Abstract
Endosomal escape remains a fundamental barrier hindering advancement of nucleic acid therapeutics. Taking inspiration from natural phospholipids that comprise biological membranes, we report the combinatorial synthesis of multi-tailed ionizable phospholipids (iPhos) capable of delivering mRNA or mRNA/sgRNA for gene editing in vivo. Optimized iPhos lipids are composed of one pH-switchable zwitterion and three hydrophobic tails, which adopt a cone shape in endosomal acidic environment to facilitate membrane hexagonal transformation and subsequent cargo release from endosomes. Structure-activity relationships reveal that iPhos chemical structure can control in vivo efficacy and organ selectivity. iPhos lipids synergistically function with various helper lipids to formulate multi-component lipid nanoparticles (iPLNPs) for Selective Organ Targeting (SORT). Zwitterionic, ionizable cationic, and permanently cationic helper lipids enable tissue-selective mRNA delivery and CRISPR/Cas9 gene editing in spleen, liver, and lungs (respectively) following intravenous administration. This rational design of functional phospholipids demonstrates significant value for gene editing research and therapeutic applications.
The CRISPR/Cas9 (clustered regularly interspaced short palindromic repeat/CRISPR-associated protein 9 (Cas9)) technology is a promising therapeutic modality for correction of DNA mutations that cause genetic diseases1–6. Systemic delivery of CRISPR/Cas cargoes using non-viral materials has attracted increasing attention because the transient nature confers increased safety over viral delivery and allows repeat dosing7–9. However, efficacious and safe in vivo gene editing using synthetic nanoparticles remains challenging, particularly due to low endosomal escape. To date, there has been a major focus on developing cationic lipids for RNA delivery10–13, with little attention to the critical roles played by zwitterionic phospholipids. Although phospholipids mimic biological membranes, aid RNA encapsulation, and are powerful at the membrane fusion step critical for endosomal escape14, 15, their chemical architectures are limited by lack of flexibility, such as pH response and tail number alteration. Inspired by biological membranes, we developed a class of pH-switchable, multi-tailed ionizable phospholipids (iPhos) with endosomal membrane destabilization and determined the underlying mechanisms of iPhos-mediated mRNA delivery and CRISPR/Cas9 gene editing.
Phospholipids are distributed throughout nature, serve as the key components of biological membranes and organelles, and are involved in cellular transport pathways16, 17. Because even the most effective carriers, including FDA-approved DLin-MC3-DMA lipid nanoparticles (LNPs), can only mediate 1–4% of RNA release into the cytoplasm, endosomal escape remains as the most daunting step for delivery18–20. Phospholipids represent an exciting opportunity to overcome a critical limitation of current carriers by inserting into naturally occurring membranes to enhance endosomal escape. Nonetheless, chemical pursuit of lipid structures to access the key physiochemical requirements to facilitate endosomal escape has been severely hampered by limitations in both reactions to access phospholipids and their inherently limited structural flexibility. For instance, the most commonly employed phospholipids, 1,2-distearoyl-sn-glycero-3-phosphocholine (DSPC)21 and 1,2-dioleoyl-sn-glycero-3-phosphoethanolamine (DOPE)22–24 possess constant one irreversible zwitterion (at least in organisms) and two hydrophobic tails, lacking chemical handles to manipulate. To enrich the diversity of phospholipids, well-tailored structures are demanded from rational chemical design.
Inspired by cationic lipid designs, we hypothesized that integration of their advantages (e.g. ionizable amines and multiple alkyl chains) into phospholipid designs with membrane integration potential would dramatically improve the resulting functionality. First, an ionizable reversible zwitterion between neutral physiological environment and acidic endosomal compartment might enable pH-triggered membrane rupture. Second, one small zwitterion head combined with over two hydrophobic tail body would adopt a cone shape more easily, inducing strong trend of membrane phase transformation. To realize these assumptions and design multifunctional phospholipids, ring-opening of dioxaphospholane oxide molecules provides a way forward. These molecules are capable of conjugating with primary, secondary, and tertiary amines, such that controllable hydrophobic tail numbers and pH switchable zwitterions can be realized through this modular synthetic strategy. Until now, most vehicles only demonstrate efficacy in liver hepatocytes15, 25, 26, and delivery outside of the liver is a major need for future correction of genetic diseases. Following optimization of material architecture, we discovered that the resulting iPhos lipids enabled establishment of structure-selectivity relationships to further define mechanisms for extrahepatic delivery. Development of tissue-selective carriers is of great significance to minimize side effects and enable targeting of cell types required for treatment of genetic diseases.
Taking inspiration from natural phospholipids that comprise biological membranes and mediate transport in cells, we report the combinatorial synthesis of 572 iPhos lipids capable of delivering mRNA or mRNA/sgRNA for gene editing in vivo via bio-inspired mechanisms. Following initial evaluation, iPhos lipids (7A1P4–13A1P16) containing one tertiary amine, one phosphate group, and three alkyl tails enabled the highest protein expression. The pH-switchable small zwitterion head and multiple tails aided endosomal membrane fusion and hexagonal phase transformation. Encouragingly, Structure-activity relationships (SAR) revealed that tail length of iPhos lipids affected both in vivo efficacy and organ selectivity. In terms of LNP designs, iPhos lipids are functional and represent the important core component in the reported delivery systems. First, top-performing iPhos 9A1P9 exhibited 40- to 965-fold higher in vivo efficacy compared to previous benchmarks DOPE and DSPC. Second, 9A1P9 demonstrated universal applicability to synergistically function with zwitterionic, ionizable cationic, and permanently cationic helper lipids to formulate multi-component lipid nanoparticles for Selective Organ Targeting (SORT)27 to mediate exclusive transfection in spleen, liver, or lungs. This phenomenon is of vital significance for the treatment of various tissue-specific diseases. The best 9A1P9–5A2-SC8 and 9A1P9-DDAB formulations mediated high mRNA expression and CRISPR/Cas9 gene editing specifically in liver and lung, respectively. We believe the rationally designed iPhos and in vivo SAR expand nanomaterial development to further the fields of gene delivery and editing and present translational potential to realize clinical applications.
Results
Rational design of iPhos for superior endosomal escape
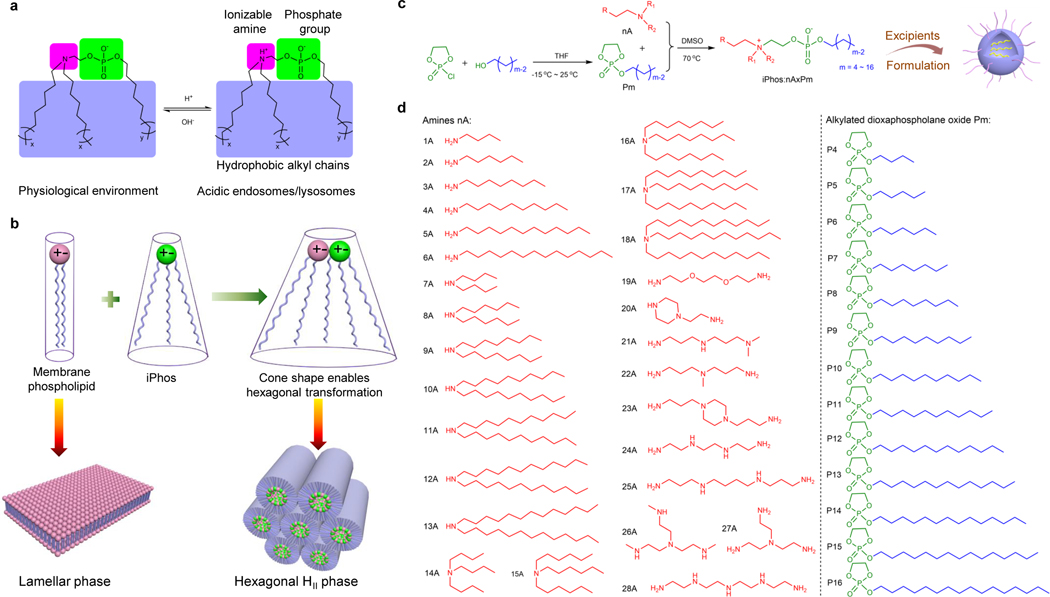
In the past decade, significant efforts have established 4-component LNP systems for RNA delivery, which are composed of cationic ionizable lipids, phospholipids, cholesterol (chol), and poly(ethylene glycol) (PEG)-lipids28, 29. Our prior work demonstrated that phospholipids assist mRNA loading into LNPs8, and increasing phospholipid content can improve delivery efficacy14. Furthermore, phospholipids show homology to biological membranes, and might enable membrane fusion readily. However, phospholipids do not natively possess a pH switch that has been shown to be essential to mediate LNP disruption and endosomal membrane fusion following cellular uptake and pH acidification in endosomes11, 30. Moreover, phospholipid species and architectures are limited to date, where common phospholipids (e.g. DSPC and DOPE) typically possess one irreversible zwitterion and two hydrophobic tails that lack the chemical features to alter shape within endosomes to address the daunting escape requirements. Previously, cationic lipids have been extensively explored28–31 and integrating their advantages (e.g. ionizable amines and multiple tails) to phospholipids represents an opportunity to tailor the structures of phospholipids. For this purpose, we rationally designed iPhos lipids, which contained an ionizable amine, a phosphate group, and three hydrophobic tails. The small zwitterion constituted by amine and phosphate group is predicted to be reversible at different pHs. In physiological pH (~7.4), the tertiary amine group will not be protonated, and the negatively charged iPhos will have difficulty fusing into the membranes. In contrast, upon entering the acidic endosomes, the tertiary amine will be protonated to form a zwitterionic head (Fig. 1a). Upon studying the substrate scope, we found that the three hydrophobic tail body was easier to mediate membrane phase transformation than that of two chains. The mechanism of action, therefore, is different from that of classic gene carriers, because synthetic iPhos lipids can overall insert into natural phospholipid membranes, with preferable small ion pairs coupled with large tail body adopting a cone shape, to facilitate hexagonal HII phase formation (Fig. 1b).
Fig. 1 ǀ. A combinatorial library of iPhos lipids was chemically synthesized and studied, which led to the elucidation of a physical mechanism of action for enhanced endosomal escape.
a, Efficacious iPhos lipids were composed of one ionizable amine, one phosphate group, and three hydrophobic alkyl tails. Upon entering acidic endosomes/lysosomes, protonation of the tertiary amine induced a zwitterionic head group, which could readily insert into membranes. b, Most biological membrane phospholipids possess a zwitterion and adopt a lamellar phase. When iPhos lipids were mixed and inserted into the endosomal membranes, the formed cone shape by small ion pair head and multiple hydrophobic tails enabled hexagonal transformation. c, Synthetic routes of iPhos: alkylated dioxaphospholane oxide molecules (Pm) were conjugated to amines (nA) to obtain iPhos (nAxPm). “x” in “nAxPm” indicated the number of Pm molecules modified on one amine molecule. d, A list of 28 amines nA and 13 alkylated dioxaphospholane oxide Pm molecules used for iPhos synthesis.
To overcome prior limitations in synthetic routes, we focused on ring-opening reactions32, 33 that could yield diverse products with chemical complexity to meet the aforementioned design guidelines. The combinatorial reaction of amines (nA) with alkylated dioxaphospholane oxides (Pm) yielded 572 iPhos lipids (termed nAxPm) (Fig. 1c) where “x” indicated the number of Pm molecules modified on one amine molecule. Pm molecules were synthesized via esterification of 2-chloro-2-oxo-1,3,2-dioxaphospholane (COP) from corresponding alcohols with different alkyl chain lengths (Supplementary Fig. 1). Primary, secondary, and tertiary amine groups could all trigger Pm ring-opening to introduce different zwitterions (Supplementary Fig. 2). To control the hydrophobic tail and zwitterion numbers, amines with different alkyl chain and amine group numbers were used (Fig. 1d and Supplementary Fig. 3). The chemical design of iPhos is unique, because, zwitterion species (pH switchable and irreversible) become available in addition to group numbers through this strategy, greatly broadening the architectures and varieties of phospholipids.
Top iPhos possessed a pH-switchable head and three tails
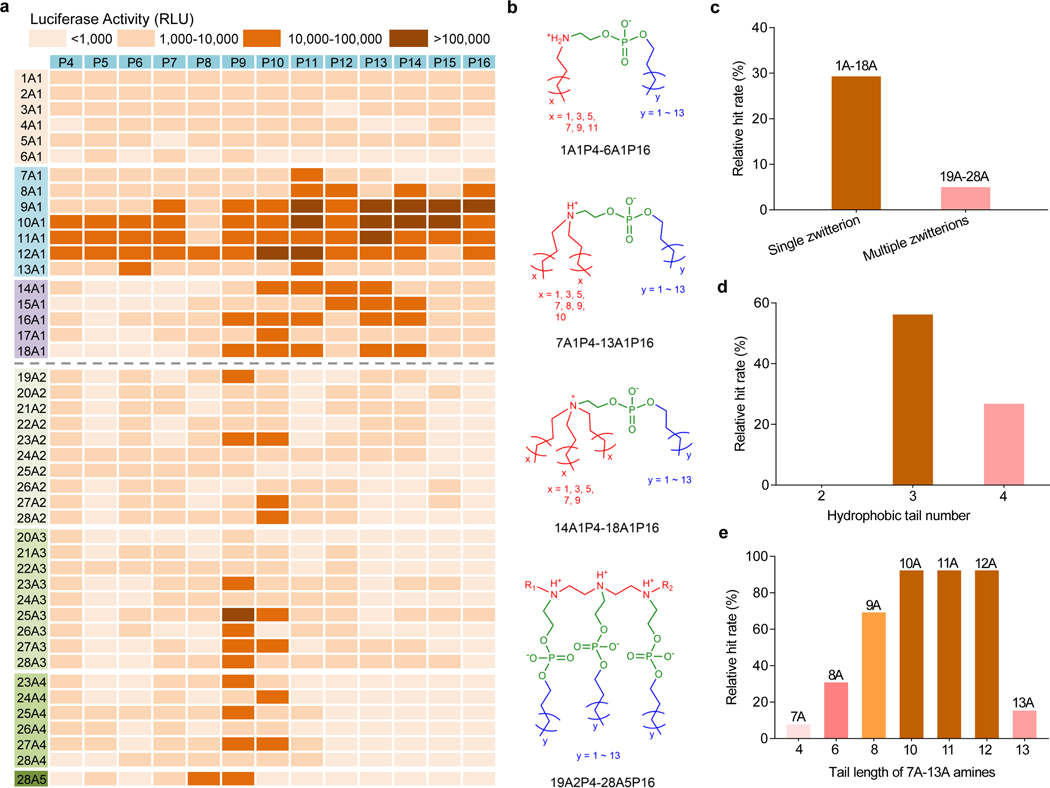
To evaluate the potential for mRNA delivery, iPhos lipid nanoparticles (iPLNPs) were used to transfect ovarian cancer cells IGROV-1. iPhos, helper lipid, chol, and 1,2-dimyristoyl-rac-glycero-3-methoxy(poly(ethylene glycol-2000)) (DMG-PEG2000) (25:30:30:1 mol/mol) were mixed with mRNA to formulate iPLNPs using the ethanol dilution method. A structurally simple lipid, N-methyldioctadecylamine (MDOA), was first employed as the helper lipid to demonstrate the iPhos function in the initial screen. iPLNPs can thus be considered a remix of traditional LNPs, where the modular emphasis is placed instead on the zwitterionic (functionally active iPhos) lipid and all other lipids become helper lipids. All initial iPhos lipids exhibited low toxicity (Supplementary Fig. 4). From the in vitro screening heat map, iPhos with a single zwitterion (1A1P4–18A1P16) showed higher mRNA efficacy than that of multiple zwitterions (19A2P4–28A5P16) (Fig. 2a-2c). The reason lied behind that multiple zwitterions constructed a larger head, making it difficult for membrane phase transformation. To further explore the SAR of iPhos with a single zwitterion, two tailed materials (1A1P4–6A1P16) exhibited far less efficacy, because the small tail body failed to formulate the cone shape with natural membrane phospholipids. iPhos 14A1P4–18A1P16 possessed a permanent zwitterion and lacked structural flexibility upon endosomal internalization. Encouragingly, iPhos (7A1P4–13A1P16) composed of one tertiary amine, one phosphate group, and three hydrophobic tails showed the highest mRNA delivery efficacy as expected, with a hit rate around 60% (Fig. 2d). The small zwitterion head and large tail body promoted membrane fusion and phase transformation from lamellar to hexagonal HII. Among these iPhos lipids, amine tail length was very important, and the hit rates of 10–12 chain lengths reached up to 92% (Fig. 2e). These observations are in contrast to previously reported ionizable amino lipid and lipidoid libraries, where efficacy generally correlated with polyamine cores and higher numbers of alkyl tails13, 34, 35. These results indicate that iPhos lipids may operate by a different mechanism that ionizable amino lipids do. Next, selected top iPhos lipids (9A1P9, 9A1P15, 10A1P10 and 10A1P16) were purified (Supplementary Fig. 5-8), and the resulting iPLNPs showed appropriate particle sizes (~ 150 nm) for endocytosis, slightly negative surface zeta potentials (~ −5 mV) for serum protein resistance, as well as suitable pKa (6.0–6.5) for in vivo assays (Supplementary Figs. 9 and 10). These capabilities impart synthetic iPhos lipids great potential for in vivo applications.
Fig. 2 ǀ. Structure-activity relationships of iPhos lipids for luciferase mRNA delivery in vitro.
a, A heat map of luciferase expression following treatment of IGROV1 cells with iPLNPs (50 ng firefly luciferase (Fluc) mRNA, n = 3 biologically independent samples). RLU > 10,000 was counted for the hit rate calculation. b, Representative chemical structures of iPhos with different numbers of zwitterions and tails in an acidic endosomal environment. c, The relative hit rate of iPhos with a single zwitterion and multiple zwitterions. d, The relative hit rate of iPhos (1A1P4–18A1P16) with a single zwitterion and different numbers of tails. e, Among the efficient iPhos (7A1P4–13A1P16), tail length of starting amines influenced the ultimate in vitro efficacy.
Model membrane studies of iPhos-mediated endosomal rupture
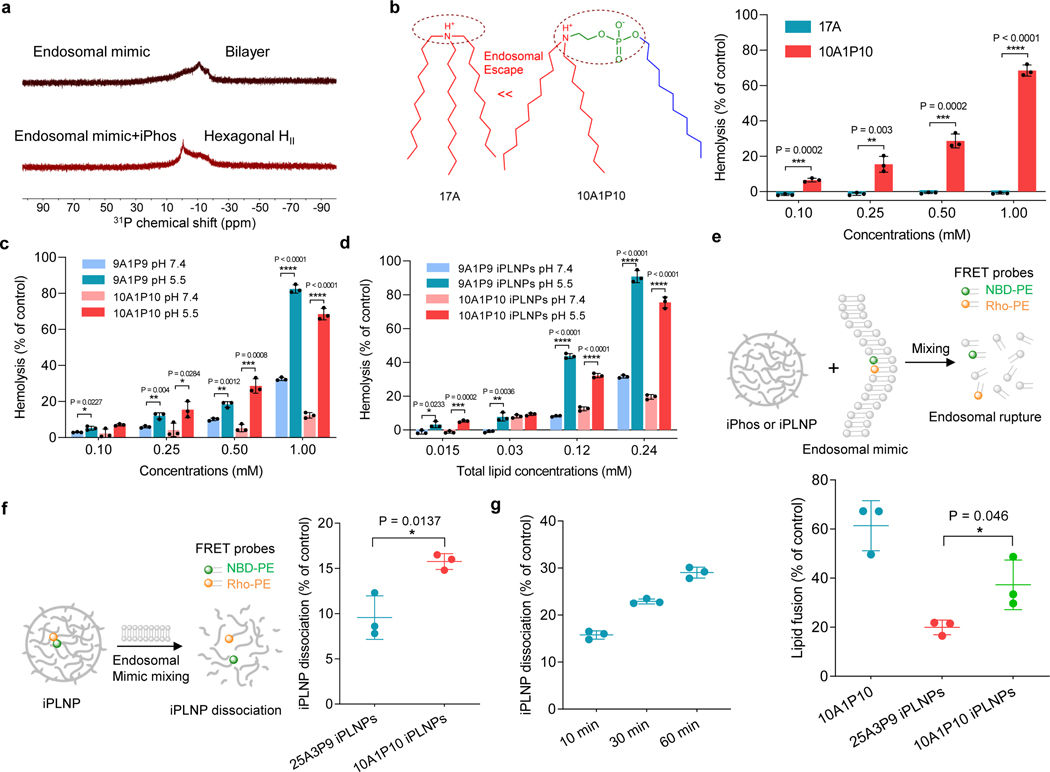
Although numerous gene delivery systems have been developed36–39, chemical designs have largely rested on cationic amines with limited pursuit of physical membrane disruption. To verify our hypothesis behind efficacious mRNA delivery, we investigated phase transformation using established 31P NMR spectroscopy techniques11, 40, 41. Endosomal mimicking liposomes adopted a bilayer structure, as evidenced by the low field shoulder. Upon mixing with iPhos lipids, the peak asymmetry was reversed and the liposomes transformed to the hexagonal phase (Fig. 3a). Next, the membrane-disruptive activity of iPhos lipids and iPLNPs was evaluated by a hemolysis model30, 42. As expected, 10A1P10 with a pH-switchable zwitterion exhibited dramatically higher hemolysis than 17A with a simple tertiary amine, validating the superiority of zwitterion in membrane fusion and rupture (Fig. 3b). Also, 9A1P9, 10A1P10, and related iPLNPs showed higher membrane-disruptive activity at acidic endosomal compartment compared to that of neutral pH (Fig. 3c,d). The high hemolysis was not due to cytotoxicity (Supplementary Fig. 11).
Fig. 3 ǀ. Model membrane studies of endosomal escape demonstrated the mechanism of iPhos lipid-mediated RNA delivery with correlation to chemical structure.
a, 31P NMR spectra of endosomal mimic and a mixture of endosomal mimic with iPhos 9A1P9. iPhos lipid mixing induced membrane hexagonal HII transformation. b, Hemolysis of 17A and 10A1P10 at pH 5.5. The zwitterion could significantly benefit the endosomal escape. c, Hemolysis of 9A1P9 and 10A1P10 at different pHs. d, Hemolysis of iPLNPs at different pHs. e, Lipid fusion and membrane rupture of 10A1P10 and iPLNPs were determined by a FRET assay at pH 5.5. f, iPLNP dissociation by FRET characterization after mixing with anionic endosomal mimics for 10 min at pH 5.5. A single zwitterion showed higher lipid fusion and iPLNP dissociation efficacy than multiple zwitterions. g, 10A1P10 iPLNP dissociation of different time intervals at pH 5.5. Data in b-g are presented as mean ± s.d. (n = 3 biologically independent samples). Statistical significance was analyzed by the two-tailed unpaired t-test: ****, P < 0.0001; ***, P < 0.001; **, P < 0.01; *, P < 0.05.
Following this, a fluorescence resonance energy transfer (FRET) assay was utilized to evaluate iPhos lipid membrane fusion and iPLNP dissociation. Two DOPE-conjugated FRET probes, 7-nitrobenzo-2-oxa-1,3-diazole (NBD-PE) and lissamine rhodamine B (Rho-PE), were formulated into a single endosomal mimicking liposome, leading to attenuated NBD fluorescence because of FRET to rhodamine. Once lipid fusion occurred, the resulting larger distance between the two probes gave rise to NBD signal increase43, 44. 10A1P10 iPLNPs exhibited higher lipid fusion than 25A3P9 iPLNPs, demonstrating that a small single zwitterion head showed stronger trend to insert and destroy endosomal membranes compared to that of multiple zwitterions (Fig. 3e). Additionally, 10A1P10 iPLNPs were easier to disassemble to release mRNA than 25A3P9 iPLNPs once mixing with endosomal mimicking liposomes (Fig. 3f,g). These results demonstrate that apart from a large tail body, a pH-switchable small zwitterion head in iPhos lipids is essential for endosomal escape.
iPhos structure controlled efficacy and organ selectivity
Due to the additional barriers for in vivo delivery, not all carriers with in vitro activity translate to animal models45, 46. Moreover, comparing siRNAs/miRNAs (18–22 bp) to long mRNAs (1,000–6,000 nt), weaker electrostatic association is required, allowing for mRNA release post cellular internalization14. Therefore, the chemistry of iPhos lipids may have an inherent advantage over cationic lipids and play a pivotal role in mRNA delivery systems.
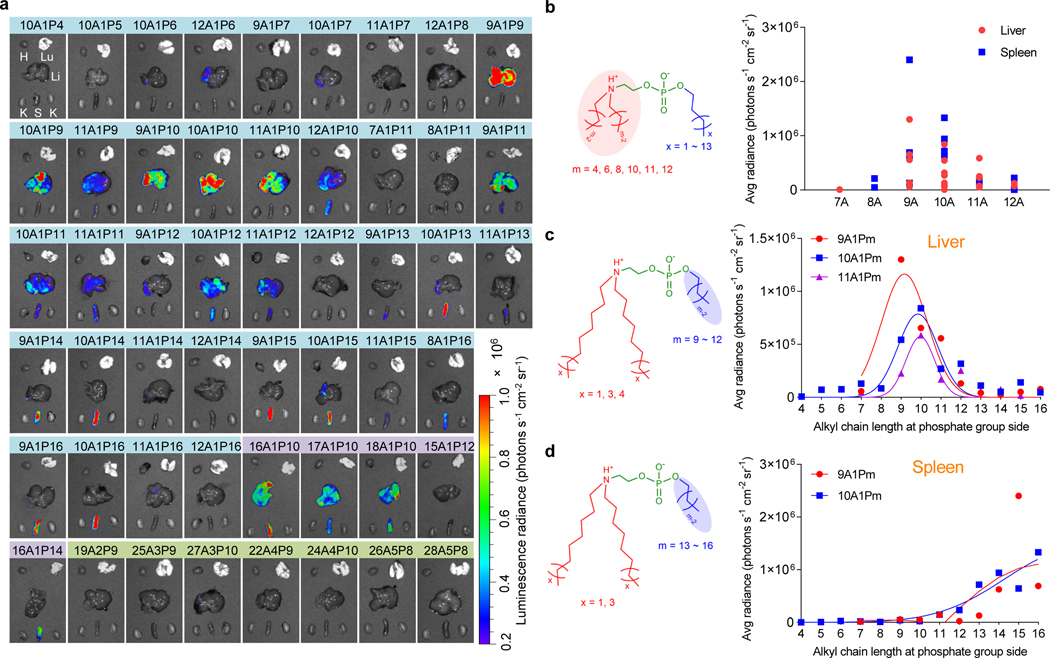
We selected 51 efficient iPhos lipids from the in vitro screen and evaluated in vivo delivery at a low mRNA dose (0.1 mg kg−1) (Fig. 4a). As expected, iPhos lipids containing multiple zwitterions failed to delivery mRNA in vivo. Further establishing SAR, iPhos lipids with one tertiary amine, one phosphate group, and three alkyl tails were the most efficacious. Interestingly, alkyl chain length tremendously affected efficacy and organ selectivity. Chain length at amine side determined efficacy, and eight to ten carbon lengths mediated high in vivo mRNA expression (Fig. 4b and Supplementary Fig. 12). Surprisingly, alkyl length beside phosphate group influenced organ selectivity (Fig. 4c,d). Shorter chains (9–12 carbons) showed mRNA translation in liver, while longer chains (13–16 carbons) would transfer protein expression to spleen. In vivo assessment of 10A1P4–10A1P16 also clearly supported this inference (Supplementary Fig. 13). Next, nanoparticle size, zeta potential, and pKa of these iPLNPs were evaluated, where no obvious differences were observed (Supplementary Fig. 14). At a higher mRNA dose (0.25 mg kg−1), organ selectivity was still achieved (Supplementary Figs. 15 and 16). While we speculate the organ selectivity relates to the chemical structure of mRNA carrier materials47, detailed mechanistic studies are still on-going. The SAR provides a guideline to develop other efficacious vector materials with organ selectivity and specificity.
Fig. 4 ǀ. Structure-activity studies revealed that iPhos lipid structure controlled in vivo efficacy and organ selectivity.
a, In vivo evaluation of 51 iPhos at a low Fluc mRNA dose (0.1 mg kg−1). Bioluminescence images of various organs were recorded 6 h after IV injection of iPLNPs into C57BL/6 mice. H: Heart, Lu: lung, Li: Liver, K: Kidney, S: Spleen. b, Among the efficacious 10A1P4–12A1P16 iPhos, hydrophobic chain length at the amine side significantly influenced the in vivo mRNA delivery efficacy. c, mRNA expression in liver by iPhos with different alkyl chain length at phosphate group side. 9 to 12 carbon lengths were the most efficient. d, mRNA expression in spleen by iPhos with different alkyl chain length at phosphate group side. 13 to 16 alkyl chain lengths were the most efficient.
iPLNPs mediated tissue-selective gene delivery and editing
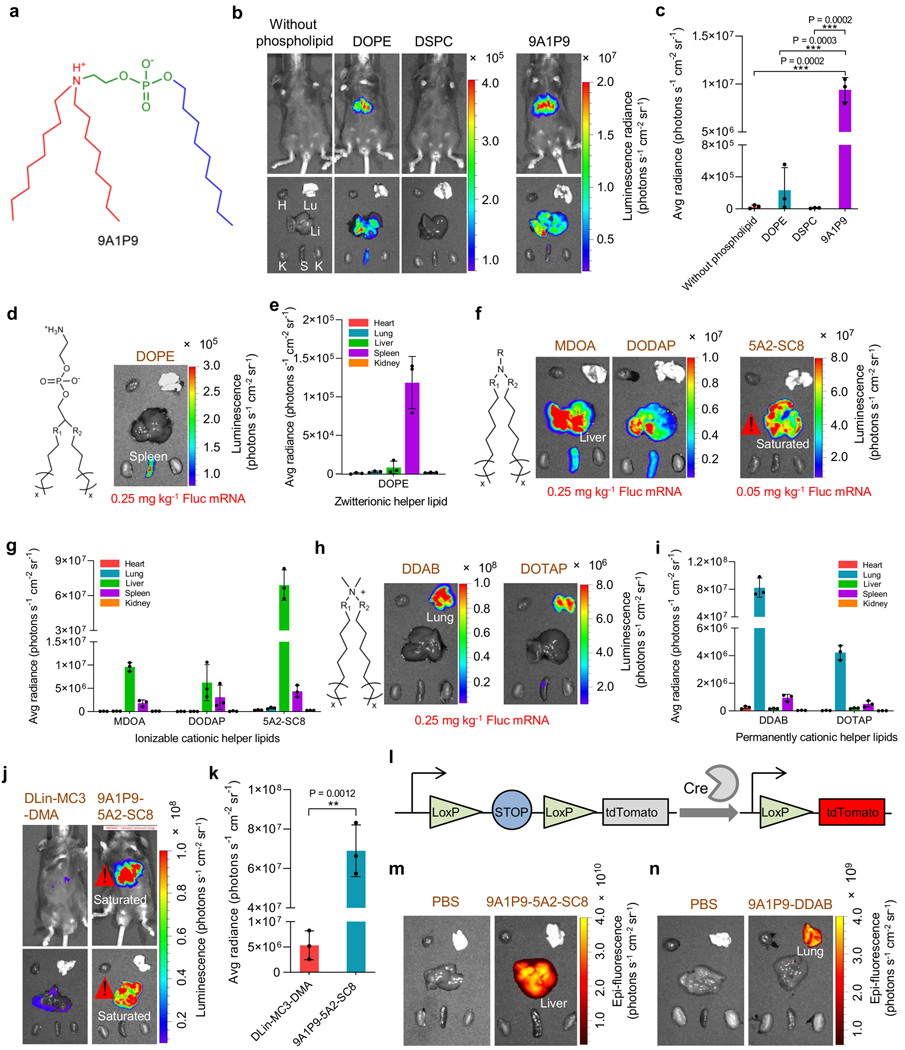
To confirm that iPhos 9A1P9 was the most important and active component of iPLNPs, we conducted a series of experiments. First, 9A1P9 exhibited 40- to 965-fold higher in vivo efficacy compared to the best currently used phospholipids DOPE and DSPC (Fig. 5a-5c). Second, a variety of other established lipids were assessed in our 9A1P9 iPLNP mRNA delivery system as helper lipids to show the broad applicability. We investigated zwitterionic lipids (DOPE), ionizable cationic lipids (MDOA, 1,2-dioleoyl-3-dimethylammonium-propane (DODAP), and 5A2-SC835), and permanently cationic lipids (dimethyldioctadecylammonium bromide salt (DDAB) and 1,2-dioleoyl-3-trimethylammonium-propane (DOTAP)) as helper lipids (Supplementary Fig. 17). The molar ratios of compositions were determined by orthogonal design methodology14, 48 and shown in Supplementary Table 1. All formulated iPLNPs exhibited appropriate diameters, zeta potentials, mRNA binding, pKa, and high in vitro mRNA delivery efficacy (Supplementary Fig. 18).
Fig. 5 ǀ. iPhos outperformed traditional phospholipids, and functioned with different helper lipids for organ selective RNA delivery.
a, Structure of iPhos 9A1P9 in the acidic endosomal environment. b,c, iPhos 9A1P9 outperformed benchmark DOPE and DSPC for mRNA delivery. Images (b) and quantification (c) of luciferase expression in liver were recorded (Fluc mRNA, 0.25 mg kg−1). H: Heart, Lu: lung, Li: Liver, K: Kidney, S: Spleen. d,e, iPLNPs containing zwitterionic helper lipid DOPE mediated mRNA expression in spleen. In vivo evaluation (d) and quantification (e) were evaluated (Fluc mRNA, 0.25 mg kg−1). f,g, iPLNPs containing ionizable cationic helper lipids mediated mRNA translation in liver. Organ selectivity (f) and quantification (g) of Fluc mRNA expression by 9A1P9 iPLNP with different ionizable cationic helper lipids were assayed (Fluc mRNA, 0.25 mg kg−1 for MDOA and DODAP; 0.05 mg kg−1 for 5A2-SC8). h,i, iPLNPs containing permanently cationic helper lipids induced mRNA transfection in lung. Organ images (h) and quantification (i) of Fluc mRNA expression by 9A1P9 iPLNP using DDAB and DOTAP were evaluated (Fluc mRNA, 0.25 mg kg−1). j,k, 9A1P9–5A2-SC8 iPLNP showed much higher mRNA delivery efficacy than positive control DLin-MC3-DMA LNPs. Images (j) and quantification (k) of luciferase expression in liver were recorded (Fluc mRNA, 0.05 mg kg−1). l-n, 9A1P9 iPLNPs enabled Cre mRNA delivery selectively in liver or lung. Schematic (l) represented a Cre-LoxP mouse model that could express tdTomato by translating Cre-recombinase mRNA to Cre protein to delete the stop. 9A1P9–5A2-SC8 iPLNP (m) and 9A1P9-DDAB iPLNP (n) mediated tdTomato expression in liver and lung, respectively (Cre mRNA, 0.25 mg kg−1). All data are presented as mean ± s.d. (n = 3 biologically independent mice). Statistical significance was analyzed by the two-tailed unpaired t-test: ****, P < 0.0001; ***, P < 0.001; **, P < 0.01; *, P < 0.05.
We next investigated whether iPhos could be used for SORT27, 49. Encouragingly, 9A1P9 iPLNPs with zwitterionic, ionizable cationic, and permanently cationic helper lipids enabled selective mRNA expression in spleen, liver, and lungs, respectively (Fig. 5d-5i). Two highly efficacious formulations were further studied and in vivo biodistribution results revealed that spherical 9A1P9–5A2-SC8 and 9A1P9-DDAB iPLNPs mediated high accumulation in liver and lung, respectively (Supplementary Figs. 19 and 20). Since iPhos lipids can enhance already efficacious formulations in a modular fashion, we determined that 9A1P9–5A2-SC8 and 9A1P9-DDAB combinations exhibited high mRNA expression in liver (~108 photons s−1 cm−2 sr−1, 0.05 mg kg−1) and lung (~108 photons s−1 cm−2 sr−1, 0.25 mg kg−1), respectively (Fig. 5f-5i and Supplementary Fig. 21). Acknowledging that DLin-MC3-DMA (used in FDA-approved Onpattro) LNPs were optimized for siRNA50 and not mRNA delivery, DLin-MC3-DMA LNPs provide value as a reference15, 51. Comparing to this established formulation, 9A1P9–5A2-SC8 iPLNPs showed 13-fold higher mRNA delivery efficacy than DLin-MC3-DMA LNPs in vivo (Fig. 5j,k and Supplementary Fig. 22). Therefore, iPLNPs are different from traditional cationic lipid LNPs, and high efficacy and controllable organ selectivity are both accomplished. Kinetic analysis revealed that protein expression occurred rapidly and peaked at around 6 h post injection (Supplementary Fig. 23).
To verify the ability of iPLNPs to induce organ-selective gene editing, we utilized an activatable Cre-LoxP mouse model that expresses Lox-stop-Lox tdTomato in all tissues. Through iPLNP delivery of Cre-recombinase mRNA (Cre mRNA), the translated Cre protein can delete the stop cassette and turn on red tdTom florescence only in successfully transfected cells (Fig. 5l). Highly efficient and organ selective gene editing was observed 2 days following Cre mRNA delivery (Fig. 5m-5n). We next used this model to quantify transfection of specific cell types in liver, lung, and spleen organs. Following delivery of Cre mRNA, liver-selective 9A1P9–5A2-SC8 iPLNPs mediated mRNA delivery to ~91% of all hepatocytes (Supplementary Fig. 24). Lung-selective 9A1P9-DDAB iPLNPs transfected ~34% of all endothelial cells, ~20% of all epithelial cells and ~13% of immune cells (Supplementary Fig. 25). Spleen-selective 10A1P16-MDOA iPLNPs transfected ~30% of all macrophages and 6% of all B cells (Supplementary Fig. 26). These results demonstrate the potential of organ selective iPLNPs for diverse therapeutic applications. iPLNP proposed here represents one of the most efficacious mRNA delivery systems and holds great potential for organ selective CRISPR/Cas9 gene editing.
Organ-specific CRISPR/Cas editing with translation potential
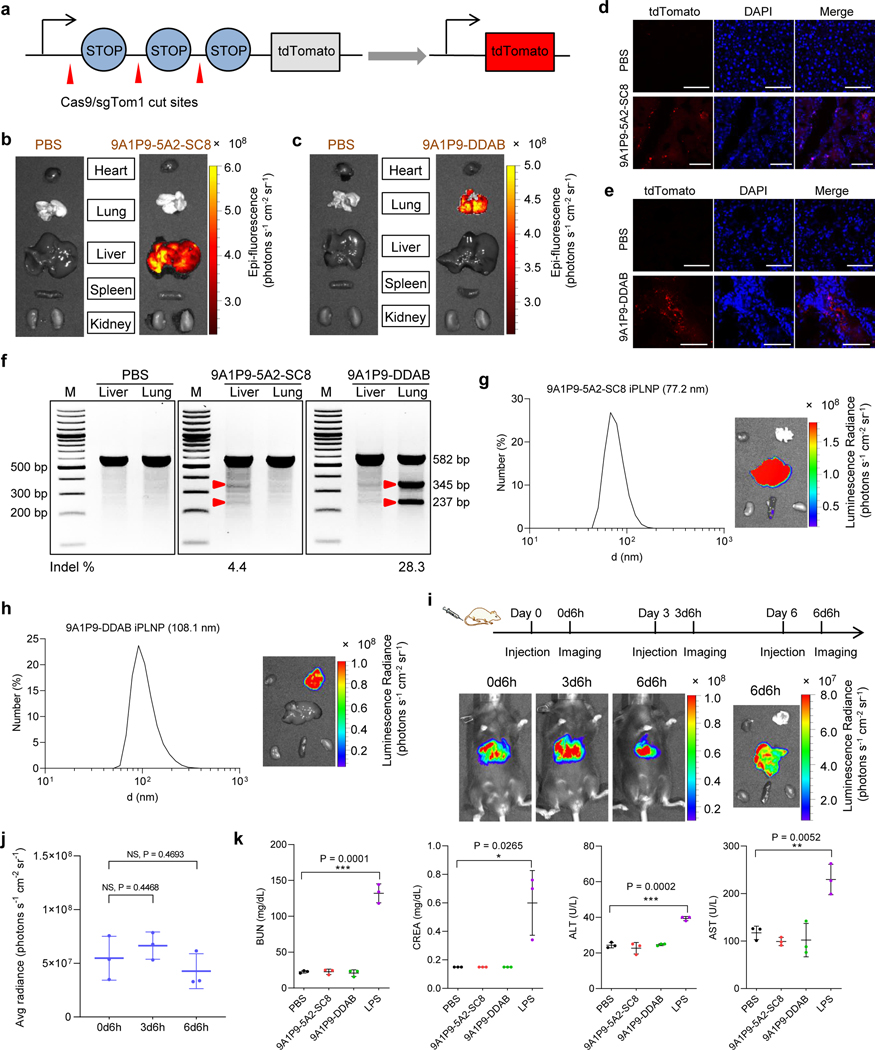
Although LNPs have been utilized to deliver mRNA, there are still few reports for successful in vivo Cas9 mRNA/sgRNA delivery for CRISPR/Cas gene editing8, 9, and even less so with precision to specific organs27. We next utilized iPLNPs to co-deliver Cas9 mRNA and sgRNA for gene editing. 9A1P9–5A2-SC8 and 9A1P9-DDAB iPLNPs containing Cas9 mRNA and Tom1 sgRNA (sgTom1) with a 4:1 weight ratio were intravenously (IV) administered into Ai9 mice at a total RNA dose of 0.75 mg kg−1, which would delete the stop cassettes and activate tdTomato protein (Fig. 6a). Fluorescent tdTomato protein was observed specifically in the liver after 9A1P9–5A2-SC8 iPLNP administration by ex vivo organ imaging (Fig. 6b). Sectioned organ analysis by confocal fluorescence microscopy showed tdTomato-positive cells in liver tissues (Fig. 6d). Similarly, 9A1P9-DDAB iPLNP induced specific gene editing in the lungs (Fig. 6c,e). Following this, PTEN sgRNA (sgPTEN) was co-delivered with Cas9 mRNA for gene editing in C57BL/6 mice (Cas9 mRNA/sgPTEN weight ratio, 4:1; total RNA dose, 0.75 mg kg−1) targeting an endogenous gene. T7E1 assay showed efficient target gene editing in liver and lung by 9A1P9–5A2-SC8 and 9A1P9-DDAB iPLNPs, respectively (Fig. 6f and Supplementary Fig. 27). CRISPR/Cas9 gene editing in specific organs has remained a long-standing challenge and the highly efficient and organ selective gene editing might broaden the iPLNP application to diverse genetic diseases.
Fig. 6 ǀ. iPLNPs enabled CRISPR/Cas9 gene editing selectively in liver and lungs and possessed potential for clinical translation.
a, Schematic of co-delivery of Cas9 mRNA and sgTom1 deletes the stop cassettes and activates tdTomato protein. b, 9A1P9–5A2-SC8 iPLNPs enabled gene editing specifically in liver. c, 9A1P9-DDAB iPLNPs enabled gene editing specifically in lung. d, Following IV administration of 9A1P9–5A2-SC8 iPLNPs containing Cas9 mRNA and sgTom1 to Ai9 mice, tdTomato-positive cells were observed in liver. Scale bars, 50 μm. e, Confocal fluorescence images showed tdTomato-positive cells in lung after administration of 9A1P9-DDAB iPLNPs. Scale bars, 50 μm. f, T7E1 assay of organ selective gene editing. 9A1P9–5A2-SC8 and 9A1P9-DDAB iPLNPs containing Cas9 mRNA and sgPTEN were IV administered into C57BL/6 mice, enabling CRISPR/Cas9 gene editing in liver and lung, respectively. For all the CRISPR/Cas9 gene editing assays, Cas9 mRNA/sgRNA weight ratio of 4:1 and total RNA dose of 0.75 mg kg−1 were used. g,h, iPLNPs were prepared by controlled microfluidic mixing, which resulted in decreased iPLNP sizes and preserved efficacy and organ selectivity. 9A1P9–5A2-SC8 iPLNPs (liver specific, Fluc mRNA, 0.05 mg kg−1) (g) and 9A1P9-DDAB iPLNPs (lung specific, Fluc mRNA, 0.25 mg kg−1) (h) demonstrated small sizes and fully retained precise organ selectivity. i,j, 9A1P9–5A2-SC8 iPLNPs (Fluc mRNA, 0.05 mg kg−1) allowed repeat dosing without loss of efficacy. Whole body imaging (i) and quantification of luciferase expression (j) was performed 6 h after each injection. k, 9A1P9–5A2-SC8 and 9A1P9-DDAB iPLNPs were well tolerated in vivo. Data are presented as mean ± s.d. and statistical significance was analyzed by the two-tailed unpaired t-test: ****, P < 0.0001; ***, P < 0.001; **, P < 0.01; *, P < 0.05; NS, P > 0.05. All data are from n = 3 biologically independent mice.
Considering potential pre-clinical activities, lead iPLNPs were manufactured at higher scale using controlled microfluidic mixing. Precise control over the mixing speed and volume ratios enabled the preparation of smaller 9A1P9–5A2-SC8 iPLNPs (77.2 nm, liver specific), 9A1P9-DDAB iPLNPs (108.1 nm, lung specific), and 10A1P16-MDOA iPLNPs (96.1 nm, spleen specific). Importantly, high in vivo mRNA delivery efficacy and precise organ selectivity were fully retained after decreasing iPLNP diameters (Fig. 6g,h and Supplementary Fig. 28). Furthermore, iPLNPs allowed repeat dosing, where high efficacy was retained after each repeat injection (Fig. 6i,j and Supplementary Fig. 29). Analysis of liver function enzymes and tissue section histology indicated that these iPLNPs showed negligible in vivo toxicity at the tested doses (Fig. 6k and Supplementary Figs. 30-31). These results highlight the potential of iPLNP system for future applications.
Discussion
The CRISPR/Cas9 gene editing system is gaining increasing interest due to its tremendous potential for genetic disease treatment4, 52, 53. Although cells build membranes and mediate transport using phospholipids, most all efficacious lipid nanoparticles for gene delivery rely on ionizable amines as the key physiochemical parameter to mediate endosomal escape via charge acquisition11–13. In carrier development, synthetic zwitterionic lipids are largely unexplored, even though they may readily enable endosomal membrane fusion and leakiness due to their homology with biological membranes.
We were therefore inspired to design and synthesize iPhos lipids to overcome these challenges. iPhos lipids include a pH-switchable small zwitterion head and a three-tail body. This unique architecture makes it easy to insert into naturally occurring membrane phospholipids and induce phase transformation for increased RNA release from endosomes. SAR revealed that iPhos chain lengths can control in vivo mRNA delivery efficacy and organ selectivity. Moreover, diverse existing zwitterionic, ionizable cationic, and permanently cationic helper lipids were evaluated in our iPLNP system, which mediated mRNA translation selectively in spleen, liver, and lungs. Ultimately, the top 9A1P9–5A2-SC8 and 9A1P9-DDAB iPLNPs were utilized to co-deliver mRNA and sgRNA to edit reporter and endogenous genes, and long-term challenging organ selective CRISPR/Cas9 gene editing was achieved. Additionally, these iPLNPs showed broad applicability to deliver other nucleic acids, including plasmid DNA and siRNA (Supplementary Fig. 32). These profiles impart synthetic ionizable phospholipids great promise for the treatment of diverse genetic diseases with minimized side effects.
Methods
Materials.
2-Chloro-2-oxo-1,3,2-dioxaphospholane (COP) and triethylamine (TEA) were purchased from Fisher Scientific. Amines, alcohols, cholesterol (chol), and N-Methyldioctadecylamine (MDOA) were purchased from Sigma-Aldrich. 1,2-dioleoyl-sn-glycero-3-phosphoethanolamine (DOPE), 1,2-dioleoyl-sn-glycero-3-phospho-L-serine (sodium salt) (DOPS), 1,2-dioleoyl-sn-glycero-3-phosphocholine (DOPC), 1,2-dioleoyl-sn-glycero-3-phosphoethanolamine-N-(7-nitro-2–1,3-benzoxadiazol-4-yl) (ammonium salt) (NBD-PE), 1,2-dioleoyl-sn-glycero-3-phosphoethanolamine-N-(lissamine rhodamine B sulfonyl) (ammonium salt) (Rho-PE), 1,2-distearoyl-sn-glycero-3-phosphocholine (DSPC), 1,2-dioleoyl-3-dimethylammonium-propane (DODAP), 1,2-dioleoyl-3-trimethylammonium-propane (chloride salt) (DOTAP) and dimethyldioctadecylammonium (Bromide Salt) (DDAB) were purchased from Avanti Lipids. 1,2-dimyristoyl-rac-glycero-3-methoxy(poly(ethylene glycol))-2000 (DMG-PEG2000) was obtained from NOF America. DLin-MC3-DMA was purchased from MedKoo Biosciences, and its LNP was prepared at the molar ratio of 50:10:38.5:1.5 (DLin-MC3-DMA: DSPC: chol: DMG-PEG2000). Dulbecco’s modified phosphate buffered saline (PBS), RPMI-1640 medium, fetal bovine serum (FBS), and trypsin-EDTA (0.25%) were purchased from Sigma-Aldrich. Firefly luciferase messenger RNA (mRNA), Cre mRNA, and Cas9 mRNA were purchased from TriLink Biotechnologies. The Quant-iT RiboGreen RNA assay kit was purchased from Life Technologies. ONE-Glo + Tox luciferase assay kit was purchased from Promega. D-Luciferin firefly, sodium salt monohydrate was purchased from Gold Biotechnology.
Synthesis of alkylated dioxaphospholane oxide molecules P4-P16.
P4-P16 were synthesized via esterification of 2-chloro-2-oxo-1,3,2-dioxaphospholane (COP) by corresponding alcohols with different alkyl chain length. For instance, to prepare P4, 1-butanol (30 mmol) and triethylamine (TEA, 30 mmol) were dissolved in 25 mL anhydrous tetrahydrofuran (THF). Then COP (30 mmol) solution in 10 mL THF was added dropwise to the mixture at −15 °C. Afterward, the reaction continued at 25 °C for 12 h. The mixture was filtered to remove the triethylamine hydrochloride and the filtrate was concentrated by rotary evaporation to give P4. P5-P10 molecules were synthesized using respective alcohols and following the general protocol described above. For P11-P16 synthesis, COP was added to corresponding alcohols at 0 °C, and other procedures were retained the same. All P4-P16 synthesis gave the yields over 90%.
General synthesis of ionizable phospholipids (iPhos) library.
iPhos (nAxPm) were synthesized through orthogonal reactions by amines (1A-28A) and alkylated dioxaphospholane oxide molecules (Pm, m = 4–16). “x” indicates the Pm molecule number modified on one amine molecule, and each Pm molecule could introduce one phosphate group and one hydrophobic alkyl chain into the iPhos. Each primary, secondary, or tertiary amine was designed to consume one equivalent of alkylated dioxaphospholane oxide molecules Pm. For amines nA (n = 1–18) with a single primary, secondary or tertiary amine, 1.1 equivalents of Pm were reacted with amines to obtain nA1Pm. For nA (n = 19–28) with multiple amine groups, each amine group was designed to introduce one zwitterion at the most. Briefly, the amines were reacted with 2.2 equivalents, 3.3 equivalents, 4.4 equivalents and 5.5 equivalents of Pm to give nA2Pm, nA3Pm, nA4Pm and nA5Pm iPhos, respectively. All the reactions were conducted in anhydrous dimethyl sulfoxide (DMSO) at the starting material concentration of 0.3 g mL-1. The mixtures were stirred at 70 °C for 3 days, then DMSO was removed through vacuum drying.
Initial mRNA delivery (in vitro and in vivo screening) experiments were conducted using crude iPhos. Selected top iPhos (e.g. 9A1P9, 10A1P10, 9A1P15, and 10A1P16) were purified by column flash chromatography and used for additional characterizations (including size, zeta potential, mRNA binding, pKa, hemolysis, FRET studies, etc.) and in vivo evaluations. The products were eluted and fractionated in silica gel column with a solvent gradient of 3% chloroform in methanol to 10% chloroform in methanol. The final iPhos were concentrated by rotary evaporation and dried under vacuum for 24 h.
In vitro iPhos nanoparticle (iPLNP) formulation and characterization.
iPLNPs were prepared by the ethanol dilution method. mRNA was diluted in citric acid/sodium citrate buffer (10 mM, pH 4.4). The lipid mixture containing synthetic iPhos, MDOA, chol, and DMG-PEG2000 was prepared in ethanol. The two solutions were rapidly mixed by pipette at a 3:1 aqueous: ethanol volumetric ratio. Post incubation for 15 minutes, the nanoparticles were diluted 3-fold with 1× PBS buffer for in vitro mRNA delivery.
For particle size and Ribogreen mRNA binding measurement, the nanoparticles were diluted 5-fold with 1× PBS buffer. Zeta potential was recorded with nanoparticles diluted by 10-fold with 1× PBS buffer. Zetasizer Nano ZS (Malvern, version 7.13) with a He-Ne laser (λ = 632 nm) was used for particle size and zeta potential measurement. For transmission electron microscopy (TEM) assay, mRNA-loaded iPLNPs were prepared and dialyzed against H2O in Pur-A-Lyzer midi dialysis chambers (Sigma-Aldrich) for 1 h (total lipid concentration, 2 mg mL−1). Then 5–8 μL of iPLNP solutions were dropped onto carbon TEM grids, allowing to deposit for 1 min before blotting with filter paper. The TEM grids were imaged with Transmission Electron Microscopy (FEI Tecnai G2 Spirit Biotwin).
Phase transformation assay by 31P NMR spectra.
Endosomal membrane mimicking liposomes were prepared by mixing DOPS: DOPC: DOPE (molar ratio 25:25:50) in chloroform, followed by rotary evaporation and overnight vacuum dry to give a thin lipid film. The films were hydrated in D2O (20 mM 4-(2-hydroxyethyl)-1-piperazineethanesulfonic acid (HEPES), pH 5.5), and the solution was vortex mixed vigorously to get a white dispersion. The dispersion was further hydrated by maintaining overnight at room temperature, and then was underwent five freeze-thaw cycles with liquid nitrogen. 31P NMR spectrum of endosomal mimics was recorded. Afterward, iPhos lipid 9A1P9 was dissolved in chloroform, rotary evaporated, and vacuum dried overnight to give a thin lipid film. Then, D2O (20 mM HEPES, pH 5.5) was added and sonicated to get the white suspended dispersion. Above endosomal mimics and 9A1P9 suspension were mixed thoroughly by sonication (9A1P9: DOPS: DOPC: DOPE molar ratio, 20:20:20:40), and the dispersion became much clearer. The solution was further underwent five freeze-thaw cycles with liquid nitrogen, and the solution became white dispersion again, which was characterized by 31P NMR. 10 μmol of total lipids was used. 31P NMR spectra were performed on a Bruker 400 MHz spectrometer at 25 °C. Acquisition parameters included 0.68 s acquisition time, 1 s relaxation delay and 7500 scans.
Hemolysis assay.
Mouse red blood cells (RBCs) were isolated from freshly collected whole blood by centrifuging at 10,000 × g for 5 min, then the RBCs were washed 5 times with PBS buffer (pH 7.4). Afterward, RBCs were suspended in PBS of pH 7.4 and 5.5, respectively. iPLNPs were formulated using the in vitro iPLNP formulation method outlined above. Lipids were dissolved in chloroform, rotary evaporated, and vacuum dried for another 2 h to give a thin lipid film. Then, PBS (pH 7.4) was added and sonicated for 20 min to obtain particle suspension. RBC suspension was added to 96-well plates, and calculated iPLNPs or lipids were added to the wells. After incubating at 37 °C for 1 h, the RBC solutions were centrifuged at 10,000 × g for 5 min and the supernatants containing hemoglobin were collected. The hemoglobin contents were evaluated at a wavelength of 540 nm with a microplate reader. That RBC suspension incubated in PBS was set as negative control, and RBC suspension incubated in Triton X-100 solutions (1 wt. %) was set as positive control. Cell viability of iPhos lipids and iPLNPs with same conditions was measured, confirming that hemolysis was not due to cytotoxicity.
Lipid fusion by fluorescence resonance energy transfer (FRET) assay.
Lipid mixing and fusion with endosomal mimicking anionic liposomes were determined by a FRET assay. The DOPE-conjugated FRET probes NBD-PE and Rho-PE were formulated into the same endosomal mimicking nanoparticle, leading to attenuated NBD fluorescence because of FRET to rhodamine. Once lipid fusion occurred, NBD signal would increase due to the larger distance between the two probes. Endosomal mimicking anionic liposomes were prepared by mixing DOPS: DOPC: DOPE: NBD-PE: Rho-PE (molar ratio 25:25:48:1:1) in chloroform, followed by rotary evaporation and another 2 h vacuum dry to give a thin lipid film. The dried film was subsequently hydrated in PBS (pH 7.4), sonicated for 20 min and total lipid concentration was fixed at 1 mM. iPLNPs were formulated using the in vitro iPLNP formulation method outlined above with an iPhos concentration of 1 mM. iPhos 10A1P10 was dissolved in chloroform, and rotary evaporated to give a thin lipid film. Then, PBS (pH 7.4) was added and sonicated for 20 min to obtain particle suspension (10 mM). 25A3P10 with multiple zwitterions failed to form particle suspension after sonication, so only lipid fusion of 25A3P10 iPLNPs was evaluated. PBS (pH 5.5) was added to black 96-well plates (100 μL/well), and 1 μL of endosomal mimicking anionic liposomes (1 mM) was added to each well. Then 10 μL iPLNPs or 1 μL lipid suspensions were added to the wells. After incubating at 37 °C for 5 min, fluorescence measurements (F) were conducted on a microplate reader at Ex/Em = 465/520 nm. Only endosomal mimicking anionic liposomes in PBS were set as negative control (Fmin). Lipids containing probes incubated with Triton X-100 solutions (2 wt. %) were set as positive control (Fmax). The lipid fusion (%) was calculated as (F-Fmin)/(Fmax-Fmin) × 100%.
In vitro screening of iPhos for mRNA delivery.
IGROV1 cells were seeded at a density of 1 × 104 cells per well in white opaque 96-well plates. 24 h later, nanoparticles with Fluc mRNA were prepared using the in vitro iPLNP formulation method outlined above in 96-well plates by rapid mixing of aqueous phase and ethanol phase (v/v = 3:1) with a multichannel pipette. iPLNPs were prepared at synthetic iPhos: mRNA molar ratio of 11622:1 and lipid mixture synthetic iPhos: MDOA: chol: DMG-PEG2000 molar ratio of 25:30:30:1. The synthetic iPhos: mRNA molar ratio of 11622:1 was fixed, where 10A1P4–12A1P16: mRNA showed an average weight ratio of 10 ± 2.5. This could ensure that each iPLNP had same moles of lipid mixture. The ratios were also used for other characterizations and in vivo evaluation, unless other noted. 50 ng mRNA per well was used. Then, 150 μL fresh cell culture media were utilized to replace the previous media, and the formulated iPLNPs were added into the cells. After another 24 h incubation, luciferase expression and cell viability were evaluated with the ONE-Glo + Tox luciferase assay kits.
In vivo iPLNP formulation and characterization.
The mRNA was diluted in citric acid/sodium citrate buffer (10 mM, pH 3.2). The lipid mixture containing synthetic iPhos, MDOA (or other help lipids), cholesterol, and DMG-PEG2000 was prepared in ethanol. The two phases were rapidly mixed by pipette at a 3:1 aqueous: ethanol volumetric ratio. Post incubation for 15 minutes, iPLNPs were dialyzed against 1× PBS in Pur-A-Lyzer midi dialysis chambers (Sigma-Aldrich) for in vivo use. Unless otherwise noted, iPLNPs were formulated by pipette.
To further decrease the sizes, iPLNPs were prepared using the NanoAssemblr microfluidic mixing system (Precision Nanosystems). Same as above, mRNA was diluted in citric acid/sodium citrate buffer (10 mM, pH 3.2), and lipid mixtures were dissolved in ethanol. The two phases were rapidly mixed at a 3:1 aqueous: ethanol volumetric ratio with a flow rate of 9 mL minute-1. A waste collection of 0.3 mL at the start and 0.1 mL in the end was set. Post incubation for 15 minutes, iPLNPs were dialyzed against 1× PBS for future use.
In vivo luciferase mRNA delivery.
For iPhos in vivo screening, nanoparticles containing Fluc mRNA were prepared as in vivo iPLNP formulation method mentioned above. Unless otherwise noted, the ratios of formulations were in accordance with that of the in vitro screening. Briefly, iPLNPs were prepared at synthetic iPhos: Fluc mRNA molar ratio of 11622:1 and lipid mixture synthetic iPhos: MDOA: chol: DMG-PEG2000 molar ratio of 25:30:30:1. Then, the nanoparticles were administered to female C57BL/6 mice (6–8 week old) via intravenous (IV) injection. 6 h later, the luciferase expression was evaluated by live animal bioluminescence imaging. Briefly, mice were anesthetized under isofluorane, and 100 μL of D-luciferin (GoldBio, 30 mg mL−1 in PBS) substrate was intraperitoneally injected. After 5 minutes under anesthesia, the luciferase activity was imaged on an IVIS Lumina system (Perkin Elmer). Afterward, the organs were isolated and imaged with the same method. The images were processed with the Living Image software version 4.3 (64-bit, Caliper Life Sciences).
iPhos 9A1P9 was used to compare with commercial phospholipids DOPE and DSPC. C57BL/6 mice were IV injected by nanoparticles at 0.25 mg kg−1 FLuc mRNA and luminescence was quantified 6 h post injection. 9A1P9: MDOA: chol: DMG-PEG2000 molar ratio of 25:30:30:1 and 9A1P9/mRNA weight ratio of 18:1 were used. For commercial phospholipid comparison, equimolar DOPE or DSPC were utilized to replace 9A1P9. Other procedures were conducted the same as mentioned above.
For 9A1P9 iPLNPs with different helper lipids, 9A1P9: DOPE: chol: DMG-PEG2000 (molar ratio) of 55:30:45:0.2, 9A1P9: MDOA (DODAP or 5A2-SC8): chol: DMG-PEG2000 (molar ratio) of 25:30:30:1 and 9A1P9: DDAB (or DOTAP): chol: DMG-PEG2000 (molar ratio) of 60:30:40:0.4 were used. For all the formulations, 9A1P9: mRNA weight ratio was fixed at 18:1. Bioluminescence imaging was conducted at different time intervals (3, 6, 12, and 24 h), following IV injection of various Fluc mRNA doses (0.05 to 0.25 mg kg−1). Other procedures were conducted the same as mentioned above.
In vivo co-delivery of Cas9 mRNA and sgTom1 for gene editing.
Nanoparticles containing Cas9 mRNA and modified sgTom1 (mRNA/sgRNA weight ratio 4:1, total RNA dose 0.75 mg kg−1) were prepared as in vivo iPLNP formulation method mentioned above (Supplementary Table 2). 9A1P9: 5A2-SC8: chol: DMG-PEG2000 (molar ratio) of 25:30:30:1 and 9A1P9: DDAB: chol: DMG-PEG2000 (molar ratio) of 60:30:40:0.4 were used for 9A1P9–5A2-SC8 iPLNP and 9A1P9-DDAB iPLNP, respectively. 9A1P9/RNA weight ratio was fixed at 18:1. Afterward, iPLNPs were administered to Ai9 mice via IV injection. PBS group was used as negative control. 10 days later, mice were sacrificed and the organs were isolated and imaged on the IVIS Spectrum in vivo imaging system (Perkin Elmer). Then, tissues were embedded in optimal cutting temperature (OCT) compound and cut into 10 μm sections. After fixing with 4% paraformaldehyde for 20 min, the sections were washed three times with PBS. Afterward, ProLong Gold Mountant with DAPI was dropped to the slides, and coverslips were covered. These slides were imaged by confocal microscopy (Zeiss LSM 700), and analyzed with ZEN 2010 software version 6.0.62 (Carl Zeiss MicroImaging GmbH).
In vivo co-delivery of Cas9 mRNA and sgPTEN for gene editing.
PTEN was selected to examine endogenous gene editing in vivo. iPLNPs containing Cas9 mRNA and modified sgPTEN (mRNA/sgRNA weight ratio 4:1, total RNA dose 0.75 mg kg−1) were prepared as in vivo iPLNP formulation method mentioned above. 9A1P9: 5A2-SC8: chol: DMG-PEG2000 (molar ratio) of 25:30:30:1 and 9A1P9: DDAB: chol: DMG-PEG2000 (molar ratio) of 60:30:40:0.4 were used for 9A1P9–5A2-SC8 iPLNP and 9A1P9-DDAB iPLNP, respectively. 9A1P9/RNA weight ratio was fixed at 18:1. Afterward, iPLNPs were administered to wild type C57BL/6 mice (6–8 week old) via IV injection. 10 days later, tissues were collected, and genomic DNA was extracted with a PureLink Genomic DNA Mini Kit (Thermo Fisher). Post obtainment of PTEN PCR products (Supplementary Table 3), the T7E1 assay was used to measure the gene editing efficacy by the standard protocol (NEB) (Supplementary Fig. 27). Furthermore, gene editing efficacy of PTEN was evaluated by Image J (1.50b Java 1.8.0_60 (64-bit), National Institutes of Health, USA). Indel (%) is calculated as 100 × (1 - (1 - fraction cleaved)^0.5), where the fraction cleaved is defined as (Fragment 1 + Fragment 2)/(Fragment 1 + Fragment 2 + Parent Fragment).
Statistical analyses.
Statistical analyses were performed using GraphPad Prism version 8 (GraphPad Software). Two-tailed unpaired Student’s t-test was used to determine the significance of the indicated comparisons. Data are expressed as mean ± s.d. P-values < 0.05 (*), P < 0.01 (**), P < 0.001 (***) and P < 0.0001 (****) were considered to be statistically significant.
Supplementary Material
Acknowledgments
D.J.S. acknowledges financial support from the National Institutes of Health (NIH) National Institute of Biomedical Imaging and Bioengineering (NIBIB) (R01 EB025192-01A1), the American Cancer Society (ACS) (RSG-17-012-01), the Welch Foundation (I-1855), and the Cystic Fibrosis Foundation (CFF) (SIEGWA18XX0). T.W. acknowledges financial support from the Cancer Prevention and Research Institute of Texas (CPRIT) Training Grant (RP160157). We acknowledge the UTSW Tissue Resource, supported in part by the National Cancer Institute (5P30CA142543), and the Moody Foundation Flow Cytometry Facility.
Footnotes
Competing interests
D.J.S., S.L., Q.C., T.W., X.Y., and the Reagents of the University of Texas System have filed a patent application on this technology.
Data availability
All relevant data supporting the findings of this study are available within the paper and Supplementary Information. The raw data is available from the corresponding author upon request.
Reporting Summary. Further information on research design is available in the Nature Research Reporting Summary linked to this article.
Additional information
Supplementary information is available in the online version of the paper.
References
- 1.Wang HX et al. CRISPR/Cas9-based genome editing for disease modeling and therapy: Challenges and opportunities for nonviral delivery. Chem. Rev 117, 9874–9906 (2017). [DOI] [PubMed] [Google Scholar]
- 2.Hajj KA & Whitehead KA Tools for translation: Non-viral materials for therapeutic mRNA delivery. Nat. Rev. Mater 2, 17056 (2017). [Google Scholar]
- 3.Jinek M. et al. A programmable dual-RNA-guided DNA endonuclease in adaptive bacterial immunity. Science 337, 816–821 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Cong L. et al. Multiplex genome engineering using CRISPR/Cas systems. Science 339, 819–823 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Mali P. et al. RNA-guided human genome engineering via Cas9. Science 339, 823–826 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Shahbazi R. et al. Targeted homology-directed repair in blood stem and progenitor cells with CRISPR nanoformulations. Nat. Mater 18, 1124–1132 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Yin H. et al. Structure-guided chemical modification of guide RNA enables potent non-viral in vivo genome editing. Nat. Biotechnol 35, 1179 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Miller JB et al. Non-viral CRISPR/Cas gene editing in vitro and in vivo enabled by synthetic nanoparticle co-delivery of Cas9 mRNA and sgRNA. Angew. Chem. Int. Ed 56, 1059–1063 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Liu J. et al. Fast and efficient CRISPR/Cas9 genome editing in vivo enabled by bioreducible lipid and messenger RNA nanoparticles. Adv. Mater 31, 1902575 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Kanasty R, Dorkin JR, Vegas A & Anderson D Delivery materials for siRNA therapeutics. Nat. Mater 12, 967–977 (2013). [DOI] [PubMed] [Google Scholar]
- 11.Semple SC et al. Rational design of cationic lipids for siRNA delivery. Nat. Biotechnol 28, 172–176 (2010). [DOI] [PubMed] [Google Scholar]
- 12.Jayaraman M. et al. Maximizing the potency of siRNA lipid nanoparticles for hepatic gene silencing in vivo. Angew. Chem. Int. Ed 51, 8529–8533 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Love K. et al. Lipid-like materials for low-dose, in vivo gene silencing. Proc. Natl. Acad. Sci. U.S.A 107, 1864–1869 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Cheng Q. et al. Dendrimer-based lipid nanoparticles deliver therapeutic FAH mRNA to normalize liver function and extend survival in a mouse model of hepatorenal tyrosinemia type I. Adv. Mater 30, e1805308 (2018). [DOI] [PubMed] [Google Scholar]
- 15.Sabnis S. et al. A novel amino lipid series for mRNA delivery: Improved endosomal escape and sustained pharmacology and safety in non-human primates. Mol. Ther 26, 1509–1519 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.van Meer G, Voelker DR & Feigenson GW Membrane lipids: Where they are and how they behave. Nat. Rev. Mol. Cell Biol 9, 112–124 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Miller JB, Kos P, Tieu V, Zhou K & Siegwart DJ Development of cationic quaternary ammonium sulfonamide amino lipids for nucleic acid delivery. ACS Appl. Mater. Inter 10, 2302–2311 (2018). [DOI] [PubMed] [Google Scholar]
- 18.Gilleron J. et al. Image-based analysis of lipid nanoparticle-mediated siRNA delivery, intracellular trafficking and endosomal escape. Nat. Biotechnol 31, 638–646 (2013). [DOI] [PubMed] [Google Scholar]
- 19.Wittrup A. et al. Visualizing lipid-formulated siRNA release from endosomes and target gene knockdown. Nat. Biotechnol 33, 870–876 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Miller JB & Siegwart DJ Design of synthetic materials for intracellular delivery of RNAs: From siRNA-mediated gene silencing to CRISPR/Cas gene editing. Nano Res. 11, 5310–5337 (2018). [Google Scholar]
- 21.Whitehead KA et al. Degradable lipid nanoparticles with predictable in vivo siRNA delivery activity. Nat. Commun 5, 4277 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Miao L. et al. Delivery of mRNA vaccines with heterocyclic lipids increases anti-tumor efficacy by STING-mediated immune cell activation. Nat. Biotechnol 37, 1174–1185 (2019). [DOI] [PubMed] [Google Scholar]
- 23.Kaczmarek JC et al. Optimization of a degradable polymer–lipid nanoparticle for potent systemic delivery of mRNA to the lung endothelium and immune cells. Nano Lett 18, 6449–6454 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Paunovska K. et al. Nanoparticles containing oxidized cholesterol deliver mRNA to the liver microenvironment at clinically relevant doses. Adv. Mater 31, 1807748 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Fenton OS et al. Bioinspired alkenyl amino alcohol ionizable lipid materials for highly potent in vivo mRNA delivery. Adv. Mater 28, 2939–2943 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Truong B. et al. Lipid nanoparticle-targeted mRNA therapy as a treatment for the inherited metabolic liver disorder arginase deficiency. Proc. Natl. Acad. Sci. U.S.A 116, 21150–21159 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Cheng Q. et al. Selective ORgan Targeting (SORT) nanoparticles for tissue specific mRNA delivery and CRISPR/Cas gene editing. Nat. Nanotechnol 15, 313–320 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Hajj KA et al. Branched-tail lipid nanoparticles potently deliver mRNA In vivo due to enhanced ionization at endosomal pH. Small 15, 1805097 (2019). [DOI] [PubMed] [Google Scholar]
- 29.Fenton OS et al. Synthesis and biological evaluation of ionizable lipid materials for the in vivo delivery of messenger RNA to B lymphocytes. Adv. Mater 29, 1606944 (2017). [DOI] [PubMed] [Google Scholar]
- 30.Alabi CA et al. Multiparametric approach for the evaluation of lipid nanoparticles for siRNA delivery. Proc. Natl. Acad. Sci. U.S.A 110, 12881–12886 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Hou X. et al. Vitamin lipid nanoparticles enable adoptive macrophage transfer for the treatment of multidrug-resistant bacterial sepsis. Nat. Nanotechnol 15, 41–46 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Menger FM & Peresypkin AV A combinatorially-derived structural phase diagram for 42 zwitterionic geminis. J. Am. Chem. Soc 123, 5614–5615 (2001). [DOI] [PubMed] [Google Scholar]
- 33.Wang D. et al. Supramolecularly engineered phospholipids constructed by nucleobase molecular recognition: upgraded generation of phospholipids for drug delivery. Chem. Sci 6, 3775–3787 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Akinc A. et al. A combinatorial library of lipid-like materials for delivery of RNAi therapeutics. Nat. Biotechnol 26, 561–569 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Zhou K. et al. Modular degradable dendrimers enable small RNAs to extend survival in an aggressive liver cancer model. Proc. Natl. Acad. Sci. U.S.A 113, 520–525 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Liu S. et al. Highly branched poly(β-amino ester) delivery of minicircle DNA for transfection of neurodegenerative disease related cells. Nat. Commun 10, 3307 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Zhou J. et al. Biodegradable poly(amine-co-ester) terpolymers for targeted gene delivery. Nat. Mater 11, 82–90 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Liu S. et al. Bioreducible zinc(II)-coordinative polyethylenimine with low molecular weight for robust gene delivery of primary and stem cells. J. Am. Chem. Soc 139, 5102–5109 (2017). [DOI] [PubMed] [Google Scholar]
- 39.Dahlman JE et al. In vivo endothelial siRNA delivery using polymeric nanoparticles with low molecular weight. Nat. Nanotechnol 9, 648–655 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Schlame M. et al. The physical state of lipid substrates provides transacylation specificity for tafazzin. Nat. Chem. Bio 8, 862–869 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Hafez IM, Maurer N & Cullis PR On the mechanism whereby cationic lipids promote intracellular delivery of polynucleic acids. Gene Ther 8, 1188–1196 (2001). [DOI] [PubMed] [Google Scholar]
- 42.Wei T. et al. Anticancer drug nanomicelles formed by self-assembling amphiphilic dendrimer to combat cancer drug resistance. Proc. Natl. Acad. Sci. U.S.A 112, 2978–2983 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Walsh CL, Nguyen J & Szoka FC Synthesis and characterization of novel zwitterionic lipids with pH-responsive biophysical properties. Chem. Commun 48, 5575–5577 (2012). [DOI] [PubMed] [Google Scholar]
- 44.Zhang Y. et al. The development of an in vitro assay to screen lipid based nanoparticles for siRNA delivery. J. Controlled Release 174, 7–14 (2014). [DOI] [PubMed] [Google Scholar]
- 45.Cheng Y, Yumul RC & Pun SH Virus-inspired polymer for efficient in vitro and in vivo gene delivery. Angew. Chem. Int. Ed 55, 12013–12017 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Zhou D. et al. The transition from linear to highly branched poly(beta-amino ester)s: Branching matters for gene delivery. Science Adv 2, e1600102 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Kowalski PS et al. Ionizable amino‐polyesters synthesized via ring opening polymerization of tertiary amino‐alcohols for tissue selective mRNA delivery. Adv. Mater, 1801151 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Li B. et al. An orthogonal array optimization of lipid-like nanoparticles for mRNA delivery in vivo. Nano Lett. 15, 8099–8107 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Wei T, Cheng Q, Min YL, Olson EN & Siegwart DJ Systemic nanoparticle delivery of CRISPR-Cas9 ribonucleoproteins for effective tissue specific genome editing. Nat. Commun 11, 3232 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Akinc A. et al. The Onpattro story and the clinical translation of nanomedicines containing nucleic acid-based drugs. Nat. Nanotechnol 14, 1084–1087 (2019). [DOI] [PubMed] [Google Scholar]
- 51.Patel S. et al. Naturally-occurring cholesterol analogues in lipid nanoparticles induce polymorphic shape and enhance intracellular delivery of mRNA. Nat. Commun 11, 983 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Xue W. et al. CRISPR-mediated direct mutation of cancer genes in the mouse liver. Nature 514, 380–384 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Staahl BT et al. Efficient genome editing in the mouse brain by local delivery of engineered Cas9 ribonucleoprotein complexes. Nat. Biotechnol 35, 431–434 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.