Abstract
Long non-coding RNAs (lncRNAs) are involved in the development and progression of a variety of diseases. However, the role of the lncRNA HLA complex group 11 (HCG11) in non-small cell lung cancer (NSCLC) remains unclear. The present study showed that the expression levels of HCG11 were reduced in tumor tissues compared with adjacent normal tissues, and similar results were obtained in experiments using lung cancer cell lines. Additionally, patients with high HCG11 expression had an increased survival rate compared with patients with low HCG11 expression. Further studies have shown that overexpression of HCG11 inhibited NSCLC cell proliferation in vitro and in vivo. Interestingly, it was observed that HCG11 expression was negatively associated with the expression levels of oncogenic microRNA-875 (miR-875) in patient specimens. Specifically, HCG11 served as a sponge of miR-875. Notably, it was determined that special AT-rich sequence-binding protein 2 (SATB2) was a direct target gene of miR-875, and overexpression of miR-875 largely abrogated the effects of HCG11 in NSCLC cells. In conclusion, HCG11 was shown to suppress the malignant properties of NSCLC cells by targeting a miR-875/SATB2 axis, and may therefore be a promising target for the treatment of NSCLC.
Keywords: long non-coding RNA, HLA complex group 11, microRNA-875, non-small cell lung cancer, special AT-rich sequence-binding protein 2
Introduction
Lung cancer is one of the most common types of cancer and is the leading cause of cancer-associated death worldwide (1–3). Lung cancer is classified into two groups based on the pathological features: Non-small cell lung cancer (NSCLC) and small cell lung cancer (SCLC) (4,5). NSCLC contributes to ~80% of lung cancer diagnoses (6,7). Additionally, ~75% of patients with lung cancer are already at the advanced stage at diagnosis as they are usually asymptomatic at an early stage. Despite improvements having been made in diagnosis and treatment, the prognosis of lung cancer remains poor and the 5-year survival rate is low (8,9). Therefore, there is an urgent need to develop effective strategies for the treatment of lung cancer.
Long non-coding RNAs (lncRNAs) are defined as non-protein coding transcripts >200 nucleotides in length. Previous studies have shown that lncRNAs serve a crucial role in a variety of biological processes, including energy metabolism, apoptosis, cell proliferation and differentiation, through transcriptional and posttranscriptional regulation (10,11). Notably, an increasing number of studies have revealed that lncRNAs are involved in the development and progression of multiple types of cancer, including breast cancer, gastric cancer, prostate cancer, lung cancer and leukemia (12,13). Although several lncRNAs, including ZEB1 antisense RNA 1 (14), breast cancer anti-estrogen resistance 4 (15), small nucleolar RNA host gene 1 (16), growth arrest specific 5 (GAS5) (17) and deleted in lymphocytic leukemia 2 (18) have been explored previously, the functions and underlying mechanisms of lncRNAs in NSCLC have not been fully revealed.
LncRNA HLA complex group 11 (HCG11) is located on chromosome 6 and is a member of the lncRNA family. It has been reported that HCG11 functions as a tumor suppressor in hepatocellular carcinoma and prostate cancer (19) by inhibiting cell proliferation, invasion and metastasis, and promoting cell apoptosis. Additionally, abnormal HCG11 expression has been observed in gastric cancer (20). In particular, downregulation of HCG11 expression is predictive of a poor prognosis in prostate cancer (21). However, whether HCG11 is involved in the development and progression of NSCLC remains unclear.
The present study primarily explored the role of HCG11 in NSCLC. First, it was identified that HCG11 expression was significantly reduced in patients with NSCLC. Further investigations revealed that low HCG11 expression was associated with a poor prognosis in patients with NSCLC. In terms of the mechanism, it was demonstrated that HCG11 could inhibit the malignant phenotype of NSCLC cells, at least partially, by targeting a microRNA-875 (miR-875)/special AT-rich sequence-binding protein 2 (SATB2) signaling pathway. Collectively, the results of the present study indicated that HCG11 is an important tumor suppressor, and thus, provides a novel potential avenue for the treatment of NSCLC.
Materials and methods
Patients and specimens
In the present study, 85 pairs of NSCLC tissues and adjacent normal tissues were collected from patients. The patients included 47 males and 38 females, with a median age of 59.5 years (range, 43–79 years), and were enrolled from The Sichuan Mianyang 404 Hospital (Mianyang, China) between January 2012 and June 2015. The patients did not receive any preoperative treatments, and patients who had received anticancer treatment were excluded from the present study. Tumor-node-metastasis (TNM) stage was determined as previously described (22). All patients provided informed consent. The present study was approved by the Ethics Committee of The Sichuan Mianyang 404 Hospital (Mianyang, China).
Cell culture and transfection
A total of five NSCLC cell lines (A549, H1299, H466, H460 and H358) and 16HBE normal lung epithelial cells were obtained from The Cell Bank of Type Culture Collection of The Chinese Academy of Sciences. All cell lines were cultured in DMEM (HyClone; Cytiva) supplemented with 10% FBS (Gibco; Thermo Fisher Scientific, Inc.), 100 U/ml penicillin and 100 µg/ml streptomycin (Gibco; Thermo Fisher Scientific, Inc.). All cells were cultured in a 37°C incubator with 5% CO2. pcDNA3.1-HCG11 (pcDNA-HCG11) and corresponding negative control (pcDNA3.1-NC; pcDNA3.1-vector) were synthesized by Shanghai Gene Pharma Co., Ltd. miR-875-mimic, as well as the corresponding negative control (miR-NC), were purchased from Guangzhou RiboBio Co., Ltd. Cells were transfected using Lipofectamine® 3000 (Invitrogen; Thermo Fisher Scientific, Inc.) according to the manufacturer's protocols.
MTT assay
A549 cells were seeded in 96-well plates (2×103 cells/well). After 24 h of culture, 500 µg/ml MTT solution (Sigma-Aldrich; Merck KGaA) was added to each well, followed by incubation at 37°C for 4 h. Subsequently, 200 µl DMSO (Sigma-Aldrich; Merck KGaA) was added to each well. Finally, the absorbance of the samples was measured at 570 nm using a microplate reader (Bio-Rad Laboratories, Inc.).
Cell invasion assay
Cell invasion assays were performed using a Transwell insert (6.5 mm in diameter) with an 8-µm pore size (Corning, Inc.). Briefly, 5×104 cells in DMEM without FBS were seeded onto the Matrigel-coated membrane matrix in the upper chamber, and the lower chamber was filled with medium supplemented with 10% FBS. Following incubation at 37°C for 24 h, cells which had not invaded were removed, and cells on the bottom of the filter, which had invaded, were fixed in 4% paraformaldehyde at room temperature and stained with crystal violet at room temperature for 30 min. The number of invaded cells were counted using a microscope.
Cell migration assay
Migration assays were performed using Matrigel-coated Transwell chambers (BD Biosciences). Briefly, 1×105 cells were seeded in the upper chamber in FBS free DMEM, and DMEM supplemented with 10% FBS was added to the lower chamber. Following incubation at 37°C for 24 h, cells in the upper chamber were removed using a cotton swab. The membrane was then soaked in 90% ethanol, fixed at 37°C for 10 min, and stained with 0.2% crystal violet at room temperature for 30 min. The cells which had migrated were counted under an inverted light microscope (magnification, ×200).
Cell apoptosis analysis
Cell apoptosis was detected using an Annexin V-FITC/7-AAD kit (BD Biosciences) according to the manufacturer's protocol, followed by flow cytometry (FACSCantoII; BD Biosciences). Flow data were analyzed using FlowJo version 10.0 (FlowJo LLC).
Luciferase reporter assay
A luciferase reporter assay was performed using a dual luciferase reporter assay kit (Promega Corporation) according to the manufacturer's protocol. Briefly, pmirGLO vector containing wild-type or mutant HCG11 or SATB2 (synthesized by Sangon Biotech Co., Ltd.) and miR-875-mimic or miR-NC were co-transfected into 293T cells obtained from The Cell Bank of Type Culture Collection of The Chinese Academy of Sciences using Lipofectamine® 3000 (Invitrogen; Thermo Fisher Scientific, Inc.). The luciferase activity was measured 48 h after transfection.
Reverse transcription-quantitative PCR (RT-qPCR)
Total RNA was extracted using TRIzol® reagent (Invitrogen; Thermo Fisher Scientific, Inc.) according to the manufacturer's protocols, and then converted to cDNA using the PrimeScript RT kit (Takara Bio, Inc.) at 37°C for 15 min followed by 85°C for 5 sec. Next, qPCR was performed using the SYBR Premix Ex Taq kit (Takara Bio, Inc.) with the following thermocycling conditions: Initial denaturation at 95°C for 3 min, followed by 40 cycles of denaturation at 95°C for 5 sec, annealing at 60°C for 30 sec and extension at 72°C for 30 sec. GAPDH was used as the internal control. The primer sequences used were: HCG11 forward, 5′-GCTCTATGCCATCCTGCTT-3′ and reverse, 5′-TCCCATCTCCATCAACCC-3′; GAPDH forward, 5′-TGTTCGTCATGGGTGTGAAC-3′ and reverse, 5′-ATGGCATGGACTGTGGTCAT-3′; and SATB2 forward, 5′-GGAGAACGACAGCGAGGAA-3′ and reverse, 5′-CCGATGTATTGCTTTGCCTAGT-3′. The expressions levels of miRNAs were measured using the mirVana RT-qPCR miRNA Detection kit, according to the manufacturer's protocol (Invitrogen; Thermo Fisher Scientific, Inc.). The miRNA primers were purchased from Guangzhou RiboBio Co., Ltd. Small RNA U6 was used as an internal reference gene. The miRNA primer sequences used were: miR-875 forward 5′-CGAATGGGCCTAAGATCCCG-3′ and reverse 5′-GGAGCCCAGCACTTTGATCT-3′; miR-522 forward, 5′-ACACTCCAGCTGGGCTCTAGAGGGAAGCGC-3′ and reverse, 5′-TGGTGTCGTGGAGTCG-3′; miR-499a forward, 5′-TGCGGTGGCAGTGTATTGTTAGC-3′ and reverse, 5′-CCAGTGCAGGGTCCGAGGT-3′; miR-224 forward, 5′-GTATACTAAGTCACTAGTGGT-3′ and reverse, 5′-GTGCAGGGTCCGAGGT-3′; miR-483 forward, 5′-GCTGACTCACTCCTCCCCTC-3′ and reverse, 5′-TATGGTTGTTCACGACTCCTTCAC-3′; miR-214 forward, 5′-TCCTTCAATCACCAAATCTGG-3′ and reverse, 5′-CCCAAGCTTTCATTCAGGCTGGGTTG-3′; miR-26a forward, 5′-GGATCCGCAGAAACTCCAGAGAGAAGGA-3′ and reverse, 5′-AAGCTTGCCTTTAGCAGAAAGGAGGTT-3′; miR-455 forward, 5′-ACACTCCAGCTGGGGCAGTCCATGGGCAT-3′ and reverse, 5′-TGGTGTCGTGGAGTCG-3′; miR-543 forward, 5′-GTGCTCGGTTTGTAGGCAGT-3′ and reverse, 5′-GTGCCTTGTTTTGATGGCAG-3′; miR-155 forward, 5′-TTAATGCTAATCGTGATAG-3′ and reverse, 5′-ACCTGAGAGTAGACCAGA-3′; miR-105 forward, 5′-GCCCTCGAGATACCATATCTATCCCCTTTTTCA-3′ and reverse, 5′-GCCGAATTCCAACCATGAAGATACGAATTGATG-3′; and U6 forward, 5′-CTCGCTTCGGCAGCACA-3′ and reverse 5′-AACGCTTCACGAATTTGCGT-3′.
Western blotting
Cells were lysed using RIPA buffer (Invitrogen; Thermo Fisher Scientific, Inc.). The concentration of proteins was determined using a Bicinchoninic Acid Protein Assay kit (Beyotime Institute of Biotechnology). Proteins (30 µg/lane) were loaded on a 10% SDS-gel, resolved using SDS-PAGE, and then electrotransferred onto PVDF membranes for 90 min. Subsequently, 5% skimmed milk was used to block the membranes at 37°C for 1 h, and the membranes were incubated with the primary antibodies at 4°C overnight. The following primary antibodies were used: Proliferation cell nuclear antigen (PCNA; cat. no. 13110S; 1:1,000; Cell Signaling Technology, Inc.), cleaved-caspase 3 (cat. no. 9664S; 1:1,000; Cell Signaling Technology, Inc.), cleaved-caspase-9 (cat. no. 9505S; 1:1,000; Cell Signaling Technology, Inc.), caspase-3 (cat. no. 14220S; 1:1,000; Cell Signaling Technology, Inc.), caspase-9 (cat. no. 9502S; 1:1,000; Cell Signaling Technology, Inc.), SATB2 (cat. no. ab92446; 1:1,000; Abcam) and β-actin (cat. no. ab8227; 1:2,000; Abcam). Signals were visualized using ECL.
Animal experiments
A total of 16 male BALB/c nude mice (4–5 weeks old, 16–20 g) were obtained from Beijing Vital River Laboratory Animal Technology Co., Ltd. All mice were housed in a SPF-grade animal room (temperature 18–22°C; humidity 40–60%; light/dark cycle 12/12 h each day) and had free access to food and water. HCG11 was cloned and inserted into pHBLV-U6-mcs-CMV-PURO lentiviral vectors (Hanbio Biotechnology Co., Ltd.). A second-generation lentiviral vector system was used. The recombinant lentivirus was generated by co-transfection of 293T cells (The Cell Bank of Type Culture Collection of The Chinese Academy of Sciences) with pHBLV lentiviral plasmid (10 µg), psPAX2 packaging plasmid (10 µg) and pMD2.G envelope plasmid (5 µg; Hanbio Biotechnology Co., Ltd.) using Lipofectamine® 3000 (Invitrogen; Thermo Fisher Scientific, Inc.). After 48 h, the virus was filtered through 0.22 µm cellulose acetate filters (Millipore Corp.) and concentrated by ultracentrifugation (50,000 × g) for 2 h at 4°C. Lentivirus carrying HCG11 or negative control was stably transfected into the A549 cell line (MOI, 20) at 37°C in the presence of 5 µg/ml polybrene (Hanbio Biotechnology Co., Ltd.). At 24 h after transfection, cultures were washed with freshly DMEM containing 10% FBS. After another 48 h, transfected cells were subcutaneously injected into BALB/c nude mice. Euthanasia was performed using pentobarbital sodium (100 mg/kg intraperitoneal injection) at the end of the study. The tumor volume was measured once every 5 days. A total of 30 days after implantation, all mice were sacrificed, and tumor tissues were collected and weighed. Finally, tumor tissues were used for hematoxylin and eosin (H&E) staining (room temperature for 10 sec) and immunohistochemical analysis of Ki-67 expression. All animal experiments performed in this study were approved by the Animal Care and Use Committee of Sichuan Mianyang 404 Hospital (Mianyang, China).
Immunohistochemical staining
Paraffin-embedded tumor tissues were sliced (0.25 µm), dewaxed using xylene and rehydrated using a series of decreasing concentrations of ethanol solutions, and then incubated with anti-Ki-67 antibody (cat. no. ab16667; 1:250; Abcam) at 4°C overnight. Subsequently, tissue sections were incubated with the goat anti-rabbit IgG secondary antibody (cat. no. A0208; 1:50; Beyotime Institute of Biotechnology) at 37°C for 1 h. After counterstaining with hematoxylin at room temperature for 3 min, color development was performed using DAB (Beyotime Institute of Biotechnology).
Bioinformatics
The prediction of the interaction between HCG11 and miR-875 was performed using stabase3.0 (starbase.sysu.edu.cn). The prediction of the interaction between miR-875 and SATB2 was performed using TargetScan (targetscan.org/vert_71/).
Statistical analysis
Statistical analysis was performed using SPSS version 19.0 (IBM Corp.) and GraphPad Prism version 6.0 (GraphPad Software, Inc.) software. A Student's t-test (paired or unpaired) or one-way ANOVA followed by a Tukey-Kramer post hoc test were used to analyze differences amongst groups. A χ2 test was used to examine the association between HCG11 expression and the clinical characteristics. Cox regression models (univariate and multivariate) were used to evaluate the factors related to overall survival. Patients were divided into high and low HCG11 expression groups based on the median expression levels; overall survival was analyzed using the Kaplan-Meier method with the log-rank test. The correlation between the expression levels of HCG11 and miR-875 were analyzed using Pearson's correlation coefficient. All experiments were independently performed at least three times. Data are presented as the mean ± standard deviation. P<0.05 was considered to indicate a statistically significant difference.
Results
HCG11 expression is downregulated in NSCLC tissues and cell lines
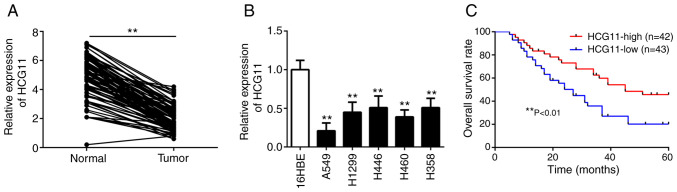
To assess the role of HCG11 in NSCLC, tumor and adjacent normal tissues from NSCLC patients were collected. Subsequently, RT-qPCR analysis showed that the expression levels of HCG11 were significantly decreased in NSCLC tissues compared with adjacent normal tissues (Fig. 1A). Additionally, the expression levels of HCG11 were significantly lower in NSCLC cell lines, particularly in A549 cells, compared with the normal lung epithelial cells (16HBE) (Fig. 1B). To assess the clinical significance of HCG11 expression in NSCLC, patients were further divided into high and low HCG11 expression groups based on the median expression levels. It was found that HCG11 expression was significantly related to tumor size and TNM stage (Table I). However, there was no significant association between HCG11 expression with age, sex, smoking history, tumor differentiation and histological subtype (Table I). In addition, Kaplan-Meier analysis showed that patients with high HCG11 expression had a higher overall survival rate than those with low HCG11 expression (Fig. 1C). In the multivariate analysis, low HCG11 expression was an independent factor in predicting a poor overall survival of NSCLC patients (Table II). These data suggest that HCG11 may serve a key role in regulating the development and progression of NSCLC.
Figure 1.
HCG11 expression is downregulated in NSCLC tissues and cell lines. (A) RT-qPCR analysis of the expression levels of HCG11 in NSCLC tissues and adjacent normal tissues obtained from 85 patients. (B) RT-qPCR analysis of the expression levels of HCG11 in NSCLC cell lines (A549, H1299, H446, H460 and H358) and 16HBE normal lung epithelial cells. (C) Overall survival rates of the 85 NSCLC patients undergoing surgery. Patients were divided into high (n=42) and low HCG11 expression group (n=43) based on the median expression levels. Data is shown as a Kaplan-Meier curve and was analyzed using a log-rank test. **P<0.01. NSCLC, non-small cell lung cancer; RT-qPCR, reverse transcription-quantitative PCR; HCG11, HLA complex group 11.
Table I.
Correlation between HLA complex group 11 expression and clinical characteristics in non-small cell lung cancer patients.
| HCG11 expression | ||||
|---|---|---|---|---|
| Variable | n | High, n=42 | Low, n=43 | P-value |
| Age | 0.452 | |||
| <60 | 37 | 20 | 17 | |
| ≥60 | 48 | 22 | 26 | |
| Sex | 0.593 | |||
| Male | 47 | 22 | 25 | |
| Female | 38 | 20 | 18 | |
| Smoking history | 0.16 | |||
| Yes | 45 | 19 | 26 | |
| No | 40 | 23 | 17 | |
| Tumor size | 0.006a | |||
| >3 cm | 35 | 11 | 24 | |
| ≤3 cm | 50 | 31 | 19 | |
| Differentiation | 0.208 | |||
| Well/moderate | 53 | 29 | 24 | |
| Poor | 32 | 13 | 19 | |
| Histological subtype | 0.622 | |||
| Adenocarcinoma | 31 | 17 | 14 | |
| Squamous cell carcinoma | 43 | 19 | 24 | |
| Others | 11 | 6 | 5 | |
| Tumor-Node-Metastasis stage | 0.003a | |||
| I/II | 51 | 32 | 19 | |
| III/IV | 34 | 10 | 24 | |
P<0.01.
Table II.
Univariate and multivariate analysis of variables for overall survival in non-small cell lung cancer patients.
| Univariate analysis | Multivariate analysis | |||
|---|---|---|---|---|
| Variables | HR (95% CI) | P-value | HR (95% CI) | P-value |
| Age | 1.085 (0.683–1.945) | 0.282 | – | – |
| Sex | 0.967 (0.544–1.876) | 0.368 | – | – |
| Smoking history | 1.636 (0.795–2.418) | 0.149 | – | – |
| Tumor size | 2.978 (1.513–4.751) | 0.004b | 2.668 (1.243–4.421) | 0.012a |
| Differentiation | 1.785 (0.832–2.735) | 0.117 | – | – |
| Histological subtype | 1.213 (0.508–2.047) | 0.456 | – | – |
| Tumor-Node-Metastasis stage | 3.254 (1.362–4.356) | 0.011a | 2.874 (1.193–4.205) | 0.025a |
| HLA complex group 11 expression | 0.356 (0.224–0.643) | 0.001b | 0.435 (0.268–0.781) | 0.008b |
P<0.01
P<0.01. HR, hazard ratio; CI, confidence interval.
HCG11 inhibits proliferation, invasion and migration, and induces apoptosis of NSCLC cells in vitro
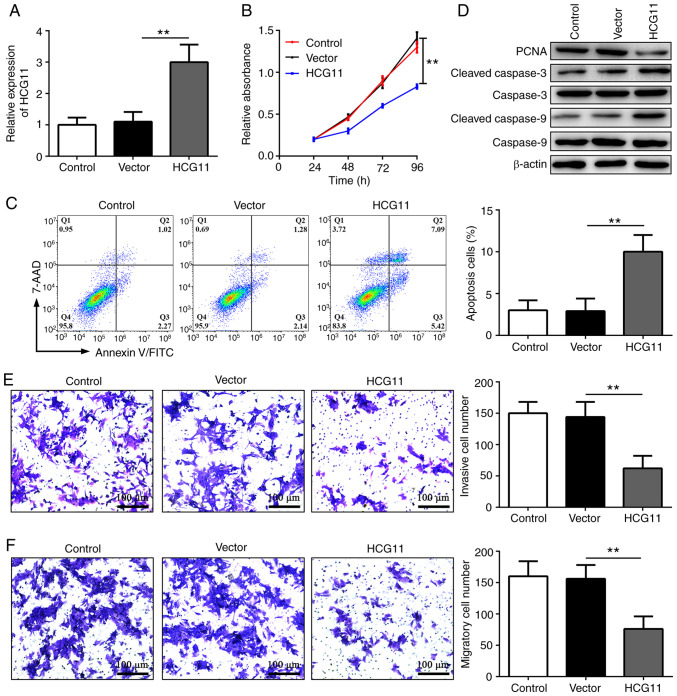
To investigate the function of HCG11 in NSCLC, a HCG11 overexpression plasmid was transfected into the A549 cell line (Fig. 2A). MTT analysis revealed that overexpression of HCG11 significantly reduced cell proliferation (Fig. 2B). Additionally, flow cytometry analysis showed that overexpression of HCG11 increased the apoptosis of NSCLC cells (Fig. 2C). Consistent with these findings, the expression levels of PCNA were decreased, and the expression levels of apoptosis-associated proteins (cleaved-caspase-3 and cleaved-caspase-9) were increased in A549 cells following overexpression of HCG11 (Fig. 2D). Furthermore, the results of Transwell assays showed that HCG11 overexpression significantly reduced the invasion and migration of NSCLC cells (Fig. 2E and F). Therefore, HCG11 may inhibit proliferation, invasion and migration, and induce apoptosis of NSCLC cells.
Figure 2.
HCG11 inhibits proliferation, invasion and migration, and induces apoptosis of non-small cell lung cancer cells in vitro. (A) Reverse transcription-quantitative PCR analysis of the expression levels of HCG11 in the Control, Vector and HCG11 groups. (B) MTT analysis of cell proliferation in the Control, Vector and HCG11 groups. (C) Flow cytometry analysis of cell apoptosis in the Control, Vector and HCG11 groups. (D) Western blot analysis of the protein expression of PCNA, cleaved-caspase-3 and cleaved-caspase-9 in the Control, Vector and HCG11 groups. (E and F) Transwell invasion assays showing (E) cell invasion and (F) migration in the Control, Vector and HCG11 groups. **P<0.01. HCG11, HLA complex group 11; PCNA, proliferation cell nuclear antigen; Vector, empty vector control.
HCG11 suppresses NSCLC tumor growth in vivo
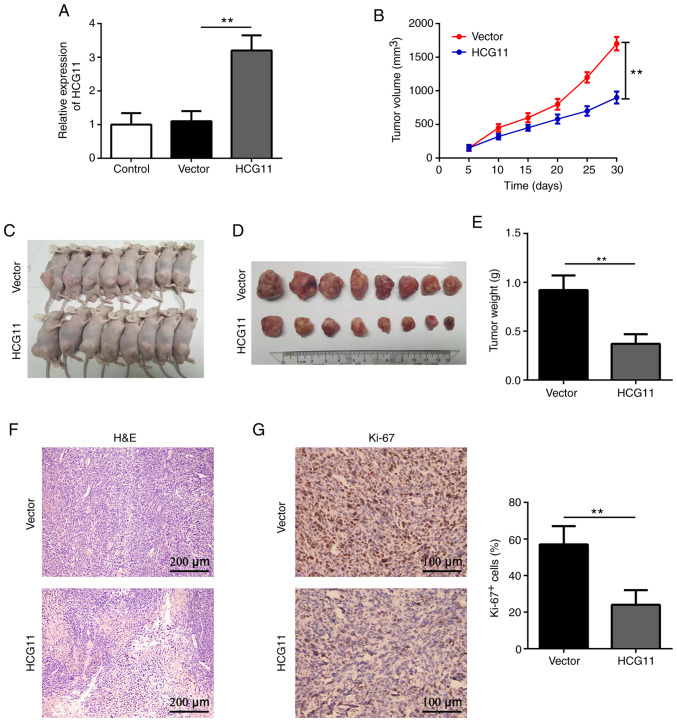
To further validate the effects of HCG11 on NSCLC, A549 cells transfected with HCG11 lentivirus were subcutaneously injected into BALB/c nude mice (Fig. 3A). The results revealed that the tumor volume was significantly decreased following HCG11 overexpression compared with the mice injected with the empty vector transfected cells (Fig. 3B and C). The tumor weight was also significantly lower in the HCG11-overexpression compared with the control group 30 days after implantation (Fig. 3D and E). Further analysis showed that HCG11-overexpressing tumors exhibited a loose tissue structure, accompanied with lower expression of Ki-67, a marker of proliferation (Fig. 3F, and G). These data suggest that HCG11 may inhibit NSCLC tumor growth in vivo.
Figure 3.
HCG11 suppresses NSCLC tumor growth in vivo. (A) Reverse transcription-quantitative PCR analysis of the expression levels of HCG11 in A549 cells after transduction with lentivirus carrying HCG11. (B-E) BALB/c nude mice were subcutaneously injected with A549 cells after transduction with a lentivirus carrying HCG11. (B) Tumor volume was measured once every 5 days. (C-E) A total of 30 days after implantation, all mice were euthanized, and tumor tissues were collected and weighed (n=8 mice per group). (F) H&E staining of the tumor tissue. (G) Immunohistochemical analysis of Ki-67 expression in tumor tissues. **P<0.01. H&E, hematoxylin and eosin; NSCLC, non-small cell lung cancer; Vector, empty vector control; HCG11, HLA complex group 11.
HCG11 functions as a sponge of miR-875 in NSCLC cells
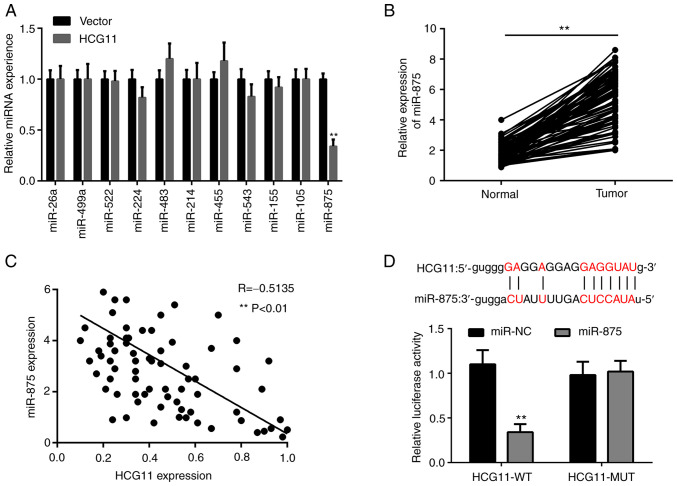
Given that lncRNAs can act as sponges of miRNAs, it was hypothesized that HCG11 may inhibit a number of oncogenic miRNAs in NSCLC cells. Bioinformatics analysis using StarBase version 3.0 (starbase.sysu.edu.cn/) revealed that several miRNAs, including miR-26a, miR-499a, miR-522, miR-224, miR-483, miR-214, miR-455, miR-543, miR-155, miR-105 and miR-875, possessed potential binding sites with HCG11. Interestingly, it was found that the expression levels of miR-875, but not other miRNAs, were significantly decreased in NSCLC cells following HCG11 overexpression (Fig. 4A). Consistent with this finding, miR-875 expression was increased in NSCLC tissues compared with adjacent normal tissues (Fig. 4B). Additionally, there was a significant negative association between HCG11 and miR-875 expression in NSCLC tissues (Fig. 4C), indicating that HCG11 suppressed NSCLC progression, potentially via inhibition of miR-875. Subsequently, a luciferase reporter assay demonstrated that HCG11 could directly bind to miR-875 (Fig. 4D). These results suggest that HCG11 may inhibit the development of NSCLC by inhibiting the expression of miR-875.
Figure 4.
HCG11 functions as a sponge of miR-875 in NSCLC cells. (A) Relative expression levels of miR-26a, miR-499a, miR-522, miR-224, miR-483, miR-214, miR-455, miR-534, miR-155, miR-105 and miR-875 in A549 cells following overexpression of HCG11. (B) Reverse transcription-quantitative PCR analysis of the expression levels of miR-875 in NSCLC tissues and adjacent normal tissues obtained from 85 patients. (C) Pearson correlation analysis of HCG11 and miR-875 expression in NSCLC tissues obtained from 85 patients. (D) Relative luciferase activity in 293T cells after co-transfection of WT or MUT HCG11 with miR-875-mimic or miR-NC. The specific binding site of miR-875 with HCG11 is shown in the top panel, based on StarBase. **P<0.01. NSCLC, non-small cell lung cancer; miR/miRNA, microRNA; NC, negative control; WT, wild-type; MUT, mutant; HCG11, HLA complex group 11.
HCG11 inhibits the malignant phenotype of NSCLC cells by regulating a miR-875/SATB2 axis
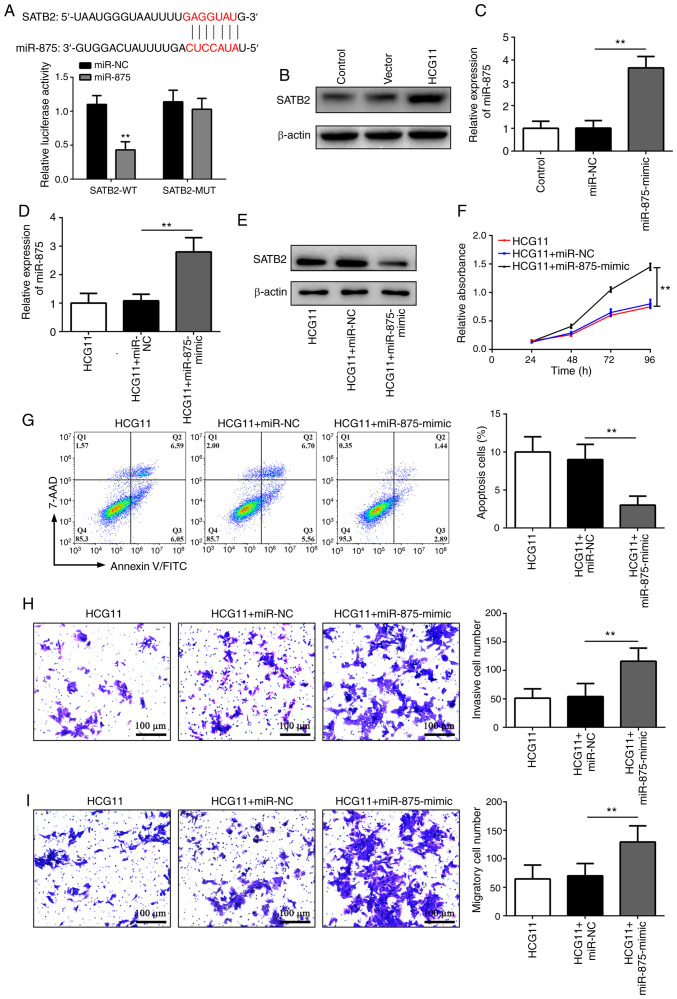
TargetScan (targetscan.org/vert_71/) was used to search for candidate targets with potential miR-875 binding sites. It was observed that there was a common binding site between miR-875 and the 3′-untranslated region of SATB2, a tumor suppressor (23), which was further verified using a luciferase reporter assay (Fig. 5A). Furthermore, it was noticed that SATB2 protein expression was significantly upregulated in HCG11-overexpressing A549 cells in which miR-875 was substantially reduced (Fig. 5B). These findings suggested that SATB2 may be a direct target of miR-875. To further confirm this hypothesis, a miR-875-mimic was used and the transfection efficiency was confirmed by RT-qPCR (Fig. 5C). Specifically, miR-875-mimic treatment could increase miR-875 levels in HCG11-overexpressing A549 cells (Fig. 5D). Overexpression of miR-875 significantly decreased the expression levels of SATB2, thereby reversing the effects of HCG11 on NSCLC cells (Fig. 5E-I). Overall, these findings show that HCG11 may serve a critical role in inhibiting the malignant phenotype of NSCLC cells by regulating a miR-875/SATB2 axis.
Figure 5.
HCG11 inhibits the malignant phenotype of NSCLC cells by regulating a miR-875/SATB2 axis. (A) Relative luciferase activity in 293T cells after co-transfection of WT or MUT SATB2 with miR-875-mimic or miR-NC. The specific binding site of miR-875 with SATB2 mRNA is shown in the top panel, based on TargetScan. (B) Western blot analysis of the expression levels of SATB2 in the Control, Vector and HCG11 groups. (C) Transfection efficiency of miR-875-mimic, as determined by RT-qPCR. (D-I) A549 cells with overexpression of HCG11 were treated with miR-NC or miR-875-mimic. Subsequently, the expression levels of (D) miR-875 were measured by RT-qPCR, and (E) SATB2 protein expression was determined by western blotting. Finally, the (F) proliferation, (G) apoptosis, (H) invasion and (I) migration were analyzed by an MTT assay, flow cytometry or Transwell invasion and migration assays, respectively. **P<0.01. NSCLC, non-small cell lung cancer; miR/miRNA, microRNA; NC, negative control; WT, wild-type; MUT, mutant; RT-qPCR, reverse transcription-quantitative PCR; SATB2, special AT-rich sequence-binding protein 2; HCG11, HLA complex group 11 Vector, empty vector control.
Discussion
Lung cancer is characterized by a high recurrence rate, high mortality rate and low 5-year survival rate, and threatens the lives of individuals worldwide (24–26). Although numerous advances have been made in our understanding of the disease, the prognosis remains unsatisfactory. Therefore, it is necessary to identify more effective strategies for early prevention, detection and treatment of lung cancer. To the best of our knowledge, the present study was the first to show that lncRNA HCG11 functions as a tumor suppressor in NSCLC by regulating a miR-875/SATB2 axis.
The role of lncRNAs in cancer is attracting increasing attention (27–29). Due to the complexity of function and heterogeneity of expression, they serve varying roles in the development and metastasis dependent on the type of cancer. For example, lncRNA H19 imprinted maternally expressed transcript is upregulated and promotes carcinogenesis and metastasis in gastric cancer (30). Additionally, downregulation of lncRNA GAS5 contributes to cell proliferation and predicts adverse consequences in cervical cancer (31). Previous studies have shown that the lncRNA HCG11 affects the progression of glioma, prostate cancer and hepatocellular carcinoma (32,33). However, to the best of our knowledge, the expression levels and role of HCG11 in NSCLC have not yet been explored. The present study revealed that the expression levels of HCG11 were significantly reduced postoperatively in tumor tissues compared with adjacent normal tissues obtained from patients with NSCLC. Reduction in HCG11 expression was observed in different lung cancer cell lines. Of note, the survival rate of patients with high HCG11 expression was higher than that of patients with low HCG11 expression. Cox regression analysis showed that HCG11 expression was an independent predictive factor for overall survival of NSCLC patients. Therefore, these data suggest that HCG11 may exert an inhibitory effect on lung cancer progression. In addition, HCG11 could be used as a prognostic marker of NSCLC.
Uncontrolled proliferation, increased invasion and migration, and decreased apoptosis are the primary causes of the development and progression of cancer (34–36). In the present study, overexpression of HCG11 significantly reduced cell proliferation, invasion and migration, and induced apoptosis in NSCLC cells. In line with these findings, the expression levels of PCNA were decreased, and the levels of cleaved-caspase 3 and cleaved-caspase-9 were increased in NSCLC cells following HCG11 overexpression. More importantly, overexpression of HCG11 also slowed NSCLC tumor growth in nude mice. These results illustrated that HCG11 can inhibit the development and progression of NSCLC.
Previous studies have revealed that HCG11 is involved in various types of cancer via targeting of miRNAs or direct regulation of gene expression (19,37,38). For instance, HCG11 can affect the growth of glioma by regulating the miR-496/cytoplasmic polyadenylation element binding protein 3 or miR-4425/metastasis associated 1 family member 3 axis. Additionally, HCG11 is capable of promoting prostate cancer progression by modulating miR-543 and the AKT/mTOR signaling pathway (33). Furthermore, HCG11 is able to suppress the apoptosis of hepatocellular carcinoma cells by regulating insulin like growth factor 2 mRNA binding protein 1 (19). The present study revealed that HCG11 overexpression resulted in a significant reduction in miR-875 expression, and HCG11 expression was negatively associated with miR-875 expression in NSCLC tissues. In particular, a luciferase reporter assay demonstrated that HCG11 could function as a competing endogenous RNA to bind to miR-875. A recent study reported that miR-875 is involved in facilitating proliferation and inhibiting apoptosis of NSCLC cells (39). Hence, the results of the present study indicated that upregulation of HCG11 can inhibit NSCLC tumor growth, potentially by downregulating miR-875.
miRNAs regulate a wide range of cell biological functions by inhibiting the translation of target genes (40–42). Previous studies have shown that SATB2 is a cancer suppressor gene and its expression is decreased in NSCLC cells, and that downregulation of SATB2 promotes the progression of NSCLC (43–45). However, how expression of SATB2 is regulated in NSCLC cells is still not fully understood. In the present study, using bioinformatics analysis and luciferase reporter assays, it was demonstrated that SATB2 was a direct target gene of miR-875. Consistently, it was noticed that SATB2 expression was significantly increased in NSCLC cells following HCG11 overexpression. miR-875-mimic treatment significantly reduced the HCG11 overexpression-induced upregulation of SATB2, thereby largely abrogating the effects of HCG11 in NSCLC cells. Taken together, these results suggested that HCG11 inhibited proliferation, invasion and migration, and facilitated apoptosis of NSCLC cells, at least in part, by regulating a miR-875/SATB2 axis. However, lncRNAs may be involved in the regulation of cell biology via numerous other mechanisms. Therefore, it is the possible that there are other mechanisms which mediate the antitumor role of HCG11 in NSCLC.
In conclusion, the present study highlighted the importance of HCG11 in alleviating the malignant phenotype of NSCLC cells by targeting a miR-875/SATB2 pathway. These data not only further improve our understanding of the mechanism of regulation of NSCLC, but also highlights a potential therapeutic target for management of NSCLC. Nevertheless, it is necessary to further explore the pathogenesis of NSCLC and treatment strategies.
Acknowledgements
Not applicable.
Funding Statement
The current study was supported by funding from the Scientific research project of Sichuan health and Family Planning Commission (grant no. 17PJ598).
Funding
The current study was supported by funding from the Scientific research project of Sichuan health and Family Planning Commission (grant no. 17PJ598).
Availability of data and materials
The datasets used and analyzed during the present study are available from the corresponding author on reasonable request.
Authors' contributions
ZS and WL conceived and designed the study. MC and RD analyzed and interpreted the results. LS and QZ performed the experiments. ZS and WL confirmed the authenticity of all the raw data. All authors have read and approved the final manuscript.
Ethics approval and consent to participate
All animal experiments in this study were approved by the Animal Care and Use Committee of Sichuan Mianyang 404 Hospital and performed in accordance with the Guide for the Care and Use of Laboratory Animals. The use of human samples was approved by the Ethics Committee of The Sichuan Mianyang 404 Hospital (Mianyang, China; approval no. 008), and each patient provided a written informed consent.
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing interests.
References
- 1.Jemal A, Bray F, Center MM, Ferlay J, Ward E, Forman D. Global cancer statistics. CA Cancer J Clin. 2011;61:69–90. doi: 10.3322/caac.20107. [DOI] [PubMed] [Google Scholar]
- 2.Carr SR, Akerley W, Cannon-Albright LA. Genetic contribution to nonsquamous, non-small cell lung cancer in nonsmokers. J Thorac Oncol. 2018;13:938–945. doi: 10.1016/j.jtho.2018.03.024. [DOI] [PubMed] [Google Scholar]
- 3.Greathouse KL, White JR, Vargas AJ, Bliskovsky VV, Beck JA, von Muhlinen N, Polley EC, Bowman ED, Khan MA, Robles AI, et al. Interaction between the microbiome and TP53 in human lung cancer. Genome Biol. 2018;19:123. doi: 10.1186/s13059-018-1501-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Velcheti V, Madison R, Ali SM, Schrock AB. WAC/RET: A novel RET oncogenic fusion variant in non-small cell lung carcinoma. J Thorac Oncol. 2018;13:e122–e123. doi: 10.1016/j.jtho.2018.03.003. [DOI] [PubMed] [Google Scholar]
- 5.Tsui DWY, Murtaza M, Wong ASC, Rueda OM, Smith CG, Chandrananda D, Soo RA, Lim HL, Goh BC, Caldas C, et al. Dynamics of multiple resistance mechanisms in plasma DNA during EGFR-targeted therapies in non-small cell lung cancer. EMBO Mol Med. 2018;10:e7945. doi: 10.15252/emmm.201707945. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Li S, Yang J, Xia Y, Fan Q, Yang KP. Long noncoding RNA NEAT1 promotes proliferation and invasion via targeting miR-181a-5p in non-small cell lung cancer. Oncol Res. 2018;26:289–296. doi: 10.3727/096504017X15009404458675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Thomas A, Rajan A, Lopez-Chavez A, Wang Y, Giaccone G. From targets to targeted therapies and molecular profiling in non-small cell lung carcinoma. Ann Oncol. 2013;24:577–585. doi: 10.1093/annonc/mds478. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Vijayvergia N, Shah PC, Denlinger CS. Survivorship in non-small cell lung cancer: Challenges faced and steps forward. J Natl Compr Canc Netw. 2015;13:1151–1161. doi: 10.6004/jnccn.2015.0140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Kudo Y, Haymaker C, Zhang J, Reuben A, Duose DY, Fujimoto J, Roy-Chowdhuri S, Solis Soto LM, Dejima H, Parra ER, et al. Suppressed immune microenvironment and repertoire in brain metastases from patients with resected non-small-cell lung cancer. Ann Oncol. 2019;30:1521–1530. doi: 10.1093/annonc/mdz207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Tang Y, Cao G, Zhao G, Wang C, Qin Q. LncRNA differentiation antagonizing non-protein coding RNA promotes proliferation and invasion through regulating miR-135a/NLRP37 axis in pancreatic cancer. Invest New Drugs. 2020;38:714–721. doi: 10.1007/s10637-019-00798-0. [DOI] [PubMed] [Google Scholar]
- 11.Chen LL. Linking Long noncoding RNA localization and function. Trends Biochem Sci. 2016;41:761–772. doi: 10.1016/j.tibs.2016.07.003. [DOI] [PubMed] [Google Scholar]
- 12.St Laurent G, Wahlestedt C, Kapranov P. The Landscape of long noncoding RNA classification. Trends Genet. 2015;31:239–251. doi: 10.1016/j.tig.2015.03.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Kopp F, Mendell JT. Functional classification and experimental dissection of long noncoding RNAs. Cell. 2018;172:393–407. doi: 10.1016/j.cell.2018.01.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Jin J, Wang H, Si J, Ni R, Liu Y, Wang J. ZEB1-AS1 is associated with poor prognosis in non-small-cell lung cancer and influences cell migration and apoptosis by repressing ID1. Clin Sci (Lond) 2019;133:381–392. doi: 10.1042/CS20180983. [DOI] [PubMed] [Google Scholar]
- 15.Yang H, Yan L, Sun K, Sun X, Zhang X, Cai K, Song T. lncRNA BCAR4 increases viability, invasion, and migration of non-small cell lung cancer cells by targeting glioma-associated oncogene 2 (GLI2) Oncol Res. 2019;27:359–369. doi: 10.3727/096504018X15220594629967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Shi SL, Zhang ZH. Long non-coding RNA SNHG1 contributes to cisplatin resistance in non-small cell lung cancer by regulating miR-140-5p/Wnt/β-catenin pathway. Neoplasma. 2019;66:756–765. doi: 10.4149/neo_2018_181218N980. [DOI] [PubMed] [Google Scholar]
- 17.Kamel LM, Atef DM, Mackawy AMH, Shalaby SM, Abdelraheim N. Circulating long non-coding RNA GAS5 and SOX2OT as potential biomarkers for diagnosis and prognosis of non-small cell lung cancer. Biotechnol Appl Biochem. 2019;66:634–642. doi: 10.1002/bab.1764. [DOI] [PubMed] [Google Scholar]
- 18.Zhou Y, Shi H, Du Y, Zhao G, Wang X, Li Q, Liu J, Ye L, Shen Z, Guo Y, Huang Y. lncRNA DLEU2 modulates cell proliferation and invasion of non-small cell lung cancer by regulating miR-30c-5p/SOX9 axis. Aging (Albany NY) 2019;11:7386–7401. doi: 10.18632/aging.102226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Xu Y, Zheng Y, Liu H, Li T. Modulation of IGF2BP1 by long non-coding RNA HCG11 suppresses apoptosis of hepatocellular carcinoma cells via MAPK signaling transduction. Int J Oncol. 2017;51:791–800. doi: 10.3892/ijo.2017.4066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Liu H, Li J, Koirala P, Ding X, Chen B, Wang Y, Wang Z, Wang C, Zhang X, Mo YY. Long non-coding RNAs as prognostic markers in human breast cancer. Oncotarget. 2016;7:20584–20596. doi: 10.18632/oncotarget.7828. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Zhang Y, Zhang P, Wan X, Su X, Kong Z, Zhai Q, Xiang X, Li L, Li Y. Downregulation of long non-coding RNA HCG11 predicts a poor prognosis in prostate cancer. Biomed Pharmacother. 2016;83:936–941. doi: 10.1016/j.biopha.2016.08.013. [DOI] [PubMed] [Google Scholar]
- 22.Costa GJ, Mello MJG, Bergmann A, Ferreira CG, Thuler LCS. Tumor-node-metastasis staging and treatment patterns of 73,167 patients with lung cancer in Brazil. J Bras Pneumol. 2020;46:e20180251. doi: 10.1590/1806-3713/e20180251. (In English, Portuguese) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Roy SK, Shrivastava A, Srivastav S, Shankar S, Srivastava RK. SATB2 is a novel biomarker and therapeutic target for cancer. J Cell Mol Med. 2020;24:11064–11069. doi: 10.1111/jcmm.15755. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Jemal A, Siegel R, Xu J, Ward E. Cancer statistics, 2010. CA Cancer J Clin. 2010;60:277–300. doi: 10.3322/caac.20073. [DOI] [PubMed] [Google Scholar]
- 25.van Rens MT, Zanen P, Brutel de La Riviere A, Elbers HR, van Swieten HA, van Den Bosch JM. Survival in synchronous vs. single lung cancer: upstaging better reflects prognosis. Chest. 2000;118:952–958. doi: 10.1378/chest.118.4.952. [DOI] [PubMed] [Google Scholar]
- 26.Marcus MW, Raji OY, Field JK. Lung cancer screening: Identifying the high risk cohort. J Thorac Dis. 2015;7(Suppl 2):S156–S162. doi: 10.3978/j.issn.2072-1439.2015.04.19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Li Y, Li L, Wang Z, Pan T, Sahni N, Jin X, Wang G, Li J, Zheng X, Zhang Y, et al. LncMAP: Pan-cancer atlas of long noncoding RNA-mediated transcriptional network perturbations. Nucleic Acids Res. 2018;46:1113–1123. doi: 10.1093/nar/gkx1311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Zhang E, He X, Zhang C, Su J, Lu X, Si X, Chen J, Yin D, Han L, De W. A novel long noncoding RNA HOXC-AS3 mediates tumorigenesis of gastric cancer by binding to YBX1. Genome Biol. 2018;19:154. doi: 10.1186/s13059-018-1523-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Khan MR, Xiang S, Song Z, Wu M. The p53-inducible long noncoding RNA TRINGS protects cancer cells from necrosis under glucose starvation. EMBO J. 2017;36:3483–3500. doi: 10.15252/embj.201696239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Yang F, Bi J, Xue X, Zheng L, Zhi K, Hua J, Fang G. Up-regulated long non-coding RNA H19 contributes to proliferation of gastric cancer cells. FEBS J. 2012;279:3159–3165. doi: 10.1111/j.1742-4658.2012.08694.x. [DOI] [PubMed] [Google Scholar]
- 31.Cao S, Liu W, Li F, Zhao W, Qin C. Decreased expression of lncRNA GAS5 predicts a poor prognosis in cervical cancer. Int J Clin Exp Pathol. 2014;7:6776–6783. [PMC free article] [PubMed] [Google Scholar]
- 32.Chen Y, Bao C, Zhang X, Lin X, Huang H, Wang Z. Long non-coding RNA HCG11 modulates glioma progression through cooperating with miR-496/CPEB3 axis. Cell Prolif. 2019;52:e12615. doi: 10.1111/cpr.12615. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Wang YC, He WY, Dong CH, Pei L, Ma YL. lncRNA HCG11 regulates cell progression by targeting miR-543 and regulating AKT/mTOR pathway in prostate cancer. Cell Biol Int. 2019 Nun; doi: 10.1002/cbin.11194. (Epub ahead of print). doi: 10.1002/cbin.11194. [DOI] [PubMed] [Google Scholar]
- 34.Cramer JD, Burtness B, Le QT, Ferris RL. The changing therapeutic landscape of head and neck cancer. Nat Rev Clin Oncol. 2019;16:669–683. doi: 10.1038/s41571-019-0227-z. [DOI] [PubMed] [Google Scholar]
- 35.Evans ER, Bugga P, Asthana V, Drezek R. Metallic Nanoparticles for Cancer Immunotherapy. Mater Today (Kidlington) 2018;21:673–685. doi: 10.1016/j.mattod.2017.11.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Turajlic S, Sottoriva A, Graham T, Swanton C. Resolving genetic heterogeneity in cancer. Nat Rev Genet. 2019;20:404–416. doi: 10.1038/s41576-019-0114-6. [DOI] [PubMed] [Google Scholar]
- 37.Long R, Liu Z, Li J, Zhang Y, Yu H. HCG11 up-regulation induced by ELK4 suppressed proliferation in vestibular schwannoma by targeting miR-620/ELK4. Cancer Cell Int. 2021;21:5. doi: 10.1186/s12935-020-01691-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Li XF, Hu DM, Zhao YX, Zhang L, Jin Y. Knockdown of lncRNA HCG11 suppresses cell progression in ovarian cancer by modulating miR-144-3p/PBX3. Eur Rev Med Pharmacol Sci. 2020;24:11032–11040. doi: 10.26355/eurrev_202011_23588. [DOI] [PubMed] [Google Scholar]
- 39.Tong JL, Wang LL, Ling XF, Wang MX, Cao W, Liu YY. MiR-875 can regulate the proliferation and apoptosis of non-small cell lung cancer cells via targeting SOCS2. Eur Rev Med Pharmacol Sci. 2019;23:5235–5241. doi: 10.26355/eurrev_201906_18189. [DOI] [PubMed] [Google Scholar]
- 40.Liang C, Liu H, Hao J, Li J, Luo L. Expression profiling and regulatory network of cucumber microRNAs and their putative target genes in response to cucumber green mottle mosaic virus infection. Arch Virol. 2019;164:1121–1134. doi: 10.1007/s00705-019-04152-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Li J, Huang L, Xiao X, Chen Y, Wang X, Zhou Z, Zhang C, Zhang Y. Photoclickable microRNA for the intracellular target identification of microRNAs. J Am Chem Soc. 2016;138:15943–15949. doi: 10.1021/jacs.6b08521. [DOI] [PubMed] [Google Scholar]
- 42.Wang Y, Luo J, Zhang H, Lu J. microRNAs in the same clusters evolve to coordinately regulate functionally related genes. Mol Biol Evol. 2016;33:2232–2247. doi: 10.1093/molbev/msw089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Kucuksayan H, Ozes ON, Akca H. Downregulation of SATB2 is critical for induction of epithelial-to-mesenchymal transition and invasion of NSCLC cells. Lung Cancer. 2016;98:122–129. doi: 10.1016/j.lungcan.2016.05.032. [DOI] [PubMed] [Google Scholar]
- 44.Tian W, Wang G, Liu Y, Huang Z, Zhang C, Ning K, Yu C, Shen Y, Wang M, Li Y, et al. The miR-599 promotes non-small cell lung cancer cell invasion via SATB2. Biochem Biophys Res Commun. 2017;485:35–40. doi: 10.1016/j.bbrc.2017.02.005. [DOI] [PubMed] [Google Scholar]
- 45.Ma YN, Zhang HY, Fei LR, Zhang MY, Wang CC, Luo Y, Han YC. SATB2 suppresses non-small cell lung cancer invasiveness by G9a. Clin Exp Med. 2018;18:37–44. doi: 10.1007/s10238-017-0464-3. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
The datasets used and analyzed during the present study are available from the corresponding author on reasonable request.