Summary
Diet and bile play critical roles in shaping gut microbiota, but the molecular mechanism underlying interplay with intestinal microbiota is unclear. Here, we showed that lemon-derived exosome-like nanoparticles (LELNs) enhance lactobacilli toleration to bile. To decipher the mechanism, we used Lactobacillus rhamnosus GG (LGG) as proof of concept to show that LELNs enhance LGG bile resistance via limiting production of Msp1 and Msp3, resulting in decrease of bile accessibility to cell membrane. Furthermore, we found that decline of Msps protein levels was regulated through specific tRNAserUCC and tRNAserUCG decay. We identified RNase P, an essential housekeeping endonuclease, being responsible for LELNs-induced tRNAserUCC and tRNAserUCG decay. We further identified galacturonic acid-enriched pectin-type polysaccharide as the active factor in LELNs to increase bile resistance and downregulate tRNAserUCC and tRNAserUCG level in the LGG. Our study demonstrates a tRNA-based gene expression regulation mechanism among lactobacilli to increase bile resistance.
Subject areas: Nanoparticles, Molecular biology, Microbiology
Graphical abstract
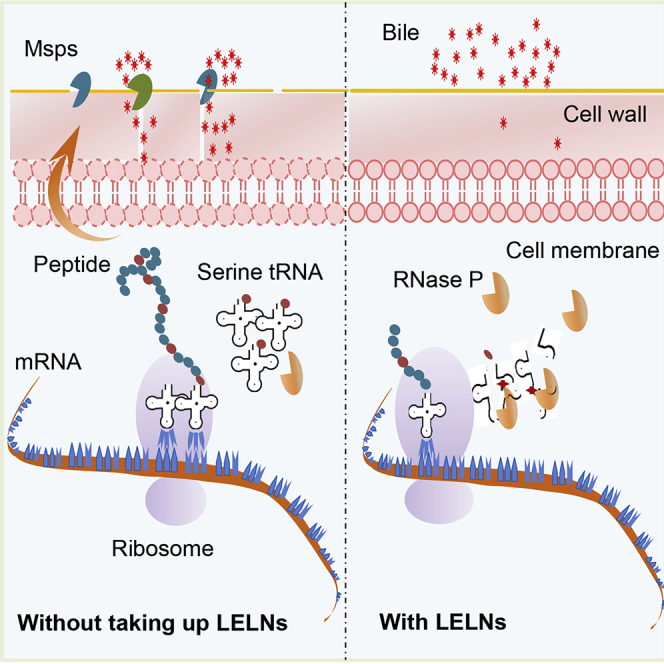
Highlights
-
•
LELN-derived pectin selectively enhances lactobacilli toleration to bile
-
•
LELNs enhance LGG bile resistance via limiting production of Msp1 and Msp3
-
•
LELNs decrease translation of Msps via specific tRNAserUCC and tRNAserUCG decay
-
•
LELNs-mediated induction of RNase P is responsible for tRNAserUCC and tRNAserUCG decay
Nanoparticles; Molecular biology; Microbiology
Introduction
It is well known that diet plays critical roles in how a host responds to a variety of stressors, such as bile, by shaping the gut microbiota composition and thus contributing to host health (Bibbo et al., 2016). Gut bacteria confront numerous stressors and insults, among which bile is one of the most challenging to the growth of bacteria in the gut. Bile kills bacteria due to its damaging effects on bacterial DNA and cell membrane integrity (Merritt and Donaldson, 2009); meanwhile, bile is also proven to be important in controlling the overgrowth of gut bacteria and shaping gut and gallbladder microbiota (Wahlstrom et al., 2016; Molinero et al., 2019). Gut bacteria develop a variety of strategies to avoid bile damage, including the expression of bile salt hydrolases, changing their subcellular membrane structure, and increasing their ability to respond to stress (Gunn, 2000; Ruiz et al., 2013). Whether diet interacts and how rapidly diet interacts with gut bacteria to regulate bile resistance at a molecular level remains elusive.
Rapid alterations of tRNA abundance were thought to be one of the widely used stress responses by both bacteria and mammalian cells (Zhong et al., 2015; Torrent et al., 2018). Modifications of tRNA are prone to affect tRNA tertiary structure and stability (Motorin and Helm, 2010) and regulate responses to stress (Gu et al., 2014). Recently published data indicated that diet can affect the tRNA modification pattern of gut bacteria (Schwartz et al., 2018), but the physiological function of this observation is not clear. Other than global tRNA changes, specific tRNA changes were also found to play critical roles in cell metabolism. Upregulation of tRNAGluUUC in breast cancer cells was found to increase cancer metastasis (Goodarzi et al., 2016).
Our previous research has shown that edible plants produce plenty of exosome-like nanoparticles (ELNs) that can have remarkable effects on both the host and gut microbiota (Teng et al., 2018). Both microRNA and lipids from ELNs have been proved to play critical roles in regulating gut bacterial gene expression (Sundaram et al., 2019; Teng et al., 2018). Here we show that lemon-derived exosome-like nanoparticles (LELNs), which could easily be included in a North American diet, selectively increase lactobacilli percentages in the small intestine of mice by increasing resistance to bile. A mechanism based on the specific tRNA decay associated with this finding is proposed.
Results
LELNs-derived pectin selectively increases bile resistance of lactobacilli
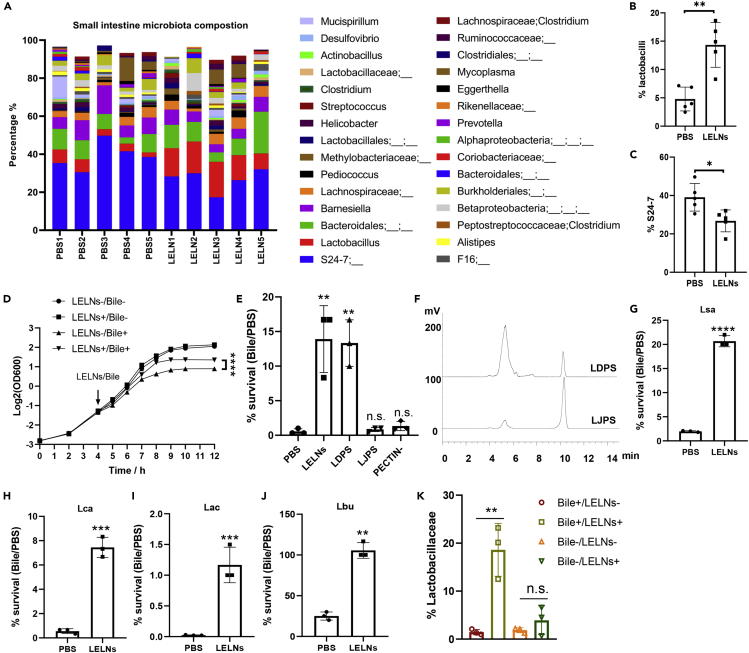
LELNs were isolated and purified through differential centrifugation (Teng et al., 2018). Sucrose gradient-purified LELNs were further characterized by electron microscopy and nanoparticle tracking analysis (Figures S1A and S1B). To test the effects of LELNs on small intestinal microbiota, mice were gavage-given LELNs and the small intestinal microbiota composition was analyzed using 16S rDNA sequencing. We found that lactobacilli were increased, whereas the unculturable Bacteroidetes family S24-7 was decreased in LELN-gavaged mice compared with the PBS control mice (Figures 1A–1C). To decipher the mechanism underlying LELNs-mediated altering of the small intestine microbiota composition, we focused on lactobacilli because they were the most significantly changed of the microbiota in the small intestine. As proof of concept, we used probiotic Lactobacillus rhamnosus GG (LGG), a well-characterized and extensively studied lactobacilli strain in human and mouse gut, to evaluate the effects of LELNs on lactobacilli. We first determined whether LELNs promote LGG growth using growth curve analysis. We found no change in growth after LELNs treatment (Figure 1D). As bile creates one of the most challenging environments for bacteria to survive in the small intestine, we tested whether LELNs promote LGG growth under bile stress. We found that LELNs remarkably enhanced LGG growth in De Man, Rogosa and Sharpe broth (MRS) in the presence of bile (Figure 1D). We thus tested the effect of LELNs on enhancing bile resistance of LGG as measured by percent survival. LELNs significantly improve percent survival of LGG under bile challenge from 0.55% ± 0.39% to 13.91% ± 4.83% (Figure 1E). To explore the active component that contributes to LGG bile resistance, LELNs components were grouped by molecular weight (cutoff 5 kDa) and underwent heat, nuclease, and protease treatments. The active component was found to be a heat-stable, nuclease and protease-resistant high-molecular-weight component (Figure S1C). Based on the aforementioned information and the viscous appearance of LELNs, we predicted that the polysaccharide in the LELNs contributed to increasing bile resistance of LGG. Polysaccharides isolated from LELNs (LDPS) increased LGG bile resistance comparable to intact LELNs (Figure 1E). We also tested and confirmed that the polysaccharides derived from lemon juice (LJPS) do not enhance LGG resistance to bile (Figure 1E). Comparison analysis of LDPS and LJPS showed that a higher molecular weight polysaccharide (LDPS-HM) was enriched in LDPS (Figure 1F). LDPS-HM was further identified using glycosyl composition and linkage analyses and pectinase digestion. The LDPS-HM was found to be a type of pectin with a molecular weight ∼450 kDa (Figure S2 and Tables S1 and S2). Pectinase-digested LELNs (PECTIN-) lose their potential for increasing LGG resistance to bile (Figure 1E), suggesting that the pectin in LELNs is a primary contributor to the resistance of LGG to bile. We also tested the effect of LELNs on resistance of other bacterial strains to bile, including four lactobacilli strains, two bacteroides strains, and one eubacterium strain. LELNs significantly improved bile resistance for all four lactobacilli strains tested but did not improve bile resistance for the bacteroides and eubacterium strains (Figures 1G–1J and S3).
Figure 1.
LELNs enhance lactobacilli survival in the small intestine by enhancing bile resistance
(A) 16S-rDNA sequencing analysis of small intestine microbiota composition in LELN-treated and PBS control C57BL/6J mice (n = 5). Top 30 genera in the small intestine were shown.
(B and C) Statistical data of Lactobacillus and S24-7 percentage in the small intestine microbiota.
(D) LGG growth curve under different treatments. LELN- and LELN+ indicate LGG growth without or with LELN treatment, and Bile- and Bile+ indicate LGG growth without or with bile treatment.
(E) LGG survival rate in a bile challenge, 1 × 1010/mL LELNs and PECTIN- or 10 mg/mL LDPS (comparable with 5 × 1010/mL LELNs) and LJPS were used in the experiments.
(F) Comparative analysis of polysaccharide composition of LDPS and LJPS by HPLC; 0.1 mg polysaccharides was injected into the HPLC system.
(G–J) Bile resistance test of four different lactobacilli strains as indicated with or without LELN treatment. Lsa represents Lactobacillus salivarius LS-33, Lca represents Lactobacillus casei LC-11, Lac represents Lactobacillus acidophilus 4356, Lbu represents Lactobacillus bulgaricus LB-87.
(K) Percentage of Lactobacillaceae in the small intestine microbiota after 24 h in vitro culture with or without LELNs treatment. The data are presented as values with standard deviation (mean ± SD). The significance is shown as ∗p ≤ 0.05; ∗∗p ≤ 0.01; ∗∗∗p ≤ 0.001 and ∗∗∗∗p ≤ 0.0001; p > 0.05 was considered not significant (n.s.).
We then tested whether bile plays a role in enhancing Lactobacillaceae percentage in general using an in vitro cultured gut microbiota (Li et al., 2019). We compared the percentage of Lactobacillaceae in the culture with or without LELNs treatment and found that LELNs treatment increased the percentage of Lactobacillaceae significantly in the presence of bile, whereas increased only slightly but with no statistical significance when bile was withdrawn from the culture media (Figure 1K).
Decline of Msp1 and Msp3 protein levels contributes to increased LGG bile resistance
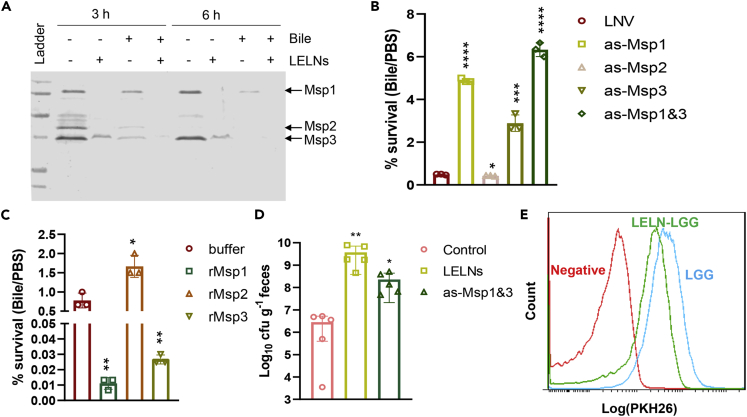
To determine the molecular basis as to how LELNs increase LGG resistance to bile, we conducted comparative proteomic analysis between LELN-treated LGG (LELN-LGG) and PBS-treated LGG as a control. We noticed that three cell surface proteins, LGG_00324 (Msp1), LGG_00031 (Msp2), and LGG_02016 (we later named Msp3) were decreased dramatically due to LELN treatment (Table S3). As Msp1 and Msp2 were reported to be secretory proteins in LGG (Claes et al., 2012), we analyzed the Msp levels by SDS-PAGE in the broth of cultured LGG. All three Msps decreased upon LELNs treatment (Figures 2A and S4). Similar to LELNs treatment, we noticed that bile treatment also decreased Msp levels in the LGG culture broth (Figure 2A). Treatment of LGG with both LELNs and bile further decreased Msp levels in the culture broth (Figure 2A). To verify whether reduction of the Msps is responsible for LGG resistance to bile, we knocked down the msp genes singly or in combination by using antisense RNA. Antisense RNAs were designed as described in Figure S5A, and knockdown efficiency was confirmed with real-time qPCR (Figure 1, Figure 2, Figure 3, Figure 4B and S5C). Knockdown of Msp1 or Msp3 genes resulted in enhancing the bile resistance of LGG (measured as survivability) from 0.49% ± 0.02% to 4.89% ± 0.10% and 2.89% ± 0.39%, respectively, whereas knockdown of the Msp2 gene decreased LGG resistance to bile (Figure 2B). Knockdown of both Msp1 and Msp3 resulted in an additive effect of enhancing LGG resistance to bile (Figure 2B). We also tested the effects of heterogeneous-expressed recombinant Msp1 (rMsp1), Msp2 (rMsp2), and Msp3 (rMsp3) on LGG bile resistance (Figure S6). rMsp1 and rMsp3 significantly decreased the bile resistance of LGG as measured by survivability from 0.79% ± 0.19% to 0.01% ± 0.004% and 0.03% ± 0.003%, respectively, whereas rMsp2 slightly increased the bile resistance (Figure 2C). We also tested whether LELN treatment or knockdown of Msp1 and Msp3 can increase LGG gut survival due to increasing bile resistance. We found that LELN treatment increased LGG survival over two orders of magnitude; knockdown of Msp1 and Msp3 also increased LGG survival over 10-fold when given to antibiotic-treated mice by gavage and analyzed by LGG ability to pass through the gastrointestinal tract (Figure 2D).
Figure 2.
Decline of Msp1 and Msp3 increase LGG bile resistance
(A) SDS-PAGE analysis of LGG secretory proteins in cultured broth. 1 × 1010/mL LELNs or/and 0.05% bile were added into LGG culture at OD600 = 0.4; supernatants were collected at two time points as indicated.
(B) The effects of LGG Msp1, Msp2, and Msp3 gene knockdown on the LGG resistance to bile. LELNs-derived Nano vectors (LNV) without RNA inclusion served as a control.
(C) The effect of Msp1, Msp2, and Msp3 on bile resistance. rMsp1, rMsp2, and rMsp3 were directly added into overnight LGG cultures 90 min before the cultured LGG was harvested. Storage buffer of recombination proteins was used as a control. To make survival rate quantifiable, LGG was incubated with bile for 30 min instead of 1 h in the bile resistance test.
(D) Survivability of LGG with indicated treatments passing through the gastrointestinal tract as evaluated by fecal LGG CFU.
(E) Cell membrane accessibility of LGG valuated by PKH26 stain. The data are presented as values with standard deviation (mean ± SD). The significance is shown as ∗p ≤ 0.05; ∗∗p ≤ 0.01; ∗∗∗p ≤ 0.001; ∗∗∗∗p ≤ 0.0001 and p > 0.05 was considered not significant (n.s.).
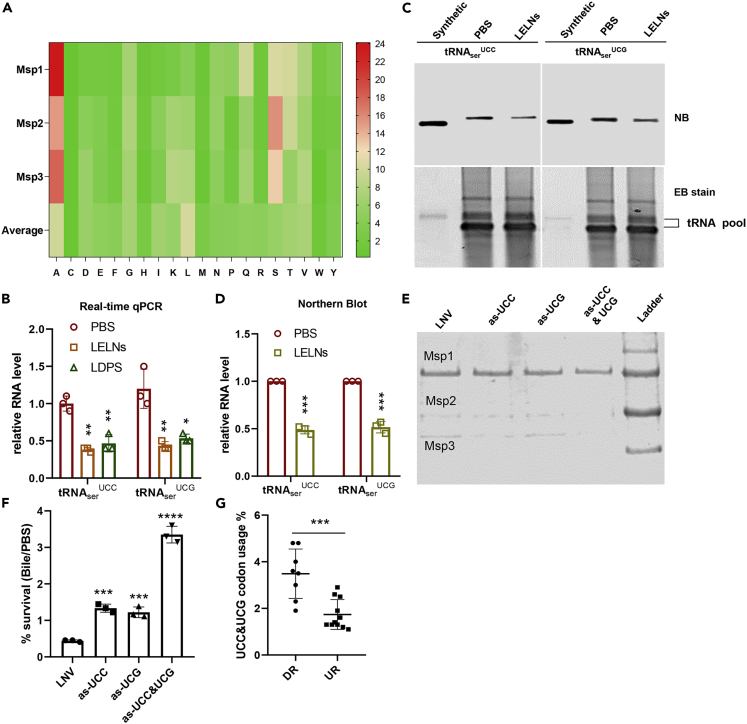
Figure 3.
Preferential utility of serine tRNA contributes to LELNs-mediated downregulation of LGG Msps
(A) Amino acid composition analysis of Msp1, Msp2, and Msp3. Average amino acid composition in whole LGG proteome was used as a control. Amino acid composition analyses were conducted using BioEdit software.
(B–D) Real-time qPCR (B) and northern blot (NB) analysis (C and D) of relative serine tRNA levels in LGG. PBS treatment served as a control, synthetic tRNA prepared by in vitro transcription was used as a positive control, and ethidium bromide (EB) stain was used as the loading control. Northern blots were quantified using ImageJ software.
(E and F) SDS-PAGE analysis of secretory proteins in the cultured broth and bile resistance tests of tRNAserUCC and tRNAserUCG knockdown strains; protein from 200 μL LGG culture supernatant were loaded on the SDS-PAGE gel. as-UCC and as-UCG indicate knockdown of either tRNAserUCC or tRNAserUCG, respectively, and as-UCC&UCG indicates knockdown of both tRNAserUCC and tRNAserUCG. LNV without RNA inclusion was used as a control. Samples were collected at 6 h (supernatant for SDS-PAGE) or 12 h (bacteria for bile resistance test) after adding antisense RNA.
(G) UCC and UCG codon usage analysis in top downregulated (DR) and upregulated (UR) genes in Table S3 due to LELNs treatment; codon usage analysis was conducted using an online codon usage tool in Sequence Manipulation Suite (Stothard, 2000). The data are presented as values with standard deviation (mean ± SD). The significance is shown as ∗p ≤ 0.05; ∗∗p ≤ 0.01; ∗∗∗p ≤ 0.001; ∗∗∗∗p ≤ 0.0001 and p > 0.05 was considered not significant (n.s.).
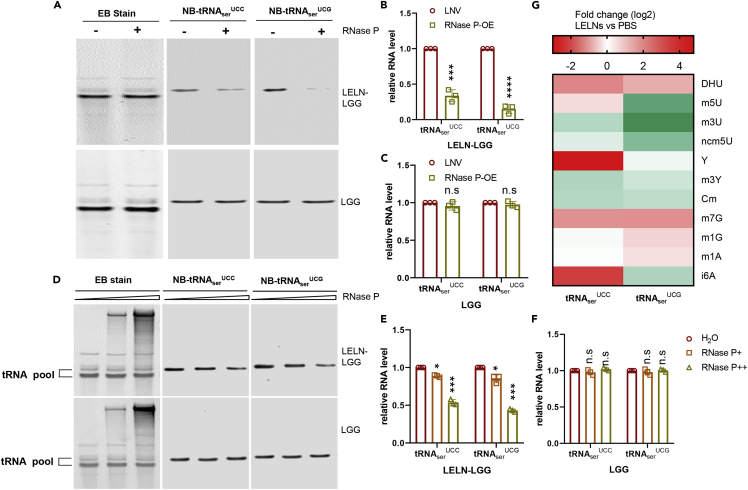
Figure 4.
RNase P is responsible for LELNs-induced tRNAserUCC and tRNAserUCG decay in LGG
(A–C) tRNAserUCC and tRNAserUCG levels in LGG after RNase P overexpression. tRNAserUCC and tRNAserUCG levels from LGG (bottom panels) or LELN-LGG (top panels) were analyzed by northern blot (A) and quantified by ImageJ software (B and C). ‘+’ indicates RNase P delivered by LNV, ‘-’ indicates LNV control without RNA included. EB stain was used as the loading control.
(D–F) In vitro RNase P cleavage assay of tRNAserUCC and tRNAserUCG derived from LGG or LELN-LGG. Two different doses of RNase P, 200 ng (middle lanes or RNase P+), and 2 μg (right lanes or RNase P++) were used in the cleavage assay. H2O was used as a control (left lanes); EB stain was used as a loading control.
(G) Nucleoside modifications analysis of tRNAserUCC and tRNAserUCG by HPLC-MASS, the nucleoside modification levels are shown as fold changes due to LELN treatment. The data are presented as values with standard deviation (mean ± SD). The significance is shown as ∗p ≤ 0.05; ∗∗p ≤ 0.01; ∗∗∗p ≤ 0.001; ∗∗∗∗p ≤ 0.0001 and p > 0.05 was considered not significant (n.s.).
Both Msp1 and Msp3 contain cell wall peptidoglycan hydrolase domains, and Msp1 has been proved to hydrolyze cell wall-derived peptidoglycan (Bauerl et al., 2010). We thus hypothesize that as a result of LELN treatment, the increased resistance of LGG to bile is likely due to decreasing accessibility of bile to the LGG cell membrane. We tested LGG membrane accessibility using the PKH26 fluorescent dye, which stains the membrane (Oh et al., 1999), and the amount of membrane-bound PKH26 dye was detected by fluorescence-activated cell sorting (FACS) analysis. FACS analysis indicates that LELNs-treated LGG had less fluorescent dye signal detected than PBS-treated LGG (Figure 2E).
LELNs downregulate Msps expression by selective tRNAserUCC and tRNAserUCG decay
To further understand the mechanism underlying LELNs regulation of Msps expression, we first analyzed msp mRNA levels using qRT-PCR. We found that msp mRNAs were only slightly decreased after LELNs treatment (Figure S7), suggesting that LELNs-mediated downregulation of Msps likely occurs at the translational or post-translational level. We then conducted protein sequence analysis of the Msps and found that alanine and serine ratios in the Msps were much higher than found in whole proteome (Figure 3A). We thus hypothesized that LELNs may target Msps in a codon usage manner. tRNA serves as the nucleic acid-decoding device that causes the insertion of codon-specific amino acids in a growing protein chain, so we analyzed the levels of serine and alanine tRNA in LGG by qRT-PCR. We found that LELNs and LDPS specifically decrease tRNAserUCC (LGG_02972) and tRNAserUCG (LGG_02998) (Figures 3B and S8A). The data generated with real-time qPCR were further supported by northern blot analysis (Figures 3C, 3D, S8B, and S8C). We then knocked down tRNAserUCC and tRNAserUCG using antisense RNA and analyzed the effects on Msp expression and bile resistance. Knockdown of tRNAserUCC or tRNAserUCG alone slightly decreased Msps detected in the broth while increasing the bile resistance of LGG from 0.44% ± 0.03% to 1.33% ± 0.11% and 1.22% ± 0.15%, respectively (Figures 3E and 3F). Knockdown of both tRNAserUCC and tRNAserUCG led to a remarkable decrease in Msps and an increase in the bile resistance of LGG to 3.35% ± 0.03% (Figures 3E and 3F). We also compared tRNAserUCC and tRNAserUCG codon usage in the top 8 upregulated genes and top 13 downregulated genes upon LELNs treatment; a negative correlation with usage of these two codons was observed (Figure 3G).
RNase P mediates LELN-induced tRNAserUCC and tRNAserUCG decay in LGG
tRNA is very stable, thus it is difficult to regulate tRNA levels instantly by transcription regulation. We found one tRNA processing enzyme, RNase P, that was increased 2-fold due to LELNs treatment according to RNA sequencing (RNA-seq)-based comparative analysis (Table S4). The results were confirmed by qRT-PCR (Figure S9A). To verify whether RNase P is responsible for the tRNAserUCC and tRNAserUCG decline, we first overexpressed RNase P in LGG and sought to detect tRNAserUCC and tRNAserUCG levels by northern blot, but found no change in the tRNAserUCC and tRNAserUCG levels (Figures 4A and 4C). To test whether RNase P decay is specific to tRNAserUCC and tRNAserUCG, we also detected the levels of three other tRNAs that are located near serine tRNA in the LGG genome. All the three tRNA are not significantly different between the control and RNase P-overexpressing group (Figures S9B–S9D). We then pretreated LGG with LELNs before overexpressing RNase P, and we found that tRNAserUCC/UCG levels were decreased in the RNase P-overexpressing group (Figures 4A and 4B). We also performed an in vitro RNase P cleavage assay directed at tRNAserUCC and tRNAserUCG. tRNA-enriched small RNA pool from LGG or LELN-LGG was incubated with RNase P in cleavage buffer and tRNAserUCC and tRNAserUCG levels were detected by northern blot. RNase P was found to specifically decay tRNAserUCC and tRNAserUCG derived from LELN-LGG under both concentrations (Figures 4D–4F), suggesting that pretreatment with LELNs is required for RNase P-mediated decay of tRNAserUCC and tRNAserUCG. tRNAserUCA was found to be decayed by a higher concentration of RNase P, and tRNAserAGC was not sensitive to RNase P in all conditions tested (Figures S9E–S9G). tRNA is reported to be one of the most modified RNAs, and the modifications have been proved to be associated with tRNA function and stability (Motorin and Helm, 2010). Therefore, we proposed that LELNs-induced tRNA epigenetic modification plays a role in RNase P-mediated tRNA decay. We thus compared nucleoside modifications of tRNAserUCC and tRNAserUCG derived from LGG and LELN-LGG by High-performance liquid chromatography–mass spectrometry. We observed dramatic changes in the pattern of tRNAserUCC and tRNAserUCG nucleoside modifications due to LELNs treatment (Figure 4G).
Discussion
In this study, our results demonstrated an elegant three-way interaction between gut microbes, bile, and diet-derived ELNs. We demonstrated that daily consumption of diet-derived nanoparticles increases the percentage of probiotic LGG in the small intestine by enhancing bile resistance. This finding is significant because most of the probiotic bacteria currently available in the market belong to the genera Lactobacillus and overcoming bile salt damage to probiotics is a great challenge. To generate beneficial health effects, these bacteria must be able to counteract the deleterious action of bile before they can transiently colonize our gut. Although diet-derived factors can modulate the survivability of gut microbiota including probiotics in general, the molecular mechanisms underlying it are still elusive. In this study, LELNs, as a proof of concept, were used to provide the molecular evidence for ELNs-mediated induction of LGG bile resistance via reduction of protein expression of Msp 1 and Msp3. Interestingly, treatment of LGG with both LELNs and bile further decreased Msp protein levels in the culture broth. This result suggests that when LGG receives an insult from bile, a survival signaling pathway that mediates inhibition of the expression of Msps protein may be activated via a feedback mechanism that attempts to rescue LGG from bile-induced cell death. This assumption was further supported by the fact that LELNs treatment increased the percentage of Lactobacillaceae significantly in an in-vitro cultured microbiota in the presence of bile, whereas there was no statistically significant increase in the percentage of Lactobacillaceae when bile was withdrawn from the culture media (Figure 1K). Whether the bile-induced survival signaling pathway in LGG is the same pathway as LELNs that we demonstrated in this study is unknown and requires further study.
It has been well documented that stress-induced tRNA decay is one of the fastest ways to modulate bacterial fitness to stress conditions (Sorensen et al., 2018; Huber et al., 2019). Our study showed that LELNs can induce specific bacterial tRNA decay and increase survivability of LGG under bile stress. We noticed that knockdown of serine tRNA by antisense RNA causes a less extensive decrease of Msp expression compared with LELNs treatment, suggesting that other factors derived from the LELNs may also contribute to inhibition of Msp expression. Another possibility is that the antisense strategy we used in this study may not be sufficient in completely inhibiting Msp expression.
We also noticed that whereas LELNs treatment increased LGG abundance of Lactobacillus, S.24-7 was decreased. Based on published data (Teng et al., 2018), orally taking edible plant ELNs could have both direct and indirect effects on the gut microbiomes. ELNs are preferentially taken up by certain species of gut bacteria leading to either promotion or inhibition of the growth of ELN recipients depending on the composition of ELNs. Additionally, metabolites released from ELN recipient cells could have either promoting or inhibiting effects on neighboring bacteria. Our finding that LELNs treatment increased LGG abundance of Lactobacillus whereas S.24-7 was decreased provides a foundation for further studying whether inhibition of S.24-7 growth in the intestine is due to direct or indirect effects.
RNase P is conserved and is found in all three kingdoms of life, and it is well known to process the 5′-end of pre-tRNA (Kazantsev and Pace, 2006). RNase P has a very promiscuous recognition sequence. Other than pre-tRNA, RNase P has also been reported to cleave 4.5s RNA, tmRNA, and m6A-containing mRNA (Coughlin et al., 2008; Park et al., 2019). tRNA contains many kinds of epigenetic modifications that can affect tRNA tertiary structure and stability (Lorenz et al., 2017). Interestingly, our data show that RNase P can only cleave serine tRNA derived from LELNs-pretreated LGG, but not PBS-treated LGG. Our data also show that LELNs can change the epigenetic modification pattern of serine tRNA; we thus propose that the LELNs-mediated epigenetic modification pattern and the special 2D structure of serine tRNA may play critical roles in substrate recognition and degradation mediated by RNase P. This is consistent with our data showing that simple overexpression of RNase P in LGG cannot induce serine tRNA decay. We speculate that LELNs or LDPS may activate the LGG stress response pathway, such as the stringent stress response signaled by ppGpp (Irving and Corrigan, 2018) to modulate epigenetic modifications of tRNA and eventually induce tRNA decay.
Reduction of peptidoglycan hydrolases decreases cell membrane accessibility, thus preventing cell membrane-associated insults. Knockdown of Msp1 and Msp3 significantly increases LGG bile resistance; however, is it not equivalent to the bile resistance induced by LELNs; the Msp1 and Msp3 double knockdown strain further enhanced LGG resistance to the bile. Our results also suggest that Msp2 exhibits the opposite effect of Msp1 and Msp3 on resistance to bile, although all Msps contain cell wall-associated hydrolase domains that may be caused by hydrolyzation of different cell wall sites. The functional divergences between Msp1 and Msp2 can also be concluded from the difference in their morphology of mutants (Claes et al., 2012). Further research is needed to identify the exact functions of Msps and their relationship with cell membrane accessibility.
We cautiously draw the conclusion that LELNs protect gut bacteria from bile damage. It is well known that gram-negative bacteria are more resistant to bile damage due to their outer membrane protection, and therefore it is conceivable that LELN treatment may have no beneficial effect on the survivability of these species of bacteria under bile stress in the gut. Different edible plant ELNs have different biological effects on our health due to different chemical compositions and because different gut bacteria are targeted by the ELNs. Our findings provide a foundation for further investigating whether other ELNs have roles in stress responses of recipient bacteria.
In addition, it is important to note why we use the ELN terminology. Edible plant nanoparticles that we and others have studied are naturally released from edible plants consumed, and like exosomes, they consist of protein, lipid, and nucleic acids and are nanosized. However, as these nanoparticles are not spontaneously released from the plants we consume, we have determined the most appropriate nomenclature for edible plant particles is “exosome-like nanoparticles.”
Limitations of study
Further study is required to decipher which specific epigenetic modification(s) of serine tRNA is responsible for RNase P recognition and cleavage.
STAR★methods
Key resource table
| REAGENT OR RESOURCE | SOURCE | IDENTIFIER |
|---|---|---|
| Bacterial strains | ||
| Lactobacillus rhamnosus GG | ATCC | Cat# 53103 |
| Lactobacillus acidophilus | ATCC | Cat# 4356 |
| Lactobacillus bulgaricus LB-87 | Custom Probiotic Inc | Cat# CP-2016 |
| Lactobacillus casei LC-11 | Custom Probiotic Inc | Cat# CP-2012 |
| Lactobacillus salivarius LS-33 | Custom Probiotic Inc | Cat# CP-2010 |
| Bacteroides thetaiotaomicron | ATCC | Cat# 29148 |
| Bacteroides fragilis | ATCC | Cat# 25285 |
| Eubacterium rectale | ATCC | Cat# 33656 |
| E. coli Top10 | Invitrogen | Cat# C404006 |
| E. coli BL21 (DE3) | Invitrogen | Cat# C600003 |
| Plasmids and primers | ||
| pET28b-MBP-TEV | Addgene | Cat# 69929 |
| pET28b-MBP-TEV-Msp1 | This study | N/A |
| pET28b-MBP-TEV-Msp2 | This study | N/A |
| pET28b-MBP-TEV-Msp3 | This study | N/A |
| Primers see Table S5 | N/A | N/A |
| Chemicals, Peptides, and Recombinant Proteins | ||
| MRS broth | Hardy Diagnostics | Cat# C5931 |
| BHI broth | Hardy Diagnostics | Cat# C5141 |
| Lemon fruits | Sam’s Club | Cat#127308 |
| Kanamycin | Sigma | Cat# K1876 |
| RNAprotect Bacteria Reagent | Qiagen | Cat# 76506 |
| Porcine bile extract | Sigma | Cat# B8631 |
| Ni-NTA Agarose | Qiagen | Cat# 30210 |
| IRDye 800cw streptavidin | VWR | Cat# 102673-342 |
| Dynabeads™ M-270 Streptavidin | Invitrogen | Cat# 65305 |
| Tskgel G5000PW column | VWR | Cat# 100368-036 |
| SuperScrip III First-Strand Synthesis System | Invitrogen | Cat#18080051 |
| RNase-free DNase І | NEB | Cat# M0303S |
| TEV protease | Sigma | Cat# T4455 |
| Nuclease P1 | NEB | Cat# M0660S |
| CIP | NEB | Cat# M0525S |
| Critical Commercial Assays | ||
| RNeasy Mini Kit | Qiagen | Cat# 74104 |
| miRNeasy Mini Kit | Qiagen | Cat# 217004 |
| RiboPure RNA Purification Kit, bacteria | Invitrogen | Cat# AM1925 |
| QuantiTect SYBR Green PCR Kits | Qiagen | Cat# 204143 |
| PKH26 Fluorescent Cell Linker Kit | Sigma | Cat# PKH26GL |
| GeneArt Seamless Cloning and Assembly Kit | Invitrogen | cat# A14606 |
| HiScribe T7 Quick High Yield RNA Synthesis Kit | NEB | Cat# E2050S |
| QIAamp DNA Stool Mini Kits | Qiagen | Cat# 51504 |
| Experimental Models: Organisms/Strains Mouse | ||
| C57BL/6 | Jackson Laboratory Stock | Cat# 000664 |
| Deposited data | ||
| Sequenced data reported in this paper is stored at Sequence Read Archive (SRA): Accession code for Deposited Data: PRJNA722680 | ||
| Software and algorithms | ||
| Prism (GraphPad Software) | GraphPad | https://www.graphpad.com |
| BioEdit | BioEdit | https://www.bioedit.com |
| Sequence manipulation suite | (Stothard, 2000) | https://www.bioinformatics.org/sms2 |
Resource availability
Lead contact
Further information and requests for resources and reagents should be directed to and will be fulfilled by the lead contact, Dr. Huang-Ge Zhang, Email: h0zhan17@louisville.edu.
Materials availability
All unique reagents generated in this study can be obtained upon reasonable request to the Lead Contact.
Data and code availability
The accession number for the sequenced data reported in this paper is NCBI Sequence Read Archive (SRA): PRJNA722680.
Experimental model and subject details
Mice
Mice 8-week-old male C57BL/6 specific-pathogen-free (SPF) mice were purchased from the Jackson Laboratory (Bar Harbor, ME) and housed under specific-pathogen-free conditions with a 12 h light-dark cycle. All mice experiments were conducted following guidelines of Institute for Laboratory Animal Research (ILAR). All protocols were approved by the University of Louisville Institutional Animal Care and Use Committee (Louisville, KY).
Bacteria
All lactobacilli strains were grown in MRS media at 37°C without shaking. E. coli strains were grown at 37°C in LB broth, 50 μg/ml of kanamycin was added when needed. Bacteroides and Eubacterium strains were cultured in BHIS media under anaerobic condition.
Method details
LELNs extraction and purification from lemon fruit
LELNs extraction and purification were performed as described previously (Teng et al., 2018). Briefly, lemon fruit were peeled and squeezed, followed with a gradient centrifugation 1,000× g for 10 min, 2,000× g for 20 min, 4,000× g for 30 min, 8,000× g for 60 min, 36,000× g for 2 hr. Collect pellet after 36,000× g centrifugation step and resuspended in ice-cold 1× PBS, further purified by sucrose gradient ultracentrifuge (8, 30, 45, and 60% sucrose in 20 mM Tris-HCl, pH 7.2). The main band located at the 30%–45% sucrose interface was collected. All centrifugation steps were performed at 4°C. LELN size distribution and quantity were determined using a Zetasizer Nano ZS (Malvern Instrument).
In vitro LGG growth test
Overnight LGG cultures were inoculated into fresh MRS media at a ratio of 1:100 and cultured at 37°C without shaking. OD600 was determined hourly to evaluate LGG growth, with exception of the first two time points, which were collected at 2-h intervals. 1 × 1010/mL LELNs or 0.05% porcine bile extract was added into LGG cultures at the indicated time point as appropriate.
Bile resistance test
Bile resistance testing was performed as previously reported with some modifications (Vinderola CG, 2003). LGG was collected by centrifuge at 5,000× g for 10 min and washed twice in ice-cold 1× PBS to remove any remaining broth. 1 × 109 CFU LGG were incubated at 37°C for 1 h with 0.2% porcine bile extract except where otherwise stated. 0.2% porcine bile extract is comparable with bile concentrations in the small intestine (Kristoffersen et al., 2007). LGG was washed twice to remove remaining bile before plating on MRS-agar and the CFU was counted using the Miles and Misra method (Miles et al., 1938). The survival percent after bile treatment was calculated as the CFU counts on MRS-agar/input CFU∗100 to evaluate bile resistance.
In vitro gut microbiota culture
In vitro gut microbiota culture was conducted according to a previous report with minor modifications (Li et al., 2019). A porcine bile extract at 0.05% was used in the microbiota culture media. Microbiota from small intestine were collected after sacrificing mice and were inoculated into microbiota culture media at a ratio of 2% (w/v) and culture for 24 h under anaerobic condition. The percentage of bacteria was analyzed by qRT-PCR using specific primers as reported (Nava et al., 2011).
Polysaccharide extraction, purification and characterization
Polysaccharides were extracted from LELNs using a sonication method (Chen et al., 2012). In brief, LELNs were sonicated with a probe for 10 min at 200 watts using a sonic dismembrater (Fisher Scientific). To prevent overheating, LELN samples were incubated in a room temperature water bath. Pelleted debris was removed by ultra-centrifugation at 100,000× g for 1 h and the supernatant containing soluble polysaccharides was collected. Five volumes of pre-cooled 95% ethanol was added to the supernatant and allowed to stand overnight at −20°C to precipitate polysaccharides. Crude polysaccharides were further purified by HPLC using the Agilent 1260 system equipped with a Tskgel G5000PW column (7.5 mm × 30 cm, 17 μm). De-ionized water was used as a mobile phase to run through the HPLC system and ELSD was used to detect polysaccharides using previously reported parameters (Condezo-Hoyos et al., 2015). Molecular weight, monosaccharide composition, and linkage analysis were conducted at the Complex Carbohydrate Research Center at the University of Georgia. The molecular weight was determined by size-exclusion chromatography (SEC) using dextrans as standards. Glycosyl composition analysis was performed by combined gas chromatography/mass spectrometry (GC/MS) of the per-O-trimethylsilyl (TMS) derivatives of the monosaccharide methyl glycosides produced from the sample by acidic methanolysis as described previously by Santander (Santander et al., 2013). For glycosyl linkage analysis, the polysaccharide sample was prepared by pipetting out 0.6 mg of sample from a solution of known concentration in water. The sample was treated with 0.5 M methanolic HCl for 20 min and then reduced for 3 h with NaBD4. Permethylation was affected by two rounds of treatment with sodium hydroxide base (15 min) and methyl iodide (45 min). Following sample workup, the permethylated material was hydrolyzed using 2 M TFA (2 h in a sealed tube at 121°C), reduced with NaBD4, and acetylated using acetic anhydride/TFA. The sample was dried under N2 stream and reconstituted in DCM for injection into a GC-MS. Partially methylated alditol acetates were analyzed on an Agilent 7890A GC interfaced to a 5975C MSD and separation was performed on a Supelco 2331 fused silica capillary column (30 m × 0.25 mm ID).
LGG proteomic analysis
1 × 1010/mL LELNs were added to LGG when an OD600 = 0.6 was reached. LGG was collected 8 h after adding LELNs and washed twice with ice-cold 1× PBS. LGG was then resuspended in lysis buffer (2% SDS, 100 mM DTT, 20 mM Tris-HCl, pH 8.8) and lysed by sonication at 200 watts using a sonic dismembrater (Fisher Scientific). Cell debris was removed by centrifuging at 12,000× g for 30 min and the supernatant was collected for HPLC-MASS analysis. HPLC-MASS and data analysis were conducted as previously described (Teng et al., 2018).
SDS-PAGE analysis of LGG secretory proteins in the broth
1 × 1010/mL LELNs or 10 mg/mL LDPS were added into LGG cultures at OD600 = 0.6. The culture supernatant was collected at indicated times after adding LELNs or LDPS. From 200 μL of supernatant, secretory proteins were enriched by precipitation using ethanol and loaded onto a 12% SDS-PAGE gel. PBS treatment was used as control for analysis.
Total RNA extraction and quantitative real-time PCR
1 × 1010/mL LELNs or 10 mg/mL LDPS were added to LGG cultures that had an OD600 = 0.6; bacteria were collected 4 hr after adding LELNs or LDPS. 1 × 109 bacteria were mixed with 2 volumes of RNAprotect Bacteria Reagent and maintained for 10 min at room temperature immediately before harvesting. Total RNA was then extracted using an RNeasy Mini Kit according to the manufacturer's instructions. Purified RNA was digested with 5 U RNase-free DNase І for 30 min at 37°C to remove remain genomic DNA, followed by incubation for 30 min at 75°C to inactivate DNase І. Reverse transcription was performed using the SuperScrip III First-Strand Synthesis System according to the manufacturer's instruction. For LGG qRT-PCR, a housekeeping gene rpoD was used as the internal reference. For the in vitro microbiota qRT-PCR, a universal 16s rRNA primer set was used to amplify all gut bacteria as input reference. Real time PCR was conducted using QuantiTect SYBR Green PCR Kits and run on the CFX96™ Real-Time PCR system (Bio-Rad).
PKH26 membrane labeling
LGG membranes were labeled using PKH26 Fluorescent Cell Linker Kits following the manufacturer's instructions. 1 × 108 CFU LGG or LELN-LGG were incubated with 1 μL PKH26 dye for 1 min at 37°C, followed with 2 washes in ice-cold 1× PBS to remove free dye. Labeled LGG was covered with aluminum foil to protect from light. The percentage of PKH26 positive and PKH26 fluorescent intensity were detected by FACs (BD Canto) according to a previous report (Teng et al., 2018).
Msps cloning and heterologous expression
Msp genes were amplified from LGG genomic DNA by primers LGG-Msp1-OE-F/R, LGG-Msp2-OE-F/R, LGG-Msp3-OE-F/R. For Msp1 expression, msp1 gene was cloned into the pET28b-MBP-TEV vector (Currinn et al., 2016). For Msp2 and Msp3 expression, msp2 and msp3 genes were cloned into the pET28b vector. msp genes were cloned into a corresponding vector using the GeneArt Seamless Cloning and Assembly Kit following the manufacturer's instruction. Positive colonies were confirmed by colony PCR and Sanger sequencing. Constructed expression vectors were then transformed into BL21 (DE3). Msps were induced by adding 0.1 mM IPTG when an OD600 = 0.8 was achieved and the bacteria were grown overnight at 18°C. Bacteria were collected by centrifuge at 5,000× g for 10 min and lysed by sonication. Msps were purified using Ni-NTA Agarose according to the manufacturer's instructions and stored in storage buffer (50 mM Tris-HCl, 150 mM NaCl, 1 mM DTT, 10% glycerol, pH 7.6). For Msp1-MBP fusion protein, the MBP tag was removed by TEV protease following the manufacturer's instruction. Purified proteins were confirmed by SDS-PAGE.
In-vitro transcription
In-vitro transcription was conducted using a HiScribe T7 Quick High Yield RNA Synthesis Kit according to the manufacturer's instruction. T7 promoter was incorporated by T7 promoter tailored primer in the 5′ terminal of template DNA. RNA was transcribed from a 1 μg template of DNA and was then purified using the miRNeasy Mini Kit following the manufacturer's instructions. The RNA was quantified using a NanoDrop 2000 (ThermoFisher scientific).
LELNs derived Nano vector (LNV) preparation and RNA encapsulation
Lipid was extracted from LELNs using the chloroform-methanol method described previously (Folch et al., 1957). Briefly, 0.2 mL LELN sample was sequentially extracted with 0.75 mL CHCl3-methanol solution (1:2), 0.25 mL methanol, 0.25 mL water, centrifuge 10 min at 2,000 rpm and collect bottom organic phase. Lipid was dried by nitrogen flow and hydrated in 1× PBS before LNV formation. LNVs were prepared using a sonication method as previously described (Teng et al., 2018). For RNA inclusion, 1 to 10 μg RNA was incubated with PEI for 5 min on ice and then added into the hydrated lipid immediately before sonication. RNA encapsulation efficiency was determined by comparison with RNA concentration in the supernatant before and after inclusion.
Gene knockdown by antisense RNA
For msps gene knockdown, antisense RNAs with length of 100 nts to 150 nts that pair with the 5′ UTR and 5′-end CDS of the target genes were designed. For tRNA knockdown, antisense RNA paired with the full length tRNA gene was designed. 1 μg of antisense RNA obtained from in vitro transcription was encapsulated into the LNV and added to LGG cultures that had reached an OD600 = 0.6. Bacteria were collected at indicated time points. LNV without RNA inclusion was used as blank control.
Northern blot analysis of tRNA
tRNA enriched small RNA was extracted by using acid phenol (pH4.5) according to a previous report (Varshney et al., 1991). 1-5 OD Bacteria were collected by centrifugation and resuspend in RNase-free miliQ water, the suspension was extracted with 1-mL hot acid phenol for 3 times by rigorously vortex. Collect and combine the aqueous phase, the RNA was precipitated by adding 3 volume of ice-cold ethanol, and wash twice with ice-cold 75% ethanol. Northern blots were performed according to a previous report with minor modifications (Hamad et al., 2017). Briefly, 1 ug tRNA enriched small RNA was loaded onto an 8 M urea-denatured PAGE gel and run in 1× TBE with a constant voltage of 80 V. The RNA was transferred to a positively charged nylon membrane and hybridized using a specific probe. The tRNA probe was designed to pair with the anticodon loop region and labeled with biotin at the 5′ end. The biotin label tRNA probe was detected using IRDye 800cw streptavidin (LI-COR) according to the manufacturer's instructions.
In-vitro RNase P cleavage assay
An in vitro RNase P cleavage assay was conducted as previously reported with minor modifications (Guerrier-Takada et al., 1983). Briefly, 1 ug of tRNA enriched small RNA was incubated for 30 min at 37°C with different amounts of RNase P in cleavage buffer (50 mM Tris-HCl, 60 mM NH4Cl, 60 mM MgCl2, 1 mM spermine, pH7.6). The reaction was terminated by adding formamide containing loading buffer and boiling for 5 min. The amounts of specific serine tRNA were detected by Northern blot using corresponding probes.
Specific tRNA isolation and purification from total tRNA pool
Specific tRNA isolation and purification were performed as previously reported with minor modifications (Yokogawa et al., 2010). 200-OD LGG were used to isolate tRNA enriched small RNA as described above. 1 μg biotin labelled probes were immobilized to magnetic streptavidin beads by incubating at room temperature for 30 min in binding buffer (10 mM Tris–HCl, 100 mM NaCl, pH 7.6). Unbound probe was removed by washing the beads twice with washing buffer (5 mM Tris–HCl, 1 M NaCl, 0.1 mM EDTA, pH 7.6). Probe bound beads were then incubated with 1 mg tRNA enriched small RNA for 1 h at 45°C in hybridization buffer (10 mM Tris–HCl, 0.9 M tetramethylammonium chloride, 0.1mM EDTA, pH 7.6) followed by two washes with washing buffer. tRNA was eluted from the beads by heating for 5 min at 65°C in H2O.
Nucleoside modifications analysis
1 μg of tRNA purified from tRNA enriched small RNA was digested with nuclease P1 for 16 h, followed by CIP dephosphorylation. Nucleosides were analyzed by HPLC-MASS following a previous report (He et al., 2019). Briefly, digested product was lyophilized and reconstituted in equal volume of 50% ACN. After centrifugation, 2 μL of the supernatant was used for LC-MS/MS analysis. All samples were randomly analyzed on a Thermo Q Exactive HF Hybrid Quadrupole-Orbitrap Mass Spectrometer coupled with a Thermo DIONEX UltiMate 3000 HPLC system (Thermo Fisher Scientific, Waltham, MA, USA). The UltiMate 3000 HPLC system was equipped with ACQUITY UPLC HSS T3 column (150 × 2.1 mm i.d., 1.8 μm) purchased from Waters (Milford, MA, USA). The temperature of the column and nucleoside separation and mass spectrometry parameters were set to be the same as previously reported (He et al., 2019). For LC-MS data analysis, Compound Discoverer software 3.0 (Thermo Fisher Scientific, Inc., Germany) was used for spectrum deconvolution, metabolite identification, and peak list alignment. To identify metabolites, the LC-MS/MS data of pooled samples were matched to the mzVault database that contains the parent ion m/z, MS/MS spectra, and retention time of 68 nucleoside standards. The threshold for the spectral similarity of the MS/MS spectra of a metabolite standard and a spectrum of the pooled sample was set as ≥ 60 with a maximum score of 100, while the thresholds of retention time difference and m/z variations window were respectively set as ≤ 0.2 min and ≤5 ppm.
16S rDNA sequencing
LELNs were given to mice at a concentration of 5 × 109/g bodyweight twice per day by gavage. Mice were sacrificed on day 7 after gavaging and microbiota were isolated from collected small intestine contents. Total genomic DNA was purified using a QIAamp DNA Microbiome Kit following the manufacturer's instruction. The V3-V4 region was amplified using primer 319F (ACTCCTACGGGAGGCAGCAG) and 806R (GGACTACHVGGGTWTCTAAT) and the amplified product was used to prepare library for sequencing.
Quantification and statistical analysis
All statistics analyses were performed using GraphPad Prism 8 software. The data are presented as values with standard deviation (mean ± SD). The significance was analyzed using t-tests for two-group analyses or ANOVA for multiple-group analyses. The significance is shown as p ≤ 0.05∗, p ≤ 0.01∗∗, p ≤ 0.001∗∗∗ and p ≤ 0.0001∗∗∗∗, p > 0.05 was considered not significant (n.s.). Otherwise as indicated, all statistical analyses were compared with control groups. Data are representative of three independent experiments.
Acknowledgments
We thank Dr. Parastoo Azadi at the Complex Carbohydrate Research Center of University of Georgia for polysaccharide analysis and Dr. J. Ainsworth for editorial assistance. Funding: This work was supported by a grant from the National Institutes of Health (NIH) (R01AT008617, R01AA023190, P20GM125504) and the Robley Rex VA Medical Center Merit Review Grants (H-GZ). H.-G. Z. is supported by a Research Career Scientist (RCS) Award. Y.T. is supported by COBRE Pilot Project, NIH (P20GM125504). W.F. is supported by R01AA023190. M.M. and X.Z. are supported by P50 AA024337 and P20 GM113226. J.P was supported by the National Institute of General Medical Sciences of the National Institutes of Health grant P20GM103436.
Author contributions
C.L. and H.-G.Z. designed the study, analyzed and interpreted the data, and prepared the manuscript; C.L. performed the experiments and interpreted the data; M.S., Y.T., and J.W.P. analyzed RNA-seq data; L.H. and X.Z. performed nucleoside modifications analysis; M.L.M. performed protein analysis; J.M., F.X., and X.Z. provided histological analysis; A.K., K.S., L.Z., S.C., S.Z., W.F., and J.Y interpreted the findings.
Declaration of interests
The authors declare no competing interests.
Published: June 25, 2021
Footnotes
Supplemental information can be found online at https://doi.org/10.1016/j.isci.2021.102511.
Supplemental information
References
- Bauerl C., Perez-Martinez G., Yan F., Polk D.B., Monedero V. Functional analysis of the p40 and p75 proteins from Lactobacillus casei BL23. J. Mol. Microbiol. Biotechnol. 2010;19:231–241. doi: 10.1159/000322233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bibbo S., Ianiro G., Giorgio V., Scaldaferri F., Masucci L., Gasbarrini A., Cammarota G. The role of diet on gut microbiota composition. Eur. Rev. Med. Pharmacol. Sci. 2016;20:4742–4749. [PubMed] [Google Scholar]
- Chen R., Li Y., Dong H., Liu Z., Li S., Yang S., Li X. Optimization of ultrasonic extraction process of polysaccharides from Ornithogalum Caudatum Ait and evaluation of its biological activities. Ultrason. Sonochem. 2012;19:1160–1168. doi: 10.1016/j.ultsonch.2012.03.008. [DOI] [PubMed] [Google Scholar]
- Claes I.J., Schoofs G., Regulski K., Courtin P., Chapot-Chartier M.P., Rolain T., Hols P., von Ossowski I., Reunanen J., de Vos W.M. Genetic and biochemical characterization of the cell wall hydrolase activity of the major secreted protein of Lactobacillus rhamnosus GG. PLoS One. 2012;7 doi: 10.1371/journal.pone.0031588. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Condezo-Hoyos L., Perez-Lopez E., Ruperez P. Improved evaporative light scattering detection for carbohydrate analysis. Food Chem. 2015;180:265–271. doi: 10.1016/j.foodchem.2015.02.039. [DOI] [PubMed] [Google Scholar]
- Coughlin D.J., Pleiss J.A., Walker S.C., Whitworth G.B., Engelke D.R. Genome-wide search for yeast RNase P substrates reveals role in maturation of intron-encoded box C/D small nucleolar RNAs. Proc. Natl. Acad. Sci. U S A. 2008;105:12218–12223. doi: 10.1073/pnas.0801906105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Currinn H., Guscott B., Balklava Z., Rothnie A., Wassmer T. APP controls the formation of PI(3,5)P(2) vesicles through its binding of the PIKfyve complex. Cell. Mol. Life Sci. 2016;73:393–408. doi: 10.1007/s00018-015-1993-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Folch J., Lees M., Sloane Stanley G.H. A simple method for the isolation and purification of total lipides from animal tissues. J. Biol. Chem. 1957;226:497–509. [PubMed] [Google Scholar]
- Goodarzi H., Nguyen H.C.B., Zhang S., Dill B.D., Molina H., Tavazoie S.F. Modulated expression of specific trnas drives gene expression and cancer progression. Cell. 2016;165:1416–1427. doi: 10.1016/j.cell.2016.05.046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gu C., Begley T.J., Dedon P.C. Trna modifications regulate translation during cellular stress. FEBS Lett. 2014;588:4287–4296. doi: 10.1016/j.febslet.2014.09.038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guerrier-Takada C., Gardiner K., Marsh T., Pace N., Altman S. The RNA moiety of ribonuclease P is the catalytic subunit of the enzyme. Cell. 1983;35:849–857. doi: 10.1016/0092-8674(83)90117-4. [DOI] [PubMed] [Google Scholar]
- Gunn J.S. Mechanisms of bacterial resistance and response to bile. Microbes Infect. 2000;2:907–913. doi: 10.1016/s1286-4579(00)00392-0. [DOI] [PubMed] [Google Scholar]
- Hamad A., Jeong H.-Y., Han J.-H., Rather I. Detection of cytosolic tRNA in mammal by Northern blot analysis. Bangladesh J. Pharmacol. 2017;12:243–250. [Google Scholar]
- He L., Wei X., Ma X., Yin X., Song M., Donninger H., Yaddanapudi K., Mcclain C.J., Zhang X. Simultaneous quantification of nucleosides and nucleotides from biological samples. J. Am. Soc. Mass Spectrom. 2019;30:987–1000. doi: 10.1007/s13361-019-02140-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huber S.M., Leonardi A., Dedon P.C., Begley T.J. The versatile roles of the tRNA epitranscriptome during cellular responses to toxic exposures and environmental stress. Toxics. 2019;7:17. doi: 10.3390/toxics7010017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Irving S.E., Corrigan R.M. Triggering the stringent response: signals responsible for activating (p)ppGpp synthesis in bacteria. Microbiology (Reading) 2018;164:268–276. doi: 10.1099/mic.0.000621. [DOI] [PubMed] [Google Scholar]
- Kazantsev A.V., Pace N.R. Bacterial RNase P: a new view of an ancient enzyme. Nat. Rev. Microbiol. 2006;4:729–740. doi: 10.1038/nrmicro1491. [DOI] [PubMed] [Google Scholar]
- Kristoffersen S.M., Ravnum S., Tourasse N.J., Okstad O.A., Kolsto A.B., Davies W. Low concentrations of bile salts induce stress responses and reduce motility in Bacillus cereus ATCC 14579. J. Bacteriol. 2007;189:5302–5313. doi: 10.1128/JB.00239-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li L., Abou-Samra E., Ning Z., Zhang X., Mayne J., Wang J., Cheng K., Walker K., Stintzi A., Figeys D. An in vitro model maintaining taxon-specific functional activities of the gut microbiome. Nat. Commun. 2019;10:4146. doi: 10.1038/s41467-019-12087-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lorenz C., Lünse C.E., Mörl M. tRNA modifications: impact on structure and thermal adaptation. Biomolecules. 2017;7:35. doi: 10.3390/biom7020035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Merritt M.E., Donaldson J.R. Effect of bile salts on the DNA and membrane integrity of enteric bacteria. J. Med. Microbiol. 2009;58:1533–1541. doi: 10.1099/jmm.0.014092-0. [DOI] [PubMed] [Google Scholar]
- Miles A.A., Misra S.S., Irwin J.O. The estimation of the bactericidal power of the blood. J. Hyg. (Lond) 1938;38:732–749. doi: 10.1017/s002217240001158x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Molinero N., Ruiz L., Milani C., Gutierrez-Diaz I., Sanchez B., Mangifesta M., Segura J., Cambero I., Campelo A.B., Garcia-Bernardo C.M. The human gallbladder microbiome is related to the physiological state and the biliary metabolic profile. Microbiome. 2019;7:100. doi: 10.1186/s40168-019-0712-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Motorin Y., Helm M. tRNA stabilization by modified nucleotides. Biochemistry. 2010;49:4934–4944. doi: 10.1021/bi100408z. [DOI] [PubMed] [Google Scholar]
- Nava G.M., Friedrichsen H.J., Stappenbeck T.S. Spatial organization of intestinal microbiota in the mouse ascending colon. ISME J. 2011;5:627–638. doi: 10.1038/ismej.2010.161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oh D.J., Lee G.M., Francis K., Palsson B.O. Phototoxicity of the fluorescent membrane dyes PKH2 and PKH26 on the human hematopoietic KG1a progenitor cell line. Cytometry. 1999;36:312–318. doi: 10.1002/(sici)1097-0320(19990801)36:4<312::aid-cyto5>3.0.co;2-v. [DOI] [PubMed] [Google Scholar]
- Park O.H., Ha H., Lee Y., Boo S.H., Kwon D.H., Song H.K., Kim Y.K. Endoribonucleolytic cleavage of m(6)A-containing RNAs by RNase P/MRP complex. Mol. Cell. 2019;74:494–507.e8. doi: 10.1016/j.molcel.2019.02.034. [DOI] [PubMed] [Google Scholar]
- Ruiz L., Margolles A., Sanchez B. Bile resistance mechanisms in lactobacillus and bifidobacterium. Front. Microbiol. 2013;4:396. doi: 10.3389/fmicb.2013.00396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Santander J., Martin T., Loh A., Pohlenz C., Gatlin D.M., Curtiss R. Mechanisms of intrinsic resistance to antimicrobial peptides of Edwardsiella ictaluri and its influence on fish gut inflammation and virulence. Microbiology. 2013;159:1471–1486. doi: 10.1099/mic.0.066639-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schwartz M.H., Wang H., Pan J.N., Clark W.C., Cui S., Eckwahl M.J., Pan D.W., Parisien M., Owens S.M., Cheng B.L. Microbiome characterization by high-throughput transfer RNA sequencing and modification analysis. Nat. Commun. 2018;9:5353. doi: 10.1038/s41467-018-07675-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sorensen M.A., Fehler A.O., Lo Svenningsen S. Transfer RNA instability as a stress response in Escherichia coli: rapid dynamics of the tRNA pool as a function of demand. RNA Biol. 2018;15:586–593. doi: 10.1080/15476286.2017.1391440. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stothard P. The sequence manipulation suite: JavaScript programs for analyzing and formatting protein and DNA sequences. Biotechniques. 2000;28:1102–1104. doi: 10.2144/00286ir01. [DOI] [PubMed] [Google Scholar]
- Sundaram K., Miller D.P., Kumar A., Teng Y., Sayed M., Mu J., Lei C., Sriwastva M.K., Zhang L., Jun Y. Plant-derived exosomal nanoparticles inhibit pathogenicity of porphyromonas gingivalis. iScience. 2019;21:308–327. doi: 10.1016/j.isci.2019.10.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Teng Y., Ren Y., Sayed M., Hu X., Lei C., Kumar A., Hutchins E., Mu J., Deng Z., Luo C. Plant-derived exosomal MicroRNAs shape the gut microbiota. Cell Host Microbe. 2018;24:637–652 e8. doi: 10.1016/j.chom.2018.10.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Torrent M., Chalancon G., de Groot N.S., Wuster A., Madan Babu M. Cells alter their tRNA abundance to selectively regulate protein synthesis during stress conditions. Sci. Signal. 2018;11 doi: 10.1126/scisignal.aat6409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Varshney U., Lee C.P., Rajbhandary U.L. Direct analysis of aminoacylation levels of tRNAs in vivo. Application to studying recognition of Escherichia coli initiator tRNA mutants by glutaminyl-tRNA synthetase. J. Biol. Chem. 1991;266:24712–24718. [PubMed] [Google Scholar]
- Vinderola C.G., Reinheimer J. Lactic acid starter and probiotic bacteria: a comparative "in vitro" study of probiotic characteristics and biological barrier resistance. Food Res. Int. 2003;36:895–904. [Google Scholar]
- Wahlstrom A., Sayin S.I., Marschall H.U., Backhed F. Intestinal crosstalk between bile acids and microbiota and its impact on host metabolism. Cell Metab. 2016;24:41–50. doi: 10.1016/j.cmet.2016.05.005. [DOI] [PubMed] [Google Scholar]
- Yokogawa T., Kitamura Y., Nakamura D., Ohno S., Nishikawa K. Optimization of the hybridization-based method for purification of thermostable tRNAs in the presence of tetraalkylammonium salts. Nucleic Acids Res. 2010;38:e89. doi: 10.1093/nar/gkp1182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhong J., Xiao C., Gu W., Du G., Sun X., He Q.Y., Zhang G. Transfer RNAs mediate the rapid adaptation of Escherichia coli to oxidative stress. PLoS Genet. 2015;11 doi: 10.1371/journal.pgen.1005302. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The accession number for the sequenced data reported in this paper is NCBI Sequence Read Archive (SRA): PRJNA722680.