ABSTRACT
The four-component meningococcal serogroup B vaccine (4CMenB) contains antigens present in the majority of meningococci causing invasive meningococcal disease (IMD) and may potentially offer protection against strains belonging to non-B serogroups.
This study aimed to evaluate the ability of 4CMenB-induced antibodies to kill, in a human serum bactericidal assay (hSBA), non-B meningococci belonging to the main genotypes responsible for IMD in Italy.
Meningococci, collected between 2015 and 2017, was characterized for PorA, FetA and sequence type, and for clonal complex. Twenty non-B isolates, representative of the most frequent genotypes, were molecularly characterized for 4CMenB antigens and tested in hSBA with sera from 4CMenB-vaccinated infants and adolescents.
Among twenty isolates, eleven were serogroup C, five were Y, two W and two X. All isolates contained genes encoding for fHbp and NHBA antigens and four harbored the NadA full-length encoding gene. Positive hSBA titers were obtained against all serogroup W, X and Y isolates and against five serogroup C isolates.
These data show that the 4CMenB vaccine can induce bactericidal antibodies against genetically representative meningococcal W, Y and X strains from Italy. For serogroup C, different susceptibilities to killing were observed for strains with similar antigenic repertoires.
KEYWORDS: Neisseria meningitides, 4CMeNB vaccine, cross-reactivity, non-B serogroups, serum bactericidal assay, Italy
Neisseria meningitidis serogroups A, B, C, W, Y and X account for the majority of invasive meningococcal disease (IMD) worldwide.1 In Italy, as in other European countries,2 serogroups B and C are the most frequent.3 During recent years they predominated alternatingly: serogroup C in 2015–2016, accounting for 43% of cases, and serogroup B in 2017–2018 with the same percentage. Since the year 2000, serogroups W and Y have shown a slow but steady increase, following the epidemiological changes in these serogroups worldwide,2 resulting in 9% and 18% of cases in 2018, respectively. Serogroups A and X are rare, with a total of 18 and 7 cases, respectively, from 2000 to 2019 (http://old.iss.it/mabi/, last access: 24 September 2020).
The Italian National Vaccination Plan 2017–20194 recommends: i) the meningococcal conjugate serogroup C vaccine during the second year of life; ii) the meningococcal quadrivalent conjugate vaccine for serogroups A, C, W, Y (MenACWY) from 12 to 18 years of age; and iii) the four-component meningococcal serogroup B vaccine (4CMenB or Bexsero). The latter is licensed for vaccination starting from 2 months of age and offered to infants aged 3 and 5 months, with a booster at 12 months. Moreover, a two-component serogroup B vaccine (Trumenba),5 based on two factor H-binding proteins (fHbp), has also been licensed in Italy for use from 10 years of age.6
The 4CMenB vaccine contains four antigens. Three are recombinant proteins, namely fHbp, Neisserial heparin binding antigen (NHBA) and Neisseria adhesin A (NadA). All three show a high level of conservation in the majority of IMD-causing meningococci. The fourth antigen is PorA, as the major component of the outer membrane vesicles (OMV).7 Since the antigens present in the 4CMenB vaccine are also harbored by meningococci belonging to other serogroups, this vaccine could potentially offer a certain level of protection against non-B serogroup strains. This has been demonstrated in a study conducted on N. meningitides W strains isolated in the UK, in which sera from 4CMenB-vaccinated infants and adolescents were able to induce complement bactericidal killing of all MenW strains tested, and in a similar study conducted on serogroup X strains, in which all African serogroup X isolates were killed by 4CMenB antisera.8,9 Moreover, sera from infants and adolescents immunized with 4CMenB were shown to exhibit bactericidal activity against a large panel of MenC, MenW and MenY clinical isolates from France, Germany, the UK and Brazil.10
The present study aimed to evaluate the ability of 4CMenB to induce antibodies in humans with bactericidal activity against a representative panel of non-B meningococcal strains responsible for IMD in Italy during the epidemiological years 2015–2017.
All meningococci sent to the National Reference Laboratory (NRL) of Istituto Superiore di Sanità (ISS), within the framework of the IMD National Surveillance System (NSS), were characterized for serogroup by slide agglutination with commercial antisera (Thermo Scientific, Waltham, Massachusetts, US) or by multiplex PCR.11 Chromosomal DNA was extracted using the QiAmp mini kit (Qiagen, Hilden, Germany) from an overnight culture. Whole genome sequencing (WGS) was performed using the Illumina MiSeq platform on each non-B isolate as previously described.12 Based on genome sequence, the clonal complex (cc), sequence type (ST), PorA-VR1 and VR2 type, FetA type and MenB vaccine antigen variants (fHbp, NHBA, NadA) were defined using the PubMLST.org database (http://pubmlst.org/neisseria/).
Pooled sera derived from infants before vaccination (n = 181, NCT00657709) or infants who received a primary series of 4CMenB at 2, 4 and 6 months of age plus a booster at 12 months of age (n = 94, NCT00847145), pooled sera derived from adolescents before or after two doses of 4CMenB administered 2 months apart (n = 39, NCT00661713) and pooled sera derived from adolescents before or after one dose of MenACWY vaccine (Menveo) (n = 24, NCT01210885) were used for the study. The use of pooled sera was due to constraints in the amount of sera available and based on previous study by Budroni et al. in which correlation between pooled serum bactericidal titers and seroresponse rate was demonstrated even at low bactericidal titer.13 hSBA assays were performed as described by Borrow et al. with minor modifications.14 Bacteria were subcultured overnight on Chocolate Agar, resuspended in Mueller Hinton Medium to an optical density (OD600) of 0.05 and grown until OD600 of 0.25 before use in the assay. Bacteria were incubated with sera and 25% of human plasma for 1 hour, and plated overnight before colony counting. hSBA titers were determined as the highest dilution that resulted in at least a 50% reduction in colony-forming units (CFU) relative to the number of CFU present in the reaction incubated without serum. No decrease in CFU was observed in the presence of heat-inactivated complement or in the presence of active complement without serum, indicating that the decrease in colony numbers observed in the presence of serum, even at a low dilution (1:2), was attributable to the presence of antibodies able to mediate complement-dependent killing. Human plasma, obtained from volunteer donors under informed consent, was selected for use as complement source with a given strain only if it did not significantly reduce CFU counts compared to T0 when added to the assay at a final concentration of 50%.
For adolescent sera, the highest serum dilution tested was 1:128 while for infant sera, the highest dilution corresponded to 1:64. Higher dilutions were not tested when a difference in titers of at least fourfold between the pre- and post-vaccination sera was achieved.
During the study period (epidemiological years 2015–2017) in Italy, 425 IMD cases were reported at NSS, with an average incidence of 0.34 cases/100,000 inhabitants. The capsular serogroup of meningococci was identified in 360 IMD cases: 143 were C (MenC), 133 were B (MenB), 58 were Y (MenY), 21 were W (MenW) and 5 were X (MenX). The proportion of serogroup B vs non-B meningococci was 37% vs 63%. The numbers of non-B vs serogroup B IMD cases in each age group are shown in Figure 1. Serogroup B prevailed among infants <1 year old, non-B serogroup cases were the majority by the age of 5. The median age of non-B cases was 29 years. Meningitis plus septicemia were the most frequent clinical picture (36%, n = 82/227); the case fatality rate was 17% (n = 29/172) (http://old.iss.it/mabi/, last access: 24 September 2020). The total number of MenC, MenY, MenW and MenX isolates suitable for analysis by WGS was 97, 44, 16 and 5, respectively. The most frequent clonal complexes for MenC were cc11 (n = 86, 89%) and cc334 (n = 8, 8%); for MenY, the majority belonged to cc23 (n = 41, 93%); for MenW, the clonal complexes recovered were cc11 (n = 14, 88%) and cc22 (n = 2, 12%); for MenX, all belonged to cc181 (n = 5, 100%).
Figure 1.
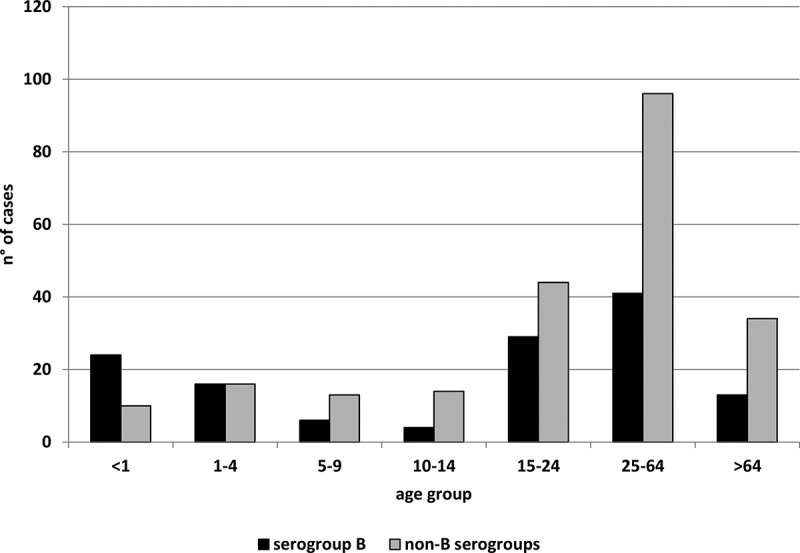
Number of invasive meningococcal diseases cases due to B and non-B serogroups by age group (years) in Italy, epidemiological years 2015–2017
A representative subsample of 20 meningococci was selected based on their genotypes (PorA-VR1 and VR2, FetA, ST, cc). As shown in Table 1, they were distributed as follows: eleven MenC (eight cc11 and three cc334), two MenW (one cc11 and one cc22), two MenX (cc181) and five MenY (all cc23).
Table 1.
Molecular characteristics of 20 isolates analyzed in this study: serogroup, genotype (cc, ST, porA-VR1, VR2, FetA) and 4CMenB antigen variants (fHbp, NHBA, NadA). hSBA titers for each isolate/pooled sera are shown. NCT01210885: Pooled sera derived from adolescents before and after one dose of MenACWY vaccine (n = 24); NCT00661713: Pooled sera derived from adolescents before and after two doses of 4CMenB vaccine administered 2 months apart (n = 39); NCT00657709: Pooled sera derived from infants before vaccination (n = 181); NCT00847145: Infants who received the primary series of 4CMenB plus the booster (n = 94)
|
NCT01210885 |
NCT00661713 |
NCT 00657709 |
NCT 00847145 |
||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Genotype |
4CMenB antigen variant |
MenACWY adolescent |
4CMenB adolescent |
4CMenB infants |
|||||||||||||
| ID | Serogroup | CC | ST | PorA VR1 | PorA VR2 | FetA | fHbp variant | NHBA peptide | NadA variant | NadA peptide | PI | post1 | PI | post2 | PI | post4 | |
| IT_C1 | C | cc11 | ST-11 | 5 | 2 | F3-3 | 2.22 | 29 | 2/3 | 121 | <4 | >512 | <4 | 4* | <2 | <2 | |
| IT_C2 | C | cc11 | ST-11 | 5 | 2 | F3-3 | 2.22 | 29 | 2/3 | 121 | <4 | >512 | <4 | 4 | <2 | <2 | |
| IT_C3 | C | cc11 | ST-11 | 5 | 2 | F3-3 | 2.22 | 29 | 2/3 | 121 | <4 | >512 | <4 | 4* | <2 | <2 | |
| IT_C4 | C | cc11 | ST-11 | 5–1 | 10–8 | F3-6 | 1.13 | 20 | nadA gene contains IS element | <4 | >512 | <4 | <4 | <2 | <2 | ||
| IT_C5 | C | cc11 | ST-11 | 5–1 | 10–8 | F3-6 | 1.808 | 20 | nadA gene contains IS element | <4 | >512 | <4 | <4 | <2 | <2 | ||
| IT_C6 | C | cc11 | ST-11 | 5–1 | 10–8 | F3-6 | 1.808 | 20 | nadA gene contains IS element | <4 | >512 | <4 | <4 | <2 | <2 | ||
| IT_C7 | C | cc334 | ST-1031 | 7–4 | 14–6 | F3-9 | 2.19 | 6 | Frameshift in the nadA gene | <4 | 512 | <4 | 16 | <2 | 2 | ||
| IT_C8 | C | cc334 | ST-1031 | 7–4 | 14–6 | F3-9 | 1.13 | 6 | Frameshift in the nadA gene | <4 | >512 | <4 | 32 | <2 | 4 | ||
| IT_C9 | C | cc334 | ST-1031 | 7–4 | 14–6 | F3-9 | 2.1297 | 6 | Frameshift in the nadA gene | <4 | 256 | <4 | <4 | <2 | <2 | ||
| IT_C10 | C | cc11 | ST-11760 | 5–1 | 10–8 | F3-6 | 2.23 | 20 | nadA gene contains IS element | <4 | 256 | <4 | <4 | <2 | <2 | ||
| IT_C11 | C | cc11 | ST-11760 | 5–1 | 10–8 | F3-6 | 1.462 | 20 | nadA gene contains IS element | <4 | 128 | <4 | <4 | <2 | <2 | ||
| IT_W1 | W | cc11 | ST-11 | 5 | 2 | F1-1 | 1.9 | 96 | 2/3 | 6 | <4 | 256 | <4 | >128 | <2 | >64 | |
| IT_W2 | W | cc22 | ST-184 | 18–1 | 3 | F4-1 | 2.16 | 20 | Frameshift in the nadA gene | <4 | 256 | <4 | >128 | 8 | >64 | ||
| IT_X1 | X | cc181 | ST-181 | 5–1 | 10–1 | F1-31 | 1.74 | 359 | nadA gene contains IS element | <4 | <16 | <4 | >128 | 2 | 64 | ||
| IT_X2 | X | cc181 | ST-181 | 5–1 | 10–1 | F1-31 | 1.74 | 359 | Frameshift in the nadA gene | <4 | <16 | <4 | >128 | <2 | >64 | ||
| IT_Y1 | Y | cc23 | ST-23 | 5–2 | 10–2 | F2-13 | 1.393 | 8 | Frameshift in the nadA gene | <4 | 256 | 32* | >128 | <2 | >64 | ||
| IT_Y2 | Y | cc23 | ST-23 | 5–2 | 10–2 | F2-13 | 2.104 | 8 | nadA gene contains IS element | <4 | 128 | <4 | >128 | 4 | >64 | ||
| IT_Y3 | Y | cc23 | ST-23 | 5–2 | 10–2 | F4-1 | 3.29 | 8 | nadA gene contains IS element | <4 | 128 | <4 | 64 | 2 | 64 | ||
| IT_Y4 | Y | cc23 | ST-1655 | 5–1 | 10–1 | F4-1 | 2.25 | 7 | nadA gene contains IS element | 16* | 512 | 16 | >128 | 4 | >64 | ||
| IT_Y5 | Y | cc23 | ST-23 | 5–2 | 10–1 | F4-1 | 2.25 | 7 | Frameshift in the nadA gene | <4 | 128 | <4 | >128 | 4 | >64 | ||
* bacteriostatic.
Eleven different genotype profiles were detected among the four serogroups as follows: four for MenC, two for MenW, one for MenX and four for MenY (Figure 2).
Figure 2.
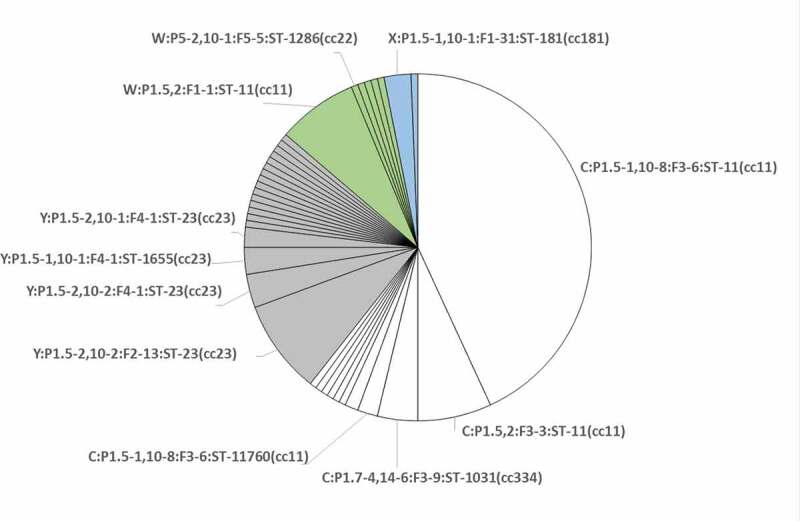
Distribution of genotypes identified within the 162 isolates characterized for this study. Each slice corresponds to a genotype. The 11 genotypes, characterizing the 20 isolates selected for this study, are shown. Serogroups are differentiated by color: white for MenC; gray for MenY; green for MenW; light blue for MenX
Fourteen fHbp peptides, of which six belonging to variant 1 (nine isolates, 45%), seven to variant 2 (ten isolates, 50%) and one to variant 3 (one isolate, 5%) were identified. The most frequent fHbp peptides were variant 2.22 (three isolates), variant 2.25 (two isolates), variant 1.74 (two isolates), variant 1.13 (two isolates) and variant 1.808 (two isolates).
Seven different NHBA peptides were identified, of which peptide 20 was the most frequent (six isolates). Regarding NadA, three isolates carried the gene encoding for variant 2/3, peptide 121, and one isolate for variant 2/3, peptide 6 (Table 1). The other isolates had mutations, such as frameshift mutations or IS insertions, that are known to abolish NadA expression. Finally, six different PorA-VR2 types were identified among the twenty isolates, of which the most frequent were 10–8 (five isolates) and 10–1 (four isolates).
No isolates expressed any of the 4CMenB vaccine antigenic variants (variant 1 fHbp peptide 1, NHBA peptide 2, NadA peptide 3.8, PorA-VR2 4).
For fHbp, the comparison between the vaccine antigen variant (peptide 1) and the variants found in the isolates highlighted two groups: peptides 9, 13, 462, 74, 808, 393 had a percentage of identity with peptide 1 greater than 90%; peptides 16, 1297, 19, 23, 22, 104, 25, 29 had identities ranging from 61.02% to 72.83%. For NHBA, all the variants presented a similarity with the vaccine antigen variant 2 higher than 80%; in particular peptides 6, 7, 8, 359 had an identity of 88.17% to 88.26%; peptides 20, 29, 96 had identities of 82.02% to 83.76%. For NadA, the two peptides identified, 6 and 121, showed an identity with the vaccine variant 3.8 of 99.01% and 98.74%, respectively.
The vaccine antigen variants were distributed diversely among different genotypes (Table 1). The NHBA peptides were identical in isolates belonging to the same genotype. The fHbp assortment was more variable, depending on the genotypes. The C:P1.5–1,10-8:F3-6:ST-11(cc11) isolates showed two different fHbp peptides: peptides 13 and 808 (variant 1), differing in seven amino acids. The MenC ST-1031(cc334) and ST-11760(cc11) isolates each expressed a different fHbp peptide. The C:P1.5,2:F3-3:ST-11(cc11) isolates shared the same combination of fHbp, NHBA and NadA peptides. The MenW cc11 and cc22 showed two different fHbp peptides. The X:P1.5–1,10-1:F1-31;ST-181(cc181) isolates expressed the same fHbp and NHBA peptides. Among the four MenY genotypes, four different fHbp and two different NHBA peptides were identified (Table 1).
All 20 isolates were tested in hSBA and results are summarized in Table 1 and Figure 3. MenW (n = 2; IT_W1-2), MenY (n = 5; IT_Y1-5) and MenX (n = 2; IT_X1-2) isolates were killed by both pooled adolescent and infant post-immune sera with bactericidal titer ≥ 4. Bactericidal titers were very high, all being ≥64 against all these strains. Killing by pre-immune adolescent and infant sera was observed against two (IT_Y1 and IT_Y4) and four strains (IT_W2, IT_Y2, IT_Y4 and IT_Y5), respectively. However, a fourfold rise in hSBA titers of post-immune sera vs pre-immune sera was measured against all strains.
Figure 3.
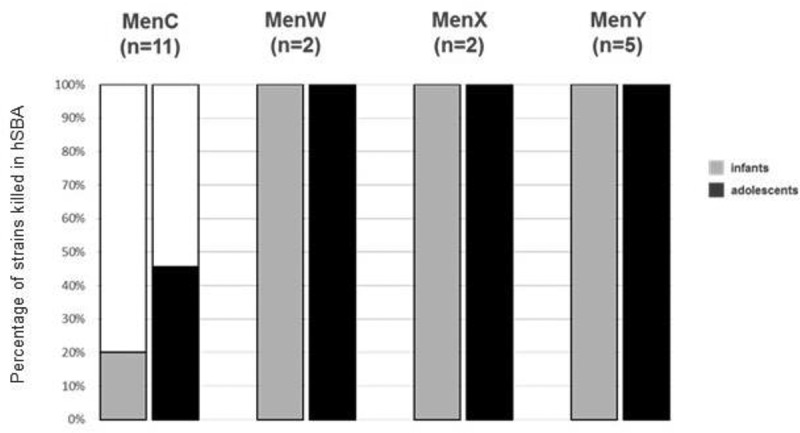
Percentage of non-MenB strains showing hSBA ≥2 with adolescent 4CMenB- (solid colored fill) and infant (dotted colored fill) 4CMenB immune sera
Among the eleven tested MenC strains, five strains were killed with hSBA titers ≥ 4 by adolescent sera and two by infant sera with hSBA titers ≥ 2. A fourfold increase in hSBA titers of post-immune sera vs pre-immune sera was observed against two strains when using adolescent sera (IT_C7 and IT_C8) and one strain when using infant sera (IT_C8), while killing by pre-immune sera was not observed against any of the strains. As expected, sera from MenACWY-vaccinated adolescents, used as positive control, were able to induce bactericidal killing of all tested MenC, MenW and MenY strains, with only one strain (IT_Y4) killed by pre-immune sera and with a fourfold increase in hSBA titers of post-immune sera vs pre-immune sera against all strains (Table 1).
The MenW, MenX and MenY isolates, killed in hSBA by 4CMenB post-immune sera, presented a wide variety of fHbp (seven different peptides, of which three variant 1 and four variant 2 or 3) and NHBA (five peptides: 7, 8, 20, 96, 359); they carried a truncated nadA gene, with the exception of the MenW/cc11 (IT_W1) isolate, harboring the NadA peptide 6, variant 2/3. The two MenC/cc334 isolates (IT_C7, IT_C8), showing a fourfold increase in hSBA titers of post-immune vs pre-immune sera, had two different fHbp peptides (variant 1.13 and variant 2.19), the same NHBA (peptide 6) and no NadA (Table 1).
Although the 4CMenB vaccine has been licensed for prevention of MenB disease, the most prevalent serogroup causing IMD in Europe and nowadays also in Italy,3 variants of the vaccine antigens are also found among non-B meningococci, independently of the capsule. Therefore, antibodies raised by these antigens may induce complement-mediated killing against other meningococcal serogroups.
During the study period, 63% of reported IMD isolates belonged to non-B serogroups, mostly from people of 5 years old and up. However, the annual incidence of non-B disease in infants (<1 year) is noteworthy, accounting for 1 per 100,000 inhabitants (http://old.iss.it/mabi/last access: 24 September 2020). A cross-reactivity by 4CMenB sera against non-B meningococci among infants may contribute to the reduction of IMD cases in this age group.
4CMenB conferred cross-reactivity against all MenW, MenY and MenX isolates analyzed in this study. In particular, the results obtained on two MenX/cc181, isolated from refugees who arrived in Italy from Bangladesh and Morocco,15 confirm the ability of 4CMenB-induced antibodies to kill the MenX strains isolated in Africa, as described by Hong et al.9 Since there is no vaccine available against MenX, the 4CMenB vaccine may represent a possible alternative. Moreover, considering that MenW, MenX and MenY IMD occur mostly among adolescents and young adults,12,15,16 vaccination with 4CMenB may also be valuable for these age groups.
The two MenW strains analyzed in the study had different genetic profiles, and were both killed by anti-4CMenB antibodies with bactericidal titers ≥ 1:64. Interestingly, the MenW/cc11 (IT_W1) isolate differed in 4CMenB antigen variants from all the MenW/UK isolates analyzed in a similar study.8 While for the strains in the UK study, bactericidal killing may have been mediated by anti-NHBA and NadA antibodies, for the IT_W1 strain in the present study, bactericidal killing may have been mediated by antibodies directed against the three antigens, NHBA, NadA and fHbp. For the strain IT_W2 belonging to cc22, killing may have been mediated by NHBA antibodies. This is consistent with the findings of an ongoing study on a collection of MenW isolates from Europe and Brazil.10
The same observation applies to MenY isolates, for which, with the exclusion of IT_Y1 harboring fHbp variant 1, only NHBA antibodies may have played a role in killing.
Considering that antibodies directed against OMV could also contribute to killing,16 we cannot exclude that the killing in hSBA might be the result of a synergistic activity of antibodies directed against the main antigens and antibodies directed against minor OMV antigens.
Overall, genotypic and serological data for MenX, MenW and MenY suggest that antibody killing activity is independent of the serogroup or clonal complex.
Antibody killing induced by 4CMenB against the 11 MenC strains was, instead, less evident compared to what was found for the other serogroups using both adolescent and infant sera.
Of note, although generally a titer of 4 is needed to consider a strain killed in hSBA, and a fourfold rise compared to baseline is considered to evaluate the effect of vaccination, in an experimental setting, a bacteriostatic titer as well as an hSBA titer of 2 or a twofold rise compared to baseline, may be indicative of functional activity probably corresponding to sub-bactericidal level of antibodies recognizing specific antigens.17,18
Interestingly, the three cc334 MenC strains showed different bactericidal results, despite their similar antigenic repertoire, probably due to the different synergistic effects of NHBA peptide 6 that, even if is not bactericidal on its own, could act in synergy with diverse fHbp subvariants or OMV minor components.19
The rationale to perform a bactericidal assay in this study was driven by the evidence that all the methods developed so far to predict coverage by 4CMenB (such as MATS,20 gMATS21 and BAST22) have been built on the basis of the correlation between antigenicity (measured by MATS as level of antigen expression and diversity) and susceptibility to bactericidal killing of a panel of meningococcal B strains. When the MATS analysis was extended to a high number of non-B strains, the correlation was not strong enough to define a positive bactericidal threshold as predictor of coverage. Therefore, MATS (as well as gMATS and BAST) should not a priori be applied to predict coverage of isolates belonging to other serogroups. Moreover, the currently available predictor tools (MATS, gMATS or BAST) do not take into account the power of contribution to killing of antibodies acting synergistically.
A limitation of the study could be the representativeness of isolates analyzed with respect to all meningococci isolated in the study period. The number of analyzed isolates for each serogroup represents a proportion of 11–12.5% of all isolates, except for MenX, because of the low total number of cases for this serogroup (n = 5). Within each serogroup, the criteria of selection were based on the clonal complex followed by the genotypic profile.
In conclusion, this study suggests that the 4CMenB vaccine is able to induce cross-reactivity against meningococci belonging to non-B serogroups and, in particular, against MenW, MenX and MenY, independently of their genotypes, but dependent on their antigenic repertoire. However, at least for MenC strains, a lower susceptibility to antibody killing was observed. Therefore, the ability of 4CMenB vaccine to induce cross-reactivity against MenC strains is more difficult to predict solely on the basis of the antigenic assortment.
Real-world evidence could provide further information on real effectiveness of 4CMenB against non-B serogroups.
Acknowledgments
The authors thank the following people for the collaboration in the National Surveillance System of Invasive Bacterial Diseases and for samples and clinical data: Milena Arghittu (Microbiology Unit, Fondazione IRCCS Ca’ Granda, Ospedale Maggiore Policlinico, Milan, Italy), Paolo Castiglia (Università di Sassari, Sassari, Italy), Laura Daprai (Microbiology and Virology Unit, Fondazione IRCCS Ca’ Grande Ospedale Maggiore, Milan, Italy), Antonino Di Caro (Microbiology section, Istituto Nazionale per le Malattie Infettive “L. Spallanzani”, Rome, Italy), Irene Alessandra Galanti (Microbiology Laboratory, Azienda USL Toscana sud est, Arezzo, Italy), Patrizia Innocenti (Microbiology and Virology Laboratory, Azienda Sanitaria dell’Alto Adige, Bolzano, Italy), Teresa Lopizzo (Microbiology and Virology Unit, Azienda Ospedaliera San Carlo, Potenza, Italy), Francesca Orecchioni (Clinical Analysis Laboratory, Section of Microbiology, Ospedali Riuniti di Ancona, Italy), Eleonora Riccobono (Microbiology and Virology Unit, Careggi University Hospital, Firenze, Italy), Gianmaria Rossolini (Microbiology and Virology Unit, Careggi University Hospital, Firenze, Italy), Teresa Spanu (Microbiology and Infectious Diseases Unit, Policlinico Gemelli, Rome, Italy), Carlo Tascini (First Division of Infectious Diseases, Cotugno Hospital, AORN dei Colli, Naples, Italy), Caterina Vocale (Unit of Clinical Microbiology, Regional Reference Centre for Microbiological Emergencies, St. Orsola Malpighi University Hospital, Bologna, Italy), Antonella Vulcano (Microbiology section, Istituto Nazionale per le Malattie Infettive “L. Spallanzani”, Rome, Italy). The authors thank Anna Anselmo, Andrea Ciammaruconi, Antonella Fortunato, Anna Maria Palozzi, Silvia Fillo and Florigio Lista (Molecular Biology Section, Army Medical and Veterinary Research Center, Rome, Italy) for the whole genome sequencing.
This publication made use of the Neisseria Multi Locus Sequence Typing website (https://pubmlst.org/neisseria/) sited at the University of Oxford (Jolley et al. Wellcome Open Res 2018, 3:124). The development of this site has been funded by the Wellcome Trust and European Union.
Funding Statement
Funding for this study was provided by GlaxoSmithKline Biologicals SA. GlaxoSmithKline Biologicals SA was provided the opportunity to review a preliminary version of this manuscript for factual accuracy, but the authors are solely responsible for final content and interpretation.
Author contributions
Marzia Monica Giuliani, Rita La Gaetana, Elena Mori, Mariagrazia Pizza, Laura Serino and Paola Stefanelli conceived the study. Alessia Biolchi and Cecilia Fazio provided insight on microbiological investigation, serological analysis and drafted the manuscript. Arianna Neri, Paola Vacca, Luigina Ambrosio, Annapina Palmieri carried out the laboratory analyses, contributed to the molecular analyses and provided insight into interpretation of results. Sara Tomei contributed to the serological analyses. All authors participated in the drafting and revision of this manuscript and gave their final approval of this version.
Disclosure of potential conflicts of interest
CF, AN, PV, LA, AP, PS do not have any conflicts of interest.
AB, ST, EM, RLG, MP, MMG, and LS are employees of the GSK group of companies. RLG, MMG, and LS hold shares in the GSK group of companies.
Trademark statement
Bexsero and Menveo are trademarks owned by or licensed to the GSK group of companies.
References
- 1.Caugant DA, Brynildsrud OB. Neisseria meningitidis: using genomics to understand diversity, evolution and pathogenesis. Nat Rev Microbiol. 2020. February;18(2):84–96. doi: 10.1038/s41579-019-0282-6. Epub 2019 Nov 8. [DOI] [PubMed] [Google Scholar]
- 2.European Centre for Disease prevention and Control (ECDC) . Annual epidemiological report for 2017. Invasive meningococcal disease. Stockholm (Sweden): ECDC; 2019. “https://www.ecdc.europa.eu/sites/default/files/documents/AER_for_2017-invasive-meningococcal-disease.pdf” [accessed 2020 Sept 24]. [Google Scholar]
- 3.Italian National Health Institute (Istituto Superiore di Sanità, ISS) . [Surveillance data on invasive bacterial diseases updated on 23 April 2019]. Italian. http://old.iss.it/binary/mabi/cont/Interim_Report_2018_finale.pdf [accessed 2020 Sept 24].
- 4.Piano Nazionale Prevenzione Vaccinale PNPV 2017-2019 [National plan for vaccine prevention NPVP 2017-2019]. Rome (Italy): Ministero della Salute; 2017. Italian. http://www.salute.gov.it/imgs/C_17_pubblicazioni_2571_allegato.pdf [Google Scholar]
- 5.Gandhi A, Balmer P, York LJ.. Characteristics of a new meningococcal serogroup B vaccine, bivalent rLP2086 (MenB-FHbp; Trumenba®). Postgrad Med. 2016;128(6):548–56. doi: 10.1080/00325481.2016.1203238. [DOI] [PubMed] [Google Scholar]
- 6.Trumenba, INN-meningococcal group B vaccine - European medicines agency. https://www.ema.europa.eu/en/documents/product-information/trumenba-epar-product-information_it.pdf [accessed 2020 Sept 24].
- 7.Serruto D, Bottomley MJ, Ram S, Giuliani MM, Rappuoli R. The new multicomponent vaccine against meningococcal serogroup B, 4CMenB: immunological, functional and structural characterization of the antigens. Vaccine. 2012;30(Suppl 2):B87–97. doi: 10.1016/j.vaccine.2012.01.033.9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Ladhani SN, Giuliani MM, Biolchi A, Pizza M, Beebeejaun K, Lucidarme J, Findlow J, Ramsay ME, Borrow R. Effectiveness of meningococcal B vaccine against endemic hypervirulent Neisseria meningitidis W Strain, England. Emerg Infect Dis. 2016. February;22(2):309–11. doi: 10.3201/eid2202.150369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Hong E, Giuliani MM, Deghmane AE, Comanducci M, Brunelli B, Dull P, Pizza M, Taha MK. Could the multicomponent meningococcal serogroup B vaccine (4CMenB) control Neisseria meningitidis capsular group X outbreaks in Africa? Vaccine. 2013. February 4;31(7):1113–16. doi: 10.1016/j.vaccine.2012.12.022. Epub 2012 Dec 20. [DOI] [PubMed] [Google Scholar]
- 10.Pizza M. 4CMenB, a multicomponent meningococcal vaccine developed for serogroup B meningococci, elicits cross-reactive immunity against serogroups C, W and Y. 15th EMGM congress; 2019. May 27-30; Lisbon, Portugal. [Google Scholar]
- 11.Zhu H, Wang Q, Wen L, Xu J, Shao Z, Chen M, Chen M, Reeves PR, Cao B, Wang L. Development of a multiplex PCR assay for detection and genogrouping of Neisseria meningitidis. J Clin Microbiol. 2012;50(1):46–51. doi: 10.1128/JCM.00918-11. PMID: 22090406 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Fazio C, Neri A, Vacca P, Ciammaruconi A, Arghittu M, Barbui AM, Vocale C, Bernaschi P, Isola P, Galanti IA, et al. Cocirculation of Hajj and non-Hajj strains among serogroup W meningococci in Italy, 2000 to 2016. Euro Surveill. 2019. January;24:4. doi: 10.2807/1560-7917.ES.2019.24.4.1800183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Budroni S, Kleinschmidt A, Boucher P, Medini D. Pooled-sera hSBA titres predict individual seroprotection in infants and toddlers vaccinated with 4CMenB. Vaccine. 2016. May 17;34(23):2579–84. doi: 10.1016/j.vaccine.2016.04.009. [DOI] [PubMed] [Google Scholar]
- 14.Borrow R, Aaberge IS, Santos GF, Eudey TL, Oster P, Glennie A, Findlow J, Høiby EA, Rosenqvist E, Balmer P, et al. Interlaboratory standardization of the measurement of serum bactericidal activity by using human complement against meningococcal serogroup b, strain 44/76-SL, before and after vaccination with the Norwegian MenBvac outer membrane vesicle vaccine. Clin Diagn Lab Immunol. 2005;12:970–76. doi: 10.1128/CDLI.12.8.970-976.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Stefanelli P, Neri A, Vacca P, Picicco D, Daprai L, Mainardi G, Rossolini GM, Bartoloni A, Anselmo A, Ciammaruconi A et al. Meningococci of serogroup X clonal complex 181 in refugee camps, Italy. Emerg Infect Dis. 2017. May;23(5):870–72. doi: 10.3201/eid2305.161713. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Fazio C, Neri A, Renna G, Vacca P, Antonetti R, Barbui AM, Daprai L, Lanzafame P, Rossi L, Santino I, et al. Persistent occurrence of serogroup Y/sequence type (ST)-23 complex invasive meningococcal disease among patients aged five to 14 years, Italy, 2007 to 2013. Euro Surveill. 2015;20:45. doi: 10.2807/1560-7917.ES.2015.20.45.30061. [DOI] [PubMed] [Google Scholar]
- 17.Goldschneider I, Gotschlich EC, Artenstein MS. Human immunity to the meningococcus. I. The role of humoral antibodies. J Exp Med. 1969. June 1;129(6):1307–26. doi: 10.1084/jem.129.6.1307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Welsch JA, Granoff D. Immunity to Neisseria meningitidis group B in adults despite lack of serum bactericidal antibody. Clin Vaccine Immunol. 2007. December;14(12):1596–602. doi: 10.1128/CVI.00341-07. Epub 2007 Oct 3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Giuliani MM, Biolchi A, Serruto D, Ferlicca F, Vienken K, Oster P, Rappuoli R, Pizza MG, Donnelly J. Measuring antigen-specific bactericidal responses to a multicomponent vaccine against serogroup B meningococcus. Vaccine. 2010;28:5023–30. [DOI] [PubMed] [Google Scholar]
- 20.Donnelly J, Medini D, Boccadifuoco G, Biolchi A, Ward J, Frasch MR, Stella M, Comanducci M, Bambini S, Muzzi A, et al. Qualitative and quantitative assessment of meningococcal antigens to evaluate the potential strain coverage of protein-based vaccines. PNAS. 2010. November;107(45):19491–95. doi: 10.1073/pnas.1013758107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Muzzi A, Brozzi A, Serino L, Bodini M, Abad R, Caugant D, Comanducci M, Lemos AP, Gorla MC, Křižová, et al. Genetic meningococcal antigen typing system (gMATS): A genotyping tool that predicts 4CMenB strain coverage worldwide. Vaccine. 2019. February 8;37(7):991–1000. doi: 10.1016/j.vaccine.2018.12.061. Epub 2019 Jan 17. [DOI] [PubMed] [Google Scholar]
- 22.Brehony C, Rodrigues CMC, Borrow R, Smith A, Cunney RE, Moxon R, Maiden MCJ. Distribution of bexsero antigen sequence types (BASTs) in invasive meningococcal disease isolates: implications for immunisation. Vaccine. 2016;34:4690–97. [DOI] [PMC free article] [PubMed] [Google Scholar]


