Abstract
Background
Sorghum, the C4 dry-land cereal, important for food, fodder, feed and fuel, is a model crop for abiotic stress tolerance with smaller genome size, genetic diversity, and bio-energy traits. The heat shock proteins/chaperonin 60s (HSP60/Cpn60s) assist the plastid proteins, and participate in the folding and aggregation of proteins. However, the functions of HSP60s in abiotic stress tolerance in Sorghum remain unclear.
Methods
Genome-wide screening and in silico characterization of SbHSP60s were carried out along with tissue and stress-specific expression analysis.
Results
A total of 36 HSP60 genes were identified in Sorghum bicolor. They were subdivided into 2 groups, the HSP60 and HSP10 co-chaperonins encoded by 30 and 6 genes, respectively. The genes are distributed on all the chromosomes, chromosome 1 being the hot spot with 9 genes. All the HSP60s were found hydrophilic and highly unstable. The HSP60 genes showed a large number of introns, the majority of them with more than 10. Among the 12 paralogs, only 1 was tandem and the remaining 11 segmental, indicating their role in the expansion of SbHSP60s. Majority of the SbHSP60 genes expressed uniformly in leaf while a moderate expression was observed in the root tissues, with the highest expression displayed by SbHSP60-1. From expression analysis, SbHSP60-3 for drought, SbHSP60-9 for salt, SbHSP60-9 and 24 for heat and SbHSP60-3, 9 and SbHSP10-2 have been found implicated for cold stress tolerance and appeared as the key regulatory genes.
Conclusion
This work paves the way for the utilization of chaperonin family genes for achieving abiotic stress tolerance in plants.
Keywords: HSP60, HSP10, abiotic stress-responsive, phylogenetic tree, gene expressions, chaperonin
1. INTRODUCTION
Plants face unavoidable environmental challenges like biotic and abiotic stresses, which drastically limit both growth and final productivity [1, 2]. Abiotic stress is the most deterrent and causes 70% yield losses in crop plants [3]. To combat this, plants have developed various mechanisms like morphological (leaf orientation), anatomical (stomatal conductance and increased leaf pubescence), phenological (changes in the developmental stages), physiological (root hydraulic conductance and photosynthesis), metabolic (accumulation of the osmolytes), hormonal balance (ABA, ethylene, and
salicylic acid) and secondary metabolites (isopropanoid, flavonoid, anthocyanin, and lignin) [4-7]. Besides these mechanisms, plants should recognize and respond to the stress at cellular and molecular levels, and repress the expression of normal proteins and induce the expression of diverse stress-associated proteins [8]. Abiotic stress generally leads to protein aggregation, which can subsequently cause metabolic dysfunction. To survive under stress conditions, it is imperative for the plants to maintain native conformation of proteins and, at the same time to reduce the accumulation of non-native proteins. To overcome such a problem, plants produce heat shock proteins called molecular chaperones, which are implicated in abiotic and biotic stress tolerance [9, 10]. They act as multi-functional proteins and maintain homeostasis by protein folding, trafficking and disaggregation under stress conditions [9, 11]. Based on their molecular weights, they are categorized into HSP100, HSP90, HSP70, HSP60, HSP40, HSP20, and HSP10 [12, 13].
HSP60 chaperonins are essential for the viability of cells in all growth conditions because they are required for the efficient folding of numerous proteins that mediate vital cellular functions. They are ubiquitously present in archaea, eubacteria, and eukaryotes. They facilitate protein folding, refolding, aggregation and transportation to chloroplasts and mitochondria [14-16]. The oligomeric protein complexes form double-ring structures, participate in protein functional conformation, folding of denatured and newly synthesized proteins through hydrolysis of ATP [17-20]. So far, two types of chaperonins, type I in bacteria (called GroEL), plastids (HSP60) and mitochondria (Hsp60), and type II in archaea and eukaryotic cytosol (CCT/TriC) [21], with a distinct, conserved crystal structure were identified [18, 22]. In plants, the nuclear genome encodes mitochondrial and plastidial chaperonin homologues [21, 23]. E. coli GroEL and mitochondrial HSP60 contain 14 identical subunits, while the plastid HSP60 contains two distinct subunits, α and β [24, 25]. The number of subunits varies in land plants; 3α and 4β subunits in Physcomitrella patens [26], 3α and only 1β in Brassica napus [27], and 2α and 4β subunits in Arabidopsis [21]. In protein folding, while type I chaperonins are assisted by co-chaperonin GroES/HSP10, type II performs by an in-built component “cap” for substrate encapsulation [11, 28, 29]. Bacterial HSP10 is encoded by a single gene GroES, in contrast to the plastid HSP10 found in algae and plants encoded by several genes [30]. In plants, they are considered as a sub-family of chaperonins [21]. The HSP10, along with HSP60, forms back-to-back double-ring assemblies consisting of closely related and rotationally symmetrical subunits, which help in folding, assembly and sorting of the proteins [17]. The two subunits containing 20-kDa homologue of bacterial HSP10 have been identified in plastids, and the subunits are linked by a TDDVKD-linker in head to tail manner [31].
The HSP60 in plant chloroplasts and E. coli [32], protects the rubisco enzyme, a prime protein that mediates CO2 fixation [12]. It has been observed that folding of plant RbcL subunits is mediated by the chaperonin HSP60, and also its cofactor HSP20 [12, 33]. Chloroplast chaperonin and the auxiliary factors when overexpressed in an E. coli strain, functional plant rubisco enzyme was noticed [33]. The results indicate that chaperonins cooperate and play pivotal roles in the functional expression of proteins encoded by transgenes. HSP60 plays an important role in RNA metabolism, RNA protection and processing, too [34]. Hsu et al. [35], noticed HSP60-mediated mitochondrial RNA splicing in Arabidopsis. Among the HSP60 gene family members, AtHSP60α1, AtHSP60β1 and AtHSP60β2 were strongly induced in comparison with others under heat stress conditions in Arabidopsis [36]. Prasad and Stewart [37] pointed out that HSP60 plays a vital role in the developmental regulation, mitochondrial biogenesis, heat stress tolerance and in assembly of oligomeric protein structures. The mRNA and protein expression levels of PtHSP60 are altered according to salinity changes and thus appear to play a key role in the regulation of the pathway associated with salinity stress [38]. When HSP60 and HSP10 were overexpressed in E. coli and yeast, they exhibited tolerance against osmotic and salt stresses [12]. Thus, the chaperonins are associated with stress tolerance and play crucial roles during the process.
Haq et al. [39], noticed a total of 16 HSP60 genes in pepper, and concluded their defence response against heat and other abiotic stresses. They act as lipo-chaperonins and participate in membrane stabilization, and refolding thus prevent irreversible thermal aggregation. HSP10 homologues were identified in Oryza sativa [40], Hordeum vulgare [41], Triticum aestivum, Phaseolus vulgaris, Pisum sativum [42], and Pennisetum glaucum [43], and these might help in ameliorating abiotic stresses.
Sorghum bicolor, the Great Millet or camel crop is a C4 crop plant with an extensive root system, waxy bloom on its leaves and culm, can survive to some extent in drought-prone areas [44, 45]. It has the potential to adapt to global warming conditions by maintaining high levels of chlorophyll, known as the stay-green phenotype [46, 47]. Its stay- green character is thus the distinctive feature for its ability to withstand drought stress [48, 49], though the growth is slowed down during drought-like other crops [50]; but it has the potential to adapt to the stress conditions by maintaining high levels of chlorophyll, known as stay-green genotypes. However, the molecular mechanisms of drought and heat stress tolerance and the underlying genes need to be validated. By sharing a double-ring-like structure, chaperonins play a key role in protein functional conformation and transport [20]. Therefore, identification of HSP60s and their expression profiling is highly crucial. They are well studied in prokaryotic systems like E. coli, but not in higher plants like sorghum. Hence, the present study aims to find out the number of chaperonins present by screening the whole genome sequence along with their in silico characterization and also the expression profiles of chaperonins (HSP60 and 10) during abiotic stress tolerance in different tissues of S. bicolor.
2. MATERIALS AND METHODS
2.1. Plant Material
Sorghum bicolor BTx623 variety procured from ICRISAT, Hyderabad, India, was used for gene expression analysis. The seedlings of BTx623 were grown in pots under glasshouse conditions. The 40-day-old plants were subjected to abiotic stress treatments as described earlier by Nagaraju et al. [51]. Drought stress was imposed by treating the S. bicolor plants with 150 mM mannitol for 4 h. Similarly, seedlings were exposed to 150 mM NaCl for 4 h, temperature stress at 40 °C in a growth chamber for 4 h, and cold stress in a refrigerator at 4 °C for 4 h. The treated root, stem, and leaf tissues, along with corresponding controls, were collected separately, frozen immediately in liquid nitrogen and stored at -80 ºC until further use.
2.2. Identification of HSP60 Genes in Sorghum
For this study, the HSP60 sequences of Oryza and Arabidopsis were retrieved from HSPIR (http://pdslab.biochem. iisc.ernet.in/hspir/) [52], and NCBI (https://www.ncbi. nlm.nih.gov/), and used as query sequences in Gramene (http://www.gramene.org/) [53] database by blasting against S. bicolor genome, with default parameters. The non-redundant putative protein sequences were subjected to the MOTIF search tool (http://www.genome.jp/tools/motif/) to identify their conserved domains and other Pfam domains [54] and the reliability was checked by employing the SMART tool (http://smart.emblheidelberg.de/). Proteins that failed to exhibit reliability were eliminated.
2.3. In silico Sequence Characterization of SbHSP60s
The chromosomal distributions of HSP60 genes were determined based on the sorghum genome annotation files at Gramene (http://www.gramene.org/) [53]. By blasting the HSP60 amino acid sequences in the ExPASy-ProtParam tool (http://web.expas y.org/ProtParam/) [55], the molecular weight (MW), isoelectric point (pI), grand average of hydropathicity (GRAVY), instability index, and aliphatic indices were calculated. The Gene Structure Display Server (GSDS2.0) (http://gsds.cbi.pku.edu.cn/) [56] was used for determining gene structures (exon/intron and utrs), WOLF PSORT (https://www.genscript.com/wolf-psort.html) [57] for sub-cellular localizations, TMHMM Server V. 2.0 [58] for trans-membrane helices, NetPhosK3 software [59] for putative phosphorylation sites, MEME software (http://meme-suite.org/) [60] for conserved motifs by setting default parameters according to Nagaraju et al. [51]. The GeneMANIA platform (https://genemania.org/), STRING database (https://string-db.org/), and STITCH v5.0 (http://stitch.embl.de/) were used to identify the interaction between gene-gene, protein-protein and protein-chemical. Using PLACE [61] and PLANTCARE [62] tools, cis-elements were detected in the upstream regions of SbHSP60s. The MEGA 6.2 software [63] was used to construct a maximum likelihood (ML) phylogenetic tree with the parameters Poisson correction, complete deletion and bootstrap value of 1,000 replicates for statistical reliability. PAL2NAL (http://www.bork.embl.de/pal2nal/) [64] was used for calculating the substitution rates for non-synonymous to synonymous sites of paralogs and orthologous gene pairs. The putative miRNAs targeting the SbHSP60s were predicted by employing the psRNATarget server [65] with default parameters.
2.4. Digital Expression Analysis of SbHSP60 Genes
In order to perform in silico expression profiling of the identified SbHSP60s, probe ids for respective genes were manually obtained from the sorghum functional genomics database (SorghumFDB) [66]. Further, these probe ids were utilized to perform expression profiling of SbHSP60s using curated whole transcriptome data embedded in the Genevestigator platform [67]. Expression analysis was performed in different anatomical parts and developmental stages under several abiotic stresses, including drought, salt, heat, and cold, with different samples embedded in the platform. Heat map that displays expression profiling was prepared using the clustering tool available on Genevestigator [68] utilizing 30 different transcriptome data sets embedded in the Genevestigator library. Clustering of expressed SbHSP genes was performed following the criteria used by Kumar et al. [69].
2.5. qRT-PCR Analysis of SbHSP60s Under Abiotic Stress Conditions
Due to the availability of limited resources, expression patterns of the identified HSP-60 genes were reduced for this study. The genes were selected based on the homology with other crops, their subcellular localizations and subclasses. From each subclass, at least one representative gene was selected for qRT-PCR analysis. Total RNAs were extracted using MACHEREY-NAGEL RNA isolation kit according to the manufacturer’s instructions, and after checking the purity, integrity and quality, 3 µg of RNA was reverse transcribed to cDNA by using Thermo Scientific first-strand synthesis kit. The NCBI PRIMER Blast (www.ncbi. nlm.nih.gov/tools/primer-blast/) [70] was used to design the gene-specific primers with the default parameters: 57-60°C annealing temperature, 18-22 bp primer length, 50-55% GC contents, and 80-140 bp amplicon length (Table S1 (1.5MB, pdf) ). SYBR Green master mix (2X) (Takara) was used for the qRT-PCR, and the analysis was performed with three biological replicates, and two technical replicates in Agilent Mx3000p with the following thermal cycles: 1 cycle at 95°C for 10 min, followed by 40 cycles alternatively at 95°C for 15 sec and 60°C for 1 min. The amplicon dissociation curves were recorded with a fluorescent lamp after 40th cycle by heating from 58 to 95°C within 20 min and by maintaining triplicates. The sorghum SbAcp (Acyl Carrier Protein 2) and SbEF-P (Elongation Factor P) genes were used as internal controls [71]. The average values ± standard error are represented. The t-test (Tukeys test) was employed to determine the statistical significance for all the values (*P ≤ 0.05). The relative gene expressions were calculated by employing Rest software [72].
3. RESULTS
3.1. Identification and In silico Characterization of HSP60 Genes in Sorghum
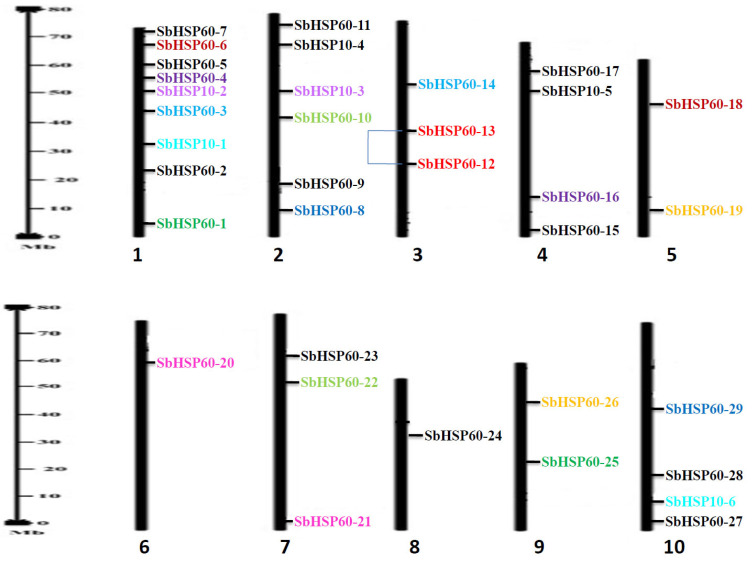
In the present study, genome-wide scanning of S. bicolor resulted in the identification of 36 HSP60 genes which are sub-divided into 2 groups; HSP60 chaperonins with 30 genes and HSP10, the co-chaperonins with 6 genes. The genes were distributed on all the chromosomes, with the highest number of 9 on chromosome 1, while only one each on chromosomes 6 and 8 (Table 1 and Fig. 1). The number of amino acids varied from 149 (SbHSP60-19) to 2056 (SbHSP60-28), with molecular weights ranging from 16563.89 (SbHSP60-19) to 229911.54 Da (HSP60-28) in HSP60s. In HSP10s, the number of amino acids ranged from 134 (SbHSP10-1) to 1844 (SbHSP10-4) and molecular weights from 14264.54 (SbHSP10-1) to 199040.62 Da (HSP10-4). The iso-electric points varied between 5.14 (SbHSP60-2) to 10.07 (SbHSP60-13). Out of 30 HSP60s, 16 were found acidic, while the remaining basic in nature. Based on the GRAVY values, SbHSP60s have been found hydrophilic, except SbHSP60-7, 16 and SbHSP10-1. Out of 36 SbHSP60s, 26 proteins displayed instability index ranging between 28.47 (SbHSP60-27) to 62.75 (SbHSP60-28), while the aliphatic index varied from 74.86 (SbHSP60-11) to 62.75 (SbHSP10-6). All the HSP10s revealed the highest index with unstable, basic, and hydrophilic nature and with less molecular weights (Table 1). Out of 30 SbHSP60s, 6 were localized in the cytosol, 9 in the nucleus, 10 in the chloroplast, 3 in mitochondria, and 2 in the plastid. Of the 6 HSP10s, 3 were localized in chloroplast and 1 each in cytosol, nucleus and vacuole (Table 1). Among the 36 HSP60s, only 4 exhibited transmembrane helices. Three helices were noticed in SbHSP60-9, one each in HSP60-3, 16 and HSP10-2.
Table 1. Characteristics of identified SbHSP60 genes in Sorghum bicolor number of amino acids, chromosomal location, isoelectric point (pI) and molecular weight (MW), DNA binding domains (DBD), number of exons, localization, GRAVY, instability index, and aliphatic index.
| Gene | Common Name | No. of a. a. | Chr. loc. | pI / MW | DBD | No. of Exon | Localization | GRAVITY | Instability Index | Aliphatic Index | |
|---|---|---|---|---|---|---|---|---|---|---|---|
| Sb01g000380 | SbHSP60-1 | 1217 | 1 | 6.22/131849.91 | 67-547 | 20 | Chl | -0.254 | 44.29 | 92.36 | |
| Sb01g020010 | SbHSP60-2 | 1077 | 1 | 5.14/119175.59 | 502-1010 | 26 | Chl | -0.410 | 41.11 | 90.27 | |
| Sb01g033120 | SbHSP60-3 | 1925 | 1 | 5.95/215246.07 | 396-646 | 15 | N | -0.456 | 48.58 | 75.67 | |
| Sb01g041170 | SbHSP60-4 | 1811 | 1 | 9.54/195624.48 | 1206-1735 | 24 | N | -0.175 | 44.74 | 87.33 | |
| Sb01g043220 | SbHSP60-5 | 688 | 1 | 8.90/74959.68 | 155-656 | 16 | C | -0.189 | 38.89* | 93.31 | |
| Sb01g047360 | SbHSP60-6 | 1180 | 1 | 8.74/131338.29 | 189-554 | 27 | C | -0.433 | 43.21 | 84.53 | |
| Sb01g049370 | SbHSP60-7 | 628 | 1 | 7.20/67781.40 | 37-592 | 5 | C | 0.041 | 42.98 | 108.77 | |
| Sb02g001450 | SbHSP60-8 | 799 | 2 | 8.73/87612.79 | 40-519 | 15 | C | -0.402 | 39.63* | 84.37 | |
| Sb02g011260 | SbHSP60-9 | 1308 | 2 | 8.53/141326.78 | 798-1273 | 22 | P | -0.157 | 46.24 | 90.32 | |
| Sb02g024300 | SbHSP60-10 | 1285 | 2 | 5.41/141642.08 | 1052-1274 | 9 | N | -0.346 | 42.05 | 79.53 | |
| Sb02g043440 | SbHSP60-11 | 569 | 2 | 7.59/62720.26 | 63-562 | 16 | M | -0.253 | 34.15* | 96.47 | |
| Sb03g009490 | SbHSP60-12 | 775 | 3 | 8.80/85157.56 | 286-676 | 19 | M | -0.385 | 40.91 | 82.76 | |
| Sb03g005190 | SbHSP60-13 | 871 | 3 | 10.07/96258.74 | 102-557 | 17 | C | -0.259 | 49.14 | 90.52 | |
| Sb03g041400 | SbHSP60-14 | 1619 | 3 | 5.21/182219.68 | 559-719 | 17 | N | -0.423 | 45.29 | 80.15 | |
| Sb04g000370 | SbHSP60-15 | 576 | 4 | 5.62/61680.65 | 52-555 | 14 | Chl | -0.143 | 28.98* | 99.51 | |
| Sb04g004030 | SbHSP60-16 | 1777 | 4 | 5.99/ 190958.85 | 37-498 | 17 | P | 0.033 | 37.71* | 101.93 | |
| Sb04g035610 | SbHSP60-17 | 380 | 4 | 9.03/40641.08 | 31-301 | 12 | C | -0.122 | 38.47* | 96.55 | |
| Sb05g022470 | SbHSP60-18 | 1706 | 5 | 5.68/189340.50 | 34-522 | 17 | N | -0.445 | 42.00 | 76.25 | |
| Sb05g008785 | SbHSP60-19 | 149 | 5 | 5.61/16563.89 | 11-51 | 5 | M | -0.125 | 39.59* | 99.53 | |
| Sb06g034080 | SbHSP60-20 | 1561 | 6 | 5.87/175336.41 | 197-477 | 12 | N | -0.470 | 45.02 | 78.31 | |
| Sb07g000590 | SbHSP60-21 | 1814 | 7 | 5.64/202460.95 | 426-566 | 16 | N | -0.503 | 44.30 | 75.51 | |
| Sb07g020930 | SbHSP60-22 | 1558 | 7 | 6.15/173645.21 | 323-407 | 15 | Chl | -0.425 | 44.15 | 79.16 | |
| Sb07g022110 | SbHSP60-23 | 1811 | 7 | 5.62/199458.67 | 431-681 | 13 | N | -0.407 | 51.36 | 76.21 | |
| Sb08g014925 | SbHSP60-24 | 1569 | 8 | 8.56/178143.90 | 126-232 | 14 | Chl | -0.484 | 50.06 | 77.23 | |
| Sb09g014430 | SbHSP60-25 | 1164 | 9 | 7.84/127154.19 | 150-630 | 16 | Chl | -0.250 | 38.22* | 92.90 | |
| Sb09g026970 | SbHSP60-26 | 1061 | 9 | 8.92/114446.97 | 102-588 | 29 | Chl | -0.321 | 34.99* | 88.75 | |
| Sb10g001120 | SbHSP60-27 | 665 | 10 | 6.92/71398.96 | 139-648 | 15 | Chl | -0.105 | 28.47* | 100.32 | |
| Sb10g009370 | SbHSP60-28 | 2056 | 10 | 6.81/229911.54 | 707-968 | 16 | N | -0.443 | 49.68 | 76.31 | |
| Sb10g022220 | SbHSP60-29 | 553 | 10 | 5.74/61136.68 | 40-549 | 9 | Chl | -0.156 | 43.37 | 98.23 | |
| Sb0011s00747 | SbHSP60-30 | 1564 | u | 9.08/177985.80 | 192-238 | 15 | Chl | -0.388 | 46.30 | 83.08 | |
| HSP-10 | |||||||||||
| Sb01g028650 | SbHSP10-1 | 134 | 1 | 7.76/14264.54 | 47-133 | 6 | Chl | 0.110 | 41.50 | 105.60 | |
| Sb01g034530 | SbHSP10-2 | 319 | 1 | 9.65/34727.58 | 123-199 | 6 | V | -0.082 | 49.74 | 105.80 | |
| Sb02g040860 | SbHSP10-3 | 169 | 2 | 9.26/17502.38 | 7-87 | 5 | C | -0.470 | 42.02 | 90.19 | |
| Sb02g025700 | SbHSP10-4 | 1844 | 2 | 8.67/199040.62 | 927-1115 | 17 | N | -0.439 | 52.49 | 74.86 | |
| Sb04g035040 | SbHSP10-5 | 487 | 4 | 9.89/52602.21 | 63-245 | 8 | Chl | -0.320 | 48.48 | 85.91 | |
| Sb10g006450 | SbHSP10-6 | 1467 | 10 | 8.43/160424.43 | 1276-1465 | 4 | Chl | -0.482 | 62.75 | 78.28 | |
Abbreviations: (a. a.: amino acids, Chrom.: Chromosome, pI: isoelectric point; MW: Molecular weight, Chl.: Chloroplast, C: Cytoplasm, N: Nucleus, P: Plastid, M: Mitochondria, V: Vacuole, GRAVY: Grand average hydropathy, * stable).
Fig. (1).
Distribution of SbHSP60 genes on 10 chromosomes with duplications. Duplications are illustrated by colours (segmental duplications in same colour) and regional duplications are linked with line. (A higher resolution / colour version of this figure is available in the electronic copy of the article).
3.2. Prediction of Phosphorylation Sites in SbHSP60s
The serine and threonine residues, known as the phosphorylation sites, have been identified in the HSP60 family and found dominant. The phosphorylation sites vary in number, 13 (SbHSP60-19) to 172 (SbHSP20-28) for serine, 2 (SbHSP60-19) to 59 (SbHSP20-28) for threonine, and 1 (SbHSP60-19) to 28 (SbHSP20-28) for tyrosine. Among the specific kinases targeting HSP60s, protein kinase C (PKC), casein kinase 2 (CK2), cell division cycle protein 2 (CDC2), protein kinase A (PKA), casein kinase 1 (CK1), P38 MAP kinase and CDK5 were noticed as dominant kinases targeting HSP60s phosphorylation, compared to Ataxia-telangiectasia mutated kinase (ATM), epidermal growth factor receptor (EGFR), insulin receptor tyrosine kinase (INSR) and glycogen synthase kinase 3 (GSK3) (Table S2 (1.5MB, pdf) ).
3.3. Gene Structure and Motif Analysis of SbHSP60s
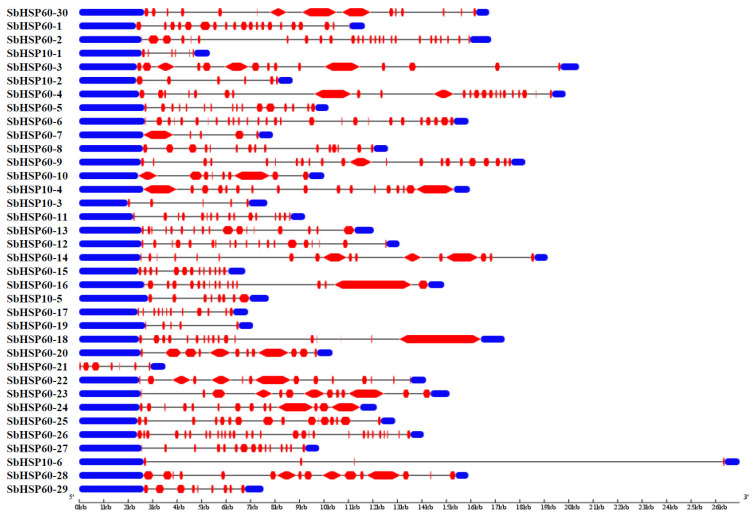
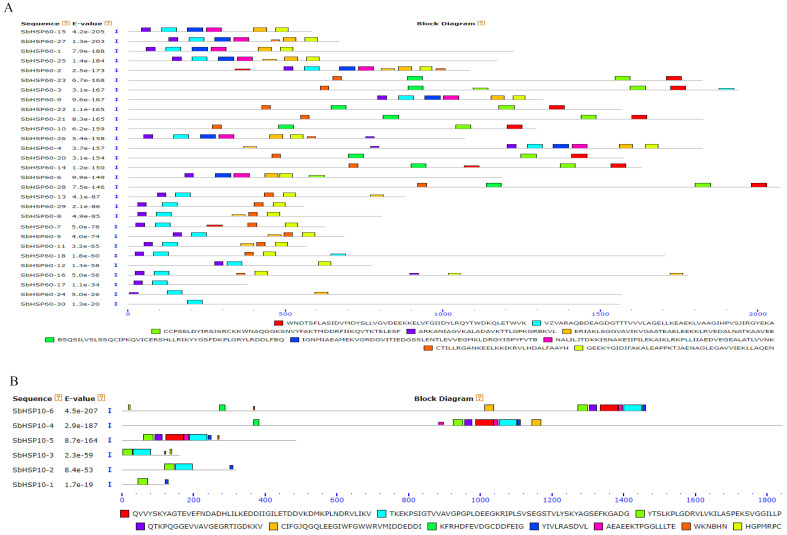
The GSDS software analysis of the SbHSP60 family revealed a large number of introns. Twenty-six (86.6%) of the 30 HSP60s have more than 10 introns, the highest number being 28 present in SbHSP60-26. On the other hand, in the HSP10 sub-group, only HSP10-4 exhibited the highest number of 16, while HSP10-6 showed 3 introns (Table 1 and Fig. 2). MEME identified 10 conserved motifs in each of the SbHSP60 and SbHSP10, with divergent distribution within each of the sub-cellular organelles. Motifs 2, 4, 7, 8, and 9 are the most conserved, commonly found in 66% of the HSP60s. Though motifs 2 and 4 are seen at N-terminus, 6 and 9 have been noticed as highly conserved. SbHSP60-3, 10, 14, 20, 21, 22, 23 and 28 have been found localized in the nucleus. They contain motif 1 at C terminus next to motif 6 repeats, and the conserved motif 2 is completely absent. They were also absent in the rest of the SbHSP60s (Fig. 3A and Fig. S1 (1.5MB, pdf) ). The amino acids in motif 1 are aromatic and acidic, responsible for nuclear localization, hence absent in other HSP60s. The stretch EEKK represents the nuclear localization signal. All the HSP10s showed similar motif distribution patterns, with 3 at N terminus and 8 at C terminus (Fig. 3B and Fig. S2 (1.5MB, pdf) ).
Fig. (2).
Structure of the SbHSP60 and HSP10s showing exons, introns and up/drown stream regions. (A higher resolution / colour version of this figure is available in the electronic copy of the article).
Fig. (3).
Conserved motifs in A) SbHSP-60s and B) SbHSP10s. The scale represents the lengths of the proteins and motifs. (A higher resolution / colour version of this figure is available in the electronic copy of the article).
3.4. Promoter Analysis of SbHSP60 Genes
Biotic and abiotic stress-responsive (drought, salt, heat, cold, light, desiccation), hormone-specific, and developmental-specific cis-elements (40-different types) were detected in the promoter regions. MYB, MYC, and HSE were dominant among the abiotic stress-responsive elements, with SbHSP60-2 and 12 exhibiting 20 ABA-responsive elements. Of the 36 HSP60s, 33 (91.6%) were noticed with ABA-responsive elements in their promoter regions and 29 of them exhibit more than 2 ABA elements. The SbHSP60-5, 27 and HSP10-5 were seen devoid of ABA-responsive but rich with heat, low temperature and salt stress-responsive elements. Interestingly, 86% of the SbHSP60s were rich in light-responsive, 41.6% phosphorous starvation-responsive and cytokinin-responsive elements (Table S3 (1.5MB, pdf) ).
3.5. Identification of miRNAs that Target SbHSP60s
A total of 48 different types of miRNAs target and regulate the SbHSP60 family genes, which participate in cleavage and translation inhibition of the RNAs. The sbi-miR6225, sbi-miR5568, miR437, miR6220, miR821, miR166, miR395, and miR5565 are some of the major miRNAs that have been found to target SbHSP60s. The miR167 plays an important role in plant developmental processes, target SbHSP60-11 and 18. The miR166 (drought stress-induced) target SbHSP60-3, 4, 5 and 25. The multiple stress-responsive miR398 targets the SbHSP10-4, SbHSP60-10 and 14. The identified miRNAs (putative) can target more than one HSP60, indicating their nature of interaction with multiple genes. For example, miR6225 interacts with SbHSP10-4, HSP10-6, HSP60-2, HSP60-3, HSP60-4, HSP60-8, HSP60-14, HSP60-18, HSP60-20, HSP60-22, and HSP60-27 (Table S4 (1.5MB, pdf) ).
3.6. Protein-protein Interaction and Chemical-protein Interaction Network of SbHSP60s
In order to predict the regulatory partners of HSP60s, the automated GeneMANIA server has been utilized. The predicted regulatory network suggests that members of the HSP60 family have various regulatory partners based on the conserved functional domains, physical interactions, and co- expression. As evident from Figure S3, the protein-protein interaction network of HSP60s revealed their complex molecular interaction with (i) 20 functional partners including T- complex protein 1 subunit alpha, (ii) 4 chaperonin proteins, (mitochondrial HSP60-3A, HSP60-2, CPN60, and 10 kDa chaperonin), (iii) 2 putative 1-phosphatidylinositol-3-phosphate 5-kinase (FAB1C, FAB1D), (iv) 4 phosphatidylinositol 4-phosphate 5-kinase (PIP5K3, PIP5K6, PIP5K10, PIP5K11), (v) 4 RING/FYVE/PHD zinc finger superfamily proteins, (vi) phosphoinositide binding protein, (vii) 2 regulators of chromosome condensation (RCC1) family proteins, (viii) GroES-like family protein, and (ix) a protein of unknown function (DUF581). The gene ontology (GO) terms analysis obtained through the GeneMANIA server suggested that predicted regulatory partners of HSP60s play key roles in different biological activity, including phosphatidylinositol phosphate kinase activity 1-phosphatidylinositol-4-phosphate 5-kinase activity, phosphatidylinositol binding, phospholipid binding, lipid binding, apical plasma membrane, and vacuole organization. The interaction between predicted regulatory partners and HSP60s can be validated using different molecular methods, including co-immunoprecipitation (Co-IP) and in situ hybridization (Fig. S3 (1.5MB, pdf) ). The protein-protein interaction analysis predicts that SbHSP60s interact with other chaperones like HSP70s, and 90s along with other proteins like phosphatidylinositol-4-phoshate 5-kinase, Mre/Mbl, ribosomal proteins, elongation factor-1, histidine kinase, glutathione S-transferase, FYVE zinc finger, dynein-related subfamily proteins and Holliday-junction DNA helicase rvuB. By interacting with the above proteins, HSP60s participate in inositol phosphate metabolism, RNA transport and degradation, phagosome, phosphatidylinositol signalling, proteosome, protein processing in endoplasmic reticulum and autophagy (Fig. S4 (1.5MB, pdf) ). Besides the prediction of the protein interaction network of HSP60s, a chemical-protein interaction network map was generated using the STITCH server. Interestingly, the HSP60s were found to interact with phosphate, ammonia, and lotion (urea or carbamide) (Fig. S5 (1.5MB, pdf) ). These results support the earlier prediction of HSP60s that interact with phosphorous-starvation-responsive elements. However, since these interactions are not validated, the data need to be taken into consideration with caution.
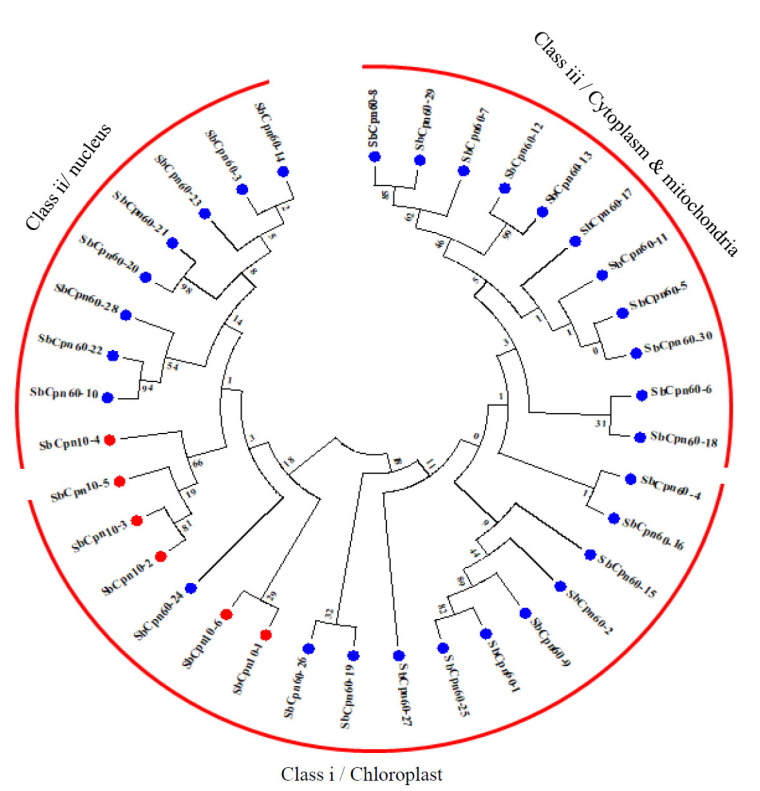
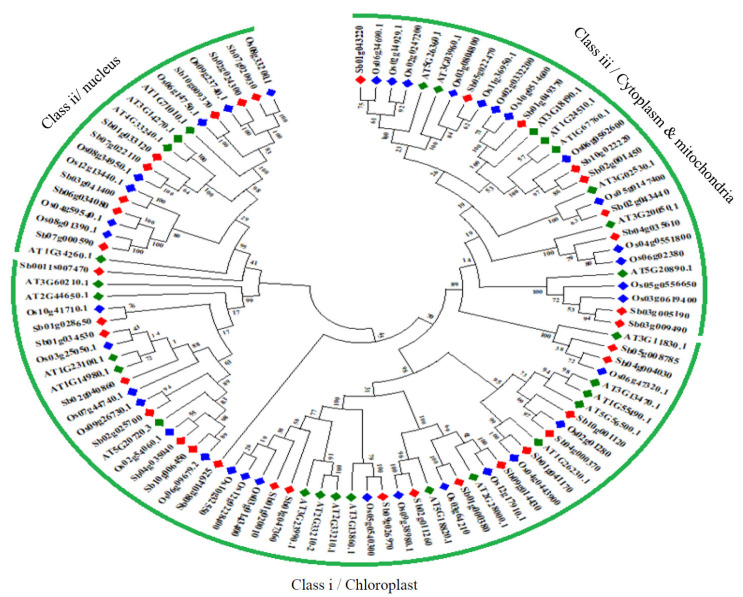
3.7. Phylogenetic Analysis of HSP60 Proteins
Phylogenetic analysis of HSP60 proteins classified them into three subclasses I to III, with a conserved HSP60_TCP1 domain. All the chloroplast and nuclear-localized proteins are clustered into class I and II, while cytosol and mitochondria localized into class III (Fig. 4). A total of 12 paralogous pairs have been detected in the SbHSP60 family, of which only one is tandem (SbHSP60-12/13 on chromosome 3) and the remaining segmental duplications (Figs. 1 and 4, Table S5 (1.5MB, pdf) ). A phylogenetic tree with 36 Sorghum, 35 rice and 28 Arabidopsis HSP60 genes was constructed to know the evolutionary relationship, exhibiting 3 subclasses according to their conserved motifs and sub-cellular localizations. Sorghum followed the trend like that of rice and Arabidopsis HSP60s sub-cellular localizations and also the cluster. It exhibited a total of 23 orthologous events with rice, as shown in Fig. (5) and Table S6 (1.5MB, pdf) .
Fig. (4).
Phylogenetic tree of SbHSP60s. The gene sub groups were classified based on their localization. (A higher resolution / colour version of this figure is available in the electronic copy of the article).
Fig. (5).
Phylogenetic tree constructed using protein sequences Sorghum, Oryza, and Arabidopsis to know the evolutionary relationship of HSP60s. (A higher resolution / colour version of this figure is available in the electronic copy of the article).
3.8. Calculation of Synonymous and Non-synonymous Substitution Rates (dN/dS)
The non-synonymous (dN) to synonymous substitution (ds) ratios for paralogous and orthologous gene pairs of HSP60 genes were estimated to know their evolution by Darwinian selection in duplication and divergence. Out of the 12 paralogs, the dN/dS ratios ranged from 0.4539 (SbHSP10-2/SbHSP10-3) to 99.0000. The SbHSP10-2/SbHSP10-3 exhibited a dN/dS ratio of 0.4539, purifying selection and the rest showed positive/Darwinian selection (Table S5 (1.5MB, pdf) ). Twenty-three orthologs exhibited ratios between 0.6926-99.0000, of which 2 showed purifying selection (ratio < 1), and the remaining were positive/Darwinian selection with >1 value (Table S6 (1.5MB, pdf) ).
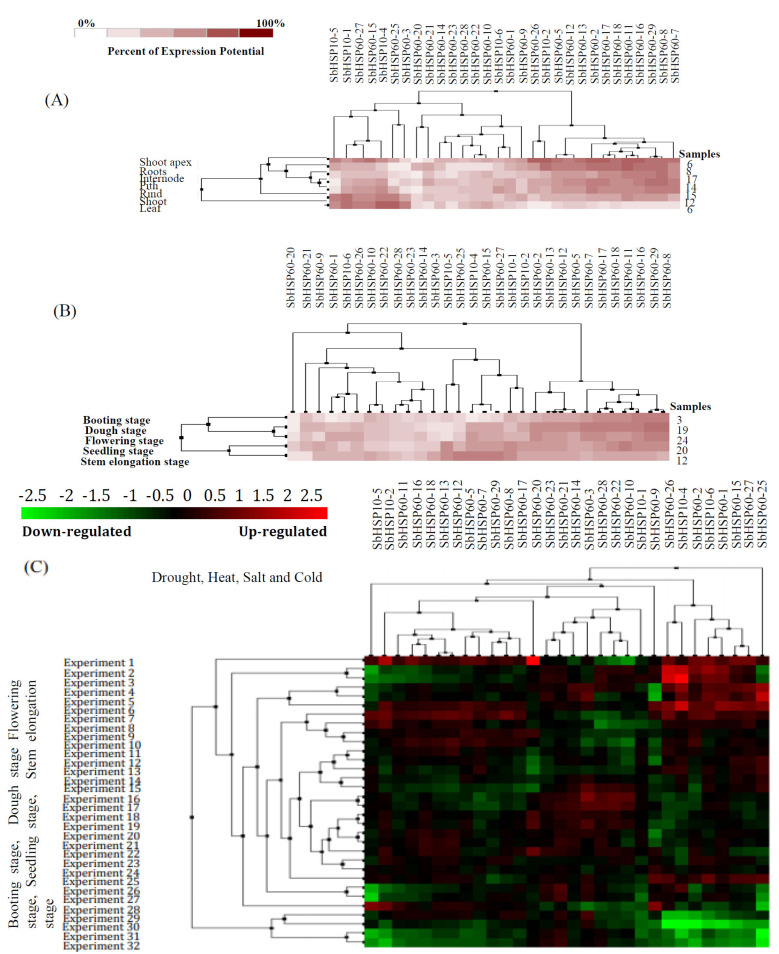
3.9. Transcriptome-based Gene Expression Profiling of SbHSP60s in Different Tissues and Developmental Stages Under Abiotic Stress Conditions
Of the 36 SbHSP60 genes, probe ids for 30 were available on the SorghumFDB. These data were utilized for gene expression profiling. Using transcriptome data, expression of these 30 SbHSP60s in seven different tissues viz., shoot apex, root, internode, pith, rind, shoot, and leaf were analyzed under normal and abiotic stress conditions such as drought, heat, salt, and cold (Fig. 6A). In the shoot apex and root tissues, a cluster of 13 SbHSP60s (SbHSP60-26, SbHSP10-2, SbHSP60-5, SbHSP60-12, SbHSP60-13, SbHSP60-2, SbHSP60-17, SbHSP60-18, SbHSP60-11, SbHSP60-16, SbHSP60-29, SbHSP60-8, and SbHSP60-7) portrayed higher expression. Five different stages like booting, dough, flowering, seedling, and stem elongation were taken for studying the expression profiles. SbHSP60s were found expressed (either upregulated or downregulated) in all developmental stages. As compared with other SbHSP60 genes, expression profiling of 11 SbHSP60 genes such as SbHSP60-2, SbHSP60-13, SbHSP60-12, SbHSP60-5, SbHSP60-7, SbHSP60-17, SbHSP60-18, SbHSP60-11, SbHSP60-16, SbHSP60-29, and SbHSP60-8 have been found higher compared to other SbHSP60 genes (Fig. 6B). High expressions of SbHSP60s during developmental stages may indicate their involvement in development-related cellular activities followed by metabolic or physiological changes that affect the gene regulation under abiotic stresses. Hierarchical clustering based on expression profiles of SbHSP60s under drought, heat, salt, and cold allowed grouping of these 30 genes in two different clusters. One of these clusters contained only one SbHSP60-25 gene, which depicts upregulation under the experimental conditions. The remaining SbHSP60s are distributed among other subclusters of the second and major cluster (Fig. 6C). The heat map of different SbHSP60s following abiotic stresses showed significantly altered expression profiling, either downregulation or upregulation up to 6.60-folds, especially in SbHSP60-20. The SbHSP10-2 and SbHSP60-20 along with the cluster of eight SbHSP60s (SbHSP60-26, SbHSP10-4, SbHSP60-2, SbHSP10-6, SbHSP60-1, SbHSP60-15, SbHSP60-27, and SbHSP60-25) displayed upregulation under abiotic stress conditions.
Fig. (6).
Hierarchical clustering of SbHSP60s based on their expression in (A): 6 different tissues; (B) 5 different developmental stages and C) under different abiotic stress conditions. (A higher resolution / colour version of this figure is available in the electronic copy of the article).
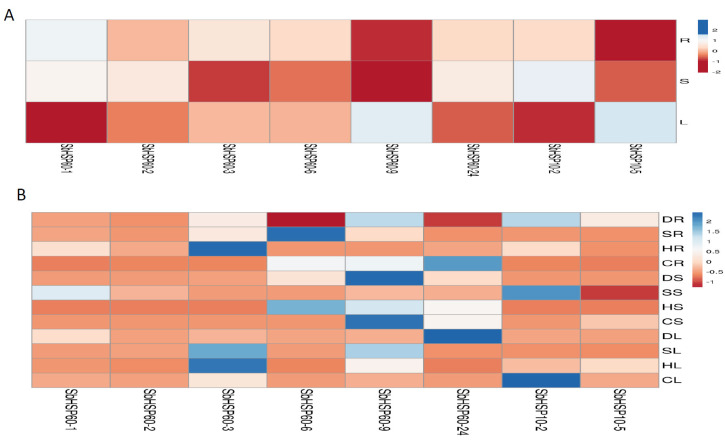
3.10. Expression Profiling of SbHSP60 Genes
Further to find out the expression levels of selected SbHSP60 genes, qRT-PCR was performed under different abiotic stress conditions. Of the 36, only 8 genes; SbHSP60-1, 2, 3, 6, 9, 24, HSP10-2, and 10-5 were selected based on the number of introns, homology with other crops, subcellular localizations and their microarray expressions. Expression profiling of 8 selected SbHSP60 genes was carried out in root, stem and leaf tissues of S. bicolor. All the 8 genes depicted uniform expressions only in the leaf tissues. All the 8 selected genes portrayed low expressions in stems (except under cold), moderate profiles in roots (but not under cold), and leaves. HSP60-1 displayed 2.38-folds higher expression under drought, 2.92- under salt, 6.68- under heat, and 0.11- folds under cold stress in roots. In stem tissues, 0.25-, 7.74-, 0.18-, and 0.43-fold higher expressions were recorded under drought, salt, heat and cold stresses, respectively. Drought exposed leaves of HSP60-1 exhibited high (12.66-folds) expressions in comparison with salt (1.32-folds), heat (0.18- folds) and cold (4.73-folds) stresses (Figs. 7, S6 and Table S7 (1.5MB, pdf) ). Root tissues of HSP60-2 exhibited 2.22- and 2.01-folds higher expressions under drought and heat stresses, respectively. Stem tissues of HSP60-2 also showed 2.81-folds higher expressions when exposed to salt stress. Both root and leaf tissues of HSP60-3 displayed higher expressions in comparison with stem and the expression levels were 4.04-, 14.32-, and 29.24-folds higher in root tissues under drought, salt and heat stresses, respectively. Leaf tissues also displayed 3.79-, 19.07-, 3.13-, and 23.97-folds higher expressions when plants were subjected to drought, salt, heat and cold stresses, respectively. Root tissues of HSP60-6 exhibited 47.17-folds under salt and 9.44-folds higher expressions in cold stress. Surprisingly, stem tissues displayed 4.16- (drought), 2.11- (salt), and 23.86-folds (heat) higher expressions than the leaf. The gene HSP60-9 was upregulated under drought (5.92-folds in roots, 17.63-folds in stems, and 3.34-folds in leaf), salt (10.05-folds in roots, 2.87-folds in stem and 16.03-folds in leaf), cold (9.98-folds in root, 224.93-folds in stem, and 5.91-folds in leaf). Stems exposed to heat stress showed 18.33-folds higher expressions, but not the other two tissues (Table S7 (1.5MB, pdf) ). Stem exhibited 4.16-, 2.11-, 23.6-, and 1.56-folds higher expressions under drought, salt, heat and cold stresses, respectively. In the case of leaf, 1.2-, 1.16-, 0.05- heat and 0.11-folds higher expressions under cold stress were noticed. Expression of root SbHSP60-24 was significantly high (18.63-folds) under cold stress, but not under other stresses. But the expression of the same gene in the stem was 3.24-, 2.66-, 12.35-, and 82.32- folds higher under drought, salt, heat and cold stresses respectively. Leaf, on the other hand, showed high expression (71.67-folds) only under drought, but not under other abiotic stresses. Expression of SbHSP10-2 was 5.96-, and 5.81- folds higher in the roots subjected to drought and heat stresses, respectively. Stem tissues displayed 11.65-folds expression under salt, and 100.65-folds in leaf under cold stress but not in other abiotic stresses. In the case of roots, the expression levels of SbHSP10-5 were 3.96-, 0.59-, 0.04- and 0.01-folds under drought, salt, heat and cold-stresses, respectively. Expression levels in the stem varied from 0.05- (drought), 0.03- (salt), 0.24- (heat) to 30.55-folds under cold stress. The gene showed 0.01-folds higher expression in drought, 0.42- under salt, 0.71- under heat and 3.79-folds under cold stress conditions in the leaf. Surprisingly, any of the HSP60s, HSP10-2 and HSP10-5 genes did not show any expressions under diverse developmental stages. From the expression analysis, HSP60-9 and HSP60-24 for drought, HSP60-3 and HSP60-6 for salt, HSP60-3 and HSP60-6 for heat, HSP60-6 and HSP10-2 for cold stress have been found as candidate genes in S. bicolor but need further validation (Figs. 7, S6 and Table S7 (1.5MB, pdf) ).
4. DISCUSSION
The HSP60 chaperonins play a critical role in folding and aggregation of other proteins present in chloroplasts and mitochondria which are often subjected to stress [14, 26]. The systematic genome-wide analysis of S. bicolor resulted in the identification of 36 genes, and subdivided into 2 groups; the HSP60 chaperonins with 30 and HSP10 co-chaperonins with 6. Under stress conditions, HSP60s interact with co-chaperones HSP10 and alleviate the aggregation of denatured proteins and step up the refolding of non-native proteins [73]. The number of chaperonins vary in plants, for example 29 in A. thaliana [21, 74, 75], 29 in rice [76], 20 in foxtail millet [77], 49 in Populus trichocarpa [78] and 16 in pepper [35]. Based on the earlier reports of their sub-cellular localization, SbHSP60 genes were subdivided into 3 groups, the cytosol/nucleus, mitochondrial and chloroplast, all of them containing the conserved HSP60_TCP1 domain. But in the present study, 2 additional subgroups were detected based on their localization in plastids (SbHSP60-9 and 16), and vacuole (SbHSP10-2). The HSP60s with the highest number of introns, and HSP10s with a smaller number of introns are consistent with the earlier records of Zhang et al. [79]. The majority of them were hydrophilic, acidic, unstable with a higher aliphatic index, indicating that these are the characteristic features of stress proteins [80]. The motifs were remarkably conserved within respective subgroups, consistent with the earlier reports [77, 79]. The duplicated genes showed a similar number of introns, conserved motifs, and properties. In the present investigation, 11 of the 12 total duplications were segmental, indicating the expansion of the SbHSP60 family through segmental duplication events [81, 82], which is in confirmation with the earlier reports in Populus and Setaria [77, 79].
The HSP60s contain several phosphorylation sites, post- translational modifications and ubiquitination, which are perhaps the reasons behind the multifaceted nature of these proteins. Phosphorylation of the HSPs under dehydration triggers the defense regulatory pathway [83]. By protein-protein interactions, HSP60s mediate the multiple stress-tolerant pathways through phosphorylation. The protein-protein interaction network analysis illustrates that SbHSP60s show interaction with phosphatidylinositol-4-phoshate 5-kinase, which plays a pivotal role in flowering, and plant growth under environmental stress. It phosphorylates phosphatidylinositol-4-phosphate to make it to phosphatidylinositol-4,5-bisphosphate, which is the precursor of inositol-1,4,5-triphosphate and diacylglycerol, and activates cellular proteins that participate in signal transduction and cytoskeletal organization [84]. HSP60s interact with histidine kinase, participate in ethylene, and cytokinin signalling, osmosensing, mega-gametophyte development, cold perception, salt and drought stress resistance [85]. It is known that glutathione S-transferase (GST) contributes to both biotic and abiotic stress tolerance [86]. There are more chances of denaturation of proteins such as GST in stress conditions and HSPs interact with GST and act as molecular chaperones and develop into an HSP-GST complex to restore and maintain its active form. Thus, the chaperonins play very critical roles during stress situations in plants. Many miRNAs are implicated in plant growth, development, metabolism, in abiotic and pathogen stress resistance [87, 88]. In the present investigation, miR171 appears to target several SbHSP60 genes. miR171 was also found in Arabidopsis, which response during all of the abiotic stress conditions [89]. miR393, and miR319 were upregulated under drought stress in rice [90], miR528 and 397 exhibited enhanced expression under arsenic stress [91], miR172c targeted AP2-like transcription factor and played key roles in flowering and abiotic stress, and its over-expression resulted in enhanced water deficit and salt tolerance in A. thaliana [92], inferring the prime roles that miRNAs play during plant development.
Promoter analysis of the SbHSP60 family genes indicated that they may be associated with abiotic and biotic stress tolerance along with other developmental-responsive elements [93-95]. The presence of ABA-responsive elements in the promoter regions indicates that these genes may work in an ABA-dependent manner. This infers that HSP60s play vital roles in multiple stresses. Chloroplast HSP60 in Arabidopsis showed enhanced expression levels under high temperature and drought stresses [96], and in pepper, almost all the candidate genes were expressed under multiple abiotic stresses [35], indicating the important roles that these genes play during stress.
In the present investigation, 6 of the HSP10 subgroup genes identified were found to have similar functions to that of the genes reported earlier in Arabidopsis [21]. Chloroplast chaperonins can physically interact with each other, and usually, assist in the refolding of two different target proteins. Also, successful partitioning to the native state-required ATP hydrolysis besides chaperonin 10 [42]. The chloroplast HSP21, the functional homologue of the mitochondrial HSP10 and HSP21 proteins, were identified in Arabidopsis and pea [97], but not noticed in S. bicolor. Sorghum contains 3 chloroplast-localized HSP10s, which exhibit their independent evolution in plants from endocytic events, while only 2 of them were reported in Arabidopsis [21]. Kim et al. [98], also found that OsHSP60α1 encodes the plastid chaperonin 60α subunit and is essential for folding of rbcL protein, inferring the importance of chaperonins in protein protection.
Earlier reports indicated the participation of Hsfs and HSPs in several biological processes like in plant development and stress tolerance [4, 76, 99]. In the present investigation, 8 selected HSP60 genes showed divergent expression levels in roots, stems and leaf tissues, majority of them with high expressions in leaf tissues in comparison with roots and stems implicating their role in leaf developmental processes or protection under stress. Downregulation of SbHSP60 genes observed in the stem tissues is contrary to the earlier reports in Populus [79]. HSPs play critical roles in conferring tolerance against multiple abiotic stresses [99, 100], indicating their overlap response which are crucial in the crosstalk of various abiotic stress conditions [76, 99]. This is akin to the implication of SbHSP60-3, 6, 9, 24 and SbHSP10-2 in various tissues under diverse stress conditions, which indicates their crosstalk response. The selected genes exhibited higher levels of upregulation under cold stress (100- and 224-folds) in comparison with drought and salt stresses (Table S7 (1.5MB, pdf) ). Cross checking of the genes implicated in developmental activities revealed that they are not involved in abiotic stress conditions and vice versa. Thus, there is no functional overlap among the genes between the two different events. But, in the case of PgHSP10, abundant expression was recorded under salt, heat and drought compared to cold stress [43]. In the present investigation, SbHSP10-2, 100-fold enhanced expression was observed under cold stress, especially in the leaf, indicating that it may be involved in the protection of low-temperature stress in the leaves. Turhan et al. [101], demonstrated upregulation of HSP60 in grafted tomato under heat stress, while Taj et al. [102], in AT1G55490 (Arabidopsis) and its homologue PtHSP60-33 by Yer et al. [78], under salt stress, indicating the involvement of HSP60s in multiple stress tolerance. In Arabidopsis and Zea, HSP60 genes showed development-mediated expression during seed germination under heat stress, and in the assembly of macromolecules necessary for mitochondrial biogenesis [33]. Thus, a functional overlap of the genes was noticed in certain plants. In the present investigation, the selected SbHSP60-1 and 3 genes exhibited high levels of expression in leaf tissues under drought and salt stresses, indicating the prominent roles they play in leaf rather than stem and root. These results are indistinguishable from the results reported in Setaria italica [77]. In the present study, HSP10-2 was found as a candidate gene, which exhibited enhanced expression under drought, salt heat and cold stress conditions. This co-chaperone may interact with HSP60s which participate in the aggregation of denatured proteins and refolding the non-native proteins, as has been pointed out by Guo et al. [73]. The majority of the selected SbHSP60 genes showed the highest expression levels under drought, and salt stresses followed by cold stress.
CONCLUSION
Taken together, our findings provide an early insight into the role of HSP60 genes in plant development and abiotic stress tolerance. The results obtained further pave the path for characterization of HSP60s in other crops as well and for understanding the mechanisms associated with stress biology, thereby crop plants can grow better under such harsh environments.
Fig. (7).
qRT-PCR expression analysis of selected SbHSP60 genes A) in root, stem, and leaf tissues. B) under drought, salt, heat and cold stress in root, stem and leaf.Abbreviations: (DR: Drought root, DS: Drought stem, DL: Drought Leaf, SR: Salt root, SS: Salt stem, SL: Salt leaf, HR: Heat root, HS: Heat stem, HL: Heat leaf, CR: Cold root, CS: Cold stem, CL: Cold leaf). (A higher resolution / colour version of this figure is available in the electronic copy of the article).
ACKNOWLEDGEMENTS
Declared none.
ETHICS APPROVAL AND CONSENT TO PARTICIPATE
Not applicable.
HUMAN AND ANIMAL RIGHTS
No Animals/Humans were used for studies that are the basis of this research.
RESEARCH INVOLVING PLANTS
All experiments on plants used in the present research were in accordance with the international guidelines.
CONSENT FOR PUBLICATION
All authors have read and approved the manuscript for publication.
AVAILABILITY OF DATA AND MATERIALS
Not applicable.
FUNDING
None.
CONFLICT OF INTEREST
The authors declare no conflict of interest, financial or otherwise.
SUPPLEMENTARY MATERIAL
Fig. S1 MEME identified motif sequences of Sorghum HSP60 proteins.
Fig. S2 MEME identified motif sequences of Sorghum HSP10 proteins.
Fig. S3 Gene-gene interaction and GO annotation analysis of SbHSP60s.
Fig. S4 STRING identified protein-protein interactions of HSP60 in Sorghum.
Fig. S5 Protein - chemical interaction network of SbHSP60s.
Fig. S6 Relative expression values of selected SbHSP60 genes in the root, stem, and leaves under drought, salt, heat and cold stress obtained through qRT-PCR analysis (DR: Drought root, DS: Drought stem, DL: Drought leaf, SR: Salt root, SS: Salt stem, SL: Salt leaf, HR: Heat root, HS: Heat stem, HL: Heat leaf, CR: Cold root, CS: Cold stem, CL: Cold leaf). Each value represents mean ± SD of three replicates. *indicates significant differences calculated by t-test (*P ≤ 0.05).
Table S1. Gene-specific primers used in the gene expression analysis of SbHSP60s.
Table S2. Protein kinases that participate in the phosphorylation of SbHSP60s.
Table S3. Cis-acting elements present in the promoter regions of Sorghum HSP60 family.
Table S4. Identification of miRNAs that target SbHSP60 family of genes.
Table S5. Non-synonymous to synonymous substitution rates (dN/dS) of SbHSP60 paralogs.
Table S6. Non-synonymous to synonymous substitutions of HSP60 orthologs of Sorghum, Oryza and Arabidopsis.
Table S7. qRT-PCR expression levels of SbHSP60s.
Supplementary material is available on the publisher’s website along with the published article.
References
- 1.Canter L.W. Environmental Impact of Agricultural Production Activities. Broken Sound Parkway: NW CRC Press: 2018. [DOI] [Google Scholar]
- 2.Vaughan M., Block A., Christensen S.A., Allen L.H., Schmelz E.A. The effects of climate change associated abiotic stresses on maize phytochemical defences. Phytochem. Rev. 2018;17:37–49. doi: 10.1007/s11101-017-9508-2. [DOI] [Google Scholar]
- 3.Mantri N., Patade V., Penna S., Ford R., Pang E. Abiotic stress responses in plants. Springer New York; 2012. Abiotic stress responses in plants: Present and future. pp. 1–19. [DOI] [Google Scholar]
- 4.Wahid A., Gelani S., Ashraf M., Foolad M.R. Heat tolerance in plants: An overview. Environ. Exp. Bot. 2007;61:199–223. doi: 10.1016/j.envexpbot.2007.05.011. [DOI] [Google Scholar]
- 5.Zhang J., Jia W., Yang J., Ismail A.M. Role of ABA in integrating plant responses to drought and salt stresses. Field Crops Res. 2006;97:111–119. doi: 10.1016/j.fcr.2005.08.018. [DOI] [Google Scholar]
- 6.Sato S., Kamiyama M., Iwata T., Makita N., Furukawa H., Ikeda H. Moderate increase of mean daily temperature adversely affects fruit set of Lycopersicon esculentum by disrupting specific physiological processes in male reproductive development. Ann. Bot. 2006;97(5):731–738. doi: 10.1093/aob/mcl037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Wang L.J., Li S.H. Salicylic acid-induced heat or cold tolerance in relation to Ca2+ homeostasis and antioxidant systems in young grape plants. Plant Sci. 2006;170:685–694. doi: 10.1016/j.plantsci.2005.09.005. [DOI] [Google Scholar]
- 8.Pareek A., Sopory S.K., Bohnert H.J. Physiological Molecular and Genomic Foundation. Springer Dordrecht; 2010. Abiotic stress adaptation in plants. [DOI] [Google Scholar]
- 9.Vierling E. The roles of heat shock proteins in plant. Annu. Rev. Plant Physiol. Plant Mol. Biol. 1991;42:579–620. doi: 10.1146/annurev.pp.42.060191.003051. [DOI] [Google Scholar]
- 10.Guo M., Liu J.H., Ma X., Luo D.X., Gong Z.H., Lu M.H. The plant heat stress transcription factors (HSFs): structure, regulation, and function in response to abiotic stresses. Front. Plant Sci. 2016;7:114. doi: 10.3389/fpls.2016.00114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Hartl F.U. Molecular chaperones in cellular protein folding. Nature. 1996;381(6583):571–579. doi: 10.1038/381571a0. [DOI] [PubMed] [Google Scholar]
- 12.Wang W., Vinocur B., Shoseyov O., Altman A. Role of plant heat-shock proteins and molecular chaperones in the abiotic stress response. Trends Plant Sci. 2004;9(5):244–252. doi: 10.1016/j.tplants.2004.03.006. [DOI] [PubMed] [Google Scholar]
- 13.Gupta S.C., Sharma A., Mishra M., Mishra R.K., Chowdhuri D.K. Heat shock proteins in toxicology: how close and how far? Life Sci. 2010;86(11-12):377–384. doi: 10.1016/j.lfs.2009.12.015. [DOI] [PubMed] [Google Scholar]
- 14.Lubben T.H., Donaldson G.K., Viitanen P.V., Gatenby A.A. Several proteins imported into chloroplasts form stable complexes with the GroEL-related chloroplast molecular chaperone. Plant Cell. 1989;1(12):1223–1230. doi: 10.1105/tpc.1.12.1223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Hartl F.U., Bracher A., Hayer-Hartl M. Molecular chaperones in protein folding and proteostasis. Nature. 2011;475(7356):324–332. doi: 10.1038/nature10317. [DOI] [PubMed] [Google Scholar]
- 16.Balchin D., Hayer-Hartl M., Hartl F.U. In vivo aspects of protein folding and quality control. Science. 2016;353(6294):aac4354. doi: 10.1126/science.aac4354. [DOI] [PubMed] [Google Scholar]
- 17.Bukau B., Horwich A.L. The Hsp70 and Hsp60 chaperone machines. Cell. 1998;92(3):351–366. doi: 10.1016/S0092-8674(00)80928-9. [DOI] [PubMed] [Google Scholar]
- 18.Ditzel L., Löwe J., Stock D., Stetter K.O., Huber H., Huber R., Steinbacher S. Crystal structure of the thermosome, the archaeal chaperonin and homolog of CCT. Cell. 1998;93(1):125–138. doi: 10.1016/S0092-8674(00)81152-6. [DOI] [PubMed] [Google Scholar]
- 19.Sigler P.B., Xu Z., Rye H.S., Burston S.G., Fenton W.A., Horwich A.L. Structure and function in GroEL-mediated protein folding. Annu. Rev. Biochem. 1998;67:581–608. doi: 10.1146/annurev.biochem.67.1.581. [DOI] [PubMed] [Google Scholar]
- 20.Saibil H. Chaperone machines for protein folding, unfolding and disaggregation. Nat. Rev. Mol. Cell Biol. 2013;14(10):630–642. doi: 10.1038/nrm3658. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Hill J.E., Hemmingsen S.M. Arabidopsis thaliana type I and II chaperonins. Cell Stress Chaperones. 2001;6(3):190–200. doi: 10.1379/1466-1268(2001)006<0190:ATTIAI>2.0.CO;2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Braig K., Otwinowski Z., Hegde R., Boisvert D.C., Joachimiak A., Horwich A.L., Sigler P.B. The crystal structure of the bacterial chaperonin GroEL at 2.8 A. Nature. 1994;371(6498):578–586. doi: 10.1038/371578a0. [DOI] [PubMed] [Google Scholar]
- 23.Peng L., Fukao Y., Myouga F., Motohashi R., Shinozaki K., Shikanai T. A chaperonin subunit with unique structures is essential for folding of a specific substrate. PLoS Biol. 2011;9(4):e1001040. doi: 10.1371/journal.pbio.1001040. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 24.Martel R., Cloney L.P., Pelcher L.E., Hemmingsen S.M. Unique composition of plastid chaperonin-60: alpha and beta polypeptide-encoding genes are highly divergent. Gene. 1990;94(2):181–187. doi: 10.1016/0378-1119(90)90385-5. [DOI] [PubMed] [Google Scholar]
- 25.Nishio K., Hirohashi T., Nakai M. Chloroplast chaperonins: evidence for heterogeneous assembly of alpha and beta Cpn60 polypeptides into a chaperonin oligomer. Biochem. Biophys. Res. Commun. 1999;266(2):584–587. doi: 10.1006/bbrc.1999.1868. [DOI] [PubMed] [Google Scholar]
- 26.Suzuki K., Nakanishi H., Bower J., Yoder D.W., Osteryoung K.W., Miyagishima S.Y. Plastid chaperonin proteins Cpn60 α and Cpn60 β are required for plastid division in Arabidopsis thaliana. BMC Plant Biol. 2009;9:38. doi: 10.1186/1471-2229-9-38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Cloney L.P., Bekkaoui D.R., Feist G.L., Lane W.S., Hemmingsen S.M. Brassica napus plastid and mitochondrial chaperonin-60 proteins contain multiple distinct polypeptides. Plant Physiol. 1994;105(1):233–241. doi: 10.1104/pp.105.1.233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Saibil H. Molecular chaperones: containers and surfaces for folding, stabilising or unfolding proteins. Curr. Opin. Struct. Biol. 2000;10(2):251–258. doi: 10.1016/S0959-440X(00)00074-9. [DOI] [PubMed] [Google Scholar]
- 29.Horwich A.L., Fenton W.A., Chapman E., Farr G.W. Two families of chaperonin: physiology and mechanism. Annu. Rev. Cell Dev. Biol. 2007;23:115–145. doi: 10.1146/annurev.cellbio.23.090506.123555. [DOI] [PubMed] [Google Scholar]
- 30.Schroda M. The Chlamydomonas genome reveals its secrets: chaperone genes and the potential roles of their gene products in the chloroplast. Photosynth. Res. 2004;82(3):221–240. doi: 10.1007/s11120-004-2216-y. [DOI] [PubMed] [Google Scholar]
- 31.Trösch R., Mühlhaus T., Schroda M., Willmund F. ATP-dependent molecular chaperones in plastids--More complex than expected. Biochim. Biophys. Acta. 2015;1847(9):872–888. doi: 10.1016/j.bbabio.2015.01.002. [DOI] [PubMed] [Google Scholar]
- 32.Hemmingsen S.M., Woolford C., van der Vies S.M., Tilly K., Dennis D.T., Georgopoulos C.P., Hendrix R.W., Ellis R.J. Homologous plant and bacterial proteins chaperone oligomeric protein assembly. Nature. 1988;333(6171):330–334. doi: 10.1038/333330a0. [DOI] [PubMed] [Google Scholar]
- 33.Wilson R.H., Hayer-Hartl M. Complex chaperone dependence of Rubisco biogenesis. Biochemistry. 2018;57(23):3210–3216. doi: 10.1021/acs.biochem.8b00132. [DOI] [PubMed] [Google Scholar]
- 34.Ruggero D., Ciammaruconi A., Londei P. The chaperonin of the archaeon Sulfolobus solfataricus is an RNA-binding protein that participates in ribosomal RNA processing. EMBO J. 1998;17(12):3471–3477. doi: 10.1093/emboj/17.12.3471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Hsu Y.W., Juan C.T., Wang C.M., Jauh G.Y. Mitochondrial heat shock protein 60s interact with what’s this factor 9 to regulate RNA splicing of ccmFC and rpl2. Plant Cell Physiol. 2019;60(1):116–125. doi: 10.1093/pcp/pcy199. [DOI] [PubMed] [Google Scholar]
- 36.Weiss C., Bonshtien A., Farchi-Pisanty O., Vitlin A., Azem A. Cpn20: siamese twins of the chaperonin world. Plant Mol. Biol. 2009;69(3):227–238. doi: 10.1007/s11103-008-9432-3. [DOI] [PubMed] [Google Scholar]
- 37.Prasad T.K., Stewart C.R. cDNA clones encoding Arabidopsis thaliana and Zea mays mitochondrial chaperonin HSP60 and gene expression during seed germination and heat shock. Plant Mol. Biol. 1992;18(5):873–885. doi: 10.1007/BF00019202. [DOI] [PubMed] [Google Scholar]
- 38.Xu Q., Qin Y. Molecular cloning of heat shock protein 60 (PtHSP60) from Portunus trituberculatus and its expression response to salinity stress. Cell Stress Chaperones. 2012;17(5):589–601. doi: 10.1007/s12192-012-0334-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Haq S.U., Khan A., Ali M., Gai W.X., Zhang H.X., Yu Q.H., Yang S.B., Wei A.M., Gong Z.H. Knockdown of CaHSP60-6 confers enhanced sensitivity to heat stress in pepper (Capsicum annuum L.). Planta. 2019;250(6):2127–2145. doi: 10.1007/s00425-019-03290-4. [DOI] [PubMed] [Google Scholar]
- 40.Jung K.H., Ko H.J., Nguyen M.X., Kim S.R., Ronald P., An G. Genome-wide identification and analysis of early heat stress responsive genes in rice. J. Plant Biol. 2012;55:458–468. doi: 10.1007/s12374-012-0271-z. [DOI] [Google Scholar]
- 41.Hartman D.J., Dougan D., Hoogenraad N.J., Høj P.B. Heat shock proteins of barley mitochondria and chloroplasts. Identification of organellar hsp 10 and 12: putative chaperonin 10 homologues. FEBS Lett. 1992;305(2):147–150. doi: 10.1016/0014-5793(92)80883-I. [DOI] [PubMed] [Google Scholar]
- 42.Viitanen P.V., Schmidt M., Buchner J., Suzuki T., Vierling E., Dickson R., Lorimer G.H., Gatenby A., Soll J. Functional characterization of the higher plant chloroplast chaperonins. J. Biol. Chem. 1995;270(30):18158–18164. doi: 10.1074/jbc.270.30.18158. [DOI] [PubMed] [Google Scholar]
- 43.Nitnavare R.B., Yeshvekar R.K., Sharma K.K., Vadez V., Reddy M.K., Reddy P.S. Molecular cloning, characterization and expression analysis of a heat shock protein 10 (Hsp10) from Pennisetum glaucum (L.), a C4 cereal plant from the semi-arid tropics. Mol. Biol. Rep. 2016;43(8):861–870. doi: 10.1007/s11033-016-4012-0. [DOI] [PubMed] [Google Scholar]
- 44.Carter P.R., Hicks D.R., Oplinger E.S., Doll J.D., Bundy L.G., Schuler R.T., Holmes B.T. Grain Sorghum (Milo). 2020 Available from: http://corn.agronomy.wisc.edu/Crops/SorghumGrain.aspx.
- 45.Premachandra G.S., Hahn D.T., Joly R.J. Leaf water relations and gas exchange in two grain sorghum genotypes differing in their pre-and post-flowering drought tolerance. J. Plant Physiol. 1994;143:96–101. doi: 10.1016/S0176-1617(11)82103-6. [DOI] [Google Scholar]
- 46.Rosenow D.T. Breeding for lodging resistance in sorghum. Proceedings of 32nd Annual corn and sorghum industry research conference. 1997:171–185. [Google Scholar]
- 47.Borrell A.K., Graeme L.H., Andrew C.L.D. Does maintaining green leaf area in sorghum improve yield under drought? I. Leaf growth and senescence. Crop Sci. 2000;40:1026. doi: 10.2135/cropsci2000.4041026x. [DOI] [Google Scholar]
- 48.Kusaba M., Tanaka A., Tanaka R. Stay-green plants: what do they tell us about the molecular mechanism of leaf senescence. Photosynth. Res. 2013;117(1-3):221–234. doi: 10.1007/s11120-013-9862-x. [DOI] [PubMed] [Google Scholar]
- 49.Borrell A.K., van Oosterom E.J., Mullet J.E., George-Jaeggli B., Jordan D.R., Klein P.E., Hammer G.L. Stay-green alleles individually enhance grain yield in sorghum under drought by modifying canopy development and water uptake patterns. New Phytol. 2014;203(3):817–830. doi: 10.1111/nph.12869. [DOI] [PubMed] [Google Scholar]
- 50.Rosenow D., Quisenberry J.E., Wendt C.E., Clark L.E. Drought tolerant sorghum and cotton germplasm. Agric. Water Manage. 1983;7:207–222. doi: 10.1016/0378-3774(83)90084-7. [DOI] [Google Scholar]
- 51.Nagaraju M., Kumar S.A., Reddy P.S., Kumar A., Rao D.M., Kavi K.P.B. Genome-scale identification, classification, and tissue specific expression analysis of late embryogenesis abundant (LEA) genes under abiotic stress conditions in Sorghum bicolor L. PLoS One. 2019;14(1):e0209980. doi: 10.1371/journal.pone.0209980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.R R.K., N S N., S P A., Sinha D., Veedin Rajan V.B., Esthaki V.K., D’Silva P. HSPIR: a manually annotated heat shock protein information resource. Bioinformatics. 2012;28(21):2853–2855. doi: 10.1093/bioinformatics/bts520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Monaco M.K., Stein J., Naithani S., Wei S., Dharmawardhana P., Kumari S., Amarasinghe V., Youens-Clark K., Thomason J., Preece J., Pasternak S., Olson A., Jiao Y., Lu Z., Bolser D., Kerhornou A., Staines D., Walts B., Wu G., D’Eustachio P., Haw R., Croft D., Kersey P.J., Stein L., Jaiswal P., Ware D. Gramene 2013: comparative plant genomics resources. Nucleic Acids Res. 2014;42(Database issue):D1193–D1199. doi: 10.1093/nar/gkt1110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Letunic I., Copley R.R., Schmidt S., Ciccarelli F.D., Doerks T., Schultz J., Ponting C.P., Bork P. SMART 4.0: towards genomic data integration. Nucleic Acids Res. 2004;32(Database issue):D142–D144. doi: 10.1093/nar/gkh088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Gasteiger E., Hoogland C., Gattiker A., Duvaud S., Wilkins M.R., Appel R.D., Bairoch A. Protein identification and analysis tools on the ExPASy Server. In: Walker J.M., editor. The Proteomics Protocols Handbook. Humana Press; 2005. pp. 571–607. [DOI] [Google Scholar]
- 56.Guo A.Y., Zhu Q.H., Chen X., Luo J.C., Chuan Y.L. [GSDS: a gene structure display server]. Yi Chuan. 2007;29(8):1023–1026. doi: 10.1360/yc-007-1023. [DOI] [PubMed] [Google Scholar]
- 57.Horton P., Park K.J., Obayashi T., Fujita N., Harada H., Adams-Collier C.J., Nakai K. WoLF PSORT: protein localization predictor. Nucleic Acids Res. 2007;35(Web Server issue):W585-7. doi: 10.1093/nar/gkm259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Möller S., Croning M.D.R., Apweiler R. Evaluation of methods for the prediction of membrane spanning regions. Bioinformatics. 2001;17(7):646–653. doi: 10.1093/bioinformatics/17.7.646. [DOI] [PubMed] [Google Scholar]
- 59.Blom N., Sicheritz-Pontén T., Gupta R., Gammeltoft S., Brunak S. Prediction of post-translational glycosylation and phosphorylation of proteins from the amino acid sequence. Proteomics. 2004;4(6):1633–1649. doi: 10.1002/pmic.200300771. [DOI] [PubMed] [Google Scholar]
- 60.Bailey T.L., Williams N., Misleh C., Li W.W. MEME: discovering and analyzing DNA and protein sequence motifs. Nucleic Acids Res. 2006;34(Web Server issue) Suppl. 2:W369-73. doi: 10.1093/nar/gkl198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Higo K., Ugawa Y., Iwamoto M., Korenaga T. Plant cis-acting regulatory DNA elements (PLACE) database: 1999. Nucleic Acids Res. 1999;27(1):297–300. doi: 10.1093/nar/27.1.297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Lescot M., Déhais P., Thijs G., Marchal K., Moreau Y., Van de Peer Y., Rouzé P., Rombauts S. PlantCARE, a database of plant cis-acting regulatory elements and a portal to tools for in silico analysis of promoter sequences. Nucleic Acids Res. 2002;30(1):325–327. doi: 10.1093/nar/30.1.325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Tamura K., Stecher G., Peterson D., Filipski A., Kumar S. MEGA6: Molecular evolutionary genetics analysis version 6. Mol. Biol. Evol. 2013;30(12):2725–2729. doi: 10.1093/molbev/mst197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Suyama M., Torrents D., Bork P. PAL2NAL: robust conversion of protein sequence alignments into the corresponding codon alignments. Nucleic Acids Res. 2006;34(Web Server issue):W609-12. doi: 10.1093/nar/gkl315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Dai X., Zhao P.X. psRNATarget: a plant small RNA target analysis server. Nucleic Acids Res. 2011;39(Web Server issue):W155-9. doi: 10.1093/nar/gkr319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Tian T., You Q., Zhang L., Yi X., Yan H., Xu W., Su Z. SorghumFDB: sorghum functional genomics database with multidimensional network analysis. Database (Oxford) 2016;2016:baw099. doi: 10.1093/database/baw099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Hruz T., Laule O., Szabo G., Wessendorp F., Bleuler S., Oertle L., Widmayer P., Gruissem W., Zimmermann P. Genevestigator v3: A reference expression database for the meta analysis of transcriptomes. Adv. Bioinformatics. 2008:420747. doi: 10.1155/2008/420747. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Grennan A.K. Genevestigator. Facilitating web-based gene-expression analysis. Plant Physiol. 2006;141(4):1164–1166. doi: 10.1104/pp.104.900198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Kumar A., Batra R., Gahlaut V., Gautam T., Kumar S., Sharma M., Tyagi S., Singh K.P., Balyan H.S., Pandey R., Gupta P.K. Genome-wide identification and characterization of gene family for RWP-RK transcription factors in wheat (Triticum aestivum L.). PLoS One. 2018;13(12):e0208409. doi: 10.1371/journal.pone.0208409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Ye J., Coulouris G., Zaretskaya I., Cutcutache I., Rozen S., Madden T.L. Primer-BLAST: a tool to design target-specific primers for polymerase chain reaction. BMC Bioinformatics. 2012;13:134. doi: 10.1186/1471-2105-13-134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Sudhakar Reddy P., Srinivas R.D., Sivasakthi K., Bhatnagar-Mathur P., Vadez V., Sharma K.K. Evaluation of sorghum [Sorghum bicolor (L.)] reference genes in various tissues and under abiotic stress conditions for quantitative real-time PCR data normalization. Front. Plant Sci. 2016;7:529. doi: 10.3389/fpls.2016.00529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Pfaffl M.W., Horgan G.W., Dempfle L. Relative expression software tool (REST) for group-wise comparison and statistical analysis of relative expression results in real-time PCR. Nucleic Acids Res. 2002;30(9):e36. doi: 10.1093/nar/30.9.e36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Guo M., Liu J.-H., Lu J.-P., Zhai Y.F., Wang H., Gong Z.H., Wang S.B., Lu M.H. Genome-wide analysis of the CaHsp20 gene family in pepper: comprehensive sequence and expression profile analysis under heat stress. Front. Plant Sci. 2015;6:806. doi: 10.3389/fpls.2015.00806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Scharf K.D., Siddique M., Vierling E. The expanding family of Arabidopsis thaliana small heat stress proteins and a new family of proteins containing alpha-crystallin domains (Acd proteins). Cell Stress Chaperones. 2001;6(3):225–237. doi: 10.1379/1466-1268(2001)006<0225:TEFOAT>2.0.CO;2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Lee U., Rioflorido I., Hong S.W., Larkindale J., Waters E.R., Vierling E. The Arabidopsis ClpB/Hsp100 family of proteins: chaperones for stress and chloroplast development. Plant J. 2007;49(1):115–127. doi: 10.1111/j.1365-313X.2006.02940.x. [DOI] [PubMed] [Google Scholar]
- 76.Hu W., Hu G., Han B. Genome-wide survey and expression profiling of heat shock proteins and heat shock factors revealed overlapped and stress specific response under abiotic stresses in rice. Plant Sci. 2009;176(4):583–590. doi: 10.1016/j.plantsci.2009.01.016. [DOI] [PubMed] [Google Scholar]
- 77.Singh R.K., Jaishankar J., Muthamilarasan M., Shweta S., Dangi A., Prasad M. Genome-wide analysis of heat shock proteins in C4 model, foxtail millet identifies potential candidates for crop improvement under abiotic stress. Sci. Rep. 2016;6:32641. doi: 10.1038/srep32641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Yer E.N., Baloglu M.C., Ayan S. Identification and expression profiling of all Hsp family member genes under salinity stress in different poplar clones. Gene. 2018;678:324–336. doi: 10.1016/j.gene.2018.08.049. [DOI] [PubMed] [Google Scholar]
- 79.Zhang J., Liu B., Li J., Zhang L., Wang Y., Zheng H., Lu M., Chen J. Hsf and Hsp gene families in Populus: genome-wide identification, organization and correlated expression during development and in stress responses. BMC Genomics. 2015;16:181. doi: 10.1186/s12864-015-1398-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Rao P.K., Roxas B.A., Li Q. Determination of global protein turnover in stressed mycobacterium cells using hybrid-linear ion trap-fourier transform mass spectrometry. Anal. Chem. 2008;80(2):396–406. doi: 10.1021/ac701690d. [DOI] [PubMed] [Google Scholar]
- 81.Cannon S.B., Mitra A., Baumgarten A., Young N.D., May G. The roles of segmental and tandem gene duplication in the evolution of large gene families in Arabidopsis thaliana. BMC Plant Biol. 2004;4:10. doi: 10.1186/1471-2229-4-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Sémon M., Wolfe K.H., Wolfe K.H. Rearrangement rate following the whole-genome duplication in teleosts. Mol. Biol. Evol. 2007;24(3):860–867. doi: 10.1093/molbev/msm003. [DOI] [PubMed] [Google Scholar]
- 83.Subba P., Barua P., Kumar R., Datta A., Soni K.K., Chakraborty S., Chakraborty N. Phosphoproteomic dynamics of chickpea (Cicer arietinum L.) reveals shared and distinct components of dehydration response. J. Proteome Res. 2013;12(11):5025–5047. doi: 10.1021/pr400628j. [DOI] [PubMed] [Google Scholar]
- 84.Mikami K., Katagiri T., Iuchi S., Yamaguchi-Shinozaki K., Shinozaki K. A gene encoding phosphatidylinositol-4-phosphate 5-kinase is induced by water stress and abscisic acid in Arabidopsis thaliana. Plant J. 1998;15(4):563–568. doi: 10.1046/j.1365-313X.1998.00227.x. [DOI] [PubMed] [Google Scholar]
- 85.Tran L.S.P., Urao T., Qin F., Maruyama K., Kakimoto T., Shinozaki K., Yamaguchi-Shinozaki K. Functional analysis of AHK1/ATHK1 and cytokinin receptor histidine kinases in response to abscisic acid, drought, and salt stress in Arabidopsis. Proc. Natl. Acad. Sci. USA. 2007;104(51):20623–20628. doi: 10.1073/pnas.0706547105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Dalton D.A., Boniface C., Turner Z., Lindahl A., Kim H.J., Jelinek L., Govindarajulu M., Finger R.E., Taylor C.G. Physiological roles of glutathione s-transferases in soybean root nodules. Plant Physiol. 2009;150(1):521–530. doi: 10.1104/pp.109.136630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Yang C., Li D., Mao D., Liu X., Ji C., Li X., Zhao X., Cheng Z., Chen C., Zhu L. Overexpression of microRNA319 impacts leaf morphogenesis and leads to enhanced cold tolerance in rice (Oryza sativa L.). Plant Cell Environ. 2013;36(12):2207–2218. doi: 10.1111/pce.12130. [DOI] [PubMed] [Google Scholar]
- 88.Xie F., Jones D.C., Wang Q., Sun R., Zhang B. Small RNA sequencing identifies miRNA roles in ovule and fibre development. Plant Biotechnol. J. 2015;13(3):355–369. doi: 10.1111/pbi.12296. [DOI] [PubMed] [Google Scholar]
- 89.Liu H.H., Tian X., Li Y.J., Wu C.A., Zheng C.C. Microarray-based analysis of stress-regulated microRNAs in Arabidopsis thaliana. RNA. 2008;14(5):836–843. doi: 10.1261/rna.895308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Zhao B., Liang R., Ge L., Li W., Xiao H., Lin H., Ruan K., Jin Y. Identification of drought-induced microRNAs in rice. Biochem. Biophys. Res. Commun. 2007;354(2):585–590. doi: 10.1016/j.bbrc.2007.01.022. [DOI] [PubMed] [Google Scholar]
- 91.Liu Q., Zhang H. Molecular identification and analysis of arsenite stress-responsive miRNAs in rice. J. Agric. Food Chem. 2012;60(26):6524–6536. doi: 10.1021/jf300724t. [DOI] [PubMed] [Google Scholar]
- 92.Li W., Wang T., Zhang Y., Li Y. Overexpression of soybean miR172c confers tolerance to water deficit and salt stress, but increases ABA sensitivity in transgenic Arabidopsis thaliana. J. Exp. Bot. 2016;67(1):175–194. doi: 10.1093/jxb/erv450. [DOI] [PubMed] [Google Scholar]
- 93.Bate N., Twell D. Functional architecture of a late pollen promoter: pollen-specific transcription is developmentally regulated by multiple stage-specific and co-dependent activator elements. Plant Mol. Biol. 1998;37(5):859–869. doi: 10.1023/A:1006095023050. [DOI] [PubMed] [Google Scholar]
- 94.Chen W., Provart N.J., Glazebrook J., Katagiri F., Chang H.S., Eulgem T., Mauch F., Luan S., Zou G., Whitham S.A., Budworth P.R., Tao Y., Xie Z., Chen X., Lam S., Kreps J.A., Harper J.F., Si-Ammour A., Mauch-Mani B., Heinlein M., Kobayashi K., Hohn T., Dangl J.L., Wang X., Zhu T. Expression profile matrix of Arabidopsis transcription factor genes suggests their putative functions in response to environmental stresses. Plant Cell. 2002;14(3):559–574. doi: 10.1105/tpc.010410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Nishiuchi T., Shinshi H., Suzuki K. Rapid and transient activation of transcription of the ERF3 gene by wounding in tobacco leaves: possible involvement of NtWRKYs and autorepression. J. Biol. Chem. 2004;279(53):55355–55361. doi: 10.1074/jbc.M409674200. [DOI] [PubMed] [Google Scholar]
- 96.Xu C., Huang B. Comparative analysis of drought responsive proteins in Kentucky blue grass cultivars contrasting in drought tolerance. Crop Sci. 2010;50:2543–2552. doi: 10.2135/cropsci2010.03.0152. [DOI] [Google Scholar]
- 97.Schlicher T., Soll J. Molecular chaperones are present in the thylakoid lumen of pea chloroplasts. FEBS Lett. 1996;379(3):302–304. doi: 10.1016/0014-5793(95)01534-5. [DOI] [PubMed] [Google Scholar]
- 98.Kim S.R., Yang J.I., An G. OsCpn60α1, encoding the plastid chaperonin 60α subunit, is essential for folding of rbcL. Mol. Cells. 2013;35(5):402–409. doi: 10.1007/s10059-013-2337-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Swindell W.R., Huebner M., Weber A.P. Transcriptional profiling of Arabidopsis heat shock proteins and transcription factors reveals extensive overlap between heat and non-heat stress response pathways. BMC Genomics. 2007;8:125. doi: 10.1186/1471-2164-8-125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Hahn A., Bublak D., Schleiff E., Scharf K.D. Crosstalk between Hsp90 and Hsp70 chaperones and heat stress transcription factors in tomato. Plant Cell. 2011;23(2):741–755. doi: 10.1105/tpc.110.076018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Turhan E., Ergin S., Aydogan C., Ozturk N. Influence of grafting on heat shock proteins of tomato (Lycopersicon esculentum Mill) plants under heat stress. J. Biotechnol. 2016;231:27. doi: 10.1016/j.jbiotec.2016.05.115. [DOI] [Google Scholar]
- 102.Taj G., Agarwal P., Grant M., Kumar A. MAPK machinery in plants: recognition and response to different stresses through multiple signal transduction pathways. Plant Signal. Behav. 2010;5(11):1370–1378. doi: 10.4161/psb.5.11.13020. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Fig. S1 MEME identified motif sequences of Sorghum HSP60 proteins.
Fig. S2 MEME identified motif sequences of Sorghum HSP10 proteins.
Fig. S3 Gene-gene interaction and GO annotation analysis of SbHSP60s.
Fig. S4 STRING identified protein-protein interactions of HSP60 in Sorghum.
Fig. S5 Protein - chemical interaction network of SbHSP60s.
Fig. S6 Relative expression values of selected SbHSP60 genes in the root, stem, and leaves under drought, salt, heat and cold stress obtained through qRT-PCR analysis (DR: Drought root, DS: Drought stem, DL: Drought leaf, SR: Salt root, SS: Salt stem, SL: Salt leaf, HR: Heat root, HS: Heat stem, HL: Heat leaf, CR: Cold root, CS: Cold stem, CL: Cold leaf). Each value represents mean ± SD of three replicates. *indicates significant differences calculated by t-test (*P ≤ 0.05).
Table S1. Gene-specific primers used in the gene expression analysis of SbHSP60s.
Table S2. Protein kinases that participate in the phosphorylation of SbHSP60s.
Table S3. Cis-acting elements present in the promoter regions of Sorghum HSP60 family.
Table S4. Identification of miRNAs that target SbHSP60 family of genes.
Table S5. Non-synonymous to synonymous substitution rates (dN/dS) of SbHSP60 paralogs.
Table S6. Non-synonymous to synonymous substitutions of HSP60 orthologs of Sorghum, Oryza and Arabidopsis.
Table S7. qRT-PCR expression levels of SbHSP60s.
Supplementary material is available on the publisher’s website along with the published article.
Data Availability Statement
Not applicable.