Abstract
The serotonin transporter (SERT, SLC6A4) is a Na+-dependent transporter that regulates the availability of serotonin (5-HT, 5-hydroxytryptamine), a key neurotransmitter and hormone in the brain and the intestine. The human SERT gene consists of two alternate promoters that drive expression of an identical SERT protein. However, there are different mRNA transcript variants derived from these two promoters that differ in their 5′ untranslated region (5′UTR), which is the region of the mRNA upstream from the protein-coding region. Two of these transcripts contain exon-1a and are abundant in neuronal tissue, whereas the third transcript contains exon-1c and is abundant in the intestine. The 3′UTR is nearly identical among the transcripts. Current studies tested the hypothesis that the UTRs of SERT influence its expression in intestinal epithelial cells (IECs) by controlling mRNA or protein levels. The SERT UTRs were cloned into luciferase reporter plasmids and luciferase mRNA and activity were measured following transient transfection of the UTR constructs into the model IEC Caco-2. Luciferase activity and mRNA abundance were higher than the empty vector for two of the three 5′UTR variants. Calculation of translation index (luciferase activity divided by the relative luciferase mRNA level) revealed that the exon-1a containing 5′UTRs had enhanced translation when compared to the exon-1c containing 5′UTR which exhibited a low translation efficiency. Compared to the empty vector, the SERT 3′UTR markedly decreased luciferase activity. In silico analysis of the SERT 3′UTR revealed many conserved potential miRNA binding sites that may be responsible for this decrease. In conclusion, we have shown that the UTRs of SERT regulate mRNA abundance and protein expression. Delineating the molecular basis by which the UTRs of SERT influence its expression will lead to an increased understanding of post-transcriptional regulation of SERT in GI disorders associated with altered 5-HT availability.
Keywords: Serotonin, Untranslated regions, Intestine, SERT, SLC6A4
1. Introduction
Serotonin (5-HT, 5-hydroxytryptamine) is a neurotransmitter and hormone with many diverse roles in the central nervous system (CNS) as well as the gastrointestinal (GI) tract. In the brain, 5-HT participates in many physiological processes including sensing of noxious stimuli (Mitchell et al., 1998), regulating the sleep-wake cycle (Boutrel et al., 1999), and controlling mood (Meltzer, 1999). Central 5-HT also is involved in the pathophysiology of depression (Barnes and Sharp, 1999), anxiety (Ramboz et al., 1998), and autism (Cook and Leventhal, 1996). The importance of 5-HT within the CNS is underscored by the fact that many commonly prescribed drugs target the central serotonergic system to treat various psychiatric disorders. Despite this importance, the majority of total body 5-HT is found in the GI tract. In the intestine, 5-HT is synthesized by specialized enteroendocrine cells called enterochromaffin cells. 5-HT released by these cells controls GI motility (Bulbring and Crema, 1958; Jin et al., 1999; Heredia et al., 2009; Kadowaki et al., 1996), activates both intrinsic and extrinsic neural reflexes (Zhu et al., 2001; Li et al., 2000; Savastano and Covasa, 2007; Vanner and Macnaughton, 2004), and modulates electrolyte and fluid homoeostasis (Gill et al., 2005; Saksena et al., 2005; Ning et al., 2004; Kaji et al., 2015). Dysregulation of the 5-HT system is notable in GI disorders including inflammatory bowel disease (IBD) (Wang et al., 2013; Tada et al., 2016; Minderhoud et al., 2007; El-Salhy et al., 1997) and irritable bowel syndrome (IBS) (Coates et al., 2004; Kerckhoffs et al., 2012; Camilleri et al., 2007; Atkinson et al., 2006). Notably, 5-HT mediates its physiological functions via acting upon specific 5-HT receptor subtypes present on neurons, as well as immune and epithelial cells. The availability of 5-HT to act upon these receptor subtypes is controlled by the serotonin transporter (SERT), which transports 5-HT across the plasma membrane in a Na+-dependent manner (Gill et al., 2008). Both preclinical and clinical studies have shown that expression of SERT is decreased in both IBD and IBS (Tada et al., 2016; Coates et al., 2004; Kerckhoffs et al., 2012; Shajib et al., 2018).
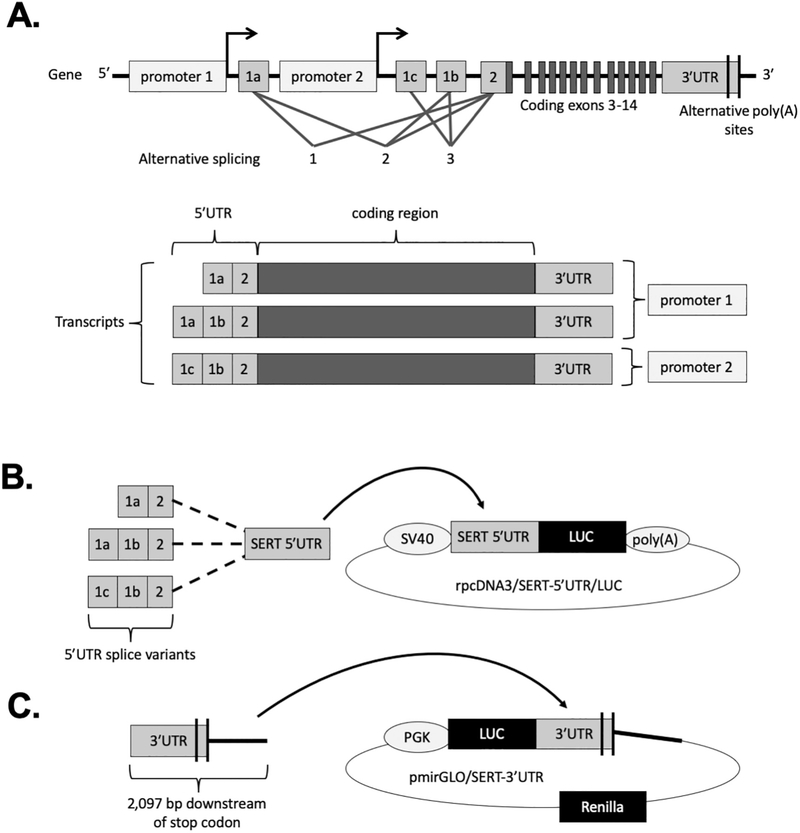
SERT is expressed in multiple tissues and has been localized various cell types, including neurons and intestinal epithelial cells (IECs). The human SERT gene consists of two promoters that drive expression of an identical SERT protein. However, there are different mRNA transcript variants derived from these two promoters (Fig. 1A) (Gill et al., 2008; Linden et al., 2009; Ozsarac et al., 2002; Bradley and Blakely, 1997). These transcript variants differ in their 5′ untranslated region (5′UTR), which is the region of the mRNA upstream from the protein-coding region. While there are three unique 5′UTR splice variants produced from the SERT gene throughout the body by alternative splicing of noncoding exons 1a, 1b, 1c, and 2, only two of these transcript variants are present in intestine (Gill et al., 2008; Linden et al., 2009). Transcription at the most upstream promoter produces transcripts with 5′UTRs composed of either exon-1a spliced to exon-2 (1a-2) or exon-1a spliced to exon-1b spliced to exon-2 (1a-1b-2) with only the former being present in the intestine. Transcription at the second promoter, which is downstream of exon-1a and upstream of exon-1c, produces a single transcript with a 5′UTR composed of exon-1c spliced to exon-1b spliced to exon-2 (1c-1b-2) which is detected at the greatest abundance in the intestine (Gill et al., 2008; Linden et al., 2009). The 5′UTR is known to regulate mRNA stability and turnover, as well as trafficking and translation efficiency (Araujo et al., 2012; Wang et al., 2005). However, the functional difference between the SERT mRNA 5′UTR transcript variants is unknown.
Fig. 1.
Human SERT gene and cloning strategy. (A) The structure of the human SERT (SLC6A4) gene and transcript variants which result from alternative splicing are shown. Transcription at promoter 1 produces two exon 1a-containing transcript variants whereas transcription at promoter 2 produces the exon 1c-containing transcript variant. (B) The rpcDNA3/SERT-5′UTR/LUC constructs were generated by inserting each of the SERT 5′UTR splice variants downstream of the SV40 promoter and immediately upstream of firefly luciferase (LUC) in the rpcDNA3 vector. (C) The pmirGLO/SERT-3′UTR construct was generated by inserting a 2097 bp fragment of the SERT gene downstream of the stop codon immediately downstream of firefly luciferase (LUC) in the pmirGLO vector.
The 3′UTR, the region of the mRNA downstream of the protein-coding region, is also known to influence protein expression (Tian and Manley, 2017). The SERT 3′UTR exists as one of two variants, which differ by approximately 130 bp, due to alternative polyadenylation sites (Yoon et al., 2013; Heils et al., 1995; Battersby et al., 1999). Among other functions, the 3′UTR is a common target for protein binding and miRNA binding. miRNAs are evolutionarily conserved, single-stranded noncoding RNAs typically 20–22 nucleotides in length that bind to multiple mRNAs to influence stability and translation of their target mRNAs (Anbazhagan et al., 2014; Valinezhad Orang et al., 2014). Differential expression of miRNAs has been shown in both IBD and IBS, which have led to functional changes in gene expression (Soroosh et al., 2017; Dalal and Kwon, 2010; Park, 2016; Martínez et al., 2017). While some studies have shown that the SERT 3′UTR harbors functional binding sites for miRNAs which influence protein expression (Liao et al., 2016; Baudry et al., 2010), the overall impact of the SERT 3′UTR in regulating protein expression in intestinal epithelial cells was not assessed.
The overarching goal of these studies was to determine the potential influence of SERT 5′ and 3′ UTRs on SERT expression in the intestinal epithelial cell line Caco-2. First, we identified how the 5′UTR variants affect mRNA abundance and protein expression utilizing luciferase reporter constructs containing the SERT 5′UTRs inserted directly upstream of the luciferase gene. Additionally, we investigated the role of the SERT 3′UTR in regulating protein expression in Caco-2 cells utilizing a luciferase reporter construct containing the SERT 3′UTR downstream of the luciferase gene and performed in silico analysis of potential miRNA binding sites. A greater understanding of how these UTRs influence the expression of SERT at the post-transcriptional level could aid in the development of therapies to treat intestinal disorders associated with decreased SERT expression. These findings may also help identify novel regulatory factors which can influence SERT expression and regulation in the intestine, as well as in other organs such as the brain.
2. Materials and methods
2.1. Cell culture
Caco-2 cells and HEK293 cells were obtained from American Type Culture Collection (ATCC). Caco-2 cells were grown routinely in Eagle’s minimum essential medium (EMEM) (ATCC) supplemented with 10% fetal bovine serum (FBS) while HEK293 cells were grown routinely in Dulbecco’s modified Eagle’s medium (DMEM) (ATCC) supplemented with 10% FBS. Caco-2 and HEK293 cells were maintained in 5% CO2–95% air at 37C. Cell lines were routinely tested for mycoplasma using a commercially available kit (Lonza) according the manufacturer’s instructions.
2.2. Cloning
Double stranded DNA fragments consisting of 1a-2, 1a-1b-2, and 1c-1b-2 5′UTR sequences with HindIII and BglII restriction sites (Table S1) were purchased as gBlocks Gene Fragments from Integrated DNA Technologies (IDT). These fragments were digested and cloned directly into digested rpcDNA3/5′UTR/LUC (generous gift from Joana Floros) (Wang et al., 2005) downstream of the SV40 promoter and upstream of the firefly luciferase reporter gene to generate rpcDNA3/1a-2/LUC, rpcDNA3/1a-1b-2/LUC, and rpcDNA3/1c-1b-2/LUC (Fig. 1B). A construct with no 5′UTR containing the SV40 promoter directly upstream of luciferase was generated by blunt ligation of the digested rpcDNA3/5′UTR/LUC, and was considered the empty vector. The ligated plasmids were transformed into competent DH5α cells (Fisher) and were plated on ampicillin agar plates. A 2097-bp fragment of the SERT gene downstream of the stop codon was amplified by PCR and purified using Qiaquick Gel extraction kit (Qiagen). Purified PCR product was cloned into the multiple cloning sire of the pmirGLO Dual Luciferase miRNA target expression vector (Promega) downstream of the firefly luciferase gene (Fig. 1C). The primer sequences flanked by Sac1 and Sal1 sites used for PCR amplification are presented in Table S2. The resulting plasmid (pmirGLO/SERT-3′UTR) was transformed into the competent JM-109 cell line (Promega) and were plated on ampicillin agar plates. Ampicillin resistant colonies were grown overnight and DNA was extracted using a plasmid miniprep kit (Qiagen). The fidelity of the constructs was confirmed by DNA sequencing.
2.3. Transient transfection and luciferase assay
Caco-2 cells and HEK293 cells were transiently transfected with reporter constructs using lipofectamine 2000 reagent (Invitrogen) as recommended by the manufacturer. For 5′UTR studies, each luciferase reporter construct was co-transfected with CMV-β, a β-gal mammalian expression plasmid (BD Biosciences). Activities of luciferase and β-gal were measured by a luminometer (Promega) utilizing kits from Promega and Clontech, respectively, according to the manufacturer’s instructions. Reporter activity was calculated as a ratio of the luciferase activity to the β-gal activity for each sample. For 3′UTR studies, pmirGLO or pmirGLO-3′UTR-SERT was transfected alone. Reporter activity was determined using the Dual Luciferase Assay Kit (Promega) measured by a luminometer (Promega). Reporter activity was calculated as a ratio of the firefly luciferase activity to the renilla luciferase activity for each sample. Each experiment was performed in quadruplicate wells and repeated at least 3 times.
2.4. RNA extraction and real time RT-PCR
RNA was extracted from cells using RNEasy column purification (Qiagen). Quantitative RT-PCR was performed using SYBR Green fluorescence (Invitrogen) as previously described (Gill et al., 2011). The gene-specific primer sequences are listed in Table S2. The relative mRNA levels were normalized to GAPDH mRNA levels using the Ct method.
2.5. Bioinformatic analysis
In silico identification of putative miRNAs targeting the SERT 3′UTR (696 bp) was performed by employing common prediction algorithms within open source online software. TargetScan (http://www.targetscan.org/) predicted miRNAs that were considered consisted of both conserved and poorly conserved sites among both vertebrates and mammals (Agarwal et al., 2015). miRanda (http://www.microrna.org/) predicted miRNAs that were considered consisted of all conserved sites with any mirSVR score (John et al., 2004). miRDB (http://mirdb.org/miRDB/) that were considered consisted of all miRNAs with a target score of 70 or higher (Wong and Wang, 2015).
3. Results
3.1. Comparison of relative luciferase activity among the SERT 5′UTR splice variants
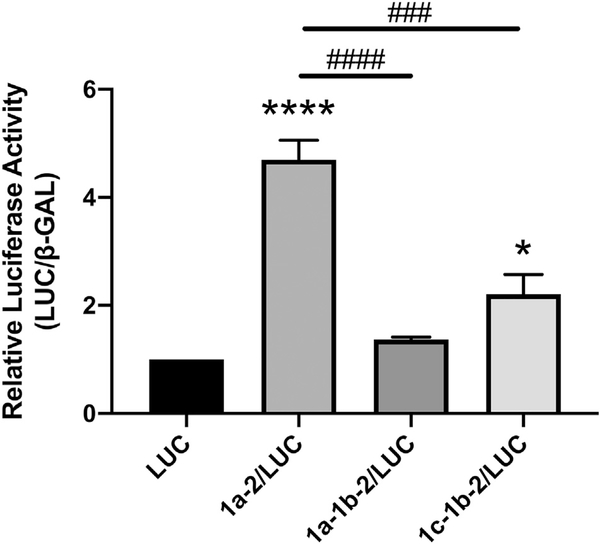
In order to assess the roles of the SERT 5′UTR splice variants on SERT protein expression, the three 5′UTR variants were cloned into rpcDNA3/LUC to generate rpcDNA3/1a-2/LUC, rpcDNA3/1a-1b-2/LUC, and rpcDNA3/1c-1b-2/LUC (Fig. 1B). Caco-2 cells were transiently transfected with rpcDNA3/1a-2/LUC, rpcDNA3/1a-1b-2/LUC, rpcDNA3/1c-1b-2/LUC, or the empty vector rpcDNA3/LUC and luciferase activity was measured 36 h post-transfection. When compared to transient transfection with the empty vector rpcDNA3/LUC, transient transfection with rpcDNA3/1a-2/LUC greatly enhanced luciferase activity approximately 4.7-fold while transient transfection with rpcDNA3/1c-1b-2/LUC slightly enhanced luciferase activity 2.2-fold (Fig. 2). Transfection of either rpcDNA3/1a-1b-2/LUC and with rpcDNA3/1c-1b-2/LUC led to significantly less luciferase activity compared to with rpcDNA3/1a-2/LUC. These data indicate that only two of the three SERT 5′UTR splice variants, specifically 1a-2 and 1c-1b-2, enhance protein expression in IECs whereas the 1a-1b-2 variant does not significantly enhance protein expression.
Fig. 2.
SERT 5′UTR variants mediate expression of luciferase reporter activity. Caco-2 cells were transiently transfected with 5′UTR/LUC reporter constructs or the LUC construct with no 5′UTR along with a β-gal mammalian expression plasmid (CMV-β) by lipofectamine. Luciferase activity was performed in quadruplicate 36 h post transfection and was normalized to β-gal activity. Results are expressed as fold change activity relative to the empty vector LUC transfected cells. Data analyzed by 1-way ANOVA followed by Tukey’s multiple comparisons test (n = 4). *P < 0.05, ****P < 0.0001 vs. LUC transfected cells. ###P < 0.001, ####P < 0.0001 between 5′UTR variants.
3.2. Comparison of luciferase mRNA content among the SERT 5′UTR splice variants
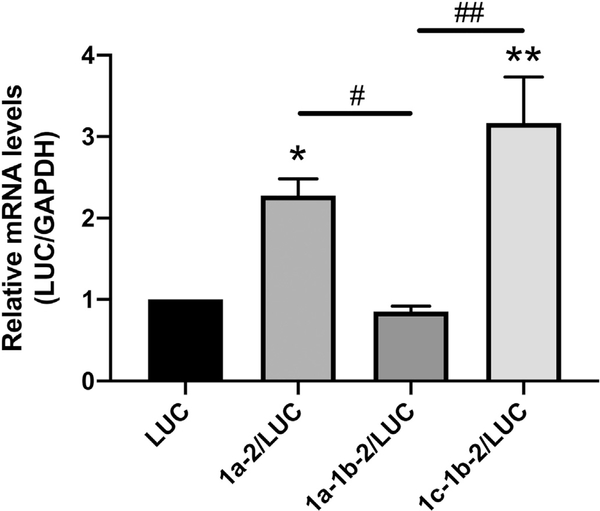
To examine the roles of the SERT 5′UTR splice variants on SERT mRNA levels, Caco-2 cells were transiently transfected with rpcDNA3/1a-2/LUC, rpcDNA3/1a-1b-2/LUC, rpcDNA3/1c-1b-2/LUC, or the empty vector rpcDNA3/LUC and luciferase mRNA content was measured 36 h post-transfection. When compared to transient transfection with the empty vector rpcDNA3/LUC, transient transfection with rpcDNA3/1a-2/LUC increased luciferase mRNA content 2.2-fold and transient transfection with rpcDNA3/1c-1b-2/LUC increased luciferase mRNA content 3.2-fold (Fig. 3). Transient transfection with rpcDNA3/1a-1b-2/LUC did not significantly alter luciferase mRNA levels and led to significantly less luciferase mRNA content than both rpcDNA3/1a-2/LUC and rpcDNA3/1c-1b-2/LUC. These data indicate that the 1a-2 and 1c-1b-2 SERT 5′UTR splice variants increase mRNA abundance in IECs whereas the 1a-1b-2 variant does not significantly alter mRNA abundance.
Fig. 3.
SERT 5′UTR variants mediate luciferase reporter mRNA levels. Caco-2 cells were transiently transfected with 5′UTR/LUC reporter constructs or the LUC construct with no 5′UTR along with a β-gal mammalian expression plasmid (CMV-β) by lipofectamine. RNA was extracted 36 h post transfection and RT-PCR for Luciferase and GAPDH was performed. Data represent Luciferase mRNA levels relative to GAPDH mRNA. Results are expressed as fold change mRNA relative to the empty vector LUC transfected cells. Data analyzed by 1-way ANOVA followed by Tukey’s multiple comparisons test (n = 4). *P < 0.05, **P < 0.01 vs. LUC transfected cells. #P < 0.05, ##P < 0.01 between 5′UTR variants.
3.3. Translation efficiency index of SERT 5′UTR variants
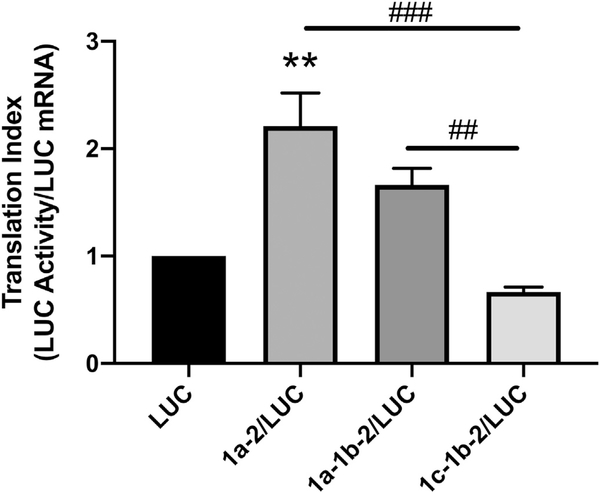
The translation efficiencies of the SERT 5′UTR variants were evaluated using the index of luciferase activity/relative mRNA content. A larger index indicates a higher translation efficiency for the 5′UTR. Only rpcDNA3/1a-2/LUC exhibited a significantly increased translation index compared to the empty vector rpcDNA3/LUC (Fig. 4). However, both rpcDNA3/1a-2/LUC and rpcDNA3/1a-1b-2/LUC exhibited significantly higher translation efficiencies compared to rpcDNA3/1c-1b-2/LUC. This is in spite of the fact that rpcDNA3/1c-1b-2/LUC led to enhanced luciferase mRNA content and activity compared to the empty vector. These results demonstrate that the 1c-1b-2 SERT 5′UTR, which is the most abundant variant found within IECs, may assume an inhibitory role in protein translation by decreasing translation efficiency and may indicate a mechanism by which the SERT 5′UTR variants modulate expression of SERT.
Fig. 4.
Comparison of 5′UTR-mediated translational indexes among SERT 5′UTR splice variants. Caco-2 cells were transiently transfected with 5′UTR/LUC reporter constructs or the LUC construct with no 5′UTR along with a β-gal mammalian expression plasmid (CMV-β) by lipofectamine. The translational index for each 5′UTR was computed by diving the relative Luciferase activity by the relative mRNA level for each construct. Translational indices are expressed relative to the translational index of the empty vector LUC. Data analyzed by 1-way ANOVA followed by Tukey’s multiple comparisons test (n = 4). **P < 0.01 vs. LUC transfected cells. ##P < 0.01, ##P < 0.001 between 5′UTR variants.
3.4. Effect of the SERT 3′UTR on luciferase reporter activity
To investigate the potential role of the SERT 3′UTR on regulating SERT protein expression, both HEK293 cells and Caco-2 cells were transiently transfected with pmirGLO/SERT-3′UTR or the empty vector pmirGLO and luciferase activity was measured after 48 h. In both cell types, there was a significant reduction in luciferase activity with pmirGLO/SERT-3′UTR compared to the empty vector (Fig. 5). However, the reduction in activity in HEK293 cells was small (12%) whereas the reduction in activity in Caco-2 cells was much greater (88%). These results suggest that there are regulatory factors, which are specifically present within IECs, that may regulate SERT expression via the 3′UTR.
Fig. 5.
SERT 3′UTR Luciferase reporter activity in different cell lines. HEK293 cells (A) and Caco-2 cells (B) were transiently transfected with pmirGLO/SERT-3′UTR or the empty vector pmirGLO. Dual luciferase assay was performed in quadruplicate 48 h post transfection. Firefly luciferase activity was normalized to renilla luciferase activity. Results are expressed as fold change activity relative to the empty vector pmirGLO transfected cells. Data analyzed by paired Student’s t-test (n = 3). *P < 0.05, **P < 0.01 vs. pmirGLO transfected cells.
3.5. Prediction of miRNAs targeting the SERT 3′UTR
TargetScan, miRDB, and MiRanda algorithms were utilized to predict miRNAs with putative binding sites within the SERT 3′UTR. The longest SERT 3′UTR variant produced by alternative polyadenylation (696 bp) was used for analysis. As shown in Table 1, TargetScan predicted 24, miRDB predicted 31, and miRanda predicted 40 miRNAs targeting the SERT 3′UTR. However, only 8 miRNAs have been experimentally validated to decrease SERT expression via the 3′UTR, and only 2 of these miRNAs (miR-24 and miR-200a) have been shown to affect the SERT 3′UTR in intestinal epithelial cells (Table 2). Therefore, it is possible that there are additional miRNAs that target the SERT 3′UTR in intestinal epithelial cells to decrease SERT expression that have not yet been discovered.
Table 1.
Number of miRNAs predicted to target the SERT 3′UTR by three different prediction algorithms.
| Algorithm | Website | Number of predicted targeting miRNAs | Reference |
|---|---|---|---|
| TargetScan | http://www.targetscan.org/ | 24 | (Agarwal et al., 2015) |
| miRDB | http://mirdb.org/miRDB/ | 31 | (Wong and Wang, 2015) |
| miRanda | http://www.microrna.org/ | 40 | (John et al., 2004) |
Table 2.
Experimentally validated miRNAs targeting SERT.
| miRNA | Cell line | Cell type | Species | Validation technique | Reference |
|---|---|---|---|---|---|
| miR-15a | JAR | Placental | Human | Reporter assay, Western blot, qPCR | (Moya et al., 2013) |
| RN46A | Neuronal | Rat | Reporter assay | (Moya et al., 2013) | |
| Primary smooth muscle cells | Vascular | Human | Western blot | (Gu et al., 2017) | |
| miR-16 | JAR | Placental | Human | Reporter assay, Western blot, qPCR | (Moya et al., 2013) |
| RN46A | Neuronal | Rat | Reporter assay | (Moya et al., 2013) | |
| 1C11 | Neuroectodermal | Mouse | Functional assay | (Baudry et al., 2010) | |
| HeLa | Cervical | Human | Reporter assay | (Baudry et al., 2010) | |
| In vivo | Neurons | Mouse | Functional assay | (Launay et al., 2011) | |
| Primary smooth muscle cells | Vascular | Human | Western blot | (Gu et al., 2017) | |
| miR-24 | NCM460 | Intestinal | Human | Reporter assay, Western blot, qPCR | (Liao et al., 2016) |
| miR-135a | HEK293 | Fibroblast | Human | Reporter assay | (Issler et al., 2014) |
| In vivo | Neurons | Mouse | Western blot, qPCR | (Issler et al., 2014) | |
| miR-135b | HEK293 | Fibroblast | Human | Reporter assay | (Issler et al., 2014) |
| miR-195 | Primary smooth muscle cells | Vascular | Human | Western blot | (Gu et al., 2017) |
| miR-200a | Primary colonic cells | Intestinal | Rat | Reporter assay, Western blot, qPCR | (Hou et al., 2018) |
| miR-322 | Primary smooth muscle cells | Vascular | Human | Western blot | (Gu et al., 2017) |
4. Discussion
Many previous studies have established that SERT is regulated via transcriptional mechanisms, including transcription factor binding to its promoters (Gill et al., 2011; García-Frigola and Herrera, 2010; Esmaili et al., 2009a), as well as post-translational mechanisms such as phosphorylation (Singhal et al., 2017; Jayanthi et al., 2005; Steiner et al., 2008; Esmaili et al., 2009b). However, little is known regarding post-transcriptional regulation of SERT mRNA transcripts. This study has established that 5′UTR variants produced by alternative splicing and transcription at alternative promoters have differential effects on SERT mRNA abundance and protein expression, as well as translation efficiency. Further, we observed that the SERT 3′UTR decreases protein expression of the reporter gene remarkably in intestinal epithelial cells but has little effect on the reporter gene expression in human embryonic kidney 293 cells. This may be due to differential expression of miRNAs targeting the SERT 3′UTR, as evidenced by bioinformatic analysis of the SERT 3′UTR, as well as a literature review of established miRNAs which have been shown to decrease SERT expression. Two of the three SERT 5′UTR variants, 1a-2 and 1c-1b-2, are expressed in intestinal epithelial cells. Previous studies have shown that the 1c-1b-2 containing variant is expressed higher than the 1a-2 variant in the intestine (Linden et al., 2009; Gill et al., 2013). Our current studies utilizing luciferase reporter constructs show that mRNA abundance for these variants is similar, suggesting that the differences observed in the intestine are likely due to increased activity at promoter 2, located upstream of exon 1c (Fig. 1), rather than differences in mRNA stability. However, the 1a-2 variant exhibited greater translation efficiency compared to the 1c-1b-2 variant. This suggests that the amount of SERT protein produced by the variants is not equivalent with the 1a-containing transcript producing SERT protein more efficiently than the 1c-containing transcript. While not detected significantly in the intestine, the 1a-1b-2 variant also exhibited a higher translation efficiency compared to the 1c-1b-2 variant.
It has been established that the 5′UTR plays an important role in the initiation of translation (Araujo et al., 2012). Two of the structural elements of the 5′UTR which determine the efficiency of translation initiation are the length of the 5′UTR and the presence of AUG codons in the 5′UTR (Araujo et al., 2012; Wang et al., 2005). The higher efficiency of 1a-2 (208 bp) compared to 1a-1b-2 (305 bp) and 1c-1b-2 (364 bp) is consistent with the idea that increased 5′UTR length enhances translation efficiency. Additionally, the SERT 5′UTR variants differ in the prevalence of AUGs. These AUGs can be categorized into either upstream open reading frames (uORFs) or upstream AUGs (uAUGs). uORFs consist of an AUG followed by an in-frame stop codon whereas an uAUG consists of an AUG that is not followed by an in-frame stop codon and are considered to be more detrimental to translation efficiency than uORFs (Araujo et al., 2012; Wang et al., 2005). The 1a-2 sequence contains one uORF, 1a-1b-2 contains one uORF and one uAUG, and 1c-1b-2 contains two uORFs and one uAUGs (Table S3). We speculate that these sequences are at least partially responsible for the differences in translation efficiency among the 5′UTR variants. Future mutation studies will confirm that these sequences are functionally active in regulating SERT expression.
Many investigations have established that the miRNA expression profile is altered during intestinal pathologies including IBS and IBD (Soroosh et al., 2017; Dalal and Kwon, 2010; Park, 2016; Martínez et al., 2017). During disease, expression of certain of miRNAs can either increase or decrease. Since our data showed that the SERT 3′UTR decreased protein expression, it is possible that miRNAs that bind to the SERT 3′UTR to decrease translation are increased in these diseases. Alternatively, it has been shown that some miRNA-mRNA interactions activate translation (Valinezhad Orang et al., 2014; Ni and Leng, 2016). Therefore, activating miRNAs targeting SERT may exist, and these miRNAs may be decreased in IBS or IBD.
Few studies have already related alterations in SERT expression to changes in miRNA levels during intestinal pathology. One intriguing study found that miR-24 was upregulated in IBS patients as well as a mouse model of IBS, experimentally validated SERT as a target of miR24, and showed that treatment with miR-24 inhibitor increased pain threshold and nociceptive threshold levels (Liao et al., 2016). Another study found that miR-200a was elevated in a mouse model of IBS-D displaying increased nociceptive visceral hypersensitivity accompanied by a decrease in SERT expression and proceeded to show that SERT was a target of miR-200a (Hou et al., 2018). In the context of IBD, it was found that mucosal miR-424 expression was decreased during pouchitis which inversely correlated to SERT expression (Sherman Horev et al., 2018). Targeting of miR-424 to the SERT mRNA was not experimentally validated which makes it unclear whether miR-424 is directly involved in regulating SERT during IBD. With only two miRNA species experimentally validated to target SERT in intestinal epithelial cells, future studies may reveal additional miRNAs which target SERT.
Alternatively, there may be other factors, including RNA binding proteins (RBPs), which bind to SERT mRNA variants to regulate protein expression. Such proteins are able to bind to both the 5′UTR the 3′UTR, as well as the coding region, and have many different functions (Hentze et al., 2018). One instance of transcript-specific RBP-mediated SERT regulation was established, which showed that heterogeneous nuclear ribonucleoprotein K (hnRNPK) bound only to the SERT 3′UTR variant produced via the downstream polyadenylation site (Yoon et al., 2013). It is possible that a phenomenon exists in the SERT 5′UTR, where certain RBPs bind exclusively to SERT mRNA transcripts containing a specified exon. Additional studies will address whether such interactions exist, and whether these interactions are altered during IBS or IBD.
One question that has not been addressed is the relative distribution of SERT mRNA variants along the length of the human intestine. In this regard, a previous study has shown that both the 1a-2 and 1c-1b-2 5′UTR-containing SERT transcripts can be amplified from human colonic mucosa cDNA (Linden et al., 2009). We have shown previously that total SERT mRNA and protein expression is highest in human small intestine, more precisely the ileum, and is lower in the colon (Gill et al., 2008). It is possible that post-transcriptional regulation of SERT mRNA variants via the 5′UTR may contribute to the regional distribution of SERT expression in the intestine as well as in different intestinal epithelial cell lines. The relative abundance of the SERT 5′UTR splice variants along the length of the intestine and among cultured cell models will be the subject of future investigation.
5. Conclusion
It can be concluded that SERT expression is controlled by both the 5′UTR as well as the 3′UTR in IECs. SERT 5′UTR splice variants differentially influence mRNA abundance, protein expression as well as translation efficiency. The SERT 3′UTR has an overall negative impact on protein expression in intestinal epithelial cells, which may be due to negative modulation by miRNAs or other factors which bind the 3′UTR. Understanding these mechanisms by which the UTRs of SERT influence its protein expression may open new avenues for targeting SERT during intestinal pathologies including IBS and IBD.
Supplementary Material
Acknowledgements
This research was supported by NIDDK grants R01 DK098170 (RKG) and Dept of Veteran Affairs BX002011 (PKD), BX000152 (WAA) and BX 002867 (SS).
Abbreviations
- 5-HT
serotonin
- CNS
central nervous system
- GI
gastrointestinal
- IBD
inflammatory bowel disease
- IBS
irritable bowel syndrome
- SLC6A4
solute carrier family 6 member 4
- SERT
serotonin transporter
- IEC
intestinal epithelial cell
- UTR
untranslated region
- bp
base-pair
- EMEM
Eagle’s essential minimum medium
- FBS
fetal bovine serum
- DMEM
Dulbecco’s modified Eagle’s medium
- uORF
upstream open reading frame
- uAUG
upstream AUG
- RBP
RNA binding proteins
- hnRNPK
heterogeneous nuclear ribonucleoprotein K
Footnotes
Declaration of competing interest
None.
Appendix A. Supplementary data
Supplementary data to this article can be found online at https://doi.org/10.1016/j.genrep.2019.100513.
References
- Agarwal V, Bell GW, Nam J-W, Bartel DP, 2015. August 12. Predicting effective microRNA target sites in mammalian mRNAs. Elife 4, 101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anbazhagan AN, Priyamvada S, Kumar A, Maher DB, Borthakur A, Alrefai WA, et al. , 2014. Translational repression of SLC26A3 by miR-494 in intestinal epithelial cells. Am. J. Physiol. Gastrointest. Liver Physiol 306 (2), G123–G131 January. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Araujo PR, Yoon K, Ko D, Smith AD, Qiao M, Suresh U, et al. , 2012. Before it gets started: regulating translation at the 5’ UTR. In: Comp. Funct. Genomics. 2012. Hindawi Publishing Corporation, pp. 475731–475738 (4). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Atkinson W, Lockhart S, Whorwell PJ, Keevil B, Houghton LA, 2006. Altered 5-hydroxytryptamine signaling in patients with constipation- and diarrhea-predominant irritable bowel syndrome. Gastroenterology 130 (1), 34–43 January. [DOI] [PubMed] [Google Scholar]
- Barnes NM, Sharp T, 1999. A review of central 5-HT receptors and their function. Neuropharmacology 38 (8), 1083–1152 August. [DOI] [PubMed] [Google Scholar]
- Battersby S, Ogilvie AD, Blackwood DH, Shen S, Muqit MM, Muir WJ, et al. , 1999. Presence of multiple functional polyadenylation signals and a single nucleotide polymorphism in the 3′ untranslated region of the human serotonin transporter gene. J. Neurochem 72 (4), 1384–1388. [DOI] [PubMed] [Google Scholar]
- Baudry A, Mouillet-Richard S, Schneider B, Launay J-M, Kellermann O, 2010. miR-16 targets the serotonin transporter: a new facet for adaptive responses to antidepressants. Science 329 (5998), 1537–1541. [DOI] [PubMed] [Google Scholar]
- Boutrel B, Franc B, Hen R, Hamon M, Adrien J, 1999. Key role of 5-HT1B receptors in the regulation of paradoxical sleep as evidenced in 5-HT1B knock-out mice. J. Neurosci 19 (8), 3204–3212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bradley CC, Blakely RD, 1997. Alternative splicing of the human serotonin transporter gene. J. Neurochem 69 (4), 1356–1367. [DOI] [PubMed] [Google Scholar]
- Bulbring E, Crema A, 1958. December. Observations concerning the action of 5-hydroxytryptamine on the peristaltic reflex. Br. J. Pharmacol. Chemother 13 (4), 444–457 Wiley-Blackwell. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Camilleri M, Andrews CN, Bharucha AE, Carlson PJ, Ferber I, Stephens D, et al. , 2007. Alterations in expression of p11 and SERT in mucosal biopsy specimens of patients with irritable bowel syndrome. Gastroenterology 132 (1), 17–25 January. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coates MD, Mahoney CR, Linden DR, Sampson JE, Chen J, Blaszyk H, et al. , 2004. Molecular defects in mucosal serotonin content and decreased serotonin reuptake transporter in ulcerative colitis and irritable bowel syndrome. Gastroenterology 126 (7), 1657–1664 June. [DOI] [PubMed] [Google Scholar]
- Cook EH, Leventhal BL. The serotonin system in autism. Curr. Opin. Pediatr 1996; 8(4):348–54. [DOI] [PubMed] [Google Scholar]
- Dalal SR, Kwon JH, 2010. The Role of MicroRNA in Inflammatory Bowel Disease. Gastroenterol Hepatol (N Y) 6(11). Millenium Medical Publishing, pp. 714–722 November. [PMC free article] [PubMed] [Google Scholar]
- El-Salhy M, Danielsson A, Stenling R, Grimelius L, 1997. Colonic endocrine cells in inflammatory bowel disease. J. Intern. Med. 242 (5), 413–419 November. [DOI] [PubMed] [Google Scholar]
- Esmaili A, Nazir SF, Borthakur A, Yu D, Turner JR, Saksena S, et al. , 2009a. Enteropathogenic Escherichia coli infection inhibits intestinal serotonin transporter function and expression. Gastroenterology 137 (6), 2074–2083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Esmaili A, Nazir SF, Borthakur A, Yu D, Turner JR, Saksena S, et al. , 2009b. Enteropathogenic Escherichia coli infection inhibits intestinal serotonin transporter function and expression. Gastroenterology 137 (6), 2074–2083 December. [DOI] [PMC free article] [PubMed] [Google Scholar]
- García-Frigola C, Herrera E, 2010. Zic2 regulates the expression of Sert to modulate eye-specific refinement at the visual targets. EMBO J. 29 (18), 3170–3183 September 15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gill RK, Saksena S, Tyagi S, Alrefai WA, Malakooti J, Sarwar Z, et al. , 2005. Serotonin inhibits Na+/H+ exchange activity via 5-HT4 receptors and activation of PKC alpha in human intestinal epithelial cells. Gastroenterology 128 (4), 962–974 April. [DOI] [PubMed] [Google Scholar]
- Gill RK, Pant N, Saksena S, Singla A, Nazir TM, Vohwinkel L, et al. , 2008. Function, expression, and characterization of the serotonin transporter in the native human intestine. Am. J. Physiol.-Gastr. L 294 (1), G254–G262 (American Physiological Society). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gill RK, Anbazhagan AN, Esmaili A, Kumar A, Nazir S, Malakooti J, et al. , 2011. April 1. Epidermal growth factor upregulates serotonin transporter in human intestinal epithelial cells via transcriptional mechanisms. Am. J. Physiol.-Gastr. L 300 (4), G627–G636 American Physiological Society. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gill RK, Kumar A, Malhotra P, Maher D, Singh V, Dudeja PK, et al. , 2013. Regulation of intestinal serotonin transporter expression via epigenetic mechanisms: role of HDAC2. Am. J. Physiol., Cell Physiol 304 (4), C334–C341 February 15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gu J, Zhang H, Ji B, Jiang H, Zhao T, Jiang R, et al. , 2017. Vesicle miR-195 derived from endothelial cells inhibits expression of serotonin transporter in vessel smooth muscle cells. Sci. Rep 7 (1), 43546 Nature Publishing Group. March 8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heils A, Teufel A, Petri S, Seemann M, Bengel D, Balling U, et al. , 1995. Functional promoter and polyadenylation site mapping of the human serotonin (5-HT) transporter gene. J. Neural Transm. Gen. Sect 102 (3), 247–254. [DOI] [PubMed] [Google Scholar]
- Hentze MW, Castello A, Schwarzl T, Preiss T, 2018. A brave new world of RNA-binding proteins. Nat. Rev. Mol. Cell Biol. 19 (5), 327–341 May. [DOI] [PubMed] [Google Scholar]
- Heredia DJ, Dickson EJ, Bayguinov PO, Hennig GW, Smith TK, 2009. Localized release of serotonin (5-hydroxytryptamine) by a fecal pellet regulates migrating motor complexes in murine colon. Gastroenterology 136 (4), 1328–1338 April. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hou Q, Huang Y, Zhang C, Zhu S, Li P, Chen X, et al. , 2018. October 1. MicroRNA-200a targets cannabinoid receptor 1 and serotonin transporter to increase visceral Hyperalgesia in diarrhea-predominant irritable bowel syndrome rats. J Neurogastroenterol Motil 24 (4), 656–668 Korean Society of Neurogastroenterology and Motility. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Issler O, Haramati S, Paul ED, Maeno H, Navon I, Zwang R, et al. , 2014. MicroRNA 135 is essential for chronic stress resiliency, antidepressant efficacy, and intact serotonergic activity. Neuron 83 (2), 344–360 July 16. [DOI] [PubMed] [Google Scholar]
- Jayanthi LD, Samuvel DJ, Blakely RD, Ramamoorthy S, 2005. Evidence for bi-phasic effects of protein kinase C on serotonin transporter function, endocytosis, and phosphorylation. Mol. Pharmacol. 67 (6), 2077–2087 June. [DOI] [PubMed] [Google Scholar]
- Jin JG, Foxx-Orenstein AE, Grider JR, 1999. Propulsion in Guinea pig colon induced by 5-hydroxytryptamine (HT) via 5-HT4 and 5-HT3 receptors. J. Pharmacol. Exp. Ther. 288 (1), 93–97 January. [PubMed] [Google Scholar]
- John B, Enright AJ, Aravin A, Tuschl T, Sander C, Marks DS, 2004. Human MicroRNA targets. PLoS Biol. 2 (11), e363 November. (Editor: Carrington James C). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kadowaki M, Wade PR, Gershon MD, 1996. November. Participation of 5-HT3, 5-HT4, and nicotinic receptors in the peristaltic reflex of Guinea pig distal colon. Am. J. Phys. 271 (5 Pt 1), G849–G857. [DOI] [PubMed] [Google Scholar]
- Kaji I, Akiba Y, Said H, Narimatsu K, Kaunitz JD, 2015. October. Luminal 5-HT stimulates colonic bicarbonate secretion in rats. Br. J. Pharmacol. 172 (19), 4655–4670 John Wiley & Sons, Ltd (10.1111). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kerckhoffs APM, Linde ter JJM, Akkermans LMA, Samsom M, 2012. May 1. SERT and TPH-1 mRNA expression are reduced in irritable bowel syndrome patients regardless of visceral sensitivity state in large intestine. Am. J. Physiol. Gastrointest. Liver Physiol 302 (9), G1053–G1060 American Physiological Society; Bethesda, MD. [DOI] [PubMed] [Google Scholar]
- Launay JM, Mouillet-Richard S, Baudry A, Pietri M, Kellermann O, 2011. November 22. Raphe-mediated signals control the hippocampal response to SRI antidepressants via miR-16. Transl. Psychiatry 1 (11), 1–8 Nature Publishing Group. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Y, Hao Y, Zhu J, Owyang C, 2000. Serotonin released from intestinal enterochromaffin cells mediates luminal non-cholecystokinin-stimulated pancreatic secretion in rats. Gastroenterology 118 (6), 1197–1207 June. [DOI] [PubMed] [Google Scholar]
- Liao X-J, Mao W-M, Wang Q, Yang G-G, Wu W-J, Shao S-X, 2016. MicroRNA-24 inhibits serotonin reuptake transporter expression and aggravates irritable bowel syndrome. Biochem. Biophys. Res. Commun Elsevier Ltd 469 (2), 288–293. [DOI] [PubMed] [Google Scholar]
- Linden DR, White SL, Brooks EM, Mawe GM, 2009. May. Novel promoter and alternate transcription start site of the human serotonin reuptake transporter in intestinal mucosa. In: Neurogastroenterology & Motility. 21(5). Blackwell Publishing Ltd, pp. 534–e11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martínez C, Rodiño-Janeiro BK, Lobo B, Stanifer ML, Klaus B, Granzow M, et al. , 2017. miR-16 and miR-125b are involved in barrier function dysregulation through the modulation of claudin-2 and cingulin expression in the jejunum in IBS with diarrhoea. Gut 66 (9), 1537–1538. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meltzer HY, 1999. The role of serotonin in antipsychotic drug action. Neuropsychopharmacology 21 (2 Suppl), 106S–115S. [DOI] [PubMed] [Google Scholar]
- Minderhoud IM, Oldenburg B, Schipper MEI, Linde ter JJM, Samsom M, 2007. Serotonin synthesis and uptake in symptomatic patients with Crohn’s disease in remission. Clin. Gastroenterol. Hepatol. 5 (6), 714–720 June. [DOI] [PubMed] [Google Scholar]
- Mitchell JM, Lowe D, Fields HL, 1998. The contribution of the rostral ventromedial medulla to the antinociceptive effects of systemic morphine in restrained and unrestrained rats. Neuroscience 87 (1), 123–133 November. [DOI] [PubMed] [Google Scholar]
- Moya PR, Wendland JR, Salemme J, Fried RL, Murphy DL, 2013. miR-15a and miR-16 regulate serotonin transporter expression in human placental and rat brain raphe cells. Int. J. Neuropsychopharmacol. 16 (3), 621–629. [DOI] [PubMed] [Google Scholar]
- Ni W-J, Leng X-M, 2016. miRNA-dependent activation of mRNA translation. Microrna 5 (2), 83–86. [DOI] [PubMed] [Google Scholar]
- Ning Y, Zhu JX, Chan HC, 2004. Regulation of ion transport by 5-hydroxytryptamine in rat colon. Clin. Exp. Pharmacol. Physiol. 31 (7), 424–428 John Wiley & Sons, Ltd (10.1111). Jul. [DOI] [PubMed] [Google Scholar]
- Ozsarac N, Santha E, Hoffman BJ, 2002. Alternative Non-coding Exons Support Serotonin Transporter mRNA Expression in the Brain and Gut. 82(2). pp. 336–344 Jul. [DOI] [PubMed] [Google Scholar]
- Park JH, 2016. Dysregulated MicroRNA expression in irritable bowel syndrome. J Neurogastroenterol Motil 22 (2), 166–167 April 30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ramboz S, Oosting R, Amara DA, Kung HF, Blier P, Mendelsohn M, et al. , 1998. Serotonin receptor 1A knockout: an animal model of anxiety-related disorder. Proc. Natl. Acad. Sci. 95 (24), 14476–14481 November 24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saksena S, Gill RK, Tyagi S, Alrefai WA, Sarwar Z, Ramaswamy K, et al. , 2005. Involvement of c-Src and protein kinase C delta in the inhibition of Cl(−)/OH- exchange activity in Caco-2 cells by serotonin. J. Biol. Chem. Am. Soc. Biochem. Mol. Biol 280 (12), 11859–11868 March 25. [DOI] [PubMed] [Google Scholar]
- Savastano DM, Covasa M, 2007. Intestinal nutrients elicit satiation through concomitant activation of CCK(1) and 5-HT(3) receptors. Physiol. Behav. 92 (3), 434–442 October 22. [DOI] [PubMed] [Google Scholar]
- Shajib MS, Chauhan U, Adeeb S, Chetty Y, Armstrong D, Halder SLS, et al. , 2018. Characterization of serotonin signaling components in patients with inflammatory bowel disease. J. Can. Assoc. Gastroenterol. 1 (7), 299 August 10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sherman Horev H, Rabinowitz KM, Elad H, Barkan R, Ben-Shachar S, Pasmanik Chor M, et al. , 2018. Increase in processing factors is involved in skewed MicroRNA expression in patients with ulcerative colitis who develop small intestine inflammation after pouch surgery. Inflamm. Bowel Dis. 24 (5), 1045–1054 April 23. [DOI] [PubMed] [Google Scholar]
- Singhal M, Manzella C, Soni V, Alrefai WA, Saksena S, Hecht GA, et al. , 2017. May 1. Role of SHP2 protein tyrosine phosphatase in SERT inhibition by enteropathogenic E. coli (EPEC). Am. J. Physiol. Gastrointest. Liver Physiol 312 (5), G443–G449 American Physiological Society. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Soroosh A, Koutsioumpa M, Pothoulakis C, Iliopoulos D, 2017. November 16. Functional role and therapeutic targeting of microRNAs in inflammatory bowel disease. Am. J. Physiol. Gastrointest. Liver Physiol 314, G256–G262 ajpgi002682017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Steiner JA, Carneiro AMD, Blakely RD, 2008. Going with the flow: trafficking-dependent and -independent regulation of serotonin transport. Traffic 9 (9), 1393–1402 September. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tada Y, Ishihara S, Kawashima K, Fukuba N, Sonoyama H, Kusunoki R, et al. , 2016. Downregulation of serotonin reuptake transporter gene expression in healing colonic mucosa in presence of remaining low-grade inflammation in ulcerative colitis. J. Gastroenterol. Hepatol. 31 (8), 1443–1452 August. [DOI] [PubMed] [Google Scholar]
- Tian B, Manley JL, 2017. Alternative polyadenylation of mRNA precursors. Nat. Rev. Mol. Cell Biol. 18 (1), 18–30 Jan. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Valinezhad Orang A, Safaralizadeh R, Kazemzadeh-Bavili M, 2014. Mechanisms of miRNA-mediated gene regulation from common downregulation to mRNA-specific upregulation. Int J Genomics. 2014 (12), 970607–970615 Hindawi. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vanner S, Macnaughton WK, 2004. April. Submucosal secretomotor and vasodilator reflexes. Neurogastroenterol. Motil. 16 (Suppl. 1), 39–43 John Wiley & Sons, Ltd (10.1111). s1. [DOI] [PubMed] [Google Scholar]
- Wang G, Wang G, Guo X, Floros J, 2005. Differences in the translation efficiency and mRNA stability mediated by 5’-UTR splice variants of human SP-A1 and SP-A2 genes. Am. J. Physiol. Lung Cell Mol. Physiol 289 (3), L497–L508 September. [DOI] [PubMed] [Google Scholar]
- Wang Y, Gong H, Lopez R, Lian L, Kiran RP, Soffer EE, et al. , 2013. Correlation between serum serotonin and endoscopy inflammation scores in patients with ileal pouches. J Crohns Colitis 7 (4), e133–e142 May. [DOI] [PubMed] [Google Scholar]
- Wong N, Wang X, 2015. miRDB: an online resource for microRNA target prediction and functional annotations. Nucleic Acids Res. 43 (Database issue), D146–D152 January. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoon Y, McKenna MC, Rollins DA, Song M, Nuriel T, Gross SS, et al. , 2013. Anxiety-associated alternative polyadenylation of the serotonin transporter mRNA confers translational regulation by hnRNPK. Proc. Natl. Acad. Sci. U.S.A. National Academy of Sciences 110 (28), 11624–11629 July 9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu JX, Zhu XY, Owyang C, Li Y, 2001. Intestinal serotonin acts as a paracrine substance to mediate vagal signal transmission evoked by luminal factors in the rat. J. Physiol. Lond 530 (Pt 3), 431–442 Wiley-Blackwell. February 1. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.