Abstract
Escherichia coli is one major cause of bacterial infections and can horizontally acquire antimicrobial resistance and virulence genes through conjugation. Because conjugative plasmids can rapidly spread among bacteria of different species, the plasmids carrying both antimicrobial resistance and virulence genes may pose a significant threat to public health. Therefore, the identification and characterization of these plasmids may facilitate a better understanding of E. coli pathogenesis and the development of new strategies against E. coli infections. Because iron uptake ability is a potential virulence trait of bacteria, we screened for E. coli conjugative plasmids able to confer both iron uptake ability and ampicillin resistance. The plasmid pEC41, which was derived from the bacteremia clinical isolate EC41, was identified. EC41, which carried the fimH27 allele, belonged to sequence type (ST) 405 and phylogroup D. According to the sequencing analyses, pEC41 was 86 kb in size, and its backbone structure was almost identical to that of another highly conjugative plasmid, pCTX-M3, in which the extended-spectrum β-lactamase gene blaCTX–M–3 was originally identified. pEC41 carried blaCTX–M–3 and blaTEM–1. The ferric citrate uptake (fec) system was identified in pEC41 and was responsible for conferring iron uptake ability. The fec system contributes to the pathogenesis of EC41 in systemic infections but not in urinary tract infections (UTIs). However, this system promoted competitive fitness of a cystitis-associated clinical isolate to colonize urinary tracts. Additionally, the distribution of the fec system was related to E. coli isolates associated with human bacteremia and UTIs. In summary, the present study identified a novel conjugative plasmid, pEC41, which conferred both antimicrobial resistance and an extra iron uptake ability to E. coli. The iron uptake ability was encoded in the fec system and contributed to E. coli pathogenesis. This study is the first to show that the fec system is a virulence factor in E. coli.
Keywords: iron uptake system, blaCTX–M–3, ESBL, bacteremia, urinary tract infections, ferric citrate, E. coli, conjugative plasmid
Introduction
Escherichia coli is one of the major Gram-negative etiological causes of bacteremia and urinary tract infections (UTIs) (Medina and Castillo-Pino, 2019; Bonten et al., 2020; Wagenlehner et al., 2020). E. coli can obtained antimicrobial resistance and virulence factors through horizontal gene transfer (HGT) (Paauw et al., 2009; Wang et al., 2011; Johnson and Lang, 2012). Conjugative plasmids, which are one of the key agents of HGT, can spread efficiently and broadly among distinct bacterial species through conjugation (Ilangovan et al., 2015; Partridge et al., 2018). Therefore, the plasmids carrying both antimicrobial resistance and virulence genes may pose a significant threat to public health (Paauw et al., 2009; Wang et al., 2011; Johnson and Lang, 2012). The identification and characterization of these plasmids would provide potential strategies to tackle the worldwide public health issues. However, identifying the virulence factors carried by the plasmids could be a challenge because some of the pathogenic effects of these factors are only observed in specific tissues, and the potential presence of other factors with similar functions in the same host bacteria may hinder the discovery of such plasmid-carried virulence factors (Cusumano et al., 2010; Garcia et al., 2011; Huang et al., 2020).
Iron uptake ability is a potential virulence trait for invading bacteria to survive in hosts, where the bioavailability of iron is restricted (Hood and Skaar, 2012). Iron uptake systems are often identified in bacterial chromosomes, while plasmid-encoded systems are relatively rare (Ramirez et al., 2014). However, based on in silico analysis, various potential iron uptake genes have been detected in plasmid sequences, suggesting that many plasmid-encoded iron uptake systems that contribute to bacterial virulence have yet to be identified. Siderophore systems, including enterobactin, salmochelin, aerobactin, and yersiniabactin, have been utilized by pathogenic bacteria to retrieve ferric iron (Fe3+) as an iron source in hosts and thus contribute to the virulence of the bacteria. On the other hand, the fecIRABCDE (fec) system also enables bacteria to retrieve Fe3+ through the uptake of ferric citrate (Braun and Mahren, 2005). However, researchers have not clearly determined whether this system contributes to any infections. This system is composed of the upstream fecIR operon and the downstream fecABCDE operons. The fecABCDE operons encode the ability to take up ferric citrate, while fecIR is responsible for regulating the transcription of fecABCDE. The transcription of fecABCDE and fecIR is repressed by iron-bound Fur. The expression of fecIR is induced under iron-limiting conditions, while the expression of fecABCDE requires both the induction of iron limitation and the presence of ferric citrate (Braun and Mahren, 2005). Given the constant presence of citrate in body fluids and the high affinity of this compound for ferric iron (Kirejczyk et al., 2014; Costello and Franklin, 2016), the fec system may contribute to the virulence of bacteria during infections.
In the present study, we aimed to identify and characterize conjugative plasmid-encoded iron uptake systems that can contribute to the pathogenesis of E. coli infections. We started by performing an in vitro screen of clinical isolate-derived conjugative plasmids that increase the iron uptake ability of bacterial hosts. Because pathogenic E. coli usually harbors multiple iron systems to cope with iron-limited environments, the coexistence of functionally redundant iron uptake factors in the bacteria may interfere with the screening. To avoid such potential interference, ampicillin-resistant conjugative plasmids in clinical strains were conjugally transferred to an iron uptake-defective commensal E. coli K12 strain whose enterobactin-dependent uptake system was inactivated. The resulting transconjugants were then subjected to a screen for the upregulated abilities to acquire iron. A plasmid encoding the fec system was identified, and this system was found to contribute to the virulence of E. coli. In addition, this plasmid carried genes encoding an extended-spectrum β-lactamase (ESBL) and a broad-spectrum β-lactamase.
Materials and Methods
Bacterial Strains, Plasmids, Growth Conditions, and Reagents
The E. coli strains utilized for genetic manipulation are shown in Table 1. Donor clinical E. coli isolates were collected from National Cheng Kung University Hospital (NCKUH), Taiwan (NCKUH IRB approval no. A-ER-104-313). For the isolates used in the molecular epidemiology study, the fecal and UTI-associated E. coli isolates were described previously (Mao et al., 2012), and bacteremia-associated E. coli were collected from the blood specimens of patients at NCKUH between October and December of 2005. The bacteria were grown on Luria-Bertani (LB) (1% tryptone, 0.5% yeast extract, and 1% NaCl) broth or agar with appropriate antimicrobial s at 37°C unless otherwise indicated. The reagents and antimicrobial s were purchased from Sigma-Aldrich, St. Louis, MO, United States unless otherwise indicated.
TABLE 1.
Bacterial strains and plasmids used in this study.
| Strains or plasmids | Description | Known AR markersa | Sources |
| Strains | |||
| EC41 | An E. coli strain isolated from the blood of a patient | Ap, Str, Km | This study |
| A487 | An E. coli strain isolated from the urine of a patient with cystitis | Mao et al., 2012 | |
| CFT073str | A spontaneous streptomycin-resistant mutant of the urosepsis-causing E. coli strain CFT073 | Str | This study |
| UTI89str | A spontaneous streptomycin-resistant mutant of the cystitis-causing E. coli strain UTI89 | Str | This study |
| MG1655str | A spontaneous streptomycin-resistant mutant of E. coli K12 MG1655 | Str | This study |
| IP10 | An entA mutant of MG1655 with an insertion of a kanamycin resistance cassette in the entA locus | Km | This study |
| MP10 | An entA mutant of MG1655str | Str | This study |
| MP10LA | A lacZ mutant of MP10 with an insertion of a chloramphenicol resistance cassette in the lacZ locus | Str, Cm | This study |
| ELA41 | MP10LA harboring pEC41 | Str, Cm, Ap | This study |
| IP10/pEC41 | IP10 harboring pEC41 | Km, Ap | This study |
| MP10/pEC41 | MP10 harboring pEC41 | Str, Ap | This study |
| Δfec-MP10 | A fecMG1655 deletion mutant of MP10 with an insertion of a gentamicin resistance cassette in the chromosomal fecMG1655 locus | Str, Gm | This study |
| MP10/pEC41Δfec | MP10 harboring pEC41Δfec | Str, Ap, Cm | This study |
| Δfec-MP10/pEC41 | Δfec-MP10 harboring pEC41 | Str, Ap, Gm | This study |
| Δfec-MP10/pEC41Δfec | Δfec-MP10 harboring pEC41Δfec | Str, Ap, Gm, Cm | This study |
| ΔfecpEC41-EC41 | A fecEC41 deletion mutant of EC41 with an insertion of chloramphenicol resistance cassette in the fecEC41 locus | Ap, Cm | This study |
| A487/pEC41 | A487 harboring pEC41 | Ap | This study |
| A487/pEC41Δfec | A487 harboring pEC41Δfec | Ap, Cm | This study |
| CFT073str/pEC41 | A spontaneous streptomycin-resistant strain of E. coli CFT073 harboring pEC41 | Str, Ap | This study |
| UTI89str/pEC41 | A spontaneous streptomycin-resistant strain of E. coli UTI89 harboring pEC41 | Str, Ap | This study |
| Plasmids | |||
| pEC41 | The plasmid isolated from EC41 | Ap | This study |
| pEC41Δfec | A fecEC41 deletion mutant of pEC41 with an insertion of chloramphenicol resistance cassette in the fecEC41 locus | Ap, Cm | This study |
aAR markers, antimicrobial resistance markers commonly used in laboratories; Ap, ampicillin; Str, streptomycin; Km, kanamycin; Cm, chloramphenicol; Gm, gentamicin.
Conjugation
To collect ampicillin-resistant conjugative plasmids from clinical E. coli isolates by using the chloramphenicol-resistant E. coli strain MP10LA (Table 1) as the recipient through conjugation, clinical isolates that exhibited ampicillin resistance and chloramphenicol susceptibility (i.e., isolates able to grow in medium containing 50 μg/ml of ampicillin, but unable to grow in medium containing 15 μg/ml of chloramphenicol) was selected as donors. Equal volumes of overnight cultures of the donor clinical isolates and the recipient strain MP10LA were mixed and spread on LB agar to capture conjugative plasmids conferring ampicillin resistance from clinical E. coli isolates. After 6 h of incubation, the bacteria were suspended in LB broth and plated on LB agar with chloramphenicol and ampicillin to select against the donor and recipient bacteria. Therefore, the resulting transconjugants, which were MP10LA strains that transconjugatively obtained ampicillin-resistant plasmids, survived and grew on LB agar.
The pEC41-harboring donor strains (EC41, ELA41, and IP10/pEC41) were mixed with recipients for the conjugative transfer of pEC41. Ampicillin and a proper antimicrobial were used to select against the recipient and donor strains, respectively and to identify the resulting transconjugants.
E. coli Typing
The sequence type (ST), phylogroup, and fimH type of EC41 were determined using the PubMLST.org website, ClermonTyping, and FimTyper, respectively, as described previously (Roer et al., 2017; Beghain et al., 2018; Jolley et al., 2018).
Mutant Construction
The E. coli mutants utilized in the present study were constructed using λ red mediated recombineering as described previously (Datsenko and Wanner, 2000; Hsu et al., 2020).
To construct the fec system deletion mutant of the plasmid pEC41, pEC41 was conjugatively transferred to MG1655red (KmR), which chromosomally encodes a λ red recombinase whose expression is induced by arabinose (Hashimoto and Kato, 2003). The mutant plasmid (pEC41Δfec; Table 1) was constructed in MG1655red (KmR) using λ red recombinase-mediated recombination to replace the fec system (fecIRABCDE) with a chloramphenicol resistance cassette. Then, pEC41Δfec was transferred from the strain to other bacterial hosts through conjugation or transformation by electroporation.
To construct ΔfecpEC41-EC41, the pEC41Δfec plasmid was conjugatively transferred into EC41 from MP10/pEC41Δfec (Table 1). The resulting transconjugant that harbored both pEC41 and pEC41Δfec was cultured in LB medium containing chloramphenicol to select for the presence of pEC41Δfec and to facilitate the expulsion of pEC41 from the bacteria. After 12 h of culture, the bacteria were spread on LB agar supplemented with chloramphenicol. ΔfecpEC41-EC41 that harbored pEC41Δfec but not pEC41 was confirmed using PCR to detect the size of the fec locus in the plasmid.
Plasmid Profile Analysis
The method reported by Kado and Liu (1981) was used to extract plasmid DNA and determine the plasmid profiles in E. coli strains, with some modifications. Briefly, one milliliter of an overnight bacterial culture was pelleted by centrifugation at 14,000 × g for 5 min at 4°C. The resulting bacterial pellet was resuspended in 60 μl of preheated lysis buffer (1.5% sodium dodecyl sulfate and 0.2 N NaOH, pH 12.8) to lyse bacterial cells, followed by mixing with an equal amount of PCI (phenol:chloroform:isoamyl alcohol 25:24:1). After centrifugation, 20 μl of the aqueous phase (the upper layer of the solution) was mixed with 4 μl of DNA dye and subjected to electrophoresis in a 0.6% agarose gel.
Iron Growth Promotion Assay
The ability of E. coli strains to utilize various iron sources was determined by performing an iron growth promotion assay. Overnight cultures (1 ml) of the transconjugants or the control strains were centrifuged, washed with PBS, and then resuspended in 10 ml of PBS. The bacterial suspension (100 μl) was spread onto 15 ml of iron-limited medium (ILM) agar composed of 15% agar and ILM medium [0.1 mM CaCl2, 50 mM Na2HPO4, 20 mM KH2PO4, 10 mM NaCl, 20 mM NH4Cl, 20% glucose, 2 mM MgSO4, 0.025 μM sodium citrate, and 100 μM 2,2′-bipyridyl (DIP)]. The resulting bacterium-containing agar plates were punched with pipette tips to create wells of approximately 4-mm diameter, and 10 μl of ferrous sulfate (10 mM), ferric chloride (10 mM), or ferric citrate (10 mM) were added to the wells as iron sources for the bacteria in the agar. The plates were incubated at 37°C for 24 h, and the diameter of growing bacteria surrounding each well was measured.
Plasmid Stability Assays Using pEC41
Because pEC41 conferred ampicillin resistance, the E. coli strains harboring pEC41 were cultured in LB medium with ampicillin (50 μg/ml) overnight. Then the cultures were transferred to fresh ampicillin-containing (50 μg/ml) LB medium in a 1:100 ratio and incubated for 2 h to ensure that all the live cells harbored the plasmid. Then, the bacteria were transferred to fresh LB medium in a 1:100 ratio. The bacterial cultures were spread on LB agar at different time points. The percentages of pEC41-harboring bacteria were determined by randomly selecting 50 colonies and determining their abilities to grow on LB agar containing ampicillin.
Plasmid DNA Extraction and Sequencing Analysis
The pEC41 plasmid was purified from MP10/pEC41 (Table 1) by using a QIAGEN Large Construction Kit (QIAGEN, United Kingdom, Cat. no. 12462) and subjected to sequencing by using the Illumina MiSeq Platform in a paired-end configuration. The sequencing data were de novo assembled into contigs using CLC Genomic Workbench. The gaps in the contigs were closed by Sanger sequencing the PCR fragment amplified between the contigs. The primers used to close the gaps are shown in Supplementary Table 1. Proteins were predicted using SnapGene and NCBI CD searches. A similarity search and pairwise alignment were performed using BLAST programs. The sequences of pEC41 are deposited in the GenBank repository, accession number MW548582.
Mouse Infection Models
For the mouse systemic infection models, the E. coli strains EC41 and ΔfecpEC41-EC41 (1 × 107 CFU/per strain) were coinoculated or independently inoculated into 6–7-week-old BALB/c mice through a tail vein injection as described previously (Subashchandrabose et al., 2013), with some modification. At 24 h postinfection (hpi), blood, hearts, livers, spleens, kidneys and lungs were collected. The solid tissue samples were weighed and homogenized in 3 ml of 0.85% NaCl. The bacterial counts in the blood and the homogenates were determined by plating the samples on LB agar containing appropriate antimicrobials. For the coinfection experiments, EC41 and ΔfecpEC41-EC41 were differentiated by chloramphenicol resistance (Table 1).
For the UTI models, 8–10-week-old female C3H/HeN mice were utilized. Equal numbers (1 × 108 CFU) of the strains harboring pEC41 or pEC41Δfec were coinoculated into the animals transurethrally as described previously (Wright et al., 2005), with some modification. At 24 hpi, the mice were sacrificed, and the bladders and kidneys were collected, weighed, and homogenized. The organ bacterial counts were then determined and differentiated by chloramphenicol resistance, because pEC41Δfec, which harbored a chloramphenicol resistance cassette, conferred chloramphenicol resistance to host bacteria, while pEC41 didn’t (Table 1). Equal volumes of the tissue homogenates were plated on LB agar without and with chloramphenicol (15 μl/ml), respectively. The colony numbers on the LB agar reflected the total bacterial counts in the samples, while the numbers on the chloramphenicol-containing LB agar reflected the counts of pEC41Δfec-harboring strains. The counts of pEC41-harboring strains were determined by subtracting the total bacterial counts by the pEC41Δfec-harboring strain counts in the samples.
The competitive indices (CIs) were calculated by dividing the ratio of pEC41Δfec-harboring to the pEC41-harboring strains in the output by the ratio of the two strains in the inoculum [(CFUpEC41Δfec/CFUpEC41)output/(CFUpEC41Δfec/CFUpEC41) input]. All experimental procedures were approved by the Institutional Animal Care and Use Committee (IACUC) of National Cheng Kung University, Tainan City, Taiwan (approval no. 108130).
Serum and Urine Survival Assay
For serum survival assays, equal numbers of bacterial strains harboring pEC41 and pEC41Δfec (2 × 105 CFU/per strain) were coincubated with 40 μl of 90% heat-inactivated pooled mouse serum (5 mice) in PBS at 37°C. After different incubation periods, the live bacterial counts of the strains were determined by spreading the culture on LB agar plates. The strains harboring pEC41 and pEC41Δfec were differentiated by chloramphenicol resistance, which was conferred by pEC41Δfec but not pEC41. Equal volumes of the serum samples were plated on LB agar without and with chloramphenicol (15 μl/ml), respectively. The colony numbers on the LB agar reflected the total bacterial counts in the samples, while the numbers on the chloramphenicol-containing LB agar reflected the counts of pEC41Δfec-harboring strains. The counts of pEC41-harboring strains were determined by subtracting the total bacterial counts by the pEC41Δfec-harboring strain counts in the samples.
As for urine survival assay, equal numbers of bacterial strains harboring pEC41 and pEC41Δfec (4 × 103 CFU/per strain) were coinoculated in 80 μl of 90% pooled human urine (5 donners) in PBS at 37°C for 4 h and the live bacterial counts of the strains were determined and differentiated by chloramphenicol resistance. Then, the bacterial culture were transferred to fresh 90% urine in a 1:250 dilution for every 4 h of incubation at 37°C and the live bacterial counts were determine right before each transfer.
RNA Isolation and Real-Time PCR
Bacteria were grown in M9 medium containing 100 μM DIP with and without supplementation of 20 μM ferric citrate for 16 h at 37°C with shaking at 200 rpm. RNA was extracted from bacteria using the TRI Reagent according to the manufacturer’s instructions (Sigma-Aldrich, St. Louis, MO, United States). To remove contaminating DNA, the resulting RNA was treated with DNase I (Roche Applied Science, Mannheim, Germany) at 37°C for 1.5 h. Then, the mixture was subjected to phenol/chloroform (1:1) extraction and ethanol precipitation. Finally, the purified RNA was dissolved in RNase-free water and stored at −80°C. The complementary DNA (cDNA) was converted from the purified RNA using random hexamer and Moloney Murine Leukemia Virus (M-MLV) reverse transcriptase according to manufacturer’s instruction (Invitrogen, Carlsbad, CA, United States). For real-time PCR, the cDNA was mixed with primers and KAPA SYBR FAST qPCR Master Mix (Kapa Biosystems, Boston, MA, United States), followed by PCR using StepOnePlus Real-Time PCR Systems with standard reaction conditions (Applied Biosystems, Carlsbad, CA, United States). The primers used in the assays were shown in Supplementary Table 1. The expression level of fecA was normalized to that of ftsZ.
Distribution of fecA
The frequencies of fecA among E. coli were determined by PCR-based analysis. The primers used in this analysis were shown in Supplementary Table 1. The PCR analyses were carried out in a 10 μl volume consisting 1.1 μl of MQ water, 1 μl of 10× Taq Buffer, 0.8 μl dNTP (2.5 mM), 5 μl of template DNA, 1 μl of each primer (20 μM), and 0.1 μl of Taq polymerase. Taq PCR condition was as follows: an initial denaturing step at 95°C for 5 min, followed by 30 cycles of 95°C of denaturation for 1 min, annealing at 55°C for 30 s, and elongation at 72°C for 1 min, and final extension at 72°C for 10 min. The PCR products were separated by gel electrophoresis on 1% agarose gel [1 g agarose in 100 ml of 1× Tris-Acetic acid-EDTA (TAE) buffer]. Agarose was dissolved by heating and cooled down before adding ethidium bromide (EtBr) to a final concentration of approximately 0.05 μg/ml. 2.5 μl of sample was typically loaded to each well along with molecular weight DNA ladder. The DNA fragments were separated by electrophoresis at 100 V for about 30 min and were visualized under UV light.
Statistical Analysis
Before statistical analysis, each set of the experiment data was tested for the normality using the Shapiro–Wilk test. The data whose normality was rejected by the test (P < 0.05) were subsequently subjected to non-parameteic statistical analyses. Otherwise, the data (P ≥ 0.05 in the normality test) were subjected to parametric statistical analyses. For non-parametric analyses, the Mann–Whitney U, Wilcoxon signed-rank, and Wilcoxon rank-sum tests were utilized to analyze the results of mouse independent infection, mouse coinfection, and some iron growth promotion assays, respectively. For the parametric analyses, the one-way ANOVA was utilized to analyze the results of plasmid stability, real-time PCR, and some iron growth promotion assays. The two-way ANOVA was utilized to analyze independent culture experiments. The paired two-tailed student’s t-test was used to analyze the results of bacterial growth in co-culture experiments. Comparisons involving the frequencies of fecA in different source groups of E. coli isolates were analyzed by the two-tailed Fisher’s exact test. A P-value of <0.05 was arbitrarily set as the threshold for statistical significance.
Results
The E. coli Plasmid pEC41 Provides Iron Uptake-Deficient Strains the Ability to Take Up Iron
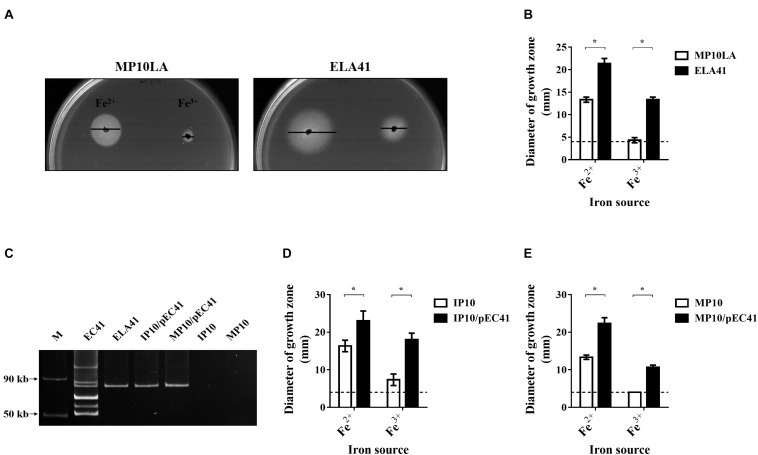
To screen for antimicrobial-resistant conjugative plasmids that provide iron uptake ability, an E. coli mutant defective in iron uptake was constructed and utilized to collect ampicillin-resistant conjugative plasmid through conjugation. The mutant strain was an E. coli MG1655 strain with an entA deletion (MP10LA; Table 1). Because the entA gene is essential for enterobactin synthesis (Liu et al., 1989), MP10LA lacked the enterobactin-dependent iron uptake system and thus exhibited growth defects in iron-deficient medium. MP10LA served as the recipient to capture ampicillin-resistant plasmids through conjugation from 50 donor E. coli clinical isolates that were associated with bacteremia or UTIs. Thirty-six transconjugants were obtained from the conjugations. The plasmids that were able to promote the growth MP10LA under iron-limited conditions are likely to harbor iron uptake-related genes. Thus, these transconjugants were subjected to growth promotion assays in iron-limited medium (ILM; see section “Materials and Methods”) agar. Among them, the transconjugant ELA41 (Table 1) showed an increased iron uptake ability (Figures 1A,B). We further investigated the plasmid patterns of ELA41 and its corresponding donor clinical isolate EC41. The plasmids were extracted from the strains using the method described previously (Kado and Liu, 1981) (please check section “Materials and Methods”) and subjected to agarose gel electrophoresis. As shown in Figure 1C, the plasmid extract of ELA41 exhibited one band with a size ranging from 50 to 90 kb on the gel, while that of EC41 showed multiple bands. The band in ELA41 showed a size similar to that of one of the bands in EC41. These findings suggest that the transconjugant ELA41 acquired one plasmid from EC41. The plasmid was named pEC41. The multiple bands in EC41 suggest that the isolate harbor multiple plasmids. However, some of the bands may be resulted from the fragmentation of plasmid DNA at the time of extraction.
FIGURE 1.
The growth of the transconjugant ELA41 under iron-limited conditions and the plasmid the transconjugant obtained through conjugation. (A) Growth promotion assays of ELA41 and MP10LA with Fe2+ (ferrous sulfate) and Fe3+ (ferric chloride). Iron-limited medium (ILM) agar was seeded with ELA41 and MP10LA. Wells with a diameter of 4 mm were punched into agar plates and filled with 10 μl of 10 mM ferrous sulfate and ferric chloride. The diameters of growth zones around the wells were determined after 24 h incubation at 37°C. Bars indicate the diameters of the growth zones. (B) Quantification of the diameters of the growth zones of ELA41 and MP10LA. Dashed lines indicate the diameter of the wells (4 mm) containing iron sources. The experiments were performed in triplicate and presented as the means ± SD. (C) The plasmid profiles of EC41, ELA41, IP10/pEC41, and MP10/pEC41. The plasmids were extracted using the method describe by Kado and Liu (1981). The plasmid extracts without any restriction enzyme digestion were subjected to agarose gel electrophoresis. Therefore, the plasmids in the gel were in a circular form. M: Fifty- and 90-kb plasmid markers. The markers were extracted from Salmonella choleraesuis OU7526 (Tzeng et al., 2012). (D,E) Quantitative analysis of the growth promotion assays of IP10 (D) and MP10 (E) strains with and without pEC41. The experiments were performed in triplicate and are presented as the means ± SD. *P < 0.05 (Wilcoxon rank-sum test).
Two sequential conjugation experiments were performed to determine whether pEC41 and the increased iron uptake ability in ELA41 were able to be conjugatively transferred to other bacterial hosts. First, ELA41 served as the donor strain, and an MG1655 entA mutant-derived strain, IP10 (Table 1), served as the recipient strain. The resulting transconjugant was designated IP10/pEC41. Then, IP10/pEC41 served as the donor strain, and another MG1655 entA mutant-derived strain, MP10 (Table 1), served as the recipient strain. The resulting transconjugant was MP10/pEC41. Similar to ELA41, IP10/pEC41, and MP10/pEC41 harbored pEC41 (Figure 1C), and these transconjugant showed increased iron uptake abilities in the growth promotion assay compared to their corresponding recipient strains (Figures 1D,E). These findings suggest that pEC41 can be transferred though conjugation independent of its original host E. coli (EC41) background and that the iron uptake ability is transferred with the plasmid.
The pEC41 Plasmid Belongs to the IncM Plasmid Groups That Have Been Shown to Carry Multiple Antimicrobial Resistance Genes, Including ESBL-Encoding Genes
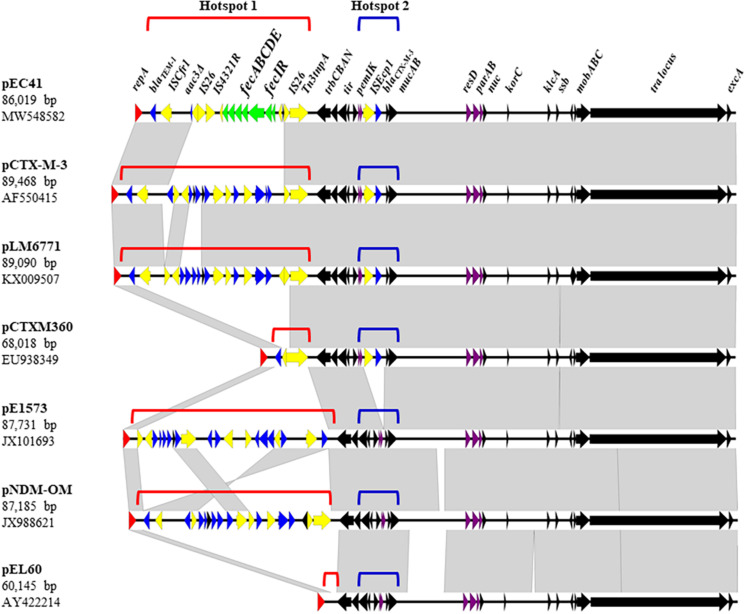
The pEC41 plasmid was purified from ELA41 and subjected to DNA sequencing. This plasmid was 86,019 bp in size with an average G + C content of 52.37%. It harbored 112 potential ORFs, which included a complete array of genes involved in plasmid replication, conjugation and stability, along with a number of mobile genetic elements (Figure 2, Supplementary Table 2, and Supplementary Figure 1). Based on an in silico analysis using the web tool PlasmidFinder-2.11 (Carattoli et al., 2014), the pEC41 plasmid belongs to the IncM plasmid groups. Based on BLAST analysis, the backbone structure of pEC41 was highly similar to the backbone structures of a group of multiple antimicrobial resistance genes (ARGs)-bearing IncM plasmids (Carattoli et al., 2015), such as pCTX-M3 from Citrobacter freundii (85% query coverage; 99% identity) (Golebiewski et al., 2007), pLM6771 from E. coli (84% query coverage; 99% identity) (Gong et al., 2019), pCTXM360 from Klebsiella. pneumoniae (77% query coverage; 99% identity) (Zhu et al., 2009), pEl1573 from Enterobacter cloacae (80% query coverage; 99% identity) (Partridge et al., 2012), and pNDM-OM from K. pneumoniae (81% query coverage; 99% identity) (Bonnin et al., 2013). The most conserved parts among these plasmids were the regions encoding the transfer loci (tra), the replicon function (repA), the partitioning factors (parA and parB), the postsegregational killing system (pemI and pemK), and a large number of hypothetical proteins (Figure 2, Supplementary Table 2, and Supplementary Figure 1) (Golebiewski et al., 2007). These plasmids exhibited two insertion hotspots (Hotspot 1 and Hotspot 2) that contained different acquired sequences. These plasmids have been proposed to have evolved from the Erwinia amylovora plasmid pEL60, which harbors no antimicrobial resistance gene and no insertion sequence in the hot spots (Bonnin et al., 2013) (Figure 2).
FIGURE 2.
Linear comparison of pEC41 with other IncM group plasmids. Black arrows indicate the common regions shared among the plasmids. The tra locus contains tra genes involved in conjugal transfer. The partition system (parAB) and postsegregational killing system (pemIK) are indicated by purple arrows. Resistance genes are indicated by blue arrows. Insertion sequences and transposon components are indicated by yellow arrows. The fec system on pEC41 is indicated with green arrows. The replicase gene repA is indicated by red arrows. The direction of arrows indicates the direction of the ORF. Gray shading denotes regions of shared homology among different plasmids. Diagrams were drawn from sequences available under the following GenBank accession numbers: pEC41 (MW548582), pCTX-M-3 (AF550415), pLM6771 (KX009507), pCTXM360 (EU938349), pEl1573 (JX101693), pNDM-OM (JX988621), and pEL60 (AY422214). The comparison map was generated using Easyfig software (Sullivan et al., 2011).
Hotspot 1 was located between the replication locus (rep) and the Trb transfer operon (trbCBAN) (Figure 2). In pEC41, a fec system (fecIRABCDE) was identified in this locus, which may be responsible for the iron uptake ability. The fec system encoded in this plasmid was designated fecpEC41 in this study. In pCTXM360, a Tn3 transposon that carries the broad-spectrum β-lactamase-encoding gene blaTEM–1 (Ghafourian et al., 2015) was located in the hotspot. In pEC41, pCTX-M3, pLM6771, and pNDM-OM, IS-mediated insertions (IS26 and/or ISCfr1) occurred in Tn3, leading to the truncation of a part of the Tn3 structure with the blaTEM–1 genes remaining in the loci. The IS insertions brought extra antimicrobial resistance genes into pCTX-M3, pLM6771, and pNDM-OM and brought fecpEC41 into pEC41.
Hotspot 2 was located between pemIK and the UV resistance genes (mucAB), in which the insertion sequence ISEcp1with the ESBL-encoding gene blaCTX–M–3 (Golebiewski et al., 2007) was inserted in four of the six multiple ARGs-bearing plasmids (Figure 2). The mobilization of blaCTX–M–3 gene may occur with the assistance of the ISEcp1 element, which were commonly found 127 bp upstream of the antimicrobial resistance gene (Literacka et al., 2009; Zong et al., 2010). On the other hand, pEl1573 and pNDM-OM lacked the ISEcp1-blaCTX–M–3 sequence at this locus.
Taken together, the plasmid pEC41 carried fecpEC41 and blaTEM–1 in the hot spot 1 and harbored blaCTX–M–3 in the hot spot 2.
The fec System in pEC41 Enhances the Iron Uptake Ability of Iron Uptake-Deficient Strains
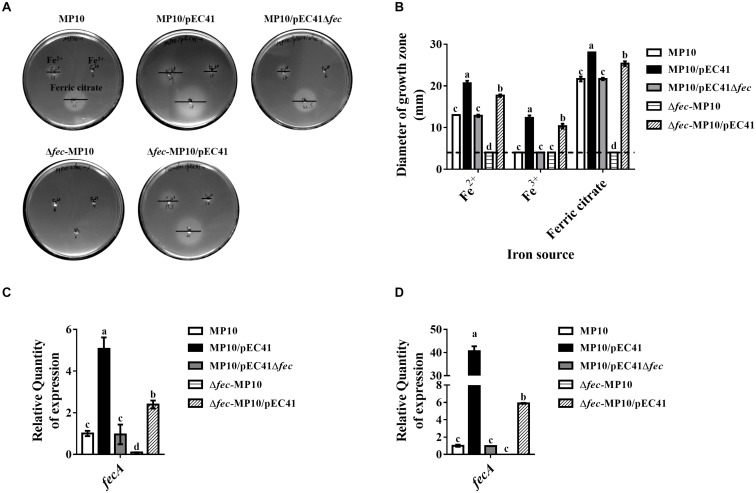
The fecpEC41 sequence was deleted in pEC41 (pEC41Δfec) and the effect of the deletion on iron uptake by bacteria was evaluated to investigate whether fecpEC41 is responsible for conferring the iron uptake ability. Since MG1655 and its derived strains chromosomally encode a copy of the fec system that shares 94% identity with the fecpEC41 system (data not shown), the chromosomal copy of the fec system in the MG1655-derived strains was designated as fecMG1655 to differentiate it from fecpEC41. The E. coli strains MP10, MP10/pEC41, MP10/pEC41Δfec, Δfec-MP10/pEC41, and Δfec-MP10/pEC41Δfec (Table 1), which harbored fecMG1655, fecpEC41/fecMG1655, fecMG1655, fecpEC41, and no fec system, respectively, were evaluated for their abilities to utilize ferrous sulfate (Fe2+), ferric chloride (Fe3+) and ferric citrate. MP10/pEC41 exhibited larger growth zones around the iron sources than MP10/pEC41Δfec (Figures 3A,B), indicating that fecpEC41 is responsible for the iron uptake ability encoded in pEC41. MP10/pEC41, which harbored 2 copies of the fec system, exhibited the largest growth zones around the iron sources, while the strains harboring one copy of the fec system (MP10, MP10/pEC41Δfec, and Δfec-MP10/pEC41) showed larger growth zones around ferrous sulfate and ferric citrate than the strain without any fec system (Δfec-MP10; Table 1). Based on these results, fecMG1655 and fecpEC41 exert additive effects on the iron uptake ability.
FIGURE 3.
The fec system of pEC41 provides iron uptake abilities. (A) Iron growth promotion assays of MP10 strains harboring pEC41 or pEC41Δfec with and without the chromosomal deletion of fecMG1655. ILM agar plates were seeded with the indicated strains, and three punched wells on each plate were loaded with 10 μl of 10 mM ferrous sulfate (Fe2+; top left panel), ferric chloride (Fe3+; top right panel) and ferric citrate (bottom panel). The diameters of growth zones around the wells were determined after 24 h of incubation at 37°C. Bars indicated the diameters of the growth zones. (B) Quantification of the diameters of the growth zones of the strains in (A). Dashed lines indicate the diameter of the wells (4 mm) of iron sources. (C,D) The mRNA levels of fecA in strains grown under iron-limited conditions without or with supplementation with ferric citrate. The bacteria were grown in M9 medium containing 100 μM DIP without (C) and with (D) supplementation of 20 μM ferric citrate overnight at 37°C. Levels of the fecA transcript were determined with real-time PCR using primers able to recognize the fecA genes in fecpEC41 and fecMG1655. The fecA transcript level in each strain was normalized to the ftsZ level and presented as the relative level compared to that of MP10. The experiments were performed in triplicate and presented as the means ± SD. Different letters marked above the bars indicate groups with significant differences (P < 0.05, one-way ANOVA).
We investigated whether the expression levels of the fecABCDE operons in these strains were correlated with their iron uptake abilities. Under iron-depleted conditions (M9 medium with 100 μM DIP) with or without ferric citrate, the total fecA mRNA levels, including the mRNA derived from fecpEC41 and fecMG1655, in MP10/pEC41 were significantly higher than those in the strains harboring either fecMG1655 or fecpEC41 (Figures 3C,D). The Δfec-MP10/pEC41 strain showed higher levels of expression than MP10/pEC41Δfec and MP10, suggesting that fecpEC41 may provide a higher level of fecA expression than fecMG1655. In addition, supplementation with ferric citrate significantly increased fecA expression, especially in MP10/pEC41 cells (Figures 3C,D). Therefore, the expression of the fecABCDE genes is consistent with the iron uptake abilities of these strains.
In addition, to investigate whether fecpEC41 contributes to EC41 survival in ILM, the fecpEC41 deletion mutant of EC41 (ΔfecpEC41-EC41) was constructed by conjugatively transferring pEC41Δfec into EC41 to compete with and thus replace pEC41 (Table 1). Then, the EC41 strains with and without fecpEC41 were subjected to iron growth promotion assays. Unlike the MP10 strains, both EC41 and ΔfecpEC41-EC41 grew in ILM agar without additional iron supplementation (data not shown), suggesting that fecpEC41 may not play a major role in EC41 growth in ILM.
The pEC41 Plasmid Is Stable in EC41, MP10, and the Cystitis-Associated Isolate A487, but Not the UPEC Strains CFT073str and UTI89str
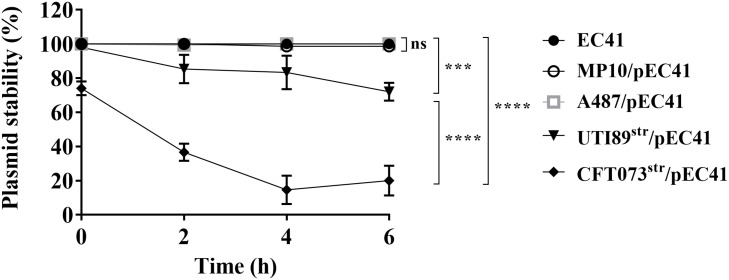
To determine whether pEC41 is stably maintained in different E. coli strains, the stabilities of the plasmid in EC41, MP10, the prototype UPEC strains UTI89 and CFT073 (Welch et al., 2002; Chen et al., 2006), and a cystitis-associated clinical isolate, A487 (Table 1), were assessed. A487 acquired pEC41 through transformation to become A487/pEC41. CFT073 and UTI89 encode no known antimicrobial resistance genes as markers for conjugation experiments, we utilized spontaneous streptomycin-resistant mutants of the strains (CFT073str and UTI89str; Table 1) to conjugatively acquire pEC41 from EC41, resulting in CFT073str/pEC41 and UTI89str/pEC41 (Table 1). After an incubation in LB medium for different periods, the percentages of pEC41-harboring cells in EC41, MP10/pEC41, A487/pEC41, CFT073str/pEC41, and UTI89str/pEC41 cultures were determined. As shown in Figure 4, pEC41 was stably maintained in MP10 and the clinical E. coli isolates EC41 and A487. However, the percentages of pEC41-containing cells in CFT073str/pEC41 and UTI89str/pEC41 cultures decreased significantly during incubation. These findings suggest that the stability of pEC41 depends on its host backgrounds. In addition, since CFT073 harbors no plasmid and UTI89 harbors an IncFIB/IIA plasmid that is not in the same incompatibility group as pEC41 (Welch et al., 2002; Chen et al., 2006), the instability of pEC41 in the strains was not likely due to plasmid interference from the bacterial hosts.
FIGURE 4.
The stability of pEC41 in different E. coli host strains. The stability of pEC41 in EC41, MP10, A487, UTI89str, and CFT073str was evaluated. The percentage of pEC41-containing cells was determined by measuring the percentage of ampicillin-resistant cells at the indicated time points. The experiments were performed in triplicate and are presented as the means ± SD. ***P < 0.001, ****P < 0.0001; ns, no significant difference (one-way ANOVA).
fecpEC41 Contributes to the Virulence of EC41 in Systemic Infections
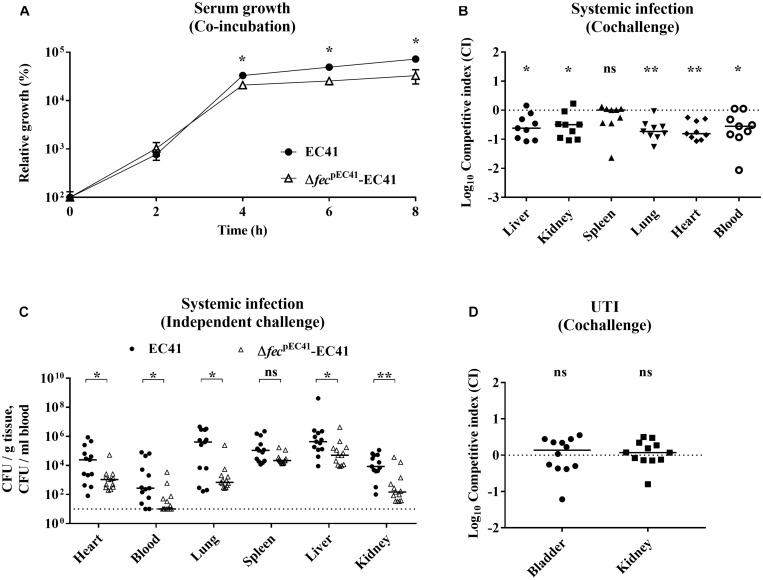
EC41, which was assigned to sequence type (ST) 405, phylogroup D, and fimH27, was isolated from the bloodstream of a patient, suggesting that this pathogen causes systemic infections. We evaluated whether fecpEC41 contributes to EC41 survival in serum, in which the bioavailability of iron is restricted (Hood and Skaar, 2012). In coincubation experiments, EC41 outcompeted ΔfecpEC41-EC41 in heat-inactivated serum (Figure 5A). Based on these results, fecpEC41 confers a fitness advantage in terms of the serum survival of EC41 and fecpEC41 may contribute to the pathogenesis of E. coli.
FIGURE 5.
The roles of fecpEC41 in EC41 growth in heat-inactivated serum and in the pathogenesis of systemic infections and urinary tract infections. (A) The growth of EC41 and ΔfecpEC41-EC41 cocultured in 90% heat-inactivated mouse serum. The results are presented as the means ± SD. The data are shown as the relative growth rates compared to the bacterial counts of the original inoculums. The data are representative of three independent experiments performed in triplicate. (B) Cochallenge of EC41 and ΔfecpEC41-EC41 in a mouse model of systemic infection. Equal numbers of EC41 and ΔfecpEC41-EC41 were coinoculated into BALB/c mice via the tail vein (N = 9). At 24 h postinfection, the bacterial numbers of each strain in the liver, kidney, spleen, lung, heart, and blood were determined, and the competitive indices (CI) of ΔfecpEC41-EC41 vs. EC41 in these tissues were calculated. (C) Independent challenge with EC41 and ΔfecpEC41-EC41 in the mouse model of systemic infection. The bacterial counts in the organs and blood were determined at 24 h postinfection. (N = 13 for the animals infected with each bacterial strain). (D) Cochallenge of EC41 and ΔfecpEC41-EC41 in a mouse model of UTIs. Equal numbers of the two strains were transurethrally coinoculated into female C3H/HeN mice (N = 12). At 24 h postinfection, the bacterial counts and the CI of ΔfecpEC41-EC41 vs. EC41 in bladders and kidneys were determined. The horizontal bars indicate median values. For (C), the dashed line represents the detection limit. For (B,D), the dashed lines represent the log10 scale of CI = 1. The CI in the cochallenge experiments was calculated using bacterial counts with the equation [(ΔfecpEC41-EC41 CFU/EC41 CFU)output/(ΔfecpEC41-EC41 CFU/ΔfecpEC41-EC41 CFU)input]. The log10 scale of CI values less than 0 (CI = 1) indicates a comparative defect in ΔfecpEC41-EC41. The paired two-tailed student’s t test and The Mann–Whitney U test was used for the statistical analyses of (A,C). The Wilcoxon signed-rank test was used in (B,D). *P < 0.05, **P < 0.01; ns, no significant difference.
The ability of ΔfecpEC41-EC41 to cause systemic infection was compared to that of EC41 to further determine whether fecpEC41 facilitates E. coli infection. In cochallenge experiments, equal numbers of EC41 and ΔfecpEC41-EC41 were intravenously coinoculated into mice. As shown in Figure 5B, at 24 h postinfection, ΔfecpEC41-EC41 was outcompeted by EC41 in the liver, kidneys, lungs, heart, and blood. Similarly, in independent challenge experiments, mice infected with EC41 showed significantly higher bacterial burdens in the organs and blood than those infected with ΔfecpEC41-EC41 (Figure 5C). Thus, fecpEC41 contributes to the pathogenesis of EC41 in systemic infections.
In addition, we examined whether fecpEC41 contributed to UTIs. EC41 and ΔfecpEC41-EC41 were intraurethrally coinoculated into mice. However, similar bacterial counts of the wild-type and mutant strains were observed in the bladders and kidneys at 24 h postinfection (Figure 5D), suggesting that fecpEC41 may not play a significant role in EC41-mediated UTIs.
fecpEC41 Promotes the Fitness of A487 During Urinary Tract Infections
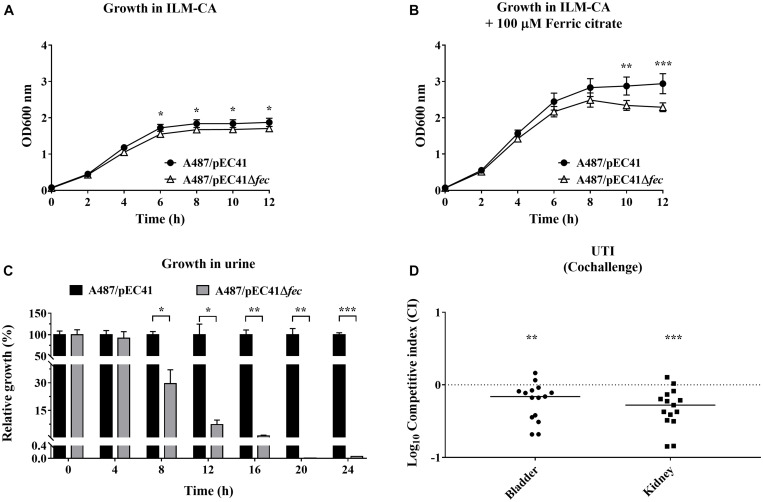
pEC41 could be stably maintained by A487 cells. We further investigated whether fecpEC41 contributes to the host’s abilities to grow in an iron-limited condition and to cause infection when A487 obtains pEC41. The growth of A487/pEC41was compared with that of A487/pEC41Δfec. Since these strains were unable to grow on ILM agar with and without iron supplementation (data not shown), we measured their growth in ILM broth supplemented with 1% casamino acid as a carbon source (ILM-CA). A487/pEC41 showed higher levels of growth than A487/pEC41Δfec in ILM-CA and in ILM-CA supplemented with ferric citrate (Figures 6A,B). Supplementation with ferric citrate increased the difference in growth between the strains with and without fecpEC41. These findings suggested that fecpEC41 significantly contributes to A487 growth under iron-limited conditions.
FIGURE 6.
The effect of fecpEC41 on the growth of A487 under iron-limited conditions and in urine and on the ability of the bacteria to colonize urinary tracts. (A) The growth of A487/pEC41 and A487/pEC41Δfec cultured in ILM-CA. (B) The growth of A487/pEC41 and A487/pEC41Δfec cultured in ILM-CA supplemented with 100 μM ferric citrate. (C) The growth of A487/pEC41 and A487/pEC41Δfec cocultured in urine. The results are shown as the relative growth rates compared to A487/pEC41 at each indicated time point. For (A–C), the results are presented as the means ± SD. The data are representative of three independent experiments performed in triplicate. (D) Cochallenge of A487/pEC41 and A487/pEC41Δfec in a mouse model of UTI. Equal numbers of the two strains were transurethrally coinoculated into female C3H/HeN mice (N = 15). At 24 h postinfection, the bacterial counts and the CI of A487/pEC41Δfec vs. A487/pEC41 in bladders and kidneys were determined. The horizontal bars indicate median values. The dashed line represents the log10 scale of CI = 1. The two-way ANOVA was used in the statistical analysis of (A,B). The paired two-tailed student’s t-test was used in the statistical analysis of (C), while the Wilcoxon signed-rank test was used in (D). *P < 0.05, **P < 0.01, ***P < 0.001.
Because A487 was associated with UTIs, the growth of A487/pEC41 and A487/pEC41Δfec in the urine was evaluated. Equal amounts of A487/pEC41 and A487/pEC41Δfec were coinoculated in 90% human urine and incubated at 37°C. At intervals of 4 h, the bacterial counts of the two strains were determined, and the cultures were transferred to fresh 90% urine in a 1:250 dilution for the next 4 h of incubation. As shown in Figure 6C, A487/pEC41 significantly outcompeted A487/pEC41Δfec after 12 h of incubation in urine. Based on these findings, the fec system optimizes the fitness of A487 growing in urine.
Furthermore, we investigated whether fecpEC41 facilitates A487 colonization of the urinary tract. A487/pEC41 and A487/pEC41Δfec were coinoculated into the urinary tract of mice. At 24 h postinfection, the bacterial loads of A487/pEC41 outcompeted those of A487/pEC41Δfec in bladders and kidneys (Figure 6D), suggesting that fecpEC41 increases the fitness of A487 to infect urinary tracts.
The Distribution of the fec System in Fecal and Clinical E. coli Isolates
Because the fec system increased the virulence or fitness of E. coli to cause infections (Figures 5C, 6D), we scrutinized whether the distribution of the system was associated with E. coli infections. We determined the distribution of the fec system by performing PCR analysis with primers that probe the fecA gene (the first gene in the fecABCDE operon of the fec system). The primers were designed to target the conserved region of the fecA gene. Therefore, the fecA alleles of fecpEC41 and fecMG1655 were both detected in the PCR-based analysis. The fecA frequencies in commensal fecal E. coli and UTI- and bacteremia- associated clinical isolates were investigated. As shown in Table 2, the cystitis-, pyelonephritis-, and bacteremia- associated E. coli groups showed significantly higher frequencies of fecA than the fecal isolates. These findings suggest that the fec system is positively associated with E. coli infections in humans and thus suggest their pathogenic roles in bacterial infections in humans.
TABLE 2.
Distribution of fecA in different source groups of E. coli isolates.
| No. (%) of E. coli isolates |
P-valuea |
||||||
| Gene | Fecal isolates (N = 87) | Cystitis isolates (N = 67) | Pyelonephritis isolates (N = 72) | Bacteremia isolates (N = 43) | Fecal vs. cystitis isolates | Fecal vs. pyelonephritis isolates | Fecal vs. bacteremia isolates |
| fecA | 22 (25.29) | 37 (55.22) | 40 (55.56) | 22 (51.62) | <0.001 | <0.001 | 0.005 |
aFisher’s exact test.
Discussion
The present study identified a novel conjugative plasmid, pEC41, which was able to promote the iron uptake ability of E. coli, thus contributing to the pathogenesis of systemic E. coli infections and conferring fitness to pathogenic E. coli during UTIs. The pEC41 plasmid belongs to the IncM plasmid groups that have been shown to carry multiple ARGs, including ESBL-encoding genes (Figure 2) (Carattoli et al., 2015). To our knowledge, pEC41 is the first IncM group plasmid identified to be able to confer virulence and multiple ARGs (Carattoli et al., 2015). The fec system encoded on pEC41 was responsible for promoting the iron uptake ability and virulence of host bacteria. Notably, this study is the first to show that the fec iron uptake system contributes to bacterial infections, although it has been previously known to be involved in iron uptake (Braun and Mahren, 2005). Consistent with these findings, our epidemiological investigation showed the association of this system with human bacteremia and UTIs, which supported its role as a bacterial virulence factor in human infections. Thus, the novel plasmid pEC41, which can disseminate virulence and antimicrobial resistance genes though conjugation among bacterial hosts, is a potential threat to public health.
The pEC41 plasmid is highly similar to the well-studied broad-host-range IncM plasmid pCTX-M3, a plasmid isolated from C. freundii (Gniadkowski et al., 1998; Baraniak et al., 2002; Golebiewski et al., 2007; Mierzejewska et al., 2007), in which the ESBL-encoding gene blaCTX–M–3 was originally identified (Golebiewski et al., 2007). pCTX-M3 is a highly conjugative plasmid that can be transferred to solid and liquid media at the same frequency (Golebiewski et al., 2007). This plasmid is proposed to be responsible for the broad dissemination of blaCTX–M–3 in clinical isolates of Enterobacteriaceae in Poland (Baraniak et al., 2002; Golebiewski et al., 2007; Carattoli, 2009). The conjugative transfer system of pCTX-M3 allows plasmid transfer between the soil bacterium Agrobacterium tumefaciens and E. coli (Golebiewski et al., 2007; Mierzejewska et al., 2007), suggesting that environmental bacteria may serve as a reservoir of these plasmids. The backbone used for the transfer and replication of pEC41 is approximately 100% identical to that of pCTX-M3, suggesting that pEC41 is a plasmid with a broad host range that can effectively transfer among clinically relevant and environmental bacteria. An investigation of the distribution of pEC41 among distinct bacterial pathogens and environmental bacteria would facilitate the evaluation of the plasmid’s threat to public health.
The contribution of the fec system to virulence and fitness of E. coli is considered niche-dependent and strain-specific because fecpEC41 facilitated EC41 virulence in systemic infections but not UTIs (Figures 5C,D), while it conferred fitness to A487 in UTIs (Figure 6D). This discrepancy may be because bacterial pathogens always harbor multiple distinct iron uptake systems that may unequally contribute to infections in a specific environment or tissue (functional hierarchy) (Garcia et al., 2011) and because different E. coli strains may harbor distinct combinations of iron uptake systems (Welch et al., 2002; Brzuszkiewicz et al., 2006). Iron bioavailability is restricted to invading bacterial pathogens in blood, inner organ tissues, and urinary tracts (Snyder et al., 2004; Skaar, 2010). Although fecpEC41 plays an important role in systemic EC41 infections, some other iron uptake system(s) in EC41 may play a more dominant role than fecpEC41 during UTIs, concealing the contribution of the fec system. On the other hand, fecpEC41 may have a relatively higher functional hierarchy in A487 cells and thus confers competitive fitness in the urinary tract.
The finding that the fec system contributed to the survival of E. coli in the serum and urine suggests that invading pathogenic bacteria may utilize ferric citrate as an iron source in the body. Accordingly, an increase in the availability of ferric citrate in the host may increase the risk of infections caused by bacteria that harbor the fec system. The dissemination of the fec system would be a potential clinical concern because citrate-based compounds have been utilized as medicines to manage some symptoms associated with bacterial infections. For example, ferric citrate has been utilized to treat hyperphosphatemia in patients with chronic kidney diseases (CKDs) who are easily complicated with UTIs (Naqvi and Collins, 2006; Pennoyer and Bridgeman, 2015). Additionally, citrate compounds have been prescribed as alkalinizers for patients with UTIs to relieve the pain of cystitis (Ueda et al., 2014). Citrate-based medicine may interact with ferric iron and form ferric citrate to serve as an iron source for pathogens in the body. Consequently, treatment with citrate-based medicines may facilitate the survival of invading bacterial pathogens that harbor the fec system and thus facilitate the spread of fec-encoding plasmids, such as pEC41. The impact of the usage of citrate-based medicine on bacterial infections and the spread of these plasmids must be monitored.
The MP10 strain solely harboring fecpEC41 (Δfec-MP10/pEC41) exhibited higher levels of iron uptake and fecA transcripts than the strains solely harboring fecMG1655 (MP10 and MP10/pEC41Δfec) (Figure 3). These findings suggest that fecpEC41 confers a higher level of the fecA transcript (i.e., a higher transcript level of the fecABCDE operons) and thus contributes a higher level of iron uptake ability than fecMG1655. Because fecpEC41 is encoded on pEC41, while fecMG1655 is located on the chromosome, the copy numbers of pEC41 are likely higher than those of the chromosome, consequently resulting in the presence of multiple copies of fecpEC41 and a higher level of fecA expression in the bacteria. Alternatively, since the sequences of fecpEC41 and fecMG1655 are not entirely identical (94% identity), the minor difference in their sequences may cause differences in the promoter activities and/or mRNA stability of the fecABCDE operons in the two fec system variants, thus resulting in different fecA mRNA levels and iron uptake abilities.
Conclusion
In the present study, we identified a novel conjugative plasmid, pEC41, whose backbone was almost identical to the multiple ARGs-bearing plasmid pCTX-M3, which is highly conjugative and is potentially transmissible between clinical and environmental bacterial species. pEC41 harbored a fec iron uptake system that was absent in pCTX-M3. The fec system increases the virulence and competitive fitness of E. coli in systemic infections and UTIs, respectively. Due to its properties of efficient and broad spread of both antimicrobial resistance and virulence genes, pEC41 deserves further study on its distribution in environmental and clinical bacteria and on the impact of these plasmids on public health.
Data Availability Statement
The sequences of pEC41 are deposited in the GenBank repository, accession number MW548582.
Ethics Statement
All of the animal experimental procedures were reviewed and approved by The Institutional Animal Care and Use Committee (IACUC) of National Cheng Kung University, Tainan City, Taiwan (approval no. 108130). The studies involving human participants and bacteria collection were reviewed and approved by The Institutional Reviewer Board (IRB) of National Cheng Kung University Hospital, Tainan City, Taiwan (approval no. A-ER-107-403 and A-ER-104-313). The patients/participants provided their written informed consent to participate in this study.
Author Contributions
W-CH, M-YW, and S-HW carried out the experiments in this study. MH, Y-LC, and C-HT contributed to the study conception, planning experiments, data analysis, and interpretation. M-HL, M-FL, and Y-LC contributed the materials and technical supports of the animal experiments. J-JW, S-LJ, and J-LW participated in conceptualization and interpretation of the data. M-CW, W-HL, and C-HT contributed materials and technical support. W-CH, M-YW, S-HW, Y-LC, and C-HT wrote the manuscript. All authors contributed to the article and approved the submitted version.
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
We thank the Center of Allergy and Clinical Immunology Research (ACIR) and the Headquarters of University Advancement at the National Cheng Kung University, which is sponsored by the Ministry of Education in Taiwan, for assisting the publication of this study.
Funding. This study was supported by the Ministry of Science and Technology, Taiwan (MOST 106-2320-B-006-032, MOST 108-2320-B-017-002-MY3, MOST 108-2320-B-006-034-MY3, and MOST 109-2320-B-006-060). The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fmicb.2021.667782/full#supplementary-material
References
- Baraniak A., Fiett J., Sulikowska A., Hryniewicz W., Gniadkowski M. (2002). Countrywide spread of CTX-M-3 extended-spectrum beta-lactamase-producing microorganisms of the family Enterobacteriaceae in Poland. Antimicrob. Agents Chemother. 46 151–159. 10.1128/aac.46.1.151-159.2002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beghain J., Bridier-Nahmias A., Le Nagard H., Denamur E., Clermont O. (2018). ClermonTyping: an easy-to-use and accurate in silico method for Escherichia genus strain phylotyping. Microb. Genom. 4:e000192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bonnin R. A., Nordmann P., Carattoli A., Poirel L. (2013). Comparative genomics of IncL/M-type plasmids: evolution by acquisition of resistance genes and insertion sequences. Antimicrob. Agents Chemother. 57 674–676. 10.1128/aac.01086-12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bonten M., Johnson J. R., Van Den Biggelaar A. H. J., Georgalis L., Geurtsen J., De Palacios P. I., et al. (2020). Epidemiology of Escherichia coli bacteremia: a systematic literature review. Clin. Infect. Dis. 72 1211–1219. 10.1093/cid/ciaa210 [DOI] [PubMed] [Google Scholar]
- Braun V., Mahren S. (2005). Transmembrane transcriptional control (surface signalling) of the Escherichia coli Fec type. FEMS Microbiol. Rev. 29 673–684. 10.1016/j.femsre.2004.10.001 [DOI] [PubMed] [Google Scholar]
- Brzuszkiewicz E., Bruggemann H., Liesegang H., Emmerth M., Olschlager T., Nagy G., et al. (2006). How to become a uropathogen: comparative genomic analysis of extraintestinal pathogenic Escherichia coli strains. Proc. Natl. Acad. Sci. U.S.A. 103 12879–12884. 10.1073/pnas.0603038103 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carattoli A. (2009). Resistance plasmid families in Enterobacteriaceae. Antimicrob. Agents Chemother. 53 2227–2238. 10.1128/aac.01707-08 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carattoli A., Seiffert S. N., Schwendener S., Perreten V., Endimiani A. (2015). Differentiation of IncL and IncM plasmids associated with the spread of clinically relevant antimicrobial resistance. PLoS One 10:e0123063. 10.1371/journal.pone.0123063 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carattoli A., Zankari E., Garcia-Fernandez A., Voldby Larsen M., Lund O., Villa L., et al. (2014). In silico detection and typing of plasmids using PlasmidFinder and plasmid multilocus sequence typing. Antimicrob. Agents Chemother. 58 3895–3903. 10.1128/aac.02412-14 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen S. L., Hung C. S., Xu J., Reigstad C. S., Magrini V., Sabo A., et al. (2006). Identification of genes subject to positive selection in uropathogenic strains of Escherichia coli: a comparative genomics approach. Proc. Natl. Acad. Sci. U.S.A. 103 5977–5982. 10.1073/pnas.0600938103 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Costello L. C., Franklin R. B. (2016). Plasma citrate homeostasis: how it is regulated; and its physiological and clinical implications. an important, but neglected, relationship in medicine. HSOA J. Hum. Endocrinol. 1:5. [PMC free article] [PubMed] [Google Scholar]
- Cusumano C. K., Hung C. S., Chen S. L., Hultgren S. J. (2010). Virulence plasmid harbored by uropathogenic Escherichia coli functions in acute stages of pathogenesis. Infect. Immun. 78 1457–1467. 10.1128/iai.01260-09 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Datsenko K. A., Wanner B. L. (2000). One-step inactivation of chromosomal genes in Escherichia coli K-12 using PCR products. Proc. Natl. Acad. Sci. U.S.A. 97 6640–6645. 10.1073/pnas.120163297 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garcia E. C., Brumbaugh A. R., Mobley H. L. (2011). Redundancy and specificity of Escherichia coli iron acquisition systems during urinary tract infection. Infect. Immun. 79 1225–1235. 10.1128/iai.01222-10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ghafourian S., Sadeghifard N., Soheili S., Sekawi Z. (2015). Extended spectrum Beta-lactamases: definition. classification and epidemiology. Curr. Issues Mol. Biol. 17 11–21. [PubMed] [Google Scholar]
- Gniadkowski M., Schneider I., Palucha A., Jungwirth R., Mikiewicz B., Bauernfeind A. (1998). Cefotaxime-resistant Enterobacteriaceae isolates from a hospital in Warsaw, Poland: identification of a new CTX-M-3 cefotaxime-hydrolyzing beta-lactamase that is closely related to the CTX-M-1/MEN-1 enzyme. Antimicrob. Agents Chemother. 42 827–832. 10.1128/aac.42.4.827 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Golebiewski M., Kern-Zdanowicz I., Zienkiewicz M., Adamczyk M., Zylinska J., Baraniak A., et al. (2007). Complete nucleotide sequence of the pCTX-M3 plasmid and its involvement in spread of the extended-spectrum beta-lactamase gene blaCTX-M-3. Antimicrob. Agents Chemother. 51 3789–3795. 10.1128/aac.00457-07 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gong J., Zeng X., Zhang P., Zhang D., Wang C., Lin J. (2019). Characterization of the emerging multidrug-resistant Salmonella enterica serovar Indiana strains in China. Emerg. Microbes Infect. 8 29–39. 10.1080/22221751.2018.1558961 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hashimoto M., Kato J. (2003). Indispensability of the Escherichia coli carbonic anhydrases YadF and CynT in cell proliferation at a low CO2 partial pressure. Biosci. Biotechnol. Biochem. 67 919–922. 10.1271/bbb.67.919 [DOI] [PubMed] [Google Scholar]
- Hood M. I., Skaar E. P. (2012). Nutritional immunity: transition metals at the pathogen-host interface. Nat. Rev. Microbiol. 10 525–537. 10.1038/nrmicro2836 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hsu P. C., Chen C. S., Wang S., Hashimoto M., Huang W. C., Teng C. H. (2020). Identification of MltG as a Prc protease substrate whose dysregulation contributes to the conditional growth defect of Prc-deficient Escherichia coli. Front. Microbiol. 11:2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang W. C., Liao Y. J., Hashimoto M., Chen K. F., Chu C., Hsu P. C., et al. (2020). cjrABC-senB hinders survival of extraintestinal pathogenic E. coli in the bloodstream through triggering complement-mediated killing. J. Biomed. Sci. 27:86. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ilangovan A., Connery S., Waksman G. (2015). Structural biology of the gram-negative bacterial conjugation systems. Trends Microbiol. 23 301–310. 10.1016/j.tim.2015.02.012 [DOI] [PubMed] [Google Scholar]
- Johnson T. J., Lang K. S. (2012). IncA/C plasmids: an emerging threat to human and animal health? Mob. Genet. Elements 2 55–58. 10.4161/mge.19626 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jolley K. A., Bray J. E., Maiden M. C. J. (2018). Open-access bacterial population genomics: BIGSdb software, the PubMLST.org website and their applications. Wellcome Open Res. 3:124. 10.12688/wellcomeopenres.14826.1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kado C. I., Liu S. T. (1981). Rapid procedure for detection and isolation of large and small plasmids. J. Bacteriol. 145 1365–1373. 10.1128/jb.145.3.1365-1373.1981 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kirejczyk J. K., Porowski T., Konstantynowicz J., Kozerska A., Nazarkiewicz A., Hoppe B., et al. (2014). Urinary citrate excretion in healthy children depends on age and gender. Pediatr. Nephrol. 29 1575–1582. 10.1007/s00467-014-2806-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Literacka E., Bedenic B., Baraniak A., Fiett J., Tonkic M., Jajic-Bencic I., et al. (2009). blaCTX-M genes in Escherichia coli strains from Croatian hospitals are located in new (blaCTX-M-3a) and widely spread (blaCTX-M-3a and blaCTX-M-15) genetic structures. Antimicrob. Agents Chemother. 53 1630–1635. 10.1128/aac.01431-08 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu J., Duncan K., Walsh C. T. (1989). Nucleotide sequence of a cluster of Escherichia coli enterobactin biosynthesis genes: identification of entA and purification of its product 2,3-dihydro-2,3-dihydroxybenzoate dehydrogenase. J. Bacteriol. 171 791–798. 10.1128/jb.171.2.791-798.1989 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mao B. H., Chang Y. F., Scaria J., Chang C. C., Chou L. W., Tien N., et al. (2012). Identification of Escherichia coli genes associated with urinary tract infections. J. Clin. Microbiol. 50 449–456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Medina M., Castillo-Pino E. (2019). An introduction to the epidemiology and burden of urinary tract infections. Ther. Adv. Urol. 11:1756287219832172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mierzejewska J., Kulinska A., Jagura-Burdzy G. (2007). Functional analysis of replication and stability regions of broad-host-range conjugative plasmid CTX-M3 from the IncL/M incompatibility group. Plasmid 57 95–107. 10.1016/j.plasmid.2006.09.001 [DOI] [PubMed] [Google Scholar]
- Naqvi S. B., Collins A. J. (2006). Infectious complications in chronic kidney disease. Adv. Chronic Kidney Dis. 13 199–204. 10.1053/j.ackd.2006.04.004 [DOI] [PubMed] [Google Scholar]
- Paauw A., Caspers M. P., Leverstein-Van Hall M. A., Schuren F. H., Montijn R. C., Verhoef J., et al. (2009). Identification of resistance and virulence factors in an epidemic Enterobacter hormaechei outbreak strain. Microbiology 155 1478–1488. 10.1099/mic.0.024828-0 [DOI] [PubMed] [Google Scholar]
- Partridge S. R., Ginn A. N., Paulsen I. T., Iredell J. R. (2012). pEl1573 Carrying blaIMP-4, from Sydney, Australia, is closely related to other IncL/M plasmids. Antimicrob. Agents Chemother. 56 6029–6032. 10.1128/aac.01189-12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Partridge S. R., Kwong S. M., Firth N., Jensen S. O. (2018). Mobile genetic elements associated with antimicrobial resistance. Clin. Microbiol. Rev. 31:e00088-e17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pennoyer A., Bridgeman M. B. (2015). Ferric citrate (auryxia) for the treatment of hyperphosphatemia. P T 40 329–339. [PMC free article] [PubMed] [Google Scholar]
- Ramirez M. S., Traglia G. M., Lin D. L., Tran T., Tolmasky M. E. (2014). Plasmid-mediated antibiotic resistance and virulence in gram-negatives: the Klebsiella pneumoniae paradigm. Microbiol. Spectr. 2 1–15. 10.14302/issn.2690-4721.ijcm-19-3154 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roer L., Tchesnokova V., Allesoe R., Muradova M., Chattopadhyay S., Ahrenfeldt J., et al. (2017). Development of a web tool for Escherichia coli subtyping based on fimH Alleles. J. Clin. Microbiol. 55 2538–2543. 10.1128/jcm.00737-17 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Skaar E. P. (2010). The battle for iron between bacterial pathogens and their vertebrate hosts. PLoS Pathog. 6:e1000949. 10.1371/journal.ppat.1000949 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Snyder J. A., Haugen B. J., Buckles E. L., Lockatell C. V., Johnson D. E., Donnenberg M. S., et al. (2004). Transcriptome of uropathogenic Escherichia coli during urinary tract infection. Infect. Immun. 72 6373–6381. 10.1128/iai.72.11.6373-6381.2004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Subashchandrabose S., Smith S. N., Spurbeck R. R., Kole M. M., Mobley H. L. (2013). Genome-wide detection of fitness genes in uropathogenic Escherichia coli during systemic infection. PLoS Pathog. 9:e1003788. 10.1371/journal.ppat.1003788 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sullivan M. J., Petty N. K., Beatson S. A. (2011). Easyfig: a genome comparison visualizer. Bioinformatics 27 1009–1010. 10.1093/bioinformatics/btr039 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tzeng J. I., Chu C. H., Chen S. W., Yeh C. M., Chiu C. H., Chiou C. S., et al. (2012). Reduction of Salmonella enterica serovar Choleraesuis carrying large virulence plasmids after the foot and mouth disease outbreak in swine in southern Taiwan, and their independent evolution in human and pig. J. Microbiol. Immunol. Infect. 45 418–425. 10.1016/j.jmii.2011.12.029 [DOI] [PubMed] [Google Scholar]
- Ueda T., Yoshida T., Tanoue H., Ito M., Tamaki M., Ito Y., et al. (2014). Urine alkalization improves the problems of pain and sleep in hypersensitive bladder syndrome. Int. J. Urol. 21 512–517. 10.1111/iju.12324 [DOI] [PubMed] [Google Scholar]
- Wagenlehner F. M. E., Bjerklund Johansen T. E., Cai T., Koves B., Kranz J., Pilatz A., et al. (2020). Epidemiology, definition and treatment of complicated urinary tract infections. Nat. Rev. Urol. 17 586–600. [DOI] [PubMed] [Google Scholar]
- Wang P., Xiong Y., Lan R., Ye C., Wang H., Ren J., et al. (2011). pO157_Sal, a novel conjugative plasmid detected in outbreak isolates of Escherichia coli O157:H7. J. Clin. Microbiol. 49 1594–1597. 10.1128/jcm.02530-10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Welch R. A., Burland V., Plunkett G., Redford P., Roesch P., Rasko D., et al. (2002). Extensive mosaic structure revealed by the complete genome sequence of uropathogenic Escherichia coli. Proc. Natl. Acad. Sci. U.S.A. 99 17020–17024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wright K. J., Seed P. C., Hultgren S. J. (2005). Uropathogenic Escherichia coli flagella aid in efficient urinary tract colonization. Infect. Immun. 73 7657–7668. 10.1128/iai.73.11.7657-7668.2005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu W. H., Luo L., Wang J. Y., Zhuang X. H., Zhong L., Liao K., et al. (2009). Complete nucleotide sequence of pCTX-M360, an intermediate plasmid between pEL60 and pCTX-M3, from a multidrug-resistant Klebsiella pneumoniae strain isolated in China. Antimicrob. Agents Chemother. 53 5291–5293. 10.1128/aac.00032-09 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zong Z., Partridge S. R., Iredell J. R. (2010). ISEcp1-mediated transposition and homologous recombination can explain the context of bla(CTX-M-62) linked to qnrB2. Antimicrob. Agents Chemother. 54 3039–3042. 10.1128/aac.00041-10 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The sequences of pEC41 are deposited in the GenBank repository, accession number MW548582.