Summary
Soybean, one of the most valuable oilseed crops, is under constant pressure from pathogens. bZIP transcription factors (TFs) composing one of the largest TF families in plants have diverse functions. Biochemical and physiological analyses were performed to characterize the regulatory roles of soybean bZIP TF GmbZIP15 in response to pathogens. We found that transgenic soybean plants overexpressing GmbZIP15 has increased resistance against Sclerotinia sclerotiorum and Phytophthora sojae. Besides, GmbZIP15 regulates pathogen response by modulating the antioxidant defense system and phytohormone signaling. In addition, we performed chromatin immunoprecipitation sequencing to identify the downstream genes of GmbZIP15 in response to S. sclerotiorum and found that GmbZIP15 can activate or repress the expression of defense-related genes through direct promoter binding. Taken together, these results indicate that GmbZIP15 plays a positive role in pathogen resistance in soybean, and this activity may be dependent on phytohormone signaling.
Subject area: Epigenetics, Plant Biology, Plant Pathology
Graphical abstract
Highlights
-
•
GmbZIP15 improves resistance against pathogen
-
•
GmbZIP15 modulates the antioxidant defense system
-
•
GmbZIP15 regulates phytohormone signaling
-
•
GmbZIP15 can direct bind to G-box
Epigenetics; Plant biology; Plant pathology;
Introduction
Environmental signals elicit cellular responses in living organisms, and these responses are crucial for survival. Soybean (Glycine max), a major oilseed crop and source of plant proteins, hosts a wide variety of pathogens that cause significant yield loss (Ranjan et al., 2019). The agronomical importance of soybean has led to research focused on its interactions with pathogens, including Phytophthora sojae, Phakopsora pachyrhizi, and Sclerotinia sclerotiorum (Hoffman et al., 1998; Whitham et al., 2016). The oomycete pathogen P. sojae belongs to the kingdom Stramenopiles, and the stem and root rot caused by this destructive pathogen costs the soybean industry millions of dollars each year (Tyler, 2007; Tyler et al., 2006). The necrotrophic ascomycete S. sclerotiorum is the causative agent of Sclerotinia stem rot or white mold, which causes significant yield loss and economic damage to soybean production (Westrick et al., 2019; Zhang et al., 2014).
Plant-pathogen interactions are a two-way communication process: plants recognize pathogens and induce different defense mechanisms, while pathogens threaten plant functional physiology and counterattack plant defense strategies (Cui et al., 2018; Silva et al., 2018). Invasion by most pathogens is recognized through transmembrane plant proteins called pattern recognition receptors that detect microbe-derived molecules known as pathogen-associated molecular pattern molecules (PAMPs). PAMP-triggered immunity (PTI) is activated by endogenous plant signals released during pathogen attack (Boutrot and Zipfel, 2017; Jones and Dangl, 2006). Another plant strategy for recognizing virulence effectors or their actions is effector-triggered immunity (ETI) (Cui et al., 2015; Mine et al., 2018). Activation of ETI leads to transcriptional upregulation of defense-related genes and is often associated with rapid localized cell death at the infection site, known as the hypersensitive response (HR) (Betsuyaku et al., 2018; Dodds and Rathjen, 2010).
Phytohormones, including abscisic acid (ABA), jasmonic acid (JA), ethylene (ET), and salicylic acid (SA), play vital roles in the regulation of plant-pathogen recognition and signal transduction pathways (Adie et al., 2007; Betsuyaku et al., 2018; Cui et al., 2018; Guo and Stotz, 2007; Liu et al., 2015; Ranjan et al., 2019). In plants under pathogen attack, PTI and ETI activate various hormone signaling pathways (Pieterse et al., 2012), resulting in a complex network of both complementary and antagonistic activities that ultimately fine-tune the defense response to pathogens (Adie et al., 2007; Betsuyaku et al., 2018; Di et al., 2017). A previous study has revealed that ABA can affect JA biosynthesis in the activation of defenses against Pythium irregulare in Arabidopsis (Adie et al., 2007). AcERF2, a halophyte Atriplex canescens ethylene-responsive factor (ERF), induced transcript accumulation of plant defense-related genes and increased Arabidopsis resistance to tomato Botrytis cinerea (Sun et al., 2018). BnWRKY33, as an S. sclerotiorum-responsive gene, positively regulates resistance to this pathogen by enhancing the expression of SA and JA-regulated genes in oilseed rape (Liu et al., 2018). Besides, overexpression of GmKR3 in soybean was found to enhance viral resistance in part through ABA signaling (Xun et al., 2019).
Effective signal transduction leads to the activation of transcription factors (TFs) that regulate the bioprocesses responsible for plant defense (Schluttenhofer and Yuan, 2015; Tsuda and Somssich, 2015). The bZIP genes encode plant-specific TFs composing one of the largest TF families in plants (Baloglu et al., 2014; Zhang et al., 2018) and plant bZIP TFs can be classified into three groups based on their DNA-binding specificity to G-box (CACGTG) or C-box (GACGTC) elements. It has reported that groupⅠ proteins exhibit a stronger binding affinity for G-box elements and group Ⅱ proteins bind G-box and C-box elements with comparable binding affinity, whereas group Ⅲ proteins display a stronger binding affinity for C-box motif (Izawa et al., 1993). The bZIP TFs of all three groups play diverse and critical roles in abiotic stress responses, flower development, pathogen defense, and seed maturation (Chang et al., 2019; Dong et al., 2019; Eleblu et al., 2019; Gaguancela et al., 2016). For example, StbZIP61 and StNPR3L cooperatively regulate the temporal activation of SA biosynthesis, which contributes to SA-mediated immunity against Phytophthora infestans infection in potato (Zhou et al., 2018). Overexpression of MebZIP3 and MebZIP5 enhanced callose deposition and improved disease resistance against cassava bacterial blight (Li et al., 2017). OsbZIP79 overexpression in rice resulted in suppression of the elicitor-inducible diterpenoid phytoalexin biosynthesis genes, leading to reduced phytoalexin accumulation in rice cells (Miyamoto et al., 2015). However, the roles of bZIP genes in soybean are still poorly understood, especially in response to pathogens.
Based on the gene structure and phylogenetic analysis, we previously divided the bZIP family in soybean into 12 groups and group K includes only one member named GmbZIP15 (Zhang et al., 2018). In this study, we isolated soybean GmbZIP15 (Glyma.02G161100) gene and found that the encoded protein contains a typical bZIP domain and activates transcription in yeast cells. Furthermore, biochemical and physiological analyses were performed to reveal the regulatory roles of GmbZIP15 in pathogen response.
Results
Sequence and domain analyses of GmbZIP15
Sequence analysis of GmbZIP15 was performed by aligning the amino acid sequence of 12 GmbZIP proteins with one member per subgroup. As shown in Figure S1, they shared the bZIP domain including a conserved 40–80 amino acid with two structural features, a basic DNA-binding region and a leucine zipper dimerization motif. The basic region contains an invariant N-×7-R/K-×9 motif of about 18 amino acid residues and the leucine zipper domain contains an L-×6-L-×6-L motif (Figures S1A and S1B). Surprisingly, only the member of the subgroup K GmbZIP15 contains a K1 domain, which is a transmembrane domain and has a conserved sequence QESAVL (Figures S1B and S1C). To further confirm whether K1 domain was conserved from different species or not, sequence alignment was performed with the six top-scoring matches of GmbZIP15 from basic local alignment search tool. Seven sequences from various plant species, including soybean, grape, poplar, rice, Arabidopsis, Thellungiella salsuginea, and Brachypodium distachyon display high similarity, including a bZIP domain and a K1 domain (Figure S1C), indicating that subgroup K own a specific K1 domain indeed. People have found this domain exists in Arabidopsis, rice, and black cottonwood (Corrêa et al., 2008), and we also found that this domain exists in grape, poplar, T. salsuginea, and B. distachyon.
Transcriptional activation activity and subcellular localization of GmbZIP15
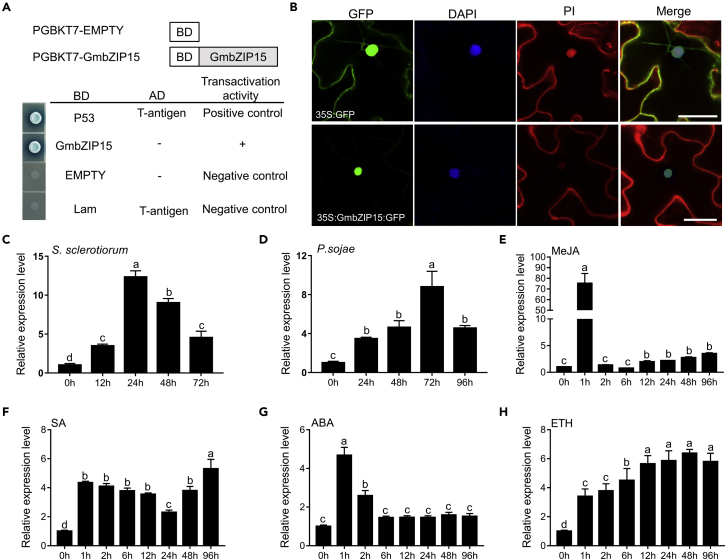
The bZIP domain is a highly conserved structural motif involved in transcriptional regulation (Hobo et al., 1999; Kim et al., 2004; Zhou et al., 2018). To test the transcription activation function of GmbZIP15, we performed a transient expression assay in yeast cells using a GAL4-responsive reporter system. As shown in Figure 1A, transformed yeast cells harboring DBD-P53+T-antigen (pGBKT7-53 + pGADT7-T, positive control) and DBD-GmbZIP15 (pGBKT7-GmbZIP15) grew well in synthetic dropout medium without tryptophan, histidine, and adenine [SD (-Trp/-His/-Ade)] and had α-galactosidase (α-gal) activity. Yeast cells containing empty pGBKT7 (negative control) exhibited no α-gal activity. This result indicated that GmbZIP15 protein has a transcriptional activation activity.
Figure 1.
Transcriptional activation and expression pattern analysis of GmbZIP15 in soybean
(A) Transcriptional activation analysis of GmbZIP15.
(B) Subcellular localization of GmbZIP15 in tobacco.
(C and D) Response of GmbZIP15 to pathogens infection in WT and GmbZIP15 transcript levels were detected by qPCR with S. sclerotiorum (C) and P. sojae (D) infection at different time point in soybean.
(E–H) GmbZIP15 transcript levels in soybean in response to hormone treatment. The relative expression level of GmbZIP15 in WT soybean was determined by qRT-PCR at 0, 1, 2, 6, 12, 24, 48, and 96 h after different phytohormone treatments. (E) 100 μM MeJA (methyl jasmonate). (F) 1 mM SA (salicylic acid). (G) 100 μM ABA (abscisic acid). (H) 100 μM ETH (ethephon). Errors bars indicate ±SD of three biological replicates. Significant differences between samples were determined by one-way ANOVA, P < 0.05.
To determine the subcellular location of GmbZIP15, we generated a 35S:GmbZIP15-GFP fusion construct and expressed the construct in Nicotiana benthamiana leaves. As shown in Figure 1B, GmbZIP15-GFP was exclusively localized in the nucleus, whereas the 35S-promoter-driven GFP control was observed in multiple subcellular compartments including the cytoplasm and nucleus. These results indicated that GmbZIP15 is a typical transcription factor.
Expression pattern of GmbZIP15 in response to various treatments
To evaluate whether GmbZIP15 is involved in response to pathogens infection, we performed quantitative polymerase chain reaction (qPCR) and examined the transcript levels of GmbZIP15 after S. sclerotiorum or P. sojae infection in wild-type soybean C03-3. The results showed that the expression of GmbZIP15 was significantly enhanced within 72 h postinoculation (hpi) with the S. sclerotiorum (Figure 1C). The transcript levels of GmbZIP15 were also elevated within 96 hpi with the P. sojae (Figure 1D). These results indicate that GmbZIP15 is differentially expressed upon S. sclerotiorum and P. sojae infection in soybean.
Because soybean defense against pathogens is known to be mediated by plant hormones, such as SA, JA, ET, and ABA (Xun et al., 2019; Yang et al., 2019a, 2019b; Zhang et al., 2019), we quantified GmbZIP15 transcript levels in response to hormone treatment by qPCR. GmbZIP15 was induced rapidly by jasmonic acid methyl ester (MeJA), SA, ABA, and ethephon (ETH) (Figures 1E–1H). The GmbZIP15 expression was rapidly induced by SA and ETH and kept a high expression level after 1-h treatment. GmbZIP15 expression in response to MeJA and ABA peaked at 1 h then decreased rapidly. These results suggest that the expression of GmbZIP15 is differentially regulated by different hormone treatments.
Functional analysis of GmbZIP15 in response to S. sclerotiorum
To determine the function of GmbZIP15 in response to S. sclerotiorum, we transformed soybean plants with a construct harboring GmbZIP15-GFP driven by the 35S promoter (OX-GmbZIP15) and a functional repressor form of GmbZIP15 (35S:bZIP15-SRDX) via Agrobacterium-mediated transformation and two lines with higher expression levels were selected for further research, respectively (Zhang et al., 2020). Hypocotyl-wound inoculation and detached-leaf inoculation were used to assess the response to S. sclerotiorum. As shown in Figure 2A, hypocotyl-wound inoculation was performed in 15-day-old wild-type (WT), OX-GmbZIP15 (OE), and 35S:GmbZIP15-SRDX (SRDX) seedlings with 48 plants per sample, which led to complete wilting of about 80% WT and SRDX plants at about 5 days postinoculation (dpi), while about 70%–80% of two OE lines developed less-severe disease symptoms and showed less tissue damage than WT plants. Besides, the detached-leaf inoculation assay was performed with the first pair ternate palmate leaves, which showed that two OE line leaves display increased resistance to S. sclerotiorum at 24 hpi, with smaller lesions and relative biomass of S. sclerotiorum on the leaves; the degree of resistance was similar between the SRDX lines and WT (Figures 2B–2D).
Figure 2.
Phenotype of GmbZIP15 in response to S. sclerotiorum
(A)15-day-old WT and GmbZIP15 transgenic soybean plants after about 5 days of S. sclerotiorum inoculation. Numbers in the panels denote the frequencies of the phenotypes shown. Bar = 1 cm.
(B) S. sclerotiorum lesion progression on soybean leaves.
(C and D) Lesion areas and relative S. sclerotiorum biomass measurement at 24 and 48 hpi. Errors bars indicate ±SD of three biological replicates. Significant differences between samples were determined by one-way ANOVA, P < 0.05.
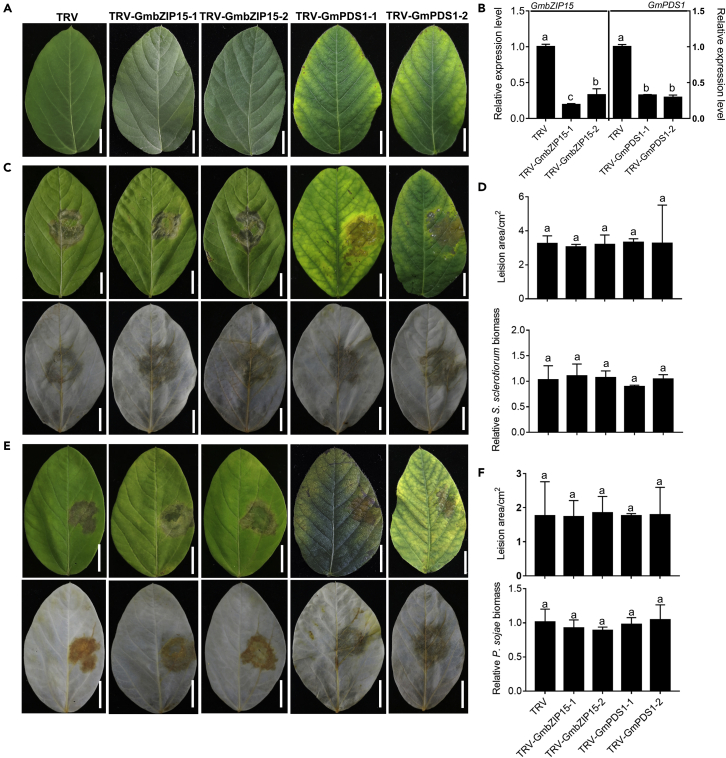
To further confirm the function of GmbZIP15, virus-induced gene silencing (VIGS) was used to knock down the expression of GmbZIP15. The silencing efficiency was determined by qPCR and compared with empty vector control (TRV). To evaluate the efficacy of our VIGS system in soybean, we silenced the soybean phytoene desaturase (GmPDS1), a gene involved in carotenoid biosynthesis (Zhang et al., 2010), and obtained consistent photobleaching of the host plants (Figure 3A), which was used as an additional control to determine the success of gene silencing. TRV-PDS as an additional control in the VIGS system has been reported in many species, such as pepper (Cai et al., 2015) and tomato (Naing et al., 2019). After about 18–24 dpi, the TRV-GmPDS1-infected leaves began to become albino (Figure 3A) and the expression of the target genes (GmbZIP15 and GmPDS1) were reduced to 20%–30% compared with the expression of these genes in empty vector control (Figure 3B). Compared with TRV-infected controls, plants infiltrated with TRV-GmPDS1 and TRV-GmbZIP15 displayed similar resistance to S. sclerotiorum (Figure 3C), with detached-leaf assay showed all plant leaves have similar lesion areas and relative biomass of S. sclerotiorum at 48 hpi (Figure 3D), which implies that the TRV construction did not affect plant resistance to S. sclerotiorum and knock down of GmbZIP15 did not change the resistance of soybean against S. sclerotiorum. Taken together, these results indicate that overexpression of GmbZIP15 improved resistance to S. sclerotiorum in soybean.
Figure 3.
Virus-induced gene silencing (VIGS) of GmbZIP15 showed similar resistance to wild type
(A) Soybean plants infected with TRV, TRV-GmbZIP15 or TRV-GmPDS1.
(B) Transcript level detection of GmbZIP15 and GmPDS1 in the first pair ternate palmate leaves of soybean.
(C) Phenotype of soybean plants with S. sclerotiorum inoculation for 48 h.
(D) Lesion areas and relative S. sclerotiorum biomass measurement at 48 h.
(E) Phenotype of soybean plants with P. sojae inoculation for 72 h.
(F) Lesion areas and relative P. sojae biomass measurement at 72 h. Errors bars indicate ±SD of three biological replicates. Significant differences between samples were determined by one-way ANOVA, P < 0.05.
Functional analysis of GmbZIP15 in response to P. sojae
To examine whether GmbZIP15 also functions against P. sojae, we inoculated soybean plants with P. sojae. After hypocotyl-wound inoculation, about 80% WT and SRDX plants exhibited similar wilting symptoms and chlorosis at about 7 dpi, and most (~85%) OE plants were healthier than WT (Figure 4A). The detached-leaf inoculation result showed that the lesion areas and relative biomass of P. sojae were similar on the leaves of SRDX and WT plants (Figures 4B–4D). At 48 hpi, the OE lines had less lesion areas and relative biomass of P. sojae than WT, the difference was more obvious at 72 hpi (Figures 4C and 4D). In addition, TRV-GmPDS1 and TRV-GmbZIP15 displayed similar resistance to P. sojae compared with TRV-infected controls (Figures 3E and 3F). These results indicate that GmbZIP15 also contributes resistance to P. sojae in soybean.
Figure 4.
Phenotype of GmbZIP15 in response to P. sojae
(A) Phenotypes of 15-day-old WT and GmbZIP15 transgenic soybean plants after about 7 days of P. sojae inoculation. Numbers in the panels denote the frequencies of the phenotypes shown.
(B) P. sojae lesion progression on soybean leaves. Bar = 1 cm.
(C and D) Lesion areas and relative P. sojae biomass measurement at 48 and 72 hpi. Errors bars indicate ±SD of three biological replicates. Significant differences between samples were determined by one-way ANOVA, P < 0.05.
GmbZ1P15-mediated pathogen defense involves the antioxidant defense system
Reactive oxygen (ROS) are key signaling molecules produced under biotic and abiotic stress conditions and trigger a variety of plant defense responses (Huckelhoven and Kogel, 2003). To better understand the resistance mechanism of GmbZIP15 transgenic soybean plants, we collected S. sclerotiorum- or P. sojae-inoculated leaves of WT and transgenic plants after 48 hpi or 72 hpi. We then stained them with 3,3-diaminobenzidine (DAB) to visualize H2O2 accumulation. After pathogen infection, more staining was observed in leaves of SRDX and WT plants, while less in leaves of OE plants exhibiting enhanced resistance to the pathogen (Figures 2B and 4B).
Malondialdehyde (MDA) is a typical product of lipid peroxidation, and its content indirectly reflects the degree of damage and antioxidant capacity (Yu et al., 2016). After infection with S. sclerotiorum and P. sojae, the MDA level was significantly elevated but that was distinctly lower in OE plants than that of WT plants, and the MDA concentration did not significantly differ between WT and SRDX plants (Figures S2A and S2B). To study whether the increased lipid peroxidation and ROS were caused by altered antioxidant activities, the activities of superoxide dismutase (SOD), peroxidase (POD), and catalase (CAT) were detected. These enzymes are important to plants for eliminating ROS (Alscher et al., 2002; Manju and Nair, 2006). Under mock and pathogen infection conditions, SOD, POD, and CAT activities were much higher in OE plants than in WT plants (Figures S2A and S2B). These results suggest that GmbZIP15 may reduce ROS accumulation in soybean by increasing antioxidant enzymatic activities.
GmbZ1P5-mediated pathogen defense involves phytohormone signaling
To further investigate how GmbZIP15 promotes resistance to S. sclerotiorum and P. sojae infection, we analyzed the expression of defense-related genes in WT and GmbZIP15 transgenic soybean plants. First, two HR-related genes (GmNPR1 and GmRAR1) were significantly elevated after pathogens infection (Figures S3 and S4). In addition, the expression of phytohormone-responsive genes, including ABA response genes (GmAA O 3A and GmNCED1), ET and JA response gene (GmERF1), and SA response gene (GmPR2), was also induced in OE lines compared with WT plants by S. sclerotiorum and P. sojae infection in a different degree, while the transcript level of these marker genes in SRDX plants was similar or slightly lower than that in WT plants (Figures S3 and S4). In addition, the expression of auxin-related gene GmWUS showed no significantly difference in all plants after pathogens infection. We also detected the biosynthetic genes of SA and JA and found that the transcript level of JA biosynthetic gene GmAOS was significantly higher in OE plants and that was similar or lower in SRDX plants compared with WT plants after S. sclerotiorum infection and was higher in SRDX plants than in WT plants after S. sclerotiorum and P. sojae infection. The expression of SA biosynthetic gene GmPAL showed no obvious difference in all plants (Figures S3 and S4).
Conservation of the GmbZIP15-mediated response to S. sclerotiorum in soybean and Arabidopsis
To investigate the functional conservation of GmbZIP15 in S. sclerotiorum plant interactions, we generated OX-GmbZIP15 and 35S:GmbZIP15-SRDX transgenic Arabidopsis plants (Zhang et al., 2020) and evaluated the effect of GmbZIP15 on the response to S. sclerotiorum in Arabidopsis. As shown in Figure S5, the OX-GmbZIP15 plants had improved resistance to S. sclerotiorum compared with WT plant, while there was no obvious difference between 35S:GmbZIP15-SRDX plants and WT plants. The S. sclerotiorum lesion areas were about 0.6 cm2 on the leaves of WT and two 35S:GmbZIP15-SRDX lines and about 0.3 cm2 on the leaves of two OX-GmbZIP15 lines at 24 hpi, and the relative biomass of S. sclerotiorum was significantly lower in two OX-GmbZIP15 lines than in WT plants (Figure S5A). These results suggest that GmbZIP15 enhanced the resistance of Arabidopsis to S. sclerotiorum. We also analyzed the function of AtbZIP60, the Arabidopsis homolog of GmbZIP15. The Atbzip60 mutant was more sensitive to S. sclerotiorum compared to WT and had larger lesion areas and relative biomass of S. sclerotiorum than WT, which was consistent with the DAB staining assay (Figure S5A). Besides, OX-GmbZIP15 could partially restore S. sclerotiorum resistance in Atbzip60 in lines (Figure S5A).
To further understand the causal factors behind the pathogen resistance of GmbZIP15 transgenic Arabidopsis plants, the expression levels of some defense-related genes were tested in WT, Atbzip60 mutant, and GmbZIP15 transgenic Arabidopsis after S. sclerotiorum infection. The transcript levels of AtABI2, AtABI5, AtERF1, AtACS6, and AtPDF1.2 were significantly elevated, whereas the transcript levels of AtAOC3, AtICS1, AtLOX4, and AtPR1 had no noticeable difference in OX-GmbZIP15 transgenic Arabidopsis plants compared with WT plants (Figure S5B). The expression level of most marker genes in Atbzip60 mutant was lower than or similar to that of WT. Besides, the expression level of most marker genes in 35S:GmbZIP15-SRDX transgenic Arabidopsis plants was lower than or equal to that of WT, except for AtICS1 and AtPDF1.2 at 12 h and AtABI2, AtABI5, AtACS6, AtERF1, AtLOX4, and ATPR1 at 24 h (Figure S5B). On one hand, ectopic overexpression 35S:GmbZIP15-SRDX construction might affect other endogenous genes expression in Arabidopsis, and somehow, these genes might influence the marker genes at only some time point. On the other hand, this result might cause by little differences between different individuals or by a little difference in the process of pathogen inoculating. Taken together, these results suggest that GmbZIP15 confers S. sclerotiorum resistance in Arabidopsis as it does in soybean.
Identification of GmbZIP15-binding sites by chromatin immunoprecipitation sequencing
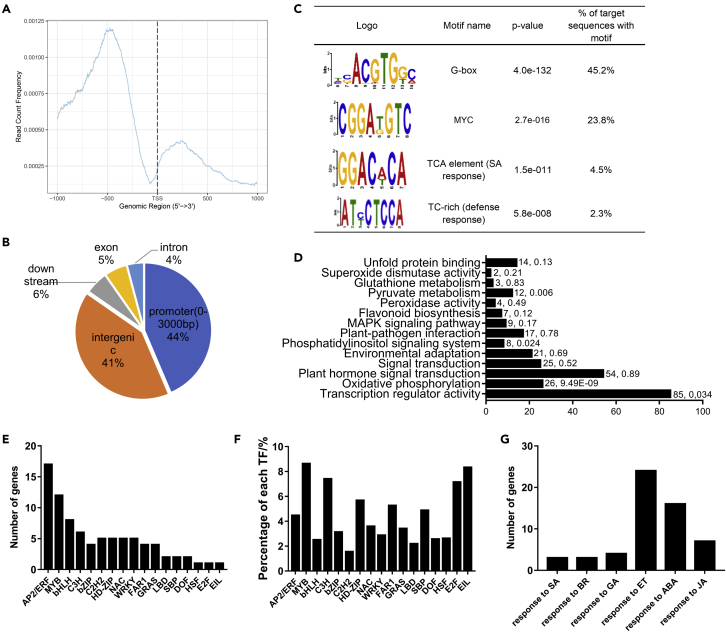
To identify the binding sites of GmbZIP15 throughout the genome, chromatin immunoprecipitation sequencing (ChIP-seq) was performed using the leaf tissues of OX-GmbZIP15-16 (35S:GmbZIP15:GFP) transgenic soybean plants under normal growth condition and a GFP antibody to pull down the putative GmbZIP15-bound DNA. After sequencing, we obtained 10,458,022 uniquely mapped reads, which were mainly located within 500 bp upstream of transcription start sites (TSS; Figure 5A). Using model-based ChIP-seq analysis software, we identified 1,865 peaks in OE plants (Table S2). As per the distribution of the GmbZIP15-binding sites in the soybean genome, 44% were within the 3,000-bp promoter regions upstream of the TSS and 41% were in the 5′ intergenic areas (Figure 5B). To analyze the binding motifs of GmbZIP15, flanking sequences (±100 bp) around the peaks were submitted to MEME-ChIP (http://meme-suite.org/tools/meme-chip) to identify consensus sequences. The most frequent sequence (about 45% of all identified peaks) in the analysis was ACGTG(G/T) (C/A) (Figure 5C), which is similar to the previously identified G-box (T/G/C/)ACGT(G/T)GC, with both containing an ACGT core (Hobo et al., 1999; Nijhawan et al., 2008; Zhang et al., 2017). The variation of the bases surrounding the ACGT core may determine the binding specificity of individual bZIP TFs. Among the peaks, we identified 1,647 candidate genes which may be directly bound by GmbZIP15. Gene ontology (GO)-based analysis of these genes revealed that the significantly enriched GO terms include the genes involved in transcriptional regulation, defense functions, including plant-pathogen interaction, plant hormone signal transduction, and some antioxidant metabolism processes (e.g., oxidative phosphorylation, phenylalanine metabolism, glutathione metabolism, SOD, and POD activity) (Figure 5D). These findings suggest that GmbZIP15 regulates the transcription of genes associated with a variety of plant defense processes (Figure 5D). As transcription factors (TFs) and phytohormones play a pivotal role in plant defense response, we picked TFs and phytohormone response genes up in detail (Table S3). We found that in total, there are 6 TF familie, including MYB, C3H, HD-ZIP, FAR1, E2F and EIL, containing more than 5% of family members were targeted by GmbZIP15 (Figures 5E and 5F and Table S3), indicating these genes are potentially regulated by GmbZIP15. Besides, there are four bZIP genes (GmbZIP60, GmABF, GmbZIP137, and GmbZIP159), accounting for about 3% of bZIP genes were targeted by GmbZIP15 (Figure 5E and Table S3). There are also many GmbZIP15 target genes response to ABA, ET and JA signaling regulation processes (Figure 5G and Table S4), such as GmABF1, GmERS1, GmJAZ8, GmLOX2 and GmLOX3.
Figure 5.
Identification of GmbZIP15-binding sites based on ChIP-seq data
(A) Enrichment of GmbZIP15 binding peaks in the promoter region. TSS, transcription start site.
(B) GmbZIP15 binding peak distribution in the soybean genome.
(C) Consensus sequence identified by MEME-ChIP among the GmbZIP15 binding peaks.
(D) Gene ontology analysis of genes targeted by GmbZIP15.
(E) Number of transcription factors targeted by GmbZIP15.
(F) The percentage of TFs in each TF family.
(G) Number of phytohormone-response genes targeted by GmbZIP15.
GmbZIP15 directly binds to the promoters of GmABF1, GmJAZ8 and GmERS1
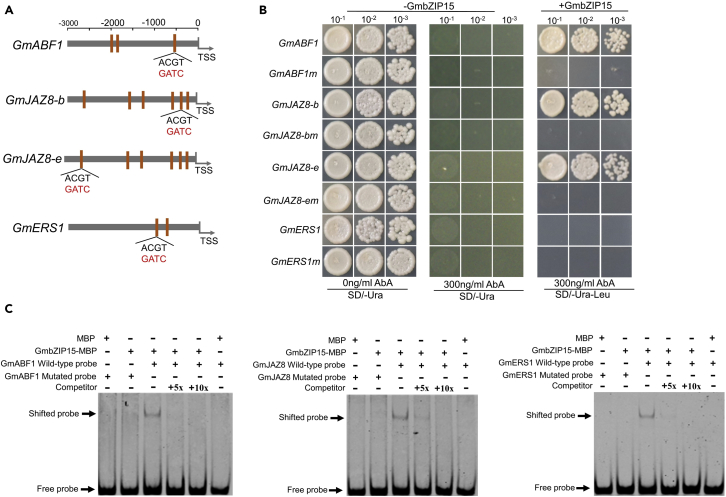
As phytohormones critically regulate the plant defense process (Lorenzo et al., 2003; Mine et al., 2018), three genes associated with ABA, JA and ET signaling (GmABF1, GmJAZ8 and GmERS1) were chosen randomly to examine their in vivo binding with GmbZIP15 and transcription profles by ChIP-qPCR. As shown in Figure S5, GmbZIP15 physically interacted with at least one site containing the G-box motif localized within the promoter region of each of the three candidate genes. A distinctive peak was detected approximately 1 kb upstream of the TSS of GmABF1(GmbZIP157) (Figure S6A), and ChIP-qPCR showed that GmbZIP15 specifically binds to the 600 bp region upstream of the GmABF1 TSS (Figure S6B). GmbZIP15 was also found to bind sites immediately upstream of the TSS of GmJAZ8 (sites b and e) and GmERS1 (sites b), as shown by ChIP-qPCR (Figure S5B). Besides, the binding specifity can also be found after S. sclerotiorum infection (Figure S6C).
To further confirm this result, yeast one-hybrid assay (Y1H) was performed. We mutated the core elements of binding site ACGT to GATC (Figure 6A), and a WT fragment and a mutated fragment of three genes were used as bait and cloned into the pAbAi vector, and GmbZIP15 was used as prey. Aureobasidin A (AbA) is an antifungal antibiotic produced by Aureobasidium pullulans R106, and AURI is a novel gene conferring aureobasidin resistance on Saccharomy cescerevisia, which can be used for screening in Y1H assay (Hashida-Okado et al., 1996; Takesako et al., 1993). As shown in Figure 6B, the yeast cells of all samples grew well on the screening medium (SD/-Ura). Moreover, cell growth of all mutated samples-GmbZIP15 prey transformats were completely inhibited by 300 ng/mL AbA, while most normal samples-GmbZIP15 prey transformats were survived except for JAZ8-e-GmbZIP15 transformat. The Y1H assay indicated that GmbZIP15 can only bind to the site b of GmJAZ8 promoter. We subsequently validated the binding regions of GmbZIP15 by performing an electrophoretic mobility shift assay (EMSA). MBP-GmbZIP15 fusion proteins could bind to the DNA probes, and the DNA-binding intensity of GmbZIP15 fusion proteins decreased due to competition from non-labeled probes, while the mutated probe was completely abolished (Figure 6C), indicating a binding specificity of GmbZIP15. Y1H and EMSA experimental evidence together suggest that GmbZIP15 can directly bind to the specific loci of GmABF1, GmJAZ8 and GmERS1 promoters.
Figure 6.
Validation of GmbZIP15 binding site and function analysis of three binding genes
(A) Diagram of the G-box distribution of GmABF1, GmJAZ8, and GmERS1 promoter fragments and wild-type G-box element (ACGT) was mutated to mG-box element (GATC).
(B)Yeast one-hybrid assay using vectors contains GmbZIP15 binding site or fragments with introduced mutations. Yeast cells carting or lacking pGAD-GmbZIP15 were grown on SD/-Ura or SD/-Ura, SD/-Ura/-Leu containing 300 ng/mL AbA.
(C) EMSA of the binding sites of GmbZIP15 protein to the promoter of GmABF1, GmJAZ8, and GmERS1. 5x or 10x competitor probe DNA (unlabeled) were added as control in the assay.
GmbZIP15 regulates the expression of GmABF1, GmJAZ8 and GmERS1 in response to S. sclerotiorum
To further investigate the function of these three genes, we detected the expression profile of these three genes by qPCR. Upon infection of OX-GmbZIP15 soybean plants with S. sclerotiorum, the transcript level of GmABF1, genes associated with ABA, was increased, suggesting direct and positive regulation by GmbZIP15 in response to pathogen infection (Figure S6D). As described above, GmbZIP15 also bound to GmERS1 and GmJAZ8, genes involved in ET and JA catabolism. Still, their transcript levels decreased in OE plant after infection (Figure S6D), indicating negative regulation by GmbZIP15 in response to pathogen infection. Subcellular localization analysis of the proteins encoded by these three genes showed that they were mainly located at the nucleus, except for GmERS1, which was detected in the cytoplasm and nucleus (Figure S7).
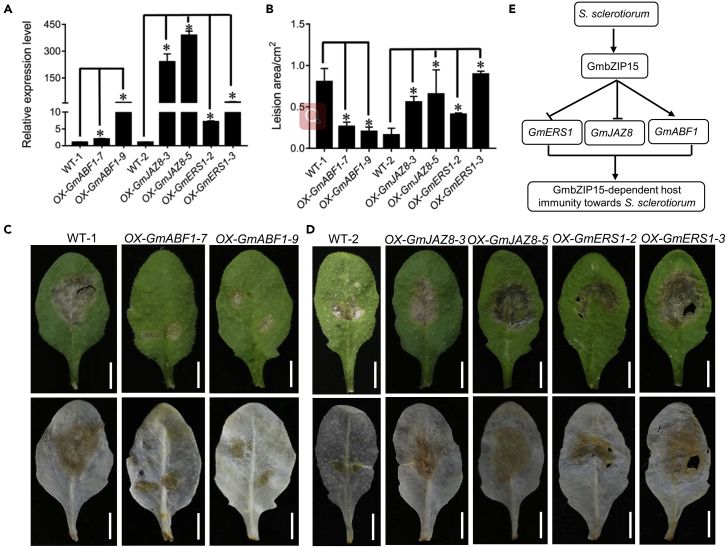
To confirm these expression changes after pathogen infection, these three genes were overexpressed in transgenic Arabidopsis, and their transcript levels were detected by qPCR in the T2 homozygous transgenic lines. As expected, the expression levels of the three genes in the corresponding transgenic lines were dramatically increased compared to WT (Figure 7A). GmABF1 overexpression in transgenic Arabidopsis conferred enhanced resistance to S. sclerotiorum, based on the smaller lesion areas and reduced DAB staining (Figures 7B and 7C), while the GmERS1 and GmJAZ8 overexpression lines showed hypersensitivity to this pathogen (Figure 7D). Therefore, in response to S. sclerotiorum, GmbZIP15 could directly activate GmABF1 expression and repress GmJAZ8 and GmERS1 by binding to their promoters (Figure 7E).
Figure 7.
Function analysis of three binding genes
(A) Transcript level of three genes overexpression in Arabidopsis.
(B) Lesion areas measurement after S. sclerotiorum infection. Errors bars indicate ±SD of three biological replicates. Significant differences between samples labeled with asterisk were determined by one-way ANOVA, P < 0.05.
(C) Phenotype observation of GmABF1 with S. sclerotiorum infection 24 h.
(D) Phenotype observation of GmJAZ8 and GmERS1 with S. sclerotiorum infection 12 h.
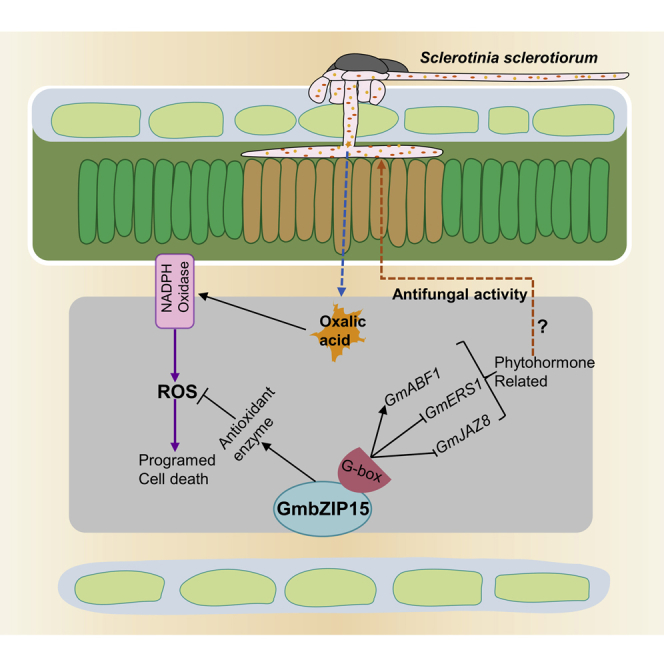
(E) Working model of GmbZIP15 responding to pathogen infection. GmbZIP15 play a positive role in the response of soybean to S. sclerotiorum infection. The expression of GmbZIP15 is induced by pathogen infection, and GmbZIP15 activates the phytohormone signaling pathways by binding to G-box of the promoter of the phytohormone-related genes, such as GmbZIP15 directly represses ET response genes such as GmERS1 and the JA signaling repressor GmJAZ8, but it directly enhances ABA signaling by activating GmABF1 to improve pathogen resistance.
Discussion
Plant resistance to pathogens, such as S. sclerotiorum, remains unclear due in part to the complex network of responses to these pathogens. bZIP TFs are involved in the regulation of various biological processes, such as plant development, abiotic stress and pathogen defense responses (Kim et al., 2004; Lee et al., 2006; Li et al., 2017). In soybean, the functions of bZIP proteins in response to pathogen infection largely remain uncharacterized. In this study, we investigated the roles of GmbZIP15 in response to pathogens infection. As a group K member, GmbZIP15 contains a K1 transmembrane domain, with a similar phenotype found in Arabidopsis and rice. Overexpression of GmbZIP15 in soybean did not cause any unexpected visible changes in agronomic traits. Notably, the OX-GmbZIP15 transgenic soybean plants have significantly increased resistance to S. sclerotiorum and P. sojae. However, the resistance of 35S:GmbZIP15-SRDX transgenic and TRV-GmbZIP15 penetration soybean plants were comparable to that of WT plants against these two pathogens.
ROS function as signaling molecules and is involved in host-pathogen interactions, including interactions with fungal and oomycete pathogens. Once plant-pathogen interaction is established, the rapid generation and accumulation of ROS, such as H2O2 or O2−, trigger programmed cell death (PCD), commonly known as the hypersensitive response (HR). HR may result in penetration failure of pathogens in their attempt to invade plant epidermal cells (Barna et al., 2003; Cheng et al., 2018). Upon pathogens challenge, the generation of H2O2 (Figures 2B and 4B) and the significantly elevated transcript levels of two HR-related genes GmNPR1 and GmRAR1 in OX-GmbZIP15 plants compared to WT (Figure S3), indicate a positive reaction of GmbZIP15 on pathogen invasion. Biotrophs feed on living host tissue, whereas necrotrophs kill host tissue and feed on the remains. However, in the case of necrotrophs, it seems that programmed cell death in the host would merely make life easier for the pathogen (Glazebrook and Jane, 2005). So in the next stage, other defense-related processes might be activited, such as POD, SOD, and CAT enzyme activities, induces to counter ROS deleterious effect and to prevent cell death (Yu et al., 2016). In this study, SOD, POD and CAT activities were much higher in OX-GmbZIP15 plants than in WT after pathogens infection (Figure S2A), which can eliminate the harmful substances produced by organisms during various metabolic processes (Choi et al., 2004). We also observed the enrichment of genes targeted by GmbZIP15 related to glutathione metabolism, SOD and POD activity (Figure 5D), as these processes are important in preventing oxidative damage imposed by pathogens (Morel et al., 2009; Ranjan et al., 2019). Thus, it is suggested that the scavenging of ROS in OX-GmbZIP15 transgenic plants is an important defense mechanism against S. sclerotiorum, which might in part be owing to the activation of antioxidant enzymatic activities in soybean.
However, defense signaling regulation pathways involved in the interaction between plants and pathogen are complex. The present study reveals that phytohormones (such as SA, JA, ET, and ABA) and their responsive marker genes play important roles in plants response to biotic stresses (Adie et al., 2007; Jing et al., 2019; Luan et al., 2019; Mazarei et al., 2007). In this study, the expression of GmbZIP15 after various hormone treatments (Figures 1E–1H) indicates its potential involvement in phytohormone pathways. Consistently, several marker genes responsive to ABA, JA, ET, and SA were significantly upregulated in the OX-GmbZIP15 transgenic soybean lines upon pathogen exposure (Figures S3 and S4). Furthermore, GmbZIP15 could directly bind to the promoter regions of GmABF1, GmERS1, and GmJAZ8, which are associated with the ABA, ET, and JA pathways, and these three genes were found to impact S. sclerotiorum resistance (Figures 7C and 7D). Moreover, GmbZIP15-dependent signaling through these pathways was demonstrated using well-characterized marker genes, such as AtAOC3, AtPDF1.2 AtERF1, AtACS6, AtABI2, AtABI5, AtICS1, and AtPR1 in Arabidopsis (Figure S5B). These results suggest that GmbZIP15 activates phytohormone signaling in response to pathogen invasion and that this pathway is conserved in soybean and Arabidopsis. The SA signaling pathway has been shown to protect against biotrophic fungi, oomycetes, and bacteria, such as Erysiphe orontii and Pseudomonas syringae, whereas JA signaling activates defense responses against many necrotrophic fungi, such as Alternaria brassicicola and B. cinerea (Glazebrook, 2005; Thomma et al., 1998). However, our study clearly indicated that the SA, ET/JA, and ABA signaling pathways were all involved in the response to S. sclerotiorum in the GmbZIP15-overexpressing soybean plants.
Some TFs also function to activate phytohormone signals upon pathogens or bacterial infection. In our study, TFs, such as ERF, bZIP, WRKY, and MYB gene families (Table S3), were targeted by GmbZIP15 and their homolog in Arabidopsis has been found to function in pathogen resistance. For example, overexpression of ethylene-responsive ERF1 in Arabidopsis confers resistance to some necrotrophic pathogens (Berrocal-Lobo et al., 2002), and ERF1 expression can be activated synergistically by ET and JA (Lorenzo et al., 2003). Overexpression of MYB102 in Arabidopsis increased susceptibility to green peach aphids (Myzus persicae) by promoting ET biosynthesis (Zhu et al., 2018). AtWRKY6 positively influenced pathogen-defense-associated PR1 promoter activity, most likely involving NPR1 function (Robatzek and Somssich, 2002). These results suggest that phytohormones play a critical role in pathogen response.
Taken together, these results suggest that altered expression of GmbZIP15 affects the pathogen-induced defense responses, including the trigger of HR response and scavenging of ROS and expression of defense genes associated with SA, ABA, JA, and ET signaling. Based on the results presented here, a working model of GmbZIP15 responding to pathogen infection can be proposed (Figure 7E). GmbZIP15 play a positive role in the response of soybean to S. sclerotiorum infection. The expression of GmbZIP15 is induced by pathogen infection, and GmbZIP15 activates the phytohormone signaling pathways by binding to G-box of the promoter of the phytohormone-related genes, such as GmbZIP15 directly represses ET response genes such as GmERS1 and the JA signaling repressor GmJAZ8, but it directly enhances ABA signaling by activating GmABF1 to improve pathogen resistance (Figure 7E). Collectively, our findings provide key insights into the pathogen response mechanisms in soybean.
Limitations of the study
In this study, we revealed that GmbZIP15 improved resistance against S. sclerotiorum and P. sojae by activating or repressing the expression of plant hormone-related genes (GmABF1, GmJAZ8, and GmERS1) through binding to the G-box of their promoters. GmbZIP15 regulates pathogen response by modulating the antioxidant enzymes activities. Additional studies are required to elucidate whether other phytohormones or antioxidant-system-related genes are involved in the downstream of GmABF1, GmJAZ8, or GmERS1 to regulate pathogen responses.
STAR★Methods
Key resources table
| REAGENT or RESOURCE | SOURCE | IDENTIFIER |
|---|---|---|
| Bacterial and virus strains | ||
| GV3101 | This paper | N/A |
| Experimental models: Organisms | ||
| Arabidopsis: OX-GmABF1 | This paper | N/A |
| Arabidopsis: OX-GmERS1 | This paper | N/A |
| Arabidopsis: OX-GmJAZ8 | This paper | N/A |
| Arabidopsis: SALK-050203C | Tair | N/A |
| Soybean: OX-GmbZIP15 | This paper | N/A |
| Soybean: 35S:GmbZIP15-SRDX | This paper | N/A |
| Software | ||
| MEME-ChIP | This paper | http://meme-suite.org/tools/meme-chip |
| Deposited data | ||
| Raw ChIP-seq data | This paper | PRJEB44708 |
| Recombinant DNA | ||
| Plasmid: pGWB605 | Cai et al., 2017 | N/A |
| TRV-GmbZIP15 | This paper | N/A |
| 35S:GmbZIP15 | This paper | N/A |
| 35S:GmbZIP15-SRDX | This paper | N/A |
Resource availability
Lead contact
Further information and requests for resources and reagents should be directed to and will be fulfilled by the lead contact, Yuan Qin (yuanqin@fafu.edu.cn).
Materials availability
This study did not generate new unique reagents.
Data and code availability
The original ChIP-seq data is available at European Nucleotide Archive (ENA) under accession number PRJEB44708.
Experimental model and subject details
Soybean plant material
Soybean ecotype C03-3 was used in this study. Soybean seeds were sown in soil and grown at 25°C with a 16-h light/8-h dark photoperiod. Agrobacterium-mediated transformation was performed with an expression construct harboring the OX-GmbZIP15 and 35S: GmbZIP15-SRDX (Yang et al., 2019b). To screen the transgenic plants progenies and obtain homozygous lines, both glufosinate spraying and polymerase chain reaction (PCR) analysis were conducted.
Pathogens infection
Fifteen-day-old soybean plants were infected with Sclerotinia sclerotiorum and Phytophthora sojae, which were collected from Fujian Agriculture and Forestry University, Fuzhou, China. The fungus S. sclerotiorum was germinated to produce hyphal inoculum on potato dextrose agar and P. sojae was cultivated at 25 °C for 7 days on V8 juice agar. The detached-leaf inoculation assay was performed based on the description reported by Yang et al. In a Petri dish, mycelial plugs were placed on the completely expanded leaves from 15-day-old soybean plants. Eight leaves were inoculated for each transgenic line and WT plants. Lesion areas on the detached leaves were monitored and photographed at 24 h or 48 h after inoculation. Hypocotyl-wound inoculation was performed based on previous research with 48 plants per samples. Briefly, a wound about 1 cm in length was cut into the hypocotyl epidermis 1 cm below the cotyledon of 15-day-old soybean plants, and then, it was inoculated with mycelial plugs. After inoculation, plants were placed in a moisturizing room at 23–25°C with 100% humidity for 48 h and then transferred to a greenhouse for further cultivation at 25°C.
Method details
Vector construction and transformation
To generate the overexpression construct, the coding DNA sequences (CDS) of GmbZIP15 (Glyma.02G161100), GmABF1 (GmbZIP157, Glyma.20G049200), GmJAZ8 (Glyma.05G141200), and GmERS1 (Glyma.03G216700) were amplified, and 35S: GmbZIP15-SRDX was generated by amplifying GmbZIP15 cDNA sequence and an SRDX motif was added to the end of the cDNA sequence (ctagatctggatctagaactccgtttgggtttcgcttaa). All the PCR fragment was cloned into the pENTR/D-TOPO vector (Invitrogen), and the pENTR clones were recombined into the destination vector pGWB605 (with 35S promoter) using LR Clonase II (Invitrogen). The resulting construct also contained the selectable marker BAR for glufosinate resistance (Cai et al., 2017; Zhao et al., 2018). All the primers used in this article are listed in Table S1.
Virus-induced gene silencing of GmbZIP15 in soybean plants
To make the GmbZIP15 silencing construct, primers (Forward: 5’-ctcacgcgtctcgaggcccCCGGGTATTACAAGCTCTTCGAG-3’, Reverse: 5’-atgtcttcgggacatgcccCACATACAACTTCTTCCTCTCCC-3’) were used to amplify a 212-bp fragment. The amplified fragment was ligated into the vector pTRV: RNA2 at BamHI and SmlI sites. Silencing was monitored using the construct pTRV: RNA2-GmPDS1, which targets the soybean phytoene desaturase (PDS), leading to photobleaching of plants. The Agrobacterium was then resuspended in agroinfiltration buffer (10 mM MgCl2, 10 mM MES pH 5.7, 100 μM acetosyringone, OD600=0.8). Each agrosuspension containing the TRV2 derivatives (TRV2-GmPDS1 and TRV2-GmbZIP15) was mixed with TRV1 and then infiltrated in ten-day-old soybean leaves. After about 18-24 dpi, the transcript levels of the first pair ternate palmate leaves were examined by qPCR.
Soybean seedling phytohormone treatments
For phytohormone treatments, SA (1 mM), MeJA (100 μM), ETH (100 μM), and ABA (100 μM) were dissolved in 0.01% Tween 20 and sprayed onto 15-day-old WT soybean young leaves, and soybean leaves were collected at 0, 1, 2, 6,12, 24, and 48 h. The control leaves were sprayed with an equal volume of 0.01% (v/v) Tween 20.
Arabidopsis plant material and pathogen infection
Arabidopsis ecotype Col-0 was used in this study. All the constructs were transformed into WT Arabidopsis (Col-0) plants by the floral dip method, and transgenic plants screening was performed as described previously. The T-DNA mutant Atbzip60 (SALK_050203) was obtained from the Arabidopsis Biological Resource Center. Arabidopsis seeds were sterilized and placed on 1/2 MS medium. After stratification at 4°C for 3 days, the plates were kept at 22°C with a 16-h light/8-h dark photoperiod for plant growth, followed by infection with S. sclerotiorum in 4-week-old plants. The detached-leaf inoculation assay was performed.
Diaminobenzidine staining
Following the method reported by Zhang et al. for hydrogen peroxide (H2O2) detection (Zhang et al., 2014), diaminobenzidine (DAB) staining was performed in the leaves of pathogen-treated plants. Briefly, the harvested leaves were vacuum-infiltrated for 20 min with Tris-HCl (pH 7.4) containing 1 % (w/v) DAB. The leaves were placed in light for 10 h and then boiled for 20 min in 75 % ethanol.
RNA extraction and quantitative real-time PCR
Total RNA was extracted using TRIzol (Invitrogen, Carlsbad, CA, USA) and reverse-transcribed using the PrimeScript RT-PCR kit (TaKaRa). Real-time PCR was performed with specific primers to analyze the relative transcript levels of selected genes (Table S1), the Bio-Rad Real-time PCR system (Foster City, CA, USA) using SYBR Premix Ex Taq II (TaKaRa Perfect Real Time). The qPCR program was 95°C for 30 s; 40 cycles of 95°C for 5 s and 60°C for 34 s; and 95°C for 15 s. GmActin was used for normalization.
Transcriptional activation analysis in yeast cells
The GmbZIP15 ORF was introduced into pGBKT7 to generate pGBKT7-GmbZIP15. The yeast strain AH109 was transformed with pGBKT7-53 + pGADT7-T, pGBKT7-GmbZIP15, and pGBKT7. The transformed cells were grown on SD (-Trp), SD (-Trp/--His/-Ade), and SD (-Trp/-His/-Ade/α-gal). The transactivation activity of proteins was detected by the growth status and α-gal activity.
Determination of antioxidant enzyme activity
Superoxide dismutase, peroxidase, catalase, and malondialdehyde enzymes were extracted from approximately 0.1 g of leaves using 1 mL extraction solution. The enzyme activities were measured as per the protocol from the Solarbio Biochemical Assay Division.
Subcellular localization
To examine GmbZIP15 localization in planta, the Agrobacteria containing the construct 35S:GmbZIP15-GFP was resuspended in infiltration media (10 mM ethanesulfonic acid [pH 5.8], 10 mM MgCl2, and 200 μM acetosyringone) before infiltration into the leaves of 30-day-old Nicotiana benthamiana plants. Two days later, leaf discs were observed under a confocal microscope (Leica TCS SP8X DLS) for GFP.
Chromatin immunoprecipitation assay
Chromatin immunoprecipitation (ChIP) was performed using a GFP antibody as previously described (Dai et al., 2018) with minor modifications. For each ChIP experiment, 3.0 g of soybean leaves was used, and three biological replicates were performed because the mapping rate of two samples was too low to conduct further analysis, so only one sample data could be used for further analysis. First, cross-linked chromatin was fragmented using 4 units of micrococcal nuclease (Sigma) in 1 ml of MNase digestion buffer (10 % sucrose, 50 mM Tris–HCl [pH 7.5], 25 mM MgCl2, 1 mM CaCl2), and then, the digestion was stopped with 5 Mm EDTA. All ChIP experiments were performed in a buffer containing 50 mM HEPES (pH 7.5), 1 mM EDTA (pH8.0), 150 mM NaCl, 1% Triton X-100, and 13 protease inhibitor cocktail (Roche). The ChIP-seq libraries were prepared as previously described (Zhang et al., 2012) then sequenced on the Illumina HiSeq 2500 platform. The ChIP-seq sequence reads were mapped to the phytozome v12.1 reference genome (https://phytozome.jgi.doe.gov/pz/portal.html) (Table S2). ChIP-Seq data processing and analysis were performed as described previously (Zong et al., 2013).
Yeast one-hybrid assays
GmbZIP15 CDS without a stop codon was amplified and then integrated into the pGADT7-T vector by an In-Fusion cloning kit (Clontech, Takara) to form a pGADT7-GmbZIP15 bait report vector. Based on the predicted binding site in our ChIP-seq data, the normal or mutational fragments were synthesized by DNA synthesis technology (Sangon Biotech, Shanghai, China; Table S1) and cloned into a pABAi vector by In-Fusion technology to form pABAi-GmABF1/pABAi-GmABF1m, pABAi-GmERS1/pABAi-GmERS1m, pABAi-GmJAZ8/pABAi-GmJAZ8m, and pABAi-GmEIX1/pABAi-GmEIX1m prey report vectors.
Yeast one-hybrid was carried out as per instructions provided by Clontech (Takara). Prey was transformed into Y1H gold yeast strain and cultured on SD/-Ura or SD/-Ura/-Leu medium with or without 300 ng/mL aureobasidin A (AbA) for 3 days. Furthermore, the yeast cells cotransformed by prey and bait were cultured on SD/-Ura/-Leu medium containing 300 ng/mL AbA for 3 days.
Electrophoretic mobility shift assays
The CDS of GmbZIP15 was amplified and inserted into the vector pMAL (NEB) to fuse with the maltose-binding protein (MBP) CDS, generating the recombinant MBP-GmbZIP15 protein. The resulting construct was expressed in Escherichia coli strain BL21(DE3) cells, the recombinant protein was expressed and purified using Amylose Resin (New England Biolabs, cat. no. E8021S). The probes were synthesized and labeled with Cy5, and different concentrations of nonlabeled probe were added to the reactions for competition. Finally, the DNA-protein complexes were electrophoresed on 6 % nondenaturing polyacrylamide gels in an ice water bath.
Quantification and statistical analysis
Analysis of variance was used to calculate the least significant differences between mean values by the t-test at P = 0.05 using SPSS software (v. 17.0). The type of statistical test, the number of replicates and whether error bars denote standard deviation or the standard error of the mean, is specified in each figure legend.
Acknowledgment
This work was supported by the Science and Technology Program of Fujian Province (2019N5008), the Science and Technology Major Project of Guangxi (Gui Ke 2018-266-Z01) and the National Natural Science Foundation of China (31970333) to Y.Q.
Author contributions
MZ performed vector construction and phenotype analysis. YL performed RNA-seq and ChIP-seq analysis. ZL, MC, and QH calculated all the data and prepared the plastic sections. ZS, YH, and FC performed qPCR analysis. HC performed yeast hybridization, SS performed soybean transformation, and HC and YQ designed the research. MZ and YQ wrote the manuscript. MA, BW, and YQ revised the manuscript. All authors have read and approved the manuscript.
Declaration of interests
The authors declare no competing interests.
Published: June 25, 2021
Footnotes
Supplemental information can be found online at https://doi.org/10.1016/j.isci.2021.102642.
Contributor Information
Bingrui Wang, Email: brwang@mail.hzau.edu.cn.
Hanyang Cai, Email: caihanyang123@163.com.
Yuan Qin, Email: yuanqin@fafu.edu.cn.
Supplemental information
References
- Adie B.A., Perez-Perez J., Perez-Perez M.M., Godoy M., Sanchez-Serrano J.J., Schmelz E.A., Solano R. ABA is an essential signal for plant resistance to pathogens affecting JA biosynthesis and the activation of defenses in Arabidopsis. Plant Cell. 2007;19:1665–1681. doi: 10.1105/tpc.106.048041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alscher R.G., Erturk N., Heath L.S. Role of superoxide dismutases (SODs) in controlling oxidative stress in plants. J. Exp. Bot. 2002;53:1331–1341. doi: 10.1093/jexbot/53.372.1331. [DOI] [PubMed] [Google Scholar]
- Baloglu M.C., Eldem V., Hajyzadeh M., Unver T. Genome-wide analysis of the bZIP transcription factors in cucumber. PLoS One. 2014;9:e96014. doi: 10.1371/journal.pone.0096014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barna B., Fodor J., Pogany M., Kiraly Z. Role of reactive oxygen species and antioxidants in plant disease resistance. Pest Manag. Sci. 2003;59:459–464. doi: 10.1002/ps.706. [DOI] [PubMed] [Google Scholar]
- Berrocal-Lobo M., Molina A., Solano R. Constitutive expression of ETHYLENE-RESPONSE-FACTOR1 in Arabidopsis confers resistance to several necrotrophic fungi. Plant J. 2002;29:23–32. doi: 10.1046/j.1365-313x.2002.01191.x. [DOI] [PubMed] [Google Scholar]
- Betsuyaku S., Katou S., Takebayashi Y., Sakakibara H., Nomura N., Fukuda H. Salicylic acid and jasmonic acid pathways are activated in spatially different domains around the infection site during effector-triggered immunity in Arabidopsis thaliana. Plant Cell Physiol. 2018;59:8–16. doi: 10.1093/pcp/pcx181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boutrot F., Zipfel C. Function, discovery, and exploitation of plant pattern recognition receptors for broad-spectrum disease resistance. Annu. Rev. Phytopathol. 2017;55:257–286. doi: 10.1146/annurev-phyto-080614-120106. [DOI] [PubMed] [Google Scholar]
- Cai H.Y., Yang Y., Xiao Z.L., Cheng J.B., Qiu A.L. CaWRKY6 transcriptionally activates CaWRKY40, regulates Ralstonia solanacearum resistance, and confers high-temperature and high-humidity tolerance in pepper. J. Exp. Bot. 2015;66:3163–3174. doi: 10.1093/jxb/erv125. [DOI] [PubMed] [Google Scholar]
- Cai H., Zhao L., Wang L., Zhang M., Su Z., Cheng Y., Zhao H., Qin Y. ERECTA signaling controls Arabidopsis inflorescence architecture through chromatin-mediated activation of PRE1 expression. New Phytol. 2017;214:1579–1596. doi: 10.1111/nph.14521. [DOI] [PubMed] [Google Scholar]
- Chang H.C., Tsai M.C., Wu S.S., Chang I.F. Regulation of ABI5 expression by ABF3 during salt stress responses in Arabidopsis thaliana. Bot. Stud. 2019;60:16. doi: 10.1186/s40529-019-0264-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheng Q., Dong L., Gao T., Liu T., Li N., Wang L., Chang X., Wu J., Xu P., Zhang S. The bHLH transcription factor GmPIB1 facilitates resistance to Phytophthora sojae in Glycine max. J. Exp. Bot. 2018;69:2527–2541. doi: 10.1093/jxb/ery103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Choi D.G., Yoo N.H., Yu C.Y., de Los Reyes B., Yun S.J. The activities of antioxidant enzymes in response to oxidative stresses and hormones in paraquat-tolerant Rehmannia glutinosa plants. J. Biochem. Mol. Biol. 2004;37:618–624. doi: 10.5483/bmbrep.2004.37.5.618. [DOI] [PubMed] [Google Scholar]
- Corrêa L., Riaño-Pachón D., Schrago C.G., Santos R., Mueller-Roeber B., Vincentz M. The role of bZIP transcription factors in green plant evolution: adaptive features emerging from four founder genes. PLoS One. 2008;3:e2944. doi: 10.1371/journal.pone.0002944. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cui H., Qiu J., Zhou Y., Bhandari D.D., Zhao C., Bautor J., Parker J.E. Antagonism of transcription factor MYC2 by EDS1/PAD4 complexes bolsters salicylic acid defense in Arabidopsis effector-triggered immunity. Mol. Plant. 2018;11:1053–1066. doi: 10.1016/j.molp.2018.05.007. [DOI] [PubMed] [Google Scholar]
- Cui H., Tsuda K., Parker J.E. Effector-triggered immunity: from pathogen perception to robust defense. Annu. Rev. Plant Biol. 2015;66:487–511. doi: 10.1146/annurev-arplant-050213-040012. [DOI] [PubMed] [Google Scholar]
- Dai X., Bai Y., Zhao L., Dou X., Liu Y., Wang L., Li Y., Li W., Hui Y., Huang X. H2A.Z represses gene expression by modulating promoter nucleosome structure and enhancer histone modifications in Arabidopsis. Mol. Plant. 2018;11:635. doi: 10.1016/j.molp.2018.03.011. [DOI] [PubMed] [Google Scholar]
- Di X., Gomila J., Takken F.L.W. Involvement of salicylic acid, ethylene and jasmonic acid signalling pathways in the susceptibility of tomato to Fusarium oxysporum. Mol. Plant Pathol. 2017;18:1024–1035. doi: 10.1111/mpp.12559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dodds P.N., Rathjen J.P. Plant immunity: towards an integrated view of plant-pathogen interactions. Nat. Rev. Genet. 2010;11:539–548. doi: 10.1038/nrg2812. [DOI] [PubMed] [Google Scholar]
- Dong Q., Xu Q., Kong J., Peng X., Zhou W., Chen L., Wu J., Xiang Y., Jiang H., Cheng B. Overexpression of ZmbZIP22 gene alters endosperm starch content and composition in maize and rice. Plant Sci. 2019;283:407–415. doi: 10.1016/j.plantsci.2019.03.001. [DOI] [PubMed] [Google Scholar]
- Eleblu J.S.Y., Haraghi A., Mania B., Camps C., Rashid D., Morin H., Dogimont C., Boualem A., Bendahmane A. The gynoecious CmWIP1 transcription factor interacts with CmbZIP48 to inhibit carpel development. Sci. Rep. 2019;9:15443. doi: 10.1038/s41598-019-52004-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gaguancela O.A., Zuniga L.P., Arias A.V., Halterman D., Flores F.J., Johansen I.E., Wang A., Yamaji Y., Verchot J. The IRE1/bZIP60 pathway and bax inhibitor 1 suppress systemic accumulation of potyviruses and potexviruses in Arabidopsis and Nicotiana benthamiana plants. Mol. Plant Microbe Interact. 2016;29:750–766. doi: 10.1094/MPMI-07-16-0147-R. [DOI] [PubMed] [Google Scholar]
- Glazebrook, Jane Contrasting mechanisms of defense against biotrophic and necrotrophic pathogens. Annu. Rev. Phytopathol. 2005;43:205–227. doi: 10.1146/annurev.phyto.43.040204.135923. [DOI] [PubMed] [Google Scholar]
- Glazebrook J. Contrasting mechanisms of defense against biotrophic and necrotrophic pathogens. Annu. Rev. Phytopathol. 2005;43:205–227. doi: 10.1146/annurev.phyto.43.040204.135923. [DOI] [PubMed] [Google Scholar]
- Guo X., Stotz H.U. Defense against Sclerotinia sclerotiorum in Arabidopsis is dependent on jasmonic acid, salicylic acid, and ethylene signaling. Mol. Plant Microbe Interact. 2007;20:1384–1395. doi: 10.1094/MPMI-20-11-1384. [DOI] [PubMed] [Google Scholar]
- Hashida-Okado T., Ogawa A., Endo M., Yasumoto R., Takesako K., Kato I. AUR1, a novel gene conferring aureobasidin resistance on Saccharomyces cerevisiae: a study of defective morphologies in Aur1p-depleted cells. Mol. Gen. Genet. 1996;251:236–244. doi: 10.1007/BF02172923. [DOI] [PubMed] [Google Scholar]
- Hobo T., Kowyama Y., Hattori T. A bZIP factor, TRAB1, interacts with VP1 and mediates abscisic acid-induced transcription. Proc. Natl. Acad. Sci. U S A. 1999;96:15348–15353. doi: 10.1073/pnas.96.26.15348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoffman D.D., Hartman G.L., Mueller D.S., Leitz R.A., Nickell C.D., Pedersen W.L. Yield and seed quality of soybean cultivars infected with Sclerotinia sclerotiorum. Plant Dis. 1998;82:826–829. doi: 10.1094/PDIS.1998.82.7.826. [DOI] [PubMed] [Google Scholar]
- Huckelhoven R., Kogel K.H. Reactive oxygen intermediates in plant-microbe interactions: who is who in powdery mildew resistance? Planta. 2003;216:891–902. doi: 10.1007/s00425-003-0973-z. [DOI] [PubMed] [Google Scholar]
- Izawa T., Foster R., Chua N.H. Plant bZIP protein DNA binding specificity. J. Mol. Biol. 1993;230:1131–1144. doi: 10.1006/jmbi.1993.1230. [DOI] [PubMed] [Google Scholar]
- Jing Y., Liu J., Liu P., Ming D., Sun J. Overexpression of TaJAZ1 increases powdery mildew resistance through promoting reactive oxygen species accumulation in bread wheat. Sci. Rep. 2019;9:5691. doi: 10.1038/s41598-019-42177-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jones J.D., Dangl J.L. The plant immune system. Nature. 2006;444:323–329. doi: 10.1038/nature05286. [DOI] [PubMed] [Google Scholar]
- Kim S., Kang J.Y., Cho D.I., Park J.H., Kim S.Y. ABF2, an ABRE-binding bZIP factor, is an essential component of glucose signaling and its overexpression affects multiple stress tolerance. Plant J. 2004;40:75–87. doi: 10.1111/j.1365-313X.2004.02192.x. [DOI] [PubMed] [Google Scholar]
- Lee S.C., Choi H.W., Hwang I.S., Choi D.S., Hwang B.K. Functional roles of the pepper pathogen-induced bZIP transcription factor, CAbZIP1, in enhanced resistance to pathogen infection and environmental stresses. Planta. 2006;224:1209–1225. doi: 10.1007/s00425-006-0302-4. [DOI] [PubMed] [Google Scholar]
- Li X., Fan S., Hu W., Liu G., Wei Y., He C., Shi H. Two cassava basic leucine zipper (bZIP) transcription factors (MebZIP3 and MebZIP5) confer disease resistance against cassava bacterial blight. Front. Plant Sci. 2017;8:2110. doi: 10.3389/fpls.2017.02110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu F., Li X., Wang M., Wen J., Yi B., Shen J., Ma C., Fu T., Tu J. Interactions of WRKY15 and WRKY33 transcription factors and their roles in the resistance of oilseed rape to Sclerotinia infection. Plant Biotechnol. J. 2018;16:911–925. doi: 10.1111/pbi.12838. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu S., Kracher B., Ziegler J., Birkenbihl R.P., Somssich I.E. Negative regulation of ABA signaling by WRKY33 is critical for Arabidopsis immunity towards Botrytis cinerea 2100. Elife. 2015;4:e07295. doi: 10.7554/eLife.07295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lorenzo O., Piqueras R., Sanchez-Serrano J.J., Solano R. ETHYLENE RESPONSE FACTOR1 integrates signals from ethylene and jasmonate pathways in plant defense. Plant Cell. 2003;15:165–178. doi: 10.1105/tpc.007468. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luan Q., Chen C., Liu M., Li Q., Wang L., Ren Z. CsWRKY50 mediates defense responses to Pseudoperonospora cubensis infection in Cucumis sativus. Plant Sci. 2019;279:59–69. doi: 10.1016/j.plantsci.2018.11.002. [DOI] [PubMed] [Google Scholar]
- Manju L., Nair R.R. Magnesium deficiency augments myocardial response to reactive oxygen species. Can. J. Physiol. Pharmacol. 2006;84:617–624. doi: 10.1139/y06-017. [DOI] [PubMed] [Google Scholar]
- Mazarei M., Elling A.A., Maier T.R., Puthoff D.P., Baum T.J. GmEREBP1 is a transcription factor activating defense genes in soybean and Arabidopsis. Mol. Plant Microbe Interact. 2007;20:107–119. doi: 10.1094/MPMI-20-2-0107. [DOI] [PubMed] [Google Scholar]
- Mine A., Seyfferth C., Kracher B., Berens M.L., Becker D., Tsuda K. The defense phytohormone signaling network enables rapid, high-amplitude transcriptional reprogramming during effector-triggered immunity. Plant Cell. 2018;30:1199–1219. doi: 10.1105/tpc.17.00970. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miyamoto K., Nishizawa Y., Minami E., Nojiri H., Yamane H., Okada K. Overexpression of the bZIP transcription factor OsbZIP79 suppresses the production of diterpenoid phytoalexin in rice cells. J. Plant Physiol. 2015;173:19–27. doi: 10.1016/j.jplph.2014.09.001. [DOI] [PubMed] [Google Scholar]
- Morel M., Ngadin A.A., Droux M., Jacquot J.P., Gelhaye E. The fungal glutathione S-transferase system. Evidence of new classes in the wood-degrading basidiomycete Phanerochaete chrysosporium. Cell Mol. Life Sci. 2009;66:3711–3725. doi: 10.1007/s00018-009-0104-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Naing A.H., Kyu S.Y., Pe P.P.W., Park K.I., Lee J.M., Lim K.B., Kim C.K. Silencing of the phytoene desaturase (PDS) gene affects the expression of fruit-ripening genes in tomatoes. Plant Methods. 2019;15:110. doi: 10.1186/s13007-019-0491-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nijhawan A., Jain M., Tyagi A.K., Khurana J.P. Genomic survey and gene expression analysis of the basic leucine zipper transcription factor family in rice. Plant Physiol. 2008;146:333–350. doi: 10.1104/pp.107.112821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pieterse C.M., Van der Does D., Zamioudis C., Leon-Reyes A., Van Wees S.C. Hormonal modulation of plant immunity. Annu. Rev. Cell Dev. Biol. 2012;28:489–521. doi: 10.1146/annurev-cellbio-092910-154055. [DOI] [PubMed] [Google Scholar]
- Ranjan A., Westrick N.M., Jain S., Piotrowski J.S., Ranjan M., Kessens R., Stiegman L., Grau C.R., Conley S.P., Smith D.L., Kabbage M. Resistance against Sclerotinia sclerotiorum in soybean involves a reprogramming of the phenylpropanoid pathway and up-regulation of antifungal activity targeting ergosterol biosynthesis. Plant Biotechnol. J. 2019 doi: 10.1111/pbi.13082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robatzek S., Somssich I.E. Targets of AtWRKY6 regulation during plant senescence and pathogen defense. Genes Dev. 2002;16:1139–1149. doi: 10.1101/gad.222702. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schluttenhofer C., Yuan L. Regulation of specialized metabolism by WRKY transcription factors. Plant Physiol. 2015;167:295–306. doi: 10.1104/pp.114.251769. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Silva M.S., Arraes F.B.M., Campos M.A., Grossi-de-Sa M., Fernandez D., Candido E.S., Cardoso M.H., Franco O.L., Grossi-de-Sa M.F. Review: potential biotechnological assets related to plant immunity modulation applicable in engineering disease-resistant crops. Plant Sci. 2018;270:72–84. doi: 10.1016/j.plantsci.2018.02.013. [DOI] [PubMed] [Google Scholar]
- Sun X., Yu G., Li J., Liu J., Wang X., Zhu G., Zhang X., Pan H. AcERF2, an ethylene-responsive factor of Atriplex canescens, positively modulates osmotic and disease resistance in Arabidopsis thaliana. Plant Sci. 2018;274:32–43. doi: 10.1016/j.plantsci.2018.05.004. [DOI] [PubMed] [Google Scholar]
- Takesako K., Kuroda H., Inoue T., Haruna F., Yoshikawa Y., Kato I., Uchida K., Hiratani T., Yamaguchi H. Biological properties of aureobasidin A, a cyclic depsipeptide antifungal antibiotic. J. Antibiot. 1993;46:1414–1420. doi: 10.7164/antibiotics.46.1414. [DOI] [PubMed] [Google Scholar]
- Thomma B.P., Eggermont K., Penninckx I.A., Mauch-Mani B., Vogelsang R., Cammue B.P., Broekaert W.F. Separate jasmonate-dependent and salicylate-dependent defense-response pathways in Arabidopsis are essential for resistance to distinct microbial pathogens. Proc. Natl. Acad. Sci. U S A. 1998;95:15107–15111. doi: 10.1073/pnas.95.25.15107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsuda K., Somssich I.E. Transcriptional networks in plant immunity. New Phytol. 2015;206:932–947. doi: 10.1111/nph.13286. [DOI] [PubMed] [Google Scholar]
- Tyler B.M. Phytophthora sojae: root rot pathogen of soybean and model oomycete. Mol. Plant Pathol. 2007;8:1–8. doi: 10.1111/j.1364-3703.2006.00373.x. [DOI] [PubMed] [Google Scholar]
- Tyler B.M., Tripathy S., Zhang X., Dehal P., Jiang R.H., Aerts A., Arredondo F.D., Baxter L., Bensasson D., Beynon J.L. Phytophthora genome sequences uncover evolutionary origins and mechanisms of pathogenesis. Science. 2006;313:1261–1266. doi: 10.1126/science.1128796. [DOI] [PubMed] [Google Scholar]
- Westrick N.M., Ranjan A., Jain S., Grau C.R., Smith D.L., Kabbage M. Gene regulation of Sclerotinia sclerotiorum during infection of Glycine max: on the road to pathogenesis. BMC Genomics. 2019;20:157. doi: 10.1186/s12864-019-5517-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Whitham S.A., Qi M., Innes R.W., Ma W., Lopes-Caitar V., Hewezi T. Molecular soybean-pathogen interactions. Annu. Rev. Phytopathol. 2016;54:443–468. doi: 10.1146/annurev-phyto-080615-100156. [DOI] [PubMed] [Google Scholar]
- Xun H., Yang X., He H., Wang M., Guo P., Wang Y., Pang J., Dong Y., Feng X., Wang S., Liu B. Over-expression of GmKR3, a TIR-NBS-LRR type R gene, confers resistance to multiple viruses in soybean. Plant Mol. Biol. 2019;99:95–111. doi: 10.1007/s11103-018-0804-z. [DOI] [PubMed] [Google Scholar]
- Yang B., Wang Y., Guo B., Jing M., Zhou H., Li Y., Wang H., Huang J., Wang Y., Ye W. The Phytophthora sojae RXLR effector Avh238 destabilizes soybean Type2 GmACSs to suppress ethylene biosynthesis and promote infection. New Phytol. 2019;222:425–437. doi: 10.1111/nph.15581. [DOI] [PubMed] [Google Scholar]
- Yang X., Yang J., Wang Y., He H., Niu L., Guo D., Xing G., Zhao Q., Zhong X., Sui L. Enhanced resistance to sclerotinia stem rot in transgenic soybean that overexpresses a wheat oxalate oxidase. Transgenic Res. 2019;28:103–114. doi: 10.1007/s11248-018-0106-x. [DOI] [PubMed] [Google Scholar]
- Yu H., Gao Q., Dong S., Zhou J., Ye Z., Lan Y. Effects of dietary n-3 highly unsaturated fatty acids (HUFAs) on growth, fatty acid profiles, antioxidant capacity and immunity of sea cucumber Apostichopus japonicus (Selenka) Fish Shellfish Immunol. 2016;54:211–219. doi: 10.1016/j.fsi.2016.04.013. [DOI] [PubMed] [Google Scholar]
- Zhang C., Bradshaw J.D., Whitham S.A., Hill J.H. The development of an efficient multipurpose bean pod mottle virus viral vector set for foreign gene expression and RNA silencing. Plant Physiol. 2010;153:52–65. doi: 10.1104/pp.109.151639. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang C., Gao H., Li R., Han D., Wang L., Wu J., Xu P., Zhang S. GmBTB/POZ, a novel BTB/POZ domain-containing nuclear protein, positively regulates the response of soybean to Phytophthora sojae infection. Mol. Plant Pathol. 2019;20:78–91. doi: 10.1111/mpp.12741. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang C., Li C., Liu J., Lv Y., Yu C., Li H., Zhao T., Liu B. The OsABF1 transcription factor improves drought tolerance by activating the transcription of COR413-TM1 in rice. J. Exp. Bot. 2017;68:4695–4707. doi: 10.1093/jxb/erx260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang H., Wu Q., Cao S., Zhao T., Chen L., Zhuang P., Zhou X., Gao Z. A novel protein elicitor (SsCut) from Sclerotinia sclerotiorum induces multiple defense responses in plants. Plant Mol. Biol. 2014;86:495–511. doi: 10.1007/s11103-014-0244-3. [DOI] [PubMed] [Google Scholar]
- Zhang M., Liu Y., Cai H., Guo M., Chai M., She Z., Ye L., Cheng Y., Wang B., Qin Y. The bZIP transcription factor GmbZIP15 negatively regulates salt- and drought-stress responses in soybean. Int. J. Mol. Sci. 2020;21 doi: 10.3390/ijms21207778. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang M., Liu Y., Shi H., Guo M., Chai M., He Q., Yan M., Cao D., Zhao L., Cai H., Qin Y. Evolutionary and expression analyses of soybean basic Leucine zipper transcription factor family. BMC Genomics. 2018;19:159. doi: 10.1186/s12864-018-4511-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang W., Wu Y., Schnable J.C., Zeng Z., Freeling M., Crawford G.E., Jiang J. High-resolution mapping of open chromatin in the rice genome. Genome Res. 2012;22:151–162. doi: 10.1101/gr.131342.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao L.H., Cai H.Y.,Z.X., Wang L.L., Huang X.Y., Zhang KLU suppresses megasporocyte cell fate through SWR1-mediated activation of WRKY28 expression in Arabidopsis. Proc. Natl. Acad. Sci. U S A. 2018 doi: 10.1073/pnas.1716054115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou X.T., Jia L.J., Wang H.Y., Zhao P., Wang W.Y., Liu N., Song S.W., Wu Y., Su L., Zhang J. The potato transcription factor StbZIP61 regulates dynamic biosynthesis of salicylic acid in defense against Phytophthora infestans infection. Plant J. 2018;95:1055–1068. doi: 10.1111/tpj.14010. [DOI] [PubMed] [Google Scholar]
- Zhu L., Guo J., Ma Z., Wang J., Zhou C. Arabidopsis transcription factor MYB102 increases plant susceptibility to aphids by substantial activation of ethylene biosynthesis. Biomolecules. 2018;8 doi: 10.3390/biom8020039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zong W., Zhong X., You J., Xiong L. Genome-wide profiling of histone H3K4-tri-methylation and gene expression in rice under drought stress. Plant Mol. Biol. 2013;81:175–188. doi: 10.1007/s11103-012-9990-2. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The original ChIP-seq data is available at European Nucleotide Archive (ENA) under accession number PRJEB44708.