Abstract
Purpose:
This study aimed to identify cases of developmental stuttering and associated comorbidities in de-identified electronic health records (EHRs) at Vanderbilt University Medical Center, and, in turn, build and test a stuttering prediction model.
Methods:
A multi-step process including a keyword search of medical notes, a text-mining algorithm, and manual review was employed to identify stuttering cases in the EHR. Confirmed cases were compared to matched controls in a phenotype code (phecode) enrichment analysis to reveal conditions associated with stuttering (i.e., comorbidities). These associated phenotypes were used as proxy variables to phenotypically predict stuttering in subjects within the EHR that were not otherwise identifiable using the multi-step identification process described above.
Results:
The multi-step process resulted in the manually reviewed identification of 1,143 stuttering cases in the EHR. Highly enriched phecodes included codes related to childhood onset fluency disorder, adult-onset fluency disorder, hearing loss, sleep disorders, atopy, a multitude of codes for infections, neurological deficits, and body weight. These phecodes were used as variables to create a phenome risk classifier (PheRC) prediction model to identify additional high likelihood stuttering cases. The PheRC prediction model resulted in a positive predictive value of 83 %.
Conclusions:
This study demonstrates the feasibility of using EHRs in the study of stuttering and found phenotypic associations. The creation of the PheRC has the potential to enable future studies of stuttering using existing EHR data, including investigations into the genetic etiology.
Keywords: Developmental stuttering, Electronic health records, Stuttering comorbidities, Machine learning
1. Introduction
The advent of large, diverse biomedical datasets has enabled agnostic, data-driven approaches in many areas of biomedical science. Electronic health records (EHRs) represent one such biomedical resource that encompasses large, readily accessible volumes of data. While the primary purpose of EHRs is to enhance individual patient care, they also provide a wealth of data useful for understanding disease patterns, treatment efficacy, and, when paired with DNA biobanks, the contribution of genetic factors in health. By combining demographic information with clinical notes, medication lists, and billing and procedural codes throughout the lifetime of a patient with DNA biobank samples, EHRs can facilitate novel approaches to questions that would be impractical to answer in traditional study designs. EHR-based studies are now commonplace in other fields, yet they represent an unexplored opportunity to expand the scope of developmental stuttering research. This project harnessed the power of data contained in EHRs to 1) identify stuttering cases via a text-mining algorithm and manual review, which in turn enabled us to 2) detect enrichments of co-occurring phenotypes (i.e., comorbidities) and 3) to create a phenome risk classifier (PheRC) to predict stuttering cases in EHR linked biobanks.
Over the past decade, hundreds of studies utilizing EHRs have been published (Denny et al., 2011; Onitilo, Engel, Greenlee, & Mukesh, 2009; Walters et al., 2020). For example, Namjou et al. (2014) investigated whether genotyped data from previously published GWAS studies (i.e., known gene variants) were associated with 539 EHR-derived phenotypes (including speech disorders) within a pediatric cohort. These studies, among many others, have validated that data collected through routine clinical care and captured in EHRs can achieve similar data quality compared to prospective study collection. Furthermore, when clinical EHR data is paired with genotyped samples in a DNA biobank, genetic studies such as genome-wide association studies are feasible and effective for disease gene discovery.
Electronic health record studies fundamentally rely on a process called phenotyping, or reliably identifying cases and controls of diseases and conditions of interest within or between EHR data sets. This usually involves creating an algorithm using International Classification of Diseases, Ninth and Tenth Revision (ICD-9 and −10) codes, Current Procedural Terminology (CPT) codes, laboratory test results, prescriptions, vital sign measurements, and/or free text keyword search, and then testing the algorithm against expert manual review. Traditionally, phenotypic characterizations within large-scale EHRs have relied on the presence of a diagnostic billing code, for example, an ICD-9 or ICD-10 code. While billing codes are adequate indicators of many clinical phenotypes, some conditions such as developmental stuttering are not always well captured by these data. For example, conditions that do not lead to hospital visits, are diagnosed outside of a hospital or outpatient setting, have broad or nonspecific billing codes, or are not covered by health insurance may be under-identified in the EHR. To demonstrate the inadequacy of using billing codes alone for identifying developmental stuttering cases, a preliminary search of de-identified EHRs at Vanderbilt University Medical Center (VUMC) revealed that ICD-9 and ICD-10 billing codes were conspicuously sparse for developmental stuttering: the billing code search returned only 90 cases out of approximately 93,000 records (0.01 % prevalence), far below the expected population 1–3 % prevalence (Pruett, Below, & Jones, 2018). For stuttering and other similar conditions, additional data beyond diagnostic codes is necessary for accurate phenotyping.
Underrepresentation of developmental stuttering using basic search methods in the EHR is likely driven by several factors. First, evaluation and treatment of communication disorders are typically performed by speech-language pathologists, not medical doctors, and many stuttering evaluations take place in public schools or in private speech-language pathology clinics. Consequently, these records regarding the evaluation and treatment of stuttering are generally not included in medical center EHRs. Second, communication disorders such as stuttering are often ancillary to the purpose of a doctor visit, so descriptions of speech and language may not be considered relevant to record in EHRs. Third, communication disorders may simply go unnoticed by medical professionals during an encounter. For example, stuttering is variable in nature and individuals may not overtly stutter during a medical consultation. In this case, unless a patient was specifically asked about his or her speech, or being seen for a speech-related reason, stuttering would not be noted in the EHR. Despite these hurdles, patients who stutter are regularly seen within medical center settings and some have sufficient notation to positively identify them as individuals who stutter.
Keyword searches and text-mining algorithms can augment diagnostic codes to identify potential cases of a given disease or disorder. In fact, depending on the condition and medical context, keywords may provide even better representation of the phenotype of interest. In our preliminary study, when “stuttering” was mentioned within VUMC EHR notes, words associated with the disorder were often used to describe conditions other than developmental stuttering (Pruett et al., 2018). In a manual review of 1,822 records returned from a keyword search, 29 % (521 records) contained exclusively non-speech related mentions of stuttering (Pruett et al., 2018). For example, the most common non-speech related usages of the keyword “stuttering” included: (1) “stuttering onset”, or a symptom that comes and goes, (2) “stuttering gait”, or a gait marked by instability and frequent halting, and (3) “stuttering angina”, or random or unstable heart pain. The widespread use of the terms provided evidence that a simple keyword search would be insufficient to define developmental stuttering cases in the EHR. Overall, the low prevalence of ICD codes combined with the high false-positive keyword search provided evidence that a more nuanced approach was necessary to define developmental stuttering within the EHR. It is not the case that information on stuttering is absent or irrelevant in these large databases, it is merely more difficult to parse than other disorders. Therefore, one purpose of the present study was to develop a valid and replicable approach to identify stuttering cases within EHRs.
Once developmental stuttering cases are identified, EHRs provide a practical and efficient method for investigating stuttering comorbidities, or the presence of one or more conditions that co-occur with developmental stuttering at a higher frequency than would be expected by random chance in a control sample. Interactions among conditions and diseases may have far-reaching effects on both personal health and the health care system at large. According to Valderas, Starfield, Sibbald, Salisbury, and Roland (2009), comorbidity is associated with worse health outcomes, more complex clinical management, and increased health care costs. Mechanisms underlying the coexistence of two or more conditions in a patient include direct or indirect causation, shared risk factors, or independence, and understanding these mechanisms can impact clinical care, epidemiology, and health services planning (Valderas et al., 2009). Greater understanding of stuttering comorbidity may not only improve clinical care, but also our understanding of causes of stuttering.
Clinical and anecdotal evidence suggests a high incidence of comorbid speech, language, and attention disorders within patients with developmental stuttering (Arndt & Healey, 2001; Donaher & Richels, 2012). For the purpose of this study, diagnosed conditions or disorders rather than between-group differences, are considered comorbidities (for examples of studies that have examined between-group differences, but not statistically tested for differences in the frequency of diagnosed conditions or disorders associated with stuttering, see Ambrose, Yairi, Loucks, Seery, & Throneburg, 2015; Kefalianos et al., 2017; Reilly et al., 2013; Watkins, Ehud, & Grinager, 1999). To date, a variety of methods have been used to examine stuttering comorbidities. For example, using the National Health Interview Survey, Briley and Ellis (2018) found the presence of at least one disabling developmental condition (from among (a) intellectual disability, (b) learning disability, (c) attention-deficit/hyperactivity disorder (ADHD)/ADD, (d) seizures, (e) autism, Asperger’s, or pervasive developmental disorder (PDD), and (f) any other developmental delay) to be 5.5 times higher in children who stutter compared to children who do not. Additionally, using the 1995 Australian Health Survey, Keating, Turrell and Ozanne (2001) found children who stutter had a higher incidence of developmental delay and emotional problems as well as asthma, allergies, and deafness from among 16 pre-selected conditions. Furthermore, a mail survey sent to practicing speech-language pathologists in the United States inquiring about clients on their caseload suggested that 63 % of young children who stutter have co-occurring speech, language, or non-speech-language disorders from among 18 pre-selected conditions (Blood, Ridenour, Qualls, & Hammer, 2003). Other studies have taken a more targeted approach and examined individual conditions comorbid with stuttering including anxiety (e. g., Iverach et al., 2016; cf. Manning & Beck, 2013), inattention and hyperactivity (e.g., Donaher & Richels, 2012), and articulation and phonological disorders (e.g., Wolk, Conture, & Edwards, 1990). These studies have identified a number of disorders and conditions comorbid with developmental stuttering but were limited by 1) a reliance on either clinician or caregiver recall (Briley & Ellis, 2018), 2) lack of a control population (Blood et al., 2003; Donaher & Richels, 2012; Manning & Beck, 2013), and/or 3) a scope limited to preselected conditions (i.e., other potential comorbidities with stuttering may not be included on a given survey).
In contrast to these approaches, EHRs offer a greater depth and breadth of examined medical conditions. Consequently, queried comorbidities are not limited to a predetermined list and are not dependent on the recall of an examiner or examinee. Additionally, the average length of medical records encompass more years than most comorbidity studies, capturing conditions across a greater portion of the lifespan. Therefore, the present investigation aimed to assess conditions associated with stuttering using a novel method with the potential to both replicate previously identified comorbidities and explore latent, unstudied, comorbidities. A greater understanding of stuttering comorbidities, especially those beyond the scope of previous studies, has the potential to impact clinical care management and reveal underlying shared etiology of associated conditions and diseases (Valderas et al., 2009).
Further, identification of comorbidities enables the creation of a phenome risk classifier. Phenome risk classifiers use the association of comorbidities to create an algorithm to identify likely cases independent of positive diagnoses (e.g., no ICD-9 or −10 code and/or no positive keyword search identification) via underlying similarities to manually reviewed cases (e.g., a similar constellation of comorbid conditions). Using this tool, we can overcome the challenges in identifying developmental stuttering cases within EHRs to greatly increase the number of high-likelihood cases for genetic analysis within the Vanderbilt EHR. More broadly, this approach may be adapted and employed in other EHRs with limited free-text search abilities to identify high-likelihood stuttering cases, or other phenotypes that are not well captured by diagnostic codes themselves. For example, any significant genetic findings resulting from work within the Vanderbilt EHR could be replicated within other biobank-paired EHRs such as the UK Biobank or the eMERGE Network (Gottesman et al., 2013; Sudlow et al., 2015). This approach has been previously used to replicate known associations between conditions and genetic variants in studies examining asthma (Zhu et al., 2018) cognition (Davies et al., 2016), depression (Howard et al., 2018), glaucoma (Verma et al., 2016), muscle strength (Tikkanen et al., 2018), and osteoarthritis (Zengini et al., 2018), among many others, demonstrating the utility and feasibility of this approach across a wide variety of diseases and conditions. For the field of stuttering, this could lead to the discovery of population level causally-related stuttering genes.
Therefore, the purpose of this project is to translate these powerful approaches to the study of developmental stuttering. Accordingly, the present study represents the first agnostic, wide-scale EHR-based study of developmental stuttering and associated conditions. The major advantages to using EHRs to investigate developmental stuttering include: 1) access to much larger sample sizes than prospective cohort studies, numbering in the thousands to hundreds of thousands, 2) access to a greater depth and breadth of nonspeech-language pathology medical history (e.g., to assess a wide range of associated comorbidities), and, when paired with DNA biobanks, 3) access to existing EHR-linked genetic data. We employed these advantages to propel novel studies of the etiology of stuttering at scale. These studies are particularly critical, because despite large heritability estimates, strong familial trends, and high population prevalence, the genetic architecture of developmental stuttering is still largely unknown (Ambrose, Cox, & Yairi, 1997; Fagnani, Fibiger, Skytthe, & Hjelmborg, 2011; Yairi, Ambrose, & Cox, 1996; Yairi, Ambrose, & Cox, 1996). Therefore, in order to begin to address this gap in our knowledge, this project harnessed the power of data contained in EHRs to: identify stuttering cases via text-mining and manual review, detect comorbidities, and create a PheRC to predict high-likelihood stuttering cases in an EHR-linked biobank. As has been done with other conditions, this nascent work may enable the application of this approach to other biobanks and related genetic analyses in the area of developmental stuttering.
2. Methods
2.1. Overall strategy
The first step to identifying conditions associated with developmental stuttering within the Vanderbilt University Medical Center (VUMC) EHR involved “defining” the developmental stuttering phenotype status by systematically labeling individuals with explicit indicators of disfluency as developmental stuttering cases and those without as population controls. Due to the dearth of developmental stuttering notation within the EHR (Pruett et al., 2018), this process involved (a) a keyword search of clinical notes followed by, (b) a text-mining algorithm, and (c) manual review. This multi-step approach was used to broadly search for developmental stuttering cases while limiting manual review in an otherwise prohibitively large (2.8 million records) clinical note set. Following identification, manually reviewed developmental stuttering cases were compared to matched population controls in a phecode enrichment analysis to reveal conditions comorbid with stuttering. Finally, these enriched phecodes were used as variables to create a phenome risk classifier (PheRC) prediction model to identify high-likelihood stuttering cases without the use of keywords and text-mining. All code used to develop the text-mining algorithm, phecode enrichment analysis, and phenome risk classifier is publicly available and open for use at https://github.com/belowlab/StutteringCART.
2.2. Data source – the Synthetic Derivative (SD) of the Vanderbilt University Medical Center EHR
Vanderbilt University Medical Center (VUMC), located in Nashville, Tennessee, is one of the largest academic medical centers in the United States and offers primary and specialty care in hundreds of adult and pediatric specialties with over 2 million patient visits each year. In addition to Vanderbilt University Hospital and Monroe Carell Jr. Children’s Hospital, VUMC comprises over 100 outpatient clinics in greater Tennessee. Furthermore, VUMC includes The Vanderbilt Bill Wilkerson Center for Otolaryngology and Communication Sciences, a multidisciplinary clinic that specializes in ear, nose, and throat diseases, and communication disorders such as hearing, speech, language, and voice problems.
The size and diversity of VUMC is reflected in the composition of the EHR. Vanderbilt University Medical Center maintains a de-identified EHR database called the Synthetic Derivative (SD) currently containing ~2.8 million patient records. The Synthetic Derivative is 52 % female, with a plurality of patients currently 18–44 years of age (33.1 %) and 15.7 % currently under 18 years of age. Current race/ethnicity estimates are: 59.8 % Caucasian, 9.6 % African American, 3.2 % Latino, 1.3 % Asian, 0.1 % Native American, 0.6 % other, 0.2 % multiple, and 25.2 % unknown. It should be noted that the recording of race is done by a third party and the procedure is not uniform, leading to a high incidence of reporting error; this cannot be directly addressed in this study but should be acknowledged as a potential, though likely minimal, source of bias.
The VUMC SD interface allows the user to search data extracted from most of the major health information databases at Vanderbilt. Specifically, the search interface provides users access to basic clinical and demographic information, such as ICD-9 and 10 codes, CPT procedure codes, medications, lab values, and free text from within medical notes, and returns de-identified data for review. Medical notes is an EHR category that contains free text, numerical, and categorical notes about an encounter. Medical note categories include admission notes, ancillary reports, discharge summaries, emergency department notes, family history, inpatient notes, general notes, nursing reports, pathology reports, outpatient notes, problem lists, and radiology reports. Medical notes make up the bulk of information on a patient and provide excellent context for understanding encounters in the EHR. Listed within medical note categories are: reason for the visit, medical history, general description of the patient, evaluation of the chief complaint, description of other relevant medical, social and familial context, test results, diagnoses, and follow-up plans. Medical notes also include transcripts of email, mail, and telephone correspondence between patients, providers, and referring providers. Psychological and psychiatric records are protected and not directly accessible, but neurology reports and psychological referral correspondence are accessible. New clinical data are added to the database bimonthly.
2.3. Identifying developmental stuttering cases in the EHR via keyword search and manual review
2.3.1. Exploratory keyword search
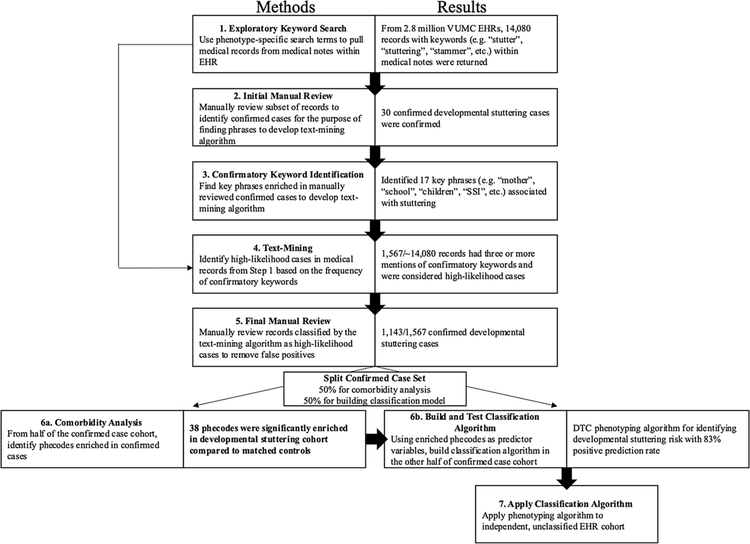
A list of exploratory keywords including descriptors of developmental stuttering in EHRs was developed based on investigator phenotypic expertise (Fig. 1, Step 1). A preliminary study of the VUMC SD examining cases with stuttering ICD 9 and 10 codes (n = 142) revealed that in free text chart notations written by doctors and nurses, stuttering was often misspelled and there were several words used to describe the condition (Pruett et al., 2018). However, in this preliminary study, every confirmed case of developmental stuttering contained at least one of these exploratory keywords, “stutter”, “studder” [sic], “stuttering”, “studdering” [sic], “stammer”, “stammering”, “disfluency”, and “dysfluency” within medical notes. Therefore, this list of exploratory keywords was used to initially filter the approximately 2.8 million records in the VUMC SD.
Fig. 1.
Flow Chart Depicting Methodological Steps with Results.
Note. All code used to develop the text-mining algorithm, phecode enrichment analysis, and phenome risk classifier is publicly available and open for use at https://github.com/belowlab/StutteringCART. Step 7 applies to future studies.
2.3.2. Initial manual review
Reviewers with expertise in stuttering then examined de-identified text files with the exploratory keywords highlighted in context to determine if the observed keywords were used to describe developmental stuttering (Fig. 1, Step 2). Medical notes with standardized test scores from a speech-language pathologist indicating developmental stuttering (e.g., Stuttering Severity Instrument, or stuttering-like disfluencies from a speech sample), description of stuttering speech combined with supporting familial/educational context (e.g., “Patient’s mother is concerned with child’s stuttering and describes teasing at school. Disfluency noted during encounter. Referral to speech-language pathologist.”) and mentions of stuttered speech without reference to confounding conditions like stroke, traumatic brain injury, seizures, and psychological/psychiatric conditions (e.g., “Patient displays stuttering in his speech, as per baseline.”) determined case status to be used in subsequent analyses. If case status could not be determined from the text surrounding keywords, the search was expanded outside the scope of the keywords (e.g., clarifying whether a disqualifying condition, such as stroke, occurred previously to the mention of stuttering). The purpose of this step is to remove cases where stuttering speech occurs as the result of (a) stroke or traumatic brain injury, termed neurogenic stuttering, (b) side effects from pharmaceuticals, termed neuropharmacological stuttering, and (c) idiopathic stuttering in conjunction with psychosis or schizophrenia, termed psychogenic stuttering. While these conditions may present similarly to developmental stuttering, with an adult onset and being often transient in nature, they likely represent a distinct pathogenesis (Theys, van Wieringen, & De Nil, 2008) and were removed from consideration as cases or controls. Similarly, ambiguous cases lacking adequate inclusion criteria (described above) were also removed from consideration as cases or controls.
Because the records used for this exploratory keyword search and context development would be lost to subsequent steps, we opted to limit our search to the first 30 positively identified stuttering cases. Due to limited sample size, there is a tradeoff between using cases to develop the text-mining algorithm and using cases for enrichment analysis and model training/testing. Therefore, the relatively limited number of cases used in this filtering step preserved cases for subsequent steps.
2.3.3. Confirmatory keyword identification
Using confirmed stuttering cases from the initial manual review, we selected all words immediately surrounding (within ten words of) an exploratory keyword instance (i.e., “stutter”, “studder”, “stuttering”, “studdering”, “stammer”, “stammering”, “disfluency”, and “dysfluency”) and calculated the frequency of observation. We then identified words whose presence was enriched in the records of developmental stuttering cases (Fig. 1, Step 3). The seventeen most strongly associated words were considered “confirmatory keywords” (i.e., “mom”, “mother”, “mom’s”, “parent”, “parents”, “school”, “preschool”, “pre-school”, “child”, “children”, “birth”, “dad”, “father”, “dad’s”, “father’s”, “SSI-3”, and “SSI”) and were selected for use in subsequent filtering steps. This data allowed us to create a text-mining algorithm, which served as a search engine for identifying high-likelihood developmental stuttering cases by the words used to describe it within medical notes.
2.3.4. Text-mining algorithm
We developed this text-mining algorithm to identify high-likelihood cases and thus reduce the number of false positive keyword hits (Fig. 1, Step 4). As a result, far fewer files required final manual review. Specifically, the algorithm tallies the number of confirmatory keywords within ten words of exploratory keyword instances to create a numerical score for each individual. After an initial trial with five or more confirmatory keyword phrases returned too few cases, the threshold was lowered to three confirmatory keyword phrases to cast a wider net of potential cases. Consequently, all patients with three or more instances of confirmatory keyword phrases (i.e. “mom”, “mother”, “mom’s”, “parent”, “parents”, “school”, “preschool”, “pre-school”, “child”, “children”, “birth”, “dad”, “father”, “dad’s”, “father’s”, “SSI-3”, and “SSI”) combined with exploratory keywords (i.e. “stutter”, “studder”, “stuttering”, “studdering”, “stammer”, “stammering”, “disfluency”, and “dysfluency”) were selected as high-likelihood developmental stuttering cases. Patients that had any direct mentions of “developmental stuttering” were also included.
2.3.5. Final manual review
An additional, final manual review (using the same procedure as the initial manual review) was conducted to remove false positive cases identified by the text-mining algorithm (Fig. 1, Step 5). Again, false positives included cases of neurogenic, neuropharmacological, and idiopathic stuttering. Specifically, this process ensured that all developmental stuttering cases were manually reviewed and confirmed by the phenotyping team.
Ultimately, the end goal of the multi-step identification process was to maximize the positive predictive value, or the proportion of confirmed cases among those identified as potential cases, in order to reduce manual review in future studies. Positive predictive value is a function of both the design of the text-mining algorithm as well as the base rate of the condition within a population. For example, a study examining phenotyping diseases within EHRs using a combination of natural language processing and structured data found that positive predictive value ranged from 88 % for rheumatoid arthritis to 98 % for Crohn’s disease (Liao et al., 2015). Given the lack of specific billing codes for developmental stuttering within the VUMC SD, we assumed our keyword-based text-mining algorithm would result in a slightly lower positive predictive value. This multi-step process accomplishes the first aim of the project: identifying stuttering cases via a text-mining algorithm and manual review within the VUMC SD.
2.4. Comorbidity analysis via phecode enrichment
Once identified using the multi-step process described above, half of the developmental stuttering cases were randomly selected for the phecode enrichment analysis (i.e., comorbidity analysis), reserving the other half for the training and testing of the phenome risk classifier as described below. Each of these cases was assigned up to five controls matched for age (< 5 years of age difference), sex, race, ancestry, and number of clinical encounters (< 5 clinical visit difference between case and control). Clinical encounters were estimated by the number of unique days a patient received a diagnostic code (i.e., phecode), an index of the frequency of medical care and thus length of record.
Phecodes represent hierarchical diagnostic groupings for EHR data derived from ICD-9 codes (Denny et al., 2010, 2013). There are 1,645 phecodes, loosely following the ICD-9 code system, but revised based on statistical co-occurrence and code frequency. For example, Phecode 315 Developmental Delays and Disorders contains the subcategory Phecode 315.2 Speech and Language Disorder, which encompasses the following ICD-9 categories: Developmental dyslexia (315.02), Other specific developmental reading disorder (315.09), Developmental speech or language disorder (315.3), Expressive language disorder (315.31), Mixed receptive-expressive language disorder (315.32), Speech and language developmental delay due to hearing loss (315.34), Childhood onset fluency disorder (315.35), and Other developmental speech disorder (315.39).
Enrichment analysis was used to compare the frequency of phecodes within the developmental stuttering cohort to selected controls. The approach uses a mathematical model (e.g., a distribution) to directly compute an empirical p value by calculating the frequency of each phecode in the stuttering case set compared to a null distribution created through multiple permutations of the randomized control set (Reimand et al., 2019). Specifically, for each permutation, one control was selected at random from each case’s matched set (i.e., 1 of the 5 controls for each stuttering case was randomly selected). By selecting exactly one control from each case’s matched set, we ensured that the overall control demographic structure (e.g., age, sex, race, etc.) remained similar to the demographics of the stuttering case set and helps account for possible confounds. We then calculated the frequency of each phecode for this randomly selected set of controls. This process was repeated for 10,000 permutations, resulting in 10,000 randomized sets of controls as well as the observed phecode frequency for each of these sets, creating a null distribution. A p-value for each phecode was then calculated by comparing the observed phecode counts in the stuttering case set to the null distribution of phecode counts created through the 10,000 permutations. Phecodes were considered significantly enriched in the stuttering case set if the observed counts exceeded the maximum observed counts in the control set distribution. For example, the maximum frequency of the Phecode 315 (Developmental delays and disorders) across all 10,000 randomized control sets was 128, whereas in our stuttering case set there were 351 patients who had this phecode; it is therefore significantly enriched in the stuttering case set. For further comparison, for Phecode 315, the 50th percentile of the observed counts across all 10,000 randomized control sets was 104 codes and the 99th percentile was 123 codes. For all significant phecodes, the stuttering case set had more observed codes than the maximum codes found across all 10,000 randomized control sets, corresponding to a conservative p-value threshold (p ~ 0). Phecodes which were observed in less than 3.5 % of developmental stuttering cases were excluded regardless of significance because these were deemed to be too infrequently occurring for comparison. For a sensitivity analysis, we repeated this methodology while restricting the developmental stuttering dataset to include only cases with Stuttering Severity Instrument (SSI) scores above “sub-clinical”, indicating that stuttering diagnosis was clinically measured and confirmed by speech-language pathologists. Overall, these steps accomplish the second aim of the project: to utilize a novel approach to detect enrichments of co-occurring phenotypes to examine developmental stuttering comorbidities across the breadth and depth of the VUMC SD.
2.5. Phenome risk classifier (PheRC) using machine learning
All significantly enriched phecodes (i.e., comorbidities) were used to model a decision tree classifier algorithm to create a phenome risk classifier (PheRC) to predict potential developmental stuttering cases. Broadly speaking, by mapping clinical phenotypes extracted from the EHR, the PheRC expresses the degree to which clusters of symptoms, represented by phecodes, predict a condition of interest, in this case developmental stuttering (for a similar approach that used symptom clusters and phecodes for identifying cases, see Bastarache et al., 2018). For example, a person with a higher frequency of codes that match the codes enriched in our developmental stuttering cohort (e.g., Developmental delays and disorders, Sleep disorders, Allergic reaction to food, etc.), would be identified as having underlying similarities to a person who stutters. Importantly, the predictiveness of any single phecode depends on the “impurity” of the phecode as a proxy variable. Impurity is determined by how effectively a certain phecode can split the dataset into stuttering and non-stuttering cases. For example, if everyone with a given phecode stutters and everyone lacking that phecode doesn’t stutter, then that phecode has an impurity of 0. Conversely, if a phecode doesn’t predict stuttering status any better than a coin flip, then that phecode has an impurity of 0.5. Consequently, a patient could be a predicted stuttering case with 2 or 3 phecodes with very low impurities, or with 6 or 7 phecodes with higher levels of impurity. The binary outcome of the PheRC classifies individual patients within the EHR as either similar to our stuttering cohort or similar to the control cohort based on their phecodes. Those deemed similar to the stuttering cohort were considered PheRC-predicted cases based on comorbidities.
2.5.1. Build and test classification algorithm
To create the PheRC algorithm, the remaining half of the manually reviewed developmental stuttering cases not used in the comorbidity analysis were used to train and test the model. For the model training (or model creation), 80 % of stuttering cases with their accompanying phecodes were used in a gini-index based classification and regression tree classification model, using the presence or absence of each phecode as predictors, and a binary determination of developmental stuttering cases or controls as the outcome (Pedregosa et al., 2011). To avoid overfitting the model, we minimized tuning our hyperparameters and avoided re-testing and training the PheRC model. No restrictions were placed on the tree depth or leaf node counts. Minimum samples per leaf was set to five. Twenty percent of the remaining manually reviewed developmental stuttering cases were used to test (or validate) the model, comparing developmental stuttering cases identified by manual review against PheRC-predicted cases. This step accomplishes the third aim of the project: to create a PheRC to predict potential developmental stuttering cases in EHR-linked biobanks.
3. Results
3.1. Exploratory keyword search
The exploratory keyword search returned 14,080 individuals with at least one keyword mention (“stutter”, “studder”, “stuttering”, “studdering”, “stammer”, “stammering”, “disfluency”, and “dysfluency”) in their records (Fig. 1, Step 1). These keywords cast a wide net but contained false positives from non-speech related uses of the words and/or stuttering resulting from exclusionary criteria.
3.2. Initial manual review
An initial manual review identified 30 developmental stuttering cases (Fig. 1, Step 2). The 30 cases represented a sufficient number for the confirmatory keyword identification while also maintaining as many cases as possible for the subsequent steps (Pruett et al., 2018).
3.3. Confirmatory keyword identification
Based on analyses using the 30 confirmed developmental stuttering records, the top twenty confirmatory keywords associated with stuttering were chosen. Keywords that were also enriched within non-stuttering cases (“edition”, “test”, “total”) were excluded. The resulting seventeen confirmatory keywords associated with stuttering included: “mom”, “mother”, “mom’s”, “parent”, “parents”, “school”, “preschool”, “pre-school”, “child”, “children”, “birth”, “dad”, “father”, “dad’s”, “father’s”, “SSI-3”, and “SSI” (Fig. 1, Step 3). The identification of these confirmatory keywords was used to facilitate the creation of the text-mining algorithm, described subsequently.
3.4. Text-Mining
From the 14,080 individuals with stuttering-related exploratory keywords, the text-mining algorithm further reduced the number of high-likelihood cases to 1,567 (Fig. 1, Step 4). As previously mentioned, these 1,567 high-likelihood cases of developmental stuttering had: 1) at least three or more occurrences of confirmatory keyword phrases (i.e., “mom”, “mother”, “mom’s”, “parent”, “parents”, “school”, “preschool”, “pre-school”, “child”, “children”, “birth”, “dad”, “father”, “dad’s”, “father’s”, “SSI-3”, and “SSI”) combined with exploratory keywords (i.e., “stutter”, “studder”, “stuttering”, “studdering”, “stammer”, “stammering”, “disfluency”, and “dysfluency”), and/or 2) a direct mention of “developmental stuttering.”
3.5. Final manual review
Of the 1,567 high likelihood cases of developmental stuttering identified by the text-mining algorithm, 1,143 were determined to be true cases after manual review by members of the investigative team (Fig. 1, Step 5). This equates to a positive predictive value (the number of confirmed cases divided by the total number of suspected cases) of 73 %. The relatively high positive predictive value indicates the exploratory keyword search combined with the text-mining algorithm successfully identified a much higher proportion of developmental stuttering cases compared to a keyword search alone (approximately 20 % positive predictive value, see Pruett et al., 2018). The average current age of developmental stuttering cases was approximately 17 with a standard deviation of 9. Birthdates for confirmed cases ranged from 1946 to 2015. Manual review typically required 3−5 min, with approximately 10 % of cases requiring more extensive review.
As mentioned previously, positive indicators for developmental stuttering cases within the 1,567 high-likelihood records included standardized test scores from a speech-language pathologist assessing developmental stuttering, description of stuttering speech combined with supporting familial/educational context, and mentions of stuttered speech without reference to confounding conditions like stroke, traumatic brain injury, seizures, and psychological/psychiatric conditions. Approximately 43 % of manually reviewed confirmed cases had a clinically documented developmental stuttering diagnosis as determined by Stuttering Severity Instrument scores and speech-language pathologist assessment. An additional ~20 % of manually reviewed stuttering cases had a speech-language pathology referral for stuttering concerns. Common exclusions within the 1,567 high-likelihood records included records with stuttering discussed in the context of (a) stroke or traumatic brain injury, termed neurogenic stuttering, (b) side effects from pharmaceuticals, termed neuropharmacological stuttering, and (c) idiopathic stuttering in conjunction with psychosis or schizophrenia, termed psychogenic stuttering. While these conditions are similar to developmental stuttering, with an adult onset and being often transient in nature, they likely represent a distinct pathogenesis (Theys et al., 2008). Ambiguous cases were rare and represented less than 1% of cases reviewed. These cases included mentions of stuttering (e.g., “Patient stuttering”) but lacked sufficient supporting context to confirm developmental stuttering. For demographics of confirmed developmental stuttering cases, see Table 1.
Table 1.
Demographics for Confirmed Developmental Stuttering Patients Following Manual Review.
| Confirmed Developmental Stuttering Cases n (%) | |
|---|---|
| Total | 1143 |
| Male | 867 (75.3 %) |
| Female | 276 (24.7 %) |
| Demographics | Mean (SD) |
| Age (years) | 17.7 (9.8) |
| Race/Ethnicity | n (%) |
| Caucasian | 526 (46.0 %) |
| African American | 321 (28.1 %) |
| Asian | 16 (1.4 %) |
| Hispanic | 68 (5.9 %) |
| Unknown | 202 (17.7 %) |
3.6. Comorbidity analysis
According to our phecode enrichment analysis, compared to matched controls (n = 2765), developmental stuttering cases not used for the development of the phenome risk classifier (n = 572) were enriched for a variety of previously suggested comorbidities as well as potential novel comorbidities (Fig. 1, Step 6a). For demographics of cases and controls used for phecode enrichment, see Table 2.
Table 2.
Case and Control Demographics for Phecode Enrichment Analysis.
| Cases n (%) | Controls n (%) | |
|---|---|---|
| Total | 572 (17.1 %) | 2765 (82.9 %) |
| Male | 432 (75.6 %) | 2081 (75.3 %) |
| Female | 140 (24.5 %) | 684 (24.7 %) |
| Demographics | Mean (SD) | Mean (SD) |
| Age (years) | 17.4 (9.48) | 18.2 (10.2) |
| Visits | 24.64 (34.0) | 21.41 (28.7) |
| Race/Ethnicity | n (%) | n (%) |
| Caucasian | 260 (45.5 %) | 1286 (46.5 %) |
| African American | 161 (28.1 %) | 773 (28.0 %) |
| Asian | 11 (2.0 %) | 50 (1.8 %) |
| Hispanic | 30 (5.2 %) | 133 (4.8 %) |
| Unknown | 100 (17.5 %) | 489 (17.7 %) |
| Cases | Matched Controls | |
| 0 (0%) | 0 | |
| 11 (1.9 %) | 1 | |
| 6 (1.1 %) | 2 | |
| 11 (1.9 %) | 3 | |
| 11 (1.9 %) | 4 | |
| 533 (93.2 %) | 5 | |
Enriched phecodes associated with previously suggested comorbidities included Developmental Delays and Disorders (Phecode 315) (Arndt & Healey, 2001; Blood et al., 2003), Speech and Language Disorder (Phecode 315.2) (Arndt & Healey, 2001; Blood et al., 2003), Pervasive Developmental Disorders (Phecode 313) (Scott, 2015), Tics and Stuttering (Phecode 313.2) (Ooki, 2005), Hearing Loss (Phecode 389) and Conductive Hearing Loss (Phecode 389.2) (Arenas, Walker, & Oleson, 2017), Sleep Disorders (Phecode 327) (Macey et al., 2002), and a variety of codes related to the atopic triad including Acute Upper Respiratory Infections of Multiple or Unspecific Cites (Phecode 465) (Strom & Silverberg, 2016a, 2016b), Allergic Reaction to Food (Phecode 930) (Strom & Silverberg, 2016a, 2016b), Rash and Other Non-specific Skin Eruption (Phecode 687.1) (Strom & Silverberg, 2016a, 2016b), Atopic/contact Dermatitis Unspecified (Phecode 939) (Strom & Silverberg, 2016a, 2016b), and Cough (Phecode 512.8) (Strom & Silverberg, 2016a, 2016b).
Other enriched phecodes included Other Tests (Phecode 1010) and associated infections, Neurological Deficits (Phecode 292), Aphasia/Speech Disturbance (Phecode 292.1), Overweight, Obesity, and Hyperalimentation (Phecode 278), Symptoms Concerning Nutrition, Metabolism, and Development (Phecode 1002), and Lack of Normal Physiological Development (Phecode 264). All significantly enriched phecodes are presented in Table 3. For non-significant phecodes of interest (up to p = .12), see Table 4.
Table 3.
Significant Phecode Enrichments (developmental stuttering cases, n = 572; matched controls, n = 2765).
| Significantly Enriched Phecodes | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| Phecode | Description | Count | p. value | p01 | p05 | p10 | p50 | p90 | p95 | p99 | max |
| Childhood onset fluency disorder | |||||||||||
| 315 | Develomental delays and disorders | 351 | 0.000 | 81 | 87 | 89 | 104 | 117 | 121 | 123 | 128 |
| 315.2 | Speech and language disorder | 337 | 0.000 | 64 | 74 | 77 | 89 | 102 | 106 | 109 | 115 |
| Pervasive development disorders and adult onset fluency disorder | |||||||||||
| 313 | Pervasive developmental disorders | 186 | 0.000 | 66 | 69 | 72 | 80 | 90 | 93 | 95 | 95 |
| 313.2 | Tics and stuttering | 141 | 0.000 | 15 | 17 | 19 | 25 | 30 | 32 | 35 | 37 |
| Hearing loss | |||||||||||
| 389 | Hearing loss | 79 | 0.000 | 38 | 43 | 45 | 53 | 59 | 62 | 66 | 66 |
| 389.2 | Conductive hearing loss | 46 | 0.000 | 18 | 22 | 23 | 30 | 35 | 37 | 39 | 44 |
| Sleep disorders | |||||||||||
| 327 | Sleep disorders | 47 | 0.000 | 18 | 24 | 25 | 30 | 35 | 36 | 39 | 42 |
| Atopic triad | |||||||||||
| 465 | Acute upper respiratory infections of multiple or unspecified sites | 174 | 0.000 | 131 | 135 | 140 | 150 | 163 | 164 | 171 | 172 |
| 930 | Allergic reaction to food | 20 | 0.000 | 4 | 6 | 6 | 10 | 14 | 15 | 16 | 19 |
| 687.1 | Rash and other nonspecific skin eruption | 63 | 0.000 | 29 | 33 | 34 | 41 | 48 | 50 | 52 | 53 |
| 939 | Atopic/contact dermatitis due to other or unspecified | 84 | 0.000 | 37 | 42 | 44 | 51 | 59 | 60 | 62 | 68 |
| 512.8 | Cough | 120 | 0.000 | 73 | 76 | 81 | 90 | 99 | 101 | 106 | 106 |
| Diagnostic testing and infections | |||||||||||
| 1010 | Other tests | 143 | 0.000 | 15 | 20 | 21 | 27 | 33 | 34 | 37 | 40 |
| 783 | Fever of unknown origin | 152 | 0.000 | 109 | 111 | 115 | 124 | 136 | 139 | 144 | 145 |
| 79 | Viral infection | 116 | 0.000 | 67 | 70 | 76 | 85 | 95 | 98 | 102 | 109 |
| 472 | Chronic pharyngitis and nasopharyngitis | 29 | 0.000 | 10 | 13 | 14 | 19 | 24 | 25 | 28 | 29 |
| 381 | Otitis media and Eustachian tube disorders | 149 | 0.000 | 94 | 104 | 108 | 120 | 131 | 133 | 136 | 143 |
| 381.1 | Otitis media | 144 | 0.000 | 89 | 99 | 101 | 114 | 125 | 126 | 129 | 131 |
| 381.11 | Suppurative and unspecified otitis media | 134 | 0.000 | 84 | 90 | 95 | 106 | 116 | 118 | 122 | 122 |
| 110 | Dermatophytosis / Dermatomycosis | 39 | 0.000 | 17 | 20 | 21 | 26 | 31 | 33 | 35 | 35 |
| 110.1 | Dermatophytosis | 38 | 0.000 | 16 | 18 | 20 | 24 | 29 | 31 | 33 | 34 |
| 112 | Candidiasis | 33 | 0.000 | 14 | 16 | 17 | 23 | 28 | 30 | 32 | 33 |
| 369 | Infection of the eye | 58 | 0.000 | 25 | 29 | 34 | 42 | 50 | 51 | 54 | 58 |
| 369.5 | Conjunctivitis; infectious | 54 | 0.000 | 22 | 26 | 29 | 37 | 44 | 46 | 50 | 53 |
| 382 | Otalgia | 27 | 0.000 | 5 | 9 | 11 | 16 | 21 | 21 | 23 | 25 |
| 870 | Open wounds of head; neck; and trunk | 54 | 0.000 | 21 | 31 | 31 | 39 | 46 | 47 | 49 | 50 |
| 870.3 | Other open wound of head and face | 38 | 0.000 | 12 | 17 | 19 | 25 | 30 | 32 | 35 | 36 |
| Neurological deficits | |||||||||||
| 292 | Neurological disorders | 55 | 0.000 | 21 | 23 | 26 | 32 | 37 | 39 | 41 | 41 |
| 292.1 | Aphasia/speech disturbance | 40 | 0.000 | 6 | 8 | 10 | 14 | 18 | 19 | 21 | 23 |
| 350 | Abnormal movement | 53 | 0.000 | 14 | 15 | 17 | 22 | 27 | 28 | 31 | 32 |
| 350.2 | Abnormality of gait | 24 | 0.000 | 0 | 1 | 2 | 4 | 6 | 7 | 7 | 10 |
| 350.3 | Lack of coordination | 20 | 0.000 | 3 | 5 | 7 | 11 | 15 | 16 | 18 | 19 |
| 345 | Epilepsy; recurrent seizures; convulsions | 55 | 0.000 | 29 | 35 | 37 | 44 | 50 | 51 | 54 | 55 |
| 345.3 | Convulsions | 54 | 0.000 | 27 | 32 | 34 | 41 | 47 | 48 | 51 | 51 |
| Weight Control | |||||||||||
| 278 | Overweight; obesity and other hyperalimentation | 29 | 0.000 | 1 | 2 | 3 | 5 | 8 | 9 | 10 | 10 |
| 1002 | Symptoms concerning nutrition; metabolism; and development | 31 | 0.000 | 2 | 3 | 4 | 7 | 10 | 10 | 11 | 11 |
| 264 | Lack of normal physiological development | 83 | 0.000 | 53 | 55 | 56 | 65 | 73 | 74 | 79 | 82 |
| 264.9 | Lack of normal physiological development; unspecified | 43 | 0.000 | 19 | 21 | 23 | 28 | 33 | 36 | 38 | 40 |
Note. p values < .0001 indicated as 0.000 in the table. p01, p05, p10, p50, p90, p95, and p99 represent the number of phecodes found in the 1st, 5th, 10th, 50th, 90th, 95th, and 99th percentile of the 10,000 control resamplings. max represents the maximum number of phecodes found in the 10,000 control resamplings.
Table 4.
Non-Significant Phecodes of Interest (developmental stuttering cases, n = 572; matched controls, n = 2765).
| Non-Significant Phecodes of Interest | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| Phecode | Description | Count | p.value | p01 | p05 | p10 | p50 | p90 | p95 | p99 | max |
| 300 | Anxiety disorders | 34 | 0.0100 | 16 | 20 | 21 | 26 | 32 | 33 | 34 | 36 |
| 476 | Allergic rhinitis | 77 | 0.0100 | 47 | 54 | 56 | 64 | 71 | 73 | 75 | 78 |
| 512 | Other symptoms of respiratory system | 163 | 0.0100 | 120 | 128 | 132 | 140 | 151 | 154 | 157 | 165 |
| 558 | Noninfectious gastroenteritis | 47 | 0.0100 | 23 | 28 | 30 | 35 | 41 | 43 | 47 | 49 |
| 264.3 | Delayed milestones | 33 | 0.0200 | 14 | 17 | 18 | 24 | 29 | 30 | 35 | 37 |
| 479 | Other upper respiratory disease | 56 | 0.0200 | 33 | 36 | 38 | 47 | 53 | 54 | 57 | 57 |
| 481 | Influenza | 28 | 0.0200 | 12 | 13 | 15 | 21 | 24 | 26 | 30 | 30 |
| 8 | Intestinal infection | 38 | 0.0400 | 15 | 20 | 24 | 30 | 36 | 38 | 40 | 42 |
| 313.3 | Autism | 23 | 0.0400 | 9 | 11 | 12 | 18 | 22 | 23 | 25 | 26 |
| 371 | Inflammation of the eye | 22 | 0.0400 | 9 | 11 | 13 | 17 | 21 | 22 | 25 | 26 |
| 495.2 | Asthma with exacerbation | 44 | 0.0400 | 24 | 25 | 26 | 32 | 39 | 41 | 45 | 47 |
| 915 | Superficial injury without mention of infection | 27 | 0.0400 | 13 | 14 | 16 | 21 | 27 | 27 | 29 | 31 |
| 474 | Acute and chronic tonsillitis | 55 | 0.0500 | 33 | 37 | 39 | 46 | 53 | 55 | 61 | 64 |
| 474.2 | Chronic tonsillitis and adenoiditis | 52 | 0.0500 | 32 | 34 | 36 | 43 | 49 | 52 | 58 | 61 |
| 465.2 | Acute pharyngitis | 75 | 0.0600 | 47 | 53 | 55 | 63 | 73 | 76 | 82 | 82 |
| 495 | Asthma | 74 | 0.0600 | 48 | 54 | 56 | 62 | 70 | 75 | 78 | 81 |
| 772.3 | Muscle weakness | 21 | 0.0600 | 8 | 11 | 11 | 16 | 21 | 22 | 26 | 27 |
| 512.1 | Wheezing | 50 | 0.0700 | 32 | 34 | 36 | 41 | 50 | 51 | 55 | 55 |
| 658 | Maternal complication of pregnancy affecting fetus or newborn | 27 | 0.0700 | 15 | 16 | 17 | 21 | 27 | 29 | 32 | 34 |
| 381.2 | Eustachian tube disorders | 39 | 0.0800 | 20 | 24 | 26 | 31 | 39 | 40 | 43 | 45 |
| 483 | Acute bronchitis and bronchiolitis | 59 | 0.0800 | 35 | 39 | 43 | 50 | 59 | 61 | 62 | 63 |
| 803.2 | Fracture of radius and ulna | 27 | 0.0900 | 13 | 15 | 17 | 22 | 27 | 29 | 31 | 31 |
| 327.3 | Sleep apnea | 38 | 0.1000 | 17 | 25 | 26 | 32 | 38 | 39 | 42 | 47 |
| 327.32 | Obstructive sleep apnea | 28 | 0.1000 | 11 | 17 | 18 | 23 | 28 | 30 | 33 | 35 |
| 313.1 | Attention deficit hyperactivity disorder | 49 | 0.1200 | 30 | 36 | 38 | 43 | 50 | 51 | 54 | 55 |
Note. p values < .0001 indicated as 0.000 in the table. p01, p05, p10, p50, p90, p95, and p99 represent the number of phecodes found in the 1st, 5th, 10th, 50th, 90th, 95th, and 99th percentile of the 10,000 control resamplings. max represents the maximum number of phecodes found in the 10,000 control resamplings.
For the sensitivity analysis, we performed an additional phecode enrichment analysis which included only records containing Stuttering Severity Instrument (SSI) scores higher than “sub-clinical” with accompanying assessment by a speech-language pathologist (n = 243 cases, 1,173 controls). Of the 38 phecodes identified in the phecode enrichment analysis, 27 phecodes still exhibited enrichment (p < 0.05) in the sensitivity analysis. Two phenotypes that were no longer enriched, “Lack of coordination” (Phecode 350.3) and “Conductive hearing loss” (Phecode 389.2), had parent, or overarching, phecodes (“Abnormal movement” (Phecode 350) and “Hearing loss” (Phecode 389), respectively) which remained enriched in the sensitivity analysis. The other nine phecodes included: Candidiasis (Phecode 112), Lack of normal physiological development (Phecode 264), Lack of normal physiological development; unspecified (Phecode 264.9), Epilepsy; recurrent seizures; convulsions (Phecode 345), Convulsions (Phecode 345), Otitis media (Phecode 381), Otitis media and eustachian tube disorders (Phecode 381.1), Suppurative and unspecified otitis media (Phecode 381.11), and Otalgia (Phecode 382).
3.7. Phenome risk classifier (PheRC) using machine learning
From the 1,143 confirmed developmental stuttering cases, 571 randomly selected cases, with up to five matched controls (n = 2754), were used to develop the gini-index decision tree classifier. Of this set of 571 stuttering cases, 430 cases and their matched controls (n = 2070) were used to build the model, and 141 cases and their matched controls (n = 684) were used to test the model. Ultimately, of the 116 patients classified by the PheRC as high-likelihood cases, 97 were manually-reviewed positives from the case set and 19 were determined false positives from the control set, equating to a positive predictive value of >83 % (Fig. 1, Step 6b). It’s important to note that the positive predictive value rate is an estimate; because we cannot be certain of the true absence of stuttering in the control group, we cannot calculate the exact sensitivity, specificity, or negative predictive value for this model. However, the high estimated positive predictive value makes it useful for case acquisition: that is, patients identified as PheRC-predicted stuttering cases are very likely to be correctly categorized. For the confusion matrix for developmental stuttering classification, see Table 5.
Table 5.
Confusion Matrix for Developmental Stuttering Classification.
| Manually Reviewed Developmental Stuttering Cases | Matched Controls | |
|---|---|---|
| Patients classified as high likelihood stuttering cases | 97 | 19 |
| Patients classified as low likelihood stuttering cases | 44 | 665 |
Note. We cannot be certain of the true absence of stuttering in the control group; it is assumed that the prevalence of stuttering in the control group is approximately the population prevalence.
4. Discussion
The present study developed a multi-step process for identifying developmental stuttering cases within an EHR-based database. The subsequent phecode enrichment analysis of the VUMC EHR revealed phecodes enriched in developmental stuttering records, representing a variety of potential comorbid conditions. Using these phecode enrichments, a phenome risk classifier (PheRC) was developed to increase the number of likely developmental stuttering cases identified within EHRs using a prediction model to increase the number of cases and enable further genetic study using the EHR.
4.1. Identifying developmental stuttering in the synthetic derivative of the VUMC EHR
Because communication disorders such as stuttering are often ancillary to the purpose of a doctor visit at major medical centers, they are not well captured through billing codes. Thus, our keyword search and subsequent filtering steps using text-mining and manual review were needed for accurate phenotyping of cases. The first step to identifying conditions associated with developmental stuttering within the Vanderbilt University Medical Center (VUMC) EHR involved “defining” the developmental stuttering phenotype status by systematically labeling individuals with explicit indicators of disfluency as developmental stuttering cases and those without as population controls. Due to the dearth of developmental stuttering notation within the EHR (Pruett et al., 2018), this process involved (a) a keyword search of clinical notes followed by, (b) an initial manual review to identify cases to create (c) a text-mining algorithm to highlight high-likelihood cases (and further reduce the need for extensive manual review) and (d) a final manual review of all high-likelihood stuttering cases. This multi-step approach was used to broadly search for developmental stuttering cases while limiting manual review in an otherwise prohibitively large clinical note set. Through this approach, an initial database of approximately 2.8 million patients was narrowed to 14,080 patients with exploratory keyword hits, then reduced to 1,567 high-likelihood cases as determined by text-mining, and, finally, after manual review, 1,143 developmental stuttering cases were identified. While there is high confidence in manually reviewed cases, we note that descriptions of speech and language may not be considered relevant to record in EHRs, and/or communication disorders may simply go unnoticed by medical professionals during an encounter, so there may be “hidden” developmental stuttering cases in our control population, slightly impacting power.
Despite these challenges, there was sufficient notation to positively identify 1,143 cases of stuttering by expert manual review. As we’ve shown, it is not the case that information on stuttering is absent or irrelevant in these large databases, but that identifying cases requires a careful review process. The approach developed here could be applied in other EHR databases with access to medical notes and may be adapted to identify other communication disorders that also fall outside the scope of typical medical encounters. The importance of comprehensively documenting conditions, even if they’re not related to the purpose of the visit, extends beyond stuttering. As Valderas et al. (2009) states, doing so would “enhance both the precision and generalizability of [comorbidity] findings, leading to improved understanding of the causes of co-occurring diseases and their consequences for health service providers and planners.”
4.2. Comorbidity analysis via phecode enrichment
Following identification and systematic review, confirmed developmental stuttering cases were compared to matched population controls in a phecode enrichment analysis to reveal conditions comorbid with stuttering within this sample. On balance, the methods employed in this study recapitulated some previously suggested stuttering comorbidities identified via other methods and produced intriguing possibilities for future comorbidity study.
Importantly, interpreting developmental stuttering comorbidities from the phecode enrichments requires understanding how the ICD-9 codes (from which the phecodes are derived) are used clinically. While some phenotypes that co-occur with developmental stuttering in the EHR may share biological underpinnings, others may be enriched due to the manner in which information is coded and utilized in the medical setting, such as an inappropriately broad billing code. The methods utilized within the EHR are distinct from previous stuttering comorbidity studies, which rely on clinician recall or are hypothesis driven. For example, our analysis of enriched phecodes examines the entire record of the subject and was not restricted to conditions observed concurrently with a mention of stuttering in medical notes. That being said, our EHR-based approach may be differentially powered to detect some enrichments, such as those that, like stuttering, are under-documented in medical records. Therefore, the absence of a significant positive finding should not be taken as a negative finding but rather as a null finding. Additionally, the demographic characteristics of a study sample from a hospital-based population may enable the identification of phenotypic associations that would not be detectable in a study of a well population.
The two most highly enriched phecodes, Developmental Delays and Disorders (Phecode 315) and Speech and Language Disorder (Phecode 315.2), encompass developmental stuttering, the phenotype of interest, and articulation disorders (Arndt & Healey, 2001; Blood et al., 2003). These enrichments provide evidence that developmental stuttering cases identified within the VUMC EHR contained expected speech conditions.
Pervasive Developmental Disorders (Phecode 313) and Tics and Stuttering (Phecode 313.2) were the next most enriched phecodes. These phecodes include the ICD-9 code for Adult Onset Fluency Disorder (307.0). This billing code was designated for acquired stuttering (i.e., neurogenic or psychogenic stuttering); however, according to experienced speech-language pathologists, the code was also commonly used for adult patients with developmental stuttering persisting into adulthood. Interestingly, while Pervasive Developmental Disorders was enriched, the autism-specific phecode Autism (313.3) was not significantly enriched (Table 4). There is increasing clinical interest in the interplay between autism and stuttering, especially autism-specific speech disfluencies (Sisskin & Wasilus, 2014; Sisskin, 2006). Further study will be necessary to illuminate possible links between autism and developmental stuttering.
The enrichment of Hearing Loss (Phecode 389) and Conductive Hearing Loss (Phecode 389.2) was particularly intriguing considering the history of stuttering, audition, and hearing loss. For decades, the prevailing consensus was that stuttering prevalence was lower among children with hearing loss, suggesting hearing loss may be a protective factor against stuttering (Backus, 1938; Harms & Malone, 1939; Montgomery & Fitch, 1988). However, more recent studies have challenged that assumption, finding preschool-aged children with mild to severe hearing loss have an increased stuttering prevalence (Arenas et al., 2017). In addition to hearing loss, studies of delayed auditory feedback, frequency shifted feedback, choral speech, and noise masking have shown that each of these aural phenomena can temporarily increase fluency in some people who stutter (Bloodstein & Bernstein Ratner, 2008). While the underlying mechanisms leading to increased fluency are not fully understood, some have hypothesized that people who stutter have disrupted sensory feedback during speech production, and that decreased hearing may counteract this disrupted auditory feedback (Hutchinson & Ringel, 1975; Tourville, Reilly, & Guenther, 2008; van Lieshout, Peters, Starkweather, & Hulstijn, 1993). The varying directionality of findings related to hearing loss combined with known aural phenomena that increase fluency suggests the interaction of stuttering and hearing loss warrants further investigation.
While not commonly studied in combination with stuttering, emerging evidence suggests possible sleep differences in people who stutter. For example, a study examining structural changes in the brain resulting from obstructive sleep apnea found that, compared to the control group, the experimental group had significantly more individuals who stutter (Macey et al., 2002). Additionally, a recent study found a difference in the likelihood of sleep problems and daily consequences of sleep deprivation among children who stutter (Briley, 2019). Yet another study found that children who stutter, as a group, appear to exhibit more irregular biological patterns of sleep, hunger, and elimination patterns (Anderson, Pellowski, Conture, & Kelly, 2003). One hypothesis posits that sleep deprivation in childhood may interfere with memory consolidation required for early language and speech-motor mastery (Strom & Silverberg, 2016b).
Although the phecode for Asthma (495) fell short of our threshold for significance (Table 4), collectively, the significantly enriched phecodes for Acute Upper Respiratory Infections (Phecode 465), Allergic Reaction to Food (Phecode 930), Rash (Phecode 687.1), Atopic/Contact Dermatitis (Phecode 939), and Cough (Phecode 512.8) comprise elements of the asthma-allergy-eczema atopic triad. These conditions are called a triad because they frequently occur together: more than 60 % of children with eczema also have asthma and/or allergies to environmental aeroallergens or certain foods (Kapoor et al., 2008). Retrospective analyses of the National Survey of Children’s Health and the National Health Interview Survey conducted in 2016 found that a history of asthma, hay fever, food allergy, and eczema were associated with increased risk of speech disorder, and, anecdotally, some speech-language pathologists report asthma and allergies are more frequently observed among children who stutter on their caseload (Strom & Silverberg, 2016b). While the pathophysiology connecting developmental stuttering and atopy is unclear, one explanation points, again, to the effects of chronic disease and resultant chronic sleep deprivation during childhood (Strom & Silverberg, 2016a). This explanation is especially compelling since sleep disorders were also a significant comorbidity in this study.
Other significant enrichments were previously unobserved in the literature. Other Tests (Phecode 1010), an extensive category that includes billing and procedural codes for a variety of diagnostic tests encompassing viral and bacterial infection, vaccine prophylaxis, and allergy testing, was significantly enriched. Due to the breadth of codes included in this category, it is difficult to pinpoint any single condition or infectious agent associated with stuttering. However, broadly speaking, an increase in diagnostic testing suggests developmental stuttering is associated with a greater overall burden of childhood medical conditions. This association is further supported by significant phecodes for conditions including Fever of Unknown Origin (Phecode 783), Viral Infection (Phecode 79), Chronic Pharyngitis and Nasopharyngitis (Phecode 472), Otitis Media, Dermatophytosis (Phecode 110), Candidiasis (Phecode 112), Infection of the Eye (Phecode 369), Otalgia (Phecode 382), and Open Wounds of the Head, Neck, and Trunk (Phecode 870).
Additionally, phecodes associated with a range of neurological conditions affecting speech, language, and gross motor movements were enriched in the developmental stuttering group, including Neurological Deficits (Phecode 292), Aphasia/Speech Disturbance (Phecode 292.1), Abnormal Movement (Phecode 350), Abnormality of Gait (Phecode 350.2), Lack of Coordination (Phecode 350.3), and Epilepsy and Convulsions (Phecode 345). Notably, Neurological Deficits and Aphasia/Speech Disturbance include ICD-9 codes for Aphasia (784.3) and Dysarthria (784.52), conditions that can present with speech disruptions similar to developmental stuttering. While clinicians may use related terminology to describe these conditions, our manual review process required documentation of developmental stuttering and excluded cases of acquired stuttering, so overlapping terminology alone would not account for the enrichment. Additionally, while the codes for Aphasia/Speech Disturbance and Abnormality of Gait are seen more frequently in the geriatric population and Epilepsy is seen more frequently in pediatrics, our age- and length-of-record matching procedure ensured that these codes are indeed enriched in comparison to the non-stuttering control sample and not as a result of age bias. Overall, these enriched neurological conditions pose the question of whether underlying vulnerabilities in the speech-motor and language systems, and the motor system at large, may be associated with developmental stuttering.
Furthermore, at first glance, the enriched codes for Overweight, Obesity, and Hyperalimentation (Phecode 278), Symptoms Concerning Nutrition, Metabolism, and Development (Phecode 1002), and Lack of Normal Physiological Development (Phecode 264) appear contradictory in that phecodes for both excessive weight gain and weight loss are present. One interpretation is that atypical weight regulation may be associated with stuttering. Alternatively, weight loss and weight gain may represent two distinct comorbidities. For example, Symptoms Concerning Nutrition, Metabolism, and Development and Lack of Normal Physiological Development include ICD-9 codes for Feeding Difficulties and Mismanagement (783.3) and Lack of Normal Physiological Development (783.40), conditions related to failure to thrive. While failure to thrive has many causes, children in families with inadequate support or resources are especially susceptible, and nutritional deficits during key developmental stages may have far-reaching impacts on physical, intellectual, and social growth, including speech and language development. In contrast, a cogent explanation regarding the association with weight gain is less clear and may require additional investigation of mediator conditions.
Importantly, the purpose of this phenome-wide, hypothesis-free testing is to be hypothesis generating; we cannot make assumptions about causality from our data. In this analysis we detect traits associated with stuttering, but these associations could be observed for a number of reasons: 1) associated traits may exert an effect on stuttering, increasing risk of stuttering either directly or through a mediator, 2) associated traits may similarly be affected by stuttering, 3) associations may be spurious 4) associations may be due to a common cause, 5) associations may be synthetic (that is, due to correlation with a confounder that is associated with stuttering). Despite multiple possible explanations for association, this study, and others like it, are important first steps towards generating new hypotheses about how complex traits manifest clinically and lay the groundwork for future studies designed to validate novel associations and determine causality.
4.2.1. Clinical features associated with developmental stuttering not identified via comorbidity analysis
Numerous studies have examined differences in language development, temperament, and emotion between children who stutter and children who do not stutter. (e.g., Ambrose et al., 2015; Arenas et al., 2017; Eggers, Luc, & Van den Bergh, 2013; Jones, Choi, Conture, & Walden, 2014; Kefalianos et al., 2017; Singer, Walden, & Jones, 2019; Yairi & Ambrose, 2005). Despite a body of evidence showing differences within those domains, the current study did not find significantly enriched phecodes specifically related to developmental language disorders, temperament, or emotion. One possible explanation is that while these domains may, on average, differ between children who stutter and children who do not, the differences fall short of clinical diagnosis, or, like stuttering, are underreported in the EHR. Consequently, fully examining these domains within EHRs will require a different approach than the one employed in this study.
Additionally, our study used Bonferroni correction, a conservative multiple comparisons correction method, a critical step considering the number of non-independent comparisons made here. Based on our sample and methods, it is possible that our results contain false negatives. For example, anxiety (p = .01) and ADHD (p = .12), two conditions with previous evidence as stuttering comorbidities (e.g., Iverach et al., 2016; Donaher & Richels, 2012), did not reach significance after correction yet the phecodes were enriched at the 99th and 90th percentiles of that seen in the control sample, respectively, relative to the control resampling distributions (see Table 4). Again, due to potential differences in our EHR-based sample, the absence of a positive finding in this case is not a negative finding, but rather a null finding. Recognizing this, we included a list of non-significant findings of interest that, using less conservative multiple comparison correction methods, different sampling methods, or different diagnostic criteria, may be significant. Consequently, these findings warrant future consideration.
4.2.2. Sensitivity analysis
The purpose of this sensitivity analysis was to test if the phecode enrichments were still observed in the subset of cases that received clinical speech-language pathology assessments and are therefore more similar to clinical research cohorts. We performed the phecode enrichment analysis on a restricted stuttering cohort, including only subjects with Stuttering Severity Instrument (SSI) results that were above “sub-clinical” with documentation of a positive diagnosis from a speech-language pathologist. This case set included 243 stuttering cases (roughly 42.5 % of the larger case set used in our primary analyses) and SSI results ranged from “mild” to “very severe”. Of the 38 phecodes identified in the primary phecode enrichment analysis, 27 phecodes still exhibited enrichment (p < .05) in the sensitivity analysis. Additionally, two phenotypes that were no longer enriched likely due to a reduction in power, “Lack of coordination” (Phecode 350.3) and “Conductive hearing loss” (Phecode 389.2), had parent, or overarching, phecodes (“Abnormal movement” (Phecode 350) and “Hearing loss” (Phecode 389), respectively) which remained enriched in the sensitivity analysis. It’s difficult to assess whether the remaining nine phecodes did not retain significance due to increased specificity of the developmental stuttering cohort used for the sensitivity analysis, or due to the decrease in statistical power that comes with removing ~57.5 % of our case set. A PheRC built using the 27 phecodes that exhibited enrichment in our sensitivity analysis had a positive predictive value of 81.2 %, compared to 83.3 % in our primary analysis. The higher positive predictive value and largely similar enrichments support the relevance of our case definition strategy for identifying associated clinical measures and developing the phenome risk classifier.
4.3. Creation of the phenome risk classifier (PheRC)
In addition to highlighting phecode enrichments relative to a control sample, phecodes were used as variables to create a Phenome Risk Classifier (PheRC) prediction model to identify high-likelihood stuttering cases without the use of keywords and text-mining. The model identified 97 stuttering cases from the case set and 19 false positives from the control set for a positive predictive value of at least 83 % (Fig. 1, Step 6b). These results demonstrate that the PheRC successfully classified these cases of developmental stuttering independent of keywords, text-mining, and manual review. This is notable for future projects as it 1) greatly increases the number of cases phenotypically similar to stuttering available for genetic analysis within the Vanderbilt EHR and 2) provides an approach that may be adapted for use in other EHRs with limited free-text search abilities. For example, several large, EHR-linked biobanks provide limited access to individual medical notes. Using a PheRC-based approach, the lack of direct access to medical notes would not be a barrier. Even in instances where databases do allow free-text search of medical notes, the manual review process can be prohibitively time-intensive, and a PheRC-based approach could greatly assist in the identification of cases.
5. Future direction: application of phenome risk classifier (PheRC)
As mentioned, the PheRC was developed to increase the number of cases phenotypically similar to developmental stuttering identified within the EHR for further study. When this tool is applied to an EHR-linked biobank, such as BioVU at VUMC, the PheRC enables a well-powered genome-wide association study (GWAS), an approach that will help to elucidate the biological mechanism of developmental stuttering, a disorder with unknown etiology. Additionally, an approach utilizing a PheRC, rather than one using text-based search, allows for the identification of cases in both publicly available and restricted-access genetic databases, such as UK Biobank or the eMERGE network, where text search is not permitted. These databases will be essential to validate any preliminary GWAS findings from VUMC’s BioVU and will be especially important considering the advantages of multi-site EHRs (Baxter et al., 2021).
6. EHR limitations for developmental stuttering
This study has revealed previously unknown barriers to utilizing EHRs as a data source for developmental stuttering, and likely, communication disorders at large. Some of these barriers, such as keywords used to describe non-stuttering conditions, can be overcome through thoughtful research design; others, such as total lack of notation of stuttering within the EHR, are more systemic in nature. Fully overcoming these obstacles for future research will require coordination from the medical community and EHR developers to increase categorical and free-text descriptions of speech-language disorders. Refining EHRs is a continual process, and, hopefully, future iterations will move toward enhancing their research potential for communication disorders such as stuttering. Again, this highlights the importance of interdisciplinary collaboration and correspondence between academics, speech-language pathology clinicians, and the medical community.
7. Conclusion
Overall, our VUMC EHR review identified cases of developmental stuttering and revealed diverse phecode enrichments (i.e., comorbidities), some of which may suggest potential novel comorbidities. Our study also demonstrated the feasibility of a data-driven approach using a large, pre-existing dataset. Importantly, this approach depended on both clinical stuttering expertise combined with phenotyping via text-mining and machine learning.
Following the lead of other biomedical science fields, the next decade could transform how we look for patterns in data in the communication sciences. Rather than hand-selecting a set of variables, we can model using a patient’s entire chart; this study marks the first exploration of this process for stuttering. Better characterization of the enriched phecodes also has the potential to enhance patient care. Ultimately, understanding conditions associated with stuttering may help us further understand the etiology and development of stuttering through shared pathophysiology, as well as provide data needed for future characterizations such as estimating the heritability and genetic and epigenetic architecture that contribute to risk, hopefully yielding advances in our knowledge of this complex disorder.
Acknowledgements
This work was supported by National Institutes of Health (NIH) grants from the National Institute on Deafness and Other Communication Disorders (NIDCD) to Vanderbilt University Medical Center and Wayne State University (1R03DC015329-01 and R01DC017175) and to Vanderbilt University Medical Center (5R21DC016723-03). The dataset(s) used for the analyses described were obtained from Vanderbilt University Medical Center’s BioVU and Synthetic Derivative, which is supported by numerous sources: institutional funding, private agencies, and federal grants. These include the NIH funded Shared Instrumentation Grant S10RR025141; and CTSA grants UL1TR002243, UL1TR000445, and UL1RR024975. The content is solely the responsibility of the authors and does not necessarily represent the official views of the NIH.
Biography
Dillon G. Pruett is a doctorate student in the Department of Hearing and Speech Sciences at Vanderbilt University. Pruett’s primary interest relates to developmental stuttering, with a focus on utilizing electronic medical records to develop novel research approaches.
Douglas Shaw is a PhD student at Vanderbilt University working in human genetics, leveraging Vanderbilt’s electronic health system and genetic database to study population genetics. Doug applies machine-learning techniques to characterize underreported diseases such as developmental stuttering in these databases and uses these techniques to study genetic risk factors associated with developmental stuttering to better understand its underlying genetic etiology.
Hung-Hsin Chen is a student in Epidemiology PhD program at Vanderbilt University. Chen’s research interest focuses on genetic epidemiology and human population genetics, especially on methods development for cardiovascular diseases, related metabolic traits, and speech phenotypes.
Lauren E. Petty is a PhD student studying genetic epidemiology at Vanderbilt University. She is primarily interested in improving methods to identify relatedness using genetic data and leveraging distant relatedness to discover genetic factors underlying a wide spectrum of human diseases.
Hannah G. Polikowsky is a doctorate student at Vanderbilt University in human genetics. Her research focuses on population and familial based analyses of speech and language traits. Current research approaches include utilizing common variant GWAS and linkage analysis of family pedigrees to investigate the genetic architecture of developmental stuttering.
Shelly Jo Kraft’s current research focuses on the biological and behavioral genetics of stuttering, autism, SLI, SSD, and hearing loss. Other research interests include neuro-anatomical and functional features of people who stutter, auditory feedback mechanisms of speech control, autism treatment strategies, new genetic analysis techniques for modeling epigenetic complexity and exploring the relationship between cognition, temperament, and stuttering severity.
Robin M. Jones is an assistant professor in the Department of Hearing and Speech Sciences in the Vanderbilt University School of Medicine. Jones’s primary research interest relates to childhood stuttering, with a focus on emotional, cognitive, and linguistic contributions to stuttering as well as empirical assessment and treatment of stuttering.
Jennifer Below is interested in developing computational methodologies to further our understanding of the genetic basis of human disease. Specifically, development of novel strategies for identifying and confirming genetic risk factors to complex traits including speech and language pathologies, diabetes, obesity, Alzheimer’s disease, cardiovascular disease, and metabolic traits via ascertainment of dense genetic (specifically whole genome/exome sequenced and whole genome imputed datasets) and phenotypic data.
Footnotes
Declaration of Competing Interest
The authors report no declarations of interest.
References
- Ambrose NG, Cox NJ, & Yairi E (1997). The genetic basis of persistence and recovery in stuttering. Journal of Speech Language and Hearing Research, 40(3), 567–580. 10.1044/jslhr.4003.567. [DOI] [PubMed] [Google Scholar]
- Ambrose NG, Yairi E, Loucks TM, Seery CH, & Throneburg R (2015). Relation of motor, linguistic and temperament factors in epidemiologic subtypes of persistent and recovered stuttering: Initial findings. Journal of Fluency Disorders, 45, 12–26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anderson JD, Pellowski MW, Conture EG, & Kelly EM (2003). Temperamental characteristics of young children who stutter. Journal of Speech Language and Hearing Research, 46(5), 1221–1233. 10.1044/1092-4388(2003/095). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arenas RM, Walker EA, & Oleson JJ (2017). Developmental stuttering in children who are hard of hearing. Language, Speech, and Hearing Services in Schools, 48 (4), 234–248. 10.1044/2017_LSHSS-17-0028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arndt J, & Healey EC (2001). Concomitant disorders in school-age children who stutter. Language, Speech, and Hearing Services in Schools, 32(2), 68–78. 10.1044/0161-1461(2001/006). [DOI] [PubMed] [Google Scholar]
- Backus O (1938). Incidence of stuttering among the deaf. The Annals of Otology, Rhinology, and Laryngology, 47(3), 632–635. 10.1177/000348943804700304. [DOI] [Google Scholar]
- Bastarache L, Hughey JJ, Hebbring S, Marlo J, Zhao W, Ho WT, … Denny JC (2018). Phenotype risk scores identify patients with unrecognized Mendelian disease patterns. Science, 359(6381), 1233–1239. 10.1126/science.aal4043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baxter SL, Saseendrakumar BR, Paul P, Kim J, Bonomi L, Kuo T-T, … Ohno-Machado L (2021). Predictive analytics for glaucoma using data from the all of us research program. American Journal of Ophthalmology, 0(0). 10.1016/j.ajo.2021.01.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blood GW, Ridenour VJ, Qualls CD, & Hammer CS (2003). Co-occurring disorders in children who stutter. Journal of Communication Disorders, 36(6), 427–448. 10.1016/S0021-9924(03)00023-6. [DOI] [PubMed] [Google Scholar]
- Bloodstein O, & Bernstein Ratner N (2008). A handbook of stuttering. Thomson Delmar Learning. [Google Scholar]
- Briley P (2019). Sleep issues in children who stutter. November Poster presented at the American speech-language-hearing association conference. [Google Scholar]
- Briley P, & Ellis C (2018). The coexistence of disabling conditions in children who stutter: Evidence from the national health interview survey. Journal of Speech Language and Hearing Research, 61(12), 2895–2905. 10.1044/2018_JSLHR-S-17-0378. [DOI] [PubMed] [Google Scholar]
- Davies G, Marioni RE, Liewald DC, Hill WD, Hagenaars SP, Harris SE, … Deary IJ (2016). Genome-wide association study of cognitive functions and educational attainment in UK Biobank (N =112 151). Molecular Psychiatry, 21(6), 758–767. 10.1038/mp.2016.45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Denny JC, Crawford DC, Ritchie MD, Bielinski SJ, Basford MA, Bradford Y, … Carrell D (2011). Variants near FOXE1 are associated with hypothyroidism and other thyroid conditions: Using electronic medical records for genome-and phenome-wide studies. American Journal of Human Genetics, 89 (4), 529–542. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Denny JC, Bastarache L, Ritchie MD, Carroll RJ, Zink R, Mosley JD, … Basford MA (2013). Systematic comparison of phenome-wide association study of electronic medical record data and genome-wide association study data. Nature Biotechnology, 31(12), 1102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Denny JC, Ritchie MD, Basford MA, Pulley JM, Bastarache L, Brown-Gentry K, … Crawford DC (2010). PheWAS: Demonstrating the feasibility of a phenome-wide scan to discover gene–disease associations. Bioinformatics, 26(9), 1205–1210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Donaher J, & Richels C (2012). Traits of attention deficit/hyperactivity disorder in school-age children who stutter. Journal of Fluency Disorders, 37(4), 242–252. 10.1016/j.jfludis.2012.08.002. [DOI] [PubMed] [Google Scholar]
- Eggers K, Luc F, & Van den Bergh BR (2013). Inhibitory control in childhood stuttering. Journal of Fluency Disorders, 38(1), 1–13. [DOI] [PubMed] [Google Scholar]
- Fagnani C, Fibiger S, Skytthe A, & Hjelmborg JV (2011). Heritability and environmental effects for self-reported periods with stuttering: a twin study from Denmark. Logopedics Phoniatrics Vocology, 36(3), 114–120. [DOI] [PubMed] [Google Scholar]
- Gottesman O, Kuivaniemi H, Tromp G, Faucett WA, Li R, Manolio TA, … Williams MS (2013). The electronic medical records and genomics (eMERGE) network: Past, present, and future. Genetics in Medicine, 15(10), 761–771. 10.1038/gim.2013.72. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harms MA, & Malone JY (1939). The relationship of hearing acuity to stammering. The Journal of Speech Disorders, 4(4), 363–370. 10.1044/jshd.0404.363. [DOI] [Google Scholar]
- Howard DM, Adams MJ, Shirali M, Clarke T-K, Marioni RE, Davies G, … McIntosh AM (2018). Genome-wide association study of depression phenotypes in UK Biobank identifies variants in excitatory synaptic pathways. Nature Communications, 9(1), 1470. 10.1038/s41467-018-03819-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hutchinson JM, & Ringel RL (1975). The effect of oral sensory deprivation on stuttering behavior. Journal of Communication Disorders, 8(3), 249–258. [DOI] [PubMed] [Google Scholar]
- Iverach L, Jones M, McLellan LF, Lyneham HJ, Menzies RG, Onslow M, & Rapee RM (2016). Prevalence of anxiety disorders among children who stutter. Journal of Fluency Disorders, 49, 13–28. 10.1016/j.jfludis.2016.07.002. [DOI] [PubMed] [Google Scholar]
- Jones R, Choi D, Conture E, & Walden T (2014). Temperament, emotion, and childhood stuttering. In Seminars in speech and language (Vol. 35, pp. 114–131). Thieme Medical Publishers. May, No. 02. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kapoor R, Menon C, Hoffstad O, Bilker W, Leclerc P, & Margolis DJ (2008). The prevalence of atopic triad in children with physician-confirmed atopic dermatitis. Journal of the American Academy of Dermatology, 58(1), 68–73. 10.1016/j.jaad.2007.06.041. [DOI] [PubMed] [Google Scholar]
- Keating D, Turrell G, & Ozanne A (2001). Childhood speech disorders: Reported prevalence, comorbidity and socioeconomic profile. Journal of Paediatrics and Child Health, 37(5), 431–436. [DOI] [PubMed] [Google Scholar]
- Kefalianos E, Onslow M, Packman A, Vogel A, Pezic A, Mensah F, … Reilly S (2017). The history of stuttering by 7 years of age: Follow-up of a prospective community cohort. Journal of Speech Language and Hearing Research, 60(10), 2828–2839. [DOI] [PubMed] [Google Scholar]
- Liao KP, Cai T, Savova GK, Murphy SN, Karlson EW, Ananthakrishnan AN, … Kohane I (2015). Development of phenotype algorithms using electronic medical records and incorporating natural language processing. The BMJ, 350. 10.1136/bmj.h1885. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Macey PM, Henderson LA, Macey KE, Alger JR, Frysinger RC, Woo MA, … Harper RM (2002). Brain morphology associated with obstructive sleep apnea. American Journal of Respiratory and Critical Care Medicine, 166(10), 1382–1387. 10.1164/rccm.200201-050OC. [DOI] [PubMed] [Google Scholar]
- Manning W, & Beck JG (2013). Personality dysfunction in adults who stutter: Another look. Journal of Fluency Disorders, 38(2), 184–192. 10.1016/j.jfludis.2013.02.001. [DOI] [PubMed] [Google Scholar]
- Montgomery BM, & Fitch JL (1988). The prevalence of stuttering in the hearing-impaired school age population. The Journal of Speech and Hearing Disorders, 53 (2), 131–135. [DOI] [PubMed] [Google Scholar]
- Namjou B, Marsolo K, Carroll R, Denny J, Ritchie MD, Lingren T, … Kohane I (2014). Phenome-wide association study (PheWAS) in EMR-linked pediatric cohorts. Frontiers in Genetics, 5, 401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Onitilo AA, Engel JM, Greenlee RT, & Mukesh BN (2009). Breast cancer subtypes based on ER/PR and Her2 expression: Comparison of clinicopathologic features and survival. Clinical Medicine & Research, 7(1–2), 4–13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ooki S (2005). Genetic and environmental influences on stuttering and tics in Japanese twin children. Twin Research and Human Genetics, 8(1), 69–75. [DOI] [PubMed] [Google Scholar]
- Pedregosa F, Varoquaux G, Gramfort A, Michel V, Thirion B, Grisel O, … Vanderplas J (2011). Scikit-learn: Machine learning in Python. Journal of Machine Learning Research, 12, 2825–2830. [Google Scholar]
- Pruett DG, Below JE, & Jones RM (2018). Defining developmental stuttering in electronic medical records: Preliminary results. November Poster session presented at the American speech-language-hearing association convention. [Google Scholar]
- Reilly S, Onslow M, Packman A, Cini E, Conway L, Ukoumunne OC, … Wake M (2013). Natural history of stuttering to 4 years of age: A prospective community-based study. Pediatrics, 132(3), 460–467. 10.1542/peds.2012-3067. [DOI] [PubMed] [Google Scholar]
- Reimand J, Isserlin R, Voisin V, Kucera M, Tannus-Lopes C, Rostamianfar A, … Bader GD (2019). Pathway enrichment analysis and visualization of omics data using g:Profiler, GSEA, Cytoscape and EnrichmentMap. Nature Protocols, 14(2), 482–517. 10.1038/s41596-018-0103-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scott KS (2015). Dysfluency in autism spectrum disorders. Procedia–Social and Behavioral Sciences, 193, 239–245. [Google Scholar]
- Singer CM, Walden TA, & Jones RM (2019). Differences in the relation between temperament and vocabulary based on children’s stuttering trajectories. Journal of Communication Disorders, 78, 57–68. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sisskin V (2006). Speech disfluency in asperger’s syndrome: Two cases of interest. Perspectives on Fluency and Fluency Disorders, 16(2), 12–14. 10.1044/ffd16.2.12. [DOI] [Google Scholar]
- Sisskin V, & Wasilus S (2014). Lost in the literature, but not the caseload: Working with atypical disfluency from theory to practice. Seminars in Speech and Language, 35(02), 144–152. 10.1055/s-0034-1371757. [DOI] [PubMed] [Google Scholar]
- Strom MA, & Silverberg JI (2016a). Asthma, hay fever, and food allergy are associated with caregiver-reported speech disorders in US children. Pediatric Allergy and Immunology, 27(6), 604–611. 10.1111/pai.12580. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Strom MA, & Silverberg JI (2016b). Eczema is associated with childhood speech disorder: A retrospective analysis from the National Survey of Children’s Health and the National Health Interview Survey. The Journal of Pediatrics, 168, 185–192. 10.1016/j.jpeds.2015.09.066.e4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sudlow C, Gallacher J, Allen N, Beral V, Burton P, Danesh J, … Collins R (2015). UK biobank: An open access resource for identifying the causes of a wide range of complex diseases of middle and old age. PLoS Medicine, 12(3), Article e1001779. 10.1371/journal.pmed.1001779. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Theys C, van Wieringen A, & De Nil LF (2008). A clinician survey of speech and non-speech characteristics of neurogenic stuttering. Journal of Fluency Disorders, 33(1), 1–23. 10.1016/j.jfludis.2007.09.001. [DOI] [PubMed] [Google Scholar]
- Tikkanen E, Gustafsson S, Amar D, Shcherbina A, Waggott D, Ashley EA, & Ingelsson E (2018). Biological insights into muscular strength: Genetic findings in the UK biobank. Scientific Reports, 8(1), 6451. 10.1038/s41598-018-24735-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tourville JA, Reilly KJ, & Guenther FH (2008). Neural mechanisms underlying auditory feedback control of speech. Neuroimage, 39(3), 1429–1443. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Valderas JM, Starfield B, Sibbald B, Salisbury C, & Roland M (2009). Defining comorbidity: Implications for understanding health and health services. Annals of Family Medicine, 7(4), 357–363. 10.1370/afm.983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Lieshout PH, Peters HF, Starkweather CW, & Hulstijn W (1993). Physiological differences between stutterers and nonstutterers in perceptually fluent speech: EMG amplitude and duration. Journal of Speech Language and Hearing Research, 36(1), 55–63. [DOI] [PubMed] [Google Scholar]
- Verma SS, Bailey JNC, Lucas A, Bradford Y, Linneman JG, Hauser MA, … Network, eMERGE, & Consortium, N. (2016). Epistatic gene-based interaction analyses for glaucoma in eMERGE and NEIGHBOR consortium. PLoS Genetics, 12(9), Article e1006186. 10.1371/journal.pgen.1006186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walters CE Jr., Nitin R, Margulis K, Boorom O, Gustavson DE, Bush CT, … Gordon RL (2020). Automated phenotyping tool for identifying developmental language disorder cases in health systems data (APT-DLD): A new research algorithm for deployment in large-scale electronic health record systems. Journal of Speech Language and Hearing Research, 1–17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Watkins RV, Ehud Y, & Grinager AN (1999). Early childhood stuttering III. Journal of Speech Language and Hearing Research, 42(5), 1125–1135. 10.1044/jslhr.4205.1125. [DOI] [PubMed] [Google Scholar]
- Wolk L, Conture EG, & Edwards ML (1990). Comorbidity of stuttering disordered phonology in young children. South African Journal of Communication Disorders, 37(1), 15–20. 10.4102/sajcd.v37i1.284. [DOI] [PubMed] [Google Scholar]
- Yairi E, & Ambrose NG (2005). Early childhood stuttering: For clinicians, by clinicians. 8700 Shoal Creek Blvd, Austin, TX 78757: ProEd, Inc. [Google Scholar]
- Yairi E, Ambrose N, & Cox N (1996a). Genetics of stuttering: A critical review. Journal of Speech Language and Hearing Research, 39. 10.1044/jshr.3904.771. [DOI] [PubMed] [Google Scholar]
- Yairi E, Ambrose N, & Cox N (1996b). Genetics of stuttering: A critical review. Journal of Speech Language and Hearing Research, 39(4), 771–784. 10.1044/jshr.3904.771. [DOI] [PubMed] [Google Scholar]
- Zengini E, Hatzikotoulas K, Tachmazidou I, Steinberg J, Hartwig FP, Southam L, … Zeggini E (2018). Genome-wide analyses using UK Biobank data provide insights into the genetic architecture of osteoarthritis. Nature Genetics, 50(4), 549–558. 10.1038/s41588-018-0079-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu Z, Lee PH, Chaffin MD, Chung W, Loh P-R, Lu Q, … Liang L (2018). A genome-wide cross-trait analysis from UK Biobank highlights the shared genetic architecture of asthma and allergic diseases. Nature Genetics, 50(6), 857–864. 10.1038/s41588-018-0121-0. [DOI] [PMC free article] [PubMed] [Google Scholar]