptomic studies. Identified genes have been found to be strongly expressed in immunity, lipid metabolism, tau binding proteins, and amyloid precursor protein metabolism8. One recent study confirmed 20 previous risk loci (CR1, BIN1, INPP5D, HLA-DRB1, TREM2, CD2AP, NYAP1, EPHA1, PTK2B, CLU, SPI1, MS4A2, PICALM, SORL1, FERMT2) and identified five novel loci (IQCK, ACE, ADAM10, ADAMTS1, and WWOX)8. A parallel GWAS study found 29 risk loci, including previously classified risk loci and several novel loci9. In parallel, transcriptomic analyses found at least 50 network hub genes upregulated in co-expression networks of LOAD compared to normal brain tissues. For example, TYROBP was identified as a pivotal regulator of the phagocytotic pathway10, cross-validating with immunity-implicated genes found by GWAS.
While GWAS and transcriptomic studies have nominated gene candidates associated with AD pathology, the therapeutic viability of manipulating functions or levels of such targets remain largely unknown. An ideal gene target should not only show sensitivity by being implicated in AD etiology, but also specificity by which its targeting intervention would not cause widespread, off-target effects (e.g., house-keeping genes or genes serving essential cognitive functions). Genome-wide CRISPR-Cas9 knockout or RNAi knockdown screens are tools which can help prioritize therapeutic targets and can guide inhibition treatments that are both effective and potentially have reduced toxicity11. The Cancer Dependency Map Project (DepMap) has curated dependency profiles of almost 18,000 genes across more than 700 human cell lines11,12. While involving multiple cell lines that share lineages with cells implicated in AD, the dataset has not been used to evaluate AD targets. Herein, we leveraged the large-scale transcriptomic/proteomic expression and dependency data from the DepMap genome-scale CRISPR-Cas9 and RNAi screens to assess the effects of knocking out and knocking down candidate AD associated genes, revealing their potential as precision therapeutic targets.
Methods
Data source and download
Genes used in this publication were obtained from multiple papers. Genome wide associated studies (GWAS) that identified genes implicated in AD were downloaded from Kunkle et al. and Jansen et al.8,9. 42 unique genes from these two sources were compiled, and APP, PSEN1, and PSEN2 were added to the AD risk gene list13. 60 key AD network hub genes associated with AD were downloaded from Zhang et al.10. Cancer gene expression and dependency scores were downloaded from the DepMap Portal Public 20Q2 release in May 202014.
Expression-driven dependency using DepMap
The DepMap Public 20Q2 release contains the Achilles dataset and results of genome-scale CRISPR knockout screens for 19,144 genes across 1206 cell lines, including both cancer and normal cell lines. 20 tissue types from DepMap with at least 25 cell lines each were selected for further analysis from an original list of 40 tissue types. For each of the 104 genes associated with AD, gene expressions and corresponding CERES dependency scores were analyzed. CERES is a computational method developed by Meyers et al., which estimates gene dependency levels derived from CRISPR-Cas9 essentiality screens and factors in the possibility of an increase in false positives in copy number amplified locations15. For each AD-associated gene, we calculated the proportion of significantly negative CERES scores (< − 0.5) by tissue type to generate heatmaps. A negative CERES score indicates that a gene knockout results in a slower growth rate of a cell line, and a score lower than − 0.5 indicates a notable reduction15. We also calculated the Pearson correlation and corresponding p-values for each AD-associated gene after stratifying the gene expression and CERES scores by tissue type. The analysis was limited to examining expression-driven dependencies in three tissue types: hematopoietic and lymphoid tissue, central nervous system, and autonomic ganglia for their involvement in AD presentation and progression16–18. Overall, 50 gene and tissue combinations were significant at the p-value of 0.05. Volcano plots display the correlation coefficients (x-axis) against the negative log of FDR corrected p-values (y-axis). We focused on gene-tissue combinations with negative correlation coefficients as a high expression of the AD-associated gene and corresponding low CERES score indicates that the gene is needed for cancer cell survival in knockout experiments. Scatterplots of dependency versus expression were plotted for AD gene tissue combinations with the highest negative log p-values, and correlation coefficients were calculated.
The DepMap Public 20Q2 release also contains genetic dependency scores estimated from RNAi and CRISPR loss-of-function screens for 17,309 genes across 712 cell lines14. These scores were calculated using DEMETER2, a framework developed by McFarland et al. to unify results from multiple large-scale RNAi screening datasets19. Calculations of negative dependency scores, volcano plots highlighting correlations, and scatterplots of AD-associated genes were completed in a similar process as CERES scores. Analysis of genetic dependency data was performed using Python (v 3.7.6), Numpy (v 1.7.4), and SciPy (v 1.3.2).
Results
Gene list and DepMap data assembly
A curated list of 104 AD-associated genes, consisting of 44 AD risk genes and 60 AD network hub genes, were utilized for this study (see Supplementary Table S1). There were 20 tissue types with data for at least 25 cell lines each from the DepMap 2020 Q2 release which were selected for further analysis (Table 1). We specifically highlighted results from three tissue types most relevant to AD etiology: hematopoietic and lymphoid tissue (HL, given the now established role of immune response and phagocytotic pathway in AD), central nervous system (CNS), and autonomic ganglia (AG)16–18.
Table 1.
Overview of tissue types and number of unique cell lines used for cell line genetic dependency analysis.
| Lineage | Dependency CERES | Dependency DEMETER2 | mRNA |
|---|---|---|---|
| Autonomic ganglia | 20 | 9 | 28 |
| Biliary tract | 28 | 1 | 39 |
| Bone | 28 | 15 | 37 |
| Breast | 34 | 82 | 57 |
| Central nervous system | 60 | 55 | 84 |
| Endometrium | 24 | 20 | 33 |
| Haematopoietic and lymphoid tissue | 82 | 61 | 205 |
| Kidney | 21 | 29 | 34 |
| Large intestine | 37 | 44 | 67 |
| Liver | 23 | 18 | 25 |
| Lung | 97 | 129 | 192 |
| Oesophagus | 25 | 24 | 32 |
| Ovary | 42 | 38 | 58 |
| Pancreas | 34 | 33 | 51 |
| Pleura | 9 | 6 | 17 |
| Skin | 51 | 46 | 67 |
| Soft tissue | 38 | 25 | 65 |
| Stomach | 26 | 25 | 41 |
| Upper aerodigestive tract | 31 | 18 | 43 |
| Urinary tract | 29 | 12 | 36 |
20 tissue types from the Cancer Dependency Map (DepMap) project with data for at least 25 cell lines were utilized for this analysis.
Pan-tissue genetic dependency
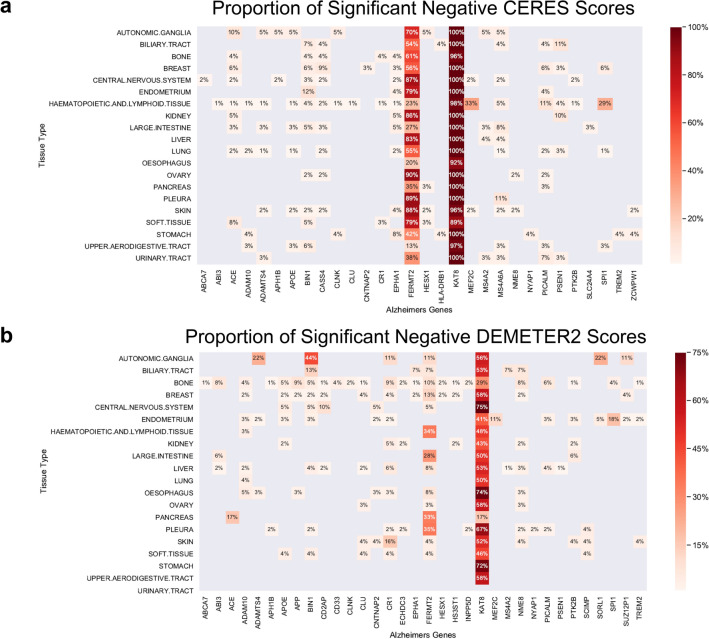
To examine the genetic dependency of AD-associated genes across tissues, we identified the proportion of cell lines within each tissue type showing significant negative dependency scores (< − 0.5), as lower scores indicate that a gene is required for cell survival and proliferation. From 44 AD risk genes found through GWAS, KAT8 showed predominant negative dependency scores in the CRISPR knockout data (CERES score (< − 0.5), with nearly 100% negative dependency scores across all 22 examined tissue types (Fig. 1a). FERMT2 had varying levels of negative CERES scores across tissues with ovary, pleura, and skin cell lines having the highest proportion of negative scores. MEF2C and SPI1 have negative CERES scores localized to the hematopoietic and lymphoid tissue (HL) cell lineage, with 33% and 29%, respectively. We also identified the proportion of cell lines with significant negative genetic dependency scores (DEMETER2 score < − 0.5) utilizing data from large-scale RNAi screens. From the list of AD risk genes, KAT8 had a range of significant negative DEMETER2 scores from 17 to 75% across tissue types, with a peak of 75% in endometrium cell lines (Fig. 1b). ADAMTS4, BIN1, SORL1, and SUZ12P1 had their highest levels of negative DEMETER2 scores in autonomic ganglia (AG) cell lines. SPI1 had its highest proportion of negative DEMETER2 scores in HL cell lines.
Figure 1.
Genetic dependency of AD risk genes in CRISPR knockout and knockdown screens across tissue lineages. (a) The proportion of pass-DepMap-threshold negative dependency scores (< − 0.5) in cell lines by tissue types based on the CERES scores derived from CRISPR knockout screens. (b) The proportion of pass-DepMap-threshold negative dependency scores (< − 0.5) in cell lines by tissue types based on the DEMETER2 scores derived from RNAi knockdown screens. For both (a) and (b), 20 tissue types from the Cancer Dependency Map (DepMap) project with data for at least 25 cell lines were utilized to evaluate 44 AD risk genes. A negative dependency score indicates a gene’s necessity for a cell line’s growth and survival.
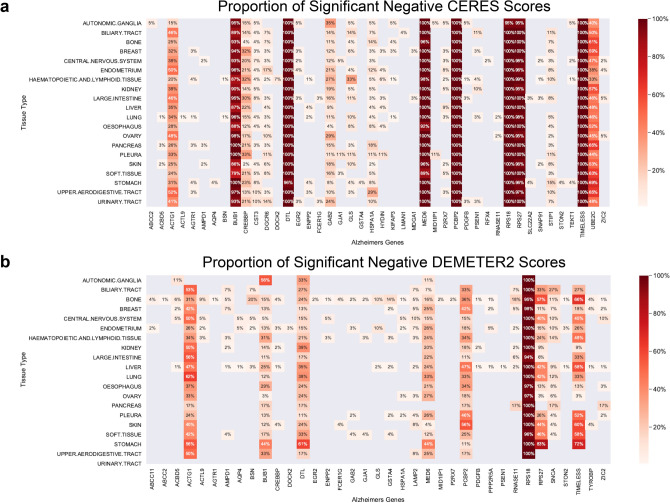
Among the 60 AD network hub genes, BUB1, DTL, MED6, PCBP2, RPS18, RPS27, and TIMELESS had significant negative CERES scores of approximately 100% across tissue types (Fig. 2a). UBE2C, ACTG1, CREBBP had varying levels of negative CERES scores ranging from 20 to 60% across tissue types. GLS and GAB2 showed highest proportion of negative CERES scores in HL (33%) and AG (35%) cell lines, respectively. From AD-associated network hub genes, RPS18 had approximately 100% negative DEMETER2 scores across all tissue types (Fig. 2b). ACTG1, BUB1, DTL, MED6, PCBP2, RPS27, and TIMELESS had varying levels of negative DEMETER2 scores from 10 to 75% across tissue types. The largely concordant CRISPR/RNAi screen results suggest that knocking out/down of several genes, including KAT8 and FERM2, results in widespread consequences affecting most cells’ survival, whereas other candidates (e.g., MEF2C in HL, GAB in AG) more selectively affect fractions of AD-relevant cell types, thus may serve as better targets.
Figure 2.
Genetic dependency of AD network hub genes in CRISPR knockout and knockdown screens across tissue lineages. (a) The proportion of pass-DepMap-threshold negative dependency scores (< − 0.5) in cell lines by tissue types based on the CERES scores derived from CRISPR knockout screens. (b) The proportion of pass-DepMap-threshold negative dependency scores (< − 0.5) in cell lines by tissue types based on the DEMETER2 scores derived from RNAi knockdown screens. For both (a) and (b), 20 tissue types from the Cancer Dependency Map (DepMap) project with data for at least 25 cell lines were utilized to evaluate 60 AD network hub genes. A negative dependency score indicates a gene’s necessity for a cell line’s growth and survival.
Expression-driven cellular dependencies of AD risk genes
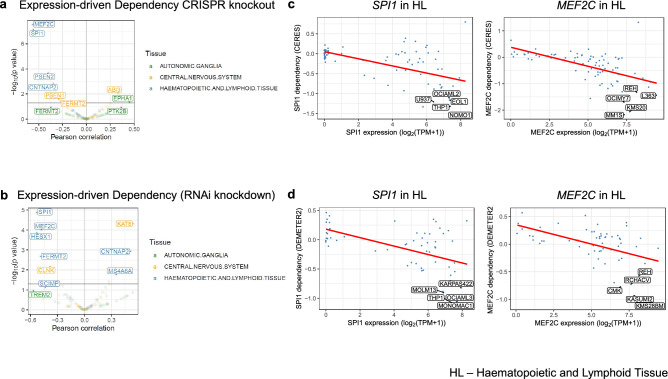
We reasoned that AD-associated genes may show aberrant expression in disease-driving/affected cells. Thus, the candidate genes would likely serve as better targets if their knockout or knockdown most strongly influenced the cells showing aberrant expression of the targeted genes. We next sought to further filter for genes whose expression is significantly correlated with dependencies of cell lines within these tissue types, i.e., expression-driven dependency. We conducted a systematic Pearson correlation analysis to identify such genes of interest, and identified four genes whose expression was significantly associated with cellular dependencies (FDR < 0.05), including MEF2C in HL cell lines (R = − 0.6, FDR = 8.13e−06), SPI1 in HL cell lines (R = − 0.6, FDR = 9.38e−06), PSEN2 in HL cell lines (R = − 0.4, FDR = 0.0131), and CNTNAP2 in HL cell lines (R = − 0.3, FDR = 0.0433) (Fig. 3a). We further conducted the correlation analysis using RNAi knockdown-based DEMETER2 scores to identify similarities and differences to CERES results. We found five genes with significant expression-driven dependency (FDR < 0.05), including SPI1 in HL cell lines (R = − 0.5, FDR = 0.00147), MEF2C in HL cell lines (R = − 0.5, FDR = 0.00545), HESX1 in HL cell lines (R = − 0.5, FDR = 0.00578), CNTNAP2 in HL cell lines (R = 0.5, FDR = 0.0424), KAT8 in CNS cell lines (R = 0.5, FDR = 0.00348) (Fig. 3b).
Figure 3.
Expression-driven Dependency of AD Risk Genes. (a) A volcano plot showing expression-driven dependency of AD risk genes based on CRISPR knockout screen data. (b) A volcano plot showing expression-driven dependency of AD risk genes based on RNAi knockdown screen data. For (a) and (b), Pearson correlation coefficients were calculated for AD risk genes using a gene’s expression versus the dependency scores for three tissue types (color-coded) relevant to AD etiology: hematopoietic and lymphoid tissue (HL), central nervous system (CNS), and autonomic ganglia (AG). The correlation plots of the expression-driven dependency were shown for associations found in the HL cell lines, including (c) SPI1 and MEF2C expression with their respective CERES dependency scores, as well as (d) with their respective DEMETER2 dependency scores. The best-fitted regression lines are shown in red and the cell lines with the highest expression and dependency are further labeled in each plot.
We next highlighted the cell lines that were most affected by knockout/knockdowns; given the challenge of functionally modeling AD in human systems20, these lines may provide alternatives that show aberrantly high expression of the selected AD-associated genes. For SPI1 in HL cell lines, these include NOMO1, THP1, MONOMAC1, THP1, and EOL1. For MEF2C in HL cell lines, these were MM1S, KMS20, OCIMY7, KMS28BM, KASUMI2, and L363 (Fig. 3c,d).
Expression-driven cellular dependencies of AD network hub genes
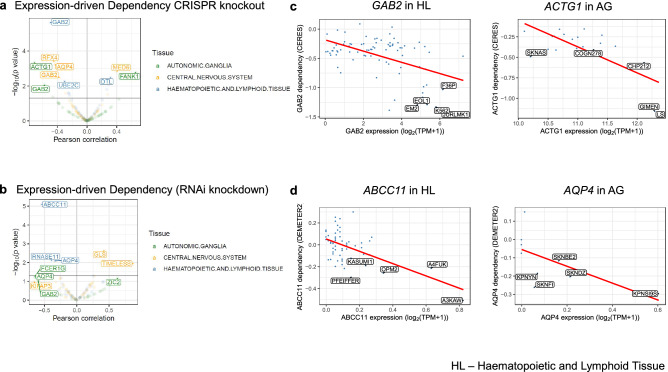
We applied the same expression-driven dependency analysis for the AD network hub genes. Based on the CRISPR screen data, we identified six genes whose expression was significantly associated with cellular dependencies (FDR < 0.05), including GAB2 in HL cell lines (R = − 0.5, FDR = 0.000221), ACTG1 in AG cell lines (R = − 0.7, FDR = 0.0203), RFX4 in CNS cell lines (R = − 0.4, FDR = 0.0203), AQP4 in CNS cell lines (R = − 0.4, FDR = 0.0222), FANK1 in AG cell lines (R = 0.7, FDR = 0.0436), MED6 in CNS cell lines (R = 0.4, FDR = 0.043254) (Fig. 4a). Using the RNAi screen data, we identified one gene with significant expression-driven dependency (FDR < 0.05), ABCC11 in HL cell lines (R = − 0.5, FDR = 0.00147) (Fig. 4b).
Figure 4.
Expression-driven Dependency of AD Network Hub Genes. (a) A volcano plot showing expression-driven dependency of AD network hub genes based on CRISPR knockout screen data. (b) A volcano plot showing expression-driven dependency of AD network hub genes based on RNAi knockdown screen data. For (a) and (b), Pearson correlation coefficients were calculated for AD risk genes using a gene’s expression versus the dependency scores for three tissue types (color-coded) relevant to AD etiology: hematopoietic and lymphoid tissue (HL), central nervous system (CNS), and autonomic ganglia (AG). The correlation plots of the expression-driven dependency were shown for associations found for (c) GAB2 in HL cell lines and ACTG1 in AG cell lines against CERES scores (d) ABCC11 in HL cell lines and AQP4 in AG cell lines against DEMETER2 scores. The best-fitted regression lines are shown in red and the cell lines with the highest expression and dependency are further labeled in each plot.
To highlight potential cell lines that can help study the implication of targeting aberrant expressions of these genes, JURLMK1 showed the lowest CERES dependency score and high expression for GAB2 in HL cell lines (Fig. 4c). For ACTG1 in AG cell lines, the cell lines of interest were LS, GIMEN, CHP212, SKNAS, and COGN278. Based on DEMETER dependency scores, for ABCC11 in HL cell lines, A3KAW, A4FUK, OPM2, KASUMI1, and PFEIFFER were highlighted (Fig. 4d). For AQP4 in AG, we noted that the cell lines KPNSI9S, SKNFI, SKNDZ, KPNYN, and SKNBE2 show expression. For GLS in CNS, the cell lines were LN235, U178, KNS60, LN215, and HS683 (see Supplementary Fig. S1).
Discussion
We conducted a comprehensive genetic dependency analysis of 104 AD-associated genes using expression and genetic screen data from over 700 cell lines. The analysis enabled us to identify whether genetic knockdown or knockout of these AD-associated genes may affect cellular survival. Our results show that the knockdown or knockout of multiple AD-associated genes (KAT8, FERMT2, BUB1, DTL, MED6, PCBP2, RPS18, RPS27, TIMELESS, ACTG1, and UBE2C) significantly reduced survival of cell lines across multiple tissues. Thus, down-regulating or inhibiting these genes could lead to pervasive damaging effects and may have limited therapeutic viability.
Meanwhile, several genes showed dependency that is correlated with gene expression within cell lineages. After limiting our analysis to three tissue types implicated in AD, hematopoietic and lymphoid tissue, central nervous system, and autonomic ganglia, we demonstrated that SPI1, MEF2C, GAB2, and ABCC11 showed expression-driven dependencies in hematopoietic and lymphoid tissue cell lines. SPI1 and MEF2C were consistently identified in both CRISPR knockout and RNAi screen data, further highlighting their potential as AD treatment targets.
Dysregulated microglial response is a hallmark of AD21,22, and the candidates showing expression-driven dependency in the hematopoietic and lymphoid tissue analysis may present targets whose intervention may only affect disease-associated microglia (DAM). For example, SPI1 (PU.1) is a transcription factor encoded by SPI1; it is considered as a master regulator of macrophages necessary for the development and differentiation of microglia. GWAS, eQTL, and epigenetic analyses have recently implicated SPI1 and its regulated network in AD23. SPI1 specifies regulatory regions and establishes the chromatin landscape24, and its target genes include TYROBP, a key regulator upregulated in LOAD10. In a study by Olmos-Alonso, et al., inhibition of Colony Stimulating Factor 1 Receptor (Csf1r), another target gene of SPI1, led to decreased expression of SPI1 and microglial proliferation in APPswe/PSEN1dE9 mice, a transgenic model of AD-like pathology25. As another example, Myocyte-enhancer factor 2C (MEF2C) has been identified as a candidate regulator in establishing the chromatin landscape of microglia24. MEF2C restrains microglial inflammatory response26, which is associated with AD pathology in multiple recent studies, and its expression is lost in aging brains. The expression of MEF2C in microglia is negatively regulated by interferon type I (IFN-1) expressed chronically during aging14. Chronically expressed IFN-1 downregulates MEF2C, and loss of MEF2C leads to a ‘priming’ state as microglia become more sensitive to immune stimuli. A study by Sao et al. demonstrated that MEF2C mRNA expression in AD subjects was significantly lower than expression in control subjects27. Given that AD is a neurodegenerative disease with a chronic pro-inflammatory state, downregulation of MEF2C expression may contribute to exaggerated pro-inflammatory conditions of the disease27. Our results support the importance of SPI1 and MEF2C in HL cells expressing the respective genes (Fig. 3). While these genes may be inhibited or augmented in a targeted manner, the effects of various dosing and timing needs to be thoroughly examined due to their regulatory functions.
A limitation of our study is that most cell lines screened are not derived from AD-patient or brain tissues. Cell lines in the DepMap project, as most of the currently established human cell lines, are derived from cancer samples that may show properties not relevant to AD etiologies. Further, our analysis may miss insights from several critical cell types in AD not represented in DepMap, such as astrocytes and oligodendrocytes, and results for microglia are approximated using hematopoietic and lymphoid cell data. A wide array of cell lines derived from such tissues is unlikely to be available soon. Meanwhile, the gene candidates nominated herein to affect proximal cell types may be further investigated using relevant models, such as patient-derived iPSC or further derived cell lines. Data from genetic screen strategies targeting AD etiology could also strengthen findings. For example, Chiu et al. (2020) used a CRISPR-Cas9 screen to identify regulators of amyloid β peptide (Aβ) synthesis and described Cib1 as a novel negative regulator in this process28. Lastly, our expression-driven dependency analysis identifies targets whose knockout/knockdown/inhibition shows specificity in disease-aberrant cells, and even with limited use of AD models, our findings help rule out genes that are essential for cell lines across tissues or show unpredictable essentiality patterns that may be undesirable for a treatment target.
Conclusions
This study utilizes CRISPR knockout/RNAi knockdown screen data from DepMap to highlight potential candidates, from a list of AD-associated genes, to be further investigated as AD treatment targets. The expression-driven dependency analyses uniquely identify SPI1, MEF2C, GAB2, ABCC11, and ACTG1, to have desirable on- vs. off-target effects. These results provide a basis for applying these methods to newly identified genes in AD and evaluating targets in other genetic disorders.
Supplementary Information
Acknowledgements
The authors acknowledge the team from the DepMap project for generating the genetic screen data. This work was supported by NIH R35GM138113 and Mount Sinai seed fund to K.H.
Abbreviations
- AD
Alzheimers’ disease
- GWAS
Genome-wide association studies
- DepMap
The Cancer Dependency Map Project
- EOAD
Early-onset Alzheimer’s disease
- LOAD
Late-onset Alzheimer’s disease
- Aβ
Amyloid β peptide
- HL
Hematopoietic and lymphoid tissue
- CNS
Central nervous system
- AG
Autonomic ganglia
Author contributions
S.K.J. and A.E. conducted the bioinformatic analyses. K.H. conceived the research and designed the analyses. All authors wrote, read, and approved the manuscript.
Data availability
The datasets generated and analyzed in the current study are available from the DepMap project under the DepMap Public 20Q2 release https://doi.org/10.6084/m9.figshare.12280541.v3(14).
Competing interests
The authors declare no competing interests.
Footnotes
Publisher's note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
These authors contributed equally: Suraj K. Jaladanki and Abdulkadir Elmas.
Supplementary Information
The online version contains supplementary material available at 10.1038/s41598-021-91713-2.
References
- 1.Vos T, et al. Global, regional, and national incidence, prevalence, and years lived with disability for 328 diseases and injuries for 195 countries, 1990–2016: A systematic analysis for the Global Burden of Disease Study 2016. Lancet. 2017;390:1211–1259. doi: 10.1016/S0140-6736(17)32154-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Weller J, Budson A. Current understanding of Alzheimer's disease diagnosis and treatment. F1000 Res. 2018;7:1161. doi: 10.12688/f1000research.14506.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.van Dyck CH. Anti-amyloid-β monoclonal antibodies for Alzheimer’s disease: Pitfalls and promise. Biol. Psychiat. 2018;83:311–319. doi: 10.1016/j.biopsych.2017.08.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Rygiel K. Novel strategies for Alzheimer's disease treatment: An overview of anti-amyloid beta monoclonal antibodies. Indian J. Pharmacol. 2016;48:629–636. doi: 10.4103/0253-7613.194867. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Apter JT, Shastri K, Pizano K. Update on disease-modifying/preventive therapies in Alzheimer's disease. Curr. Geriatr. Rep. 2015;4:312–317. doi: 10.1007/s13670-015-0141-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Vaz M, Silvestre S. Alzheimer's disease: Recent treatment strategies. Eur. J. Pharmacol. 2020;887:173554. doi: 10.1016/j.ejphar.2020.173554. [DOI] [PubMed] [Google Scholar]
- 7.Howard R, Liu KY. Questions EMERGE as Biogen claims aducanumab turnaround. Nat. Rev. Neurol. 2020;16:63–64. doi: 10.1038/s41582-019-0295-9. [DOI] [PubMed] [Google Scholar]
- 8.Kunkle BW, et al. Genetic meta-analysis of diagnosed Alzheimer’s disease identifies new risk loci and implicates Aβ, tau, immunity and lipid processing. Nat. Genet. 2019;51:414–430. doi: 10.1038/s41588-019-0358-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Jansen IE, et al. Genome-wide meta-analysis identifies new loci and functional pathways influencing Alzheimer’s disease risk. Nat. Genet. 2019;51:404–413. doi: 10.1038/s41588-018-0311-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Zhang B, et al. Integrated systems approach identifies genetic nodes and networks in late-onset Alzheimer’s disease. Cell. 2013;153:707–720. doi: 10.1016/j.cell.2013.03.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Behan FM, et al. Prioritization of cancer therapeutic targets using CRISPR–Cas9 screens. Nature. 2019;568:511–516. doi: 10.1038/s41586-019-1103-9. [DOI] [PubMed] [Google Scholar]
- 12.Tsherniak A, et al. Defining a cancer dependency map. Cell. 2017;170:564–576. doi: 10.1016/j.cell.2017.06.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Giau VV, et al. Genetic analyses of early-onset Alzheimer’s disease using next generation sequencing. Sci. Rep. 2019;9:8368. doi: 10.1038/s41598-019-44848-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.DepMap, B. DepMap 20Q2 Public. 10.6084/m6089.figshare.12280541.v12280543, 10.6084/m9.figshare.12280541.v3 (2020).
- 15.Meyers RM, et al. Computational correction of copy number effect improves specificity of CRISPR–Cas9 essentiality screens in cancer cells. Nat. Genet. 2017;49:1779–1784. doi: 10.1038/ng.3984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Solana C, Tarazona R, Solana R. Immunosenescence of natural killer cells, inflammation, and Alzheimer's disease. Int. J. Alzheimers Dis. 2018 doi: 10.1155/2018/3128758. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Negi N, Das BK. CNS: Not an immunoprivilaged site anymore but a virtual secondary lymphoid organ. Int. Rev. Immunol. 2018;37:57–68. doi: 10.1080/08830185.2017.1357719. [DOI] [PubMed] [Google Scholar]
- 18.Issac TG, et al. Autonomic dysfunction: A comparative study of patients with Alzheimer's and frontotemporal dementia: A pilot study. J. Neurosci. Rural Pract. 2017;8:84–88. doi: 10.4103/0976-3147.193545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.McFarland JM, et al. Improved estimation of cancer dependencies from large-scale RNAi screens using model-based normalization and data integration. Nat. Commun. 2018;9:4610. doi: 10.1038/s41467-018-06916-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Cenini G, et al. Dissecting Alzheimer's disease pathogenesis in human 2D and 3D models. Mol. Cell Neurosci. 2020;1:103568. doi: 10.1016/j.mcn.2020.103568. [DOI] [PubMed] [Google Scholar]
- 21.Satoh J-I, Asahina N, Kitano S, Kino Y. A comprehensive profile of ChIP-Seq-Based PU.1/Spi1 target genes in Microglia. Gene Regul. Syst. Biol. 2014;8:127–139. doi: 10.4137/GRSB.S19711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Mathys H, et al. Single-cell transcriptomic analysis of Alzheimer's disease. Nature. 2019;570:332–337. doi: 10.1038/s41586-019-1195-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Huang KL, et al. A common haplotype lowers PU.1 expression in myeloid cells and delays onset of Alzheimer's disease. Nat. Neurosci. 2017;20:1052–1061. doi: 10.1038/nn.4587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Lavin Y, et al. Tissue-resident macrophage enhancer landscapes are shaped by the local microenvironment. Cell. 2014;159:1312–1326. doi: 10.1016/j.cell.2014.11.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Olmos-Alonso A, et al. Pharmacological targeting of CSF1R inhibits microglial proliferation and prevents the progression of Alzheimer's-like pathology. Brain. 2016;139:891–907. doi: 10.1093/brain/awv379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Deczkowska A, et al. Mef2C restrains microglial inflammatory response and is lost in brain ageing in an IFN-I-dependent manner. Nat. Commun. 2017;8:717. doi: 10.1038/s41467-017-00769-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Sao T, et al. MEF2C mRNA expression and cognitive function in Japanese patients with Alzheimer's disease. Psychiatry Clin. Neurosci. 2018;72:160–167. doi: 10.1111/pcn.12618. [DOI] [PubMed] [Google Scholar]
- 28.Chiu YW, et al. Identification of calcium and integrin-binding protein 1 as a novel regulator of production of amyloid beta peptide using CRISPR/Cas9-based screening system. FASEB J. 2020;34:7661–7674. doi: 10.1096/fj.201902966RR. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The datasets generated and analyzed in the current study are available from the DepMap project under the DepMap Public 20Q2 release https://doi.org/10.6084/m9.figshare.12280541.v3(14).