Abstract
Background
Our previous studies have revealed the roles of ribosomal protein (RP) genes in the abiotic stress responses of rice.
Methods
In the current investigation, we examine the possible involvement of these genes in insect stress responses. We have characterized the RP genes that included both Ribosomal Protein Large (RPL) and Ribosomal Protein Small (RPS) subunit genes in response to infestation by two economically important insect pests, the brown planthopper (BPH) and the Asian rice gall midge (GM) in rice. Differential transcript patterns of seventy selected RP genes were studied in a susceptible and a resistant genotype of indica rice: BPT5204 and RPNF05, respectively. An in silico analyses of the upstream regions of these genes also revealed the presence of cis-elements that are associated with wound signaling.
Results
We identified the genes that were up or downregulated in either one of the genotypes, or both of them after pest infestation. The transcript patterns of a majority of the genes were found to be temporally-regulated by both the pests. In the resistant RPNF05, BPH infestation activated RPL15, L51 and RPS5a genes while GM infestation induced RPL15, L18a, L22, L36.2, L38, RPS5, S9.2 and S25a at a certain point of time. These genes that were particularly upregulated in the resistant genotype, RPNF05, but not in BPT5204 suggest their potential involvement in plant resistance against either of the two pests studied.
Conclusion
Taken together, RPL15, L51, L18a, RPS5, S5a, S9.2, and S25a appear to be the genes with possible roles in insect resistance in rice.
Keywords: Ribosomal protein genes, brown planthopper, rice, gall midge pests, transcriptional response, resistance
1. INTRODUCTION
The global population is growing exponentially and is expected to exceed nine billion by 2050 [1]. One of the important ways of ensuring food security and feeding the burgeoning population is through enhanced and sustained productivity of staple crops. Rice is a principal source of nutrition and a staple cereal for more than two billion population in Asia and thus, accounts for more than 40% of the calorie consumption. Increasing demand necessitates a proportionate increase in production of rice, which is alarmingly affected by biotic and abiotic stresses that result in unstable productivity.
Biotic stress comprises attacks by insect pests, bacterial, fungal and viral pathogens, and weed competition, while abiotic stresses such as salinity, drought and extreme climates cause a huge impediment towards sustainable rice production across the world [2]. The major biotic stresses include bacterial leaf blight (BLB), fungal blast diseases and attack by insect pests like BPH, gall midge and stem borers that together account for a major share of losses across the globe [3]. Out of this, approximately 21% of the losses are solely attributed to insect pests. Among the three major groups of insect pests listed above, BPH represents sucking pests that mainly feed on phloem sap depriving the plant of nutrients, which results in plant mortality called “hopper burn”.
Gall midges are unique gall formers that do not kill the plant but hijack its normal developmental processes by inducing the formation of galls and sterility. Though these two pests are managed by the farmers through the application of insecticides, these are also amenable for management through host-plant resistance. Extensive research has identified scores of rice germplasm lines that are resistant to the attack by these pests [4]. Intensive studies have unveiled genetics of resistance leading to tagging and mapping plant resistance (R) genes and the development of linked molecular markers that are being used in marker-assisted selection (MAS). MAS has made breeding rice for insect resistance more precise with rapid pyramiding of genes for multiple and durable resistance. Concomitant research over decades has tried to understand insect resistance in rice at genetic, anatomical, physiological, biochemical and molecular levels [5]. These studies, in summary, highlight a two-tier defence system in rice against insects as in the case of pathogen attack. The first line of defence is an innate immune response of general nature triggered by molecular patterns associated with herbivory (HAMP) and or damage (DAMP) [6]. Plants recognize these molecules through receptors on the cell membrane and trigger an immune response called pattern triggered immunity (PTI). This defence is race non-specific. In response, insects secrete specialized chemicals called effectors that nullify PTI and allow the insect to feed and develop on the plants [7-10].
The second level of defence is induced by plants in response to the effectors and is termed as effector-triggered immunity (ETI) [10]. This defence is race-specific and involves both constitutive and induced responses with or without hypersensitive reaction (HR), oxidative burst, signal transduction, hormone regulation and systemic acquired resistance [11, 12]. Some of these involve secondary metabolism of synthesis of defence chemicals that are not essential for the regular growth, development and reproduction of plant. Thus, there is a compromise between primary and secondary metabolism [12].
Insect resistance has long been proposed to consist of components like nonpreference or antixenosis, antibiosis resulting in reduced fitness of the infesting insect and tolerance [13]. While the first two components constitute the active plant defence system detailed above, the tolerance component accounts for the ability of the plant to compensate for the damage inflicted by the insect and maintain near normal growth, development and yield. This component has been least studied in greater detail. This primary metabolism-mediated insect resistance in rice may involve basic functions like photosynthesis and protein synthesis. In such a situation, the ribosomal proteins (RPs) might play a role here. The present investigation is aimed at this angle of insect resistance through monitoring a subset of RP coding genes.
RPs are known to play an integral role in rRNA structure and formation of protein synthesizing machinery in cells. These are also crucial for the growth and development of all the organisms. The formation of a ribosomal complex is a compendium process requiring co-transcription, coordinated expression and assembly of hundreds of proteins [14]. The ribosomal DNA (rDNA) exists as tandemly arrayed repeats separated by spacers at nucleolus organizing regions (NORs). The nucleolus is the site of transcription of rDNA, processing of primary transcripts into different rRNAs and assembly of preribosomal subunits, which together form the linchpins of ribosome biogenesis [15-17]. The rRNAs and RPs together maintain the structural stability of ribosomes that perform diverse cellular activities required for organismal growth and development [18]. Thus, to cope up with the changing growth conditions and continuous exposure to external stimuli, which occur at different stages of the life cycle in an organism, cells must normalize the ribosome biosynthesis for its normal functioning [15, 19]. Any imbalance in ribosome biosynthesis or mutations in rRNA and RP-coding genes results in perturbed plant phenotypes, which might also be lethal to cells. Mutations in different RP genes caused varying consequences from morphological changes [20-25] to embryonic lethality [26]. For example, RPL23aA exists as two paralogs, RPL23aA and aB. While the knockout of aA resulted in an overall reduction in ribosome biogenesis producing phenotypes characterized by growth retardation and reduced fertility, the knockout of aB showed no obvious phenotypic consequences [27]. Also, RPL23aB was found to be the only paralog that did not show any phenotypic change upon knockout [28]. However, these variations in aA and aB knockout mutations have been attributed to their native expression levels, pointing to the fact that these two paralogs are functionally equivalent [29].
It has been observed that upregulation of RP genes in plants under abiotic stress conditions might suggest a differential reconstruction of protein-synthesizing machinery by the incorporation of different paralogous members of RPs in different situations [30-36]. This heterogeneity of ribosomes formed by different combinations of RP paralogs is controlled by specific cell types, developmental stages and environmental factors [34, 37]. The high ribosomal heterogeneity, a characteristic feature of sessile plants has been attributed to their potential to adapt under different environmental signals [30, 35]. Although there are no significant reports on the contribution of RP genes in conferring resistance to pests, we have identified potential Ribosomal Protein Large (RPL) subunit genes (RPL23a and RPL6) to be significantly upregulated in studies for enhanced water-use efficiency (WUE) in indica rice following gain of function mutagenesis through the activation tagging approach [38]. Subsequently, we studied the regulatory roles of all the RPL and RPS genes under different external stimuli and observed the significant up-regulation of a considerable number of these genes under several stresses including abiotic and biotic conditions in rice [39, 40]. In the present investigation, we made a transcript analysis of selected 70 out of 255 RP genes in susceptible (BPT5204) and resistant (RPNF05) genotypes of indica rice challenged by two major insect pests, BPH and GM. We have also tried to identify the potential candidate RP genes that are specifically or commonly upregulated in one or both the above genotypes.
2. MATERIALS AND METHODS
2.1. Nucleotide Sequence Retrieval of RP Genes
The nucleotide sequences of all the RP genes were retrieved from MSU Rice Genome Annotation Project data base (RGAP-DB)1 under putative function search tool using a keyword ‘ribosomal’. This search provided a list of a total of 428 genes. Because plants also contain chloroplast ribosomes, the 50S and 30S RP genes were excluded and only cytosolic RP genes were selected. The large and small subunit genes were differentiated based on their prefixes. The genes starting with a prefix 60S or ‘L’ (for large) were considered as RPL genes and those starting with 40S or ‘S’ (for small) were categorized as RPS genes. According to the latest update of RGAP-DB, the cytosolic ribosomal proteins of rice are encoded by at least 70 genes out of a family of 255 RP genes that include paralogs [38-40]. In the current study, these 70 RP candidate genes (35 RPL and 35 RPS) excluding the paralogs were selected for transcript analysis. After sequence retrieval, the 70 genes were validated by a BLAST search in other databases like NCBI2, RAP-DB3 and Phytozome4.
2.2. In silico Putative Promoter Analysis of RP Genes
To determine the presence of cis-elements that respond to wound signaling resulting from insect attack, about 1 kb nucleotide sequence upstream from each of 70 RP genes was retrieved from RGAP-DB and submitted to the PlantCARE5 database. The complete list of cis-elements in promoter regions of RP genes has been reported in the published literature [39, 40] and those concerning insect attack responses have been emphasized here.
2.3. Plant Growth Conditions and Infestation with Insect Pests
Two rice genotypes, BPT5204 and RPNF05 were used in this study. BPT5204 is susceptible to both pests and is obtained from the Indian Institute of Rice Research, Hyderabad, India. RPNF05, which has been shown to exhibit moderate to a high level of resistance against BPH and GM, respectively is collected from the Agri Biotech Foundation, Hyderabad, India [41, 42] and is used as a positive control. It is a derivative of BPT5204 obtained by systematic introgression of bacterial blight (Xa21, xa13 and xa5), fungal blast (Pi54) and GM resistance genes (Gm1, Gm3, Gm4 and Gm8) [42]. Hence, the background of both the genotypes used in this study is similar except for the introgression of the desirable genes governing pest stress tolerance in RPNF05.
The growth of BPT5204 and RPNF05 genotypes and their infestation with gall midge (GM) and brown planthopper (BPH) were performed as mentioned earlier [42]. In brief, rice seeds of both the genotypes were sown in plastic trays (60 X 40 X 7 cm) and plants were raised under green house conditions (30 ± 2°C, 16 hour (h) light/ 8 h dark photoperiods, (Supplementary Fig. 1a). Three-week-old plants were challenged with gall flies belonging to gall midge biotype 1 (GMB1) for 24-48 h, following which the plants were shifted to a humidity chamber for the next 48 h to facilitate hatching of eggs. On the fifth day (120 h), some of the plants were observed for the presence of maggots at the apical meristem region through plant dissection to confirm the infestation. Stem samples of 2 cm length above the soil level were cut at 24 and 120 hours after infestation (HAI) and stored in liquid nitrogen for RNA isolation. Samples collected from uninfested plants of similar age groups grown under similar conditions were used as corresponding controls for double normalization. All the samples were collected in biological (independent experiments) and technical triplicates for transcript analysis. Likewise, three-week-old test plants were infested with BPH nymphs, and lower leaf sheath samples were collected at 6 and 12 HAI (Supplementary Fig. 1b (854.6KB, pdf) ).
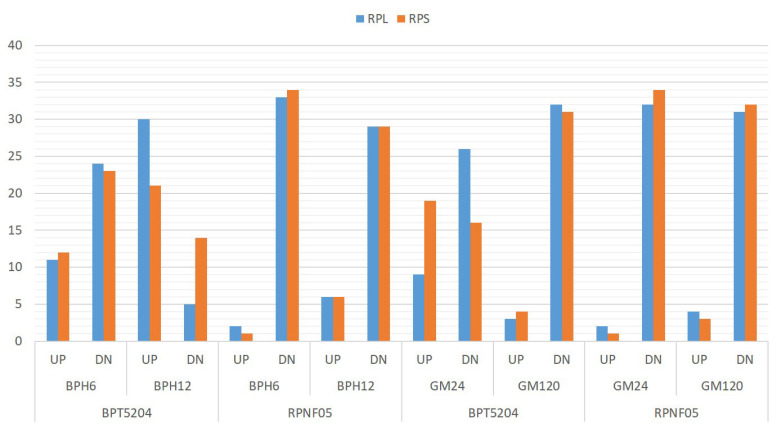
Fig. (1).
Transcript profiles of RPL and RPS genes. Overall, transcript profiles of 35 RPL and RPS genes in both the susceptible BPT5204 and the resistant RPNF05 following infestation with either BPH or GM revealed a predominant number of genes to be downregulated in comparison to the number that was upregulated. The numbers on the y-axis correspond to the mean of the total number of RP genes that were up or downregulated after technical and biological triplicate experiments. The blue bars are RPL and the orange bars indicate RPS genes. *UP, upregulated; DN, downregulated BPH6, brown planthopper 6 HAI; BPH12, brown planthopper 12 HAI; GM24, gall midge 24 HAI; GM120, gall midge 120 HAI. (A higher resolution / colour version of this figure is available in the electronic copy of the article).
2.4. Total RNA Extraction, cDNA Synthesis and Quantitative-PCR (qRT-PCR)
Total RNA was isolated from shoot samples of infested and uninfested BPT5204 and RPNF05 plants using the Trizol (Sigma-Aldrich, US) method. About 2 μg of total RNA was used to synthesize the first-strand cDNA with reverse transcriptase (Takara Bio, Clontech, USA). The cDNA was diluted in 1:7 ration with sterile Milli-Q water and 2 μl of this diluted cDNA was used for analyzing the transcript levels of RPL and RPS genes in the two genotypes. The rice-specific RPL and RPS primers, designed through the primer-36 tool was used in qRT-PCR. Primer sequences used in the study are listed in Supplementary Table 2 (854.6KB, pdf) . The qRT-PCR was performed using SYBR Green R Premix (Takara Bio, USA). The qRT-PCR cyclic conditions included an initial denaturation at 94 °C for 2 min, followed by 40 cycles of 94 °C for 15 s, an annealing temperature specific to each RP gene for 25 s and an amplicon extension step at 72 °C for 30 s. This was followed by a melting curve step to analyze the specificity of each amplicon. The qRT-PCR was performed as three technical and biological repeats and the fold change was calculated using the standard ΔΔCT double normalization method [43].
2.5. Statistical Analysis
The fold change in each BPH and GM infested sample was normalized with the corresponding uninfested samples using two reference genes, act-1 and β-tub that are specific to rice. The mean fold change of each replicate obtained after normalization with these two genes was considered as the final fold change. In this study, genotype and the infestation time were the major experimental factors. A two-way analysis of variance (ANOVA) was employed for each gene on all replicates to look for effects of genotype and infestation time and their interactions. Tukey’s pairwise comparisons were used to calculate the statistical significance. Probability of P < 0.05 was considered as statistically significant. SigmaPlot v.11 and R program v.3.6.3 were used for statistical analyses. The P values obtained from two-way ANOVA were adjusted with a Bonferroni correction in Microsoft excel v.2013 following the instructions available on web tool7. The transcript patterns of RP genes were also represented in the form of heatmaps constructed using the mean of fold change values. These maps were generated with the online program, Morpheus8.
3. RESULTS
3.1. In silico Analysis of RP Gene Upstream Promoter Sequences
The upstream nucleotide sequences up to 1 kb from the transcription start site of each of 70 genes were assessed for the presence of cis-elements that were shown to be associated with wound signaling in earlier reports. A majority of the promoter sequences were found to have elements that responded to phytohormones such as salicylic acid (TCA-motif), ethylene (Ethylene-responsive element) and methyl jasmonate (TGACG and CGTCA motifs). In addition, the motifs associated with wound signalling like W-box and WUN-motif are also present. Nine RPL and four RPS genes have W-box motif; one RPL and two RPS genes have WUN-motif, 11 RPL and 15 RPS genes have one TCA element, three RPL and four RPS genes have one ERE-element, 21 RPL and 26 RPS genes have the TGACG motif, 14 RPL and 25 RPS genes exhibited the CGTCA motif in their upstream regions. The list of these cis-elements in the promoter regions of RP genes has been summarized in Supplementary Table 2 (854.6KB, pdf) .
3.2. Differential Transcriptional Regulation of RPL Genes in Response to BPH and GM
The cytoplasmic RPs of rice are encoded by atleast 70 genes out of a family of 255 RP genes including paralogs. These 70 genes (35 RPL and 35 RPS) excluding the paralogs were selected in the present study to investigate their response to infestation by the BPH and GM pests at two different time points. Those genes that exhibited ≥2fold transcript levels were considered as upregulated. The original P values obtained from two-way ANOVA were adjusted with a Bonferroni correction. The term significance hereafter refers to statistical significance at P < 0.05 from P adjusted values. Overall, transcript profiles of 35 RPL genes in both the susceptible BPT5204 and the resistant RPNF05 following infestation by either BPH or GM revealed that a predominant number of genes were downregulated in comparison to the number that was upregulated. These pests induced the downregulation of a large number of RP genes (>80%) in RPNF05 than in BPT5204 (Fig. 1). In BPT5204, BPH infestation at 6 HAI resulted in 11 upregulated RPL genes, while 24 were downregulated. At 12 hai with BPH, 30 of the 35 RPL genes were upregulated as against the remaining five genes that were either downregulated or unaffected.
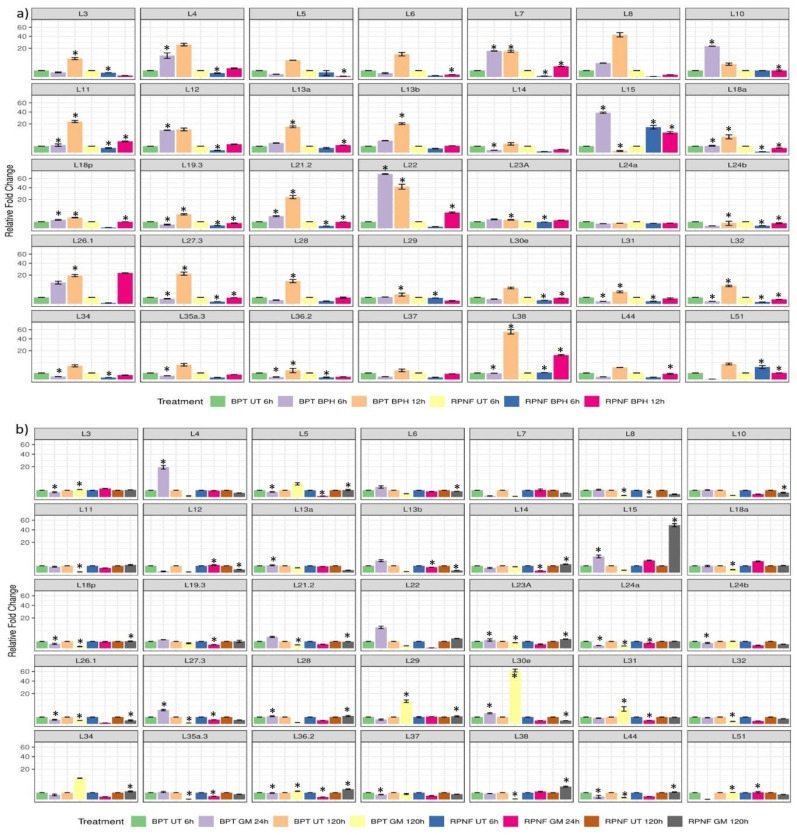
Further, six genes viz., RPL4, L7, L10, L12, L15 and L22 among the 11 upregulated RPL genes registered significant transcript levels of >10-fold at 6 hai with BPH in BPT5204. Among those that were downregulated, RPL14, L19.3, L27.3, L31, L32, L34, L35a.3 and L36.2 were significant. At 12 hai, 30 genes were upregulated, of which 14 genes (RPL4, L6, L7, L8, L11, L12, L13a, L13b, L21.2, L22, L26.1, L27.3, L28 and L38) were of >10-fold magnitude and upregulation of 19 genes (RPL3, L7, L11, L13a, L13b, L18a, L19p, L19.3, L21.2, L22, L23a, L26.1, L27.3, L28, L29, L31, L32, L36.2, L38) was significant. At both, the time points, three genes (RPL7, L12 and L22) had higher transcript levels, with the upregulation of RPL7 and L22 being significant. In RPNF05, only RPL15 (15-fold) and L51 (4-fold) were significantly upregulated at 6 hai and transcripts of the remaining 33 genes were downregulated. Among these, expression of 18 genes (RPL3, L4, L7, L11, L12, L18a, L19.3, L21.2, L23A, L24b, L27.3, L29, L30e, L31, L32, L34, L36.2 and L51) became significantly declined. At 120 hai, four genes, RPL15 (10-fold), L22 (6-fold), L26.1 (23-fold) and L38 (14-fold) showed high upregulation, whereas RPL7 and L11 were moderately induced (3-fold) (Fig. 2a). The upregulation of all these transcripts except RPL26.1 was significant. Interestingly, RPL51 was significantly upregulated in RPNF05 and downregulated in BPT5204 at 6 hai. Likewise, RPL15 was significantly upregulated in RPNF05 but significantly downregulated in BPT5204 at 12 HAI.
Fig. (2).
Graphical representation of the differential transcriptional response of individual RPL genes. To understand the transcript patterns of individual RPL genes upon infestation with (a) BPH and (b) GM pests, bar diagrams were constructed for each RPL gene separately. The data on the y-axis is the relative fold change calculated using the ΔΔCT method, whereas treatments are indicated on the x-axis. The asterisks in bar diagrams represent statistical significance at P < 0.05 obtained after adjusting probability values of two-way ANOVA with Bonferroni correction. Different colored grids at the bottom of each figure correspond to the type of treatment. Bar diagrams were generated using statistical program R v.3.6.3, two-way ANOVA was calculated in SigmaPlot v.11 and p values were adjusted in Microsoft excel v.2013. *UT, untreated; BPH, brown planthopper; GM, gall midge. (A higher resolution / colour version of this figure is available in the electronic copy of the article).
Of the seven upregulated genes (RPL4, L6, L13b L21.2, L22, L27.3 and L30e) in BPT5204 at 24 hai following GM infestation, L22 and L4 exhibited 10 and 19-fold induction in transcript levels, respectively. The upregulation of RPL4, L27.3 and L30e and the downregulation of RPL3, L5, L18p, L24a, L24b, L26.1, L36.2, L37 and L44 were significant. At 120 hai, L30e recorded highest and significant (62-fold) upregulation and the transcripts of RPL5, L29, L31 and L34 were moderately induced. In RPNF05, only RPL15 and L18a were moderately upregulated at 24 hai with GM. The upregulation of L18a was specifically noticed in RPNF05, which was downregulated in BPT5204. A significant downregulation was observed in transcripts of RPL8, L11, L18a, L18p, L21.2, L23a, L24a, L26.1, L27.3, L32, L35a.3, L38, L44 and L51. At 120 hai following GM infestation, 16 genes were significantly downregulated. Among the upregulated ones, RPL15, L36.2 and L38 were significantly induced with the transcript level of L15 showing the highest upregulation (50-fold), but these were downregulated in BPT5204 (Fig. 2b). Also, RPL15 was significantly and highly activated in RPNF05 at 6 hai with BPH and 120 hai with GM but was downregulated at 120 hai with GM in BPT5204. The transcripts of RPL14, L23a, L24a and L24b were commonly downregulated in response to BPH and GM infestations in both BPT5204 and RPNF05 genotypes. Of these, the downregulation of RPL14 at 6 hai and L24b at 12 hai with BPH in BPT5204 was significant, which was also significantly noticed at both the time points with GM infestation in RPNF05.
3.3. Differential Transcriptional Regulation of RPS Genes
The overall response of RPS genes was similar to that noted for RPL genes with the majority of them being either downregulated or unaltered in response to either BPH or GM infestation in both susceptible BPT5204 and resistant RPNF05 genotypes. Notable exceptions were at 24 hai with GM in BPT5204 wherein a slightly higher number of RPS genes were upregulated than those downregulated, in contrast to RPL genes. But, as in the case of RPL genes, a higher number of RPS genes were also upregulated in BPT5204 at 6 hai following BPH infestation.
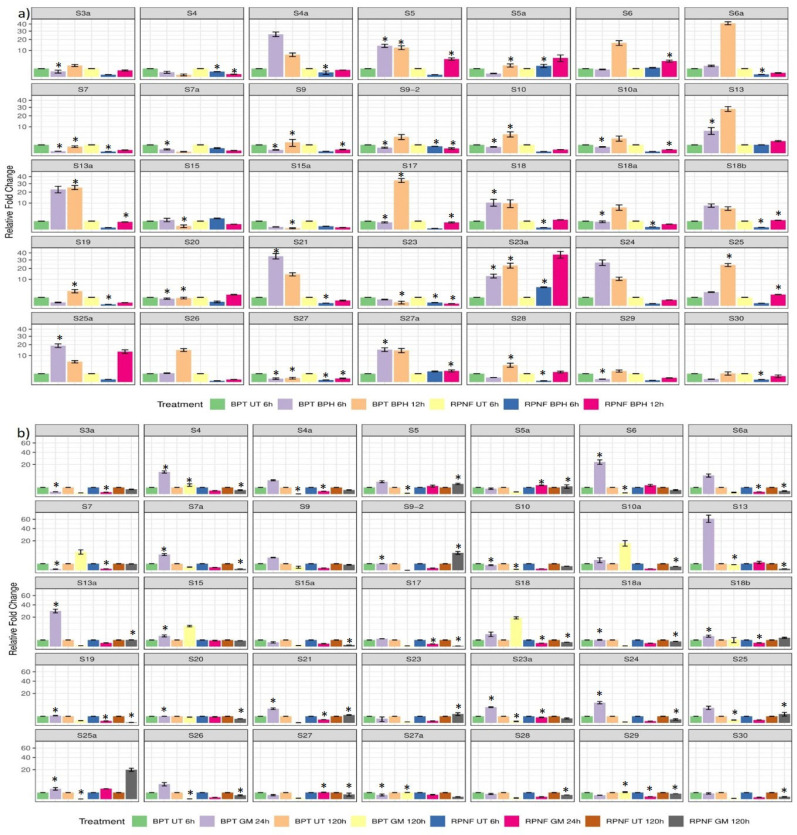
Among the induced genes, more than 10-fold upregulation was noted in 12 genes (RPS4a, S5, S6, S6a, S13, S13a, S18, S21, S23a, S24, S25a and S27a) at 6 hai with BPH in BPT5204 and a significant upregulation was observed in RPS5, S13, S18, S21, S23a, S25a and S27a. At 12 hai, the transcripts of RPS5, S10, S13a, S17, S23a, and S25 were significantly upregulated, whereas RPS4, S4a, S5a, S6a, S7, S9-2, S18, S18a, S18b, S19, S21, S23, S27, S28 and S30 were significantly downregulated. In RPNF05, only RPS23a became significantly upregulated (5-fold) at 6 hai, whereas at 12 hai, six genes such as RPS5, S5a, S6, S13, S23a and S25a were upregulated. Among these, the highest induction was noticed in the transcripts of S23a (37-fold) and RPS25a (13-fold) and a significant upregulation was observed in RPS5 and S6 (Fig. 3a). The transcript levels of RPS5a (5-fold) were specifically upregulated at 12 hai with BPH in RPNF05 but was downregulated in BPT5204 suggesting its possible role in resistance.
Fig. (3).
Differential transcript patterns of individual RPS genes. The bar diagrams were constructed separately for each RPS gene upon infestation with (a) BPH and (b) GM pests. These were prepared using the statistical program ‘R’ v.3.6. The statistical significance at P < 0.05 obtained after adjusting the original p values of two-way ANOVA with Bonferroni correction was represented with asterisks in bar diagrams. The data on x and y-axis corresponds to the treatment and the relative fold change, respectively. Statistical program R v.3.6.3 and SigmaPlot v.11 were used to draw the bar diagrams and calculate two-way ANOVA, respectively. *UT, untreated; BPH, brown planthopper; GM, gall midge. (A higher resolution / colour version of this figure is available in the electronic copy of the article).
In response to GM at 24 HAI in BPT5204, a total of 19 genes were induced. Among them, high upregulation (more than 10-fold) was noted in four genes (RPS4, S6, S13 and S13a) and a significant upregulation was observed in 10 genes (RPS4, S6, SS7a, S13a, S15, S18b, S21, SS23a, S24 and S25a). At 120 HAI, only RPS7, S10a, S15 and S18 were upregulated (Fig. 3b). RPS15 was consistently upregulated at both the time points. In RPNF05, the transcript level of only RPS25a was induced at 24 hai, whereas three genes (RPS5, S9.2 and S25a) were upregulated specifically at 120 hai but were downregulated in BPT5204. The up and downregulation of RPS5 in RPNF05 and BPT5204, respectively was significant.
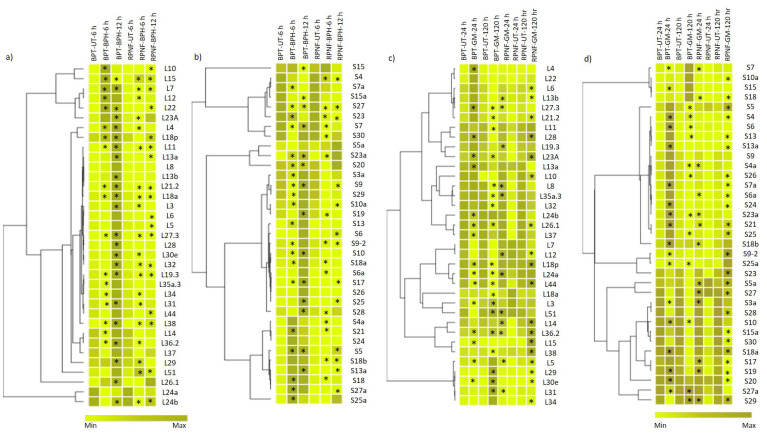
In summary, seven of the RP genes viz., RPL51, RPL15, RPL18, RPS5, RPS5a, RPS9.2 and RPS25a were shortlisted for their possible involvement in insect resistance. These genes were selected because of their specific upregulation in resistant genotype at a given time point in either of the two pests. The differential transcript levels of RPL and RPS genes were also depicted in the form of heatmaps (Fig. 4). Each grid in the map corresponds to a particular RP gene, with the dark-colored ones exhibiting higher transcript levels whereas, the pale-colored grids represent weak or no induction. We have also shortlisted the genes that exhibited an overlap in transcript levels or those that were commonly up
Fig. (4).
Heatmap depiction of differential transcriptional regulation of RP genes. Heatmaps were used to represent the transcript levels of RPL and RPS genes infested with (a, b) BPH and (c, d) GM pests. These were generated by incorporating the mean of fold change values obtained from three biological and three technical replicates normalized by the ΔΔCT method. The hierarchical clustering of genes with one minus Pearson correlation and average linkage method was used to group the genes with similar transcript patterns. A color scale is provided at the left and right bottom of the figure that denotes an increase in transcript levels from a gradient of light to dark color. The light-colored grids indicate a decrease in transcripts and the dark-colored grids correspond to an increase in transcript levels. The asterisks in the grids represent statistical significance at P < 0.05 calculated through two-way ANOVA using SigmaPlot v.11 and adjusted with Bonferroni correction. Heatmaps were developed using an online program, Morpheus. *UT, untreated; BPH, brown planthopper; GM, gall midge. (A higher resolution / colour version of this figure is available in the electronic copy of the article).
or downregulated. Venn diagrams were used to describe this overlap in transcript patterns (Fig. 5) and the list of these genes was provided in Supplementary Table 3 (854.6KB, pdf) . The P values of each gene under a given insect treatment calculated through two-way ANOVA were adjusted with a Bonferroni correction and the statistical significance of RPL and RPS genes was represented in Supplementary Tables 4 (854.6KB, pdf) and 5 (854.6KB, pdf) , respectively.
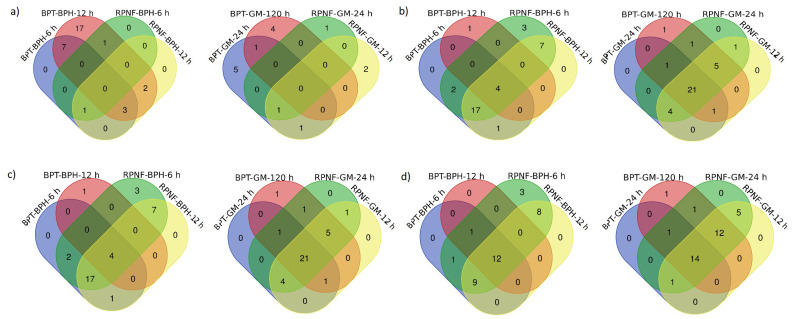
Fig. (5).
Venn diagrams representing the inducible up and downregulation of RP genes. The Venn diagrams were used to depict the total number of commonly (a) up and (b) downregulated RPL and (c) up and (d) downregulated RPS genes at 6 and 12 h after BPH and 24 and 120 h of GM infestation in BPT5204 and RPNF05 genotypes of rice. *BPH, brown planthopper; GM, gall midge. (A higher resolution / colour version of this figure is available in the electronic copy of the article).
4. DISCUSSION
Plants, being sessile, respond to changing environmental conditions by maintaining an equilibrium between their growth and development and stress adaptation mechanisms [44]. One of the important cellular changes that occur during the onset of stress is the regulation of protein turnover starting from transcription through translation to post-translational modifications including protein ubiquitination [45, 46]. Phytohormones are also the critical regulators of tolerance to biotic and abiotic stresses besides their role in growth and development [47, 48]. While abiotic stresses induce the modulation of ABA, auxin, methyl Jasmonate (MeJa), and salicylic acid (SA) responsive genes, the biotic stresses also involve genes responsive to the same phytohormones along with cytokinin and ethylene [49, 50]. MeJa and/or SA are found to be positive regulators of resistance to certain insect pests or pathogen attacks whereas, ABA generally acts as a negative regulator [51, 52].
There have been extensive studies on understanding the morphological, anatomical, biochemical, physiological and molecular basis of insect resistance in rice [5]. One approach for these studies is based on functional genomics with the genetic characterization of resistance (R) genes, their cloning and understanding the function of these cloned genes [53] while the other approach has been through the studies on a set of resistant versus susceptible varieties and following their responses to insect infestation [54, 55]. These two approaches are yet to merge and provide a vivid understanding of the three basic components of insect resistance viz., antixenosis, antibiosis and tolerance [13]. Our current understanding of antixenosis or non-preference is through green leaf volatiles like (Z)3-hexanol that attracts BPH [56] and/or natural enemies of the pest through rice hydroperoxide lyase (OsHPL3) gene function [57]. Antibiosis component in terms of insect development, survival and fecundity could be the result of reduced feeding due to mechanical obstruction or due to the presence of toxins or lack of essential nutrients in the plant [58]. The tolerance component of planthopper resistance is not well studied. This may be because it is not a true defence pathway, but represents general vigor and enhanced photosynthesis under stress conditions [59, 60]. The underlying genes may be mostly constitutively expressed and difficult to capture by over-expression or omics studies.
Ribosomes, being the essential cellular moieties, respond to the environmental cues likely by the co-ordinated transcriptional upregulation of a few selected or a group of RP genes [39, 40]. The upregulation of a majority of RP genes was also observed in response to macro element deficiency in Arabidopsis [61]. This transcriptional induction of genes involved in ribosome biogenesis is a strategy adapted under stresses [17, 62]. The induction might also help in maintaining or improving the synthesis of not only of its own proteins involved in inducing structural stability but also of the other transcription factors or signal transduction proteins under the conditions of stress [63]. In response to both abiotic stresses and insect pests, we noticed the upregulation of a large number of RP genes. The transcripts of some of these genes responded by immediate upregulation after the onset of stress and some were elevated to high fold levels. This co-ordinated high and instantaneous activation of RP genes in response to invaders might function as an immediate defence. In rice, the RPs are encoded by at least 70 genes, of which, the proteins of large and small subunits are encoded by 35 genes each. A majority of these RP genes further exist as 2-3 paralogs in the rice genome taking the total number of RP-encoding genes to 255. Thus, the same RP is encoded by several paralogous genes. The presence of a large number of RP genes suggests that the RPs in a ribosomal complex are heterogeneous. The RP synthesized from a member of a paralogous group is incorporated into a given ribosome in a particular tissue or under a given condition [64]. The existence of a large number of RP gene paralogs in plants compared to other biological systems means that some thousands of RP combinations and hence, the formation of functionally dedicated ribosomes are possible [33-37, 65]. Environmental stress plays a major role in modifying ribosomal composition by differential expression of RP genes [66, 67]. The likely existence of dedicated ribosomes and multiple divergent paralogs provides a platform for mining individual RP gene functions, particularly in stress responses. Therefore, the identification of specific RP genes that respond to environmental signals is the first step in this direction.
Our studies of activation tagged gain-of-function mutants revealed the activation of two RPL genes, RPL6 and RPL23a, for enhanced water use efficiency (WUE) trait in rice, indicating their involvement in extra-ribosomal activities apart from their basic cellular functions [38]. Subsequent transcript analysis of the entire RP gene family indicated that they were differentially regulated under various abiotic stresses [39, 40]. Further, functional validation of one candidate RPL gene, RPL23a, under multiple abiotic stress treatments highlighted the role of RP genes in the amelioration of abiotic stresses in rice [68]. However, the response of RP genes under pest attack is less investigated and no direct report is hitherto available to suggest their role in insect resistance.
In the current study, we have conducted a differential transcript analysis of RP genes in response to infestation by two divergent insect pests representing gall-forming ones and phloem feeders. Here, we have also shortlisted the genes that were highly induced under a given insect challenge in each genotype. These included the genes that were common to both the genotypes and also the genes that were specifically upregulated in the resistant genotype, RPNF05. Taken together, we noticed the upregulation of a large number of RPL genes in the susceptible genotype, BPT5204, compared to RPNF05 against BPH. Therefore, it is unlikely that such genes are involved in resistance against BPH but instead contribute to susceptibility in BPT5204. Modulation or overlap in the upregulation of a few RPL genes in both BPT5204 and RPNF05 under infestation with GM suggests that they have no possible role either in susceptibility or resistance. Unlike RPL, the RPS genes, in general, were low responding in numbers against either BPH or GM infestation in either of the genotypes. When the response of individual gene(s) was considered, some of the common genes like RPL7, L12 or RPS6, S13 and S23a against BPH or RPS23a against GM were activated independently of genotype or stress indicating that they might not have a specific role in insect-plant interactions. In contrast, the genes, including RPL15, L51, L18a, RPS5, S5a, S9.2 and S25a which were specifically upregulated in the resistant genotype, RPNF05, but not in BPT5204 either against BPH or GM suggest their possible role in plant resistance against the corresponding pest. RPL15 was also found to be highly upregulated in response to bacterial blight [39]. RPL15 also contains a MeJa-responsive motif (CGTCA) in its promoter region. The upstream sequence of RPL18a was found to have SA (TCA-element) and MeJa-responsive (CGTCA and TGACG) cis-elements that responded to wounding or mechanical damage induced by pests. This gene was also highly upregulated under abiotic stress treatments [39]. Among the RPS genes, the transcript levels of RPS5a were specifically upregulated at 12 hai with BPH whereas RPS5, S9.2 and S25a were upregulated at 120 hai with GM in RPNF05. The promoter regions of these three genes carry both MeJa-responsive motifs (CGTCA and TGACG). In our previous study, several RPL genes including RPL6, L10, L11, L15 and L24a also showed considerable upregulation in rice plants infected with bacterial blight causing Xanthomonas oryzae pv. oryzae [39]. RPL6 and L10 were also activated in samples treated with MeJA and SA, the key hormones involved in plant defence mechanisms against insects and pathogens [39]. The proteins encoded by wound-responsive genes are involved in either direct repairing of damage site, inhibiting growth of the pest, eliciting the defence signaling cascade or modulating the plant metabolism to compensate the nutritional loss [69].
In addition to forming dedicated ribosomes to translate specific mRNAs [34-37, 70], the involvement of RPs in defence-related signals might also occur by individually interacting with a network of other proteins to form a functional circuit [71]. In humans, RPL6, L8 and RPS14 were found to be important members of repair signaling cascade that are recruited at sites of DNA damage induced by environmental stresses [71]. The temporal and resistant genotype-specific upregulation of these RP genes upon challenge with BPH or GM along with the presence of related stress-responsive elements and their repeats on their respective putative promoter regions further provides a strong basis for their possible involvement in providing resistance to these pests. The differential regulation of RP transcripts in response to infestation by two pests at two different time points might have occurred co-ordinately to change the canonical ribosomal composition to translate specific stress-related mRNAs. In addition to the exchange of paralogs, ribosome heterogeneity also includes sequence variation of rRNAs, absence of specific RPs, posttranscriptional or posttranslational modifications of rRNA or RPs [36]. For instance, the up and downregulation, respectively of 11 and 24 RPL genes at 6 hai with BPH; 30 and five genes at 12 hai in BPT5204 in the current study can be correlated with ribosome heterogeneity for adaptation under different stress levels. Alternatively, the upregulated genes might also have specialized functions in inducing resistance [72-74]. This should be investigated in future studies. The genes that were specifically upregulated (RPL15, L51, L18a, RPS5, S5a, S9.2 and S25a) in resistant genotype might directly participate in defence-related signal transduction pathways or probably involved in primary metabolism-mediated tolerance component of insect resistance such as enhancing overall photosynthesis and vigor of the plant under stress.
The tolerance component of resistance is important in rice against BPH [75]. It refers to the genetic ability of the genotype that can compensate for the loss inflicted by the insect through feeding. The physiological basis of tolerance covers an increased rate of photosynthesis and primary metabolism as studied in rice varieties like Triveni, Utri Rajapan and Kenchana [75]. Increased activities of the majority of both RPL and RPS genes in BPT5204, but not in RPNF05 at 6 and 12 hai with BPH strongly suggest enhanced protein synthesis to gear up production and transport of primary metabolites, especially, sugars. Among these, RPL22 displayed more than 40-fold upregulation in BPT5204 at both the time points while it was either not induced or poorly induced in RPNF05 at the corresponding time points. Likewise, RPS6a, RPS17 and RPS21 among RPS genes registered more than 25-fold upregulation in BPT5204 with little or no induction in RPNF05. While induction of these genes suggests a strong component of tolerance in BPT5204 against BPH, more greenhouse studies are needed to confirm this observation. Interestingly, one of the RPS genes, RPS23 was highly induced in RPNF05 compared to BPT5204 against BPH at 12 hai. The observations from this study on differential induction of RP genes under pest treatments with specific upregulation of a few of them in resistant genotype clearly point to their involvement in extra-ribosomal functions like stress responses and possibly in inducing resistance.
CONCLUSION
The information provided in this study on the differential transcriptional analyses of selected RP genes in response to two economically important pests in rice needs to be exploited further by independent functional characterization of the selected genes. In particular, investigations on the RPL and RPS genes that are specifically upregulated in the resistant genotype is of importance and such genes require further focus in the direction of developing transgenic crop plants resistant to the respective pests.
ACKNOWLEDGEMENTS
Mazahar Moin is grateful to the Department of Science and Technology (DST), Government of India for providing research grants and fellowship in the form of INSPIRE-faculty award. Achala Bakshi is thankful to the Department of Biotechnology for Research Associate (DBT-RA) fellowship. Authors also acknowledge Dr. J.S. Bentur for helping in conducting pest treatments on rice plants. Mazahar Moin acknowledges Pritam Saha for help in ‘R’ statistical program. Kirti PB acknowledges the Senior Scientist position awarded by the National Academy of Sceinces-India (NASI). Authors acknowledge the facilities obtained from the Biotechnology Division, ICAR-Indian Institute of Rice Research, Hyderabad, India.
Footnotes
1http://rice.plantbiology.msu.edu/ 2https://blast.ncbi.nlm.nih.gov/Blast.cgi 3https://rapdb.dna.affrc.go.jp/index.html 4https://phytozome.jgi.doe.gov/pz/portal.html 5http://bioinformatics.psb.ugent.be/webtools/plantcare/html/ 6http://bioinfo.ut.ee/primer3-0.4.0/ 7https://www.statisticssolutions.com/bonferroni-correction/ 8https://software.broadinstitute.org/morpheus/
AUTHORS’ CONTRIBUTIONS
Mazahar Moin, Kirti P.B. and Madhav M.S. designed the experiments. Mazahar Moin performed all the experiments. Anusree Saha and Achala Bakshi helped in performing a few qRT-PCR experiments. Divya D. performed the insect infestations on rice. Mazahar Moin and Kirti P.B. prepared the manuscript. All the authors read and approved the manuscript.
ETHICS APPROVAL AND CONSENT TO PARTICIPATE
Not applicable.
HUMAN AND ANIMAL RIGHTS
No Animals/Humans were used for studies that are the basis of this research.
CONSENT FOR PUBLICATION
Not applicable.
AVAILABILITY OF DATA AND MATERIALS
Not applicable.
FUNDING
The funding for the current work has been obtained through a grant sponsored by the Department of Science and Technology (DST), Government of India, in the form of DST-INSPIRE faculty award to Mazahar Moin. The grant number is IFA17-LSPA67.
CONFLICT OF INTEREST
The authors declare no conflict of interest, financial or otherwise.
SUPPLEMENTARY MATERIAL
Supplementary material is available on the publisher’s website along with the published article.
REFERENCES
- 1.Ehrlich P.R., Harte J. Opinion: To feed the world in 2050 will require a global revolution. Proc. Natl. Acad. Sci. USA. 2015;112:14743–14744. doi: 10.1073/pnas.1519841112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Das G., Patra J.K., Baek K.H. Corrigendum: Insight into MAS: a molecular tool for development of stress resistant and quality of rice through gene stacking. Front. Plant Sci. 2017;8:1321. doi: 10.3389/fpls.2017.01321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Prahalada G.D., Shivakumar N., Lohithaswa H.C., Gowda D.S., Ramkumar G., Kim S.R., Ramachandra C., Hittalmani S., Mohapatra T., Jena K.K. Identification and fine mapping of a new gene, BPH31 conferring resistance to brown planthopper biotype 4 of India to improve rice, Oryza sativa L. Rice (N. Y.) 2017;10:41. doi: 10.1186/s12284-017-0178-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bentur J.S., Rawat N., Divya D., Sinha D.K., Agarrwal R., Atray I., Nair S. Rice–gall midge interactions: battle for survival. J. Insect Physiol. 2016;84:40–49. doi: 10.1016/j.jinsphys.2015.09.008. [DOI] [PubMed] [Google Scholar]
- 5.Du B., Chen R., Guo J., He G. Current understanding of the genomic, genetic, and molecular control of insect resistance in rice. Mol. Breed. 2020;40:24. [Google Scholar]
- 6.Abdul Malik N.A., Kumar I.S., Nadarajah K. Elicitor and Receptor Molecules: Orchestrators of Plant Defense and Immunity. Int. J. Mol. Sci. 2020;21(3):963. doi: 10.3390/ijms21030963. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Jones J.D., Dangl J.L. The plant immune system. Nature. 2006;444(7117):323–329. doi: 10.1038/nature05286. [DOI] [PubMed] [Google Scholar]
- 8.Dodds P.N., Rathjen J.P. Plant immunity: towards an integrated view of plant–pathogen interactions. Nat. Rev. Genet. 2010;11(8):539–548. doi: 10.1038/nrg2812. [DOI] [PubMed] [Google Scholar]
- 9.Boller T., He S.Y. Innate immunity in plants: an arms race between pattern recognition receptors in plants and effectors in microbial pathogens. Science. 2009;324(5928):742–744. doi: 10.1126/science.1171647. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Tsuda K., Katagiri F. Comparing signaling mechanisms engaged in pattern-triggered and effector-triggered immunity. Curr. Opin. Plant Biol. 2010;13(4):459–465. doi: 10.1016/j.pbi.2010.04.006. [DOI] [PubMed] [Google Scholar]
- 11.Cui H., Tsuda K., Parker J.E. Effector-triggered immunity: from pathogen perception to robust defense. Annu. Rev. Plant Biol. 2015;66:487–511. doi: 10.1146/annurev-arplant-050213-040012. [DOI] [PubMed] [Google Scholar]
- 12.Hatsugai N., Igarashi D., Mase K., Lu Y., Tsuda Y., Chakravarthy S., et al. A plant effector-triggered immunity signaling sector is inhibited by pattern-triggered immunity. EMBO J. 2017;36(18):2758–2769. doi: 10.15252/embj.201796529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Cartier J.J., Painter R.H. Differential reactions of two biotypes of the corn leaf aphid to resistant and susceptible varieties, hybrids and selections of sorghums. J. Econ. Entomol. 1956;49:498–508. [Google Scholar]
- 14.Moss T., Langlois F., Gagnon-Kugler T., Stefanovsky V. A housekeeper with power of attorney: the rRNA genes in ribosome biogenesis. Cell. Mol. Life Sci. 2007;64(1):29–49. doi: 10.1007/s00018-006-6278-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Tschochner H., Hurt E. Pre-ribosomes on the road from the nucleolus to the cytoplasm. Trends Cell Biol. 2003;13:255–263. doi: 10.1016/s0962-8924(03)00054-0. [DOI] [PubMed] [Google Scholar]
- 16.Grummt I. The nucleolus—guardian of cellular homeostasis and genome integrity. Chromosoma. 2013;122(6):487–497. doi: 10.1007/s00412-013-0430-0. [DOI] [PubMed] [Google Scholar]
- 17.Sáez-Vásquez J., Delseny M. Ribosome biogenesis in plants: from functional 45S ribosomal DNA organization to ribosome assembly factors. Plant Cell. 2019;31(9):1945–1967. doi: 10.1105/tpc.18.00874. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Ferreira-Cerca S., Hurt E. Arrest by ribosome. Nature. 2009;459:46–47. doi: 10.1038/459046a. [DOI] [PubMed] [Google Scholar]
- 19.Kressler D., Hurt E., Baßler J. Driving ribosome assembly. Biochimica Et Biophysica Acta (BBA)-. Molecular Cell Research. 1803;2010:673–683. doi: 10.1016/j.bbamcr.2009.10.009. [DOI] [PubMed] [Google Scholar]
- 20.Ito T., Kim G.T., Shinozaki K. Disruption of an Arabidopsis cytoplasmic ribosomal protein S13- homologous gene by transposon-mediated mutagenesis causes aberrant growth and development. Plant J. 2000;22(3):257–264. doi: 10.1046/j.1365-313x.2000.00728.x. [DOI] [PubMed] [Google Scholar]
- 21.Yao Y., Ling Q.H., Wang H., Huang H. Ribosomal proteins promote leaf adaxial identity. Development. 2008;135(7):1325–1334. doi: 10.1242/dev.017913. [DOI] [PubMed] [Google Scholar]
- 22.Szakonyi D., Byrne M.E. Ribosomal protein L27a is required for growth and patterning in Arabidopsis thaliana. Plant J. 2011;65(2):269–281. doi: 10.1111/j.1365-313X.2010.04422.x. [DOI] [PubMed] [Google Scholar]
- 23.Ferreyra M.L.F., Pezza A., Biarc J., Burlingame A.L., Casati P. Plant L10 ribosomal proteins have different roles during development and translation under ultraviolet-B stress. Plant Physiol. 2010;153:1878–1894. doi: 10.1104/pp.110.157057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kakehi J.I., Kawano E., Yoshimoto K., Cai Q., Imai A., Takahashi T. Mutations in ribosomal proteins, RPL4 and RACK1, suppress the phenotype of a thermospermine-deficient mutant of Arabidopsis thaliana. PLoS One. 2015;10:e0117309. doi: 10.1371/journal.pone.0117309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Zheng M., Wang Y., Liu X., Sun J., Wang Y., Xu Y. The RICE MINUTE-LIKE1 (RML1) gene, encoding a ribosomal large subunit protein L3B, regulates leaf morphology and plant architecture in rice. J. Exp. Bot. 2016;67(11):3457–3469. doi: 10.1093/jxb/erw167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Byrne M.E. A role for the ribosome in development. Trends Plant Sci. 2009;14(9):512–519. doi: 10.1016/j.tplants.2009.06.009. [DOI] [PubMed] [Google Scholar]
- 27.Degenhardt R.F., Bonham-Smith P.C. Arabidopsis ribosomal proteins RPL23aA and RPL23aB are differentially targeted to the nucleolus and are disparately required for normal development. Plant Physiol. 2008;147(1):128–142. doi: 10.1104/pp.107.111799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Degenhardt R.F., Bonham-Smith P.C. Evolutionary divergence of ribosomal protein paralogs in Arabidopsis. Plant Signal. Behav. 2008;3(7):493–495. [Google Scholar]
- 29.Xiong W., Chen X., Zhu C., Zhang J., Lan T. Arabidopsis ribosomal proteins RPL23aA and RPL23aB are functionally equivalent. BMC Plant Biol. 2020 doi: 10.1186/s12870-020-02672-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Giavalisco P., Wilson D., Kreitler T., Lehrach H., Klose J., Gobom J., Fucini P. High heterogeneity within the ribosomal proteins of the Arabidopsis thaliana 80S ribosome. Plant Mol. Biol. 2005;57(4):577–591. doi: 10.1007/s11103-005-0699-3. [DOI] [PubMed] [Google Scholar]
- 31.Carroll A.J., Heazlewood J.L., Ito J., Millar A.H. Analysis of the Arabidopsis cytosolic ribosome proteome provides detailed insights into its components and their post-translational modification. Mol. Cell. Proteomics. 2008;7(2):347–369. doi: 10.1074/mcp.M700052-MCP200. [DOI] [PubMed] [Google Scholar]
- 32.Sugihara Y., Honda H., Iida T., Morinaga T., Hino S., Okajima T., et al. Proteomic analysis of rodent ribosomes revealed heterogeneity including ribosomal proteins L10-like, L22-like 1, and L39-like. J. Proteome Res. 2010;9:1351–1366. doi: 10.1021/pr9008964. [DOI] [PubMed] [Google Scholar]
- 33.Xue S., Barna M. Specialized ribosomes: a new frontier in gene regulation and organismal biology. Nat. Rev. Mol. Cell Biol. 2012;13:355–369. doi: 10.1038/nrm3359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Hummel M., Cordewener J.H., de Groot J.C., Smeekens S., America A.H., Hanson J. Dynamic protein composition of Arabidopsis thaliana cytosolic ribosomes in response to sucrose feeding as revealed by label free MS E proteomics. Proteomics. 2012;12(7):1024–1038. doi: 10.1002/pmic.201100413. [DOI] [PubMed] [Google Scholar]
- 35.Carroll A.J. The Arabidopsis cytosolic ribosomal proteome: from form to function. Front. Plant Sci. 2013;4:32. doi: 10.3389/fpls.2013.00032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Martinez-Seidel F., Beine-Golovchuk O., Hsieh Y.C., Kopka J. Systematic review of plant ribosome heterogeneity and specialization. Front. Plant Sci. 2020;11:948. doi: 10.3389/fpls.2020.00948. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Komili S., Farny N.G., Roth F.P., Silver P.A. Functional specificity among ribosomal proteins regulates gene expression. Cell. 2007;131:557–571. doi: 10.1016/j.cell.2007.08.037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Moin M., Bakshi A., Saha A., Udaya Kumar M., Reddy A.R., Rao K.V., Siddiq E.A., Kirti P.B. Activation tagging in indica rice identifies ribosomal proteins as potential targets for manipulation of water-use efficiency and abiotic stress tolerance in plants. Plant Cell Environ. 2016;39:2440–2459. doi: 10.1111/pce.12796. [DOI] [PubMed] [Google Scholar]
- 39.Moin M., Bakshi A., Saha A., Dutta M., Madhav S.M., Kirti P.B. Rice ribosomal protein large subunit genes and their spatio-temporal and stress regulation. Front. Plant Sci. 2016;7:1284. doi: 10.3389/fpls.2016.01284. b. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Saha A., Das S., Moin M., Dutta M., Bakshi A., Madhav M.S., Kirti P.B. Genome-wide identification and comprehensive expression profiling of Ribosomal Protein Small Subunit (RPS) genes and their comparative analysis with the Large Subunit (RPL) genes in rice. Front. Plant Sci. 2017;8:1553. doi: 10.3389/fpls.2017.01553. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Sama V.S.A.K., Rawat N., Sundaram R.M., Himabindu K., Naik B.S., Viratamath B.C., Bentur J.S. A putative candidate for the recessive gall midge resistance gene gm3 in rice identified and validated. Theor. Appl. Genet. 2014;127:113–124. doi: 10.1007/s00122-013-2205-7. [DOI] [PubMed] [Google Scholar]
- 42.Divya D., Madhavi K.R., Dass M.A., Maku R.V., Mallikarjuna G., Sundaram R.M., Laha G.S., Padmakumari A.P., Patel H.K., Prasad M.S., Sonti R.V. Expression profile of defense genes in rice lines pyramided with resistance genes against bacterial blight, fungal blast and insect gall midge. Rice (N.Y.) 2018;11:40. doi: 10.1186/s12284-018-0231-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Livak K.J., Schmittgen T.D. Analysis of relative gene expression data using real-time quantitative PCR and the 2− ΔΔCT method. Methods. 2001;25:402–408. doi: 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]
- 44.Claeys H., Inzé D. The agony of choice: how plants balance growth and survival under water-limiting conditions. Plant Physiol. 2013;162(4):1768–1779. doi: 10.1104/pp.113.220921. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Mazzucotelli E., Mastrangelo A.M., Crosatti C., Guerra D., Stanca A.M., Cattivelli L. Abiotic stress response in plants: when post-transcriptional and post-translational regulations control transcription. Plant Sci. 2008;174:420–431. [Google Scholar]
- 46.Nelson C.J., Millar A.H. Protein turnover in plant biology. Nat. Plants. 2015;1:15017. doi: 10.1038/nplants.2015.17. [DOI] [PubMed] [Google Scholar]
- 47.Pieterse C.M., Van der Does D., Zamioudis C., Leon-Reyes A., Van Wees S.C. Hormonal modulation of plant immunity. Annu. Rev. Cell Dev. Biol. 2012;28:489–521. doi: 10.1146/annurev-cellbio-092910-154055. [DOI] [PubMed] [Google Scholar]
- 48.Cohen S.P., Leach J.E. Abiotic and biotic stresses induce a core transcriptome response in rice. Sci. Rep. 2019;18:1–1. doi: 10.1038/s41598-019-42731-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Bari R., Jones J.D. Role of plant hormones in plant defence responses. Plant Mol. Biol. 2009;69(4):473–488. doi: 10.1007/s11103-008-9435-0. [DOI] [PubMed] [Google Scholar]
- 50.Li N., Han X., Feng D., Yuan D., Huang L.J. Signaling crosstalk between salicylic acid and ethylene/jasmonate in plant defense: do we understand what they are whispering? Int. J. Mol. Sci. 2019;20(3):671. doi: 10.3390/ijms20030671. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Smith C.M., Boyko E.V. The molecular bases of plant resistance and defense responses to aphid feeding: current status. Entomol. Exp. Appl. 2007;122:1–6. [Google Scholar]
- 52.Klessig D.F., Choi H.W., Dempsey D.M.A. Systemic acquired resistance and salicylic acid: past, present and future. Mol. Plant Microbe Interact. 2018;31:871–888. doi: 10.1094/MPMI-03-18-0067-CR. [DOI] [PubMed] [Google Scholar]
- 53.Deng Y., Zhu X., Shen Y., He Z. Genetic characterization and fine mapping of the blast resistance locus Pigm (t) tightly linked to Pi2 and Pi9 in a broad-spectrum resistant Chinese variety. Theor. Appl. Genet. 2006;113(4):705–713. doi: 10.1007/s00122-006-0338-7. [DOI] [PubMed] [Google Scholar]
- 54.Liu C., Hao F., Hu J., Zhang W., Wan L., Zhu L., et al. Revealing different systems responses to brown planthopper infestation for pest susceptible and resistant rice plants with the combined metabonomic and gene-expression analysis. J. Proteome Res. 2010;9(12):6774–6785. doi: 10.1021/pr100970q. [DOI] [PubMed] [Google Scholar]
- 55.Du B., Wei Z., Wang Z., Wang X., Peng X., Du B., et al. Phloem-exudate proteome analysis of response to insect brown plant-hopper in rice. J. Plant Physiol. 2015;183:13–22. doi: 10.1016/j.jplph.2015.03.020. [DOI] [PubMed] [Google Scholar]
- 56.Wang H., Ye S., Mou T. Molecular breeding of rice restorer lines and hybrids for brown planthopper (BPH) resistance using the Bph14 and Bph15 genes. Rice (N.Y.) 2016;9:53. doi: 10.1186/s12284-016-0126-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Tong X., Qi J., Zhu X., Mao B., Zeng L., Wang B., Li Q., Zhou G., Xu X., Lou Y., He Z. The rice hydroperoxide lyase OsHPL3 functions in defense responses by modulating the oxylipin pathway. Plant J. 2012;71:763–775. doi: 10.1111/j.1365-313X.2012.05027.x. [DOI] [PubMed] [Google Scholar]
- 58.Pathak M.D., Saxena R.C. INSECT RESISTANCE IN CROP PLANTS. Commentaries Plant Sci. 2013;2:2–61. [Google Scholar]
- 59.Wei Z., Hu W., Lin Q., Cheng X., Tong M., Zhu L., et al. Understanding rice plant resistance to the Brown Planthopper (Nilaparvata lugens): A proteomic approach. Proteomics. 2009;9(10):2798–2808. doi: 10.1002/pmic.200800840. [DOI] [PubMed] [Google Scholar]
- 60.Douglas A.E. Strategies for enhanced crop resistance to insect pests. Annu. Rev. Plant Biol. 2018;69:637–660. doi: 10.1146/annurev-arplant-042817-040248. [DOI] [PubMed] [Google Scholar]
- 61.Wang J., Lan P., Gao H., Zheng L., Li W., Schmidt W. Expression changes of ribosomal proteins in phosphate-and iron-deficient Arabidopsis roots predict stress-specific alterations in ribosome composition. BMC Genomics. 2013;14(1):783. doi: 10.1186/1471-2164-14-783. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Ferreyra M.L.F., Casadevall R., Luciani M.D., Pezza A., Casati P. New evidence for differential roles of l10 ribosomal proteins from Arabidopsis. Plant Physiol. 2013;163(1):378–391. doi: 10.1104/pp.113.223222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Kim K.Y., Park S.W., Chung Y.S., Chung C.H., Kim J.I., Lee J.H. Molecular cloning of low-temperature-inducible ribosomal proteins from soybean. J. Exp. Bot. 2004;55(399):1153–1155. doi: 10.1093/jxb/erh125. [DOI] [PubMed] [Google Scholar]
- 64.Genuth N.R., Barna M. The discovery of ribosome heterogeneity and its implications for gene regulation and organismal life. Mol. Cell. 2018;71(3):364–374. doi: 10.1016/j.molcel.2018.07.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Whittle C.A., Krochko J.E. Transcript profiling provides evidence of functional divergence and expression networks among ribosomal protein gene paralogs in Brassica napus. Plant Cell. 2009;21:2203–2219. doi: 10.1105/tpc.109.068411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Sormani R., Masclaux-Daubresse C., Daniele-Vedele F., Chardon F. Transcriptional regulation of ribosome components are determined by stress according to cellular compartments in Arabidopsis thaliana. PLoS One. 2011:6. doi: 10.1371/journal.pone.0028070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Ghulam M.M., Catala M., Abou Elela S. Differential expression of duplicated ribosomal protein genes modifies ribosome composition in response to stress. Nucleic Acids Res. 2020;48(4):1954–1968. doi: 10.1093/nar/gkz1183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Moin M., Bakshi A., Madhav M.S., Kirti P.B. Expression profiling of ribosomal protein gene family in dehydration stress responses and characterization of transgenic rice plants overexpressing RPL23A for water-use efficiency and tolerance to drought and salt stresses. Front Chem. 2017;5:97. doi: 10.3389/fchem.2017.00097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.León J., Rojo E., Sánchez-Serrano J.J. Wound signalling in plants. J. Exp. Bot. 2001;52(354):1–9. doi: 10.1093/jexbot/52.354.1. [DOI] [PubMed] [Google Scholar]
- 70.Gerst J.E. Pimp my ribosome: ribosomal protein paralogs specify translational control. Trends Genet. 2018;34(11):832–845. doi: 10.1016/j.tig.2018.08.004. [DOI] [PubMed] [Google Scholar]
- 71.Yang C., Zang W., Ji Y., Li T., Yang Y., Zheng X. Ribosomal protein L6 (RPL6) is recruited to DNA damage sites in a poly (ADP-ribose) polymerase–dependent manner and regulates the DNA damage response. J. Biol. Chem. 2019;294:2827–2838. doi: 10.1074/jbc.RA118.007009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Yu J., Hu S., Ma K., Sun L., Hu H., Zou F., et al. Ribosomal protein S29 regulates metabolic insecticide resistance through binding and degradation of CYP6N3. PLoS One. 2014;9(4):e94611. doi: 10.1371/journal.pone.0094611. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Liu X.D., Xie L., Wei Y., Zhou X., Jia B., Liu J., Zhang S. Abiotic stress resistance, a novel moonlighting function of ribosomal protein RPL44 in the halophilic fungus Aspergillus glaucus. Appl. Environ. Microbiol. 2014;80(14):4294–4300. doi: 10.1128/AEM.00292-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Zhu F., Zhou Y.K., Ji Z.L., Chen X.R. The plant ribosome-inactivating proteins play important roles in defense against pathogens and insect pest attacks. Front. Plant Sci. 2018;9:146. doi: 10.3389/fpls.2018.00146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Panda N., Heinrichs E.A. Levels of tolerance and antibiosis in rice varieties having moderate resistance to the brown planthopper, Nilaparvata lugens (Stål)(Hemiptera: Delphacidae). Environ. Entomol. 1983;12:1204–1214. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary material is available on the publisher’s website along with the published article.
Data Availability Statement
Not applicable.