Abstract
Parasitic nematodes are a major cause of human health problems with an estimated 1 billion people infected worldwide by these organisms. Identifying biochemical targets that differ between the parasite and host species is essential for finding effective new anti-parasitic molecules. The free-living nematode Caenorhabditis elegans is a powerful model system for experiments in genetics and developmental biology needed to achieve this goal; however, in-depth understanding of metabolic processes in this organism is limited as it still contains unexplored biochemical pathways. Eukaryotes. including nematodes and humans, share many similar metabolic pathways, which makes specific targeting of nematode parasites challenging. Recent studies suggest that C. elegans and other nematodes may use a plant-like pathway as the major biosynthetic route to phosphatidylcholine. In this pathway, a pair of phosphoethanolamine methyltransferases (PMT) catalyze the sequential methylation of phosphoethanolamine to phosphocholine, which can be incorporated into phosphatidylcholine. RNAi experiments demonstrate that both PMT are required for normal growth and development of C. elegans. Because the PMT are highly conserved across nematode parasites of humans, livestock, and plants, as well as in protozoan parasites, understanding how these enzymes function and the identification of inhibitors will aid in the development of new anti-parasite compounds of potential medical, veterinary, and agricultural value.
Keywords: Caernorhabditis elegans, nematode, phosphobase methylation, phospholipids, methyltransferase, phosphoethanolamine, phosphocholine
INTRODUCTION
Caenorhabditis elegans is a powerful model system for genetics and developmental biology, and for identifying biochemical targets for the development of drugs active against parasitic nematodes. Eukaryotes, including nematodes and humans, share many similar metabolic pathways, which makes specific targeting of parasites challenging. Recent studies suggest that C. elegans and other nematodes, unlike other animals, may use a plant-like pathway as the major biosynthetic route to phosphatidylcholine. In this pathway, a pair of phosphoethanolamine methyltransferases (PMT) catalyze the sequential methylation of phosphoethanolamine to phosphocholine, which can be incorporated into phosphatidylcholine. Although RNAi experiments demonstrate that both PMT are required for normal growth and development of C. elegans, the structural and molecular basis for how these enzymes function and their role in phospholipid metabolism remain to be established.
OVERVIEW OF PHOSPHOLIPID SYNTHESIS PATHWAYS IN EUKARYOTES
Phosphatidylcholine is a major phospholipid component in the cellular membranes of eukaryotes [1–2]. Phosphatidylcholine synthesis occurs through three metabolic routes in different organisms (Fig. 1). In mammals, the de novo choline or Kennedy pathway is the major synthetic route from choline to phosphatidylcholine [1–4]. Yeast and mammalian liver cells primarily use the Bremer-Greenberg pathway to methylate phosphatidylethanolamine to phosphatidylcholine [5–6]. In plants, metabolic flux into the Kennedy pathway occurs through a third route that involves S-adenosy 1-methionine (SAM)-dependent methylation of phosphoethanolamine to phosphocholine [7–8]. Recent work suggests that C. elegans and other nematodes [9–11], along with the protozoan parasite Plasmodium falciparum (the causative agent of malaria), use the plant-like phosphobase pathway as the primary route to provide phosphocholine for the synthesis of phosphatidylcholine [12–13].
Fig. 1. Overview of Pathways.
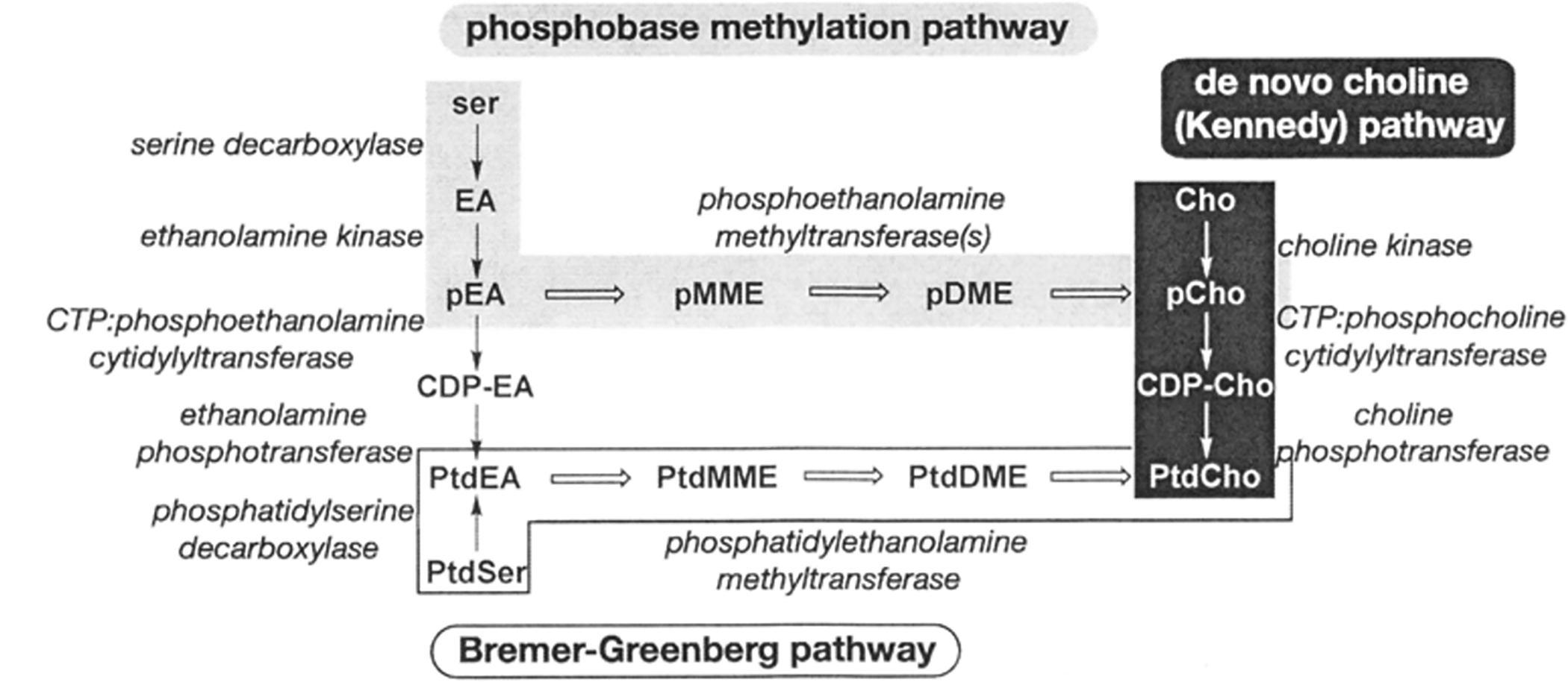
The de novo choline, Bremer-Greenbcrg, and phosphobase methylation pathways are shown. Conversions between the base, phosphobase, CDP, and phospholipid substrates arc shown as vertical arrows. Horizontal arrows correspond to SAM-dependent methylations. Names of metabolites consist of a prefix (p, phospho: CDP-. cytidine 5’-diphosphate; or Ptd, phosphatidyl) and a core name (EA, ethanolamine: MME, monomethylethanolamine; DME, dimethylcthanolamine; Cho, choline).
KNOWN ROUTES TO PHOSPHATIDYLCHOLINE IN C. elegans
The C. elegans genome contains the genes for both the de novo choline and Bremer-Greenberg pathways for the synthesis of phosphatidylcholine [11]; however, molecular studies suggest the presence of an alternate route to this phospholipid. In C. elegans, both the de novo choline and Bremer-Greenberg pathways have been examined to varying degrees by biochemical and high-throughput RNA interference (RNAi) approaches. These studies highlight how little is known about phospholipid synthesis in this model organism at the molecular level. RNAi screens suggest that later enzymes in the de novo choline pathway are important for worm growth and development, that the Bremer-Greenberg pathway plays a lesser role, and that an alternate pathway may exist in C. elegans to provide metabolites for the synthesis of phosphatidylcholine.
Biochemically, the best-studied parts of these pathways are the initial steps of the Kennedy pathway catalyzed by choline kinase and CTP:phosphocholine cytidylyltransferase (CCT). Choline kinase catalyzes the first step in the Kennedy pathway (Fig. 1). C. elegans encodes seven isoforms of this enzyme, which cluster into three families A, B, and C [14]. The A-family isoforms (cka-1 and −2) are 35% identical to the mammalian choline kinases, while the B-family (ckb-1, −2, −3, and −4) and C-family (ckc-1) share <25% sequence identity [14]. Biochemical analysis of CKA-1, CKA-2, CKB-2, and CKB-4 demonstrates a 10- to 20-fold preference for choline versus ethanolamine as a substrate [13], The remaining isoforms are biochemically uncharacterized, but postulated to function as ethanolamine kinases [13]. In the next reaction, CCT catalyzes the synthesis of CDP-choline [1–2], The genome of C. elegans contains five genes sharing roughly 60% sequence identity with mammalian CCT [14], but only one of these genes functions as a CCT [15], The last enzyme of the pathway, diacylglycerol choline phosphotransferase, converts CDP-choline and 1,2-diacylglycerol to CMP and phosphatidylcholine. It is a transmembrane protein that has not been purified to homogeneity from any source [1], and there are no reports describing this enzyme or its activity in C. elegans.
Genome-wide RNAi experiments indicate that certain isoforms of the enzymes in the de novo choline pathway are critical for normal growth and development [16–23]. Abnormal embryo and post-embryo developmental phenotypes were observed for ckb-1 and ckb-2 in some studies [17–19]; however, other genome-wide studies describe no effect resulting from silencing of any ckb gene [16, 20–23]. This difference may result from functional redundancy of the isoforms, alternate routes for phosphatidylcholine synthesis, or different growth conditions in these studies. It is unclear if C. elegans utilizes an external supply of choline for synthesis of phosphatidylcholine, as growth phenotypes require the addition of choline at concentrations (10–30 mM) far exceeding physiological levels [9–10]. Of the possible CCT isoforms, only RNAi of the verified CCT showed larva arrest in some experiments [16–17, 21], RNAi of the diacylglycerol choline phosphotransferase results in phenotypes ranging from the arrest of larva growth to embryo lethality [16–17, 21, 23]. Importantly, these studies indicate that the last two steps of the pathway are essential for normal growth and development. It should be noted that genome-wide RNAi screens are initial efforts to broadly define a role for these genes, and supporting studies are required to elucidate the contribution of various enzymes in the Kennedy pathway of nematodes.
The metabolic contribution of the Bremer-Greenberg pathway to phosphatidylcholine synthesis in nematodes appears less important for normal growth. The presence of genes encoding phosphatidylserine decarboxylase and phosphatidylethanolamine N-methyltransferase implies a functional Bremer-Greenberg pathway in C. elegans (Fig. 1). The putative C. elegans phosphatidylserine decarboxylase shares less than 40% identity with the mammalian enzyme [11]. RNAi of the decarboxylase results in abnormal development at the embryo and post-embryo stages [17–19, 21], suggesting that metabolism of ethanolamine to phosphatidylethanolamine cannot substitute for phosphatidylserine decarboxylase during development [24], In contrast RNAi of the phosphatidylethanolamine N-methyltransferase genes in C. elegans yields no observable phenotype [16–17], It is unclear if these results indicate that the methylation of phosphatidylethanolamine is not essential or if other pathways compensate for their loss.
PHOSPHOBASE METHYLATION IS REQUIRED FOR GROWTH AND DEVELOPMENT IN C. elegans
Unlike animals, plants use a pathway for the endogenous synthesis of phosphatidylcholine that effectively bypasses the need for either exogenous or recycled choline (Fig. 1). In the initial reaction of the phosphobase pathway, plants generate ethanolamine by direct decarboxylation of serine [25]. There is no information on the corresponding enzyme in C. elegans, but at least four genes encode biochemically uncharacterized proteins sharing ~40% sequence similarity with the plant serine decarboxylase.
The core of the phosphobase methylation pathway involves the sequential methylation of phosphoethanolamine into phosphocholine (Fig. 2). Early reports on the plant and Plasmodium PMT described the presence of two hypothetical genes in C. elegans (ZK622.3a/b; pmtl and F54D11.1; pmt2) sharing less than 20% identity with the PMT from plants and protozoa [12, 26–28], These observations suggested that C. elegans might use a plant-like phosphobase methylation pathway for phosphoeholine synthesis. Each of the C. elegans PMT are more closely related to the plant enzymes than they are to each other, as they share only 12% sequence identity [9–10].
Fig. 2. The Phosphobase Methylation Pathway.
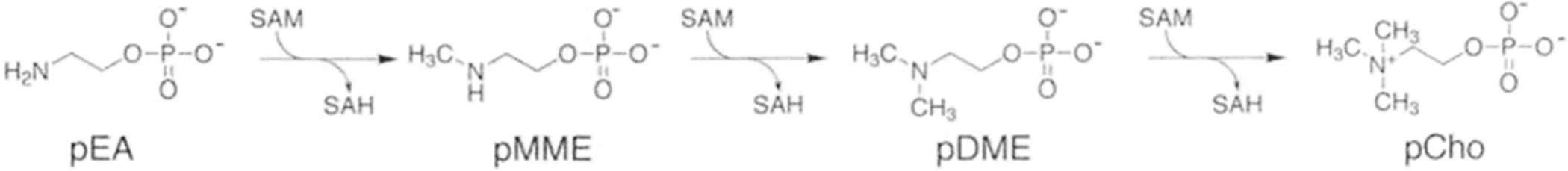
The sequential methylation of phosphoethanolamine (pEA) to phosphomonomethyletha-nolamine (pMME), phosphodimethylethanolamine (pDME), and phosphocholine (pCho) catalyzed by the PMT provides an entry point into the de novo choline pathway.
Biochemical analysis of each potential PMT from C. elegans showed that each enzyme is a SAM-dependent methyltransferase with distinct reaction specificity [9–10]. CePMTl catalyzes only the first methylation reaction (phosphoethanolamine -> phosphomonomethylethanolamine; pMME) in the pathway [10], whereas, CePMT2 completes the pathway by converting pMME to phosphodimethylethanolamine (pDME) and pDME to phosphoeholine [9].
Targeted gene silencing of either pmtl or pmt2 by RNAi yields pronounced phenotypes. Expression of each gene is required for normal growth and development of C. elegans at the LI (early larva), L4 (late larva), and dauer (stasis) stages of its lifecycle [9–10], For RNAi of pmtl,when the parental worm (P0) exposure began either as an LI or dauer larva, the phenotype was complete or highly penetrant (>95%) P0 sterility [10]. When exposure began as a P0 L4 larva, the phenotype was arrested development and lethality in the progeny (F1) at the L1 /L2 or the L3 larva stage. The worms show large lipid droplet accumulation in the intestines, suggesting pmt-l is also involved in the homeostasis of triacylglycerol and phosphatidylcholine [29], Since the pmtl RNAi experiments show that different larva stages were all developmentally impaired, pmtl is necessary at multiple stages in the life cycle. To determine if providing pathway metabolites reverses the pmtl RNAi phenotype, growth media were supplemented with ethanolamine, MME, DME, or choline [10]. Addition of ethanolamine did not reverse the pmtl phenotype for the LI, dauer, or L4 worms. Supplementing the media with MME (10 mM), DME (10 mM), or choline (30 mM) rescued the P0 sterility and FI larval arrest. These results confirm that CePMTl functions at a step before MME enters the pathway [10].
Similar experiments were performed using RNAi of pmt2 [9]. LI and dauer larva grown in the presence of E. coli expressing double-stranded RNA (dsRNA) from the pmt2 gene yielded sterile progeny. Likewise, feeding RNAi to L4 larva resulted in arrested development and lethality in the L1/L2 larva [10], Chemical complementation of the pmt2 phenotype provides information on the biological function of the protein [10], Growing C. elegans in media supplemented with choline (30 mM) reversed the pmt2 RNAi-mediated phenotype; however, chemical rescue of the phenotype did not occur in media supplemented with ethanolamine, MME, or DME. This indicates that CePMT2 is upstream of phosphocholine entry into the Kennedy pathway and is required for the utilization of pMME and pDME [10],
The observed phenotypes of both pmtl and pmt2 are similar to those reported in RNAi experiments targeting phosphatidylcholine metabolism in C. elegans that resulted in drastic reductions in fertility [16–23], which suggests a role of phosphatidylcholine in embryo development and worm growth. To provide insight on temporal and spatial expression pattern of PMT, several localization studies of PMT1 have been conducted. Transgenic worms containing a pmtlr::gfp construct showed strong expression of PMT1 in the hypodermis in the all larval stages and in seam cells in the adult stage [29]. Using wild-type worms, immunolocalization of PMT 1 suggests the presence of PMT 1 in the whole body of larva (data not shown) and in the vulva of adult worms (Fig. 3). Although RNAi experiments demonstrate a critical role for both PMT and recent localization studies examine the expression patterns of PMT1, tissue-specific expression patterns of PMT2 and co-expression patterns of both PMT in C. elegans remain to be examined.
Fig. 3. lmmunolocalization of PMT1 in C. elegans.

The permeabilized adult worms were incubated with primary PMT1 antibodies, and then with Alexa Fluor dye-conjugated anti-rabbit antibodies.
THREE TYPES OF PMT IN NEMATODES, PROTOZOANS, AND PLANTS
Although organisms ranging from nematodes to protozoans to plants utilize PMT for the SAM-dependent methylation of phosphoethanolamine to phosphocholine, the physical organization of the methyltransferasc domains in plants (type 1), protozoa (type 2), and nematodes (type 3) differs dramatically (Fig. 4). All of the PMT share four consensus sequence motifs (I-IV; Fig. 5) that define canonical SAM-binding domains in various methyltransferases [30].
Fig. 4. Methvltransferase Domain Organization in the Three Types of PMT.
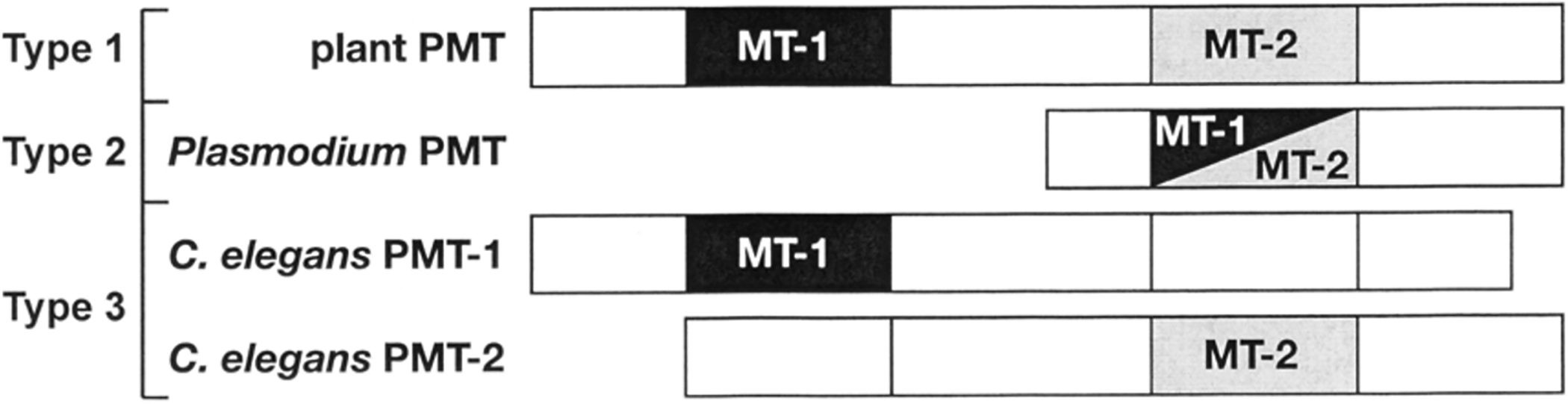
Plant PMT contain two methyltransferasc domains (MT) n tandem. MT-1 (black) catalyzes the conversion ofpEA to pMME. MT-2 (grey) catalyzes the methylation ofpMME to pDME and pDME o pCho. P. falciparum PMT contains a single bifunctional domain that catalyzes all three methylation reactions. Nematodes, like C. elegans, mcodc monofunctional PMT that catalyze either the MT-1 or MT-2 reaction. Nematode PMT also contain a vestigial MT domain (white jox).
Fig. 5. SAM-binding Motifs in the Two Domains of the PMT.
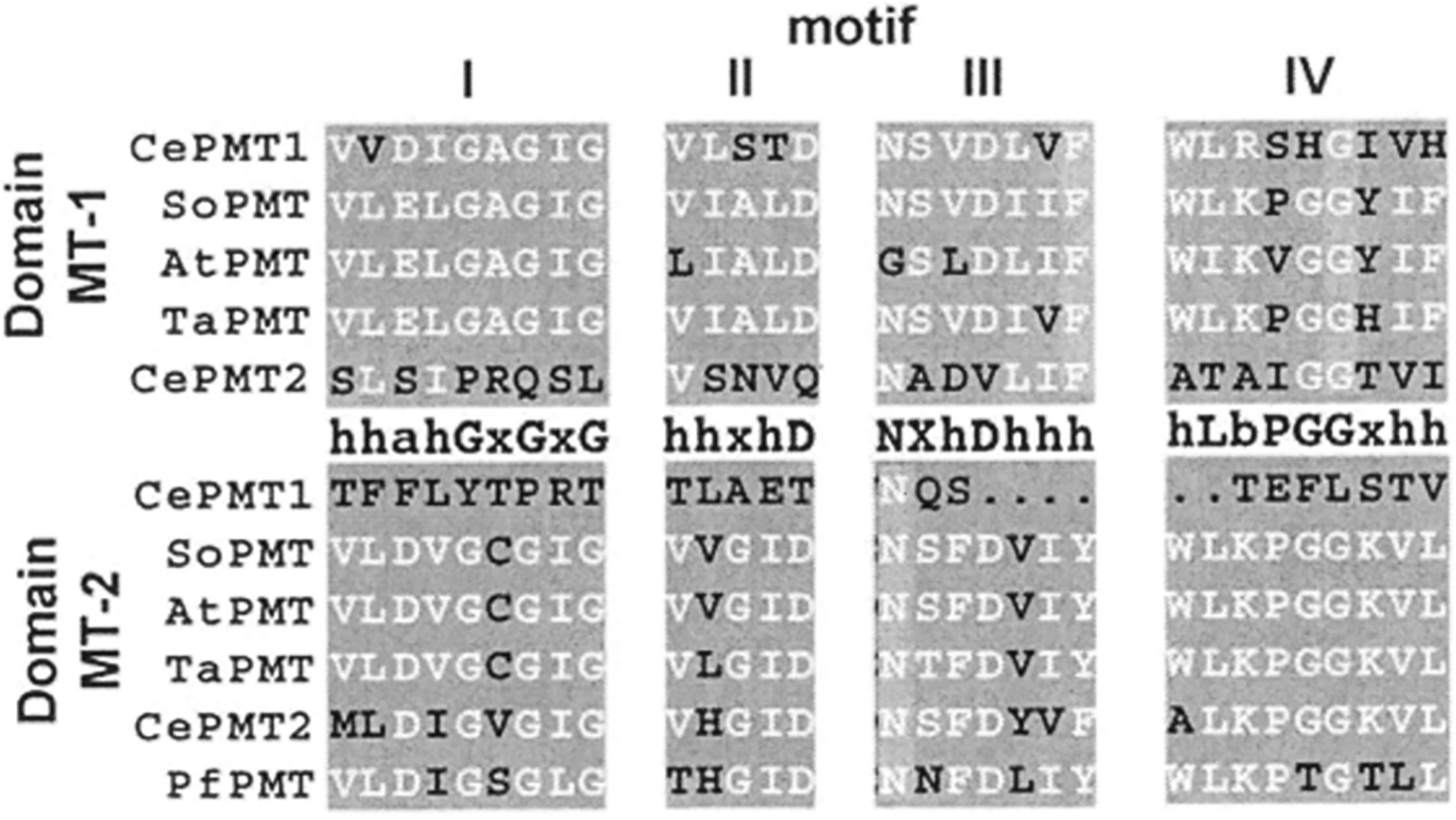
The canonical sequence is shown between the MT-1 (dark grey) and MT-2 (light grey) sequences (h=hydrophobic, a=acidic, b=basic, x=variable). The CePMT, SoPMT, AtPMT, TaPMT, and PfPMT are from C. elegans, spinach, Arabidopsis, wheat, and P. falciparum, respectively.
The plant PMT contain tandem methyltransferase domains in a single polypeptide [26–28] (Fig. 4). Generation of a truncated plant PMT variant lacking the C-terminal domain resulted in an N-terminal methyltransferase domain protein that only catalyzed the initial methylation of phosphoethanolamine to pMME, suggesting that the C-terminal methyltransferase domain performs the next two reactions to yield phosphocholinc [27],
In contrast to the plant PMT, the enzyme from the malarial parasite P. falciparum is half the length and consists of a single methyltransferase domain that acccpts all three phosphobase substrates [12–13] (Fig. 4). As with C. elegans, the PfPMT is required for normal growth and development of this protozoan parasite [13].
The PMT in C. elegans reveal a third scheme for organization of the methyltransferasc active sites (Fig. 4). Although the C. elegans PMT are distantly related to the plant enzymes, the dual methyltransferase domain architecture, as defined by the SAM-binding motifs, is not found in the C. elegans PMT [30]. The C. elegans genes encode proteins containing either the N-terminal methyltransferase domain (CePMT 1) or the C-tcrminal methyltransferase domain (CcPMT2). In each C. elegans PMT, the methyltransferasc consensus motifs in the other half of each protein shows major sequence differences (Fig. 5), suggesting that the biochemical function of these domains is lost. Our published work on the steady-state kinetics of these proteins shows that CePMT 1 indeed only catalyzes the conversion of phosphoethanolamine to pMME and that CePMT2 methylates pMME to pDME and pDME to phosphocholine with each enzyme using a random bi bi kinetic mechanism [9–10]. In C. elegans, gene duplication and divergence into separate enzymes that perform the initial and subsequent methylation reactions, respectively, has occurred.
Although the phosphobase methylation pathway is found in three different kingdoms - animals (C. elegans and other nematodes), protists (P. falciparum), and plants (such as Arabidopsis, spinach, and wheat) - the molecular basis for the structural evolution of the three types of PMT remains unresolved. Sequence examination of the genome of multiple parasitic nematodes of humans, including Ascaris lumhricoides (intestinal roundworm), Trichuris trichiura (whipworm), Necator americanus (hookworm) and Ancylostoma duode-nale (hookworm), as well as parasites of livestock and plants, indicates that these organisms all encode highly homologous (>50% sequence identity) type 3 PMT with shared methyltransferase domain architectures [9–11]. Moreover, the type 2 PMT are also conserved (>75% sequence identity) in all four Plasmodium species associated with malaria [12]. Overall, the PMT are not found in humans or other mammals and are essential for normal growth and development. Thus, these proteins are new targets for the development of anti-parasitic inhibitors.
CONCLUSIONS
Parasitic nematodes are a major cause of mortality and morbidity worldwide with an estimated 1 billion people infected by each species of major nematode parasites (i.e., Ascaris, Trichuris, and hookworms) [31–32], Moreover, increasing population levels and urbanization correlate with a rising prevalence of helminth infections [31]. Parasitic nematodes also result in considerable losses in livestock and domestic animals and more than $125 billion in crop damage annually [32–35]. The existing pharmacopoeia targeting nematode parasites is chemically varied [11], but the primary impetus to develop new broad-spectrum anthelmintics, much like the drive for novel antibiotics, is drug resistance. Currently, reports describe resistance to all compounds of the few anthelmintic classes available [36–37], The development of new anti-parasitic molecules requires better understanding of the biochemistry of nematodes and the identification of new pathways in these organisms that differ from the host species.
Genome science has transformed parasitology as sequence information on free-living and parasitic nematodes, including C. elegans, Brugia malayi (lymphatic filariasis), Onchocerca volvulus (river blindness), N. americanus (human hookworm), A. lumhricoides (human intestinal roundworm), and Trichinella spiralis (human-infective muscle cell nematode), and protozoans (Babesia, Eimeria, Giardia, Leishmania, Plasmodium, Toxoplasma, Trichomonas/Try-panosomes) allows for testing of hypothesis developed by comparative bioinformatics [32–35, 38], Exploration of these organisms is essential to provide fundamental insights on eukaryote and pathogen biology and to open opportunities to identify and exploit novel and essential metabolic features for drug discovery targeted at neglected diseases endemic in the developing regions of Africa, Asia, and the Americas.
For example, the free-living nematode C. elegans serves as a useful model system for studying nematode biology and as a platform for biochemical discovery and anti-parasitic drug development [32–35]. Using C. elegans as a model system provides a myriad of advantages: the worms are easily cultured, rapidly and prolifically reproduce, they are transparent and allow easy visualization of molecular, cellular, and organelle structure, molecular methods are well established, the developmental lineage of each cell is known, ongoing functional annotation of the genome continues through the WormBase (www.wormbase.org) consortium, and RNAi provides a valuable tool to analyze gene function [32–35]. Moreover, C. elegans shares physiological and pharmacological properties with many nematode parasites [32–35]. In contrast to the depth of developmental biology and genetic insight on C. elegans; however, the biochemical understanding of metabolism in C. elegans is limited in scope. This promises continued surprises about how this organism functions.
REFERENCES
- [1].Kent C Eukaryotic phospholipid biosynthesis. Annu Rev Biochem 1995; 64: 314–43. [DOI] [PubMed] [Google Scholar]
- [2].Kent C Regulatory enzymes of phosphatidylcholine biosynthesis: a personal perspective. Biochm Biophys Acta 2005; 1733: 53–66. [DOI] [PubMed] [Google Scholar]
- [3].Carman GM, Henry SA. Phospholipid biosynthesis in yeast. Annu Rev Biochem 1989; 58: 635–69. [DOI] [PubMed] [Google Scholar]
- [4].Sohlenkamp C, Lopez-Lara IM, Geiger O. Biosynthesis of phosphatidylcholine in bacteria. Prog Lipid Res 2003; 42: 115–62. [DOI] [PubMed] [Google Scholar]
- [5].Kanipes MI, Henry SA. The phospholipid methyltransferases in yeast. Biochim Biophys Acta 1997; 1348: 138–41. [DOI] [PubMed] [Google Scholar]
- [6].Vance DE, Walkey CJ., Cui Z. Phosphatidylcthanolamine N-methyltransferase from liver. Biochim Biophys Acta 1997; 1348: 142–50. [DOI] [PubMed] [Google Scholar]
- [7].Mudd SH, Datko AH. Phosphoethanolaminc bases as intermediates in phosphatidylcholine synthesis by l.emna. Plant Physiol 1986; 82: 126–35. ‘ [DOI] [PMC free article] [PubMed] [Google Scholar]
- [8].Datko AH, Mudd SH. Phosphatidylcholine synthesis: ditfering patterns in soybean and carrot. Plant Physiol 1988; 88: 854–61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [9].Palavalli LH, Brendza KM, Haakenson W, et al. Defining the role of phosphomethylethanolamine N-methyltransferase from Caenorhabditis elegans in phosphocholine biosynthesis by biochemical and kinetic analysis. Biochemistry 2006; 45: 6056–65. [DOI] [PubMed] [Google Scholar]
- [10].Brendza KM, Haakenson W. Cahoon RE, et al. Phosphoethanolamine N-methyltransferasc (PMT-1) catalyzes the first reaction of a new pathway for phosphocholine biosynthesis in Caenorhabditis elegans. Biochem J 2007; 404: 439–48. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [11].Jez JM. Phosphatidylcholine biosynthesis as a potential target for inhibition of metabolism in parasitic nematodes. Curr Enz Inhib 2007; 3: 133–42. [Google Scholar]
- [12].Pessi G, Kociubinski G, Mamoun CB. A pathway for phosphatidylcholine biosynthesis in Plasmodium falciparum involving phosphoethanolamine methylation. Proc Natl Acad Sci USA 2004; 101: 6206–11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [13].Pessi G, Choi JY, Reynolds JM, Voelker DR, Mamoun CB. In vivo evidence for the specificity of Plasmodium falciparum phosphoethanolamine methyltransferase and its coupling to the Kennedy pathway. J Biol Chem 2005; 280: 12461–6. [DOI] [PubMed] [Google Scholar]
- [14].Gee P, Kent C. Multiple isoforms of choline kinase from Caenorhabditis elegans: cloning, expression, purification, and characterization. Biochim Biophys Acta 2003; 1648: 33–42. [DOI] [PubMed] [Google Scholar]
- [15].Friesen JA, Liu MF, Kent C. Cloning and characterization of a lipid-activated CTP:phosphocholine cytidylyltransferase from Caenorhabditis elegans: identification of a 21-residue segment critical for lipid activation. Biochim Biophys Acta 2001; 1533: 8698. [DOI] [PubMed] [Google Scholar]
- [16].Sonnichsen B, Koski LB, Walsh A, et al. Full-genome RNAi profiling of early embryogencsis in Caenorhabditis elegans. Nature 2005; 434: 462–9. [DOI] [PubMed] [Google Scholar]
- [17].Kamath RS, Fraser AG, Dong Y, et al. Systematic functional analysis of the Caenorhabditis elegans genome using RNAi. Nature 2003; 421: 231–7. [DOI] [PubMed] [Google Scholar]
- [18].Rual JF, Ccron J, Korcth J, et al. Toward improving Caenorhabditis elegans phenomc mapping with an ORFeome-based RNAi library. Genome Res 2004; 14: 2162–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [19].GOnczy P, Echeverri C, Oegema K, et al. Functional genomic analysis of cell division in C. elegans using RNAi of genes on chromosome III. Nature 2000; 408: 331–6. [DOI] [PubMed] [Google Scholar]
- [20].Piano F, Schetter AJ, Mangone M, Stein L, Kemphues KJ. RNAi analysis of genes expressed in the ovary of Caenorhabditis elegans. CurrBiol 2000; 10: 1619–22. [DOI] [PubMed] [Google Scholar]
- [21].Simmer F, Moorman C, van der Linden AM, et al. Genome-wide RNAi of C. elegans using the hypersensitive rrf-3 strain reveals novel gene functions. PLoS Biol 2003; 1: E12.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [22].Maeda I, Kohara Y, Yamamoto M, Sugimoto A. Large-scale analysis of gene function in Caenorhabditis elegans by high-throughput RNAi. CurrBiol 2001; 11: 171–6. [DOI] [PubMed] [Google Scholar]
- [23].Fraser AG, Kamath RS, Zipperlcn P, Martinez-Campos M, Sohrmann M, Ahringer J. Functional genomic analysis of C. elegans chromosome I by systematic RNA interference. Nature 2000; 408: 325–30. [DOI] [PubMed] [Google Scholar]
- [24].Steenbergen R, Nanowski TS, Beigneux A, Kulinski A, Young SG, Vance JE. Disruption of the phosphatidylserine decarboxylase gene in mice causes embryonic lethality and mitochondrial defects. J Biol Chem 2005; 280: 40032–40. “ [DOI] [PMC free article] [PubMed] [Google Scholar]
- [25].Rontein D, Nishida I, Tashiro G, et al. Plants synthesize ethanolamine by direct decarboxylation of serine using a pyridoxal phosphate enzyme. J Biol Chem 2001; 276: 35523–9. [DOI] [PubMed] [Google Scholar]
- [26].Bolognese CP, McGraw P. The isolation and characterization in yeast of a gene for Arabidopsis S-adenosylmethionine:phospho-ethanolamine N-methyltransferase. Plant Physiol 2000; 124: 180013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [27].Nuccio ML, Ziemak MJ, Henry SA, Weretilnyk EA, Hanson AD. cDNA Cloning of phosphoethanolamine N-mcthyltransferase from spinach by complementation in Schizosaccharomyces pombe and characterization of the recombinant enzyme. J Biol Chem 2000; 275: 14095–101. [DOI] [PubMed] [Google Scholar]
- [28].Charron JB, Breton G. Danyluk J, Muzac I, Ibrahim RK, Sarhan F. Molecular and biochemical characterization of a cold-regulated phosphoethanolamine N-methyltransferase from wheat. Plant Physiol 2002; 129: 363–73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [29].Li Y, Na K, Lee HJ, Lee EY. Paik YK. Contribution of sams-l and pmt-l to lipid homoeostasis in adult Caenorhabditis elegans. J Biochem 2010; 149: 529–38. [DOI] [PubMed] [Google Scholar]
- [30].Kagan RM, Clarke S. Widespread occurrence of three sequence motifs in diverse S-adenosylmethionine-dependent methyltransferases suggests a common structure for these enzymes. Arch Biochem Biophys 1994; 310: 417–27. [DOI] [PubMed] [Google Scholar]
- [31].Chan WS. The global burden of intestinal nematode infections -fifty years on. Parasitol Today 1997; 13: 438–43. [DOI] [PubMed] [Google Scholar]
- [32].Jones AK, Buckingham SD, Sattelle DB. Chcmistry-to-gene screens in Caenorhabditis elegans. Nat Rev Drug Discov 2005; 4: 321–30. [DOI] [PubMed] [Google Scholar]
- [33].Hashmi S, Tawe W, Lustigman S. Caenorhabditis elegans and the study of gene function in parasites. Trends Parasitol 2001; 17: 38793. [DOI] [PubMed] [Google Scholar]
- [34].Kaletta T, Hengartncr MO. Finding function in novel targets: C. elegans as a model organism. Nat Rev Drug Discov 2008: 5: 38798. [DOI] [PubMed] [Google Scholar]
- [35].Renslo AR, McKcrrow JH. Drug discovery and development for neglected parasitic diseases. Nature Chem Biol 2006; 2: 701–10. [DOI] [PubMed] [Google Scholar]
- [36].Kaminsky R Drug resistance in nematodes: a paper tiger or a real problem? Curr Opin Infect Dis 2003; 6: 559–64, [DOI] [PubMed] [Google Scholar]
- [37].van den Enden E Pharmacotherapy of helminth infection. Expert Opin Pharma 2009; 10: 435–51. [DOI] [PubMed] [Google Scholar]
- [38].Hunter WN. Structure-based ligand design and the promise held for antiprotozoan drug discovery. J Biol Chem 2009; 284: 11749–53. [DOI] [PMC free article] [PubMed] [Google Scholar]


