SUMMARY
We report here cryo-electron microscopy reconstructions of Type IV pili (T4P) from two important human pathogens, Pseudomonas aeruginosa and Neisseria gonorrhoeae, at ~ 8 Å and 5 Å resolution, respectively. The two structures reveal distinct arrangements of the pilin globular domains on the pilus surfaces, which impart different helical parameters, but similar packing of the conserved N-terminal α-helices, α1, in the filament core. In contrast to the continuous α-helix seen in the x-ray crystal structures of the P. aeruginosa and N. gonorrhoeae pilins, α1 in the pilus filaments has a melted segment located between conserved helix-breaking residues Gly14 and Pro22, as seen for the Neisseria meningitidis T4P. Using mutagenesis we show that Pro22 is critical for pilus assembly, as are Thr2 and Glu5, which are positioned to hydrogen bond in the hydrophobic filament core. These structures provide a framework for understanding T4P assembly, function and biophysical properties.
Keywords: Type IV pili, cryoelectron microscopy, bacterial pathogenesis, membrane protein, macromolecular machine, IHRSR
eTOC blurb
Wang et al. report cryo-EM reconstructions of Type IV pili from P. aeruginosa and N. gonorrhoeae. These structures reveal that a melted portion of the pilin α-helical N-terminus is a common feature of Type IVa pili and is necessary for packing into the pilus filament.
INTRODUCTION
The Type IV pili of pathogens such as Pseudomonas aeruginosa and Neisseria gonorrhoeae have received intense scrutiny due to their critical roles in pathogenesis. The P. aeruginosa T4P mediate twitching motility and adhesion, allowing this opportunistic microbe to colonize the lungs of cystic fibrosis patients, causing potentially fatal pneumonia (Anyan et al., 2014; Bradley, 1980; Burrows, 2012; Engel, 2003; Leighton et al., 2015; Mattick, 2002; Woods et al., 1980). N. gonorrhoeae T4P are required for bacterial aggregation, twitching motility, natural transformation, adhesion and host cell signaling (Merz and So, 2000; Naumann et al., 1999). Antigenic variation of the pilin subunits contributes to immune escape for this invasive sexually transmitted pathogen, which is responsible for the sexually transmitted disease gonorrhea, with complications that include ectopic pregnancies, pelvic inflammatory disease and infertility. No vaccines are available for either P. aeruginosa or N. gonorrhoeae and both organisms exhibit resistance to multiple antibiotics, which severely limits treatment options. An atomic level understanding of the critical surface exposed T4P will contribute to the design of both vaccines and therapeutics for these important human pathogens.
T4P are polymers comprised primarily of a single major pilin subunit. The T4P of P. aeruginosa (Pa) and N. gonorrhoeae (Ng) are classified as Type IVa pili based on the amino acid sequences of the pilin subunits, which are generally smaller, with shorter signal peptides and more complex assembly machineries compared to those of the Type IVb pili. Crystal structures have been determined for the full length pilin subunits PilA from P. aeruginosa strain K (PaK) (Craig et al., 2003) and PilE from N. gonorrhoeae (Craig et al., 2006; Parge et al., 1995) revealing a canonical pilin fold, with an extended curving N-terminal α-helix, α1, the C-terminal half of which is packed against a 4-stranded β-sheet, forming the globular domain. A disulfide bond connects the β-sheet strand β4 to the most C-terminal loop and delineates the D-region of the pilin. The curvature of α1 seen in crystals is due to three helix-breaking residues: a glycine at position 14, a proline at 22 and glycine at 42. The N-terminal 25 residues are highly conserved in the Type IVa pilins. This α-helical segment is hydrophobic with the exception of a threonine or serine at position 2 and glutamic acid at position 5, and anchors the pilin subunits in the bacterial inner membrane prior to filament assembly. PaK PilA and Ng PilE differ primarily in the structures of the D-region and the αβ-loop, which connects α1 to the globular domain β-sheet. The αβ-loop lies on one side of the β-sheet is an irregular loop in both pilins but is bulkier in PilA, with a small β-sheet subdomain inserted near its N-terminus. In both pilins the D-regions are irregular loops that flank the β-sheet on the side opposite the αβ-loop, but Ng PilE has a β-hairpin insert between strand β4 and this loop, that sits atop the β-sheet. This β-hairpin contains the hypervariable region of Ng PilE (residues 127–142). Thus, despite the canonical pilin fold for both proteins, these two insertions, the β-sheet subdomain in the αβ-loop of PaK PilA and the hypervariable β-hairpin in the D-region of Ng PilE, result in substantially different shapes for the two globular domains.
The structure of Ng T4P has been solved by cryo-electron microscopy (cryo-EM) at 12.5 Å resolution (Craig et al., 2006). Ng T4P has a right-handed 1-start [(+)1] helix with a pitch of ~ 37 Å generated by an axial rise per subunit of ~ 10.5 Å and an azimuthal rotation of ~ 100.8°. Although secondary structures were not resolved at this resolution, the globular domain of the Ng PilE crystal structure was easily docked into the cryo-EM density, aided by its asymmetric structure and protruding surface-exposed β-hairpin. This rigid body fitting resulted in the N-terminal α-helices wrapping around each other in a helical array in the filament core. Though the precise positions of these helices were not resolved in the cryo-EM map, they nonetheless fit well, requiring only small adjustments in the N-terminal two residues, which were modeled in an extended conformation to minimize steric clashes. This arrangement placed the conserved Glu5 in a position to form a salt bridge with the N-terminal amine of the adjacent α1N of the (+)1 helix, neutralizing these charges in the otherwise hydrophobic core of the filament. Glu5 is critical for T4P assembly (Aas et al., 2007a; Horiuchi and Komano, 1998; Li et al., 2012; Pasloske and Paranchych, 1988; Strom and Lory, 1991), presumably because it contributes to docking of pilin subunits into the growing pilus (Craig et al., 2006). The globular domains fit neatly together on the pilus surface and almost completely bury the N-terminal α-helical core, but it is the extensive interactions among the mostly hydrophobic α1s that are thought to provide the remarkable stability of the Ng T4P, which are resistant to proteolysis, heat and chemical denaturation and require detergent to dissociate them into pilin subunits (Li et al., 2012; Watts et al., 1982).
A substantially higher quality ~ 6 Å resolution structure was recently determined for the closely related T4P from N. meningitidis (Kolappan et al., 2016), whose pilin subunit, PilE, is homologous to Ng PilE in amino acid sequence (~78% identity) and structure. Nm T4P is similar to Ng T4P in helical symmetry, with an average rise per subunit of ~ 10.3 Å and a rotation of ~ 100.8°, and in the arrangement of the globular domains on the pilus surface. Importantly, the higher quality Nm T4P structure revealed rod-like density defining α-helical secondary structure in the filament core, but only weak and extended density for residues ~ 15–23 indicating partial melting of the α-helix in the pilus filament. This melted segment, which is flanked by the conserved helix-breaking residues Gly14 and Pro22, was modeled in an extended conformation. This loss of α-helical structure results in a substantially different positioning of the α1:1–14 helices relative to their predicted orientation in the lower resolution Ng pilus structure, lying almost parallel to each other in the Nm filament core. This melting of α1:15–23 was proposed to occur as pilin subunits dock into the pilus to optimize the packing of the subunits in the pilus, to provide flexibility for the pilus, and to accommodate the force-induced stretching observed for both Nm and Ng T4P (Biais et al., 2010; Brissac et al., 2012). The conservation of the N-terminal residues in the Type IVa pilins, particularly the helix-breaking residues Gly14 and Pro22, suggest that helical melting is a conserved feature of the Type IVa pili.
While no empirical structures have yet been published for the P. aeruginosa T4P, a computational model for the filament was determined for PaK pili (Craig et al., 2004) based upon the PilA crystal structure, a filament diameter of 52 Å and fiber diffraction data supporting a 1-start helix with a 41 Å pitch and near parallel α-helices (Folkhard et al., 1981). The PaK T4P computational model has a subunit rise of 10.25 Å and a rotation of 90°, resulting in subunits stacking directly above one another along the 4-start helices. The much smaller diameter of the PaK T4P is due to its smaller globular domain compared with the Neisseria pilins. Here we report cryo-EM reconstructions for the PaK T4P at ~ 8 Å resolution and the Ng T4P at ~ 5 Å resolution, which share the melted central portion of α1 seen for Nm T4P. We describe mutational analysis for the closely related Nm PilE, providing further insights into the role of this melted segment in pilus assembly and function.
RESULTS
Cryo-EM reconstruction and filament model for the P. aeruginosa Type IV pilus
Pili were sheared from the surface of P. aeruginosa strain K (PaK) cells and resuspended in an alkaline buffer (ethanolamine, pH 10.5), which disrupts pilus bundles (Craig et al., 2003). Pili were purified and concentrated and plunge-frozen in vitreous ice on lacey carbon grids. Cryo-EM images were collected using a direct electron detector (Figure 1A) and filaments were boxed and segmented into overlapping segments. Filament segments were sorted revealing considerable structural polymorphism, with a rise per pilin subunit ranging from 8.9 Å to 11.9 Å and rotations ranging from 85.3° to 89.3° (Figure S1A, Movie M1). A reconstruction was obtained from the largest bin containing 22% of the segments using the Iterative Helical Real Space Reconstruction (IHRSR) method (Egelman, 2000). The reconstruction shows a smooth undulating filament surface with gently rounded protrusions that represent the PaK pilin globular domains (Figure 1B, C). These protrusions delineate a right-handed (+) 1-start helix with a rise of 10.5 Å and a rotation of 87.3° between subunits. The protrusions also follow left-handed (−)3- and (−)4-start helices. Cavities at the intersection of three globular domain densities reveal internal rod-like density corresponding to the N-terminal α-helices in the filament core. The rod-like density is well-defined but weakens before joining the globular domain density, indicating melting of a central portion of α1 (Figure 1D, E), as seen for the N. meningitidis cryo-EM reconstruction (Kolappan et al., 2016). A bridge of density connects the end of each rod to the side of an adjacent rod near its end (see Figure 2D).
Figure 1. Cryo-EM reconstruction and filament model of the P. aeruginosa T4P.
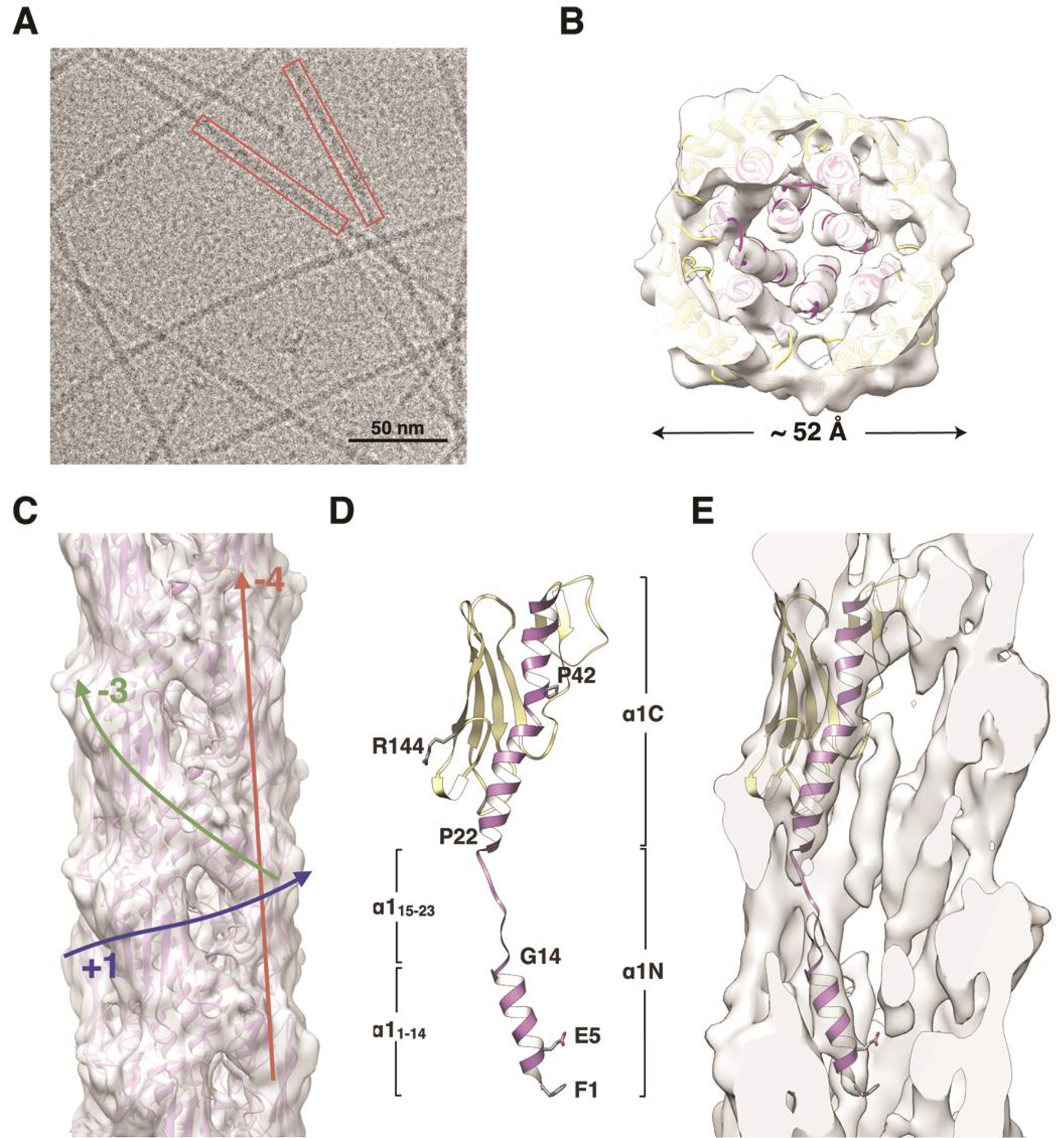
(A) Cryo-electron micrograph of P. aeruginosa T4P. (B) Slice of the PaK pilus reconstruction and filament model viewed along the filament axis. (C) Side view of the PaK pilus reconstruction and filament model. Colored arrows indicate the paths of the (+)1-, (−)3- and (−)4-start helices. (D) Ribbon representation of the PilA subunit model generated by fitting the protein into the cryo-EM density in three segments: the globular domain and α1:1–14 were fit separately as rigid bodies from the PilA crystal structure 1OQW, and α1:15–23 was modeled in an extended conformation. α1 is purple and the remainder of the globular domain is yellow. This subunit model was used to generate the PaK pilus filament model. (E) Slice through the cryo-EM density map showing a single PilA subunit. See also Figure S1A, Movie S1.
Figure 2. Interactions among subunits in the cryo-EM-derived PaK pilus model.
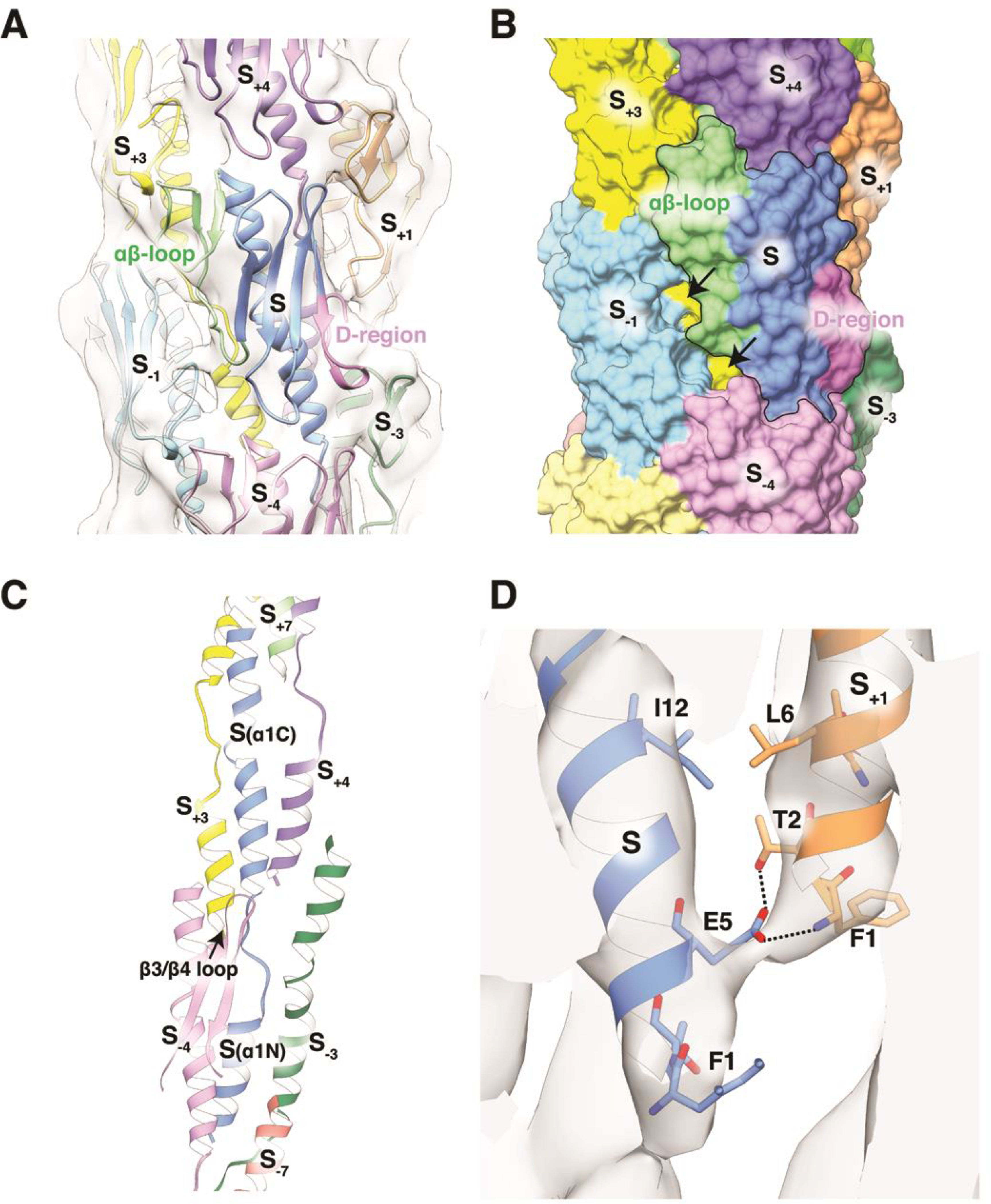
(A) Ribbon representation of the PaK pilus showing interactions among the globular domains. The central subunit, S, is shown in blue with its αβ-loop colored green and its D-region colored magenta. Other subunits have a single color. Subunits S−1, S and S+1 lie along the (+)1 helix; subunits S−3, S and S+3 lie along the (−)3 helix; S−4, S and S4 lie along the (−)4 helix. (B) Surface representation of the PaK pilus model colored as in (A), with the central subunit outlined. Arrows indicate where gaps between subunits in the (+)1 helix expose the α-helical core of the pilus. (C) Packing of α1s in the filament core. (D) Close-up of the cryo-EM map and PaK pilus model showing the bridging density representing an electrostatic bond between Glu5 and Phe1 in an adjacent subunit in the (+)1-helix. This density is also consistent with a Glu5:Thr2 hydrogen bond.
The crystal structure of full length PaK PilA (1OQW, (Craig et al., 2003)) was fit as a rigid body into the globular density of the cryo-EM map, which placed its N-terminus outside the rod-like density. Thus, N-terminal α-helical segment of PilA (α1:1–14) was fit separately into the rod-like density, rotating it about its long axis to fit the Glu5 side chain into the bridging density between adjacent rods (Figure 1E). Next, residues 15–23 were modeled in an extended conformation to approximately match the weak density for this region and to connect α1:1–14 with α1C (residues 24–53) in the globular domain. A filament model was generated from this single subunit by applying the helical symmetry parameters from the reconstruction and cycles of refinement and fitting were performed to optimize the fit of the filament model to the cryo-EM map. The resulting filament model is shown in Figure 1B and C. Refinement statistics for the model are shown in Table 1. The resolution of the reconstruction is estimated at 8 Å overall and 6.8 Å for the filament core based on Fourier shell correlation (FSC) of the cryo-EM density map with the resulting pilus model (Figure S2A, B). The resolution is likely limited by the variability in both rise and rotation despite the sorting process.
Table 1:
Refinement statistics for the Ng and PaK T4P filament model
| PaK T4P filament model | Ng T4P filament model | |
|---|---|---|
| Clash score, all atoms | 15.7 | 11.8 |
| Protein geometry | ||
| Ramachandran favored (%) | 87.3 | 89.0 |
| Ramachandran outliers (%) | 0 | 0 |
| Rotamer outliers (%) | 0 | 0.8 |
| Cβ deviations > 0.25 Å | 0 | 0 |
| RMS deviations | ||
| Bonds (Å) | 0.01 | 0.01 |
| Angles (°) | 1.44 | 1.23 |
| MolProbity score | 2.31 (99th, 3.25 Å – 8.25 Å) | 2.16 (100th, 3.25 Å – 5.35 Å) |
| PDB ID | XXXX | XXXX |
The diameter of the PaK pilus model is ~ 52 Å (Figure 1B). The globular domains comprise the outer shell of the filament and each have unique interaction interfaces with adjacent globular domains in the (+)1-, (−)3- and (−)4-start helices (Figure 1C, 2A, B). Minimal interactions along the (+)1-start helix, between parts of the αβ-loop of each subunit, S, and the β3/4 loop and the most C-terminal tail of subunit S−1, partially expose the α1 helices in the filament core. The near-90° rotation per subunit along the (+)1 helix results in the globular domains being stacked almost directly on top of each other along the (−)4 helices, which run almost parallel to the filament axis. A shallow depression at the top of each globular domain, formed by the tip of α1C and the β1/β2- and β3/β4 loops, is filled by the β2/β3 loop of the subunit directly above it (S → S+4). And along the (−)3 helix, the edge of the β-sheet subdomain within the αβ-loop of subunit S contacts a turn in the D-region loop of S+3, delineated by the disulfide-bonded cysteines Cys129 and Cys142.
In the filament core extensive hydrophobic interactions occur among the α1 helices, which run approximately parallel to each other and tilted slightly to the filament axis (Figure 2C). The shallow S-shaped curve seen for α1 in the PaK PilA crystal structure is disrupted by the non-helical segment, α1:15–23, which lies between the two helix-breaking residues Gly14 and Pro22 (Figure 1D). The α1N segments from subunits in each turn of the (+)1 helix interdigitate between the α1C segments of subunits the next turn down. Thus, each α1N, which is comprised of the helical segment α1:1–14 and the non-helical segment α1:15–23 (Figure 1D), is sandwiched between the globular domain α1Cs of two subunits in the next turn down of the (+)1 helix (Figure 2C). Conversely, each α1C is wedged between the α1Ns of two subunits in the next turn up in the (+)1 helix (Figure 2C). In addition to the hydrophobic interactions that mediate α1:α1 contacts, the bridge of density that connects the ends of the rod-like densities is consistent with a salt bridge between the Glu5 side chain of subunit S and the Phe1 amino nitrogen of S+1 (Figure 2D). Glu5 is also positioned to hydrogen bond with Thr2 of S+1. An end-on view of the filament shows that the helical segments of α1, α1:1–14 and α1C, and the globular domain β-sheets form concentric rings in the filament: the α1:1–14 helices occupy the core of the filament; the α1C helices form a ring that encircles the α1:1–14 core; and the β-sheets form the outermost shell of the filament (Figure 5D).
Figure 5. Comparison of the P. aeruginosa, N. gonorrhoeae and N. meningitidis Type IV pilus structures.
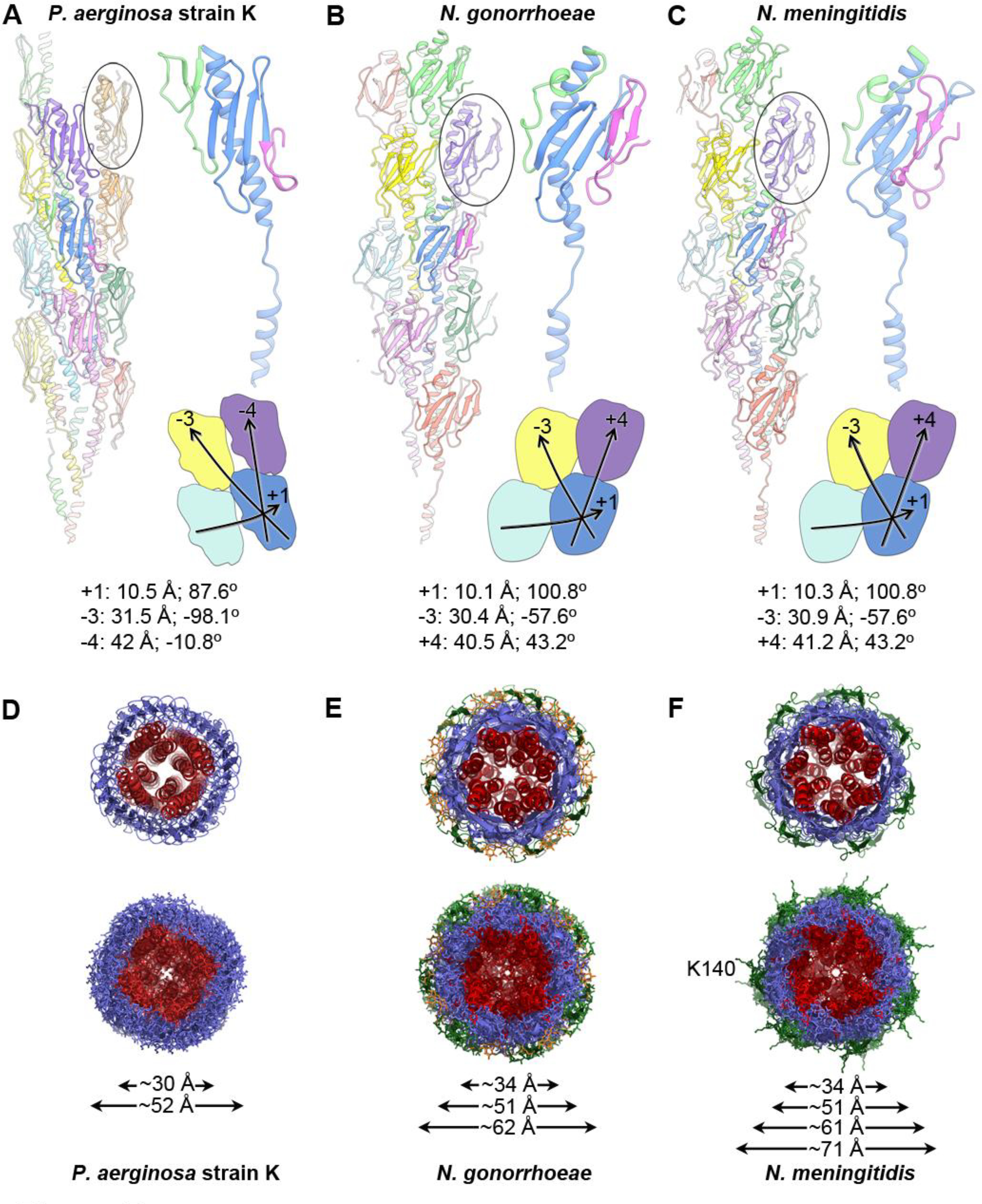
(A) A ribbon representation of the P. aeruginosa strain K T4P is shown on the left with the globular domain of a pilin subunit shown in side view circled. The central blue subunit is enlarged on the right, with its αβ-loop colored green and its D-region colored magenta. The general shape and packing of the globular domains is shown below, as are the symmetry parameters for the reconstruction. (B, C) The N. gonorrhoeae and N. meningitidis T4P structures, packing and symmetry are shown as in (A). The shape and packing of the globular domains is substantially different for PaK T4P than for the Neisseria pili. (D-F) End views of the PaK (D), N. gonorrhoeae (E) and previously determined N. meningitidis (F) T4P structures show similar diameters for the α-helix/β-sheet components for all three pili, with the β-hairpins and post-translational modifications accounting for the larger diameters of the Neisseria pili. The α1s are colored red, the hypervariable β-hairpins present on the Neisseria pili are shown in green, and the post-translational modifications at Ser63 and Ser68 of N. gonorrhoeae are colored orange. Lys140 protrudes from the β-hairpins in N. meningitidis T4P. Diameter measurements are the maximum Cα-Cα distances, with the exception of the largest diameter reported for Nm T4P, which includes the protruding Lys140 side chains.
Cryo-EM reconstruction and filament model for the N. gonorrhoeae Type IV pilus
To obtain a cryo-EM structure of the N. gonorrhoeae T4P, the pili were sheared from the surfaces of N. gonorrhoeae C30 cells and purified using a protocol similar to that for the PaK pili. Cryo-EM images of the frozen-vitrified pili were collected (Figure 3A), boxed and segmented. Filament segments were sorted by rise and twist, which ranged from 9.6 Å to 10.6 Å, and 99.8° to 101.8°, respectively (Figure S1B, Movie M2). Segments in the largest bin, representing 25% of the total segments, were processed using IHRSR. The resulting cryo-EM map has an axial rise per subunit of 10.1 Å and an azimuthal rotation of 100.8°, generating a (+)1 helix with a pitch of 36 Å (Figure 3B, C). The cryo-EM density map of the globular domain is well-defined, with rod-like density for the α-helical spine (α1C, residues 24–53) and extended density for the β-strands of the central β-sheet, allowing unambiguous fitting of this region, residues 24–158, using the full-length Ng PilE structure (2HI2, (Craig et al., 2006)). However, as with the PaK T4P reconstruction, rigid body fitting of PilE into the globular domain density resulted in a poor fit for the N-terminal half of α1N (residues 1–14) into the rod-like density corresponding to this segment. Thus, α1:1–14 was fit separately, and the remaining α1N segment, residues 15–23, was modeled in a partially extended conformation to align with the weak density connecting α1N(1–14) with the globular domain (Figure 3D, E). After optimizing the fit of a single subunit using RosettaCM protocol (Song et al., 2013), filament models were generated by applying the determined symmetry parameters. A single best filament model was selected based on its overall fit with the experimental cryo-EM map (Figure 3B, C, Table 1). The overall resolution of the Ng T4P reconstruction is estimated at ~ 5.1 Å, with ~ 4.3 Å for the helical core based on FSC calculations between the refined atomic model and the map (Sup. Figure S2C, D). Though less heterogeneous than PaK pili, the polymorphism of Ng pili likely also limited the resolution of this reconstruction.
Figure 3. Cryo-EM density map and filament model for the N. gonorrhoeae T4P.
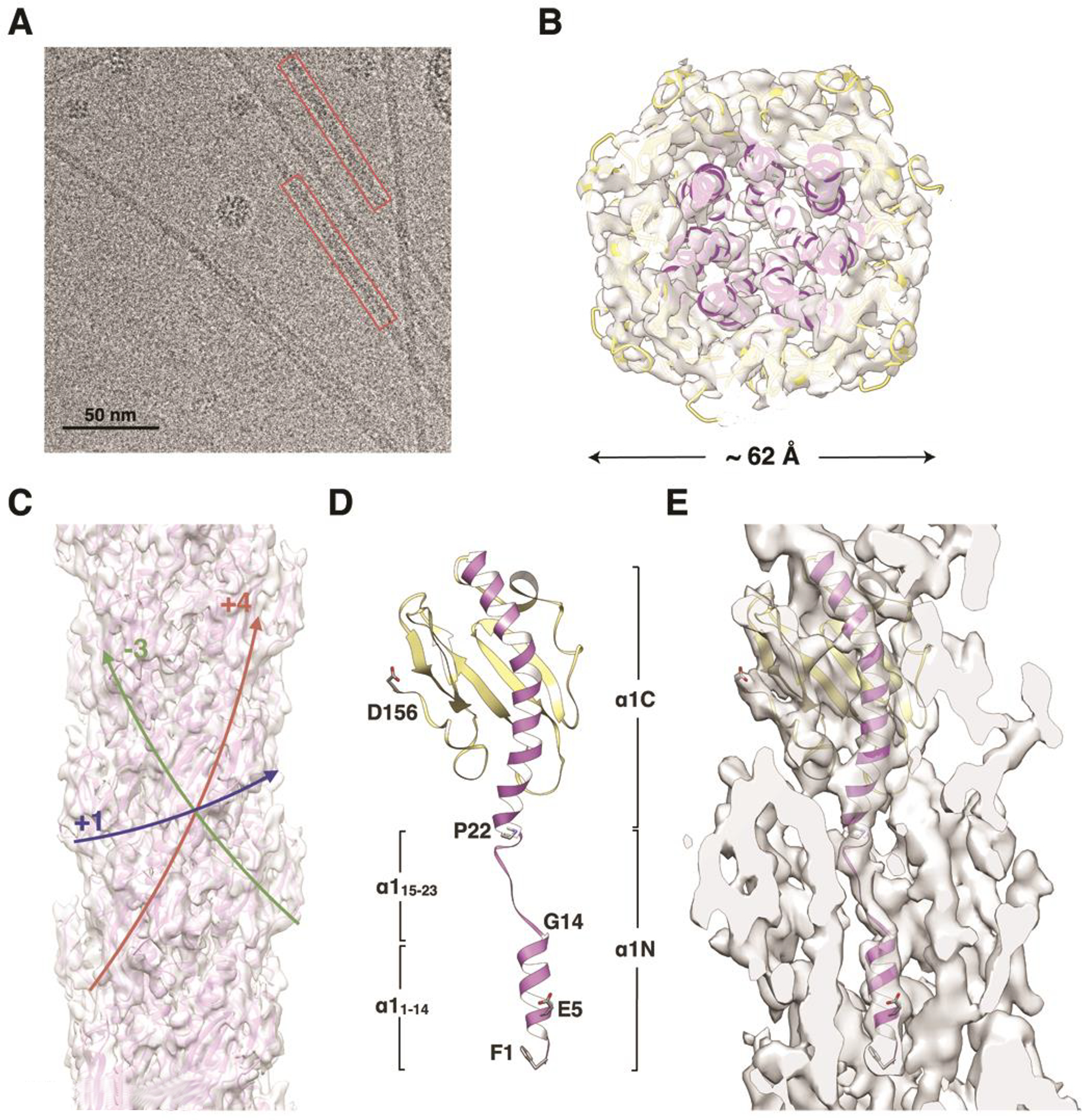
(A) Cryo-electron micrograph of N. gonorrhoeae T4P. (B) Slice of the Ng pilus reconstruction and filament model viewed along the filament axis. (C) Side view of the PaK pilus reconstruction and filament model. Colored arrows indicate the paths of the (+)1-, (−)3- and (+)4-start helices. (D) Ribbon representation of the Ng PilE model generated by fitting the protein into the cryo-EM density in three segments: the globular domain and α1:1–14 were fit separately as rigid bodies from the PilE crystal structure 2HI2, and α1:15–23 was modeled in an extended conformation. α1 is purple and the remainder of the globular domain is yellow. This subunit model was used to generate the Ng pilus filament model. (E) Slice through the cryo-EM density map showing a single PilE subunit. See also Figure S1B, Movie S2.
The overall architecture of the Ng pilus structure is very similar to that of the 12.5 Å resolution Ng pilus structure (Craig et al., 2006) and the recently published ~ 6 Å resolution N. meningitidis pilus structure (Kolappan et al., 2016), yet provides the highest resolution and most detailed T4P structure available to date. Individual polypeptide strands and structural features can be resolved when the map is contoured at a high level (Figure 4A): the αβ-loop (residues 54–77), the β-strands of the central β-sheet, the β2/β3-loop, the hypervariable β-hairpin within the D-region, much of the C-terminal loop, and the proximal sugar of the glycosyl moiety attached to Ser63. This sugar has been identified by mass spectrometry as hexose linked to a proximal 2,4-diacetamido-2,4,6-trideoxyhexose sugar (HexDATDH) (Aas et al., 2007a). The density for this sugar moiety was not present in the original map and thus the post-translational modifications were initially omitted from the model. Upon appearance of extra density at Ser63 after refinement of the model against the map, HexDATDH and phosphoethanolamine moieties were added to the filament model at Ser63 and Ser68, respectively and the model refined further (Figure 4A). No density is observed for the distal hexose of the disaccharide or the phosphoethanoloamine attached to Ser68, likely due to greater flexibility for these groups. Nonetheless, the positions of Ser63 and Ser68 as well as the density for the proximal sugar on Ser63, indicate that the post-translational modifications are displayed on the surface of the pilus. The D-region of PilE, delineated by the disulfide bond between Cys121 and Cys151, is well-defined in the map as a dominant protrusion from the globular domain density, corresponding to the β-hairpin that contains the hypervariable region, and the bulging loop that immediately follows this feature. Density is also apparent for the remainder of the D-region loop, which folds back into the pilus and reconnects with the β-sheet via the disulfide bond. Following the disulfide bonded Cys151 the most C-terminal segment of PilA re-emerges on the pilus surface. Terminal residues Ala157 and Lys158 are not resolved in the cryo-EM map and were thus omitted from the model.
Figure. 4. Interactions among subunits in the Ng T4P model.
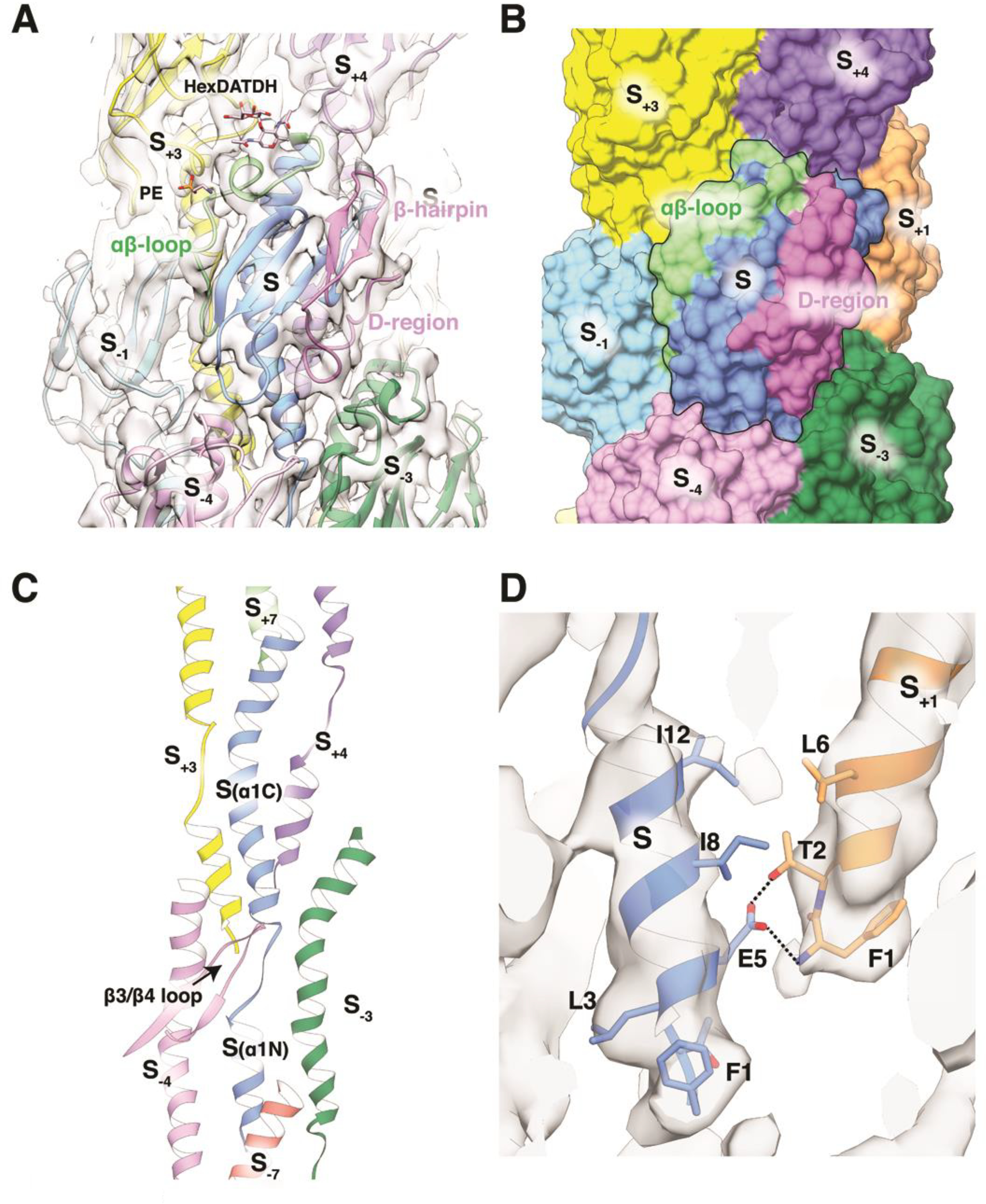
(A) Ng T4P map and model shown in ribbon representation from the outer face of the pilus. The central subunit S is colored blue with the αβ-loop shown in green and the D-region in magenta. The map is thresholded at a high level to show that portions of the αβ-loop, β-strands and the hypervariable β-hairpin within the D-region are resolved. The post-translational modifications Hex-DATDH at Ser63 and PE at Ser68 are shown as sticks. (B) Surface representation of the Ng pilus model colored as in (A) to show the interactions between the central subunit, S (outlined), and its neighboring subunits in the (+)1, (−)3 and (+)4 helices. (C) Packing of α1s in the filament core. (D) Close-up of the salt bridge and hydrogen bond between Glu5 on one subunit and Phe1 and Thr2 on an adjacent subunit in the (+)1 helix.
Direct interactions between the globular domains of Ng PilE occur at three unique interfaces (Figure 4A, B): along the (+)1 helix the C-terminal half of the αβ-loop of subunit S with its bulky side chains (Lys74, Lys76 and Tyr77) fits perfectly into the curved surface created by the β3/β4-loop, β4 and the D-region C-terminus of subunit S−1; along the (−)3 helices between the C-terminal residues of α1 and the beginning of the αβ-loop of subunit S and the D-region loop of S+3; and along the (+)4 helices between the tip of α1 of subunit S and the β2/β3 loop of S−4. The stereochemical complementarity between these interfaces allows close packing of the globular domains on the surface of the Ng pilus model, which almost completely encloses the N-terminal α-helices in the core of the pilus (Figure 4B).
Within the filament core rod-like density defines both the C-terminal half of α1, α1C (~24–53), and its most N-terminal segment, α1:1–14 (Figure 3E). α1:1–14 of each subunit, S−1, is positioned within the rod-like density such that its Glu5 forms a salt bridge and a hydrogen bond with the N-terminal amino nitrogen and Thr2, respectively, of the neighboring subunit in the (+)1 helix (subunit S) (Figure 4D), though the bridge of density representing this interaction is not as well resolved as in the PaK T4P reconstruction, and is only visible at lower contouring of the map. Between α1:1–14 and α1C the density is weaker, indicating melting of the α-helix between the helix-breaking residues Gly14 and Pro22 (Figure 3D, E). This segment was modeled as a partially extended polypeptide, though the positions of individual residues are not well-defined due to the weak density in this region. Nonetheless, connections are evident between side chains in this melted segment and the β3/β4-loop of subunit S−4, and α1C of subunit S−3 (Figure 4C). Such detail has not been observed previously for a T4P cryo-EM reconstruction. The N-terminal α-helices run approximately parallel to the filament axis, with the N-terminal halves (α1N) interdigitating between α1Cs of two subunits in the next turn down of the (+)1 helix, as seen for the PaK pilus. But unlike Pak PilA, which has a relatively straight α1C, α1C of Nm PilE is curved as a result of a glycine at position 42, with its C-terminal end bending out toward the C-terminal edge of the subunit and toward the filament surface (Figure 5A, B). This segment is flanked by two α1Ns from the next turn up in the (+)1 helix, as well as by the most N-terminal segment of a subunit (S+7) two turns up (Figure 4C).
Comparison of the PaK, Ng and Nm T4P structures
The new PaK and Ng pilus cryo-EM structures and the recently determined Nm T4P structure share similar architectures, including a melted α1 between Gly14 and Pro22, extensive hydrophobic interactions among their N-terminal α-helices in the core of the filament, and smaller interaction interfaces between the globular domains along the (+)1, (−)3 and (+/−4)-start helices (Figure 5A-C)). Yet significant architectural differences are also apparent between PaK pilus and the highly similar Ng and Nm pili, primarily due to differences in the shapes of their globular domains and the curvature of α1C. PaK PilA has an elongated almost cylindrical globular domain, and incomplete contacts between these globular domains in the (+)1 helix leave gaps that expose part of α1N. The Ng and Nm PilE globular domains are more egg-shaped and pack together more compactly in the filament. The long axis of the PilA globular domain is tilted ~ −15° relative to the filament axis. α1C follows this tilt, whereas the β-strands run almost parallel to this axis (Figure 5A). In contrast, the long axes of the Ng and Nm PilE globular domains are oriented almost parallel to the filament axis with their α1Cs curved away from the axis and in line with the β-strands, angled ~ +15° to this axis (Figure 5B, C). The curvature in the Ng and Nm PilE spine also pushes the top half of α1C out toward the surface of the filament.
The differences in the shape and packing of the globular domains produce different helical symmetries for the filaments. The PaK pilin subunits are rotated by ~ 90° along the (+)1 helix (87.3°), which places the globular domains almost directly on top of each other in subsequent turns of this helix (Figure 5A). This arrangement gives (−)4-start helices with very small rotations of −10.8 ° per subunit along each strand. In contrast the 100.8° rotation for subunits along the (+)1 helix of Ng and Nm T4P means they are staggered along the length of the filament, with globular domains in each turn of the helix partially intercalated between globular domains in subsequent turns of the helix (Figure 5B, C). This symmetry produces left-handed (−)4-start helices. Packing differences between the PaK and Neisseria T4P are also evident in the core of the pilus. Although the conformation of α1 N-terminal to residue 42 and its orientation relative to the filament axis are similar in all three pili, and they have similar neighboring α1 segments (Figure 2, 4) they are staggered throughout the filament core in the Ng and Nm T4P compared to the PaK pilus where they are stacked on top of each other (Figure 5D-F). Thus, although the α1s interact more extensively than do the globular domains, the packing of the bulky globular domains nonetheless dictates the arrangement of the α1 segments. Interestingly, the cryo-EM-based PaK pilus model is remarkably similar to the computational model that was built based on the full length PilA structure and fiber diffraction data (Craig et al., 2004). This earlier structure predicted a (+)1 helix with a 10.3 Å rise and a 90° rotation, with a similar orientation for the globular domains.
Views of the cross-sections of these T4P show similar arrangements of concentric rings (Figure 5D-E), where the N-terminal α-helices fill the filament core with diameters of 30–34 Å (max. Cα-Cα distances); the β-sheets and αβ-loops form the next ring, comprising the outer layer for PaK pili (Figure 5D), with diameters of 51–52 Å. The hypervariable β-hairpin in Ng and Nm PilE and the post-translational modifications on Ng PilE increases the diameter of these pili to 62 Å, although these features do not coat the entire pilus surface (Figure 5E). N. meningitidis T4P is similarly modified, with a trisaccharide at Ser63 and a phosphoglycerol at Ser69 and Ser93 (Chamot-Rooke et al., 2011; Chamot-Rooke et al., 2007; Marceau et al., 1995), although these modifications are not resolved in the cryo-EM reconstruction. Lys140 protrudes from the hypervariable loop of Nm PilE, extending the Nm pilus diameter to 71 Å (measured atom-to-atom, Figure 5F).
Role of the pilin N-terminal residues in pilus assembly and functions
The identification of the melted helix in the Nm T4P (Kolappan et al., 2016), which is also seen here for the PaK and Ng pilus structures, prompted us to test the importance of backbone flexibility in pilus assembly. The α1 sequences of Ng and Nm PilE are identical to each other and 77% identical to PaK in the first 30 residues (Figure 6A). Gly14 and Pro22 flank the melted segment and may destabilize the α-helix to allow melting. Gly14 is largely conserved in both the Type IVa and IVb pilins, though some possess another small amino acid, alanine, at this position (Figure 6A). Pro22 is conserved in the Type IVa pilins. In addition, there are several conserved alanines within and adjacent to the melted region that may contribute to flexibility and packing of this unfolded region in the pilus due to their small side chains. To test whether these conserved residues are important for pilus assembly we introduced point mutations in the N. meningitidis pilE gene and assessed the mutants for their ability to assemble functional pili. Gly14 and Pro22 were changed to alanine, which are common in α-helices, and alanines were changed to the bulkier hydrophobic valine. We also mutated codons for Thr2 and Glu5, changing these to alanine, to test the requirements for the putative Thr2:Glu5 hydrogen bond and the Phe1N:Glu5 salt bridge for pilus assembly (Figure 6B). The mutants were evaluated based on pilin expression, pilus assembly and pilus functions in bacterial aggregation.
Figure 6. Conserved residues in the Type IV pilin α1 are critical for pilus assembly and functions.
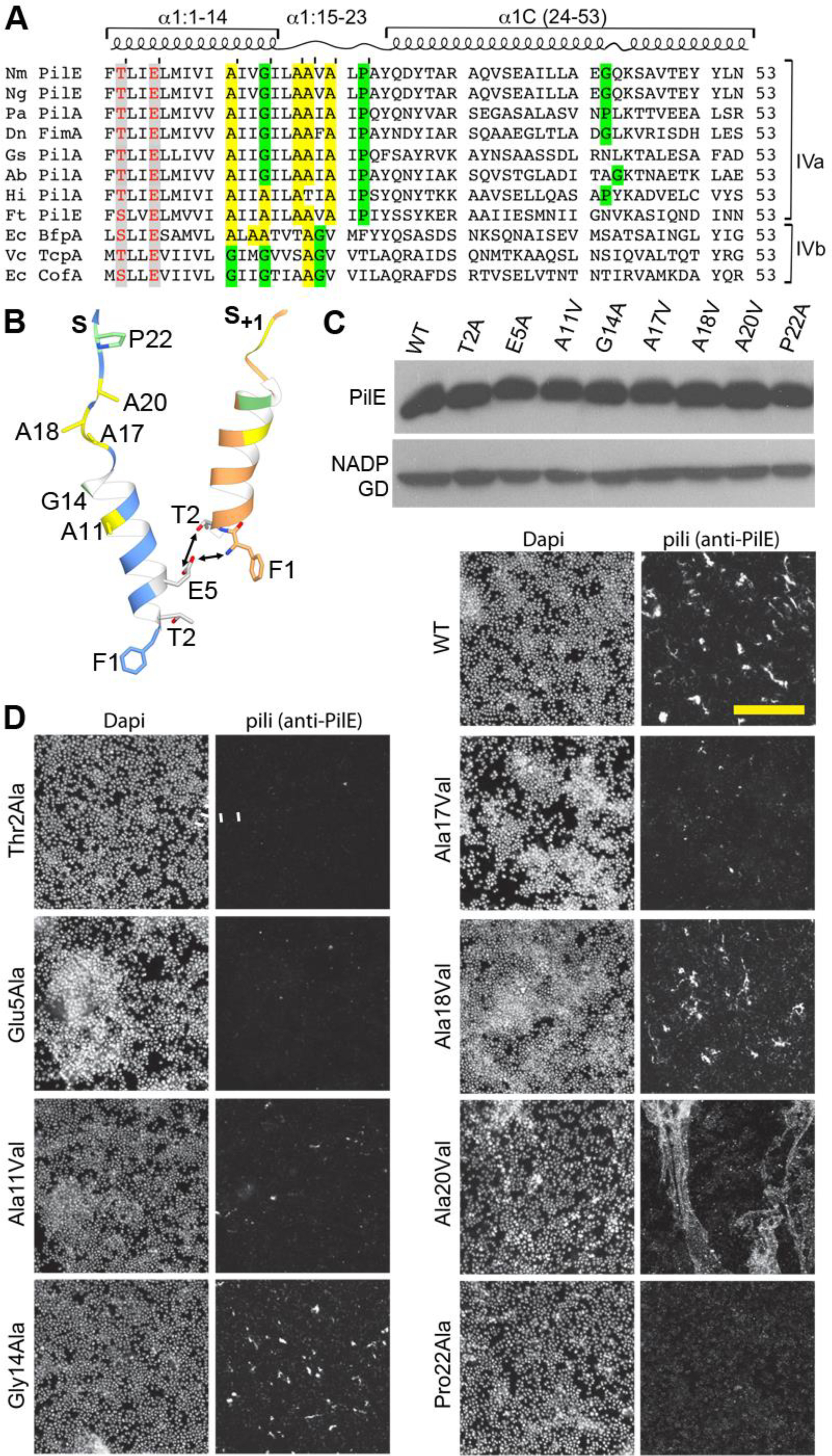
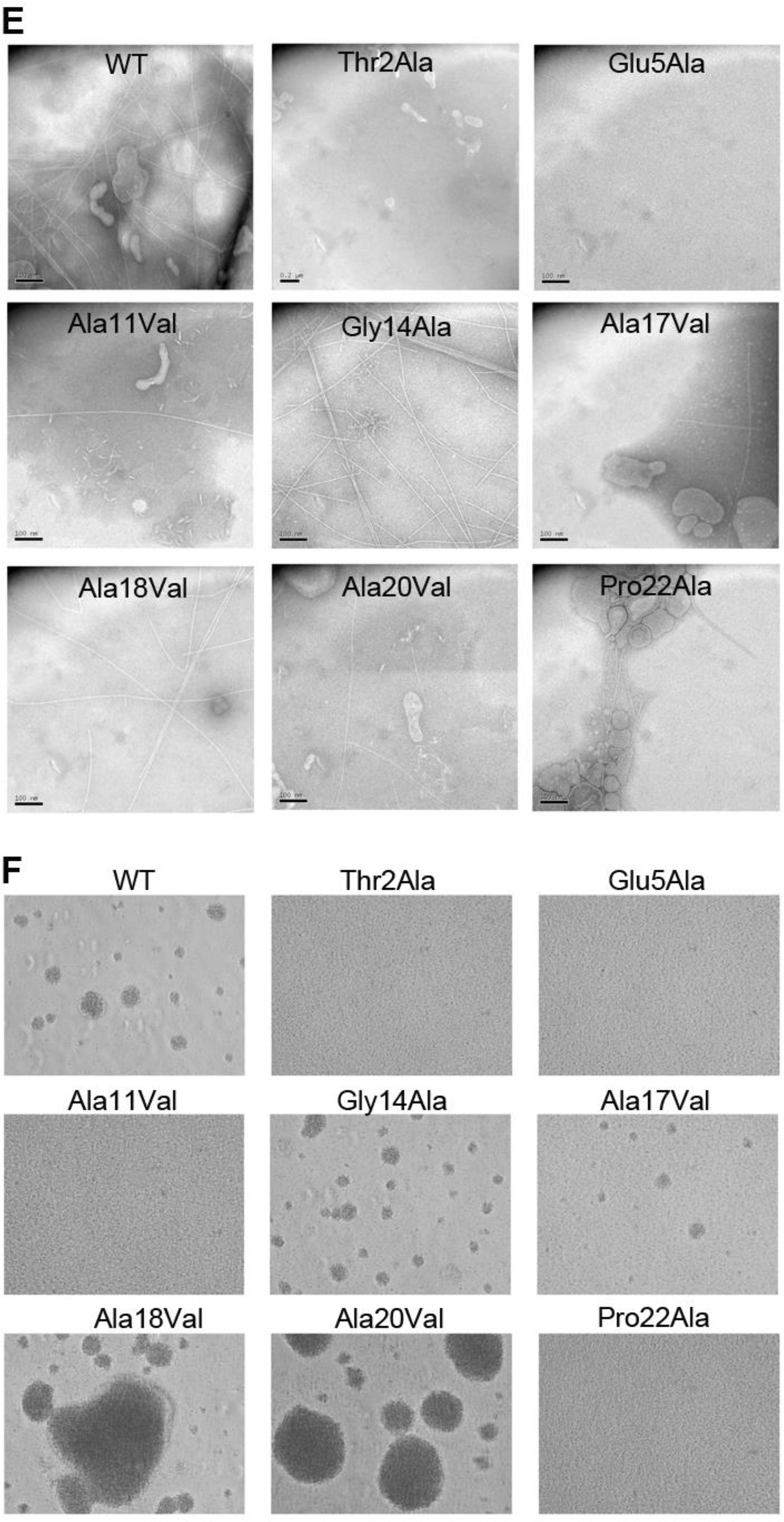
(A) Amino acid sequence alignment of α1 (residues 1–53) from Type IVa and Type IVb pilins. The α-helical portions of this segment are indicated based on the cryo-EM density maps for Nm, Ng and PaK T4P, and the melted region is boxed. Thr/Ser2 and Glu5 are highlighted in grey with red text. Conserved alanines within or near the melted region are highlighted yellow and the conserved helix-breaking residues proline and glycine are highlighted green. Nm PilE, N. meningitidis PilE (GenBank accession number WP_014573675); Ng PilE, N. gonorrhoeae PilE (for 2HI2, P02974); PaK PilA, P. aeruginosa PilA (P02973); Dn FimA, Dichelobacter nodosus (X52403), Gs PilA, Geobacter sulfurreducins PilA (2M7G_A); Ab PilA, Acinetobacter baumannii ACICU (ACC58690); Hi, non-typeable Haemophilus influenzae (AAX87353); Ft PilE1, Francisella tularensis Schu S4 PilE1 (CAG45522); Ec BfpA, enteropathogenic Escherichia coli BfpA (Z68186); Vc TcpA, Vibrio cholerae TcpA (ABQ19609); Ec CofA, enterotoxigenic E. coli (CofA, BAA07174) (B) Close-up of the N-terminus of two PilE subunits in the (+)1 helix of Nm T4P model (Kolappan et al., 2016) showing the melted α1:15–23 segment and the interactions between Glu5 of subunit S and Phe1 and Thr2 in subunit S+1 in the (+)1 helix. (C) Immunoblot of whole cell lysates of N. meningitidis SB-Kn (WT) and Sb-Kn pilE mutants probed with antibodies against PilE and a cytosolic marker protein, NADP glutamate dehydrogenase. (D) Immunofluorescent labeling of N. meningitidis WT and pilE mutant strains stained with DAPI to visualize the bacteria and anti-PilE and goat anti-mouse Alexa 488 antibody to visualize the pili. The scale bar is 20 μm. (E) Aggregation of N. meningitidis WT and pilE mutant strains imaged on an inverted microscope after overnight growth and resuspension in fetal bovine serum. (F) TEM imaging of pili sheared from N. meningitidis WT and pilE mutant strains.
As seen from immunoblots of whole cell lysates, PilE is expressed at approximately wild type levels in all of the pilE mutants (Figure 6C), demonstrating that the amino acid substitutions do not disrupt the pilin fold. Pilus assembly was assessed by immunofluorescence of N. meningitidis cells using anti-PilE antibody clone 20D9 (Pujol et al., 1999). As predicted, pili staining levels are substantially reduced in the Thr2Ala, Glu5Ala and Pro22Ala variants (Figure 6D), suggesting these residues are critical for pilus assembly. Pili were sheared from the cells and imaged by TEM to assess their morphologies. No pili were observed for Thr2Ala and Glu5Ala and few were observed for Pro22Ala (Figure 6E). Consistent with these results, no bacterial aggregation was observed for these strains (Figure 6F). Thus, the PilE Thr2Ala, Glu5Ala and Pro22Ala variants are defective in pilus assembly. These results suggest that the Thr2:Glu5 hydrogen bond and the Phe1N:Glu5 salt bridge both contribute to pilus stability. The kink induced by Pro22 likely destabilizes the N-terminal α-helix, allowing it to unfold and pack into the pilus core, whereas an alanine at this position may stabilize the α-helical conformation. In contrast, despite being conserved, Gly14 could be changed to alanine without affecting pilus assembly. The alanine substitution may be too conservative a change at this site to detect altered piliation.
Replacement of the conserved alanines with the bulky hydrophobic valines disrupted pilus assembly at only one site, Ala11, as shown by reduced piliation and aggregation (Figure 6). The Ala17Val variant is comparable in phenotype to the WT strain. And surprisingly, the Ala18Val and Ala20Val variants appear to be hyperpiliated, as shown by immunofluorescence and aggregation assays (Figure 6).
DISCUSSION
The structures of the PaK and Nm T4P described here highlight features that appear to be broadly conserved among the Type IV pili, such as the N1:Glu5 and Thr2:Glu5 interactions and the melted helix between Gly14 and Pro22, as well as features unique to each pilus. The conservation of Thr/Ser2 and Glu5 in both Type IVa and IVb pilins and the importance of these residues in pilus assembly suggest that these polar interactions are conserved among the Type IV pilin class in general (Figure 6A). The requirement for Glu5 for efficient pilus assembly is well documented in both T4P classes (Aas et al., 2007b; Horiuchi and Komano, 1998; Li et al., 2012; Pasloske and Paranchych, 1988; Strom and Lory, 1991) but to our knowledge this is the first demonstration of a role for Thr2. The conserved Gly14 and Pro22 flank the melted segment of α1 and may destabilize the α-helix in this region. The Gly14Ala variant has a WT phenotype, implying that a helix-breaking amino acid is not necessary at position 14. Since alanine is present at this position in some Type IV pilins, the preference may be for a small side chain rather than glycine specifically. Pro22 is unique to the Type IVa pilins, but the Type IVb pilins possess several glycines in the segment that is melted in the PaK, Ng and Nm T4P structures, which may also destabilize the helix. We expected that changing the conserved alanines within the melted segment to the bulkier side chain valine might impose restraints on this segment, but in fact these changes resulted in more efficient pilus assembly in the case of Ala18Val and Ala20Val. These variants showed wild type levels of DNA transformation (data not shown) indicating that their hyperpiliation is not due to loss of retraction. Since alanine is found more frequently than valine in α-helices, it may be that the valines at these positions make the melting transition more favorable. Nonetheless, the selection of alanines at these positions presumably means these residues are optimal for pilus assembly and functions in vivo.
We have proposed that α1N is helical when the pilin in anchored in the inner membrane, but melts as the pilin subunit incorporates into the pilus filament in order to pack the α-helices in the core of the pilus and prevent their clashing (Kolappan et al., 2016). In PaK, Ng and Nm T4P, the melted segment serves to straighten out α1 such that the most N-terminal segment, α1:1–14, lies almost parallel to the filament axis and to adjacent α1s, optimizing their interactions instead of clashing with them. It is these N-terminal hydrophobic interactions and not the polar globular domain interactions that hold the subunits together, as detergent is required to dissociate pili into pilin subunits without denaturing them (Li et al., 2012). Interestingly, detergent solubilization increases the α-helical content of PaK PilA by ~ 6%, as measured by circular dichroism (Watts et al., 1982), consistent with the fully α-helical α1N for the PaK and Ng crystal structures obtained from pili dissociated with octyl β-D-glucopyranoside (Craig et al., 2003; Craig et al., 2006; Parge et al., 1995). The contribution of α1Ns to filament formation is exemplified in the Type IVa pilus from Geobacter sulfurreducens, whose 61 amino acid pilin subunits are comprised almost exclusively of α1. The G. sulfurreducens pilins possess Gly14, Pro22 and several of the alanines conserved in the Type IVa pilins (Figure 6A), suggesting that these pilins adopt a packing arrangement similar to that of PaK, Ng and Nm T4P with no constraints imposed by fitting the globular domains onto the pilus surface. Nonetheless, it is curious why the proline/glycine-induced curvature is conserved in the Type IV pilins, when straighter α-helices appear to fit better in the filament. In fact, the Ignicoccus hospitalis Iho670 flagellar–like filament subunits have a hydrophobic N-terminal segment homologous to the Type IV pilins, but these segments form straight continuous α-helices that are well-resolved in the ~ 4 Å cryo-EM reconstruction of Iho670 (Braun et al., 2016). Notably, the packing and symmetry are quite different from that observed in any true T4P. The I. hospitalis subunits lack the helix-breaking Gly14 and Pro22 and glycines between these positions, which may prove to be predictors of helix melting. Conservation of these residues would allow α1 to adopt an α-helical conformation when acting as an inner membrane anchor prior to pilus assembly, and to partially unfold to incorporate and pack into the growing pilus. The α-helical conformation would shield the backbone amide nitrogen and carbonyl carbon from the hydrophobic lipid bilayer, and the melted conformation would facilitate packing of α1N in the filament core and impart flexibility and extensibility on the pilus, allowing it to adhere to surfaces and resist high sheer forces.
The curvature of α1 is further enhanced by Gly42 in α1C of Ng and Nm pilins, to a greater degree than for Pro42 in PaK PilA, which has a straighter α1C. This subtle structural difference profoundly impacts the orientation of the globular domain and hence its packing on the surface of the pilus. The Dichelobacter nosodus Type IVa pilin FimA and Acinetobacter baumanni PilA have curved α1Cs due to glycines at positions 42 and 43 respectively (Hartung et al., 2011; Piepenbrink et al., 2016), but other Type IV pilins, including G. sulfurreducens PilA and the Type II secretion pseudopilins, have neither a glycine nor a proline at this position and have straight a1Cs (Alphonse et al., 2010; Craig et al., 2006; Kolappan et al., 2012; Korotkov et al., 2009; Lim et al., 2010; Piepenbrink et al., 2015)
The new PAK pilus structure is markedly different from the Ng and Nm structures due to its smaller globular domain, which lacks the hypervariable hairpin loop that protrudes from its exposed face, and to its straight α1C, which places the globular domain in a different orientation relative to the PilE globular domains. PaK pili function in adhesion to glycans on the surfaces of epithelial cells (Bucior et al., 2012; Comolli et al., 1999; Irvin et al., 1989; Krivan et al., 1988; Saiman and Prince, 1993; Wall and Kaiser, 1999), though the identity of these receptors is somewhat controversial (Emam et al., 2006; Schroeder et al., 2001). Residues in the C-terminal D-region are implicated in receptor binding (Wong et al., 1995) despite there being considerable sequence variation in this region. The D-region forms a tight loop at the edge of the β-sheet that is stabilized by the disulfide bond between Cys129 and Cys142. The D-region contacts neighboring subunits in the (−)3 and (+)4 helices and thus is not particularly accessible to host receptors, except at the pilus tip, which has been shown to act as an adhesion (Lee et al., 1994). However, PaK pili, like Ng and Nm pili, are very flexible and extensible, with spring-like properties and the ability to withstand forces up to 250 pN (Beaussart et al., 2014; Lu et al., 2015; Touhami et al., 2006). Thus, PaK pili may undergo reversible force-induced conformational changes that disrupt the weaker subunit:subunit interactions, exposing the D-region, while maintaining the more extensive hydrophobic α1:α1 interactions. This mechanism was proposed (Kolappan et al., 2016) to explain the force-induced exposure of the EYYLN epitope in Ng and Nm T4P (Biais et al., 2010; Brissac et al., 2012). PaK T4P also bind to hydrophobic abiotic surfaces, allowing P. aeruginosa to colonize surfaces such as indwelling catheters and contact lenses (Beaussart et al., 2014; Giltner et al., 2010; Miller and Ahearn, 1987; Nickel et al., 1989). These interactions occur all along the length of the pilus (Lu et al., 2015) despite their surface-exposed residues being predominantly polar. Hydrophobic residues in the melted segment of α1N are partially exposed in the gap between pilin subunits, but this gap is no more than 10 Å at its broadest point and as deep, making these residues accessible only to long thin receptors. It is conceivable that stress on the pilus might enlarge this gap and make the residues in the melted α1:14–23 segment more accessible to bind hydrophobic surfaces.
The PaK and Ng T4P structures reported here help to explain the remarkable mechanical and biological properties that make these filaments essential virulence factors for so many human pathogens. Insights derived from these structures can contribute to the development of pilus-based vaccines and therapeutics, which may help to address the growing problem of antibiotic resistance. Virulence factors such as the T4P are highly attractive as targets for vaccines due to their surface exposure, although antigenic variability prohibits their use in pathogens such as Neisseria. Antibiotics directed at T4P and their assembly machinery have strong potential due to their specificity, which would prevent wholesale destruction of the beneficial microbiota, and may reduce the selective pressure to evolve resistance mechanisms because they would disarm rather than kill select microbes. A molecular understanding of these filaments, their assembly and functions, is invaluable in these efforts.
STAR METHODS
Bacterial strains and growth media
The hyperpiliated P. aeruginosa strain PAK/2pfs (Bradley, 1974) was a gift from Dr. Randal Irvin, University of Alberta. The N. gonorrhoeae strain C30, a variant of MS11 (Segal et al., 1985) was a gift from Dr. John Tainer, The Scripps Research Institute, La Jolla CA. pilE mutations were introduced at the pilE expression locus and expressed in the high adhesive SB variant of N. meningitidis 8013 as previously described (Nassif et al., 1993).
Purification of P. aeuginosa strain K pili
P. aeruginosa strain PAK/2pfs (Bradley, 1974) was grown on a Tryptic soy agar (3% TS broth, 1.5% agar) plates at 30 °C overnight. A single colony was passed onto a new plate and the cells were grown overnight at 30°C. Approximately 20 colonies were harvested and resuspended in 2 ml 3% TS broth and 50 μl of the resuspension was spread over each of the thirty TS agar plates that had been sitting for 3 days at room temperature and one day at 30 °C at ~20% humidity to reduce their moisture content. Cells were grown for 24 hr at 30°C, 20% humidity. The bacterial lawns were scraped off of the plates and resuspended in 25 ml of chilled Buffer 1 (150 mM ethanolamine, 1 mM DTT, pH 10.5). Pili were sheared from the cells by three cycles of 1 min bursts of vigorous vortexing followed by 1 min on ice. The cells were removed from the pilus suspension by centrifugation (8000 xg, 30 min) at 4°C. The supernatant containing the pili was centrifuged again to pellet residual cells. The supernatant was dialyzed against 4 l Buffer 2 (150 mM NaCl, 50 mM Tris, 1 mM DTT, 0.02% NaN3, pH 8.0) three times, each time for at least four hours, at 4 °C using a 6000–8000 MWCO dialysis membrane to promote aggregation of the pili. Pili were concentrated by centrifugation 24000 xg for 30 min at 4 °C. The supernatant was discarded and the pellet was resuspended in 1 ml cold Buffer 1 and dialyzed against 4 l Buffer 2 three times, each time for at least four hours at 4 °C using a 3500 MWCO dialysis membrane. The dialysate was centrifuged at 10 000 xg for 30 min at 4 °C to pellet the pili, which were resuspended in 200 μl cold Buffer 1 and stored at 4°C.
Purification of N. gonorrhoeae T4P
N. gonorrhoeae strain C30 cells (Segal et al., 1985) were grown on a GC plate at 37 °C with 5% CO2 for 24 h. To prepare GC plates, 54.38 g BD Difco™ GC medium base and 1.88 g Agar Noble were added to 1.5 l of H20. The solution was autoclaved and cooled to 65 °C and 15 ml of Supplement I (2.22 M glucose, 68 mM L-glutamine, 66.5 μM thiamine) and 1.5 ml of Supplement II (20.7 mM Fe(NO3)3) were added. Three pilus producing (P+) colonies, identified using light micrcospy by their almost spherical morphology, were passed onto a new GC plate in a triad formation. The plate was incubated at 37°C, with 5% CO2 for 24 h. Three new P+ colonies were passed onto two GC plates in a triad formation. The plates were incubated at 37°C with 5% CO2 for 20 h. After confirming that the colonies are P+, they were swabbed off of the two plates and resuspended in 5 ml GC Broth (1.5% (w/v) BD Bacto™ Proteose Peptone No. 3, 23 mM K2HPO4, 7.3 mM KH2PO4, 86 mM NaCl). Fifty GC plates were inoculated with 100 μl of the cell suspension and the cells were evenly spread over the plates. Cells were grown at 37 °C, with 5% CO2 for 24 h. The bacterial lawns were scraped off of the plates and resuspended in 20 ml CHES Buffer (50 mM CHES, 1 mM DTT, pH 9.5) at 4 °C. To shear pili off, the cells were vortexed vigorously 3 times for 1-min bursts, returning them to ice for 1 min between the bursts. Cells were removed from the pilus suspension by centrifugation at 10 000 xg for 20 min at 4 °C. The supernatant containing the pili was dialyzed against 4 l cold Pili Buffer (50 mM Tris, 150 mM NaCl, 1 mM DTT, 0.02% NaN3, pH 7.5) twice, each time for at least 2 hours at 4 °C using a 6000–8000 MWCO dialysis membrane. Aggregated pili were pelleted by centrifugation at 10 000 xg for 20 min at 4 °C and resuspended in 3 ml cold CHES Buffer. The protein suspension was centrifuged at 3500 xg for 20 min at 4 °C to remove contaminants then dialyzed against 4 l cold Pili Buffer twice, each time for at least 2 hours at 4 °C using a 6000–8000 MWCO dialysis membrane. Purified pili were pelleted from the dialysate by centrifugation at 17 500 xg for 30 min and resuspended in 200 μl cold CHES Buffer without DTT and stored at 4 °C.
Cryo-electron microscopy and image processing of pilus filaments
Samples (2 μl) were applied to lacey carbon grids and vitrified using a Vitrobot Mark IV (FEI). The grids were imaged in a Titan Krios operating at 300 keV using a Falcon II camera with 1.05 Å/px sampling, with the imaging controlled by the EPU software. Images were collected using a defocus range of 0.5 to 3.0 μm. The CTFFIND3 program (Mindell and Grigorieff, 2003) was used for determining the actual defocus of the images.
PaK pili.
A total of 440 images of PaK pili were selected and 3,453 filaments of varying lengths were boxed from these using the e2helixboxer program within EMAN2 (Tang et al., 2007). The SPIDER software package (Frank et al., 1996) was used for most other operations. Images were corrected for the contrast transfer function (CTF) by multiplying them by the theoretical CTF function, both reversing phases where needed and boosting the SNR. Overlapping 384 px long boxes (with a shift of 15 px, ~ 1.5 times the axial rise per subunit) were cut from the long filament boxes, yielding 154,641 segments. A reference-based sorting procedure was used to bin the segments based on the axial rise and azimuthal rotation. After sorting, 31,231 segments from the largest bin, representing 22% of the total segments, were processed by IHRSR (Egelman, 2000) to produce the final reconstruction.
Ng pili.
A total of 225 images were selected for image processing, from which 1,715 filaments boxes were cut. Overlapping 384 px long boxes (with a shift of 14 px) were cut from the long filament boxes, yielding 39,197 boxes segments. After sorting by both the twist and axial rise, 9,855 segments representing 25% of the total segments, were used in the final reconstruction.
Model Building
For each reconstruction a single pilin subunit was segmented from the map in Chimera (Pettersen et al., 2004). Model building began by docking x-ray crystal structures of the full length pilin subunits (1OQW and 2HI2, for the PaK and Ng maps, respectively) into the experimental density data using Chimera’s “fit in map” tool. Since these fits resulted in poor correspondence between the N-terminal segment, the N-terminal α-helix (α1:1–14) was fit separately and the melted loop (α1:15–23) was built with RosettaCM protocols (Wang et al., 2015). A total of 2500 models were generated for both pilin subunits using the segmented map, and 15 models selected based on Rosetta’s energy function, were combined into one model by manual editing in Coot (Emsley and Cowtan, 2004) to yield the best overall fit to the density map. This model was used as the starting model for a whole filament RosettaCM rebuilding restrained by the determined helical symmetry and the filament map. A total of 500 filament models were generated for both pili, and the best 15 models were combined again into a single model in Coot. Further refinements and editing were carried by Phenix real-space refinement (Adams et al., 2010) and Coot. Ser63 and Ser68 in the final Ng T4P model were replaced with the post-translationally modified serines from the Ng PilE crystal structure (2HI2). MolProbity (Chen et al., 2010) was used to evaluate the quality of the models. The MolProbity scores for the PaK and Ng T4P filament models compare favorably (99th and 100th percentile, respectively) with structures of similar resolution.
Mutagenesis of N. meningitidis pilE
To introduce mutations at the pilE expression locus, a transcriptional fusion of the pilE coding sequence with the aph-3’ gene (confers resistance to kanamycin) was engineered and cloned into the pMiniT plasmid. ppilE(Kn)-pMiniT plasmids were linearized for transformation of N. meningitidis strain 2C4.3, whereby the pilE(Kn) fusion replaces the existing pilE gene by homologous recombination (Nassif et al., 1993). Selection on kanamycin ensures that the pilE gene has not recombined with the promoterless pilE genes. The plasmid, ppilE(Kn)-pMiniT was prepared by overlap PCR. Briefly, DNA segments encoding the pilESB promoter and the pilESB gene and downstream sequence were amplified from genomic DNA using the primer pairs pilE_5’UTR_FW and pilE_5’UTR_RV, and pilE_3’UTR_FW and pilE_3’UTR_RV (Table 2). The aph-3’ coding sequence was amplified from the TOPO 2.1 vector using the primers Kn_FW and Kn_RV. The three fragments were assembled by overlapping PCR and the resulting product was cloned into the pMiniT plasmid following the NEB PCR Cloning Kit protocol. This construct was transformed into TOP10 chemically competent E. coli cells (Life Technologies) and the ppilE(Kn)-pMiniT vector was isolated from kanamycin-resistant colonies by miniprep (PureLink miniprep, Life Technologies). The correct vector was confirmed by sequencing (GATC-Biotech). Mutations were introduced into pilE within the plasmid by PCR Site Directed Mutagenesis using the AA primers containing the desired mutations, listed in Table 2. PCR products were purified, ligated and transformed into TOP10 chemically competent E. coli cells (Life Technologies). Plasmids were isolated from colonies grown on ampicillin and sequenced to verify the correct mutation. Linearized plasmids were transformed into N. meningitidis strain 2C4.3 and three kanamycin-resistant transformants were isolated for each mutant and sequenced to confirm the presence of the correct pilE-Kn gene fusion. Genomic DNA from these clones was purified and used to transform N. meningitidis 8013 SB, which were grown overnight on GCB-Kn plates. The pilE-Kn gene was verified by sequencing some of the Kn-resistant bacteria and the whole population was collected and frozen at −80 °C in 15% glycerol.
Table 2:
N. meningitidis pilE cloning and mutagenesis primers
| Primer Name | Sequence |
|---|---|
| pilE_5’UTR_FW | CCACCAAAACGGACGACCCCG |
| pilE_5’UTR_RV | TGAATTTTCAGACGGCGTAAATTATACCATAAATTTTAAATAAATCAAGCGGTAAGTGA |
| Kn_FW | GCCGTCTGAAAATTCAGAAGAACTCGTCAAGAAGGCGATAGAA |
| Kn_RV | CTGCGTGCAATCCATCTTGTTCAATCATGCGAAACGATCCTC |
| pilE_3’UTR_FW | CATGCGAAACGATCCTCTTAGCTGGCAGATGAATCATCG |
| pilE_3’UTR_RV | CCTCAAAACACAAACTTGATACCAATC |
| 2AA_FW | GATAAGagcAAAACCTTTTTGAAGGGTGTTCA* |
| 2AA_RV | GAGCTGATGATTGTGATTGCCATCGTC |
| 5AA_FW | CATCAGagcGATAAGGGTAAAACCTTTTTGA |
| 5AA_RV | ATTGTGATTGCCATCGTCGGCATTTTG |
| 11AA_FW | CATTTTGGCGGCAGTCGCCCTTC |
| 11AA_RV | CATTTTGGCGGCAGTCGCCCTTC |
| 14AA_FW | GCCAAAATagcGACGATGGCAATCACAATCATCA |
| 14AA_RV | GGCAGTCGCCCTTCCTGCTTATCA |
| 17AA_FW | CGACTGCaacCAAAATGCCGACGATGGC |
| 18AA_FW | CGACaacCGCCAAAATGCCGACGATGGC |
| 17_18AA_RV | CCCTTCCTGCTTATCAAGACTACACAGCCC |
| 20AA_FW | GGAAGaacGACTGCCGCCAAAATGCCGAC |
| 20AA_RV | GATAAGCagcAAGGGCGACTGCCGCCAAAATGCCGACG |
| 22AA_FW | GATAAGCagcAAGGGCGACTGCCGCCAAAATGCCGACG |
| 22AA_RV | AAGACTACACAGCCCGCGCACAAGTTTCCG |
Lower case letters indicate the mutations.
Immunofluorescence imaging of N. meningitidis pilE mutants
Bacteria were grown overnight on agar plates. Several meningococcal colonies were coated on glass slides, incubated in 4% paraformaldehyde for 30 minutes and dried for 2 hours before immunostaining. Pili were visualized using mouse anti-PilE antibodies (clone 20D9 (Pujol et al., 1999)) in phosphate-buffered saline containing 0.3% BSA; bacteria were visualized using DAPI staining of DNA and pili were visualized using goat anti-mouse Alexa 488 (Life Technologies). Slides were mounted in Mowiol (Citifluor Ltd) before image acquisition with a laser-scanning confocal microscope (Leica SP5). Confocal images were collected and processed using the Leica Application Suite AF lite software (Leica Microsystems).
Negative stain imaging of N. meningitidis pili from pilE mutants
N. meningitidis PilE mutants were grown overnight on GCB agar plates and resuspended in 20 mM ethanolamine, pH 10.5 and vortexed to shear the pili from the cells. Cells were removed by two rounds of centrifugation at 4000 xg and the pili in the supernatant fraction were resuspended in ammonium sulfate to 10% saturation and incubated for 16 h at 16 °C to precipitate the pili, which were collected by centrifugation (13000 x g, 60 min, 4°C) and resuspended in 20 mM ethanolamine, pH 10.5. Pili were negatively stained with 1% phosphotungstic acid on glow-discharged copper grids and imaged on a Hitachi 8100 STEM.
Aggregation assay for N. meningitidis pilE mutants
Aggregation of each strain was assessed in 24 wells plate in DMEM, 10% FBS. Briefly, the day of infection, a suspension of bacteria from an overnight culture grown on a GCB agar plate was adjusted to OD600 of 0.05 and incubated for 1h at 37°C in 24 wells plate in a pre-warmed DMEM, 10% fetal bovine serum under agitation. Aggregates were mechanically disrupted by pipetting up and down ten times. The bacteria were then incubated for 1 more hour without agitation and images of aggregates were acquired on an inverted microscope.
Supplementary Material
Movie S1. Animation showing the averaged power spectra computed from PaK pilus segments in the histogram bins 1, bin 5 and bin 9 shown in Figure S1A.
Movie S2. Animation showing the averaged power spectra computed from Ng pilus segments in the histogram bins 1, bin 5 and bin 9 shown in Figure S1B.
Highlights.
Cryo-EM structures were obtained for P. aeruginosa and N. gonorrhoeae Type IV pili
A portion of the pilin N-terminal α-helix is melted in both filament structures
The conserved Pro22 flanking the melted segment is necessary for pilus assembly
Pro22 destabilizes the α-helix to allow subunit packing and filament flexibility
ACKNOWLEDGEMENTS
This work was supported by a research grants of the Agence Nationale de la Recherche of France (ANR-14-CE14–0010-02) and the Fondation pour la Recherche Médicale (to XN), by NIH AI114902 (to EHE), by CIHR MOP125959 (to LC). Pilus sample screening and negative stain imaging of Nm mutant pili were performed at the SFU Nanoimaging Facility, part of the 4D LABS shared facilities supported by the Canada Foundation for Innovation (CFI), British Columbia Knowledge Development Fund, Western Economic Diversification Canada and Simon Fraser University. The cryo-EM work was conducted at the Molecular Electron Microscopy Core facility at the University of Virginia, which is supported by the School of Medicine and built with NIH grant (G20-RR31199). The Titan Krios and Falcon II direct electron detector within the Core were purchased with NIH SIG S10-RR025067 and S10-OD018149, respectively.
Footnotes
DATA AVAILABILITY
The PaK and Ng T4P reconstructions were deposited in the Electron Microscopy Data Bank under accession numbers EMD-XXXX and EMD-YYYY, respectively. Filament models were deposited in the Protein Data Bank under accession numbers XXX and YYY.
REFERENCES
- Aas FE, Vik A, Vedde J, Koomey M, and Egge-Jacobsen W (2007a). Neisseria gonorrhoeae O-linked pilin glycosylation: functional analyses define both the biosynthetic pathway and glycan structure. Mol Microbiol 65, 607–624. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aas FE, Winther-Larsen HC, Wolfgang M, Frye S, Lovold C, Roos N, van Putten JP, and Koomey M (2007b). Substitutions in the N-terminal alpha helical spine of Neisseria gonorrhoeae pilin affect Type IV pilus assembly, dynamics and associated functions. Mol Microbiol 63, 69–85. [DOI] [PubMed] [Google Scholar]
- Adams PD, Afonine PV, Bunkoczi G, Chen VB, Davis IW, Echols N, Headd JJ, Hung LW, Kapral GJ, Grosse-Kunstleve RW, et al. (2010). PHENIX: a comprehensive Python-based system for macromolecular structure solution. Acta crystallographica. Section D, Biological crystallography 66, 213–221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alphonse S, Durand E, Douzi B, Waegele B, Darbon H, Filloux A, Voulhoux R, and Bernard C (2010). Structure of the Pseudomonas aeruginosa XcpT pseudopilin, a major component of the type II secretion system. J Struct Biol 169, 75–80. [DOI] [PubMed] [Google Scholar]
- Anyan ME, Amiri A, Harvey CW, Tierra G, Morales-Soto N, Driscoll CM, Alber MS, and Shrout JD (2014). Type IV pili interactions promote intercellular association and moderate swarming of Pseudomonas aeruginosa. Proc Natl Acad Sci U S A 111, 18013–18018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beaussart A, Baker AE, Kuchma SL, El-Kirat-Chatel S, O’Toole GA, and Dufrene YF (2014). Nanoscale adhesion forces of Pseudomonas aeruginosa type IV Pili. ACS nano 8, 10723–10733. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Biais N, Higashi DL, Brujic J, So M, and Sheetz MP (2010). Force-dependent polymorphism in type IV pili reveals hidden epitopes. Proc Natl Acad Sci U S A 107, 11358–11363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bradley DE (1974). The adsorption of Pseudomonas aeruginosa pilus-dependent bacteriophages to a host mutant with nonretractile pili. Virology 58, 149–163. [DOI] [PubMed] [Google Scholar]
- Bradley DE (1980). A function of Pseudomonas aeruginosa PAO polar pili: twitching motility. Can J Microbiol 26, 146–154. [DOI] [PubMed] [Google Scholar]
- Braun T, Vos M, Kalisman N, Sherman NE, Rachel R, Wirth R, Schroder GF, and Egelman E (2016). Archaeal flagellin combines a bacterial type IV pilin domain with an Ig-like domain. Proc Natl Acad Sci U S A In press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brissac T, Mikaty G, Dumenil G, Coureuil M, and Nassif X (2012). The meningococcal minor pilin PilX is responsible for type IV pilus conformational changes associated with signaling to endothelial cells. Infect Immun 80, 3297–3306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bucior I, Pielage JF, and Engel JN (2012). Pseudomonas aeruginosa pili and flagella mediate distinct binding and signaling events at the apical and basolateral surface of airway epithelium. PLoS Pathog 8, e1002616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burrows LL (2012). Pseudomonas aeruginosa twitching motility: Type IV pili in action. Annual review of microbiology 66, 493–520. [DOI] [PubMed] [Google Scholar]
- Chamot-Rooke J, Mikaty G, Malosse C, Soyer M, Dumont A, Gault J, Imhaus AF, Martin P, Trellet M, Clary G, et al. (2011). Posttranslational modification of pili upon cell contact triggers N. meningitidis dissemination. Science 331, 778–782. [DOI] [PubMed] [Google Scholar]
- Chamot-Rooke J, Rousseau B, Lanternier F, Mikaty G, Mairey E, Malosse C, Bouchoux G, Pelicic V, Camoin L, Nassif X, et al. (2007). Alternative Neisseria spp. type IV pilin glycosylation with a glyceramido acetamido trideoxyhexose residue. Proc Natl Acad Sci U S A 104, 14783–14788. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen VB, Arendall WB 3rd, Headd JJ, Keedy DA, Immormino RM, Kapral GJ, Murray LW, Richardson JS, and Richardson DC (2010). MolProbity: all-atom structure validation for macromolecular crystallography. Acta crystallographica. Section D, Biological crystallography 66, 12–21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Comolli JC, Waite LL, Mostov KE, and Engel JN (1999). Pili binding to asialo-GM1 on epithelial cells can mediate cytotoxicity or bacterial internalization by Pseudomonas aeruginosa. Infect Immun 67, 3207–3214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Craig L, Pique ME, and Tainer JA (2004). Type IV pilus structure and bacterial pathogenicity. Nat Rev Microbiol 2, 363–378. [DOI] [PubMed] [Google Scholar]
- Craig L, Taylor RK, Pique ME, Adair BD, Arvai AS, Singh M, Lloyd SJ, Shin DS, Getzoff ED, Yeager M, et al. (2003). Type IV pilin structure and assembly: X-ray and EM analyses of Vibrio cholerae toxin-coregulated pilus and Pseudomonas aeruginosa PAK pilin. Mol Cell 11, 1139–1150. [DOI] [PubMed] [Google Scholar]
- Craig L, Volkmann N, Arvai AS, Pique ME, Yeager M, Egelman EH, and Tainer JA (2006). Type IV pilus structure by cryo-electron microscopy and crystallography: implications for pilus assembly and functions. Mol Cell 23, 651–662. [DOI] [PubMed] [Google Scholar]
- Egelman EH (2000). A robust algorithm for the reconstruction of helical filaments using single-particle methods. Ultramicroscopy 85, 225–234. [DOI] [PubMed] [Google Scholar]
- Emam A, Yu AR, Park HJ, Mahfoud R, Kus J, Burrows LL, and Lingwood CA (2006). Laboratory and clinical Pseudomonas aeruginosa strains do not bind glycosphingolipids in vitro or during type IV pili-mediated initial host cell attachment. Microbiology 152, 2789–2799. [DOI] [PubMed] [Google Scholar]
- Emsley P, and Cowtan K (2004). Coot: model-building tools for molecular graphics. Acta crystallographica. Section D, Biological crystallography 60, 2126–2132. [DOI] [PubMed] [Google Scholar]
- Engel JN (2003). Molecular pathogenesis of acute Pseudomonas aeruginosa infections. In Severe Infections Caused by Pseudomonas aeruginosa, Hauser A, and Rello J, eds. (New York: Kluwer Academic/Plenum Press; ), pp. 201–230. [Google Scholar]
- Folkhard W, Marvin DA, Watts TH, and Paranchych W (1981). Structure of polar pili from Pseudomonas aeruginosa strains K and O. J Mol Biol 149, 79–93. [DOI] [PubMed] [Google Scholar]
- Frank J, Radermacher M, Penczek P, Zhu J, Li Y, Ladjadj M, and Leith A (1996). SPIDER and WEB: processing and visualization of images in 3D electron microscopy and related fields. J Struct Biol 116, 190–199. [DOI] [PubMed] [Google Scholar]
- Giltner CL, Habash M, and Burrows LL (2010). Pseudomonas aeruginosa minor pilins are incorporated into type IV pili. J Mol Biol 398, 444–461. [DOI] [PubMed] [Google Scholar]
- Hartung S, Arvai AS, Wood T, Kolappan S, Shin DS, Craig L, and Tainer JA (2011). Ultra-high resolution and full-length pilin structures with insights for filament assembly, pathogenic functions and vaccine potential. J Biol Chem 286, 44254–44265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Horiuchi T, and Komano T (1998). Mutational analysis of plasmid R64 thin pilus prepilin: the entire prepilin sequence is required for processing by type IV prepilin peptidase. J Bacteriol 180, 4613–4620. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Irvin RT, Doig P, Lee KK, Sastry PA, Paranchych W, Todd T, and Hodges RS (1989). Characterization of the Pseudomonas aeruginosa pilus adhesin: confirmation that the pilin structural protein subunit contains a human epithelial cell-binding domain. Infect Immun 57, 3720–3726. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kolappan S, Coureuil M, Xiong Y, Nassif X, Egelman E, and Craig L (2016). Structure of the Neisseria meningitidis Type IV pilus. Nature Communications 7, 13015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kolappan S, Roos J, Yuen AS, Pierce OM, and Craig L (2012). Structure of CFA/III and longus Type IV pili from enterotoxigenic Escherichia coli. J Bacteriol 194, 2725–2735. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Korotkov KV, Gray MD, Kreger A, Turley S, Sandkvist M, and Hol WG (2009). Calcium is essential for the major pseudopilin in the type 2 secretion system. J Biol Chem 284, 25466–25470. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krivan HC, Roberts DD, and Ginsburg V (1988). Many pulmonary pathogenic bacteria bind specifically to the carbohydrate sequence GalNAc beta 1–4Gal found in some glycolipids. Proc Natl Acad Sci U S A 85, 6157–6161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee KK, Sheth HB, Wong WY, Sherburne R, Paranchych W, Hodges RS, Lingwood CA, Krivan H, and Irvin RT (1994). The binding of Pseudomonas aeruginosa pili to glycosphingolipids is a tip-associated event involving the C-terminal region of the structural pilin subunit. Mol Microbiol 11, 705–713. [DOI] [PubMed] [Google Scholar]
- Leighton TL, Buensuceso RN, Howell PL, and Burrows LL (2015). Biogenesis of Pseudomonas aeruginosa type IV pili and regulation of their function. Environ Microbiol 17, 4148–4163. [DOI] [PubMed] [Google Scholar]
- Li J, Egelman EH, and Craig L (2012). Structure of the Vibrio cholerae Type IVb pilus and stability comparison with the Neisseria gonorrhoeae Type IVa pilus. J Mol Biol 418, 47–64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lim MS, Ng D, Zong Z, Arvai AS, Taylor RK, Tainer JA, and Craig L (2010). Vibrio cholerae El Tor TcpA crystal structure and mechanism for pilus-mediated microcolony formation. Molecular Microbiology 77, 755–770. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu S, Giuliani M, Harvey H, Burrows LL, Wickham RA, and Dutcher JR (2015). Nanoscale Pulling of Type IV Pili Reveals Their Flexibility and Adhesion to Surfaces over Extended Lengths of the Pili. Biophys J 108, 2865–2875. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marceau M, Beretti JL, and Nassif X (1995). High adhesiveness of encapsulated Neisseria meningitidis to epithelial cells is associated with the formation of bundles of pili. Mol Microbiol 17, 855–863. [DOI] [PubMed] [Google Scholar]
- Mattick JS (2002). Type IV pili and twitching motility. Annual review of microbiology 56, 289–314. [DOI] [PubMed] [Google Scholar]
- Merz AJ, and So M (2000). Interactions of pathogenic Neisseriae with epithelial cell membranes. Annu Rev Cell Dev Biol 16, 423–457. [DOI] [PubMed] [Google Scholar]
- Miller MJ, and Ahearn DG (1987). Adherence of Pseudomonas aeruginosa to hydrophilic contact lenses and other substrata. J Clin Microbiol 25, 1392–1397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mindell JA, and Grigorieff N (2003). Accurate determination of local defocus and specimen tilt in electron microscopy. J Struct Biol 142, 334–347. [DOI] [PubMed] [Google Scholar]
- Nassif X, Lowy J, Stenberg P, O’Gaora P, Ganji A, and So M (1993). Antigenic variation of pilin regulates adhesion of Neisseria meningitidis to human epithelial cells. Mol Microbiol 8, 719–725. [DOI] [PubMed] [Google Scholar]
- Naumann M, Rudel T, and Meyer TF (1999). Host cell interactions and signalling with Neisseria gonorrhoeae. Current opinion in microbiology 2, 62–70. [DOI] [PubMed] [Google Scholar]
- Nickel JC, Downey JA, and Costerton JW (1989). Ultrastructural study of microbiologic colonization of urinary catheters. Urology 34, 284–291. [DOI] [PubMed] [Google Scholar]
- Parge HE, Forest KT, Hickey MJ, Christensen DA, Getzoff ED, and Tainer JA (1995). Structure of the fibre-forming protein pilin at 2.6 Å resolution. Nature 378, 32–38. [DOI] [PubMed] [Google Scholar]
- Pasloske BL, and Paranchych W (1988). The expression of mutant pilins in Pseudomonas aeruginosa: fifth position glutamate affects pilin methylation. Mol Microbiol 2, 489–495. [DOI] [PubMed] [Google Scholar]
- Pettersen EF, Goddard TD, Huang CC, Couch GS, Greenblatt DM, Meng EC, and Ferrin TE (2004). UCSF Chimera - A Visualization System for Exploratory Research and Analysis. J. Comput. Chem. 25, 1605–1612. [DOI] [PubMed] [Google Scholar]
- Piepenbrink KH, Lillehoj E, Harding CM, Labonte JW, Zuo X, Rapp CA, Munson RS Jr., Goldblum SE, Feldman MF, Gray JJ, et al. (2016). Structural Diversity in the Type IV Pili of Multidrug-resistant Acinetobacter. J Biol Chem 291, 22924–22935. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Piepenbrink KH, Maldarelli GA, Martinez de la Pena CF, Dingle TC, Mulvey GL, Lee A, von Rosenvinge E, Armstrong GD, Donnenberg MS, and Sundberg EJ (2015). Structural and evolutionary analyses show unique stabilization strategies in the type IV pili of Clostridium difficile. Structure 23, 385–396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pujol C, Eugene E, Marceau M, and Nassif X (1999). The meningococcal PilT protein is required for induction of intimate attachment to epithelial cells following pilus-mediated adhesion. Proc Natl Acad Sci U S A 96, 4017–4022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saiman L, and Prince A (1993). Pseudomonas aeruginosa pili bind to asialoGM1 which is increased on the surface of cystic fibrosis epithelial cells. The Journal of clinical investigation 92, 1875–1880. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schroeder TH, Zaidi T, and Pier GB (2001). Lack of adherence of clinical isolates of Pseudomonas aeruginosa to asialo-GM(1) on epithelial cells. Infect Immun 69, 719–729. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Segal E, Billyard E, So M, Storzbach S, and Meyer TF (1985). Role of chromosomal rearrangement in N. gonorrhoeae pilus phase variation. Cell 40, 293–300. [DOI] [PubMed] [Google Scholar]
- Song Y, DiMaio F, Wang RY, Kim D, Miles C, Brunette T, Thompson J, and Baker D (2013). High-resolution comparative modeling with RosettaCM. Structure 21, 1735–1742. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Strom MS, and Lory S (1991). Amino acid substitutions in pilin of Pseudomonas aeruginosa. Effect on leader peptide cleavage, amino-terminal methylation, and pilus assembly. J Biol Chem 266, 1656–1664. [PubMed] [Google Scholar]
- Tang G, Peng L, Baldwin PR, Mann DS, Jiang W, Rees I, and Ludtke SJ (2007). EMAN2: an extensible image processing suite for electron microscopy. J Struct Biol 157, 38–46. [DOI] [PubMed] [Google Scholar]
- Touhami A, Jericho MH, Boyd JM, and Beveridge TJ (2006). Nanoscale characterization and determination of adhesion forces of Pseudomonas aeruginosa pili by using atomic force microscopy. J Bacteriol 188, 370–377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wall D, and Kaiser D (1999). Type IV pili and cell motility. Mol Microbiol 32, 1–10. [DOI] [PubMed] [Google Scholar]
- Wang RY, Kudryashev M, Li X, Egelman EH, Basler M, Cheng Y, Baker D, and DiMaio F (2015). De novo protein structure determination from near-atomic-resolution cryo-EM maps. Nature Methods 12, 335–338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Watts TH, Scraba DG, and Paranchych W (1982). Formation of 9-nm filaments from pilin monomers obtained by octyl-glucoside dissociation of Pseudomonas aeruginosa pili. J Bacteriol 151, 1508–1513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wong WY, Campbell AP, McInnes C, Sykes BD, Paranchych W, Irvin RT, and Hodges RS (1995). Structure-function analysis of the adherence-binding domain on the pilin of Pseudomonas aeruginosa strains PAK and KB7. Biochemistry 34, 12963–12972. [DOI] [PubMed] [Google Scholar]
- Woods DE, Straus DC, Johanson WG Jr., Berry VK, and Bass JA (1980). Role of pili in adherence of Pseudomonas aeruginosa to mammalian buccal epithelial cells. Infect Immun 29, 1146–1151. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Movie S1. Animation showing the averaged power spectra computed from PaK pilus segments in the histogram bins 1, bin 5 and bin 9 shown in Figure S1A.
Movie S2. Animation showing the averaged power spectra computed from Ng pilus segments in the histogram bins 1, bin 5 and bin 9 shown in Figure S1B.


