Abstract
The data presented in this article support the accompanying research article “Identification of harmine and β-carboline analogs from a high-throughput screen of an approved drug collection; profiling as differential inhibitors of DYRK1A and monoamine oxidase A and for in vitro and in vivo anti-cancer studies” [1]. As DYRK1A (dual-specificity tyrosine phosphorylation-regulated kinase 1a) plays a role in the pathophysiology of a number of diseases including diabetes, cancer and neurodegeneration [2], [3], [4], the identification of DYRK1A inhibitors is of significant interest. This data article details the hits identified from a DYRK1A high-throughput screen of a small molecule compound library containing over 95% approved drugs. Twenty-two compounds were identified with >50% inhibition, including harmine and four of its analogs. Subsequent profiling of these harmine analogs using glioma cancer cell lines and high-content image analysis identified those with effects on growth and cytotoxicity.
Keywords: DYRK1A, High-throughput screening, High-content imaging, Harmine, Cytotoxicity, Glioma, Cancer cells
Specifications Table
| Subject | Cell Biology |
| Specific subject area | High-throughput screening and high-content image analysis |
| Type of data | Table Figure |
| How data were acquired | PHERAstar plate reader (BMG Labtech; Cary, NC, USA) and CellInsight NXT (Thermo Fisher Scientific) |
| Data format | Raw Analyzed |
| Parameters for data collection | Compounds were screened at room temperature at 6.7 μM against DYRK1A. Cancer cells were treated with harmine analogs over a range of concentrations at 37 °C for 3 days. |
| Description of data collection | Compounds from the Prestwick library were screened against DYRK1A using a TR-FRET assay format, and percent inhibition values were determined. Cancer cells were treated with harmine analogs and then assessed for effects on proliferation by Presto Blue assay for effects on cell growth and cytotoxicity by high-content imaging. |
| Data source location | Institution: North Carolina Central University City/Town/Region: Durham, North Carolina Country: U.S.A. Latitude and longitude (and GPS coordinates, if possible) for collected samples/data: 35.97630, -78.90378 |
| Data accessibility | Williams, Kevin (2021), “Raw data in article submitted to Data in Brief (Tarpley et al., 2021)”, Mendeley Data, V1, https://doi.org/10.17632/3r9r4s5yvm.1 https://data.mendeley.com/datasets/3r9r4s5yvm/1 |
| Related research article | Tarpley, M. Oladapo, H.O. Strepay, D. Caligan,T.B. Chdid, L. Shehata, H. Roques, J.R. Thomas, R. Laudeman, C.P. Onyenwoke, R.U. Darr, D.B. Williams, K.P. Identification of harmine and β-carboline analogs from a high-throughput screen of an approved drug collection; profiling as differential inhibitors of DYRK1A and monoamine oxidase A and for in vitro and in vivo anti-cancer studies, Eur. J. Pharm. Sci. 162 (2021) 105,821. https://doi.org/10.1016/j.ejps.2021.105821 |
Value of the Data
-
•
A small scale chemical screen identified small molecule inhibitors of DYRK1A such as harmine along with several of its analogs, which had differential activity in cancer cell models.
-
•
These raw data will benefit researchers in the fields of drug discovery and cancer biology.
-
•
These data will be of interest to those researchers exploring the harmine chemical scaffold for its anti-cancer potential.
1. Data Description
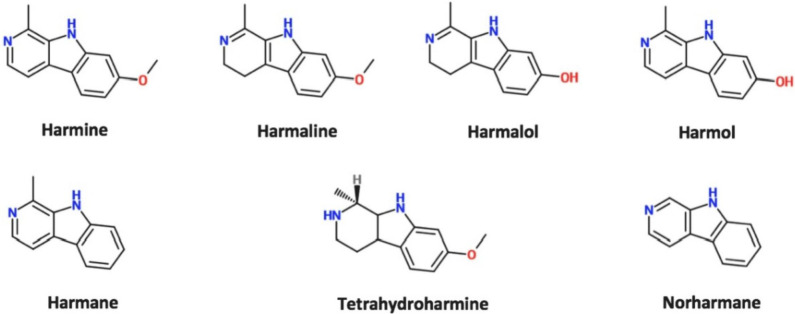
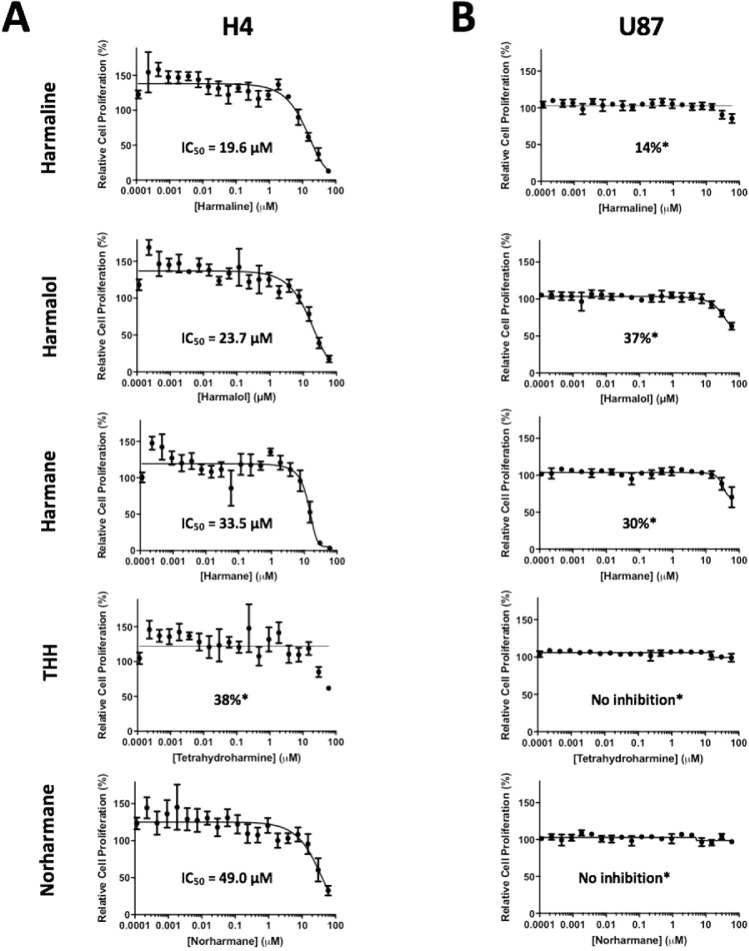
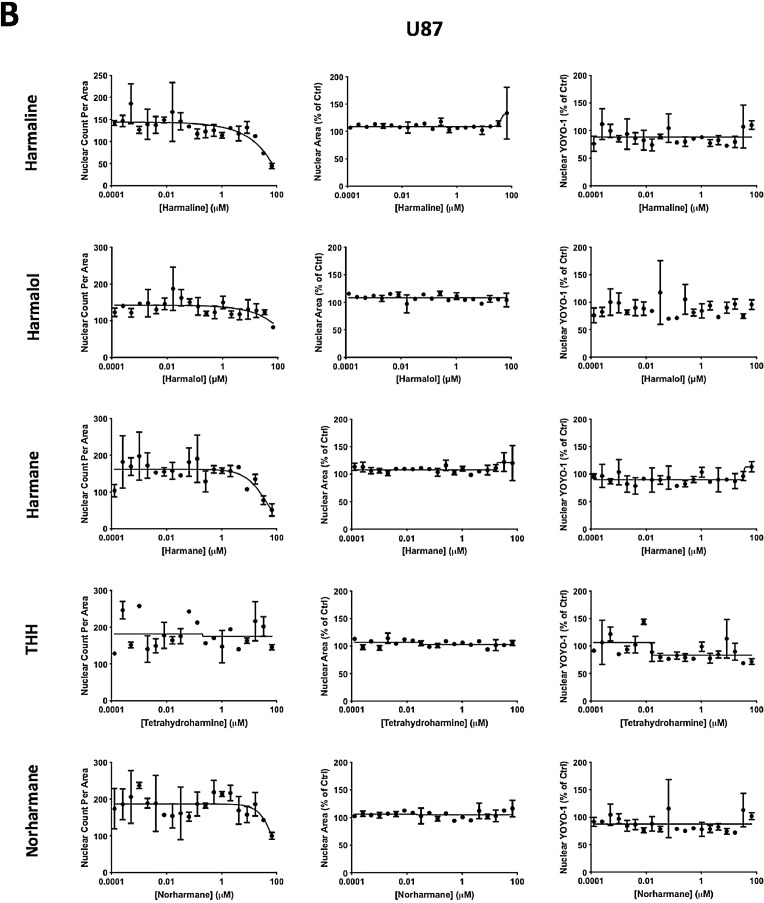
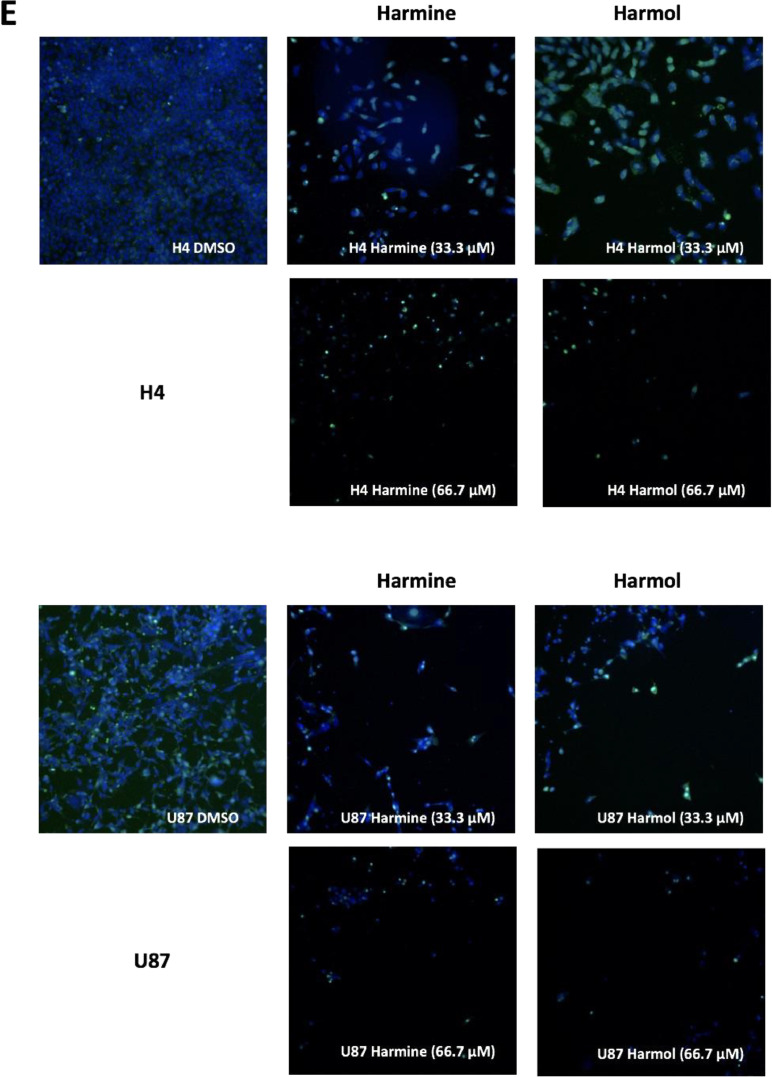
The data reported in this paper provide additional data to our recently published article “Identification of harmine and β-carboline analogs from a high-throughput screen of an approved drug collection; profiling as differential inhibitors of DYRK1A and monoamine oxidase A and for in vitro and in vivo anti-cancer studies” (Eur. J. Pharm. Sci. 162 (2021) 105821. doi: 10.1016/j.ejps.2021.105821) [1]. Table 1 lists a dataset for all 22 hits (Prestwick ID, chemical structure and name) from a pilot high-throughput screen of a library of approved compounds versus the kinase DYRK1A and lists those compounds having percent inhibition values >50%, including harmine and 4 of its analogs (harmol, harmaline, harmane and harmalol). Harmine, the 4 analogs from HTS and 2 additional purchased analogs (norhamane and tetrahydroharmine) (structures shown in Fig. 1) were assessed for effects on growth and cytotoxicity of two glioma cancer cell lines (H4 and U87). Fig. 2 shows the analyzed data and the effect of the harmine analogs on the proliferation of glioma cell lines H4 and U87, as assessed by PrestoBlue assay. Fig. 3 shows analyzed high-content imaging data and the effects of harmine anlogs on nuclear count and size (Hoechst) and plasma membrane integrity (nuclear YOYO-1). Fig. 3 also includes representative images for these data. Cell data on harmine and harmol are primarily included in [1]. The raw data are deposited at https://doi.org/10.17632/3r9r4s5yvm.1.
Table 1.
Hits from Prestwick Chemical Library Screen of DYRK1A.
| No. | Prestwick ID Identifier a | Initial Screen (% Inhibition) | Structure | Chemical Name | Repeat Screen (% Inhibition) b |
|---|---|---|---|---|---|
| 1 | PWK-433484 | 74 | 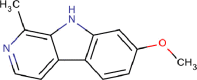 |
Harmine hydrochloride | 99 |
| 2 | PWK-433483 | 78 | 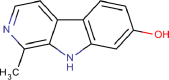 |
Harmol hydrochloride monohydrate | 98 |
| 3 | PWK-433297 | 92 |  |
Chicago sky blue 6B | 95 |
| 4 | PWK-433337 | 88 |  |
Myricetin | 93 |
| 5 | PWK-433379 | 83 |  |
Quercetine dihydrate | 93 |
| 6 | PWK-433658 | 66 | 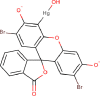 |
Merbromin | 91 |
| 7 | PWK-433485 | 74 |  |
Ellipticine | 91 |
| 8 | PWK-432974 | 99 |  |
Nocodazole | 91 |
| 9 | PWK-433481 | 81 | 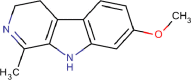 |
Harmaline hydrochloride dihydrate | 86 |
| 10 | PWK-433740 | 55 |  |
Luteolin | 84 |
| 11 | PWK-433491 | 71 |  |
Harmane hydrochloride | 78 |
| 12 | PWK-433833 | 52 | 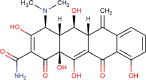 |
Methacycline hydrochloride | 77 |
| 13 | PWK-433286 | 94 |  |
Apigenin | 76 |
| 14 | PWK-433482 | 79 | 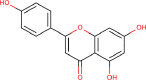 |
Harmalol hydrochloride dihydrate | 74 |
| 15 | PWK-433257 | 94 | 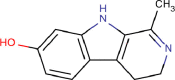 |
Mitoxantrone dihydrochloride | 73 |
| 16 | PWK-433759 | 55 | 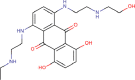 |
Chrysin | 72 |
| 17 | PWK-433328 | 88 | 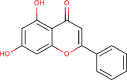 |
Meclocycline sulfosalicylate | 71 |
| 18 | PWK-433091 | 98 |  |
Mebendazole | 71 |
| 19 | PWK-433966 | 51 | 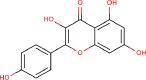 |
Kaempferol | 58 |
| 20 | PWK-433805 | 54 | 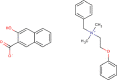 |
Bephenium hydroxynaphthoate | 55 |
| 21 | PWK-433471 | 82 | 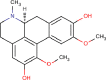 |
Boldine | 54 |
| 22 | PWK-433706 | 59 | 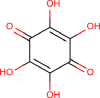 |
Tetrahydroxy-1,4-quinone monohydrate | 51 |
Chemical identifier number provided by Prestwick Corporation.
Hits defined as >50% inhibition in two independent replicate screens.
Fig. 1.
Chemical structures of harmine and anlaogs identified from DYRK1A inhibition screen. Structures were downloaded using JChem database.
Fig 2.
Cell proliferation assay data for hamine analogs. Glioma cancer cells H4 (A) and U87 (B) were incubated with harmine analogs in dose response (0.0001 – 25 µM) with proliferation assessed using PrestoBlue. For each concentration, percent inhibition values were calculated, and data were normalized to DMSO vehicle. Dose response curves were plotted, and IC50 values were determined using Prism GraphPad 7.0.
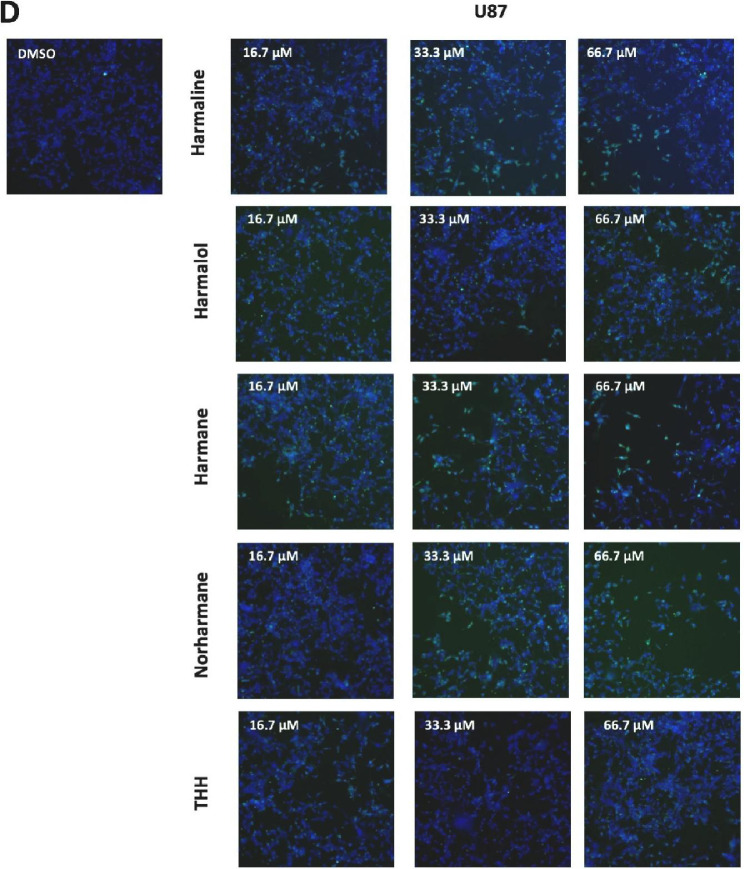
Fig 3.
High-content cell imaging data for hamine analogs. For high-content imaging, H4 and U87 cells were incubated with the indicated harmine analogs in dose response (0.1 – 66 µM) for 72 h. Cells were stained with Hoechst and YOYO-1, and nuclear count, nuclear area and nuclear YOYO-1 staining, measured and imaged as described in method 2.4 below. Dose response data for harmine analogs on H4 (A) and U87 (B) glioma cells were plotted, and IC50 values were determined using Prism GraphPad 7.0. Representative high-content images (10x; overlay of Hoechst=blue, YOYO-1=green) for vehicle (DMSO) and harmine analogs (at 16.7, 33.3. and 66.7 μM) on H4 (C) and U87 (D) glioma cells. (E) Representative high-content images for harmine and harmol (33.3 and 66.7 µM) on H4 (upper panel) and U87 (lower panel) glioma cells. Images for 16.7 μM are included in the accompanying article [1].
2. Experimental Design, Materials and Methods
2.1. Reagents and cells
Reagents for the DYRK1A screen (DYRK1A-GST, anti-GST antibody (Ab) and Kinase Tracer 236) were from Life Technologies (Carlsbad, CA). Glioma cell lines H4 and U87 were from ATCC. The Prestwick chemical library, which contains >95% FDA-approved drugs, was from Prestwick (Prestwick Chemical, Washington, DC). Tetrahydroharmine was purchased from Cayman Chemical (Ann Arbor, MI) and norharmane from Toronto Research Chemicals (North York, ON, Canada).
2.2. Hit list from high-throughput screen of DYRK1A with Prestwick drug library
The DYRK1A time-resolved fluorescence energy transfer (TR-FRET) assay was optimized and validated for HTS as described in detail in the accompanying MethodsX article [5]. Briefly, the assay is carried out as follows. DYRK1A-GST, anti-GST Ab and Kinase Tracer 236 were combined and dispensed into low volume black 384-well plates using a NanoScreen NSX-1536 equipped with a 384 head (Beckman Coulter; Brea, CA, USA). For compound addition, 50 nL of each drug from the Prestwick library (1124 compounds) was added using a Biomek NX workstation (Beckman-Coulter) equipped with a Pintool array (VP Scientific; San Diego CA) into a final assay volume of 7.5 µL to give a final compound concentration of 6.7 µM. Plates were covered, incubated for 1 h at RT and then read on a PHERAstar plate reader (BMG Labtech; Cary, NC, USA) using 665 /620 nm. Two independent screens were carried out.
2.3. Cell proliferation assay
Cell proliferation assay was carried out essentially as in [6]. Using a multidrop dispenser (Thermo), cells were plated in clear 384-well tissue culture plates at 1000 cells per well for U87 and 800 cells per well for H4. Cells were allowed to attach overnight at 37 °C and 5% CO2. Compounds were then added to cells in duplicate 20-point 2-fold concentration response (0.0001 µM to 25 µM) using a Biomek NX and incubated for 72 h. PrestoBlue in PBS was then added and after 90 min fluorescence was determined using a PHERAstar plate reader at λex 560 nm / λem 590 nm. Based on DMSO-treated controls, relative cell proliferation was calculated using raw RFU values. Data were plotted to generate dose response curves, and IC50 values were determined by non-linear regression using GraphPad Prism 7.
2.4. High-content imaging analysis
Dyes Hoechst 33342 for nuclear count and YOYO-1 to assess membrane integrity were added to glioma cancer cells (H4 and U87) for live imaging combined with quantitative multiparametric analysis of cell morphology [7]. Cells were grown and treated with compounds as in Method 2.3 (above). For cell imaging, fluorescence quantification was determined using a Thermo Fisher CellInsight NXT and HCS Screen software (Thermo Fisher Scientific), and images were captured as described in [6]. Excitation wavelengths were 386 nm and 485 nm for Hoechst 33342 and YOYO-1, respectively. A nuclear mask was established (Hoechst 33342) to determine nuclear characteristics: nuclear count (a measure of cell count) and nuclear size (a measure of nuclear morphology); as well as establishing area inside nuclear mask for YOYO-1 staining (a measure of plasma membrane integrity) [7]. For quantitation, data were plotted and analyzed in Prism GraphPad 7.0.
Ethics Statement
The authors declare that the work described is original and has not been submitted elsewhere for publication. No conflict of interest exists in this submission.
CRediT Author Statement
Michael Tarpley: Conceptualization, Methodology, Data curation, Formal analysis, Investigation, Validation, Writing - original draft, Writing - review & editing; Helen Oladapo: Conceptualization, Methodology, Data curation, Formal analysis, Investigation, Validation; Thomas Brent Caligan: Methodology, Investigation, Validation; Rob U. Onyenwoke: Data curation, Validation, Writing - original draft, Writing - review & editing; Kevin P. Williams: Funding acquisition, Project administration, Resources, Supervision, Conceptualization, Methodology, Data curation, Formal analysis, Validation, Writing original draft, Writing - Review & Editing.
Declaration of Competing Interest
The authors declare that they have no known competing financial interests or personal relationships which have or could be perceived to have influenced the work reported in this article.
Acknowledgement
This study was supported in part by funding from NIH/NCI awards R15CA208651 and P20CA202924; NIH/NIMHD RCMI U54MD012392 (K.P.W.), and project support from NIH award U54CA137844 (K.P.W., R.U.O.), and Komen Graduate Training in Disparities Research award GTDR16377604 (K.P.W. and H.O.H.) with additional support from the Golden LEAF Foundation and the BIOIMPACT Initiative of the State of North Carolina.
References
- 1.Tarpley M., Oladapo H.O., Strepay D., Caligan T.B., Chdid L., Shehata H., Roques J.R., Onyenwoke R.U., Darr D.B., Williams K.P. Identification of harmine and β-carboline analogs from a high-throughput screen of an approved drug collection; profiling as differential inhibitors of DYRK1A and monoamine oxidase A and for in vitro and in vivo anti-cancer studies. Eur. J. Pharm. Sci. 2021;162 doi: 10.1016/j.ejps.2021.105821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Ionescu A., Dufrasne F., Gelbcke M., Jabin I., Kiss R., Lamoral-Theys D. DYRK1A kinase inhibitors with emphasis on cancer. Mini Rev. Med. Chem. 2012;12:1315–1329. doi: 10.2174/13895575112091315. [DOI] [PubMed] [Google Scholar]
- 3.Rachdi L., Kariyawasam D., Guez F., Aiello V., Arbones M.L., Janel N., Delabar J.M., Polak M., Scharfmann R. Dyrk1a haploinsufficiency induces diabetes in mice through decreased pancreatic beta cell mass. Diabetologia. 2014;57:960–969. doi: 10.1007/s00125-014-3174-3. [DOI] [PubMed] [Google Scholar]
- 4.Wegiel J., Gong C.X., Hwang Y.W. The role of DYRK1A in neurodegenerative diseases. FEBS J. 2011;278:236–245. doi: 10.1111/j.1742-4658.2010.07955.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Tarpley M., Caligan T.B., Onyenwoke R., Williams K.P. Optimization and validation of a DYRK1A TR-FRET assay for high-throughput screening. MethodsX. 2021;8:101383. doi: 10.1016/j.mex.2021.101383. In press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Oladapo H.O., Tarpley M., Sauer S.J., Addo K.A., Ingram S.M., Strepay D., Ehe B.K., Chdid L., Trinkler M., Roques J.R., Darr D.B., Fleming J.M., Devi G.R., Williams K.P. Pharmacological targeting of GLI1 inhibits proliferation, tumor emboli formation and in vivo tumor growth of inflammatory breast cancer cells. Cancer Lett. 2017;411:136–149. doi: 10.1016/j.canlet.2017.09.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Abraham V.C., Towne D.L., Waring J.F., Warrior U., Burns D.J. Application of a high-content multiparameter cytotoxicity assay to prioritize compounds based on toxicity potential in humans. J. Biomol. Screen. 2008;13:527–537. doi: 10.1177/1087057108318428. [DOI] [PubMed] [Google Scholar]