Abstract
Long non-coding RNA ILF3 divergent transcript (ILF3-AS1) displays a tumor-suppressing effect. StarBase predicted that the potential target microRNA (miR) of ILF3-AS1 was miR-454-3p; therefore, the present study investigated the effect of ILF3-AS1 and its target miR-454-3p on cervical cancer (CC). Gene Expression Profiling Interactive Analysis was used to predict the expression of ILF3-AS1 in CC and the overall survival rate of patients. The present study demonstrated that ILF3-AS1 expression was significantly downregulated in human CC tissues and cells compared with adjacent tissues (ANTs) and normal cervical epithelial cells (NCEs), respectively. Patients with CC with high ILF3-AS1 expression displayed higher survival rates compared with patients with low ILF3-AS1 expression. Cell viability, apoptosis, migration and invasion were detected by performing Cell Counting Kit-8, flow cytometry, wound healing and Transwell assays, respectively. Compared with the negative control (NC) group, ILF3-AS1 overexpression significantly inhibited CC cell viability and migration, but significantly increased CC cell apoptosis. Moreover, ILF3-AS1 overexpression significantly upregulated E-Cadherin expression levels, but significantly downregulated N-Cadherin and snail family transcriptional repressor 1 expression levels compared with the NC group. miR-454-3p expression was negatively correlated with ILF3-AS1, and highly expressed in CC tissues and cells compared with ANTs and NCEs, respectively. PTEN, which was predicted and verified as the target gene for miR-454-3p, was significantly downregulated in CC tissues and cells compared with ANTs and NCEs, respectively. ILF3-AS1 expression was positively correlated with PTEN expression, and ILF3-AS1 overexpression partially reversed the inhibitory effect of miR-454-3p on PTEN expression. In conclusion, the present study indicated that ILF3-AS1 inhibited CC cell proliferation and migration, and promoted CC cell apoptosis by inhibiting epithelial-mesenchymal transition, and ILF3-AS1 overexpression partially reversed the inhibitory effect of miR-454-3p on PTEN expression.
Keywords: cervical cancer, lncRNA ILF3 divergent transcript, microRNA-454-3p, PTEN
Introduction
Cervical cancer (CC) is the second most common malignant tumor among women in Latin America (1,2). The mortality of CC in developing countries is ~80%, with ~106,000 new cases and ~48,000 deaths in China in 2018 (3). A large number of studies have demonstrated that CC is induced by several factors, including bacterial infections, sexual disorders, multiple pregnancies and long-term oral contraceptives (4–6). Harald zur Hausen demonstrated for the first time that human papilloma virus (HPV) infection is a key cause of CC (7,8). Although the development and application of HPV vaccines has increased in popularity, the occurrence and development of tumors is a complex regulation process involving multiple genes (9); therefore, activation of proto-oncogenes and inactivation of tumor suppressor genes may serve as a promising therapeutic strategy in CC.
Long non-coding RNAs (lncRNAs) are a type of RNA that lack protein-encoding functions, and are widely involved in cell proliferation, differentiation and apoptosis (10). lncRNAs are >200 nucleotides in length (11) and the majority of non-protein coding sequences are transcribed into lncRNAs (12). lncRNAs are tissue- and cell-specific, and can regulate the growth and development of single-cell eukaryotes, embryonic stem cells and adult stem cells (13). Abnormal expression or functions of lncRNAs are closely related to the occurrence and development of cancer (14–16). An integrated analysis of lncRNA-associated networks in CC indicated that several lncRNAs might serve as novel molecular markers for the diagnosis and prognosis of CC (17). Increasing evidence demonstrates that lncRNAs, such as XLOC_006390 (18), small nucleolar RNA host gene 1 (19), are important regulators of CC progression. However, the functions of lncRNAs in CC are not completely understood.
ILF3 divergent transcript (ILF3-AS1) is an lncRNA located on chromosome 19p 13.2 (20). lncRNA ILF3-AS1 is highly expressed in melanoma cells, promoting cell proliferation, migration and invasion via negative regulation of microRNA (miR/miRNA)-200b/a/429, which suggests that ILF3-AS1 may serve as a potential prognostic biomarker and therapeutic target for melanoma (20,21). In addition, ILF3-AS1 expression is significantly upregulated in osteosarcoma tissues and cells, which promotes osteosarcoma development by regulating the miR-212/SOX5 axis (22). Moreover, ILF3-AS1 is a potential biomarker for colon (23) and prostate (24) cancer. Similarly, previous studies reported that ILF3-AS1 might serve as a prognostic biomarker for CC (25,26). However, the specific regulatory mechanism underlying ILF3-AS1 in CC is not completely understood, thus, the present study investigated the effects and mechanisms underlying ILF3-AS1 in CC.
Materials and methods
Ethics statement
All patients provided written informed consent. The study was approved by the Ethics committee of Guangdong Clifford Hospital (approval no. 2014007XHK).
CC specimen collection and survival analysis
CC tissues (15 stage I/II cases and 17 stage III/IV cases) and adjacent tissues (ANTs; distance from tumor margin, 2 cm; n=32) were collected during CC (age, 37–55 years; female patients) resection at Guangdong Clifford Hospital (Guangzhou, China) between January 2017 and December 2018. Patients with CC were categorized into stages I–IV according to the criteria of the International Society of Oncology and the International Association of Obstetricians and Gynecologists (27). The exclusion criteria were as follows: i) Combined with other malignancies; ii) receiving radiotherapy and chemotherapy for CC; and iii) and acute inflammation of the reproductive tract. Patients diagnosed with cervical cancer via pathological examination were included in the present study. Tissues were washed with sterile saline, snap-frozen in liquid nitrogen for 15 min and stored at −80°C.
Database analysis
The expression and pathological stage plots of ILF3-AS1 in cervical and endocervical cancer were obtained from the Gene Expression Profiling Interactive Analysis (GEPIA) database (gepia.cancer-pku.cn). Kaplan-Meier analysis was performed to assess the survival of patients with CC based on data obtained from the GEPIA database. Survival curves were compared using log-rank tests.
Cell culture
Normal cervical epithelial cell line (NCE; PCS-480-011) and CC cell lines, including C33A (HTB-31), ME-180 (HTB-33), SiHa (HTB-35), HeLa (CCL-2) and CaSki (CRM-CRL-1550), were purchased from American Type Culture Collection. Cells were cultured in DMEM (cat. no. 12800017; Gibco; Thermo Fisher Scientific, Inc.) supplemented with 10% FBS (cat. no. 10437085; Gibco; Thermo Fisher Scientific, Inc.) at 37°C with 5% CO2.
Transfection
The overexpression vector pcDNA 3.1 (cat. no. V86020; Invitrogen; Thermo Fisher Scientific, Inc.) containing ILF3-AS1 was constructed. SiHa and HeLa cells were transfected with 2 µg ILF3-AS1, miR-454-3p mimic, ILF3-AS1 + miR-454-3p mimic or the corresponding controls [empty vector pcDNA 3.1 plasmid and mimic control (scrambled), 5′-UAGGUCUAAAUGUCUAUUGAUGG-3′] using Lipofectamine® 3000 (cat. no. L3000015; Thermo Fisher Scientific, Inc.). Briefly, 1 µg ILF3-AS1 overexpression vector or 30 pmol miR-454-3p mimic (cat. no. 4464066; Thermo Fisher Scientific, Inc.; 5′-UAGUGCAAUAUUGCUUAUAGGGU-3′) was added to Opti-MEM® medium (cat. no. 31985062; Invitrogen; Thermo Fisher Scientific, Inc.). Subsequently, 3 µl Lipofectamine 3000 reagent was added to Opti-MEM medium. Following gentle mixing of the two mixtures at room temperature, CC cells (5×105 cells/well) were incubated with the combined mixture. Blank cells were treated with transfection reagent alone. Cells in the negative control (NC) group were transfected with empty pcDNA 3.1 plasmid. Cells in the control group were co-transfected with empty pcDNA 3.1 plasmid and mimic control. The following mimic control was used: 5′-UAGCAAGGAGGUCUAUGUUAUUU-3′. Transfection was performed at room temperature for 15 min. At 48 h post-transfection, subsequent experiments were performed.
Target gene prediction
The target miRNA of ILF3-AS1 was predicted using StarBase (starbase.sysu.edu.cn) (28). TargetScan (version 7.2; www.targetscan.org) was used to analyze miRNA-mRNA interactions (29).
Luciferase assay
The dual-luciferase reporter assay was performed to verify predictions. The following four reporter plasmids were constructed using the pmirGLO plasmid (cat. no. CL414-01; Beijing Biomed Gene Technology Co., Ltd.): i) Wild-type (WT) ILF3-AS1 (ILF3-AS1-WT); ii) mutant (MUT) ILF3-AS1 (ILF3-AS1-MUT); iii) WT PTEN (PTEN-WT); and iv) mutant PTEN (PTEN-MUT). The four reporter plasmids had the following sequences: i) ILF3-AS1-WT, 5′-AGCCGAGAUUGCUCCAUUGCACUC-3′; ii) ILF3-AS1-MUT, 5′-AGGGGUGUUUCGUCCUAACGUGAC-3′; iii) PTEN-WT, 5′-CAUUAUAAUGGGCUUUUGCACUG-3′; and iv) PTEN-MUT, 5′-CAUCGAUGUGGGCUUCCUUGACG-3′. Cells were seeded (5×105 cells/well) into 24-well plates and cultured overnight. Subsequently, SiHa and HeLa cells were co-transfected with 50 ng ILF3-AS1-WT, ILF3-AS1-MUT, PTEN-WT or PTEN-MUT reporter plasmid and 100 pmol miR-454-3p mimic or mimic control using Lipofectamine® 3000 reagent (Invitrogen; Thermo Fisher Scientific, Inc.) according to the manufacturer's instructions for 48 h at 37°C. At 48 h post-transfection, firefly and Renilla luciferase activities were detected using the Dual-luciferase Reporter Assay System (cat. no. E1910; Promega Corporation). Firefly luciferase activities were normalized to Renilla luciferase activities.
Reverse transcription-quantitative PCR (RT-qPCR)
Total RNA was extracted from tissues and cells using TRIzol® reagent (cat. no. 15596026; Invitrogen; Thermo Fisher Scientific, Inc.). Total RNA concentrations were detected using a NanoDrop 2000 spectrophotometer (Thermo Fisher Scientific, Inc.). Total RNAs were reverse transcribed into cDNA using the Revert Aid First Strand cDNA Synthesis Kit (cat. no. k1622; Thermo Fisher Scientific, Inc.). For detection of miR-454-3p expression, reverse transcription was performed using the All-in-One™ miRNA First-Reverse Transcription, Strand cDNA Synthesis Kit (cat. no. QP056; GeneCopoeia, Inc.). Both RT kits were used according to the manufacturer's protocol. Subsequently, qPCR was performed using a Veriti™ 96-Well Fast Thermal Cycler (cat. no. 4375305; Thermo Fisher Scientific, Inc.) and PowerUp™ SYBR™ Green Master Mix (cat. no. A25742; Applied Biosystems; Thermo Fisher Scientific, Inc.). The following thermocycling conditions were used for qPCR: Initial denaturation at 95°C for 10 min; 40 cycles of denaturation at 95°C for 15 sec, annealing at 60°C for 1 min and elongation at 95°C for 10 sec; and extension at 65°C for 60 sec. The following primers were used for qPCR: lncRNA ILF3-AS1 forward, 5′-TAAACCCCACTGTCTTCC-3′ and reverse, 5′-TTCCTTGCTCTTCTTGCTC-3′; PTEN forward, 5′-TTTGAAGACCATAACCCACCAC-3′ and reverse, 5′-ATTACACCAGTTCGTCCCTTTC-3′; GAPDH forward, 5′-AATGGACAACTGGTCGTGGAC-3′ and reverse, 5′-CCCTCCAGGGGATCTGTTTG-3′; miR-454-3p forward, 5′-GGGTGTCGTATCCAGTGCAA-3′ and reverse, 5′-GTCGTATCCAGTGCGTGTCG-3′; and U6 forward, 5′-AGTAAGCCCTTGCTGTCAGTG-3′ and reverse, 5′-CCTGGGTCTGATAATGCTGGG-3′. miRNA and mRNA expression levels were quantified using the 2−∆∆Cq method (30) and normalized to the internal reference genes U6 and GAPDH, respectively.
Cell Counting Kit-8 (CCK-8) assay
SiHa and HeLa cell viability was assessed by performing CCK-8 assays. Cells were seeded (5×103 cells/ml; 100 µl) into 96-well plates, with six replicate wells for each experimental group. Following incubation for 24, 48 and 72 h at 37°C, 10 µl CCK-8 reagent (cat. no. PA584814; Thermo Fisher Scientific, Inc.) was added to each well and incubated for 4 h. The absorbance was measured at a wavelength of 450 nm using an iMARK microplate absorbance reader (Bio-Rad Laboratories, Inc.).
Colony formation assay
SiHa and HeLa cells were seeded (1×102 cells/well) into 6-well plates and incubated at 37°C with 5% CO2. After 2 weeks, cells were thoroughly washed with PBS, fixed with 5 ml 100% methanol (cat. no. M116119-4L; Shanghai Aladdin Biochemical Technology Co., Ltd.) for 15 min at room temperature and stained with Giemsa stain (cat. no. 32884-250ML; Sigma-Aldrich; Merck KGaA) for 20 min at room temperature. Following drying at room temperature, stained cells were counted manually. The colony formation rate (%) was calculated according to the following formula: (Number of clones/number of cells inoculated) ×100.
Flow cytometry
The Annexin V-FITC Apoptosis Detection Kit (cat. no. C1062M; Beyotime Institute of Biotechnology) was used to assess cell apoptosis. SiHa or HeLa cell (3×105 cells) suspensions were prepared using PBS. Subsequently, cells were incubated with 5 µl Annexin V-FITC and 10 µl PI (cat. no. C1062M; Beyotime Institute of Biotechnology) for 20 min in the dark at room temperature. Apoptotic cells (early and late apoptotic cells) were analyzed using a CytoFLEX flow cytometer (Beckman Coulter, Inc.) and analyzed using FlowJo (version 10.0; FlowJo LLC).
Wound healing assay
SiHa and HeLa cell migration was detected by performing wound healing assays. Cells were seeded (5×105 cells/well) into a 6-well plate and incubated overnight at 37°C. At 80% confluence, a pipette was used to create a single wound in the cell monolayer. Following washing using PBS, cells were incubated with serum-free DMEM medium at 37°C with 5% CO2. SiHa and HeLa cell migration was observed at 0 and 24 h using a TS100 light microscope (Nikon Corporation; magnification, ×100). The relative migration distance was calculated according to the following formula: (wound width at 0 h-wound width at 24 h)/wound width at 0 h. Relative migration rates were calculated as the ratio of the relative migration distance of cells in the ILF3-AS1 group to the relative migration distance of cells in the blank group.
Transwell invasion assay
Transwell invasion assays were performed to assess SiHa and HeLa cell invasion. The upper chambers of Transwell inserts (pore size, 8 µm; BD Biosciences) were coated with Matrigel for 30 min at 37°C. Subsequently, cells were seeded (1×104 cells/well) into the upper chamber with serum-free DMEM. DMEM supplemented with 10% FBS was added into the lower chamber. Following incubation for 48 h at 37°C, invading cells were fixed with 4% paraformaldehyde (cat. no. C104188-500g; Shanghai Aladdin Biochemical Technology Co., Ltd.) at room temperature for 5 min and stained with 0.1% crystal violet (cat. no. RBG1019-1; Guangzhou Rolls Biotechnology Co., Ltd.) for 15 min at room temperature. Invading cells were observed using a light microscope (magnification, ×100).
Western blotting
Following washing with PBS, total protein was isolated from SiHa and HeLa cells using 100 µl cell lysis buffer (RIPA buffer; cat. no. 9806; Cell Signaling Technology, Inc.). Proteins were obtained from the cell lysate by centrifugation at 4°C for 10 min at 1,600 × g. Protein concentrations were determined using the BCA method. Proteins (100 µg) were separated via 10% SDS-PAGE and transferred to PVDF membranes. Following washing three times with 1X TBST (0.5% Tween-20) for 10 min, the membranes were blocked with 5% skimmed milk for 1.5 h at room temperature. Subsequently, the membranes were incubated overnight at 4°C with primary antibodies targeted against the following: E-Cadherin (E-Cad; 1:1,000; cat. no. 14472; Cell Signaling Technology, Inc.), N-Cadherin (N-Cad; 1:1,000; cat. no. 14215; Cell Signaling Technology, Inc.), snail family transcriptional repressor 1 (Snail; 1:10,000; cat. no. ab53519; Abcam) and GAPDH (1:10,000; cat. no. ab8245; Abcam). Following primary incubation, the membranes were incubated with anti-goat (HRP-conjugated; 1:5,000; cat. no. ab6885; Abcam) and anti-mouse (HRP-conjugated; 1:10,000; cat. no. ab6728; Abcam) secondary antibodies for 1 h at 37°C. The membranes were washed three times. Protein bands were visualized using ECL luminescent liquid (cat. no. RBG2019-1; Guangzhou Rolls Biotechnology Co., Ltd.) and observed using a GelDoc XR gel imager (Bio-Rad Laboratories, Inc.). Protein expression levels were semi-quantified using ImageJ software (version 1.47; National Institutes of Health) with GAPDH as the loading control.
Statistical analysis
Statistical analyses were performed using SPSS software (version 23.0; IBM Corp.). Data are presented as the mean ± SD. All experiments were repeated three times. For normally distributed data, comparisons between two groups were analyzed using an unpaired Student's t test, whereas comparisons among multiple groups were analyzed using one-way ANOVA followed by Bonferroni's post hoc test. Pearson's correlation coefficient was used to analyze correlations. P<0.05 was considered to indicate a statistically significant difference.
Results
ILF3-AS1 expression is downregulated in CC tissues
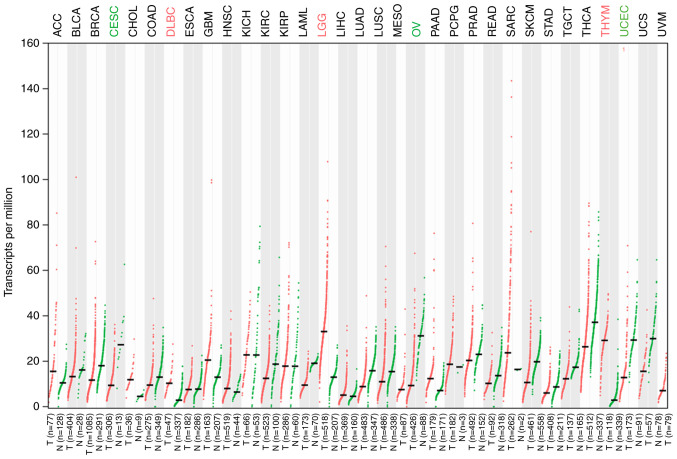
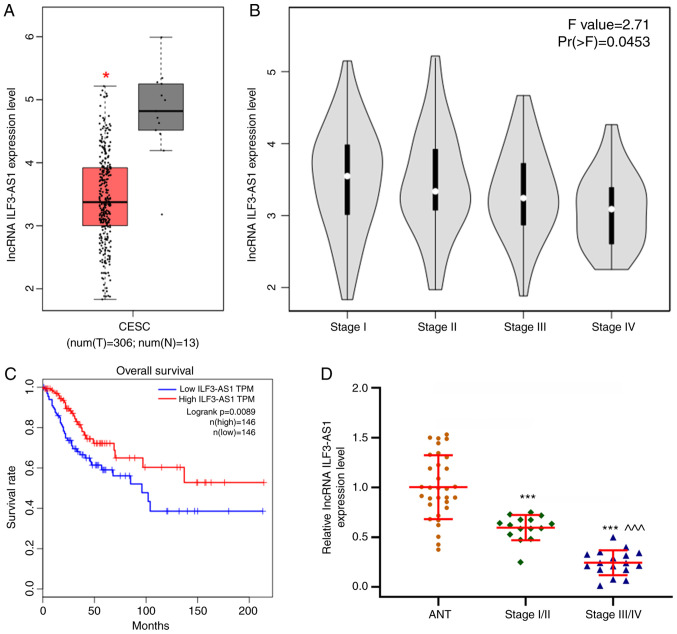
ILF3-AS1 expression in tumor tissues was detected using the GEPIA database (Fig. 1). The results demonstrated that ILF3-AS1 was expressed at lower levels in tumor tissues compared with healthy tissues. In addition, the GEPIA database predicted that ILF3-AS1 expression levels were significantly lower in CC tissues compared with healthy tissues (P<0.05; Fig. 2A). Moreover, low ILF3-AS1 expression levels were significantly associated with more advanced stages of CC (F=2.71; Pr(>F)=0.0453; Fig. 2B). The overall survival of patients with low ILF3-AS1 expression was significantly worse compared with patients with high ILF3-AS1 expression (P=0.0089; |log2FC| cutoff=1; Fig. 2C). In addition, ILF3-AS1 expression levels in CC tissues of different stages and ANTs were detected via RT-qPCR (P<0.001; Fig. 2D). ILF3-AS1 expression was significantly downregulated in CC tissues compared with ANTs, and was significantly lower in stage III/IV CC tissues compared with stage I/II CC tissues (P<0.001). The results demonstrated that lncRNA ILF3-AS1 was expressed at lower levels in CC tissues compared with ANTs, and that ILF3-AS1 expression in stage III/IV CC tissues was decreased compared with stage I/II CC tissues.
Figure 1.
Gene Expression Profiling Interactive Analysis database analysis of ILF3-AS1 expression levels in tumor tissues. Red dots represent the transcripts levels of ILF3-AS1 in tumor tissues. Green dots represent the transcripts levels of ILF3-AS1 in healthy tissues. Green text indicates the tumours with the lowest ILF3-AS1 expression. Red text indicates the tumours with the highest ILF3-AS1 expression. ILF3-AS1, ILF3 divergent transcript; T, tumor; N, healthy.
Figure 2.
ILF3-AS1 expression is downregulated in CC tissues. (A) GEPIA database analysis demonstrated that ILF3-AS1 expression was downregulated in CC tissues compared with healthy tissues. The red box represents CC tissues and the gray box represents healthy tissues. (B) Low ILF3-AS1 expression levels were associated with the stage of CC. (C) GEPIA database analysis of overall survival in patients with CC. (D) Reverse transcription-quantitative PCR was performed to measure ILF3-AS1 expression levels in CC tissues (stage I/II=15; stage III/IV=17) and ANTs (n=32). All experiments were repeated three times. *P<0.05 and ***P<0.001 vs. healthy tissues or ANTs; ^^^P<0.001 vs. stage I/II. ILF3-AS1, ILF3 divergent transcript; CC, cervical cancer; GEPIA, Gene Expression Profiling Interactive Analysis; ANT, adjacent tissue; lncRNA, long non-coding RNA; CESC, cervical and endocervical cancer; T, tumor; N, healthy; TPM, transcripts per million.
ILF3-AS1 inhibits CC cell proliferation and promotes CC cell apoptosis
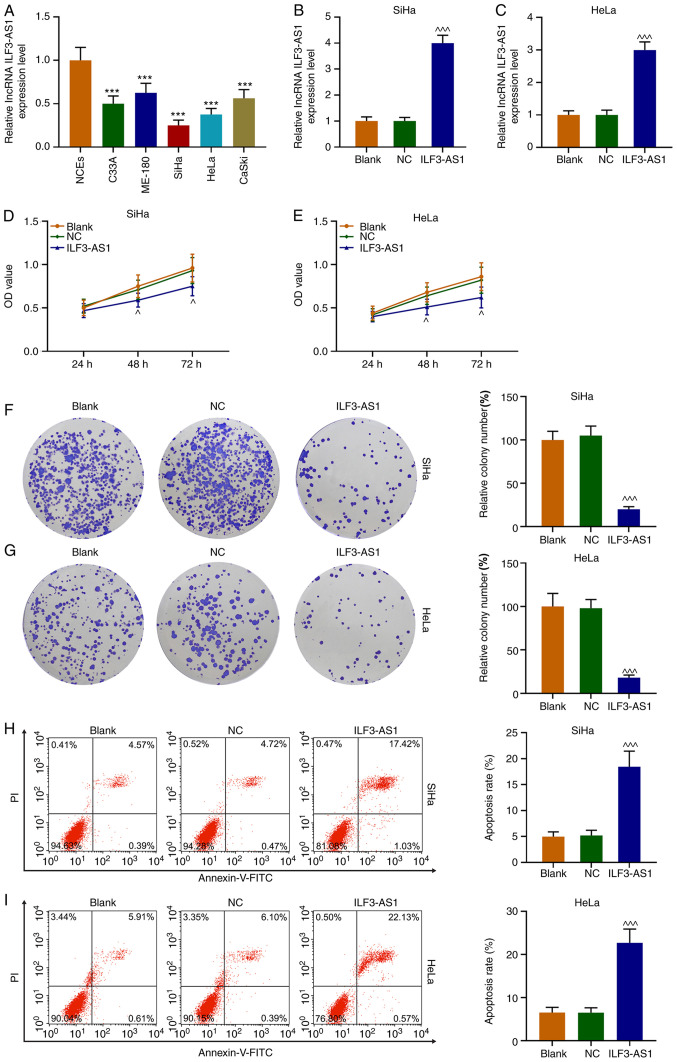
ILF3-AS1 expression levels in CC cells were detected via RT-qPCR. The results demonstrated that ILF3-AS1 expression was significantly downregulated in CC cells compared with NCEs (P<0.001; Fig. 3A). Compared with NCEs, SiHa and HeLa cells displayed the lowest expression levels of ILF3-AS1 among the CC cell lines, thus were used for subsequent experiments. Following transfection with ILF3-AS1, ILF3-AS1 expression levels were significantly increased in SiHa and HeLa cells compared with the NC group (P<0.001; Fig. 3B and C). Compared with the NC group, ILF3-AS1 overexpression significantly inhibited SiHa and HeLa cell viability at 48 and 72 h (P<0.05, Fig. 3D and E). Similarly, ILF3-AS1 overexpression significantly decreased SiHa and HeLa cell proliferation compared with the NC group (P<0.001; Fig. 3F and G). The flow cytometry results indicated that ILF3-AS1 overexpression significantly increased cell apoptosis compared with the NC group (P<0.001; Fig. 3H and I). Thus, the results demonstrated that ILF3-AS1 was expressed at lower levels in CC cells compared with NCEs, and ILF3-AS1 overexpression inhibited cell viability and promoted cell apoptosis compared with the NC group.
Figure 3.
ILF3-AS1 inhibits CC cell proliferation and promotes CC cell apoptosis. (A) ILF3-AS1 expression levels in NCEs and CC cells were detected via RT-qPCR. Transfection efficiency of ILF3-AS1 in (B) SiHa and (C) HeLa cells. (D) SiHa and (E) HeLa cell viability were assessed by performing Cell Counting Kit-8 assays. (F) SiHa and (G) HeLa cell proliferation were assessed by conducting cell colony formation assays (magnification, ×1). (H) SiHa and (I) HeLa cell apoptosis was assessed via flow cytometry. All experiments were repeated three times. ***P<0.001 vs. NCEs; ^P<0.05 and ^^^P<0.001 vs. NC. ILF3-AS1, ILF3 divergent transcript; CC, cervical cancer; NCE, normal cervical epithelial cell line; RT-qPCR, reverse transcription-quantitative PCR; NC, negative control; lncRNA, long non-coding RNA; OD, optical density.
ILF3-AS1 inhibits CC metastasis by inhibiting epithelial-mesenchymal transition (EMT)
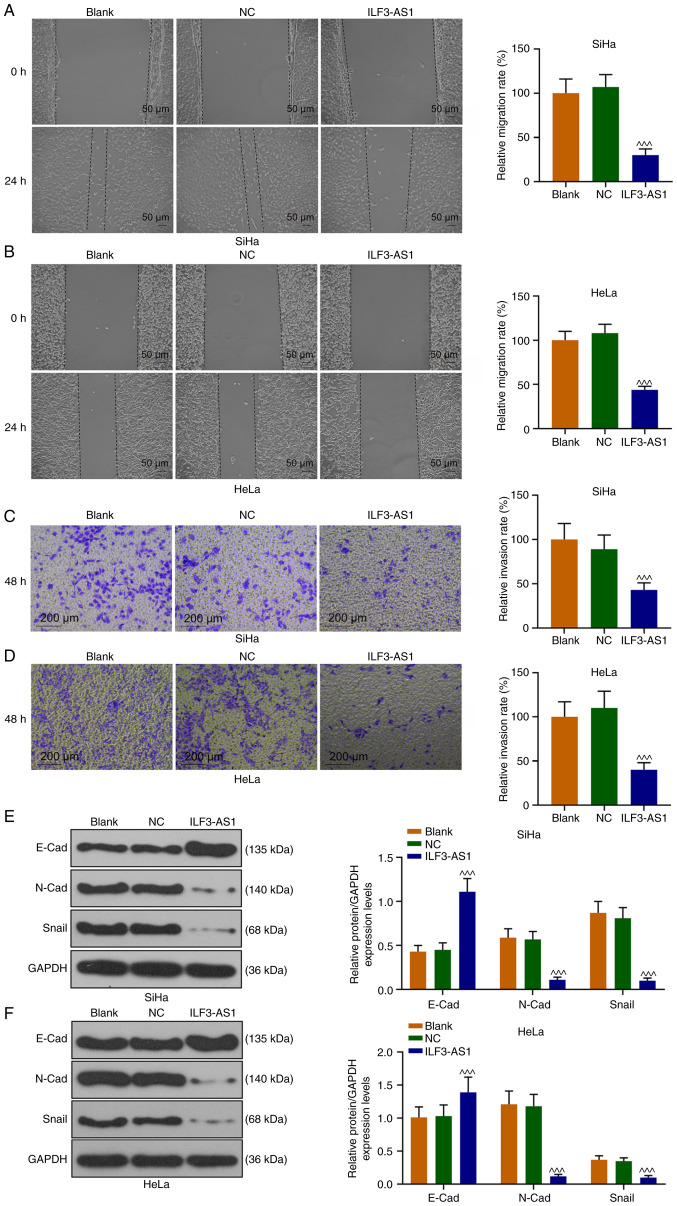
SiHa and HeLa cell migration and invasion were also assessed (Fig. 4A-D). In ILF3-AS1-overexpression cells, cell migration and invasion were significantly inhibited compared with the NC group (P<0.001; Fig. 4A-D). To further explore the mechanism underlying ILF3-AS1-mediated alterations, the expression levels of EMT-associated proteins were detected. ILF3-AS1 overexpression significantly upregulated E-Cad expression levels, and significantly downregulated N-Cad and Snail expression levels in SiHa and HeLa cells compared with the NC group (P<0.001; Fig. 4E and F). The aforementioned results indicated that ILF3-AS1 overexpression inhibited EMT in CC cells.
Figure 4.
ILF3-AS1 inhibits CC metastasis by inhibiting EMT. (A) SiHa and (B) HeLa cell migration were assessed by performing the wound healing assay. (C) SiHa and (D) HeLa cell invasion were assessed by conducting the Transwell invasion assay. Expression levels of EMT-associated proteins in (E) SiHa and (F) HeLa cells were determined via western blotting. All experiments were repeated three times. ^^^P<0.001 vs. NC. ILF3-AS1, ILF3 divergent transcript; CC, cervical cancer; EMT, epithelial-mesenchymal transition; NCE, normal cervical epithelial cell line; NC, negative control; E-cad, E-cadherin; N-cad, N-cadherin; Snail, snail family transcriptional repressor 1.
miR-454-3p is highly expressed in CC cells and is negatively correlated with ILF3-AS1
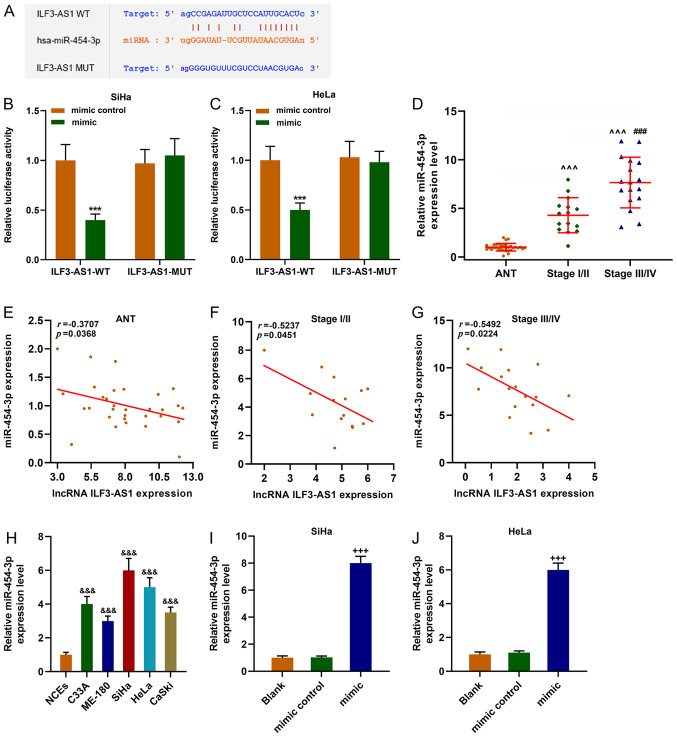
StarBase predicted that miR-454-3p was a target miRNA of ILF3-AS1, which was verified by performing dual-luciferase reporter assays (P<0.001; Fig. 5A-C). miR-454-3p expression was significantly upregulated in CC tissues and cells compared with ANTs and NCEs, respectively (P<0.001; Fig. 5D and H). miR-454-3p expression levels were significantly higher in stage III/IV CC tissues compared with stage I/II CC tissues (P<0.001; Fig. 5D). Moreover, the results indicated that ILF3-AS1 and miR-454-3p expression levels were negatively correlated (P<0.05; r<0; Fig. 5E-G). miR-454-3p mimic significantly upregulated miR-454-3p expression levels in SiHa and HeLa cells compared with the mimic control group (P<0.001; Fig. 5I and J).
Figure 5.
miR-454-3p expression is upregulated in CC cells and is negatively correlated with ILF3-AS1. (A) StarBase predicted that miR-454-3p was the targeted miRNA of ILF3-AS1. The sequences of ILF3-AS1 WT, ILF3-AS1 MUT and miR-454-3p. Dual-luciferase reporter assays were performed to verify the relationship between ILF3-AS1 and miR-454-3p in (B) SiHa and (C) HeLa cells. (D) RT-qPCR was performed to measure miR-454-3p expression levels in ANTs and CC tissues. Pearson's correlation coefficient was used to analyze the correlation between miR-454-3p and ILF3-AS1 expression in (E) ANTs, (F) stage I/II CC tissues and (G) stage III/IV CC tissues. (H) RT-qPCR was performed to measure miR-454-3p expression levels in NCEs and CC cells. Transfection efficiency of miR-454-3p mimic in (I) SiHa and (J) HeLa cells. All experiments were repeated three times. ***P<0.001 vs. mimic control; ^^^P<0.001 vs. ANT; ###P<0.001 vs. stage I/II; &&&P<0.001 vs. NCEs; +++P<0.001 vs. blank. miR/miRNA, microRNA; CC, cervical cancer; ILF3-AS1, ILF3 divergent transcript; WT, wild-type; MUT, mutant; RT-qPCR, reverse transcription-quantitative PCR; ANT, adjacent tissue; NCE, normal cervical epithelial cell line.
PTEN is expressed at low levels in CC cells, and is negatively correlated with miR-454-3p and positively correlated with ILF3-AS1
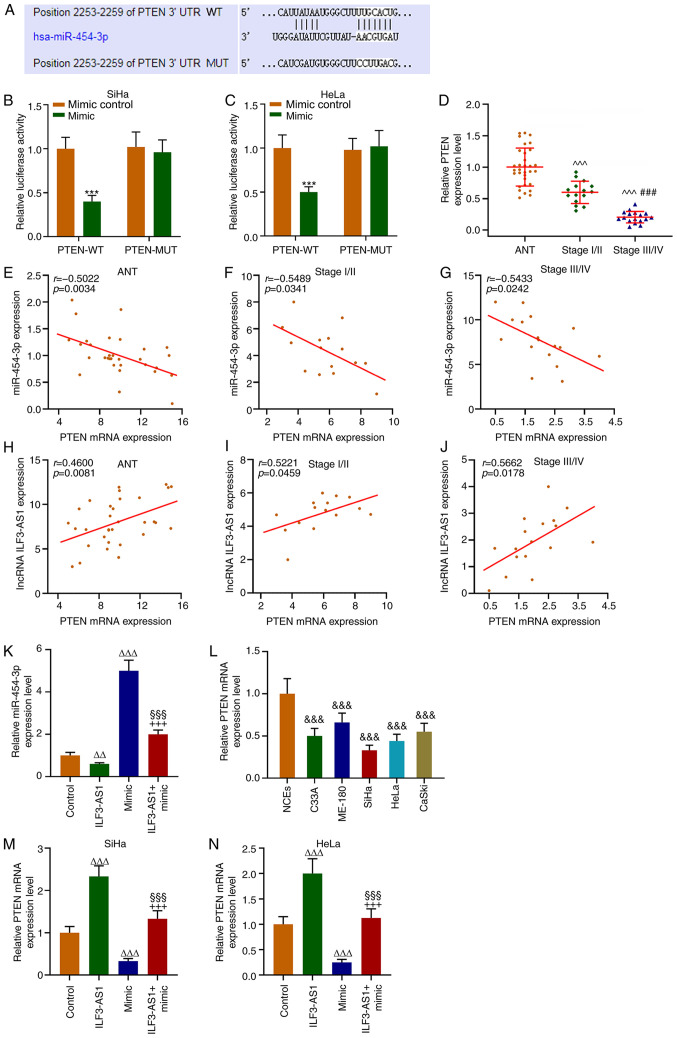
TargetScan predicted that the target gene for miR-454-3p was PTEN, which was further verified by performing dual-luciferase reporter assays (P<0.001; Fig. 6A-C). The RT-qPCR results demonstrated that PTEN expression levels were significantly lower in CC tissues and cells compared with ANTs and NECs, respectively (P<0.001; Fig. 6D and L). In addition, PTEN expression levels in stage III/IV CC tissues were significantly lower compared with stage I/II CC tissues (P<0.001; Fig. 6D). Correlation analysis demonstrated that PTEN expression was negatively correlated with miR-454-3p expression (P<0.05; r=−0.5022; Fig. 6E-G), but positively correlated with ILF3-AS1 expression (P<0.05; r=0.4600; Fig. 6H-J). In addition, CC cells were transfected with ILF3-AS1, miR-454-3p mimic or ILF3-AS1 + miR-454-3p mimic. miR-454-3p expression levels in SiHa cells were significantly decreased by ILF3-AS1 overexpression compared with the control group, which was significantly reversed by co-transfection miR-454-3p mimic (P<0.01; Fig. 6K). Compared with the control group, miR-454-3p mimic significantly downregulated PTEN expression levels, but ILF3-AS1 overexpression significantly upregulated PTEN expression levels, which was significantly reversed by co-transfection with miR-454-3p mimic (P<0.001; Fig. 6M and N). The results indicated that ILF3-AS1 upregulated PTEN gene expression by negatively regulating miR-454-3p.
Figure 6.
PTEN is a target gene for miR-454-3p and is correlated with miR-454-3p and ILF3-AS1 expression. (A) TargetScan was used to predict the target gene of miR-454-3p. Dual-luciferase reporter assays were performed to verify the relationship between PTEN and miR-454-3p in (B) SiHa and (C) HeLa cells. (D) RT-qPCR was performed to measure miR-454-3p expression levels in ANTs and CC tissues. Pearson's correlation coefficient was used to analyze the correlation between miR-454-3p and PTEN expression in (E) ANT, (F) stage I/II CC tissues and (G) stage III/IV CC tissues. Pearson's correlation coefficient was used to analyze the correlation between PTEN and ILF3-AS1 in (H) ANTs, (I) stage I/II CC tissues and (J) stage III/IV CC tissues. RT-qPCR was performed to measure (K) miR-454-3p expression levels in SiHa cells following transfection, (L) PTEN expression levels in NCEs and CC cells, and PTEN expression levels in (M) SiHa and (N) HeLa cells following transfection. All experiments were repeated three times. ***P<0.001 vs. mimic control; ^^^P<0.001 vs. ANT; ###P<0.001 vs. stage I/II; &&&P<0.001 vs. NCEs; ∆∆P<0.01; ∆∆∆P<0.001 vs. control; §§§P<0.001 vs. ILF3-AS1; +++P<0.001 vs. miR-454-3p mimic. miR, microRNA; ILF3-AS1, ILF3 divergent transcript; RT-qPCR, reverse transcription-quantitative PCR; ANT, adjacent tissue; CC, cervical cancer; NCE, normal cervical epithelial cell line; WT, wild-type; MUT, mutant; UTR, untranslated region.
Discussion
It has been reported that ILF3-AS1 can be used to predict survival in patients with CC (25,26). However, the functions and mechanism underlying ILF3-AS1 in CC are not completely understood. The present study demonstrated that ILF3-AS1 overexpression significantly inhibited CC cell migration and invasion compared with the NC group. Therefore, the expression levels of EMT-related factors were measured. E-Cad, N-Cad and Snail are important regulators of EMT (31). Downregulation or deletion of E-Cad protein can cause tumor cell release contact inhibition, and the conversion of E-Cad to N-Cad can enhance tumor cell migration and invasion, which promotes EMT (32–34). Snail downregulates the expression levels of epithelial phenotype genes by binding to the E-box sequence of the target gene (35). The present study demonstrated that compared with the NC group, ILF3-AS1 overexpression significantly upregulated E-Cad protein expression levels, and significantly downregulated N-Cad and Snail expression levels, which indicated that ILF3-AS1 overexpression inhibited CC cell EMT. EMT, a biological process by which epithelial cells are transformed into mesenchymal cells (36), serves an important role in epithelial-derived cancer cell migration and invasion (37). Therefore, investigating the molecular mechanism underlying EMT in the occurrence, development and metastasis of tumors, and developing diagnostic and molecular therapeutic strategies based on EMT may improve CC treatment.
lncRNAs affect tumor cell proliferation by inhibiting miRNA expression (38). lncRNAs can compete with miRNAs for binding to genes, and can also cleave precursors of miRNAs in cells or directly inhibit miRNA expression (39). In the present study, starBase predicted that miR-454-3p was a target miRNA of ILF3-AS1. miR-454-3p serves a regulatory role in a variety of neoplastic diseases, including breast and bladder cancer (40,41). Wang et al (42) reported that miR-454-3p is expressed at low levels in bladder cancer cells, which negatively regulated cell invasion and migration by targeting ZEB2 antisense RNA 1. Shao et al (43) demonstrated that miR-454-3p is an exosome biomarker, and miR-454-3p overexpression inhibits glioma cell proliferation, migration, invasion and autophagy. However, Ren et al (40) reported that inhibition of regulation of nuclear pre-mRNA domain containing 1A by miR-454-3p activates the Wnt/β-catenin signaling pathway, thereby promoting breast cancer metastasis and growth (40). In addition, miR-454-3p expression is upregulated in colorectal cancer tissues and participates in the development of colorectal cancer by promoting liver cancer cell proliferation, invasion and migration (44). The aforementioned studies suggested that miR-454-3p might display different effects in different types of cancer. To explore the role of miR-454-3p in CC, miR-454-3p expression levels in CC tissues and cells were measured. The results demonstrated that miR-454-3p expression levels were significantly increased in CC tissues and cells compared with ANTs and NCEs, respectively. Moreover, miR-454-3p was identified as a target miRNA of ILF3-AS1, and its expression was negatively regulated by ILF3-AS1.
In the present study, TargetScan also identified an interaction between miR-454-3p and PTEN. PTEN, which is 200 kb in length and is located on the human chromosome 10q23.3 (45), displays a variety of biological activities and encodes a dual phosphatase with lipid and protein phosphatase activities (46). Previous studies demonstrated that PTEN inhibits cell proliferation, migration and adhesion, induces apoptosis, and participates in embryonic development and other physiological functions via multiple signaling pathways, including the Akt signaling pathway (47,48). PTEN is a tumor suppressor gene and one of the most susceptible genes in tumors (46). By inhibiting FAK expression activity, PTEN reduces integrin-mediated cell proliferation and local adhesion, thereby inhibiting tumor cell migration and invasion (49). In addition, PTEN can suppress the phosphorylation of ERK, RAS and SHC adaptor protein 1, which are upstream of the MAPK/ERK signaling pathway, thereby inhibiting tumor cell proliferation (50). Therefore, the aforementioned studies indicated that increased PTEN expression levels display a significant inhibitory effect on tumor cell migration and invasion. The results of the present study demonstrated that PTEN expression was significantly decreased in CC cells compared with NCEs, which was partially reversed by ILF3-AS1 overexpression. Moreover, miR-454-3p negatively regulated ILF3-AS1.
In conclusion, the present study demonstrated that ILF3-AS1 was expressed at significantly lower levels in CC tissues and cells compared with ANTs and NCEs, respectively. Compared with the NC group, ILF3-AS1 overexpression significantly inhibited CC cell viability, reduced CC cell migration and invasion, and promoted CC cell apoptosis by inhibiting EMT. Moreover, compared with the control group, ILF3-AS1 overexpression significantly upregulated the expression levels of the tumor suppressor gene PTEN by negatively regulating miR-454-3p. Therefore, the results of the present study may aid with improving the treatment of CC.
Acknowledgements
Not applicable.
Glossary
Abbreviations
- CC
cervical cancer
- lncRNA
long non-coding RNA
- RT-qPCR
reverse transcription-quantitative PCR
- CCK-8
Cell Counting Kit-8
Funding Statement
The present study was supported by the Medical Scientific Research Foundation of Guangdong Province (grant no. B2020211), the Scientific Research Project of Guangdong Provincial Bureau of Traditional Chinese Medicine (grant no. 20201284) and the Guangzhou Health Science and Technology Project (grant no. 20202A010024).
Funding
The present study was supported by the Medical Scientific Research Foundation of Guangdong Province (grant no. B2020211), the Scientific Research Project of Guangdong Provincial Bureau of Traditional Chinese Medicine (grant no. 20201284) and the Guangzhou Health Science and Technology Project (grant no. 20202A010024).
Availability of data and materials
The datasets analyzed during the current study are available from the corresponding author on reasonable request.
Authors' contributions
LZ and RC substantially contributed to the conception and design of the present study. CJ, QX, WZ, XG and HH acquired, analyzed and interpreted the data. LZ and RC drafted the manuscript and critically revised it for important intellectual content. All authors read and approved the final manuscript.
Ethics approval and consent to participate
All patients provided written informed consent. The study was approved by Guangdong Clifford Hospital (approval no. 2014007XHK).
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing interests.
References
- 1.Burki TK. Novel mutations in cervical cancer. Lancet Oncol. 2017;18:e137. doi: 10.1016/S1470-2045(17)30080-3. [DOI] [PubMed] [Google Scholar]
- 2.Bychkovsky BL, Ferreyra ME, Strasser-Weippl K, Herold CI, de Lima Lopes G, Jr, Dizon DS, Schmeler KM, Del Carmen M, Randall TC, Nogueira-Rodrigues A, et al. Cervical cancer control in Latin America: A call to action. Cancer. 2016;122:502–514. doi: 10.1002/cncr.29813. [DOI] [PubMed] [Google Scholar]
- 3.Arbyn M, Weiderpass E, Bruni L, de Sanjosé S, Saraiya M, Ferlay J, Bray F. Estimates of incidence and mortality of cervical cancer in 2018: a worldwide analysis. Lancet Glob Health. 2020;8:e191–e203. doi: 10.1016/S2214-109X(19)30482-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Fang J, Zhang H, Jin S. Epigenetics and cervical cancer: From pathogenesis to therapy. Tumour Biol. 2014;35:5083–5093. doi: 10.1007/s13277-014-1737-z. [DOI] [PubMed] [Google Scholar]
- 5.Kanyina EW, Kamau L, Muturi M. Cervical precancerous changes and selected cervical microbial infections, Kiambu County, Kenya, 2014: A cross sectional study. BMC Infect Dis. 2017;17:647. doi: 10.1186/s12879-017-2747-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Tavakoli F, Khatami SS, Momeni F, Azadbakht J, Ghasemi F. Cervical cancer diagnosis: Insights into biochemical biomarkers and Imaging techniques. Comb Chem High Throughput Screen. 2020 Aug 31; doi: 10.2174/1386207323666200901101955. (Epub ahead of print) [DOI] [PubMed] [Google Scholar]
- 7.zur Hausen H. Papillomaviruses and cancer: From basic studies to clinical application. Nat Rev Cancer. 2002;2:342–350. doi: 10.1038/nrc798. [DOI] [PubMed] [Google Scholar]
- 8.zur Hausen H. Papillomaviruses in the causation of human cancers-a brief historical account. Virology. 2009;384:260–265. doi: 10.1016/j.virol.2008.11.046. [DOI] [PubMed] [Google Scholar]
- 9.Gillison ML. Human papillomavirus-related diseases: Oropharynx cancers and potential implications for adolescent HPV vaccination. J Adolesc Health. 2008;43(Suppl 4):S52–S60. doi: 10.1016/j.jadohealth.2008.07.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Mattick JS, Makunin IV. Non-coding RNA. Hum Mol Genet. 2006;15:R17–R29. doi: 10.1093/hmg/ddl046. [DOI] [PubMed] [Google Scholar]
- 11.Hombach S, Kretz M. Non-coding RNAs: Classification, biology and functioning. Adv Exp Med Biol. 2016;937:3–17. doi: 10.1007/978-3-319-42059-2_1. [DOI] [PubMed] [Google Scholar]
- 12.Cabianca DS, Casa V, Gabellini D. A novel molecular mechanism in human genetic disease: A DNA repeat-derived lncRNA. RNA Biol. 2012;9:1211–1217. doi: 10.4161/rna.21922. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Jarroux J, Morillon A, Pinskaya M. History, discovery, and classification of lncRNAs. Adv Exp Med Biol. 2017;1008:1–46. doi: 10.1007/978-981-10-5203-3_1. [DOI] [PubMed] [Google Scholar]
- 14.Kim J, Piao HL, Kim BJ, Yao F, Han Z, Wang Y, Xiao Z, Siverly AN, Lawhon SE, Ton BN, et al. Long noncoding RNA MALAT1 suppresses breast cancer metastasis. Nat Genet. 2018;50:1705–1715. doi: 10.1038/s41588-018-0252-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Wang Y, He L, Du Y, Zhu P, Huang G, Luo J, Yan X, Ye B, Li C, Xia P, et al. The long noncoding RNA lncTCF7 promotes self-renewal of human liver cancer stem cells through activation of Wnt signaling. Cell Stem Cell. 2015;16:413–425. doi: 10.1016/j.stem.2015.03.003. [DOI] [PubMed] [Google Scholar]
- 16.Hu X, Feng Y, Zhang D, Zhao SD, Hu Z, Greshock J, Zhang Y, Yang L, Zhong X, Wang LP, et al. A functional genomic approach identifies FAL1 as an oncogenic long noncoding RNA that associates with BMI1 and represses p21 expression in cancer. Cancer Cell. 2014;26:344–357. doi: 10.1016/j.ccr.2014.07.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Wu WJ, Shen Y, Sui J, Li CY, Yang S, Xu SY, Zhang M, Yin LH, Pu YP, Liang GY. Integrated analysis of long non-coding RNA competing interactions revealed potential biomarkers in cervical cancer: Based on a public database. Mol Med Rep. 2018;17:7845–7858. doi: 10.3892/mmr.2018.8846. [DOI] [PubMed] [Google Scholar]
- 18.Luan X, Wang Y. LncRNA XLOC_006390 facilitates cervical cancer tumorigenesis and metastasis as a ceRNA against miR-331-3p and miR-338-3p. J Gynecol Oncol. 2018;29:e95. doi: 10.3802/jgo.2018.29.e95. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Liu Y, Yang Y, Li L, Liu Y, Geng P, Li G, Song H. LncRNA SNHG1 enhances cell proliferation, migration, and invasion in cervical cancer. Biochem Cell Biol. 2018;96:38–43. doi: 10.1139/bcb-2017-0188. [DOI] [PubMed] [Google Scholar]
- 20.Chen X, Liu S, Zhao X, Ma X, Gao G, Yu L, Yan D, Dong H, Sun W. Long noncoding RNA ILF3-AS1 promotes cell proliferation, migration, and invasion via negatively regulating miR-200b/a/429 in melanoma. Biosci Rep. 2017;37:BSR20171031. doi: 10.1042/BSR20171031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Gao G, Li W, Liu S, Han D, Yao X, Jin J, Han D, Sun W, Chen X. The positive feedback loop between ILF3 and lncRNA ILF3-AS1 promotes melanoma proliferation, migration, and invasion. Cancer Manag Res. 2018;10:6791–6802. doi: 10.2147/CMAR.S186777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Hu XH, Dai J, Shang HL, Zhao ZX, Hao YD. SP1-mediated upregulation of lncRNA ILF3-AS1 functions a ceRNA for miR-212 to contribute to osteosarcoma progression via modulation of SOX5. Biochem Biophys Res Commun. 2019;511:510–517. doi: 10.1016/j.bbrc.2019.02.110. [DOI] [PubMed] [Google Scholar]
- 23.Zhou M, Hu L, Zhang Z, Wu N, Sun J, Su J. Recurrence-associated long non-coding RNA signature for determining the risk of recurrence in patients with colon cancer. Mol Ther Nucleic Acids. 2018;12:518–529. doi: 10.1016/j.omtn.2018.06.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Ye G, Guo L, Xing Y, Sun W, Yuan M. Identification of prognostic biomarkers of prostate cancer with long non-coding RNA-mediated competitive endogenous RNA network. Exp Ther Med. 2019;17:3035–3040. doi: 10.3892/etm.2019.7277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Wu W, Sui J, Liu T, Yang S, Xu S, Zhang M, Huang S, Yin L, Pu Y, Liang G. Integrated analysis of two-lncRNA signature as a potential prognostic biomarker in cervical cancer: A study based on public database. PeerJ. 2019;7:e6761. doi: 10.7717/peerj.6761. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Mao X, Qin X, Li L, Zhou J, Zhou M, Li X, Xu Y, Yuan L, Liu QN, Xing H. A 15-long non-coding RNA signature to improve prognosis prediction of cervical squamous cell carcinoma. Gynecol Oncol. 2018;149:181–187. doi: 10.1016/j.ygyno.2017.12.011. [DOI] [PubMed] [Google Scholar]
- 27.Tsikouras P, Zervoudis S, Manav B, Tomara E, Iatrakis G, Romanidis C, Bothou A, Galazios G. Cervical cancer: Screening, diagnosis and staging. J BUON. 2016;21:320–325. [PubMed] [Google Scholar]
- 28.Li JH, Liu S, Zhou H, Qu LH, Yang JH. starBase v2.0: Decoding miRNA-ceRNA, miRNA-ncRNA and protein-RNA interaction networks from large-scale CLIP-Seq data. Nucleic Acids Res. 2014;42((Database Issue)):D92–D97. doi: 10.1093/nar/gkt1248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Agarwal V, Bell GW, Nam JW, Bartel DP. Predicting effective microRNA target sites in mammalian mRNAs. Elife. 2015;4:e05005. doi: 10.7554/eLife.05005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) method. Methods. 2001;25:402–408. doi: 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]
- 31.Qu BL, Yu W, Huang YR, Cai BN, Du LH, Liu F. 6-OH-BDE-47 promotes human lung cancer cells epithelial mesenchymal transition via the AKT/Snail signal pathway. Environ Toxicol Pharmacol. 2015;39:271–279. doi: 10.1016/j.etap.2014.11.022. [DOI] [PubMed] [Google Scholar]
- 32.Wong SHM, Fang CM, Chuah LH, Leong CO, Ngai SC. E-cadherin: Its dysregulation in carcinogenesis and clinical implications. Crit Rev Oncol Hematol. 2018;121:11–22. doi: 10.1016/j.critrevonc.2017.11.010. [DOI] [PubMed] [Google Scholar]
- 33.van Roy F. Beyond E-cadherin: Roles of other cadherin superfamily members in cancer. Nat Rev Cancer. 2014;14:121–134. doi: 10.1038/nrc3647. [DOI] [PubMed] [Google Scholar]
- 34.Angst BD, Marcozzi C, Magee AI. The cadherin superfamily: Diversity in form and function. J Cell Sci. 2001;114:629–641. doi: 10.1242/jcs.114.4.629. [DOI] [PubMed] [Google Scholar]
- 35.Osorio LA, Farfán NM, Castellón EA, Contreras HR. SNAIL transcription factor increases the motility and invasive capacity of prostate cancer cells. Mol Med Rep. 2016;13:778–786. doi: 10.3892/mmr.2015.4585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Lamouille S, Xu J, Derynck R. Molecular mechanisms of epithelial-mesenchymal transition. Nat Rev Mol Cell Biol. 2014;15:178–196. doi: 10.1038/nrm3758. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Diepenbruck M, Christofori G. Epithelial-mesenchymal transition (EMT) and metastasis: Yes, no, maybe? Curr Opin Cell Biol. 2016;43:7–13. doi: 10.1016/j.ceb.2016.06.002. [DOI] [PubMed] [Google Scholar]
- 38.Cheng N, Wu J, Yin M, Xu J, Wang Y, Chen X, Nie Z, Yin J. LncRNA CASC11 promotes cancer cell proliferation in hepatocellular carcinoma by inhibiting miRNA-188-5p. Biosci Rep. 2019;39:BSR20190251. doi: 10.1042/BSR20190251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Slaby O, Laga R, Sedlacek O. Therapeutic targeting of non-coding RNAs in cancer. Biochem J. 2017;474:4219–4251. doi: 10.1042/BCJ20170079. [DOI] [PubMed] [Google Scholar]
- 40.Ren L, Chen H, Song J, Chen X, Lin C, Zhang X, Hou N, Pan J, Zhou Z, Wang L, et al. MiR-454-3p-mediated Wnt/β-catenin signaling antagonists suppression promotes breast cancer metastasis. Theranostics. 2019;9:449–465. doi: 10.7150/thno.29055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Song Y, Guo Q, Gao S, Hua K. miR-454-3p promotes proliferation and induces apoptosis in human cervical cancer cells by targeting TRIM3. Biochem Biophys Res Commun. 2019;516:872–879. doi: 10.1016/j.bbrc.2019.06.126. [DOI] [PubMed] [Google Scholar]
- 42.Wang S, Zhang G, Zheng W, Xue Q, Wei D, Zheng Y, Yuan J. MiR-454-3p and miR-374b-5p suppress migration and invasion of bladder cancer cells through targetting ZEB2. Biosci Rep. 2018;38:BSR20181436. doi: 10.1042/BSR20181436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Shao N, Xue L, Wang R, Luo K, Zhi F, Lan Q. miR-454-3p is an exosomal biomarker and functions as a tumor suppressor in glioma. Mol Cancer Ther. 2019;18:459–469. doi: 10.1158/1535-7163.MCT-18-0725. [DOI] [PubMed] [Google Scholar]
- 44.Li W, Feng Y, Ma Z, Lu L. Expression of miR-454-3p and its effect on proliferation, invasion and metastasis of colon cancer. Nan Fang Yi Ke Da Xue Xue Bao. 2018;38:1421–1426. doi: 10.12122/j.issn.1673-4254.2018.12.04. (In Chinese) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Malaney P, Uversky VN, Davé V. PTEN proteoforms in biology and disease. Cell Mol Life Sci. 2017;74:2783–2794. doi: 10.1007/s00018-017-2500-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Chen CY, Chen J, He L, Stiles BL. PTEN: Tumor suppressor and metabolic regulator. Front Endocrinol (Lausanne) 2018;9:338. doi: 10.3389/fendo.2018.00338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Ortega-Molina A, Serrano M. PTEN in cancer, metabolism, and aging. Trends Endocrinol Metab. 2013;24:184–189. doi: 10.1016/j.tem.2012.11.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Nitulescu GM, Van De Venter M, Nitulescu G, Ungurianu A, Juzenas P, Peng Q, Olaru OT, Grădinaru D, Tsatsakis A, Tsoukalas D, et al. The Akt pathway in oncology therapy and beyond (Review) Int J Oncol. 2018;53:2319–2331. doi: 10.3892/ijo.2018.4597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Milella M, Falcone I, Conciatori F, Cesta Incani U, Del Curatolo A, Inzerilli N, Nuzzo CM, Vaccaro V, Vari S, Cognetti F, Ciuffreda L. PTEN: Multiple functions in human malignant tumors. Front Oncol. 2015;5:24. doi: 10.3389/fonc.2015.00024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Ye Z, Li Q, Guo Q, Xiong Y, Guo D, Yang H, Shu Y. Ketamine induces hippocampal apoptosis through a mechanism associated with the caspase-1 dependent pyroptosis. Neuropharmacology. 2018;128:63–75. doi: 10.1016/j.neuropharm.2017.09.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
The datasets analyzed during the current study are available from the corresponding author on reasonable request.