Abstract
The Klebsiella pneumoniae carbapenemase-2 (KPC-2) is a common source of antibiotic resistance in Gram-negative bacterial infections. KPC-2 is a class A β-lactamase that exhibits a broad substrate profile and hydrolyzes most β-lactam antibiotics including carbapenems owing to rapid deacylation of the covalent acyl-enzyme intermediate. However, the features that allow KPC-2 to deacylate substrates more rapidly than non-carbapenemase enzymes are not clear. The active-site residues in KPC-2 are largely conserved in sequence and structure compared with non-carbapenemases, suggesting that subtle alterations may collectively facilitate hydrolysis of carbapenems. We utilized a nonbiased genetic approach to identify mutants deficient in carbapenem hydrolysis but competent for ampicillin hydrolysis. Subsequent pre–steady-state enzyme kinetics analyses showed that the substitutions slow the rate of deacylation of carbapenems. Structure determination via X-ray diffraction indicated that a F72Y mutant forms a hydrogen bond between the tyrosine hydroxyl group and Glu166, which may lower basicity and impair the activation of the catalytic water for deacylation, whereas several mutants impact the structure of the Q214-R220 active site loop. A T215P substitution lowers the deacylation rate and drastically alters the conformation of the loop, thereby disrupting interactions between the enzyme and the carbapenem acyl-enzyme intermediate. Thus, the environment of the Glu166 general base and the precise placement and conformational stability of the Q214-R220 loop are critical for efficient deacylation of carbapenems by the KPC-2 enzyme. Therefore, the design of carbapenem antibiotics that interact with Glu166 or alter the Q214-R220 loop conformation may disrupt enzyme function and overcome resistance.
Keywords: beta-lactamase, carbapenemase, antibiotics, antibiotic resistance, carbapenems, enzyme, enzyme structure, enzyme kinetics
Abbreviations: CRE, carbapenem-resistant Enterobacteriaceae; KPC-2, Klebsiella pneumoniae carbapenemase-2; MIC, minimum inhibitory concentration
Bacterial resistance to antibiotics is an emerging global health threat. β-Lactam antibiotics are the most widely prescribed class of antibiotics, accounting for roughly 65% of all antibiotic prescriptions. They are used to treat a wide range of bacterial infections (1, 2). Resistance to β-lactam antibiotics has steadily increased since the discovery of penicillin. In Gram-negative bacteria, the molecular basis of resistance to β-lactam antibiotics is a pathogen-produced enzyme, known as β-lactamase, which inactivates the drug by hydrolysis (2). β-Lactamases are grouped into four classes (A, B, C, and D) based on their sequence homology (3, 4). Classes A, C, and D utilize a catalytic serine to hydrolyze the β-lactam ring, whereas class B metalloenzymes require one to two zinc ions to hydrolyze β-lactam antibiotics (5, 6, 7). The evolution of these resistance enzymes as a result of widespread antibiotic use has led to the emergence of extended-spectrum β-lactamases and carbapenemases, which hydrolyze a broad range of β-lactam antibiotics (8, 9, 10). The class A enzyme, Klebsiella pneumoniae carbapenemase (KPC-2), has one of the broadest substrate profiles, hydrolyzing nearly all β-lactam antibiotics, including carbapenems (11, 12).
KPC-2 was originally isolated from an infected patient in North Carolina in 1996 and is frequently associated with carbapenem-resistant Enterobacteriaceae (CRE) (11, 12). The global dissemination of carbapenemases is of particular concern, as carbapenem antibiotics have been used as a last resort treatment for multidrug-resistant clinical infections (13). Carbapenemases of classes D (Oxa), B (NDM-1), and A (KPC-2) have established regional and endemic footholds across the globe, resulting in both the Centers for Disease Control and Prevention and World Health Organization classifying CRE as a level-one urgent public health threat (14, 15). CRE infections due to strains expressing KPC-2 have a high mortality rate and their dissemination is on the rise in the United States. In 2012 they were the prevalent strains in 42 states and are prevalent in all 50 states today (16, 17).
The kinetic model for hydrolysis of β-lactam antibiotics by serine β-lactamases is a two-step acylation–deacylation mechanism as shown in Scheme 1, where E is the enzyme, S is the substrate, EAc is the acyl-enzyme intermediate, and P is the hydrolyzed product (8, 18). Upon substrate binding (k1), a catalytic serine (Ser70) in the active site attacks the carbonyl carbon of the β-lactam ring, cleaving the ring and forming the acyl-enzyme intermediate (k2) (5). Glu166 then acts as a general base, activating a catalytic water that carries out a second nucleophilic attack that hydrolyzes the acyl-enzyme intermediate and releases the inactive product (k3) (5, 6).
Most clinically relevant class A β-lactamases such as CTX-M-14, SHV-1, and TEM-1 excel at hydrolyzing most β-lactams, yet they fail to hydrolyze and are, in fact, inhibited by carbapenems owing to very slow deacylation of the covalent acyl-enzyme intermediate (6, 19, 20).
The molecular basis for how KPC-2 can rapidly catalyze deacylation of carbapenem covalent intermediates, whereas other class A enzymes such as TEM-1 cannot remains unclear. However, carbapenems are thought to be poor substrates for non-carbapenemase enzymes owing to the presence of the pyrroline ring adjacent to the β-lactam ring and the small R1 6α-hydroxyethyl moiety (19, 20, 21, 22) (Fig. S1). For example, the 6α-hydroxyethyl group of the carbapenem can form a hydrogen bond with the deacylating water, reduce its nucleophilicity, and slow the deacylation rate (20). In contrast, the SFC-1 class A carbapenemase constrains the position of the hydroxyethyl group away from the water, which results in a faster deacylation rate due to enhanced nucleophilicity of the water. In addition, the pyrroline ring undergoes tautomerization upon acylation by the catalytic serine to form two isomers, Δ1 and Δ2, with the Δ1 isomer thought to be deacylated very slowly (22, 23, 24). Finally, structural studies of acyl-enzyme intermediates of the TEM-1 and SHV-1 enzymes with bound carbapenems have shown a conformational change of the acylated substrate where the carbonyl oxygen of the β-lactam ring is no longer in the oxyanion hole, thereby impeding the deacylation reaction (19, 20).
We recently used pre–steady-state kinetic analysis to show that the acylation rate of imipenem by the KPC-2 enzyme is fast (k2 = 209 s−1), whereas the deacylation rate is somewhat slower and rate limiting (k3 = 56 s−1) (25). However, the deacylation rate of imipenem by KPC-2 is orders of magnitude faster than that observed for noncarbapenemase class A β-lactamases such as TEM-1, which essentially does not deacylate carbapenems (26). However, the question of what structural features of KPC-2 allow it to rapidly deacylate carbapenems compared with noncarbapenemases remains ill-defined. The majority of the active-site residues in KPC-2 are also found in other class A β-lactamases and the structure of the KPC-2 and noncarbapenemase active sites are similar. Thus, it has been suggested that multiple subtle alterations, rather than gross distortions of the active site, may collectively be responsible for the rapid hydrolysis of carbapenems (Fig. 1) (27, 28). Thus, it is possible that multiple residues, including those not directly in the active site, may influence the structure and dynamics of the enzyme to facilitate deacylation of carbapenems.
Figure 1.
Structural alignment of class A β-lactamases.A, ribbon diagram structures of class A enzymes with KPC-2 colored tan (Protein Data Bank [PDB] id: 5UL8), CTX-M-14 colored green (PDB id: 4UA6), TEM-1 colored blue (PDB id: 1M40), and SHV-1 colored purple (PDB id: 4FH4). B, structural alignment of the active site region of class A β-lactamases. Color scheme as in A.
Because residues that contribute to carbapenem hydrolysis may reside anywhere in the enzyme, we reasoned that a nonbiased genetic approach might be effective in identifying residues that contribute to carbapenem hydrolysis. In particular, we set out to identify mutants that exhibit reduced carbapenem hydrolysis while retaining the ability to hydrolyze ampicillin. This approach can identify substitutions at residues that contribute specifically to carbapenem hydrolysis without a significant effect on hydrolysis of other β-lactam classes. For this purpose, we performed random mutagenesis by error-prone PCR to create a library of single-base substitutions in the KPC-2 gene and initially selected for growth in the presence of ampicillin. We then screened ampicillin-resistant clones for loss of imipenem resistance and identified several mutants including F72Y, Q128H, T215P, R220H, and T237A. Subsequent characterization by steady-state and pre–steady-state enzyme kinetics and X-ray crystallography demonstrated the substitutions act by slowing the deacylation rate of carbapenems. The F72Y mutant forms a new hydrogen bond between the tyrosine hydroxyl group and the carboxylate of the general base Glu166. This should lower basicity and impair the activation of the water for deacylation. The Q128H, T215P, and R220H are all located in or interact with the Q214-R220 loop, and the T216P substitution drastically alters the conformation of the loop. The T237A substitution is in the β-3 sheet bordering the active site and has been previously shown to impair carbapenem but not ampicillin hydrolysis (25). Therefore, the environment of Glu166 and the catalytic water and the conformation and dynamics of the Q214-R220 loop are key factors necessary for the rapid deacylation of carbapenem antibiotics by KPC-2.
Results
Identification of KPC-2 mutants deficient for carbapenem but not ampicillin hydrolysis
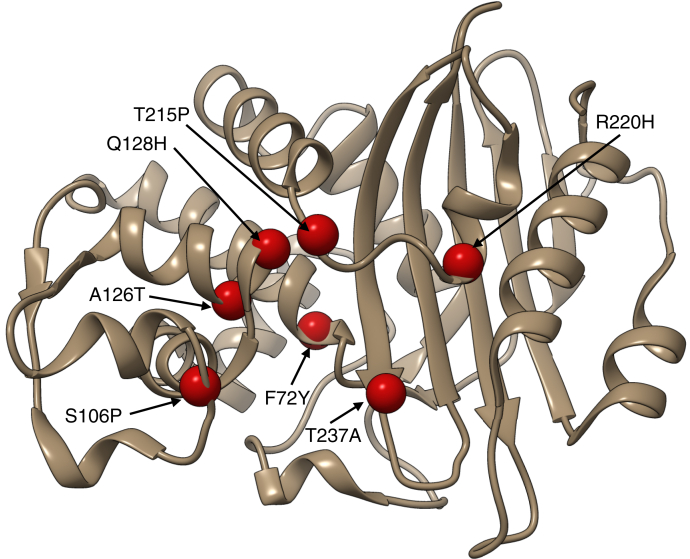
To identify mutations in KPC-2 that impair carbapenem but not penicillin hydrolysis, a library of KPC-2 mutant clones was generated using error-prone PCR. After PCR of the KPC-2 gene, the products were ligated into a bacterial protein expression plasmid (29). A total of 105 transformant colonies were pooled to create the library. Ampicillin was used as a representative penicillin, and imipenem and meropenem were used as representative carbapenems. To select for mutants that retain high penicillin resistance, the naive mutant library was transformed into Escherichia coli cells and spread on agar plates containing a concentration of ampicillin (100 μg/ml) that selects for high levels of KPC-2 catalytic activity. A total of 1764 surviving colonies were picked and individually grown in LB broth and subsequently diluted into medium containing a concentration of imipenem (0.4 μg/ml) that selects for wildtype levels of KPC-2 activity. The 103 ampicillin-resistant clones that did not grow in the medium containing imipenem were chosen for further study. DNA sequencing of imipenem-deficient mutants revealed a range of mutations, with most encoding single amino acid substitutions. The minimum inhibitory concentration (MIC) for E. coli containing each unique mutant was determined for ampicillin, imipenem, and meropenem to confirm the screening results (Table 1). The most promising mutants retained near-wildtype resistance levels toward ampicillin but exhibited increased susceptibility toward both imipenem and meropenem. Although many mutants retained high-level ampicillin resistance, several clones exhibited reductions in MIC for both ampicillin and carbapenems, despite the initial selection of the library for ampicillin resistance. Based on the MIC results, we selected the F72Y, Q128H, S106P, A126T, T215P, T216P, and T237A mutants for further characterization (Fig. 2).
Table 1.
Minimum inhibitory concentrations of E. coli containing empty vector control, wildtype, and mutant KPC-2 enzymes
| KPC mutants | Ampicillin (μg/ml) | Imipenem (μg/ml) | Meropenem (μg/ml) |
|---|---|---|---|
| E. coli XL-1 Blue pTP123 | 1.5 | 0.19 | 0.023 |
| KPC-2 | 64 | 0.75 | 0.19 |
| F72Y | 64 | 0.25 | 0.032 |
| S106P | 48 | 0.5 | 0.094 |
| A126T | 64 | 0.38 | 0.125 |
| Q128H | 64 | 0.5 | 0.125 |
| K212N | 16 | 0.25 | 0.064 |
| T215P | 64 | 0.25 | 0.064 |
| T216P | 16 | 0.19 | 0.047 |
| R220H | 64 | 0.38 | 0.064 |
| I221T | 6 | 0.19 | 0.032 |
| T237A | 48 | 0.25 | 0.047 |
| I259T | 32 | 0.5 | 0.094 |
Figure 2.
Location of amino acid residue positions substituted in KPC-2 mutants identified from carbapenem susceptibility screening. The wildtype KPC-2 β-lactamase structure (Protein Data Bank id: 5UL8) is shown as a tan ribbon. The location of residues substituted in the mutants is shown as red spheres.
Mutant enzymes identified in the genetic screen are deficient in carbapenem but not ampicillin hydrolysis
Each of the mutant enzymes chosen for further study was expressed and purified from E. coli for steady-state kinetic analysis (Table 2). The KPC-2 mutant enzymes can be placed into three groups based on the magnitudes of the observed kinetic parameters for penicillin, cephalosporin, and carbapenem hydrolysis. However, all of these mutants displayed enhanced ampicillin hydrolysis and decreased carabapenem hydrolysis, consistent with the method of selection.
Table 2.
Steady-state kinetic parameters for wildtype and mutant KPC-2 enzymes
| KPC mutants | AMP | PenG | CEP | IMI | MER | |
|---|---|---|---|---|---|---|
| KPC-2 | kcat (s−1) | 65 ± 2 | 19 ± 1 | 170 ± 10 | 48 ± 1 | 3.4 ± 0.1 |
| Km (μM) | 380 ± 30 | 46 ± 5 | 120 ± 10 | 220 ± 20 | 24 ± 1 | |
| kcat/Km (μM−1s−1) | 0.17 ± 0.02 | 0.41 ± 0.06 | 1.42 ± 0.13 | 0.22 ± 0.04 | 0.14 ± 0.04 | |
| F72Y | kcat (s−1) | 100 ± 10 | 9.8 ± 0.3 | 1.3 ± 0.1 | 0.15 ± 0.01 | 0.01 ± 0.002 |
| Km (μM) | 90 ± 10 | 27 ± 3 | 5.2 ± 1 | 0.51 ± 0.05 | 1.8 ± 0.1 | |
| kcat/Km (μM−1s−1) | 1.1 ± 0.17 | 0.36 ± 0.05 | 0.25 ± 0.07 | 0.29 ± 0.04 | 0.006 ± 0.001 | |
| T215P | kcat (s−1) | 460 ± 20 | 200 ± 10 | 190 ± 10 | 0.4 ± 0.01 | 0.16 ± 0.01 |
| Km (μM) | 460 ± 40 | 480 ± 50 | 380 ± 40 | 1.8 ± 0.2 | 11 ± 2 | |
| kcat/Km (μM−1s−1) | 1.0 ± 0.15 | 0.42 ± 0.06 | 0.50 ± 0.07 | 0.23 ± 0.02 | 0.015 ± 0.003 | |
| Q128H | kcat (s−1) | 340 ± 20 | 45 ± 2 | 270 ± 10 | 8.8 ± 0.2 | 0.89 ± 0.01 |
| Km (μM) | 550 ± 70 | 160 ± 20 | 240 ± 10 | 16.6 ± 1.3 | 4.1 ± 0.3 | |
| kcat/Km (μM−1s−1) | 0.62 ± 0.10 | 0.28 ± 0.05 | 1.13 ± 0.08 | 0.53 ± 0.05 | 0.22 ± 0.02 | |
| R220H | kcat (s−1) | 360 ± 10 | 130 ± 10 | 660 ± 20 | 20 ± 0.3 | 0.51 ± 0.03 |
| Km (μM) | 290 ± 20 | 180 ± 20 | 290 ± 30 | 33 ± 2 | 2.5 ± 0.4 | |
| kcat/Km (μM−1s−1) | 1.2 ± 0.11 | 0.72 ± 0.12 | 2.3 ± 0.07 | 0.61 ± 0.04 | 0.20 ± 0.04 | |
| T237Aa | kcat (s−1) | 150 ± 10 | 12 ± 1 | 47 ± 1 | 8.9 ± 0.3 | 0.11 ± 0.01 |
| Km (μM) | 17 ± 2 | 19 ± 2 | 51 ± 4 | 19 ± 2 | 1.3 ± 0.24 | |
| kcat/Km (μM−1s−1) | 8.8 ± 0.1 | 0.63 ± 0.09 | 0.92 ± 0.08 | 0.47 ± 0.06 | 0.09 ± 0.02 | |
| S106P | kcat (s−1) | 120 ± 10 | 12 ± 1 | 64 ± 2 | 22 ± 0.3 | 2.0 ± 0.1 |
| Km (μM) | 1150 ± 80 | 50 ± 6 | 210 ± 20 | 220 ± 10 | 28 ± 2 | |
| kcat/Km (μM−1s−1) | 0.10 ± 0.01 | 0.24 ± 0.04 | 0.30 ± 0.04 | 0.10 ± 0.01 | 0.07 ± 0.01 | |
| A126T | kcat (s−1) | 180 ± 10 | 16 ± 1 | 93 ± 2 | 27 ± 1 | 2.5 ± 0.1 |
| Km (μM) | 1550 ± 60 | 60 ± 10 | 240 ± 10 | 200 ± 20 | 30 ± 2 | |
| kcat/Km (μM−1s−1) | 0.12 ± 0.01 | 0.27 ± 0.06 | 0.39 ± 0.03 | 0.14 ± 0.02 | 0.08 ± 0.01 |
AMP, ampicillin; CEP, cephalothin; IMI, imipenem; MER, meropenem; PenG, benzylpenicillin.
T237A kinetic parameters for ampicillin, cephalothin, imipenem an meropenem are from Mehta et al. (2020).
The first group consists of F72Y and T237A (Table 2). Phe72 resides at the base of the active site pocket near the general base Glu166, and thus the F72Y substitution could directly impact catalysis (Fig. 2). This enzyme exhibits a modest (<2-fold) increase in kcat and a 4-fold decrease in KM, resulting in a 7-fold increase in catalytic efficiency (kcat/KM) for ampicillin hydrolysis (Table 2, Fig. S2). In the β-lactamase kinetic scheme, kcat is dependent on the magnitude of the acylation and deacylation rates (k2, k3) and kcat/KM reports on the rates up to the formation of the acyl-enzyme intermediate including binding affinity (k−1/k1 = KD) and the acylation rate (k2) (Fig. S2) (8, 18, 25). Therefore, the 7-fold increase in kcat/KM for ampicillin hydrolysis indicates an increased binding affinity (lower KD) and/or a faster acylation rate (k2) by the F72Y enzyme compared with wildtype KPC-2. In contrast, kcat for imipenem and meropenem hydrolysis by the F72Y enzyme is 320- and 340-fold slower, respectively (Table 2, Figs. S4 and S5). The F72Y enzyme also exhibits sharp reductions in KM for imipenem and meropenem (430- and 13-fold), which results in kcat/KM values that are similar to the wildtype KPC-2 enzyme. The decreased kcat values, coupled with the unchanged kcat/KM values, suggest that the major effect is due to a reduction in the deacylation rate (k3).
The effects of the F72Y substitution extend to other penicillins. The kinetics of penicillin G hydrolysis show modest effects for the F72Y enzyme, with kcat and KM each reduced 2-fold and kcat/KM unchanged relative to wildtype KPC-2 (Table 2, Fig. S6). Thus, although the F72Y substitution does not increase kcat and kcat/KM as observed for ampicillin, the effects on penicillin G hydrolysis are much smaller than the effects on carbapenem hydrolysis. We also determined if the F72Y substitution impacts cephalosporin hydrolysis by measuring kinetic parameters with cephalothin as substrate. The F72Y substitution has a similar effect on cephalothin hydrolysis as that observed for carbapenems, with kcat reduced 130-fold, KM reduced 20-fold, and kcat/KM reduced 5-fold (Table 2, Fig. S7). As with carbapenems, the large reduction in kcat and modest effect on kcat/KM suggests a decreased deacylation rate (k3) for cephalothin hydrolysis. Therefore, the F72Y substitution greatly decreases turnover of carbapenems and cephalosporins but has little impact on penicillin hydrolysis.
The T237A enzyme displays similar trends in kinetic parameters as F72Y, although the effects on carbapenem hydrolysis are not as extreme. Thr237 is located on the β-3 strand bordering the active site pocket, and the side chain Oγ has been shown to hydrogen bond to the C-3 carboxylate in structures of KPC-2 with faropenem and SFC-1 with meropenem (27, 30). Similar to F72Y, the T237A enzyme exhibits enhanced ampicillin hydrolysis and decreased carbapenem hydrolysis. For ampicillin there is a 2-fold increase in kcat and a 23-fold decrease in KM, resulting in a 50-fold increase in catalytic efficiency (kcat/KM) (Table 2, Fig. S3). Similar to F72Y, the T237A enzyme also displays lower kcat values for imipenem (5-fold) and meropenem (30-fold) along with lower KM values, which results in small changes in kcat/KM (Table 2, Figs. S4 and S5). As with F72Y, the decreased kcat values coupled with the unchanged kcat/KM values suggest that the T237A substitution reduces the deacylation rate (k3) for carbapenem turnover. Finally, the T237A enzyme also exhibits reduced kcat and KM and an unchanged kcat/KM for cephalothin hydrolysis indicating that, similar to F72Y, the substitution impacts carbapenem and cephalosporin hydrolysis similarly but enhances penicillin hydrolysis (Table 2, Fig. S7).
The second group of KPC-2 mutants includes single amino acid substitutions Q128H, T215P, and R220H that increase penicillin and cephalosporin hydrolysis but substantially decrease carbapenemase activity. Residues Thr215 and Arg220 are located in the 214 to 220 loop that is adjacent to the active site. Gln128 is located near the loop and makes a hydrogen bond to the main chain O of Trp210 and the main chain N and side chain Oγ of Thr215 to stabilize the loop (Fig. 2). The mutant enzymes in this group display increases in kcat (3- to 7-fold) for ampicillin hydrolysis as well as 4- to 10-fold increases in kcat/KM (Table 2, Fig. S3). The matched increases in kcat and kcat/KM suggest an increased rate of acylation of ampicillin by this group of mutants. Furthermore, these enzymes display a similar effect on penicillin G hydrolysis, suggesting the impact of the substitutions is general to penicillins (Table 2, Fig. S6). In addition, the Q128H, T215P, and R220H enzymes display moderate to 3-fold increased kcat, 2- to 3-fold increased KM, and similar kcat/KM values as wildtype KPC-2 for cephalothin hydrolysis (Table 2, Fig. S7). Therefore, as opposed to the F72Y and T237A substitutions, the Q128H, T215P, and R220H substitutions enhance the turnover of cephalosporins.
The Q128H, T215P, and R220H enzymes display decreased kcat values for carbapenem hydrolysis highlighted by T215P, which shows 120- and 26-fold decreased kcat values for imipenem and meropenem, respectively (Table 2, Figs. S4 and S5). Mutant enzymes in this group also exhibit 10- to 100-fold reduced KM values for carbapenem hydrolysis, resulting in kcat/KM values comparable with wildtype KPC-2. As with the F72Y enzyme, the decreased kcat values coupled with unchanged kcat/KM values suggest that the major effect is a reduction in the deacylation rate (k3) for carbapenem hydrolysis. The sharp reduction in kcat observed for the Q128H, T215P, and R220H mutants is restricted to carbapenem hydrolysis, a different kinetic signature from the F72Y and T237A enzymes where kcat is reduced for both carbapenem and cephalosporin substrates.
The third group of KPC-2 mutants includes substitutions at S106P and A126T. These substitutions have modest effects on the steady-state kinetics of both ampicillin and carbapenem hydrolysis. Residue Ser106 is in the 103 to 106 loop adjacent to the active site, and Ala126 is in the α helix near the loop containing the catalytic active-site residue Ser130 (Fig. 2). The S106P and A126T mutants display 2- to 3-fold increases in kcat for ampicillin hydrolysis indicating an increased k2 and/or k3 rate (Table 2, Fig. S3). In addition, these mutants exhibit 2- to 3-fold increases in KM and display kcat/KM values that are similar to wildtype KPC-2 (Table 2). These mutant enzymes display a modest 2-fold reduced kcat value for imipenem and meropenem hydrolysis indicating lower k2 and/or k3 rates. The KM values for carbapenems are unchanged and kcat/KM values are reduced 2-fold. Therefore, this group of mutants displays an increased turnover of ampicillin but a modestly reduced carbapenem turnover.
F72Y and T216P mutant enzymes display sharply reduced deacylation rates for carbapenems
Steady-state kinetics experiments, although informative, cannot provide rate constants for individual reaction steps such as acylation and deacylation. We chose the F72Y and T215P mutants from groups 1 and 2 described above for further characterization because they showed the most drastic reductions in carbapenem turnover (Table 2). We determined the acylation rate constant (k2) for imipenem hydrolysis for these mutants using stopped-flow kinetics under single turnover conditions in the presence of excess enzyme over substrate (25, 31, 32, 33) (Figs. 3 and 4, Table 3). The F72Y and T215P enzymes were found to have acylation constants (k2) that were much larger than the corresponding kcat values (500-fold for F72Y and 450-fold for T215P) indicating that acylation is not rate limiting for these mutants (Table 3, Figs. 3 and 4). The deacylation rate constant (k3) was calculated using the equation for kcat based on the kinetic scheme (Equation 3, Experimental procedures), and values of 0.2 and 0.4 s−1 were obtained for F72Y and T215P, respectively (Table 3). The values for k3 closely match the corresponding kcat values, as expected for a strongly rate-limiting step. We previously determined the k2 and k3 rate constants for imipenem turnover for the wildtype KPC-2 enzyme to be 220 and 60 s−1, respectively (25). The F72Y and T215P substitutions reduce the acylation rate by a relatively modest 3-fold and <2-fold, respectively. The major effect of the F72Y and T215P substitutions, however, is to reduce the rate constant for deacylation of imipenem by 300- and 150-fold, respectively. Although we did not determine rate constants for meropenem turnover, the similarities in steady-state kinetic parameters suggest that a reduction in the deacylation rate constant is also the major effect of the mutations. Furthermore, individual rate constants were not determined for ampicillin turnover for the F72Y and T215P mutants but the kcat values of 100 and 460 s−1 indicate that k3 must be at least this high. Therefore, the rate of deacylation of ampicillin by the mutants is at least two orders of magnitude faster than that for carbapenems.
Figure 3.
Single turnover kinetic analysis of KPC-2 F72Y mutant enzyme hydrolysis of imipenem. Imipenem, 10 μM, was used with increasing concentrations of F72Y enzyme as indicated below each plot. Absorbance is shown on the y-axis and time in seconds on the x-axis. The kobs obtained from fitting a single or double exponential equation is indicated for each plot. For fits to a double exponential equation both the fast (kobs-f) and slow (kobs-s) values are indicated. At the bottom right is the fit of the kobs values versus the F72Y enzyme concentrations to a hyperbola to obtain the acylation rate (k2).
Figure 4.
Single turnover kinetic analysis of KPC-2 T215P mutant enzyme hydrolysis of imipenem. Imipenem, 10 μM, was used with increasing concentrations of T215P enzyme as indicated below each plot. Absorbance is shown on the y-axis and time in seconds on the x-axis. The kobs values obtained from fitting a single or double exponential equation is indicated for each plot. For fits to a double exponential equation both the fast (kobs-f) and slow (kobs-s) values are indicated. At the bottom is the fit of the kobs values versus the F72Y enzyme concentrations to a hyperbola to obtain the acylation rate (k2).
Table 3.
Pre–steady-state kinetic parameters for acylation and deacylation rates for imipenem hydrolysis
| KPC mutants | k2 (s−1) | k3 (s−1) |
|---|---|---|
| Wildtypea | 210 ± 70 | 60 ± 20 |
| F72Y | 74 ± 7 | 0.2 ± 0.05 |
| T215P | 180 ± 30 | 0.4 ± 0.20 |
k2 and k3 values from Mehta et al., 2020.
F72Y substitution results in a new hydrogen bond to the Glu166 general base
Steady-state and pre–steady-state kinetic analyses indicate that the F72Y substitution significantly reduces kcat for carbapenem substrates as a direct result of a slower deacylation rate constant. To assess the molecular mechanism by which the F72Y substitution alters the carbapenem deacylation rate, X-ray crystallography was employed to determine the structures of the mutant enzyme with and without carbapenem bound in the active site (Table 4).
Table 4.
X-ray crystallography data collection and refinement statistics for KPC-2 mutant enzymes
| F72Y | F72Y/IMP | T215P | S70G/T215P/IMP | S70G/T215P/MPM | |
|---|---|---|---|---|---|
| Data Collection | |||||
| Space group | P1 | P1 | P212121 | P1 | P1 |
| a, b, c (Å) | 34.33, 37.31, 82.18 | 34.61, 37.12, 83.65 | 67.84, 71.66, 92.2 | 34.67, 37.74, 82.71 | 34.49, 37.29, 82.13 |
| a, β, γ (deg) | 92.43, 90.41, 94.62 | 88.12, 88.38, 85.48 | 90.0, 90.0, 90.0 | 91.54, 90.37, 93.84 | 91.39, 90.37, 93.79 |
| Resolution range (Å) | 37.15–1.81 (1.88–1.81) | (2.18–2.1) | 46.1–1.43 (1.48–1.43) | 37.64–1.82 (1.89–1.82) | 37.2–1.67 (1.73–1.67) |
| Rmerge (%) | 5.7 (7.6) | 3 (12.7) | 2.6 (30.81) | 4.6 (6.2) | 3.1 (8.0) |
| Rpim (%) | 5.2 (7.0) | 2.3 (10.1) | 2.6 (30.81) | 3.1 (4.2) | 2.2 (6.3) |
| I/σ | 10.84 (7.41) | 17.7 (6.32) | 11.12 (1.78) | 14.9 (12.5) | 16.3 (7.6) |
| CC(1/2) | 0.986 (0.98) | 0.999 (0.981) | 0.879 (0.999) | 0.997 (0.992) | 0.998 (0.986) |
| Multiplicity | 1.8(1.7) | 2.5 (2.5) | 1.9 (1.9) | 3 (3) | 2.6 (2.3) |
| Completeness (%) | 88.9 (87.63) | 98 (97.1) | 98.23 (93.8) | 96.6 (96.1) | 96.7 (92.2) |
| Wilson B-factor (Å2) | 6.98 | 27.85 | 14.67 | 10.87 | 12.53 |
| No. of unique reflections | 32,888 (3244) | 23,701 (2374) | 82,170 (7735) | 36,229 (3598) | 45,834 (4370) |
| Refinement | |||||
| Rwork, Rfree (%) | 15.8, 19.7 | 18.2, 21.9 | 15.73, 18.61 | 15.23, 18.71 | 17.08, 19.26 |
| No. of protein residues | 526 | 522 | 520 | 528 | 528 |
| No. of water molecules | 394 | 129 | 518 | 581 | 560 |
| Ramachandran favored (%) | 98.85 | 97.48 | 97.67 | 98.66 | 98.09 |
| Ramachandran outliers (%) | 0 | 0.39 | 0 | 0.0 | 0.19 |
| Root-mean-square deviation | |||||
| Bond lengths (Å) | 0.007 | 0.004 | 0.005 | 0.01 | 0.004 |
| Bond angles (deg) | 0.91 | 0.81 | 0.082 | 0.96 | 0.8 |
| Average B-factor (Å2) | 10.75 | 41.56 | 20.25 | 14.09 | 16.96 |
| Protein | 9.48 | 41.32 | 18.24 | 11.93 | 15.34 |
Values in parentheses in the body of the table indicate the highest-resolution shell.
The structure of wildtype KPC-2 has previously been determined with and without various substrates and inhibitors (30, 34, 35, 36). We utilized KPC-2 with a three amino acid truncation at the C terminus, which is known to facilitate crystallization (37), for our structure determinations. The F72Y apoenzyme structure was determined at 1.81-Å resolution in space group P1. The structure is very similar to the wildtype KPC-2 apoenzyme structure with the active-site residues superimposable (Fig. 5). The catalytic water molecule forms hydrogen bonds with side chains of the general base Glu166 as well as Ser70 and Asn170 (Figs. 5 and 6, A and B). A water is also present in the oxyanion hole, forming hydrogen bonds to the main chain nitrogens of Ser70 and Thr237. The most significant difference from the wildtype structure is the hydroxyl group on Tyr72 of the mutant enzyme. There is a small movement of the benzyl group of Tyr72 and the carboxylate of Glu166 to accommodate the Tyr72 hydroxyl group (Figs. 5 and 6, A and B). The hydroxyl group forms a new hydrogen bond with good geometry and distance (2.65 Å) to the side chain Oε2 of Glu166.
Figure 5.
Schematic illustration of the KPC-2 F72Y mutant enzyme.A, active site region of the F72Y enzyme. Hydrogen bonds are shown as thin black lines and water is shown as a red sphere. Carbon atoms are shown in tan, oxygen in red, and nitrogen in blue. B, structural alignment of the KPC-2 wildtype and F72Y enzyme active sites. Carbon atoms of wildtype KPC-2 (Protein Data Bank id: 5UL8) and F72Y are shown in light green and tan, respectively. Water molecules from the wildtype KPC-2 structure are shown in green and those from the F72Y structure are shown in red.
Figure 6.
Schematic illustration of the active site structures of KPC-2 F72Y apoenzyme and acyl-enzyme with imipenem.A, active site of KPC-2 F72Y apoenzyme from chain A. The deacylation water (HOH388) is in position to hydrogen bond with multiple active-site residues including Ser70, Glu166, and Asn170. A water is also found in the oxyanion hole (HOH258). The hydroxyl group of Tyr72 forms a hydrogen bond with Glu166 carboxylate Oε1. Carbon atoms are shown in tan, oxygen in red, and nitrogen in blue. B, KPC-2 F72Y apoenzyme chain B. The structure is nearly identical to chain A and makes the same interactions. C, KPC-2 F72Y imipenem acyl-enzyme structure, chain A. Imipenem (gray) is covalently linked to Ser70. The deacylation water is in position to hydrogen bond with multiple groups including the Ser70/imipenem ester oxygen, Glu166, Asn170, and the imipenem 6α-1R-hydroxyethyl oxygen. As in the apoenzyme, the hydroxyl group of Tyr72 forms a hydrogen bond with Glu166 carboxylate Oε1. D, KPC-2 F72Y imipenem acyl-enzyme structure, chain B. The structure is similar to that described for chain A except the 6α-1R-hydroxyethyl group is rotated such that the hydroxyethyl oxygen is not in position to form a hydrogen bond to the deacylation water. E, KPC-2 F72Y imipenem acyl-enzyme chain A showing the deacylation water and indicating the Burgi–Dunitz angle (θy) of 112.8° is not optimal for nucleophilic attack. F, KPC-2 F72Y imipenem acyl-enzyme chain B with θy = 113.1°.
We also determined the structure of the F72Y mutant acyl-enzyme complex with imipenem by soaking the crystal with the substrate. The structure was determined in space group P1 with two monomers in the crystallographic asymmetric unit and refined to 2.1-Å resolution. The ability to capture imipenem acylated to the catalytic Ser70 further underscores the impaired deacylation rate of the F72Y enzyme. The structure reveals minimal changes compared with the wildtype apoenzyme with the exception of the Tyr72 hydroxyl, which is hydrogen bonded to the side chain Oε2 of Glu166 in both chains in the asymmetric unit (Fig. 6, C and D). Imipenem is also covalently attached to the Ser70 Oγ in both chains in the asymmetric unit, although the conformation of the 6α-hydroxyethyl group is different between the chains, with the hydroxyethyl oxygen hydrogen bonded to the catalytic water in chain A and rotated away from the water in chain B (Fig. 6, C and D). The catalytic water is present in both chains and is in position to form hydrogen bonds with multiple groups including Ser70 Oγ, Glu166 Oε2 and Oε1, Asn170 Oδ1 and Nδ2, and Lys73 Nζ in chain A and the imipenem O in chain B. The water present in the oxyanion hole in the apoenzyme is displaced and occupied by the carbonyl oxygen of the ester bond of acylated imipenem. Note that, although the deacylation water is present, the ester bond of the acyl-enzyme is intact in both chains A and B (Fig. 6, C and D). The water is too distant, at 3.2 Å, for efficient nucleophilic attack on the carbonyl carbon of the ester. In addition, the Burgi–Dunitz angle (θy) of attack on the ester carbonyl (O7-C7-HOH) is 113° for the water in chains A and B while the optimal angle is 105° (38), indicating that it is not in optimal position for the deacylation reaction. The suboptimal distance and angle of nucleophilic attack of the water is consistent with the slow deacylation rate (38, 39, 40) (Fig. 6, E and F).
T215P substitution alters the conformation of the Q214-R220 active site loop to slow deacylation
The T215P substitution also drastically lowers kcat for carbapenem hydrolysis owing to a decreased deacylation rate. To understand how the T215P substitution lowers the deacylation rate, we determined the structure of the T215P apoenzyme and the S70G/T215P mutant enzyme with bound imipenem and meropenem. The T215P apoenzyme structure was determined at 1.43-Å resolution in space group P212121 with two monomers in the asymmetric unit. The structure reveals a large conformational change of the 214 to 220 loop that is adjacent to the active site (Fig. 7). The loop moves up and away from the active-site catalytic residues (Fig. 7). As an illustration, the Oγ of Thr216 is 2.67 Å from and forms a hydrogen bond with the Oγ of Thr235 in the wildtype KPC-2 structure. In contrast, the Oγ of Thr216 is 5.6 and 6.3 Å from the Oγ of Thr235 in chains A and B of the T215P structure, respectively. The different distances also reflect the fact that the loop conformations in chains A and B are different (Fig. 7). The altered conformations of the loop in chains A and B suggest that the loop is flexible in the T215P mutant. We, therefore, assessed the flexibility of the 214 to 220 loop by comparing the Wilson B factors between wildtype KPC-2 and the T216P mutant and found that the B-factors were much higher in the T215P mutant, consistent with increased flexibility relative to the wildtype enzyme (Fig. S8).
Figure 7.
Structure of the KPC-2 Q214-R220 loop and interactions with active-site residues.A, structure of the 214 to 220 loop in wildtype KPC-2 (light green). Oxygen and nitrogen are colored red and blue, respectively. Hydrogen bonds are shown as thin black lines. B, structure of the KPC-2 T216P apoenzyme 214 to 220 loop (pink). Chains A and B are shown in a structural alignment that indicated the loop is in different conformations in the chains. C, structural alignment of the 214 to 220 loop in wildtype KPC-2 (light green) versus chains A and B of the KPC-2 T215P apoenzyme. The 214 to 220 loop is in an altered, open conformation compared with that in wildtype KPC-2.
We attempted to determine the structure of the T215P enzyme with acylated imipenem or meropenem by rapid soaking of crystals but were not successful in trapping the intermediate. Therefore, we utilized an S70G/T215P mutant enzyme, where the substitution of Ser70 with Gly blocks the catalytic reaction (41). The structure of the S70G/T215P enzyme with imipenem bound was determined at 1.82-Å resolution in space group P1 with imipenem present in both chains of the asymmetric unit (Fig. 8, A and B). In both chains, the imipenem is hydrolyzed, showing the product complex. This is likely due to the time period of soaking that leads to slow hydrolysis. The 214 to 220 loop is found in different conformations in chains A and B (Fig. 8, A and B). In chain B, the loop is in an open position similar to the T215P apoenzyme structure, whereas in chain A the loop has shifted back to a closed conformation similar to that in wildtype KPC-2 (Fig. 8, A and B). In chain A, the hydrogen bond between the Oγ of Thr216 and Oγ of Thr235 is restored. In addition, both oxygens of the imipenem C3 carboxylate form hydrogen bonds to the Oγ of Thr235. The oxygens from the C7 carboxylate formed by hydrolysis make hydrogen bonds to Lys73 and the oxyanion hole nitrogens of Ser70 and Thr237. In contrast, in chain B the Oγ of Thr216 is 6.14 Å from the Oγ of Thr235 (Fig. 8B). However, the hydrolyzed imipenem makes similar contacts to the enzyme in chain B, with both oxygens of the imipenem C3 carboxylate forming hydrogen bonds to the Oγ of Thr235 and the oxygens from the carboxylate formed by hydrolysis making hydrogen bonds to Lys73 and the oxyanion hole nitrogens of Ser70 and Thr237 (Fig. 8B). Imipenem is in slightly different positions in the chains with the hydroxyethyl oxygen hydrogen bonding to Thr237 in chain A but not in chain B where the group is flipped in orientation. The near-wildtype conformation of the Q214-H220 loop in chain A suggests the presence of bound substrate may shift the equilibrium of the loop from the open conformation found in the T215P apoenzyme to the closed conformation of the S70G/T215P/imipenem chain A structure. However, the presence of the closed conformation in chain A and open conformation in chain B suggests both of the conformations are significantly populated. Coupled with the enzyme kinetics results, the structural findings also suggest that the conformational heterogeneity of the Q214-H220 loop hinders the deacylation of carbapenems but not penicillins or cephalosporins.
Figure 8.
Structure of the Q214-R220 loop region of KPC-2 S70G/T215P with hydrolyzed imipenem and meropenem bound in the active site.A, structure of the 214 to 220 loop in chain A of KPC-2 S70G/T215P. Imipenem product is shown in gray and carbons of the S70G/T215P enzyme are shown in salmon. Oxygen and nitrogen atoms are shown in red and blue, respectively. Hydrogen bonds are indicated by thin black lines. B, structure of the 214 to 220 loop for chain B of KPC-2 S70G/T215P with hydrolyzed imipenem. C, structural alignment of chains A and B of S70G/T215P with hydrolyzed imipenem and the KPC-2 wildtype apoenzyme (light green). D, structure of the 214 to 220 loop in chain A of KPC-2 S70G/T215P. Hydrolyzed meropenem product is shown in gray, and carbons of the S70G/T215P enzyme are shown in sienna. E, structure of the 214 to 220 loop for chain B of KPC-2 S70G/T215P with hydrolyzed meropenem. F, structural alignment of chains A and B of S70G/T215P with bound meropenem and the KPC-2 wildtype apoenzyme (light green).
We further determined the structure of the S70G/T215P enzyme with meropenem bound at 1.62-Å resolution in the P1 space group. The substrate meropenem is present in both chains of the crystallographic asymmetric unit (Fig. 8, D and E). As with the imipenem structure, meropenem is hydrolyzed and represents a product complex. The 214 to 220 loop is in the closed form similar to wildtype KPC-2 in both chains A and B in the S70G/T215P/meropenem structure. The hydrogen bond between the Oγ of Thr216 and Oγ of Thr235 is present in both chains (Fig. 8, D and E). The interactions between the S70G/T215P enzyme and meropenem are very similar to those observed in the S70G/T215P/imipenem structure, with both oxygens of the meropenem C3 carboxylate forming hydrogen bonds to the Oγ of Thr235 and the oxygens from the C7 carboxylate formed by hydrolysis making hydrogen bonds to Lys73 and the oxyanion hole nitrogens of Ser70 and Thr237 (Fig. 8, D and E). In addition, the Oγ of Thr216 in chain B directly forms a hydrogen bond with an oxygen of the C3 carboxylate group. It is also of note that there are differences in meropenem binding in chain A versus chain B, with the C3 carboxylate group shifted toward Thr216 and the 6α-hydroxyethyl group oxygen rotated away from Thr237 in chain B.
Taken together, the structural results show that, in the apoenzyme, the 214 to 220 loop undergoes a conformational change due to the T215P substitution such that it moves away from the active site and assumes multiple conformations with a larger B-factor than wildtype. Upon carbapenem binding, the loop closes to a conformation similar to that seen in the wildtype KPC-2 enzyme. The closed form of the loop is also less flexible; the B-factors of the loop residues in the closed form with substrate bound are much lower than in the open form and are similar to those observed in the wildtype enzyme (Fig. S8). However, despite the change in the 214 to 220 loop in the presence of substrate to a conformation similar to that of wildtype, the T215P enzyme displays a greatly reduced deacylation rate for carbapenem hydrolysis. However, based on the T215P apoenzyme and S70G/T215P/imipenem structures, the loop may exhibit significant conformational heterogeneity in solution that is not captured in the S70G/T215P/meropenem structure. We suggest this conformational heterogeneity leads to a loss of key contacts with the acylated carbapenem, leading to slower deacylation.
Discussion
We have identified mutations in KPC-2 β-lactamase that selectively block carbapenem hydrolysis while maintaining rapid hydrolysis of penicillins, a specificity profile that is similar to noncarbapenemase class A enzymes. The mutants could be placed in three groups based on enzyme kinetic parameters. The F72Y and T237A enzymes of the first group are characterized by enhanced turnover of ampicillin and sharply reduced turnover of both carbapenems and cephalosporins. Pre–steady-state single-turnover kinetic analysis showed that the deacylation rate for carbapenem hydrolysis is greatly reduced for the F72Y mutant. The structure of the F72Y enzyme with acylated impenem revealed that, in both chains in the asymmetric unit, the tyrosine substitution results in a new hydrogen bond to Oε2 of the general base, Glu166. This additional hydrogen bond would reduce its basicity and reduce activation of the deacylation water, slowing the deacylation rate for both carbapenems and cephalosporins (24, 42). In addition, the deacylation water is 3.3 Å from the ester carbonyl carbon of the acyl-enzyme and the Burgi–Dunitz angle (θy) of attack on the ester carbonyl (O7-C7-HOH) is 113° for the water in chains A and B, whereas the optimal angle is 105°, thereby slowing the deacylation rate of carbapenems (38). Although no structural data are available, a similar rationale could explain the slow turnover of cephalosporins. Furthermore, hydrogen bonding between the 6α-1R-hydroxyethyl oxygen of carbapenems and the deacylation water has been proposed to lower the nucleophilicity of the water and impair the deacylation reaction (19, 20, 27). The 6α-1R-hydroxyethyl substituent is in two orientations in the F72Y/imipenem structure with the hydroxyethyl oxygen in chain A positioned to make a hydrogen bond to the deacylation water but oriented away from the water in chain B. Therefore, if the chain A conformation is significantly populated in solution, it would also contribute to the reduced deacylation rate. Note, however, that this mechanism does not apply to the reduced turnover of cephalosporins, which lack the 6α-1R-hydroxyethyl group.
The various points described above provide a rationale for the slow deacylation of carbapenems by the F72Y enzyme but do not explain why ampicillin is hydrolyzed rapidly. The rates of acylation and deacylation of ampicillin by KPC-2 are not known, although they must be at least as fast as the kcat value of 65 s−1. It is formally possible that acylation of ampicillin is rate limiting and deacylation is much faster in the wildtype enzyme. In this scenario, the deacylation rate could actually be reduced by the F72Y substitution but still not be rate limiting and therefore not affect kcat. If the deacylation rate is not reduced by F72Y, it may be due to differences in the acyl-amide side chain. Ampicillin has an acyl-amide side chain appended to the β-lactam ring rather than a hydroxyethyl group (Fig. S1), which removes the possibility of the hydroxyethyl oxygen hydrogen bonding to the deacylation water and slowing the reaction. Because of the differences in structure between carbapenems and ampicillin, we suggest the water may be closer to the ester carbonyl of the ampicillin intermediate and at a better angle for nucleophilic attack, thereby compensating for the decreased basicity of Glu166. Note that we recently showed that alanine substitution of Asn170 in KPC-2, which forms hydrogen bonds to both the Glu166 carboxylate Oε1 and the deacylation water (Fig. 5), reduces the deacylation rate for imipenem by 2800-fold versus wildtype KPC-2 while reducing kcat for ampicillin hydrolysis by only 4-fold. Thus, carbapenem turnover is sensitive to the environment of Glu166, whereas ampicillin turnover is not (25). In aggregate, the results for the F72Y mutant suggest that deacylation of carbapenem acyl-enzyme intermediates by KPC-2 is highly sensitive to the environment and precise positioning of Glu166 and the deacylation water and thus it may be possible to modify carbapenems (or other β-lactams) to engage Glu166 or alter the hydrogen bonding network with Glu166 to impair deacylation.
Based on the F72Y mutant, we conclude that the environment around Glu166 is important for carbapenemase activity. Therefore, we examined the sequence conservation at residue 72 among class A carbapenemases and noncarbapenemases (Fig. S9A). Phenylalanine is found at position 72 in all 14 class A carbapenemases examined (Fig. S9A). However, Phe72 is also found in noncarbapenemase class A enzymes such as TEM-1 and SHV-1. As seen in Figure 5, Glu166 Oε1 and Oε2 in KPC-2 form hydrogen bonds with Lys73, Asn170, and the deacylation water. However, the environment also includes hydrophobic residues that are in van der Waals contact with the Glu166 carboxylate oxygens including Phe72, Leu167, and Leu169 (Fig. S9, B and C). This hydrophobic pocket could influence the pKa and enhance basicity of Glu166. For example, in triosphosphate isomerase, a hydrophobic interaction of an active-site isoleucine with the glutamate general base results in a 2-unit change in the pKa of the side chain of glutamate to enhance basicity for the isomerization reaction (43, 44). Of the hydrophobic residues surrounding Glu166 in KPC-2, Leu167 is unique to the carbapenemases. Position 167 is proline in most class A enzymes, which would reduce hydrophobicity and may influence the basicity of Glu166.
Similar to F72Y, the Thr237A mutant exhibits enhanced turnover of ampicillin but reduced turnover of carbapenems and cephalosporins. Substrate, acyl-enzyme, and product structures of carbapenems in complex with KPC-2 and SFC-1, including those presented here, show Oγ of Thr237 makes hydrogen bonds to the C3 carboxyl of carbapenems (27, 30). The T237A substitution eliminates the hydrogen bond to the C3 carboxyl of carbapenems. We previously showed that the T237A substitution decreases both k2 and k3 values for imipenem hydrolysis, compared with wildtype KPC-2 so that neither step is rate limiting (25). This suggests that the hydrogen bond to the imipenem C3 carboxylate by Thr237 is important for both acylation and deacylation rates.
The second group of mutants includes Q128H, T215P, and R220H. These residues are positioned to impact the structure and dynamics of the 214 to 220 loop. Enzyme kinetic analysis shows that the mutants in this group significantly increase kcat and kcat/KM for ampicillin hydrolysis and decrease kcat but not kcat/KM for carbapenem hydrolysis. Single turnover experiments indicate that the decrease in kcat for T215P is due to a large reduction in the deacylation (k3) rate, and structural studies indicate that the substitution results in an open 214 to 220 loop and substantial conformational heterogeneity. In the presence of imipenem or meropenem, the 214 to 220 loop closes to a conformation similar to that observed in wildtype KPC-2. However, the fact that one of the chains for the S70G/T215P imipenem structure retains the open conformation suggests that there is still conformational heterogeneity in the loop. Examination of the structures suggests that the conformational heterogeneity may impact interactions of the loop residue Thr216 with the carbapenem. Note that, in both the S70G/T215P imipenem and meropenem structures, the main chain O of Thr216 makes an interaction with the C3 carboxylate through a bridging water (Fig. S10). The Thr216 side chain Oγ also makes a direct hydrogen bond to the C3 carboxylate in the S70G/T215P meropenem structure (Fig. 8E). The Thr216 main chain O also makes a hydrogen bond with the C3 carboxylate through the same bridging water in the structure of a faropenem hydrolysis product with KPC-2 (PDB id: 5UJ4) (30). In addition, the Thr216 main chain O forms a hydrogen bond with the functional equivalent of the C3 carboxylate via the same water in the structure of KPC-2 with the boronic acid inhibitor vaborbactam (PDB id: 6TD0) and with the diazabicyclooctane inhibitor avibactam (PDB id: 4BZE) (36) (Fig. S10). Furthermore, the hydrogen bond of the Thr216 main chain O with the C3 carboxylate occurs through a similarly placed water in structures of the SFC-1 carbapenemase S70A mutant with bound meropenem substrate and an E166A mutant with acylated meropenem (PDB ids: 4EUZ, 4EV4) (27). These observations suggest that the interaction between the Thr216 main chain O with the C3 carboxylate through the conserved water is an important stabilizing force for bound carbapenems. Furthermore, the conformational heterogeneity induced by the T215P substitution disrupts this interaction. Note that, since the Thr216 interaction with carbapenems is through the main chain O, substitution of this residue would not necessarily change enzyme activity. However, the T215P substitution alters the structure of the Q214-R220 loop, which is, in effect, a main chain mutation of Thr216. Finally, since the T215P substitution disrupts the deacylation reaction, we propose that the Thr216 interaction is most important for stabilizing the acyl-enzyme intermediate. This model is consistent with the acyl-enzyme structures of the noncarbapenemase TEM-1 and SHV-1 β-lactamases with imipenem and meropenem where the acylated carbapenem has moved out of the oxyanion hole to a position not consistent with deacylation (19, 20). Of interest, the 214 to 220 loop is in an altered position in the noncarbapenemase TEM-1 and SHV-1 enzymes relative to that in KPC-2 and SFC-1, such that an interaction between the main chain O of residue 216 with the carbapenem C3 carboxyl is not possible.
The Q128H and R220H mutants exhibit similar kinetic profiles for ampicillin and carbapenem hydrolysis as T215P, although with ∼10-fold higher kcat values for carbapenem turnover (Table 2). Gln128 is located near the 214 to 220 loop and makes hydrogen bonds to the main chain O of Trp210 and main chain N and side chain Oγ of Thr215 to stabilize the loop. Therefore, the Q128H mutation may act through a similar mechanism as that described for T215P, where destabilizing the conformation of the loop disrupts the interaction between Thr216 and the C3 carboxylate of the carbapenem acyl-enzyme intermediate. In addition to its placement in the 214 to 220 loop, the guanidinium group of Arg220 also makes hydrogen bonds to stabilize the position of Thr237. Since disruption of the Thr237 Oγ/carbapenem interaction reduces turnover, the R220H substitution may indirectly do the same. Active site loop structures have been shown to play an important role in β-lactamase substrate specificity (8, 45). It has been extensively documented that changes in the 164 to 179 omega loop, which contributes to the floor of the active site and contains Glu166, result in altered substrate specificity of class A β-lactamases (8, 45, 46, 47, 48, 49, 50, 51). Changes in the conformation of the 103 to 106 loop are also associated with changes in substrate specificity (41). The identification and characterization of the group 2 mutants, Q128H, T215P, and R220H, suggests that the 214 to 220 loop is an important contributor to KPC-2 substrate specificity in that changes in the conformation of the loop reduce carbapenem turnover but do not affect hydrolysis of penicillins or cephalosporins.
The S106P and A126T mutants from the third group display increased ampicillin hydrolysis but moderately decreased levels of carbapenem hydrolysis. Ser106 is part of the 103 to 106 loop that contains Trp105. The structures of KPC-2 in complex with hydrolyzed faropenem and substrate along with the structure of the acyl-enzyme complex of the SFC-1 carbapenemase with meropenem reveal that Trp105 makes hydrophobic interactions with the pyrrole ring and hydroxyethyl group of carbapenems (27, 30). The S106P substitution would eliminate a hydrogen bond between the Ser106 Oγ and the main chain O of Val103, which could impact the conformation of the 103 to 106 loop and the position of Trp105. It has been shown previously that the N106S natural variant of CTX-M β-lactamases changes the conformation of the loop and thereby alters substrate specificity (41). In contrast, Ala126 lies in the hydrophobic core of the α-helical domain of KPC-2 and class A β-lactamases. Modeling suggests that the introduction of threonine would destabilize the core, which could impact the positioning of the loop containing Ser130 and Asn132. However, it is unclear how this would impact carbapenem hydrolysis.
It has been suggested that the pyrroline ring form of a carbapenem in the acyl-enzyme intermediate influences the rate of the deacylation reaction in serine β-lactamases (21, 22, 23, 24). Three forms of the pyrroline ring have been observed in carbapenem acyl-enzyme structures, including a 2-pyrroline enamine (Δ2) form and two epimeric 1-pyrroline imine forms ((R)-Δ1 and (S)-Δ1) (24, 27, 52, 53, 54). Structures of BlaC acyl-enzyme intermediates with doripenem after a short soaking time revealed the Δ2 form, whereas a longer soaking time yielded a structure with the Δ1 form (24). In the Δ1 form, the hydroxyethyl group was in an altered position and formed a hydrogen bond to Glu166, reducing its basicity and consistent with slower deacylation (24). Recent NMR studies suggest that the Δ2 and (R)-Δ1 forms are preferentially produced in the active site and the (S)-Δ1 form is then produced nonenzymatically (55). Therefore, we examined the KPC-2 F72Y imipenem acyl-enzyme and S70G/T215P product structures to determine the tautomeric form of the pyrroline ring. The structures are not of sufficient resolution to determine if the C2 position is protonated, although the position of the R2 side chain sulfur relative to the plane of pyrroline ring does provide an indication of tautomeric state (Fig. S11). All of the structures have two chains in the asymmetric unit. For the KPC-2 F72Y imipenem acyl-enzyme, the R2 group sulfur is clearly below the plane of the pyrroline ring for both chains, with angles of 49° and 55° from the plane defined by the C5-C3-C2 atoms indicating the (S)-Δ1 form (Fig. S11, A and B). The R2 group sulfur atom in the S70G/T215P impenem product complex is below the pyrroline plane in chain A (41°) and near the plane for chain B (13°), suggesting that chain A is in the (S)-Δ1 form and chain B is in the enamine Δ2 form (Fig. S11, C and D). The S70G/T215P meropenem product complex reveals that the angle of the sulfur is 29° and 23° below the plane for chains A and B, which is consistent with the (S)-Δ1 form (Fig. S11, E and F). Note that the hydroxyethyl group of the F72Y imipenem-acyl-enzyme structure is in different conformations in chains A and B (Fig. 6, C and D), but this is not correlated to the tautomer of the pyrroline ring as it is in the (S)-Δ1 form for both chains A and B (Fig. S11, A and B). In addition, the hydroxyethyl group of the S70G/T215P meropenem product structure is different in chains A and B but is also not correlated with the pyrroline ring form, which is (S)-Δ1 for both chains. In contrast, the altered conformation of the hydroxyethyl group in chains A and B for the S70G/T215P imipenem product structure is matched by a change in the pyrroline ring form ((S)-Δ1 and Δ2) (Fig. 8 and Fig. S11, C and D). However, it is noteworthy that the (S)-Δ1 form is produced nonenzymatically after acyl-enzyme or product formation and it is possible that (S)-Δ1 is produced during the time frame of soaking and data collection for structure determination and is not relevant to the form undergoing the deacylation reaction (55).
Based on the similar structures of the active sites of KPC-2 and noncarbapenemases such as TEM-1 and SHV-1, it has been suggested that multiple subtle structural changes could be responsible for the carbapenemase activity of KPC-2 (27, 28). A key finding from this work is that deacylation of carbapenems is more sensitive to the environment of Glu166, possibly owing to altered basicity, than is deacylation of penicillins. This is consistent with previous studies on noncarbapenemase enzymes showing that Δ1 tautomer of carbapenems is associated with the hydroxyethyl group by forming a hydrogen bond to the Glu166 to reduce its basicity and slowing the deacylation rate (20, 24). In addition, we show that the conformation of the Q214-R220 loop is critical for efficient deacylation of carbapenems but not penicillins by KPC-2 and interpret this as being due to disruption of an interaction between Thr216 and the carbapenem C-3 carboxylate. The identification of the T237A mutant also supports a role for a hydrogen bond between Thr237 and the C-3 carboxylate as an additional contribution to carbapenem hydrolysis (25, 56). Clearly, these are not the only determinants of carbapenem hydrolysis by KPC-2 but represent additional factors that together add up to efficient hydrolysis of carbapenems.
Experimental procedures
Bacterial strains and plasmids
The KPC-2 gene was inserted into the previously constructed pTP123 plasmid vector, as described (29). The KPC-2-pTP123 plasmid was introduced into E. coli XL1-Blue (57) for construction of a random mutant library of KPC-2. Library selection, library screening, and MICs were performed in XL1-Blue E. coli cells, and wildtype KPC-2 and mutant KPC-2 enzymes were transformed into E. coli RB791 for protein purification (58).
Minimum inhibitory concentration determinations
The E-test strip (Biomerieux) method was used to determine the MICs for ampicillin, imipenem, and meropenem. The wildtype, mutant KPC-2 enzymes, and empty pTP123 plasmid were transformed into E. coli XL1-Blue cells. Transformed colonies were grown overnight in 5 ml LB broth containing 12.5 μg/ml chloramphenicol. The cultures were then diluted to an absorbance of 0.05 and spread onto LB agar plates, and the E-test strip containing either ampicillin, imipenem, or meropenem was placed onto the agar surface. The plates were then incubated at 37 °C overnight, and the concentration of antibiotic sufficient to inhibit bacterial cell growth at the markings denoted on the E-test strip was reported.
Identification of KPC-2 mutants resistant to ampicillin but sensitive to imipenem
To select for KPC-2 mutants that retain high penicillin resistance and poor carbapenem resistance, the naive mutant library was transformed into E. coli XL1-Blue cells and spread on LB agar plates containing a concentration of 100 μg/ml of ampicillin. The 100-μg/ml concentration of ampicillin was chosen based on pilot experiments examining the plating efficiency of E. coli containing the wildtype KPC-2 gene whereby 100 μg/ml allows colonies to form, whereas higher concentrations reduce colony-forming units. Therefore, 100 μg/ml ampicillin selects for mutants with near-wildtype levels of KPC-2 β-lactamase activity toward ampicillin. Colonies that grew under ampicillin selection were then picked to inoculate a 96 deep well plate containing LB supplemented with 12.5 mg/ml chloramphenicol and incubated overnight at 37 °C with shaking at 150 rpm. The absorbance of the culture at 600 nm was measured to evaluate growth. Next, the cultures were tested for carbapenem resistance by diluting 1:100,000 into LB broth containing 0.4 μg/ml imipenem in sterile, clear-bottom 96-well plates and incubating overnight at 37 °C and 150 rpm. The absorbance at 600 nm was measured to evaluate growth. Plasmid DNA was isolated from cultures that failed to grow in LB-imipenem broth, and the identity of the mutations was determined by DNA sequencing.
Protein expression and purification
The wildtype KPC-2 and KPC-2 mutant enzymes were encoded on the pTP123 plasmid and transformed into E. coli RB791 cells for protein expression and purification. Cells were grown overnight at 37 °C in 10 ml LB broth containing 12.5 μg/ml chloramphenicol (LB-CMP), then back diluted into 1 l LB-CMP. The cultures were then grown at 37 °C until A600 ∼0.8, at which time the cells were induced by the addition of 200 μl 1 M IPTG and grown overnight at 23 °C. The cultures were then collected and pelleted by centrifugation at 4 °C for 15 min at 5000 rcf.
The cell pellet was resuspended in 10 mM Tris pH:8.0, 20% (w/v) sucrose and shaken at 4 °C for 30 min. A periplasmic release was performed by the addition of an equal volume of ice-cold water and incubation by shaking at 4 °C for 1 h. The periplasmic fraction was isolated by two centrifugations at 10,000 rcf for 15 min. To remove impurities, the fraction was flowed over a SP-Sepharose fast-flow (SP-FF, GE Healthcare) cation-exchange column. To bind free KPC-2 enzyme, the supernatant pH was adjusted to 6.0 with 1 M MES buffer and passed through an SP-FF column. A NaCl gradient was used to elute the bound protein. Eluted samples containing KPC-2 were pooled, concentrated, and further purified to homogeneity by size-exclusion chromatography (GE S75 increase Sepharose column, Akta pure HPLC). Fractions were assessed for purity by SDS-PAGE and samples of ≥90% pure were pooled and concentrated for further use.
Steady-state enzyme kinetic analysis
In vitro kinetic parameters for β-lactam antibiotic hydrolysis were determined as described (25). Steady-state kinetic parameters were determined for the β-lactam substrates ampicillin, imipenem, and meropenem by monitoring hydrolysis for each substrate using a DU800 spectrophotometer at 235, 295, and 295 nm, respectively. Extinction coefficients used for ampicillin, imipenem, and meropenem were −900, −9000, and −10,940 M−1 cm−1 respectively. Experiments were performed at 25 °C in 50 mM phosphate pH 7.0, 0.1 mg/ml BSA with a minimum of two replicates per substrate concentration tested. The average and standard deviation of the replicates was used with GraphPad Prism 8 to fit initial velocity measurements to the Michaelis–Menten equation v = Vmax [S]/(KM+[S]) to determine kcat and KM values.
Determination of acylation (k2) and deacylation (k3) rate constants
The acylation rate constant (k2) was determined by monitoring the rate of imipenem acylation (Δε295: −9000 M−1 cm−1) using a Kintek SF-2001 stopped-flow apparatus. Single-turnover kinetic conditions were established by using excess amounts of enzyme and low concentration of imipenem (1–10 μM). The reactions were performed at 25 °C in 50 mM phosphate pH 7.0. The enzyme concentration was increased until the rate of acylation was saturated. For each kinetic experiment, 1000 data points were collected and at least three kinetic traces were collected to ensure high-quality data. The observed data were fit to either a single or double exponential equation to determine the rate of observed acylation, where At is the absorbance at time t, A1 and A2 are absorbance at At0 and Atplateau, kobs is the apparent rate from the reaction, and biphasic fast and slow phases are f and s, respectively.
| (1) |
| (2) |
The individual kobs data at each enzyme concentration were then fit to a hyperbola to determine k2. From there, the deacylation rate constant (k3) was calculated using the determined values of kcat and k2 using Equation 3.
| (3) |
Solving Equation 3 for kcat in terms of k3 yields k3 = (k2∗kcat)/(kcat-k2). The error in the calculated value of k3 was assessed by propagating the errors in kcat and k2 through the equation by adding the absolute errors for the sum or difference in the denominator to obtain the absolute and the percentage error in kcat-k2. The percent error of the numerator, (kcat∗k2), was calculated by adding the percent error for kcat and k2, and the final, percent, and absolute errors on k3 were obtained by adding the percent error of the numerator and denominator to yield the percentage error in k3.
X-ray crystallography
A truncation to the C terminus of the KPC-2 enzyme that facilitates crystallization was introduced via site-directed mutagenesis using the following oligonucleotides (34).
Δ292 to 295: 5’-GAGGGATTGGGCTAAAACGGGCAGTAATCTAGAGTCGACC
S70G: 5’-CCCACTGTGCGGCTCATTCAAG
The F72Y and S70G/T215P mutant enzymes were purified as described above and were crystallized by mixing 200 nl 25% PEG 8000, 0.1 M KsCN, 0.1 M sodium acetate, pH 4.5 with 10 mg/ml of protein. The T215P mutant was crystallized by mixing 2.5 M NaCl, 0.1 M sodium acetate pH 5.1 with 10 mg/ml of protein. Crystals were soaked with 10 mM imipenem or meropenem in their respective mother liquor for 30 min to 1 h before being flash frozen in liquid nitrogen. The data for all enzyme structures were collected at the Berkeley Center for Structural Biology at the Advanced Light Source on synchrotron beamlines 5.0.1, 8.2.1, and 8.2.2. Data were processed using xia2, xia2/DIALS, and iMOSFLM (59). Molecular replacement was performed using expertMR phaser or MOLREP (60) in the CCP4i Suite (61). Coot was used to fit the model to the density, and phenix.refine was used to refine the data (62, 63, 64). All figures were modeled and created in the UCSF Chimera program (65).
Data availability
Accession codes: Coordinates and structure factors have been deposited in the Protein Data Bank under accession codes 7LJK, 7LLH, 7LK8, 7LLB, and 7LNL. All relevant data associated with the paper are available upon request from the corresponding author.
Supporting information
This article contains supporting information.
Conflict of interest
The authors declare they have no conflicts of interest with the contents of this article.
Acknowledgments
The authors thank Hiram Gilbert for discussions and comments on the manuscript. ALS-ENABLE beamlines are supported in part by the National Institutes of Health, National Institute of General Medical Sciences, grant P30 GM124169-01. The Advanced Light Source is a Department of Energy Office of Science User Facility under Contract No. DE-AC02-05CH11231. ALS beamlines 5.0.1, 8.2.1, and 8.2.2 were used in this study.
Author contributions
I. M. F., L. H., B. V. V. P., and T. P. formal analysis; I. M. F., L. H., and T. P. visualization; I. M. F., S. C. M., B. S., L. H., B. V. V. P., and T. P. methodology; I. M. F., B. V. V. P., and T. P. writing - original draft; I. M. F., S. C. M., B. S., L. H., B. V. V. P., and T. P. writing - review and editing; B. V. V. P. and T. P. funding acquisition.
Funding and additional information
This work was supported by NIH R01 AI32956 to T. P., NIH T32 AI055449 to I. M. F., and Welch Foundation Q1279 to B. V. V. P. The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institutes of Health.
Edited by Chris Whitfield
Supporting information
References
- 1.Livermore D.M. The β-lactamase threat in Enterobacteriaceae, Pseudomonas and Acinetobacter. Trends Microbiol. 2006;14:413–420. doi: 10.1016/j.tim.2006.07.008. [DOI] [PubMed] [Google Scholar]
- 2.Bush K., Bradford P.A. β-Lactams and β-lactamase inhibitors: An overview. Cold Spring Harb. Perspect. Med. 2016;6 doi: 10.1101/cshperspect.a025247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Ambler R.P. The structure of beta-lactamases. Philos. Trans. R. Soc. Lond. B Biol. Sci. 1980;289:321–331. doi: 10.1098/rstb.1980.0049. [DOI] [PubMed] [Google Scholar]
- 4.Ambler R.P., Coulson F.W., Frere J.-M., Ghuysen J.-M., Joris B., Forsman M., Levesque R.C., Tiraby G., Waley S.G. A standard numbering scheme for the class A β-lactamases. Biochem. J. 1991;276:269–272. doi: 10.1042/bj2760269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Strynadka N.C.J., Adachi H., Jensen S.E., Johns K., Sielecki A., Betzel C., Sutoh K., James M.N.G. Molecular structure of the acyl-enzyme intermediate in β-lactam hydrolysis at 1.7 A resolution. Nature. 1992;359:700–705. doi: 10.1038/359700a0. [DOI] [PubMed] [Google Scholar]
- 6.Fisher J.F., Mobashery S. Three decades of the class A beta-lactamase acyl-enzyme. Curr. Protein Pept. Sci. 2009;10:401–407. doi: 10.2174/138920309789351967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Palzkill T. Metallo-beta-lactamase structure and function. Ann. N.Y. Acad. Sci. 2013;1277:91–104. doi: 10.1111/j.1749-6632.2012.06796.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Palzkill T. Structural and mechanistic basis for extended-spectrum drug-resistance mutations in altering the specificity of TEM, CTX-M, and KPC β-lactamases. Front. Mol. Biosci. 2018;5:16. doi: 10.3389/fmolb.2018.00016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Bush K. Alarming beta-lactamase-mediated resistance in multidrug-resistant Enterobacteriaceae. Curr. Opin. Microb. 2010;13:558–564. doi: 10.1016/j.mib.2010.09.006. [DOI] [PubMed] [Google Scholar]
- 10.Bush K. Past and present perspectives on β-lactamases. Antimicrob. Agents Chemother. 2018;62 doi: 10.1128/AAC.01076-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Queenan A.M., Bush K. Carbapenemases: The versatile beta-lactamases. Clin. Microbiol. Rev. 2007;20:440–458. doi: 10.1128/CMR.00001-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Yigit H., Queenan A.M., Anderson G.J., Domenech-Sanchez A., Biddle J.W., Steward C.D., Alberti S., Bush K., Tenover F.C. Novel carbapenem-hydrolyzing beta-lactamase, KPC-1, from a carbapenem- resistant strain of Klebsiella pneumoniae. Antimicrob. Agents Chemother. 2001;45:1151–1161. doi: 10.1128/AAC.45.4.1151-1161.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Bonomo R.A. β-Lactamases: A focus on current challenges. Cold Spring Harb. Perspect. Med. 2017;7 doi: 10.1101/cshperspect.a025239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.CDC . Antibiotic Resistance Threats in the United States, 2019. US Department of Health and Human Services, CDC; Atlanta, GA: 2019. [Google Scholar]
- 15.World Health Organization . WHO Press; Geneva, Switzerland: 2014. Antimicrobial Resistance: Global Report on Surveillance. [Google Scholar]
- 16.Logan L.K., Weinstein R.A. The epidemiology of carbapenem-resistant Enterobacteriaceae: The impact and evolution of a global menace. J. Infect. Dis. 2017;215:S28–S36. doi: 10.1093/infdis/jiw282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Long S.W., Olsen R.J., Eagar T.N., Beres S.B., Zhao P., Davis J.J., Brettin T., Xia F., Musser J.M. Population genomic analysis of 1,777 extended-spectrum beta-lactamase-producing Klebsiella pneumoniae isolates, Houston, Texas: Unexpected Abundance of Clonal Group 307. mBio. 2017;8 doi: 10.1128/mBio.00489-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Galleni M., Frere J.-M. Kinetics of β-lactamases and penicillin-binding proteins. In: Bonomo R.A., Tolmasky M.E., editors. Enzyme-Mediated Resistance to Antibiotics: Mechanisms, Dissemination, and Prospects for Inhibition. ASM Press; Washington, DC: 2007. pp. 67–79. [Google Scholar]
- 19.Nukaga M., Bethel C.R., Thomson J.M., Hujer A.M., Distler A., Anderson V.E., Knox J.R., Bonomo R.A. Inhibition of class A β-lactamases by carbapenems: Crystallographic observation of two conformations of meropenem in SHV-1. J. Am. Chem. Soc. 2008;130:12656–12662. doi: 10.1021/ja7111146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Maveyraud L., Mourey L., Kotra L.P., Pedelacq J., Guillet V., Mobashery S., Samama J. Structural basis for clinical longevity of carbapenem antibiotics in the face of challenge by the common class A β-lactamases from the antibiotic resistant bacteria. J. Am. Chem. Soc. 1998;120:9748–9752. [Google Scholar]
- 21.Charnas R., Knowles J. Inhibition of the RTEM beta-lactamase from Escherichia coli. Interaction of enzyme with derivatives of olivanic acid. Biochemistry. 1981;20:2732–2737. doi: 10.1021/bi00513a005. [DOI] [PubMed] [Google Scholar]
- 22.Kalp M., Carey P.R. Carbapenems and SHV-1 beta-lactamase form different acyl-enzyme populations in crystals and solution. Biochemistry. 2008;47:11830–11837. doi: 10.1021/bi800833u. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Easton C.J., Knowles J.R. Inhibition of the RTEM beta-lactamase from Escherichia coli. Interaction of the enzyme with derivatives of olivanic acid. Biochemistry. 1982;21:2857–2862. doi: 10.1021/bi00541a008. [DOI] [PubMed] [Google Scholar]
- 24.Tremblay L.W., Fan F., Blanchard J.S. Biochemical and structural characterization of Mycobacterium tuberculosis β-lactamase with the carbapenems ertapenem and doripenem. Biochemistry. 2010;49:3766–3773. doi: 10.1021/bi100232q. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Mehta S.C., Furey I.M., Pemberton O.A., Boragine D.M., Chen Y., Palzkill T. KPC-2 β-lactamase enables carbapenem antibiotic resistance through fast deacylation of the covalent intermediate. J. Biol. Chem. 2020;296:100155. doi: 10.1074/jbc.RA120.015050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Zafaralla G., Mobashery S. Facilitation of the D2 Æ D1 pyrroline tautomerization of cabapenam antibiotics by the highly conserved arginine-244 of class A β-lactamases during the course of turnover. J. Am. Chem. Soc. 1992;114:1505–1506. [Google Scholar]
- 27.Fonseca F., Chudyk E.I., van der Kamp M.W., Correia A., Mulholland A.J., Spencer J. The basis for carbapenem hydrolysis by class A β-lactamases: A combined investigation using crystallography and simulations. J. Am. Chem. Soc. 2012;134:18275–18285. doi: 10.1021/ja304460j. [DOI] [PubMed] [Google Scholar]
- 28.Majiduddin F.K., Palzkill T. Amino acid residues that contribute to substrate specificity of class A beta-lactamase SME-1. Antimicrob. Agents Chemother. 2005;49:3421–3427. doi: 10.1128/AAC.49.8.3421-3427.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Mehta S.C., Rice K., Palzkill T. Natural variants of the KPC-2 carbapenemase have evolved increased catalytic efficiency for ceftazidime hydrolysis at the cost of enzyme stability. PLoS Pathog. 2015;11 doi: 10.1371/journal.ppat.1004949. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Pemberton O.A., Zhang X., Chen Y. Molecular basis of substrate recognition and product release by the Klebsiella pneumoniae carbapenemase (KPC-2) J. Med. Chem. 2017;60:3525–3530. doi: 10.1021/acs.jmedchem.7b00158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Bicknell R., Waley S.G. Single-turnover and steady-state kinetics of hydrolysis of cephalosporins by beta-lactamase I from Bacillus cereus. Biochem. J. 1985;231:83–88. doi: 10.1042/bj2310083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Hollaway M.R., Antonini E., Brunori M. The ficin-catalysed hydrolysis of p-nitrophenyl hippurate. Detailed kinetics including the measurement of the apparent dissociation constant for the enzyme-substrate complex. FEBS Lett. 1969;4:299–306. doi: 10.1016/0014-5793(69)80261-9. [DOI] [PubMed] [Google Scholar]
- 33.Soeung V., Lu S., Hu L., Judge A., Sankaran B., Prasad B.V.V., Palzkill T. A drug-resistant β-lactamase variant changes the conformation of its active site proton shuttle to alter substrate specificity and inhibitor potency. J. Biol. Chem. 2020;295:18239–18255. doi: 10.1074/jbc.RA120.016103. [DOI] [PubMed] [Google Scholar]
- 34.Ke W., Bethel C.R., Thomson J.M., Bonomo R.A., van den Akker F. Crystal structure of KPC-2: Insights into carbapenemase activity in class A β-lactamases. Biochemistry. 2007;46:5732–5740. doi: 10.1021/bi700300u. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Petrella S., Ziental-Gelus N., Mayer C., Renard M., Jarlier V., Sougakoff W. Genetic and structural insights into the dissemination potential of the extremely broad-spectrum class A beta-lactamase KPC-2 identified in an Escherichia coli strain and an Enterobacter cloacae strain isolated from the same patient in France. Antimicrob. Agents Chemother. 2008;52:3725–3736. doi: 10.1128/AAC.00163-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Krishnan N.P., Nguyen N.Q., Papp-Wallace K.M., Bonomo R.A., van den Akker F. Inhibition of Klebsiella β-lactamases (SHV-1 and KPC-2) by avibactam: A structural study. PLoS One. 2015;10 doi: 10.1371/journal.pone.0136813. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Ke W., Bethel C.R., Papp-Wallace K.M., Pagadala S.R.R., Nottingham M., Fernandez D., Buynak J.D., Bonomo R.A., van den Akker F. Crystal structures of KPC-2 β-lactamase in complex with 3-nitrophenyl boronic acid and the penam sulfone PSR-3-226. Antimicrob. Agents Chemother. 2012;56:2713–2718. doi: 10.1128/AAC.06099-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Burgi H.B., Dunitz J.D., Shefter E. Geometrical reaction coordinates. II. Nucleophilic addition to a carbonyl group. J. Am. Chem. Soc. 1973;95:5065–5067. [Google Scholar]
- 39.Radisky E.S., Koshland D.E. A clogged gutter mechanism for protease inhibitors. Proc. Natl. Acad. Sci. U. S. A. 2002;99:10316–10321. doi: 10.1073/pnas.112332899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Radisky E.S., Lee J.M., Lu C.-J.K., Koshland D.E. Insights into the serine protease mechanism from atomic resolution structures of trypsin reaction intermediates. Proc. Natl. Acad. Sci. U. S. A. 2006;103:6835–6840. doi: 10.1073/pnas.0601910103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Patel M.P., Hu L., Brown C.A., Sun Z., Adamski C.J., Stojanoski V., Sankaran B., Prasad B.V.V., Palzkill T. Synergistic effects of functionally distinct substitutions in β-lactamase variants shed light on the evolution of bacterial drug resistance. J. Biol. Chem. 2018;293:17971–17984. doi: 10.1074/jbc.RA118.003792. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Zhai X., Reinhardt C.J., Malabanan M.M., Amyes T.L., Richard J.P. Enzyme architecture: Amino acid side-chains that function to optimize the basicity of the active site glutamate of triosephosphate isomerase. J. Am. Chem. Soc. 2018;140:8277–8286. doi: 10.1021/jacs.8b04367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Malabanan M.M., Nitsch-Velasquez L., Amyes T.L., Richard J.P. Magnitude and origin of the enhanced basicity of the catalytic glutamate of triosephosphate isomerase. J. Am. Chem. Soc. 2013;135:5978–5981. doi: 10.1021/ja401504w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Richard J.P., Amyes T.L., Malabanan M.M., Zhai X., Kim K.J., Reinhardt C.J., Wierenga R.K., Drake E.J., Gulick A.M. Structure-function studies of hydrophobic residues that clamp a basic glutamate side chain during catalysis by triosephosphate isomerase. Biochemistry. 2016;55:3036–3047. doi: 10.1021/acs.biochem.6b00311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Dellus-Gur E., Toth-Petroczy A., Elias M., Tawfik D.S. What makes a protein fold amenable to functional innovation? Fold polarity and stability trade-offs. J. Mol. Biol. 2013;425:2609–2621. doi: 10.1016/j.jmb.2013.03.033. [DOI] [PubMed] [Google Scholar]
- 46.Dellus-Gur E., Elias M., Caselli E., Prati F., Salverda M.L., de Visser J.A., Fraser J.S., Tawfik D.S. Negative epistasis and evolvability in TEM-1 β-lactamase-the thin line between and enzyme’s conformational freedom and disorder. J. Mol. Biol. 2015;427:2396–2409. doi: 10.1016/j.jmb.2015.05.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Hart K.M., Ho C.M.W., Dutta S., Gross M.L., Bowman G.R. Modelling proteins’ hidden conformations to predict antibiotic resistance. Nat. Commun. 2016;7:12965. doi: 10.1038/ncomms12965. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Brown C.A., Hu L., Sun Z., Patel M.P., Singh S., Porter J.R., Sankaran B., Prasad B.V.V., Bowman G.R., Palzkill T. Antagonism between substitutions in β-lactamase explains a path not taken in the evolution of bacterial drug resistance. J. Biol. Chem. 2020;295:7376–7390. doi: 10.1074/jbc.RA119.012489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Patel M.P., Hu L., Stojanoski V., Sankaran B., Prasad B.V., Palzkill T. The drug-resistant variant P167S expands the substrate profile of CTX-M β-lactamases for oxyimino-cephalosporin antibiotics by enlarging the active site upon acylation. Biochemistry. 2017;56:3443–3453. doi: 10.1021/acs.biochem.7b00176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Vakulenko S.B., Taibi-Tronche P., Toth M., Massova I., Lerner S.A., Mobashery S. Effects on substrate profile by mutational substitutions at positions 164 and 179 of the class A TEMpUC19 b-lactamase from Escherichia coli. J. Biol. Chem. 1999;274:23052–23060. doi: 10.1074/jbc.274.33.23052. [DOI] [PubMed] [Google Scholar]
- 51.Palzkill T., Le Q.-Q., Venkatachalam K.V., LaRocco M., Ocera H. Evolution of antibiotic resistance: Several different amino acid substitutions in an active site loop alter the substrate profile of ß-lactamase. Mol. Microbiol. 1994;12:217–229. doi: 10.1111/j.1365-2958.1994.tb01011.x. [DOI] [PubMed] [Google Scholar]
- 52.Schneider K.D., Karpen M.E., Bonomo R.A., Leonard D.A., Powers R.A. The 1.4 A crystal structure of the class D beta-lactamase OXA-1 complexed with doripenem. Biochemistry. 2009;48:11840–11847. doi: 10.1021/bi901690r. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Pozzi C., Luca F.D., Benvenuti M., Poirel L., Nordmann P., Rossolini G.M., Mangani S., Docquier J.-D. Crystal structure of the Pseudomonas aeruginosa BEL-1 extended-spectrum β-lactamase and its complexes with moxalactam and imipenem. Antimicrob. Agents Chemother. 2016;60:7189–7199. doi: 10.1128/AAC.00936-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Stewart N.K., Smith C.A., Antunes N.T., Toth M., Vakulenko S.B. Role of the hydrophobic bridge in the carbapenemase activity of class D β-lactamases. Antimicrob. Agents Chemother. 2019;63 doi: 10.1128/AAC.02191-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Lohans C.T., Freeman E.I., van Groesen E., Tooke C.L., Hinchliffe P., Spencer J., Brem J., Schofield C.J. Mechanistic insights into β-lactamase-catalysed carbapenem degradation through product characterisation. Sci. Rep. 2019;9:13608. doi: 10.1038/s41598-019-49264-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Papp-Wallace K.M., Taracila M., Hornick J.M., Hujer A.M., Hujer K.M., Distler A.M., Endimiani A., Bonomo R.A. Substrate selectivity and a novel role in inhibitor discrimination by residue 237 in the KPC-2 beta-lactamase. Antimicrob. Agents Chemother. 2010;54:2867–2877. doi: 10.1128/AAC.00197-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Bullock W.O., Fernandez J.M., Short J.M. XL1-Blue: A high efficiency plasmid transforming recA Escherichia coli strain with beta-galactosidase selection. Biotechniques. 1987;5:376–379. [Google Scholar]
- 58.Amann E., Ochs B., Abel K.J. Tightly regulated tac promoter vectors useful for the expression of unfused and fused proteins in Escherichia coli. Gene. 1988;69:301–315. doi: 10.1016/0378-1119(88)90440-4. [DOI] [PubMed] [Google Scholar]
- 59.Battye T.G., Kontogiannis L., Johnson O., Powell H.R., Leslie A.G. iMOSFLM: A new graphical interface for diffraction-image processing with MOSFLM. Acta Crystallogr. D Biol. Crystallogr. 2011;67:271–281. doi: 10.1107/S0907444910048675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Vagin A., Teplyakov A. Molecular replacement with MOLREP. Acta Crystallogr. D Biol. Crystallogr. 2010;66:22–25. doi: 10.1107/S0907444909042589. [DOI] [PubMed] [Google Scholar]
- 61.Winn M.D., Ballard C.C., Cowtan K.D., Dodson E.J., Emsley P., Evans P.R., Keegan R.M., Krissinel E.B., Leslie A.G., McCoy A., McNicholas S.J., Murshudov G.N., Pannu N.S., Potterton E.A., Powell H.R. Overview of the CCP4 suite and current developments. Acta Crystallogr. D Biol. Crystallogr. 2011;67:235–242. doi: 10.1107/S0907444910045749. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Emsley P., Cowtan K. Coot: Model-building tools for molecular graphics. Acta Crystallogr. D Biol. Crystallogr. 2004;60:2126–2132. doi: 10.1107/S0907444904019158. [DOI] [PubMed] [Google Scholar]
- 63.Emsley P., Lohkamp B., Scott W.G., Cowtan K. Features and development of Coot. Acta Crystallogr. D Biol. Crystallogr. 2010;66:486–501. doi: 10.1107/S0907444910007493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Adams P.D., Afonine P.V., Bunkoczi G., Chen V.B., Davis I.W., Echols N., Headd J.J., Hung L.W., Kapral G.J., Grosse-Kunstleve R.W., McCoy A.J., Moriarty N.W., Oeffner R., Read R.J., Richardson D.C. PHENIX: A comprehensive Python-based system for macromolecular structure solution. Acta Crystallogr. D Biol. Crystallogr. 2010;66:213–221. doi: 10.1107/S0907444909052925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Pettersen E.F., Goddard T.D., Huang C.C., Couch G.S., Greenblatt D.M., Meng E.C., Ferrin T.E. UCSF Chimera--a visualization system for exploratory research and analysis. J. Comput. Chem. 2004;25:1605–1612. doi: 10.1002/jcc.20084. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Accession codes: Coordinates and structure factors have been deposited in the Protein Data Bank under accession codes 7LJK, 7LLH, 7LK8, 7LLB, and 7LNL. All relevant data associated with the paper are available upon request from the corresponding author.