Abstract
An abdominal aortic aneurysm (AAA) is a ballooning of the abdominal aorta, that if not treated tends to grow and rupture. Computed Tomography Angiography (CTA) is the main imaging modality for the management of AAAs, and segmenting them is essential for AAA rupture risk and disease progression assessment. Previous works have shown that Convolutional Neural Networks (CNNs) can accurately segment AAAs, but have the limitation of requiring large amounts of annotated data to train the networks. Thus, in this work we propose a methodology to train a CNN only with images generated with a synthetic shape model, and test its generalization and ability to segment AAAs from new original CTA scans. The synthetic images are created from realistic deformations generated by applying principal component analysis to the deformation fields obtained from the registration of few datasets. The results show that the performance of a CNN trained with synthetic data to segment AAAs from new scans is comparable to the one of a network trained with real images. This suggests that the proposed methodology may be applied to generate images and train a CNN to segment other types of aneurysms, reducing the burden of obtaining large annotated image databases.
Keywords: Abdominal aortic aneurysm, Segmentation, Convolutional Neural Network, Synthetic images, Principal component analysis
1. Introduction
An abdominal aortic aneurysm (AAA) is a focal dilation of the aorta, which in most of the cases requires surgical treatment to prevent its rupture. Computed Tomography Angiography (CTA) is the preferred imaging modality for the management of AAAs, from the diagnosis to the pre-operative planning of the intervention and the post-operative follow-up. Specifically, the diagnosis consists in evaluating the rupture risk by measuring the size of the AAA from a CTA scan. The follow-up aims at evaluating the progression of the AAA, i.e. the post-operative shrinkage of the AAA or its expansion if the intervention is unsuccessful. Hence, automatic AAA segmentation tools are essential to aid the clinicians in these tasks and they open up the opportunity to perform complex analyses of AAA morphology and biomechanics.
Convolutional Neural Networks (CNNs) have shown the ability to automatically and accurately segment infra-renal AAAs from pre-operative and postoperative CTA scans [1,2]. CNNs efficiently learn implicit object representations at different scales from input annotated data. However, the need for such annotations is one of the main limitations of CNN-based segmentation approaches, since obtaining them is time-consuming and requires expert knowledge. Here is where synthetic image generation and data augmentation play an important role in order to train a network from few annotated input data.
Thus, the aim of this study is to evaluate the ability of a 3D CNN trained only with images generated with a synthetic shape model to segment aneurysms from real CTA scans. The employed synthetic image generation approach [3] is based on realistic deformations computed from the Principal Component Analysis (PCA) of deformation fields extracted from the registration of few real CTA scans. Furthermore, we run several experiments to evaluate the performance and generalization of the network when gradually reducing the amount of input real CTA images used for synthetic image generation.
2. State-of-the-art
Segmentation of the AAA thrombus from CTA images is challenging since it appears as a non-contrasted structure with fuzzy borders, and its shape varies among patients, which makes it difficult to develop robust automatic segmentation approaches. Traditionally proposed methods combine intensity information with shape constraints to minimize a certain energy function using level-sets [9], graph-cuts [4,5,8] or deformable models [6,7]. However, these algorithms require user interaction and/or prior lumen segmentation along with centerline extraction and their performance highly depends on multiple parameter tuning, affecting their robustness and clinical applicability.
More recently, CNNs have gained attention in the scientific community for solving complex segmentation tasks, surpassing the previous state-of-the-art performance in many problems. CNN-based approaches have shown state-of-the-art results in infra-renal AAA segmentation from both pre-operative and post-operative CTA scans in a fully automatic manner and without the need of any additional prior segmentation from the scans [1,2]. The main limitation of these approaches is the need of large amounts of annotated datasets to improve the network generalization when having different types of aneurysms.
Obtaining annotated images to train CNNs is a time-consuming task and requires expert knowledge. Hence, researchers try to alleviate the burden by using data augmentation techniques. Applying rotation, translation, scaling, reflection and other random elastic deformations is the most common approach to increase the number of annotated images [10]. Realistic deformations to increase the image database have also been proposed [3,11], as well as generative adversarial networks [12]. In this work, we leverage realistic elastic deformations to generate a synthetic shape model in order to train a CNN only with data created with this model, and to test its ability to segment aneurysms from new, real CTA scans. The proposed approach could be used in the future to extend the segmentation to aneurysms at other sites of the aorta, such as thoracic aneurysms, or cerebral aneurysms without the need of large annotated databases.
3. Methods
Hereby, we aim at evaluating the generalization ability of a 3D CNN trained on data generated with a synthetic shape model to segment infra-renal AAAs from real CTA scans. The followed procedure consists in 3 main steps: synthetic image generation, CNN training with synthetic data, and validation using real data. Synthetic images are created using a method based on the principal component analysis (PCA) of deformation fields extracted from the registration of real CTA scans to a reference real CTA scan, as explained in Sect.3.2. In a second step, we train the 3D CNN designed in [2] with the generated synthetic data. Finally, we evaluate the performance of the network to segment AAAs from real CTA scans and compare the results with the baseline model presented in [2] for the same testing datasets. This baseline network was trained with more than double real input scans, plus rotations and translations.
3.1. Abdominal Aortic Aneurysm Datasets
The employed CTA volumes have been provided by Donostia University Hospital. They have been obtained with scanners of different manufacturers with an in-plane spatial resolution in the range of 0.725 mm to 0.977 mm in x and y, and a slice thickness of 0.625 mm to 1 mm in the z direction. We work with 28 postoperative CTA datasets of patients with an infra-renal AAA and that have been treated with EVAR. We use a maximum of 12 CTAs to generate synthetic images, and the remaining 16 are saved for testing the CNN model. Ground truth annotations have been obtained semi-automatically using the segmentation method developed in [1] and manually correcting the results.
3.2. Synthetic Shape Model Generation Using Realistic Deformations
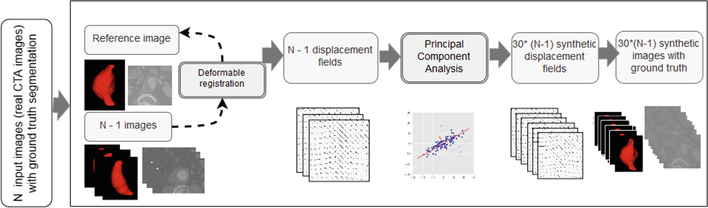
Elastic deformation-based data augmentation has been widely applied in medical imaging to increase the amount of input data to train a network. In [3], a novel approach to obtain synthetic images from realistic deformations was proposed to aid the segmentation of the pulmonary artery from CT scans. We aim at applying this method to generate synthetic AAA CTA scans, following the procedure shown in Fig. 1. The next steps are performed:
Register real AAA ground truths to a reference mask using the BSpline registration method implemented in 3D Slicer and extract the 3D deformation fields corresponding only to the elastic transform. The registration is done using the ground truth aneurysm masks.
Run the PCA from the fields and extract the eigenvectors and eigenvalues. The PCA components, together with the mean deformation extracted from the input fields, characterize the main AAA deformation directions.
Generate new deformation fields by weighting the eigenvectors with random values between 0 to ± the square root of the corresponding eigenvalue.
-
Generate synthetic volumes by applying these deformation fields to each original CTA volume, following Eq. 1.
(1) where is the new synthetic image, n is the number of eigenvectors, wi are the weights generated from the eigenvalues, Bi are the eigenvectors, and μ is the mean deformation extracted from the original deformation fields.
Fig. 1.
Synthetic image generation using realistic deformations.
With this procedure, we create 6 different datasets to run 6 experiments, consisting in training the CNN using a different amount of input real CTA and reference scans. Table 1 summarizes the number of images used in each experiment. The amount of applied translations and rotations are selected to approximate the total number of images used to train the baseline model in [2]. From experiment 1 to 3, we use a single CTA scan as reference and gradually reduce the amount of additional scans to register and create the shape model. In experiments 4 to 6, 2 reference scans are used. Figure 2 shows some created synthetic images and segmentations.
Table 1.
Summary of the employed data used to train the network in each experiment.
| Experiment | 1 | 2 | 3 | 4 | 5 | 6 |
|---|---|---|---|---|---|---|
| Total number of scans | 12 | 8 | 4 | 12 | 8 | 4 |
| Number of scans used as reference | 1 | 1 | 1 | 2 | 2 | 2 |
| Number of scans registered to each reference | 11 | 7 | 3 | 11 | 7 | 3 |
| Number of synthetic images | 330 | 210 | 90 | 660 | 420 | 180 |
| Applied translations | 2 | 4 | 9 | 2 | 2 | 4 |
| Applied rotations | 3 | 4 | 9 | 3 | 2 | 5 |
| Total number of images to train the network | 1650 | 1680 | 1620 | 1650 | 1680 | 1620 |
Fig. 2.
Examples of generated synthetic images.
3.3. 3D Convolutional Neural Network for AAA Segmentation
Once the images generated with the synthetic shape model are created, we proceed to train the 3D CNN proposed in [2], specifically designed to segment AAAs from CTA scans. The net combines fine edge detection with global information about the shape and location of the aneurysm thanks to the fusion of features maps at different scales. Figure 3 shows the architecture of the CNN, for which more details can be found in [2].
Fig. 3.
Proposed 3D convolutional neural network for aneurysm segmentation.
The network is trained using the aforementioned datasets, and to reduce the impact of the amount of images used to train the network in each experiment and to better evaluate the influence of the amount of input real CTA scans and number of reference scans, we apply a different number of rotations and translations in each case. Table 1 shows a summary of the total number of images employed to train the network, and how they have been created.
We set the initial learning rate to 1e-4, a batch size of 2 and Adam optimization are employed. A weighted Dice loss function is minimized [2]. The network is trained on a TITAN X Pascal (NVIDIA) GPU card with 11.91GB memory.
4. Results
The created CNN models are tested with the same 16 CTA scans used during the inference step in the baseline model from [2], which allows to study the generalization of the network when trained on synthetic data as compared to the network trained on real CTA scans. Furthermore, this baseline model is created from 28 real CTA scans, whereas our proposed approach requires a maximum of 12 annotated scans to generate the shape model. The total number of images after augmentation used in our experiments and to train the model in [2] is very similar, to minimize the impact of the amount of images used for training.
We compute the Dice and Jaccard scores between the ground truth annotations and the automatic segmentations as in [2]. Table 2 shows the obtained results for each of the experiments and the baseline values presented in [2].
Table 2.
Mean Dice and Jaccard scores for the testing datasets, for the networks trained on synthetic data and the baseline model trained on real CTA scans
| Baseline [2] | Exp 1 | Exp 2 | Exp 3 | Exp 4 | Exp 5 | Exp 6 | |
|---|---|---|---|---|---|---|---|
| Dice | 0.88 ± 0.05 | 0.84 ± 0.01 | 0.75 ± 0.05 | 0.57 ± 0.08 | 0.85 ± 0.01 | 0.76 ± 0.06 | 0.71 ± 0.06 |
| Jaccard | 0.78 ± 0.08 | 0.73 ± 0.01 | 0.64 ± 0.05 | 0.44 ± 0.06 | 0.75 ± 0.01 | 0.65 ± 0.05 | 0.59 ± 0.05 |
The obtained Dice and Jaccard scores for the testing datasets when training the network with synthetic data created from 12 original CTA scans (experiment 1 and 4) are close to the values achieved in [2], in which the network was trained with 28 real CTA scans. This concludes that our proposed synthetic image generation approach allows to create realistic images that can be used to effectively train a CNN with a reduced amount of input annotated data.
Furthermore, when comparing the performance of the network when using more datasets as reference (experiments 4,5 and 6), an improvement in the Dice and Jaccard scores is achieved. This improvement is more notable when comparing experiment 3 and experiment 6, in which only 4 datasets are used to train the network. For example, if a t-test between the Jaccard coefficients of experiments 1 and 4 is computed, a p-value of 3.65e-6 is achieved, meaning that the difference is significant. Having more reference datasets to extract deformation fields and create the synthetic shape model allows to account for a larger variability of the input data, which improves the generalization of the CNN. This result suggests that even having less input data, using the proposed technique with multiple reference scans can help creating a representative image database to train an AAA segmentation network that can be used to segment real CTA scans.
Finally, to further evaluate the value of the proposed augmentation method, we run a test in which we compare the performance of the network when trained (1) only with the original 12 CTA scans, (2) only applying augmentation via rotations and translations, and (3) including the synthetically generated images from experiment 1. Table 3 shows the results, which indeed confirm that using the images generated with our approach boosts the performance of the CNN.
Table 3.
Influence of adding the synthetic images in the performance of the network. T: translations, R: rotations.
| Training data | (I) 12 real CTA | (II) I + 2 T + 3 R | (III) II + 330 synthetic images |
|---|---|---|---|
| Dice | 0.22 ± 0.07 | 0.42 ± 0.02 | 0.84 ± 0.01 |
| Jaccard | 0.13 ± 0.00 | 0.27 ± 0.01 | 0.73 ± 0.01 |
5. Conclusions
This work aimed at evaluating the ability of a CNN trained only with images generated with a synthetic shape model to segment infra-renal aneurysms from real CTA scans. Having large annotated image databases to train a network is one of the main limitations of deep learning-based approaches, and thus, applying a proper methodology to generate synthetic images that account for a large variability of the input data is essential. Obtaining these images with the proposed synthetic shape model allows to generate plausible, realistic scans, controlling the final shape of the generated AAA scan, contrarily to using a fully random elastic deformation.
Our experiments have shown that the described synthetic image generation approach allows to create a wide variety of CTA images to train a CNN. Using less than a half of the original CTA scans employed in [2], the 3D CNN trained with synthetic data is able to accurately segment real aneurysm with comparable Dice and Jaccard coefficients as when the network is trained with real scans. Additionally, we have proved that using more reference datasets to extract deformation fields can further improve the segmentation results.
Hence, the obtained results suggest that the proposed methodology may be applied to generate images and train a CNN to segment other arterial or cerebral aneurysms, without the need of large annotated image databases. This is the aim of our future work, as well as including other synthetic image generation methods, such as generative adversarial networks.
References
- 1.López-Linares K, et al. : Fully automatic detection and segmentation of abdominal aortic thrombus in post-operative CTA images using deep convolutional neural networks. Med. Image Anal 46, 202–214 (2018) [DOI] [PubMed] [Google Scholar]
- 2.López-Linares K, García I, García-Familiar A, Macía I, González Ballester MA: 3D convolutional neural network for abdominal aortic aneurysm segmentation. arXiv preprint arXiv:1903.00879 (2019) [Google Scholar]
- 3.López-Linares K, et al. : 3D pulmonary artery segmentation from CTA scans usingdeep learning with realistic data augmentation. In: Image Analysis for Moving Organ, Breast, and Thoracic Images, pp. 225–237 (2018) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Duquette AA, Jodoin PM, Bouchot O, Lalande A: 3D segmentation of abdominal aorta from CT-scan and MR images. Comput. Med. Imaging Graph 36(4), 294–303 (2012) [DOI] [PubMed] [Google Scholar]
- 5.Freiman M, Esses SJ, Joskowicz L, Sosna J: An iterative model-constrained graph-cut algorithm for abdominal aortic aneurysm thrombus segmentation. In: IEEE International Symposium on Biomedical Imaging: From Nano to Macro, pp. 672–675(2010) [Google Scholar]
- 6.Demirci S, Lejeune G, Navab N: Hybrid deformable model for aneurysm segmentation. In: IEEE International Symposium on Biomedical Imaging: From Nano to Macro, pp. 33–36 (2009) [Google Scholar]
- 7.Lalys F, Yan V, Kaladji A, Lucas A, Esneault S: Generic thrombus segmentation from pre and postoperative CTA. Int. J. Comput. Assist. Radiol. Surg 12(9), 1–10 (2017) [DOI] [PubMed] [Google Scholar]
- 8.Siriapisith T, Kusakunniran W, Haddawy P: Outer wall segmentation of abdominal aortic aneurysm by variable neighborhood search through intensity and gradient spaces. J. Digital Imaging 31(4), 490–504 (2018) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Zohios C, Kossioris G, Papaharilaou Y: Geometrical methods for level set based abdominal aortic aneurysm thrombus and outer wall 2D image segmentation. Comput. Methods. Program. Biomed 107(2), 202–217 (2012) [DOI] [PubMed] [Google Scholar]
- 10.Çiçek Ö, Abdulkadir A, Lienkamp SS, Brox T, Ronneberger O: 3D U-Net: Learning Dense Volumetric Segmentation from Sparse Annotation. In: Ourselin S, Joskowicz L, Sabuncu MR, Unal G, Wells W (eds.) MICCAI 2016. LNCS, vol. 9901, pp. 424–432. Springer, Cham: (2016). 10.1007/978-3-319-46723-8_49 [DOI] [Google Scholar]
- 11.Roth HR, et al. : DeepOrgan: multi-level deep convolutional networks for automated pancreas segmentation. In: Navab N, Hornegger J, Wells WM, Frangi AF (eds.) MICCAI 2015. LNCS, vol. 9349, pp. 556–564. Springer, Cham: (2015). 10.1007/978-3-319-24553-9_68 [DOI] [Google Scholar]
- 12.Kazeminia S, et al. : GANs for medical image analysis. arXiv preprint arXiv:1809.06222 (2018) [Google Scholar]