Abstract
RNA-binding proteins (RBPs) play essential roles in diverse cellular processes through post-transcriptional regulation of RNAs. The subcellular localization of RBPs is thus under tight control, the breakdown of which is associated with aberrant cytoplasmic accumulation of nuclear RBPs such as TDP-43 and FUS, well-known pathological markers for amyotrophic lateral sclerosis and frontotemporal dementia (ALS/FTD). Here, we report in Drosophila model for ALS/FTD that nuclear accumulation of a cytoplasmic RBP Staufen may be a new pathological feature. We found that in Drosophila C4da neurons expressing PR36, one of the arginine-rich dipeptide repeat proteins (DPRs), Staufen accumulated in the nucleus in Importin- and RNA-dependent manner. Notably, expressing Staufen with exogenous NLS—but not with mutated endogenous NLS—potentiated PR-induced dendritic defect, suggesting that nuclear-accumulated Staufen can enhance PR toxicity. PR36 expression increased Fibrillarin staining in the nucleolus, which was enhanced by heterozygous mutation of stau (stau+/−), a gene that codes Staufen. Furthermore, knockdown of fib, which codes Fibrillarin, exacerbated retinal degeneration mediated by PR toxicity, suggesting that increased amount of Fibrillarin by stau+/− is protective. stau+/− also reduced the amount of PR-induced nuclear-accumulated Staufen and mitigated retinal degeneration and rescued viability of flies expressing PR36. Taken together, our data show that nuclear accumulation of Staufen in neurons may be an important pathological feature contributing to the pathogenesis of ALS/FTD.
Introduction
Post-transcriptional regulation of RNAs by RNA-binding proteins (RBPs) is essential for expanding proteome diversity and targeting RNAs to specific places through their splicing (1) and transport (2,3), respectively. Upon arriving at a specific location, a particular mRNA isoform is translated by ribosomes and RBPs (3). Because these processes require RBPs to be in specific places at a particular moment, subcellular localization of RBPs needs to be tightly controlled. Thus, it is not surprising that a breakdown of this tight control of RBPs is associated with various diseases, particularly neurodegenerative diseases such as amyotrophic lateral sclerosis and frontotemporal dementia (ALS/FTD) (4).
Staufen is a double-stranded RBP (dsRBP) that plays a role in localization, translation, stability and decay of mRNA (5) as well as alternative splicing of pre-mRNA (6). More recently, its function has been extended to include aberrant mRNA processing as well as amplification of unfolded protein response and apoptosis upon induction of stress or expression of ALS-linked genes (7,8). However, our understanding on the involvement of Staufen in ALS/FTD pathogenesis remains incomplete.
ALS and FTD are two diseases on a single pathogenic spectrum with overlapping genetic etiologies (9). GGGGCC (G4C2) repeat expansion mutation in the intron of C9orf72 is the most common cause of both familial and sporadic cases of ALS and FTD (C9-ALS/FTD) (10,11). G4C2 repeat expansion produces both sense (G4C2) and antisense (G2C4) RNAs that can either form toxic nuclear foci (12,13) or undergo repeat-associated non-ATG translation (RANT) in the cytoplasm to generate five types of dipeptide repeat proteins (DPRs) (14,15): glycine–alanine (GA), glycine–proline (GP), glycine–arginine (GR), proline–arginine (PR) and proline–alanine (PA). Among them, arginine-rich DPRs have been shown to be most toxic in fly and mammalian models for ALS (16).
In recent years, nucleocytoplasmic transport defect, which includes decreases in protein import into and mRNA export from the nucleus, has been identified as one of the prominent pathogenic features of C9-ALS/FTD (17–20), leading to accumulation of proteins in the cytoplasm and mRNA in the nucleus, respectively. In addition, recent studies showed nuclear accumulation of double-stranded RNAs (dsRNAs) in tdp-1(TDP-43)-deleted C. elegans (21) and in PR-expressing mouse cortical neurons (22). Currently, potentially toxic consequences of decreased protein import are actively being explored, one of which is cytoplasmic mislocalization of TDP-43 (23), a pathological hallmark of ALS/FTD (24). However, potentially toxic consequences of RNA accumulation in the nucleus remain largely underexplored.
RBPs such as hnRNPA1 have been shown to egress out of the nucleus to the cytoplasm when there is a decreased level of nuclear RNA (25,26). These studies lead to a hypothesis that increased RNA levels in the nucleus might induce nuclear retention of certain RBPs, resulting in their subsequent gain of nuclear toxicity in ALS. Thus, to test our hypothesis, we used C9-ALS/FTD Drosophila model system to screen for and identify RBPs whose nuclear accumulation leads to neurotoxicity and to understand its underlying mechanisms.
Results
Arginine-rich DPR proteins increase nuclear accumulation of Staufen in neurons
Drosophila class IV dendritic arborization (C4da) neurons are larger than most neurons in the brain and are localized in the peripheral areas in which neuronal population is sparse. These characteristics render C4da neurons amenable for in vivo single-cell imaging, which is desirable when examining localization pattern of RBPs. Furthermore, these neurons have been used to study neurodegenerative diseases (27–32), including ALS (33,34). Thus, to identify RBPs that show increased nuclear localization in C9-ALS/FTD Drosophila model, we screened in Drosophila C4da neurons for RBPs whose accumulation in the nucleus was increased compared to the controls. First, to identify the RBPs whose localization is cytoplasmic, we prescreened 11 RBPs the transgenic lines for which were readily available and examined their localization in larval C4da neurons. Among them, we identified seven RBPs whose localization is mostly cytoplasmic. Next, for the actual screen of these seven RBPs, we used as C9-ALS/FTD Drosophila model the flies that expressed PR repeat proteins (V5-PR36) and not GR repeat proteins, because recent studies showed that between the two arginine-rich DPRs, PR is more closely associated with nucleocytoplasmic transport defects (18,35,36). In addition, by using V5-PR36, which is expressed from an alternative codon different from repeated G4C2 sequence, we were able to preclude detection of RBPs mislocalized by G4C2 RNA from the screen.
The genetic screen (Fig. 1A and Supplementary Material, Fig. S1) identified Staufen as the RBP whose puncta size and fluorescence intensity in the nucleus increased most significantly (**P = 0.0022). In control (CTRL) C4da neurons, GFP-tagged Staufen showed mostly cytoplasmic localization with small puncta in the nucleus, whereas in C4da neurons expressing V5-PR36, Staufen-GFP puncta were much more prominent in the nucleus (Fig. 1B). Unfortunately, the detected signal for endogenous Staufen using the anti-Staufen antibody in C4da neurons was too faint to apply any proper analyses (data not shown). However, a previous study showed that the overexpressed GFP-tagged Staufen in Drosophila cells behaved in a similar manner to the endogenous Staufen in its localization (37), providing a rationale for our using Staufen-GFP for further analyses.
Figure 1 .
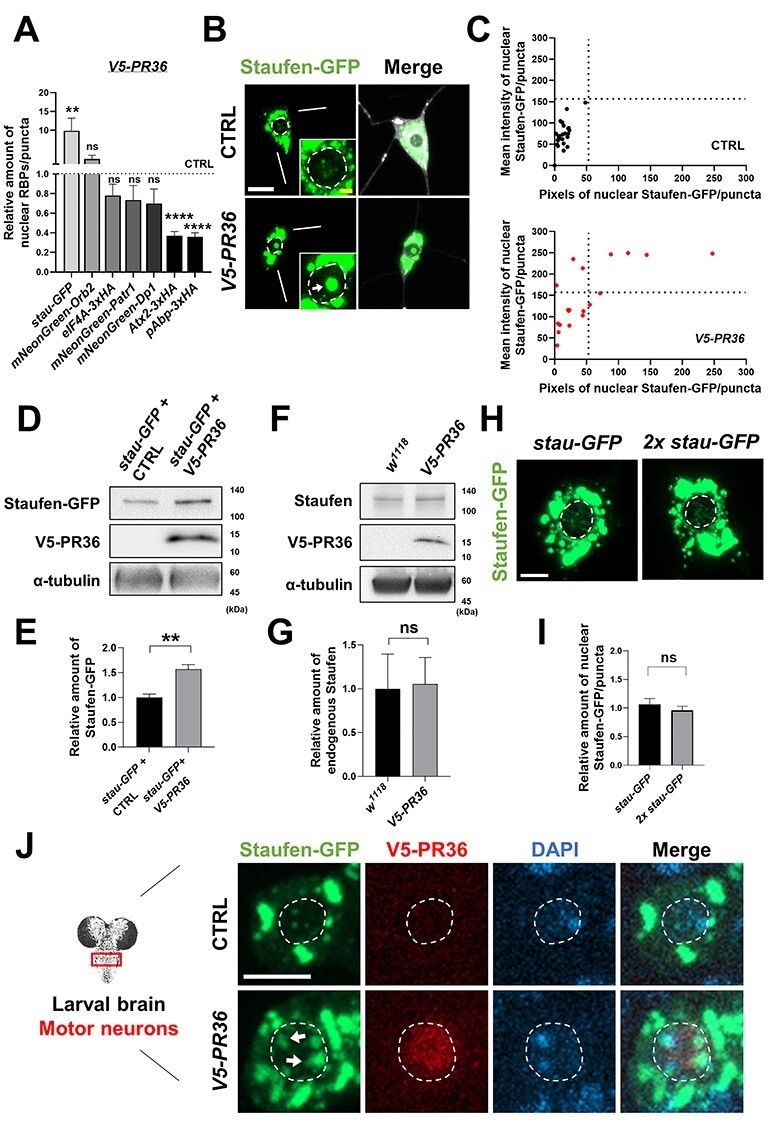
PR toxicity increases nuclear accumulation of Staufen in neurons. (A) Quantification of nuclear RBPs in C4da neurons expressing V5-PR36 compared to controls. The pixel number and the mean intensity of nuclear RBPs were multiplied to provide an estimate of the amount of nuclear RBPs. **P = 0.0026, ****P < 1.0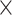 10−4 by two-tailed student’s t-test; error bars, ± SEM; n ≥ 13 RBP puncta; n = 3 larvae. (B) Subcellular localization of Staufen-GFP in larval C4da neurons of controls (CTRL) or expressing V5-PR36 (Genotypes. CTRL: UAS-stau-GFP/+; PPK1a-gal4 > UAS-mCD8RFP/+, V5-PR36: UAS-stau-GFP/+; PPK1a-gal4 > UAS-mCD8RFP/UAS-V5-PR36). Merge: merged with plasma membrane marker. Bottom right insets are magnified images of the nucleus. Dashed circular lines (white) outline the nuclei. Arrow (white) indicates increased nuclear Staufen-GFP puncta. Scale bars (yellow), 2 μm; (white), 10 μm. (C) Individual nuclear Staufen-GFP puncta in C4da neurons with denoted genotypes were counted and plotted. y-axis: the mean intensity of Staufen-GFP puncta in the nucleus; x-axis: the pixel number of Staufen-GFP puncta in the nucleus. n ≥ 19 Staufen-GFP puncta; n = 3 larvae. (D) Western blot showing the expression level of Staufen-GFP in fly heads (Genotypes. stau-GFP + CTRL: UAS-stau-GFP; elavGS-gal4 > UAS-pACU2-empty, stau-GFP + V5-PR36: UAS-stau-GFP; elavGS-gal4 > UAS-V5-PR36). α-tubulin was used as a loading control. The transgenic expression was induced by feeding 100 μM of RU486 for 10 days after eclosion at 27°C. (E) Quantification of the amount of Staufen-GFP normalized to the control. **P = 0.0024 by paired two-tailed student’s t-test; error bars, ± SEM; n = 3 with 2 technical repeats. (F) Western blot showing the expression level of endogenous Staufen in fly heads (Genotypes. w1118: +/+, V5-PR36: +; elavGS-gal4 > UAS-V5-PR36). α-tubulin was used as a loading control. The transgenic expression was induced by feeding 100 μM of RU486 for 10 days after eclosion at 27°C. (G) Quantification of the amount of Staufen-GFP normalized to the control. ns: not significant by paired two-tailed student’s t-test; error bars, ± SEM; n = 3 with 2 technical repeats. (H) Images of C4da neurons expressing either one or two copies of stau-GFP. Merge: merged with plasma membrane marker. Scale bar (white), 10 μm. (I) Quantification of the amount of nuclear Staufen-GFP normalized to the control. ns: not significant by two-tailed student’s t-test; error bars, ± SEM; n ≥ 58 Staufen-GFP puncta; n = 3 larvae. (J) IHC of dissected third instar larval motor neurons shows that V5-PR36 expression increased nuclear accumulation of Staufen-GFP (Genotypes. CTRL: UAS-stau-GFP/+; D42-Gal4 > UAS-mCD8RFP/+, V5-PR36: UAS-stau-GFP/+; D42-Gal4 > UAS-mCD8RFP/UAS-V5-PR36). Dashed circular lines (white) outline the nuclei. Arrows (white) indicate stronger nuclear Staufen-GFP signal compared to the control. Scale bar (white), 5 μm.
10−4 by two-tailed student’s t-test; error bars, ± SEM; n ≥ 13 RBP puncta; n = 3 larvae. (B) Subcellular localization of Staufen-GFP in larval C4da neurons of controls (CTRL) or expressing V5-PR36 (Genotypes. CTRL: UAS-stau-GFP/+; PPK1a-gal4 > UAS-mCD8RFP/+, V5-PR36: UAS-stau-GFP/+; PPK1a-gal4 > UAS-mCD8RFP/UAS-V5-PR36). Merge: merged with plasma membrane marker. Bottom right insets are magnified images of the nucleus. Dashed circular lines (white) outline the nuclei. Arrow (white) indicates increased nuclear Staufen-GFP puncta. Scale bars (yellow), 2 μm; (white), 10 μm. (C) Individual nuclear Staufen-GFP puncta in C4da neurons with denoted genotypes were counted and plotted. y-axis: the mean intensity of Staufen-GFP puncta in the nucleus; x-axis: the pixel number of Staufen-GFP puncta in the nucleus. n ≥ 19 Staufen-GFP puncta; n = 3 larvae. (D) Western blot showing the expression level of Staufen-GFP in fly heads (Genotypes. stau-GFP + CTRL: UAS-stau-GFP; elavGS-gal4 > UAS-pACU2-empty, stau-GFP + V5-PR36: UAS-stau-GFP; elavGS-gal4 > UAS-V5-PR36). α-tubulin was used as a loading control. The transgenic expression was induced by feeding 100 μM of RU486 for 10 days after eclosion at 27°C. (E) Quantification of the amount of Staufen-GFP normalized to the control. **P = 0.0024 by paired two-tailed student’s t-test; error bars, ± SEM; n = 3 with 2 technical repeats. (F) Western blot showing the expression level of endogenous Staufen in fly heads (Genotypes. w1118: +/+, V5-PR36: +; elavGS-gal4 > UAS-V5-PR36). α-tubulin was used as a loading control. The transgenic expression was induced by feeding 100 μM of RU486 for 10 days after eclosion at 27°C. (G) Quantification of the amount of Staufen-GFP normalized to the control. ns: not significant by paired two-tailed student’s t-test; error bars, ± SEM; n = 3 with 2 technical repeats. (H) Images of C4da neurons expressing either one or two copies of stau-GFP. Merge: merged with plasma membrane marker. Scale bar (white), 10 μm. (I) Quantification of the amount of nuclear Staufen-GFP normalized to the control. ns: not significant by two-tailed student’s t-test; error bars, ± SEM; n ≥ 58 Staufen-GFP puncta; n = 3 larvae. (J) IHC of dissected third instar larval motor neurons shows that V5-PR36 expression increased nuclear accumulation of Staufen-GFP (Genotypes. CTRL: UAS-stau-GFP/+; D42-Gal4 > UAS-mCD8RFP/+, V5-PR36: UAS-stau-GFP/+; D42-Gal4 > UAS-mCD8RFP/UAS-V5-PR36). Dashed circular lines (white) outline the nuclei. Arrows (white) indicate stronger nuclear Staufen-GFP signal compared to the control. Scale bar (white), 5 μm.
Next, we quantitatively measured the mean fluorescence intensity (y-axis) and counted pixel numbers (x-axis) for nuclear Staufen-GFP puncta in controls and in C4da neurons expressing V5-PR36 and plotted their values (Fig. 1C). V5-PR36-expressing C4da neurons showed puncta with substantially higher mean fluorescence intensity (i.e. higher density) and pixel number (i.e. larger size) compared to those in controls, suggesting that PR toxicity induces nuclear accumulation of Staufen. To test whether this increase in the nuclear accumulation of Staufen is due to an overall increase in the protein amount, we examined the amount of Staufen-GFP in fly heads with or without V5-PR36 expression. Interestingly, we found a significant (**P = 0.0024) increase in the amount of Staufen-GFP in flies expressing V5-PR36 compared to controls (Fig. 1D and E). In contrast, the amount of endogenous Staufen was not significantly different between controls and V5-PR36-expressing flies (Fig. 1F and G). The difference in the level of Staufen-GFP and endogenous Staufen upon V5-PR36 expression might be due to different promoters being used to express them: Staufen-GFP is expressed via GAL4/UAS system and not its endogenous promoter. We next tested whether merely increasing the amount of overall Staufen-GFP is sufficient to increase the amount of its nuclear localization. To this end, we expressed two copies of stau-GFP (stau gene encodes Staufen) in C4da neurons and compared the amount of Staufen-GFP in the nucleus to controls expressing only one copy (Fig. 1H). We noticed no difference in the amount of nuclear Staufen-GFP between the two (Fig. 1I), suggesting that merely increasing the dosage of Staufen is not enough to increase its nuclear localization.
To investigate whether nuclear accumulation of Staufen is specific to V5-PR36 expression, we examined in C4da neurons the localization pattern of Staufen-GFP after expressing other C9-ALS/FTD constructs: G4C236, G4C236-RNA only, Myc-PA36, and HA-GR36. Among them, G4C236 and HA-GR36 expression led to increased nuclear accumulation of Staufen (Supplementary Material, Fig. S2A and B), whereas G4C236-RNA only and Myc-PA36 expression did not. These results suggest that only the constructs that produce arginine-rich DPRs (i.e. G4C236, V5-PR36, and HA-GR36), but not the others (i.e. G4C236-RNA only and Myc-PA36), induce nuclear accumulation of Staufen. On the other hand, C4da neurons expressing pathogenic polyQ proteins (MJDtr-78Q) that predominantly accumulate in the nucleus showed no change in the amount of Staufen-GFP in the nucleus compared to controls (Supplementary Material, Fig. S2C), suggesting that nuclear accumulation of Staufen is not a general feature observed in neurons exhibiting protein toxicity.
Next, we questioned whether V5-PR36 expression can increase nuclear accumulation of Staufen in neurons of another type. To this end, we expressed V5-PR36 in motor neurons using D42-gal4 driver. Consistent with our results from C4da neurons, expression of V5-PR36 in Drosophila motor neurons showed an increase in nuclear accumulation of Staufen (Fig. 1J), compared to controls. We wondered whether Staufen also accumulates in the nucleus of non-neuronal cells upon arginine-rich DPR expression. To test this, we expressed V5-PR36 in epithelial cells using 69B-gal4 and examined the localization of Staufen. Unexpectedly, we did not observe any punctated form of Staufen in both controls and V5-PR36-expressing epithelial cells (Supplementary Material, Fig. S3), thus precluding us from quantifying the number, size and density of Staufen puncta. Instead, we measured the nuclear/cytoplasmic (Nuc/Cyt) ratio of Staufen-GFP. We found that V5-PR36 expression did not change the ratio (Supplementary Material, Fig. S3B), suggesting that PR repeat proteins do not induce nuclear accumulation of Staufen in epithelial cells. These data collectively suggest that arginine-rich DPRs may induce a neuron-specific accumulation of Staufen in the nucleus.
Imp α2 and Imp ß3 mediate nuclear accumulation of Staufen upon PR36 expression in C4da neurons
A previous study identified Exportin 5 as the nuclear transport receptor (NTR) that exports Staufen2 out of the nucleus in mammals (38). Thus, we hypothesized that a reduced function or expression of Exportin 5 may be responsible for the nuclear accumulation of Staufen upon arginine-rich DPR expression. To test this, we knocked down Exportin 5 in C4da neurons and analyzed the amount of Staufen in the nucleus. Contrary to our expectation, knockdown of Exportin 5 failed to mimic PR toxicity and did not induce nuclear accumulation of Staufen in C4da neurons (Fig. 2A and B). However, Exportin 5 knockdown enhanced the nuclear accumulation of Staufen induced by PR36 expression (Fig. 2C and D). These results suggest that PR36 expression most likely promotes nuclear import of Staufen instead of blocking its export.
Figure 2 .
Exportin- and Importin-dependent changes of nuclear Staufen-GFP upon V5-PR36 expression. (A) A subcellular distribution of Staufen-GFP in C4da neurons with or without Exportin 5 knockdown. Merge: merged with plasma membrane marker (Genotypes. CTRL: UAS-stau-GFP/+; PPK1a-Gal4,UAS-mCD8RFP/UAS-luciferase, Exportin 5 RNAi: UAS-stau-GFP/+; PPK1a-Gal4,UAS-mCD8RFP/UAS-Exportin 5 RNAi). Dashed circular lines (white) outline the nuclei. Scale bar (white), 10 μm. (B) The pixel number and the mean intensity of nuclear Staufen-GFP puncta were multiplied to provide an estimate of the amount of nuclear Staufen-GFP. ns: not significant by two-tailed student’s t-test; error bars, ± SEM; n ≥ 24 Staufen-GFP puncta; n = 3 larvae. (C) A subcellular distribution of Staufen-GFP in C4da neurons expressing V5-PR36 with or without knockdown of Exportin 5 (Genotypes. CTRL: UAS-stau-GFP/+; PPK1a-Gal4,UAS-mCD8RFP, UAS-V5-PR36/UAS-luciferase, Exportin 5 RNAi: UAS-stau-GFP/+;PPK1a-Gal4,UAS-mCD8RFP, UAS-V5-PR36/UAS-Exportin 5 RNAi). Arrow (white) indicates increased nuclear Staufen-GFP puncta. Scale bar (white), 10 μm. (D) The pixel number and the mean intensity of nuclear Staufen-GFP puncta were multiplied to provide an estimate of the amount of nuclear Staufen-GFP. Arrow (white) indicates conjunction of nuclear-accumulated Staufen-GFP. Dashed circular lines (white) outline the nuclei. Scale bar (white), 10 μm. **P = 0.0071 by two-tailed student’s t-test; error bars, ± SEM; n ≥ 29 Staufen-GFP puncta; n = 3 larvae. (E) Quantification of the amount of nuclear Staufen-GFP per puncta in C4da neurons expressing the denoted transgenes (see Fig. S4A for details). *P < 0.05; **P < 0.01; ***P < 0.001 by two-tailed student’s t-test; error bars, ±SEM; n ≥ 9 Staufen-GFP puncta; n = 3 larvae.
Although a functional bipartite nuclear localization signal (NLS) has been identified in yeast and mammalian Staufen (38,39), no specific NTRs that mediate its nuclear import has been identified. Therefore, we performed an RNAi or mutant screen of 13 NTRs involved in nuclear import (Fig. 2E and Supplementary Material, Fig. S4A) and examined their ability to block nuclear accumulation of Staufen upon V5-PR36 expression. Among the 13 NTRs, knockdown of Importin alpha 2 (Imp α2) and Ran and heterozygous mutation of Importin beta 3 (Imp β3) significantly reduced nuclear accumulation of Staufen. On the other hand, knockdown or mutation of the NTRs by themselves had no effect on nuclear accumulation of Staufen (Supplementary Material, Fig. S4B and C). These results suggest that PR repeat proteins do not disrupt nuclear export of Staufen but rather induces its nuclear import in Imp α2-, Imp β3- and Ran-dependent manner.
PR-induced nuclear accumulation of Staufen is neurotoxic
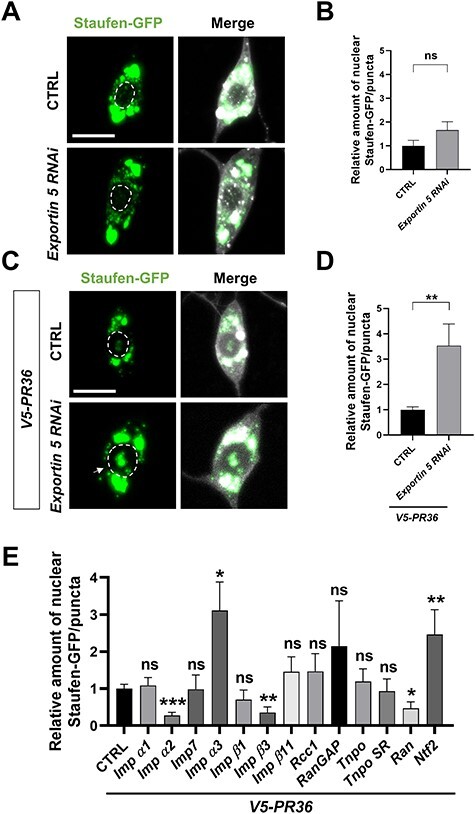
The importin-mediated nuclear accumulation of Staufen may potentiate PR toxicity. To test whether PR-induced nuclear accumulation of Staufen is toxic, we first generated a transgenic construct that express HA-tagged Staufen with exogenous NLS sequence (HA-stau-NLS) and another with mutant endogenous NLS sequence (HA-stau-∆NLS). Then, we tested whether V5-PR36 expression can induce nuclear localization of those two modified Staufen proteins in C4da neurons. As expected, HA-Staufen-∆NLS did not accumulate in the nucleus (Fig. 3A and B), whereas HA-Staufen-NLS showed strong nuclear localization upon V5-PR36 expression (Fig. 3A and B).
Figure 3 .
Nuclear Staufen enhances PR-induced dendritic defect. (A) A subcellular localization of HA-Staufen in C4da neurons (Genotype. HA-stau-NLS: UAS-HA-stau-NLS/+; PPK1a-Gal4,UAS-V5-PR36,UAS-CD4-tdTOM, HA-stau-∆NLS: UAS-HA-stau-∆NLS/+; PPK1a-Gal4,UAS-V5-PR36,UAS-CD4-tdTOM). Scale bar (white), 5 μm. (B) Quantification of the amount of HA-Staufen normalized to controls. *P = 0.0142 by paired two-tailed student’s t-test; error bars, ± SEM; n ≥ 22 HA-Staufen puncta; n = 3 larvae. (C) Skeletonized dendritic images of C4da neurons expressing the denoted transgenes described in Figure 3A. Scale bar (black), 100 μm. (D) Quantification of total dendritic length of C4da neurons expressing the denoted transgenes. ns: not significant, P = 0.0721; ***P = 0.0003; ****P < 1.0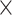 10−4 by paired two-tailed student’s t-test; error bars, ± SEM; n = 9 neurons; n = 3 larvae.
10−4 by paired two-tailed student’s t-test; error bars, ± SEM; n = 9 neurons; n = 3 larvae.
Neuronal dendrites are quite susceptible to degeneration and their lengths are often shortened in neurodegenerative diseases including ALS/FTD (40–43). Thus, we decided to use dendritic length to approximate neurotoxicity in C4da neurons. Expression of V5-PR36 led to an average dendritic length similar to those observed in C4da neurons expressing another PR36 transgene (43) (Fig. 3C and D). Compared to when V5-PR36 is expressed alone, V5-PR36 expressed in combination with HA-stau-NLS led to a significantly shorter dendritic length, whereas the length did not change with HA-stau-∆NLS (Fig. 3C and D). These data show that nuclear-accumulated Staufen potentiates PR-induced dendritic defect and suggest that PR-induced nuclear accumulation of Staufen is toxic.
Nuclear-accumulated Staufen partially colocalizes with heterochromatin in PR36-expressing neurons
A recent study reported that PR repeat proteins colocalize with heterochromatin and disrupt heterochromatin protein 1α (HP1α) liquid droplets (22). This disruption in mouse cortical neurons expressing PR repeat proteins leads to misexpression of repetitive elements (RE) that form dsRNAs, which accumulates within the nucleus (22). We hypothesized based on this previous study that Staufen, which is a dsRNA-binding protein (dsRBP) (44), may be recruited to the heterochromatin region, a site of dsRNA production, in neurons presenting PR toxicity. To test this hypothesis, we expressed RFP-tagged HP1, a Drosophila ortholog of HP1α, and compared its colocalization pattern with Staufen-GFP with or without PR36 expression in C4da neurons (Fig. 4A and B). C4da neurons with PR36 expression showed that greater fraction of HP1 puncta colocalized with Staufen-GFP puncta than in controls (P = 0.0339) (Fig. 4C). Consistently, Staufen-GFP partially colocalized with 4′, 6-diamidino-2-phenylindole (DAPI), which labels heterochromatin, in C4da neurons expressing V5-PR36 (Fig. 4D and E). These data suggest that PR toxicity can augment heterochromatin distribution of Staufen in Drosophila neurons.
Figure 4 .
Nuclear-accumulated Staufen partially colocalizes with heterochromatin in C4da neurons expressing PR36. (A) Nuclear-accumulated Staufen-GFP partially colocalizes with RFP-HP1 in V5-PR36-expressing C4da neurons (Genotypes. CTRL: UAS-stau-GFP/+; PPK1a-gal4/RFP-HP1, PR36: UAS-stau-GFP/UAS-PR36; PPK1a-gal4/RFP-HP1). Dashed circular lines (white) outline the nuclei. Arrow (white) indicates colocalization between nuclear Staufen-GFP and RFP-HP1. Scale bar (white), 5 μm. (B) 3D images of Figure 4A are presented to better visualize the overlap of Staufen-GFP and RFP-HP1 in the nucleus. Arrows (white) indicate colocalization between nuclear Staufen-GFP and RFP-HP1 (white). (C) The proportion of RFP-HP1 puncta that colocalize with Staufen-GFP were normalized to controls. *P = 0.0339 by two-tailed student’s t-test; error bars, ± SEM; n ≥ 25 RFP-HP1 puncta; n = 3 larvae. (D) IHC of dissected third instar larvae shows that nuclear-accumulated Staufen-GFP partially colocalizes with DAPI in V5-PR36-expressing C4da neurons (Genotypes. CTRL: UAS-stau-GFP/+; PPK1a-gal4 > UAS-mCD8RFP/+, V5-PR36: UAS-stau-GFP/+; PPK1a-gal4 > UAS-mCD8RFP/UAS-V5-PR36). DAPI (blue) staining labels heterochromatin. Dashed circular lines (white) outline the nuclei. Arrow (white) indicates colocalization between nuclear Staufen-GFP and DAPI. Scale bar (white), 2 μm. (E) 3D images of Figure 4D are presented to better visualize the overlap of Staufen-GFP and DAPI in the nucleus. Arrows (white) indicate colocalization between nuclear Staufen-GFP and DAPI (white).
PR-induced nuclear accumulation of Staufen is mediated by TBPH
We asked how PR toxicity might induce nuclear accumulation of dsRNAs in C4da neurons. Recent studies showed that loss of nuclear TDP-43, a prominent pathological feature of C9-ALS/FTD (45), can derepress RE transcription from heterochromatin (21,46,47), leading to the accumulation of dsRNAs in the nucleus. Thus, we tested whether decreasing the amount of nuclear TBPH, a Drosophila ortholog of TDP-43, is sufficient to increase accumulation of Staufen in the nucleus. To this end, we knocked down TBPH and examined the localization pattern of Staufen in the nucleus of C4da neurons (Fig. 5A). Compared to the controls, TBPH RNAi led to an increased accumulation of Staufen in the nucleus of C4da neurons (Fig. 5A and B, Supplementary Material, Fig. S5A and B).
Figure 5 .
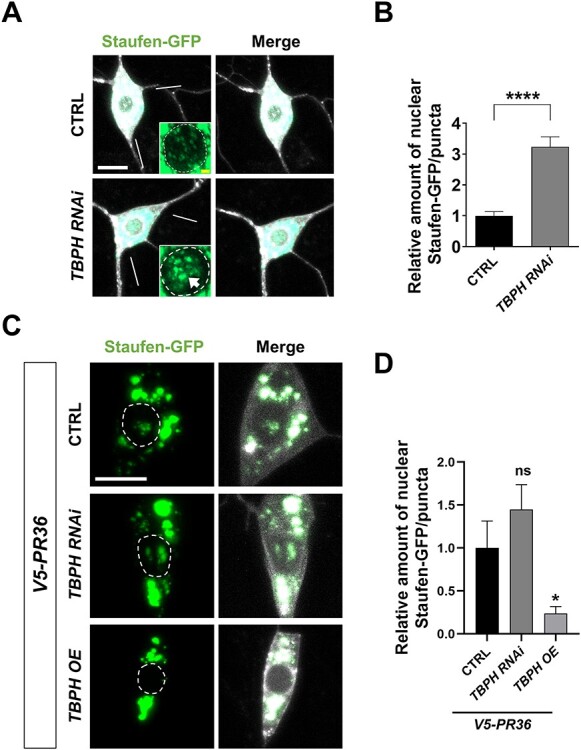
PR-induced nuclear accumulation of Staufen is mediated by TBPH. (A) A subcellular localization of Staufen-GFP upon knockdown of TBPH in C4da neurons (Genotypes. CTRL: UAS-Stau-GFP/+; PPK1a-gal4 > UAS-mCD8RFP/UAS-Luciferase, TBPH RNAi: UAS-Stau-GFP/+; PPK1a-gal4 > UAS-mCD8RFP/UAS-TBPH RNAi). Arrow indicates the increased nuclear accumulation of Staufen-GFP. Scale bar (yellow), 2 μm; (white), 10 μm. (B) Quantification of the amount of Staufen-GFP normalized to controls. ****P < 1.0 10−4 by paired two-tailed student’s t-test; error bars, ± SEM; n ≥ 57 Staufen-GFP puncta; n = 3 larvae. (C) A subcellular localization of Staufen-GFP upon knockdown or overexpression of TBPH in combination with V5-PR36 expression in C4da neurons (Genotypes. CTRL: UAS-Stau-GFP/+; PPK1a-gal4 > UAS-V5-PR36, UAS-mCD8RFP/UAS-Luciferase, TBPH RNAi: UAS-Stau-GFP/+; PPK1a-gal4 > UAS-V5-PR36, UAS-mCD8RFP/UAS-TBPH RNAi, TBPH OE: UAS-Stau-GFP/UAS-TBPH OE; PPK1a-gal4 > UAS-V5-PR36, UAS-mCD8RFP/+). Scale bar (white), 10 μm. (D) Quantification of the amount of Staufen-GFP normalized to controls. ns: not significant; *P = 0.0248 by paired two-tailed student’s t-test; error bars, ± SEM; n ≥ 17 Staufen-GFP puncta; n = 3 larvae.
10−4 by paired two-tailed student’s t-test; error bars, ± SEM; n ≥ 57 Staufen-GFP puncta; n = 3 larvae. (C) A subcellular localization of Staufen-GFP upon knockdown or overexpression of TBPH in combination with V5-PR36 expression in C4da neurons (Genotypes. CTRL: UAS-Stau-GFP/+; PPK1a-gal4 > UAS-V5-PR36, UAS-mCD8RFP/UAS-Luciferase, TBPH RNAi: UAS-Stau-GFP/+; PPK1a-gal4 > UAS-V5-PR36, UAS-mCD8RFP/UAS-TBPH RNAi, TBPH OE: UAS-Stau-GFP/UAS-TBPH OE; PPK1a-gal4 > UAS-V5-PR36, UAS-mCD8RFP/+). Scale bar (white), 10 μm. (D) Quantification of the amount of Staufen-GFP normalized to controls. ns: not significant; *P = 0.0248 by paired two-tailed student’s t-test; error bars, ± SEM; n ≥ 17 Staufen-GFP puncta; n = 3 larvae.
Next, we tested whether the loss of TBPH is necessary for PR-induced nuclear accumulation of Staufen. We knocked down and overexpressed TBPH in C4da neurons expressing V5-PR36 and examined the Staufen-GFP localization pattern. We found that the V5-PR36-induced nuclear localization of Staufen was significantly decreased by TBPH overexpression but remained unchanged with TBPH knockdown (Fig. 5C and D). We suspect that TBPH knockdown did not further increase the nuclear accumulation of Staufen because V5-PR36 likely has reduced the nuclear function of TBPH to the point where it is negligible. These data suggest that the loss of nuclear TBPH (e.g. in the V5-PR36 expression or TBPH knockdown condition) is both sufficient and necessary for the nuclear accumulation of Staufen in PR-expressing neurons.
Increased nuclear accumulation of Staufen by PR toxicity is RNA-dependent
It is possible that Staufen can accumulate in the nucleus by its binding to RNAs, such as dsRNAs. To test whether PR-induced nuclear accumulation of Staufen is dependent on RNA, we dissected larvae with V5-PR36 expression in C4da neurons and treated them with RNase A for 1 h at 37°C before fixing them for imaging analysis. The RNase A treatment protocol we used effectively reduced RNA levels in C4da neurons (Supplementary Material, Fig. S6A and B). Reducing RNA levels led to a substantial decrease in the V5-PR36-induced nuclear accumulation of Staufen (Fig. 6A). We then measured the mean fluorescence intensity (y-axis) and counted pixel numbers (x-axis) of nuclear Staufen-GFP puncta in C4da neurons expressing V5-PR36 with or without RNase A treatment and plotted their values (Fig. 6B). RNase A treatment led to decreases in density and size of Staufen-GFP puncta. The amount of Staufen-GFP proteins in the nucleus was calculated by multiplying the mean fluorescence intensity with the pixel number of Staufen-GFP puncta. RNase A-treated C4da neurons expressing V5-PR36 exhibited significantly (P = 0.0334) lower amount of Staufen-GFP proteins in the nucleus compared to the controls (Fig. 6C). These results suggest that increased accumulation of Staufen in the nucleus by PR toxicity is RNA-dependent.
Figure 6 .
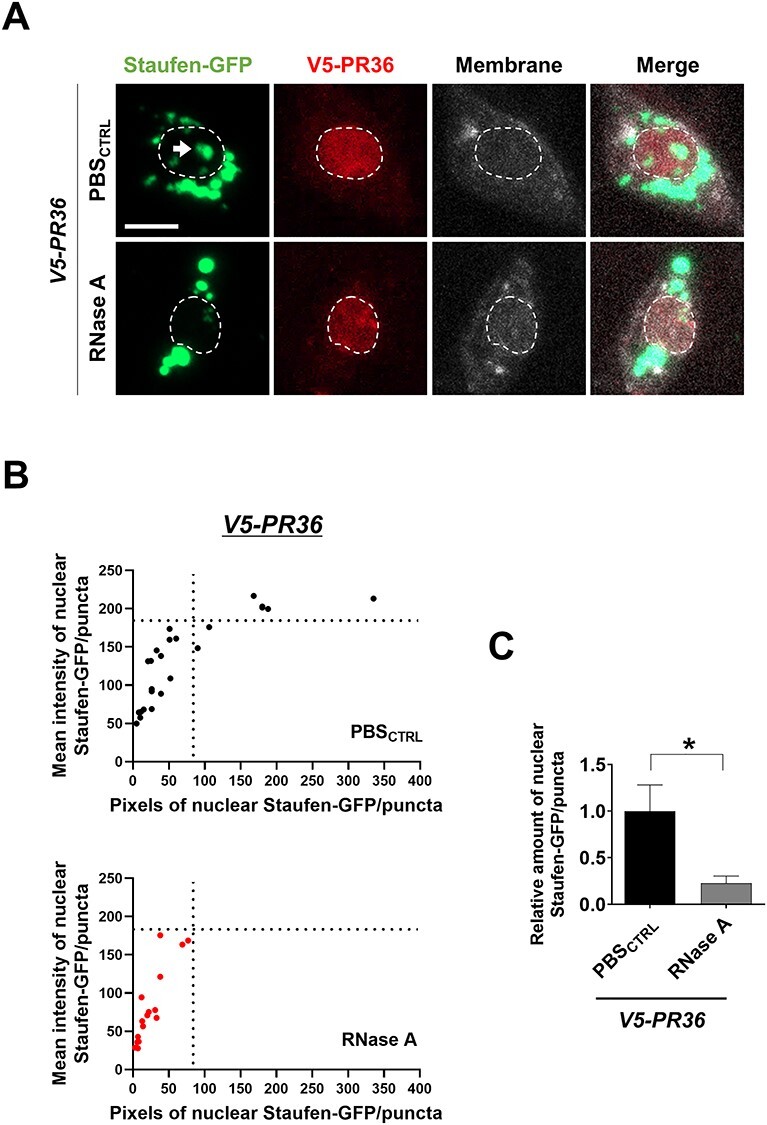
PR toxicity in C4da neurons induces RNA-dependent accumulation of Staufen in the nucleus. (A) RNase A treatment decreased nuclear accumulation of Staufen-GFP induced by PR toxicity in C4da neurons (Genotypes. V5-PR36: UAS-stau-GFP/+; PPK1a-gal4 > UAS-mCD8RFP/UAS-V5-PR36). Dissected third instar larvae were treated for 1 h at 37°C with (RNase A) or without (PBSCTRL) 20 μg/ml of RNase A. Dashed lines (white) indicate the outlines of the nuclei. Arrow (white) indicates faint nuclear Staufen-GFP signals remained after RNase A treatment. Scale bar (white), 5 μm. (B) Nuclear Staufen-GFP puncta in C4da neurons with denoted genotypes from Figure 6A were counted and plotted. y-axis: the mean intensity of Staufen-GFP puncta in nuclei; x-axis: the pixel number of Staufen-GFP puncta in nuclei. n ≥ 16 Staufen-GFP puncta. (C) The pixel number and the mean intensity of nuclear Staufen-GFP puncta (obtained from Figure 6B) were multiplied to provide an estimate of the amount of nuclear Staufen-GFP. *P = 0.0334 by two-tailed student’s t-test; error bars, ± SEM; n ≥ 16 Staufen-GFP puncta; n = 3 larvae.
Increased nucleolar expression of Fibrillarin by reduced stau dosage is a compensatory response to PR toxicity in Drosophila
A large portion of heterochromatin, which consists of inactive ribosomal DNA (rDNA), decorates the outer region of the nucleolus within which actively transcribed rDNA are situated (48). In addition, previous studies have shown that Staufen can localize to the nucleolus (49–51), the dysfunction of which has been reported in C9-ALS/FTD models and patients (52). These data suggest a possibility that in addition to the heterochromatin, PR toxicity may also induce Staufen to accumulate in or influence the activity of the neighboring nucleolus. To test this possibility, we expressed V5-PR36 and stau-GFP in C4da neurons and immunostained for Fibrillarin, which is localized in the dense fibrillar component of the nucleolus (53). Staufen-GFP and Fibrillarin did not colocalize but were oftentimes localized adjacent to each other (Supplementary Material, Fig. S7A and B). Interestingly, we noticed that Fibrillarin staining was considerably stronger (P = 0.0195) in C4da neurons expressing V5-PR36 than in controls (Supplementary Material, Fig. S8A and B). These data are in line with a previous report showing increased nucleolar size determined by Fibrillarin staining in U2OS and NSC-34 cells expressing PR20 (54).
Although Staufen and Fibrillarin did not colocalize with each other, abnormal accumulation of Staufen in the nucleus may still contribute to the increased staining of Fibrillarin in C4da neurons expressing V5-PR36. To test this possibility, we examined the amount of Fibrillarin upon V5-PR36 expression in flies with heterozygous mutation of stau (stau+/−) and compared it to the controls. Surprisingly, stau+/− actually increased (P = 0.0371) nucleolar staining of Fibrillarin in C4da neurons expressing V5-PR36 (Fig. 7A and B). Of note, about 15.6% (5/32) of those neurons showed more than 3.5-fold increase in nucleolar staining of Fibrillarin compared to V5-PR36 only controls.
Figure 7 .
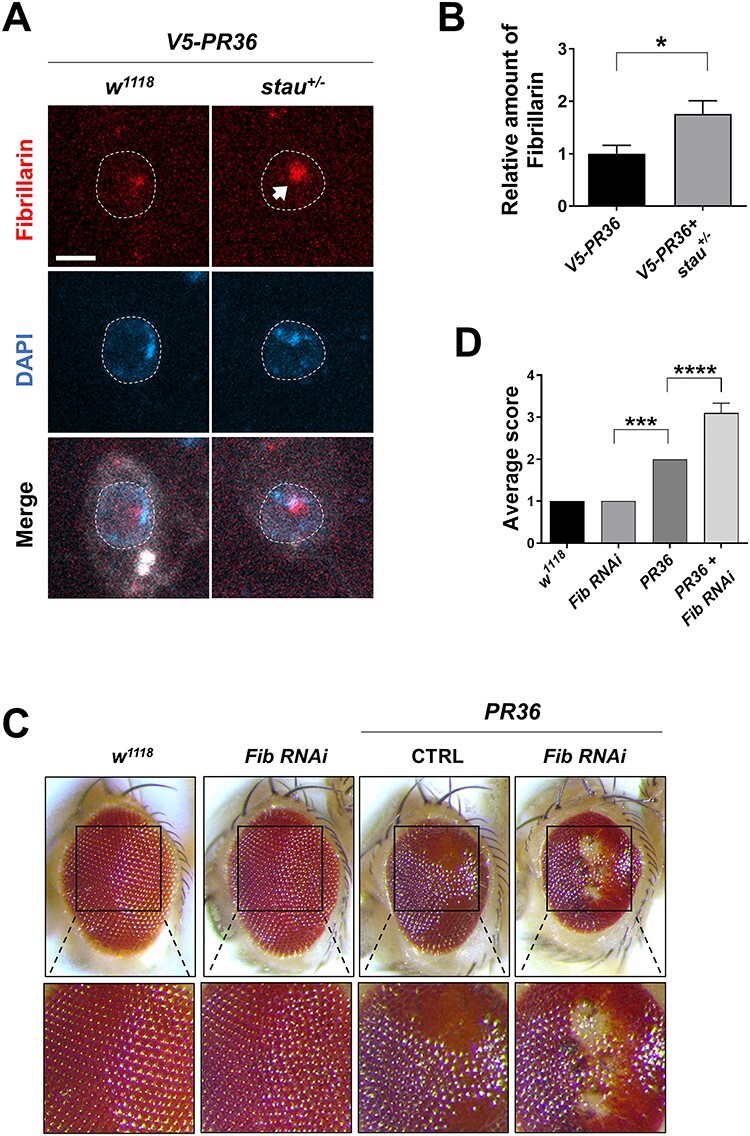
Increased Fibrillarin expression by reduced stau dosage is a compensatory response to PR toxicity in Drosophila. (A) IHC of dissected third instar larvae shows that heterozygous loss of stau increased nucleolar expression of Fibrillarin in V5-PR36-expressing C4da neurons (Genotypes. w1118 + V5-PR36: +/+; PPK1a-gal4 > UAS-mCD8RFP/UAS-V5-PR36, V5-PR36 + stau+/−: staury9/+; PPK1a-gal4 > UAS-mCD8RFP/UAS-V5-PR36). Dashed lines (white) indicate the outlines of the nuclei. Arrow (white) indicates increased Fibrillarin signal after reducing stau dosage. Scale bar (white), 5 μm. (B) The pixel number was multiplied by the mean intensity of Fibrillarin puncta to estimate the amount of Fibrillarin in C4da neurons with the denoted genotypes from Figure 7A. *P = 0.0371 by two-tailed student’s t-test; error bars, ± SEM; n ≥ 18 Fibrillarin puncta; n = 3 larvae. (C) Knockdown of Fib exacerbated retinal degeneration induced by PR toxicity (Genotypes. w1118: GMR-gal4/+, Fib RNAi: GMR-gal4/UAS-Fib RNAi, CTRL + PR36: GMR-gal4 > UAS-PR36/UAS-40D, Fib RNAi + PR36: GMR-gal4 > UAS-PR36/UAS-Fib RNAi). Fly eyes were imaged at 4 days after eclosion. Images were acquired using 160× objective lens. (D) Quantification of the eye defect for the flies with genotypes mentioned in Figure 7C. We scored the eyes in the following way: (1) normal, (2) some ommatidial disruptions, (3) small regions of depigmentation and/or necrosis and (4) large regions of depigmentation and/or necrosis. ***P = 0.0003; ****P < 1.0 10−4 by one-way ANOVA; error bars, ± SEM; n ≥ 6.
10−4 by one-way ANOVA; error bars, ± SEM; n ≥ 6.
Increased level of Fibrillarin in the nucleolus may be either a toxic feature of or a compensatory response to PR toxicity. To test which is the case, we decreased the level of Fibrillarin by a knockdown of the gene encoding Fibrillarin (Fib) in Drosophila eyes using GMR-gal4 and examined whether PR36-induced retinal degeneration was exacerbated or mitigated. Knockdown of Fib exacerbated the PR36-induced retinal degeneration, whereas the knockdown by itself showed no toxicity (Fig. 7C and D). Interestingly, a previous study showed that knockdown of Fib can also exacerbate GR-induced retinal degeneration in Drosophila (55). These data suggest that a partial loss of stau increases the level of Fibrillarin, which may be a compensatory response to, and not a toxic feature of, PR toxicity.
Heterozygous loss of stau mitigates PR toxicity in Drosophila
We showed above that in neurons expressing V5-PR36, Staufen partially overlaps with Fibrillarin in the nucleolus and that reducing the dosage of stau increases the amount of Fibrillarin, which may be a compensatory response to PR toxicity. Furthermore, we have shown that PR-induced nuclear accumulation of Staufen can cause dendritic shortening. Based on these data, we hypothesized that reducing stau gene dosage might mitigate PR toxicity. To test this hypothesis, we expressed PR36 in the Drosophila eyes via GMR-gal4 and compared their external morphologies to the controls. By four days after eclosion, flies expressing PR36 displayed a considerable retinal degeneration with loss of pigments and gain of necrotic spots (Fig. 8A). Consistent with our hypothesis, stau+/− was able to significantly suppress PR36-induced retinal degeneration (Fig. 8B). To see whether this suppression was specific to stau+/−, we tested whether knockdown or heterozygous mutant of the other six RBPs screened could also suppress PR36-induced retinal degeneration (Supplementary Material, Fig. S9A and C). Interestingly, knockdown of Patr-1, orb2 and Dp1 exacerbated the PR-induced degeneration (Supplementary Material, Fig. S9B); only Atx2 heterozygous mutant suppressed the degenerative phenotype, albeit only slightly (Supplementary Material, Fig. S9D).
Figure 8 .
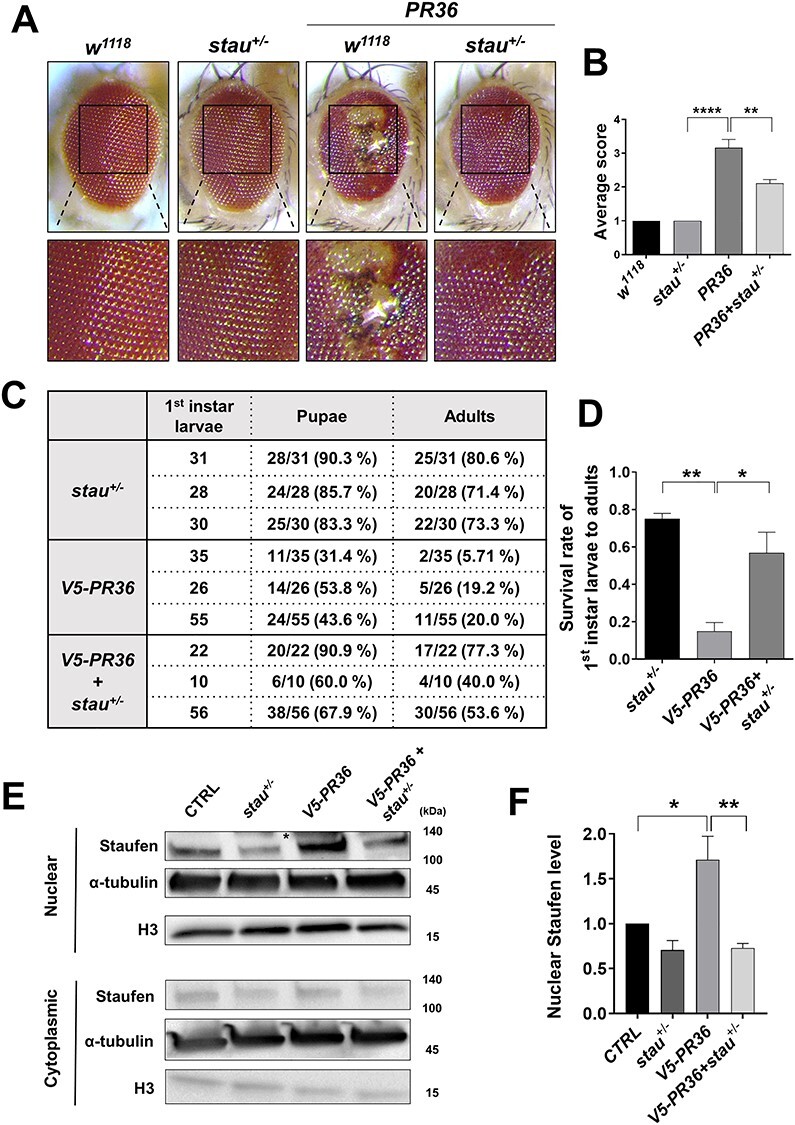
Reduction of stau dosage suppresses PR toxicity in Drosophila. (A) Heterozygous loss of stau restored PR36-induced retinal degeneration (Genotypes. w1118: GMR-gal4/+, stau+/−: GMR-gal4/staury9, w1118 + PR36: GMR-gal4 > UAS-PR36, PR36 + stau+/−: GMR-gal4 > UAS-PR36/staury9). Fly eyes were imaged at 4 days after eclosion. Images were acquired using 160× objective lens. (B) Quantification of the eye defect for the flies with genotypes mentioned in Figure 8A. **P = 0.012; ****P < 0.0001 by one-way ANOVA; error bars, ± SEM; n ≥ 9. (C) Heterozygous loss of stau restored viability of flies expressing V5-PR36 (Genotypes. stau+/−: staury9/+; elav-gal4/+, V5-PR36: elav-gal4/UAS-V5-PR36, V5-PR36 + stau+/−: staury9/+; elav-gal4/UAS-V5-PR36). First instar larvae were collected separately per each line at 25°C. Surviving pupae and eclosed adult flies were tallied and listed. Percentages of first instar larvae-to-pupae and first instar larvae-to-adults are denoted in parentheses. (D) The percentages of first instar larva-to-adult viability obtained from three independent experiments were plotted and compared: stau+/− to V5-PR36 and V5-PR36 to V5-PR36 + stau+/−. **P = 0.0022; *P = 0.0131 by one-way ANOVA; error bars, ± SEM; n = 3 vials. (E) Western blot after nuclear and cytoplasmic fractionation of heads of flies with the following genotypes (Genotypes. CTRL: elavGS-gal4/+, stau+/−: staury9/+; elavGS-gal4/+, V5-PR36: elavGS-gal4/UAS-V5-PR36, V5-PR36 + stau+/−: staury9/+; elavGS-gal4/UAS-V5-PR36). The transgenic expression was induced by feeding 100 μM of RU486 for 10 days after eclosion at 27°C. H3 and α-tubulin were used as loading controls. An asterisk marks a nonspecific band. (F) Quantification of Figure 8E. *P = 0.0316, **P = 0.0054; by one-way ANOVA; error bars, ± SEM; n = 3.
Next, we asked whether stau+/− could increase the viability of flies expressing V5-PR36. We measured the percentage of larvae (Genotypes. stau+/−: staury9/+; elav-gal4/+, V5-PR36: elav-gal4/UAS-V5-PR36, V5-PR36 + stau+/−: staury9/+; elav-gal4/UAS-V5-PR36) reaching the pupal and adult stages and compared them with one another. Most of the stau+/− larvae reached the pupal (86.46%) and adult (75.14%) stages, whereas only 42.97% and 14.98% of V5-PR36 larvae reached the pupal and adult stages, respectively (P = 0.0022) (Fig. 8C and D). Remarkably, the larva-to-pupa (72.92%) and larva-to-adult (56.95%; P = 0.0131) viabilities were largely rescued in stau+/− + V5-PR36 larvae (Fig. 8C and D). Importantly, heads of flies expressing V5-PR36 showed an increased (P = 0.0316) level of endogenous Staufen in the nucleus, and stau+/− reduced its level back near to that of control (P = 0.0054; Fig. 8E and F). Taken together, these results suggest that PR toxicity induces aberrant accumulation of Staufen in the neuronal nucleus and that reducing the stau gene dosage is sufficient to rescue PR toxicity in Drosophila.
Discussion
Most ALS/FTD pathology can be defined by nuclear loss and cytoplasmic accumulation of disease-associated proteins, such as TDP-43, FUS, TAF15, EWSR1, hnRNPA1 or hnRNPA2/B1, thereby leading to their loss of nuclear function and gain of toxic cytoplasmic function (56). On the other hand, to our knowledge, no cytoplasmic protein has been characterized thus far in ALS/FTD that translocates into the nucleus and accumulates therein. In this study, we identified Staufen, a cytoplasmic dsRBP, as a potential disease-associated protein in C9-ALS/FTD Drosophila model that exhibits nuclear accumulation in Importin- and RNA-dependent manner. We found that increased nuclear—but not cytoplasmic—Staufen potentiates PR-induced dendritic defects. Once inside the nucleus, Staufen partially interacted with heterochromatin and nucleolus. Reducing the dosage of stau mitigated retinal degeneration in flies expressing V5-PR36 and rescued their viability.
Recently, nucleocytoplasmic transport defect has been identified in numerous neurodegenerative diseases including ALS (17–20,57). Moreover, a study identified via LC–MS/MS the proteins whose nucleocytoplasmic distribution was altered in a C9-ALS/FTD model (58). This study identified 126 proteins, 44% of which shifted to the nucleus from cytoplasm. Staufen, however, was not among them. This apparent discrepancy may be accounted for by the following major differences between the abovementioned study and ours: we used in vivo Drosophila neurons, they HEK293 cells; we used mostly PR36 as the disease construct, they G4C258; we experimented in flies with manifest toxicity, they in cells without overt toxicity. Because of these differences, studies such as ours complement—not undermine—studies such as those mentioned above by overcoming issues regarding false negatives and false positives.
Among the seven RBPs screened in this study, only Staufen was identified as a dsRBP. This is a significant point, because what most distinguishes Staufen from other RBPs screened is its ability to bind to dsRNA (44), a toxic byproduct of disrupted heterochromatin caused by PR toxicity (22) and nuclear loss of TDP-43 (21,46,47). In this study, we showed that PR expression led to increased colocalization of Staufen with heterochromatin and that degrading RNAs via RNase led to decreased nuclear accumulation of Staufen. These data support the notion that upon PR expression in neurons, Staufen accumulates around heterochromatin and binds to dsRNA the degradation of which disperses nuclear-accumulated Staufen, leading to its nuclear egress. Whether other dsRBPs also localize near the heterochromatin upon induction of PR toxicity is difficult to ascertain, because those dsRBPs (Zn72D and pasha) we tested (which were included in the prescreen) originally localized to the nucleus even without PR expression (data not shown).
Staufen1 and Staufen2 have a bipartite NLS at the end of double-stranded RNA-binding domain 3 (dsRBD3) that can deliver Staufen to nucleolus in yeast and mammals (38,39). However, Staufen normally localizes mainly to the cytoplasm in mammalian and Drosophila cells (59,60), potentially due to its anchoring to the ER and cytoskeleton (39), its binding to RNAs that may mask NLS (38) or its active nuclear export. Although the specific Importin receptors for Staufen remained unidentified, a nuclear export receptor Exportin 5 has been shown to export Staufen2 out of the nucleus in BHK and HeLa cells (38). In Drosophila neurons, we observed that Staufen normally localizes also to the cytoplasm but accumulates in the nucleus upon arginine-rich DPR expression. Lack of nuclear accumulation of Staufen in C4da neurons upon Exportin 5 knockdown seems to suggest that Staufen is not actively being imported into the nucleus under normal conditions. However, knockdown of Exportin 5 in combination with V5-PR36 expression enhanced nuclear accumulation of Staufen, suggesting that PR repeat proteins may stimulate nuclear import of Staufen. As anticipated, PR-induced nuclear accumulation of Staufen can be largely prevented by a knockdown or mutation of Imp α2, Imp ß3 or Ran (Fig. 2E and Supplementary Material, Fig. S4A). These data suggest that instead of blocking nuclear export of Staufen, PR repeat proteins likely promote nuclear import of Staufen. Further biochemical analysis is needed to test our prediction that Imp α2 and Imp ß3 form a complex together to mediate Staufen import into the nucleus in a Ran-dependent manner.
In this study, we found that expression of PR repeat proteins led to an increased staining of Fibrillarin. What might best account for the increased amount of Fibrillarin by PR toxicity? A previous study identified p53 as a repressor of FBL (which codes Fibrillarin protein in humans) promoter, thereby regulating the amount of Fibrillarin, in various cancer cell lines (61). Interestingly, another study showed that dsRNA induces reduction of p53 in HT1080 cells (62). These pieces of data suggest that increased dsRNA production by PR toxicity may reduce p53, thereby disinhibiting Fibrillarin production. Consistent with this notion, our western blot data showed a significantly (P = 0.0321) reduced level of p53 protein in heads of flies expressing V5-PR36 compared to controls (Supplementary Material, Fig. S10A and B). In addition, we showed that in C4da neurons V5-PR36 expression increased the amount of immunostained Fibrillarin compared to the controls, and this amount was further increased by knockdown of p53 (Supplementary Material, Fig. S10C). In contrast to our data, a previous study showed that treatment of a high dose (10 μmol/L) of PR20 in SH-SY5Y cells led to increased level of p53, albeit with a significant amount of cell death (63). Thus, further studies are required for understanding a potential role of p53 in mediating PR-induced Fibrillarin induction.
Similar to p53 RNAi, stau+/− enhanced the level of Fibrillarin in C4da neurons expressing V5-PR36 (Fig. 7A and B), which appears to be a compensatory mechanism against PR toxicity (Fig. 7C and D). Moreover, stau+/− by itself also increased the level of Fibrillarin in fly heads (Supplementary Material, Fig. S11A and B). However, how stau+/−increases the level of Fibrillarin remains unknown. Interestingly, a previous study showed that a knockdown of STAU1 led to a reduced half-life of TP53 (a human homolog of p53) mRNA in H1299 cells upon actinomycin D treatment (64). However, western blot of p53 (Supplementary Material, Fig. S11C and D) and its potential downstream targets, Pp2A-29B and Bicaudal-D (65) (Supplementary Material, Fig. S11E and F), showed no significant difference in their level in fly heads between the w1118 and stau+/−. We surmise that V5-PR36 and stau+/− up-regulate Fibrillarin via two distinct mechanisms: p53-dependent increase in Fibrillarin level by V5-PR36, and p53-independent increase by stau+/−.
In many neurodegenerative diseases, such as Huntington’s disease, abnormal nuclear accumulation of proteins is known to be highly toxic (66,67). Based on our results, we propose that nuclear-accumulated Staufen may also be toxic in a fly model of C9-ALS/FTD. Our results show that (1) increasing nuclear—but not cytoplasmic—localization of Staufen potentiates PR toxicity, (2) reducing stau dosage in PR-expressing neurons increased the amount of Fibrillarin, which may be a compensatory response to PR toxicity and (3) reducing stau dosage decreases the level of nuclear Staufen and rescues retinal degeneration and viability of flies. In addition, a recent proteomics study identified Stau2, an ortholog of Drosophila Staufen (68), to be commonly enriched in both PR and GR interactome in rat primary neurons (69). This raises the possibility that Staufen may interact with PR in the nucleus in an RNA-dependent manner to contribute to toxicity. These evidence, however, do not exclude the possibility that stau+/− may confer neuroprotection via reducing cytoplasmic level of Staufen. Further investigation will be needed to elucidate the potential role of cytoplasmic Staufen in the disease pathogenesis.
A recent study showed that Spinocerebellar ataxia type 2 (SCA2) mouse model and human patients presented elevated level of Stau1 protein whose interaction with polyQ-expanded ATXN2 caused neurotoxicity (7). Reducing STAU1 in SCA2 mouse improved motor behaviors and reduced polyQ-expanded ATXN2 aggregation. In the same study, they also showed an elevated level of Stau1 in fibroblasts from ALS patients with TDP-43 G298S mutation (7). In a follow-up study, they showed that up-regulated STAU1 induced the unfolded protein response pathway, thereby contributing to toxicity in fibroblasts from ALS patients (8). In our study, we showed that Staufen accumulates in the nucleus, that its accumulation enhances PR toxicity, and that reducing stau suppressed retinal degeneration and rescued viability of flies exhibiting PR toxicity. Taken together, we surmise that aberrant accumulation of Staufen in neurons is toxic and that its reduction might be a suitable therapeutic option for a spectrum of diseases including SCA2, ALS and FTD.
Materials and Methods
Drosophila stocks
All flies were maintained at 25°C and 60% humidity. The following lines were obtained from Bloomington Drosophila Stock Center (Indiana, USA): w1118 (3605), UAS-pasha-GFP (36506), P{PTT-GA}Zn72DCA07703 (50830), Mi{MIC}eIF4BMI14258 (61727), UAS-(G4C2)36 (58688), UAS-(G4C2)36-RNA only (58689), UAS-PR36 (58694), RFP-HP1 (30562), staury9/+ (10742), UAS-TBPH RNAi (29517), UAS-MJDtr-78Q (8141), UAS-CD4-tdTOM (35841), UAS-Luciferase (35788), elav-GeneSwitch(GS)-gal4 (43642), D42-gal4 (8816), 69B-gal4 (1774), GMR-gal4 (1104), elav-gal4 (8760), UAS-Imp α1 RNAi (27523), UAS-Imp α2 RNAi (27692), UAS-Imp β1 RNAi (31242), UAS-Imp 7 RNAi (33626), UAS-Imp β11 RNAi (55142), UAS-Tnpo RNAi (50732), UAS-Tnpo-SR RNAi (56974), UAS-Ran RNAi (42482) and UAS-Ntf-2 RNAi (28633). The following lines were obtained from Vienna Drosophila Resource Center (VDRC): UAS-Imp α3 RNAi (106249), UAS-RanGAP RNAi (108264), UAS-Exportin 5 RNAi (31706), UAS-Rcc1 RNAi (110321), UAS-Fib RNAi (104372KK), fTRG00916.sfGFP-TVPTBF (318270) and UAS-40D (60101). The following lines were obtained from FlyORF (70): UAS-eIF4A-3xHA (F001132), UAS-Atx2-3xHA (F001031), and UAS-pAbp-3xHA (F000753). UAS -stau-GFP was provided by Andrea Brand (University of Cambridge, UK); UAS-mCD8RFP and PPK1a-gal4 were provided by Yuh Nung Jan (UCSF, USA); UAS-TBPH was provided by Brian D.McCabe (EPFL, Switzerland). elavGS-gal4 expression was induced by feeding adult flies 100 μM of RU486 (Mifepristone) (Sigma. M8046) for 10 days after eclosion at 27°C.
Molecular cloning and generation of transgenic flies
V5-PR36, HA-GR36 and Myc-PA36 were synthesized and subcloned (Genscript, Inc.) into pACU2 vectors (from Chun Han, Cornell University) using BglIII and XhoI, and the epitopes were added at the N-terminus. Alternative codons were used to construct the abovementioned DPR lines. HA-stau-NLS, and HA-stau-∆NLS were also synthesized and subcloned into the pACU2 vector in the same manner but with different enzymes: NOTI and XbaI. For HA-stau-NLS, we added PY-NLS from HNRNPA1 at the C-terminus. For HA-stau-∆NLS, we first identified the bipartite NLS in fly stau by comparing its sequence to that of NLS in human Stau1 (39). Then, we mutated all the lysines into alanines within the putative NLS to generate HA-stau-∆NLS. HA epitopes were added to the N-terminus. Dp1 (LD21677), Patr-1 (LD27979) and orb2 (LP05645) cDNAs were obtained from Drosophila Genomics Resource Center. These genes were subcloned (using NheI and KpnI for Dp1; NheI and xhoI for orb2; and NotI and XbaI for Patr-1) into a modified pACU2 vector that has mNeonGreen (Allele biotechnology) before the multicloning site so that the transgenes are tagged at the N-terminus. Transgenic flies were generated by BestGene, Inc. Nucleotide sequences are available upon request.
Western blot
Fly heads were suspended in lysis buffer (50 mM Tris-buffered saline (Tris–HCL) pH 7.5, 150 mM NaCl, 1% Triton ×100) with protease inhibitor cocktail (Thermo Scientific, #87786; 1:100) in which they were homogenized and then centrifuged at 13 300 rpm for 10–15 min at 4°C. Supernatant was collected and its protein amount was measured via Bradford protein assay. Laemmli buffer (Bio-Rad, #161-0747) to 2-mercaptoethanol (BIOSESANG, #60-24-2) in 9:1 ratio was then added and boiled for 5 min at 95°C and then loaded onto Mini-PROTEAN TGX Stain-Free, 4–15% gel (#BR456-8083, Bio-Rad). After the transfer, the membranes were incubated in 5% skim milk diluted in 1% TBST (blocking buffer) for 45 min or 1 h at room temperature (RT), and then in blocking buffer with primary antibodies overnight at 4°C. The primary antibodies used are as follows: mouse anti-GFP (Santa Cruz, sc-9996) (1:2000); mouse anti-V5 (Thermo Scientific, R960–25) (1:1000); mouse anti-p53 (DSHB, 7A4) (1:200); and mouse anti-α-tubulin (DSHB, E7-s) (1:2000 or 1:4000). After washing five times in 1% TBST at RT, the membranes were incubated in blocking buffer with secondary antibody mouse anti-IgGκ BP-HRP (Santa Cruz, sc-516 102) (1:2000) for 1 h at RT. After washing five times in 1% TBST at RT, the membranes were incubated in ECL solution prior to detection with chemiluminescence (#BR170-5060, Bio-Rad) in ChemiDoc™ XRS+.
Nuclear/cytoplasmic fractionation and western blot
Nuclear and cytoplasmic fractions of whole protein lysate were isolated by using Nuclear and Cytoplasmic Extraction Reagents (NE-PER) (Thermo Scientific, #77835). Protein lysates were extracted from adult fly head samples collected at 10 days after eclosion. Experimental procedures of nuclear/cytoplasmic fractionation were carried out according to the manufacturer’s standard protocol. Western blot analysis was then performed as previously described (29). 4× Laemmli buffer and β-mercaptoethanol were added to each nuclear and cytoplasmic extract to make the final concentration be 1× and 2.5%, respectively. Mini-PROTEAN TGX Stain-Free, 4–15% gel (Bio-Rad, #BR456-8084) was used to run the gel electrophoresis, and the proteins were subsequently transferred to nitrocellulose membrane (Bio-Rad, #BR170-4270). Then, nitrocellulose membrane was incubated for 1 h in the blocking buffer (5% skim milk solution in TBST (50 mM Tris–HCl pH 7.4, 150 mM NaCl, and 0.1% Tween-20)). The following primary antibodies were used in this experiment: rabbit anti-Staufen (gifted by Dr St Johnston, Cambridge, UK) (71) (1:1000 dilution) to detect endogenous Staufen proteins; mouse anti-V5 (Thermo Fisher Scientific, R960-25) (1:1000 dilution) to detect V5-PR36 protein; and rabbit anti-Histone 3 (Abcam, Ab1791) (1:1000 dilution) and mouse anti-α-tubulin (DSHB, 12G10) (1:1000 dilution) as loading controls. To detect these primary antibodies, the following secondary antibodies were used: goat anti-rabbit IgG-HRP (Santa Cruz, sc-3837) (1:3000) and anti-mouse IgGκ BP-HRP (Santa Cruz, sc-516 102) (1:3000).
Immunohistochemistry
Third instar larvae were dissected and immediately fixed in 3.7% formaldehyde (JUNSEI, #50-00-0) for 20 min at RT. Formaldehyde was diluted in 1× phosphate buffered saline (PBS) (MENTOS). After washing three times at RT in 0.3% PBST (0.3% triton X-100 in 1× PBS), the samples were incubated in the 5% normal donkey serum (NDS) diluted in washing buffer (blocking buffer) for 1 h at RT. Next, the samples were incubated in primary antibodies diluted in blocking buffer overnight at 4°C. To detect Staufen-GFP, V5-PR36, HA-Staufen, Staufen and Fibrillarin, the following primary antibodies were used respectively: rabbit anti-GFP (Abcam, ab183734) (1:500); mouse anti-V5 (Thermo Scientific, R960–25) (1:500); rabbit anti-HA (Cell Signaling, C29F4) (1:500); rabbit anti-Staufen (gifted by Dr. St Johnston) (1:100); and mouse anti-Fibrillarin (Abcam, ab4566) (1:200). After washing five times for 10 min in washing buffer at RT, the samples were incubated with secondary antibodies diluted in blocking buffer for 2–4 h at RT. The following secondary antibodies were used: goat anti-rabbit Alexa 488 (Invitrogen, A11034) (1:1000); goat anti-mouse Alexa 647 (Invitrogen, A21236) (1:1000 or 1:400); and goat anti-HRP Cy3 (Jackson Immunoresearch Laboratories, #123–165-021) (1:200). The anti-HRP antibody was used to label the whole neuronal membranes of soma and dendrites. The samples were then washed five times for 5 min in washing buffer and mounted using mounting medium with DAPI (4′,6-diamidino-2-phenylindole) (VECTASHIELD, H-1200) for imaging. DAPI was used for visualizing heterochromatin located in the nucleus. For immunohistochemistry (IHC) of adult brains and larval brains, all primary antibodies were diluted 1:200 and all secondary antibodies were diluted 1:400 in blocking buffer. All procedures in IHC experiments were performed in the dark.
Imaging analysis
Confocal images were acquired using LSM 780 (ZEISS) or LSM 800 (ZEISS) microscope. In vivo neuronal images of third instar larvae were taken at 200× magnification using 20× objective with the confocal microscope. Neuronal images from staining experiments in third instar larvae were taken at 400× magnification using the 40× water immersion objective with the confocal microscope. All C4da neurons were acquired from the abdominal segments A4-A6. Retinal images were acquired using Leica SP5. Fly eyes (left eyes only) were taken at 160× magnification immediately upon dissection. For quantification, we arbitrarily defined the retinal phenotypes into four categories: (1) normal, (2) some ommatidial disruptions, (3) small regions of depigmentation and/or necrosis and (4) large regions of depigmentation and/or necrosis.
Dendrite analysis
Dendritic images taken from Confocal microscope were first converted into skeletal images by ImageJ. These skeletonized images were used to measure dendritic length.
Ribonuclease A treatment
The protocol for ribonuclease A (RNase A) treatment in Drosophila embryos was slightly modified (72) to suit C4da neurons of third instar larval fillet. Samples were incubated in 20 μg/ml of RNase A (Sigma, R6513) suspended in 1× PBS for 1 h at 37°C before fixation. As controls, samples were incubated in 1× PBS instead of RNase A for 1 h at 37°C before fixation. The next steps of IHC were performed as described above.
RNA fluorescent in situ hybridization
The protocol for protein-RNA double labeling in Drosophila ovaries (73) was slightly modified to apply fluorescent in situ hybridization (FISH) to C4da neurons of third instar larval fillet. To confirm mRNA reduction in RNase A-treated C4da neurons of third instar larvae, a Cy 5-tagged poly(A)-tail-targeting (Cy5-oligo-dT) probe was designed as follows: 5′-Cy5-TTT TTT TTT TTT TTT TTT TT-3′ (6555.6 g/mol MW, 37.9°C Tm) (Macrogen Inc.). All procedures in FISH experiments were performed in the dark. In FISH experiments, IHC was first performed using dissected third instar larvae, but with different solutions from IHC described above: 0.1% tween-20 (Biotech, #9005-64-5) diluted in 1× PBS (PBT); 3.7% formaldehyde in PBT; and 1% skim milk in PBT. Then, FISH procedures were applied to the samples as previously described (73). 3 μM (tests for 0.1–5 μM) of a Cy5-oligo-dT probe was hybridized to the samples at 37.9°C (Tm) overnight. The following FISH solutions were used: EtOH (Merk, #64-17-5); Xylenes (Sigma, #1330-20-7); RIPA (Biosesang, R2002); Formamide (Sigma, F9037); 5× SSC (Biosesang, S2012); Heparin (Sigma, H4784); DEPC-treated water (MENTOS, M1409). Finally, the samples were mounted for imaging.
Image processing and statistical analysis
Obtained confocal images were further analyzed using Image J for quantification of pixel number and mean intensity. Microsoft Excel was used to plot puncta patterns on the graph. For statistical analyses, GraphPad Prism 6.01 was used for two-tailed student’s t-test, one-way ANOVA, and the production of all the other graphs. Mean intensities and pixel values were calculated and used for analyses. Error bars show SEM and all P values were summarized with asterisks: *P < 0.05; **P < 0.01; ***P < 0.001; ****P < 1.0 10−4.
10−4.
Supplementary Material
Acknowledgements
We would like to thank Dr. St Johnston (Cambridge, UK) for the anti-Staufen antibody. The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
Contributor Information
Eun Seon Kim, Department of Brain & Cognitive Sciences, DGIST, Daegu 42988, Republic of Korea; Dementia research group, Korea Brain Research Institute (KBRI), Daegu 41068, Republic of Korea.
Chang Geon Chung, Department of Brain & Cognitive Sciences, DGIST, Daegu 42988, Republic of Korea.
Jeong Hyang Park, Department of Brain & Cognitive Sciences, DGIST, Daegu 42988, Republic of Korea.
Byung Su Ko, Department of Brain & Cognitive Sciences, DGIST, Daegu 42988, Republic of Korea.
Sung Soon Park, Department of Brain & Cognitive Sciences, DGIST, Daegu 42988, Republic of Korea.
Yoon Ha Kim, Department of Brain & Cognitive Sciences, DGIST, Daegu 42988, Republic of Korea.
In Jun Cha, Department of Brain & Cognitive Sciences, DGIST, Daegu 42988, Republic of Korea.
Jaekwang Kim, Dementia research group, Korea Brain Research Institute (KBRI), Daegu 41068, Republic of Korea.
Chang Man Ha, Research Division and Brain Research Core Facilities of Korea Brain Research Institute (KBRI), Daegu 41068, Republic of Korea.
Hyung-Jun Kim, Dementia research group, Korea Brain Research Institute (KBRI), Daegu 41068, Republic of Korea.
Sung Bae Lee, Department of Brain & Cognitive Sciences, DGIST, Daegu 42988, Republic of Korea; Dementia research group, Korea Brain Research Institute (KBRI), Daegu 41068, Republic of Korea.
Funding
National Research Foundation of Korea (NRF) funded by the Ministry of Education (2020R1A6A3A01096351 to CGC); the KBRI Research Program of the Ministry of Science, ICT & Future Planning (21-BR-02-16 to JK), (21-BR-03-02 to H-JK, CMH and SBL); and Basic Science Research Program through the NRF, funded by the Ministry of Science and ICT (2019R1A4A1024278 to SBL).
Conflict of Interest Statement
None declared.
References
- 1. Liu, Y., Pelham-Webb, B., Di Giammartino, D.C., Li, J., Kim, D., Kita, K., Saiz, N., Garg, V., Doane, A., Giannakakou, P. et al. (2017) Widespread mitotic bookmarking by histone marks and transcription factors in pluripotent stem cells. Cell Rep., 19, 1283–1293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Medioni, C., Mowry, K. and Besse, F. (2012) Principles and roles of mRNA localization in animal development. Development, 139, 3263–3276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Glock, C., Heumuller, M. and Schuman, E.M. (2017) mRNA transport & local translation in neurons. Curr. Opin. Neurobiol., 45, 169–177. [DOI] [PubMed] [Google Scholar]
- 4. Conlon, E.G. and Manley, J.L. (2017) RNA-binding proteins in neurodegeneration: mechanisms in aggregate. Genes Dev., 31, 1509–1528. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Heraud-Farlow, J.E. and Kiebler, M.A. (2014) The multifunctional Staufen proteins: conserved roles from neurogenesis to synaptic plasticity. Trends Neurosci., 37, 470–479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Ravel-Chapuis, A., Belanger, G., Yadava, R.S., Mahadevan, M.S., DesGroseillers, L., Cote, J. and Jasmin, B.J. (2012) The RNA-binding protein Staufen1 is increased in DM1 skeletal muscle and promotes alternative pre-mRNA splicing. J. Cell Biol., 196, 699–712. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Paul, S., Dansithong, W., Figueroa, K.P., Scoles, D.R. and Pulst, S.M. (2018) Staufen1 links RNA stress granules and autophagy in a model of neurodegeneration. Nat. Commun., 9, 3648. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Gandelman, M., Dansithong, W., Figueroa, K.P., Paul, S., Scoles, D.R. and Pulst, S.M. (2020) Staufen 1 amplifies proapoptotic activation of the unfolded protein response. Cell Death Differ., 27, 2942–2951. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Taylor, J.P., Brown, R.H., Jr. and Cleveland, D.W. (2016) Decoding ALS: from genes to mechanism. Nature, 539, 197–206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. DeJesus-Hernandez, M., Mackenzie, I.R., Boeve, B.F., Boxer, A.L., Baker, M., Rutherford, N.J., Nicholson, A.M., Finch, N.A., Flynn, H., Adamson, J. et al. (2011) Expanded GGGGCC hexanucleotide repeat in noncoding region of C9ORF72 causes chromosome 9p-linked FTD and ALS. Neuron, 72, 245–256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Renton, A.E., Majounie, E., Waite, A., Simon-Sanchez, J., Rollinson, S., Gibbs, J.R., Schymick, J.C., Laaksovirta, H., van Swieten, J.C., Myllykangas, L. et al. (2011) A hexanucleotide repeat expansion in C9ORF72 is the cause of chromosome 9p21-linked ALS-FTD. Neuron, 72, 257–268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Gendron, T.F., Bieniek, K.F., Zhang, Y.J., Jansen-West, K., Ash, P.E., Caulfield, T., Daughrity, L., Dunmore, J.H., Castanedes-Casey, M., Chew, J. et al. (2013) Antisense transcripts of the expanded C9ORF72 hexanucleotide repeat form nuclear RNA foci and undergo repeat-associated non-ATG translation in c9FTD/ALS. Acta Neuropathol., 126, 829–844. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Lee, Y.B., Chen, H.J., Peres, J.N., Gomez-Deza, J., Attig, J., Stalekar, M., Troakes, C., Nishimura, A.L., Scotter, E.L., Vance, C. et al. (2013) Hexanucleotide repeats in ALS/FTD form length-dependent RNA foci, sequester RNA binding proteins, and are neurotoxic. Cell Rep., 5, 1178–1186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Ash, P.E., Bieniek, K.F., Gendron, T.F., Caulfield, T., Lin, W.L., Dejesus-Hernandez, M., van Blitterswijk, M.M., Jansen-West, K., Paul, J.W., 3rd, Rademakers, R. et al. (2013) Unconventional translation of C9ORF72 GGGGCC expansion generates insoluble polypeptides specific to c9FTD/ALS. Neuron, 77, 639–646. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Kanamori, T., Kanai, M.I., Dairyo, Y., Yasunaga, K., Morikawa, R.K. and Emoto, K. (2013) Compartmentalized calcium transients trigger dendrite pruning in Drosophila sensory neurons. Science, 340, 1475–1478. [DOI] [PubMed] [Google Scholar]
- 16. Freibaum, B.D. and Taylor, J.P. (2017) The role of dipeptide repeats in C9ORF72-related ALS-FTD. Front. Mol. Neurosci., 10, 35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Freibaum, B.D., Lu, Y., Lopez-Gonzalez, R., Kim, N.C., Almeida, S., Lee, K.H., Badders, N., Valentine, M., Miller, B.L., Wong, P.C. et al. (2015) GGGGCC repeat expansion in C9orf72 compromises nucleocytoplasmic transport. Nature, 525, 129–133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Jovicic, A., Mertens, J., Boeynaems, S., Bogaert, E., Chai, N., Yamada, S.B., Paul, J.W., 3rd, Sun, S., Herdy, J.R., Bieri, G. et al. (2015) Modifiers of C9orf72 dipeptide repeat toxicity connect nucleocytoplasmic transport defects to FTD/ALS. Nat. Neurosci., 18, 1226–1229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Rossi, S., Serrano, A., Gerbino, V., Giorgi, A., Di Francesco, L., Nencini, M., Bozzo, F., Schinina, M.E., Bagni, C., Cestra, G. et al. (2015) Nuclear accumulation of mRNAs underlies G4C2-repeat-induced translational repression in a cellular model of C9orf72 ALS. J. Cell Sci., 128, 1787–1799. [DOI] [PubMed] [Google Scholar]
- 20. Zhang, K., Donnelly, C.J., Haeusler, A.R., Grima, J.C., Machamer, J.B., Steinwald, P., Daley, E.L., Miller, S.J., Cunningham, K.M., Vidensky, S. et al. (2015) The C9orf72 repeat expansion disrupts nucleocytoplasmic transport. Nature, 525, 56–61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Saldi, T.K., Ash, P.E., Wilson, G., Gonzales, P., Garrido-Lecca, A., Roberts, C.M., Dostal, V., Gendron, T.F., Stein, L.D., Blumenthal, T. et al. (2014) TDP-1, the Caenorhabditis elegans ortholog of TDP-43, limits the accumulation of double-stranded RNA. EMBO J., 33, 2947–2966. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Zhang, Y.J., Guo, L., Gonzales, P.K., Gendron, T.F., Wu, Y., Jansen-West, K., O'Raw, A.D., Pickles, S.R., Prudencio, M., Carlomagno, Y. et al. (2019) Heterochromatin anomalies and double-stranded RNA accumulation underlie C9orf72 poly(PR) toxicity. Science, 363, eaav2606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Zhang, K., Grima, J.C., Rothstein, J.D. and Lloyd, T.E. (2016) Nucleocytoplasmic transport in C9orf72-mediated ALS/FTD. Nucleus-Phila, 7, 132–137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Neumann, M., Sampathu, D.M., Kwong, L.K., Truax, A.C., Micsenyi, M.C., Chou, T.T., Bruce, J., Schuck, T., Grossman, M., Clark, C.M. et al. (2006) Ubiquitinated TDP-43 in frontotemporal lobar degeneration and amyotrophic lateral sclerosis. Science, 314, 130–133. [DOI] [PubMed] [Google Scholar]
- 25. Lichtenstein, M., Guo, W. and Tartakoff, A.M. (2001) Control of nuclear export of hnRNP A1. Traffic, 2, 261–267. [DOI] [PubMed] [Google Scholar]
- 26. Ayala, Y.M., Zago, P., D'Ambrogio, A., Xu, Y.F., Petrucelli, L., Buratti, E. and Baralle, F.E. (2008) Structural determinants of the cellular localization and shuttling of TDP-43. J. Cell Sci., 121, 3778–3785. [DOI] [PubMed] [Google Scholar]
- 27. Chung, C.G., Kwon, M.J., Jeon, K.H., Hyeon, D.Y., Han, M.H., Park, J.H., Cha, I.J., Cho, J.H., Kim, K., Rho, S. et al. (2017) Golgi outpost synthesis impaired by toxic polyglutamine proteins contributes to dendritic pathology in neurons. Cell Rep., 20, 356–369. [DOI] [PubMed] [Google Scholar]
- 28. Clark, S.G., Graybeal, L.L., Bhattacharjee, S., Thomas, C., Bhattacharya, S. and Cox, D.N. (2018) Basal autophagy is required for promoting dendritic terminal branching in Drosophila sensory neurons. PLoS One, 13, e0206743. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Kwon, M.J., Han, M.H., Bagley, J.A., Hyeon, D.Y., Ko, B.S., Lee, Y.M., Cha, I.J., Kim, S.Y., Kim, D.Y., Kim, H.M. et al. (2018) Coiled-coil structure-dependent interactions between polyQ proteins and Foxo lead to dendrite pathology and behavioral defects. Proc. Natl. Acad. Sci. USA, 115, E10748–E10757. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Lee, S.B., Bagley, J.A., Lee, H.Y., Jan, L.Y. and Jan, Y.N. (2011) Pathogenic polyglutamine proteins cause dendrite defects associated with specific actin cytoskeletal alterations in Drosophila. Proc. Natl. Acad. Sci. USA, 108, 16795–16800. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Niehues, S., Bussmann, J., Steffes, G., Erdmann, I., Kohrer, C., Sun, L., Wagner, M., Schafer, K., Wang, G., Koerdt, S.N. et al. (2015) Impaired protein translation in Drosophila models for Charcot-Marie-Tooth neuropathy caused by mutant tRNA synthetases. Nat. Commun., 6, 7520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Woolums, B.M., McCray, B.A., Sung, H., Tabuchi, M., Sullivan, J.M., Ruppell, K.T., Yang, Y., Mamah, C., Aisenberg, W.H., Saavedra-Rivera, P.C. et al. (2020) TRPV4 disrupts mitochondrial transport and causes axonal degeneration via a CaMKII-dependent elevation of intracellular Ca(2). Nat. Commun., 11, 2679. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Han, M.H., Kwon, M.J., Ko, B.S., Hyeon, D.Y., Lee, D., Kim, H.J., Hwang, D. and Lee, S.B. (2020) NF-kappaB disinhibition contributes to dendrite defects in fly models of neurodegenerative diseases. J. Cell Biol., 219, e202004107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Machamer, J.B., Woolums, B.M., Fuller, G.G. and Lloyd, T.E. (2018) FUS causes synaptic hyperexcitability in Drosophila dendritic arborization neurons. Brain Res., 1693, 55–66. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Boeynaems, S., Bogaert, E., Michiels, E., Gijselinck, I., Sieben, A., Jovicic, A., De Baets, G., Scheveneels, W., Steyaert, J., Cuijt, I. et al. (2016) Drosophila screen connects nuclear transport genes to DPR pathology in c9ALS/FTD. Sci. Rep., 6, 20877. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Shi, K.Y., Mori, E., Nizami, Z.F., Lin, Y., Kato, M., Xiang, S., Wu, L.C., Ding, M., Yu, Y., Gall, J.G. et al. (2017) Toxic PRn poly-dipeptides encoded by the C9orf72 repeat expansion block nuclear import and export. Proc. Natl. Acad. Sci. USA, 114, E1111–E1117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Barbee, S.A., Estes, P.S., Cziko, A.M., Hillebrand, J., Luedeman, R.A., Coller, J.M., Johnson, N., Howlett, I.C., Geng, C., Ueda, R. et al. (2006) Staufen- and FMRP-containing neuronal RNPs are structurally and functionally related to somatic P bodies. Neuron, 52, 997–1009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Macchi, P., Brownawell, A.M., Grunewald, B., DesGroseillers, L., Macara, I.G. and Kiebler, M.A. (2004) The brain-specific double-stranded RNA-binding protein Staufen2 - Nucleolar accumulation and isoform-specific exportin-5-dependent export. J. Biol. Chem., 279, 31440–31444. [DOI] [PubMed] [Google Scholar]
- 39. Martel, C., Macchi, P., Furic, L., Kiebler, M.A. and DesGroseillers, L. (2006) Staufen1 is imported into the nucleolus via a bipartite nuclear localization signal and several modulatory determinants. Biochem. J., 393, 245–254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Ferrer, I. (1999) Neurons and their dendrites in frontotemporal dementia. Dement. Geriatr. Cogn. Disord., 10(Suppl 1), 55–60. [DOI] [PubMed] [Google Scholar]
- 41. Kweon, J.H., Kim, S. and Lee, S.B. (2017) The cellular basis of dendrite pathology in neurodegenerative diseases. BMB Rep., 50, 5–11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Nakano, I. and Hirano, A. (1987) Atrophic cell processes of large motor neurons in the anterior horn in amyotrophic lateral sclerosis: observation with silver impregnation method. J. Neuropathol. Exp. Neurol., 46, 40–49. [DOI] [PubMed] [Google Scholar]
- 43. Park, J.H., Chung, C.G., Seo, J., Lee, B.H., Lee, Y.S., Kweon, J.H. and Lee, S.B. (2020) C9orf72-Associated Arginine-Rich dipeptide repeat proteins reduce the number of golgi outposts and dendritic branches in Drosophila neurons. Mol Cells, 43, 821–830. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. St Johnston, D., Brown, N.H., Gall, J.G. and Jantsch, M. (1992) A conserved double-stranded RNA-binding domain. Proc. Natl. Acad. Sci. USA, 89, 10979–10983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Balendra, R. and Isaacs, A.M. (2018) C9orf72-mediated ALS and FTD: multiple pathways to disease. Nat. Rev. Neurol., 14, 544–558. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Saldi, T.K., Gonzales, P., Garrido-Lecca, A., Dostal, V., Roberts, C.M., Petrucelli, L. and Link, C.D. (2018) The Caenorhabditis elegans Ortholog of TDP-43 Regulates the Chromatin Localization of the Heterochromatin Protein 1 Homolog HPL-2. Mol. Cell. Biol., 38, e00668–e00617. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Liu, E.Y., Russ, J., Cali, C.P., Phan, J.M., Amlie-Wolf, A. and Lee, E.B. (2019) Loss of Nuclear TDP-43 Is Associated with Decondensation of LINE Retrotransposons. Cell Rep., 27, 1409–1421 e1406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Pontvianne, F., Blevins, T., Chandrasekhara, C., Mozgova, I., Hassel, C., Pontes, O.M., Tucker, S., Mokros, P., Muchova, V., Fajkus, J. et al. (2013) Subnuclear partitioning of rRNA genes between the nucleolus and nucleoplasm reflects alternative epiallelic states. Genes Dev., 27, 1545–1550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Macchi, P., Brownawell, A.M., Grunewald, B., DesGroseillers, L., Macara, I.G. and Kiebler, M.A. (2004) The brain-specific double-stranded RNA-binding protein Staufen2: nucleolar accumulation and isoform-specific exportin-5-dependent export. J. Biol. Chem., 279, 31440–31444. [DOI] [PubMed] [Google Scholar]
- 50. Kiebler, M.A., Jansen, R.P., Dahm, R. and Macchi, P. (2005) A putative nuclear function for mammalian Staufen. Trends Biochem. Sci., 30, 228–231. [DOI] [PubMed] [Google Scholar]
- 51. Martel, C., Macchi, P., Furic, L., Kiebler, M.A. and Desgroseillers, L. (2006) Staufen1 is imported into the nucleolus via a bipartite nuclear localization signal and several modulatory determinants. Biochem. J., 393, 245–254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Herrmann, D. and Parlato, R. (2018) C9orf72-associated neurodegeneration in ALS-FTD: breaking new ground in ribosomal RNA and nucleolar dysfunction. Cell Tissue Res., 373, 351–360. [DOI] [PubMed] [Google Scholar]
- 53. Hernandez-Verdun, D., Roussel, P., Thiry, M., Sirri, V. and Lafontaine, D.L.J. (2010) The nucleolus: structure/function relationship in RNA metabolism. Wires Rna, 1, 415–431. [DOI] [PubMed] [Google Scholar]
- 54. Corman, A., Jung, B., Haggblad, M., Brautigam, L., Lafarga, V., Lidemalm, L., Huhn, D., Carreras-Puigvert, J. and Fernandez-Capetillo, O. (2019) A chemical screen identifies compounds limiting the toxicity of C9ORF72 Dipeptide repeats. Cell Chem Biol, 26, 235–243 e235. [DOI] [PubMed] [Google Scholar]
- 55. Goodman, L.D., Prudencio, M., Kramer, N.J., Martinez-Ramirez, L.F., Srinivasan, A.R., Lan, M., Parisi, M.J., Zhu, Y., Chew, J., Cook, C.N. et al. (2019) Toxic expanded GGGGCC repeat transcription is mediated by the PAF1 complex in C9orf72-associated FTD. Nat. Neurosci., 22, 863–874. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Kapeli, K., Martinez, F.J. and Yeo, G.W. (2017) Genetic mutations in RNA-binding proteins and their roles in ALS. Hum. Genet., 136, 1193–1214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Moore, S., Rabichow, B.E. and Sattler, R. (2020) The Hitchhiker's Guide to Nucleocytoplasmic Trafficking in Neurodegeneration. Neurochem. Res., 45, 1306–1327. [DOI] [PubMed] [Google Scholar]
- 58. Ortega, J.A., Daley, E.L., Kour, S., Samani, M., Tellez, L., Smith, H.S., Hall, E.A., Esengul, Y.T., Tsai, Y.H., Gendron, T.F. et al. (2020) Nucleocytoplasmic Proteomic Analysis Uncovers eRF1 and Nonsense-Mediated Decay as Modifiers of ALS/FTD C9orf72 Toxicity. Neuron, 106, 90–107 e113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Fuerstenberg, S., Peng, C.Y., Alvarez-Ortiz, P., Hor, T. and Doe, C.Q. (1998) Identification of Miranda protein domains regulating asymmetric cortical localization, cargo binding, and cortical release. Mol. Cell. Neurosci., 12, 325–339. [DOI] [PubMed] [Google Scholar]
- 60. Wickham, L., Duchaine, T., Luo, M., Nabi, I.R. and DesGroseillers, L. (1999) Mammalian staufen is a double-stranded-RNA- and tubulin-binding protein which localizes to the rough endoplasmic reticulum. Mol. Cell. Biol., 19, 2220–2230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Marcel, V., Ghayad, S.E., Belin, S., Therizols, G., Morel, A.P., Solano-Gonzalez, E., Vendrell, J.A., Hacot, S., Mertani, H.C., Albaret, M.A. et al. (2013) p53 Acts as a safeguard of translational control by regulating fibrillarin and rRNA methylation in cancer. Cancer Cell, 24, 318–330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. Marques, J.T., Rebouillat, D., Ramana, C.V., Murakami, J., Hill, J.E., Gudkov, A., Silverman, R.H., Stark, G.R. and Williams, B.R. (2005) Down-regulation of p53 by double-stranded RNA modulates the antiviral response. J. Virol., 79, 11105–11114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63. Wang, R., Xu, X., Hao, Z., Zhang, S., Wu, D., Sun, H., Mu, C., Ren, H. and Wang, G. (2019) Poly-PR in C9ORF72-related amyotrophic lateral sclerosis/frontotemporal dementia causes neurotoxicity by Clathrin-dependent endocytosis. Neurosci. Bull., 35, 889–900. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64. Siprashvili, Z., Webster, D.E., Kretz, M., Johnston, D., Rinn, J.L., Chang, H.Y. and Khavari, P.A. (2012) Identification of proteins binding coding and non-coding human RNAs using protein microarrays. BMC Genomics, 13, 633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65. Heraud-Farlow, J.E., Sharangdhar, T., Li, X., Pfeifer, P., Tauber, S., Orozco, D., Hormann, A., Thomas, S., Bakosova, A., Farlow, A.R. et al. (2013) Staufen2 regulates neuronal target RNAs. Cell Rep., 5, 1511–1518. [DOI] [PubMed] [Google Scholar]
- 66. Chung, C.G., Lee, H. and Lee, S.B. (2018) Mechanisms of protein toxicity in neurodegenerative diseases. Cell. Mol. Life Sci., 75, 3159–3180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67. Woulfe, J.M. (2007) Abnormalities of the nucleus and nuclear inclusions in neurodegenerative disease: a work in progress. Neuropathol. Appl. Neurobiol., 33, 2–42. [DOI] [PubMed] [Google Scholar]
- 68. Buchner, G., Bassi, M.T., Andolfi, G., Ballabio, A. and Franco, B. (1999) Identification of a novel homolog of the Drosophila staufen protein in the chromosome 8q13-q21.1 region. Genomics, 62, 113–118. [DOI] [PubMed] [Google Scholar]
- 69. Hartmann, H., Hornburg, D., Czuppa, M., Bader, J., Michaelsen, M., Farny, D., Arzberger, T., Mann, M., Meissner, F. and Edbauer, D. (2018) Proteomics and C9orf72 neuropathology identify ribosomes as poly-GR/PR interactors driving toxicity. Life Sci Alliance, 1, e201800070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70. Bischof, J., Bjorklund, M., Furger, E., Schertel, C., Taipale, J. and Basler, K. (2013) A versatile platform for creating a comprehensive UAS-ORFeome library in Drosophila. Development, 140, 2434–2442. [DOI] [PubMed] [Google Scholar]
- 71. St Johnston, D., Beuchle, D. and Nusslein-Volhard, C. (1991) Staufen, a gene required to localize maternal RNAs in the Drosophila egg. Cell, 66, 51–63. [DOI] [PubMed] [Google Scholar]
- 72. Mills, W.K., Lee, Y.C.G., Kochendoerfer, A.M., Dunleavy, E.M. and Karpen, G.H. (2019) RNA from a simple-tandem repeat is required for sperm maturation and male fertility in Drosophila melanogaster. elife, 8, e48940. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73. Zimmerman, S.G., Peters, N.C., Altaras, A.E. and Berg, C.A. (2013) Optimized RNA ISH, RNA FISH and protein-RNA double labeling (IF/FISH) in Drosophila ovaries. Nat. Protoc., 8, 2158–2179. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


