Abstract
Escherichia coli levels in recreational waters are often used to predict when fecal-associated pathogen levels are a human health risk. The reach of the Chattahoochee River that flows through the Chattahoochee River National Recreation Area (CRNRA), located in the Atlanta-metropolitan area, is a popular recreation area that frequently exceeds the U.S. Environmental Protection Agency beach action value (BAV) for E. coli. A BacteriALERT program has been implemented to provide real-time E. coli estimates in the reach and notify the public of potentially harmful levels of fecal-associated pathogens as indicated by surrogate models based on real-time turbidity measurements from continuous water quality monitoring stations. However, E. coli does not provide information about the sources of fecal contamination and its accuracy as a human health indicator is questionable when sources of contamination are non-human. The objectives of our study were to investigate, within the Park and surrounding watersheds, seasonal and precipitation-related patterns in microbial source tracking marker concentrations of possible sources (human, dog, and ruminant), assess correlations between source contamination levels and culturable E. coli levels, determine which sources best explained model-based E. coli estimates above the BAV and detection of esp2 (a marker for the esp gene associated with pathogenic strains of Enterococcus faecium and Enterococcus faecalis), and investigate associations between source contamination levels and land use features. Three BacteriALERT sites on the Chattahoochee River were sampled six times per season in the winter and summer from December 2015 through September 2017, and 11 additional stream sites (synoptic sites) from the CRNRA watershed were sampled once per season. Samples were screened with microbial source tracking (MST) quantitative PCR (qPCR) markers for humans (HF183 Taqman), dogs (DogBact), and ruminants (Rum2Bac), the esp2 qPCR marker, and culturable E. coli. At the BacteriALERT sites, HF183 Taqman concentrations were higher under wet conditions DogBact concentrations were greater in the winter and under wet conditions, and Rum2Bac concentrations were comparatively low throughout the study with no difference across seasons or precipitation conditions. Concentrations of HF183 Taqman, DogBact, and Rum2Bac were positively correlated with culturable E. coli concentrations; however, DogBact had the largest R2 value among the three markers, and the forward stepwise regression indicated it was the best predictor of culturable E. coli concentrations at the BacteriALERT sites. Recursive partitioning indicated that BAV exceedances of model-based E. coli estimates were best explained by DogBact concentrations ≥3 gene copies per mL (CN/mL). Detections of esp2 at BacteriALERT sites were best explained by DogBact concentrations ≥11 CN/mL, while detections of esp2 at synoptic sites were best explained by HF183 Taqman ≥29 CN/mL. At the synoptic sites, HF183 Taqman levels were associated with wastewater treatment plant density. However, this relationship was driven primarily by a single site, suggesting possible conveyance issues in that catchment. esp2 detections at synoptic sites were positively associated with development within a 2-km radius and negatively associated with development within the catchment, suggesting multiple sources of esp2 in the watershed. DogBact and Rum2Bac were not associated with the land use features included in our analyses. Implications for Park management include: 1) fecal contamination levels were highest during wet conditions and in the off season when fewer visitors are expected to be participating in water-based recreation, 2) dogs are likely contributors to fecal contamination in the CRNRA and may be sources of pathogenic bacteria indicating further investigation of the origins of this contamination may be warranted as would be research to understand the human health risks from exposure to dog fecal contamination, and 3) high levels of the human marker at one site in the CRNRA watershed suggests more extensive monitoring in that catchment may locate the origin of human fecal contamination detected during this study.
Keywords: BacteriALERT, Quantitative PCR, Bacteroidales, Fecal pollution, Septic systems
1. Introduction
Monitoring for fecal indicator bacteria (FIB) as a proxy for human health outcomes in recreational freshwater bodies is often conducted by collecting water samples, culturing Escherichia coli in the samples, and enumerating E. coli colonies (U.S. Environmental Protection Agency, 2012). However, this method for monitoring FIB has an 18e24 h minimum delay between potentially harmful conditions at the time of sample collection and notification to the public. While not yet widely implemented, surrogate models for E. coli from continuous water-quality monitoring data can solve the delay issue between sample collection and public notification of a potential human health risk (Lawrence, 2012).
The Chattahoochee River National Recreation Area (CRNRA) has been a case study of the use of surrogate models for predicting E. coli concentrations in recreational waters. Models of E. coli concentrations based on water quality condition (i.e., turbidity) were developed for two USGS continuous water-quality monitoring stations (USGS 02335000 and USGS02336000; Fig. 1) on the Chattahoochee River reach within the CRNRA by Lawrence (2012) as part of the collaborative BacteriALERT program between the National Park Service (NPS), Chattahoochee Riverkeeper, and U.S. Geological Survey (USGS). The purpose of BacteriALERT is to monitor E. coli levels in the recreational waters of the CRNRA and inform the Park and public when levels exceed the U.S. Environmental Protection Agency (USEPA) beach action value (BAV) for primary contact (>235 colony forming units per 100-mL; U.S. Environmental Protection Agency, 2012). When model-based estimates of E. coli concentrations exceed 235 cfu/100 mL, the Park cancels all NPS-sponsored water-based activities (e.g., rafting, interpretive events, river-based trash clean ups). Water-based recreational park concessionaires (e.g., tubing and kayaking companies) are also required to post the health advisory until the notice is lifted, which can deter patrons from their establishments. Model-based estimates of E. coli concentrations are updated every 15 min on the BactertiALERT website (https://www2.usgs.gov/water/southatlantic/ga/bacteria/, accessed Oct. 15, 2019) for the public to make informed decisions about recreating in the Park.
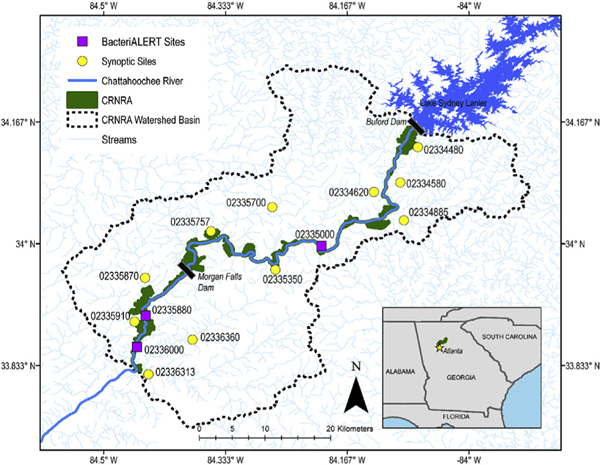
Fig. 1.
Map of microbial source tracking sample sites within the Chattahoochee River National Recreation Area (CRNRA) watershed. Twenty-four samples were collected from BacteriALERT sites and three to four samples were collected from each synoptic site between December 2015 and September 2017 for microbial source-tracking analysis.
While model-based estimates of E. coli in recreational waters can provide a general indication when a human health risk exists based on USEPA recommendations they do not provide source or spatial origins information for directing mitigation efforts. Further, if the primary sources of fecal contamination are non-human, the reliability of FIB as indicators of the risk to human health can be inconsistent (Calderon et al.,1991; McBride et al.1998; Colford et al. 2007). Studies used by the USEPA to develop the BAV for E. coli were based on gastrointestinal illness rates in people exposed to waters contaminated primarily by human waste from wastewater treatment plants (U.S. Environmental Protection Agency, 2012). Due inpart to the host specificity of most viruses (Webby et al., 2004; Medina and García-Sastre, 2011) there the possibility that exposure to human fecal contamination is a greater human-health risk than exposure to fecal contamination from non-human sources as some studies have indicated (Colford et al. 2007; Schoen and Ashbolt, 2010; Soller et al. 2010b; Soller et al. 2014, but see McBride et al., 1998). However, few studies have directly investigated human morbidity rates associated with exposure to fecal contamination from specific non-human sources (but see Soller et al. 2015). Escherichia coli are shed in the feces of many different warmblooded animals, including wildlife and domesticated species. Therefore, the health risk from exposure to contaminated waters with E. coli concentrations above the BAV may be different if the primary sources of fecal contamination are non-human versus human. Determining the sources of fecal contamination in the CRNRA, and sources associated with E. coli levels above the BAV may indicate if further research is warranted to accurately assess the human health risk from exposure to fecal contamination when recreating in the CRNRA.
Microbial source tracking (MST) is an approach for determining sources of fecal contamination by detecting bacteria, often by quantitative PCR (qPCR), that occur in the gastrointestinal tract of particular species, which allows for the assessment of the presence or absence of enteric bacteria as well as identification of probable animal sources (Bradshaw et al.; Bernhard and Field, 2000a; Meays et al. 2004; Beversdorf et al., 2007; Byappanahalli et al. 2012a; Byappanahalli et al., 2012b; Boehm et al. 2013). The intestinal systems of different animals such as humans, dogs, and deer have distinct physiological and dietary signatures. Because of these differences, the characteristics of enteric bacterial populations differ as well. Quantitative PCR is a sensitive molecular technique for detecting DNA sequences of interest and, when combined with fluorescently-labeled probes, can add additional specificity to an assay. The Species of the order Bacteroidales have been established as useful indicator microorganisms for fecal contamination (Allsop and Stickler, 1985) because they are restricted to warm-blooded animals, make up a significant portion of the bacteria present in mammalian feces, can be highly specific to particular species or taxonomic groups, and are strict anaerobes and therefore indicative of recent contamination. As most E. coli is non-pathogenic to humans, the inclusion in an MST study of a molecular marker associated with a pathogen (from here on referred to as a pathogenic marker) can provide further insight to associations between MST markers and human health risks.
The goals of this project were to investigate spatial and temporal trends in sources of fecal contamination in the CRNRA, identify potential primary sources (humans, dogs, and deer) and spatial or land use origins of contamination, and determine sources and MST marker levels associated with exceedances of the BAV for E. coli and presence of a pathogenic marker. The objectives of this study were to (1) compare levels of qPCR MST markers at BacteriALERT sites in the CRNRA by season (summer: high visitation; and winter: low visitation) and precipitation condition (wet: precipitation within the 24 h prior to sample collection, and dry: little to no precipitation 24 h prior to sample collection) to assess trends in sources of fecal contamination; (2) correlate culturable E. coli concentrations with qPCR MST marker levels to test for evidence of primary sources of E. coli; (3) investigate which qPCR MST markers best explain model-based estimates of E. coli concentrations above the BAV and pathogenic marker detection; and (4) investigate qPCR MST marker levels and pathogenic marker detection in tributaries throughout the CRNRA watershed to assess associations between marker levels and surrounding land use characteristics.
Understanding contamination sources and spatial origins will enable Park managers target outreach efforts within the CRNRA and facilitate discussions with local communities from which runoff originates that can impact waterways located within Park boundaries. Further, determining which markers best predict E. coli BAV exceedances from model-based concentration estimates may help assess if the current USEPA BAV for E. coli is an appropriate indicator of a human health risk in the CRNRA, or if further research is warranted to determine more effective indicators of human health risk from exposure to contamination of non-human sources.
2. Methods
2.1. Study locations
This study took place in the CRNRA watershed in Georgia in the greater Atlanta-metropolitan area. A total of 114 samples were collected over the duration of the study. Different sampling designs were used to study three BacteriALERT sites on the Chattahoochee River (USGS 02335000, USGS 02335880, and USGS 02336000; Fig. 1) versus 11 synoptic sites located throughout the CRNRA watershed on tributaries to the Chattahoochee River (Fig. 1). BacteriALERT sites were sampled six times in the summer (June–September) and winter (December–March) of water years (WY) 2016 (October 1, 2015–September 30, 2016) and WY 2017 (October 1, 2016–September 30, 2017), totaling 72 BacteriALERT samples. Synoptic sites across the CRNRA watershed were sampled once in the summer and winter in WY2016 and WY2017, with the exception of USGS02336313 and USGS02336360, which were not sampled in winter WY2016, totaling 42 synoptic samples. Synoptic sample locations were determined based on the presence of a USGS gaging station (current or historic), and with an approximately even distribution along the 48-mile stretch of the CRNRA, on both sides of the River (Fig. 1). Water samples were collected from the approximated middle of the stream or river channel by hand-dipping or with a bridge sampler in sterile 1-L bottles (U.S. Geological Survey variously dated).
2.2. Water sample collection and bacterial enrichment and enumeration
Water samples were processed within 48 h of sample collection, with samples kept away from light and refrigerated in the interim. To collect bacterial DNA, 50 to 100-mL (McKee, 2019) of water were filtered on a vacuum manifold through a 47-mm polycarbonate filters with 0.4-μm pore size (Millipore, Bedford, MA, USA). Six filter replicates were collected per sample, two of which were selectively enriched for Enterococci with mEI agar as described by USEPA Method 1600 (U.S. Environmental Protection Agency, 2006). DNA from filters enriched for Enterococci were subsequently screened for a gene associated with pathogenic strains of Enterococcus faecium and Enterococcus faecalis (see section 2.3.2). Filters were stored in sterile tubes at 80°C until DNA extraction. To investigate correlations between MST marker and culturable E. coli concentrations at the three BacteriALERT sites, we used the most probable number (MPN) method with IDEXX Colilert-18 (IDEXX Laboratories, Inc. Westbrook, ME1) for enumerating E. coli, following Myers et al. (2014). Sterile de-ionized water controls were filtered in conjunction with each sample event to test for contamination during sample collection or laboratory processing and were treated identically to filtered samples in subsequent processing.
Model-based estimates of E. coli were only available for two of the three BacteriALERT sites (USGS 02335000 and USGS 02336000) over the duration of the study. Model-based E. coli concentration estimates were calculated for the time of sample collection at the associated site from models by Lawrence (2012) with turbidity data from continuous water-quality monitoring stations. As turbidity measurements are recorded every 15 min, turbidity values for times measurements were interpolated for samples collected between measurements.
2.3. Molecular methods
2.3.1. DNA extraction
For non-enriched filters, two filter replicates per sample were extracted, and remaining filter replicates were stored as backups. For enriched filters, DNA was extracted from one replicate with the other stored as a backup. Based on equipment availability, DNA was extracted using the Qiagen DNeasy PowerLyzer PowerSoil DNA Isolation Kit (Catalog No. 12855–100) with bead-beater homogenization, or the Qiagen DNeasy PowerSoil DNA Isolation Kit (Catalog No. 12888–100) with sample homogenization via a vortex adapter following manufacturer’s instructions (S1 Table). These kits isolate genomic DNA from environmental samples and can help remove PCR inhibitors that interfere with quantitative qPCR (qPCR) accuracy. Extraction blanks were processed with all batches of DNA extractions to test for contamination during the DNA extraction process.
2.3.2. Quantitative polymerase chain reaction for microbial-source tracking markers
Samples were tested for PCR inhibition following Bradshaw et al. (2016) using the Sketa22 assay (Haugland et al. 2010), where a 1.5 Ct difference or more between the mean Ct values for the qPCR no template control (NTC, PCR-grade water) and sample was indicative of PCR inhibition. PCR inhibition was not detected in any of the samples and therefore no further action to mitigate inhibition was necessary. Microbial source-tracking markers HF183 Taqman (human associated marker, Table 1, Green et al. 2014a), Rum2Bac (ruminant associated marker suggestive of deer contamination in this study, Table 1, Mieszkin et al. 2010), and DogBact (canine associated marker, Table 1, Dick et al., 2005; Shibata et al. 2010) were screened to detect possible sources of fecal contamination within the CRNRA. We also screened the mEIenriched filters for presence of the esp gene associated pathogenic strains of Enterococcus faecium and E. faecalis (Table 1, Scott et al. 2005), which can also be suggestive of human fecal material (Shankar et al. 1999; Scott et al. 2005). mEI-enrichments began starting with samples collected on February 23, 2016 (McKee, 2019). All markers were screened using qPCR on a QuantStudio 3 (Applied Biosystems). Extracted DNA from non-enriched duplicate filters were screened for the MST markers HF183 Taqman, DogBact, and Rum2Bac. Each extracted DNA sample was run with two qPCR replicates, for a total of four qPCR replicates per sample (with the exception of one sample for HF183 Taqman (two qPCR replicates) and DogBact (3 qPCR replicates), and the Sketa22 assay which was screened on two qPCR replicates from one DNA sample; McKee 2019). Because DogBact has demonstrated low specificity to dogs (Boehm et al. 2013), samples that tested positive for DogBact were screened with a second dog source-tracking marker, DG3 (Green et al. 2014b) to verify DogBact results. DNA from filters enriched with mEI agar were screened for esp2 (Table 1), a probe-based qPCR marker for the esp gene. Two qPCR replicates were run for DNA from one mEI-enriched filter replicate, resulting in two esp2 qPCR replicates per sample.
Table 1.
Quantitative PCR Markers. LOQ is the limit of quantification (gene copy numbers from 4 mL DNA per qPCR reaction), which was the lowest copy number with a coefficient of variation of estimated gene copies less than 35%.
| Marker Name | Primer and Probe Sequences 5’→3’ | Target Organism | Amplicon Length (base pairs) | Annealing Temp ( C) | LOQ (CN/qPCR reaction) | Reference |
|---|---|---|---|---|---|---|
| HF183 Taqman | ||||||
| HF183 Taqman-F | ATCATGAGTTCACATGTCCG | Human-associated microbial populations | 126 | 60 | 40 | Green et al. (2014a) |
| HF183 Taqman-R | CTTCCTCTCAGAACCCCTATCC | |||||
| HF183 Taqman-P | FAM-CTAATGGAACGCATCCC-MGB | |||||
| DogBact | ||||||
| DF475F | CGCTTGTATGTACCGGTACG | Dog-associated microbial populations | 251 | 60 | 100 | (Dick et al., 2005; Shibata et al. 2010) |
| Bac708R | CAATCGGAGTTCTTCGTG | |||||
| DogBactP | FAM-ATTCGTGGTGTAGCGGTGAAAT GCTTAG -TAMRA | |||||
| Rum2Bac | ||||||
| BacB2–590F | ACAGCCCGCGATTGATACTGGTAA | Ruminant-associated microbial populations | 99 | 60 | 40 | Mieszkin et al. (2010) |
| Bac708Rm | CAATCGGAGTTCTTCGTGAT | |||||
| BacB2–626P | FAM-ATGAGGTGGATGGAATTCGTGGTGT TAMRA | |||||
| esp2 | esp gene associated with pathogenic strains of Enterococcus faecium and E. faecalis | 113 | 60 | NA | (Scott et al. 2005; Adapated for qPCR by Yolanda Brook, USGS contractor FY2015 (Joe Duris, personal communication)) | |
| esp2-F | TTACCCCATTAGGAGCGGTTT | |||||
| esp2-R | TGATTCGCTGGGTAGACCTACA | |||||
| esp2-P | FAM-CGTATCGGTTGTTTCTGCCCCAGC-TAMRA | |||||
Reactions were carried out in 96-well qPCR plates, sealed with optical 8 strip caps. Quantitative PCRs were carried out in 20-mL reaction volumes. Final concentrations of reagents in the assays were 1-mM forward and reverse primers; 80-nM 6-carboxy-fluorescein FAM-labeled TaqManTM probe, 0.02-mg/mL BSA (Life Technologies), and 1x DNA TaqMan™ Fast Universal PCR Master Mix (HF183 Taqman, Rum2Bac, esp2, and DG3; Life Technologies) or 1x TaqMan™ Environmental Master Mix (DogBact; Life Technologies) and 4-μL of template (genomic DNA, PCR-grade water as no template controls), standard concentrations of 10 to 106 target sequence copies of HF183 Taqman, Rum2Bac, or DogBact, or positive genomic DNA control for E. faecium). No template controls were run in duplicate with all qPCR plates, as were standard curves for all markers except esp2, for which positive controls were run with every plate. Quantification of target sequence copies for HF183 Taqman, Rum2Bac, and DogBact was determined from a serial dilution standard curve of linearized control plasmid DNA containing the targeted amplicon sequence. Minimum R2 values for standard curves was 0.99 for all markers except DogBact, which had a minimum R2 value of 0.96. Quantitative PCR efficiencies for all markers were between 80 and 110%, with slopes ranging from 3.76 to 3.05 (McKee, 2019).
HF183 Taqman, Rum2Bac, and esp2 were run with a qPCR protocol of a 20 s 95°C hold followed by 40 cycles of a 95°C for 3 s and 60°C for 30 s. DogBact was run with a qPCR protocol of a 2-min 50°C hold, followed by a 10-min 95°C hold, followed by 40 cycles of a 95°C for 15 s and 60°C for 1 min. Guidelines for determining the limits of quantification (LOQ) are based on Forootan et al. (2017). LOQ was determined by the lowest copy number with a coefficient of variation (CV = 100*s/m; where s is the standard deviation, and m is the mean number of estimated gene copies) less than 35% (Table 1). Gene copy estimates were based on the equation where X0 is the initial number of target copies in the qPCR; EAMP is the exponential amplification value, which is 1 + the amplification efficiency (e.g. if the amplification efficiency is 0.94, EAMP = 1.94); b is the y-intercept of the standard curve; and CT is the cycle number when the amplification curve crosses the threshold line that distinguishes fluorescence intensity of a reaction from background levels.
2.3.3. Estimating microbial source-tracking marker concentrations
Quantitative PCR data provide an estimate of the initial number of marker copies present in the qPCR reaction (also referred to as the copy number per reaction; CN/rxn), which is then used to calculate an estimated concentration of the bacterial marker in the original water sample (CN/mL). The calculation from CN/rxn to CN/ mL is a function of the volume of sample that was filtered, the estimated percent of DNA from the filter that was captured during the extraction process (we assumed capture of approximately 50% of the total sample DNA sample based on a 50% volume recovery after the homogenization step in the DNA extraction), the volume of solution in which the DNA was eluted during the extraction process, and the volume of DNA added to the qPCR reaction. When CN/rxn estimates of qPCR replicates were less than LOQ, including when the marker was not detected (CN/rxn = 0), we accounted for the error in the CN/rxn estimate by randomly selecting a value from a normal distribution centered around the CN/rxn estimate, the standard error of which was a function of the qPCR-based estimate (S1–S3 Files). When the randomly selected value was negative or greater than LOQ, the distribution was resampled until a value between 0 and LOQ was obtained (S1–S3 Files). The value selected from the distribution (S1 Table) was used in subsequent analyses and treated as an accurate estimate of concentration. Because of the variation in volumes of water filtered across several filter replicates, CN/mL was calculated for both subsamples of the qPCR replicate for HF183 Taqman, DogBact, and Rum2Bac, then averaged by sample (four reaction average) and rounded to the nearest integer to obtain an estimated CN/mL value for each sample. To facilitate interpretation and discussion of results, we used the threshold of ≥10 CN/mL as a general determinant of high marker concentrations for all three MST markers as HF183 Taqman concentrations ≥≥10 CN/mL may indicate sewage contamination (Templar et al. 2016). The esp2 results were reported as detected/not-detected because of the enrichment with mEI agar, where amplification at any CT in either qPCR replicate was indicative of esp2 detection for the sample.
2.4. Land use characterization
Factors affecting bacterial levels at Chattahoochee River BacteriALERT sites may differ from those affecting bacterial levels at synoptic stream sites. Buford dam releases can affect flow and resuspension of sediments at the BacteriALERT sites, which may mask possible land use associations with bacterial levels. Analyses of land use associations with MST marker concentrations and esp2 detections were therefore limited to synoptic sites. The land use features investigated for associations with a given marker were based on hypotheses about the relationship between the source and the land use feature (Table 2).
Table 2.
Description of land use variables and hypothesized relevance to possible sources (and associated microbial source-tracking markers) of fecal contamination. NA indicates the variable was not included in model selection.
| Land use variable abbreviation | Variable description (units) | Human (HF183 Taqman and esp2) | Dog (DogBact) | Deer (Rum2Bac) |
|---|---|---|---|---|
| WWTP | Wastewater treatment | WWTP density may indicate sewer line density plant density (#/km2) | NA | NA |
| SEPTIC | Septic system density (#/km2) | Septic systems may be source of contamination | NA | NA |
| DEVEL | Developed land (%) | May be a combined representation of septic and sewer line density | Dog ownership may correlate with human population density | NA |
| OPEN | Open developed land (%) | NA | Possible recreational areas for people to take their dogs | Possible foraging habitat for deer |
| FOREST | Forested land (%) | NA | Possible recreational areas for people to take their dogs | Possible deer habitat |
We investigated land use features at three different spatial scales: 1-km, 2-km, and catchment-wide. MST marker correlations with land use proximal to the stream channel (1 and 2-km buffers) would support detection of recent contamination from nearby inputs. In contrast, MST marker correlations with catchment-scale land use may indicate more broad scale sources of contamination. Catchment areas were delineated for each synoptic site using StreamStats (https://streamstats.usgs.gov, accessed September 12, 2018). We created 1 and 2-km buffers around our study sites in ArcMap (v. 10.4.1; Environmental Systems Research Institute, Redlands, CA) and used the Intersecting Layers Mask tool with our site-specific catchment areas to create GIS layers of 1 and 2-km buffers within the catchment for each site (Long and Plummer, 2004). For each buffer and catchment, we calculated septic system density (Clarke and Painter, 2014); percent developed land (DEVEL; Homer et al. 2015, Classes 21–24, 30-m resolution); percent forested land (FOREST; Homer et al. 2015, Classes 41–43, 30-m resolution); and percent open developed land, which includes parks, lawns, and vegetation planted in recreational areas (OPEN; Homer et al. 2015, Class 21, 30-m resolution). Percentage variables were arcsin-square root transformed for linear modeling. Wastewater discharge density, which was used as a surrogate for sewer infrastructure density, was calculated for each catchment area as the number of wastewater treatment plants divided by the area of the catchment (WWTP), as was septic system density. Land use variables are heretofore presented as VARIABLEscale.
Within a river basin, downstream USGS Station IDs are numerically higher than upstream USGS Station IDs. While USGS Station ID was not included as an independent variable in land use models of MST marker concentrations, it was included as a numerical variable with Spearman Rank correlations against independent variables to investigate if land use features demonstrated an upstream-downstream spatial pattern across the CRNRA watershed.
2.5. Statistical analysis
HF183 Taqman, DogBact, Rum2Bac, and E. coli concentration estimates were log10 transformed to reduce data skew for linear regression. Numerous qPCR marker concentration estimates were 0 or 1 CN/mL for HF183 Taqman, DogBact, and Rum2Bac after the random number generation for values below LOQ as described in section 2.3.3. To include qPCR marker estimates of 0 or 1 CN/mL and E. coli estimates of 0 or 1 colonies/100-mL in subsequent analyses, we converted log10 (1) to 0.001 and log10 (0) to 0.0001. Antecedent precipitation values for each sample were obtained through the USGS National Water Information System (NWIS; https://waterdata.usgs.gov/nwis, Accessed August 29, 2018). For sites without precipitation gages, we used values from the nearest USGS precipitation gage (S1 Table).
2.5.1. Seasonal and precipitations patterns in MST concentrations
To investigate seasonal and precipitation-related patterns of HF183 Taqman, Rum2Bac, and DogBact concentrations across the three Chattahoochee River BacteriALERT sites, we used precipitation within 24 h prior to of sample collection as our indicator of precipitation conditions. Any sites that received more than 0.01 inches of rain within 24 h prior to collection were termed wet samples, versus dry samples. We performed a three factor ANOVA and Student’s T pairwise comparisons in JMP® (v 14.2.0, SAS Institute Inc.) to determine how bacterial concentrations and sources varied by site, season, and precipitation. Forward stepwise regression with Bayesian Information Criterion (BIC; Schwarz, 1978; Aho et al., 2014) in JMP was used to determine which source tracking marker concentrations were most closely correlated with E. coli concentrations at the three BacteriALERT sites individually and combined.
2.5.2. Recursive partitioning of E. coli beach action value exceedances
To investigate which MST marker best explained BAV exceedances based on real-time gaging station estimates of E. coli concentrations (S1 Table) and the presence of esp2 we used the Partition platform in JMP® with a minimum split size of five data points. Recursive partitioning is a nonparametric data-mining analysis that splits the response variable into two categories based on an explanatory variable cutting value. The cutting value is determined with a partition algorithm that finds the explanatory variable split that maximizes the difference in the response frequencies between the two nodes of the split. Recursive partitioning of model-based E. coli estimates only included samples for which associated turbidity data were available at the time of sample collection, resulting in 19 records for USGS 02335000 and 17 records for USGS 02336000 (S1 Table). Recursive partitioning of esp2 was conducted separately for samples from BacteriALERT (N = 63) and synoptic sites (N = 42). Explanatory variables included in the analysis were untransformed concentrations of the three source tracking markers (HF183 Taqman, DogBact, and Rum2Bac).
2.5.3. Land use associations with MST markers
Seasonal variation and the magnitude and timing of rainfall can affect patterns of bacterial contamination in streams and rivers (Gentry et al. 2006; Shanks et al. 2006; St Laurent and Mazumder, 2014). To determine land use factors associated with bacteria levels across synoptic sites within the CRNRA watershed, we first accounted for potential variations in bacteria levels due to season and precipitation. We used generalized linear modeling to test combinations of antecedent precipitation (12, 24, 48, and 72 h; PRECIPtime) and season to determine which were most strongly associated with abundances of each MST marker at synoptic sites. HF183 Taqman, DogBact, and Rum2Bac CN/mL values were modeled with log-normal distributions using the glm function in R (v.3.4.1; R Core Team, 2017), and esp2 detections were modeled with a negative binomial distribution using the glm.nb function from the MASS package (Venables and Ripley, 2002) in R. Top models were determined as the model with the lowest BIC.
Residuals from the top model for each marker were used to calculate a mean residual value (MRV) for each synoptic site, which represented MST marker concentrations after accounting for the effects of season and precipitation. MRVs were used as the dependent variables in forward stepwise linear regression in JMP®, with a minimum of one explanatory variable, to determine which selected land use factors (Table 2) were most closely associated with bacterial concentrations (HF183 Taqman, DogBact, and Rum2Bac) or presence (esp2) (Table 2) at synoptic sites. The lowest small-sample-size corrected version of Akaike information criterion (AICc; Burnham and Anderson, 2004) for models with at least one explanatory variable was used as the stopping rule. All tests were considered significant at p < 0.05.
3. Results
A total of 114 samples were collected over the two-year study period, 72 of which were from the 3 Chattahoochee River BacteriALERT sites, and 42 samples from 11 synoptic sites across the CRNRA watershed (S2 Table). From the 3 BacteriALERT sites combined, there were 19 summer-dry samples, 17 summer-wet samples, 17 winter-dry samples, and 19 winter-wet samples. The number of samples with concentrations greater than the LOQ (HF183 Taqman = 40 CN/rxn; DogBact = 100 CN/rxn; Rum2Bac = 40 CN/rxn) for HF183 Taqman, DogBact, and Rum2Bac were 12, 17, and 7, respectively. esp2 was detected in 18 of 105 samples, 8 of which were from synoptic sites (McKee, 2019). Model-based E. coli concentration estimates indicated BAV exceedances associated with for 15 of 36 samples from USGS 02335000 and USGS 02336000 (S1 Table).
Catchment sizes among sites were variable, ranging in size from 7 to 257 km2. WWTP density within catchments ranged from 0 to 0.11 WWTP/km2, and estimated catchment-wide septic system density ranged from 1 to 164 septic systems/km2 (S2 Table). Greater variability was seen at proximal scales of 1 and 2-km buffers than at the catchment scale for land cover variables DEVEL, OPEN, and FOREST (e.g. DEVEL1km ranged from 40 to 93% versus DEVELcatchment, which ranged from 55 to 89%; S2 Table). DEVEL and FOREST variables were negatively correlated. Correlations between land use variables and USGS Station ID indicated upstream-downstream land cover spatial trends. USGS Station ID was positively correlated with all spatial scales of DEVEL and was negatively correlated with FORESTcatchment, indicating trends of increasing development downstream (towards Atlanta) and increasing forest cover upstream.
3.1. Verification of DogBact results
We screened samples that were positive for DogBact with a less sensitive second dog MST marker, DG3, which was found to be highly specific, including the ability to distinguish dog from human fecal contamination (Green et al., 2014b). Of the 37 samples that tested positive for DogBact, 25 also tested positive for DG3 (S1 Figure), and DogBact and DG3 mean CN/rxn for qPCR duplicates from corresponding filter replicates were strongly correlated (Pearson r = 0.909, p < 0.001; S1 Figure). DogBact and HF183 Taqman mean CN/rxn for qPCR duplicates were not correlated (Pearson r = 0.005, p = 0.969; S1 Figure), nor were DG3 and HF183 Taqman (Pearson r = 0.018, p = 0.882; S1 Figure), further supporting the validity of DogBact.
3.2. Microbial source-tracking marker and E. coli concentrations at BacteriALERT sites
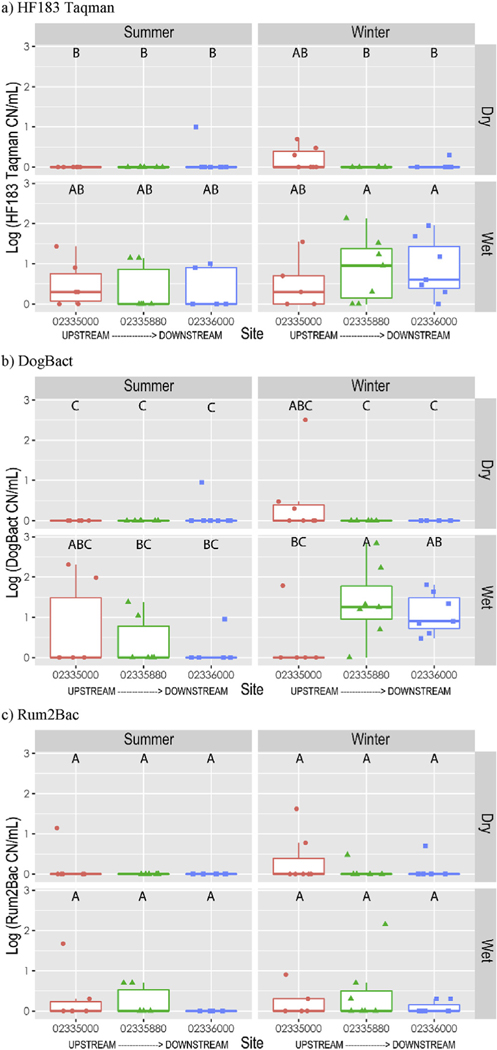
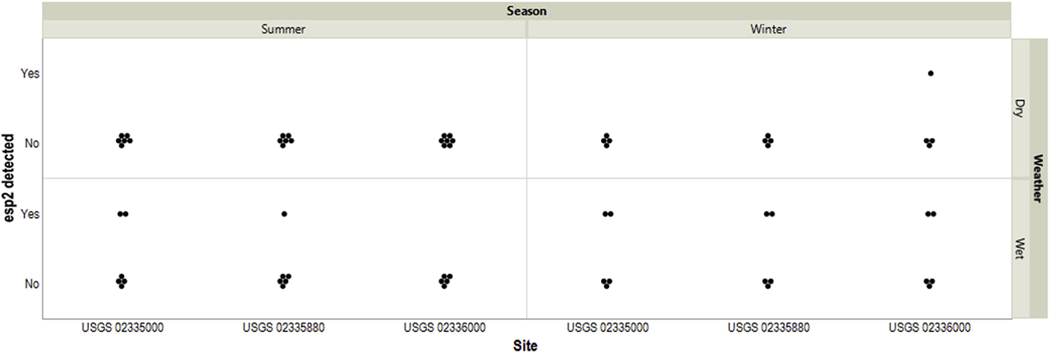
Concentrations ≥10 CN/mL of HF183 Taqman, DogBact, and Rum2Bac were detected in 12, 14, and 4 samples out of 72 samples from BacteriALERT sites, respectively (S1 Table). HF183 Taqman concentrations were higher in wet samples (S3 Table, Fig. 2a). DogBact concentrations were greater in winter and wet samples overall, with significantly higher levels at the middle and downstream BacteriALERT sites under winter-wet conditions compared to the most upstream BacteriALERT site (S3 Table, Fig. 2b). Rum2Bac concentrations were not different across seasons, precipitation condition, or by site (S3 Table, Fig. 2c). The low number of samples with Rum2Bac concentrations ≥10 CN/mL suggested that ruminant contamination was generally low. esp2 was detected in 10 of 63 samples from BacteriALERTsites, 9 of which were in wet conditions. During the summer, esp2 was only detected under wet conditions (Fig. 3).
Fig. 2.
Concentrations of fecal source tracking markers a) HF183 Taqman, b) DogBact, c) Rum2Bac, at three USGS stream gaging stations in the Chattahoochee River National Recreation Area by season and precipitation conditions (Wet: ≥ 0.01 inches of rain within 24 h prior to sample collection; Dry: < 0.01 inches of precipitation 24 h of samples collection). Box indicates first and third quartiles, upper whisker extends to the largest value within 150% of the inter-quartile range. Letters over box and whiskers indicate pairwise differences as determined by the Student’s T test with p < 0.01.
Fig. 3.
Detections of a molecular marker for pathogenic strains of Enterococcus faecium and Enterococcus faecalis in samples from three BacteriALERT sites by season and precipitation condition. Samples were considered to have been collected under wet conditions when the site received more than 0.01 inches of rain within 24 h prior to sample collection.
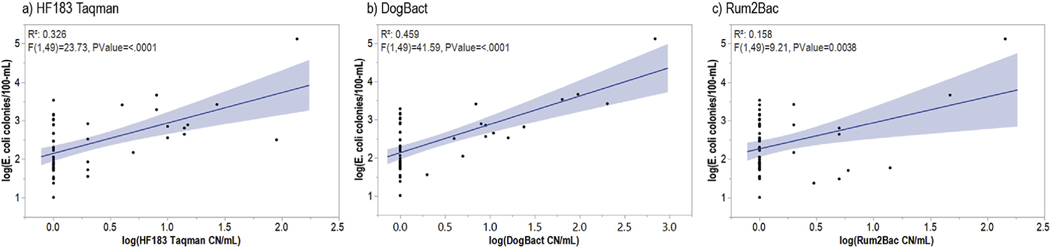
Concentrations of all three MST markers were positively correlated with E. coli (Fig. 4). DogBact was the most strongly correlated MST marker with E. coli and was the only MST marker in the best predictive model of E. coli concentrations (S4 Table). DogBact was also the best predictor of E. coli concentrations at each individual BacteriALERT site (S4 Table) indicating the strong correlation between DogBact and E. coli was not driven by a single site.
Fig. 4.
Correlations between E. coli and microbial source tracking marker concentrations for a) humans, b) dogs, and c) ruminants from three BacteriALERT sites in the Chattahoochee River National Recreation Area. Blue shading represents the 95% confidence interval for predicted values. (For interpretation of the references to colour in this figure legend, the reader is referred to the Web version of this article.)
3.3. Recursive partitioning of human health indicators
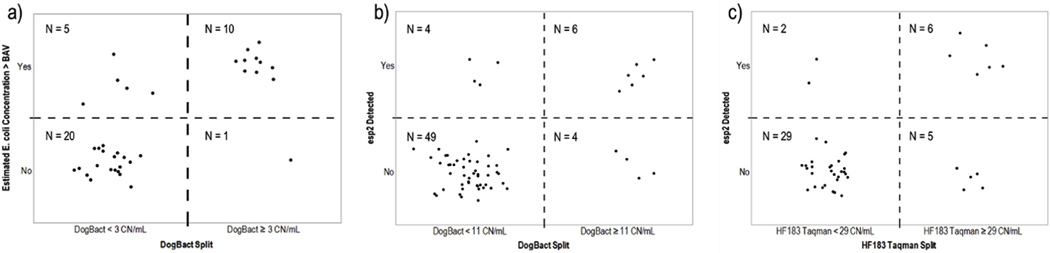
Recursive partitioning showed that DogBact was the MST marker that best explained model-based E. coli BAV exceedances (R2 = 0.347). Samples with concentrations of DogBact greater than or equal to 3 CN/mL at USGS 02335000 and USGS 02336000 had an 87% probability of exceeding the E. coli BAV, whereas samples with DogBact concentrations less than 3 CN/mL had a 21% probability of exceeding the E. coli BAV. The DogBact split correctly identified 10 of 15 instances of estimated E. coli concentrations exceeding BAV and 20 of 21 instances of E. coli estimates below BAV (Fig. 5a) totaling 83% correct predictions.
Fig. 5.
Recursive partitioning confusion matrices for a) model-based estimates of E. coli concentrations exceeding the U.S. Environmental Protection Agency beach action value (BAV, 235 colonies/100-mL) concurrently timed with sample collection at two BacteriALERT sites, b) esp2 (pathogenic Enterococcus) detection in samples from three BacteriALERT sites, and c) esp2 detection in samples from 11 synoptic sites. Possible explanatory variables included microbial source tracking (MST) markers for humans (HF183 Taqman), dogs (DogBact), and ruminants (Rum2Bac). The splitting value for the best explanatory marker was determined by a partitioning algorithm that finds the explanatory variable split that maximizes the difference in the response frequencies between the two nodes of the split. MST marker concentrations units are copies per mL.
At BacteriALERT sites, esp2 detection was also best explained by DogBact, but with poor predictive power (R2 = 0.240). DogBact concentrations 11 CN/mL in samples from BacteriALERT sites had a 56% probability of esp2 detection, whereas the probability of esp2 detection in samples with DogBact concentrations <11 CN/mL was only 8%. The DogBact split at 11 CN/mL for samples from the three BacteriALERT sites resulted in correct classification of esp2 detection for 55 of 63 samples, 49 of which were for correct classification of non-detects. False-positives and false-negatives were split for the remaining samples (Fig. 5b). At synoptic sites, esp2 detection was best explained by HF183 Taqman, but with poor predictive power (R2 = 0.266) and a misclassification rate of 17% (Fig. 5c). There was essentially no difference in the likelihood of esp2 detection or non-detection for HF183 Taqman concentrations greater than or equal to 29 CN/mL (52% probability of esp2 detection for HF183 Taqman ≥29 CN/mL) at synoptic sites; however, HF183 Taqman concentrations less than 29 CN/mL were associated with a 93% probability of not detecting esp2.
3.4. Synoptic sample associations with surrounding land use
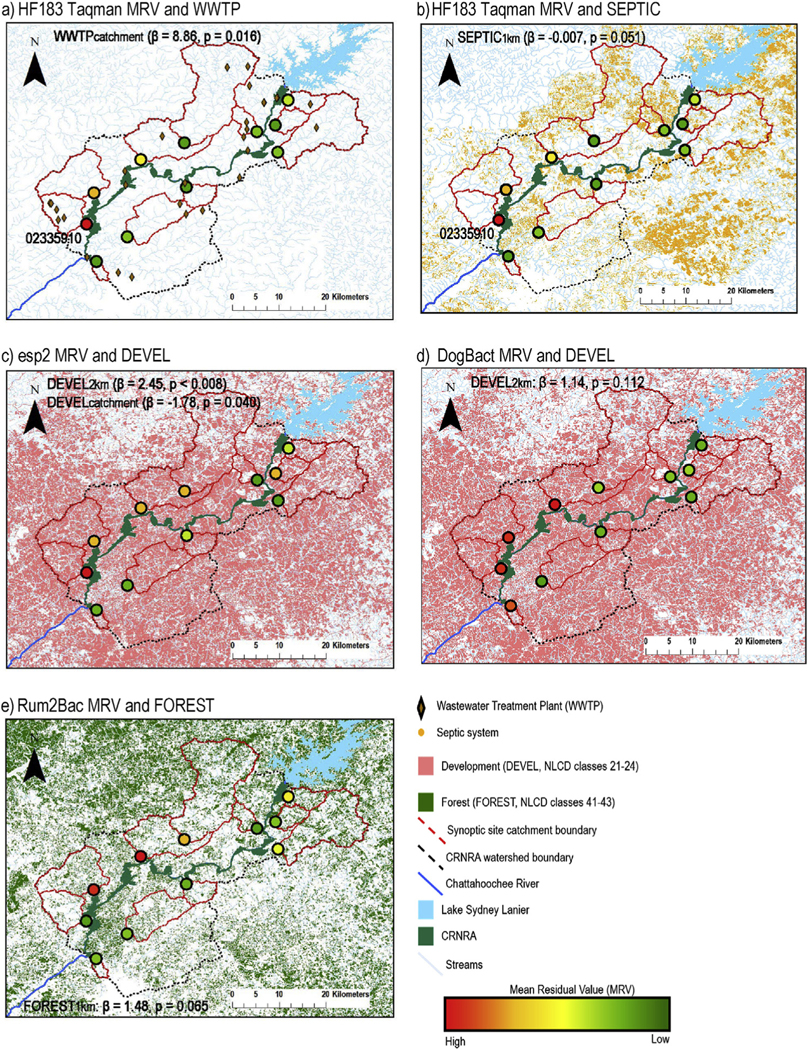
The top season and precipitation models differed for all MST markers at synoptic sites and indicated higher HF183 Taqman levels and esp2 detections in the winter and increasing Rum2Bac concentrations with increasing 72-h antecedent precipitation (S5 Table). For all markers, the sites with the highest marker level were in the downstream half of the CRNRA and on the west side of the CRNRA (Fig. 6). The top land use model for HF183 Taqman was WWTPcatchment (β = 8.86, p = 0.016; Fig. 6a) + SEPTIC1km (β=0.007, p = 0.051; Fig. 6b, S6 Table). The relationships between HF183 Taqman MRV and the two explanatory variables in the top model were largely driven by a single site, USGS 02335910 (Fig. 6a). The top model for esp2 included DEVEL at two spatial scales (S6 Table), but with opposite directional effects between the two scales (DEVEL2km: β= 2.45, p = 0.008, Fig. 6c; and DEVELcatchment: β = 1.78, p = 0.040, Fig. 6c). The directions of association for explanatory variables in the top models of DogBact (DEVEL2km: β = 1.13, p = 0.112, Fig. 6d) and Rum2Bac (FOREST1km: β = 1.48, p = 0.065, Fig. 6e) were in line with our hypotheses (Table 2); however, the associations were not significant.
Fig. 6.
Synoptic site locations in the Chattahoochee River National Recreation Area Watershed, land use, and mean residuals values (MRVs) of microbial source tracking (MST) marker concentrations after modeling the effects of season and precipitation on MST marker concentrations. Land use maps are for explanatory variables included in the top models (β indicates the effect size and direction of the variable) of MRVs for the MST markers a-b) HF183 Taqman, c) esp2, d) DogBact, e) and Rum2Bac. The HF183 Taqman MRV (a–b) at site 02335910 was an outlier that drove the relationships with WWTP and DEVEL in the top model.
4. Discussion
Escherichia coli is a commonly used indicator of the human health risk from exposure to fecal-associated pathogens in recreational waters, but standard methods for quantifying E. coli have an 18e24 h delay between sample collection and public notification of potentially harmful conditions. Model-based estimates of E. coli from continuous water quality monitoring can reduce this delay by providing the public with notification of harmful conditions in real-time. However, E. coli occurs across a wide range of animal species and does not provide information about sources of fecal contamination, which is critical information for developing effective mitigation and public outreach plans. Further, while E. coli has been demonstrated to be a relatively accurate surrogate of morbidity rates when the primary source of fecal contamination is human, the health risk from exposure to non-human contamination is poorly understood (U.S. Environmental Protection Agency, 2012). Viral pathogens, which are often host-specific (Webby et al., 2004), are believed to be the primary causes of GI illness for swimmers in wastewater impacted recreational waters swimmers (Sinclair et al., 2009; Soller et al. 2010a), Given the host-specificity of many viral pathogens, exposure to non-human contamination in recreational waters may be a lower human health risk than exposure to waters impacted by human fecal contamination. Therefore, assessing sources of contamination in recreational waters is important for directing outreach and mitigation efforts as well as determining whether further research is needed to accurately assess human health risks if the primary source of fecal contamination is not human. Our study has indicated that MST analysis can be combined with model-based estimates of E. coli concentrations to determine sources of contamination and MST marker concentrations predictive of E. coli estimates that suggest a human health risk.
We investigated the frequency and magnitude of concentrations of three MST markers and one pathogenic marker across the CRNRA watershed to assess seasonal and precipitation-associated patterns in the sources of fecal contamination impacting the watershed. In addition, we identified land use characteristics associated with the presence of MST markers in tributaries of the Chattahoochee River located within the CRNRA. A common challenge with MST markers is the lack of specificity or sensitivity. We addressed this issue by using Bacteriodales markers determined to be sensitive and specific for humans (HF183MGB, Green et al. 2014a) and ruminants (Rum2Bac, Boehm et al. 2013), and supplementing a dog marker that has demonstrated high sensitivity and low specificity (DogBact, Boehm et al. 2013) with a more specific and potentially less sensitive dog marker (DG3, Green et al. 2014b). While we cannot rule out cross-amplification with other sources by DogBact, the strong correlation between DG3 and DogBact estimates suggest that DogBact was primarily detecting dog fecal material (although neither marker has been tested on coyotes, which do occur in the Atlanta metropolitan area). We also screened for presence of the esp gene associated with pathogenic strains of E. faecium and E. faecalis, which is also generally associated with human contamination but has been detected in other species, including pigs and dog, at lower frequencies than human sewage or septage (Whitman et al. 2007).
Among the three sources we tested for, humans and dogs were detected most frequently. The human and dog markers were detected at levels ≥10 CN/mL at all but two study sites, indicating that the origins of human and dog contamination are not specific to single locations in the CRNRA watershed. Dogs in particular appeared to be a primary contributor of fecal contamination in the CRNRA as indicated by a strong correlations with culturable E. coli and as the best explanatory variable for E. coli BAV exceedances from model-based estimates. We found no relationship between land use variables and DogBact concentrations at the synoptic sites, suggesting the variables we analyzed were not representative of the number or density of dogs within a 1-km or 2-km buffer, or within the catchment. More direct indicators of dog presence, such dog park density, may provide better representation of dog density in a given area.
Dog contamination was higher in the winter and under wet conditions, which is consistent with contamination caused by runoff. We also saw higher levels of dog contamination at the middle and downstream BacteriALERT sites in winter-wet conditions. Riedel et al. (2015) also found elevated levels of dog contamination in the winter at Topanga Beach, CA and attributed it to changes in owner behavior. Dogs were not allowed at Topanga Beach; however the authors suggested that illegal dog walking increased in the winter as a result decreased lifeguard hours or lessened social pressure from fewer visitors to the Beach in the winter. While dogs are allowed in the CRNRA, they are required to be kept on leash. Further investigation of the origins (e.g. within the CRNRA, dog parks, or residential areas) of dog fecal contamination may be warranted to determine if contamination is originating from within or outside the Park.
Human contamination levels were higher in samples collected during wet conditions. When groundwater levels are above septic systems, they are more likely to fail (Cogger and Carlile, 1984), and when groundwater levels are above sewer lines, groundwater infiltration to sewer systems can occur (Wittenberg and Aksoy, 2010; Thorndahl et al., 2016), resulting in capacity exceedences for the systems as well as for treatment processes at WWTPs. Modeling of land use features associated with the human marker suggested sewer infrastructure as plausible origins of human contamination in this study. After accounting for the seasonal and precipitation effects on HF183 Taqman concentrations at synoptic sites, the human MST marker was significantly associated with increasing WWTP density in the catchment, which we used as a surrogate for sewer line density. This relationship with WWTP density was primarily driven by one site, suggesting possible conveyance issues in that catchment. While not significant, it is worth noting that the top land use model for HF183 Taqman indicated a near significant (p = 0.051) negative correlation between HF183 Taqman and septic system density within 1 km. Peed et al. (2011) found a strong negative correlation between septic and sewer line densities, suggesting the negative correlation between HF183 Taqman and septic system density may indicate a positive correlation with sewer line density. HF183 Taqman was frequently detected at levels below LOQ in samples collected during dry conditions, suggesting chronic low levels of human contamination due to leaking sewer lines across the CRNRA watershed.
Recursive partitioning results for esp2 at synoptic sites suggested that humans are a source of esp2 in tributaries in the CRNRA watershed. While the correlation strength was relatively low, the predictive split for esp2 detection at synoptic sites of HF183 Taqman 29 CN/mL is similar to the 32.2 to 42 3220 to 4200 HF183 gene copies (GC) per 100 mL (3220 to 4200 GC/100 mL; Bernhard and Field, 2000b; Seurinck et al. 2005) determined by quantitative microbial risks assessments to be a benchmarks for 30 to 36 gastrointestinal (GI) illnesses/1000 swimmers/swimming event in recreational waters contaminated with untreated and secondary treated sewage (Boehm et al., 2015; Ahmed et al. 2018). This QMRA-based benchmark is essentially the same as the benchmark GI illness rates (36 illnesses/1000 swimmers/swimming event) used to determine the E. coli BAV threshold (U.S. Environmental Protection Agency, 2012), suggesting further research is warranted to determine if and under what conditions do HF183 Taqman concentrations 29 CN/mL indicate a human health risk similar to the E. coli BAV.
Humans as a source of esp2 at synoptic sites is further supported by the positive association between esp2 and development within 2-km upstream of the sampling location. The negative correlation between esp2 and development at the catchment scale is counterintuitive as values of DEVEL2km and DEVELcatchment were positively correlated. However, the inverse relationship between esp2 and development at the 2-km and catchment scales may indicate that multiple sources are contributing to esp2 presence at synoptic sites. While the esp gene occurs at only about a 30% rate in septic waste (Whitman et al. 2007), the negative correlation between development and septic system density at the catchment scale supports the possibility that human contamination originating from septic systems further upstream in the watershed may be sources of esp2. The esp gene is even less frequent in dog waste (~21%; Whitman et al. 2007); however, recursive partitioning of esp2 detection at BacteriALERT sites suggests dogs were a source of esp2 in the CRNRA therefore also possibly sources of esp2 in streams within the CRNRA watershed.
While levels of Rum2Bac were generally lower than HF183 Taqman or DogBact, infrequent detections of Rum2Bac ≥10 CN/mL suggests that deer are occasional sources of fecal contamination in the CRNRA. Cha et al. (2016) found that fecal indicator bacteria levels in forested areas increased in response to temperature under both wet and dry conditions, potentially because of an increase in wildlife activity. Given that deer contamination in the Chattahoochee River likely would be from surface water runoff, we would expect to see an increase in contamination levels in wet samples. If deer activity increased with warmer temperatures, we would expect that Rum2Bac contamination levels would be highest in summer-wet samples. This pattern did not appear to be the case for deer in our study. While Rum2Bac concentrations were positively associated with rainfall within the antecedent 72 h at synoptic sites, they did not significantly differ across seasons or rain conditions at the three BacteriALERT sites.
Of the three MST markers tested, the dog MST marker was the best predictor of BAV exceedances, demonstrating that dog fecal contamination is in part periodically responsible for Park cessation of water-based activities. The error in DogBact predictions of BAV exceedances tended towards false negatives (i.e., BAV was exceeded but DogBact concentrations were below 3 CN/mL) indicating that when DogBact concentrations are greater than or equal to 3 CN/mL, E. coli concentrations are likely greater than 235 cfu/100 mL. The association between DogBact concentrations and esp2 detections in the CRNRA indicate that swimming in recreational waters contaminated by dog waste is potentially a human health risk. Further research into the human health risks from exposure to dog contamination would help the Park determine if the E. coli BAV is an appropriate surrogate of human health risk when dogs are primary sources of contamination, and if not, to assess potential alternative surrogate indicators that provide more relevant indices.
5. Conclusions
While several samples from sites within the CRNRA had human marker concentrations indicative of a risk to human health, these levels were only detected in the off season for the park, indicating a lower likelihood of guests being exposed to unsafe levels of contamination in the water than if these levels had occurred in samples collected during peak visitation season in the summer.
For sites outside the CRNRA, human MST marker concentrations were associated with wastewater treatment plant density. However, this relationship was dictated primarily by one site, suggesting possible conveyance issues in the associated catchment. Future studies may target this catchment to assess possible point or distributed sources of sewage impact.
Because the dog MST marker was the best predictor of E. coli concentrations in the CRNRA, we assume that dog fecal material contributes to a significant portion of the fecal contamination at these locations. While a supplementary assay provided evidence of specificity of the dog marker in this study, previous studies have demonstrated low marker specificity indicating a need to be cautious about designating dogs as the major source of E. coli in the CRNRA.
Dog fecal contamination was the best predictor of USEPA E. coli beach action value exceedances, indicating dogs are in part, periodically responsible for Park cessation of water-based activities. Determining the human health outcomes from primary contact with dog contamination in recreational waters would assist the development of more appropriate indicators of human health risk in recreational waters for which dogs are the primary sources of fecal contamination.
Supplementary Material
Acknowledgements
We are grateful for the assistance of numerous people throughout the duration of this project. Alan Cressler, Cristal Younker, Daniel Calhoun, Daniel McCay, Elliot Stahl, Kristina Bowen, Marcella Cruz, Robert Forde, Robert Everett, Russell Mclester, Samantha Kephart, and Skylar McHenry assisted with sample collection and processing. John Joiner, Chris Smith, Javier Spencer, and Paul Ankcorn assisted with coordinating personnel and field equipment. Jessica Sterling and Jason Ulseth provided input on sample site locations and assisted with sampling logistics. Joseph Duris was an invaluable advisor for the molecular work and provided a thoughtful and thorough review. Ourania Georgacopoulos, Blake Snyder, and Robert Sowah provided advice and assisted with troubleshooting for the molecular laboratory work. This project was supported by the U.S. Geological Survey-National Park Service Water Quality Partnership Program. This document has been reviewed in accordance with U.S Environmental Protection Agency policy and approved for publication. Any mention of trade names, manufacturer or products does not imply an endorsement by the United States Government or the U.S. Environmental Protection Agency. USEPA, USGS, NPS and their employees do not endorse any commercial products. We thank three anonymous reviewers, Dan Calhoun, and Sandra Cooper for their thoughtful feedback that helped improve this manuscript.
Footnotes
Data availability
Quantitative PCR and E. coli data collected for this study are available in the USGS ScienceBase-Catalog (https://doi.org/10.5066/P957P46S), which the authoritative copy of these data. Supplementary Material S2 File contains redundant qPCR data, formatted as an input file for the R code (File S1). Quantitative PCR marker concentration estimates in Table S1 were calculated from the output of File S1. Additional data presented in the Supplementary Material obtained through other sources are noted in the file descriptions.
Declaration of competing interest
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Appendix A. Supplementary data
Supplementary data to this article can be found online at https://doi.org/10.1016/j.watres.2019.115435.
Any use of trade, firm, or product names is for descriptive purposes only and does not imply endorsement by the U.S. Government.
References
- Ahmed W, Hamilton KA, Lobos A, Hughes B, Staley C, Sadowsky MJ, Harwood VJ, 2018. Quantitative microbial risk assessment of microbial source tracking markers in recreational water contaminated with fresh untreated and secondary treated sewage. Environ. Int 117, 243–249. [DOI] [PubMed] [Google Scholar]
- Aho K, Derryberry D, Peterson T, 2014. Model selection for ecologists: the worldviews of AIC and BIC. Ecology 95, 631–636. [DOI] [PubMed] [Google Scholar]
- Allsop K, Stickler J, 1985. An assessment fo Bacteroides fragilis group organisms as indicators of human faecal pollution. J. Appl. Bacteriol 58, 95–99. [DOI] [PubMed] [Google Scholar]
- Bernhard A, Field K, 2000a. A PCR assay to discriminate human and ruminant feces on the basis of host differences in Bacteroidese-Prevotella genes encoding 16S rRNA. Appl. Environ. Microbiol 66, 4571–4574. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bernhard AE, Field KG, 2000b. A PCR assay to discriminate human and ruminant feces on the basis of host differences in Bacteroides-Prevotella genes encoding 16S rRNA. Appl. Environ. Microbiol 66, 4571–4574. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beversdorf LJ, Bornstein-Forst SM, McLellan SL, 2007. The potential for beach sand to serve as a reservoir for Escherichia coli and the physical influences on cell die-off. J. Appl. Microbiol 102, 1372–1381. [DOI] [PubMed] [Google Scholar]
- Boehm AB, Soller JA, Shanks OC, 2015. Human-associated fecal quantitative polymerase chain reaction measurements and simulated risk of gastrointestinal illness in recreational waters contaminated with raw sewage. Environ. Sci. Technol. Lett 2, 270–275. [Google Scholar]
- Boehm AB, Van De Werfhorst LC, Griffith JF, Holden PA, Jay JA, Shanks OC, Wang D, Weisberg SB, 2013. Performance of forty-one microbial source tracking methods: a twenty-seven lab evaluation study. Water Res. 47, 6812–6828. [DOI] [PubMed] [Google Scholar]
- Bradshaw JK, Snyder BJ, Oladeinde A, Spidle D, Berrang ME, Meinersmann RJ, Oakley B, Sidle RC, Sullivan K & Molina M Characterizing relationships among fecal indicator bacteria, microbial source tracking markers, and associated waterborne pathogen occurrence in stream water and sediments in a mixed land use watershed. Water Res.. [DOI] [PubMed] [Google Scholar]
- Bradshaw JK, Snyder BJ, Oladeinde A, Spidle D, Berrang ME, Meinersmann RJ, Oakley B, Sidle RC, Sullivan K, Molina M, 2016. Characterizing relationships among fecal indicator bacteria, microbial source tracking markers, and associated waterborne pathogen occurrence in stream water and sediments in a mixed land use watershed. Water Res. 101, 498–509. [DOI] [PubMed] [Google Scholar]
- Burnham KP, Anderson DR, 2004. Multimodel inference: understanding AIC and BIC in model selection. Sociol. Methods Res 33, 261–304. [Google Scholar]
- Byappanahalli MN, Nevers MB, Korajkic A, Staley ZR, Harwood VJ, 2012a. Enterococci in the environment. Microbiol. Mol. Biol. Rev.: MMBR 76, 685–706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Byappanahalli MN, Roll BM, Fujioka RS, 2012b. Evidence for occurrence, persistence, and growth potential of Escherichia coli and Enterococci in Hawaii’s soil environments. Microb. Environ 27, 164–170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Calderon RL, Mood EW, Dufour AP, 1991. Health effects of swimmers and nonpoint sources of contaminated water. Int. J. Environ. Health Res 1, 21–31. [DOI] [PubMed] [Google Scholar]
- Cha Y, Park M-H, Lee S-H, Kim JH, Cho KH, 2016. Modeling spatiotemporal bacterial variability with meteorological and watershed land-use characteristics. Water Res. 100, 306–315. [DOI] [PubMed] [Google Scholar]
- Clarke JS, Painter JA, 2014. Influence of Septic Systems on Stream Base Flow in the Apalachicolae-Chattahoocheee-Flint River Basin Near Metropolitan Atlanta, Georgia, 2012. U.S. Geological Survey Scientific Investigations, p. 68. Report 2014–5144. [Google Scholar]
- Cogger CG, Carlile BL, 1984. Field performance of conventional and alternative septic systems in wet Soils1. J. Environ. Qual 13, 137–142. [Google Scholar]
- Colford JMJ, Wade TJ, Schiff KC, Wright CC, Griffith JF, Sandhu SK, Burns S, Sobsey M, Lovelace G, Weisberg SB, 2007. Water quality indicators and the risk of illness at beaches with nonpoint sources of fecal contamination. Epidemiology 18, 27–35. [DOI] [PubMed] [Google Scholar]
- Dick LK, Simonich MT, Field KG, 2005. Microplate subtractive hybridization to enrich for Bacteroidales genetic markers for fecal source identification. Appl. Environ. Microbiol 71, 3179–3183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Forootan A, Sjo€back R, Bjo€rkman J, Sjo€green B, Linz L, Kubista M, 2017. Methods to determine limit of detection and limit of quantification in quantitative real-time PCR (qPCR). Biomol. Detect. Quantification 12, 1–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gentry RW, McCarthy J, Layton A, McKay LD, Williams D, Koirala SR, Sayler GS, 2006. Escherichia coli loading at or near base flow in a mixed-use watershed. J. Environ. Qual 35, 2244–2249. [DOI] [PubMed] [Google Scholar]
- Green HC, Haugland RA, Varma M, Millen HT, Borchardt MA, Field KG, Walters WA, Knight R, Sivaganesan M, Kelty CA, 2014a. Improved HF183 quantitative real-time PCR assay for characterization of human fecal pollution in ambient surface water samples. Appl. Environ. Microbiol 80, 3086–3094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Green HC, White KM, Kelty CA, Shanks OC, 2014b. Development of rapid canine fecal source identification PCR-based assays. Environ. Sci. Technol 48, 11453–11461. [DOI] [PubMed] [Google Scholar]
- Haugland RA, Varma M, Sivaganesan M, Kelty C, Peed L, Shanks OC, 2010. Evaluation of genetic markers from the 16S rRNA gene V2 region for use in quantitative detection of selected Bacteroidales species and human fecal waste by qPCR. Syst. Appl. Microbiol 33, 348–357. [DOI] [PubMed] [Google Scholar]
- Homer CG, Dewitz JA, Yang L, Jin S, Danielson P, Xian G, Coulston J, Herold ND, Wickham JD, Megown K, 2015. Completion of the 2011 National Land Cover Database for the conterminous United States-Representing a decade of land cover change information. Photogramm. Eng. Remote Sens 81, 345–354. [Google Scholar]
- Lawrence SJ, 2012. Escherichia coli Bacteria Density in Relation to Turbidity, Streamflow Characteristics, and Season in the Chattahoochee River Near Atlanta, Georgia, October 2000 through September 2008dDescription, Statistical Analysis, and Predictive Modeling. U.S. Geological Survey Scientific Investigations. Report, pp. 99. [Google Scholar]
- Long SC, Plummer JD, 2004. Assessing land use impacts on water quality using microbial source tracking. J. Am. Water Resour. Assoc 40, 1433–1448. [Google Scholar]
- McBride GB, Salmond CE, Bandaranayake DR, Turner SJ, Lewis GD, Till DG, 1998. Health effects of marine bathing in New Zealand. Int. J. Environ. Health Res 8, 173–189. [Google Scholar]
- McKee AM, 2019. Microbial Source Tracking Marker Concentrations in the Chattahoochee River National Recreation Area Watershed in 2015–2017. U.S. Geological Survey data release, Georgia, USA 10.5066/P957P46S. [DOI] [Google Scholar]
- Meays C, Broersma K, Nordin R, Mazumder A, 2004. Source tracking fecal bacteria in water: a critical review of current methods. J. Environ. Manag 73, 71–79. [DOI] [PubMed] [Google Scholar]
- Medina RA, García-Sastre A, 2011. Influenza A viruses: new research developments. Nat. Rev. Microbiol 9, 590–603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mieszkin S, Yala JF, Joubrel R, Gourmelon M, 2010. Phylogenetic analysis of Bacteroidales 16S rRNA gene sequences from human and animal effluents and assessment of ruminant faecal pollution by real-time PCR. J. Appl. Microbiol 108, 974–984. [DOI] [PubMed] [Google Scholar]
- Myers DN, Stoeckel DM, Bushon RN, Francy DS, Brady AMG, 2014. Fecal Indicator Bacteria (V. 2.1). U.S. Geological Survey Techniques of Water-Resources Investigations. Book 9, chap. A7, section 7.1, May 2014. [Google Scholar]
- Peed LA, Nietch CT, Kelty CA, Meckes M, Mooney T, Sivaganesan M, Shanks OC, 2011. Combining land use information and small stream sampling with PCR-based methods for better characterization of diffuse sources of human fecal pollution. Environ. Sci. Technol 45, 5652–5659. [DOI] [PubMed] [Google Scholar]
- R Core Team, 2017. R: A Language and Environment for Statistical Computing. R Foundation for Statistical Computing, Vienna, Austria. URL. https://www.Rproject.org/. [Google Scholar]
- Riedel TE, Thulsiraj V, Zimmer-Faust AG, Dagit R, Krug J, Hanley KT, Adamek K, Ebentier DL, Torres R, Cobian U, 2015. Long-term monitoring of molecular markers can distinguish different seasonal patterns of fecal indicating bacteria sources. Water Res. 71, 227–243. [DOI] [PubMed] [Google Scholar]
- Schoen ME, Ashbolt NJ, 2010. Assessing pathogen risk to swimmers at nonsewage impacted recreational beaches. Environ. Sci. Technol 44, 2286–2291. [DOI] [PubMed] [Google Scholar]
- Schwarz G, 1978. Estimating the dimension of a model. Ann. Stat 6, 461–464. [Google Scholar]
- Scott TM, Jenkins TM, Lukasik J, Rose JB, 2005. Potential use of a host associated molecular marker in Enterococcus faecium as an index of human fecal pollution. Environ. Sci. Technol 39, 283–287. [PubMed] [Google Scholar]
- Seurinck S, Defoirdt T, Verstraete W, Siciliano SD, 2005. Detection and quantification of the human-specific HF183 Bacteroides 16S rRNA genetic marker with real-time PCR for assessment of human faecal pollution in freshwater. Environ. Microbiol 7, 249–259. [DOI] [PubMed] [Google Scholar]
- Shankar V, Baghdayan AS, Huycke MM, Lindahl G, Gilmore MS, 1999. Infection-derived Enterococcus faecalis strains are enriched in esp, a gene encoding a novel surface protein. Infect. Immun 67, 193–200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shanks OC, Nietch C, Simonich M, Younger M, Reynolds D, Field KG, 2006. Basin-wide analysis of the dynamics of fecal contamination and fecal source identification in Tillamook Bay, Oregon. Appl. Environ. Microbiol 72, 5537–5546. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shibata T, Solo-Gabriele HM, Sinigalliano CD, Gidley ML, Plano LRW, Fleisher JM, Wang JD, Elmir SM, He G, Wright ME, Abdelzaher AM, Ortega C, Wanless D, Garza AC, Kish J, Scott T, Hollenbeck J, Backer LC, Fleming LE, 2010. Evaluation of conventional and alternative monitoring methods for a recreational marine beach with nonpoint source of fecal contamination. Environ. Sci. Technol 44, 8175–8181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sinclair RG, Jones EL, Gerba CP, 2009. Viruses in recreational water-borne disease outbreaks: a review. J. Appl. Microbiol 107, 1769–1780. [DOI] [PubMed] [Google Scholar]
- Soller J, Bartrand T, Ravenscroft J, Molina M, Whelan G, Schoen M, Ashbolt N, 2015. Estimated human health risks from recreational exposures to stormwater runoff containing animal faecal material. Environ. Model. Softw 72, 21–32. [Google Scholar]
- Soller JA, Bartrand T, Ashbolt NJ, Ravenscroft J, Wade TJ, 2010a. Estimating the primary etiologic agents in recreational freshwaters impacted by human sources of faecal contamination. Water Res. 44, 4736–4747. [DOI] [PubMed] [Google Scholar]
- Soller JA, Schoen ME, Bartrand T, Ravenscroft JE, Ashbolt NJ, 2010b. Estimated human health risks from exposure to recreational waters impacted by human and non-human sources of faecal contamination. Water Res. 44, 4674–4691. [DOI] [PubMed] [Google Scholar]
- Soller JA, Schoen ME, Varghese A, Ichida AM, Boehm AB, Eftim S, Ashbolt NJ, Ravenscroft JE, 2014. Human health risk implications of multiple sources of faecal indicator bacteria in a recreational waterbody. Water Res. 66, 254–264. [DOI] [PubMed] [Google Scholar]
- St Laurent J, Mazumder A, 2014. Influence of seasonal and inter-annual hydrometeorological variability on surface water fecal coliform concentration under varying land-use composition. Water Res. 48, 170–178. [DOI] [PubMed] [Google Scholar]
- Templar HA, Dila DK, Bootsma MJ, Corsi SR, McLellan SL, 2016. Quantification of human-associated fecal indicators reveal sewage from urban watersheds as a source of pollution to Lake Michigan. Water Res. 100, 556–567. [DOI] [PubMed] [Google Scholar]
- Thorndahl S, Balling JD, Larsen UBB, 2016. Analysis and integrated modelling of groundwater infiltration to sewer networks. Hydrol. Process 30, 3228–3238. [Google Scholar]
- U.S. Environmental Protection Agency, 2006. Method 1600: Enterococci in Water by Membrane Filtration Using Membrane-Enterococcus Indoxyl-SS-D-Glucoside Agar (mEI). U.S. Environmental Protection Agency. Report 821-R-06–009. [Google Scholar]
- U.S. Environmental Protection Agency, 2012. Recreational Water Quality Criteria. Health and Ecological Criteria Division, Office of Science and Technology. U.S. Environmental Protection Agency. EPA 820-F-12–058. [Google Scholar]
- National Field Manual for the Collection of Water-Quality Data: U.S. Geological Survey Techniques of Water-Resources Investigations, book 9, chaps. A1-A10, available online at: http://pubs.water.usgs.gov/twri9A. [Google Scholar]
- Venables WN, Ripley BD, 2002. Modern Applied Statistics with S, fourth ed. Springer, New York. [Google Scholar]
- Webby R, Hoffmann E, Webster R, 2004. Molecular constraints to interspecies transmission of viral pathogens. Nat. Med 10, S77–S81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Whitman RL, Przybyla-Kelly K, Shively DA, Byappanahalli MN, 2007. Incidence of the enterococcal surface protein (esp) gene in human and animal fecal sources. Environ. Sci. Technol 41, 6090–6095. [DOI] [PubMed] [Google Scholar]
- Wittenberg H, Aksoy H, 2010. Groundwater intrusion into leaky sewer systems. Water Sci. Technol 62, 92–98. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.