Five equine herpesvirus 1 (EHV-1) genome sequences with links to an EHV-1 outbreak with neurological disorders after a horse gathering in Valencia, Spain, in February 2021, were determined. All strains showed the closest relationships to strains from Belgium and the United Kingdom, indicating a common source of infection.
ABSTRACT
Five equine herpesvirus 1 (EHV-1) genome sequences with links to an EHV-1 outbreak with neurological disorders after a horse gathering in Valencia, Spain, in February 2021, were determined. All strains showed the closest relationships to strains from Belgium and the United Kingdom, indicating a common source of infection.
ANNOUNCEMENT
One of the most serious equine herpesvirus 1 (EHV-1) outbreaks in Europe was reported following the International CES Valencia Spring Tour (Spain) in February 2021, which was attended by 752 horses. As 17 dead horses and neurological disorders were reported, quarantine regulations were implemented quickly in Spain and other European Union countries to prevent further spread (19 March 2021). Nevertheless, the aggressive EHV-1 strain escaped Spain with outbreaks in nine countries, including Belgium and France (FEI Updates 2021, https://inside.fei.org/fei/ehv-1/department-updates?year=). Here, we report five genomes from EHV-1 isolates from affected horses in Belgium and France with links to the Spanish Tour in 2021, as obtained through rapid long-read sequencing.
EHV-1 was isolated on rabbit kidney (RK-13) cells from nasal swab samples or peripheral blood mononuclear cells (PBMCs) from Belgian and French horses with neurological symptoms after attending the Spanish Sunshine Tour in February 2021 (Table 1).
TABLE 1.
Overview of clinical data and sequencing output for three Belgian and two French EHV-1 isolates
| Strain | Origin | Horse age (yr) | Horse gendera | Horse type | Site in Spain (sampling timeb) | PCR test result forc: |
Virus isolation from: |
Sequencing output |
|||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Nose sample | Blood sample | Nose sample | PBMCs | Length (bp) | GC content (%) | Coverage (×) | Read N50 (bp) | ||||||
| BE/21P40/2021 | Belgium | 8 | F | Jumping | Valencia (2 wk) | + | + | + | − | 149,513 | 56.6 | 230 | 4,058 |
| BE/21P41/2021 | Belgium | 12 | F | Jumping | Vejer (<24 h) | + | + | + | + | 154,163 | 56.5 | 140 | 4,169 |
| BE/21P43_BD5/2021 | Belgium | 11 | F | Jumping | Valencia (2 wk) | + | + | − | + | 161,236 | 56.6 | 592 | 1,748 |
| FR/Valencia1/2021 | France | 6 | F | Jumping | Valencia (<24 h) | + | ND | + | ND | 150,964 | 56.6 | 311 | 400 |
| RF/Valencia2/2021 | France | 9 | F | Jumping | Valencia (<24 h) | + | ND | + | ND | 156,482 | 56.5 | 309 | 417 |
F, female.
The sampling time indicated is the time between the return from Spain and sampling.
+, positive result; −, negative result; ND, not determined (test was not performed).
Viral DNA was extracted from the cell culture supernatant using the Quick-DNA/RNA viral kit (Zymo Research) at PathoSense BV. Native EHV-1 DNA was sequenced on a MinION R.9.4 flow cell (FLO-MIN106; Oxford Nanopore Technologies [ONT]) using the ONT ligation sequencing protocol (LSK-109; ONT). Raw data were processed using an in-house pipeline. In short, raw data were base called using the high-accuracy algorithm, demultiplexed and trimmed, and filtered using Guppy (v3.6; ONT) (-c dna_r9.4.1_450bps_hac.cfg), qcat (v1.1.0; ONT), and NanoFilt (v2.7.1) (1), respectively. Filtered reads were used for de novo EHV-1 genome assembly using Canu (v2.0) (2). Consensus genomes were obtained after read alignment using GraphMap (v0.5.2) (3) and polishing using medaka (v1.0.0; ONT). Downstream analyses included multiple-sequence alignment, pairwise identity determination, and phylogenetic analysis using MAFFT (v7.471) (4), BLASTN (v2.10.1+), and FastTree (v2) (5) (-nt -gamma -gtr), respectively. All software was run using default settings. Values are represented as means ± standard deviation.
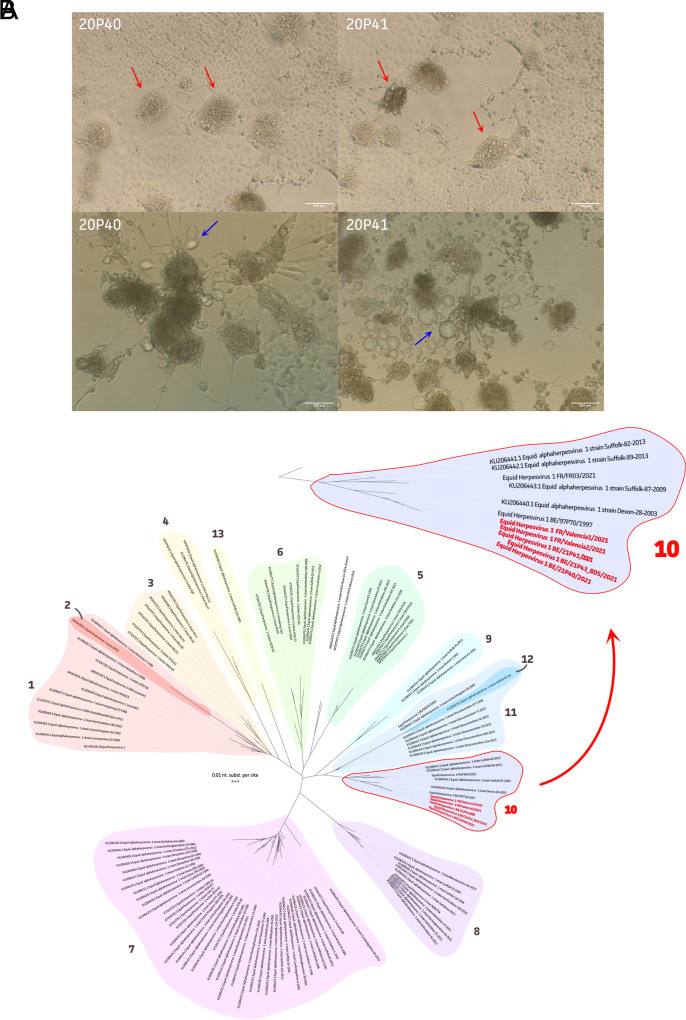
The three Belgian isolates showed comparable cytopathic effect (CPE) on RK-13 cells. As shown in Fig. 1A, formation of typical EHV-1 syncytia was observed. Multiple balloon-shaped structures were present close to these syncytia.
FIG 1.
Phenotypic characterization and clade determination of new EHV-1 strains. (A) Phenotypic characterization of two Belgian isolates (21P40 and 21P41) on RK-13 cells, showing the formation of syncytia (red arrows) and balloon-shaped structures (blue arrows). Bar, 500 μm. (B) Phylogenetic tree of all available EHV-1 genomes, highlighting the new Belgian (n = 3) and French (n = 2) EHV-1 strains in red within clade 10.
The EHV-1 genomes (154,472 ± 4,658 bp) were phylogenetically classified based on clades previously defined by Bryant et al. (6). All Belgian and French outbreak strains were closely related to each other (nucleotide identities of 99.91 ± 0.03%) and belonged to clade 10. This suggests the spread of a single EHV-1 strain during the outbreak in Spain. Interestingly, clade 10 comprised a total of four abortive EHV-1 strains from Belgium (n = 1; 1997) and the United Kingdom (n = 3; 2009 to 2013) (Fig. 1B). We further analyzed whether G2254/D752, among other neuropathogenic markers, was present in open reading frame 30 (ORF30) (6–8). All isolates demonstrated an H250/N752/Y753/K990 genotype. In the past, the N752 (A2254) marker was shown to be predominant (52% to 97%) in abortion cases in different countries (8–13). While mostly partial genomes are available (<80% nucleotides of ∼150 kbp), we encourage more complete high-quality genome sequences and clinical data to be made available in order to establish a clear genetic EHV-1 context (6, 8).
Data availability.
The EHV-1 genome sequences were deposited in the NCBI database, and raw reads (Nanopore) are available in the ENA (BioProject number PRJEB43980). The accession numbers are MW855958 (BE/21P40/2021), MW855959 (BE/21P41/2021), MW855960 (BE/21P43_BD5/2021), MW855961 (FR/Valencia1/2021), and MW855962 (FR/Valencia2/2021).
ACKNOWLEDGMENTS
We are thankful for the support received from Carine Boone, Marthe Pauwels, Gauthier Danneels, Camille Normand, and Christine Fortier.
N.V. is supported by a grant from the Flemish Agency for Innovation and Entrepreneurship (Baekeland Mandate HBC.2020.2889). F.C. is supported by a grant from the Fonds Eperon (grant N39-2019) and by GIS CENTAURE. The funders had no role in study design, data collection and interpretation, or the decision to submit the work for publication.
S.T. and H.N. are cofounders and co-owners of PathoSense.
REFERENCES
- 1.De Coster W, D'Hert S, Schultz DT, Cruts M, Van Broeckhoven C. 2018. NanoPack: visualizing and processing long-read sequencing data. Bioinformatics 34:2666–2669. doi: 10.1093/bioinformatics/bty149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Koren S, Walenz BP, Berlin K, Miller JR, Bergman NH, Phillippy AM. 2017. Canu: scalable and accurate long-read assembly via adaptive k-mer weighting and repeat separation. Genome Res 27:722–736. doi: 10.1101/gr.215087.116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Sović I, Šikić M, Wilm A, Fenlon SN, Chen S, Nagarajan N. 2016. Fast and sensitive mapping of Nanopore sequencing reads with GraphMap. Nat Commun 7:11307. doi: 10.1038/ncomms11307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Katoh K, Standley DM. 2013. MAFFT multiple sequence alignment software version 7: improvements in performance and usability. Mol Biol Evol 30:772–780. doi: 10.1093/molbev/mst010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Price MN, Dehal PS, Arkin AP. 2010. FastTree 2: approximately maximum-likelihood trees for large alignments. PLoS One 5:e9490. doi: 10.1371/journal.pone.0009490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Bryant NA, Wilkie GS, Russell CA, Compston L, Grafham D, Clissold L, McLay K, Medcalf L, Newton R, Davison AJ, Elton DM. 2018. Genetic diversity of equine herpesvirus 1 isolated from neurological, abortigenic and respiratory disease outbreaks. Transbound Emerg Dis 65:817–832. doi: 10.1111/tbed.12809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Nugent J, Birch-Machin I, Smith KC, Mumford JA, Swann Z, Newton JR, Bowden RJ, Allen GP, Davis-Poynter N. 2006. Analysis of equid herpesvirus 1 strain variation reveals a point mutation of the DNA polymerase strongly associated with neuropathogenic versus nonneuropathogenic disease outbreaks. J Virol 80:4047–4060. doi: 10.1128/JVI.80.8.4047-4060.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Sutton G, Garvey M, Cullinane A, Jourdan M, Fortier C, Moreau P. 2019. Molecular surveillance of EHV-1 strains circulating in France during and after the major 2009 outbreak in Normandy involving respiratory infection, neurological disorder, and abortion. Viruses 11:916. doi: 10.3390/v11100916. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Smith KL, Allen GP, Branscum AJ, Cook RF, Vickers ML, Timoney PJ, Balasuriya UBR. 2010. The increased prevalence of neuropathogenic strains of EHV-1 in equine abortions. Vet Microbiol 141:5–11. doi: 10.1016/j.vetmic.2009.07.030. [DOI] [PubMed] [Google Scholar]
- 10.Perkins GA, Goodman LB, Tsujimura K, Van de Walle GR, Kim SG, Dubovi EJ, Osterrieder N. 2009. Investigation of the prevalence of neurologic equine herpes virus type 1 (EHV-1) in a 23-year retrospective analysis (1984–2007). Vet Microbiol 139:375–378. doi: 10.1016/j.vetmic.2009.06.033. [DOI] [PubMed] [Google Scholar]
- 11.Stasiak K, Rola J, Ploszay G, Socha W, Zmudzinski JF. 2015. Detection of the neuropathogenic variant of equine herpesvirus 1 associated with abortions in mares in Poland. BMC Vet Res 11:102. doi: 10.1186/s12917-015-0416-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Latorre-Pérez A, Villalba-Bermell P, Pascual J, Vilanova C. 2020. Assembly methods for Nanopore-based metagenomic sequencing: a comparative study. Sci Rep 10:13588. doi: 10.1038/s41598-020-70491-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Tsujimura K, Oyama T, Katayama Y, Muranaka M, Bannai H, Nemoto M, Yamanaka T, Kondo T, Kato M, Matsumura T. 2011. Prevalence of equine herpesvirus type 1 strains of neuropathogenic genotype in a major breeding area of Japan. J Vet Med Sci 73:1663–1667. doi: 10.1292/jvms.11-0140. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
The EHV-1 genome sequences were deposited in the NCBI database, and raw reads (Nanopore) are available in the ENA (BioProject number PRJEB43980). The accession numbers are MW855958 (BE/21P40/2021), MW855959 (BE/21P41/2021), MW855960 (BE/21P43_BD5/2021), MW855961 (FR/Valencia1/2021), and MW855962 (FR/Valencia2/2021).