Abstract
Objective
To investigate the potential mechanism of action of Yi-Qi-Huo-Xue-Tong-Luo formula (YQHXTLF) in the treatment of diabetic peripheral neuropathy (DPN).
Methods
Network pharmacology and molecular docking techniques were used in this study. Firstly, the active ingredients and the corresponding targets of YQHXTLF were retrieved using the Traditional Chinese Medicine Systems Pharmacology (TCMSP) platform; subsequently, the targets related to DPN were retrieved using GeneCards, Online Mendelian Inheritance in Man (OMIM), Pharmgkb, Therapeutic Target Database (TTD) and Drugbank databases; the common targets of YQHXTLF and DPN were obtained by Venn diagram; afterwards, the “YQHXTLF Pharmacodynamic Component-DPN Target” regulatory network was visualized using Cytoscape 3.6.1 software, and Gene Ontology (GO) enrichment analysis and Kyoto Encyclopedia of Genes and Genomes (KEGG) pathway analysis were performed on the potential targets using R 3.6.3 software. Finally, molecular docking of the main chemical components in the PPI network with the core targets was verified by Autodock Vina software.
Results
A total of 86 active ingredients and 229 targets in YQHXTLF were screened, and 81 active ingredients and 110 targets were identified to be closely related to diabetic peripheral neuropathy disease. PPI network mapping identified TP53, MAPK1, JUN, and STAT3 as possible core targets. KEGG pathway analysis showed that these targets are mostly involved in AGE-RAGE signaling pathway in diabetic complications, TNF signaling pathway, and MAPK signaling pathway. The molecular docking results showed that the main chemical components of YQHXTLF have a stable binding activity to the core pivotal targets.
Conclusion
YQHXTLF may act on TP53, MAPK1, JUN, and STAT3 to regulate inflammatory response, apoptosis, or proliferation as a molecular mechanism for the treatment of diabetic peripheral neuropathy, reflecting its multitarget and multipathway action, and providing new ideas to further uncover its pharmacological basis and mechanism of action.
1. Introduction
Diabetic peripheral neuropathy (DPN) is one of the most common and serious microvascular complications of diabetes which is characterized by pain, sensory abnormalities, and loss of sensation [1, 2]. It has been shown that DPN affects approximately 40% to 60% of people with diabetes [3], and if not well treated, it increases the risk of disability and mortality [4]. Currently, there is no specific treatment for DPN in modern medicine, which consists mainly of improving metabolic disorders and pain management [5, 6]. With the continuous improvement in medical care and the increasing demand for health, the main issue facing us today is how to prevent and control the progression of DPN and improve the quality of survival of patients. Traditional Chinese Medicine (TCM) has a long history of treating diabetes mellitus and its complications [7, 8]. In TCM, DPN is often classified as “paralysis,” “impotence,” and “blood paralysis” [9]. It is often caused by prolonged thirst, depletion of Qi and blood, deficiency of both yin and yang, and loss of nourishment for the tendons and veins, resulting in coldness, numbness, and muscle atrophy [10, 11]. Treatment is mostly based on benefiting Qi and nourishing Yin, invigorating blood circulation and relieving pain, emphasizing the treatment of both the symptoms and the root cause, and overall regulation [12]. A large number of studies have shown that Chinese medicine is effective in treating DPN, significantly improving the clinical symptoms of patients and delaying the development of the disease, with few toxic side effects [13]. Therefore, exploring the regulatory mechanisms of TCM can help develop new therapeutic approaches to improve the treatment of DPN.
Yi-Qi-Huo-Xue-Tong-Luo formula (YQHXTLF) is an in-hospital preparation for the prevention and control of diabetic peripheral neuropathy at Anhui Provincial Hospital of Traditional Chinese Medicine. It consists of 7 Chinese herbal medicines: Astragalus (Huangqi), Radix Angelicae Sinensis (Danggui), Radix et Rhizoma Dioscoreae (Dihuang), Radix et Rhizoma Yanhusuo (Yanhusuo), Radix et Rhizoma Puerariae (Gegen), Radix et Rhizoma Chrysanthemum (Jixueteng), and Radix et Rhizoma Weilingensis (Weilingxian). The combination of all the herbs in this formula can treat both the symptoms and the root cause of pain by treating Qi and blood together, which can benefit Qi and invigorate blood, as well as promote circulation and relieve pain. Preliminary clinical studies have shown that this formula can alleviate abnormal skin sensation, numbness, and tingling in the limbs and improve motor nerve conduction speed and sensory nerve conduction speed in DPN patients [14]. Basic research found that this formula can reduce islet cell damage in db/db mice, improve blood flow in the sciatic nerve area, and promote the repair and regeneration of damaged nerves, and the mechanism of action may be related to the improvement of diabetic inflammatory lesions and inhibition of excessive activation of the MAPK signaling pathway [15]. However, due to the multicomponent and multitarget nature of the Chinese medicine formula, the exact mechanism of action of the formula is still unclear.
Network pharmacology is a new discipline involving the analysis of drug-target-disease network associations [16]. It provides a systematic approach to the analysis of complex drug mechanisms of action and potential disease interventions by identifying the core targets shared by drugs and diseases [17, 18]. Through the use of network pharmacology, we can not only explore the complex active molecular components and potential molecular targets in Chinese medicine formulations but also understand the molecular relationships between different components in a compound formula and between the components and complex diseases and extract possible pathways for drug interventions to target diseases [19]. With the rapid development of network pharmacology, the mechanisms of TCM in the treatment of many serious diseases have been successfully predicted, and the multitarget integrated prevention and treatment approach has been applied to cancer [20], arthritis [21], diabetes, and other diseases with certain results [22, 23].
In this study, we use network pharmacology as a tool to further analyze the possible targets, molecular mechanisms, biological processes, and pathways of YQHXTLF for the treatment of DPN. We modeled the interrelationship between the targets of DPN and elucidated the synergistic mechanism between the active components of Chinese medicine, providing insights into the interrelationship and changes between Chinese medicine and diseases from the perspective of biological networks, which provided new possibilities and directions for the treatment of DPN. The flow chart of this study is shown in Figure 1.
Figure 1.
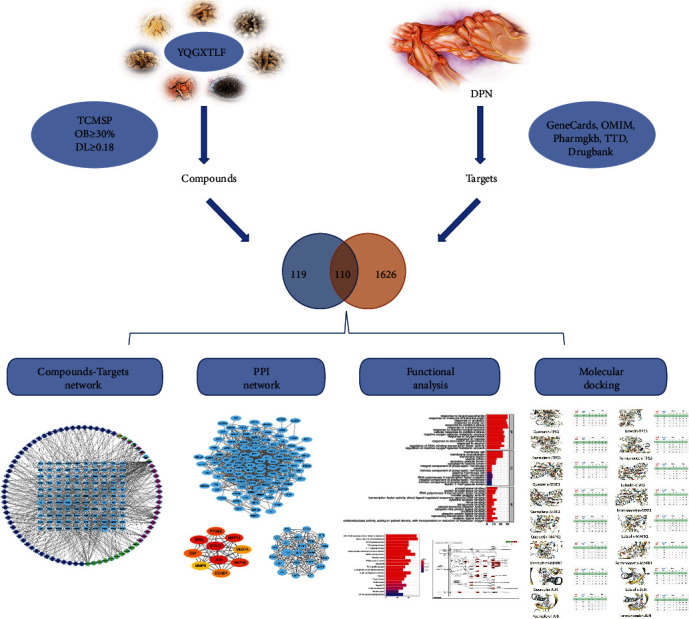
Workflow of the network pharmacology of YQHXTLF in the treatment of DPN. First, the effective active compounds of YQHXTLF were screened from the TCMSP. Relevant targets of DPN were summarized by searching databases. The intersection targets of compound targets and disease targets were obtained soon. Secondly, the interaction net between the compounds and the filtered targets was established. These key targets were analyzed by PPI analysis, functional analysis, and molecular docking verification. Finally, the key genes were used to find the biologic pathway and explain the therapeutic mechanism by network pharmacology analysis.
2. Materials and Methods
2.1. Screening of Active Ingredients of YQHXTLF
The Traditional Chinese Medicine Systems Pharmacology (TCMSP, https://www.tcmspw.com) analysis platform [24] was used to screen the chemical components of YQHXTLF. TCMSP is a unique systemic pharmacology platform for Chinese herbal medicines that captures the relationship between drug, target, and disease. Oral bioavailability (OB) is one of the most important pharmacokinetic parameters in drug absorption, distribution, metabolism, and excretion (ADME) [25]. Drug-like properties (DL) refer to the similarity of a compound to a known drug. In drug development, drug-like studies are based on lead compounds, and drug-like molecules can be considered as high-quality lead compounds [26]. In this study, compounds with OB ≥ 30% and DL ≥ 0.18 were selected as potential active ingredients. The target information of these active ingredients was standardized by using the Uniprot database (https://www.uniprot.org/) with the species “homo sapiens.”
2.2. Exploring Potential DPN Targets
We searched several important databases with the keyword "diabetic peripheral neuropathy", including the GeneCards database (https://www.genecards.org/), the OMIM database (https://www.omim), the Pharmgkb database (https://www.pharmgkb.org/), the TTD database (http://bidd.nus.edu.sg/group/cjttd/), and the Drugbank data (https://www.drugbank.ca/). For the targets in GeneCards, only those with a score ≥ 10 were chosen. The targets obtained from the above four databases were then integrated to construct a DPN-related target set.
2.3. Constructing a Regulatory Network of Targets for the Treatment of DPN with YQHXTLF
The overlapping targets were considered to be the common targets of YQHXTLF and DPN. Cytoscape (version 3.6.1; https://www.cytoscape.org/) [27] was then used to visualize the complex relationships between the active chemical components and the potential target genes to construct a regulatory network of targets for the treatment of DPN with YQHXTLF. The layout tool was used to quantify the degree of each node; the larger the node in the network, the higher the degree value of the node.
2.4. Protein-Protein Interaction Analysis and Core Gene Screening
The intersecting gene targets of YQHXTLF and DPN were imported into the STRING protein interaction analysis platform, and the protein classification was set to “homo sapiens” with a maximum confidence level of ≥0.7, hiding the unlinked nodes in the network. Protein interaction network analysis was performed, and the TSV file was downloaded. Cytoscape 3.6.1 software was then imported to construct protein-protein interaction (PPI) network maps. The hub targets in the PPI network were screened using the cytoHubba plugin, with darker node colors representing higher scores [28]. The Molecular Complex Detection (MCODE) plugin was used to discover closely linked regions in the PPI network [29]. The score value of a module reflects how dense the module is to the surrounding nodes, with higher scores indicating that the nodes are becoming more important.
2.5. Gene Ontology and KEGG Pathway Enrichment Analysis
Gene Ontology (GO) functional analysis is mainly used to describe the function of gene targets, including biological process, cellular component, and molecular function. KEGG enrichment analysis can obtain the signal pathways enriched by the common targets of YQHXTLF and DPN. By using R 3.6.3 software-related R packages (Colorspace, Stringi, GGPLOt2, BiocManager, Dose, clusterProfiler, Enrichplot, Pathview), GO enrichment analysis and KEGG pathway enrichment analysis were conducted for the intersection target genes of YQHXTLF-DPN. According to Fisher's test, a p value < 0.05 and q value < 0.05 were considered statistically significant.
2.6. Molecular Docking Validation
Molecular docking validation is achieved through the CB-Dock server [30], a docking tool that predicts the binding site of a given protein and calculates the center and size using a novel curvature-based lumenal detection method. The 2D structure of the compound's small-molecule ligand was first obtained from the PubChem online database (https://pubchem.ncbi.nlm.nih.gov/) as the small-molecule ligand file for molecular docking. The core target protein receptor structure file was obtained from the PDB online database (https://www.rcsb.org). The CB-Dock software was used for molecular docking, and the score of the molecular docking complexes was calculated using the Vina program to evaluate their binding activity.
3. Results
3.1. Active Ingredients in YQHXTKF
The active ingredients were screened by searching the TCMSP database with a threshold value of OB ≥ 30% and DL ≥ 0 : 18. A total of 86 active ingredients derived from YQHXTLF were screened. Among them, 20 ingredients were from Astragalus, 2 from Radix Angelicae Sinensis, 4 from Radix Puerariae, 24 from Spatholobi Caulis, 2 from Rehmanniae Radix, 7 from Radix Clematidis, and 49 from Corydalis Rhizoma. The search for their corresponding targets showed that 86 compounds acted on 229 targets. The basic information of the 86 active compounds is shown in Table 1.
Table 1.
Detailed information of the 86 active compounds from YQHXTLF.
| Mol Id | Mol name | Structure | OB (%) | DL |
|---|---|---|---|---|
| MOL000358 | Beta-sitosterol |
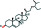
|
36.91 | 0.75 |
| MOL000449 | Stigmasterol |
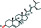
|
43.83 | 0.76 |
| MOL000392 | Formononetin |
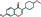
|
69.67 | 0.21 |
| MOL002959 | 3′-Methoxydaidzein |
|
48.57 | 0.24 |
| MOL003629 | Daidzein-4,7-diglucoside |
|
47.27 | 0.67 |
| MOL000211 | Mairin |
|
55.38 | 0.78 |
| MOL000239 | Jaranol |
|
50.83 | 0.29 |
| MOL000296 | Hederagenin |
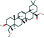
|
36.91 | 0.75 |
| MOL000033 | (3S,8S,9S,10R,13R,14S,17R)-10,13-Dimethyl-17-[(2R,5S)-5-propan-2-yloctan-2-yl]-2,3,4,7,8,9,11,12,14,15,16,17-dodecahydro-1H-cyclopenta[a]phenanthren-3-ol |
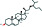
|
36.23 | 0.78 |
| MOL000354 | Isorhamnetin |
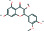
|
49.6 | 0.31 |
| MOL000371 | 3,9-Di-O-methylnissolin |
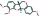
|
53.74 | 0.48 |
| MOL000378 | 7-O-Methylisomucronulatol |
|
74.69 | 0.3 |
| MOL000379 | 9,10-Dimethoxypterocarpane -3-O-β-D-glucoside |
|
36.74 | 0.92 |
| MOL000380 | (6aR,11aR)-9,10-Dimethoxy-6a,11a-dihydro-6H-benzofurano[3,2-c]chromen-3-ol |
|
64.26 | 0.42 |
| MOL000387 | Bifendate |
|
31.1 | 0.67 |
| MOL000417 | Calycosin |

|
47.75 | 0.24 |
| MOL000422 | Kaempferol |
|
41.88 | 0.24 |
| MOL000433 | FA |
|
68.96 | 0.71 |
| MOL000439 | Isomucronulatol-7,2′-di-O-glucosiole |
|
49.28 | 0.62 |
| MOL000442 | 1,7-Dihydroxy-3,9-dimethoxy pterocarpene |
|
39.05 | 0.48 |
| MOL000098 | Quercetin |
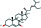
|
46.43 | 0.28 |
| MOL000461 | 3,7-Dihydroxy-6-methoxy-dihydroflavonol |
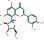
|
43.8 | 0.26 |
| MOL000468 | 8-O-Methylreyusin |
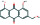
|
70.32 | 0.27 |
| MOL000469 | 3-Hydroxystigmast-5-en-7-one |
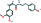
|
40.93 | 0.78 |
| MOL000470 | 8-C-α-L-Arabinosylluteolin |
|
35.54 | 0.66 |
| MOL000471 | Aloe-emodin |
|
83.38 | 0.24 |
| MOL000483 | (Z)-3-(4-Hydroxy-3-methoxy-phenyl)-N-[2-(4-hydroxyphenyl)ethyl]acrylamide |
|
118.35 | 0.26 |
| MOL000490 | Petunidin |
|
30.05 | 0.31 |
| MOL000492 | (+)-Catechin |
|
54.83 | 0.24 |
| MOL000493 | Campesterol |

|
37.58 | 0.71 |
| MOL000497 | Licochalcone a |
|
40.79 | 0.29 |
| MOL000500 | Vestitol |
|
74.66 | 0.21 |
| MOL000501 | Consume close grain |
|
68.12 | 0.27 |
| MOL000502 | Cajinin |
|
68.8 | 0.27 |
| MOL000503 | Medicagol |
|
57.49 | 0.6 |
| MOL000506 | Lupinidine |
|
61.89 | 0.21 |
| MOL000507 | Psi-baptigenin |
|
70.12 | 0.31 |
| MOL000006 | Luteolin |
|
36.16 | 0.25 |
| MOL000359 | Sitosterol |

|
36.91 | 0.75 |
| MOL005603 | Diheptyl phthalate |
|
42.26 | 0.31 |
| MOL001454 | Berberine |
|
36.86 | 0.78 |
| MOL001458 | Coptisine |

|
30.67 | 0.86 |
| MOL001460 | Cryptopine |

|
78.74 | 0.72 |
| MOL001461 | Dihydrochelerythrine |
|
32.73 | 0.81 |
| MOL001463 | Dihydrosanguinarine |
|
59.31 | 0.86 |
| MOL001474 | Sanguinarine |
|
37.81 | 0.86 |
| MOL000217 | (S)-Scoulerine |
|
32.28 | 0.54 |
| MOL002670 | Cavidine |
|
35.64 | 0.81 |
| MOL002903 | (R)-Canadine |
|
55.37 | 0.77 |
| MOL004071 | Hyndarine |
|
36.91 | 0.75 |
| MOL004190 | (-)-Alpha-N-methylcanadine |
|
73.94 | 0.64 |
| MOL004191 | Capaurine |

|
45.06 | 0.8 |
| MOL004193 | Clarkeanidine |
|
62.91 | 0.69 |
| MOL004195 | CORYDALINE |
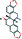
|
86.65 | 0.54 |
| MOL004196 | Corydalmine |
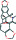
|
65.84 | 0.68 |
| MOL004197 | Corydine |
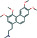
|
52.5 | 0.59 |
| MOL004198 | 18797-79-0 |

|
37.16 | 0.55 |
| MOL004199 | Corynoloxine |
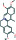
|
46.06 | 0.85 |
| MOL004200 | Methyl-[2-(3,4,6,7-tetramethoxy-1-phenanthryl)ethyl]amine |
|
38.12 | 0.6 |
| MOL004202 | Dehydrocavidine |
|
61.15 | 0.44 |
| MOL004203 | Dehydrocorybulbine |
|
38.99 | 0.81 |
| MOL004204 | Dehydrocorydaline |
|
46.97 | 0.63 |
| MOL004205 | Dehydrocorydalmine |
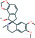
|
41.98 | 0.68 |
| MOL004208 | Demethylcorydalmatine |
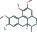
|
43.9 | 0.59 |
| MOL004209 | 13-Methyldehydrocorydalmine |
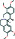
|
38.99 | 0.54 |
| MOL004210 | (1S,8′R)-6,7-Dimethoxy-2-methylspiro[3,4-dihydroisoquinoline-1,7′-6,8-dihydrocyclopenta[g][1,3]benzodioxole]-8′-ol |
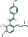
|
35.94 | 0.63 |
| MOL004763 | Izoteolin |
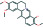
|
43.95 | 0.72 |
| MOL004214 | Isocorybulbine |
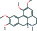
|
39.53 | 0.51 |
| MOL004215 | Leonticine |
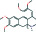
|
40.18 | 0.66 |
| MOL004216 | 13-Methylpalmatrubine |
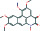
|
45.79 | 0.26 |
| MOL004220 | N-Methyllaurotetanine |
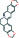
|
40.97 | 0.63 |
| MOL004221 | Norglaucing |
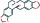
|
41.62 | 0.56 |
| MOL004224 | Pontevedrine |
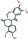
|
30.35 | 0.56 |
| MOL004225 | Pseudocoptisine |

|
30.28 | 0.71 |
| MOL004226 | 24240-05-9 |
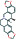
|
38.97 | 0.86 |
| MOL004228 | Saulatine |
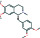
|
53.75 | 0.83 |
| MOL004230 | Stylopine |
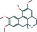
|
42.74 | 0.79 |
| MOL004231 | Tetrahydrocorysamine |

|
48.25 | 0.85 |
| MOL004232 | Tetrahydroprotopapaverine |
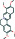
|
34.17 | 0.86 |
| MOL004233 | ST057701 |

|
36.86 | 0.78 |
| MOL004234 | 2,3,9,10-Tetramethoxy-13-methyl-5,6-dihydroisoquinolino[2,1-b]isoquinolin-8-one |

|
30.67 | 0.86 |
| MOL000785 | Palmatine |
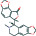
|
78.74 | 0.72 |
| MOL000787 | Fumarine |
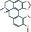
|
32.73 | 0.81 |
| MOL000790 | Isocorypalmine |
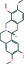
|
59.31 | 0.86 |
| MOL000791 | Bicuculline |
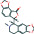
|
37.81 | 0.86 |
| MOL000793 | C09367 |
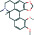
|
32.28 | 0.54 |
Mol Id: molecule Id; Mol name: molecule name; OB: oral bioavailability; DL: drug-like.
3.2. DPN-Related Gene Targets
Using the keyword “Diabetic peripheral neuropathy,” the GeneCards website screened 1521 genes related to diabetic peripheral neuropathy, the OMIM database screened 118, the Pharmgkb database retrieved 198, the TTD online website retrieved 32, and the Drugbank database retrieved 20. After removing duplicate targets and standardized gene names, a total of 1736 DPN potential action targets were screened.
3.3. Intersection of TQHXTLF and DPN Target
The Hiplot research data visualization platform (https://hiplot.com.cn) was used to show the intersection of TQHXTLF and DPN targets, and Venn diagrams were drawn to obtain a total of 110 candidate targets for TQHXTLF and DPN, as shown in Figure 2.
Figure 2.
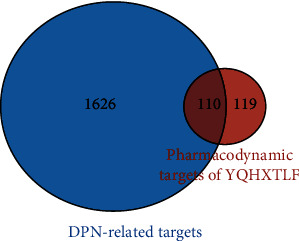
Venn diagram of targets of DPN treated by YQHXTLF.
3.4. Regulatory Network Analysis of YQHXTLF and DPN Targets
The “YQHXTLF Pharmacodynamic Component-DPN Target” regulatory network was mapped by using Cytoscape 3.6.1 software to match the YQHXTLF pharmacodynamic components with the 110 action targets obtained from the screening. As shown in Figure 3, the network contained 191 nodes and 696 relationships. From the overall characteristics of the network, it can be found that among the 81 compounds in YQHXTLF, there is one compound corresponding to multiple targets and one target corresponding to multiple compounds; the larger the node, the larger the network value of the target. From the regulatory network, it can be seen that the chemical with the highest degree of connectivity is quercetin, which interacts with 75 targets, followed by luteolin, which interacts with 31 targets; kaempferol, which interacts with 28 targets; and formononetin, which interacts with 18 targets.
Figure 3.
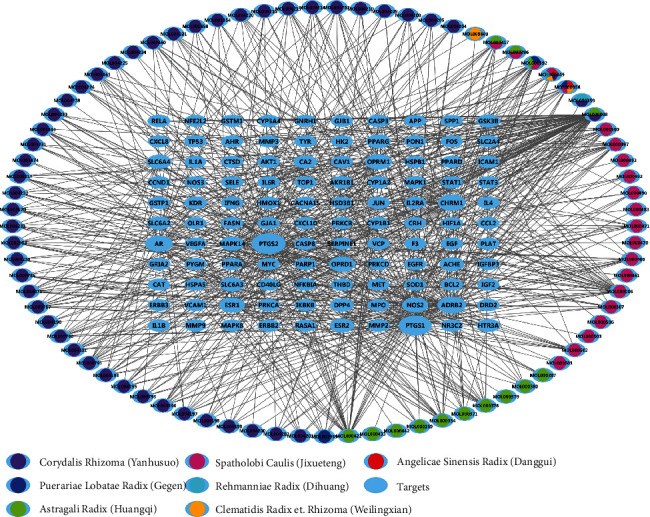
Construction of a target modulation network for DPN treatment with YQHXTLF. The blue ellipse represents the target site, the other coloured nodes represent the different herbal compounds, and the connecting lines represent the interaction between the compound and the target site.
3.5. PPI Network Construction and Core Module Analysis
We imported common targets into the STRING 11.0 platform and visualized the PPI network using Cytoscape 3.6.1 software, as shown in Figure 4(a). 105 nodes and 797 edges were included in the network. Five targets did not interact with any other targets and were therefore not included in the PPI network. The cytoHubba plugin was applied to calculate the degree of degree connectivity for each target (Figure 4(b)). The top 10 hub target genes ranked by node degree were AKT1, MAPK8, TP53, MAPK1, STAT3, VEGFA, JUN, EGFR, and EGF. These target genes may play a key role in the network. The MCODE plugin was analyzed to filter out the most significant modules, with an MCODE score of 17.238, containing 22 nodes and 181 edges. The top ten targets in the MOCDE score were TP53, ESR1, ICAM1, JUN, AR, STAT3, CXCL8, MAPK1, and FOS (Figure 4(c)). The results showed that TP53, MAPK1, JUN, and STAT3 were the key targets under different algorithms.
Figure 4.
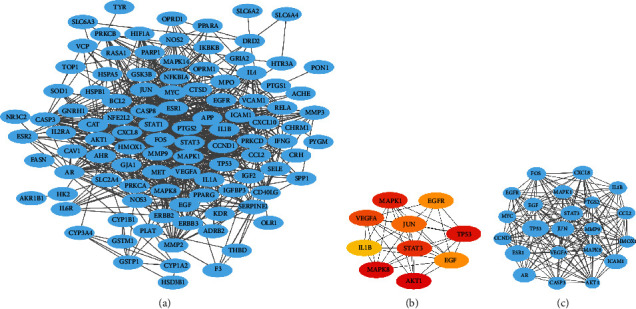
PPI network construction and core module: (a) YQHXTLF pharmacodynamic composition-DPN target PPI network; (b) top 10 hub target genes ranked by node degree; (c) the most significant modules analyzed by the MCODE plugin.
3.6. Gene Ontology and KEGG Pathway Enrichment Analysis
We intercepted the first 15 terms from the smallest to the largest according to the adj. p value. The results of the biological process analysis showed that the intersection targets were mostly enriched in response to lipopolysaccharide, response to molecule of bacterial origin, response to antibiotic, response to metal ion response to oxidative stress, and response to nutrient levels. The results of cell composition showed that the intersection targets were mostly enriched in membrane raft, membrane microdomain, membrane region, vesicle lumen, secretory granule lumen, cytoplasmic vesicle lumen, etc. Molecular functions mainly include phosphatase binding, protein phosphatase binding, RNA polymerase II transcription factor binding, nuclear receptor activity, transcription factor activity, direct ligand regulated sequence-specific DNA binding, and cytokine receptor binding, as shown in Figure 5(a).
Figure 5.
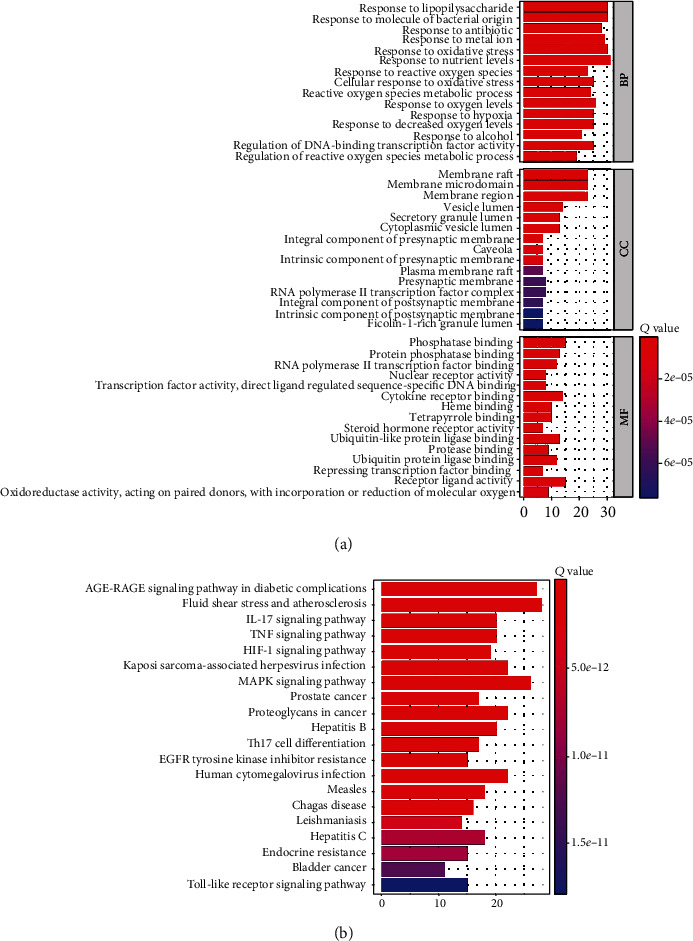
GO and KEGG pathway enrichment analysis: (a) the top 15 significantly enriched GO terms of BP, CC, and MF; (b) the top 20 significantly enriched KEGG pathways.
We intercepted the top 20 KEGG pathways from the smallest to the largest based on the p value. The analysis showed that these targets were mostly enriched in the AGE-RAGE signaling pathway in diabetic complications, fluid shear stress and atherosclerosis, TNF signaling pathway, MAPK signaling pathway, etc., as shown in Figure 5(b).
3.7. Molecular Docking Validation
The four active ingredients quercetin, luteolin, kaempferol, and formononetin, which have a high number of targets in YQHXTKF, were selected for molecular docking validation with the core targets TP53, MAPK1, JUN, and STAT3 in the PPI network map. The binding energy (Vina score) was used to evaluate the bonding strength between the docked molecules, and the value of the Vina score indicated some binding activity between the proteins and the compounds. The smaller the binding energy (Vina score), the higher the affinity of the receptor and ligand and the more stable the binding of the compound to the target site. The binding energies of the four active ingredients and the corresponding target proteins were all less than -5.0, indicating good docking and high binding activity [31]. Molecular docking simulations showed a stable point docking structure for the binding of small-molecule ligands and protein receptors (Figure 6).
Figure 6.

The docking model diagram of the active ingredient of the drug and the core target molecule: (a–d) the action mode of TP53 and quercetin, luteolin, kaempferol, and formononetin; (e–h) the action mode of STAT3 and quercetin, luteolin, kaempferol, and formononetin; (i–l) the action mode of MAPK1 and quercetin, luteolin, kaempferol, and formononetin; (m–p) the action mode of JUN and quercetin, luteolin, kaempferol, and formononetin.
4. Discussion
In this study, we used network pharmacology and molecular docking techniques to analyze the potential molecular biological mechanisms of YQHXTLF for the treatment of DPN. We firstly screened the active ingredients of YQHXTLF using the criteria of OB ≥ 30% and DL ≥ 0.18 and obtained 94 compounds, 229 potential targets, and 110 targets overlapping with DPN to construct a compound-disease target regulatory network. Through the network analysis, 81 components in this prescription may act on 110 targets to exert the therapeutic effect. These identified compounds, particularly the four compounds including quercetin, luteolin, kaempferol, and formononetin, were linked to more than 10 targets, indicating that these compounds might play a vital role in the process of DPN treatment. Quercetin is a flavonoid which is widely found in nature and has been proved to have a great effect on antibacterial, anti-inflammatory, and antiallergic therapy [32]. Quercetin reduces neuronal loss and inhibits neuronal apoptosis by improving neurotrophic factor levels [33, 34]. It has been reported that quercetin exerts neuroprotective effects on DRG neuronal cells in a high-glucose environment, possibly by activating Nrf-2/HO-1 and inhibiting NF-κB to reduce apoptosis [35]. Quercetin also has neuroprotective effects in diabetic peripheral neuropathy by inducing autophagy to reduce the damage to neuronal cells from high glucose and improving the antioxidant status [36, 37]. Luteolin significantly upregulates the protein levels of NRF2 and HO-1 in diabetic nerves and improved nerve conduction velocity and nerve blood flow [38]. Luteolin has been shown to improve blood glucose, glycosylation, insulin, and HOMR-IR levels in diabetic model mice, with positive effects against diabetes and its complications [39]. Kaempferol has excellent antioxidant properties and can correct hyperglycemia in DM rats by regulating oxidative stress and reducing AGE accumulation, thus preventing the risk of complications [40, 41]. It reduces the expression of IL-1β and TNF-α, thereby inhibiting the neuroimmune activation of microglia and alleviating the progression of diabetic neuropathy [42]. Formononetin is an isoflavone that is known to prevent and slow the progression of long-term diabetic complications by reducing hyperglycemia and highlighting neuroprotective effects [43]. Formononetin protects diabetic animals from hyperglycemia-induced neuronal damage by increasing the expression of SIRT1 and NGF in neural tissue [44, 45]. Therefore, quercetin, luteolin, kaempferol, and formononetin may be the most important components of YQHXTLF for the treatment of DPN, as shown in Table 2.
Table 2.
The potential mechanisms of the four main compounds in the treatment of DPN.
| Compound | Mechanism | Model | Reference |
|---|---|---|---|
| Quercetin | Increased BDNF, NGF, and Bcl-2, inhibited caspase-3 | Diabetic rats | [33] |
| Maintained the density of the general neuronal population, reduced the loss of interosseous neurons, antioxidant | Diabetic rats | [34] | |
| Activated the Nrf-2/HO-1 pathway, inhibited the NF-κB pathway, and inhibited iNOS, COX-2, IL-6, and TNF-α | DRG cells | [35] | |
| Upregulated Beclin-1 and LC3 protein expression levels, increased cell proliferation, and upregulated autophagy | Schwann cells | [36] | |
| Reduced total cholesterol and TBARS levels, increased HDL-cholesterol, SOD, CAT, and GSH-Px activity | Db/db mice | [37] | |
| Luteolin | Upregulated protein levels of Nrf2 and HO-1, improved nerve conduction velocity and nerve blood flow | Diabetic rats | [38] |
| Improved the levels of blood glucose, HbA 1c, insulin, and HOMR-IR | KK-Ay mice | [39] | |
| Reduced mRNA expression of SREBP-1c, TNF-α | |||
| Kaempferol | Regulated oxidative and nitrosative stress and reduced the formation of AGEs | Diabetic rats | [40] |
| Reduced ROS production and inhibited caspase-3 activation | PC12 cells | [41] | |
| Reduction IL-1β, TNF-α, IC, and ROS and inhibited neuroimmune activation of microglia | Diabetic mice | [42] | |
| Formononetin | Inhibited islet B cell apoptosis and promoted islet B cell regeneration, insulin secretion, hepatic glycogen synthesis, and hepatic glycolysis | Diabetic mice | [43] |
| Controlled hyperglycemia and increased expression of SIRT1 and NGF | Diabetic rats | [44] | |
| Increased SIRT1 expression and reduced blood glucose | Diabetic rats | [45] |
The exact etiology of DPN is still unclear, and its pathogenesis may be related to mitochondrial dysfunction and oxidative stress, polyol pathway activation, advanced glycosylation end products, and endoplasmic reticulum stress due to prolonged and severe hyperglycaemia [46, 47]. In addition, metabolic inflammation, neurotrophic vasculopathy, insulin resistance, and neurotrophic factors are all involved, creating a complex and interrelated pathogenesis [48]. Studies have shown that elevated levels of RAGE expression have been found in skin biopsy specimens from DPN patients [49]. Advanced glycosylation end products (AGEs) are a complex group of compounds. The primary receptor for AGEs (RAGE or AGER), which belongs to the immunoglobulin superfamily, has been described as a pattern recognition receptor [50]. AGE/RAGE signaling causes activation of multiple intracellular signaling pathways involving NADPH oxidase, protein kinase C (PKC), and MAPKs [51] and promotes the expression of multiple proinflammatory cytokines, leading to segmental demyelination of peripheral neurons (Figure 7). The interaction between AGE and RAGE leads to increased diacylglycerol (DAG) synthesis and excessive activation of PKC, which increases NF-κB and tissue-type fibrinogen activator inhibitor-1 (PAI-1) expression, further activating inflammatory factors, transforming growth factor-β (TGF-β), and vascular endothelial growth factor (VEGF), leading to altered vascular function and peripheral neurological microangiopathy [52, 53]. Numerous studies have shown that AGEs bound to their receptors rapidly activate NADPH oxidase, increasing the level of mitochondrial oxidative stress, generating large amounts of ROS, and promoting a signal transduction cascade to induce apoptosis [54, 55]. STAT3 and MAPK pathways are important signaling pathways involved in inflammatory responses, cell proliferation, and apoptosis. STAT3, signal transducer and activator of transcription 3, a member of the signal transducer and activator of transcription family, is closely associated with central cell growth, proliferation and survival, and immune response [56]. Activated STAT3 can be implicated in the activity of downstream mediators such as p27kip1, P16INK4A, and p21kip1 proteins, which regulate cell growth, differentiation, and angiogenesis and are involved in the pathogenesis of diabetes [57]. MAPK is a mitogen-activated protein kinase involved in a variety of cellular functions, including cell proliferation, differentiation, and migration [58]. MAPKs cause neuronal apoptosis, impaired neuronal regeneration, and neuropathy through the direct action of glucose and glucose-induced oxidative stress [59]. Tumor necrosis factor-α (TNF-α) is a proinflammatory factor involved in peripheral nerve injury. It stimulates monocytes and endothelial cells to secrete IL-1β and IL-6 and other inflammatory factors, which have toxic effects on neurons and glial cells and lead to demyelination [60]. TNF-α inhibits nitric oxide synthase (NOS) activity in vascular endothelial cells, resulting in reduced NO-induced vasodilation, which leads to endothelial dysfunction and neurotrophic vascular damage and induces neuropathy [61]. In addition, TNF-α can activate the c-jun amino-terminal kinase JNK signaling pathway, leading to apoptosis [62]. TP53, cellular tumor antigen p53, is a stress-sensitive transcription factor responsible for controlling cell survival and death to prevent tumor formation [63]. TP53 has been found to inhibit glycolysis and to be involved in oxidative stress, the TP53 gene polymorphism marker Pro72Arg has been associated with DPN pathogenesis [64], and TP53 serum levels are significantly increased in patients with T2DM [65], so it is hypothesized that TP53 may play an important role in metabolic diseases such as diabetes. The molecular docking results showed that the active ingredients of the key compounds in YQHXTLF were able to bind stably to TP53, MAPK1, JUN, and STAT3. Thus, these results also confirm that our screened targets are consistent with literature reports, suggesting that YQHXTLF can play a therapeutic role in DPN by regulating apoptosis or proliferation and mediating inflammatory responses or oxidative stress.
Figure 7.
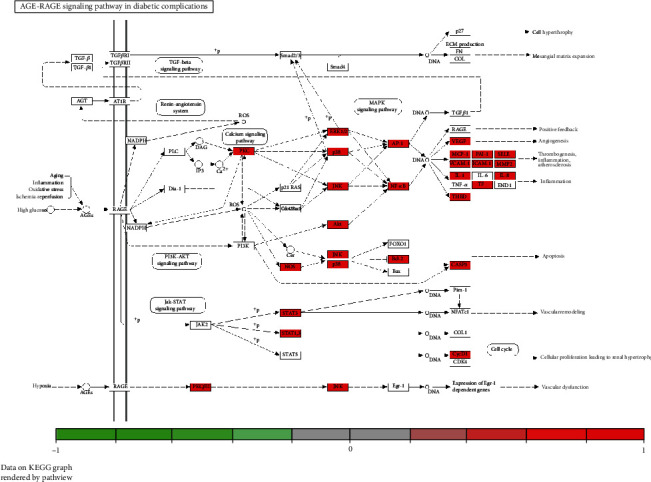
Pathway map of YQHXTLF in the treatment of DPN. AGE/RAGE signaling causes activation of multiple intracellular signaling pathways involving NADPH oxidase, protein kinase C (PKC), and MAPKs and promotes the expression of multiple proinflammatory cytokines, leading to segmental demyelination of peripheral neurons.
5. Conclusion
In summary, this study analyzed the potential molecular biological mechanisms of YQHXTLF in the treatment of DPN throughnetwork pharmacology and molecular docking approach. The results showed that the active components of YQHXTLF in DPN treatment were composed of 81 compounds, among which quercetin, luteolin, kaempferol, and formononetin were the important active components. Moreover, a total of 110 target genes were screened, among which TP53, MAPK1, JUN, and STAT3 are possible core targets. The pathways involved in the treatment of DPN by YQHXTLF may be related to the AGE-RAGE signaling pathway, TNF signaling pathway, and MAPK signaling pathway, reflecting the multicomponent, multitarget, and multipathway biological properties of YQHXTLF. However, the network pharmacology is only a reasonable prediction of the mechanism of action of the herbal compound for the treatment of DPN based on the data mining perspective, which can provide a reference for the study of its therapeutic mechanism. We will be followed by animal and clinical experiments to validate the screened targets based on this prediction analysis, to provide more scientific evidence for its clinical application.
Acknowledgments
This study was supported by the National Natural Science Foundation of China (No. 81603574, No. 81774286), Major Natural Science Research Projects of Colleges and Universities in Anhui Province (No. KJ2018ZD029), and the second batch of scientific research projects for the construction of the national traditional Chinese medicine clinical research base business (JDZX2015123).
Contributor Information
Zhaohui Fang, Email: fangzhaohui9097@163.com.
Guoming Shen, Email: shengm_66@163.com.
Data Availability
We have presented all our main data in the form of figures and additional files. The data used to support the conclusions of this study are available from the authors.
Conflicts of Interest
The authors declare that they have no conflicts of interest.
Supplementary Materials
Ingredients: potential active ingredients and action targets of the herbal compound were obtained through screening.
Diseases: DPN-related targets were obtained.
References
- 1.Singh R., Kishore L., Kaur N. Diabetic peripheral neuropathy: current perspective and future directions. Pharmacological Research. 2014;80:21–35. doi: 10.1016/j.phrs.2013.12.005. [DOI] [PubMed] [Google Scholar]
- 2.Boulton A. J., Gries F. A., Jervell J. A. Guidelines for the diagnosis and outpatient management of diabetic peripheral neuropathy. Diabetic Medicine. 1998;15(6):508–514. doi: 10.1002/(SICI)1096-9136(199806)15:6<508::AID-DIA613>3.0.CO;2-L. [DOI] [PubMed] [Google Scholar]
- 3.Tesfaye S., Selvarajah D. Advances in the epidemiology, pathogenesis and management of diabetic peripheral neuropathy. Diabetes/Metabolism Research and Reviews. 2012;28(Supplement 1):8–14. doi: 10.1002/dmrr.2239. [DOI] [PubMed] [Google Scholar]
- 4.Waldfogel J. M., Nesbit S. A., Dy S. M., et al. Pharmacotherapy for diabetic peripheral neuropathy pain and quality of life: a systematic review. Neurology. 2017;88(20):1958–1967. doi: 10.1212/WNL.0000000000003882. [DOI] [PubMed] [Google Scholar]
- 5.Callaghan B. C., Cheng H. T., Stables C. L., Smith A. L., Feldman E. L. Diabetic neuropathy: clinical manifestations and current treatments. Lancet Neurology. 2012;11(6):521–534. doi: 10.1016/S1474-4422(12)70065-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Iqbal Z., Azmi S., Yadav R., et al. Diabetic peripheral neuropathy: epidemiology, diagnosis, and pharmacotherapy. Clinical Therapeutics. 2018;40(6):828–849. doi: 10.1016/j.clinthera.2018.04.001. [DOI] [PubMed] [Google Scholar]
- 7.Yang D., Liang X. C. Strategies and research progress of Chinese medicine in prevention and treatment of diabetic peripheral neuropathy. Chinese Journal of Integrative Medicine. 2018;24(10):794–800. doi: 10.1007/s11655-018-3051-x. [DOI] [PubMed] [Google Scholar]
- 8.Xiao E., Luo L. Alternative therapies for diabetes: a comparison of western and traditional Chinese medicine (TCM) approaches. Current Diabetes Reviews. 2018;14(6):487–496. doi: 10.2174/1573399813666170519103230. [DOI] [PubMed] [Google Scholar]
- 9.Li M., Su H., Xiang Y., Zhang J., Zhao J., Lin L. Features analysis on traditional Chinese medicine syndromes in patients with diabetic peripheral neuropathy. Journal of Traditional Chinese Medicine. 2013;33(5):637–641. doi: 10.1016/S0254-6272(14)60034-0. [DOI] [PubMed] [Google Scholar]
- 10.Piao Y., Liang X. Chinese medicine in diabetic peripheral neuropathy: experimental research on nerve repair and regeneration. Evidence-based Complementary and Alternative Medicine. 2012;2012:13. doi: 10.1155/2012/191632.191632 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Xu J. Y., Gao H. Y. Combined treatment of traditional Chinese medicine and western medicine in the treatment of diabetic peripheral neuropathy. Journal of Biological Regulators and Homeostatic Agents. 2018;32(4):945–949. [PubMed] [Google Scholar]
- 12.Fu Q., Yang H., Zhang L., et al. Traditional Chinese medicine foot bath combined with acupoint massage for the treatment of diabetic peripheral neuropathy: a systematic review and meta-analysis of 31 RCTs. Diabetes/metabolism research and reviews. 2020;36(2, article e3218) doi: 10.1002/dmrr.3218. [DOI] [PubMed] [Google Scholar]
- 13.Kang S., Zhong Y., Liu D., Li W. Traditional Chinese medicine fumigation as auxiliary treatment of diabetic peripheral neuropathy: a protocol for systematic review and meta-analysis. Medicine (Baltimore) 2021;100(4, article e24200) doi: 10.1097/MD.0000000000024200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Lin Y., Wang F., Li J., Ruan N., Fang Z., Shen G. Therapeutic effect of Yiqi Huoxue Tongluo Fang on diabetic peripheral neuropathy. Clinical Journal of Traditional Chinese Medicine. 2019;31(11):2135–2138. [Google Scholar]
- 15.Yao Y., Shen G., Jiang A., Wang L., Ye S., Wang L. Influence of Yiqi Huoxue Tongluo Fang on JNK signal pathway and inflammatory factors in mice with diabetic peripheral neuropathy. Journal of Beijing University of Traditional Chinese Medicine. 2017;40(11):923–927. [Google Scholar]
- 16.Hopkins A. L. Network pharmacology: the next paradigm in drug discovery. Nature Chemical Biology. 2008;4(11):682–690. doi: 10.1038/nchembio.118. [DOI] [PubMed] [Google Scholar]
- 17.Berger S. I., Iyengar R. Network analyses in systems pharmacology. Bioinformatics. 2009;25(19):2466–2472. doi: 10.1093/bioinformatics/btp465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Hao D. C., Xiao P. G. Network pharmacology: a Rosetta Stone for traditional Chinese medicine. Drug Development Research. 2014;75(5):299–312. doi: 10.1002/ddr.21214. [DOI] [PubMed] [Google Scholar]
- 19.Jiao X., Jin X., Ma Y., et al. A comprehensive application: molecular docking and network pharmacology for the prediction of bioactive constituents and elucidation of mechanisms of action in component-based Chinese medicine. Computational Biology and Chemistry. 2021;90, article 107402 doi: 10.1016/j.compbiolchem.2020.107402. [DOI] [PubMed] [Google Scholar]
- 20.Lee H. S., Lee I. H., Kang K., et al. A network pharmacology study on the molecular mechanisms of FDY003 for breast cancer treatment. Evidence-based Complementary and Alternative Medicine. 2021;2021:18. doi: 10.1155/2021/3919143.3919143 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Xie B., Lu H., Xu J., et al. Targets of hydroxychloroquine in the treatment of rheumatoid arthritis. A network pharmacology study. Joint Bone Spine. 2021;88(2, article 105099) doi: 10.1016/j.jbspin.2020.105099. [DOI] [PubMed] [Google Scholar]
- 22.Zhou J., Wang Q., Xiang Z., et al. Network pharmacology analysis of traditional Chinese medicine formula Xiao Ke Yin Shui treating type 2 diabetes mellitus. Evidence-based Complementary and Alternative Medicine. 2019;2019:15. doi: 10.1155/2019/4202563.4202563 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Hu R. F., Sun X. B. Design of new traditional Chinese medicine herbal formulae for treatment of type 2 diabetes mellitus based on network pharmacology. Chinese Journal of Natural Medicines. 2017;15(6):436–441. doi: 10.1016/S1875-5364(17)30065-1. [DOI] [PubMed] [Google Scholar]
- 24.Ru J., Li P., Wang J., et al. TCMSP: a database of systems pharmacology for drug discovery from herbal medicines. Journal of Cheminformatics. 2014;6(1):p. 13. doi: 10.1186/1758-2946-6-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Xu X., Zhang W., Huang C., et al. A novel chemometric method for the prediction of human oral bioavailability. International Journal of Molecular Sciences. 2012;13(6):6964–6982. doi: 10.3390/ijms13066964. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Di L., Kerns E. H. Profiling drug-like properties in discovery research. Current Opinion in Chemical Biology. 2003;7(3):402–408. doi: 10.1016/S1367-5931(03)00055-3. [DOI] [PubMed] [Google Scholar]
- 27.Shannon P., Markiel A., Ozier O., et al. Cytoscape: a software environment for integrated models of biomolecular interaction networks. Genome Research. 2003;13(11):2498–2504. doi: 10.1101/gr.1239303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Chin C. H., Chen S. H., Wu H. H., Ho C. W., Ko M. T., Lin C. Y. cytoHubba: identifying hub objects and sub-networks from complex interactome. BMC systems biology. 2014;8(Supplement 4):p. S11. doi: 10.1186/1752-0509-8-S4-S11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Saito R., Smoot M. E., Ono K., et al. A travel guide to Cytoscape plugins. Nature Methods. 2012;9(11):1069–1076. doi: 10.1038/nmeth.2212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Liu Y., Grimm M., Dai W. T., Hou M. C., Xiao Z. X., Cao Y. CB-Dock: a web server for cavity detection-guided protein-ligand blind docking. Acta Pharmacologica Sinica. 2020;41(1):138–144. doi: 10.1038/s41401-019-0228-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Hsin K. Y., Ghosh S., Kitano H. Combining machine learning systems and multiple docking simulation packages to improve docking prediction reliability for network pharmacology. PLoS One. 2013;8(12, article e83922) doi: 10.1371/journal.pone.0083922. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Boots A. W., Haenen G. R., Bast A. Health effects of quercetin: from antioxidant to nutraceutical. European Journal of Pharmacology. 2008;585(2-3):325–337. doi: 10.1016/j.ejphar.2008.03.008. [DOI] [PubMed] [Google Scholar]
- 33.Ola M. S., Ahmed M. M., Shams S., Al-Rejaie S. S. Neuroprotective effects of quercetin in diabetic rat retina. Saudi journal of biological sciences. 2017;24(6):1186–1194. doi: 10.1016/j.sjbs.2016.11.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Ferreira P. E., Lopes C. R., Alves A. M., et al. Diabetic neuropathy: an evaluation of the use of quercetin in the cecum of rats. World Journal of Gastroenterology. 2013;19(38):6416–6426. doi: 10.3748/wjg.v19.i38.6416. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Shi Y., Liang X. C., Zhang H., Wu Q. L., Qu L., Sun Q. Quercetin protects rat dorsal root ganglion neurons against high glucose-induced injury in vitro through Nrf-2/HO-1 activation and NF-κB inhibition. Acta Pharmacologica Sinica. 2013;34(9):1140–1148. doi: 10.1038/aps.2013.59. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Qu L., Liang X., Gu B., Liu W. Quercetin alleviates high glucose-induced Schwann cell damage by autophagy. Neural Regeneration Research. 2014;9(12):1195–1203. doi: 10.4103/1673-5374.135328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Jeong S. M., Kang M. J., Choi H. N., Kim J. H., Kim J. I. Quercetin ameliorates hyperglycemia and dyslipidemia and improves antioxidant status in type 2 diabetic db/db mice. Nutrition Research and Practice. 2012;6(3):201–207. doi: 10.4162/nrp.2012.6.3.201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Li M., Li Q., Zhao Q., Zhang J., Lin J. Luteolin improves the impaired nerve functions in diabetic neuropathy: behavioral and biochemical evidences. International Journal of Clinical and Experimental Pathology. 2015;8(9):10112–10120. [PMC free article] [PubMed] [Google Scholar]
- 39.Zang Y., Igarashi K., Li Y. Anti-diabetic effects of luteolin and luteolin-7-O-glucoside on KK-Ay mice. Bioscience, Biotechnology, and Biochemistry. 2016;80(8):1580–1586. doi: 10.1080/09168451.2015.1116928. [DOI] [PubMed] [Google Scholar]
- 40.Kishore L., Kaur N., Singh R. Effect of kaempferol isolated from seeds of Eruca sativa on changes of pain sensitivity in Streptozotocin-induced diabetic neuropathy. Inflammopharmacology. 2018;26(4):993–1003. doi: 10.1007/s10787-017-0416-2. [DOI] [PubMed] [Google Scholar]
- 41.Kaeidi A., Taati M., Hajializadeh Z., Jahandari F., Rashidipour M. Aqueous extract of Zizyphus jujuba fruit attenuates glucose induced neurotoxicity in an in vitro model of diabetic neuropathy. Iranian Journal of Basic Medical Sciences. 2015;18(3):301–306. [PMC free article] [PubMed] [Google Scholar]
- 42.Al-Numair K. S., Chandramohan G., Veeramani C., Alsaif M. A. Ameliorative effect of kaempferol, a flavonoid, on oxidative stress in streptozotocin-induced diabetic rats. Redox Report. 2015;20(5):198–209. doi: 10.1179/1351000214Y.0000000117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Qiu G., Tian W., Huan M., Chen J., Fu H. Formononetin exhibits anti-hyperglycemic activity in alloxan-induced type 1 diabetic mice. Experimental Biology and Medicine (Maywood, N.J.) 2017;242(2):223–230. doi: 10.1177/1535370216657445. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Oza M. J., Kulkarni Y. A. Formononetin ameliorates diabetic neuropathy by increasing expression of SIRT1 and NGF. Chemistry & Biodiversity. 2020;17(6, article e2000162) doi: 10.1002/cbdv.202000162. [DOI] [PubMed] [Google Scholar]
- 45.Oza M. J., Kulkarni Y. A. Formononetin treatment in type 2 diabetic rats reduces insulin resistance and hyperglycemia. Frontiers in Pharmacology. 2018;9:p. 739. doi: 10.3389/fphar.2018.00739. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Selvarajah D., Kar D., Khunti K., et al. Diabetic peripheral neuropathy: advances in diagnosis and strategies for screening and early intervention. The Lancet Diabetes and Endocrinology. 2019;7(12):938–948. doi: 10.1016/S2213-8587(19)30081-6. [DOI] [PubMed] [Google Scholar]
- 47.Tang H. Y., Jiang A. J., Ma J. L., Wang F. J., Shen G. M. Understanding the signaling pathways related to the mechanism and treatment of diabetic peripheral neuropathy. Endocrinology. 2019;160(9):2119–2127. doi: 10.1210/en.2019-00311. [DOI] [PubMed] [Google Scholar]
- 48.Ristikj-Stomnaroska D., Risteska-Nejashmikj V., Papazova M. Role of inflammation in the pathogenesis of diabetic peripheral neuropathy. Open access Macedonian journal of medical sciences. 2019;7(14):2267–2270. doi: 10.3889/oamjms.2019.646. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Park S. Y., Kim Y. A., Hong Y. H., Moon M. K., Koo B. K., Kim T. W. Up-regulation of the receptor for advanced glycation end products in the skin biopsy specimens of patients with severe diabetic neuropathy. Journal of Clinical Neurology. 2014;10(4):334–341. doi: 10.3988/jcn.2014.10.4.334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Yan S. F., Ramasamy R., Schmidt A. M. Receptor for AGE (RAGE) and its ligands-cast into leading roles in diabetes and the inflammatory response. Journal of Molecular Medicine (Berlin, Germany) 2009;87(3):235–247. doi: 10.1007/s00109-009-0439-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Soman S., Raju R., Sandhya V. K., et al. A multicellular signal transduction network of AGE/RAGE signaling. Journal of cell communication and signaling. 2013;7(1):19–23. doi: 10.1007/s12079-012-0181-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Chowdhury S. K., Dobrowsky R. T., Fernyhough P. Nutrient excess and altered mitochondrial proteome and function contribute to neurodegeneration in diabetes. Mitochondrion. 2011;11(6):845–854. doi: 10.1016/j.mito.2011.06.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Yamamoto Y., Yamagishi S., Yonekura H., et al. Roles of the AGE-RAGE system in vascular injury in Diabetesa. Annals of the New York Academy of Sciences. 2000;902(1):163–172. doi: 10.1111/j.1749-6632.2000.tb06311.x. [DOI] [PubMed] [Google Scholar]
- 54.Wada R., Yagihashi S. Role of advanced glycation end products and their receptors in development of diabetic neuropathy. Annals of the New York Academy of Sciences. 2005;1043(1):598–604. doi: 10.1196/annals.1338.067. [DOI] [PubMed] [Google Scholar]
- 55.Brownlee M. Biochemistry and molecular cell biology of diabetic complications. Nature. 2001;414(6865):813–820. doi: 10.1038/414813a. [DOI] [PubMed] [Google Scholar]
- 56.Yu H., Pardoll D., Jove R. STATs in cancer inflammation and immunity: a leading role for STAT3. Nature Reviews. Cancer. 2009;9(11):798–809. doi: 10.1038/nrc2734. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Chen M., Obasanmi G., Armstrong D., et al. STAT3 activation in circulating myeloid-derived cells contributes to retinal microvascular dysfunction in diabetes. Journal of Neuroinflammation. 2019;16(1):p. 138. doi: 10.1186/s12974-019-1533-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Dewanjee S., Das S., Das A. K., et al. Molecular mechanism of diabetic neuropathy and its pharmacotherapeutic targets. European Journal of Pharmacology. 2018;833:472–523. doi: 10.1016/j.ejphar.2018.06.034. [DOI] [PubMed] [Google Scholar]
- 59.Wang K., Fu X. Y., Fu X. T., et al. DSePA antagonizes high glucose-induced neurotoxicity: evidences for DNA damage-mediated p53 phosphorylation and MAPKs and AKT pathways. Molecular Neurobiology. 2016;53(7):4363–4374. doi: 10.1007/s12035-015-9373-1. [DOI] [PubMed] [Google Scholar]
- 60.Fromont A., De Seze J., Fleury M. C., Maillefert J. F., Moreau T. Inflammatory demyelinating events following treatment with anti-tumor necrosis factor. Cytokine. 2009;45(2):55–57. doi: 10.1016/j.cyto.2008.11.002. [DOI] [PubMed] [Google Scholar]
- 61.Mu Z. P., Wang Y. G., Li C. Q., et al. Association between tumor necrosis factor-α and diabetic peripheral neuropathy in patients with type 2 diabetes: a meta-analysis. Molecular Neurobiology. 2017;54(2):983–996. doi: 10.1007/s12035-016-9702-z. [DOI] [PubMed] [Google Scholar]
- 62.Yamakawa I., Kojima H., Terashima T., et al. Inactivation of TNF-α ameliorates diabetic neuropathy in mice. American Journal of Physiology. Endocrinology and Metabolism. 2011;301(5):E844–E852. doi: 10.1152/ajpendo.00029.2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Fromentel C. C. D., Soussi T. TP53 tumor suppressor gene: a model for investigating human mutagenesis. Genes Chromosomes Cancer. 1992;4(1):1–15. doi: 10.1002/gcc.2870040102. [DOI] [PubMed] [Google Scholar]
- 64.Spitsina E. V., Iakunina NIu, Chudakova D. A., et al. Association of polymorphous markers Pro72Arg and C(-594)CC OF TP53 gene with diabetic polyneuropathy in patients with type 1 diabetes mellitus living in Moscow. Molekuliarnaia biologiia. 2007;41(6):989–993. [PubMed] [Google Scholar]
- 65.Sliwinska A., Kasznicki J., Kosmalski M., et al. Tumour protein 53 is linked with type 2 diabetes mellitus. The Indian Journal of Medical Research. 2017;146(2):237–243. doi: 10.4103/ijmr.IJMR_1401_15. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Ingredients: potential active ingredients and action targets of the herbal compound were obtained through screening.
Diseases: DPN-related targets were obtained.
Data Availability Statement
We have presented all our main data in the form of figures and additional files. The data used to support the conclusions of this study are available from the authors.


