Abstract
Lipid droplets (LDs) are dynamic fat-storage organelles that interact readily with numerous cellular structures and organelles. A prominent LD contact site is with degradative vesicles such as autophagosomes, lysosomes, autolysosomes, and late endosomes. These contacts support lipid catabolism through the selective autophagy of LDs (i.e., lipophagy) or the recruitment of cytosolic lipases to the LD surface (i.e., lipolysis). However, LD-autophagosome contacts serve additional functions beyond lipid catabolism, including the supply of lipids for autophagosome biogenesis. In this review, we discuss the molecular mediators of LD contacts with autophagosomes and other degradative organelles as well as the diverse cellular functions of these contact sites in health and disease.
Keywords: autophagosome, autophagy, cell biology, endosome, lipid droplet, lysosome
Lipid droplets (LDs) are fat-storage organelles that play diverse roles in health and disease. Present in nearly every cell type and across virtually all species, LDs are first formed at the endoplasmic reticulum (ER) where diacylglycerol, acetyl coenzyme A, lecithin retinol and other acyltransferases (DGATs, ACATs, LRATs), generate triglycerides (TAGs), and cholesterol esters (CEs) that accumulate between the two leaflets of the ER lipid bilayer (Olzmann and Carvalho, 2019). Nascent LDs extend into the cytoplasm from the ER, surrounded by a phospholipid monolayer that is distinct in membrane composition from that of the ER (Tauchi-Sato et al., 2002; Jackson, 2019). The LD monolayer recruits a distinct proteome that regulates the access of stored lipids by lipases such as adipose triglyceride lipase (ATGL), hormone sensitive lipase (HSL), lysosomal acid lipase, and many others that have varying subcellular localizations and affinities for different neutral lipid species. The LD is also host to numerous additional adaptors including an extensive network of small Rab guanosine triphosphatases (GTPases) that facilitate intracellular trafficking and contact with other organelles. In fact, many LD-organelle contacts are now appreciated to contribute to numerous functions related to lipid transport, protein degradation, and metabolism (Schuldiner and Bohnert, 2017; Bersuker and Olzmann, 2018; Henne and Hariri, 2018; Krahmer and Mann, 2019; Thiam and Dugail, 2019).
An excessive accumulation of LDs is a critical factor in many disease pathologies (i.e., fatty liver disease, obesity, atherosclerosis, diabetes), which has generated intense research interest in defining how these complex organelles are metabolized (Onal et al., 2017). Lipolysis appears to be a predominant pathway for LD breakdown through the action of soluble lipases like ATGL and HSL. However, a seminal study by Singh et al. (2009) showed an additional pathway for LD breakdown involving the autophagic machinery. The selective autophagy of LDs, termed lipophagy, is thought to require autophagosomes that engulf LDs and, upon fusion with the lysosome, hydrolyze TAG and CE through the action of lysosomal acid lipase (Zechner et al., 2017; Olzmann and Carvalho, 2019).
The concept of lipophagy has inspired considerable interest in defining how LDs interact with degradative vesicles (i.e., autophagosomes, late endosomes, multivesicular bodies [MVBs], and lysosomes). These heterotypic, organelle-organelle contacts are widespread but variable in nature. For example, LD-lysosome contacts have been defined as transient kiss-and-run events that function in protein transfer and perhaps piecemeal lipid catabolism (Kaushik and Cuervo, 2015; Schroeder et al., 2015; Schulze et al., 2017). In contrast, LDs can form stable interactions with autophagosomes, or the yeast vacuole as discussed later, and result in the complete envelopment of the LD within these vesicles (Garcia et al., 2018). These diverse LD contact sites likely comprise unique proteomes and phospholipid membrane dynamics that are yet to be fully understood. The discovery of new molecular targets that facilitate some of these interactions has uncovered a diverse set of functions in addition to lipophagy.
In this review, we highlight recent insights into the function of LD contacts with degradative vesicles during lipophagy. In addition, LD transient interactions with autophagic vesicles have also been implicated in the trafficking of cytosolic lipases to the LD surface. LDs may also serve as lipid reservoirs at sites of autophagosome biogenesis. These LD contacts are relevant to human health, particularly in liver diseases of lipid metabolism and viral infection. Later, we describe the diverse functions resulting from LD associations with autophagosomes and other degradative organelles in more detail (Figure 1).
Figure 1.
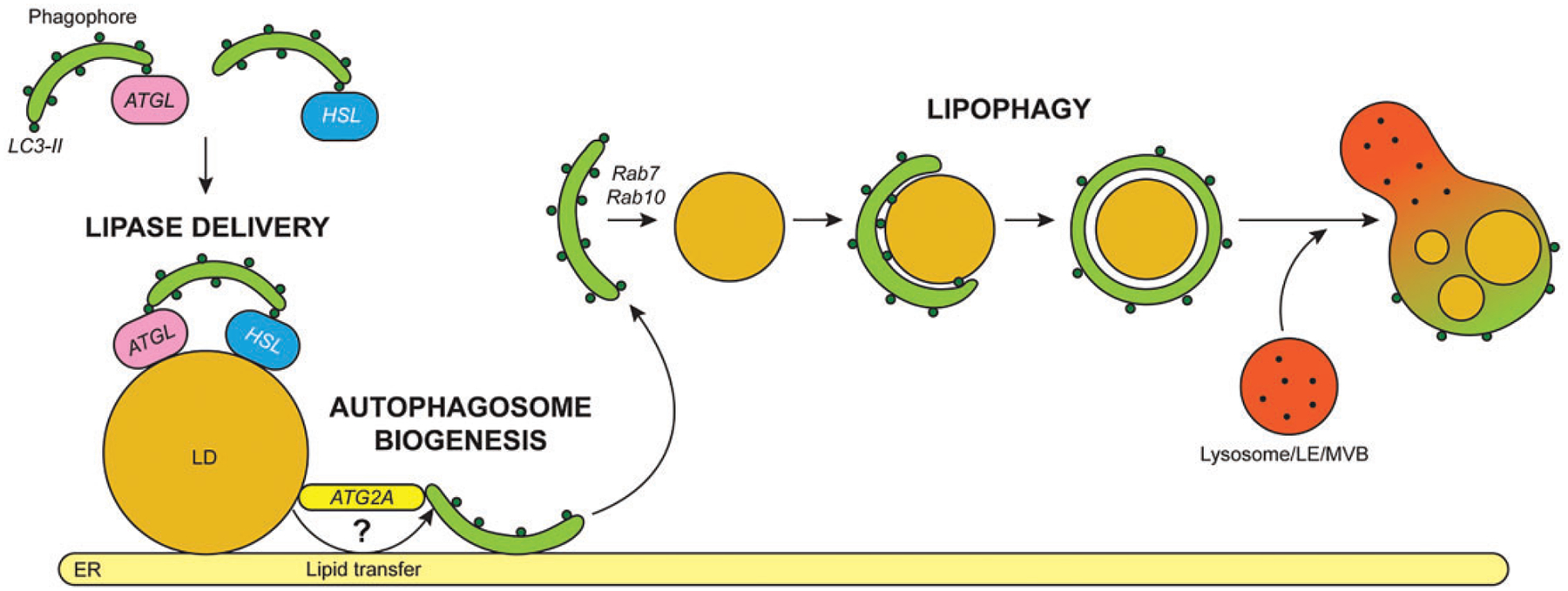
Diverse Functions of LD-Autophagosome Interactions. Traditionally, LD associations with autophagic membranes have been thought to result in LD breakdown via lipophagy. During this process, Rab7 and Rab10 assist in LD recognition and engulfment within autophagosomes, which fuse with lysosomes, LEs or MVBs containing lysosomal acid lipase. In addition to lipophagy, autophagosomes can also facilitate lipolysis by delivering ATGL and HSL to the LD surface via lipase interactions with LC3. Finally, neutral lipids stored within LDs can serve as a lipid source for autophagosome biogenesis, perhaps through the tethering and lipid transfer activity of Atg2a at the ER. LEs = late endosomes; MVBs = multivesicular bodies; ER = endoplasmic reticulum; LD = lipid droplet; ATGL = adipose triglyceride LD lipase. HSL = hormone sensitive lipase.
LD Contacts With Autophagosomes and Other Degradative Organelles During Lipophagy
Mechanisms of Selective Autophagy
Selective targeting of organelles and proteins by the autophagy machinery has been an area of intense investigation (Gatica et al., 2018). As of today, nearly 20 types of selective autophagy pathways have been described to include autophagic degradation of mitochondria (mitophagy), peroxisomes (pexophagy), lysosomes (lysophagy), ER (reticulophagy), LDs (lipophagy), aggregated and misfolded proteins (aggrephagy), and pathogens (xenophagy) to name a few (Kim et al., 2008; Singh et al., 2009; Thurston et al., 2009; Zheng et al., 2009; Deosaran et al., 2013; Hung et al. 2013; Khaminets et al., 2015; Lazarou et al., 2015; Khaminets et al., 2016). These diverse selective autophagy pathways share a common mechanism of utilizing autophagy receptors that recognize specific cargo that is tagged for degradation and provide a physical bridge with autophagosomes to ensure selective engulfment of that particular cargo (Stolz et al., 2014).
Ubiquitination is a common post-translational modification that signals for selective autophagy and can target damaged organelles, aggregated proteins or pathogens to mark them for selective autophagy, although ubiquitin-independent signals also exist (Kirkin et al., 2009; Khaminets et al., 2016). Ubiquitinated proteins are recognized by autophagy receptors such as sequestosome-1 (SQSTM1, also known as p62), optineurin (OPTN), neighbor of BRCA1 gene 1 (NBR1), and nuclear dot protein 52 kDa (NDP52) that physically associate with ubiquitin-modified cargoes through consensus ubiquitin-binding domains (UBDs; Stolz et al., 2014). In addition to binding ubiquitin, autophagy receptors also contain an LC3-interacting region (LIR) with the consensus sequence of Trp/Phe/Tyr-X-X-Leu/Ile/Val. Through this sequence, autophagy receptors bind directly to the LC3/GABARAP family of proteins that decorate autophagic membranes (Rogov et al., 2014). LC3 interactions with autophagy receptors function to recruit nascent phagophores to ubiquitinated cargo. Interestingly, this interaction also stimulates autophagosome formation around the cargo that is destined for degradation (Kamber et al., 2015b). Thus, autophagy receptors behave not only as a physical link between ubiquitinated cargo and autophagosomes, but can also engage in targeted autophagosome biogenesis around the cargo itself (Turco et al., 2019; Vargas et al., 2019). Despite the considerable advancement in our understanding of selective autophagy, how these mechanisms apply to LD autophagy is not fully understood.
A Role for Ubiquitin and Autophagy Receptors During Lipophagy
A key role for autophagy as a means to degrade cellular lipid was demonstrated roughly a decade ago using autophagy-deficient hepatocytes and mouse embryonic fibroblasts (MEFs) which accumulate excess LDs compared to control cells (Singh et al., 2009). In this ground-breaking study, Singh et al. further demonstrated that LC3-labeled autophagosomes and Lamp1-labeled lysosomes intimately associate with LDs to drive autophagic LD catabolism which previously had been thought to occur mainly through utilization of cytosolic lipases. It was proposed that autophagosomes consume LDs whole or through piecemeal sampling prior to fusion with the lysosomes. Today, lipophagy is considered to be a central LD breakdown pathway in many different cell types, although our understanding of the molecular mechanisms that mediate LD recognition by autophagy machinery is only beginning to emerge.
The LD is host to several ubiquitin adaptors as well as ubiquitinated proteins (Bersuker and Olzmann, 2019). While ubiquitination of LD-resident proteins likely serves as a signal for proteasome degradation, ubiquitin modifications may also signal autophagosome recruitment to the LD similar to other forms of selective autophagy. Consistent with this, many LD-resident proteins involved in ubiquitin conjugation, binding, and autophagy are now known to influence LD breakdown. For example, spartin, a protein involved in hereditary spastic paraplegia, localizes to LDs and recruits the E3 ubiquitin ligase AIP4 to the LD surface (Eastman et al., 2009; Edwards et al., 2009; Hooper et al., 2010). Here, spartin increases the activity of AIP4 ligase on the LD, which then ubiquitinates the LD-resident protein PLIN2. Depletion of endogenous spartin increased LD content after oleic acid loading, suggesting that spartin/AIP4-mediated ubiquitination of PLIN2 may influence LD breakdown. However, overexpression of spartin similarly increased LD content, suggesting a complex interplay between LD ubiquitination and lipid metabolism (Eastman et al., 2009). Whether spartin/AIP4 ubiquitination of LD proteins provides a stimulus for lipophagy as opposed to proteasome degradation will require future exploration. Other ubiquitin-modifying proteins such as mysterin and ancient ubiquitous protein 1 (AUP1)-E2 ubiquitin conjugase Ubc7 complex have been found to reside on the LD surface as well, although their role during lipophagy remains unclear (Klemm et al., 2011; Spandl et al., 2011; Jo et al., 2013).
The notion that ubiquitination can serve as a signal for lipophagy is supported by studies implicating autophagy receptors and other ubiquitin binding proteins in LD-autophagosome interactions. In a recent study of Drosophila hepatocyte-like oenocytes and human HepG2 hepatoma cells, nutrient starvation induced aggregates of the autophagy receptor p62 as well as the autophagy regulator histone deacetylase 6 (HDAC6), which selectively recruits LDs for autophagic degradation (Yan et al., 2019). HDAC6 depletion prevented LD breakdown, as did the overexpression of HDAC6 mutants that are not able to bind polyubiquitin and overexpression of p62 mutants that are not able to bind LC3 and ubiquitin. This suggests that HDAC6 and p62 bind cooperatively to ubiquitinated cargo on the LD surface and to LC3-labeled autophagosomes during lipophagy. In support of these findings, an additional study demonstrated that both p62 and LC3-labeled autophagosomes colocalize with LDs in AML12 mouse hepatocytes acutely exposed to alcohol, a known stimulus for autophagy (Wang et al., 2017). This further suggests that p62 may function as a selective lipophagy receptor that recruits autophagosomes to LDs in response to autophagy activation (Figure 2).
Figure 2.
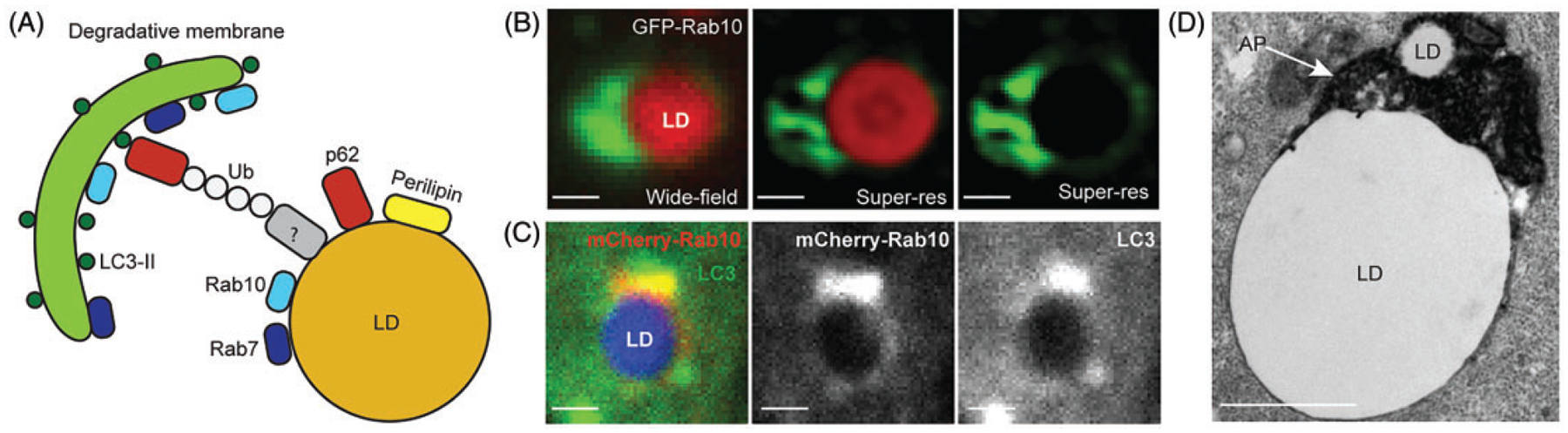
Proposed Model of LD Contacts With Degradative Organelles. Panel A: A cartoon showing proposed proteins involved in regulating LD associations with APs, lysosomes, MVBs and other degradative vesicles. Panel B: Comparison of images of a GFP-Rab10-positive LD from nutrient-starved Huh7 cells observed using wide-field epifluorescence (left) or super-resolution microscopy (middle and right). Panel C: Images from nutrient-starved Hep3B cells showing colocalization between LD-localized mCherry-Rab10 and endogenous LC3 (LD is shown in blue). Panel D: Low-magnification electron microscopy image of LD-AP or other degradative lysosomal membrane interactions in Huh7 cells that are expressing GFP-tagged active Rab10 (Q68L) form and have been starved in nutrient-depleted medium. Scale bars, 1 μm. Microscopy images (B-D) have been adapted from Li et al. (2016).
LD = lipid droplet; AP = autophagosome.
What proteins on the LD surface are recognized by the selective autophagy machinery? One possibility is PLIN1, a LD-resident protein that is highly expressed in adipose tissue, which was shown to be subjected to ubiquitin modification and recognition by p62 in response to inflammatory cytokines secreted during obesity (Ju et al., 2019). As described earlier, PLIN2 is also ubiquitinated by spartin/AIP4, but its role in p62-autophagosome association remains to be explored. Finally, Huntingtin (Htt) has been proposed to function as a selective autophagy scaffold protein on the LD that promotes p62 binding to both ubiquitinated cargos and LC3-positive autophagosomes (Rui et al., 2015). Loss of Htt expression reduced LC3-LD colocalization and increased LD content, suggesting that Htt might also aid in LD targeting by autophagosomes similar to p62 and HDAC6. Clearly, a number of different proteins with innate functions in ubiquitin modification/binding or autophagy have a potential to regulate LD associations with autophagosomes and provide interesting future avenues to explore (Figure 2 and Table 1).
Table 1.
List of Proteins Implicated in LD Contacts With Degradative Vesicles.
| Proteins | Process | Organism | Proposed role | Citation |
|---|---|---|---|---|
| AIP4 | Unclear | Mammals | Ubiquitin ligase; ubiquitinates LD-resident protein PLIN2 | Edwards et al., 2009; Hooper et al., 2010 |
| Atg14L | Autophagosome biogenesis | Mammals | Autophagy-related protein; localizes to LDs; proposed to transport lipids; knockdown increases LD levels | Pfisterer et al., 2014 |
| Atg2a/b | Autophagosome biogenesis | Mammals | Autophagy-related protein; localizes to LDs; transports lipids; knockdown increases LD levels | Velikkakath et al., 2012; Pfisterer et al., 2014; Maeda et al., 2019; Valverde et al., 2019 |
| Atg5 | Lipophagy | Mammals | Autophagy-related protein; regulates LC3 lipidation | Singh et al., 2009 |
| Atg7 | Lipophagy | Mammals | Autophagy-related protein; regulates LC3 lipidation | Singh et al., 2009 |
| ATGL | Lipolysis | Mammals | Lipase; interacts with LC3 through LIR motif | Martinez-Lopez et al., 2016 |
| AUP1 | Lipophagy | Mammals | Localizes to LDs, ubiquitinated following DENV infection; binds the E2 ubiquitin ligase Ube2g2 | Wang et al., 2017; Yan et al., 2019 |
| HDAC6 | Lipophagy | Mammals | Autophagy regulator; binds to ubiquitinated cargo on the LD surface; might cooperate with p62 | Yan et al., 2019 |
| Hsc70 | CMA | Mammals | Chaperone; recognizes and unfolds substrates | Kaushik & Cuervo, 2015 |
| HSL | Lipolysis | Mammals | Lipase; interacts with LC3 through LIR motif | Martinez-Lopez et al., 2016 |
| Huntingtin | Lipophagy | Mammals | Selective autophagy scaffold; promotes p62 binding to ubiquitinated cargo and LC3-positive autophagosomes on the LD surface | Rui et al., 2015 |
| Lamp2A | CMA | Mammals | Receptor; binds and translocates substrates across lysosomal lumen | Kaushik & Cuervo, 2015 |
| Mysterin | Unclear | Mammals | Localizes to LDs; contains a ubiquitin ligase domain; possible mediator of LD catabolism | Sugihara et al., 2019 |
| p62 | Lipophagy | Mammals | Selective autophagy receptor; binds to ubiquitinated cargo on the LD surface and recruits autophagosomes | Wang et al., 2017; Yan et al., 2019 |
| Plin1 | LD catabolism | Mammals | LD-resident protein; ubiquitinated and interacts with p62 | Ju et al., 2019 |
| Plin2 | CMA | Mammals | LD-resident protein; CMA substrate; interacts with Lamp2A; ubiquitinated by Spartin-AIP4 (unknown function) | Hooper et al., 2010; Kaushik & Cuervo, 2015 |
| Plin3 | CMA | Mammals | LD-resident protein, CMA substrate; interacts with Lamp2A | Kaushik & Cuervo, 2015 |
| PNPLA5 | Autophagosome biogenesis | Mammals | Triglyceride lipase; supplies lipids for autophagosome biogenesis | Dupont et al., 2014 |
| Rab10 | Lipophagy | Mammals | Membrane trafficking protein; mediates autophagic LD engulfment | Schroeder et al., 2015; Schulze et al., 2017 |
| Rab7 | Lipophagy | Mammals | Membrane trafficking protein; mediates LD association with lysosomes and MVBs | Schroeder et al., 2015; Li et al., 2016; Schulze et al., 2017 |
| Spartin | Unclear | Mammals | Recruits the ubiquitin ligase AIP4 to the LD surface which ubiquitinates LD-resident protein PLIN2; affects LD content | Eastman et al., 2009; Edwards et al., 2009; Hooper et al., 2010 |
| Ubc7/Ube2g2 | Unclear | Mammals | Localizes to LDs; contains ubiquitin ligase domain; associates with AUP1; possible role in lipophagy | Spandl et al., 2011; Klemm et al., 2011; Jo et al., 2013 |
| WIPI1–2 | Autophagosome biogenesis | Mammals | Early autophagy protein; localizes near LDs during autophagy induction | Velikkakath et al., 2012; Dupont et al., 2014; Pfisterer et al., 2014; Valverde et al., 2019; Maeda et al., 2019 |
| Atg1,3–10, Atg12, Atg14, Atg16 and Vps34 | Microlipophagy | Yeast | Core autophagy proteins; regulate LD uptake into the vacuole | van Zutphe et al., 2014; Wang et al., 2014; Seo et al., 2017 |
| Vps4, Vps27 | Microlipophagy | Yeast | Endosomal sorting complexes required for transport proteins; regulate LD uptake into the vacuole | Wang et al., 2014; Vevea et al., 2015; Oku et al., 2017 |
LD = lipid droplet; MVB = multivesicular body; CMA = Chaperon-mediated autophagy; HSL = hormone sensitive lipase.
Small Rab GTPases as Mediators of LD Contacts With Degradative Organelles
Several members of the vesicular trafficking machinery, particularly small Rab GTPases, have been found to reside on the LD surface and mediate various steps of LD metabolism. Rab GTPases are molecular switches that cycle between active (GTP-bound) or inactive (GDP-bound) states and regulate nearly all membrane trafficking processes in the cell (Zhen and Stenmark, 2015). A subset of these Rab GTPases is also involved in key membrane trafficking steps during autophagy such as autophagosome formation, movement, and fusion with lysosomes (Ao et al., 2014; Nakamura and Yoshimori, 2017). A significant number of human Rabs have been found to reside on the LD surface via proteomic studies, although only a few of them have been implicated in regulating LD associations with autophagosomes and other degradative organelles such as late endosomes, MVBs, and lysosomes (Kiss and Nilsson, 2014; Schroeder et al., 2015; Li et al., 2016).
For example, two well-studied Rab proteins, Rab7, and Rab10, that have been already implicated in the selective degradation of damaged mitochondria during mitophagy (Yamano et al., 2014; Jimenez-Orgaz et al., 2018; Yamano et al., 2018; Wauters et al., 2019), also perform key functions during lipophagy. Rab7 predominantly resides on late endosomes, MVBs, and lysosomes and coordinates the trafficking and degradation of both endocytic and autophagic cargo within the lysosome (Guerra and Bucci, 2016). When in an active, GTP-bound state, Rab7 associates with effectors such as motor adaptors and tethering proteins and in turn regulates maturation, transport and fusion between late endosomes, autophagosomes, MVBs, and lysosomes (Jordens et al., 2001; Gutierrez et al., 2004; Jager et al., 2004; Pankiv et al., 2010; Hyttinen et al., 2013). Rab7 also resides on the LD surface and plays an important role in LD associations with autophagosomes or other degradative organelles during lipophagy (Figure 2). In support of this idea, Schroeder et al. (2015) found that Rab7 is activated on the LD surface under nutrient-deprived conditions and mediates LD recognition by MVBs and lysosomes to induce lipophagic LD breakdown in human hepatoma cells. Loss of Rab7 expression reduced MVB/lysosome association with LDs and therefore inhibited LD breakdown during starvation-induced lipophagy. More recently, it was reported that in hepatocytes, chronic ethanol exposure inhibits lipophagy by interfering not only with lysosome morphology and motility but also with Rab7 activation, suggesting that LD recognition by the autophagic machinery could be altered (Schulze et al., 2017). Of note, Rab7 has also been implicated in 3T3-L1 adipocyte lipolysis after β-adrenergic receptor stimulation, suggesting a complex interplay between lipophagy and lipolysis in the function of Rab7 (Lizaso et al., 2013).
Rab10 has also emerged as a key player of autophagic LD engulfment during lipophagy (Figure 2). Like Rab7, Rab10 also becomes activated under nutrient starvation coincident with increased localization on LDs (Li et al., 2016). Here, Rab10 recruits degradative membranes positive for autophagic markers LC3, ATG16L, and lysosomal marker Lamp1. In its active state, Rab10 also recruits its downstream effector EH domain binding protein 1 (EHBP1) and a membrane remodeling ATPase named EH domain-containing protein 2 (EHD2) to the LD-autophagosome synapse where this protein complex drives LD engulfment by autophagic membranes. Loss of the Rab10-EHBP1-EHD2 complex reduces autophagic membrane targeting and engulfment of LDs, resulting in excess LD accumulation. This suggests that Rab10 plays a key role in degradative membrane targeting to the LD surface as well as LD engulfment within these structures during lipophagy.
LD Engulfment by the Vacuole During Microlipophagy
In addition to autophagosome-based lipophagy, an alternative pathway for LD breakdown, microlipophagy, has been described in Saccharomyces cerevisiae. Microlipophagy is the direct internalization of LDs into the yeast vacuole (similar to mammalian lysosomes) for degradation. This appears to correspond with a previously described phenomenon whereby the vacuolar membrane is dramatically reorganized during stationary phase starvation or other conditions of cellular stress (Toulmay and Prinz, 2013). During membrane reorganization, sterol-enriched liquid-ordered (Lo) microdomains akin to lipid rafts become distinctly segregated from liquid-disordered (Ld) membrane domains. These domains accommodate sites of LD docking and engulfment at Lo domains, while Ld microdomains further constrain LD movement (Wang et al., 2014). In addition, it appears that stationary phase microlipophagy itself is required to maintain the integrity of vacuolar microdomains most likely through LD-stored sterol ester catabolism.
Yeast microlipophagy has been described to occur in response to various starvation conditions. For example, nitrogen starvation was shown to induce LD uptake into the vacuole, and this also required several core macroautophagy proteins including Atg1, Atg3–10, Atg12, Atg14, Atg16, and Vps34 (Table 1). Acute glucose starvation also induces microlipophagy via AMP-activated protein kinase (AMPK) and this also required core autophagy genes (Seo et al., 2017). Currently, the requirement of stable LD-autophagosome contact sites in microlipophagy remains unclear. Two additional studies reported that gradual glucose starvation can stimulate microlipophagy and requires the endosomal sorting complexes required for transport (ESCRT) proteins Vps4 and Vps27. These studies diverged somewhat on the role of macroautophagy genes in microlipophagy, as Atg proteins seem to be required at the stationary phase, but not during the diauxic shift (Oku et al., 2017).
As mentioned earlier, the importance of autophagosome formation during microlipophagy may vary depending on the stimulus. For example, while the earlier studies utilized nitrogen or glucose starvation, Vevea et al. reported that during lipid stress (resulting from a block of phosphatidylcholine biosynthesis), yeast upregulates both LD biogenesis and LD degradation via microlipophagy, but this was independent of the core autophagy protein Atg7 and LD associations with autophagosomes. Instead, lipid stress-induced lipophagy required the ESCRT protein Vps4 (Vevea et al., 2015). These studies suggest that the ESCRT machinery may play a critical role in microlipophagy, perhaps in cooperation with canonical macroautophagy. Currently, microlipophagy has not been described in mammalian cells, although it seems likely that similar mechanisms exist given that microautophagy is conserved from yeast to mammals (Sahu et al., 2011). It would be of interest to better understand the different stimuli that could lead to microlipophagy activation as well as the requirement for autophagosomes, autophagy receptors, and ubiquitination in this pathway.
The Role of Lipophagy in Liver Pathology
Lipophagy is a critical pathway for lipid homeostasis particularly in the liver. Not surprisingly, numerous liver diseases have now been shown to impact lipophagy and drive disease pathogenesis (Schulze et al., 2017). This is particularly true of nonalcoholic fatty liver disease (NAFLD), which is characterized by excess hepatic LD accumulation and is a common manifestation of metabolic syndrome affecting nearly a third of the western world population (Kim et al., 2018). Genetic mutations can often increase patient risk for NAFLD, which may negatively impact LD breakdown by lipophagy. In 2008, Romeo et al. identified a missense sequence variant within patatin-like phospholipase domain-containing protein 3 (PNPLA3) (I48M) that is associated with increased hepatic fat content and is considered a major genetic risk factor for NAFLD. Further studies demonstrated that PNPLA3 I48M knock in mice develop fatty liver if fed high-sucrose diet and show increased accumulation of mutant PNPLA3 on LDs compared to the wild-type (WT) protein suggesting that it may play a role in LD breakdown pathways (Smagris et al., 2015). More recently, the relationship between PNPLA3 I48M variant and lipophagy was explored. In this study, HepG2 cells expressing I48M mutant showed resistance to starvation-induced LD loss accompanied with reduced autophagic flux and lipophagy compared to cells expressing WT PNPLA3 (Negoita et al., 2019). These data suggest that the accumulation of I48M PNPLA3 variant on LDs renders them resistant for lipophagic recognition leading to increased LD content, although further studies are necessary to confirm this observation.
In contrast to lipophagy being potentially defective during metabolic fatty liver disease, this selective autophagy pathway can be hijacked by viruses providing them a beneficial environment for replication within the host cell. For example, dengue virus (DENV) infection was shown to induce deubiquitination of LD-resident protein AUP1 and its binding partner Ubc7/Ube2g2 in HepG2 and HeLa cells (Zhang et al., 2018). This displaces deubiquitinated AUP1 from LDs to autophagosomes leading to increased lipophagy pathway, LD loss, and increased viral replication.
Regulation of Lipolysis by LD-Autophagosome/Lysosome Contacts
During lipolysis, cytoplasmic lipases ATGL, HSL, and MGL target LDs to hydrolyze fatty acids from triacylglycerol, diacylglycerol, and monoacylglycerol, respectively. Nutrient deprivation as well as G-protein-coupled receptor activation regulates lipase trafficking to the LD surface through signaling pathways involving AMPK, cyclic adenosine monophosphate (cAMP)/protein kinase A (PKA), and others (Zechner et al., 2017). In addition, LD-resident cofactors can also block lipase access to the LD or directly modulate lipase activity. While these mechanisms are complex and variable among different tissue types, several studies now implicate autophagosomes and lysosomal vesicles in the regulation of cytosolic lipases.
In brown adipose tissue, Martinez-Lopez et al. (2016) show that cold exposure increases autophagosome interaction with LDs, coincident with ATGL and HSL recruitment to the LD surface. Sequence analysis of ATGL and HSL revealed multiple LIR motifs, and mutation of one such LIR on ATGL (F146A, V149A) prevented starvation-induced LD loss and reduced LD recruitment of ATGL (Martinez-Lopez et al., 2016). In addition, HSL immunoprecipitation from purified LDs showed lipase association with LC3. This study implicates autophagosomes in the delivery of cytosolic lipases to the LD surface, an idea that is consistent with previous vesicle-based mechanisms of ATGL transport to the LD (Soni et al., 2009). Interestingly, the LIR motif of ATGL also appears critical for lipophagy in NIH3T3 cells, raising the possibility that this interaction may also facilitate autophagosome tethering to LDs (Martinez-Lopez et al., 2016) (Figure 1).
Lysosome-LD interactions also appear to stimulate lipolysis by removing perilipin proteins that block cytosolic lipases access to the LD. Kaushik and Cuervo (2015) propose that LD-resident PLIN2 and PLIN3 are degraded by chaperone-mediated autophagy, a process that utilizes Lamp2A, situated in the lysosome membrane, to transport specific proteins directly into the lysosome lumen for degradation. In Lamp2A-deficient cells, β-oxidation due to lipolysis was severely downregulated along with reduced ATGL trafficking to the LD. In addition, lipolysis and ATGL targeting to the LD was blocked in cells expressing a mouse PLIN2 mutant that lacks a putative pentapeptide consensus sequence for CMA degradation (414SLKVQ). This work suggests that LD-lysosome kiss and run events can aid in degrading proteins at the LD surface, clearing the way for cytosolic lipases access. Interestingly, LD-autophagosome contacts were also reduced in CMA-deficient cells, suggesting an intimate interplay between CMA, lipolysis, and lipophagy.
Regulation of Lipophagy by Cytosolic Lipases
While it is clear that autophagy regulates lipolysis, several studies now show that lipolysis can influence autophagy and LD-autophagosome interactions. Mashek and coworkers were first to show that ATGL activates the transcription of autophagy genes as well as that of fatty acid oxidation and lysosome biogenesis. In hepatocytes, ATGL activates PPARα through a mechanism involving the deacetylase SIRT1 (Khan et al., 2015; Sathyanarayan et al., 2017), suggesting that lipolysis is an important upstream mediator of autophagosome formation and lipophagy. This is supported by a recent study suggesting that ATGL targets larger sized LDs upstream of lipophagy, and that lipophagy preferentially targets smaller sized LDs <1 μm in diameter (Schott et al., 2019). Moreover, lipolysis is important for producing these small LDs, both by reducing large LD diameters and by generating nonesterified fatty acids that are repackaged into nascent LDs by DGAT1/2 enzymes (Paar et al., 2012; Schott et al., 2019).
The mechanisms that preferentially target lipophagic vesicles to smaller-sized LDs is unclear. One possibility is that the size of degradative vesicles themselves limits LD cargo. Autophagosomes, lysosomes, and late endosomes can exceed diameters >1 μm under certain conditions, although the majority of these vesicles are much smaller (Huotari and Helenius, 2011; Xu and Ren, 2015; Su et al., 2016). Interestingly, LC3 was reported to be lipidated preferentially on membranes with high curvature due to Atg3 sensing (Nath et al., 2014), raising the possibility that similar curvature-sensing mechanisms could exist for lipophagic targeting of small LDs. Along these lines, the autophagosome biogenesis protein Atg14L (first identified as Barkor; Sun et al., 2008) functions as a membrane curvature sensor (Fan et al., 2011) and can localize to LDs in a variety of cell lines. It is possible that small-LD targeting by lipophagic vesicles involves curvature-sensing machinery, but this remains to be investigated.
LDs as Lipid Reservoirs for Autophagosome Biogenesis
In addition to LD catabolism, LD-autophagosome contact sites may also mark sites of autophagosome biogenesis. Inhibition of mechanistic target of rapamycin complex 1 (mTORC1) initiates the process of autophagosome formation by activating Unc-51 Like Autophagy Activating Kinase 1 (ULK1) and Beclin 1-PI3K complexes to produce PtdIns3P (PIP3) necessary for outgrowth of early autophagosomal membranes (i.e., omegasomes, phagophores; Grasso et al., 2018; Mercer et al., 2018). Nascent autophagosomes utilize lipids from various organelles (i.e., Golgi, ER, mitochondria, plasma membrane) to supply the membrane needed for outgrowth (Shibutani and Yoshimori, 2014). Among these sources, LDs offer an abundant pool of neutral lipids that can be processed into autophagosome membrane. Consistent with this, Dupont et al. (2014) showed that oleic acid (OA)-loaded cells have a greater capacity for autophagosome formation and autophagic flux. Several members of the PNPLA family of lipases, most notably PNPLA5, support autophagosome formation by converting LD triacylglycerol into diacylglycerol that is used to make phospholipids for the budding autophagosome (Dupont et al., 2014).
One putative link between LDs and nascent autophagosomes is the autophagy protein Atg2a/b, which binds to both organelles through distinct regions at its N- and C-termini. Velikkakath et al. (2012) was first to report that Atg2a/b is involved in autophagosome biogenesis, and that this function requires amino acids 1723–1829 which are also necessary for LD localization. Interestingly, Atg2a/b also controls LD content in a reciprocal fashion, as knockdown of Atg2a/b, but not Atg5, caused an increase in total LD area per cell and appeared to cluster LDs in a majority of cells (Velikkakath et al., 2012). This study raised the possibility that LD contacts with autophagosomes may denote sites of autophagosome biogenesis in addition to lipophagic degradation. However, it remains unclear whether Atg2a functions as a definitive tether between LDs and nascent autophagosomes, as Atg2a controls autophagosome biogenesis from the ER and functions as a lipid transfer protein (Maeda et al., 2019; Valverde et al., 2019). Interestingly, Atg2a shares some sequence similarity with the protein Vps13 particularly at its N-terminal chorein domain, which is responsible for lipid transport at organelle contact sites (Kumar et al., 2018). Structural studies of this domain suggested that it adopts a scoop-like fold with a hydrophobic cavity which could accommodate multiple lipid molecules. In support of this, cryo-EM reconstruction of Atg2a also identified a cavity (or multiple cavities) within the protein which could similarly facilitate lipid transfer between the organelles (Valverde et al., 2019). Maeda et al. (2019) also demonstrated a lipid transport function by Atg2a and suggested that phagophore tethering by WIPI4 and WIPI1 (both PI3P effector proteins) aids in this process (Maeda et al., 2019). Based on these data, Atg2a/b likely functions as a lipid transporter between lipid-rich LDs and nascent autophagosomes; however, the requirement of Atg2a in LD-autophagosome tethering remains to be determined and likely involves an intimate connection with the ER (Figure 1).
Autophagosome biogenesis from LDs may also involve Atg14L/Barkor. As a subunit of the PI3K complex, Atg14L aids in delivering PIP3 to nascent phagophores and has been shown to localize to LDs along with Atg2a in cancer cells (Pfisterer et al., 2014). Like Atg2a, Atg14L gene silencing also induces LD accumulation in addition to reduced autophagy. One possibility is that these genes function to extend nascent autophagosomes around LDs for lipophagy, as has been described for cargo-induced autophagosome biogenesis in selective autophagy (Kamber et al., 2015a; Turco et al., 2019). Future work is required to define the fate of these nascent autophagosomes in lipophagy versus other forms of autophagy.
Conclusions
LD contacts with degradative vesicles serve important purposes in both LD catabolism and autophagosome biogenesis (Figure 1). Given the widespread importance of these processes to metabolism, future research into LD-autophagosome contacts will uncover important new insights into cell function and disease pathogenesis. It is particularly exciting to consider how future proteomic studies will help define new molecular tethers, lipophagy receptors, and posttranslational modifications present at the interface between LDs and degradative vesicles.
In future work, it will be critical to define LD-auto-phagosome contacts as a balance between lipophagy, lipolysis, and autophagosome biogenesis. Do these processes work simultaneously during nutrient starvation? How are different LDs selected for lipophagy versus autophagosome biogenesis? Along these lines, it will be of interest to investigate how LD heterogeneity and varying nutrient availability may dictate the different forms of lipophagy (i.e., macro, micro, kiss-and-run) in different cells and tissues. This may also require a closer investigation into posttranslational modifications at LD-autophagosome contact sites. In particular, ubiquitination of LD proteins appears to facilitate LD catabolism, but the relative contribution of ubiquitin to lipophagy as opposed to proteasome clearance of LD proteins is unclear. Future work will need to differentiate these pathways, including role of unique ubiquitin architectures as new technological advances emerge (Swatek et al., 2019).
Funding
The author(s) disclosed receipt of the following financial support for the research, authorship, and/or publication of this article: This work was supported by the National Institutes of Health Grants - the National Institute on Alcohol Abuse and Alcoholism (NIAAA) K99AA026877 (M.B. Schott), NIAAA R01AA020735 (M.A. McNiven), the National Institute of Diabetes and Digestive and Kidney Diseases (NIDDK) R01DK044650 (M.A. McNiven) and NIDDK P30DK084567 (M.A. McNiven).
Footnotes
Declaration of Conflicting Interests
The author(s) declared no potential conflicts of interest with respect to the research, authorship, and/or publication of this article.
References
- Ao X, Zou L, and Wu Y. (2014). Regulation of autophagy by the Rab GTPase network. Cell Death Differ 21, 348–358. doi: 10.1038/cdd.2013.187 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bersuker K, and Olzmann JA. (2018). In Close Proximity: The Lipid Droplet Proteome and Crosstalk With the Endoplasmic Reticulum. Contact (Thousand Oaks) 1. doi: 10.1177/2515256418768996 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bersuker K, and Olzmann JA. (2019). Identification of Lipid Droplet Proteomes by Proximity Labeling Proteomics Using APEX2. Methods in molecular biology 2008, 57–72. doi: 10.1007/978-1-4939-9537-0_5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deosaran E, Larsen KB, Hua R, Sargent G, Wang Y, Kim S, Lamark T, Jauregui M, Law K, Lippincott-Schwartz J, et al. (2013). NBR1 acts as an autophagy receptor for peroxisomes. J Cell Sci 126, 939–952. doi: 10.1242/jcs.114819 [DOI] [PubMed] [Google Scholar]
- Dupont N, Chauhan S, Arko-Mensah J, Castillo EF, Masedunskas A, Weigert R, Robenek H, Proikas-Cezanne T, and Deretic V. (2014). Neutral lipid stores and lipase PNPLA5 contribute to autophagosome biogenesis. Curr Biol 24, 609–620. doi: 10.1016/j.cub.2014.02.008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eastman SW, Yassaee M, and Bieniasz PD. (2009). A role for ubiquitin ligases and Spartin/SPG20 in lipid droplet turnover. J Cell Biol 184, 881–894. doi: 10.1083/jcb.200808041 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Edwards TL, Clowes VE, Tsang HT, Connell JW, Sanderson CM, Luzio JP, and Reid E. (2009). Endogenous spartin (SPG20) is recruited to endosomes and lipid droplets and interacts with the ubiquitin E3 ligases AIP4 and AIP5. Biochem J 423, 31–39. doi: 10.1042/BJ20082398 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fan W, Nassiri A, and Zhong Q. (2011). Autophagosome targeting and membrane curvature sensing by Barkor/Atg14 (L). Proc Natl Acad Sci U S A 108, 7769–7774. doi 10.1073/pnas.1016472108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gatica D, Lahiri V, and Klionsky DJ. (2018). Cargo recognition and degradation by selective autophagy. Nat Cell Biol 20, 233–242. doi: 10.1038/s41556-018-0037-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grasso D, Renna FJ, and Vaccaro MI. (2018). Initial Steps in Mammalian Autophagosome Biogenesis. Front Cell Dev Biol 6, 146. doi: 10.3389/fcell.2018.00146 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guerra F, and Bucci C. (2016). Multiple Roles of the Small GTPase Rab7. Cells 5. doi: 10.3390/cells5030034 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gutierrez MG, Munafo DB, Beron W, and Colombo MI. (2004). Rab7 is required for the normal progression of the autophagic pathway in mammalian cells. J Cell Sci 117, 2687–2697. doi: 10.1242/jcs.01114 [DOI] [PubMed] [Google Scholar]
- Henne WM, and Hariri H. (2018). Endoplasmic Reticulum-Vacuole Contact Sites “Bloom” With Stress-Induced Lipid Droplets. Contact (Thousand Oaks) 1. doi: 10.1177/2515256418756112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hyttinen JM, Niittykoski M, Salminen A, and Kaarniranta K. (2013). Maturation of autophagosomes and endosomes: a key role for Rab7. Biochim Biophys Acta 1833, 503–510. doi: 10.1016/j.bbamcr.2012.11.018 [DOI] [PubMed] [Google Scholar]
- Hooper C, Puttamadappa SS, Loring Z, Shekhtman A, and Bakowska JC. (2010). Spartin activates atrophin-1-interacting protein 4 (AIP4) E3 ubiquitin ligase and promotes ubiquitination of adipophilin on lipid droplets. BMC Biol 8, 72. doi: 10.1186/1741-7007-8-72 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hung YH, Chen LM, Yang JY, and Yang WY. (2013). Spatiotemporally controlled induction of autophagy-mediated lysosome turnover. Nat Commun 4, 2111. doi: 10.1038/ncomms3111 [DOI] [PubMed] [Google Scholar]
- Huotari J, and Helenius A. (2011). Endosome maturation. EMBO J 30, 3481–3500. doi: 10.1038/emboj.2011.286 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamano K, Fogel AI, Wang C, van der Bliek AM, and Youle RJ. (2014). Mitochondrial Rab GAPs govern autophagosome biogenesis during mitophagy. Elife 3, e01612. doi: 10.7554/eLife.01612 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamano K, Wang C, Sarraf SA, Munch C, Kikuchi R, Noda NN, Hizukuri Y, Kanemaki MT, Harper W, Tanaka K, et al. (2018). Endosomal Rab cycles regulate Parkin-mediated mitophagy. eLife 7. doi: 10.7554/eLife.31326 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yan Y, Wang H, Wei C, Xiang Y, Liang X, Phang CW, and Jiao R. (2019). HDAC6 regulates lipid droplet turnover in response to nutrient deprivation via p62-mediated selective autophagy. J Genet Genomics 46, 221–229. doi: 10.1016/j.jgg.2019.03.008 [DOI] [PubMed] [Google Scholar]
- Jackson CL. (2019). Lipid droplet biogenesis. Curr Opin Cell Biol 59, 88–96. doi: 10.1016/j.ceb.2019.03.018 [DOI] [PubMed] [Google Scholar]
- Jager S, Bucci C, Tanida I, Ueno T, Kominami E, Saftig P, and Eskelinen EL. (2004). Role for Rab7 in maturation of late autophagic vacuoles. J Cell Sci 117, 4837–4848. doi: 10.1242/jcs.01370 [DOI] [PubMed] [Google Scholar]
- Jimenez-Orgaz A, Kvainickas A, Nagele H, Denner J, Eimer S, Dengjel J, and Steinberg F. (2018). Control of RAB7 activity and localization through the retromer-TBC1D5 complex enables RAB7-dependent mitophagy. EMBO J 37, 235–254. doi: 10.15252/embj.201797128 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jo Y, Hartman IZ, and DeBose-Boyd RA. (2013). Ancient ubiquitous protein-1 mediates sterol-induced ubiquitination of 3-hydroxy-3-methylglutaryl CoA reductase in lipid droplet-associated endoplasmic reticulum membranes. Mol Biol Cell 24, 169–183. doi: 10.1091/mbc.E12-07-0564 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jordens I, Fernandez-Borja M, Marsman M, Dusseljee S, Janssen L, Calafat J, Janssen H, Wubbolts R, and Neefjes J. (2001). The Rab7 effector protein RILP controls lysosomal transport by inducing the recruitment of dynein-dynactin motors. Curr Biol 11, 1680–1685. doi: 10.1016/s0960-9822(01)00531-0 [DOI] [PubMed] [Google Scholar]
- Ju L, Han J, Zhang X, Deng Y, Yan H, Wang C, Li X, Chen S, Alimujiang M, Li X, et al. (2019). Obesity-associated inflammation triggers an autophagy-lysosomal response in adipocytes and causes degradation of perilipin 1. Cell Death Dis 10, 121. doi: 10.1038/s41419-019-1393-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kamber RA, Shoemaker CJ, and Denic V. (2015a). A molecular switch for selective autophagosome formation. Autophagy 11, 2132–2133. doi: 10.1080/15548627.2015.1098799 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kamber RA, Shoemaker CJ, and Denic V. (2015b). Receptor-Bound Targets of Selective Autophagy Use a Scaffold Protein to Activate the Atg1 Kinase. Mol Cell 59, 372–381. doi: 10.1016/j.molcel.2015.06.009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaushik S, and Cuervo AM. (2015). Degradation of lipid droplet-associated proteins by chaperone-mediated autophagy facilitates lipolysis. Nat Cell Biol 17, 759–770. doi: 10.1038/ncb3166 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khaminets A, Behl C, and Dikic I. (2016). Ubiquitin-Dependent And Independent Signals In Selective Autophagy. Trends Cell Biol 26, 6–16. doi: 10.1016/j.tcb.2015.08.010 [DOI] [PubMed] [Google Scholar]
- Khaminets A, Heinrich T, Mari M, Grumati P, Huebner AK, Akutsu M, Liebmann L, Stolz A, Nietzsche S, Koch N, et al. (2015). Regulation of endoplasmic reticulum turnover by selective autophagy. Nature 522, 354–358. doi: 10.1038/nature14498 [DOI] [PubMed] [Google Scholar]
- Khan SA, Sathyanarayan A, Mashek MT, Ong KT, Wollaston-Hayden EE, and Mashek DG. (2015). ATGL-catalyzed lipolysis regulates SIRT1 to control PGC-1alpha/PPAR-alpha signaling. Diabetes 64, 418–426. doi: 10.2337/db14-0325 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim D, Touros A, and Kim WR. (2018). Nonalcoholic Fatty Liver Disease and Metabolic Syndrome. Clin Liver Dis 22, 133–140. doi: 10.1016/j.cld.2017.08.010 [DOI] [PubMed] [Google Scholar]
- Kim PK, Hailey DW, Mullen RT, and Lippincott-Schwartz J. (2008). Ubiquitin signals autophagic degradation of cytosolic proteins and peroxisomes. Proc Natl Acad Sci U S A 105, 20567–20574. doi: 10.1073/pnas.0810611105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kirkin V, McEwan DG, Novak I, and Dikic I. (2009). A role for ubiquitin in selective autophagy. Mol Cell 34, 259–269. doi: 10.1016/j.molcel.2009.04.026 [DOI] [PubMed] [Google Scholar]
- Kiss RS, and Nilsson T. (2014). Rab proteins implicated in lipid storage and mobilization. J Biomed Res 28, 169–177. doi: 10.7555/JBR.28.20140029 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klemm EJ, Spooner E, and Ploegh HL. (2011). Dual role of ancient ubiquitous protein 1 (AUP1) in lipid droplet accumulation and endoplasmic reticulum (ER) protein quality control. J Biol Chem 286, 37602–37614. doi: 10.1074/jbc.M111.284794 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klionsky DJ, Abdelmohsen K, Abe A, Abedin MJ, Abeliovich H, Acevedo Arozena A, Adachi H, Adams CM, Adams PD, Adeli K, et al. (2016). Guidelines for the use and interpretation of assays for monitoring autophagy (3rd edition). Autophagy 12, 1–222. doi: 10.1080/15548627.2015.1100356 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krahmer N, and Mann M. (2019). Catching Lipid Droplet Contacts by Proteomics. Contact (Thousand Oaks) 2, 2515256419859186. doi: 10.1177/2515256419859186 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kumar N, Leonzino M, Hancock-Cerutti W, Horenkamp FA, Li P, Lees JA, Wheeler H, Reinisch KM, and De Camilli P. (2018). VPS13A and VPS13C are lipid transport proteins differentially localized at ER contact sites. J Cell Biol 217, 3625–3639. doi: 10.1083/jcb.201807019 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lazarou M, Sliter DA, Kane LA, Sarraf SA, Wang C, Burman JL, Sideris DP, Fogel AI, and Youle RJ. (2015). The ubiquitin kinase PINK1 recruits autophagy receptors to induce mitophagy. Nature 524, 309–314. doi: 10.1038/nature14893 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Z, Schulze RJ, Weller SG, Krueger EW, Schott MB, Zhang X, Casey CA, Liu J, Stockli J, James DE, and McNiven MA. (2016). A novel Rab10-EHBP1-EHD2 complex essential for the autophagic engulfment of lipid droplets. Sci Adv 2, e1601470. doi: 10.1126/sciadv.1601470 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lizaso A, Tan KT, and Lee YH. (2013). beta-adrenergic receptor-stimulated lipolysis requires the RAB7-mediated autolysosomal lipid degradation. Autophagy 9, 1228–1243. doi: 10.4161/auto.24893 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maeda S, Otomo C, and Otomo T. (2019). The autophagic membrane tether ATG2A transfers lipids between membranes. Elife 8. doi: 10.7554/eLife.45777 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martinez-Lopez N, Garcia-Macia M, Sahu S, Athonvarangkul D, Liebling E, Merlo P, Cecconi F, Schwartz GJ, and Singh R. (2016). Autophagy in the CNS and Periphery Coordinate Lipophagy and Lipolysis in the Brown Adipose Tissue and Liver. Cell Metab 23, 113–127. doi: 10.1016/j.cmet.2015.10.008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mercer TJ, Gubas A, and Tooze SA. (2018). A molecular perspective of mammalian autophagosome biogenesis. J Biol Chem 293, 5386–5395. doi: 10.1074/jbc.R117.810366 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakamura S, and Yoshimori T. (2017). New insights into autophagosome-lysosome fusion. J Cell Sci 130, 1209–1216. doi: 10.1242/jcs.196352 [DOI] [PubMed] [Google Scholar]
- Nath S, Dancourt J, Shteyn V, Puente G, Fong WM, Nag S, Bewersdorf J, Yamamoto A, Antonny B, and Melia TJ. (2014). Lipidation of the LC3/GABARAP family of autophagy proteins relies on a membrane-curvature-sensing domain in Atg3. Nat Cell Biol 16, 415–424. doi: 10.1038/ncb2940 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Negoita F, Blomdahl J, Wasserstrom S, Winberg ME, Osmark P, Larsson S, Stenkula KG, Ekstedt M, Kechagias S, Holm C, and Jones HA. (2019). PNPLA3 variant M148 causes resistance to starvation-mediated lipid droplet autophagy in human hepatocytes. J Cell Biochem 120, 343–356. doi: 10.1002/jcb.27378 [DOI] [PubMed] [Google Scholar]
- Oku M, Maeda Y, Kagohashi Y, Kondo T, Yamada M, Fujimoto T, and Sakai Y. (2017a). Evidence for ESCRT-and clathrin-dependent microautophagy. J Cell Biol 216, 3263–3274. doi: 10.1083/jcb.201611029 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oku M, Maeda Y, Kagohashi Y, Kondo T, Yamada M, Fujimoto T, and Sakai Y. (2017b). Evidence for ESCRT-and clathrin-dependent microautophagy. J Cell Biol. doi: 10.1083/jcb.201611029 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Olzmann JA, and Carvalho P. (2019). Dynamics and functions of lipid droplets. Nature reviews. Molecular cell biology 20, 137–155. doi: 10.1038/s41580-018-0085-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- Onal G, Kutlu O, Gozuacik D, and Dokmeci Emre S. (2017). Lipid Droplets in Health and Disease. Lipids Health Dis 16, 128. doi: 10.1186/s12944-017-0521-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paar M, Jungst C, Steiner NA, Magnes C, Sinner F, Kolb D, Lass A, Zimmermann R, Zumbusch A, Kohlwein SD, and Wolinski H. (2012). Remodeling of lipid droplets during lipolysis and growth in adipocytes. J Biol Chem 287, 11164–11173. doi: 10.1074/jbc.M111.316794 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pankiv S, Alemu EA, Brech A, Bruun JA, Lamark T, Overvatn A, Bjorkoy G, and Johansen T. (2010). FYCO1 is a Rab7 effector that binds to LC3 and PI3P to mediate microtubule plus end-directed vesicle transport. J Cell Biol 188, 253–269. doi: 10.1083/jcb.200907015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pfisterer SG, Bakula D, Frickey T, Cezanne A, Brigger D, Tschan MP, Robenek H, and Proikas-Cezanne T. (2014). Lipid droplet and early autophagosomal membrane targeting of Atg2A and Atg14L in human tumor cells. J Lipid Res 55, 1267–1278. doi: 10.1194/jlr.M046359 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rogov V, Dotsch V, Johansen T, and Kirkin V. (2014). Interactions between autophagy receptors and ubiquitin-like proteins form the molecular basis for selective autophagy. Mol Cell 53, 167–178. doi: 10.1016/j.molcel.2013.12.014 [DOI] [PubMed] [Google Scholar]
- Romeo S, Kozlitina J, Xing C, Pertsemlidis A, Cox D, Pennacchio LA, Boerwinkle E, Cohen JC, and Hobbs HH. (2008). Genetic variation in PNPLA3 confers susceptibility to nonalcoholic fatty liver disease. Nat Genet 40, 1461–1465. doi: 10.1038/ng.257 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rui YN, Xu Z, Patel B, Chen Z, Chen D, Tito A, David G, Sun Y, Stimming EF, Bellen HJ, et al. (2015). Huntingtin functions as a scaffold for selective macroautophagy. Nat Cell Biol 17, 262–275. doi: 10.1038/ncb3101 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sahu R, Kaushik S, Clement CC, Cannizzo ES, Scharf B, Follenzi A, Potolicchio I, Nieves E, Cuervo AM, and Santambrogio L. (2011). Microautophagy of cytosolic proteins by late endosomes. Dev Cell 20, 131–139. doi: 10.1016/j.devcel.2010.12.003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sathyanarayan A, Mashek MT, and Mashek DG. (2017). ATGL Promotes Autophagy/Lipophagy via SIRT1 to Control Hepatic Lipid Droplet Catabolism. Cell Rep 19, 1–9. doi: 10.1016/j.celrep.2017.03.026 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schott MB, Weller SG, Schulze RJ, Krueger EW, Drizyte-Miller K, Casey CA, and McNiven MA. (2019). Lipid droplet size directs lipolysis and lipophagy catabolism in hepatocytes. J Cell Biol. doi: 10.1083/jcb.201803153 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schroeder B, Schulze RJ, Weller SG, Sletten AC, Casey CA, and McNiven MA. (2015). The small GTPase Rab7 as a central regulator of hepatocellular lipophagy. Hepatology 61, 1896–1907. doi: 10.1002/hep.27667 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schuldiner M, and Bohnert M. (2017). A different kind of love - lipid droplet contact sites. Biochim Biophys Acta Mol Cell Biol Lipids 1862, 1188–1196. doi: 10.1016/j.bbalip.2017.06.005 [DOI] [PubMed] [Google Scholar]
- Schulze RJ, Drizyte K, Casey CA, and McNiven MA. (2017a). Hepatic Lipophagy: New Insights into Autophagic Catabolism of Lipid Droplets in the Liver. Hepatol Commun 1, 359–369. doi: 10.1002/hep4.1056 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schulze RJ, Rasineni K, Weller SG, Schott MB, Schroeder B, Casey CA, and McNiven MA. (2017b). Ethanol exposure inhibits hepatocyte lipophagy by inactivating the small guanosine triphosphatase Rab7. Hepatol Commun 1, 140–152. doi: 10.1002/hep4.1021 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seo AY, Lau PW, Feliciano D, Sengupta P, Gros MAL, Cinquin B, Larabell CA, and Lippincott-Schwartz J. (2017). AMPK and vacuole-associated Atg14p orchestrate mu-lipophagy for energy production and long-term survival under glucose starvation. Elife 6. doi: 10.7554/eLife.21690 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shibutani ST, and Yoshimori T. (2014). A current perspective of autophagosome biogenesis. Cell Res 24, 58–68. doi: 10.1038/cr.2013.159 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Singh R, Kaushik S, Wang Y, Xiang Y, Novak I, Komatsu M, Tanaka K, Cuervo AM, and Czaja MJ. (2009). Autophagy regulates lipid metabolism. Nature 458, 1131–1135. doi: 10.1038/nature07976 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smagris E, BasuRay S, Li J, Huang Y, Lai KM, Gromada J, Cohen JC, and Hobbs HH. (2015). Pnpla3I148M knockin mice accumulate PNPLA3 on lipid droplets and develop hepatic steatosis. Hepatology 61, 108–118. doi: 10.1002/hep.27242 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Soni KG, Mardones GA, Sougrat R, Smirnova E, Jackson CL, and Bonifacino JS. (2009). Coatomer-dependent protein delivery to lipid droplets. J Cell Sci 122, 1834–1841. doi: 10.1242/jcs.045849 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spandl J, Lohmann D, Kuerschner L, Moessinger C, and Thiele C. (2011). Ancient ubiquitous protein 1 (AUP1) localizes to lipid droplets and binds the E2 ubiquitin conjugase G2 (Ube2g2) via its G2 binding region. J Biol Chem 286, 5599–5606. doi: 10.1074/jbc.M110.190785 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stolz A, Ernst A, and Dikic I. (2014). Cargo recognition and trafficking in selective autophagy. Nat Cell Biol 16, 495–501. doi: 10.1038/ncb2979 [DOI] [PubMed] [Google Scholar]
- Su QP, Du W, Ji Q, Xue B, Jiang D, Zhu Y, Ren H, Zhang C, Lou J, Yu L, and Sun Y. (2016). Vesicle Size Regulates Nanotube Formation in the Cell. Sci Rep 6, 24002. doi: 10.1038/srep24002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sugihara M, Morito D, Ainuki S, Hirano Y, Ogino K, Kitamura A, Hirata H, and Nagata K. (2019). The AAA + ATPase/ubiquitin ligase mysterin stabilizes cytoplasmic lipid droplets. J Cell Biol 218, 949–960. doi: 10.1083/jcb.201712120 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun Q, Fan W, Chen K, Ding X, Chen S, and Zhong Q. (2008). Identification of Barkor as a mammalian autophagy-specific factor for Beclin 1 and class III phosphatidylinositol 3-kinase. Proc Natl Acad Sci U S A 105, 19211–19216. doi: 10.1073/pnas.0810452105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Swatek KN, Usher JL, Kueck AF, Gladkova C, Mevissen TET, Pruneda JN, Skern T, and Komander D. (2019). Insights into ubiquitin chain architecture using Ub-clipping. Nature 572, 533–537. doi: 10.1038/s41586-019-1482-y [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tauchi-Sato K, Ozeki S, Houjou T, Taguchi R, and Fujimoto T. (2002). The surface of lipid droplets is a phospholipid monolayer with a unique Fatty Acid composition. J Biol Chem 277, 44507–44512. doi: 10.1074/jbc.M207712200 [DOI] [PubMed] [Google Scholar]
- Thiam AR, and Dugail I. (2019). Lipid droplet-membrane contact sites - from protein binding to function. J Cell Sci 132. doi: 10.1242/jcs.230169 [DOI] [PubMed] [Google Scholar]
- Thurston TL, Ryzhakov G, Bloor S, von Muhlinen N, and Randow F. (2009). The TBK1 adaptor and autophagy receptor NDP52 restricts the proliferation of ubiquitin-coated bacteria. Nat Immunol 10, 1215–1221. doi: 10.1038/ni.1800 [DOI] [PubMed] [Google Scholar]
- Turco E, Fracchiolla D, and Martens S. (2019a). Recruitment and Activation of the ULK1/Atg1 Kinase Complex in Selective Autophagy. J Mol Biol. doi: 10.1016/j.jmb.2019.07.027 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Turco E, Witt M, Abert C, Bock-Bierbaum T, Su MY, Trapannone R, Sztacho M, Danieli A, Shi X, Zaffagnini G, et al. (2019b). FIP200 Claw Domain Binding to p62 Promotes Autophagosome Formation at Ubiquitin Condensates. Mol Cell 74, 330–346 e311. doi: 10.1016/j.molcel.2019.01.035 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Valverde DP, Yu S, Boggavarapu V, Kumar N, Lees JA, Walz T, Reinisch KM, and Melia TJ. (2019). ATG2 transports lipids to promote autophagosome biogenesis. J Cell Biol 218, 1787–1798. doi: 10.1083/jcb.201811139 [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Zutphen T, Todde V, de Boer R, Kreim M, Hofbauer HF, Wolinski H, Veenhuis M, van der Klei IJ, and Kohlwein SD. (2014). Lipid droplet autophagy in the yeast Saccharomyces cerevisiae. Mol Biol Cell 25, 290–301. doi: 10.1091/mbc.E13-08-0448 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vargas JNS, Wang C, Bunker E, Hao L, Maric D, Schiavo G, Randow F, and Youle RJ. (2019). Spatiotemporal Control of ULK1 Activation by NDP52 and TBK1 during Selective Autophagy. Mol Cell 74, 347–362 e346. doi: 10.1016/j.molcel.2019.02.010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Velikkakath AK, Nishimura T, Oita E, Ishihara N, and Mizushima N. (2012). Mammalian Atg2 proteins are essential for autophagosome formation and important for regulation of size and distribution of lipid droplets. Mol Biol Cell 23, 896–909. doi: 10.1091/mbc.E11-09-0785 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vevea JD, Garcia EJ, Chan RB, Zhou B, Schultz M, Di Paolo G, McCaffery JM, and Pon LA. (2015). Role for Lipid Droplet Biogenesis and Microlipophagy in Adaptation to Lipid Imbalance in Yeast. Dev Cell 35, 584–599. doi: 10.1016/j.devcel.2015.11.010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang CW, Miao YH, and Chang YS. (2014). A sterol-enriched vacuolar microdomain mediates stationary phase lipophagy in budding yeast. J Cell Biol 206, 357–366. doi: 10.1083/jcb.201404115 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang L, Zhou J, Yan S, Lei G, Lee CH, and Yin XM. (2017). Ethanol-triggered Lipophagy Requires SQSTM1 in AML12 Hepatic Cells. Sci Rep 7, 12307. doi: 10.1038/s41598-017-12485-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wauters F, Cornelissen T, Imberechts D, Martin S, Koentjoro B, Sue C, Vangheluwe P, and Vandenberghe W. (2019). LRRK2 mutations impair depolarization-induced mitophagy through inhibition of mitochondrial accumulation of RAB10. Autophagy, 1–20. doi: 10.1080/15548627.2019.1603548 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu H, and Ren D. (2015). Lysosomal physiology. Annu Rev Physiol 77, 57–80. doi: 10.1146/annurev-physiol-021014-071649 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zechner R, Madeo F, and Kratky D. (2017). Cytosolic lipolysis and lipophagy: two sides of the same coin. Nature reviews. Molecular cell biology 18, 671–684. doi: 10.1038/nrm.2017.76 [DOI] [PubMed] [Google Scholar]
- Zhang J, Lan Y, Li MY, Lamers MM, Fusade-Boyer M, Klemm E, Thiele C, Ashour J, and Sanyal S. (2018). Flaviviruses Exploit the Lipid Droplet Protein AUP1 to Trigger Lipophagy and Drive Virus Production. Cell Host Microbe 23, 819–831 e815. doi: 10.1016/j.chom.2018.05.005 [DOI] [PubMed] [Google Scholar]
- Zhen Y, and Stenmark H. (2015). Cellular functions of Rab GTPases at a glance. J Cell Sci 128, 3171–3176. doi: 10.1242/jcs.166074 [DOI] [PubMed] [Google Scholar]
- Zheng YT, Shahnazari S, Brech A, Lamark T, Johansen T, and Brumell JH. (2009). The adaptor protein p62/SQSTM1 targets invading bacteria to the autophagy pathway. J Immunol 183, 5909–5916. doi: 10.4049/jimmunol.0900441 [DOI] [PubMed] [Google Scholar]


