Abstract
Background
Acinetobacter baumannii is a major contributor to nosocomial infections. Extended-spectrum β-lactamase (ESBL)-producing A. baumannii is spreading worldwide. We aimed to determine the frequency of ESBL-encoding genes in clinical isolates of A. baumannii and to access their clonal relationship by repetitive extragenic palindromic-PCR (rep-PCR).
Methods
In this descriptive cross-sectional study, 203 isolates of A. baumannii were collected from Qazvin hospitals. The Identification of isolates was performed by standard laboratory methods. To verify ESBL production, all isolates were screened by disk agar diffusion and confirmed by the combined disk method. Subsequently, ESBL-encoding genes were detected by PCR and sequencing. Possible clonal association of ESBL-producing isolates was evaluated using rep-PCR.
Results
Two hundred (98.5%) isolates showed reduced susceptibility to one of the antibiotics used in the ESBL screening test, of which 127 isolates (62.6%) produced ESBL. PCR results showed blaOXA-1 (20.5%) was the most prevalent gene followed by blaTEM-1 (20%), blaGES-1 (15.7%), blaCTX-M-15 (7.9%), and blaPER-1 (1.6%). Rep-PCR results revealed that ESBL-producing isolates belonged to clones A (85%), B (13.4%), and C (1.6%).
Conclusion
Our study showed the significant presence of blaOXA-1, blaTEM-1, blaGES-1, blaCTX-M-15, and blaPER-1 genes in ESBL-producing A. baumannii isolates in the studied hospitals. This is the first report on the emergence of blaOXA-1 gene in these isolates in Iran. The use of comprehensive antimicrobial treatment guidelines based on laboratory data and appropriate infection control interventions are essential.
Keywords: Acinetobacter baumannii, ESBL, repetitive extragenic palindromic-PCR
Introduction
Acinetobacter baumannii (A. baumannii), an opportunistic pathogen involved in health care-associated infections, is a gram-negative, non-fermentative, aerobic bacterium. These organisms are major causes of infections such as urinary tract infection (UTI), bacteremia, meningitis, pneumonia, and burn infections (1,2). In the last decade, Acinetobacter strains have become resistant to most antibiotics, especially in Asia (3–5). Over use and/or misuse of antimicrobials is leading to increasing the emergence of multidrug resistant strains of A. baumannii worldwide. This organism has become resistant to several antibiotic classes including β-lactam, aminoglycoside, and quinolones (6). Due to its high ability to develop resistance and create MDR strains, the treatment of these infections has become difficult and costly and sometimes impossible (7). β-lactam antibiotics have been frequently used for treating A. baumannii infections (8). Resistance to β-lactams in A. baumannii isolates is due to intrinsic and acquired mechanisms including enzymatic alteration, mutation in target genes, alteration of outer membrane permeability and increased efflux pump activity (9). The most important mechanism of β-lactam resistance in Gram-negative bacteria is the production of β-lactamase enzymes that has posed many challenges in the antibiotic treatment of infections caused by these organisms (10). The Ambler classification divides the β-lactamases into four major molecular classes (A-D). The extended-spectrum β-lactamases (ESBLs) belong to class A and can hydrolyze first, second and third-generation of cephalosporins, penicillins, and monobactams but are inhibited by clavulanate (11). The important ESBL families in many enteric gram-negative bacilli are CTX, TEM, OXA, SHV, GES, VEB and PER (12). The ESBL-encoding genes are often found on plasmids that allow the spread of resistance factors between these isolates and other organisms that cause healthcare-associated infections (13).
Currently, given the high prevalence and rapid worldwide spread of multiple drug resistant (MDR) A. baumannii isolates, mortality in hospitalized patients, especially in the intensive care units (ICUs), has increased significantly (14). Several recent studies suggest that the release of ESBL-encoding genes could complicate the treatment of A. baumannii infections because of limited therapeutic choices (15). Therefore, we aimed to determine the prevalence of ESBLs and the genes encoding these enzymes including blaGES, blaVEB, blaSHV, blaTEM, blaCTX, blaOXA, and blaPER in clinical isolates of A. baumannii as well as to assess their clonal relationship in teaching hospitals of Qazvin, Iran.
Methods
Bacterial isolates: The current study was a descriptive cross-sectional study done during July 2017 to September 2018 in two teaching hospitals in Qazvin, Iran. In total, 203 A. baumannii isolates (one isolate per patient) were collected from different clinical specimens including urine, blood, trachea, wounds and from patients admitted to infectious diseases intensive care units, neurosurgery, and surgery wards. Identification of isolates was performed using standard microbiological and biochemical tests including gram staining and microscopic examination, culture on MacConkey agar medium, lactose fermentation, negative oxidase, immobility on SIM medium, alkaline slant/alkaline butt (ALK/ALK) pattern on triple sugar iron (TSI) and no pigment production (16). The identification was further confirmed by detecting of the intrinsic blaOXA-51-like and gltA (encoding bacterial citrate synthase) genes as previously described (17–18). All isolates were then stored at -70 °C in tryptic soy broth (TSB) medium with 20% glycerol until subsequent tests.
Ethical considerations: This study was approved by the Ethics Committee of Qazvin University of Medical Sciences (code IR.QUMS.REC.1396.696). Written informed consent was obtained from all individuals enrolled in the study.
Detection of ESBL-producing isolates by phenotypic methods: First, all isolates were prepared using standard Disk Agar Diffusion (DAD) method using cefpodoxime (30 µg), ceftazidime (30 µg), ceftriaxone (30 µg), cefotaxime (30 µg) and aztreonam (30 µg) discs (Mast Diagnostics Group Ltd, Merseyside, UK) according to Clinical and Laboratory Standard Institute (CLSI) guidelines (19). Then, isolates that reduced susceptibility to one of the used antibiotics were screened for confirmation of ESBL production using the combined disc method. In this method, discs containing ceftazidime or cefotaxime (30 µg) with and without clavulanic acid (10 µg) were used. An ≥5 mm increase in zone of inhibition for ceftazidime/clavulanate (30/10 µg) and cefotaxime/clavulanate (30/10 µg) compared to the zone diameter in the absence of clavulanate was considered to be an ESBL producer. For the quality control of susceptibility testing, A. baumannii standard isolate American Type Culture Collection (ATCC) 19606 was used.
Detection of ESBL-encoding genes by PCR and Sequencing: The amplification of blaOXA, blaGES-1, blaVEB-1, blaSHV, blaTEM, blaCTX-M, and blaPER-1 genes was performed using specific primers through PCR (Table 1). Total DNAs was first extracted by the extraction kit (Bioneer Company, South Korea) according to the manufacturer's instructions. Then, the PCR was done in a total reaction volume of 20 µl, containing 8 µl of DNA polymerase 2x Master Mix (Ampliqon, Odense, Denmark), 0.5 µM were prepared from each reverse and forward primer, 10 µl distilled water and 1µL DNA template (50 g concentration). PCR amplification of each of the studied genes was done using a thermocycler (Applied Biosystems, USA) under the following program: initial denaturation temperature of 96 °C for 5 min, 30 cycles including denaturation temperature of 95 °C for 1 min, specific annealing temperature for each primer for 1 minutes, extension temperature of 72 °C for 1 min.
Table 1.
Sequences of primers used in this study
| Target genes |
Primer sequence (5′-3′) | Annealing temperature (°C) |
References |
| blaSHV | F: GGTTATGCGTTATATTCGCC R: TTAGCGTTGCTTGTGCTC |
50 | (20) |
| blaTEM | F: ATGAGTATTCAACATTTCCG R: CTGACAGTTACCAATGCTTA |
50 | (21) |
| blaVEB-1 | F: CGACTTCCATTTCCCGATGC R: GGACTCTGCAACAAATACGC |
55 | (22) |
| blaPER-1 | F: AATTTGGGCTTAGGGCAGAA R: ATGAATGTCATTATAAAAGC |
53 | (23) |
| blaGES-1 | F: ATGCGCTTCATTCACGCAC R: CTATTTGTCCGTGCTCAGG |
53 | (24) |
| blaOXA-4 | F: TCAACAGATATCTCTACTG TT R: TTTATCCCATTTGAATATGGT |
50 | (25) |
|
blaOXA-10 (group I) |
F: AGCCGTTAAAATTAAGCCC R: CTTGATTGAAGGGTTGGGCG |
56 | (22) |
|
blaOXA-2 (group II) |
F: GCCAAAGGCACGATAGTTGT R: GCGTCCGAGTTGACTGCCGG |
62 | (26) |
|
blaOXA-1 (group III) |
F: AGCCGTTAAAATTAAGCCC R: CTTGATTGAAGGGTTGGGCG |
53 | (27) |
| blaCTX-M-1 | F: ATGGTTAAAAAATCACTGCGTC R: TTGGTGACGATTTTAGCCGC |
55 | (28) |
| blaCTX-M-2 | F: ATGATGACTCAGAGCATTCG R: TGGGTTACGATTTTCGCCGC |
55 | (28) |
| blaCTX-M-8 | F: ACTTCAGCCACACGGATTCA R: CGAGTACGTCACGACGACTT |
55 | (28) |
| blaCTX-M-9 | F: ATGGTGACAAAGAGAGTGCA R: CCCTTCGGCGATGATTCTC |
55 | (28) |
| rep-PCR | F: IIIGCGCCGICATCAGGC R: ACGTCTTATCAGGCCTAC |
45 | (29) |
A final extension step of 8 minutes at 72ºC was also performed. PCR products were visualized by electrophoresis on agarose gel (1%) using gel documentation system (UVtec, UK). The purified PCR products sent to the Macrogen Company (Seoul, Korea) for sequencing and then the data were analyzed online on The Basic Local Alignment Search Tool (BLAST) available at National Center for Information Biotechnology.
Rep-PCR analysis: Rep-PCR was done in a final volume of 20 µl, including 10 µl of distilled water, 8 µl of Master Mix (Ampliqon, Odense, Denmark), 0.5 µl of each forward and reverse primers (Table 1), 1 µl of the template DNA. The conditions of the amplification were as follows: initial denaturation of 95 °C for 3 minutes; then 35 cycles including denaturation temperature of 92 °C for 1 min, specific annealing temperature of 36 °C for 1 min, extension time of 72 °C for 10 min. A final extension step of 16 minutes at 72ºC was also performed. The PCR products were then analyzed by gel electrophoresis using 2% agarose gel and the results were visualized and the isolates with similar patterns (less than two bands difference) were assigned to the same clonal groups (30).
Results
Patients and isolates data: In this study, A. baumannii isolates were collected from the trachea (109, 53.8%), urine (33, 16.4%), blood (32, 15.8%) and wound (12, 5.9%), and sputum (17, 4.8%), respectively. The isolates were obtained from 122(60.1%) patients admitted to ICUs: 39(19.2%) to internal medicine, 25(12.3%) to infectious diseases, 11(5.4%) to neurosurgery and 6(3%) to surgery wards. One hundred and twenty-seven (58%) strains were isolated from females and 76(42%) from males. Patients' mean age was 54±12 years (range: 26–81 years).
ESBLs and prevalence ESBL-encoding genes: Of the 203 isolates, 200(98.5%) showed reduced susceptibility to one of the five cephalosporins used in the primary screening. By performing confirmatory test, 127(62.6%) isolates were positive for ESBL production. Twenty-six (20.5%) isolates harbored the blaOXA-1 gene which was the most frequent gene, followed by 25(20%), 20(15.7%), 10(7.9%) and 2(1.6%) isolates carrying the blaTEM-1, blaGES-1, blaCTX-M-15, and blaPER-1 genes, respectively. The studied isolates were negative for the presence of blaVEB-1 and blaSHV-1 genes. Also, simultaneous presence of blaOXA-1-blaGES-1 genes was seen in 17(13.4%) isolates, blaTEM-1-blaOXA-1 genes in 7(5.5%) isolates, and blaOXA-blaTEM-1 genes in 4 (3.1%) isolates (Table 2).
Table 2.
Repetitive-sequence-based PCR (rep-PCR) results of the ESBL-positive A. baumannii isolates
| No. of isolates (%) | ||||
| Genes | Type A | Type B | Type C | Total |
| blaGES-1+blaOXA-1 | 15 (11.8) | 1 (0.8) | 1 (0.8) | 17 (13.4) |
| blaTEM-1+blaOXA-1 | 6 (4.7) | 1 (0.8) | - | 7 (5.5) |
| blaTEM-1+blaGES-1+blaOXA-1 | 7 (5.5) | - | - | 7 (5.5) |
| blaGES-1+blaOXA-1+blaCTX-M | 5 (3.9) | 1 (0.8) | - | 6 (4.7) |
| blaOXA-1+blaCTX-M | 4 (3.1) | - | - | 4 (3.1) |
| blaTEM-1+blaGES-1+blaOXA-1+blaCTX-M | 2 (1.6) | - | - | 2 (1.6) |
| blaTEM-1+blaOXA-1+blaPER-1 | 1 (0.8) | 1 (0.8) | - | 2 (1.6) |
Rep-PCR: Rep-PCR results showed that ESBL-producing isolates belonged to 3 clones: A (85%), B (13.4%), and C (1.6%) indicating clonal spread of these resistant isolates among patients between different wards (Figure 1). As shown in Table 2, the presence of the blaOXA-1 gene alone or in combination was often shown in isolates belonging to clone A (50.3%) followed by clones B (5.6%) and C (0.8%).
Figure 1.
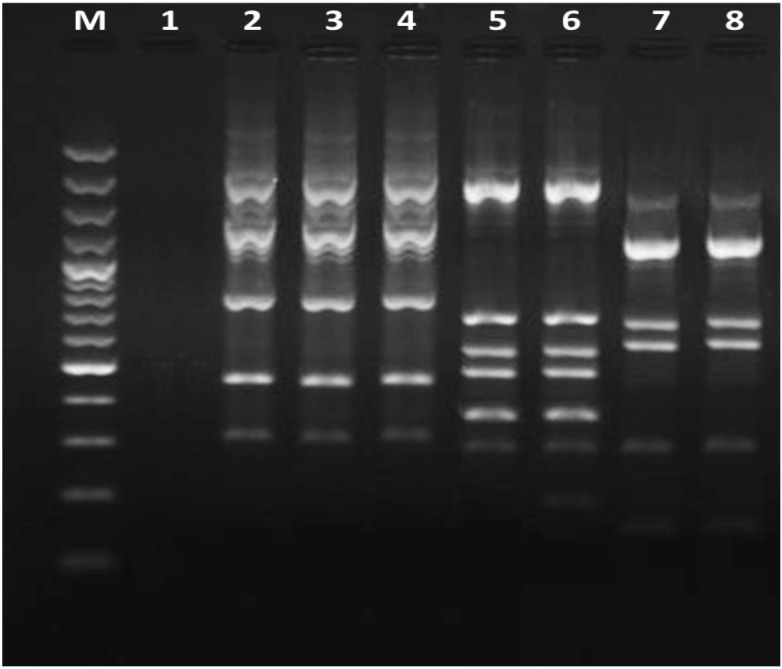
Representative REP-PCR fingerprints of Acinetobacter baumannii isolated from Qazvin hospitals. Lane M, 100 bp DNA ladder; lane 1, negative control (without template DNA), lanes 2 to 4, genotype A; lanes 5 and 6, genotype B; lanes 7 and 8, genotype C.
Discussion
Health care-associated infection caused by A. baumannii isolates is currently one of the major causes of patient mortality, especially in ICUs. Several reports from different geographical regions of Iran showed the presence of ESBLs in Gram negative bacteria (31–33). However, there was no study that investigated the presence of these enzymes in A. baumannii isolates in Qazvin province, central Iran.
In the current study, 62.6% of A. baumannii isolates were ESBL producers. Our results were consistent with two other studies in Iran in 2013 in Tabriz and in 2017 in Kermanshah on A. baumannii isolates which showed that 70% and 53.8% of the isolates produced ESBL, respectively (32,34). However, we found a higher prevalence of ESBL-producing A. baumannii compared with other studies in Iran in Hamedan (7%) and Tehran (16%) (35,36). The present study showed that ESBL-producing isolates were seen more in patients admitted to ICUs, internal and infectious wards and in trachea and urine specimens. Prolonged hospitalization and critical conditions for patients admitted to ICUs, the use of invasive procedures and devices, and prolonged exposure to broad-spectrum antibiotics may facilitate infection with these resistant organisms.
It seems that the presence of these resistant organisms in the studied hospitals may be due to inadequate management of infection control and lack of proper and rational medication administration. Our data emphasize the need to establish an antimicrobial resistance monitoring system locally and nationally and can effectively monitor the possible occurrence of this resistant organism in our healthcare system. However, it should be noted that the distribution of these resistant hospitalized organisms varies in different regions of the world. Factors such as age, severity of infection, patient immune response, method of administering broad-spectrum antibiotics, length of hospital stay and aggressive diagnostic and treatment methods considered for the patient may affect the incidence and prevalence of these infections.
In the present study, 20.5%, 20%, 15.7%, 7.9%, and 1.6% of isolates carried the blaOXA-1, blaTEM-1, blaGES-1, blaCTX-M-15 and blaPER-1 genes, respectively. In one study in Iran, the prevalence of blaTEM and blaCTX genes was 52.1% and 43.4%, respectively, in ESBL-producing A. baumannii isolates (36). Two other studies also reported the presence of blaCTX-M and blaTEM genes in clinical isolated of ESBL-producing A. baumannii isolates (34,37). In 2019, 42% of A. baumannii strains isolated from patients admitted to hospitals in Tehran were positive for the blaTEM gene and negative for the presence of the blaSHV gene (33). Rezaee et al. in 2013 found that 37% of the A. baumannii isolates harbored at least one of the blaTEM-1 and blaPER-1 genes, and none carried the blaSHV gene (38). We previously showed the presence of blaOXA-1, blaGES-1, and blaVEB-1 in clinical isolates of Pseudomonas aeruginosa (39). In Iraq, Ghaima and colleagues found that 75% and 45% of A. baumannii isolates carried blaTEM and blaCTX-M genes, respectively (40). In another study, the prevalence of blaTEM and blaSHV genes was 8.8% and 25%, respectively (41). In Turkey, the frequencies of TEM, SHV and GES in ESBL-producing A. baumannii isolates were 55.7%, 7.7% and 1.5%, respectively (42). Taken together, these results confirm the presence of different ESBLs-encoding genes in A. baumannii isolated from medical centers worldwide. Several recent studies indicate an increasing presence of less prevalent ESBLs genes (blaGES, blaPER, and blaSHV) in A. baumannii isolates (38,43).
In this study, rep-PCR results showed that ESBL-producing A. baumannii isolates belonged to three separate clones which was strongly influenced by the clonal spread of these resistant isolate as well as patient-to-patient transmission. Moreover, we found that the dissemination of ESBL-producing isolates in different parts of the hospital as well as the dissemination of ESBLs-encoding genes occur in a clonal manner. This issue necessitates the use of appropriate infection control tools, especially in ICUs.
Our findings showed the significant presence of ESBL-producing A. baumannii isolates in the studied medical centers. This is the first report of the presence of blaOXA-1 gene in these isolates in Iran. Since the ESBL-producing isolates are often resistant to other antibiotics, they have created serious limitations and difficulties in the treatment of infected patients. Given the plasmid nature and rapid and widespread dissemination of ESBLs in treatment centers, early identification of these resistant organisms, the use of comprehensive antimicrobial treatment guidelines based on laboratory data and the use of appropriate infection control tools to prevent further infection are essential.
Acknowledgments
The authors thank the staff of the Medical Microbiology Research Center, Qazvin University of Medical Sciences.
References
- 1.Visca P, Seifert H, Towner KJ. Acinetobacter infection-an emerging threat to human health. IUBMB life. 2011;63(12):1048–1054. doi: 10.1002/iub.534. [DOI] [PubMed] [Google Scholar]
- 2.Valentine SC, Contreras D, Tan S, Real LJ, Chu S, Xu HH, et al. Phenotypic and molecular characterization of Acinetobacter baumannii clinical isolates from nosocomial outbreaks in Los Angeles County, California. J Clin Microbiol. 2008;46(8):2499–2507. doi: 10.1128/JCM.00367-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Peymani A, Farajnia S, Nahaei MR, Sohrabi N, Abbasi L, Ansarin K, et al. Prevalence of class 1 integron among multidrug-resistant Acinetobacter baumannii in Tabriz, northwest of Iran. Pol J Microbiol. 2012;61(1):57–60. [PubMed] [Google Scholar]
- 4.Hoang Quoc C, Nguyen Thi Phuong T, Nguyen Duc H, Tran Le T, Tran Th Thu H, Nguyen Tuan S, et al. Carbapenemase genes and multidrug resistance of Acinetobacter baumannii: A cross sectional study of patients with pneumonia in southern vietnam. Antibiotics (Basel) 2019;8(3):148. doi: 10.3390/antibiotics8030148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Tada T, Uchida H, Hishinuma T, Watanabe S, Tohya M, Kuwahara-Arai K, et al. Molecular epidemiology of multidrug-resistant Acinetobacter baumannii isolates from hospitals in Myanmar. J Glob Antimicrob Resist. 2020:2213–7165. doi: 10.1016/j.jgar.2020.02.011. [DOI] [PubMed] [Google Scholar]
- 6.Chen F, Wang L, Wang M, Xie Y, Xia X, Li X, et al. Genetic characterization and in vitro activity of antimicrobial combinations of multidrug-resistant Acinetobacter baumannii from a general hospital in China. Oncol Lett. 2018;15(2):2305–2315. doi: 10.3892/ol.2017.7600. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Hanlon GW. The emergence of multidrug resistant Acinetobacter species: a major concern in the hospital setting. Lett Appl Microbiol. 2005;41(5):375–378. doi: 10.1111/j.1472-765X.2005.01791.x. [DOI] [PubMed] [Google Scholar]
- 8.Fournier PE, Richet H. The epidemiology and control of Acinetobactr baumannii in health care facilities. Clin Infect Dis. 2006;42(5):692–699. doi: 10.1086/500202. [DOI] [PubMed] [Google Scholar]
- 9.Poirel L, Nordmann P. Carbapenem resistance in Acinetobacter baumannii: mechanisms and epidemiology. Clin Microbiol Infect. 2006;12(9):826–836. doi: 10.1111/j.1469-0691.2006.01456.x. [DOI] [PubMed] [Google Scholar]
- 10.Litake GM, Ghole VS, Niphadkar KB, Joshi SG. Phenotypic ESBL detection in Acinetobacter baumannii: a real challenge. Am J Infect Dis. 2015;11(3):48–53. [Google Scholar]
- 11.Shah AA, Hasan F, Ahmed S, Hameed A. Extended-spectrum beta-lactamases (ESBLs): characterization, epidemiology and detection. Crit Rev Microbio. 2004;30(1):25–32. doi: 10.1080/10408410490266429. [DOI] [PubMed] [Google Scholar]
- 12.Bush K, Bradford PA. Interplay between β-lactamases and new β-lactamase inhibitors. Nature Rev Microbiol. 2019;17(5):295–306. doi: 10.1038/s41579-019-0159-8. [DOI] [PubMed] [Google Scholar]
- 13.Pfeifer Y, Cullik A, Witte W. Resistance to cephalosporins and carbapenems in Gram-negative bacterial pathogens. Int J Med Microbiol. 2010;300(6):371–379. doi: 10.1016/j.ijmm.2010.04.005. [DOI] [PubMed] [Google Scholar]
- 14.Ren J, Li X, Wang L, Liu M, Zheng K, Wang Y, et al. Risk factors and drug resistance of the MDR Acinetobacter baumannii in pneumonia patients in ICU. Open Med (Wars) 2019;14:772–777. doi: 10.1515/med-2019-0090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Simhon A, Rahav G, Shazberg G, Block C, Bercovier H, Shapiro M, et al. Acinetobacter baumannii at a tertiary-care teaching hospital in Jerusalem. J Clin Microbiol. 2001;39(1):389–391. doi: 10.1128/JCM.39.1.389-391.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Mahon CR, Lehman DC, Manuselis G. Textbook of diagnostic microbiology. 4rd ed. Maryland Heights: Mo Saunders/Elsevier; 2011. 1395 p. [Google Scholar]
- 17.Wen H, Wang K, Liu Y, Tay M, Lauro FM, Huang H, et al. Population dynamics of an Acinetobacter baumannii clonal complex during colonization of patients. J Clin Microbiol. 2014;52(9):3200–3208. doi: 10.1128/JCM.00921-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Mendes RE, Kiyota KA, Monteiro J, Castanheira M, Andrade SS, Gales AC, et al. Rapid detection and identification of metallobeta-lactamase-encoding genes by multiplex real-time PCR assay and melt curve analysis. J Clin Microbiol. 2007;45(2):544–547. doi: 10.1128/JCM.01728-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Wayne PA CLSI. Clinical and laboratory standards institute performance standards for antimicrobial susceptibility testing. Twenty-Fifth informational supplement 2015; M100-S23. [Google Scholar]
- 20.Biendo M, Manoliu C, Laurans G, Castelain S, Canarelli B, Thomas D, et al. Molecular typing and characterization of extended-spectrum blaTEM, blaSHV and blaCTX-M β-lactamases in clinical isolates of Enterobacter cloacae. Res Microbiol. 2008;159(9–10):590–594. doi: 10.1016/j.resmic.2008.09.003. [DOI] [PubMed] [Google Scholar]
- 21.Naiemi NA, Duim B, Savelkoul PH, Spanjaard L, de Jonge E, Bart A, et al. Widespread transfer of resistance genes between bacterial species in an intensive care unit: implications for hospital epidemiology. J Clin Microbiol. 2005;43(9):4862–4864. doi: 10.1128/JCM.43.9.4862-4864.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Aubert D, Poirel L, Chevalier J, Leotard S, Pages JM, Nordmann P. Oxacillinase-mediated resistance to cefepime and susceptibility to ceftazidime in Pseudomonas aeruginosa. Antimicrob Agents Chemother. 2001;45(6):1615–1620. doi: 10.1128/AAC.45.6.1615-1620.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Poirel L, Le Thomas I, Naas T, Karim A, Nordmann P. Biochemical sequence analyses of GES-1, a novel class A extended-spectrum β-lactamase, and the class 1 integron In52 from Klebsiella pneumoniae. Antimicrob Agents Chemother. 2000;44(3):622–632. doi: 10.1128/aac.44.3.622-632.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Shahcheraghi F, Nikbin VS, Feizabadi MM. Prevalence of ESBLs genes among multidrug-resistant isolates of Pseudomonas aeruginosa isolated from patients in Tehran. Microb Drug Resist. 2009;15(1):37–39. doi: 10.1089/mdr.2009.0880. [DOI] [PubMed] [Google Scholar]
- 25.Naas T, Benaoudia F, Massuard S, Nordmann P. Integron-located VEB-1 extended- spectrum β-lactamase gene in a Proteus mirabilis clinical isolate from Vietnam. J Antimicrob Chemother. 2000;46(5):703–711. doi: 10.1093/jac/46.5.703. [DOI] [PubMed] [Google Scholar]
- 26.De Champs C, Poirel L, Bonnet R, Sirot D, Chanal C, Sirot J, et al. Prospective survey of β-lactamases produced by ceftazidime-resistant Pseudomonas aeruginosa isolated in a French hospital in 2000. Antimicrob Agents Chemother. 2002;46(9):3031–3034. doi: 10.1128/AAC.46.9.3031-3034.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Kim YR, Kim SI, Lee JY, Park YJ, Lee KY, Kang MW, et al. Nosocomial transmission of CTX-M-15 and OXA-30 beta-lactamase-producing Escherichia coli in a neurosurgical intensive care unit. Ann Clin Lab Sci. 2005;35(3):297–301. [PubMed] [Google Scholar]
- 28.Ben Achour N, Mercuri PS, Power P, Belhadj C, Ben Moussa M, Galleni M, et al. First detection of CTX-M-28 in a Tunisian hospital from a cefotaxime-resistant Klebsiella Pneumoniae strain. Pathol Biol. (Paris) 2009;57(5):343–348. doi: 10.1016/j.patbio.2008.07.016. [DOI] [PubMed] [Google Scholar]
- 29.Bou G, Cervero G, Dominguez MA, Quereda C, Martinez-Beltran J. PCR-based DNA fingerprinting (REP-PCR, AP-PCR) and pulsed-field gel electrophoresis characterization of a nosocomial outbreak caused by imipenem-and meropenem-resistant Acinetobacter baumannii. Clin Microbiol Infect. 2000;6(12):635–643. doi: 10.1046/j.1469-0691.2000.00181.x. [DOI] [PubMed] [Google Scholar]
- 30.Chen CH, Huang CC. Molecular epidemiological study of clinical Acinetobacter baumannii isolates: phenotype switching of antibiotic resistance. Ann Clin Microbiol Antimicrob. 2013;12(1):21. doi: 10.1186/1476-0711-12-21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Peymani A, Naserpour-Farivar T, Zare E, Azarhoosh KH. Distribution of blaTEM, blaSHV, and blaCTX-M genes among ESBL-producing P. aeruginosa isolated from Qazvin and Tehran hospitals, Iran. J Prev Med Hyg. 2017;58(2):E155–E160. [PMC free article] [PubMed] [Google Scholar]
- 32.Farajnia S, Azhari F, Alikhani MY, Hosseini MK, Peymani A, Sohrabi N, et al. Prevalence of PER and VEB type extended spectrum betalactamases among multidrug resistant Acinetobacter baumannii isolates in North-West of Iran. Iran J Basic Med Sci. 2013;16(6):751–755. [PMC free article] [PubMed] [Google Scholar]
- 33.Abdar MH, Taheri-Kalani M, Taheri K, Emadi B, Hasanzadeh A, Sedighi A, et al. Prevalence of extended-spectrum beta-lactamase genes in Acinetobacter baumannii strains isolated from nosocomial infections in Tehran, Iran. GMS Hyg Infect. 2019;14 doi: 10.3205/dgkh000318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Azizi M, Mortazavi SH, Etemadimajed M, Gheini S, Vaziri S, Alvandi A, et al. Prevalence of extended-spectrum β-Lactamases and antibiotic resistance patterns in Acinetobacter baumannii isolated from clinical samples in Kermanshah, Iran. Jundishapur J Microbiol. 2017;10(12):61522. [Google Scholar]
- 35.Safari M, Nejad AS, Bahador A, Jafar R, Alikhani MY. Prevalence of ESBL and MBL encoding genes in Acinetobacter baumannii strains isolated from patients of intensive care units (ICU) Saudi J Biol Sci. 2015;22(4):424–429. doi: 10.1016/j.sjbs.2015.01.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Ranjbar R, Tolon SS, Zayeri S, Sami M. The frequency of antibiotic resistance and ESBLs among clinically Acinetobacter baumannii strains isolated from patients in a Major Hospital in Tehran, Iran. Open Microbiol J. 2018;12:254–260. doi: 10.2174/1874285801812010254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Khosroshahi SA, Farajnia S, Azhari F, Hosseini MK, Khanipour F, Farajnia H, et al. Antimicrobial susceptibility pattern and prevalence of extended-spectrum β-lactamase genotypes among clinical isolates of Acinetobacter baumannii in Tabriz, North-West of Iran. Jundishapur J Microbiol. 2017;10(6):13368. [Google Scholar]
- 38.Rezaee MA, Pajand O, Nahaei MR, Mahdian R, Aghazadeh M, Ghojazadeh M, et al. Prevalence of ambler class A beta-lactamases and ampC expression in cephalosporin-resistant isolates of Acinetobacter baumannii. Diagn Microbiol Infect Dis. 2013;76(3):330–334. doi: 10.1016/j.diagmicrobio.2013.04.003. [DOI] [PubMed] [Google Scholar]
- 39.Amirkamali S, Naserpour-Farivar T, Azarhoosh K, Peymani A. Distribution of the blaOXA, blaVEB-1, and blaGES-1 genes and resistance patterns of ESBL-producing Pseudomonas aeruginosa isolated from hospitals in Tehran and Qazvin, Iran. Rev Soc Bras Med Trop. 2017;50(3):315–320. doi: 10.1590/0037-8682-0478-2016. [DOI] [PubMed] [Google Scholar]
- 40.Ghaima KK. Distribution of extended spectrum beta-lactamase (ESBL) genes among Acinetobacter baumannii isolated from burn infections. MOJ Cell Sci Rep. 2018;5(3):42–46. [Google Scholar]
- 41.AL-Thahab AA. Molecular detection of extended-spectrum beta-lactamases in clinical isolates of Acinetobacter baumannii. J Biol Agric Health. 2013;3(7):32–38. [Google Scholar]
- 42.Beriş FŞ, Budak EE, Gülek D, Uzun A, Cizmeci Z, Mengeloğlu FZ, et al. Investigation of the frequency and distribution of betalactamase genes in the clinical isolates of Acinetobacter baumannii collected from different regions of Turkey: a multicenter study. Mikrobiyol Bul. 2016;50(4):511–521. doi: 10.5578/mb.29176. [DOI] [PubMed] [Google Scholar]
- 43.Poirel L, Menuteau O, Agoli N, Cattoen C, Nordmann P. Outbreak of extended-spectrum beta-lactamase VEB-1-producing isolates of Acinetobacter baumannii in a French hospital. J Clin Microbiol. 2003;41(8):3542–3547. doi: 10.1128/JCM.41.8.3542-3547.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]


