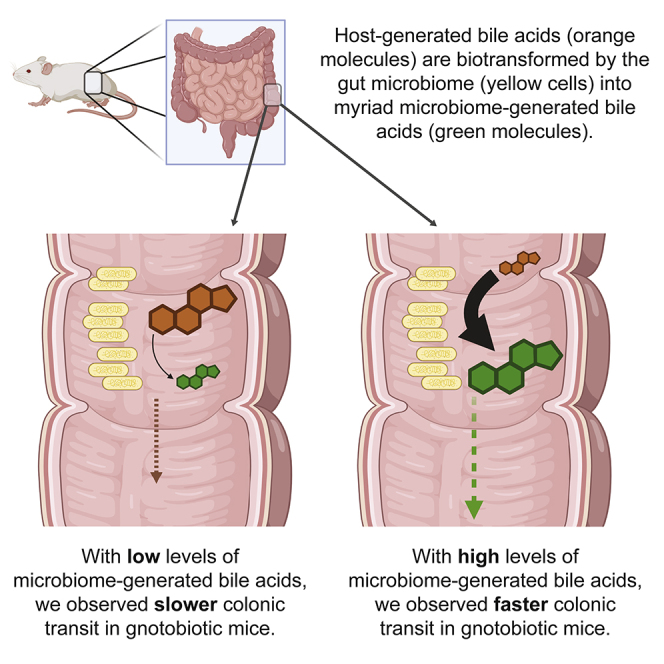
Summary
Gut motility is regulated by the microbiome via mechanisms that include bile acid metabolism. To localize the effects of microbiome-generated bile acids, we colonized gnotobiotic mice with different synthetic gut bacterial communities that were metabolically phenotyped using a functional in vitro screen. Using two different marker-based assays of gut transit, we inferred that bile acids exert effects on colonic transit. We validated this using an intra-colonic bile acid infusion assay and determined that these effects were dependent upon signaling via the bile acid receptor, TGR5. The intra-colonic bile acid infusion experiments further revealed sex-biased bile acid-specific effects on colonic transit, with lithocholic acid having the largest pro-motility effect. Transcriptional responses of the enteric nervous system (ENS) were stereotypic, regional, and observed in response to different microbiota, their associated bile acid profiles, and even to a single diet ingredient, evidencing exquisite sensitivity of the ENS to environmental perturbations.
Subject areas: Microbiology, Microbiome, Gastroenterology
Graphical abstract
Highlights
-
•
Gut microbiome-generated bile acids regulate colonic transit via TGR5.
-
•
Lithocholic acid had the largest colonic pro-motility effect.
-
•
Bile acids exert sex-biased effects on gut transit times.
-
•
Enteric nervous system transcriptional responses are regional and microbiome-specific.
Microbiology; Microbiome; Gastroenterology
Introduction
Disturbances and disorders of gastrointestinal motility, a physiologic parameter critical to optimizing digestion and nutrition, form the basis for a shared set of common human experiences. Interactions between diet, the gut microbiome, and the enteric nervous system (ENS) regulate gut motility (Abrams and Bishop, 1967; Husebye et al., 1994; Anitha et al., 2012; Kashyap et al., 2013; Dey et al., 2015; Bhattarai et al., 2018). Bile acids are metabolites that lie at the intersection of all these factors, metabolically at the interface between the host and its gut microbiome. Produced from cholesterol in the liver, host-generated primary bile acids are secreted into the first part of the small intestine. As they transit through the gut, they are subjected to gut bacterial metabolism, beginning with deconjugation (i.e., removal of glycine or taurine) via bile salt hydrolases (BSH). BSHs set the stage for myriad subsequent microbiome-mediated biotransformations of primary bile acids into secondary bile acids, each varying in their effects on host and microbial biology via incompletely understood pathways. We previously showed in a gnotobiotic model that unconjugated bile acids generated by bacterial BSHs correlated with faster gut transit (Dey et al., 2015). The correlation between BSHs and motility implicates unconjugated primary and secondary bile acids, consistent with observations in individuals with diarrhea-predominant irritable bowel syndrome (Zhao et al., 2020). Bile acid-mediated effects on gut transit times require RET signaling in the ENS and can be modulated by dietary ingredients that stimulate bile secretion (e.g., turmeric) (Dey et al., 2015). Whether bile acids exert regional or global motility effects throughout the gut is unknown, as are the ENS transcriptional responses underpinning the observed physiologic responses. In the present study, we sought to elucidate the biogeography of bile acid-regulated motility phenotypes and to characterize ENS-specific gene expression in the context of different microbiomes (plus germ-free controls) and their associated bile acids.
Results
Design of synthetic gut microbial communities
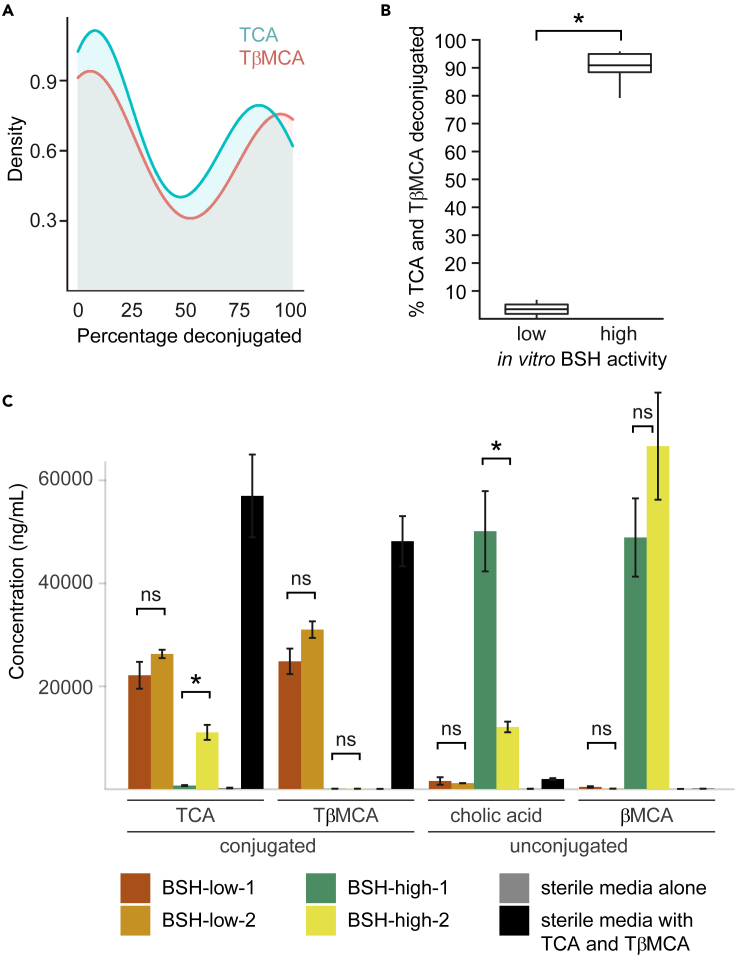
Germ-free (GF) animals are known to exhibit slower gut motility and less complex bile acid profiles than colonized animals (Husebye et al., 2001; Sayin et al., 2013; Kashyap et al., 2013; Dey et al., 2015). To decouple gut microbial colonization from bile acid metabolism, we designed synthetic bacterial communities based on an in vitro functional screen of bile acid metabolism. We anaerobically cultured 30 consortia, each comprising 2-21 gut bacterial strains selected from a pool of 30 type strains (Table S1), in media containing conjugated primary bile acid substrates (taurocholic acid [TCA], tauro-beta-muricholic acid [TβMCA]). We made in silico genome-based predictions of which bile acids the bacterial consortia would generate, and we tested our predictions using liquid chromatography mass spectrometry (LC-MS) to quantitate bile acids in culture media collected after 48 hr. We found that deconjugation, which was predicted in all co-cultures (i.e., all co-cultures contained a bsh-encoding organism), was observed in 67% of instances (Table S1), consistent with our prior observation that genome-based predictions of BSH activities are imperfect (Dey et al., 2015). A bimodal distribution of BSH activities was observed, thereby permitting classification of consortia as “BSH-low” or “BSH-high” (Figure 1A). BSH-high consortia deconjugated TCA and TβMCA to a significantly greater degree than BSH-low consortia (91% ± 2% vs 4% ± 3% [mean ± SEM], p< 0.006, two-tailed t-test; Figure 1B), mirroring what we observed in fecal samples from gnotobiotic mice colonized with fecal microbiota from conventionally housed specific pathogen-free (SPF) mice (92.7% ± 2.4%) compared to GF mice (1.9% ± 0.6%) (Table S2).
Figure 1.
Bile acid metabolism of synthetic bacterial communities (See also Figure S1 and Table 1)
(A) Bimodal distribution of in vitro low or high BSH activities of 30 synthetic communities.
(B) Percent deconjugation of conjugated primary bile acid substrates of “BSH-low” and “BSH-high” consortia.
(C) Bile acid profiles of 4 synthetic consortia, along with controls.
Statistical significance was determined using a Student's two-tailed t-test; ∗p< 0.05.
In (B), the horizontal lines within boxes denote the mean values, while lower and upper boundaries of boxes represent the 25th and 75th percentiles, respectively, and whiskers represent 1.5 times the interquartile range. In (C), bile acid concentrations are represented by mean values ± SEM.
Co-cultures containing Bacteroides vulgatus, Dorea longicatena, or Clostridium asparagiforme were consistently associated with robust bile salt deconjugation, whereas co-cultures containing Bacteroides finegoldii were associated with poor deconjugation (consistent with B. finegoldii encoding a bsh phylotype associated with low enzyme activity (Song et al., 2019)). Co-cultures containing Clostridium sporogenes were associated with highly variable deconjugation (5-98% TCA deconjugation, 1-98% TβMCA deconjugation), suggesting that interspecific interactions may regulate its bile acid metabolism. From these consortia, we identified two pairs of 6-member consortia that were divergent in BSH activity level, one that was genus-matched (“BSH-low-1” and “BSH-high-1”) and another that was not (“BSH-high-2” and “BSH-low-2”) (Table 1). While the unconjugated primary bile acids βMCA and cholic acid were the most abundant bile acids detected in BSH-high-1 cultures, βMCA predominated in BSH-high-2 cultures (Figure 1C). Thus, these two “BSH-high” consortia presented an opportunity to compare and contrast the gut motility phenotypes associated with two synthetic communities that have similarly high but slightly divergent BSH activities.
Table 1.
| Microbiota | Community members | Vendor | Catalog number |
|---|---|---|---|
| Complete mouse microbiota | Fecal microbiota suspension | NA | NA |
| BSH-low-1 | Bacteroides caccae | ATCC | 43185 |
| Bacteroides finegoldii | DSM | 17565 | |
| Clostridium leptum | DSM | 753 | |
| Clostridium scindens | ATCC | 35704 | |
| Dorea formicigenerans | ATCC | 27755 | |
| Ruminococcus torques | ATCC | 27756 | |
| BSH-high-1 | Bacteroides thetaiotaomicron | VPI | 5482 |
| Bacteroides vulgatus | ATCC | 8482 | |
| Clostridium asparagiforme | DSM | 15981 | |
| Clostridium sporogenes | ATCC | 15579 | |
| Dorea longicatena | DSM | 13814 | |
| Ruminococcus obeum | ATCC | 29174 | |
| BSH-low-2 | Citrobacter youngae | ATCC | 29220 |
| Dorea formicigenerans | ATCC | 27755 | |
| Enterobacter cancerogenus | ATCC | 35316 | |
| Escherichia fergusonii | ATCC | 35469 | |
| Megamonas funiformis | DSM | 19343 | |
| Ruminococcus torques | ATCC | 27756 | |
| BSH-high-2 | Bacteroides vulgatus | ATCC | 8482 |
| Clostridium asparagiforme | DSM | 15981 | |
| Clostridium sporogenes | ATCC | 15579 | |
| Lactobacillus ruminus | ATCC | 25644 | |
| Ruminococcus gnavus | ATCC | 29149 | |
| Ruminococcus obeum | ATCC | 29174 |
Membership of synthetic communities possessing high or low BSH activities that were used in gnotobiotic studies.
Validation of serial deployment of two assays quantifying gut transit
We characterized gut transit in a gnotobiotic model using two methods: a fecal marker-based assay (which permits quantification of whole gut transit times (Dey et al., 2015) via oral gavage with a red carmine dye solution followed by timing of the interval to fecal passage) and a gut fluorescence-based assay (in which fluorescein isothiocyanate [FITC] is administered via oral gavage, with characterization of the spatial resolution of fluorescence throughout the gut [Samuel et al., 2008]). In the FITC-based assay, we euthanized mice at a defined interval after gavage with an FITC-dextran solution (typically 2 hr, except in one experiment described below in which a 4-hr interval was used), partitioned gastrointestinal tracts into 12 segments numbered 1-12 from proximal to distal gut, and quantified FITC detected in each segment using a fluorometer. We then calculated geometric mean (g) using the following equation:
in which i indicates the gut segment number (1 is stomach, 2-9 are small intestine, and 10-12 are colon) and FITCi represents the proportion of total gut FITC detected in that gut segment.
To establish reference ranges, we deployed these two assays in GF wild-type Swiss Webster mice and age-matched gnotobiotic mice colonized with an SPF fecal microbiota (Figure S1A; Table S3). Compared to colonized mice, GF mice had significantly longer whole gut transit times (5.2 ± 0.3 hr in GF mice vs 2.7 ± 0.2 hr in colonized mice, p = 0.0006, two-tailed t-test), lower geometric means of fluorescence (6.7 ± 0.3 in GF mice vs 9.2 ± 0.3 in colonized mice, p = 0.0004, two-tailed t-test), and lower percent of total gut FITC detected in the colon (1.1% ± 0.2% in GF mice vs 61.3% ± 7.6% in colonized mice, p = 0.004, two-tailed t-test).
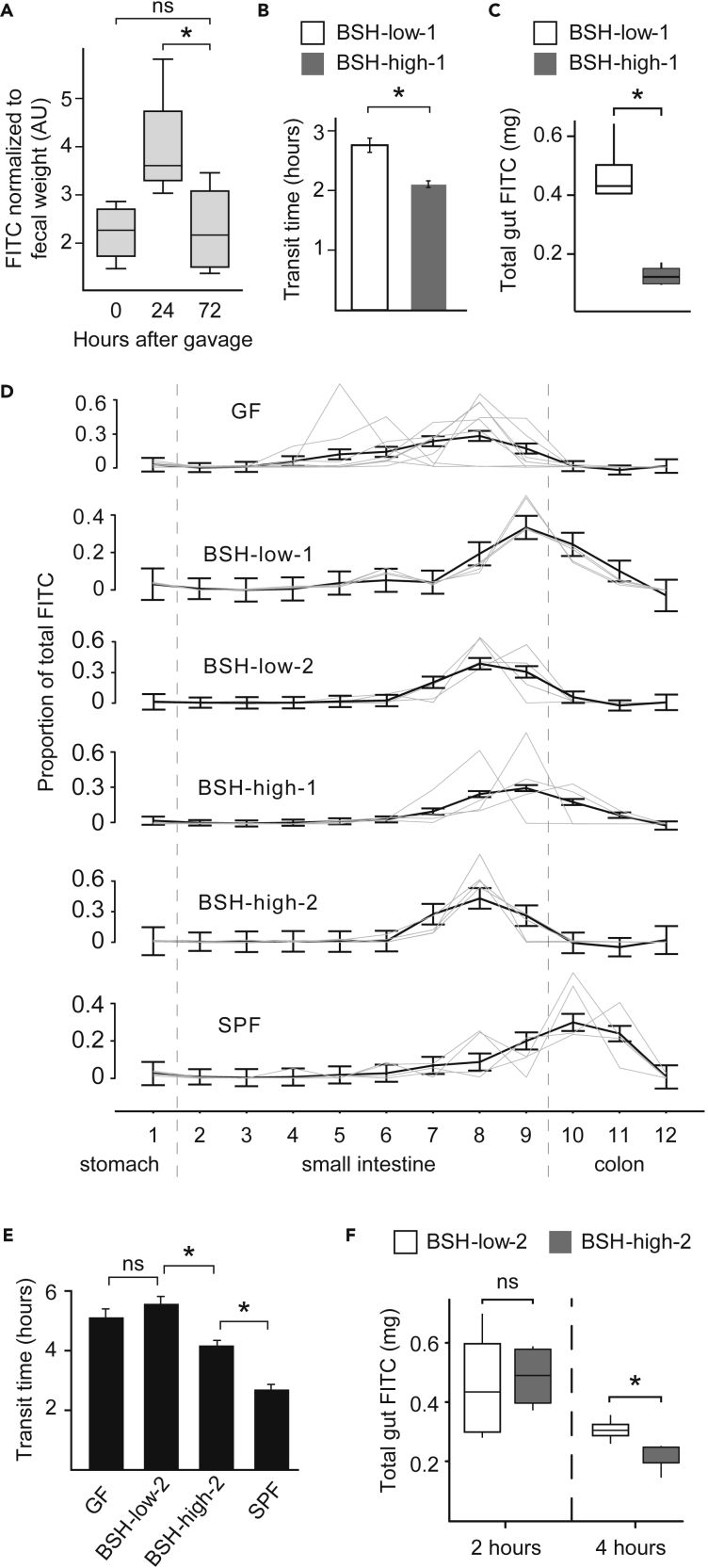
To determine whether these assays may be used in tandem in a given mouse, we first estimated the duration for which motility markers persist in the guts of gnotobiotic mice. One week after colonization of gnotobiotic mice with the 12 strains comprising the BSH-high-1 and BSH-low-1 consortia, we gavaged mice with FITC and measured fluorescence in fecal pellets collected at baseline and then 24 hr and 72 hr later (n = 4 mice). Fluorescence at 72 hr was significantly lower than at 24 hr (p< 0.04, paired two-tailed Student’s t-test; Figure 2A) but not significantly different from baseline levels (p = 0.9, paired two-tailed Student’s t-test). We validated this 3-day interval in age-matched gnotobiotic mice colonized with the BSH-high-1 or BSH-low-1 consortia (n = 7–8 mice/consortium). Approximately half of each cohort was subjected to the carmine dye assay 3 days prior to the FITC assay, which was performed in all mice at the time of euthanasia. Neither total gut FITC nor geometric means significantly differed if carmine had been previously administered (p> 0.2, two-tailed t-test), suggesting that carmine did not interfere with FITC readouts 3 days later. Indeed, carmine dye was no longer visible in fecal pellets 3 days after gavage.
Figure 2.
Differential gut bacterial BSH activity regulates colonic motility (See also Figures S3 and S7 and Table 1)
(A) Clearance of a FITC-labeled marker of gut transit administered via oral gavage by 72 hr.
(B) Whole gut transit times in gnotobiotic mice colonized by BSH-low-1 or BSH-high-1 communities.
(C) Residual gut FITC is lower in gnotobiotic mice colonized with the BSH-high-1 consortium than the BSH-low-1 consortium.
(D) Intestinal FITC distributions in gnotobiotic mice either maintained as GF or harboring different microbiota 2 hr after gavage.
(E) Whole gut transit times in BSH-high-2- and BSH-low-2-colonized mice, along with GF and SPF microbiota-colonized gnotobiotic mice.
(F) Residual gut FITC in mice colonized with the BSH-high-2 or BSH-low-2 consortia at 2 hr and at 4 hr after gavage.
Statistical significance was determined using a Student's two-tailed t-test; ∗p< 0.05. In (A), (C), and (F), the horizontal lines within boxes denote the mean values, while lower and upper boundaries of boxes represent the 25th and 75th percentiles, respectively, and whiskers represent 1.5 times the interquartile range. In (B), (D), and (E), mean values ± SEM are shown.
Gut bacterial bile acid metabolism regulates colonic transit
We had previously observed that turmeric, which has cholekinetic properties (Rasyid and Lelo, 1999; Rasyid et al., 2002; Marciani et al., 2013), could be administered to mice in order to stimulate bile secretion and thereby elicit bile acid-mediated microbiome-dependent physiologic responses (Dey et al., 2015). Consistent with our prior experience, we found that gnotobiotic Swiss Webster mice colonized with the BSH-high-1 consortium experienced significantly faster whole gut transit times compared to age-matched mice harboring the BSH-low-1 consortium when fed a turmeric-containing diet (n = 4 mice/group; 2.2 ± 0.1 vs 2.9 ± 0.1 hr, p< 0.02, two-tailed t-test; Figures 2B and S1B), but this difference was not significant in the same mice in the setting of a bland diet (Table S3). In separate cohorts of gnotobiotic mice fed a monotonous turmeric-containing diet and then subjected to the FITC motility assay, we observed that total gut FITC was significantly lower in BSH-high-1-colonized than in BSH-low-1-colonized mice (n = 4 mice/group; p< 0.05, two-tailed t-test; Figures 2C and S1A), evidencing greater fecal passage of this fluorophore marker 2 hr after oral gavage in BSH-high-1-colonized mice. However, the distribution of gut FITC was not skewed toward the more distal gut in BSH-high-1-colonized mice; rather, FITC intensity peaked in the distal small intestine in both treatment groups (Figure 2D). Together, these observations suggest that the FITC motility marker passed through the gastrointestinal tract at a relatively even pace from the stomach to the distal small intestine regardless of gut bacterial BSH activity; once past the cecum, however, the marker passed through the colons of mice harboring gut microbiota with high BSH activity more quickly.
To test whether this phenomenon was robust to different microbiota, we then colonized gnotobiotic mice with either the BSH-high-2 or BSH-low-2 consortium and administered a monotonous turmeric-containing diet throughout the experiment (n = 4 mice/group; Figure S1A). Similar to our observations in mice colonized with the first pair of consortia, mice harboring BSH-high-2 microbes had significantly faster whole gut transit times compared to BSH-low-2 mice (4.2 ± 0.2 hr vs 5.6 ± 0.3 hr, p< 0.006, two-tailed t-test) (Figure 2E). However, both intestinal FITC distributions and total gut FITC at 2 hr were similar in these two cohorts (Figures 2D and 2F). We then assessed FITC distributions 4 hr after oral gavage (rather than 2 hr after) in separate cohorts of gnotobiotic mice (n = 4–5 mice/group), reasoning that sufficient FITC may not have transited to the colon within 2 hr given the overall slower transit times compared to mice colonized with the first pair of consortia. Indeed, while intestinal FITC distributions were similar in both cohorts of mice, total gut FITC was significantly lower in BSH-high-2 than BSH-low-2 mice (10 ± 1 vs 15 ± 1 μg/mL, p = 0.01, two-tailed t test; Figure 2F), again indicating that the digesta passes through the stomach and small intestine at similar rates, with motility diverging in the colon on the basis of gut bacterial BSH activity. Both sets of comparisons of gnotobiotic mice colonized with bacterial communities exhibiting high or low BSH activity suggested that the colon is the site at which bacterial BSHs regulate gut transit.
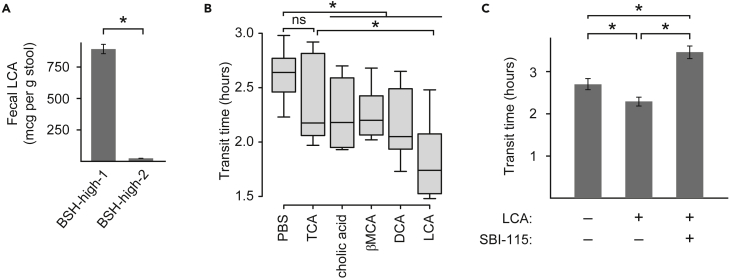
As predicted by our in vitro functional screen, fecal concentrations of unconjugated bile acids were greater in mice colonized with gut microbes possessing high BSH activity (p = 0.02, F1,26 = 5.9, one-way ANOVA). Total unconjugated bile acid concentrations correlated with faster transit in vivo across all treatment groups (Spearman rho −0.66, p< 10−5). BSH-high-1-colonized mice, which had faster transit times than BSH-high-2-colonized mice, had significantly greater fecal lithocholic acid (LCA) concentrations (890 ± 40 vs 20 ± 0.5 mcg/g feces, p< 0.0002, two-tailed t-test; Figure 3A). This suggested that in addition to BSH activity, variations in gut bacterial secondary bile acid metabolism further regulated gut transit. Given the association between LCA production and fast gut transit in BSH-high-1-colonized mice, we further hypothesized that LCA is a pro-motility bile acid in the colon.
Figure 3.
Unconjugated bile acids, notably LCA, expedite colonic transit (See also Figures S2,S4, and S7 and Table 1)
(A) Fecal LCA concentrations differ between mice colonized with the two BSH-high consortia.
(B) Intra-colonic bile acid infusions modulate gut transit.
(C) The pro-motility effect of lithocholic acid is inhibited by the TGR5 inhibitor SBI-115.
Statistical significance was determined using a two-tailed Student’s t-test (A) or paired two-tailed Student's t-test (B and C) as indicated in the text; ∗p< 0.05.
In (B), the horizontal lines within boxes denote the mean values, while lower and upper boundaries of boxes represent the 25th and 75th percentiles, respectively, and whiskers represent 1.5 times the interquartile range. In (A) and (C), mean values ± SEM are shown.
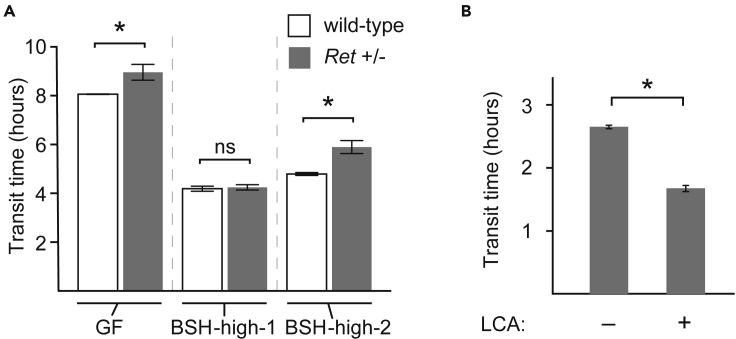
To validate our model that microbiome-generated bile acids regulate colonic transit, we next directly quantified the effects of intra-colonic infusions of cholic acid, βMCA, DCA, and LCA in comparison with the conjugated primary bile acid TCA as well as a buffer-only negative control (n = 8 wild-type SPF mice; Table S4). We performed intra-colonic infusions per rectum rather than oral gavage because unconjugated bile acids are passively absorbed in the small intestine, and therefore orally administered unconjugated bile acids may not reach the colon. Mice received intra-colonic bile acid (or buffer-only) infusions serially (with infusions ≥48 hr apart), and transit times were measured concurrently via the carmine assay. In repeated measures ANOVA in which transit time was the dependent variable, the specific bile acid infused was a significant determinant (p = 0.001); in addition, sex was found to be a significant variable (p = 0.0001), as discussed in the following section. Compared to the negative control, unconjugated bile acids (i.e., bile acids requiring microbiome-encoded enzymes for generation) exerted significant pro-motility effects in the colon (p< 0.04, paired two-tailed t-tests; Figure 3B). In contrast, the transit time effect of the conjugated bile acid TCA was not significant in comparison to the negative control (p = 0.2, paired two-tailed t-test; Figure 3B), suggesting that bile acid effects on gut transit may be more attributable to pro-motility effects of unconjugated bile acids rather than anti-motility effects of conjugated bile acids. Colonic LCA infusion induced the fastest transit times, consistent with our observations in BSH-high-1-colonized mice.
We hypothesized that bile acid-mediated effects on transit times were transmitted by TGR5, a bile acid receptor previously reported to be expressed by enteric neurons (Poole et al., 2010). To test this hypothesis, we performed intra-colonic infusions of LCA ± SBI-115 (a TGR5 inhibitor) in a separate cohort of wild-type SPF mice (n = 6). Compared to placebo, LCA infusion again resulted in significantly faster transit (p = 0.004, paired two-tailed t-test). Consistent with our hypothesis, concomitant administration of SBI-115 was associated with significantly slower transit than seen with LCA alone (p = 0.002, paired two-tailed t-test; Figure 3C), suggesting that TGR5 inhibition blocked LCA’s effects. Transit times observed following SBI-115 infusion were significantly slower than with placebo (p< 0.02, paired two-tailed t-test), suggesting blockade of signals transmitted by endogenous colonic bile acids generated by the SPF mouse microbiota. Together, these data support the model that microbiome-encoded bile acid metabolism modulates colonic motility in a TGR5-dependent fashion.
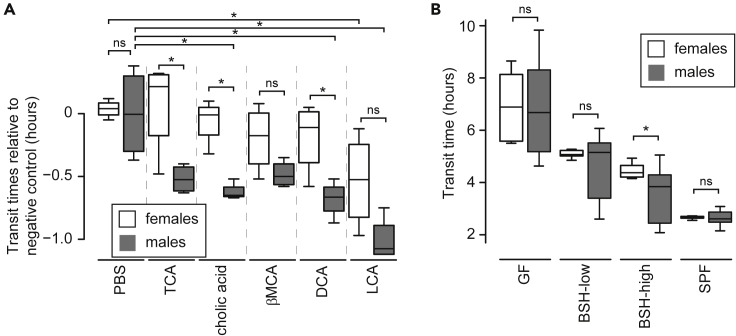
Bile acids have sex-biased effects on gut transit times
In our intra-colonic bile acid infusion experiment above, we found sex to be a significant variable determining transit time. In males, cholic acid, DCA, and LCA induced significantly faster transit compared to the negative control, whereas in females, LCA was the only bile acid that induced significantly faster transit (p< 0.05, paired two-tailed t-test; Figure 4A). Pro-motility effects were generally stronger in males than females for each bile acid tested (Figure 4A). This was true even for TCA, a conjugated primary bile acid produced by the host without any microbial contribution, suggesting that a sex-biased response to bile acids exists at baseline. Since diet can determine bile secretion, diet alone could theoretically mediate differential responses in males versus females. Nonetheless, the sex-biased effect seen with TCA was modest in comparison to the effects of some microbiome-encoded bile acids.
Figure 4.
Sex bias in bile acid effects on gut transit (See also Figures S3,S4, and S7 and Table 1)
(A) Sex bias in transit times following intra-colonic bile acid infusions. In order to normalize recorded transit times (Table S4) to a single value, each recorded transit time was subtracted from the mean transit time of males after intra-colonic infusion with the buffer-only negative control.
(B) Sex-biased transit time differences in mice colonized with BSH-high consortia but not in mice harboring BSH-low consortia or SPF microbiota (including conventionally raised mice; transit times measured following intra-colonic infusion of PBS included), or in GF controls.
Statistical significance was determined using a Student's two-tailed t-test; ∗p< 0.05. In (A) and (B), the horizontal lines within boxes denote the mean values, while lower and upper boundaries of boxes represent the 25th and 75th percentiles, respectively, and whiskers represent 1.5 times the interquartile range.
In light of these observations, we revisited the transit times recorded in the gnotobiotic experiments above to ascertain whether sex biases were associated with our synthetic communities. We found that males had lower transit times (i.e., faster motility) than females in cohorts colonized with BSH-high consortia (p = 0.009, two-tailed t-test) (Figure 4B). Significant differences were not seen in GF or BSH-low cohorts, consistent with the notion that microbiome-generated bile acids exert more profound sex-biased effects than host-generated bile acids. However, no significant difference was seen in the SPF cohort either, suggesting that other microbiome effects on transit time are able to overcome the BSH-associated sex-bias.
Interactions between the ENS and microbiota regulate gut transit
We next asked whether the motility phenotypes transmitted by BSH-high consortia were dependent upon a functional ENS. We transplanted these synthetic communities into gnotobiotic C57BL/6 mice harboring a null allele of Ret, a gene critical in ENS development and maintenance that is most commonly implicated in Hirschsprung’s disease (Edery et al., 1994; Romeo et al., 1994; Tsuzuki et al., 1995; Enomoto et al., 2001; Gianino et al., 2003), as well as wild-type littermate controls. Ret+/- mice have equivalent numbers of enteric neurons as wild-type mice but exhibit ENS dysfunction (Gianino et al., 2003). Intriguingly, we observed that the baseline difference in transit times between GF Ret+/- and GF wild-type mice could be mitigated by colonization with BSH-high-1 but not BSH-high-2 (microbiota: p< 10−12, F2,18 = 210; Ret genotype: p< 10−3, F1,18 = 19; two-way ANOVA; Figure 5A). Putting this together with our observations above that the BSH-high-1 consortium produces abundant LCA and that LCA is a potent pro-motility bile acid, we hypothesized that LCA mediates this effect. Indeed, intra-colonic infusion of LCA induced significant acceleration of colonic transit in Ret+/- mice just as in wild-type mice (Figure 5B).
Figure 5.
Interactions between the ENS and microbiota regulate gut transit (See also Figures S3,S4, and S7 and Table 1)
(A) Transit times in Ret+/- mice (and wild-type littermates) colonized with BSH-high consortia compared to GF controls.
(B) LCA has significant pro-motility effects following intra-colonic infusion.
Statistical significance was determined using a Student's two-tailed t-test; ∗p< 0.05. Mean values ± SEM are shown.
The differential effects of these two BSH-high consortia suggest that interactions between the gut microbiota and RET signaling can modulate gut transit in a microbiota-specific manner. In a recent study of conventionally raised mice (harboring a complete mouse microbiota), it was reported that motility differences between Ret+/- and wild-type mice are age-dependent (Kulkarni et al., 2020); in addition, we propose a microbiota-dependency.
Design of a multiplexed host gene expression panel
Building upon our prior observation that BSH-mediated motility effects are mediated by the ENS (Dey et al., 2015), we hypothesized that bacterial BSH activity and associated motility phenotypes would be associated with distinct transcriptional signatures of the ENS. We quantified gene expression in gnotobiotic mice using a multiplexed gene expression NanoString panel designed on the basis of two reports identifying ENS-specific genes (Heanue and Pachnis, 2006; Vohra et al., 2006). We included probes targeting genes whose expression was estimated to be at least 10-fold enriched in the ENS in either study; in addition, a 25% error factor was used to compensate for any technical variability in these studies (McIntyre et al., 2011; Hicks et al., 2018), effectively lowering the threshold for inclusion to 7.5-fold enrichment in the ENS. Ultimately, our panel comprised of 91 ENS-specific genes (broadly classified as involved in signaling, in development and maintenance, or both), as well as 10 host genes involved in bile acid metabolism, 4 other genes of interest, and 7 housekeeping genes (Table S5A).
To better understand the effects of different normalization strategies, we compared normalized datasets generated using different subsets (minimum 3) of the 7 housekeeping genes (Rpl19, Rpl10, Hprt1, Tubb5, G6pdx, Gusb, and G6pdx). Normalized values were robust to normalization strategy in this dataset: for example, a comparison of normalization using 3 (Rpl19, Rpl10, and Hprt1) versus all 7 housekeeping genes demonstrated concordance (statistics not calculated given pseudo-replication, as each gene is represented multiple times, once per gut segment per mouse) (Figure S2A). We ultimately normalized our data using 3 housekeeping genes, rationalizing that we did not observe a systematic bias of any one normalization strategy.
We also considered the degree to which estimated fold-changes (FCs) of ENS-specific genes in our panel (z) might deviate from true FCs in ENS cells (y) as a function of the percent of non-ENS cells expressing a given gene (x), the percent of ENS cells expressing this gene (w), the FC in gene expression by non-ENS cells (v), and the total number of cells expressing this gene (n). In our model, we assumed that ENS cells comprise 1% of cells in full-thickness intestinal sections (Drokhlyansky et al., 2020). We found that
We then simulated a dataset in which we systematically altered each of these biological variables to assess effects on z (Table S5D). In the edge case where x = 0 (i.e., 100% ENS specificity of gene expression), z = y, meaning that experimental readouts from our panel exactly mirror actual changes in gene expression. However, with gene expression by non-ENS cells, z precipitously declines, leading to the perception that FCs in ENS cells are much lower than in actuality (Figure S2B). For example, in the scenario where all cells evenly express a gene at baseline, a 100-fold upregulation in the 1% of ENS cells with unchanged expression in non-ENS cells would appear merely as a 1% upregulation. Given that the genes in our panel are likely variably expressed by non-ENS cells, we suspect that some of the FCs we report below underestimate true values.
With these considerations in mind, we then utilized this targeted gene expression panel to characterize transcription in each of the 4 intestinal sections (proximal small intestine, distal small intestine, proximal colon, and distal colon) harvested from 76 gnotobiotic mice, for a total of 304 samples (Table S5B and S5C).
Stereotypic regional ENS transcriptional responses to gut microbes, metabolites, and diet
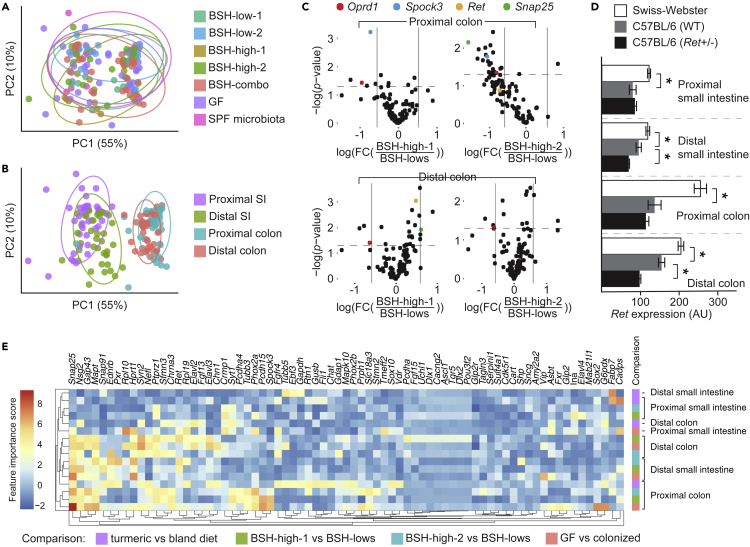
ENS transcriptomic signatures were unique to each microbiota (p = 0.002, PERMANOVA [permutational multivariate analysis of variance using distance matrices] using Bray-Curtis dissimilarities) and reproducible in gnotobiotic mice colonized with the same microbiota (Figure 6A). However, biogeography (i.e., anatomical location of the gut segment profiled) was by far the predominant factor explaining variance in gene expression (adjusted R2 0.58, p = 0.001, PERMANOVA; Figure 6B). The explained variance in global ENS transcriptomic signatures was lower and non-significant for bacterial BSH activity level and sex. (All 4 of these factors were included in the same statistical model.)
Figure 6.
Gene expression signatures were microbiota-specific, highly reproducible, and distinct between gut segments (See also Figures S5 and S7 and Table 1)
In an analysis of Bray-Curtis dissimilarities between ENS-specific gene expression signatures of intestinal samples in our dataset, we observed significant clustering by (A) microbiota and (B) biogeography.
(C) Volcano plots for BSH-high vs BSH-low in the colon. The y axis reflects log10-transformed p values, and the x axis shows log2-transformed fold-changes.
(D) Ret expression as a function of genotype.
(E) The most discriminatory features in comparisons of 4 dietary and microbial exposures as determined using a Random Forest classifier, filtered for genes found to have an importance score of >1 in at least 1 comparison, with hierarchical clustering applied across rows (specific comparison) and columns (genes).
In (D), mean values ± SEM are shown.
Genes that were differentially expressed between BSH-high- and BSH-low-colonized mice play roles in ENS signaling (e.g., colonic expression of Oprd1, which encodes opioid receptor delta 1, was lower in BSH-high mice) and in neuronal development and maintenance (e.g., Spock3 was lower in the proximal colon of BSH-high mice), and a subset of these genes were upregulated in one BSH-high consortium but downregulated in the other (e.g., Ret and Snap25), highlighting both microbiota-specific effects and common effects potentially attributable to shared metabolic traits (Figure 6C).
Genes that were significantly differentially expressed between males and females were associated with more modest fold-changes, although it is possible that our gene expression panel underestimated fold-changes as described above. Several of these genes have precedence for sex-based bias in the central nervous system (e.g., Penk (Corchero et al., 2002) and Gad2 (Colic et al., 2018)). Expression of genes involved in bile acid sensing (Fxr, Pxr, Vdr, and Tgr5) were not significantly different between sexes.
In a comparison of wild-type Swiss-Webster and C57BL/6 gnotobiotic mice, we found that microbiota explained the variability of ENS gene expression signatures to a greater extent than genotype (microbiota: p = 0.002, adjusted partial R2 0.04; genotype: p = 0.001, adjusted partial R2 0.001; biogeography: p = 0.001, adjusted partial R2 0.57; PERMANOVA in which all variables were included in the same model). Notably, Ret expression was significantly lower throughout the small intestine and colon in C57BL/6 mice compared to Swiss-Webster mice, and this was more pronounced in the Ret+/- mice (gut location: p< 10−32, F3,264 = 70; Ret genotype: p< 10−34, F2,264 = 107; two-way ANOVA; Figure 6D). These results suggest that while the effect sizes of the microbiota and biogeography are greater than that of genotype, there are significant genotype-based differences that may have biological implications.
Given turmeric’s limited absorption during intestinal passage (Ghosh et al., 2018), we hypothesized that it may have effects on ENS gene expression. Indeed, turmeric consumption alone was associated with several reproducible regional effects in germ-free mice (Figure S1C, Table S5C), including significant upregulation of two genes involved in neuronal development and maintenance, Elavl2 and Ptprz1, throughout the colon, and of Fabp7 (fatty acid binding protein 7, also involved in neuronal development) throughout the small intestine. Turmeric consumption alone did not have a significant effect on gut transit times (Table S3), supporting our model of its microbiota-dependent effect on motility.
Most ENS-specific genes in our panel were correlated with one another to varying degrees (Figure S3). The majority of genes involved in bile acid sensing clustered separately; of these, several genes (Pxr and Vdr, which encode bile acid receptors pregnane X receptor and vitamin D receptor, respectively, plus Fgf15 and Shp) were strongly anti-correlated with ENS-specific genes. This cluster of genes was directly correlated with Ednrb (endothelin receptor type B; regulates ENS growth during development). On the other hand, expression of Fxr and Asbt, which were correlated, was largely independent of ENS-specific genes, as was Fabp7 (fatty acid binding protein 7, involved in neuronal development). Tgr5 was only detectable at near-background levels and was strongly correlated with other genes similarly expressed at low levels.
Given the predominance of biogeography in explaining the variability of gene expression, we deployed a supervised machine learning algorithm, Random Forest, to identify regional gene expression signatures. If microbial colonization, bacterial bile acid metabolism, or turmeric consumption had uniform effects along the length of the gastrointestinal tract, then one would expect clustering by these factors. However, hierarchical clustering (Figure 6E) revealed that sets of discriminatory genes were most similar within gut regions, suggesting that regional responses to dietary, microbial, and metabolomic perturbations are stereotypic. Within the set of discriminatory genes identified in this model, Snap25 (which encodes a component of membrane fusion complexes critical to neurotransmission) was notable for its consistently high importance scores in the distal small intestine, proximal colon, and distal colon, suggesting that it may mediate ENS responses to environmental cues.
Effects of gut microbial colonization on ENS gene expression
In a comparison of gene expression between GF and gnotobiotic mice colonized with SPF microbiota, FCs ranged from −7.7 to 5.2, consistent with prior publications which have used this technology to assess microbial effects on the host (Dheer et al., 2020; Sweeney et al., 2016). We observed broad effects of gut microbial colonization on expression of ENS-specific genes involved in signaling as well as development and maintenance.
In the small intestine, the effects of colonization on gene expression often varied between proximal and distal aspects (Figure S4A). In the proximal small intestine, colonization was associated with upregulation of a number of genes involved in ENS signaling: Vip (as in the colon); Snap91 (synaptosome-associated protein 91; involved in neurotransmission); Cart (cocaine and amphetamine regulated transcript; involved in appetite and addiction); Sult4a1 (sulfotransferase family 4A, member 1; involved in metabolism of neurotransmitters); and Sncg (synuclein, gamma; implicated in neurodegenerative diseases). In the distal small intestine, a different set of genes involved in ENS signaling were upregulated: Nos2, Glp2r (glucagon-like peptide 2 receptor; expressed by enteric neurons, mediates growth signals to intestinal epithelial cells, receptor for teduglutide), and Syn2 (synapsin II; binds to vesicles in pre-synaptic nerve terminal). In addition, differential expression of ENS genes involved in development was observed: Fabp7 was upregulated with colonization in both proximal and distal small intestine; and Ednrb, which was downregulated in the colon with colonization, was similarly downregulated in the proximal small intestine but upregulated in the distal small intestine, a finding of unclear significance. Finally, consistent with published reports on the effects of colonization, several genes involved in bile acid metabolism were downregulated with colonization: Asbt, Fgf15, and Vdr. Colonization likely drives downregulation of Asbt and Fgf15 via Fxr-dependent feedback regulation of bile acid synthesis (Sayin et al., 2013).
Two genes were expressed at significantly higher levels in the distal small intestine than in the proximal small intestine: Asbt and Fgf15 (Figure S4B). Asbt encodes an apical sodium-dependent bile acid transporter that is critical in active intestinal transport of bile acids (Oelkers et al., 1997), while Fgf15 encodes a microbiota-sensitive component of the feedback loop that regulates bile acid production (Degirolamo et al., 2014). Both are known to be expressed at high levels in the ileum. Asbt was also expressed in the proximal colon, consistent with a prior report (Craddock et al., 1998), this was more pronounced in GF mice compared to SPF microbiota-colonized gnotobiotic mice, suggesting that the gut microbiota drives downregulation (5456 ± 972 arbitrary units [AU] in GF vs 657 ± 301 AU in colonized, p< 0.0002, two-tailed t-test). Fxr, which is critical in mediating negative feedback from the ileum to the liver regulating bile acid production, has been reported to be upregulated with colonization; consistent with these reports, we found that distal small intestinal Fxr expression was greater in colonized mice (4,413 ± 404 AU in colonized vs 3,209 ± 162 AU in GF, p = 0.05, two-tailed t-test; Figure S4B).
In the colon, a number of genes involved in ENS development and maintenance were differentially expressed in SPF-colonized mice compared to GF mice, suggesting that the gut microbiome may, in fact, influence postnatal ENS homeostasis (Figure S4C). Most of these differentially expressed genes were downregulated with colonization: Elavl2 (embryonic lethal abnormal visual system-like RNA binding protein 2; involved in neuronal development and maintenance); Mapk10 (mitogen-activated protein kinase 10; involved in intracellular signal transduction cascades that regulate several biological processes, including apoptosis); Gap43 (growth-associated protein 43; involved in neuronal development and axonal regeneration); and Ednrb. In addition, a gene involved in neuronal signaling, Snap25 (synaptosome-associated protein 25, a component of the protein machinery that facilitates pre-synaptic vesicle fusion in neurotransmission), and a gene involved in bile acid sensing, Asbt (described above; higher expression in the proximal colon than distal colon (Craddock et al., 1998), consistent with our findings [Figure S4D]), were significantly downregulated with colonization. In contrast, genes that were upregulated in the colon with colonization included Mapt (microtubule-associated protein tau; involved in microtubule dynamics); Shp (a regulator of host bile acid production); and two genes implicated in gut motility, Vip (vasoactive intestinal peptide; a pro-motility neuropeptide) and Nos2 (nitric oxide synthase 2; involved in both motility and immunity).
Finally, we asked whether our results mirrored previously published reports (El Aidy et al., 2012; Larsson et al., 2012), to which we compared differential gene expression that we observed in gnotobiotic mice colonized with SPF microbiota versus GF controls. Interestingly, we found disagreements in both small intestinal and colonic gene expression, not only with our findings but also between these two studies (Pearson correlation rho< 0.1 in all comparisons; Figure S5A). We hypothesized that this discrepancy in intestinal gene expression was attributable to heterogeneity in the SPF microbiome (O'Rourke et al., 1988; Dobson et al., 2019; Sivan et al., 2015). To further evaluate this, we generated shotgun sequencing data profiling the fecal microbiota of gnotobiotic mice colonized with SPF microbiota (Table S6). Phylum-level taxonomy was reported in the two cited studies; even at this broad taxonomic rank, we found that our SPF microbiota harbored a greater proportional abundance of Firmicutes and lower abundances of Proteobacteria and TM7 genera (Figure S5B). Methodological differences may have contributed to these discrepancies (e.g., use of microarray (El Aidy et al., 2012) and 454 pyrosequencing (Larsson et al., 2012) versus Illumina sequencing in our study; use of conventional versus “conventionalized” mice, which impacts intestinal gene expression (Camp et al., 2014); precise location of intestinal tract profiled; and use of exclusively male mice, whereas we studied males and females).
Discussion
Our observations in a gnotobiotic mouse model suggest that gut bacterial bile acid metabolism regulates colonic transit in a sex-biased manner: greater gut bacterial BSH activity drives faster colonic transit, with greater pro-motility effects in males. ENS transcriptomic signatures were microbiota-specific, exquisitely sensitive to environmental perturbations (including to a single diet ingredient), and regionally stereotypic, consistent with two recent studies (Muller et al., 2020; Obata et al., 2020). Differences in observed effects of gut microbial colonization on intestinal gene expression between studies (El Aidy et al., 2012; Larsson et al., 2012) underscore the point that because the microbiome is not a singular entity with a uniform effect, the state of being colonized is therefore not a singular thing. This highlights the challenges with reproducibility in microbiome studies and underscores the value of gnotobiotic models and defined microbial consortia that have been carefully phenotyped and genotyped.
Motility is often slower in females, both in health (Lampe et al., 1993) and in irritable bowel syndrome (Adeyemo et al., 2010; Klem et al., 2017; Kim and Kim, 2018). Sex-biased differences in gut motility may have broader implications for health (Burkitt et al., 1972; Bjerknes and Cheng, 2001; Maruti et al., 2008). Our findings are consistent with human studies that have demonstrated (1) faster gut motility in males that correlates with differences in bile acid profiles (Lampe et al., 1993) and (2) motility effects of bile acid administration (Odunsi-Shiyanbade et al., 2010; Rao et al., 2010). Together, our results suggest that strategies for treating or preventing gastrointestinal diseases may need to be tailored to sex and to biogeography of the gut. While targeting the microbiome and the ENS is justified, our observation of significant transcriptional responses to defined interventions in a highly controlled gnotobiotic setting also highlights challenges to clinical translation. The human experience—which reflects the aggregate effects of the innumerable dietary ingredients that we consume daily, the hugely diverse metabolically dynamic microbes that inhabit our guts, our own digestive processes, and the interactions of all of the above that result in thousands of gut metabolites—entails significantly more complex and variable transcriptional responses to environmental cues.
Limitations of the study
There are several limitations of our study. First, sex-biased gut motility phenotypes, which we could reproduce via intra-colonic bile acid infusions, could not be explained by sex-biased differential expression of bile acid receptors (e.g., Tgr5, which encodes the bile acid receptor known to be expressed by enteric neurons (Poole et al., 2010)) or by differential bile acid production by the gut microbiome, both of which we found to be equivalent in males and females. Similar to our findings, another recent study did not identify sex-biased gene expression by enteric neurons (Obata et al., 2020). Uncovering the underlying host biology may require more granular ENS gene expression profiling at single-cell resolution (Drokhlyansky et al., 2020) combined with mouse models in which hormones and hormone receptors are manipulated. Second, our use of gnotobiotic mice is almost certainly an imperfect model of humans, but this system permitted us to carefully and precisely control colonization and our experimental interventions. Third, our differential gene expression calculations likely underestimate true fold-changes for some of these genes due to non-specific expression (as illustrated in Figure S2B). Fourth, our reductionist model overlooks differences between the two BSH-low communities and between the two BSH-high communities that are unrelated to BSH, including metabolic differences that may influence gut transit. Nonetheless, our study establishes gut microbial bile acid metabolism as a driver of sex-biased motility phenotypes, and we demonstrate potential modulation of motility via colonic administration of bile acids. Our results should help inform the design and interpretation of clinical studies testing the efficacy of bile acid-based approaches in treating motility disorders in humans.
STAR★Methods
Key resources table
| REAGENT or RESOURCE | SOURCE | IDENTIFIER |
|---|---|---|
| Bacterial and virus strains | ||
| Bacteroides caccae | ATCC | 43185 |
| Bacteroides finegoldii | DSM | 17565 |
| Bacteroides thetaiotaomicron | VPI | 5482 |
| Bacteroides vulgatus | ATCC | 8482 |
| Citrobacter youngae | ATCC | 29220 |
| Clostridium asparagiforme | DSM | 15981 |
| Clostridium leptum | DSM | 753 |
| Clostridium scindens | ATCC | 35704 |
| Clostridium sporogenes | ATCC | 15579 |
| Dorea formicigenerans | ATCC | 27755 |
| Dorea longicatena | DSM | 13814 |
| Enterobacter cancerogenus | ATCC | 35316 |
| Escherichia fergusonii | ATCC | 35469 |
| Lactobacillus ruminis | ATCC | 25644 |
| Megamonas funiformis | DSM | 19343 |
| Ruminococcus gnavus | ATCC | 29149 |
| Ruminococcus obeum | ATCC | 29174 |
| Ruminococcus torques | ATCC | 27756 |
| Chemicals, peptides, and recombinant proteins | ||
| SBI-115 | MedChemExpress | HY-111534 |
| Fluorescein isothiocyanate-dextran (FITC-dextran; 70,000 MW) | Sigma-Aldrich | 46945 |
| Carmine powder | Sigma-Aldrich | C1022 |
| Methylcellulose | Sigma-Aldrich | M0512 |
| Phosphate buffered saline tablets (PBS) | Fisher Scientific | BP2944100 |
| RNAlater | Qiagen | 76106 |
| Deoxycholic acid | Sigma-Aldrich | 30960 |
| Lithocholic acid | Sigma-Aldrich | L6250 |
| Cholic acid | Sigma-Aldrich | C1129 |
| β-Muricholic Acid | Sigma-Aldrich | SML2372 |
| Taurocholic acid | Sigma-Aldrich | T4009 |
| RNeasy Mini Kit | Qiagen | 74106 |
| AIN-93M irradiated mouse chow | Envigo | TD.170815 |
| AIN-93M irradiated mouse chow with turmeric | Envigo | TD.170816 |
| Deposited data | ||
| Bacterial shotgun sequencing datasets | European Nucleotide Archive (ENA) | ENA: PRJEB38547 |
| Mouse gene expression data | El Aidy et al., 2012 | https://www.nature.com/articles/mi201232 |
| Mouse gene expression data | Larsson et al., 2012 | https://gut.bmj.com/content/61/8/1124 |
| Experimental models: organisms/strains | ||
| Mouse: Wild-type Swiss-Webster | http://www.uwgnotobiotics.org | N/A |
| Mouse: Ret+/- and wild-type littermates | Enomoto et al., 2001 | N/A |
| Software and algorithms | ||
| nSolver™ Analysis Software | NanoString Technologies | https://www.nanostring.com/products/analysis-solutions/ncounter-advanced-analysis-software/ |
| Custom R scripts | Dey Lab | https://github.com/DeyLab |
| Kraken2 | Wood and Salzberg, 2014 | https://github.com/DerrickWood/kraken2/wiki |
| MetaPhlAn2 | Segata et al., 2012 | https://huttenhower.sph.harvard.edu/metaphlan2/ |
| USEARCH | Edgar and Flyvbjerg, 2015 | https://www.drive5.com/usearch/ |
Resource availability
Lead contact
Further information and requests for resources and reagents should be directed to and will be fulfilled by the lead contact, Dr. Neelendu Dey (ndey@fredhutch.org).
Materials availability
This study did not develop new unique reagents.
Data and code availability
The accession number for the bacterial shotgun sequencing data reported in this paper is ENA: PRJEB38547. Experimental data associated with this study are available in supplemental information. Custom scripts used for data analysis available at https://github.com/DeyLab.
Experimental model and subject details
Animal husbandry
Gnotobiotic mouse experiments were performed on male and female (1) wild-type Swiss-Webster mice and (2) Ret+/- mice and wild-type littermates (Enomoto et al., 2001) using protocols approved by the Institutional Animal Care and Use Committee of the University of Washington. Mice were 6-8 weeks old at the start of experiments. The intra-colonic bile acid infusion experiment was conducted in conventionally raised mice using protocols approved by the Fred Hutchinson Cancer Research Center. Mice were housed in environmentally controlled rooms of an AALAC accredited facility under 6am/8pm light/dark cycles. Breeding mice were maintained in sterile, flexible, plastic gnotobiotic isolators (Class Biologically Clean Ltd., Madison, WI), while experimental animals were housed in Tecniplast cages (Tecniplast Group, West Chester, PA). Prior to experiments, sterility was ensured by collecting fecal pellets from GF mice and subjecting them to 16S rDNA PCR, aerobic culturing, and anaerobic culturing. To minimize variability in composition between diets, the turmeric-containing and bland diets were identical in composition with the exception of 0.1% turmeric by weight: a batch of bland diet (Envigo TD.170815; AIN-93M diet; 12.4% protein and 4.1% fat by weight) was prepared and split into two, and 0.1% turmeric was added to one portion (Envigo TD.170816). Diets were sterilized via irradiation with 20-50 kGy, with documented minimum delivered dose of 27.2 kGy. Mice were fed ad libitum starting 3-4 days prior to day 0 (the day of colonization or, in the case of GF mice, the official experimental start date). The fecal microbiota and bacterial consortia used for colonization were prepared in an anaerobic chamber; the fecal microbiota suspension was derived from fecal pellets collected from conventionally housed wild-type SPF mice. All oral gavages (for purposes of microbiota transplantation and for motility assays) were performed by the same individual within a consistent time frame (between 08:00 and 09:00 local time) in all mice in order to minimize variability. Fresh fecal pellets were collected from mice throughout experiments in sterile tubes, snap-frozen in liquid nitrogen, and stored at −80°C until use.
Method details
Gut motility measurements
FITC assay. 200 μL per mouse of a sterilized 5 mg/mL solution of fluorescein isothiocyanate-dextran (FITC-dextran; 70,000 MW; Sigma-Aldrich, St. Louis, MO) prepared in PBS was delivered via oral gavage. Mice were euthanized 2 hr later (except in a single experiment in which a 4-hr interval was studied, as described in the text), and gastrointestinal tracts were immediately harvested and placed in ice-cold PBS for 30 s to inhibit motility, after which they were cut into a total of 12 segments: stomach, small intestine (partitioned into 8 equal-sized segments), and colon (with cecum first dissected out; the remainder was then partitioned into two equal-sized segments). Each of the 12 segments was flushed with 2 mL of PBS. After flushing with PBS, gut segments were stored in RNAlater (Thermo Fisher Scientific Inc., Waltham, MA) at 4°C for 24 hr and then transferred to −20°C for storage until use. The intestinal flushes were then subjected to serial dilutions, prepared in triplicate. Fluorescence was measured using a BioTek Synergy HTX (BioTek, Winooski, VT), with absolute fluorescence levels estimated using a dilution series of an FITC-dextran solution of known concentration. Carmine red dye assay. 150 μL per mouse of a sterilized 6% (w/v) solution of carmine red (Sigma-Aldrich) in 0.5% methylcellulose (Sigma-Aldrich) was delivered via oral gavage. Fecal output was monitored every 30 min or more frequently if stool was passed spontaneously. The time from gavage to appearance of bright red dye was recorded as the whole gut transit time. In the intra-colonic bile acid experiment, carmine gavage was preceded by rectal administration of 400 μL of a sterilized 4 mM bile acid solution with the aid of a mouse restraining device. Intra-colonic infusions were performed in the following order, with intervals of ≥48 hr between infusions: vehicle control, bMCA, DCA, cholic acid, TCA, and LCA. In the experiment using the TGR5 inhibitor SBI-115 (MedChemExpress, Monmouth Junction, NJ), the order of infusions was LCA, SBI-115 + LCA (100 μL of SBI-115 was administered 10 min prior to LCA infusion), and vehicle control.
Functional in vitro assay of bile acid metabolism
We anaerobically co-cultured gut bacterial strains in media containing conjugated primary bile acid substrates (taurocholic acid [TCA], tauro-beta-muricholic acid [TbMCA]). After 48 hr, sterile supernatant from bacterial cultures were subjected to bile acid profiling via LC-MS using methods described in the following section. In silico genome-based predictions of bile acid production by different bacterial consortia were binary: Individual bile acid metabolites were either predicted to be detectable or not, without consideration for concentrations.
Bile acid profiling
Bile acid extractions were performed on ice using 50 mg of each fecal sample. Samples were placed into a tube containing one 4 mm steel ball, 1 mL of ice-cold water:methanol (1:10 v/v), and 10 μL of a mixture of internal standards (10 μM each of d4-GCDCA [glyco-chenodeoxycholic acid], d4-LCA [lithocholic acid], d4-CDCA [chenodeoxycholic acid], d4-DCA, and d4-CA [cholic acid] in 1:1 water:methanol) before mechanical disruption using a TissueLyser II sonication in an ice bath for 10 min, and agitation via a vortex mixer. Samples were then centrifuged at 15,000 x g at 4°C for 15 min. 800 μL of the supernatant was transferred to a new tube and dried in a speed vacuum at room temperature. Samples were resuspended in 100 μL of 50% methanol and stored at −80°C until LC-MS. Ten working standards (2.5–10,000 ng/mL) and 3 quality control samples with replicates were prepared using the identical protocol as was used for experimental samples. Before injecting into the LC-MS, samples were transferred into 0.2 μm Costar Spin-X HPLC microcentrifuge filters (Corning Inc., Corning, NY) and centrifuged at 15,000 x g for 10 min. 5 μL filtrate per sample was injected into the Agilent G6460 (version A.00.07.32) ultra-performance liquid chromatography coupled with mass spectrometry in tandem system (UPLC-MS/MS) combined with a triple quadrupole mass spectrometer via an electrospray ionization interface. Chromatographic separation was performed using a ZORBAX Eclipse Plus C18 analytical column (2.1 × 100 mm; id: 1.8μm). The gradient profile for the LC pump under the final chromatography conditions was 0 min, 95:5; 5 min, 95:5; 14 min, 86:14; 14.5 min, 75:25; 17.50 min, 75:25; 18 min, 50:50; 22 min, 50:50; 22.50 min, 20:80; 24.50 min, 20:80; 25-28 min, 95:5 (A:B, v/v). Column temperature was 45°C; sample tray temperature was 9°C. MS/MS spectra were produced in negative ionization mode.
RNA isolation
20 mg of each sample was placed into a tube containing 0.1 mm zirconium beads, a 4 mm steel ball, and homogenization buffer before mechanical disruption using a TissueLyser II (Qiagen, Hilden, Germany). RNA was purified using RNeasy Mini kits (Qiagen).
Gene expression quantification
RNA samples were run on a custom NanoString nCounter panel in the Fred Hutch Genomics Core using manufacturer protocols. To minimize batch effects, samples were pseudo-randomized across different runs. Raw counts were imported into NanoString nSolver for normalization. Spiked-in negative control probes were used to determine the minimum threshold for detection (i.e., background thresholding): first, the geometric mean of the negative control probes was calculated, and this was used to set the threshold for background levels; then, normalized values less than background were set to this background level. Spiked-in positive control probes were used to adjust for lane-to-lane and run-to-run variations: the geometric mean of the positive control probes in combination with the expression levels of housekeeping genes and 4 samples included in multiple runs (2 colonized, 2 GF) were used to calculate normalization factors. Normalized data were then analyzed in R.
Assessment of ENS transcriptomic signatures
Principal coordinates analysis was performed on Bray-Curtis dissimilarities between samples that were calculated using the subsets of normalized NanoString data consisting of ENS-specific genes.
Supervised machine learning analysis
Discriminatory genes were identified using Random Forest as implemented in the randomForest package (version 4.6–14), with importance scores averaged and aggregated after 100 iterations and 1,000 trees per iteration.
Multiplex sequencing for gut microbiota profiling
Genomic DNA was derived from fecal pellets using combined physical (bead beating) and chemical (SDS) disruption, phenol/chloroform extraction, and purification using QIAquick spin columns (Qiagen). Genomic DNA was sheared using a Covaris LE220 ultrasonicator (Covaris, Woburn, MA). Sample-specific barcoded adapters were ligated to end-repaired DNA fragments, after which libraries were prepared for shotgun sequencing on the Illumina MiniSeq platform. An earlier sequencing run was performed using a 75 nt single-end strategy (369,980 ± 251,360 high-quality reads per sample [mean ± SEM], range 59,862 - 952,873); a subsequent run used a 150 nt paired-end strategy (76,661 ± 49,279, range 12,536 - 171,548). Reads mapping to the mouse genome (UCSC mm10; Bowtie2, version 2.2.5 (Langmead and Salzberg, 2012)) or estimated to have >1 error (USEARCH, version 11 (Edgar and Flyvbjerg, 2015)) were removed; paired-end reads were joined and quality-filtered with USEARCH's fastq_merge function using default parameters. Datasets were rarefied to 22,356 (single-end) or 7,101 (paired-end) randomly selected high-quality reads (or pairs of reads) per sample. Taxonomic classifications for samples corresponding to synthetic consortia were performed with Kraken2 (Wood and Salzberg, 2014) using custom reference databases for each defined consortium; classifications of reads generated from SPF microbiota-colonized mice were performed with MetaPhlAn2 (Segata et al., 2012) using default parameters.
Comparing prior studies
Datasets from two prior studies (El Aidy et al., 2012; Larsson et al., 2012) were downloaded from publisher websites. Shared genes (n = 1,170 between El Aidy et al. and Larsson et al.; n = 16 between El Aidy et al. and the present study; n = 29 between Larsson et al. and the present study) were identified by exact gene name matching.
Quantification and statistical analysis
Statistical comparisons were performed in R (version 4.0.0). Two-tailed t-tests and ANOVA were used for comparisons of datasets with normal distributions. PERMANOVA was used as a non-parametric multivariate test to compare dissimilarities in gene expression profiles of different samples. Factors such as diet, age, and genotype were controlled to the greatest extent possible in the reported comparisons. p values of ≤0.05 were considered significant. Plots were generated in R (using native functions as well as the ggplot2 [version 3.3.0] and pheatmap [version 1.0.12] packages) and assembled in Adobe Illustrator.
Acknowledgments
We thank our colleagues for helpful suggestions in the preparation of this paper. This work was supported by grants from the NIH (NIDDK K08 DK111941 and NCI Cancer Center Support Grant P30 CA015704 to N.D.) and the Fred Hutchinson Cancer Research Center (Pathogen-Associated Malignancies Integrated Research Center pilot funds and start-up funds to N.D.). Graphical abstract created using images from BioRender.com.
Author contributions
Conceptualization, N.L., S.T.K, D.M.L., and N.D.; Data curation, N.L., S.T.K, D.M.L., M.D., J.Y.C, and N.D.; Formal analysis, N.L., S.T.K, D.M.L., M.D., J.Y.C, and N.D.; Funding acquisition, N.D.; Investigation, N.L., S.T.K, D.M.L., and N.D.; Methodology, N.L., S.T.K, D.M.L., and N.D.; Project administration, N.L., S.T.K, D.M.L., and N.D.; Resources, J.Y.C, and N.D.; Software, S.T.K, D.M.L., and N.D.; Supervision, J.Y.C, and N.D.; Validation, N.L., S.T.K, D.M.L., M.D., J.Y.C, and N.D.; Visualization, N.L., S.T.K, D.M.L., and N.D.; Writing - original draft, N.L., S.T.K, D.M.L., and N.D.; Writing - review & editing, N.L., S.T.K, D.M.L., M.D., J.Y.C, and N.D.
Declaration of interests
The authors declare no competing interests.
Published: June 25, 2021
Footnotes
Supplemental information can be found online at https://doi.org/10.1016/j.isci.2021.102508.
Supplemental information
BSH activities of 30 bacteria consortia subjected to an in vitro functional screen.
Unconjugated and conjugated bile acids in gnotobiotic mice, as measured through a targeted liquid chromatography mass spectrometry analysis. LOD: limit of detection.
Results of an FITC-based gut motility assay. Each row summarizes data for an individual mouse. Fluorescence in a given gut segment is represented as a proportion of total fluorescence measured throughout the gut.
(A). Serial transit time measurements following intra-colonic infusions of different bile acids.
(B). Effect of a TGR5 inhibitor on transit times.
(C). Effect of intra-colonic infusion of LCA on transit times in Ret+/- mice.
(A). List of genes included in a custom NanoString nCounter CodeSet panel, classifications, target sequences, and NCBI accession numbers.
(B). Raw gene expression datafrom NanoString panel.
(C). Normalized gene expression based on normalization of raw data using negative control probes, positive control probes, and expression levels of housekeeping genes.
(D). Modeling the impact of non-specific gene expression on results of a custom NanoString panel. Fold-changes in ENS gene expression may be underestimated because of non-specific gene expression by non-ENS cells. Different “true” fold-changes were considered, ranging from 1 (no change) to 10; these values represent the unknown actual gene expression fold-changes in ENS cells, whereas the values in the cell represent fold-changes “estimated” by the gene expression panel we used.
Barcoding and accession numbers for all samples that underwent shotgun sequencing are listed.
Shotgun sequencing of genomic DNA from fecal pellets revealed that Bacteroides was the predominant taxon in mice colonized with the BSH-low-1, BSH-high-1, and BSH-high-2 consortia; Clostridia comprised the next most prevalent taxon. Citrobacter youngae, Escherichia fergusonii, and Enterobacter cancerogenus approximately equally abundant species in mice colonized with the BSH-low-2 consortium, which did not contain any Bacteroides or Clostridia.
References
- Abrams G.D., Bishop J.E. Effect of the normal microbial flora on gastrointestinal motility. Proc. Soc. Exp. Biol. Med. 1967;126:301–304. doi: 10.3181/00379727-126-32430. [DOI] [PubMed] [Google Scholar]
- Adeyemo M.A., Spiegel B.M.R., Chang L. Meta-analysis: Do irritable bowel syndrome symptoms vary between men and women?: meta-analysis: gender variance in irritable bowel syndrome. Aliment. Pharmacol. Ther. 2010;32:738–755. doi: 10.1111/j.1365-2036.2010.04409.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anitha M., Vijay-Kumar M., Sitaraman S.V., Gewirtz A.T., Srinivasan S. Gut microbial products regulate murine gastrointestinal motility via toll-like receptor 4 signaling. Gastroenterology. 2012;143:1006–1016.e4. doi: 10.1053/j.gastro.2012.06.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bhattarai Y., Williams B.B., Battaglioli E.J., Whitaker W.R., Till L., Grover M., Linden D.R., Akiba Y., Kandimalla K.K., Zachos N.C. Gut microbiota-produced tryptamine activates an epithelial G-protein-coupled receptor to increase colonic secretion. Cell Host Microbe. 2018;23:775–785.e5. doi: 10.1016/j.chom.2018.05.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bjerknes M., Cheng H. Modulation of specific intestinal epithelial progenitors by enteric neurons. Proc. Natl. Acad. Sci. U S A. 2001;98:12497–12502. doi: 10.1073/pnas.211278098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burkitt D.P., Walker A.R.P., Painter N.S. Effect of dietary fibre on stools and the transit-times, and its role in the causation of disease. The Lancet. 1972;300:1408–1411. doi: 10.1016/s0140-6736(72)92974-1. [DOI] [PubMed] [Google Scholar]
- Camp J.G., Frank C.L., Lickwar C.R., Guturu H., Rube T., Wenger A.M., Chen J., Bejerano G., Crawford G.E., Rawls J.F. Microbiota modulate transcription in the intestinal epithelium without remodeling the accessible chromatin landscape. Genome Res. 2014;24:1504–1516. doi: 10.1101/gr.165845.113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Colic L., Li M., Demenescu L.R., Li S., Müller I., Richter A., Behnisch G., Seidenbecher C.I., Speck O., Schott B.H. GAD65 promoter polymorphism rs2236418 modulates harm avoidance in women via inhibition/excitation balance in the rostral ACC. J. Neurosci. 2018;38:5067–5077. doi: 10.1523/JNEUROSCI.1985-17.2018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Corchero J., Fuentes J.A., Manzanares J. Gender differences in proenkephalin gene expression response to delta9-tetrahydrocannabinol in the hypothalamus of the rat. J. Psychopharmacol. Oxf. Engl. 2002;16:283–289. doi: 10.1177/026988110201600401. [DOI] [PubMed] [Google Scholar]
- Craddock A.L., Love M.W., Daniel R.W., Kirby L.C., Walters H.C., Wong M.H., Dawson P.A. Expression and transport properties of the human ileal and renal sodium-dependent bile acid transporter. Am. J. Physiol. 1998;274:G157–G169. doi: 10.1152/ajpgi.1998.274.1.G157. [DOI] [PubMed] [Google Scholar]
- Degirolamo C., Rainaldi S., Bovenga F., Murzilli S., Moschetta A. Microbiota modification with probiotics induces hepatic bile acid synthesis via downregulation of the fxr-fgf15 Axis in mice. Cell Rep. 2014;7:12–18. doi: 10.1016/j.celrep.2014.02.032. [DOI] [PubMed] [Google Scholar]
- Dey N., Wagner V.E., Blanton L.V., Cheng J., Fontana L., Haque R., Ahmed T., Gordon J.I. Regulators of gut motility revealed by a gnotobiotic model of diet-microbiome interactions related to travel. Cell. 2015;163:95–107. doi: 10.1016/j.cell.2015.08.059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dheer R., Davies J.M., Quintero M.A., Damas O.M., Deshpande A.R., Kerman D.H., Sawyer W.P., Pignac-Kobinger J., Ban Y., Fernandez I. Microbial signatures and innate immune gene expression in Lamina propria phagocytes of inflammatory bowel disease patients. Cell. Mol. Gastroenterol. Hepatol. 2020;9:387–402. doi: 10.1016/j.jcmgh.2019.10.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dobson G.P., Letson H.L., Biros E., Morris J. Specific pathogen-free (SPF) animal status as a variable in biomedical research: have we come full circle? EBioMedicine. 2019;41:42–43. doi: 10.1016/j.ebiom.2019.02.038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Drokhlyansky E., Smillie C.S., Van Wittenberghe N., Ericsson M., Griffin G.K., Eraslan G., Dionne D., Cuoco M.S., Goder-Reiser M.N., Sharova T. The human and mouse enteric nervous system at single-cell resolution. Cell. 2020;182:1606–1622.e23. doi: 10.1016/j.cell.2020.08.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Edery P., Lyonnet S., Mulligan L.M., Pelet A., Dow E., Abel L., Holder S., Nihoul-Fékété C., Ponder B.A., Munnich A. Mutations of the RET proto-oncogene in Hirschsprung’s disease. Nature. 1994;367:378–380. doi: 10.1038/367378a0. [DOI] [PubMed] [Google Scholar]
- Edgar R.C., Flyvbjerg H. Error filtering, pair assembly and error correction for next-generation sequencing reads. Bioinformatics. 2015;31:3476–3482. doi: 10.1093/bioinformatics/btv401. [DOI] [PubMed] [Google Scholar]
- El Aidy S., van Baarlen P., Derrien M., Lindenbergh-Kortleve D.J., Hooiveld G., Levenez F., Doré J., Dekker J., Samsom J.N., Nieuwenhuis E.E.S. Temporal and spatial interplay of microbiota and intestinal mucosa drive establishment of immune homeostasis in conventionalized mice. Mucosal Immunol. 2012;5:567–579. doi: 10.1038/mi.2012.32. [DOI] [PubMed] [Google Scholar]
- Enomoto H., Crawford P.A., Gorodinsky A., Heuckeroth R.O., Johnson E.M., Milbrandt J. RET signaling is essential for migration, axonal growth and axon guidance of developing sympathetic neurons. Development. 2001;128:3963–3974. doi: 10.1242/dev.128.20.3963. [DOI] [PubMed] [Google Scholar]
- Ghosh S.S., He H., Wang J., Gehr T.W., Ghosh S. Curcumin-mediated regulation of intestinal barrier function: the mechanism underlying its beneficial effects. Tissue Barriers. 2018;6:e1425085. doi: 10.1080/21688370.2018.1425085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gianino S., Grider J.R., Cresswell J., Enomoto H., Heuckeroth R.O. GDNF availability determines enteric neuron number by controlling precursor proliferation. Development. 2003;130:2187–2198. doi: 10.1242/dev.00433. [DOI] [PubMed] [Google Scholar]
- Heanue T.A., Pachnis V. Expression profiling the developing mammalian enteric nervous system identifies marker and candidate Hirschsprung disease genes. Proc. Natl. Acad. Sci. U S A. 2006;103:6919–6924. doi: 10.1073/pnas.0602152103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hicks S.C., Townes F.W., Teng M., Irizarry R.A. Missing data and technical variability in single-cell RNA-sequencing experiments. Biostatistics. 2018;19:562–578. doi: 10.1093/biostatistics/kxx053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Husebye E., Hellström P.M., Midtvedt T. Intestinal microflora stimulates myoelectric activity of rat small intestine by promoting cyclic initiation and aboral propagation of migrating myoelectric complex. Dig. Dis. Sci. 1994;39:946–956. doi: 10.1007/BF02087542. [DOI] [PubMed] [Google Scholar]
- Husebye E., Hellström P.M., Sundler F., Chen J., Midtvedt T. Influence of microbial species on small intestinal myoelectric activity and transit in germ-free rats. Am. J. Physiol. Gastrointest. Liver Physiol. 2001;280:G368–G380. doi: 10.1152/ajpgi.2001.280.3.G368. [DOI] [PubMed] [Google Scholar]
- Kashyap P.C., Marcobal A., Ursell L.K., Larauche M., Duboc H., Earle K.A., Sonnenburg E.D., Ferreyra J.A., Higginbottom S.K., Million M. Complex interactions among diet, gastrointestinal transit, and gut microbiota in humanized mice. Gastroenterology. 2013;144:967–977. doi: 10.1053/j.gastro.2013.01.047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim Y.S., Kim N. Sex-Gender differences in irritable bowel syndrome. J. Neurogastroenterol. Motil. 2018;24:544–558. doi: 10.5056/jnm18082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klem F., Wadhwa A., Prokop L.J., Sundt W.J., Farrugia G., Camilleri M., Singh S., Grover M. Prevalence, risk factors, and Outcomes of irritable bowel syndrome after infectious enteritis: a systematic review and meta-analysis. Gastroenterology. 2017;152:1042–1054.e1. doi: 10.1053/j.gastro.2016.12.039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kulkarni S., Saha M., Becker L., Wang Z., Liu G., Leser J., Kumar M., Bakhshi S., Anderson M., Lewandoski M. Neural crest-derived neurons are replaced by a newly identified mesodermal lineage in the post-natal and aging enteric nervous system. bioRxiv. 2020:1–16. [Google Scholar]
- Lampe J.W., Fredstrom S.B., Slavin J.L., Potter J.D. Sex differences in colonic function: a randomised trial. Gut. 1993;34:531–536. doi: 10.1136/gut.34.4.531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Langmead B., Salzberg S.L. Fast gapped-read alignment with Bowtie 2. Nat. Methods. 2012;9:357–359. doi: 10.1038/nmeth.1923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Larsson E., Tremaroli V., Lee Y.S., Koren O., Nookaew I., Fricker A., Nielsen J., Ley R.E., Bäckhed F. Analysis of gut microbial regulation of host gene expression along the length of the gut and regulation of gut microbial ecology through MyD88. Gut. 2012;61:1124–1131. doi: 10.1136/gutjnl-2011-301104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marciani L., Cox E.F., Hoad C.L., Totman J.J., Costigan C., Singh G., Shepherd V., Chalkley L., Robinson M., Ison R. Effects of various food ingredients on gall bladder emptying. Eur. J. Clin. Nutr. 2013;67:1182–1187. doi: 10.1038/ejcn.2013.168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maruti S.S., Lampe J.W., Potter J.D., Ready A., White E. A prospective study of bowel motility and related factors on Breast cancer risk. Cancer Epidemiol. Biomark. Prev. 2008;17:1746–1750. doi: 10.1158/1055-9965.EPI-07-2850. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McIntyre L.M., Lopiano K.K., Morse A.M., Amin V., Oberg A.L., Young L.J., Nuzhdin S.V. RNA-seq: technical variability and sampling. BMC Genomics. 2011;12:293. doi: 10.1186/1471-2164-12-293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muller P.A., Matheis F., Schneeberger M., Kerner Z., Jové V., Mucida D. Microbiota-modulated CART+ enteric neurons autonomously regulate blood glucose. Science. 2020;370:314–321. doi: 10.1126/science.abd6176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Obata Y., Castaño Á., Boeing S., Bon-Frauches A.C., Fung C., Fallesen T., de Agüero M.G., Yilmaz B., Lopes R., Huseynova A. Neuronal programming by microbiota regulates intestinal physiology. Nature. 2020;578:284–289. doi: 10.1038/s41586-020-1975-8. [DOI] [PubMed] [Google Scholar]
- Odunsi-Shiyanbade S.T., Camilleri M., McKinzie S., Burton D., Carlson P., Busciglio I.A., Lamsam J., Singh R., Zinsmeister A.R. Effects of chenodeoxycholate and a bile acid sequestrant, colesevelam, on intestinal transit and bowel function. Clin. Gastroenterol. Hepatol. 2010;8:159–165. doi: 10.1016/j.cgh.2009.10.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oelkers P., Kirby L.C., Heubi J.E., Dawson P.A. Primary bile acid malabsorption caused by mutations in the ileal sodium-dependent bile acid transporter gene (SLC10A2) J. Clin. Invest. 1997;99:1880–1887. doi: 10.1172/JCI119355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- O’Rourke J., Lee A., McNeill J. Differences in the gastrointestinal microbiota of specific pathogen free mice: an often unknown variable in biomedical research. Lab. Anim. 1988;22:297–303. doi: 10.1258/002367788780746142. [DOI] [PubMed] [Google Scholar]
- Poole D.P., Godfrey C., Cattaruzza F., Cottrell G.S., Kirkland J.G., Pelayo J.C., Bunnett N.W., Corvera C.U. Expression and function of the bile acid receptor GpBAR1 (TGR5) in the murine enteric nervous system. Neurogastroenterol. Motil. 2010;22 doi: 10.1111/j.1365-2982.2010.01487.x. e227–228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rao A.S., Wong B.S., Camilleri M., Odunsi-Shiyanbade S.T., McKinzie S., Ryks M., Burton D., Carlson P., Lamsam J., Singh R. Chenodeoxycholate in females with irritable bowel syndrome-constipation: a pharmacodynamic and pharmacogenetic analysis. Gastroenterology. 2010;139:1549–1558. doi: 10.1053/j.gastro.2010.07.052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rasyid A., Lelo A. The effect of curcumin and placebo on human gall-bladder function: an ultrasound study. Aliment. Pharmacol. Ther. 1999;13:245–249. doi: 10.1046/j.1365-2036.1999.00464.x. [DOI] [PubMed] [Google Scholar]
- Rasyid A., Rahman A.R.A., Jaalam K., Lelo A. Effect of different curcumin dosages on human gall bladder. Asia Pac. J. Clin. Nutr. 2002;11:314–318. doi: 10.1046/j.1440-6047.2002.00296.x. [DOI] [PubMed] [Google Scholar]
- Romeo G., Ronchetto P., Luo Y., Barone V., Seri M., Ceccherini I., Pasini B., Bocciardi R., Lerone M., Kääriäinen H. Point mutations affecting the tyrosine kinase domain of the RET proto-oncogene in Hirschsprung’s disease. Nature. 1994;367:377–378. doi: 10.1038/367377a0. [DOI] [PubMed] [Google Scholar]
- Samuel B.S., Shaito A., Motoike T., Rey F.E., Backhed F., Manchester J.K., Hammer R.E., Williams S.C., Crowley J., Yanagisawa M. Effects of the gut microbiota on host adiposity are modulated by the short-chain fatty-acid binding G protein-coupled receptor, Gpr41. Proc. Natl. Acad. Sci. U S A. 2008;105:16767–16772. doi: 10.1073/pnas.0808567105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sayin S.I., Wahlström A., Felin J., Jäntti S., Marschall H.-U., Bamberg K., Angelin B., Hyötyläinen T., Orešič M., Bäckhed F. Gut microbiota regulates bile acid metabolism by reducing the levels of tauro-beta-muricholic acid, a naturally occurring FXR antagonist. Cell Metab. 2013;17:225–235. doi: 10.1016/j.cmet.2013.01.003. [DOI] [PubMed] [Google Scholar]
- Segata N., Waldron L., Ballarini A., Narasimhan V., Jousson O., Huttenhower C. Metagenomic microbial community profiling using unique clade-specific marker genes. Nat. Methods. 2012;9:811–814. doi: 10.1038/nmeth.2066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sivan A., Corrales L., Hubert N., Williams J.B., Aquino-Michaels K., Earley Z.M., Benyamin F.W., Lei Y.M., Jabri B., Alegre M.-L. Commensal Bifidobacterium promotes antitumor immunity and facilitates anti-PD-L1 efficacy. Science. 2015;350:1084–1089. doi: 10.1126/science.aac4255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Song Z., Cai Y., Lao X., Wang X., Lin X., Cui Y., Kalavagunta P.K., Liao J., Jin L., Shang J. Taxonomic profiling and populational patterns of bacterial bile salt hydrolase (BSH) genes based on worldwide human gut microbiome. Microbiome. 2019;7:9. doi: 10.1186/s40168-019-0628-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sweeney T.E., Wong H.R., Khatri P. Robust classification of bacterial and viral infections via integrated host gene expression diagnostics. Sci. Transl. Med. 2016;8:346ra91. doi: 10.1126/scitranslmed.aaf7165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsuzuki T., Takahashi M., Asai N., Iwashita T., Matsuyama M., Asai J. Spatial and temporal expression of the ret proto-oncogene product in embryonic, infant and adult rat tissues. Oncogene. 1995;10:191–198. [PubMed] [Google Scholar]
- Vohra B.P.S., Tsuji K., Nagashimada M., Uesaka T., Wind D., Fu M., Armon J., Enomoto H., Heuckeroth R.O. Differential gene expression and functional analysis implicate novel mechanisms in enteric nervous system precursor migration and neuritogenesis. Dev. Biol. 2006;298:259–271. doi: 10.1016/j.ydbio.2006.06.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wood D.E., Salzberg S.L. Kraken: ultrafast metagenomic sequence classification using exact alignments. Genome Biol. 2014;15:R46. doi: 10.1186/gb-2014-15-3-r46. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao L., Yang W., Chen Y., Huang F., Lu L., Lin C., Huang T., Ning Z., Zhai L., Zhong L.L.D. A Clostridia-rich microbiota enhances bile acid excretion in diarrhea-predominant irritable bowel syndrome. J. Clin. Invest. 2020;130:438–450. doi: 10.1172/JCI130976. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
BSH activities of 30 bacteria consortia subjected to an in vitro functional screen.
Unconjugated and conjugated bile acids in gnotobiotic mice, as measured through a targeted liquid chromatography mass spectrometry analysis. LOD: limit of detection.
Results of an FITC-based gut motility assay. Each row summarizes data for an individual mouse. Fluorescence in a given gut segment is represented as a proportion of total fluorescence measured throughout the gut.
(A). Serial transit time measurements following intra-colonic infusions of different bile acids.
(B). Effect of a TGR5 inhibitor on transit times.
(C). Effect of intra-colonic infusion of LCA on transit times in Ret+/- mice.
(A). List of genes included in a custom NanoString nCounter CodeSet panel, classifications, target sequences, and NCBI accession numbers.
(B). Raw gene expression datafrom NanoString panel.
(C). Normalized gene expression based on normalization of raw data using negative control probes, positive control probes, and expression levels of housekeeping genes.
(D). Modeling the impact of non-specific gene expression on results of a custom NanoString panel. Fold-changes in ENS gene expression may be underestimated because of non-specific gene expression by non-ENS cells. Different “true” fold-changes were considered, ranging from 1 (no change) to 10; these values represent the unknown actual gene expression fold-changes in ENS cells, whereas the values in the cell represent fold-changes “estimated” by the gene expression panel we used.
Barcoding and accession numbers for all samples that underwent shotgun sequencing are listed.
Shotgun sequencing of genomic DNA from fecal pellets revealed that Bacteroides was the predominant taxon in mice colonized with the BSH-low-1, BSH-high-1, and BSH-high-2 consortia; Clostridia comprised the next most prevalent taxon. Citrobacter youngae, Escherichia fergusonii, and Enterobacter cancerogenus approximately equally abundant species in mice colonized with the BSH-low-2 consortium, which did not contain any Bacteroides or Clostridia.
Data Availability Statement
The accession number for the bacterial shotgun sequencing data reported in this paper is ENA: PRJEB38547. Experimental data associated with this study are available in supplemental information. Custom scripts used for data analysis available at https://github.com/DeyLab.