Abstract
The Nicotine Metabolite Ratio (NMR, 3-hydroxycotinine/cotinine), a highly heritable index of nicotine metabolic inactivation by the CYP2A6 enzyme, is associated with numerous smoking behaviors and diseases, as well as unique cessation outcomes. However, the NMR cannot be measured in non-, former- or intermittent-smokers, for example in evaluating tobacco-related disease risk. Traditional pharmacogenetic groupings based on CYP2A6 * alleles capture a modest portion of NMR variation. We previously created a CYP2A6 weighted genetic risk score (wGRS) for European-ancestry populations (EUR) by incorporating independent signals from genome-wide association studies to capture a larger proportion of NMR variation. However, CYP2A6 genetic architecture is unique to ancestral populations. In this study we developed and replicated an African-ancestry (AFR) wGRS which captured 30–35% of the variation in NMR. We demonstrated model robustness against known environmental sources of NMR variation. Furthermore, despite the vast diversity within AFR populations, we showed that the AFR wGRS was consistent between different US geographical regions and unaltered by fine AFR population substructure. The AFR and EUR wGRSs can distinguish slow from normal metabolizers in their respective populations, and were able to reflect unique smoking cessation pharmacotherapy outcomes previously observed for the NMR. Additionally, we evaluated the utility of a cross-ancestry wGRS, and the capacity of EUR, AFR, and cross-ancestry wGRSs to predict the NMR within stratified or admixed AFR-EUR populations. Overall, our findings establish the clinical benefit of applying ancestry-specific wGRSs, demonstrating superiority of the AFR wGRS in AFRs.
Keywords: Pharmacogenetics, CYP2A6, genetic-risk-score, drug-metabolism, nicotine, smoking-cessation
Introduction
Cigarette smoking is the leading cause of preventable death in the USA(1). Smoking behaviours and related disease risk differ between ancestral populations. Although Americans of African ancestry (AFR) have a comparable smoking prevalence to those of European ancestry (EUR), they have a higher incidence of lung cancer(2, 3) yet paradoxically smoke fewer cigarettes per day (CPD)(4–6). However, CPD is not an accurate measure of tobacco consumption, owing to differences in smoking intensity; quantification of nicotine intake biomarkers would better reflect differences in smoking consumption than self-reported measures(7). AFR smokers are also more likely to make a quit attempt, but less likely to succeed, than EUR smokers(8, 9). Identifying factors contributing to unique smoking behaviours in AFR may assist in tailoring treatment.
Nicotine, the primary psychoactive component in cigarettes,(10) is metabolized to cotinine and then to 3-hydroxycotinine in reactions mediated by CYP2A6(11, 12). The 3-hydroxycotinine/cotinine ratio, or nicotine metabolite ratio (NMR), is a biomarker of CYP2A6 activity(13). The NMR is highly correlated with nicotine clearance(13) thus altering smoking behaviours, including acquisition(14), quantity(15–17), topography(18), dependence(16, 17, 19), and related diseases(17).
The NMR is useful for personalizing smoking cessation(20–24).For example, the Pharmacogenetics of Nicotine Addiction and Treatment 2 (PNAT2, [NCT01314001]) trial prospectively randomized smokers by their baseline NMR: normal metabolizers (i.e. higher NMR) had greater quit rates on varenicline versus the nicotine patch, whereas slow metabolizers (i.e. lower NMR) had similar quit rates(22).
Ad-libitum NMR is measured during regular smoking from a saliva or blood sample; NMRs derived from saliva, plasma, and whole blood are highly correlated(25). This requirement makes it difficult to assess the impact of CYP2A6 activity in intermittent- and non-smokers in the pathogenesis of several diseases (e.g. COPD and lung cancer)(26, 27) and on the metabolism of other clinical drugs (e.g. Tegafur, Letrozole, and Efavirenz)(28).
The NMR is highly heritable (h2~0.60–0.80)(29), thus it should be predictable using a genetic risk score. An NMR genetic risk score would be important for non/irregular smokers and in studies where only DNA is available. In EUR smokers approximately 20% of NMR variation is captured by CYP2A6 * alleles(30). The * allele nomenclature system (see pharmvar.org) describes unique haplotypes mostly indicative of protein coding and structural variants in pharmacogenes(31–33). The relatively low portion of variation explained by CYP2A6 * alleles prompted several genome-wide association studies (GWASs) of the NMR, conducted predominantly in EURs(29, 34–36). Over 96% of genome-wide significant variants identified in a meta-analysis of >5,000 EUR smokers are around the CYP2A6 gene locus(37).
The genetic architecture (linkage disequilibrium (LD) structure and allele frequencies) of CYP2A6 differs by ancestry, necessitating the identification of population-specific genetic variants influencing the NMR. In our NMR GWAS performed in AFRs(35), a high (97%) proportion of variants were found in/around CYP2A6(35) as found in EUR(37). However, ~60% of the CYP2A6 variants that reached genome-wide significance in AFR were not among those found in EUR(35), emphasizing the unique ancestral CYP2A6 architecture. Few studies have assessed the influence of population substructure on CYP2A6 variant associations beyond broad continental categorizations. Thus, there is little knowledge in how ancestral subpopulations may affect these associations, and whether this influences allele functional heterogeneity. Additionally, the genetic architecture of AFR-EUR admixed populations, mostly excluded in ancestry-stratified GWASs, is largely unknown.
The CYP2A6 gene shares ~95% homology with the pseudogene CYP2A7. Due to high homology and the presence of structural variants, inaccurate sequencing in this region leads to poor imputation with current reference panels. Consequently, it is difficult to accurately capture several low frequency (MAF<5%) (*20, *23, *25, *28, *31, *35) and structural (*1X2, *1B, *4, *12) * alleles in the CYP2A6 gene through GWAS microarrays. Integrating these difficult-to-genotype * alleles via targeted sequencing or qPCR approaches, with the independent signals identified by GWASs, could capture a larger portion of NMR variability.
A multiplicative gene scoring model was published(38) based on a phenotype of first pass metabolism of oral nicotine (cotinine/cotinine+nicotine)(38) however it did not capture variation in the NMR(39). We previously developed an additive weighted genetic risk score (wGRS) approach for the NMR in EURs, where seven CYP2A6 variants (independent signals from GWAS and * alleles) captured ~34% of NMR variation(39), but only 20% of the NMR variation in AFRs as genetic risk scores do not transfer between ancestral populations(40, 41). Thus, the clinical utility of genetic risk scores generated from existing GWASs (conducted mostly in EUR) will continue to disproportionally benefit EUR(40, 41).
We sought to (1) develop and validate a wGRS to predict the NMR in AFR, (2) understand whether smoking characteristics or population substructure would confound the application of this wGRS, (3) examine the clinical utility of the wGRS in predicting smoking cessation outcomes, (4) create and evaluate an AFR-EUR cross-ancestry wGRS and compare it to the ancestry-specific wGRSs, and (5) evaluate the applicability of the EUR, AFR, and cross-ancestry wGRSs within an admixed AFR-EUR population.
Methods
Study Populations:
The studies were approved by institutional review boards at all participating sites.
Training Set: AFR smokers (N=954) screened for two smoking cessation clinical trials: the PNAT2 [NCT01314001] and the Kick It at Swope 3 (KIS3) [NCT00666978] trials (Table S1), where ancestry was both self-reported and confirmed by principal component (PC) clustering to the HapMap3 AFR population (predominately the African ancestry in Southwest USA (ASW) and the Yoruba in Ibadan, Nigeria (YRI) subpopulations). Of note, the ASW HapMap3 subpopulation consists of a narrow degree of EUR admixture, herein we define these individuals as AFR. The PNAT2 participants were recruited from 4 different sites: Philadelphia, Buffallo, Houston, and Toronto. The KIS3 participants were recruited exclusively from Kansas City. PNAT2 included only heavy smokers (≥10 CPD), whereas KIS3 included only light smokers (≤10). Clinical trial details are described elsewhere(22, 42).
Replication Set: AFR smokers (N=216) screened for the Quit-2-Live (Q2L) clinical trial [NCT01836276], where ancestry was self-reported. The Q2L participants were recruited exclusively from Kansas City. Q2L included a combination of heavy and light smokers. Clinical trial details are described elsewhere(9).
Admixed Set: Individuals of admixed AFR and EUR ancestry (N=68) excluded from the AFR and EUR subsets in the training set because they were on the cline between the AFR and EUR HapMap3 reference populations as determined by PCA, representing approximately equal admixture of AFR and EUR ancestry.
Genotyping:
A variety of array imputation, targeted sequencing, and qPCR approaches were used to genotype all variants across the three studies. Details are described in the supplementary material.
Principal Components Analysis (PCA):
Ancestry clustering for the training set was performed through PC clustering with the HapMap3 project reference populations as described(35): 98.5% and 96.6% of African-ancestry smokers in the PNAT2 and KIS3 trials, respectively, had genetic ancestries concordant with self-reported ancestry(35). Refined AFR population substructure was assessed by merging the training set with the AFR subpopulations included in the phase 3 release of 1000 Genomes (1KG).
AFR wGRS:
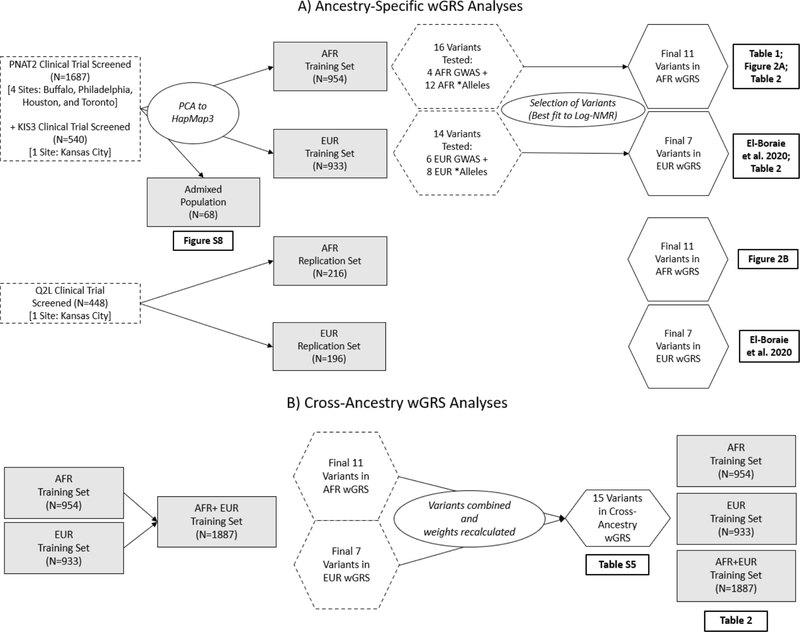
16 variants were tested from two sets. Set 1 included 4 GWAS independent signals identified in conditional analyses in a meta-analysis comprised of exclusively AFR smokers: rs12459249, rs111645190, rs185430475, and rs11878604(35). This did not include variants from NMR GWASs in multi-ethnic or EUR cohorts, such as rs56112850(39, 43), as it was not identified as a GWAS independent signal among exclusively AFR smokers, although these were tested later in a cross-ancestry wGRS discussed below(35). Set 2 included 12 CYP2A6 * alleles common in AFR populations (MAF>1%) listed on pharmvar.org; set 2 represent functional variants that could be missed in GWAS analyses owing to their low frequency (most MAF<5%). These include single nucleotide, insertion/deletion, and structural variants that are often excluded from microarrays: *1X2 (gene duplication), *1B (58 base-pair gene conversion in the 3′ UTR of CYP2A6), *4 (gene deletion), *9 (rs28399433), *12 (gene hybrid), *17 (rs28399454), *20 (rs568811809), *23 (rs56256500), *25/*26/*27 (all tagged by rs28399440), *28 (rs28399463), *31 (rs72549432), and *35 (rs143731390). The analysis outline including the populations used and the variant selection process is highlighted in (Figure 1).
Figure 1.
Flowchart for the A) Ancestry-specific and B) Cross-ancestry wGRS analyses. The training sets consisted of individuals screened from two trials (PNAT2 and KIS3) and where ancestry was determined through PC clustering analysis to the HapMap3 populations. Variants tested included GWAS independent signals and * alleles from the specified population. For the cross-ancestry wGRS, all ancestry-specific wGRS variants were merged.
An additive wGRS was developed based on the variation in the log-transformed NMR (log-NMR) in the training set. Variants were selected by backward stepwise regression after inputting all sixteen variants (sets 1 and 2). Additional models were assessed using either all the variants from set 1, or set 2, as a base model and assessing the contribution to the variance (R2) captured after entering additional variants stepwise. Scores were created by summing the number of risk alleles weighted by their unstandardized effect sizes. The use of unstandardized betas was used to retain the unit of measurement from the GWAS analysis, whereas standardized variables invite bias due to sampling error. Betas were estimated from frequentist additive linear regression models (using SNPTEST, version 2.5.2)(44) of square-root transformed NMR (sqrt-NMR) in a meta-analysis of the training set, adjusted for PCs 1 and 2, sex, age, and body mass index (BMI), and unstandardized through multiplying betas by the standard deviation (SD) of the sqrt-NMR in the training set (SD=0.181). Details on how to evaluate an individual’s wGRS are included in the supplementary material.
EUR wGRS: A CYP2A6 wGRS defined for EUR is described elsewhere(39), where the EUR training set included the EUR subset of the PNAT2 trial, where ancestry was self-reported and confirmed by PC clustering to the HapMap3 EUR population (exclusively the Utah residents with Northern and Western European ancestry (CEU) subpopulation). Briefly the final wGRS included 7 variants: independent signals from EUR GWAS (rs56113850, rs2316204, rs113288603) and CYP2A6 * alleles common in EUR populations (*2, *4, *9, *12). The analysis outline including the populations used and the variant selection process is highlighted in (Figure 1).
Cross-ancestry wGRS:
To create a cross-ancestry wGRS, a meta-analysis was performed including all EUR and AFR CYP2A6 wGRS variants, adjusting for population substructure and demographic covariates (using META, version 1.7). Both the inverse-variance method based on a fixed effects model, and a random effects model were tested. The analysis outline including the populations used and the variant selection process is highlighted in (Figure 1).
Statistical Analysis, Phenotype, and Covariates:
Beyond calculating variant betas (SNPTEST, version 2.5.2), all other statistical analyses were performed using SPSS version 20 (IBM Corporation) and MedCalc version 17.4 (MedCalc Software). The NMR (cotinine and 3-hydroxycotinine metabolites) were measured from whole blood(25) collected at intake in each study while participants were smoking ad-libitum. The NMR was not normally distributed (by the Shapiro-Wilk test) and was therefore log-transformed, which best represents the nicotine clearance rate(45). Linear regression assessed log-transformed NMR (log-NMR) variation accounted for by the wGRS models, with and without the addition of demographic covariates known to alter NMR (i.e. sex, age, and BMI)(46). Additional factors were also evaluated (i.e. mentholated cigarette use and nicotine intake). Nicotine intake was assessed as self-reported CPD and as a biological measure consisting of the sum of nicotine’s major metabolites, free cotinine and 3-hydroxycotinine (COT+3HC)(15, 47). Receiver Operating Characteristic (ROC) curve analyses were conducted with an NMR definition of normal metabolizers (≥0.31) which was used to randomize PNAT2 participants to treatment(22, 39). The Youden’s J index was used to determine the criterion for the optimal cut-point in the wGRS to dichotomize slow (<0.31) and normal metabolizers (≥0.31). Logistic regression was used to evaluate end-of-treatment quit rates (nicotine patch vs. varenicline) within slow and normal metabolizers defined by NMR or by the wGRS. An interaction between treatment and metabolizer group was also evaluated as the ratio of odds ratios (ORRs)(22).
Results
AFR Weighted Genetic Risk Score
Sixteen variants were evaluated. Backward stepwise regression identified a final set of 11 variants that provided optimal prediction of the log-NMR phenotype (Table 1). Versions of the model including rs11878604 or other common CYP2A6 * alleles (*1B, *23, *28, and *31) (Table 1) yielded poorer fit to the NMR and were consequently eliminated by stepwise regression.
Table 1.
The sixteen variants tested, with the final eleven variants included and the five variants excluded in the African-ancestry (AFR) CYP2A6 weighted genetic risk score (wGRS).
| Reference AlleleB | Risk Allele | Location with respect to CYP2A6 gene | Beta per Risk Allele | Weight per Risk AlleleC | |
|---|---|---|---|---|---|
| Included Variants | |||||
|
| |||||
| rs12459249 A | T | C | 10KB 3’ | +0.591 | +0.107 |
| rs111645190 A | G | A | 5KB 5’ | −0.624 | −0.113 |
| rs185430475 A | C | G | 10MB 3’ | +0.287 | +0.052 |
| *1×2 (CYP2A6 Duplication) | - | Duplication | Full Gene Duplication | +0.361 | +0.065 |
| *4 (CYP2A6 Deletion) | - | Deletion | Full Gene Deletion | −0.819 | −0.148 |
| *9 (rs28399433) | A | C | Promoter (TATA Box) | −0.457 | −0.083 |
| *12 (CYP2A6/2A7 Hybrid) | - | Hybrid | Translocation of Exons 1–2 | −0.538 | −0.097 |
| *17 (rs28399454) | C | T | Exon 7 (V365M) | −0.683 | −0.124 |
| *20 (rs568811809) | TT | - | Exon 4 (196 Frameshift) | −0.862 | −0.156 |
| *25/*26/*27 (rs28399440) | A | G | Exon 3 (F118L) | −0.714 | −0.129 |
| *35 (rs143731390) | T | A | Exon 9 (N438Y) | −0.312 | −0.057 |
| *1 (No wGRS Variants) | - | - | - | - | 0.000 |
|
| |||||
| Excluded Variants | |||||
|
| |||||
| rs11878604 A | T | C | 16KB 3’ | −0.654 | −0.118 |
| *1B (58 BP Conversion) | - | Conversion | UTR 3’ | +0.186 | +0.034 |
| *23 (rs56256500) | G | A | Exon 4 (R203C) | +0.051 | +0.009 |
| *28 (rs28399463) | T | C | Exon 8 (N418D) | +0.037 | +0.007 |
| *31 (rs72549432) | T | G | Exon 1 (M6L) | +0.414 | +0.075 |
KB: kilobases. BP: base pair. UTR: untranslated region.
Independent signals identified from conditional analyses in the NMR meta-GWAS of PNAT2 and KIS3 AFR smokers (training set)
Reference alleles are in relation to the positive strand of the GRCh37 genome orientation
The change in NMR, and thus the ‘weight per risk allele’ was estimated by accounting the standard deviation (SD) of NMR in the training set sample (SD=0.181)
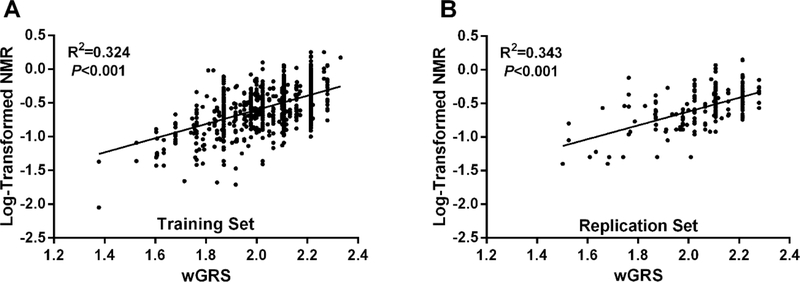
The final wGRS model explained 32.4% of log-NMR variance in the training set (Figure 2A). In the replication set (N=216), the wGRS explained 34.3% of the variance in log-NMR (Figure 2B). When sex, age, and BMI were included in the model, the overall log-NMR variance captured was 36.6% in the training set, and 39.3% in the replication set. When stratifying by sex, the wGRS explained 35.0% of the variation in males (N=401) and 30.5% in females (N=553) in the training set.
Figure 2.
Linear regression analyses of the relationship between the wGRS and log-transformed NMR in the A) training set (N=954; combined PNAT2 and KIS3 trials), and B) replication set (N=216; Q2L trial). The wGRS explained A) 32.4% of the variance in log-NMR in the training set and B) 34.3% in the replication set
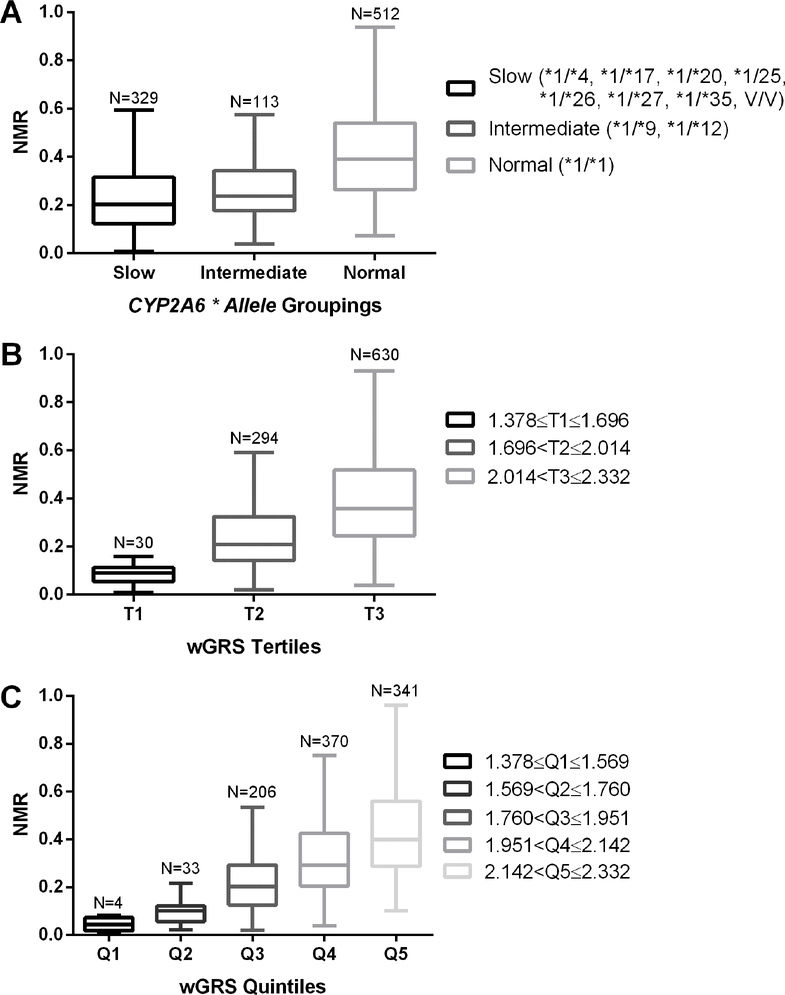
The wGRS captured more variation in the log-NMR in the training and replication sets than previous methods used to classify individuals by CYP2A6 * alleles into slow, intermediate, or normal metabolizers(48) (Figure 3A). The semi-continuous range of values of the wGRS can be used directly or grouped (e.g. by splitting the wGRS scale into tertiles (Figure 3B) or quintiles (Figure 3C)).
Figure 3.
Tukey box-and-whisker plots of nicotine metabolite ratio (NMR) distributions by genotype grouping. Data from the training set (N=954) is grouped as a function of A) * allele groupings; SM, slow metabolizers; IM, intermediate metabolizers; NM, normal metabolizers(48), V/V, two variant * alleles, B) wGRS scale (1.378–2.332) split into tertiles (T, tertile) and C), weighted genetic risk score (wGRS) scale (1.378–2.332) split into quintiles (Q, quintile)
Generalizability Across Different Subpopulations
The log-NMR variation captured by the wGRS was similar between the two clinical trials within the training set (PNAT2: 30.7% and KIS3: 35.8%) despite their baseline differences in smoking characteristics. PNAT2 was restricted to those smoking ≥10 CPD while KIS3 was restricted to those smoking ≤10 CPD. Likewise, the log-NMR variation captured was in a similar range in the replication set clinical trial (Q2L, 34.3%; Figure 2B) which included equal proportions of those smoking ≤10 and ≥10 CPD.
The wGRS was also compared between the geographical recruitment sites (Figure S1). The baseline demographics did not differ between the PNAT2 (Buffalo, Philadelphia, and Houston) and KIS3 (Kansas City) recruitment sites, apart from self-reported CPD in Kansas City as discussed above(42) (Table S1). The log-NMR variation captured by the wGRS was similar across most of the geographical sites (Table S2). Likewise, similar proportions of variation were captured by the EUR CYP2A6 wGRS(39) among EUR, after stratifying by recruitment site (Table S3). After controlling for population substructure (PCs 1+2) there was a negligible influence on the variance captured by the AFR wGRS across the geographical recruitment sites in the training set (Table S2), (PCs 1+2 yielded similar results to controlling for PCs 1–10; data not shown). In addition, demographic covariates such as sex, age, BMI, smoking levels and mentholated cigarette use did not reduce the ability of the wGRS to capture log-NMR variation (Table S2), as seen before with the EUR wGRS(39). Furthermore, no significant differences among allele frequencies were observed (Table S4).
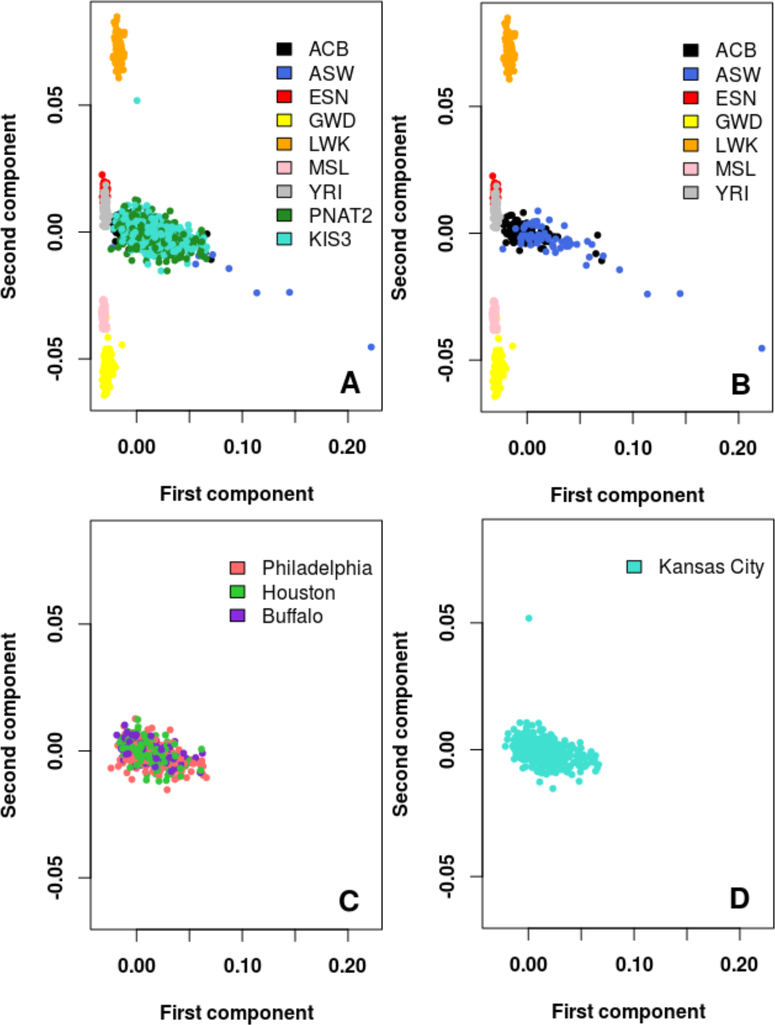
Due to the genetic diversity among North American AFR populations, fine population substructure may affect the utility of an AFR wGRS. Thus, PCs were revaluated in combination with the phase 3 release of 1KG AFR subpopulations to determine whether different US regional zones (Buffalo, Philadelphia, Houston, and Kansas City) clustered with different AFR subpopulations in 1KG (Figure 4). Most participants from the PNAT2 and KIS3 trials, either together (Figure 4A) or split by the multiple geographical recruitment sites (Figures 4C&D), overlapped with the African Caribbean (ACB) and/or the Southwest American (ASW) subpopulations from the 1KG project (Figure 4B), suggesting that the AFR populations from these recruitment sites are relatively homogenous. Similar findings were observed when restricting the PCA to the most similar 1KG subpopulations (ACB, ASW, and YRI) (Figure S2).
Figure 4.
Principal component analyses showing fine AFR population substructure. Principal components were computed from merging the 1KG AFR subpopulations and the training set (PNAT2+KIS3 AFR) samples. A) 1KG AFR subpopulations and training set (PNAT2+KIS3 AFR) participants plotted. B) 1KG AFR subpopulations plotted only. C) PNAT2 participants split by geographic recruitment sites plotted only. D) KIS3 participants plotted only.
(ACB: 1KG African Caribbeans; ASW: 1KG Southwest Americans; LWK: 1KG Luhya; ESN: 1KG Esan; GWD: 1KG Gambian; MSL: 1KG Mende; YRI: 1KG Yoruba).
Note: The PNAT2 Toronto site was excluded from the analyses in this Figure to the small sample size (N=14)
Clinical Utility of the wGRS
The wGRS model showed fair diagnostic ability to discriminate between slow (NMR<0.31) and normal (NMR≥0.31) metabolizers, yielding a significant area under the curve (AUC) of 0.73 (95% confidence interval (CI), 0.70–0.76) in the training set (Figure S3A), and 0.77 (95% CI, 0.71–0.84) in the replication set (Figure S3B). The Youden index J statistic indicated an optimal cut-point wGRS≥2.089 to best identify normal metabolizers (NMR≥0.31) in both the replication and training sets.
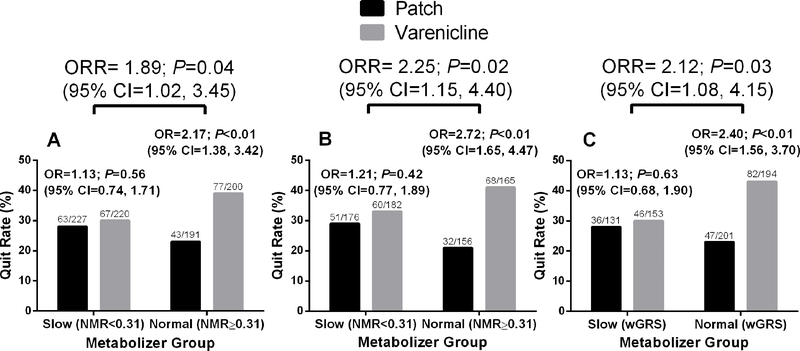
In the primary analysis of the published PNAT2 clinical trial, 838 multiracial smokers were randomized to treatment based on pre-treatment NMR(22). Normal metabolizers (NMR≥0.31) experienced significantly higher end-of-treatment quit rates on varenicline compared with the nicotine patch, while slow metabolizers (NMR<0.31) had similar quit rates on varenicline and the nicotine patch(22). This resulted in a significant NMR-by-treatment interaction (ratio of odds ratio, ORR=1.89; 95% CI, 1.02–3.45; Figure 5A). Of the 838 smokers, 404 were EUR and 275 were AFR. Of note, while 504 PNAT2 AFR smokers were assessed for baseline NMR at the recruitment screen and included in the training set described above, 229 were not randomized to treatment, leaving 275 AFR randomized to active treatment. In the combined EUR and AFR subset (N=679) that were randomized to the varenicline or nicotine patch treatment arms, a similar NMR-by-treatment interaction on quitting was observed (ORR=2.25; 95% CI, 1.15–4.45; Figure 5B). Using the wGRS previously published for EUR(39) and the wGRS described here for AFR, the pooled EUR+AFR stratified analyses reproduced a similar wGRS metabolism group-by-treatment interaction on quitting (ORR=2.12; 95% CI, 1.08–4.15; Figure 5C). The relative treatment effects within the normal metabolizer group were comparable between the three approaches: normal metabolizers showed significantly higher quit rates on varenicline versus nicotine patch defined by the NMR in the N=838 multi-racial dataset (OR=2.17, P<0.01; Figure 5A), defined by the NMR in the N=679 EUR+AFR subset (OR=2.72, P<0.01; Figure 5B), and defined by the respective wGRSs in the N=679 EUR+AFR subset (OR=2.40, P<0.01; Figure 5C). Likewise, the lack of differences between the treatment groups within slow metabolizers were observed using the three respective approaches: (OR=1.13, P=0.56; Figure 5A; OR=1.21, P=0.42; Figure 5B and OR=1.13, P=0.63; Figure 5C). When examining just the subset of AFR smokers in the trial (N=275), similar treatment effects were observed to the overall group (Figure 5) by the NMR (Figure S4A) and by the AFR wGRS (Figure S4B). A summary of the populations used and the wGRS applied for each clinical (smoking cessation) analysis is summarized in (Figure S5)
Figure 5.
End-of-treatment quit rates in the PNAT2 trial by treatment and metabolizer group. Odds ratios (OR) with 95% confidence intervals (CI) comparing the efficacy of varenicline versus the nicotine patch. Metabolizer-by-treatment interaction effects --were evaluated by the ratio of odds ratios (ORR) with 95% CI. (A) NMR stratification (slow: NMR<0.31, normal: NMR≥0.31) in the complete varenicline and nicotine patch treatment arms from intent-to-treat dataset (N=838)(22). (B) NMR stratification (slow: NMR<0.31, normal: NMR≥0.31) in the genetically determined AFR and EUR subset of the varenicline and nicotine patch treatment arms (N=679), and (C) AFR wGRS + EUR wGRS stratification (slow: wGRS<2.089 in AFR or 2.140 in EUR, normal: wGRS ≥2.089 in AFR or 2.140 in EUR) in the genetically determined AFR and EUR subset of the varenicline and nicotine patch treatment arms (N=679)
Ancestry-specific vs Combined Cross-ancestry wGRSs
Genetic risk scores incorporating ancestry-specific variants rarely function across populations(40, 41). We examined whether the CYP2A6 wGRS designated for one population worked for the other. Our AFR wGRS explained 32.4% and 20.0% of the log-NMR variance in the AFR and EUR training sets, respectively (Table 2). The EUR wGRS(39) explained 33.8% and 18.2% of the log-NMR variance in the EUR and AFR training sets, respectively (Table 2). Next, a cross-ancestry wGRS that may function across ancestral populations was evaluated. The variants from both the AFR and EUR wGRSs were meta-analyzed through a fixed-effects model and combined to create a 15-variant cross-ancestry wGRS (Table S5). The random effects model yielded similar effect sizes to the fixed-effects model. The cross-ancestry wGRS explained 33.2% of the variance in log-NMR when merging the EUR (N=933) and AFR (N=954) populations (total N=1887) (Table 2). Moreover, the cross-ancestry wGRS explained 25.0% of the variance in EUR (N=933), and 34.7% of the variance in AFR (N=954) (Table 2). However, the cross-ancestry wGRS did not yield a significant metabolizer by treatment interaction (Figure S6A), unlike when using the ancestry-specific wGRSs together (Figure 5C). Furthermore, the cross-ancestry wGRS was unable to distinguish treatment differences within the AFR normal metabolizers (Figure S6B).
Table 2.
Linear regression analysis of the relationship between respective wGRSs (European-ancestry specific, African-ancestry specific, predicted log-NMR calculated from the respective ancestry specific wGRSs, and a cross-ancestry specific) and log-transformed NMR in different ancestral populations in the PNAT2 and KIS3 clinical trials (i.e. training sets for AFR and EUR wGRSs). Values represent unique variance explained by the wGRS. P<0.001 unless otherwise indicated.
| Variance uniquely explained by wGRS | |||
|---|---|---|---|
|
| |||
| Predictor | AFR Training Set PNAT2+KIS3 (N=954) | EUR Training Set PNAT2 (N=933) | Merged AFR+EUR Training Set PNAT2+KIS3 (N=1887) |
| AFR wGRS | 0.324 | 0.200 | 0.300 |
| EUR wGRS | 0.182 | 0.338 | 0.261 |
| Predicted log-NMR (AFR or EUR wGRS) | - | - | 0.359 |
|
| |||
| Cross-ancestry wGRS | 0.347 | 0.250 | 0.332 |
To improve integration, we tested the utility of transforming the respective ancestry-specific wGRSs onto the same scale. We calculated the predicted log-NMR from the respective equations of the lines of best fit in the training set. For AFR, the predicted log-NMR = 1.0502 (AFR wGRS score) - 2.7042 (Figure 2A), and for EUR, the predicted log-NMR = 0.684 (EUR wGRS) - 1.9417(39). Using this predicted log-NMR approach, 35.9% and 35.5% of NMR variance was captured in the merged AFR+EUR training (Table 2) and replication sets (Figure S7), respectively.
AFR-EUR Admixed populations
Sixty-eight participants were deemed AFR-EUR admixed (Figure S8). The CYP2A6 wGRS developed in EUR captured 42.4% of the variation in log-NMR in the admixed group, while the AFR and cross-ancestry wGRSs captured 33.1% and 22.5% of the variance, respectively. However, these values are likely imprecise due to the small sample size of admixed participants in our study (N=68).
Discussion
We created a CYP2A6 genetic risk score specifically for AFR, based on a set of independent signals identified from an NMR GWAS conducted in AFR smokers with CYP2A6 * alleles found in AFR (Table 1). The increase in variation captured by the wGRS (30–35%) reflects the benefit of merging these two unique variant sets. The variation explained by the AFR wGRS was not reduced when accounting for several known and established demographic sources of variation in the NMR (Table S2). Furthermore, the wGRS captured a similar proportion of variation in a replication set (a group independent of the training set) (Figure 1B), when splitting by sex, by variation in smoking levels, and by most US geographical regions (Table S2), indicating that the effect size estimates are precise and robust. This suggests that this AFR wGRS should be applicable to the majority of AFR populations in the US.
The samples in our training set represented a narrow range of population substructure relative to the 1KG AFR reference subpopulations, mostly overlapping with the ACB and ASW 1KG subpopulations (Figure 4; Figure S2). This suggests that AFR substructure within the US does not vary substantially by the geographical locations included in our training dataset, or alternatively that the PCA along the AFR subpopulations included in 1KG are not sufficient to distinguish population substructure if it exists. It would be useful, going forward, to test the wGRS in other AFR world subpopulations. Less log-NMR variation captured by the wGRS in one recruitment site (Houston) of the training set in comparison to all other sites; this did not appear to be due to known demographic factors, population substructure, or CYP2A6 allele frequency differences (Table S2, Figure 4, Table S4). In contrast, among the EUR training set the variation captured by the EUR wGRS was consistent among all recruitment sites, including Houston, (Table S3). This unexplained difference may be due to a gene-environment interaction, or unknown demographic or environmental influences, that affects the AFR sample in Houston. Specific gene-environment interactions among AFR, that do not occur among EUR, have been previously published(49).
A semi-continuous metric, like our wGRS, is advantageous because it replicates previous metabolizer groupings (Figure 3B) and creates more refined groupings for different purposes (e.g. studying slow and fast CYP2A6 metabolizer extremes) (Figure 3C). Through ROC analyses, we demonstrated that the AFR wGRS shows fair diagnostic ability to distinguish normal from slow CYP2A6 metabolizers, based on an NMR cut-point (i.e. 0.31) that has been implicated in unique smoking cessation clinical trial outcomes (Figure S3).
When the EUR and AFR populations in PNAT2 were combined using their respective wGRSs, similar interaction effect sizes (ORRs) were observed (Figure 5); further, the same treatment effects were observed when splitting into EUR(39) and AFR subgroups, demonstrating that normal metabolizers benefit from varenicline over nicotine patch treatment (Figure S4). The ability to replicate smoking cessation outcomes indicate that the wGRSs could be extended to identify other clinical relationships relating to the CYP2A6 gene, including metabolism of several clinical substrates and risk for tobacco-related diseases.
Like polygenic/genetic risk scores described for other phenotypes(40, 41), there is limited ability to utilize the ancestry-specific EUR and AFR CYP2A6 wGRSs in other ancestries (Table 2). The lack of transferability of the wGRSs is likely explained by the differences in the genetic architecture between ancestral populations: there are differences in the frequencies and effect sizes of CYP2A6 variants, both * alleles and GWAS hits (Table S5).
We created a cross-ancestry wGRS by meta-analyzing the two training sets with equal numbers of AFR and EUR. This approach captured sufficient NMR variation yet showed weak predictive power when stratifying by ancestry (Table 2), and did not recapitulate the clinical outcomes as well as the ancestry-specific wGRSs did (Figure S6). While the cross-ancestry wGRS captured a similar fraction of variation in the AFR group, it was not as effective for EUR and thus was weak as a cross-ancestry wGRS. For both populations fewer variants are required, and more or equal variation is captured, using the ancestry-specific wGRS. The similarity in the variation captured by the AFR and cross-ancestry wGRS suggests that the included independent signals from the AFR GWAS already capture the variation that would have been explained by variants from multi-ethnic or EUR GWASs (e.g. rs56113850). A better approach to integrate analyses including both ancestral populations would be to convert the ancestry-specific wGRSs on to the same scale (i.e. the predicted log-NMR). This captured more of the variation in log-NMR compared to the cross-ancestry wGRS in both the merged AFR-EUR training and replication sets (Table 2; Figure S7).
In the admixed AFR-EUR population, more of the variation was captured by the EUR wGRS, than by the AFR or cross-ancestry wGRS, suggesting that the independent signals in the EUR wGRS are the most universal at tagging causal variants. Indeed, the top SNP (rs56113850) found in NMR GWASs in EUR was also the top SNP in a multi-ethnic NMR GWAS(36). However, the small sample of admixed individuals in our study (n=68) calls for increased inclusion and comprehensive analyses to extend these findings. Further, identifying causal variants unhindered by LD structure differences will likely lead to universal genetic risk scoring models with wider applicability.
Several limitations should be noted. The fraction of log-NMR variation explained by the wGRS is less than the heritability estimates for the NMR (60–80%), suggesting unaccounted for genetic variation. Nighty-eight hits were identified in the NMR GWAS in AFRs(35), mostly in, or around, the CYP2A6 gene - thus genetic risk score approaches more sensitive to high LD between variants may improve variant weight calculation(50). Nevertheless, our current approach was more successful than standard polygenic risk score (PRS) approaches, capturing 34% of the variation in NMR(39) compared to a standard PRS model which captured 9.2–16%(51). The lack of PC-determined ancestry in the replication cohort is another limitation, however a similar proportion of variation was captured suggesting minimal hindrance to the application of the wGRS. The smaller portion of PNAT2 which was AFR also reduced statistical power for the interaction effect (ORR) within the population, although the ORR was of similar magnitude. Finally, the very small number of admixed individuals limited interpretation of this data.
In conclusion, we have derived an AFR CYP2A6 genetic risk score to complement one developed for EUR(39). The models replicate into external samples and were unaffected by known demographic factors. These metrics can adequately distinguish metabolizer groups and reflected clinical outcomes captured by the NMR. Currently, ancestry-specific CYP2A6 wGRSs are more reflective of the NMR than a cross-ancestry wGRS, so we recommend using ancestry-specific wGRSs and aligning the scores by calculating a predicted NMR where necessary. The AFR wGRS for NMR can advance our understanding of the role of CYP2A6 variation in differences in smoking behaviours and related diseases in AFR populations, as well as metabolic influences on other CYP2A6 substrates. More broadly, developing population-specific genetic risk scores are an important future direction to help investigate population differences in the susceptibility for disease, and for differential responses to pharmacotherapies.
Supplementary Material
Study Highlights.
What is the current knowledge on the topic?
CYP2A6 * alleles capture a modest portion of the variation in CYP2A6 activity. Incorporation of GWAS analyses to develop genetic risk scores (GRSs) have enhanced the fraction explained in European populations, however, limited data exists on how these GRSs extend to other populations including African and admixed African-European.
What question did this study address?
How transferable is a European-specific GRS to African or admixed populations in predicting metabolizer status and reflecting unique metabolizer clinical outcomes? If not, would an African-specific GRS be more effective?
What does this study add to our knowledge?
Highlights the importance of ancestry-specific genetic risk scores and adds a new genetic scoring algorithm specifically for African populations.
How might this change clinical pharmacology or translational science?
Careful consideration of the ancestral makeup of the population is important before applying GRSs which will be a valuable tool in improving the prediction of metabolizer status. An African-specific GRS will help investigate population differences in disease susceptibility and pharmacotherapy responses.
Acknowledgements
Computations were performed on the Centre for Addiction and Mental Health (CAMH) Specialized Computing Cluster (SCC), funded by the Canada Foundation for Innovation Research Hospital Fund. We acknowledge additional members of the PGRN-PNAT Research Group including Frank Leone, Henry Glick, Angela Pinto, Paul Sanborn, Peter Gariti, Richard Landis (University of Pennsylvania); Maria Novalen, Bin Zhao, Ewa Hoffmann, Qian Zhou, Adel Aziziyeh (CAMH/University of Toronto); Martin Mahoney (Roswell Cancer Center, University of Buffalo); Maher Karam-Hage (The University of Texas M.D. Anderson Cancer Center); David Conti (University of Southern California); and Andrew Bergen (SRI International). This publication was made possible by the Pharmacogenomics Research Network-RIKEN Global Alliance (PGRN-RIKEN), which is supported by the RIKEN Center for Integrative Medical Science and the NIH Pharmacogenomics Research Network (GM115370).
Funding: Canada Research Chair in Pharmacogenomics (RFT); National Institutes of Health (NIH) grants PGRN DA020830 (R.F.T. and C.L.), R01-DA031815 (N.L.N.), and CA091912 (L.S.C.); Canadian Institutes of Health Research (CIHR) foundation grant FDN-154294 (R.F.T.) and CIHR project grant PJY-159710 (M.J.C., R.F.T., and J.K.); the Campbell Family Mental Health Research Institute of the Centre for Addiction and Mental Health (CAMH); the CAMH Foundation.
Footnotes
Conflict of Interest: R.F.T. has consulted for Quinn Emanuel and Ethismos on unrelated topics. All other authors declared no competing interests for this work.
Clinical Trial Registrations (Date of Registration): NCT00666978 (April 25, 2008), NCT01314001 (March 14, 2011), and NCT01836276 (April 19, 2013)
References
- (1).National Center for Chronic Disease Prevention and Health Promotion (US) Office on Smoking and Health. The Health Consequences of Smoking—50 Years of Progress: A Report of the Surgeon General. (Centers for Disease Control and Prevention, Atlanta, 2014). [PubMed] [Google Scholar]
- (2).Cunningham TJ, Croft JB, Liu Y, Lu H, Eke PI & Giles WH Vital Signs: Racial Disparities in Age-Specific Mortality Among Blacks or African Americans - United States, 1999–2015. MMWR Morb Mortal Wkly Rep 66, 444–56 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- (3).Haiman CA et al. Ethnic and racial differences in the smoking-related risk of lung cancer. N Engl J Med 354, 333–42 (2006). [DOI] [PubMed] [Google Scholar]
- (4).Jamal A, King BA, Neff LJ, Whitmill J, Babb SD & Graffunder CM Current Cigarette Smoking Among Adults - United States, 2005–2015. MMWR Morb Mortal Wkly Rep 65, 1205–11 (2016). [DOI] [PubMed] [Google Scholar]
- (5).Trinidad DR, Perez-Stable EJ, Emery SL, White MM, Grana RA & Messer KS Intermittent and light daily smoking across racial/ethnic groups in the United States. Nicotine Tob Res 11, 203–10 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- (6).Trinidad DR, Perez-Stable EJ, White MM, Emery SL & Messer K A nationwide analysis of US racial/ethnic disparities in smoking behaviors, smoking cessation, and cessation-related factors. Am J Public Health 101, 699–706 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- (7).Benowitz NL, Dains KM, Dempsey D, Wilson M & Jacob P Racial differences in the relationship between number of cigarettes smoked and nicotine and carcinogen exposure. Nicotine Tob Res 13, 772–83 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- (8).Goren A, Annunziata K, Schnoll RA & Suaya JA Smoking cessation and attempted cessation among adults in the United States. PLoS One 9, e93014 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- (9).Nollen NL et al. Factors That Explain Differences in Abstinence Between Black and White Smokers: A Prospective Intervention Study. J Natl Cancer Inst 111, 1078–87 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- (10).Benowitz NL Nicotine addiction. N Engl J Med 362, 2295–303 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- (11).Nakajima M et al. Role of human cytochrome P4502A6 in C-oxidation of nicotine. Drug Metab Dispos 24, 1212–7 (1996). [PubMed] [Google Scholar]
- (12).Nakajima M et al. Characterization of CYP2A6 involved in 3’-hydroxylation of cotinine in human liver microsomes. J Pharmacol Exp Ther 277, 1010–5 (1996). [PubMed] [Google Scholar]
- (13).Dempsey D et al. Nicotine metabolite ratio as an index of cytochrome P450 2A6 metabolic activity. Clin Pharmacol Ther 76, 64–72 (2004). [DOI] [PubMed] [Google Scholar]
- (14).O’Loughlin J et al. Genetically decreased CYP2A6 and the risk of tobacco dependence: a prospective study of novice smokers. Tob Control 13, 422–8 (2004). [DOI] [PMC free article] [PubMed] [Google Scholar]
- (15).Malaiyandi V, Lerman C, Benowitz NL, Jepson C, Patterson F & Tyndale RF Impact of CYP2A6 genotype on pretreatment smoking behaviour and nicotine levels from and usage of nicotine replacement therapy. Mol Psychiatry 11, 400–9 (2006). [DOI] [PubMed] [Google Scholar]
- (16).Schoedel KA, Hoffmann EB, Rao Y, Sellers EM & Tyndale RF Ethnic variation in CYP2A6 and association of genetically slow nicotine metabolism and smoking in adult Caucasians. Pharmacogenetics 14, 615–26 (2004). [DOI] [PubMed] [Google Scholar]
- (17).Wassenaar CA, Dong Q, Wei Q, Amos CI, Spitz MR & Tyndale RF Relationship between CYP2A6 and CHRNA5-CHRNA3-CHRNB4 variation and smoking behaviors and lung cancer risk. J Natl Cancer Inst 103, 1342–6 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- (18).Strasser AA et al. Nicotine metabolite ratio predicts smoking topography and carcinogen biomarker level. Cancer Epidemiol Biomarkers Prev 20, 234–8 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- (19).Sofuoglu M, Herman AI, Nadim H & Jatlow P Rapid nicotine clearance is associated with greater reward and heart rate increases from intravenous nicotine. Neuropsychopharmacology 37, 1509–16 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- (20).Schnoll RA, Patterson F, Wileyto EP, Tyndale RF, Benowitz N & Lerman C Nicotine metabolic rate predicts successful smoking cessation with transdermal nicotine: a validation study. Pharmacol Biochem Behav 92, 6–11 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- (21).Patterson F et al. Toward personalized therapy for smoking cessation: a randomized placebo-controlled trial of bupropion. Clin Pharmacol Ther 84, 320–5 (2008). [DOI] [PubMed] [Google Scholar]
- (22).Lerman C et al. Use of the nicotine metabolite ratio as a genetically informed biomarker of response to nicotine patch or varenicline for smoking cessation: a randomised, double-blind placebo-controlled trial. Lancet Respir Med 3, 131–8 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- (23).Lerman C et al. Genetic variation in nicotine metabolism predicts the efficacy of extended-duration transdermal nicotine therapy. Clin Pharmacol Ther 87, 553–7 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- (24).Lerman C et al. Nicotine metabolite ratio predicts efficacy of transdermal nicotine for smoking cessation. Clin Pharmacol Ther 79, 600–8 (2006). [DOI] [PubMed] [Google Scholar]
- (25).St.Helen G et al. Reproducibility of the Nicotine Metabolite Ratio in Cigarette Smokers. Cancer Epidemiol Biomarkers Prev 21, 1105–14 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- (26).Cho MH et al. A genome-wide association study of COPD identifies a susceptibility locus on chromosome 19q13. In: Hum Mol Genet, Vol. 21947–57 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- (27).Liu T et al. Interaction between heavy smoking and CYP2A6 genotypes on type 2 diabetes and its possible pathways. Eur J Endocrinol 165, 961–7 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- (28).McDonagh EM et al. PharmGKB summary: very important pharmacogene information for cytochrome P-450, family 2, subfamily A, polypeptide 6. Pharmacogenet Genomics 22, 695–708 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- (29).Loukola A et al. A Genome-Wide Association Study of a Biomarker of Nicotine Metabolism. PLoS Genet 11, e1005498 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- (30).Swan GE, Lessov-Schlaggar CN, Bergen AW, He Y, Tyndale RF & Benowitz NL Genetic and environmental influences on the ratio of 3’hydroxycotinine to cotinine in plasma and urine. Pharmacogenet Genomics 19, 388–98 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- (31).Ho MK, Mwenifumbo JC, Zhao B, Gillam EM & Tyndale RF A novel CYP2A6 allele, CYP2A6*23, impairs enzyme function in vitro and in vivo and decreases smoking in a population of Black-African descent. Pharmacogenet Genomics 18, 67–75 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- (32).Piliguian M et al. Novel CYP2A6 variants identified in African Americans are associated with slow nicotine metabolism in vitro and in vivo. Pharmacogenet Genomics 24, 118–28 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- (33).Fukami T et al. A novel polymorphism of human CYP2A6 gene CYP2A6*17 has an amino acid substitution (V365M) that decreases enzymatic activity in vitro and in vivo. Clin Pharmacol Ther 76, 519–27 (2004). [DOI] [PubMed] [Google Scholar]
- (34).Baurley JW et al. Genome-Wide Association of the Laboratory-Based Nicotine Metabolite Ratio in Three Ancestries. Nicotine Tob Res 18, 1837–44 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- (35).Chenoweth MJ et al. Genome-wide association study of a nicotine metabolism biomarker in African American smokers: impact of chromosome 19 genetic influences. Addiction 113, 509–23 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- (36).Patel YM et al. Novel Association of Genetic Markers Affecting CYP2A6 Activity and Lung Cancer Risk. Cancer Res 76, 5768–76 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- (37).Buchwald J et al. Genome-wide association meta-analysis of nicotine metabolism and cigarette consumption measures in smokers of European descent. Mol Psychiatry, (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- (38).Bloom J et al. The contribution of common CYP2A6 alleles to variation in nicotine metabolism among European-Americans. Pharmacogenet Genomics 21, 403–16 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- (39).El-Boraie A et al. Evaluation of a weighted genetic risk score for the prediction of biomarkers of CYP2A6 activity. Addict Biol 25, e12741 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- (40).Duncan L et al. Analysis of polygenic risk score usage and performance in diverse human populations. Nat Commun 10, 3328 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- (41).Martin AR et al. Human Demographic History Impacts Genetic Risk Prediction across Diverse Populations. Am J Hum Genet 100, 635–49 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- (42).Cox LS et al. Bupropion for smoking cessation in African American light smokers: a randomized controlled trial. J Natl Cancer Inst 104, 290–8 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- (43).Park SL, Murphy SE, Wilkens LR, Stram DO, Hecht SS & Le Marchand L Association of CYP2A6 activity with lung cancer incidence in smokers: The multiethnic cohort study. PLoS One 12, e0178435 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- (44).Marchini J, Howie B, Myers S, McVean G & Donnelly P A new multipoint method for genome-wide association studies by imputation of genotypes. Nat Genet 39, 906–13 (2007). [DOI] [PubMed] [Google Scholar]
- (45).Tanner JA et al. Nicotine metabolite ratio (3-hydroxycotinine/cotinine) in plasma and urine by different analytical methods and laboratories: implications for clinical implementation. Cancer Epidemiol Biomarkers Prev 24, 1239–46 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- (46).Chenoweth MJ et al. Known and novel sources of variability in the nicotine metabolite ratio in a large sample of treatment-seeking smokers. Cancer Epidemiol Biomarkers Prev 23, 1773–82 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- (47).Benowitz NL, Dains KM, Dempsey D, Yu L & Jacob P 3rd. Estimation of nicotine dose after low-level exposure using plasma and urine nicotine metabolites. Cancer Epidemiol Biomarkers Prev 19, 1160–6 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- (48).Ho MK et al. Association of nicotine metabolite ratio and CYP2A6 genotype with smoking cessation treatment in African-American light smokers. Clin Pharmacol Ther 85, 635–43 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- (49).Tsai HJ et al. Role of African ancestry and gene-environment interactions in predicting preterm birth. Obstet Gynecol 118, 1081–9 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- (50).Vilhjálmsson BJ et al. Modeling Linkage Disequilibrium Increases Accuracy of Polygenic Risk Scores. Am J Hum Genet 97, 576–92 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- (51).Chen LS, Hartz SM, Baker TB, Ma Y, N LS & Bierut LJ Use of polygenic risk scores of nicotine metabolism in predicting smoking behaviors. Pharmacogenomics 19, 1383–94 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.