Abstract
Background:
The ability to form enduring social bonds is characteristic of human nature and impairments in social affiliation are central features of severe neuropsychiatric disorders including autism spectrum disorders and schizophrenia. Due to its ability to form long-term pair-bonds, the socially monogamous prairie vole has emerged as an excellent model to study the neurobiology of social attachment. Despite the enduring nature of the bond, however, surprisingly few genes have been implicated in the pair-bonding process in either sex.
Methods:
Male and female prairie voles (Microtus ochrogaster) were cohabitated with an opposite-sex partner for 24 hrs or 3 weeks (3W) and transcriptomic regulations in the nucleus accumbens (NAc) were measured by RNA-sequencing.
Results:
We found sex-specific response patterns despite similar behavioral indicators of pair-bond establishment. Indeed, 24 hrs of cohabitation with an opposite-sex partner induced widespread transcriptomic changes that remained sustained to some extent in females after 3W but returned to baseline before a second set of regulations in males. This led to a highly sexually-biased NAc transcriptome at 3W related to processes such as neurotransmission, protein turnover, and DNA transcription. In particular, we found sex-specific alterations of mitochondrial dynamics following cohabitation, with a shift towards fission in males.
Conclusions:
In addition to identifying the genes, networks, and pathways involved in the pair-bonding process in the NAc, our work illustrates the vast extent of sex differences in the molecular mechanisms underlying pair-bonding in prairie voles, and paves the way to further our understanding of the complex social bonding process.
Keywords: Prairie vole, pair-bonding, selective aggression, nucleus accumbens, gene expression, mitochondria
Introduction
Social attachment is a critical characteristic of human behavior and impairments in social affiliation are thus common features in several major neuropsychiatric disorders such as autism spectrum disorders, schizophrenia, and depression (1). Despite this importance, our understanding of the neurobiology underlying social attachment has been limited by the fact that social monogamy can only be found in 3–5% of mammals (3). In this context, the socially monogamous prairie vole (M. ochrogaster) represents an excellent animal model for the investigation of neurobiology of social bonding due to its ability to establish long-term pair-bonds (5,7,8). Indeed, prairie voles cohabitated with an opposite-sex partner for 24 hrs display strong preferential affiliative behaviors towards their partner over an unfamiliar opposite-sex stranger—termed partner preference—indicative of the formation of a pair-bond. Following additional weeks of cohabitation, both male and female prairie voles develop selective aggression towards unfamiliar intruders, indicative of pair-bond maintenance (9-12).
The study of the neurocircuitry underlying social bonding in prairie voles revealed a complex network of interactions between several brain structures reflecting the complexity and multi-faceted nature of social affiliative behaviors (13,14). A key area of the brain reward pathway, the nucleus accumbens (NAc) plays a central role in the regulation of pair-bonding in prairie voles where its activity modulates the development of affiliative behaviors towards the partner (13-16). At the molecular level, a variety of neurotransmitters and their receptors, including the oxytocin, vasopressin, and dopamine systems, have been involved in partner preference formation (17), although with sex-specific alterations of gene expression in the NAc. Indeed, while 24 hrs of cohabitation with mating up-regulate oxytocin receptors (OTR) and vasopressin V1a receptors (V1aR) in females, males show an increase in OTR but not V1aR (18,19). Notably, neither OTR nor V1aR mRNA levels show changes in the ventral striatum of male and female prairie voles pair-bonded for 2 weeks, suggesting distinct mechanisms underlying pair-bond formation and its maintenance (20).
In addition to being affected by pair-bonding, specific variations in gene expression levels in the NAc relate to the levels of social affiliative behaviors in prairie voles. In females, the viral-mediated expression of OTR in the NAc facilitates partner preference formation (21). Moreover, natural variations in OTR expression levels resulting from non-coding polymorphisms are associated with greater social affiliative behaviors in males (22). Altogether, these data indicate that the transcriptomic landscape in the prairie vole NAc can not only reflect the enduring changes in social behaviors, but also participate to mediate both the formation and maintenance of the bond.
Despite the variety of neurotransmitter systems involved and the enduring nature of the bond, however, surprisingly few genes have been associated with the maintenance phase of the bond. In this study, we thus aimed at identifying by RNA-sequencing the specific transcriptomic regulations underlying the maintenance of the bond, while including both males and females to pinpoint regulations common or specific to each sex.
Methods and Materials
Detailed methods are available as supplemental information.
Animals, cohabitation, and resident-intruder test
Male and female prairie voles (Microtus ochrogaster) were paired and cohabitated with an aged-matched opposite-sex partner for 3 weeks (“3W”) or 24 hours (“24H”), or a same-sex littermate for 3 weeks (Sexually-Naive, “SN”). Selective aggression was then assessed using a 10-min resident-intruder test (RIT) as previously described (23). Experimental procedures were approved by the Institutional Animal Care and Use Committee at Florida State University.
RNA extraction, library preparation, sequencing, and bioinformatic analyses
Immediately after RIT, subjects were killed by rapid decapitation, their brain dissected out, snap-frozen, and stored at −80C until further processing. Total RNA was extracted from NAc tissue punches and a total of 46 libraries were prepared as previously described (21): 16 SN voles (6 males, 10 females), 16 24H voles (6 males, 10 females), and 11 3W voles (5 males, 6 females). All libraries were pooled and sequenced (2x50bp, NovaSeq 6000, S2 lane) at the Translational Sciences Laboratory at Florida State University.
Raw reads were quality-filtered (fastp, v0.14.1)(24), before quantification (Salmon, v0.12)(25), and differential expression analysis (DESeq2, v1.20.0)(26,27). Gene-Sets Enrichment Analyses (GSEA, v3.0)(28) were performed using publicly-available gene-sets (http://download.baderlab.org, September_01_2018 release), whereas gene ontologies and Kyoto Encyclopedia of Genes and Genomes (KEGG) pathways enrichment was tested using the Bioconductor (27) package clusterProfiler (v3.10.1) (29).
A weighted gene co-expression network analysis (WGCNA, R package WGCNA (v1.68) (30-32)) was conducted, followed by gene-gene interaction networks reconstruction (ARACNe-AP (33)) and key driver analysis (Bioconductor package Mergeomics (v1.16)) as previously described (34-36).
Results
Behavioral characterization
While the transition from affiliative to aggressive behaviors is a well-established characteristic of pair-bonded prairie voles, existing studies generally focused either on the early or the later phases of the social bond process, or only within one sex (8,11,12,20,37). We thus first aimed at characterizing the behaviors related to selective aggression in the early and later phases of pair-bonding in both sexes. To do so, adult sexually-naive male and female prairie voles (from eight different cohorts, representing a total of 194 voles: cohort 1: 29, cohort 2: 29, cohort 3: 30, cohort 4: 16, cohort 5: 16, cohort 6: 20, cohort 7: 33, cohort 8: 21) were paired for 24 hrs (24H, early phase) or 3 weeks (3W, later phase), before being subjected to a resident-intruder test (RIT) as a proxy for the verification and evaluation of the pair-bond.
While 3 weeks of pair-bonding increased aggressive behaviors in both sexes, only males exhibited aggression levels higher than their same-sex controls (SN) at 24H (Fig. 1A, Table S1). Simultaneously, a reduction in nose-to-nose sniffing was observed similarly in males and females at both 24H and 3W (Fig. 1B, Table S1). Combined with the observation of similar effects of cohabitation between males and females on the other behaviors recorded (Fig. S1, Table S1) and their evolution within the RIT session (Fig. S2, Supplemental Results), this indicates that 24 hrs or 3 weeks of cohabitation with an opposite-sex partner leads to the development of selective aggression characterized by an increase in aggression at the expense of non-aggressive social interactions (anogenital and nose-to-nose sniffing) similarly between males and females despite a slight delay in developing aggression in females.
Figure 1.
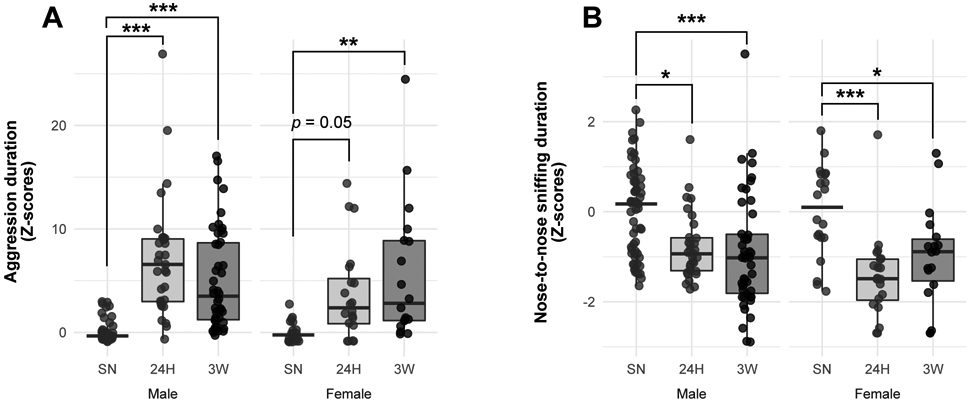
Characterization of selective aggression in male and female prairie voles during the resident-intruder test (RIT). Adult male and female prairie voles were cohabitated with a same-sex littermate (SN), or an opposite-sex partner for 24 hrs (24H) or 3 weeks (3W) before being subjected to a 10-min RIT. Scorings for the time spent in aggression (A) or nose-to-nose sniffing (B) is detailed after normalization by z-scoring within cohorts and then between sexes. Data are depicted as mean ± SEM in (A). In (A,B), each dot represents an individual vole. *p < 0.05, **p < 0.01, ***p < 0.001 vs. SN group within same sex, Tukey’s post-hoc test.
Time-specific alteration of gene expression in the NAc following cohabitation with a partner
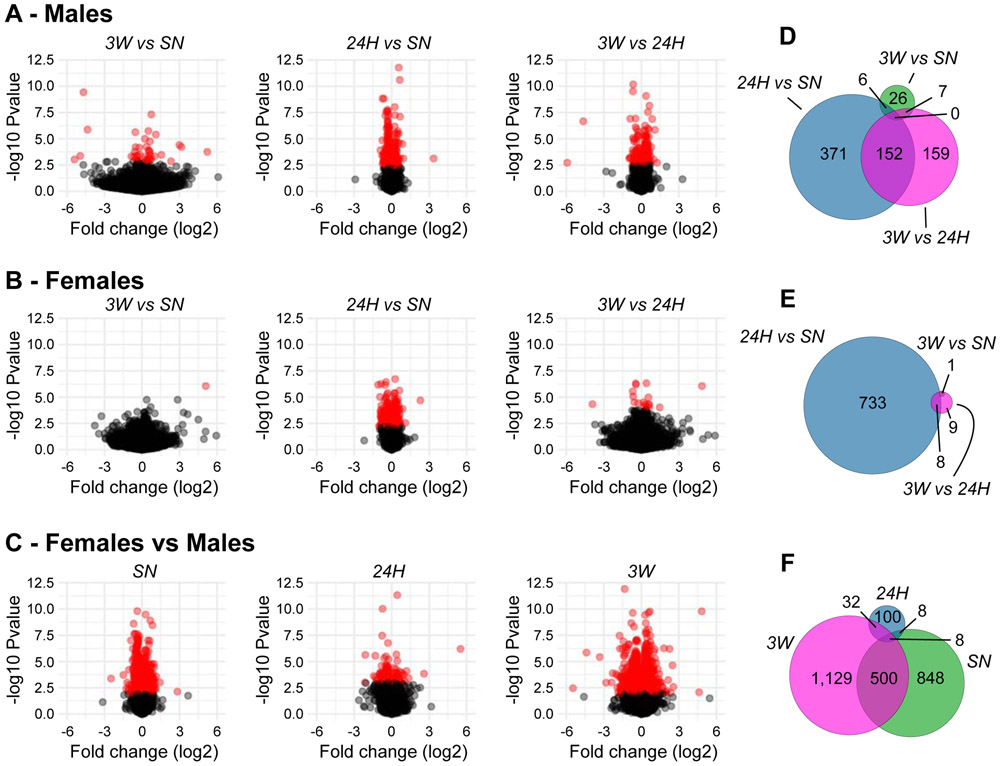
Relatively few genes survived the false discovery rate threshold to be qualified as differentially expressed (DE) at 3W when compared to SN controls (39 in males, 1 in females, Fig. 2A,B, Table S2). At 24H, however, 529 and 741 genes were DE in males and females, respectively, when compared to SN controls (Fig. 2A,B, Table S2), denoting a substantial impact of a novel social encounter on the NAc transcriptome in prairie voles. Only 37 genes were DE at 24H in both sexes, however, 35 of which sharing directionality (14 down-regulated, 21 up-regulated), indicating distinct sets of regulations between sexes. In males, 152 of these (29%) were also DE when comparing 3W to 24H (Fig. 2A,D, Table S2, Fig. S3A), which, combined to the relative high number of DE genes between the two cohabitation durations (3W vs. 24H), suggests distinct transcriptional regulations at 24H and 3W when compared to SN controls. In females, however, only 8 of the 741 DE genes in the 24H vs. SN comparison were also DE between the 24H and 3W group, which, in light of the relatively small number of DE genes in the latter comparison (Fig. 2B,E, Table S2, Fig. S3A), denotes similar profiles of regulation between the two timepoints when compared to SN controls.
Figure 2.
Differential expression in the nucleus accumbens of male and female prairie voles following cohabitation. In (A–C), volcano plots depict the log2 fold-change against the −log10 of the uncorrected p-value for each gene in each pairwise comparison in males (A), females (B), or between-sexes (C). Red dots correspond to differentially expressed genes (surviving the 0.1 false-discovery rate). In (D–F), Euler diagrams illustrate the degree of overlap between differentially-expressed genes in males (D), females (E), or between-sexes (F). The area of each circle is proportional to the number of differentially expressed genes it contains. SN: sexually-naive, 24H: 24 hrs of cohabitation, 3W: 3 weeks of cohabitation.
Surprisingly, the number of sexually-biased genes was large at baseline and 3W (1,371 and 1,678 DE genes, respectively), but relatively limited at 24H (155 DE genes, Fig. 2C) likely resulting from the large transcriptomic response observed then in either sex. Consistent with their high number, DE genes were distributed throughout the genome (Fig. S3B), indicating that sex differences in the prairie vole NAc cannot solely be explained by sex chromosomes. Moreover, no bias towards either up- or down-regulation was detected in any comparison (Table S2). Notably, only 508 (37%) out of the 1,371 DE genes in SN voles were also sexually-biased at 3W (Fig. 2F). Altogether, this suggests that despite vast sex-differences at baseline, after 3 weeks of pair-bonding, males and females retain different sexually-biased transcriptomes.
Functional enrichment following cohabitation
To analyze the biological pathways associated with the changes in gene expression following cohabitation with an opposite-sex partner and compare the early (24H) and later phases (3W) of the pair-bonding process, we conducted a threshold-free GSEA within each sex, and then visualized the resulting gene-sets using an enrichment map.
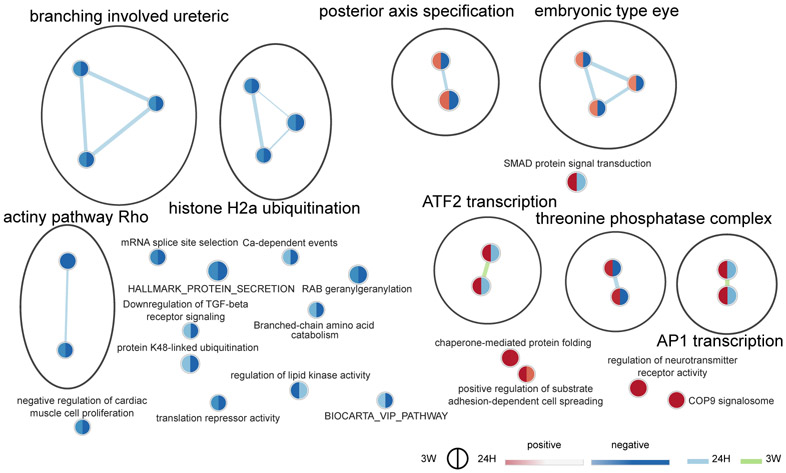
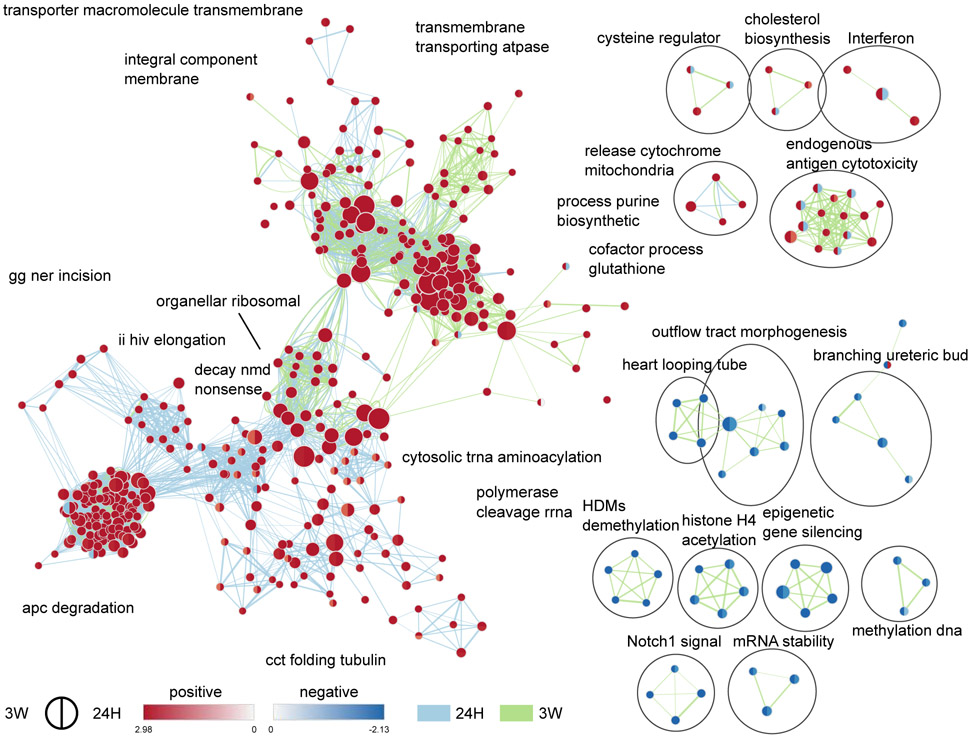
In males, we observed a relatively weak enrichment of a few gene-sets denoting a down-regulation of pathways related to chromatin modifications, actin polymerization, RNA translation, and protein degradation, but an up-regulation of those related to protein maturation and neurotransmitter receptors. Nevertheless, while most enriched gene-sets shared directionality of regulation between the two cohabitation durations when compared to SN controls, gene-sets related to AP1 and ATF2 transcription factors as well as protein phosphatases were down-regulated at 24H, but up-regulated at 3W (Fig. 3). Interestingly, these regulations were carried by several genes such as Fos, Duspl, or Nts, with known involvement in dopaminergic transmission and motivated behaviors (Supplemental Results). Combined with the partial overlap in DE genes between the 24H vs. SN and 3W vs. 24H comparisons (Fig. 2D, Fig. S3A), this supports the presence of distinct sets of regulations occurring after 24 hrs or 3 weeks of cohabitation in males. Females, however, displayed a distinct pattern with widespread enrichments of gene-sets related to all aspects of cellular activity. Gene-sets related to epigenetic regulation of gene expression were down-regulated, whereas those related to DNA transcription, RNA splicing and translation, RNA stability, protein degradation, as well as mitochondrial organization and activity were up-regulated (Fig. 4). In a neuronal context such as the NAc, this suggests a coordinated alteration of most systems required for a sustained regulation of neurotransmission—from DNA transcription to protein turnover as well as energy production. In contrast to males, all but one cluster of gene-sets—related to immune response (“interferon”, and “endogenous antigen cytotoxicity”)—share directionality of changes between the two timepoints when compared to SN controls. In combination with the very small overlap in DE genes between the 24H vs. SN and 3W vs. 24H comparisons (Fig. 2E, Fig. S3A), this supports a widespread reprogramming of gene expression in the early phase of pair-bonding in the female NAc that remains sustained, to some extent, following 3 weeks of cohabitation.
Figure 3.
Time-specific functional enrichment following cohabitation with an opposite-sex partner in male prairie voles. Enrichment map depicting the clusters of differentially modulated pathways following cohabitation when compared to sexually-naive controls (SN) in males, identified by gene-set enrichment analyses. The area of each node, representing a gene-set (functional pathway), corresponds to the number of genes of the gene-set it contains, and its color depicts the direction of enrichment (red: positive, blue: negative) with the color intensity representing the enrichment score, when compared to the SN group either in the 24H dataset (right half), or 3W dataset (left half). As a result, a gene-set with different colors in their two halves display opposite directions of regulation in the early (24H) and later (3W) phases of the bonding process when compared to SN controls. Edge thickness is proportional to the number of genes overlapping between the two connected nodes. Blue and green edges correspond to the 24H vs SN, and 3W vs SN datasets, respectively. 24H: 24 hrs of cohabitation, 3W: 3 weeks of cohabitation.
Figure 4.
Time-specific functional enrichment following cohabitation with an opposite-sex partner in female prairie voles. Enrichment maps depicting the clusters of differentially modulated pathways following cohabitation when compared to sexually-naive controls (SN) in females, identified by gene-set enrichment analyses. The area of each node, representing a gene-set (functional pathway), corresponds to the number of genes of the gene-set it contains, and its color depicts the direction of enrichment (red: positive, blue: negative) with the color intensity representing the enrichment score, when compared to the SN group either in the 24H dataset (right half), or 3W dataset (left half). As a result, a gene-set with different colors in their two halves display opposite directions of regulation in the early (24H) and later (3W) phases of the bonding process when compared to SN controls. Edge thickness is proportional to the number of genes overlapping between the two connected nodes. Blue and green edges correspond to the 24H vs SN, and 3W vs SN datasets, respectively. 24H: 24 hrs of cohabitation, 3W: 3 weeks of cohabitation.
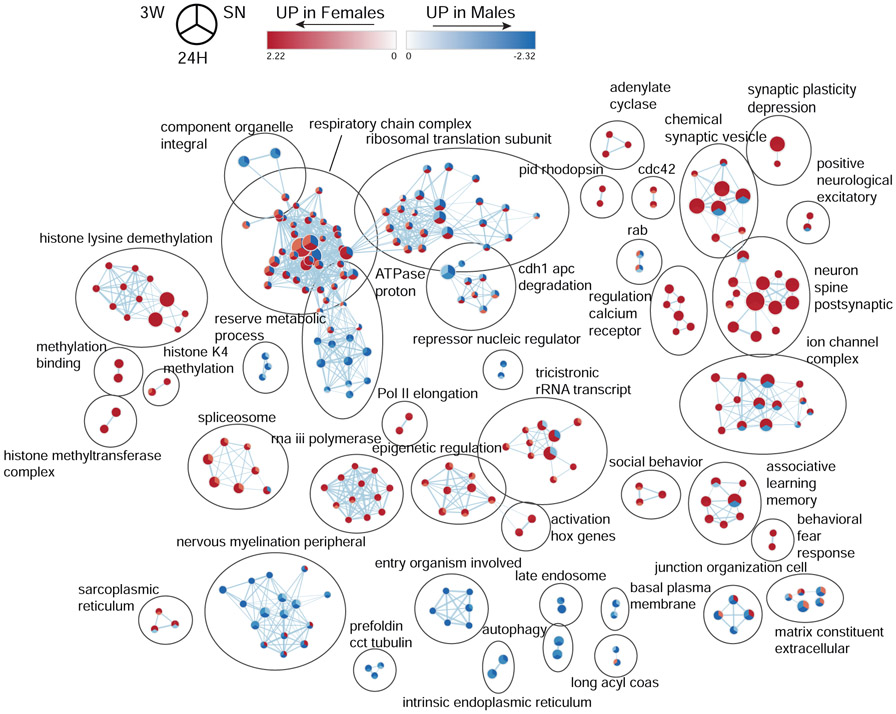
In line with the high number of sexually-biased genes at baseline and 3W (Fig. 2D,F), we observed a widespread enrichment of gene-sets between sexes at baseline (Fig. 5, Supplemental Results). Following cohabitation, three different response patterns emerged: (1) the sex bias is independent of cohabitation status, (2) the baseline sex bias is reversed at 24H but returns to its baseline state after 3 weeks, and (3) the baseline sex bias is reversed at 24H and is maintained after 3 weeks. In the first group, we found gene-sets related to epigenetic regulation of gene expression, dendritic spines, post-synaptic compartment, as well as synaptic plasticity. In the second group, we found clusters of gene-sets related to ion channels, synaptic vesicles, RNA translation, and protein degradation. Finally, the third group includes a cluster of gene-sets related to the electron transport chain and oxidative phosphorylation that, in line with the widespread upregulation of genes associated with mitochondrial function in females (Fig. 4), switches from a marked bias towards males at baseline, to a strong bias towards females at 24H that remains present at 3W (Fig. 5). In this context, mitochondrial activity emerges as an interesting candidate in underlying pair-bond maintenance in females.
Figure 5.
Sex-differences in functional enrichment following cohabitation with an opposite-sex partner in prairie voles. Enrichment map depicting the clusters of differentially modulated pathways between male and female prairie voles at baseline (SN) and following 24 hrs (24H) or 3 weeks (3W) of cohabitation, identified by gene-set enrichment analyses. The area of each node, representing a gene-set (functional pathway), corresponds to the number of genes of the gene-set it contains, and its color depicts the direction of enrichment (red: female-biased, blue: male-biased) with the color intensity representing the enrichment score, either in the SN, (top right sector), 24H (bottom sector), or 3W (top left sector) datasets. As a result, a gene-set with different colors between its three sectors display a change in sex-bias following cohabitation with an opposite-sex partner, whereas a gene-set with three similarly-colored sectors show the same sex-bias regardless of the cohabitation status. Edge thickness is proportional to the number of genes overlapping between the two connected nodes.
Cohabitation with a partner recruits different gene networks in males and females
To identify the genes underlying the regulations observed above, we conducted a weighted gene co-expression network analysis (WGCNA) to highlight clusters of genes with similar expression (modules) and extract modules of interest based on their relation to our behavioral profiling.
Across all cohabitation states, we found a consensus network of 26 modules of correlated gene expression—ranging from 108 to 1,756 genes—between males and females. Notably, the relation of each module to behavioral performances during the RIT session differs between sexes, as evidenced by the limited consensus between the male and female module–trait relationships (Fig. S4). In particular, extracting the modules of interest based on behavioral indexes of pair-bonding reveals that only 40–43% of the modules eigengenes found significantly correlated with any behavioral trait are common between males and females (Fig. S5, Supplemental Results). Interestingly, albeit common between sexes, the plum1 module shows an opposite relation to main RIT behaviors, thereby further underlining sex-specificities in underlying gene networks. Alongside these common modules, others show a sex-specific association with behavior during the RIT (Fig. S5, Supplemental Results). Supporting the functional relevance of this clustering, the underlying genes relate to ontologies and pathways distinct between groups of modules, and related to mitochondrial function, RNA translation, epigenetic mechanisms, protein degradation, as well as synaptic transmission (Supplemental Results, Fig. S6). In addition to further supporting the involvement of these biological pathways in pair-bonding, these observations do highlight sex- and modules-specific associations.
Sex-specific alterations of mitochondria
Throughout our analyses of gene expression profiles or modules of co-expression, we repeatedly found alterations related to multiple aspects of the mitochondria. To further investigate mitochondria-related regulation in the NAc following pair-bonding, mitochondrial density was first estimated by measuring copy numbers for the mitochondrial genes Nd1 and Nd4l, normalized against the nuclear gene Gapdh.
When compared to SN voles, Nd1 gDNA levels increased with cohabitation in males and reached significance at the 3W timepoint (p = 0.047), whereas females showed lower levels than males following cohabitation at both 24H (p = 0.014) and 3W (p = 0.024, Fig. 6A, Table S3). A similar pattern was observed for Nd4l gDNA levels, which tend to increase in males following cohabitation, whereas females displayed overall lower levels than males that reached significance at 3W (p = 0.003, Fig. 6B, Table S3). As these observations suggest an increase in mitochondrial density in males but not in females following pair-bonding, we then questioned whether genes controlling mitochondrial fusion (Mfn1 and Mfn2) and fission (Fis1 and Dnm1l also known as Drp1) dynamics would be altered. Despite lower levels in 3W females than in 3W males (p = 0.016), cohabitation did not affect Mfn1 mRNA levels in either sex (Fig. 6C, Table S3). Similarly, Mfn2 mRNA levels in the NAc did not differ between groups (Fig. 6D, Table S3). Fission- controlling genes, however, displayed a sex- and cohabitation-specific regulation. Indeed, Fis1 mRNA levels showed an overall sex-specific regulation by cohabitation (Fig. 6E, Table S3), whereas Dnm1l mRNA levels increased with cohabitation in males but not in females (Fig. 6F, Table S3), with higher levels in 3W males than in sexually-naive males (p = 0.013) and pair-bonded females (p = 0.034). Altogether, these observations indicate that the later phase of the pair-bonding process (3 weeks) is associated with an alteration of mitochondrial dynamics promoting fission in male but not female prairie voles, which could, at least in part, explain the increase in mitochondrial density in males suggested by their higher mitochondrial copy numbers.
Figure 6.
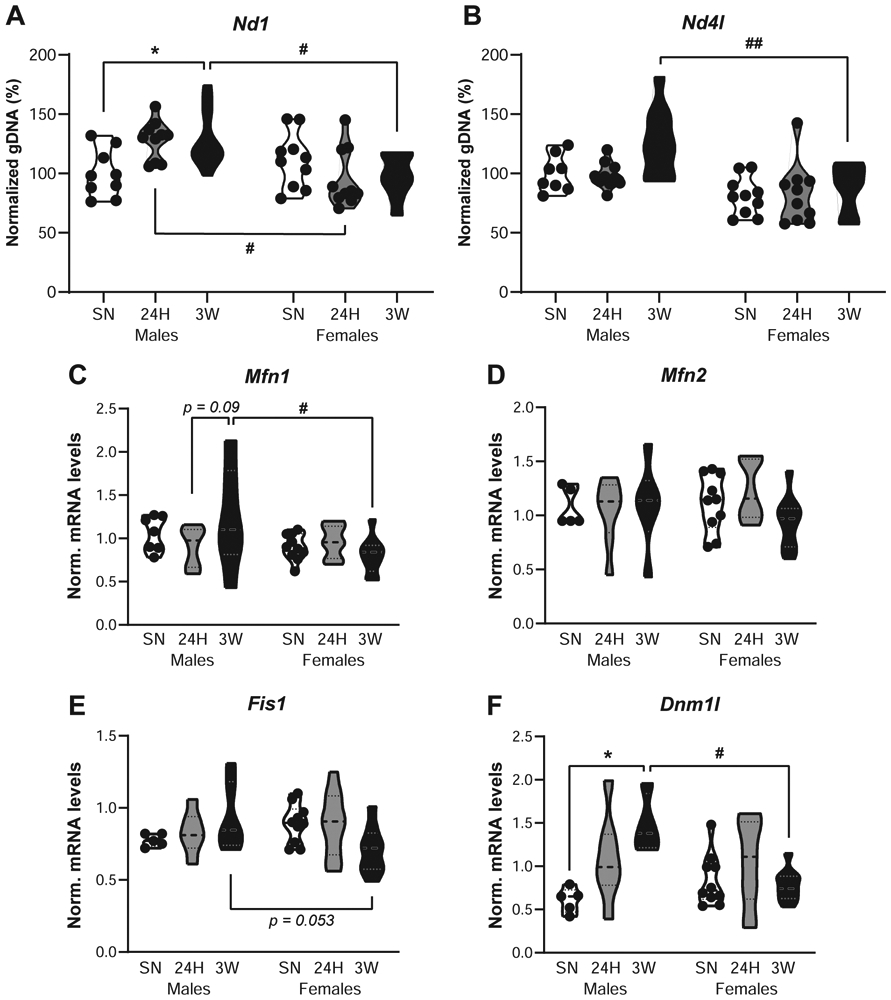
Sex-specific alterations in mitochondrial dynamics following cohabitation with an opposite-sex partner in prairie voles. Measurement of genomic DNA (gDNA) copy number for the mitochondrial genes Nd1 (A) and Nd4l (B) in the nucleus accumbens of male and female prairie voles cohabitated with a same-sex littermate (SN), or an opposite-sex partner for 24 hrs (24H) or 3 weeks (3W). Each measurement was normalized to the nuclear gene Gapdh and depicted as a percentage of controls with the “Males SN” set as 100%. In addition, normalized mRNA levels for the fusion-promoting genes Mfn1 (C) and Mfn2 (D), as well as the fission-promoting genes Fis1 (E) and Dnm1l (F) are depicted. In all panels, each dot represents an individual vole. *p < 0.05 vs. SN group within same sex; #p < 0.05, ##p < 0.01 vs. same cohabitation group in males, Sidak’s or Tukey’s post-hoc tests.
Discussion
In this study, we found a time- and sex-specific reprogramming of the NAc transcriptome in prairie voles following cohabitation with an opposite-sex partner. Indeed, while both males and females displayed selective aggression following cohabitation, males exhibited a distinct transcriptomic response from females in the NAc. When compared to sexually-naive controls, we found few pathways enriched in cohabitated voles but distinct sets between the early and later phase of the bond in males whereas in females, changes were widespread at 24H and remained sustained—to some extent—at 3W. Further highlighted by the observation of distinct modules of co-expression and underlying gene networks between males and females, this sex-specific response of the NAc transcriptome following pair-bonding involved a wide-range of systems required for a sustained regulation of neurotransmission. Interestingly, we found a modulation of mitochondrial dynamics in males but not in females following cohabitation, together with sex-specific alterations in mitochondrial activity.
Although we observed a slight delay in the development of selective aggression in females (Fig. 1A) consistent with longer duration of cohabitation required in females than males to reach maximum levels of aggression reported in the literature (12,38), both males and females displayed similar levels of aggression at 3W. This is in contradiction, however, with the previously-reported lower levels of selective aggression in females than in males pair-bonded for 2 weeks (11,20). Interestingly, this sex difference is only observed in conditions optimal for the pregnancy of the female partner (20), which could explain the absence of sex differences in aggressive behaviors we observed at 3W. Nevertheless, we did observe high levels of aggression in cohabitated animals (Fig. 1, Fig. S1) that, together with the reduction in positive social interactions towards strangers, confirm the proper establishment of a pair-bond (11,37,39).
In accordance with such profound behavioral changes, transcriptomic changes were widespread in the NAc as early as 24 hrs after the beginning of the cohabitation with an opposite-sex partner. Surprisingly, the number of DE genes was relatively limited at 3W, likely resulting from a substantial inter-individual variability—commonly reported in prairie voles at the gene and behavioral levels (22,40-43) and also observed during the RIT (Fig. S1)—impeding a traditional DE analysis with a set false-discovery rate threshold. Nevertheless, we could still detect widespread regulations of gene expression at 24H, reflecting the impact of a new social encounter in the highly social prairie voles. In this context, we used a threshold-free approach (GSEA) and found distinct sets of regulation between the early (24H) and later (3W) phases of pair-bonding in males further supported by existing evidence. For instance, mRNA levels for the oxytocin receptor (Oxtr) and vasopressin V1a receptor (Avpr1a) are upregulated in the NAc after 24 hrs (18,19), but not 2 weeks (20) of pair-bonding. Similarly, while μ-opioid receptors blockade in the NAc prevents partner preference formation following 24 hrs of cohabitation (44), κ-receptors but not μ-receptor antagonism reduces selective aggression in prairie voles pair-bonded for 2 weeks (11,20). Finally, while dopamine D1 receptors (D1R) promote the maintenance but inhibit the formation of the pair-bond, dopamine D2 receptors (D2R) inhibit its maintenance but promote its formation (37,45). Our current study thus uncovers the extent of the transcriptomic regulations specific to each phase of the pair-bonding process and highlights its preponderance in the NAc of males over females. Notably, our study is limited in the ability to differentiate the contribution of the exposure to a novel conspecific from those linked to the early stage of pair-bonding among the regulations observed at the 24H timepoint. It is important to note, however, that most functional enrichments observed at 24H in females were maintained at 3W, a timepoint at which exposure to social novelty is unlikely to substantially contribute. Another limitation results from the salient nature of the RIT, reported to induce transcriptomic changes following 20-30 mins in a variety of species (46-48). Although our SN controls were exposed to the RIT identically to pair-bonded voles, we cannot rule out the possibility that part of the transcriptomic regulations observed at 24H and 3W reflect a different response of pair-bonded animals to the social stress resulting from the RIT exposure. It is important to note, however, that few RIT-responsive genes (48) or immediate early genes, whose mRNA levels generally peak 30 min post-stimulation (46,47), are DE following cohabitation in our dataset (Table S4), which would tend to suggest that such RIT interference with the transcriptomic regulations we observed would be limited.
Alongside widespread alterations in gene expression encompassing processes required for a sustained regulation of neurotransmission (Supplemental Results), our analyses suggest modulations of mitochondria and energy production by pair-bonding. While this effect was observed at the transcript level in males, females exhibit a widespread up-regulation of related gene-sets detected as early as 24H and pronounced enough to reverse the baseline sex-bias. Indeed, consistent with higher mitochondrial respiration in males than females in the mouse cortex (49), mitochondria-related gene-sets were biased towards males at baseline, but switched towards females at 24H (Fig. 5). Surprisingly, however, cohabitation with a male did not alter Mfn1, Mfn2, Fis1, or Dnm1l mRNA levels in female prairie voles, but led to a sex-specific alteration of mitochondrial respiration (Fig. S7, Table S5, Supplemental Results). In contrast, males showed an increase in fission-controlling genes but not fusion-controlling genes at 3W that, consistent with their higher mitochondrial DNA copy numbers, would indicate an alteration of mitochondrial dynamics balance towards fission. While a link between mitochondrial dynamics and prosocial behaviors remains unclear, they constitute a critical component of neuronal plasticity in a variety of systems (50,51). The fission-promoting factor DNM1L (DRP1), for instance, regulates mitochondrial distribution to dendrites, increases spines and synapses density, and mediates a fusion-to-fission shift following KCl-induced depolarization in hippocampal neurons (52). Furthermore, while fission-promoting genes relate to D1R- and D2R-containing MSN identity at baseline (53), DRP1 in D1R-containing MSN is up-regulated by and mediates drug seeking behavior following cocaine self-administration (54). In combination with the alterations in factors—including DNM1L—controlling mitochondrial dynamics in the autistic brain and other neuropsychiatric disorders (50,51,55), our observations thus highlight mitochondrial fission in the NAc and DMM1L in particular as interesting candidates underlying pair-bond maintenance in male prairie voles.
In addition to vast sex differences at baseline, we found a sex-specific reprogramming of gene expression in the prairie vole NAc following 3 weeks of cohabitation with an opposite-sex partner. Indeed, despite similarities in functional enrichment, distinct sets of gene networks were recruited in males than in females, leading to a greater extent of sexually-biased genes at 3W than at baseline (Fig. 2C,F). As a result, even though pair-bonded male and female prairie voles show similar behaviors during the RIT, the transcriptomic mechanisms at play differ. Previously, we provided evidence supporting sex-specific regulations in the early phase of pair-bonding, in which 24 hrs of cohabitation with mating increases OTR and V1aR mRNA levels in the NAc of females, but only V1aR mRNA levels in males (18,19). Although similar regulations were not detected here, our study reveals a male-specific gene-network linked to selective aggression under the control of V1aR, as well as an enrichment of dopamine- and OTR-signaling pathways in pair-bonded females consistent with known sex-differences in these neurotransmissions underlying pair-bonding (Supplemental Results). Our current study thus not only expands the involvement of neuropeptidic and dopaminergic receptors in pair-bonding to their downstream signaling pathways, but also highlights the recruitment of distinct functional processes between males and females pair-bonded for 3 weeks (see also Supplemental Results). It is for instance particularly interesting to note the enrichment of pathways associated with response to reward and addiction in the set of genes sexually-biased at 3W only (Supplemental Results, Fig. S8, Table S6). Consistent with a sex-specific modulation of neurotransmission in the NAc following pair-bonding, female prairie voles show greater dopamine release in the NAc shell than males following two weeks of pair-bonding but not at baseline (20). This also draws an interesting parallel with known sex differences in reward processing in the NAc in rodents, where male and female rats and mice differ in sensitivity to reward as evidenced by different activation of the dopaminergic neurotransmission (56). As expected, these differences are accompanied by transcriptomic reprogramming in the NAc, but while male and female mice share common responsive genes, each sex exhibits a distinct set of regulations (57). Notably, this sex-specific response pattern of the NAc transcriptome is particularly pronounced following exposure to chronic stress in mice (58,59) and in major depressive disorder in humans (60). Although the extent and exact nature of similarities and differences between these different conditions remain to be determined, these data do confirm the marked degree of sex-specificity in transcriptomic regulations following NAc activation. By highlighting the overall pathways and gene networks at play in each sex, our transcriptomic analyses pave the way to further our elucidation of the complex social bonding process.
Supplementary Material
Acknowledgments
This work was supported by the National Institute of Mental Health grants R01-MH109450 to M.K., R01-MH058616 and R01-MH108527 to Z.W, as well as by the National Institute on Aging R01-AG057716 to P.K. The authors would like to thank Dr. Roger Mercer, Dr. Cynthia Vied, and Dr. Yanming Yang from the Florida State University Translational Science Laboratory for the generation and demultiplexing of sequencing data. Some of the computing for this manuscript was performed on the HPC/Spear clusters at the Research Computing Center at the Florida State University (FSU).
Footnotes
Disclosures
The authors report no biomedical financial interests or potential conflicts of interest.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Bora E, Yucel M, Allen NB (2009): Neurobiology of human affiliative behaviour: implications for psychiatric disorders. Curr Opin Psychiatry 22: 320–325. [DOI] [PubMed] [Google Scholar]
- 2.Duclot F, Kabbaj M (2015): The estrous cycle surpasses sex differences in regulating the transcriptome in the rat medial prefrontal cortex and reveals an underlying role of early growth response 1. Genome Biol 16: 256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Kleiman DG (1977): Monogamy in mammals. Q Rev Biol 52: 39–69. [DOI] [PubMed] [Google Scholar]
- 4.Clayton DF (2000): The genomic action potential. Neurobiol Learn Mem 74: 185–216. [DOI] [PubMed] [Google Scholar]
- 5.Thomas JA, Birney EC (1979): Parental Care and Mating System of the Prairie Vole, Microtus ochrogaster. Behav Ecol Sociobiol 5: 171–186. [Google Scholar]
- 6.Tur G, Georgieva EI, Gagete A, López-Rodas G, Rodríguez JL, Franco L (2010): Factor binding and chromatin modification in the promoter of murine Egr1 gene upon induction. Cell Mol Life Sci 67: 4065–4077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Getz LL, Hofmann JE (1986): Social organization in free-living prairie voles, Microtus ochrogaster. Behav Ecol Sociobiol 18: 275–282. [Google Scholar]
- 8.Williams JR, Catania KC, Carter CS (1992): Development of partner preferences in female prairie voles (Microtus ochrogaster): the role of social and sexual experience. Horm Behav 26: 339–349. [DOI] [PubMed] [Google Scholar]
- 9.Wang Z, Young LJ, Liu Y, Insel TR (1997): Species differences in vasopressin receptor binding are evident early in development: comparative anatomic studies in prairie and montane voles. J Comp Neurol 378: 535–546. [PubMed] [Google Scholar]
- 10.Winslow JT, Hastings N, Carter CS, Harbaugh CR, Insel TR (1993): A role for central vasopressin in pair bonding in monogamous prairie voles. Nature 365: 545–548. [DOI] [PubMed] [Google Scholar]
- 11.Resendez SL, Kuhnmuench M, Krzywosinski T, Aragona BJ (2012): κ-Opioid receptors within the nucleus accumbens shell mediate pair bond maintenance. J Neurosci 32: 6771–6784. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Carter CS, DeVries AC, Getz LL (1995): Physiological substrates of mammalian monogamy: the prairie vole model. Neurosci Biobehav Rev 19: 303–314. [DOI] [PubMed] [Google Scholar]
- 13.Lieberwirth C, Wang Z (2016): The neurobiology of pair bond formation, bond disruption, and social buffering. Curr Opin Neurobiol 40: 8–13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Young LJ, Wang Z (2004): The neurobiology of pair bonding. Nat Neurosci 7: 1048–1054. [DOI] [PubMed] [Google Scholar]
- 15.Amadei EA, Johnson ZV, Jun Kwon Y, Shpiner AC, Saravanan V, Mays WD, et al. (2017): Dynamic corticostriatal activity biases social bonding in monogamous female prairie voles. Nature 546: 297–301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Scribner JL, Vance EA, Protter DSW, Sheeran WM, Saslow E, Cameron RT, et al. (2020): A neuronal signature for monogamous reunion. Proc Natl Acad Sci U S A. 10.1073/pnas.1917287117 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Young KA, Gobrogge KL, Liu Y, Wang Z (2011): The neurobiology of pair bonding: insights from a socially monogamous rodent. Front Neuroendocrinol 32: 53–69. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Duclot F, Wang H, Youssef C, Liu Y, Wang Z, Kabbaj M (2016): Trichostatin A (TSA) facilitates formation of partner preference in male prairie voles (Microtus ochrogaster). Horm Behav 81: 68–73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Wang H, Duclot F, Liu Y, Wang Z, Kabbaj M (2013): Histone deacetylase inhibitors facilitate partner preference formation in female prairie voles. Nat Neurosci 16: 919–924. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Resendez SL, Keyes PC, Day JJ, Hambro C, Austin CJ, Maina FK, et al. (2016): Dopamine and opioid systems interact within the nucleus accumbens to maintain monogamous pair bonds. Elife 5. 10.7554/eLife.15325 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Ross HE, Freeman SM, Spiegel LL, Ren X, Terwilliger EF, Young LJ (2009): Variation in oxytocin receptor density in the nucleus accumbens has differential effects on affiliative behaviors in monogamous and polygamous voles. J Neurosci 29: 1312–1318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.King LB, Walum H, Inoue K, Eyrich NW, Young LJ (2016): Variation in the Oxytocin Receptor Gene Predicts Brain Region-Specific Expression and Social Attachment. Biol Psychiatry 80: 160–169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Gobrogge KL, Liu Y, Jia X, Wang Z (2007): Anterior hypothalamic neural activation and neurochemical associations with aggression in pair-bonded male prairie voles. J Comp Neurol 502: 1109–1122. [DOI] [PubMed] [Google Scholar]
- 24.Chen S, Zhou Y, Chen Y, Gu J (2018): fastp: an ultra-fast all-in-one FASTQ preprocessor. Bioinformatics 34: i884–i890. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Patro R, Duggal G, Love MI, Irizarry RA, Kingsford C (2017): Salmon provides fast and biasaware quantification of transcript expression. Nat Methods 14: 417–419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Love MI, Huber W, Anders S (2014): Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol 15: 550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Huber W, Carey VJ, Gentleman R, Anders S, Carlson M, Carvalho BS, et al. (2015): Orchestrating high-throughput genomic analysis with Bioconductor. Nat Methods 12: 115–121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Subramanian A, Tamayo P, Mootha VK, Mukherjee S, Ebert BL, Gillette MA, et al. (2005): Gene set enrichment analysis: a knowledge-based approach for interpreting genomewide expression profiles. Proc Natl Acad Sci U S A 102: 15545–15550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Yu G, Wang L-G, Han Y, He Q-Y (2012): clusterProfiler: an R package for comparing biological themes among gene clusters. OMICS 16: 284–287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Zhang B, Horvath S (2005): A general framework for weighted gene co-expression network analysis. Stat Appl Genet Mol Biol 4: Article17. [DOI] [PubMed] [Google Scholar]
- 31.Langfelder P, Horvath S (2008): WGCNA: an R package for weighted correlation network analysis. BMC Bioinformatics 9: 559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Langfelder P, Horvath S (2012): Fast R Functions for Robust Correlations and Hierarchical Clustering. J Stat Softw 46. Retrieved from https://www.ncbi.nlm.nih.gov/pubmed/23050260 [PMC free article] [PubMed] [Google Scholar]
- 33.Lachmann A, Giorgi FM, Lopez G, Califano A (2016): ARACNe-AP: gene network reverse engineering through adaptive partitioning inference of mutual information. Bioinformatics 32: 2233–2235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Bagot RC, Cates HM, Purushothaman I, Lorsch ZS, Walker DM, Wang J, et al. (2016): Circuit-wide Transcriptional Profiling Reveals Brain Region-Specific Gene Networks Regulating Depression Susceptibility. Neuron 90: 969–983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Zhang B, Gaiteri C, Bodea L-G, Wang Z, McElwee J, Podtelezhnikov AA, et al. (2013): Integrated systems approach identifies genetic nodes and networks in late-onset Alzheimer’s disease. Cell 153: 707–720. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Shu L, Zhao Y, Kurt Z, Byars SG, Tukiainen T, Kettunen J, et al. (2016): Mergeomics: multidimensional data integration to identify pathogenic perturbations to biological systems. BMC Genomics 17: 874. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Aragona BJ, Liu Y, Yu YJ, Curtis JT, Detwiler JM, Insel TR, Wang Z (2006): Nucleus accumbens dopamine differentially mediates the formation and maintenance of monogamous pair bonds. Nat Neurosci 9: 133–139. [DOI] [PubMed] [Google Scholar]
- 38.Firestone KB, Thompson KV, Carter CS (1991): Female-female interactions and social stress in prairie voles. Behav Neural Biol 55: 31–41. [DOI] [PubMed] [Google Scholar]
- 39.Carter CS, DeVries AC, Taymans SE, Roberts RL, Williams JR, Getz LL (1997): Peptides, steroids, and pair bonding. Ann N Y Acad Sci 807: 260–272. [DOI] [PubMed] [Google Scholar]
- 40.Hammock EAD, Young LJ (2005): Microsatellite instability generates diversity in brain and sociobehavioral traits. Science 308: 1630–1634. [DOI] [PubMed] [Google Scholar]
- 41.Barrett CE, Keebaugh AC, Ahern TH, Bass CE, Terwilliger EF, Young LJ (2013): Variation in vasopressin receptor (Avpr1a) expression creates diversity in behaviors related to monogamy in prairie voles. Horm Behav 63: 518–526. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Young LJ (1999): Frank A. Beach Award. Oxytocin and vasopressin receptors and species-typical social behaviors. Horm Behav 36: 212–221. [DOI] [PubMed] [Google Scholar]
- 43.Vogel AR, Patisaul HB, Arambula SE, Tiezzi F, McGraw LA (2018): Individual Variation in Social Behaviours of Male Lab-reared Prairie voles (Microtus ochrogaster) is Non-heritable and Weakly Associated with V1aR Density. Sci Rep 8: 1396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Resendez SL, Dome M, Gormley G, Franco D, Nevárez N, Hamid AA, Aragona BJ (2013): μ-Opioid receptors within subregions of the striatum mediate pair bond formation through parallel yet distinct reward mechanisms. J Neurosci 33: 9140–9149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Gingrich B, Liu Y, Cascio C, Wang Z, Insel TR (2000): Dopamine D2 receptors in the nucleus accumbens are important for social attachment in female prairie voles (Microtus ochrogaster). Behav Neurosci 114: 173–183. [DOI] [PubMed] [Google Scholar]
- 46.Sanogo YO, Band M, Blatti C, Sinha S, Bell AM (2012): Transcriptional regulation of brain gene expression in response to a territorial intrusion. Proc Biol Sci 279: 4929–4938. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Shpigler HY, Saul MC, Murdoch EE, Cash-Ahmed AC, Seward CH, Sloofman L, et al. (2017): Behavioral, transcriptomic and epigenetic responses to social challenge in honey bees. Genes Brain Behav 16: 579–591. [DOI] [PubMed] [Google Scholar]
- 48.Rittschof CC, Bukhari SA, Sloofman LG, Troy JM, Caetano-Anollés D, Cash-Ahmed A, et al. (2014): Neuromolecular responses to social challenge: common mechanisms across mouse, stickleback fish, and honey bee. Proc Natl Acad Sci U S A 111: 17929–17934. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Arias-Reyes C, Losantos-Ramos K, Gonzales M, Furrer D, Soliz J (2019): NADH-linked mitochondrial respiration in the developing mouse brain is sex-, age- and tissue-dependent. Respir Physiol Neurobiol 266: 156–162. [DOI] [PubMed] [Google Scholar]
- 50.Belenguer P, Duarte JMN, Schuck PF, Ferreira GC (2019): Mitochondria and the Brain: Bioenergetics and Beyond. Neurotox Res 36: 219–238. [DOI] [PubMed] [Google Scholar]
- 51.Bertholet AM, Delerue T, Millet AM, Moulis MF, David C, Daloyau M, et al. (2016): Mitochondrial fusion/fission dynamics in neurodegeneration and neuronal plasticity. Neurobiol Dis 90: 3–19. [DOI] [PubMed] [Google Scholar]
- 52.Li Z, Okamoto K-I, Hayashi Y, Sheng M (2004): The importance of dendritic mitochondria in the morphogenesis and plasticity of spines and synapses. Cell 119: 873–887. [DOI] [PubMed] [Google Scholar]
- 53.Chandra R, Calarco CA, Lobo MK (2019): Differential mitochondrial morphology in ventral striatal projection neuron subtypes. J Neurosci Res 97: 1579–1589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Chandra R, Engeln M, Schiefer C, Patton MH, Martin JA, Werner CT, et al. (2017): Drp1 Mitochondrial Fission in D1 Neurons Mediates Behavioral and Cellular Plasticity during Early Cocaine Abstinence. Neuron 96: 1327–1341.e6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Tang G, Gutierrez Rios P, Kuo S-H, Akman HO, Rosoklija G, Tanji K, et al. (2013): Mitochondrial abnormalities in temporal lobe of autistic brain. Neurobiol Dis 54: 349–361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Becker JB (2016): Sex differences in addiction. Dialogues Clin Neurosci 18: 395–402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.LaRese TP, Rheaume BA, Abraham R, Eipper BA, Mains RE (2019): Sex-Specific Gene Expression in the Mouse Nucleus Accumbens Before and After Cocaine Exposure. J Endocr Soc 3: 468–487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Pfau ML, Purushothaman I, Feng J, Golden SA, Aleyasin H, Lorsch ZS, et al. (2016): Integrative Analysis of Sex-Specific microRNA Networks Following Stress in Mouse Nucleus Accumbens. Front Mol Neurosci 9: 144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Hodes GE, Pfau ML, Purushothaman I, Ahn HF, Golden SA, Christoffel DJ, et al. (2015): Sex Differences in Nucleus Accumbens Transcriptome Profiles Associated with Susceptibility versus Resilience to Subchronic Variable Stress. J Neurosci 35: 16362–16376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Labonté B, Engmann O, Purushothaman I, Menard C, Wang J, Tan C, et al. (2017): Sex-specific transcriptional signatures in human depression. Nat Med 23: 1102–1111. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.