Abstract
The assessment of a patient’s immune function is critical in many clinical situations. In complex clinical immune dysfunction like sepsis, which results from a loss of immune homeostasis due to microbial infection, a plethora of pro- and anti-inflammatory stimuli may occur consecutively or simultaneously. Thus, any immunomodulatory therapy would require in-depth knowledge of an individual patient’s immune status at a given time. Whereas lab-based immune profiling often relies solely on quantification of cell numbers, we used an ex vivo whole-blood infection model in combination with biomathematical modeling to quantify functional parameters of innate immune cells in blood from patients undergoing cardiac surgery. These patients experience a well-characterized inflammatory insult, which results in mitigation of the pathogen-specific response patterns towards Staphylococcus aureus and Candida albicans that are characteristic of healthy people and our patients at baseline. This not only interferes with the elimination of these pathogens from blood, but also selectively augments the escape of C. albicans from phagocytosis. In summary, our model could serve as a valuable functional immune assay for recording and evaluating innate responses to infection.
Subject terms: Computational models, Infection, Inflammation, Innate immunity, Computational biology and bioinformatics, Immunology
Introduction
Critical illness may be associated with significant alterations in immune function, including both hypo- and hyper-inflammatory states. While impairment of immunity significantly increases the risk of infection, systemic inflammation promotes the development of multiple organ dysfunction syndrome. Therefore, assessment of the immune function of patients is desirable in clinical practice. However, most laboratory tests rely solely on quantification of immune cell populations. For example, the risk of infection during neutropenia is assessed by determining the number of neutrophils found in peripheral blood1. Similarly, the CD4+ T-cell count is used to quantify the degree of immunosuppression in HIV infection2. Although these assays are highly useful in clinical routine, they use quantitative thresholds and do not provide any information about immune cell function3. To improve understanding of a patient’s immune status, functional immune monitoring has been attempted using quantification of released proteins as indirect markers, such as IL-6, IL-8, procalcitonin and C-reactive protein4. All of these are derived from different immune cells, are mostly pleiotropic and may antagonize each other, all of which limits their clinical use. Sepsis-induced immunosuppression can be measured by reduced expression of the major histocompatibility (MHC) class II molecule HLA-DR on monocytes, which is associated with a diminished antigen-presenting capacity and a shift from pro- to anti-inflammatory cytokine production (reviewed in5,6). Similarily, neutrophil surface expression of high-affinity Fcγ receptor I (CD64) has been shown to increase in patients during the early immune response to bacterial infection and in systemic inflammatory response syndrome7–9. Several studies have demonstrated that CD64 measured as an index may be useful for the detection and management of sepsis and bacterial infection in neonatal intensive care units and in adult hospital patients10,11. However, while surface markers reflect the activation status of immune cells, they do not provide a direct functional readout. Therefore, in a more complex setting, ex vivo stimulation of whole blood with LPS and quantification of cellular cytokine release has been used to quantify immune function. LPS-induced TNF-α release by monocytes in whole blood tends to be less in immunosuppressed patients compared to healthy individuals12,13. Nevertheless, this approach relies on a secreted cytokine as an indirect marker of cellular immune function. The most functional read-out described so far is the assessment of neutrophil dysfunction in critically ill patients by measuring neutrophil capacity to clear zymosan particles ex vivo3,14. However, these assays selectively involve neutrophils and require prior isolation of the cells, which may alter their function, especially in patients with activated neutrophils15.
In previous studies, we applied a systems biology approach to investigate the immune response to pathogens in blood from healthy individuals16–18. Using ex vivo whole-blood infection in combination with biomathematical modeling enabled the calculation of functional parameters for blood immune cells, such as kinetic rates for cell migration and pathogen uptake. Based on these results, we have performed a pilot study to investigate whether a systems biology approach allows quantification of immune function in a clinical setting. We selected patients undergoing cardiac surgery with extracorporeal circulation. Such patients receive a standardized anesthesia regimen and operation procedure and have a well-characterized inflammatory response (e.g. increase of blood lipopolysaccharide and β-D-glucan levels), which enables analysis of immune function before and after the insult19–21. By determining functional parameters of innate immune cell populations after ex vivo whole-blood bacterial (Staphylococcus aureus) and fungal (Candida albicans) infection, we show that post-surgery inflammation results in attenuation of inter-patient and inter-pathogen differences in immune response patterns. Moreover, our model revealed enhanced immune escape by C. albicans that was not observed for S. aureus, indicating pathogen-specific adaptation to an altered immune status.
Results
Pathogen-specific immune response patterns during bacterial and fungal whole-blood infection
Prior analyses in whole blood have been carried out using viable pathogens16. However, quantification of immune cell function in patient blood samples requires the use of inactivated stimuli to avoid the effects of pathogen inactivation by antibiotics in patient blood. Thus, we quantified immune cell functional parameters for two inactivated pathogens, Staphylococcus aureus (bacterial) and Candida albicans (fungal). During a 4-h time course, neutrophils were the predominant immune cell type to interact with both pathogens (Fig. 1a). Both pathogens showed a similar association with neutrophils after 60 min of blood infection. However, the early 10 min time point revealed much faster association of S. aureus with neutrophils and monocytes (70.8 ± 10.2% and 7.6 ± 1.8%, respectively) compared with C. albicans (45.0 ± 13.3% and 3.1 ± 0.5%, respectively) (Fig. 1a,b). Consistent with the different association kinetics, C. albicans was cleared more slowly than S. aureus (10 min p.i.: extracellular C. albicans 52.9 ± 13.6%, extracellular S. aureus 21.6 ± 10.5%, Fig. 1c). Notably, 4 h after infection, a larger population of C. albicans cells remained extracellular compared to S. aureus (extracellular C. albicans 13.0 ± 4.3%, extracellular S. aureus 4.0 ± 1.5%). Comparison of these data with previous studies and a new set of experiments using viable C. albicans showed similar patterns overall, but also revealed a significantly higher number of inactivated fungal cells associated with monocytes (60 min p.i.: viable C. albicans 4.9 ± 2.3%, inactivated C. albicans 13.4 ± 2.5%, P < 0.001) (Fig. 1f and16). Association with lymphocytes could not be detected for either pathogen regardless of their viability. Whole-blood infection with C. albicans in either an active or inactive state induced a strong and comparable secretion of monocyte-derived cytokines (TNF-α, IL-1β and IL-6), whereas only low cytokine levels could be detected in mock-infected blood (Fig. 1g).
Figure 1.
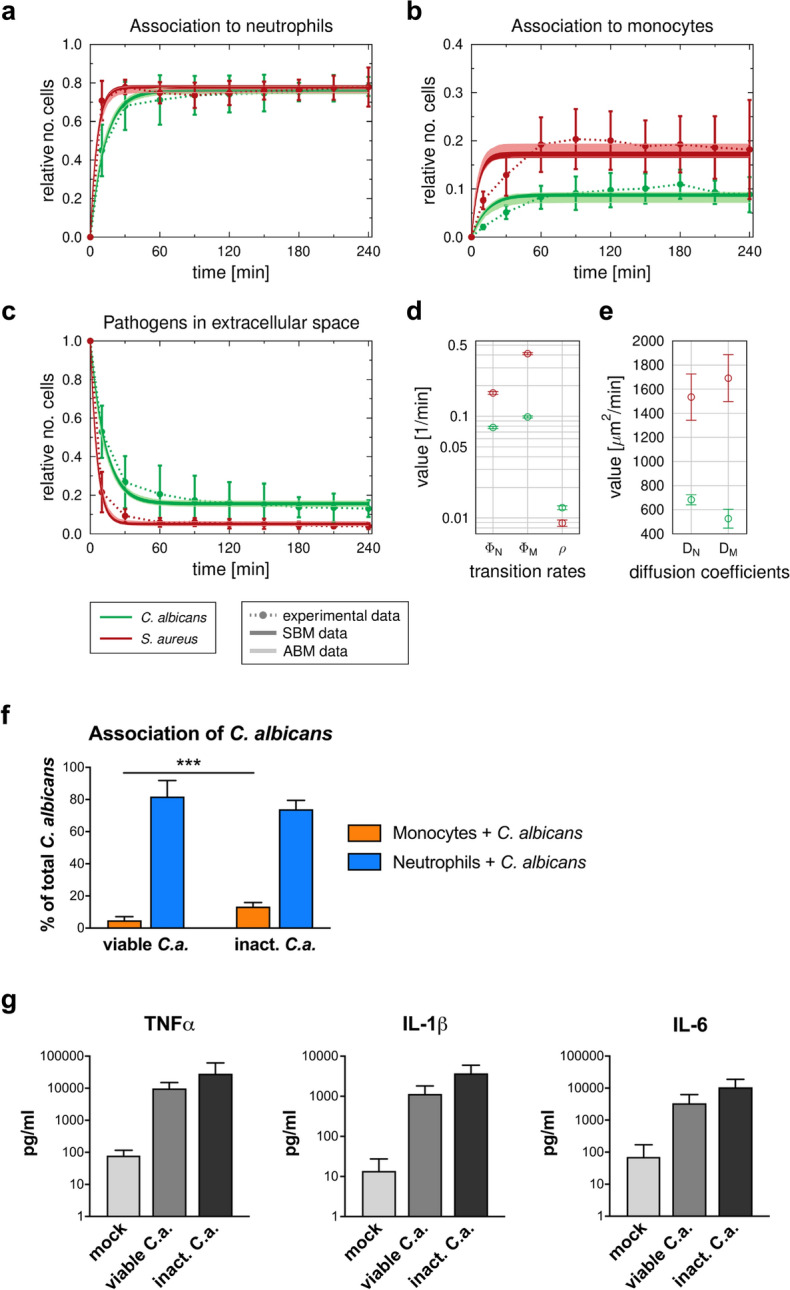
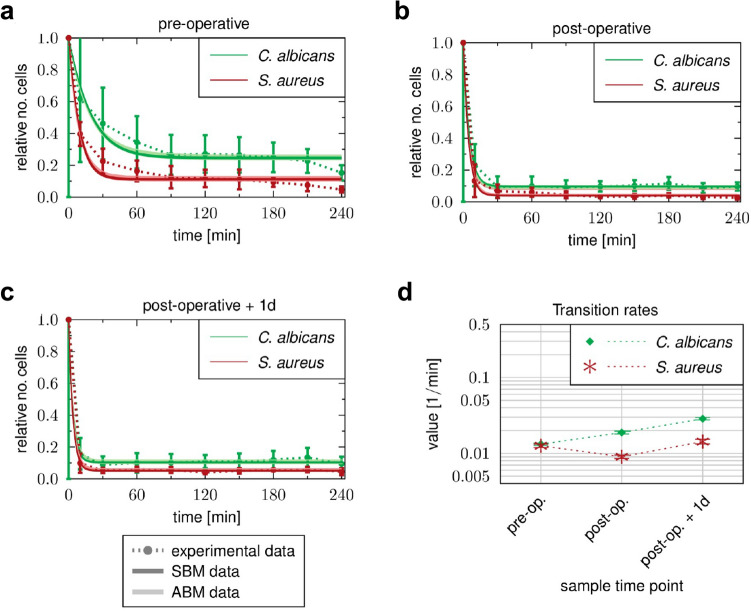
Comparison of the dynamics of host–pathogen interaction during C. albicans and S. aureus infection in healthy blood. (a–c) Results of fitting the state-based model (SBM) and the agent-based model (ABM) to experimental data. Simulated dynamics of the combined units (solid line) were obtained by fitting the SBM (dark color) and the ABM (light color) to the experimentally measured association kinetics (dotted line). Experimental data were gained from whole-blood infection assays with either C. albicans (green, n = 10) or S. aureus (red, n = 7). SBM: line thickness represents SD obtained by 50 simulations with transition rate values sampled within their corresponding SD. ABM: line thickness represents the standard deviations obtained from 30 stochastic simulations with the estimated diffusion coefficients. (a) and (b), dynamics of the combined units PN and PM, which correspond to the experimental data on pathogens associated with neutrophils and monocytes. (c) kinetics of the combined unit PE together with experimentally measured kinetics of either fungal or bacterial cells in extracellular space. (d) Mean values (± SD) of transition rate values obtained by fitting the SBM to experimental data using simulated annealing. The rate of phagocytosis by neutrophils () and by monocytes () as well as the rate for immune escape () are depicted for infection scenarios with either C. albicans (green circle) or S. aureus (red circle). (e) Diffusion coefficients for neutrophils and monocytes were estimated by fitting the ABM to the experimental data for C. albicans (green circle) and S. aureus (red circle), respectively. Mean and SD are calculated from all parameter sets with a mean LSE within the SD of the optimal parameter set. (f) Association of viable and inactivated C. albicans with blood monocytes and neutrophils after 60 min quantified using flow cytometry. Significance was estimated using the unpaired, two-sided Student t test (***P < 0.001). (g) Release of monocyte-derived cytokines (TNF-α, IL-1β, IL-6) in plasma samples generated from 4 h whole-blood infection experiments in response to viable and inactivated C. albicans cells was investigated. Bars are shown as means ± SD of at least 3 independent experiments with whole blood from different donors.
In order to comparatively quantify the functional properties of immune cells we calibrated the state-based virtual infection model (SBM) to the experimental data (see “Methods” section, Table 1, Fig. 1d). The coefficients of variation of the resulting transition rates are very small (< 7%) and the corresponding dynamics of the three combined units are similar to the mean values of the respective experimental data, indicating a good match to the experimental data (see Fig. 1a–c). By comparing the resulting transition rate values, we observed significantly higher phagocytosis rates for S. aureus than for C. albicans (for neutrophils with , for monocytes with ). The opposite ratio was found with respect to the rate of immune escape (, ( with ). These parameter differences lead to an overall faster removal of S. aureus cells from the extracellular space and a higher amount of immune-evading C. albicans at 240 min after infection (Fig. 1c).
Table 1.
Transition rate values of the SBM for C. albicans and S. aureus infection of blood samples taken from either healthy volunteers or HLM patients before cardiac surgery (pre-operative), immediately after surgery (post-operative) and one day after admission to intensive care (post-operative + 1d).
| Transition rate | Mean SD (CV ) | |||||||
|---|---|---|---|---|---|---|---|---|
| Healthy | Pre-operative | Post-operative | Post-operative + 1d | |||||
| C. albicans | S. aureus | C. albicans | S. aureus | C. albicans | S. aureus | C. albicans | S. aureus | |
| 7.75 ± 0.20 (2.6) | 16.94 ± 0.55 (3.3) | 4.45 ± 0.14 (3.1) | 10.65 ± 0.36 (3.3) | 16.67 ± 0.36 (2.2) | 20.86 ± 0.54 (2.6) | 23.24 ± 0.77 (3.3) | 25.20 ± 0.72 (2.9) | |
| 9.87 ± 0.23 (2.4) | 41.19 ± 9.45 (2.3) | 9.29 ± 0.28 (3.0) | 24.74 ± 0.64 (3.3) | 94.41 ± 1.98 (2.1) | 97.94 ± 0.98 (3.3) | 72.28 ± 2.23 (3.1) | 84.60 ± 2.24 (3.3) | |
| 1.26 ± 0.06 (4.4) | 0.89 ± 0.06 (7.0) | 1.32 ± 0.05 (3.8) | 1.26 ± 0.07 (5.3) | 1.88 ± 0.08 (4.3) | 0.90 ± 0.05 (5.4) | 2.86 ± 0.10 (3.5) | 1.43 ± 0.10 (7.0) | |
The transition rate values are given by the phagocytosis rate of neutrophils, the phagocytosis rate of monocytes and the rate for acquiring immune escape .
Based on the estimated parameters of the SBM, we also simulated the infection scenario with both pathogens in an agent-based model (ABM) (see “Methods” section). Similar to the SBM dynamics, the resulting dynamics of the ABM are in agreement with the experimental data (see Fig. 1a–c). Diffusion coefficients for neutrophils and monocytes are two to four times higher for S. aureus infection compared to C. albicans . Furthermore, the diffusion coefficient for neutrophils is higher than for monocytes during C. albicans infection and vice versa for S. aureus infection (Fig. 1e).
Cardiac surgery results in neutrophil mobilization and immune activation
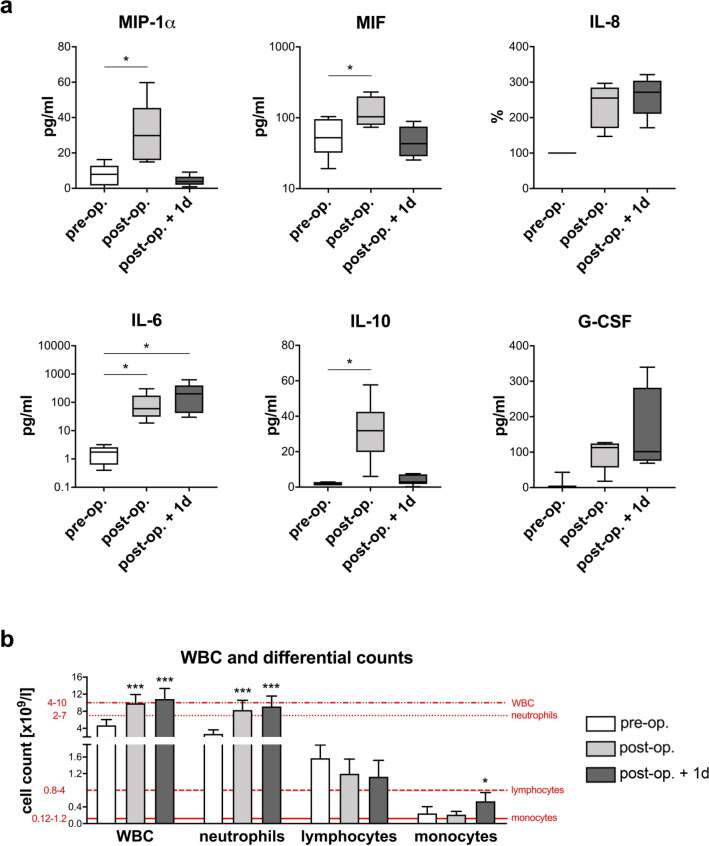
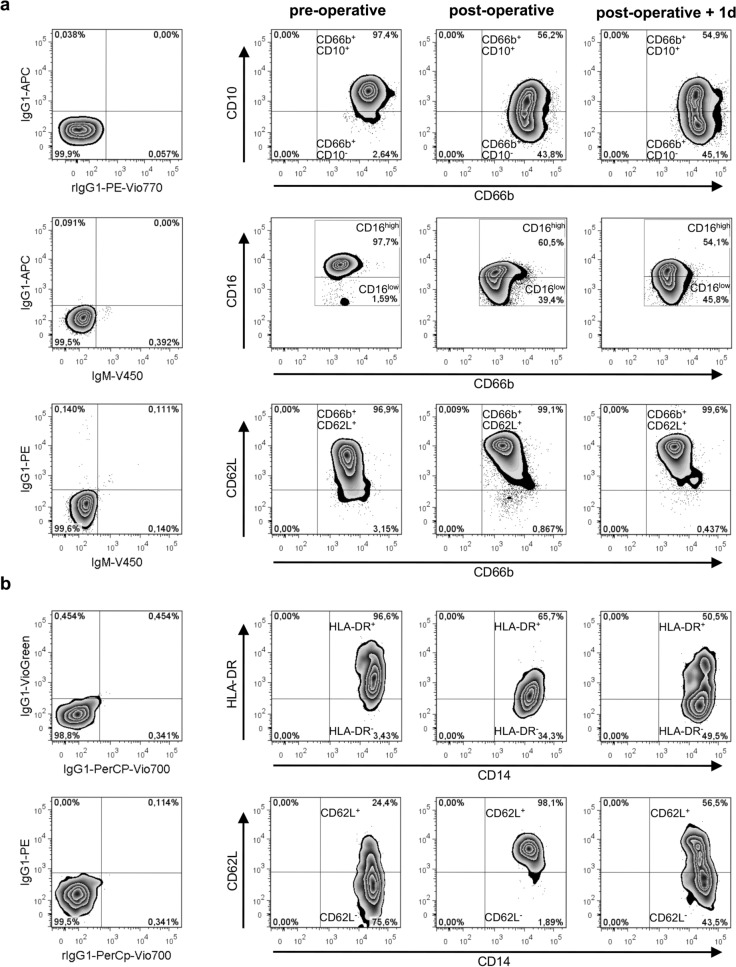
The data generated so far clearly show that a combination of ex vivo blood infection and virtual infection modelling allows quantification of immune cell functions. To address the feasibility of this approach in a clinical setting, we used blood samples from patients undergoing cardiac surgery for mitral valve insufficiency (heart lung machine (HLM) patients). Six patients (4 female, 2 male) aged between 52 and 74 years were recruited. All received minimally invasive mitral valve replacement or reconstruction surgery via lateral thoracotomy on cardiopulmonary bypass (CPB). Anesthesia was induced by Propofol, Sufentanyl and Rocuronium and sustained by Sufentanyl and Sevofluran before and Propofol during CPB. Three patients underwent additional surgery such as tricuspid valve surgery, cryoablation, atrial appendage closure and atrial septal defect closure. Bypass time ranged from 99 to 220 min, the average cumulative fluid balance on the day of surgery was 3316 ± 2486 ml (see Supplementary Table S1 for individual patient data). All patients were extubated on the day of surgery and none had any major complications. The only post-operative infection was a urinary tract infection more than two weeks after surgery. After informed consent, blood samples were taken before cardiac surgery (pre-operative), directly after surgery (post-operative) and one day after admission to intensive care (post-operative + 1d). These defined time points together with the low variance in surgical time and duration of extracorporeal circulation as well as the time-defined, strong and homogeneous inflammatory stimulus allowed us to clearly distinguish between inter-individual differences and the effects of inflammation. Levels of the pro-inflammatory cytokines IL-6, IL-8, MIP-1α and MIF and the anti-inflammatory cytokine IL-10 were increased in post-operative blood (Fig. 2a), whereas TNF-α and IL-1β were not detectable in plasma samples before and after surgery. Elevated levels of IL-6 and IL-8 were sustained until one day after surgery. Furthermore, the mean total white blood cell count increased after surgery (pre-operative: 4.70 ± 1.37 × 109/L, post-operative: 9.87 ± 2.05 × 109/L, post-operative + 1d: 10.89 ± 2.45 × 109/L) (Fig. 2b, see Supplementary Table S1 for individual patient data). Quantitative analysis of white blood cells revealed a significant increase in neutrophils (both post-operative and post-operative + 1d) and monocytes (post-operative + 1d), whereas lymphocyte counts remained stable. Neutrophilia following cardiac surgery is mediated by granulocyte colony-stimulating factor (G-CSF)-induced bone marrow neutrophil release22. G-CSF was markedly higher in plasma samples obtained directly and one day after surgery compared to pre-operative blood (Fig. 2a). Furthermore, whereas pre-operative blood was characterized by a homogenous neutrophil population that expressed abundant CD10 and high CD16 surface expression (mature neutrophils), the neutrophil population after surgery contained a CD10-negative subpopulation with reduced CD16 and increased L-selectin (CD62L) expression, indicating recruitment of immature neutrophils (Fig. 3a). Post-operatively, the CD10neg neutrophil subpopulation accounted for 49.3 ± 4.1% of the total neutrophil population and remained fairly stable after one day (46.7 ± 2.6%). Whereas no changes in monocyte counts could be observed between the pre- and post-operative time points, total monocyte numbers were significantly increased one day after surgery (pre-operative: 0.24 ± 0.16 × 109/L, post-operative + 1d: 0.54 ± 0.21 × 109/L, P < 0.05) (Fig. 2b, see Supplementary Table S1 for individual patient data). Expression of the MHC class II antigen HLA-DR on monocytes was markedly decreased after surgery (Fig. 3b). Together with the increased CD62L expression, this suggests that surgery induced an immature monocyte state.
Figure 2.
Blood after surgery shows changes in cytokine profiles and peripheral differential cell counts. Blood samples from six HLM patients taken before cardiac surgery (pre-operative), immediately after surgery (post-operative) and one day after admission to intensive care (post-operative + 1d) were analyzed for (a) cytokine levels (MIP-1α, MIF, IL-8, IL-6, IL-10, G-CSF) using Luminex technology and (b) white blood cell (WBC) as well as neutrophil, lymphocyte and monocyte counts using an automated hematology analyzer. Reference ranges of leukocytes are indicated in red. The plasma concentrations of cytokines are presented in pg/ml, except for IL-8. Levels for IL-8 in post-operative and post-operative + 1d blood samples were normalized to respective plasma levels within pre-operative blood (set to 100%) for each donor and means ± SD of the calculated percentages are shown. Significance was estimated using the unpaired, two-sided Student t test and shown as *P < 0.05, ***P < 0.001.
Figure 3.
Surface phenotypes of monocytes and neutrophils after surgery indicate an increased number of immature cells in blood of HLM patients. Flow cytometry analysis of CD66b+ neutrophils (a) and CD14+ monocytes (b) from whole blood of HLM patients taken before cardiac surgery (pre-operative), immediately after surgery (post-operative) and one day after admission to intensive care (post-operative + 1d) are shown. (a) Immature phenotype of CD66b+ blood neutrophils was analyzed by surface expression of CD10, CD16 and CD62L. Representative zebra plots show a change of the surface phenotype by the shift of the population to the lower right quadrant for CD10 and CD16 and to the higher right quadrant for CD62L. (b) Surgery-induced changes on CD14+ blood monocytes are represented by zebra plots of HLA-DR and CD62L expression patterns. Left plots show proper gate setting defined by isotype control stainings.
Pro-inflammatory stimulus during cardiopulmonary bypass mitigates inter-patient and inter-pathogen differences in immune response patterns
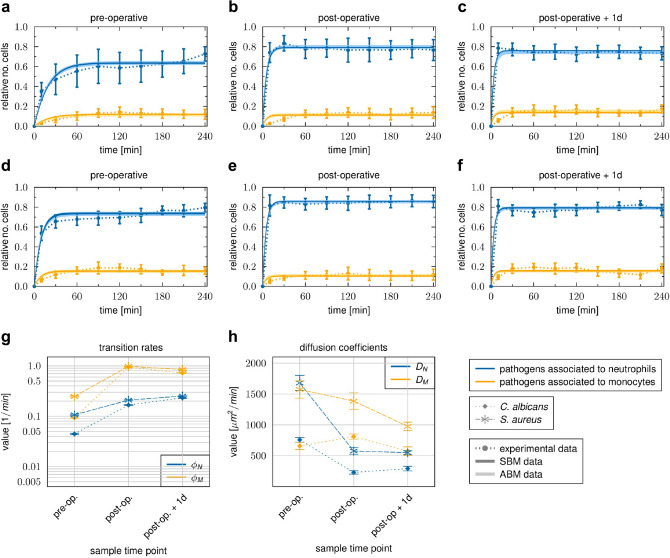
To quantify functional immune parameters in whole blood after cardiopulmonary bypass, we perfomed whole-blood infection with inactivated S. aureus or C. albicans as described above. These analyses revealed clear changes in innate immune response patterns against C. albicans and S. aureus. In blood collected after surgery, association of the pathogens with neutrophils and monocytes was increased in comparison to the pre-operative time point and occurred more quickly (Fig. 4). For instance, 10 min post infection, 35.1 ± 8.8% of C. albicans and 53.7 ± 7.2% of S. aureus cells, respectively, were associated with neutrophils in pre-operative blood (Fig. 4a,d), compared to 78.5 ± 5.1% for C. albicans and 81.1 ± 6.7% for S. aureus one day after surgery (Fig. 4c,f). To test whether the altered association kinetics could be explained by changes in leukocyte numbers or indicate functional differences, we fitted the SBM to the association kinetics and used the means of the measured immune cell counts as input for the model simulations. These analyses clearly showed that the faster association kinetics after surgery could only be explained by a significant increase in the phagocytosis rates of neutrophils and monocytes and not solely by increased immune cell counts (Fig. 4g). Importantly, pathogen-specific response patterns that were clearly visible in blood from healthy donors and in samples taken before surgery were absent after the pro-inflammatory stimulus. More specifically, S. aureus had increased and faster association with neutrophils and monocytes than C. albicans in blood taken before surgery (Fig. 4a,d). Comparing the transition rates for phagocytosis by neutrophils ( and monocytes ( between both pathogens revealed significantly higher values for S. aureus (, ) (Fig. 4g). In contrast, association kinetics for C. albicans and S. aureus as well as the corresponding phagocytosis rates for neutrophils and monocytes were almost similar in blood after surgery (e.g. post-operative + 1d: , ; Fig. 4g).
Figure 4.
Time courses of pathogen association to immune cells observed in whole-blood samples of HLM patients. Blood samples were taken before cardiac surgery (pre-operative), immediately after surgery (post-operative) and one day after admission to intensive care (post-operative + 1d). Time-resolved experimental data (dotted line) were obtained by whole-blood infection assays with either C. albicans (a–c) or S. aureus (d–f). Data points and error bars refer to the means and SD of blood samples from six HLM patients. The simulated dynamics of the combined units (solid line) were obtained by fitting the state-based model (SBM, dark color) and the agent-based model (ABM, light color) to the experimental data. The thickness of the results from the SBM represents the SD obtained by 50 simulations with transition rate values that were sampled within their corresponding SD. The thickness of the results from the ABM represents the SD obtained from 30 stochastic simulations of the ABM with the estimated diffusion coefficients. (g) Transition rate values of the SBM resulting from fitting the model to experimental data of either C. albicans or S. aureus infection in blood samples from HLM patients. The transition rate values are given for the phagocytosis rate of neutrophils and the phagocytosis rate of monocytes. (h) The diffusion coefficients are given for neutrophils and monocytes . Mean and SD are calculated from all parameter sets with a mean LSE that lies within the SD of the optimal parameter set.
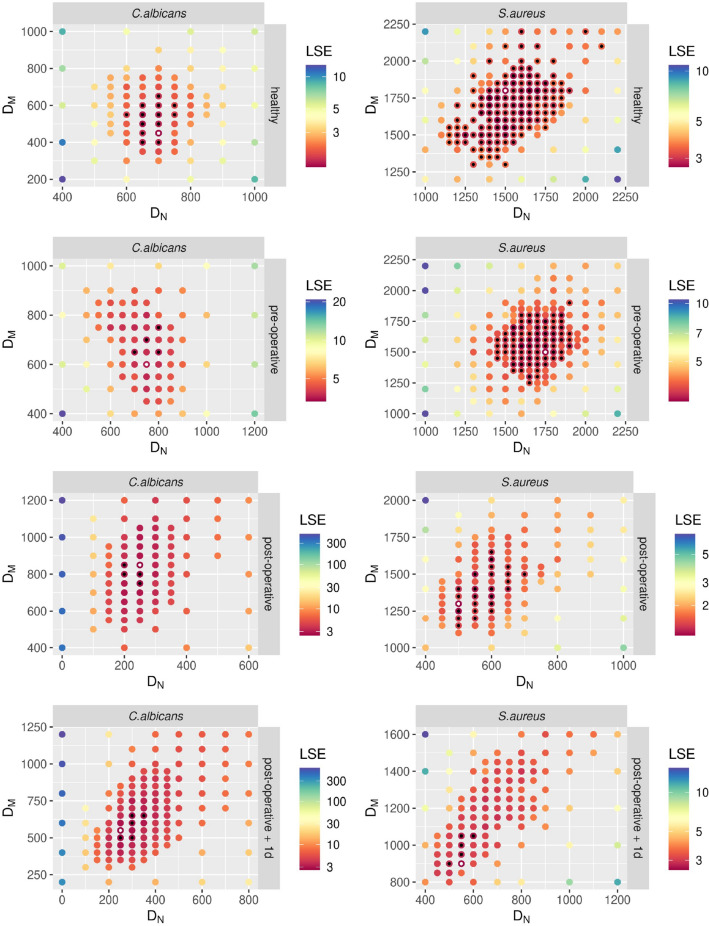
In addition, we investigated immune cell migration in blood after surgery compared to the pre-operative time point using the ABM. As in healthy donors, diffusion coefficients for monocytes and neutrophils during S. aureus infection exceeded diffusion coefficients during C. albicans infection by a factor of two to three (Figs. 4h, 5). Infection of post-operative blood with C. albicans resulted in a decrease in by a factor of and an increase in by a factor of (Figs. 4h, 5). Similarly, was decreased by a factor of during S. aureus infection. Contrary to confrontation with C. albicans, was not increased in S. aureus-infected post-operative blood, but rather slightly decreased (Figs. 4h, 5). Furthermore, did not change from the post-operative time point to one day after surgery for both species. However, in the case of we observed a decrease for both species by a factor of approximately . Comparing the values for and between the pathogens for each sample time point showed that both diffusion constants were higher in the presence of S. aureus compared to C. albicans for all time points. Similar to the immune cell association and phagocytosis rates, differences in neutrophil and monocyte diffusion between the fungal and bacterial pathogen decreased after surgery (Fig. 4h).
Figure 5.
Results of fitting the agent-based model (ABM) to the experimental data from C. albicans and S. aureus infection using the method of adaptive regular grid search. The parameter space is shown for fitting the ABM to experimental data, where blood samples from healthy donors and HLM patients before surgery (pre-operative), immediately after surgery (post-operative) and one day after admission to intensive care (post-operative + 1d) were infected with either C. albicans (left column) or S. aureus (right column). Colors of the points refer to the weighted least squares error for each parameter set . The optimal parameter set is marked with a white dot. All parameter sets with a mean LSE that lies within the SD of the optimal parameter set are marked with a black dot.
In addition to reducing differences in the responses to the two pathogens, the inflammatory stimulus decreased inter-individual differences in the association of pathogens with immune cells. For instance, the percentage standard deviation of pathogens that interacted with neutrophils decreased from 32 to 10% for C. albicans (Fig. 4a,c) and 13.5% to 8% for S. aureus infection (Fig. 4d,f) between samples collected pre-operatively and one day after surgery.
Taken together, the kinetics of immune responses were accelerated after the pro-inflammatory stimulus independent of the microbial trigger. This resulted not only from the large increase in immune cell numbers, but also from increased transition rates (see Fig. 4g). As a consequence, pathogen-specific and inter-individual differences were less pronounced after surgery.
Monocyte activation by C. albicans and S. aureus is dampened after cardiopulmonary bypass
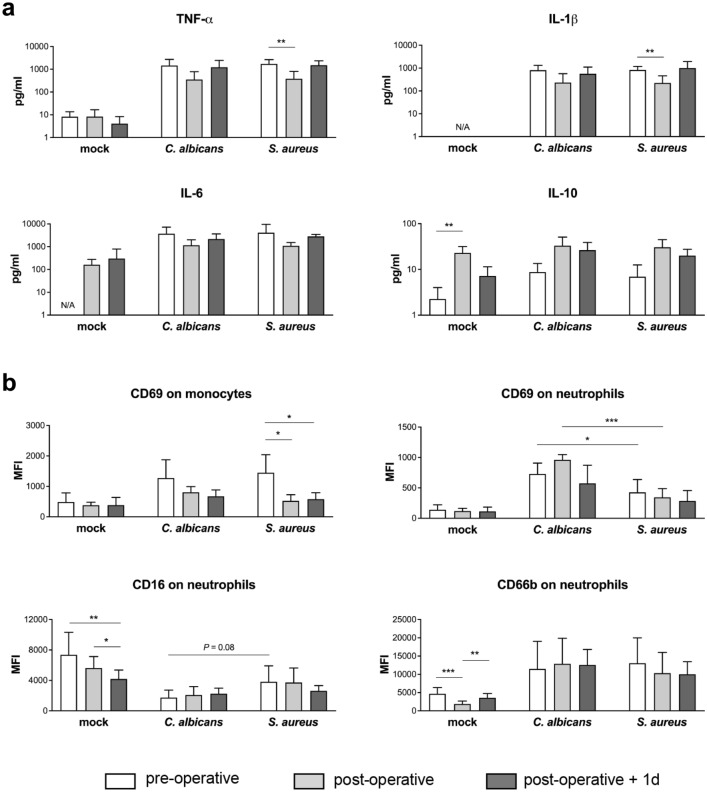
We further investigated consequences of cardiopulmonary bypass surgery-induced inflammation on activation of innate immune cells during infection. Activation of monocytes was analyzed by their surface expression of early activation antigen CD69 and cytokine secretion in response to microbial confrontation, and was reduced after cardiac surgery. The presence of C. albicans or S. aureus induced increased release of the monocytic cytokines TNF-α, IL-1β and IL-6 in blood from all three time points tested in HLM patients compared to the corresponding mock-treated samples (Fig. 6a). However, the induced levels were markedly lower in post-operative blood during both C. albicans and S. aureus infection (C. albicans: TNF-α: pre-operative 1456 ± 1248 pg/ml, post-operative 355 ± 428 pg/ml, P = 0.068; S. aureus: TNF-α: pre-operative 1730 ± 903 pg/ml, post-operative 380 ± 427 pg/ml, P < 0.01). The reduced pro-inflammatory cytokine release suggests down-regulation of monocyte activity, which could have been mediated by increased IL-10 levels detected in blood obtained directly after surgery (see Fig. 2a). Neither C. albicans nor S. aureus infection was able to induce further IL-10 secretion compared to mock-treated post-operative samples (Fig. 6a). Plasma concentrations of IL-10 almost returned to pre-operative values after one day in the ICU, and accordingly, release of monocytic cytokines was increased again in response to both pathogens (TNF-α: C. albicans 1229 ± 1234 pg/ml, S. aureus 1507 ± 852 pg/ml). In line with the cytokine data, elevated CD69 surface levels could be detected on monocytes 4 h after inoculation of C. albicans (MFI 1276 ± 598) or S. aureus (MFI 1447 ± 597) compared to mock-infection (MFI 488 ± 298) in pre-operative blood without any quantitative differences between the pathogens (Fig. 6b). A less pronounced response was detected in blood after surgery (S. aureus: post-operative MFI 524 ± 202, post-operative + 1d MFI 580 ± 212). Together with reduced monocytic cytokine release, these data indicate decreased monocyte stimulation following infection of blood after surgery. Analysis of the surface phenotype of neutrophils revealed significant differences in the expression of CD16 and CD66b between mock-infected control samples in blood taken before and after surgery as well as after one day in the ICU that could be explained by the inflammation-dependent recruitment of immature neutrophils (Fig. 6b). Activation was largely restricted to those neutrophils which had phagocytosed either C. albicans or S. aureus, and was stronger in response to C. albicans, shown by a more pronounced increase in CD69 (pre-operative: mock-infected MFI 141 ± 82, C. albicans-infected MFI 730 ± 178, S. aureus-infected MFI 427 ± 211) and down-regulation of CD16 (pre-operative: mock-infected MFI 7359 ± 2962, C. albicans-infected MFI 1721 ± 998, S. aureus-infected MFI 3812 ± 2108). However, the decrease in surface CD16 and up-regulation of the degranulation marker CD66b on pathogen-associated neutrophils was equal in blood from all three time points for the respective infections, even though surface levels of both activation markers were different on neutrophils in pre- and post-operative blood. Comparable results were observed for CD69 expression.
Figure 6.
Changes in cytokine secretion and innate immune cell activation in whole blood from HLM patients after surgery. Blood samples from HLM patients were taken before cardiac surgery (pre-operative, non-filled bars), directly after surgery (post-operative, light grey bars) and one day after admission to intensive care (post-operative + 1d, dark grey bars) and either mock-infected, treated with C. albicans or S. aureus for 4 h. (a) Plasma levels of TNF-α, IL-1β, IL-6 and IL-10 were quantified and bars are shown as means ± SD. Results are presented as pg/ml; N/A stands for values not available. (b) Surface marker expression was analyzed on the total monocyte population and on pathogen-associated neutrophils by flow cytometry. Data shown are mean fluorescence intensity (MFI) ± SD. Significance is shown as *P < 0.05; **P < 0.01; ***P < 0.001, unpaired, two-sided Student t test.
Inflammatory stimulus during cardiopulmonary bypass induces enhanced C. albicans immune escape
Despite the quantitative increase in leukocyte numbers and their functional changes, a fraction of C. albicans and S. aureus cells was not phagocytosed by neutrophils or monocytes even after 4 h of infection, and remained extracellular (Fig. 7). Comparing between different sample time points, we observed for both pathogens that the percentages of extracellular pathogens were decreased after surgery and the slope of the reaction curve was increased during the initial phase. Specifically, 60 min after inoculation of total C. albicans cells were not associated with immune cells in pre-operative blood (Fig. 7a). However, in blood samples collected immediately (post-operative) and one day after surgery, only and C. albicans cells, respectively, were not associated with immune cells at 60 min post infection (Fig. 7b,c). Although these results indicated that elimination of pathogens from the extracellular space was more efficient after surgery, pathogen-specific differences were present in the blood under the three tested conditions. In every case, the percentage of extracellular cells was higher for C. albicans than for S. aureus following 240 min of infection (Fig. 7a–c). Our model predicted that at 240 min all extracellular cells of both pathogens would have undergone immune escape (). Interestingly, despite increased neutrophil numbers and neutrophil phagocytosis rates in blood collected after surgery, estimated transition rates for acquiring immune escape () were twofold higher for C. albicans in post-operative compared to pre-operative samples, which was in contrast to S. aureus, which had almost equal values for the different sample time points (Fig. 7d, Table 1). In fact, this resulted in a changed ratio of between S. aureus and C. albicans infection in post-operative blood. While values were similar for C. albicans and S. aureus in pre-operative blood (), larger differences could be detected in blood samples taken after surgery (post-operative: , post-operative + 1d: ).
Figure 7.
Time courses of extracellular pathogens observed in whole-blood samples of HLM patients. Blood samples were taken (a) before cardiac surgery (pre-operative), (b) immediately after surgery (post-operative) and (c) one day after admission to intensive care (post-operative + 1d). Time-resolved experimental data (dotted line) were obtained by whole-blood infection assays with either C. albicans (green) or S. aureus (red). Data points and error bars refer to the means and SD of blood samples from six HLM patients. The simulated dynamics of the combined units (solid line) were obtained by fitting the state-based model (SBM, dark color) and the agent-based model (ABM, light color) to the experimental data. The thickness of the results from the SBM represents the SD obtained by 50 simulations with transition rate values that were sampled within their corresponding SD. The thickness of the results from the ABM represents the SD obtained from 30 stochastic simulations of the ABM with the estimated diffusion coefficients. (d) Transition rate values for immune evasion of the SBM resulting from fitting the model to experimental data of either C. albicans or S. aureus infection in blood samples from HLM patient.
Discussion
In this study, we applied a systems biology approach, where experimental whole-blood infection assays were combined with virtual infection modeling to investigate the effect of immune activation on the immune defense against pathogens in whole blood. In order to analyze the immune response of patients that suffer from imbalanced immune homeostasis, we used blood samples of patients who underwent cardiac surgery with extracorporeal circulation. The surgical intervention and the cardiopulmonary bypass are known to trigger a systemic inflammatory response. Although this transitory inflammation is not usually related to infection or sepsis, it can contribute to post-operative complications such as organ dysfunction or bleeding23. Pathophysiologically, a multitude of triggers that include surgical trauma, ischemia and reperfusion injury as well as endotoxinemia and glucanemia results in an acute phase reaction largely but not exclusively mediated by activation of NF-κB20,24. In particular, the inflammatory stimulus causes a surplus of the complement protein C5a, which serves as an important chemoattractant for several innate immune cells, including neutrophils and monocytes25,26. C5a molecules activate neutrophils and induce neutrophil degranulation and superoxide generation26–31. Due to this pro-inflammatory stimulus, patients show leukocytosis after cardiac surgery, including mobilization of neutrophils from the bone marrow. Furthermore, their monocytes express reduced levels of HLA-DR and increased CD62L32. Thus, transient hyperinflammation can be studied well in elective cardiac bypass surgery patients.
In accordance with this, we observed leukocyte recruitment after surgery in our study. In particular, the number of neutrophils increased more than twofold due to the recruitment of CD10neg immature neutrophils. In addition, monocytes were increased at day one, but already showed an immature phenotype in blood taken directly after surgery. Despite the higher abundance of immature neutrophils and monocytes, pathogen association with immune cells occured faster and to a larger extent in post-operative compared to pre-operative blood samples. By calibrating the state-based virtual infection model to the association kinetics of samples taken before and after surgery, we found that this was not solely due to increased immune cell numbers. Rather, phagocytosis rates of neutrophils and monocytes increased by twofold after surgery, indicating an enhanced activation state. These quantitative results showing the increase in immune cell activity could not be detected by selectively quantifying surface activation marker expression, such as the neutrophil degranulation marker CD66b. Despite the high abundance of immature neutrophils recruited in response to transient hyperinflammation, these cells showed the same maximum of CD66b and CD69 up-regulation and decrease in surface CD16 expression upon fungal phagocytosis as mature neutrophils in pre-operative blood. Immature neutrophils contain three types of granules and can produce reactive oxygen species33,34, indicating that these cells are a functional subset that is released under certain conditions. Studies by Leliefeld et al. and van Grinsven et al. using experimentally-induced acute inflammation in healthy subjects by intravenous administration of bacterial LPS have demonstrated that the different neutrophil subsets not only differ in phenotype but also in function35,36. Immature neutrophils were shown to have even higher antibacterial capacity in vitro than mature neutrophils, and exhibit efficient migration, which led to the hypothesis that immature neutrophils are not released as bystanders but rather as cells that are more efficient in pathogen killing, in line with the observed increased immune cell activity in our study. Other previous studies have reported an increase in immune cell activation for monocytes and neutrophils after surgery, by evaluating the expression of immune cell specific activation markers and the cytokine profile37–39. However, they did not investigate quantitative changes in the immune cell functionality during the response to different pathogens serving as stimuli. The observed functional activation also mitigates pathogen-specific patterns of immune activation. Prior to the inflammatory insult and as observed in samples from healthy control donors, S. aureus infection induced a faster immune response than C. albicans infection. In contrast, after surgery, the inter-pathogen differences were substantially reduced.
Additionally, we analyzed the concentration of pro- and anti-inflammatory cytokines in blood samples of HLM patients before and after surgery. Interestingly, the pro-inflammatory cytokines IL-1β and TNF-α were not present in higher levels after surgery, whereas an increase was observed for others, such as IL-6. In addition, the anti-inflammatory cytokine IL-10 was also significantly increased at the end of surgery, indicating the activation of anti-inflammatory pathways. These findings are in line with several studies reporting the surgery-induced release of both pro- and anti-inflammatory mediators27,40–43. During ex vivo infection, the release of monocytic cytokines and the up-regulation of early activation antigen CD69 on monocytes were markedly lower in post-operative blood. Together with the immature phenotype of blood monocytes, this points towards a regulatory effect of increased IL-10 levels in blood taken directly after surgery. Among the mediators released during surgery, IL-10 has already been shown to contribute to the down-regulation of HLA-DR on CD14+ cells44. IL-10 levels decreased after one day, which consequently resulted in the partial recovery of HLA-DR on monocytes and secretion of cytokines IL-1β, IL-6 and TNF-α during both C. albicans and S. aureus infection.
Similar to results of our previous studies on the immune response in whole-blood samples upon C. albicans infection16, we observed a specific population of pathogens that could not be phagocytosed by the immune cells, but were still present in the extracellular space at four hours post-infection. Therefore, these pathogens must escape the immune defense. Although the evasive pathogens could be detected in stimulated blood samples at each sample time point before and after surgery, we found that the amount of evasive C. albicans cells and S. aureus cells decreased after surgery, while the rate for this process increased for C. albicans infection and remained almost unchanged for S. aureus infection. In a previous study, we investigated the mechanism of C. albicans immune evasion in whole blood by testing potential evasion mechanisms using mathematical modeling. By simulating the infection in whole blood under neutropenic conditions, we could suggest future experimental measurements that would most likely enable us to accept or reject one of the two tested mechanisms, i.e. spontaneous evasion or neutrophil-mediated immune evasion45. Since we observed larger evasion rates after surgery, where the immune cells are more active in terms of phagocytosis, our study provides clues that the evasion of pathogens could be dependent on immune cell activity. Moreover, the change in the immune evasion rate after surgical insult is an additional characteristic property describing the changes of the immune response in whole-blood samples of patients who experienced an inflammatory stimulus.
Taken together, we show that a combination of experimental assays in ex vivo blood samples and biomathematical monitoring is a feasible approach for immune profiling in patients. Clearly, at this stage this cannot be used for diagnostic purposes and requires both further optimization and standardization. However, using this approach in its current state, we can already reveal important pathophysiological properties governing virulence of microorganisms, as in the case of immune escape of C. albicans.
Methods
Experimental methods
Patients and ethics statement
Human peripheral blood was collected from healthy volunteers and heart lung machine (HLM) patients with written informed consent. This study was conducted in accordance with the Declaration of Helsinki and all protocols were approved by the Ethics Committee of the University Hospital Jena (permit number: 4643-12/15).
Patients included in this study received standardized minimally invasive cardiac surgery for mitral valve insufficiency with a heart lung machine and same anesthesia regimen. Blood was taken from inserted catheters before cardiac surgery (pre-operative), immediately after surgery (post-operative) and one day after admission to the intensive care unit (post-operative + 1d).
Strains and culture
Candida albicans (SC5314) was grown overnight in YPD medium (2% D-glucose, 1% peptone, 0.5% yeast extract, in water) at 30 °C to stationary phase. Fungal cells were reseeded in YPD medium, grown for 3 h at 30 °C, stained with CellTracker™ Green (Invitrogen) for 1 h, and harvested in PBS. Subsequently, cells were inactivated with 0.1% thimerosal at 37 °C for 1 h and then rinsed extensively.
Staphylococcus aureus (ATCC25923) was cultivated overnight in lysogeny broth (LB) medium (10 g/l tryptone, 5 g/l yeast extract, 10 g/l sodium chloride, pH 7) at 37 °C. Bacterial cells were reseeded in LB medium and grown at 37 °C to reach the exponential growth phase (OD600 of 0.6–0.7) followed by staining with CellTracker™ Green for 30 min and inactivation with 50% ethanol for 4 h at 37 °C. Both pathogen stocks were stored at − 20 °C until use.
Ex vivo whole-blood infection assay
Peripheral blood from healthy volunteers and from patients undergoing cardiac surgery with extracorporeal circulation was collected in S-monovettes® (Sarstedt) containing recombinant Hirudin as anti-coagulant. Differential blood cell counts were measured with an auto hematology analyzer (BC-5300, Mindray).
For whole-blood infection, PBS as mock-infected control (mock) or 1 × 106 killed pathogens per ml whole blood were incubated for various time points (as indicated) at 37 °C on a rolling mixer (5 rpm). After incubation, the samples were immediately placed on ice. To collect plasma samples, whole-blood aliquots were centrifuged for 10 min at 10,000 × g and 4 °C, and the resulting plasma was stored at − 20 °C until further analysis.
Flow cytometry
Differential staining and flow cytometry was applied to identify distinct immune cell populations and to measure activation of immune cells and their association to pathogens. For surface antigen staining on the different immune cells, whole blood was stained with mouse anti-human CD3 (clone SK7, T cells), CD19 (clone HIB19, B cells), CD56 (clone MEM-188, NK cells), CD66b (clone G10F5, neutrophils), obtained from BioLegend, and CD69 (clone L78, early activation antigen, BD Biosciences). Monocytes were labelled with anti-human CD14 antibody (clone REA599, Miltenyi Biotec). Immature phenotype of neutrophils was assessed by surface CD10, CD16 and CD62L expression using mouse anti-human CD10 (clone HI10a, BioLegend), CD16 (clone 3G8, BioLegend) and CD62L (clone REA615, Miltenyi Biotec) antibodies. Surgery-induced changes on monocyte surface antigen expression were analysed for HLA-DR (anti-human HLA-DR, clone REA805, Miltenyi Biotec) and CD62L. Red blood cells were lysed with BD FACS Lysing solution followed by washing and harvesting cells in BD CellWASH solution. Acquisition was performed with the BD FACSCanto II flow cytometer and data was processed with FlowJo 7.6.4 software.
The strategy used to evaluate the association of microorganisms to immune cells in human blood was shown for GFP-expressing C. albicans in Hünniger et al., 2014 and used in the same way for CellTracker™ Green-labeled C. albicans and S. aureus in this study16.
Quantification of cytokines
The secretion of cytokines was assessed in plasma samples using Luminex technology (ProcartaPlex™ Multiplex Immunoassay, Thermo Fisher Scientific). The analyses were performed according to the instructions from the manufacturer.
Statistical analyses
Data are presented as arithmetic means ± standard deviation (SD). Statistical analysis was performed by applying the following steps. First, the Shapiro–Wilk test was applied to test whether the underlying data is normally distributed. For normally distributed data the unpaired t-test was used to test for significant differences, since the respective data was unpaired. If the data was not normally distributed, either the Wicoxon signed-rank test was applied to test paired samples, or the Mann–Whitney U test was applied to test unpaired samples for significant differences. Afterwards, a multiple comparison correction (Bonferroni’s correction) was performed if comparisons were made between several data sets. The corrected P-value is given by Significance is shown as *P < 0.05, **P < 0.01, ***P < 0.001.
Biomathematical modeling
State-based virtual infection modelling
The state-based model (SBM) is derived from our previous models of whole-blood infection16–18 and simulates the immune defence in whole blood that was infected with killed pathogens. As depicted in Supplementary Fig. S1a, the SBM contains several states for distinct cell populations in the system. Killed pathogens in extracellular space are represented by the state and pathogens that acquired immune escape are described by the state . Furthermore, the model contains the states and that represent neutrophils and monocytes, respectively, where the index denotes the number of phagocytosed pathogens. In order to compare the dynamics of the model with experimental data, we defined combined units that are specific combinations of measurable model states. Pathogens in extracellular space are represented by the combined unit :
The combined units and refer to pathogens that have been phagocytosed by either neutrophils or monocytes:
The transitions between model states represent biological processes during whole-blood infection (see the connections between states in Supplementary Fig. S1a). In the SBM, we defined three transition rates characterising specific transitions. The rate refers to the phagocytosis of killed pathogens () by neutrophils with phagocytosed pathogens ( and characterises the state transition
The phagocytosis of killed pathogens () by monocytes with phagocytosed pathogens ( is characterised by the rate and is described by
The immune escape of killed pathogens () is quantified by the rate for state transition
The transition rates enables us to quantify the dynamics of the SBM, where a transition from any state to state within the time step will be performed with probability : . The SBM dynamics was calculated by applying the random selection method46 and the flow chart of this simulation algorithm is depicted in Supplementary Fig. S1b.
Agent-based virtual infection modelling
In order to investigate also spatial aspects of host–pathogen interactions we applied a previously developed agent-based model (ABM)47,48 that was adjusted to the context of whole-blood infection assays17,18. This spatial counterpart of the SBM simulates the different immune cell types of neutrophils and monocytes as well as the killed pathogens as distinct spherical objects, i.e. the agents (Supplementary Fig. S2a). These agents can migrate and interact within an environment in a rule-based fashion, where the environment is a continuous three-dimensional representation of a section of of the whole-blood infection assay in which cells perform random walk migration due to the high density of erythrocytes17. Modeling the active migration of neutrophils and monocytes by a diffusion process with coefficients and , respectively, the passive migration of pathogens is estimated to have a small diffusion coefficient of 17. Based on the random selection method46, each stochastic simulation was repeated 30 times to obtain significant mean and standard deviation of simulation dynamics (Supplementary Fig. S2b). The migration parameters of immune cells were estimated from a minimization of least-squares error (LSE) between experiment and simulation using the method of adaptive regular grid search49.
Parameter estimation
In order to estimate the a priori unknown values of transition rates, the state-based model (SBM) is fitted to experimental data of the whole-blood infection assays by applying the algorithm of simulated annealing50 based on the Metropolis Monte Carlo scheme51. For a detailed description of the parameter estimation in the SBM we refer to our previous work17,18,29,45 and to the flowchart of this parameter fitting algorithm in Supplementary Fig. S3.
To calibrate the model parameters of the agent-based model (ABM) to the experimental data, we previously developed a bottom-up approach that combines the SBM and ABM such that estimated rates from the SBM can be used in the ABM by a self-consistent conversion of these parameters17. Therefore, the parameter search space reduces to the migration parameters of the immune cell types and . For a detailed description of the parameter estimation in the ABM we refer to our previous work17,18,29,45 and the flowchart of this algorithm in Supplementary Fig. S4.
Supplementary Information
Acknowledgements
This study was funded by the Federal Ministry for Education and Science (BMBF) within the Center for Sepsis Control and Care and by the Deutsche Forschungsgemeinschaft within the Collaborative Research Center Transregio 124 FungiNet "Pathogenic fungi and their human host: Networks of Interaction", DFG Project Number 210879364 (Projects B4 to MTF and C3 to OK). We are grateful to Prof. Torsten Doenst from the Department of Cardiothoracic Surgery for his support of the project and to Petra Bloos, Anja Haucke, Karina Knuhr-Kohlberg, Steffi Kolanos and Katrin Schwope for organizing blood draws on the ICU. We thank Carl-Magnus Svensson for scientific discussions throughout the project and Natalie Nieuwenhuizen for critical reading of the final manuscript.
Author contributions
Conceptualization and Ideas: O.K., M.T.F., K.H.; Formal Analysis including application of statistical, mathematical, computational, or other formal techniques: T.L., S.T., M.T.F.; Funding Acquisition: O.K., M.T.F.; Investigation: T.L., I.L., S.T., K.H.; Resources: D.T.R., F.B., C.S., Writing: T.L., I.L., O.K., M.T.F., K.H.
Competing interests
The authors declare no competing interests.
Footnotes
Publisher's note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
These authors contributed equally: Teresa Lehnert and Ines Leonhardt.
These authors jointly supervised this work: Marc Thilo Figge and Kerstin Hünniger.
Contributor Information
Marc Thilo Figge, Email: thilo.figge@leibniz-hki.de.
Kerstin Hünniger, Email: kerstin.huenniger@leibniz-hki.de.
Supplementary Information
The online version contains supplementary material available at 10.1038/s41598-021-91362-5.
References
- 1.Pizzo PA. Management of patients with fever and neutropenia through the arc of time. Ann. Intern. Med. 2019;170:389–397. doi: 10.7326/M18-3192. [DOI] [PubMed] [Google Scholar]
- 2.Ford N, Meintjes G, Vitoria M, Greene G, Chiller T. The evolving role of CD4 cell counts in HIV care. Curr. Opin. HIV AIDS. 2017;12:123–128. doi: 10.1097/COH.0000000000000348. [DOI] [PubMed] [Google Scholar]
- 3.Pfortmueller CA, Meisel C, Fux M, Schefold JC. Assessment of immune organ dysfunction in critical illness: utility of innate immune response markers. Intensive Care Med. Exp. 2017;5:49. doi: 10.1186/s40635-017-0163-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Reinhart K, Bauer M, Riedemann NC, Hartog CS. New approaches to sepsis: molecular diagnostics and biomarkers. Clin. Microbiol. Rev. 2012;25:609–634. doi: 10.1128/CMR.00016-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Monneret G, Venet F, Pachot A, Lepape A. Monitoring immune dysfunctions in the septic patient: a new skin for the old ceremony. Mol. Med. 2008;14:64–78. doi: 10.2119/2007-00102.Monneret. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Schefold JC. Measurement of monocytic HLA-DR (mHLA-DR) expression in patients with severe sepsis and septic shock: assessment of immune organ failure. Intensive Care Med. 2010;36:1810–1812. doi: 10.1007/s00134-010-1965-7. [DOI] [PubMed] [Google Scholar]
- 7.Qureshi SS, et al. Increased distribution and expression of CD64 on blood polymorphonuclear cells from patients with the systemic inflammatory response syndrome (SIRS) Clin. Exp. Immunol. 2001;125:258–265. doi: 10.1046/j.1365-2249.2001.01596.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Wagner C, et al. Expression patterns of the lipopolysaccharide receptor CD14, and the FCgamma receptors CD16 and CD64 on polymorphonuclear neutrophils: data from patients with severe bacterial infections and lipopolysaccharide-exposed cells. Shock. 2003;19:5–12. doi: 10.1097/00024382-200301000-00002. [DOI] [PubMed] [Google Scholar]
- 9.Mayer FL, Wilson D, Hube B, Article M. Candida albicans pathogenicity mechanisms. Virulence. 2013;4:119–128. doi: 10.4161/viru.22913. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Bhandari A, Schramm CM, Kimble C, Pappagallo M, Hussain N. Effect of a short course of prednisolone in infants with oxygen-dependent bronchopulmonary dysplasia. Pediatrics. 2008;121:e344–e349. doi: 10.1542/peds.2006-3668. [DOI] [PubMed] [Google Scholar]
- 11.Icardi M, et al. CD64 index provides simple and predictive testing for detection and monitoring of sepsis and bacterial infection in hospital patients. J. Clin. Microbiol. 2009;47:3914–3919. doi: 10.1128/JCM.00628-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Rigato O, Salomao R. Impaired production of interferon-gamma and tumor necrosis factor-alpha but not of interleukin 10 in whole blood of patients with sepsis. Shock. 2003;19:113–116. doi: 10.1097/00024382-200302000-00004. [DOI] [PubMed] [Google Scholar]
- 13.Munoz C, et al. Dysregulation of in vitro cytokine production by monocytes during sepsis. J. Clin. Invest. 1991;88:1747–1754. doi: 10.1172/JCI115493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Morris AC, et al. C5a-mediated neutrophil dysfunction is RhoA-dependent and predicts infection in critically ill patients. Blood. 2011;117:5178–5188. doi: 10.1182/blood-2010-08-304667. [DOI] [PubMed] [Google Scholar]
- 15.Duggan S, Leonhardt I, Hünniger K, Kurzai O. Host response to Candida albicans bloodstream infection and sepsis. Virulence. 2015;6:316–326. doi: 10.4161/21505594.2014.988096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Hünniger K, et al. A virtual infection model quantifies innate effector mechanisms and Candida albicans immune escape in human blood. PLoS Comput. Biol. 2014;10:e1003479. doi: 10.1371/journal.pcbi.1003479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Lehnert T, et al. Bottom-up modeling approach for the quantitative estimation of parameters in pathogen-host interactions. Front. Microbiol. 2015;6:1–15. doi: 10.3389/fmicb.2015.00608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Timme S, et al. Quantitative simulations predict treatment strategies against fungal infections in virtual neutropenic patients. Front. Immunol. 2018;9:1–14. doi: 10.3389/fimmu.2018.00667. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Clive Landis R, et al. Attenuating the systemic inflammatory response to adult cardiopulmonary bypass: a critical review of the evidence base. J. Extra Corporeal Technol. 2014;46:197–211. [PMC free article] [PubMed] [Google Scholar]
- 20.Otto GP, et al. Limitation of (1→3)-β-D-glucan monitoring in major elective surgery involving cardiopulmonary bypass. Crit. Care. 2013;17:1–2. doi: 10.1186/cc12718. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Bronicki RA, Hall M. Cardiopulmonary bypass-induced inflammatory response. Pediatr. Crit. Care Med. 2016;17:S272–S278. doi: 10.1097/PCC.0000000000000759. [DOI] [PubMed] [Google Scholar]
- 22.Quiroga MM, et al. The effect of body temperature on leukocyte kinetics during cardiopulmonary bypass. J. Thorac. Cardiovasc. Surg. 1985;90:91–96. doi: 10.1016/S0022-5223(19)38668-4. [DOI] [PubMed] [Google Scholar]
- 23.Permanyer E, et al. Mini-extracorporeal circulation surgery produces less inflammation than off-pump coronary surgery. Eur. J. Cardio-Thoracic Surg. 2019;57:496–503. doi: 10.1093/ejcts/ezz291. [DOI] [PubMed] [Google Scholar]
- 24.Paparella D, Yau TM, Young E. Cardiopulmonary bypass induced inflammation: pathophysiology and treatment. An update. Eur. J. Cardio-Thoracic Surg. 2002;21:232–244. doi: 10.1016/S1010-7940(01)01099-5. [DOI] [PubMed] [Google Scholar]
- 25.Guo R-F, Riedemann NC, Ward PA. Role of C5a–C5aR interaction in sepsis. Shock. 2004;21:1–7. doi: 10.1097/01.shk.0000105502.75189.5e. [DOI] [PubMed] [Google Scholar]
- 26.Day JRS, Taylor KM. The systemic inflammatory response syndrome and cardiopulmonary bypass. Int. J. Surg. 2005;3:129–140. doi: 10.1016/j.ijsu.2005.04.002. [DOI] [PubMed] [Google Scholar]
- 27.Ortega Loubon, C. et al. Extracorporeal circulation in cardiac surgery inflammatory response, controversies and future directions. Int. Arch. Med.8 (2015).
- 28.Edmunds LH. Inflammatory response to cardiopulmonary bypass. Ann. Thoracic Surg. 1998;66:S715–S720. doi: 10.1016/S0003-4975(98)00967-9. [DOI] [PubMed] [Google Scholar]
- 29.Hünniger K, et al. A second stimulus required for enhanced antifungal activity of human neutrophils in blood is provided by Anaphylatoxin C5a. J. Immunol. 2014;194:1199–1210. doi: 10.4049/jimmunol.1401845. [DOI] [PubMed] [Google Scholar]
- 30.Levy JH, Tanaka KA. Inflammatory response to cardiopulmonary bypass. Ann. Thorac. Surg. 2003;75:S715–S720. doi: 10.1016/S0003-4975(02)04701-X. [DOI] [PubMed] [Google Scholar]
- 31.Ozao J. Personal and surgical experiences during a rotation in san cristobal dominican republic. J. Surg. Educ. 2007;64:128–129. doi: 10.1016/j.cursur.2006.08.003. [DOI] [PubMed] [Google Scholar]
- 32.Fink R, Al-Obaidi M, Grewal S, Winter M, Pepper J. Monocyte activation markers during cardiopulmonary bypass. Perfusion. 2003;18:83–86. doi: 10.1191/0267659103pf645oa. [DOI] [PubMed] [Google Scholar]
- 33.Cowland JB, Borregaard N. Granulopoiesis and granules of human neutrophils. Immunol. Rev. 2016;273:11–28. doi: 10.1111/imr.12440. [DOI] [PubMed] [Google Scholar]
- 34.Runge MS, Haber E. Importance of experimental models in study of thrombosis and thrombolysis. Circulation. 1991;83:IV1-2. [PubMed] [Google Scholar]
- 35.van Grinsven E, et al. Immature neutrophils released in acute inflammation exhibit efficient migration despite incomplete segmentation of the nucleus. J. Immunol. 2019;202:207–217. doi: 10.4049/jimmunol.1801255. [DOI] [PubMed] [Google Scholar]
- 36.Leliefeld PHC, et al. Differential antibacterial control by neutrophil subsets. Blood Adv. 2018;2:1344–1355. doi: 10.1182/bloodadvances.2017015578. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Sbrana S, Parri MS, De Filippis R, Gianetti J, Clerico A. Monitoring of monocyte functional state after extracorporeal circulation: a flow cytometry study. Cytometry. 2004;58B:17–24. doi: 10.1002/cyto.b.10061. [DOI] [PubMed] [Google Scholar]
- 38.Franke A, Lante W, Kollig E, Markewitz A. A comparison of monocyte counts and ex vivo and in vitro monocyte cytokine production after major surgical trauma. J. Surg. Res. 2009;154:91–98. doi: 10.1016/j.jss.2008.06.003. [DOI] [PubMed] [Google Scholar]
- 39.Morse DS, Adams D, Magnani B. Platelet and neutrophil activation during cardiac surgical procedures: impact of cardiopulmonary bypass. Ann. Thorac. Surg. 1998;65:691–695. doi: 10.1016/S0003-4975(97)01425-2. [DOI] [PubMed] [Google Scholar]
- 40.Khabar KSA, et al. Circulating endotoxin and cytokines after cardiopulmonary bypass: differential correlation with duration of bypass and systemic inflammatory response/multiple organ dysfunction syndromes. Clin. Immunol. Immunopathol. 1997;85:97–103. doi: 10.1006/clin.1997.4413. [DOI] [PubMed] [Google Scholar]
- 41.Franke A, et al. Pro-inflammatory cytokines after different kinds of cardio-thoracic surgical procedures: is what we see what we know? Eur. J. Cardio-Thoracic Surg. 2005;28:569–575. doi: 10.1016/j.ejcts.2005.07.007. [DOI] [PubMed] [Google Scholar]
- 42.Sablotzki A, et al. The systemic inflammatory response syndrome following cardiac surgery: different expression of proinflammatory cytokines and procalcitonin in patients with and without multiorgan dysfunctions. Perfusion. 2002;17:103–109. doi: 10.1191/0267659102pf543oa. [DOI] [PubMed] [Google Scholar]
- 43.Diab M, et al. Changes in inflammatory and vasoactive mediator profiles during valvular surgery with or without infective endocarditis: a case control pilot study. PLoS ONE. 2020;15:e0228286. doi: 10.1371/journal.pone.0228286. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Fumeaux T, Pugin J. Role of interleukin-10 in the intracellular sequestration of human leukocyte antigen-DR in monocytes during septic shock. Am. J. Respir. Crit. Care Med. 2002;166:1475–1482. doi: 10.1164/rccm.200203-217OC. [DOI] [PubMed] [Google Scholar]
- 45.Prauße MTE, et al. Predictive virtual infection modeling of fungal immune evasion in human whole blood. Front. Immunol. 2018;9:560. doi: 10.3389/fimmu.2018.00560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Figge MT. Stochastic discrete event simulation of germinal center reactions. Phys. Rev. E. 2005;71:51907. doi: 10.1103/PhysRevE.71.051907. [DOI] [PubMed] [Google Scholar]
- 47.Pollmächer J, Figge MT. Agent-based model of human alveoli predicts chemotactic signaling by epithelial cells during early Aspergillus fumigatus infection. PLoS ONE. 2014;9:e111630. doi: 10.1371/journal.pone.0111630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Pollmächer J, Figge MT. Deciphering chemokine properties by a hybrid agent-based model of Aspergillus fumigatus infection in human alveoli. Front. Microbiol. 2015;6:503. doi: 10.3389/fmicb.2015.00503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Powell MJD. Direct search algorithms for optimization calculations. Acta Numer. 1998 doi: 10.1017/S0962492900002841. [DOI] [Google Scholar]
- 50.Kirkpatrick S, Gelatt CD, Vecchi MP. Optimization by simulated annealing. Science (80-.) 1983;220:671–680. doi: 10.1126/science.220.4598.671. [DOI] [PubMed] [Google Scholar]
- 51.Metropolis N, Rosenbluth AW, Rosenbluth MN, Teller AH, Teller E. Equation of state calculations by fast computing machines. J. Chem. Phys. 1953;21:1087. doi: 10.1063/1.1699114. [DOI] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.