Highlights
-
•
Naproxen inhibits the adenocarcinoma by 64% in KrasG12V mice.
-
•
Naproxen inhibits serum PGE2/CXCR4 levels in KrasG12V mice.
-
•
Naproxen inhibits the progression of adenocarcinoma in KrasG12V mice.
Keywords: Lung cancer, Prevention, Naproxen, KrasG12V, Lung adenocarcinoma, NSAID
Abstract
Lung cancer is the leading cause of cancer related deaths worldwide. The present study investigated the effects of naproxen (NSAID) on lung adenocarcinoma in spontaneous lung cancer mouse model. Six-week-old transgenic KrasG12V mice (n = 20; male + female) were fed modified AIN-76A diets containing naproxen (0/400 ppm) for 30 wk and euthanized at 36 wk of age. Lungs were evaluated for tumor incidence, multiplicity, and histopathological stage (adenoma and adenocarcinoma). Lung tumors were noticeable as early as 12 wk of age exclusively in the KrasG12V mice. By 36 wk age, 100% of KrasG12V mice on control diet developed lung tumors, mostly adenocarcinomas. KrasG12V mice fed control diet developed 19.8 ± 0.96 (Mean ± SEM) lung tumors (2.5 ± 0.3 adenoma, 17.3 ± 0.7 adenocarcinoma). Administration of naproxen (400 ppm) inhibited lung tumor multiplicity by ∼52% (9.4 ± 0.85; P < 0001) and adenocarcinoma by ∼64% (6.1 ± 0.6; P < 0001), compared with control-diet-fed mice. However, no significant difference was observed in the number of adenomas in either diet, suggesting that naproxen was more effective in inhibiting tumor progression to adenocarcinoma. Biomarker analysis showed significantly reduced inflammation (COX-2, IL-10), reduced tumor cell proliferation (PCNA, cyclin D1), and increased apoptosis (p21, caspase-3) in the lung tumors exposed to naproxen. Decreased serum levels of PGE2 and CXCR4 were observed in naproxen diet fed KrasG12V mice. Gene expression analysis of tumors revealed a significant increase in cytokine modulated genes (H2-Aa, H2-Ab1, Clu), which known to further modulate the cytokine signaling pathways. Overall, the results suggest a chemopreventive role of naproxen in inhibiting spontaneous lung adenocarcinoma formation in KrasG12V mice.
Introduction
Lung cancer tops the chart in terms of cancer incidence and mortality worldwide in both sexes, with approximately 235,760 new cases and 131,880 deaths in the US during 2021 [1]. Smoking increases the risk for lung cancer by 25 times compared with non-smoking, resulting in ninety percent of lung cancer cases. The 5-y survival rate of lung cancer is only 19.7% due to the high frequency of late diagnosis, resulting in non-resectable tumors, compared with the 67.1% average of all site cancers [2]. Smoking cessation has been shown to have a positive effect, decreasing the risk of death by half in patients with early lung cancer. However, the risk of developing this malignancy is still high for millions of active and former smokers. As a result of continued efforts towards smoking cessation and novel interventions, there has been an improvement in lung cancer prevention and survival over the years. Although recent data indicate a decline in death rates for lung cancer between 2008-2017 in both men and in women, as of 2021 lung cancer continues to be the leading cause of cancer-related deaths [1,3].
KRAS mutations are common in lung cancer and represent about 30% of non-small cell lung cancer (NSCLC) patients. Many studies have shown that mutant KRASG12V NSCLC patients had poor survival rates [4]. KRASG12V NSCLC patients in particular exhibited lower overall survival and time to recurrence compared than did the entire cohort (Overall survival: KRASG12V, 26 mo vs Cohort, 60 mo; Disease free Survival: KRASG12V, 15 mo vs Cohort, 24 mo) [5]. Higher metastatic and poor survival rates with KRASG12V were also observed in colorectal and other cancers [6]. The exact mechanisms by which KRASG12V mutation drives aggressive lung tumor growth are not fully understood. Some studies suggested that KRASG12V upregulates expression of CXCR4 and other proinflammatory genes, leading to immune suppression [7,8]. It has been established that prostaglandin E2 induces the overexpression of CXCR4 in the tumor microenvironment and promotes tumor growth and metastasis [9].
At present, there are no FDA-approved CXCR4 inhibitors for lung cancer patients with KRASG12V mutation. A few studies have shown that nonsteroidal anti-inflammatory drugs (NSAIDs) may block the expression of CXCR4 and suppress tumor growth [10]. So far, no preclinical studies with KrasG12V mutant lung tumor model interventions have been established using NSAIDs. It is therefore necessary to develop intervention strategies aiming at KrasG12V-driven lung tumorigenesis by NSAIDs.
Over the last few decades, numerous studies have shown that NSAIDs, alone or in combination with other agents inhibit lung cancer in experimental models, epidemiological observations, and randomized clinical trials [11,12]. The inverse association of NSAIDs use with lung cancer risk is well-studied [13]; however, NSAIDs have not been evaluated in KrasG12V mutant lung cancers. Among all NSAIDs, the commonly used medication naproxen, has been shown to be highly effective in preventing tumor development [14]. Importantly, naproxen has demonstrated overall better cardiovascular safety than other NSAIDs [15]. Therefore, in the present study, we examined the effectiveness of naproxen on lung adenoma progression to adenocarcinoma in the KrasG12V mouse model and assessed key biomarkers of tumor growth.
Materials and methods
Chemicals, antibodies, and reagents
Naproxen (>99% purity; TCI America, Oregon, USA), EDTA, EGTA, Sodium orthovanadate, Igepal CA-630, Triton X-100,) Complete Protease inhibitor tablet and PhosSTOP phosphate inhibit tablet (Sigma) were purchased. Primary antibodies against PCNA (ab-29), cyclin D1 (ab-134175), caspase-9 (ab-52298), and IL-10 (ab-133575) were purchased from Abcam (MA, USA). Antibodies against caspase-3 (cs-9662), p21 (cs-2947), PARP (cs-9542), actin (cs-4970), cre (cs-15036s), RalA (cs-4799s), PI3K (cs-4292s), and horseradish-peroxidase-conjugated secondary antibodies (anti-rabbit, -goat, and –mouse) were purchased from Cell Signaling Technology (Beverly, MA, USA). Primary antibody against p21 (sc-397), caspase-3 (sc-397; for immunohistochemistry [IHC]), and COX-2 (sc-7951) was purchased from Santa Cruz Biotechnology (Santa Cruz, CA, USA). Prostaglandin E2 ELISA kit (Cat. No. 514010) and mouse CXC-Chemokine receptor 4 (CXCR4) ELISA kit (Cat. No. MBS701736) were purchased from Cayman Chemical Company (Ann Arbor, MI) and MyBiosource.com (San Diego, USA). The Histostain-Plus 3rd Gen IHC Detection Kit (Life Technologies, NY, USA) and DeadEnd Colorimetric TUNEL system (Promega, Wisconsin, USA) were purchased.
Animals, diet, and care
All animal experiments were performed in the pathogen-free OU Rodent Barrier Facility as per the National Institutes of Health and University of Oklahoma HSC's Institutional Animal Care and Use Committee guidelines. Six-week-old male and female LSL-KrasG12V mouse strain (B6.129-Krastm1Bbd/ J; Jax Stock No 026924) carrying the KrasG12V point mutation were provided by Dr. Logsdon from the MD Anderson Cancer Center. Diets (control and experimental) were prepared based on the modified AIN-76A diet, as described earlier [16] and stored in a cold room. Naproxen was premixed with a small amount of casein and then mixed with the appropriate amount of control diet using a Hobart Mixer. Mice had unrestricted access to the diets and automated tap water purified by reverse osmosis. Food cups were refilled with diet weekly twice throughout the experiment. Mice were regularly checked for indications of weight reduction, toxicity, or any variation from the norm.
Transgenic KrasG12V mice and lung tumor development
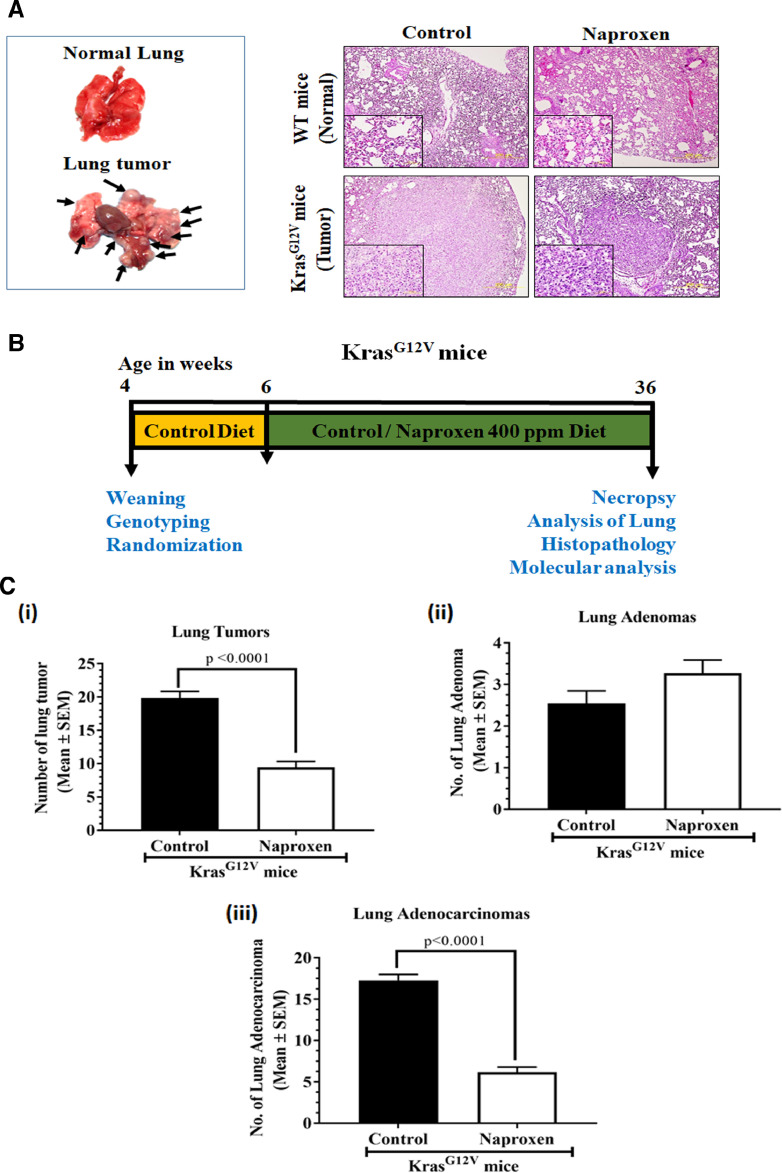
KRasG12V mice were euthanized at various ages and lungs were evaluated for tumor incidence. These mice developed spontaneous lung tumors that were visually distinguishable by 12 wk of age. The number of tumors in each mouse increased as they aged and progressed to adenocarcinoma by 24 wk. By 36 wk of age, lung tumor incidence was observed in 100% of KrasG12V mice, while wild-type mouse lungs were free from any abnormalities or tumors (Fig. 1A).
Fig. 1.
Effect of naproxen on spontaneous lung tumor development in a KrasG12V mouse model.
(A) Gross morphology of the lungs from wild-type mice and lungs with tumors in KrasG12V mice. Representative photomicrograph of hematoxylin and eosin-stained lung sections from mice belonging to control and naproxen treatment groups (magnification 10X; inset shows 60X magnification). Note the reduction in tumor size upon treatment. (B) Experimental design for evaluating the chemopreventive effect of dietary naproxen 400 ppm in a KrasG12V lung cancer mouse model. Three to 4 wk-old KrasG12V mice were weaned, genotyped, and randomized. At 6 wk of age, mice were shifted onto experimental diets containing naproxen (0 ppm or 400 ppm) for 30 wk until the end of the study. At 36 wk of age, mice were euthanized and lungs were harvested for evaluation of tumors. (C) Effect of naproxen (400 ppm) on number of (i) Lung tumors, (ii) Lung adenomas, and (iii) Lung adenocarcinomas (bottom). Data represent Mean ± SEM. Differences between groups were analyzed by unpaired student's t test with Welch's Correction.
Experimental bioassay
The experiment was intended to assess the long-term effects of naproxen on spontaneous lung adenoma and adenocarcinoma development in KrasG12V mice. Male and female KrasG12V-positive mice were identified by genotyping and randomly assigned to control and experimental groups (n = 20). Mice were fed with powdered AIN-76A experimental diets containing naproxen (0 ppm or 400 ppm) starting at 6 wk of age, and were weighed weekly until study termination. The rationale to use 400 ppm naproxen was based on previous published studies [17]. Mice were euthanized by CO2 asphyxiation after 30 wk of exposure to the test agent (Fig. 1B). Lungs were lavaged, perfused, and assessed for the tumor occurrence, number, and tumor size by 2 readers blinded to treatments. Lungs with tumors were dissected and either snap-frozen in liquid nitrogen followed by storage at -80°C for biomarker analysis or fixed in buffered formalin for histopathological identification of adenoma and adenocarcinoma.
Tumor histology
Lung tissues fixed in 10% buffered formalin were embedded in paraffin, sectioned, and stained with hematoxylin and eosin. Multiple sections of each lung tissue were evaluated by a board-certified pathologist blinded to treatment info. Tumors were categorized to adenomas and adenocarcinomas according to the criteria of the Mouse Models of Human Cancers Consortium [18].
Protein immunoblotting
For analysis of different markers, frozen lung tumors were used to prepare tissue lysates based on established protocol [19]. Equal concentration (50−100µg) of lysates were separated on SDS-PAGE (8%−12%), transferred to a polyvinylidene difluoride membrane, and probed with specific antibodies (PCNA [1:1000], cyclin D1 [1:2000], p21 [1:1000], caspase-9 [1:1000], caspase-3 [1:1000], PARP [1:1000], and Actin [1:1000]) overnight at 4°C. The next day, membranes were washed and incubated with horseradish peroxidase-conjugated secondary antibody (1:5000). Membranes were developed using chemiluminescence reagent and visualized with Biorad ChemiDoc Imaging system. Densities of various analyte proteins were calculated relative to β-actin (loading control) using ImageJ 1.52 (National Institutes of Health) software.
Immunohistochemical staining
Formalin-fixed paraffin-embedded (FFPE) normal lung tissues and adenocarcinomas were evaluated for expression of various proteins by IHC. Briefly, 5-μm-thick tissue sections were prepared. Antigen retrieval was carried out in boiling citrate buffer (10 mM, pH 6.0). Endogenous peroxidase activity quenching and nonspecific binding sites blocking were performed using a Histostain-Plus 3rd Gen IHC Detection Kit. Sections were then incubated with specific antibodies (PCNA [1:1000], cyclin D1 [1:50], p21 [1:50], IL-10 [1:50], COX-2 [1:100], caspase-9 [1:100], caspase-3 [1:250] and Cre [1:1000]) or PBS as negative controls overnight at 4°C in a humidified chamber. Later, slides were washed in PBS, incubated with biotinylated secondary antibody for 1 h, and then incubated with streptavidin-peroxidase conjugate for 30 min. After washing with PBS, slides were developed using diaminobenzidine substrate and counterstained with Mayor's hematoxylin. These slides were viewed under an Olympus microscope IX71 to capture digital images that were used to determine relative expression using IHC profiler following a semi-automated analysis protocol [20].
Terminal Deoxynucleotidyl Transferase Biotin-dUTP Nick End Labelling (TUNEL) assay
Apoptosis was assayed in FFPE sections using an in situ TUNEL assay kit (Promega, Madison, WI, USA) according to the manufacturer's protocol. Nuclei stained brown in color indicated the apoptized cells. Digital images of tumor sections (at multiple random fields per slide) from a minimum of 3 mice per group were used to calculate the apoptotic index using IHC profiler as described earlier [20].
Prostaglandin E2 measurement
Prostaglandin E2 (PGE2) levels were determined in mouse serum of wild-type, control, and naproxen treated KrasG12V mice with a PGE2 ELISA Kit from Cayman Chemical Company (Ann Arbor, MI) according to the manufacturer's protocol. The calculation was performed as per the manufacturer's protocol to determine the PGE2 levels in different treatment groups.
CXC-Chemokine receptor 4 (CXCR4) measurement
CXCR4 levels were determined in mouse serum of wild-type, control, and naproxen treated KrasG12V mice with a CXCR4 ELISA Kit from MyBiosource.com (San Diego, USA) according to the manufacturer's protocol. The calculation was performed as per the manufacture's protocol to determine the CXCR4 levels in different treatment groups.
Gene expression analysis
Mouse lung FFPE tissue samples with and without tumors were sent to HTG Molecular Diagnostics, Inc. for molecular analysis using the standard protocol described earlier [21,22]. RNA expression analysis was performed using the HTG EdgeSeq Mouse mRNA Tumor Response Panel of 1659 mRNA targets related to core signaling pathways and drug response and immune response mechanisms in oncology and other disease states. Sequencing was performed using an Illumina NextSeq sequencer and the assay was validated using negative and positive process controls. Gene expression data from the NGS instrument were processed and reported through the HTG host system software. Expression data plot were generated using HTG EdgeSeq Reveal application (3.0).
To determine the status of altered genes in lung tumor of KrasG12V mice in human, the significantly altered genes of KrasG12V mice were checked in human lung adenocarcinoma by TCGA analysis using UALCAN [23]. To understand the cellular pathways that were involved in the development of lung tumors in KrasG12V mice vs. human lung cancer, expression data were compared and included in a pathway analysis of the Kyoto Encyclopedia of Genes and Genomes (KEGG) and Reactome pathway using DAVID [24,25].
Statistical analysis
All data are presented as Mean ± SEM. Male and female data were pooled for tumors. Tumor multiplicity (average number of tumors per mice) and differences between body and organ weights were analyzed using unpaired student's t test with Welch's Correction. Differences were considered significant when the “P” value was less than 0.05. Statistical analysis was performed using GraphPad Prism Software 8.0 (GraphPad Software, Inc., San Diego, CA).
Results
General observations
No significant changes were observed in body weight gain of mice fed with control or experimental diets (naproxen 400 ppm) throughout the study (data not shown). At termination, there were no observable abnormalities in the gross findings, such as size, shape, color, and texture of major organs. Except for lungs, there were no significant differences in major organ weights (liver, kidneys, spleen, pancreas, heart, and colon) between control and naproxen-fed mice was observed (Supplementary Fig. 1).
Naproxen significantly inhibits KrasG12V-driven lung adenocarcinoma formation
To study the effect of naproxen on KrasG12V mutant tumors, lungs of the control and naproxen-treated transgenic mice were evaluated for tumors and the data were compared. Control-diet-fed KrasG12V mice developed 19.8 ± 0.96 (Mean ± SEM) lung tumors per mouse at termination. Administration of naproxen significantly suppressed total lung tumor formation by ∼52% (9.4 ± 0.85; P < 0001) compared with the control diet (Fig. 1Ci). Histopathologic analysis showed that control-mice had on average 2.5 ± 0.3 adenomas and 17.3 ± 0.7 lung adenocarcinomas, reflecting 12.6% of adenomas and 87.4% of adenocarcinomas (Figs. 1Cii and 1Ciii). Administration of 400 ppm of naproxen significantly suppressed adenocarcinoma by ∼64% (6.1 ± 0.6; P < 0001) (Fig. 1Ciii). However, no significant difference was found in the number of adenomas (Fig. 1Cii). The decrease in tumor multiplicity was also reflected in lung weight in naproxen-fed mice. Lungs of the naproxen-fed mice weighed significantly less (230.6 ± 12.2 mg, P < 0.02) than lungs of the control group (384.0 ± 68.1 mg) (Supplementary Fig. 1).
Gene expression analysis to identify pathways dysregulated in lung tumors of KrasG12V mice
Comparison of tumor related genes expression in normal lung (wild type mice) and lung tumors (KrasG12V mice) was carried out to identify significantly altered genes and cellular pathways in the lung tumors. A total of 226 genes were upregulated and 90 genes were downregulated in lung tumors of KrasG12V mice (Supplementary Table 1 and 2). Analysis of significantly altered genes (as input gene identifiers) with KEGG and Reactome pathway using DAVID identified the cytokine-cytokine receptor interaction, PI3K-Akt signaling pathway, pathways in cancer, MAPK signaling pathway, and Ras signaling pathway as the top 5 altered pathways in lung tumors of KrasG12V mice (Supplementary Figure 2A). Further, TCGA analysis of KrasG12V mouse lung tumor genes (226 genes-upregulated; 90 genes-downregulated) using the lung adenocarcinoma dataset by UALCAN software showed that 90 common genes were upregulated and 51 common genes were downregulated in patients (Supplementary Figure 2B). Furthermore, the pathways in cancer, PI3K-Akt signaling pathway, cytokine-cytokine receptor interaction pathway, focal adhesion, and Ras signaling pathway were the top 5 pathways altered in lung adenocarcinoma patients as per the analysis of the common set of KrasG12V mouse and lung adenocarcinoma patient genes with KEGG and Reactome pathway using DAVID (Supplementary Figure 2C). In the lung tumors of KrasG12V mice, we found a significant upregulation of pro-inflammatory interleukins (e.g., IL-6, IL-18RAP, IL-1B, IL-18, IL-10, IL-11), immune-oncology genes (e.g., Pdcd1lg2, Sele, Crp), MAP signaling pathway (e.g., Ccne1, Ccna1, Hras), and Fgfr pathway (e.g., Fgf5-9, Fgf13-14, Fgf16-17) which are linked to cancer-promoting inflammation (Supplementary Table 3). Further, histocompatibility 2, class II antigen A, alpha (H2-Aa, >78 fold) and beta 1 (H2-Ab1, >40 fold) genes critical for antigen presentation to CD4+ T lymphocytes are upregulated in lung tumors of KrasG12V mice. These genes play a critical role in lung cancer diagnosis, prognosis, and treatment (Supplementary Table 4). Intelectin 1 (Itln1, <24 fold), mannan-binding lectin serine peptidase 2 (Masp2, <9 fold), and prolactin receptor (Prlr, <7 fold) were downregulated in lung tumors of KrasG12V mice and lung adenocarcinoma patients (Supplementary Table 5).
Increased expression of cell proliferation, and pro-inflammation markers with PGE2 and CXCR4 levels and decreased expression of apoptotic markers in KrasG12V lung tumors
KrasG12V lung tumors were analyzed for the expression of different biomarkers of inflammation, proliferation, and apoptosis. IHC showed strong positivity for PCNA and cyclin D1 proteins, suggesting an increase in cell proliferation in lung tumors when compared with normal lung tissue (Supplementary Fig. 2D). There was also decreased expression of the apoptotic markers p21, caspase-3, and apoptotic index (Supplementary Fig. 2E) and increased expression of pro-inflammation markers COX-2 and IL-10 in the lung tumors (Supplementary Fig. 2F). Compared with normal or wild-type mice, serum samples of control-diet-fed KrasG12V mice showed significantly high expression of PGE2 (pg/ml) and CXCR4 (pg/ml) levels (Supplementary Fig. 2G).
Inflammation in KrasG12V lung tumors is reduced upon naproxen treatment
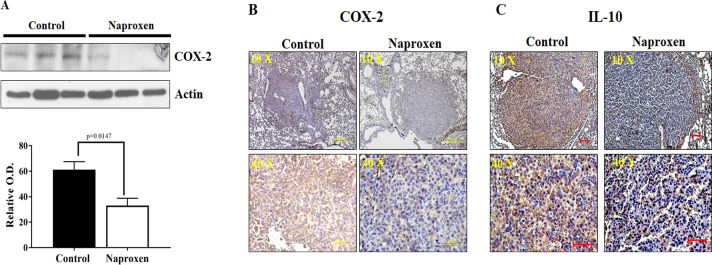
The effects of naproxen on inflammation-modulating proteins, i.e., COX-2 and IL-10, were determined using western immunoblotting and IHC. Western blot analysis suggested that naproxen significantly decreased the COX-2 protein expression in lung tumor tissues of KrasG12V mice (Fig. 2A) when compared with untreated tumors. These results were further complemented by determining the expression of COX-2 and IL-10 measured by immunohistochemical staining. IHC analysis suggested that naproxen treatment also decreased the expression of IL-10 in lung tumor tissues of KrasG12V mice (Figs. 2B and 2C). Together, these results suggest that naproxen inhibits the inflammation in lung tumors of KrasG12V mice, leading to reduced tumor formation.
Fig. 2.
Effect of naproxen on cell-inflammation markers in KrasG12V mouse lung tumors. (A) Representative western blot and optical density of COX-2 measured in total cell protein of tumor. Differences between control and treatment groups were analyzed by one-tailed t test with Welch's correction and 95% confidence interval. Data represent Mean ± SEM. P ≤ 0.05 was considered statistically significant. (B-C) Representative photomicrographs showing immunohistochemical detection of COX-2 and IL-10 (magnification 10X and 40X).
Naproxen inhibits proliferation and increases apoptosis in lung tumors of KrasG12V mice
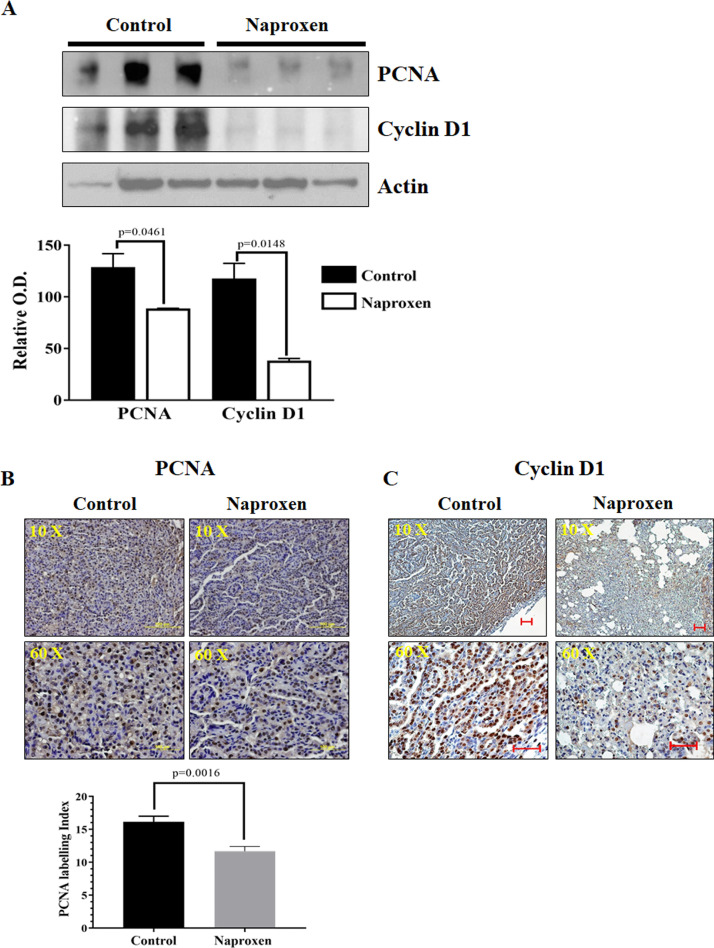
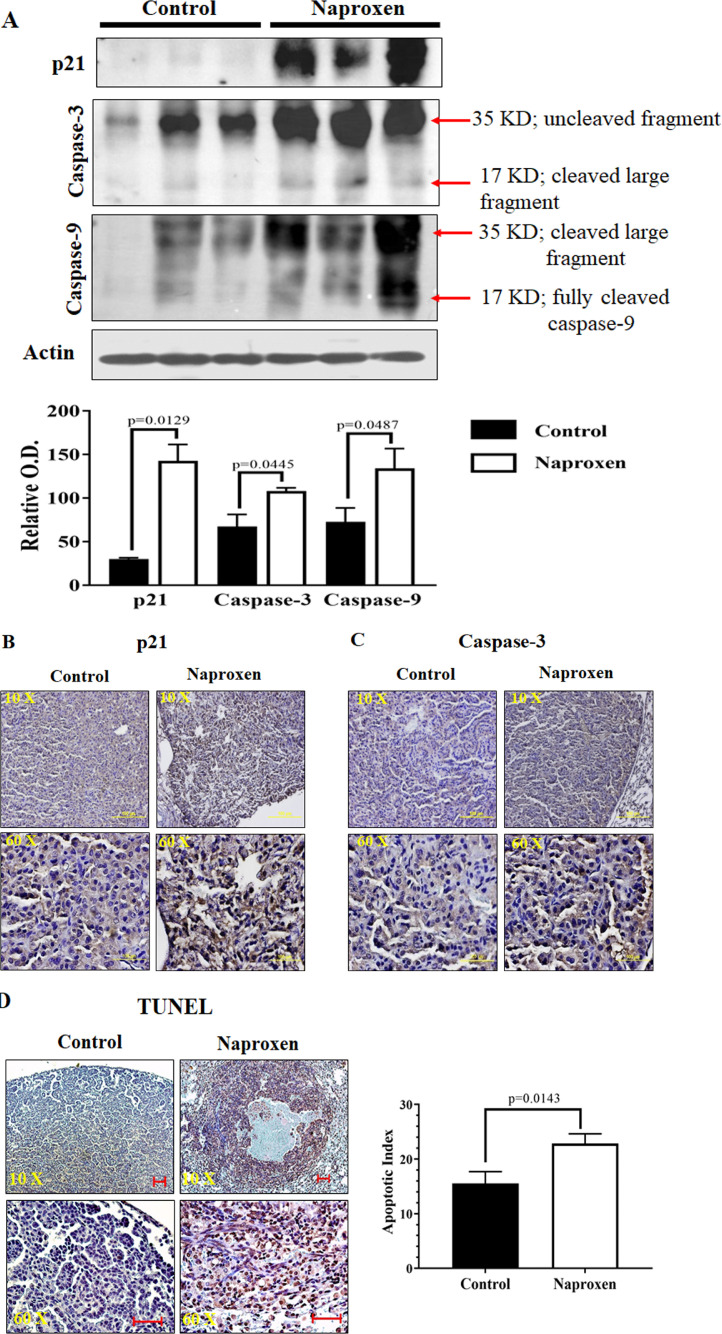
Cell proliferation markers were analyzed to study their role in decreased lung tumor multiplicity in KrasG12V mice fed with naproxen diet. For this, total cell lysates prepared from lung tumor tissues of control and naproxen-fed KrasG12V mice were used. The western blot and IHC analysis showed that the lung tumors in the control group had strong expression of proliferation markers PCNA, and cyclin D1. Naproxen significantly decreased the expression of PCNA and cyclin D1 in KrasG12V mice (Figs. 3A, 3B, and 3C). Further, naproxen treatment led to a significant increase in the expression of apoptotic markers, such as p21, caspase-3, and caspase-9, in the mouse lung tumor tissue (Figs. 4A, 4B, and 4C). These cellular effects of naproxen treatment leading to increased apoptosis were further reflected by an increase in TUNEL-positive cells in lung tumor tissues (Fig. 4D).
Fig. 3.
Effect of naproxen on cell proliferation markers (PCNA and cyclin D1) in KrasG12V mouse lung tumors. (A) Representative western blot and optical density of PCNA and cyclin D1 measured in total cell protein of tumors. (B-C) Representative photomicrographs showing immunohistochemical detection of PCNA labeling index and cyclin D1 (magnification 10X and 60X). Differences between control and treatment groups were analyzed by 1-tailed t test with Welch's correction and 95% confidence interval. Data represent Mean ± SEM. P ≤ 0.05 was considered statistically significant.
Fig. 4.
Effect of naproxen on apoptosis in KrasG12V mouse lung tumors. (A) Representative western blot of p21, caspase-3, and caspase-9 measured in total cell protein of tumors. (B-C) Representative photomicrographs showing immunohistochemical detection of p21 and caspase-3 (magnification 10X and 60X). (D) Representative photomicrographs showing calorimetric detection of apoptotic index through TUNEL assay. Differences between control and treatment groups were analyzed by 1-tailed t test with Welch's correction and 95% confidence interval. Data represent Mean ± SEM. P ≤ 0.05 was considered statistically significant.
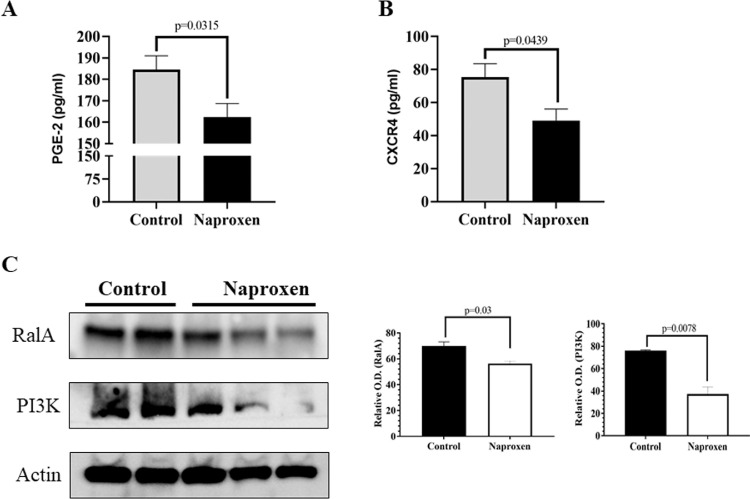
Naproxen inhibits the PGE2 and CXCR4 levels and protein expression of RalA and PI3K in lung tumors of KrasG12V mice
Levels of PGE2 and CXCR4 were determined in serum samples of control and naproxen-fed KrasG12V mice. Naproxen treatment significantly reduced PGE2 and CXCR4 levels in the KrasG12V mice (Figs. 5A and 5B). RalA and PI3K-Akt pathways, downstream of RAS has been shown to play an important part in RAS-mediated cell survival and proliferation [26]. Therefore, the effects of naproxen on RalA and PI3K were determined. Naproxen treatment led to a significant decrease in the expression of RalA and PI3K in the mouse lung tumor tissue (Fig 5C).
Fig. 5.
Effect of naproxen on levels of (A) PGE2, and (B) CXCR4, in KrasG12V mice serum samples. Data represent Mean ± SEM of at least 3 serum samples for PGE2 and CXCR4 ELISA, respectively. (C) Effect of naproxen on RalA and PI3K in KrasG12V mouse lung tumors. Representative western blot and optical density of RalA and PI3K measured in total cell protein of tumors. Differences between control and treatment groups were analyzed by 2-tailed unpaired t test with Welch's correction and 95% confidence interval. Data represent Mean ± SEM. P ≤ 0.05 was considered statistically significant.
Discussion
KRAS mutations represent one of the most common oncogenic driver mutations in lung cancer. These alterations occur in approximately 30% of lung adenocarcinomas [27]. KRAS mutations are common in pancreatic ductal adenocarcinoma (80.0%), and colorectal adenocarcinoma (47%) [28]. Of all mutations, G12C (40%), G12V (22%), and G12D (16%) are the common alterations found predominantly in codon 12 and shown to affect patient survival, and response to treatments [29]. Lung cancer patients with KRAS mutant tumors are associated with decreased progression-free survival compared with those with wild-type KRAS tumors [30].
In the present study, KrasG12V mice develop spontaneous lung tumors and progress to adenocarcinomas. One possible explanation for spontaneous lung tumor development is the gain of hyperproliferative properties of lung bronchiolo-alveolar cells that progress into adenocarcinoma [31]. This activation of KrasG12V in lungs produces less control of tumor initiation and is shown to have undesirable effects in many transgenic models. Further, transgene expression of the inducible system in the turn off stage; also known as leakiness, may also account for the development of spontaneous lung tumors [32]. To check for the presence of Cre, we performed PCR with Cre-specific primers and tested the mouse tissues for protein expression using IHC. Cre expression was absent from adult KrasG12V and wild-type mouse lung tissue. Furthermore, Cre expression was absent in lung tissues of KrasG12V mice when compared with the pancreas of p48Cre/+/LSL-KrasG12D/+ mice and salivary gland of Ela-CreERT.CAG-lox-KrasG12V (ECKG12V) mice (Supplementary Fig. 3). This data suggests the possibility of the leaky gene expression in the embryonic/early stages of KrasG12V mice lung tissues.
Mutant Kras activation plays an important role in multiple downstream signaling pathways. Mutant Kras (G12C and G12V) preferentially bind to Ral guanine nucleotide dissociation stimulator (RalGDS), a Ral GTPase-specific GEF. However, Kras (G12D) has a higher affinity for phosphatidylinositol 3-kinase (PI3K) [33]. NSCLC cell lines with KrasG12D mutation have activated PI3K and MEK signaling, whereas cell lines with KrasG12C/G12V mutations have activated Ral signaling and decreased growth factor-dependent Akt activation [34]. In present study, we also observed an almost 3-fold decreased Akt expression in KrasG12V mouse lung tissues compared with normal lung, where naproxen has shown to decrease the induced RalA expression.
Despite being the most prevalent oncogenic event, efforts towards mutant RAS targeted therapies are yet to yield clinically approved drugs. Hence, it is imperative to develop alternate strategies to prevent Kras-driven cancers to minimize the cancer mortality. Although G12V mutation makes up almost one-fifth of the total Kras mutation and is associated with poor survival, very limited data is available on treating lung cancers carrying KrasG12V mutation. While NSAID use has been shown to decrease the 28% lung cancer risk [35], this is the first study to our knowledge that specifically aims to show the efficacy of naproxen in transgenic KrasG12V mice with spontaneous lung tumors.
Gene expression analysis of the lung tumors revealed upregulated genes involved in cytokine signaling, suggesting the existence of a pro-inflammatory environment in KrasG12V mice lung tumors. Gene expression results and protein expression analysis also suggested increased expression of cell inflammation markers COX-2 and IL-10 in KrasG12V lung tumors as shown in tumor-associated macrophages, which further correlate with lung cancer stages [36,37]. Protein expression of COX-2 and IL-10 was decreased upon naproxen treatment as shown by western blot and immunohistochemical analysis. Clusterin, a stress-associated protein with an antiapoptotic role and contributes to resistance against chemotherapy, was observed to be significantly upregulated in KrasG12V lung tumor. A phase I/II trial of Clusterin protein production inhibitor in combination with gemcitabine/platinum showed higher median survival in advanced NSCLC patients [38]. Interleukins and fibroblast growth factor (FGF) are also modulated in lung tumor tissue suggesting their role in tumor cell growth, survival, differentiation, and angiogenesis. Studies have shown that FGF works synergistically with vascular endothelial growth factor to amplify tumor angiogenesis [39]. Together, interleukins, FGF, and the immune system can be used for new drug target and can play an important role in inhibition of tumor development.
Aspirin and naproxen have been shown to inhibit cigarette smoke induced lung tumors in female A/J mice [12]. The mode of action of naproxen to inhibit prostaglandin synthesis is well-established. Although reports of the effects of naproxen on lung cancer are limited, it is well studied in other cancers. Naproxen (320 mg/kg diet) significantly inhibited the formation of lung tumors in mainstream cigarette smoke-exposed neonatal mice [11]. Both naproxen and nitric oxide-naproxen have been shown to be effective in colon, urinary bladder, and mammary rodent model cancers [14,40]. Results from our laboratory have shown that continuous and intermittent dosing (1 wk-on/off) regimens of naproxen (200 or 400 ppm) inhibit azoxymethane-induced colon adenoma progression to invasive adenocarcinoma [17]. In the present study, we found that administration of naproxen (400 ppm) inhibits total lung tumor formation by ∼52% and adenocarcinoma by ∼64% in KrasG12V mice. This observation along with increased adenomas multiplicity in the KrasG12V mice fed with naproxen diet, suggests that naproxen delays the progression of lung adenoma to adenocarcinoma.
Consistent with many studies, increased cell proliferation (PCNA and cyclin D1) with decreased expression of apoptosis markers (p21 and caspase-3) were observed in mouse lung tumors compared with normal lung [[41], [42], [43]]. The present study demonstrated that the dietary administration of naproxen decreased tumor size, cell proliferation markers as compared with controls. Our results showed that naproxen treatment leads to increased apoptosis in lung tumor tissue. These results agree with previously published in vivo and in vitro studies in lung or other cancers [17,44] that showed that NSAIDs inhibit proliferation and/or induce apoptosis in a variety of cell lines, irrespective of COX-1/2 expression [45,46]. Naproxen was also found to induce cell-cycle arrest and apoptosis by targeting PI3K in bladder cancer cells [44]. The role of PGE2 in tumor formation and impact of NSAIDS on tumor development has been demonstrated in different cancer animal models and in vitro in cell lines [47]. Together, our results and published literature suggest that naproxen inhibits COX‐2-derived PGE2-mediated processes that altogether inhibit CXCR4 expression, resulting in the inhibition of lung tumor growth.
In conclusion, this is the first report of the chemopreventive effects of naproxen blocking spontaneous lung adenocarcinoma formation in the KrasG12V mouse model. We found that naproxen use was associated with a diminished occurrence of lung cancer, with a strong protective effect against the progression of adenoma to adenocarcinoma in vivo. Significantly, the outcomes are reliable and consistent with the underlying biological mechanisms of the COX-2 pathway and lung adenocarcinoma. Although our outcomes are promising, the known risks and advantages for current and former smokers while using different NSAIDs must be assessed before these medications can be prescribed as chemoprevention operators for lung malignancy.
Acknowledgments
Acknowledgments
The authors thank the following: University of Oklahoma Health Sciences Center Rodent Barrier Facility and Ms. Kathy Kyler for editorial help. Dr. Craig Logsdon (MD Anderson Cancer Center) for sharing the mouse model. HTG Molecular Diagnostics, Inc. for gene expression analysis using Mouse MRNA Tumor Response Panel. Ms. Taylor McCoy for administrative help in preparation of manuscript.
Authors’ contributions
Conception and design: G. Kumar, V. Madka, A. Mohammed, C.V. Rao
Data curation, formal analysis, methodology, and data acquisition: G. Kumar, V. Madka, A. Singh, M. Farooqui, A. Mohammed, C.V. Rao
Analysis and interpretation of data: G. Kumar, V. Madka, A. Singh, M. Farooqui, S. Lightfoot, C.V. Rao
Writing, review, and editing: G. Kumar, V. Madka, A. Mohammed, C.V. Rao
Study supervision: V. Madka, C.V. Rao
Footnotes
Funding: This study was supported by the Kerley-Cade Chair Endowment & partly by the NCI-PREVENT program HHSN261201500038i; VA Merit Award I01BX003198
✩✩Conflicts of interest: The authors have no conflicts of interest to declare.
Supplementary material associated with this article can be found, in the online version, at doi:10.1016/j.neo.2021.05.010.
Appendix. Supplementary materials
References
- 1.Siegel R.L., Miller K.D., Fuchs H.E., Jemal A. Cancer Statistics, 2021. CA Cancer J Clin. 2021;71(1):7–33. doi: 10.3322/caac.21654. [DOI] [PubMed] [Google Scholar]
- 2.Howlader N., Noone A., Krapcho M., Miller D., Brest A., Yu M., Ruhl J., Tatalovich Z., Mariotto A., Lewis D. National Cancer Institute. Bethesda; MD: 2019. SEER Cancer Statistics Review, 1975-2016. [Google Scholar]
- 3.Blandin Knight S., Crosbie P.A., Balata H., Chudziak J., Hussell T., Dive C. Progress and prospects of early detection in lung cancer. Open biology. 2017;7(9) doi: 10.1098/rsob.170070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Meng D., Yuan M., Li X., Chen L., Yang J., Zhao X., Ma W., Xin J. Prognostic value of K-RAS mutations in patients with non-small cell lung cancer: a systematic review with meta-analysis. Lung Cancer. 2013;81(1):1–10. doi: 10.1016/j.lungcan.2013.03.019. [DOI] [PubMed] [Google Scholar]
- 5.Renaud S., Falcoz P.E., Schaëffer M., Guenot D., Romain B., Olland A., Reeb J., Santelmo N., Chenard M.P., Legrain M. Prognostic value of the KRAS G12V mutation in 841 surgically resected Caucasian lung adenocarcinoma cases. Br J Cancer. 2015;113(8):1206–1215. doi: 10.1038/bjc.2015.327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Alamo P., Gallardo A., Di Nicolantonio F., Pavón M.A., Casanova I., Trias M., Mangues M.A., Lopez-Pousa A., Villaverde A., Vázquez E. Higher metastatic efficiency of KRas G12V than KRas G13D in a colorectal cancer model. FASEB J. 2015;29(2):464–476. doi: 10.1096/fj.14-262303. [DOI] [PubMed] [Google Scholar]
- 7.Eckert F., Schilbach K., Klumpp L., Bardoscia L., Sezgin E.C., Schwab M., Zips D., Huber S.M. Potential role of CXCR4 targeting in the context of radiotherapy and immunotherapy of cancer. Front Immunol. 2018;9:3018. doi: 10.3389/fimmu.2018.03018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Santagata S., Napolitano M., D'Alterio C., Desicato S., Di Maro S., Marinelli L., Fragale A., Buoncervello M., Persico F., Gabriele L. Targeting CXCR4 reverts the suppressive activity of T-regulatory cells in renal cancer. Oncotarget. 2017;8(44):77110. doi: 10.18632/oncotarget.20363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Obermajer N., Muthuswamy R., Odunsi K., Edwards R.P., Kalinski P. PGE2-induced CXCL12 production and CXCR4 expression controls the accumulation of human MDSCs in ovarian cancer environment. Cancer Res. 2011;71(24):7463–7470. doi: 10.1158/0008-5472.CAN-11-2449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Kuo C.-N., Pan J.-J., Huang Y.-W., Tsai H.-J., Chang W.-C. Association between nonsteroidal anti-inflammatory drugs and colorectal cancer: a population-based case–control study. Cancer Epidemiol Biomarkers Prev. 2018;27(7):737–745. doi: 10.1158/1055-9965.EPI-17-0876. [DOI] [PubMed] [Google Scholar]
- 11.Balansky R., Ganchev G., Iltcheva M., Micale R.T., La Maestra S., D'Oria C., Steele V.E., De Flora S. Selective inhibition by aspirin and naproxen of mainstream cigarette smoke-induced genotoxicity and lung tumors in female mice. Arch Toxicol. 2016;90(5):1251–1260. doi: 10.1007/s00204-015-1550-5. [DOI] [PubMed] [Google Scholar]
- 12.La Maestra S., D'Agostini F., Izzotti A., Micale R.T., Mastracci L., Camoirano A., Balansky R., Trosko J.E., Steele V.E., De Flora S. Modulation by aspirin and naproxen of nucleotide alterations and tumors in the lung of mice exposed to environmental cigarette smoke since birth. Carcinogenesis. 2015;36(12):1531–1538. doi: 10.1093/carcin/bgv149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Brasky T.M., Baik C.S., Slatore C.G., Potter J.D., White E. Non-steroidal anti-inflammatory drugs and small cell lung cancer risk in the VITAL study. Lung Cancer. 2012;77(2):260–264. doi: 10.1016/j.lungcan.2012.04.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Steele V.E., Rao C.V., Zhang Y., Patlolla J., Boring D., Kopelovich L., Juliana M.M., Grubbs C.J., Lubet R.A. Chemopreventive efficacy of naproxen and nitric oxide–naproxen in rodent models of colon, urinary bladder, and mammary cancers. Cancer Prev Res. 2009;2(11):951–956. doi: 10.1158/1940-6207.CAPR-09-0080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Angiolillo D.J., Weisman S.M. Clinical pharmacology and cardiovascular safety of naproxen. Am J Cardiovasc Drugs. 2017;17(2):97–107. doi: 10.1007/s40256-016-0200-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Patlolla J.M., Kopelovich L., Qian L., Zhang Y., Kumar G., Madka V., Mohammed A., Biddick L., Sadeghi M., Lightfoot S. Early and delayed intervention with Rapamycin prevents NNK-induced lung adenocarcinoma in A/J mice. Oncol Rep. 2015;34(6):2925–2934. doi: 10.3892/or.2015.4277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Mohammed A., Janakiram N.B., Madka V., Zhang Y., Singh A., Biddick L., Li Q., Lightfoot S., Steele V.E., Lubet R.A. Intermittent dosing regimens of aspirin and naproxen inhibit azoxymethane-induced colon adenoma progression to adenocarcinoma and invasive carcinoma. Cancer Prev Res. 2019;12(11):751–762. doi: 10.1158/1940-6207.CAPR-19-0312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Nikitin A.Y., Alcaraz A., Anver M.R., Bronson R.T., Cardiff R.D., Dixon D., Fraire A.E., Gabrielson E.W., Gunning W.T., Haines D.C. Classification of proliferative pulmonary lesions of the mouse: recommendations of the mouse models of human cancers consortium. Cancer Res. 2004;64(7):2307–2316. doi: 10.1158/0008-5472.can-03-3376. [DOI] [PubMed] [Google Scholar]
- 19.Kumar G., Tajpara P., Bukhari A.B., Ramchandani A.G., De A., Maru G.B. Dietary curcumin post-treatment enhances the disappearance of B(a) P-derived DNA adducts in mouse liver and lungs. Toxicol Rep. 2014;1:1181–1194. doi: 10.1016/j.toxrep.2014.11.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Varghese F., Bukhari A.B., Malhotra R., De A. IHC Profiler: an open source plugin for the quantitative evaluation and automated scoring of immunohistochemistry images of human tissue samples. PLoS One. 2014;9(5):e96801. doi: 10.1371/journal.pone.0096801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kwak M., Erdag G., Slingluff C.L. Gene expression analysis in formalin fixed paraffin embedded melanomas is associated with density of corresponding immune cells in those tissues. Sci Rep. 2020;10(1):1–8. doi: 10.1038/s41598-020-74996-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Sinn B., Loibl S., Kronenwett R., Furlanetto J., Krappmann K., Karn T., Kerns B., Weber K., Schmidt M., Denkert C. Targeted mRNA sequencing of small formalin-fixed and paraffin-embedded breast cancer samples for the quantification of immune and cancer-related genes. Ann Oncol. 2017;28:i12. [Google Scholar]
- 23.Chandrashekar D.S., Bashel B., Balasubramanya S.A.H., Creighton C.J., Ponce-Rodriguez I., Chakravarthi B.V., Varambally S. UALCAN: a portal for facilitating tumor subgroup gene expression and survival analyses. Neoplasia. 2017;19(8):649–658. doi: 10.1016/j.neo.2017.05.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Huang D.W., Sherman B.T., Lempicki R.A. Bioinformatics enrichment tools: paths toward the comprehensive functional analysis of large gene lists. Nucleic Acids Res. 2009;37(1):1–13. doi: 10.1093/nar/gkn923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Sherman B.T., Lempicki R.A. Systematic and integrative analysis of large gene lists using DAVID bioinformatics resources. Nat Protoc. 2009;4(1):44. doi: 10.1038/nprot.2008.211. [DOI] [PubMed] [Google Scholar]
- 26.Castellano E., Downward J. RAS interaction with PI3K: more than just another effector pathway. Genes Cancer. 2011;2(3):261–274. doi: 10.1177/1947601911408079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Shepherd F.A., Domerg C., Hainaut P., Jänne P.A., Pignon J.-P., Graziano S., Douillard J.-Y., Brambilla E., Chevalier T.Le, Seymour L. Pooled analysis of the prognostic and predictive effects of KRAS mutation status and KRAS mutation subtype in early-stage resected non–small-cell lung cancer in four trials of adjuvant chemotherapy. J Clin Oncol. 2013;31(17):2173. doi: 10.1200/JCO.2012.48.1390. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Consortium A.P.G. AACR Project GENIE: powering precision medicine through an international consortium. Cancer Discov. 2017;7(8):818–831. doi: 10.1158/2159-8290.CD-17-0151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Liu P., Wang Y., Li X. Targeting the untargetable KRAS in cancer therapy. Acta Pharm Sin B. 2019;9(5):871–879. doi: 10.1016/j.apsb.2019.03.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Ihle N.T., Byers L.A., Kim E.S., Saintigny P., Lee J.J., Blumenschein G.R., Tsao A., Liu S., Larsen J.E., Wang J. Effect of KRAS oncogene substitutions on protein behavior: implications for signaling and clinical outcome. J Natl Cancer Inst. 2012;104(3):228–239. doi: 10.1093/jnci/djr523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Guerra C., Mijimolle N., Dhawahir A., Dubus P., Barradas M., Serrano M., Campuzano V., Barbacid M. Tumor induction by an endogenous K-ras oncogene is highly dependent on cellular context. Cancer Cell. 2003;4(2):111–120. doi: 10.1016/s1535-6108(03)00191-0. [DOI] [PubMed] [Google Scholar]
- 32.Delerue F., White M., Ittner L.M. Inducible, tightly regulated and non-leaky neuronal gene expression in mice. Transgenic Res. 2014;23(2):225–233. doi: 10.1007/s11248-013-9767-7. [DOI] [PubMed] [Google Scholar]
- 33.Román M., Baraibar I., López I., Nadal E., Rolfo C., Vicent S., Gil-Bazo I. KRAS oncogene in non-small cell lung cancer: clinical perspectives on the treatment of an old target. Mol Cancer. 2018;17(1):33. doi: 10.1186/s12943-018-0789-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Roberts P.J., Stinchcombe T.E. KRAS mutation: should we test for it, and does it matter? J Clin Oncol. 2013;31(8):1112–1121. doi: 10.1200/JCO.2012.43.0454. [DOI] [PubMed] [Google Scholar]
- 35.Bittoni M.A., Carbone D.P., Harris R.E. Ibuprofen and fatal lung cancer: a brief report of the prospective results from the Third National Health and Nutrition Examination Survey (NHANES III) Mol Clin Oncol. 2017;6(6):917–920. doi: 10.3892/mco.2017.1239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Wang R., Lu M., Zhang J., Chen S., Luo X., Qin Y., Chen H. Increased IL-10 mRNA expression in tumor-associated macrophage correlated with late stage of lung cancer. J Exp Clin Cancer Res. 2011;30(1):62. doi: 10.1186/1756-9966-30-62. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Vahl J.M., Friedrich J., Mittler S., Trump S., Heim L., Kachler K., Balabko L., Fuhrich N., Geppert C.I., Trufa D.I. Interleukin-10-regulated tumour tolerance in non-small cell lung cancer. Br J Cancer. 2017;117(11):1644–1655. doi: 10.1038/bjc.2017.336. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Laskin J.J., Nicholas G., Lee C., Gitlitz B., Vincent M., Cormier Y., Stephenson J., Ung Y., Sanborn R., Pressnail B. Phase I/II trial of custirsen (OGX-011), an inhibitor of clusterin, in combination with a gemcitabine and platinum regimen in patients with previously untreated advanced non-small cell lung cancer. J Thorac Oncol. 2012;7(3):579–586. doi: 10.1097/JTO.0b013e31823f459c. [DOI] [PubMed] [Google Scholar]
- 39.Korc M., Friesel R.E. The role of fibroblast growth factors in tumor growth. Curr Cancer Drug Targets. 2009;9(5):639–651. doi: 10.2174/156800909789057006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Madka V., Mohammed A., Li Q., Zhang Y., Lightfoot S., Wu X.-R., Steele V., Kopelovich L., Rao C.V. Nitric oxide-releasing naproxen prevents muscle invasive bladder cancer. Proceedings: AACR 107th Annual Meeting 2016; New Orleans, LA; 2016. April 16-20, 201676(14 Supplement) [Google Scholar]
- 41.Kučera M., Rousek M. Evaluation of thermooxidation stability of biodegradable recycled rapeseed-based oil NAPRO-HO 2003. Res Agric Eng. 2008;54(4):163–169. [Google Scholar]
- 42.Suh N., Reddy B.S., DeCastro A., Paul S., Lee H.J., Smolarek A.K., So J.Y., Simi B., Wang C.X., Janakiram N.B. Combination of atorvastatin with sulindac or naproxen profoundly inhibits colonic adenocarcinomas by suppressing the p65/β-catenin/cyclin D1 signaling pathway in rats. Cancer Prev Res. 2011;4(11):1895–1902. doi: 10.1158/1940-6207.CAPR-11-0222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Kumar G., Patlolla J.M.R., Madka V., Mohammed A., Li Q., Zhang Y., Biddick L., Singh A., Gillaspy A., Lightfoot S. Simultaneous targeting of 5-LOX-COX and ODC block NNK-induced lung adenoma progression to adenocarcinoma in A/J mice. Am J Cancer Res. 2016;6(5):894. [PMC free article] [PubMed] [Google Scholar]
- 44.Kim M.-S., Kim J.-E., Huang Z., Chen H., Langfald A., Lubet R.A., Grubbs C.J., Dong Z., Bode A.M. Naproxen induces cell-cycle arrest and apoptosis in human urinary bladder cancer cell lines and chemically induced cancers by targeting PI3K. Cancer Prev Res. 2014;7(2):236–245. doi: 10.1158/1940-6207.CAPR-13-0288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Vogt T., McClelland M., Jung B., Popova S., Bogenrieder T., Becker B., Rumpler G., Landthaler M., Stolz W. Progression and NSAID-induced apoptosis in malignant melanomas are independent of cyclooxygenase II. Melanoma Res. 2001;11(6):587–599. doi: 10.1097/00008390-200112000-00005. [DOI] [PubMed] [Google Scholar]
- 46.Elder D., Halton D.E., Hague A., Paraskeva C. Induction of apoptotic cell death in human colorectal carcinoma cell lines by a cyclooxygenase-2 (COX-2)-selective nonsteroidal anti-inflammatory drug: independence from COX-2 protein expression. Clin Cancer Res. 1997;3(10):1679–1683. [PubMed] [Google Scholar]
- 47.Nakanishi M., Rosenberg D.W. Multifaceted roles of PGE2 in inflammation and cancer. Semin Immunopathol. 2013;35(2):123–137. doi: 10.1007/s00281-012-0342-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.