SUMMARY
During meiosis, the maintenance of genome integrity is critical for generating viable haploid gametes.1 In meiotic prophase I, double-strand DNA breaks (DSBs) are induced and a subset of these DSBs are repaired as interhomolog crossovers to ensure proper chromosome segregation. DSBs not resolved as crossovers with the homolog must be repaired by other pathways to ensure genome integrity.2 To determine if alternative repair templates can be engaged for meiotic DSB repair during oogenesis, we developed an assay to detect sister and/or intra-chromatid repair events at a defined DSB site during Caenorhabditis elegans meiosis. Using this assay, we directly demonstrate that the sister chromatid or the same DNA molecule can be engaged as a meiotic repair template for both crossover and noncrossover recombination, with noncrossover events being the predominant recombination outcome. We additionally find that the sister or intra-chromatid substrate is available as a recombination partner for DSBs induced throughout meiotic prophase I, including late prophase when the homolog is unavailable. Analysis of noncrossover conversion tract sequences reveals that DSBs are processed similarly throughout prophase I. We further present data indicating that the XPF-1 nuclease functions in late prophase to promote sister or intra-chromatid repair at steps of recombination following joint molecule processing. Despite its function in sister or intra-chromatid repair, we find that xpf-1 mutants do not exhibit severe defects in progeny viability following exposure to ionizing radiation. Overall, we propose that C. elegans XPF-1 may assist as an intersister or intrachromatid resolvase only in late prophase I.
Graphical abstract
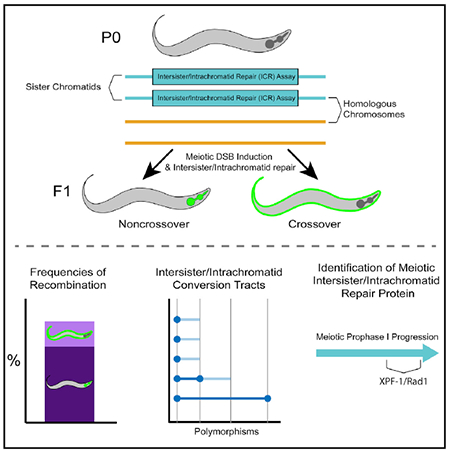
In brief
Toraason et al. develop an assay that both establishes and characterizes repair of a single induced DNA double-strand break with the sister chromatid or same DNA molecule in C. elegans. This study also demonstrates a role for the XPF-1 nuclease in promoting homolog-independent repair during late meiotic prophase I.
RESULTS AND DISCUSSION
Engagement of the sister chromatid in meiotic DSB repair
During meiotic prophase I, double-strand DNA breaks (DSBs) are induced across the genome.1 A subset of DSBs must be repaired as interhomolog crossovers to ensure accurate chromosome segregation, and the remaining DSBs are repaired through other mechanisms.3 While the homolog is the preferred recombination template in meiotic prophase I,3 access to the homolog is shut down in mid-late pachytene stage.4 Several studies have hypothesized that after access to the homolog is shut down, there is a regulated switch in template preference from the homolog to the sister chromatid during late meiotic prophase I to ensure the repair of any remaining DSBs prior to the meiotic divisions.4–6 Studies in Saccharomyces cerevisiae indicate the sister chromatid can be engaged during meiosis,7 and multiple lines of evidence have suggested that the sister chromatid may be engaged as a meiotic DSB repair template to repair these remaining DSBs in metazoan meiosis,4,5,8 but the perfect sequence identity shared between sister chromatids has precluded direct testing of this hypothesis in metazoans.
To determine whether the sister chromatid or the same chromatid can be engaged as a repair template during C. elegans meiosis, we developed a non-allelic intersister/intrachromatid repair assay (ICR assay; Figures 1A and S1) that utilizes controlled excision of a Mos1 transposon to induce a single DSB within a genetic reporter that detects repair events using a non-allelic truncated cassette on the sister chromatid or same chromatid as a template. Similar to other repair assays in S. cerevisiae meiosis9,10 and mammalian mitosis,11–13 our ICR assay is composed of two tandem reporter sequences. In our assay, the upstream copy encodes a truncated GFP allele driven by a myo-3 promoter (body wall expression). The downstream copy is driven by a myo-2 promoter (pharynx expression) and is disrupted with the Drosophila Mos1 transposable element.14 Upon heat shock-induced expression of Mos1 transposase,15 excision of the Mos1 transposon produces a single DSB.4 Previous studies determined the frequency of Mos1 excision with this heat-induced method to be 28% of C. elegans germ cell nuclei and that Mos1 is likely not to excise in both sister chromatids (see supplement of Rosu et al.4).4,16–18 Repair of the Mos1-induced DSB via nonallelic intersister or intrachromatid recombination yields restoration of functional GFP sequence and GFP+ progeny. The tissue-specific expression of the resultant functional GFP indicates which recombination pathway is engaged: (1) an intersister or intrachromatid noncrossover will generate functional pmyo-2::GFP expressed in the pharynx and (2) a deletion product indicative of an intersister or intrachromatid crossover will producepmyo-3::GFP expressed in the body wall muscle. While homology-directed single-strand annealing (SSA) could also generate a deletion product, 1,367 bp of sequence would need to be resected for this mechanism to occur and data in this manuscript indicate that SSA is likely not responsible for these products. Further, intersister crossover recombination has been demonstrated in C. elegans.19 Although the ICR assay cannot definitively distinguish between an intersister and an intrachromatid event, we suggest the assay is very likely detecting intersister events based on evidence for intersister repair in S. cerevisiae7 and strong evidence indicating use of intersister repair in C. elegans.4,5,19–22 Since allelic recombination will not restore functional GFP sequence, the ICR assay will not detect every sister chromatid or intrachromatid repair event, but it does detect nonallelic recombination outcomes, thereby enabling direct detection of such events in C. elegans.
Figure 1. Intersister/intrachromatid repair can be engaged to resolve DSBs in meiotic prophase I.
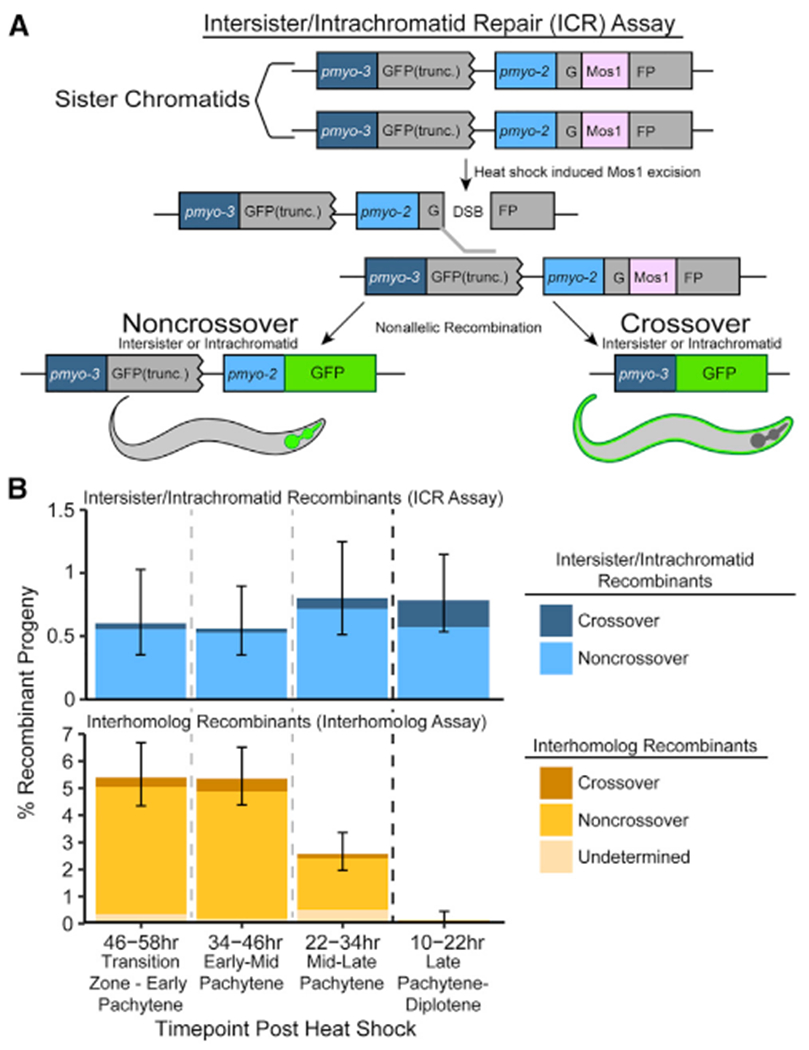
(A) Cartoon diagram of the intersister/intrachromatid repair (ICR) assay. The ICR assay is composed of two tandem GFP cassettes. The upstream GFP is driven by a pmyo-3 (body wall) promoter and is truncated, while the downstream GFP is driven by a pmyo-2 (pharynx) promoter and is interrupted by a Mos1 Drosophila transposon. Excision of Mos1 yields a single DSB. Repair of this DSB by intersister or intrachromatid recombination will yield GFP+ progeny. Figure S1A depicts how intrachromatid repair could be engaged within the ICR assay. See Figures S1B–S1D and S4 for confirmation of both ICR assay integration and noncrossover progeny genotypes.
(B) Frequency of recombinant progeny identified in the ICR assay (top) and interhomolog assay (bottom).4 Total progeny scored, n = ICR assay/interhomolog assay; 10–22 h,n = 3,317/1,625; 22–34 h,n = 2,372/1,989; 34–46 h, n = 3,032/1,721; 46–58 h, n = 2,159/1,477 (Table S1). Stacked bar plots represent the overall percent of living progeny that exhibit the indicated recombinant phenotype within a specific time point following heat shock. Error bars represent 95% binomial confidence intervals. Dashed vertical lines delineate between time points scored, while the dark black dashed line delineates between the “interhomolog window” (22–58 h post-heat shock) and “non-interhomolog window” (10–22 h post-heat shock).
The ICR assay was performed in hermaphrodites heterozygous for the assay at a locus previously assessed for interhomolog repair4 (exon 6 of unc-5) (STAR Methods). Since there is no GFP sequence on the homolog in this context, recombination repair of the Mos1-induced DSB is restricted to sister chromatid or intrachromatid events. With this assay, we observed both noncrossover and crossover GFP+ recombinants at an overall frequency of 0.69% of all progeny (including progeny that did not experience a Mos-1-induced DSB; 95% confidence interval [CI] 0.550%–0.863%), which represents the frequency of nonallelic recombination at this locus in oocytes at meiotic stages from leptotene/zygotene (transition zone) through late pachytene and diplotene at the time of DSB induction by Mos1 transposition (Figure 1B, top). Notably, noncrossover events were the predominant repair outcome from the ICR assay (85.3% of GFP+ recombinants; Table S1, top). These data directly demonstrate that the sister chromatid or same DNA molecule can be engaged as a DSB repair template in C. elegans meiosis and enable the assessment of these meiotic DNA repair pathways in a metazoan.
We next wanted to test for the hypothesized switch in template bias from the homolog to the sister chromatid during late meiotic prophase I.4–6 Similar to a previous C. elegans assay that assessed interhomolog repair during meiosis (interhomolog repair assay; Figure 1B, bottom; Table S1, bottom),4 the ICR assay can determine the stages of meiotic prophase I in which the sister chromatid can be engaged as a repair template. Given the established timing of meiotic prophase progression for C. elegans oogenesis, progeny laid in the 22–58 h time points were derived from oocytes spanning entry into meiotic prophase I through mid-pachytene at the time of heat shock (Mos1 excision), while the oocytes yielding progeny at the 10–22 h time point were at late pachytene/diplotene.4,23 While neither the interhomolog assay nor ICR assay detect whether a DSB is repaired within the same meiotic stage it was induced, we can still determine the latest window in which a repair template is available. Specifically, DSBs induced during the 22+ h time points (“interhomolog window”) and not the 10–22 h time point (“non-interhomolog window”) can be repaired with the homolog (Figure 1B, bottom; Table S1).4 The ICR assay demonstrates that DSBs induced at different times throughout meiotic prophase can be repaired using the sister chromatid or same DNA molecule, and that such repair occurs at similar frequencies regardless of the timing of DSB induction (Figure 1B, top; Table S1). Thus, while engagement of the homolog is restricted to a specific window of meiotic prophase I, the sister chromatid or same chromatid may be engaged as a repair template for DSBs induced throughout meiotic prophase I. These data further demonstrate that intersister or intrachromatid repair becomes the preferred recombination pathway in late meiotic prophase I when the homolog is no longer readily engaged for repair (10–22 h post-heat shock; Figure 1B). Moreover, we observed both noncrossover and crossover recombinant progeny at all time points (Figure 1B, top), indicating that DSBs induced throughout meiotic prophase I may be repaired by intersister/intrachromatid crossover and noncrossover recombination pathways. Crossover recombinants are specifically enriched in the non-interhomolog window (10–22 h post-heat shock) compared to the interhomolog window (Fisher’s exact test, p = 0.041). These results indicate that a late pachytene transition increases DSB resolution by intersister/intrachromatid crossover recombination.
XPF-1 nuclease promotes intersister/intrachromatid repair
We next investigated the role of the resolvase XPF-1 in intersister and intrachromatid recombination. XPF-1 is the C. elegans homolog of the XPF/RAD1 nuclease and acts semi-redundantly with other nucleases to resolve meiotic interhomolog crossovers.24–27 XPF-1 is also required for SSA, a mutagenic homology-directed repair pathway that may be engaged upon exposure of >30 bp of repeated sequence on each resected ssDNA strand of a damaged chromosome and results in deletion of sequences between tandem repeats.20,28,29 As the ICR assay contains tandem GFP cassettes (Figure 1A), engagement of SSA to resolve Mos1-induced DSBs could yield progeny with a phenotype that may be interpreted as an intersister/intrachromatid crossover event. To both assess the role of XPF-1 in intersister/intrachromatid repair and determine whether our assay is identifying SSA-mediated DSB repair, we performed the ICR assay in an xpf-1(tm2842) mutant, which exhibits normal rates of ovulation and likely does not significantly affect the timing of meiotic prophase progression (Figure S2A).
Neither the overall recombinant frequency nor the proportion of crossover progeny in xpf-1 mutants differed from wild type within the interhomolog window (Figure 2B; Tables S1 and S2; Fisher’s exact test, p > 0.05). However, there was a decrease in the total frequency of recombinants in the non-interhomolog window at 10–22 h post-heat shock (Figure 2; Fisher’s exact test, p = 0.004), including crossover recombinant progeny. If the ICR assay was primarily detecting SSA repair, then ablation of xpf-1 should result in a severe reduction of “crossover” progeny without altering observed frequencies of noncrossover progeny. Therefore, the occurrence of crossover recombinants in the xpf-1 mutant suggests SSA does not significantly contribute to the detected ICR assay repair outcomes. This result is not surprising, as multiple C. elegans studies demonstrate that mutagenic DNA repair pathways, including SSA, are only frequently utilized for meiotic DSB repair in mutants where homologous recombination is impeded.6,8,21,30,31 Notably, intersister crossovers were also cytologically observed in the accompanying publication,19 reinforcing the model that the crossover progeny we observe are likely derived from bona fide intersister crossovers. Overall, our data suggest that XPF-1 promotes meiotic sister chromatid and/or intrachromatid repair specifically in late meiotic prophase I. Since XPF-1 functions to resolve interhomolog joint molecules in C. elegans meiosis,24–27 XPF-1 may also act to cleave intersister/intrachromatid joint molecules to yield crossover and noncrossover products at this late meiotic stage.
Figure 2. XPF-1 promotes intersister/intrachromatid repair in late meiotic prophase I.
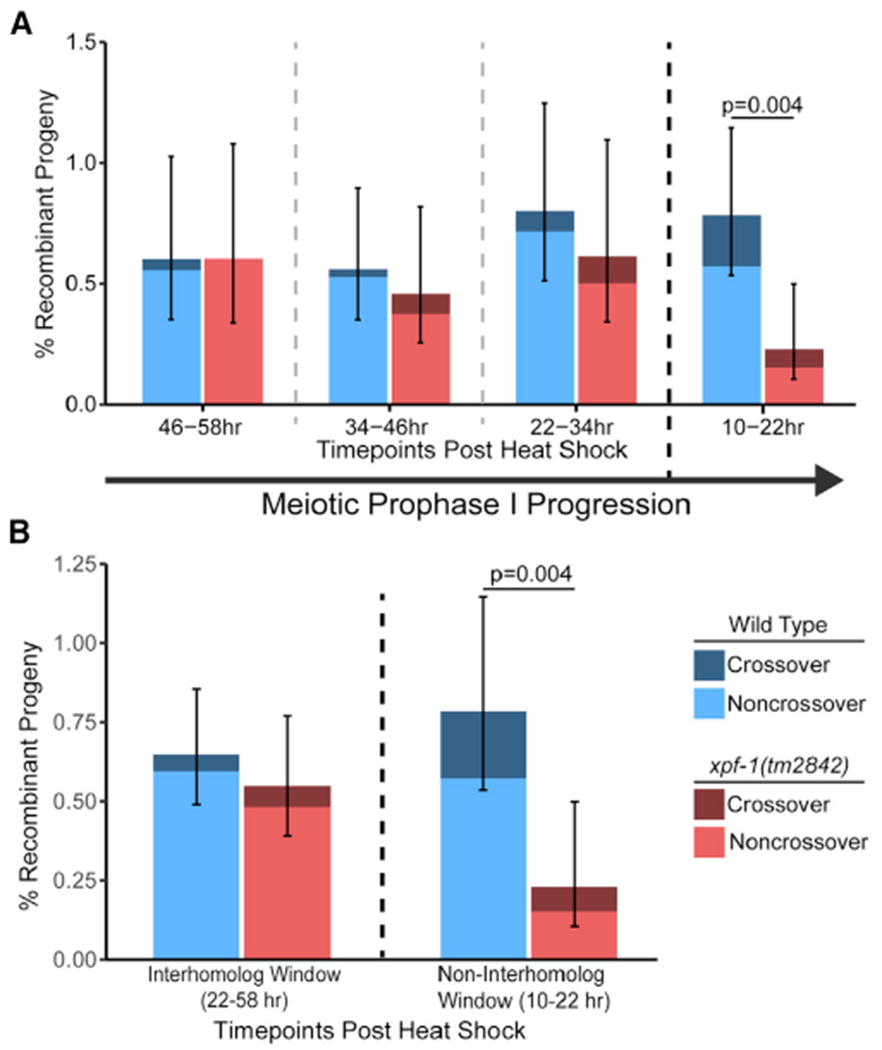
(A) Frequency of ICR assay recombinant progeny in wild-type and xpf-1(tm2842) mutants at each scored time point following heat shock. Total progeny scored, n = wild-type/xpf-1; 10–22 h, n = 3,317/2,618; 22–34 h, n = 2,372/1,793; 34–46 h,n = 3,032/2,400; 46–58 h,n = 2,159/1,819 (Tables S1 and S2). Both wild-type and xpf-1(tm2842) have similar rates of meiotic prophase progression (Figure S2A).
(B) Frequency of recombinant progeny identified in the ICR assay within binned windows of prophase I defined by observation of recombinants in the interhomolog assay. n = wild-type/xpf-1; interhomolog window, n = 7,563/ 6,012; non-interhomolog window, n = 3,317/2,618 (Tables S1 and S2). Stacked bars represent the overall percent of living progeny that exhibit the indicated recombinant phenotype within the labeled time interval following heat shock. Error bars represent 95% binomial confidence intervals. p values were calculated by Fisher’s exact test. Dashed vertical lines delineate between time points scored, while the dark black dashed line delineates between the “interhomolog window” (22–58 h post-heat shock) and “non-interhomolog window” (10–22 h post-heat shock).
Mechanisms of intersister/intrachromatid recombination
Recombination mechanisms can be inferred from gene conversion tracts, which are DNA sequence changes that arise from nonreciprocal exchanges during recombination repair with a polymorphic template. To reveal mechanisms of meiotic intersister/intrachromatid repair, we engineered polymorphisms in the two tandem GFP cassettes within the ICR assay, thereby enabling detection of conversion tracts from recombination between nonallelic GFP sequences (Figure 3A). Wild-type intersister/intrachromatid noncrossover events displayed tracts ranging from a single to multiple polymorphism conversions spanning 567 bp of sequence (Figure 3B). In all of these tracts, the polymorphism most proximal to the site of Mos1 excision (12 bp downstream) was always converted, indicating that recombination intermediates remain local to the site of DSB induction and/or that this marker is frequently incorporated within the resection area (Figure 3B). This result is also reminiscent of S. cerevisiae mitotic repair of HO-mediated DSBs where there is preferential conversion of markers proximal to the DSB site,32 likely due to the proofreading activity of polymerase delta. With this polymorphism density, we did not observe restoration tracts arising from recombination in wild-type animals, which are unconverted polymorphisms flanked by conversion events indicative of multiple template engagement, heteroduplex DNA mismatch correction, or nucleotide excision of joint molecules during recombination33–35 (Figure 3B). Although interhomolog conversion tracts in other organisms suggest frequent joint molecule migration and strand switching,35,36 our results suggest intersister/intrachromatid noncrossover repair in C. elegans possibly may not involve extensive migration from the DSB site. Future experiments in a mismatch repair mutant (e.g., msh-2 mutant) or an ICR assay with a higher density of polymorphisms could reveal additional molecular signatures and evidence of template switching during these events.
Figure 3. XPF-1 does not influence intersister/intrachromatid conversion tract length.
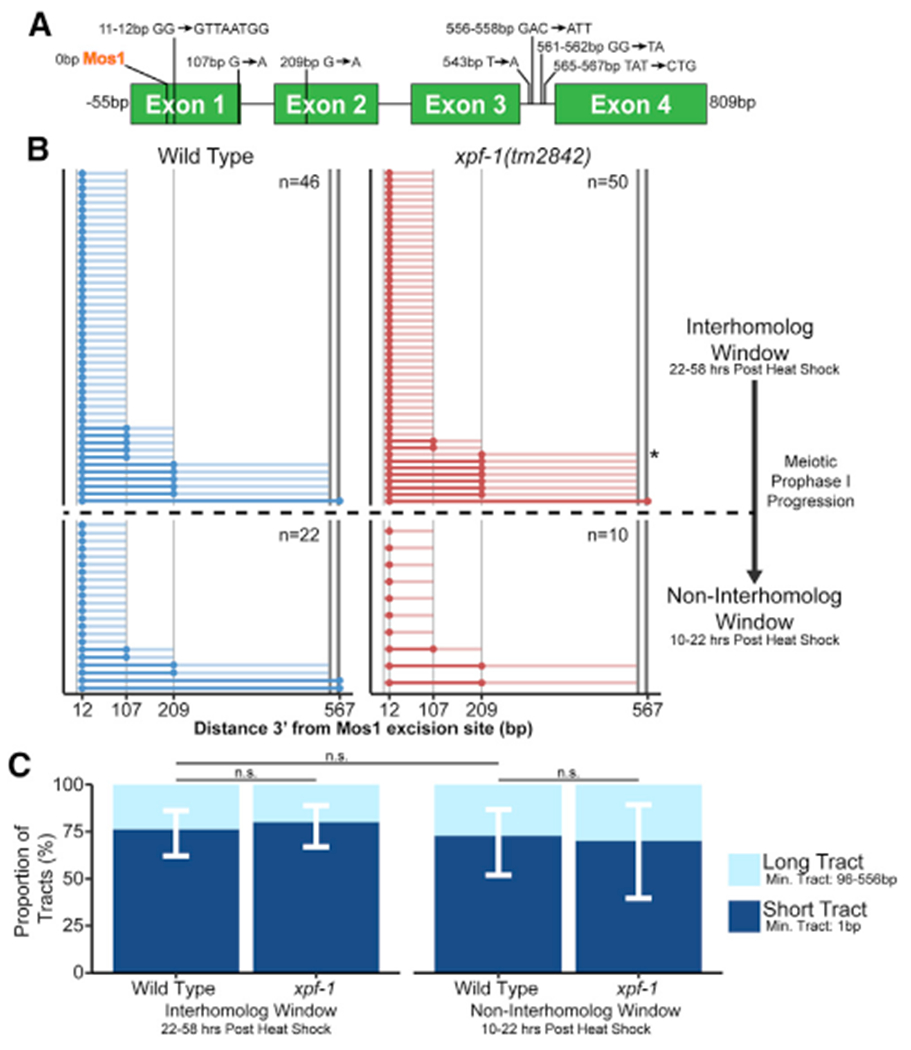
(A) Scale cartoon of ICR assay GFP cassette with annotated polymorphisms. The polymorphisms of thepmyo-2::GFP sequence are listed tothe left ofeach arrow, while the sequence of the pmyo-3::GFP polymorphism is listed to the right of each arrow. Positions of polymorphisms in bp are relative to the site of Mos1 excision.
(B) Converted polymorphisms within wild-type and xpf-1(tm2842) ICR assay noncrossover recombinant loci. Each horizontal line represents the sequenced locus of a single recombinant. High-opacity lines connect contiguous converted polymorphisms within a single tract and represent minimum tract length, while the low-opacity lines represent the range between converted and the most proximal non-converted polymorphism. See Figure S3 for crossover conversion tract data.
(C) Stacked bar plots showing the proportion of “short” (1 bp minimum tract length) and “long” (>96 bp minimum tract length). Error bars represent 95% binomial confidence intervals. p values calculated by Fisher’s exact test.
To assess whether processing of intersister/intrachromatid recombination intermediates changes during meiotic progression, we compared tracts generated at different stages of prophase I (Figures 3B and 3C). The length of a conversion tract can be influenced by 5′ strand resection, joint molecule migration, extent of strand synthesis, and mismatch repair of heteroduplex sequences.37,38 Comparing the minimum conversion tract lengths of our wild-type intersister/intrachromatid noncrossover tracts, we note that the proportion of “short” tracts converted only at one polymorphism and “long” tracts ≥96 bp in length are similar within both the interhomolog and non-interhomolog windows (interhomolog window, 76.1% “short” tracts, 95% CI 62.1%–86.1%; non-interhomolog window, 72.7% “short” tracts, 95% CI 51.8%–86.1%; Figure 3C; Fisher’s exact test, p > 0.05), suggesting that DSB processing during intersister/intrachromatid noncrossover recombination repair is likely similar throughout prophase I.
XPF-1 does not influence intersister/intrachromatid noncrossover conversion tracts
Similar to wild type, the most DSB-proximal polymorphism remained converted in every xpf-1 mutant intersister/intrachromatid noncrossover tract we sequenced (Figure 3B). However, we identified a single restoration tract arising from an interhomolog window noncrossover tract in our xpf-1 mutant dataset (Figure 3B, asterisk). While this single event is not sufficient evidence that restoration tracts are specific to or enriched in xpf-1 mutants, our identification of this tract demonstrates that complex recombination events occur in C. elegans meiosis. The proportion of “short” (1 bp) and “long” (≥96 bp) noncrossover conversion tracts arising from xpf-1 mutants was similarly indistinguishable from wild type (interhomolog window, 80.0% “short” tracts, 95% CI 67.0%–88.8%; non-interhomolog window, 70% “short” tracts, 95% CI 39.7%–89.2%; Figure 3C; Fisher’s exact test, interhomolog and non-interhomolog windows, p > 0.05). While our limited sample in the non-interhomolog window limits our interpretation of tract length proportions at this time point, the similar proportion of “short” and “long” tracts in the interhomolog window in both wild-type and xpf-1 mutants suggests that XPF-1 may function after joint molecule processing by acting as a resolvase to promote intersister/intrachromatid repair in late meiotic prophase I.
XPF-1 is not required for progeny viability following irradiation
To establish whether defects in intersister/intrachromatid recombination at specific stages of meiotic prophase I are required for fertility, we exposed young adult xpf-1 mutant hermaphrodites to ionizing radiation, which induces DSBs, and performed a reverse time course to assess effects on brood viability of damage induced at specific meiotic stages. Mutants for xpf-1 exhibited a mild but significant reduction in brood viability upon exposure to 5,000 Rads of ionizing radiation only within the interhomolog window (22–46 h time point; Figure 4; Mann-Whitney U test, p = 0.037). While our ICR assay demonstrates that XPF-1 promotes intersister/intrachromatid repair in the non-interhomolog window, xpf-1 mutants were not radiation-sensitive in late meiotic prophase I.
Figure 4.
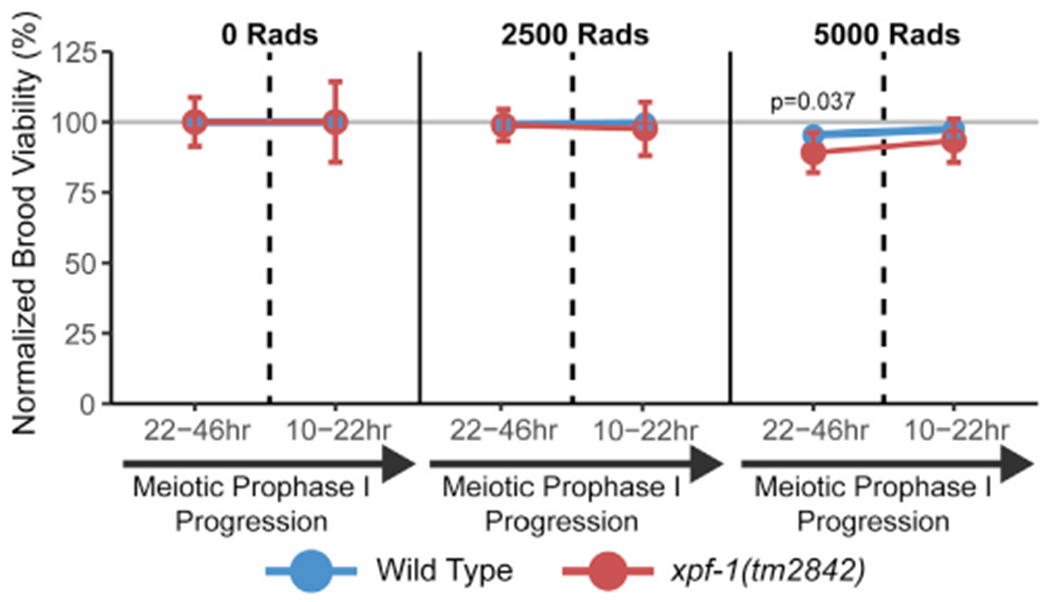
XPF-1 is not required for brood viability in response to ionizing radiation Mean brood viability of young adult hermaphrodites exposed to 0, 2,500, or 5,000 Rads of ionizing radiation, normalized to the mean brood viability for each genotype and time point scored in the absence of ionizing radiation (0 Rads treatment). Broods of n = 15 parent hermaphrodites of each respective genotype were scored for each irradiation treatment dose. Vertical dashed lines delineate between time points representing damage induced during the interhomolog window (22–46 h) and time points representing damage induced during the non-interhomolog window (10–22 h). Error bars represent SD. p values were calculated by Mann-Whitney U test. Brood viabilities of each condition without normalization are displayed in Figure S2B.
One possibility is that XPF-1 might distinguish between DSBs generated from transposition versus ionizing irradiation. Alternately, this discrepancy in ionizing radiation sensitivity and intersister/intrachromatid repair frequencies in xpf-1 mutants could reflect that defects late in meiotic recombination intermediate resolution do not necessarily impact progeny viability. A recent study found that during Drosophila and mammalian mitosis, theta-mediated end joining (TMEJ) can process joint molecules in a resolvase-deficient background.39 TMEJ is active during C. elegans meiosis and is the primary mechanism responsible for the formation of small deletions in the C. elegans germline.40 A study that profiled mutations in xpf-1 mutants demonstrated that nematode germ cells deficient in XPF-1 are susceptible to incurring small deletions in response to ionizing radiation.41 Thus, mutants in DSB repair components such as XPF-1 that affect intersister/intrachromatid recombination late in joint molecule resolution may be less impactful on fertility due to DSB resolution by alternative and error-prone repair pathway(s). Notably, sensitivity to ionizing radiation has been utilized as an important indicator for mutants deficient in intersister repair.22,42 Our data indicate that the ICR assay can further elucidate functions of proteins in meiotic DSB repair.
Conclusions
In this study, we detected recombination between sister chromatids and/or the same chromatid, thereby demonstrating that intersister/intrachromatid DNA repair can be engaged during meiosis. Additionally, we generated and analyzed intersister/intrachromatid conversion tracts to assess mechanisms of these types of events. We further show that the XPF-1 nuclease is differentially engaged within meiotic prophase I to promote intersister/intrachromatid repair. From our data, we propose that XPF-1 nuclease acts downstream of recombination intermediate processing to promote intersister/intrachromatid repair during late meiotic prophase I. Multiple repair pathways likely work with or in parallel to XPF-1 to promote meiotic intersister/intrachromatid recombination, and our ICR assay enables future elucidation of these interactions.
STAR★METHODS
RESOURCE AVAILABILITY
Lead Contact
Requests for further information, reagents, or resources should be directed to the lead contact, Diana E. Libuda (dlibuda@uoregon. edu).
Materials Availability
All strains and reagents generated for this dataset are available upon request.
Data and Code Availability
The published article includes all ICR assay datasets generated or analyzed in this study. Datasets detailing the per-hermaphrodite brood viability and ovulation counts used to generate Figures 4 and S2 are available upon request.
EXPERIMENTAL MODEL AND SUBJECT DETAILS
Caenorhabditis elegans
C. elegans strains used in this study were maintained at 15°C or 20°C on nematode growth medium (NGM) plates and were fed the OP50 Escherichia coli bacterial strain. Experiments were performed only on C. elegans strains that had been maintained at 20°C for a minimum of two generations.
C. elegans strains used in this study were obtained from the Caenorhabditis Genetics Center (CGC) or were generated by crossing and/or CRISPR/Cas9 genome editing. Genetic crosses were performed by placing L4 stage male and hermaphrodite nematodes on NGM plates with OP50 at 20°C and screening for cross progeny after 3–4 days. Genotypes of strains generated by crossing were confirmed by PCR. DLW23 was generated by crossing YE57 males to DLW14 hermaphrodites. DLW82 was generated by crossing TG1660 hermaphrodites were crossed to YE57 males to generate males carrying the xpf-1(tm2842) allele balanced by the mIn1 balancer. These F1 xpf-1/mIn1 males were then crossed to DLW14.
Strains generated by CRISPR/Cas9 genome editing were backcrossed to remove any off-target mutations that may have been incurred. The strain carrying the integrated intersister/intrachromatid repair (ICR) assay sequence, DLW14, was backcrossed three times to EN909.
METHOD DETAILS
Intersister/intrachromatid Repair (ICR) Assay Construction
The intersister/intrachromatid (ICR) assay plasmid pMG1 was constructed by integrating pmyo-3 sequence from pCFJ104 (Jorgensen Lab) into the synthetic plasmid pDL23 (GenScript) by Gibson assembly (SGI-DNA) and PCR stitching. Plasmids pMG3 and pMG14 expressing Cas9 and CRISPR guide RNAs (pMG3 protospacer 5′-GAGUAGUUCAGGAUCUGG-3′, pMG14 protospacer 5′-GUUGUUGAAUGUGGUAGAGG-3′) targeting unc-5 were generated by modifying pJW1285 (Jorgensen Lab) using PCR stitching. All plasmid sequences were confirmed by Sanger sequencing (Sequetech).
CRISPR/Cas9 C. elegans Genome Editing
CRISPR/Cas9 genome editing to integrate the ICR assay into the unc-5 locus on Chromosome IV of the C. elegans genome was performed by injecting the germlines of adult N2 hermaphrodites with a plasmid mix (100ng/μL pMG1,30ng/μL pMG3,30ng/μL pMG14). F1 progeny of injected hermaphrodites were screened for uncoordinated movement (Unc) phenotypes, indicating editing at the unc-5 locus. Integration of the ICR assay was confirmed by PCR, and the entire integrant construct sequence was confirmed by Sanger sequencing (Sequetech).
ICR assay copy number verification
DNA was isolated from adult DLW14 hermaphrodites using a DNeasy Blood and Tissue Kit (QIAGEN) following a modified version of the Kaganovich Lab Genomic DNA Isolation using QIAGEN kit protocol. 100 adult DLW14 hermaphrodites were placed into 200 μL of M9 buffer and were washed 2x by centrifuging at 2500xg for 1 min, removing ~150μL of supernatant, and then resuspending the pelleted hermaphrodites in an additional ~150μL of M9 buffer. Following washes, the hermaphrodites were centrifuged at 2500xg for 1 min, ~180μL of supernatant was removed, and the hermaphrodites were resuspended in 200μL ATL buffer (DNeasy kit) and then were frozen at −80°C overnight. The next day, the hermaphrodites were freeze thawed 3x using liquid Nitrogen and a 65°C water bath. 20μL of Proteinase K (New England Biolabs) was added and the lysed worm solution was incubated at 56°C for 2 h. 8uL of RNase A (Sigma Aldrich) was added and the solution was incubated at room temperature for 5 min. 200μL of AL buffer (DNeasy kit) and the solution was incubated for 10 min at 56°C. 200μL of 100% ethanol was then added, and the solution was vortexed. Remaining steps of the protocol followed the published QIAGEN kit instructions. All DNA was eluted from a single column in 50μL of ddH2O and was stored at −20°C until used.
The schematic to detect duplications is outlined in Figure S1. The primers used are as follows: Primer 1 (5′- GCGGACTCCTCTCGGATAGT-3′), Primer 2 (5′- GGGCGTGGAACTCCTTATCA-3′), Primer 3 (5′- TGAGGTACCAGTTCAGAGGA-3′), Primer 4 (5′- TGAAGTC CGCTATTACAATGAAGT-3′). Primer 1 and 2 amplifies the left genome-construct junction, while primers 3 and 4 amplifies the right genome construct junction. The annealing temperatures were determined empirically. Each 8 mL reaction PCR mix contained: 1x buffer (10x: 450mM Tris-HCl pH 8.8, 110mM (NH4)2SO4, 45mM MgCl2, 67mM b-mercaptoethanol, 44mM EDTA, 10mM each: dATP, dTTP, dGTP, and dCTP, and 1.13mg/mL non-acetylated BSA), 12.5mM Tris base, 0.2mM of each primer, 0.25U of Taq, and 0.05U of Pfu polymerase. The reaction mixture was then subjected to extension times, as denoted in Figure S1. In independent assays, this DNA-PCR reaction mix produce amplicons of > 10kb reliably.
Intersister/intrachromatid Repair (ICR) Assay
Parent (P0) hermaphrodites for ICR assays were generated by crossing. L4 stage P0 hermaphrodites were picked 16-18 h before heat shock and incubated overnight at 15°C. To improve progeny yields at later time points, where the abundance of hermaphrodite sperm limits brood size, N2 young adult males were added to these plates in some replicates of the ICR assay. Heat shock was performed by placing P0 hermaphrodites in an air incubator (refrigerated Peltier incubator, VWR Model VR16P) at 34°C for one h. Following heat shock, hermaphrodites were incubated at 20°C for 10 h and then were picked to individual NGM plates seeded with OP50. Data in Tables S1 and S2 contain data for time courses with 12 h time points and pooled times points (details outlined below).
For the time course with 12 h time points (10-22 h, 22-34 h, 34-46 h, 46-58 h, and 58-70 h): After 12 h (for which the plate contained F1 progeny for the 10-22 h time point), each P0 hermaphrodite was transferred to a new NGM plate. P0 hermaphrodites were similarly passaged to new NGM plates every 12 h for a total of 6 transfers. NGM plates with P0 hermaphrodites were maintained at 20°C, while NGM plates with F1 progeny only were placed at 15°C.
For time course with pooled time points encompassing the non-interhomolog window (10-22 h) and the interhomolog window (22-58 h): After 12 h (for which the plate contained F1 progeny for the 10-22 h time point), each P0 hermaphrodite was transferred to a new NGM plate for 36 h. After this 36 h (for which the plate contained F1 progeny for the 22-58 h time point) P0 hermaphrodites were discarded. NGM plates with P0 hermaphrodites were maintained at 20°C, while NGM plates with F1 progeny only were placed at 15° C.
F1 progeny were maintained at 15°C for 36-48 h. ~18 h before scoring for fluorescence, F1 progeny were placed in a 25°C incubator to enhance GFP expression. F1 progeny were scored for fluorescence using an Axio Zoom V16 fluorescent dissection microscope (Zeiss). F1s that expressed GFP in the pharynx, body wall, or both were transferred to individual plates for single worm lysis (as described in Intersister/intrachromatid Repair (ICR) Assay Conversion Tract Analysis methods). All other progeny were removed from the plate and discarded. If all F1 progeny were in larval developmental stages at the time of scoring, dead eggs and unfertilized oocytes on the plates were additionally quantified.
We noted that the majority of recombinant progeny with pmyo-2 (pharynx) GFP expression also exhibited pmyo-3 (body wall) GFP fluorescence. To determine if this expression pattern arose from a single locus, we assayed the segregation of GFP phenotypes in F2 progeny arising from pharynx and body wall GFP expressing ICR assay progeny (Figure S4). The ratios of segregation were consistent with Mendelian inheritance of a single locus (Figure S4B). PCR genotyping of progeny with both pmyo-2 (pharynx) and pmyo-3 (body wall) GFP expression produced products consistent with the presence of noncrossover/intrachromatid recombination events specifically (Figure S4A). Previous work has demonstrated that both the pmyo-2 and pmyo-3 promoters contain enhancers that alter the specificity of the other respective promoter’s expression pattern.43 We therefore suggest that recombinants with both pharynx and body wall GFP expression patterns arise from the enhancer activity of the upstream myo-3 promoter in noncrossover recombinants (Figure S4C). Progeny exhibiting both pharynx and body-wall GFP expression were scored as noncrossover/chromatid recombinants in all recombination frequency calculations.
We also found that a fraction of F1 ICR assay progeny exhibited weak fluorescence phenotypes only in a portion of the pharynx, body wall, or both tissues. These progeny were transferred to individual plates and maintained at 20°C. F2 progeny were visually screened for inheritance of a fluorescent phenotype. No partial tissue fluorescent F1 was ever observed to produce fluorescent progeny, indicating that these fluorescent phenotypes are a product of somatic Mos1 excision and subsequent DNA repair and are not the result of bona fide meiotic recombination. Partially fluorescent F1s were categorized as nonrecombinant when determining frequencies of meiotic sister chromatid recombination.
The ICR assay was replicated a minimum of three times for each genotype.
While performing ICR assays in N2 and xpf-1(tm2842) backgrounds in which the unc-5(lib1) and KrIs14 transgenes were inherited from a hermaphrodite, we observed a spontaneous change in results encompassing: (1) reduced recombinants at the 10-22 h time point following heat shock; and, (2) severe embryonic lethality among progeny laid 22+ h following heat shock. We were able to successfully restore function of the ICR assay by performing cross schemes to ensure that the parent hermaphrodites heat shocked in the ICR assay inherited their unc-5(lib1) allele and KrIs14 transgene from a male. We therefore recommend that future ICR assays only be performed on parent hermaphrodites who inherit these transgenes from a male. For descriptions of both cross schemes, see ‘Crosses to Generate Strains to Perform the ICR Assay’.
Crosses to Generate Strains to Perform the ICR Assay
N2 (wild-type) with ICR assay transgenes inherited from hermaphrodite: Parent hermaphrodites were generated by crossing: (1) DLW14 hermaphrodites x N2 males to generate F1 unc-5(lib1)/+ IV; KrIs14/+ V hermaphrodites.
N2 (wild-type) with ICR assay transgenes inherited from male: Parent hermaphrodites were generated by crossing: (1) N2 males x DLW14 hermaphrodites to generate F1 unc-5(lib1)/+ IV; KrIs14/+ V males, (2) F1 males x CB791 hermaphrodites to generate unc-5(lib1)/unc-5(e791) IV; KrIs14/+ V hermaphrodites.
xpf-1 with ICR assay transgenes inherited from male: Parent hermaphrodites were generated by crossing: (1) YE57 males x TG1660 hermaphrodites to generate xpf-1(tm2842)/mIn1 II males, (2) F1 males x DLW75 hermaphrodites to generate xpf-1(tm2842)/mIn1 II; unc-5(lib1)/+ IV; KrIs14/+ V males, (3) F2 males x DLW82 hermaphrodites to generate xpf-1(tm2842)/xpf-1(tm2842) II; unc-5(lib1)/unc-5(e791) IV; KrIs14/+ V hermaphrodites.
Intersister/intrachromatid Repair (ICR) Assay Conversion Tract Analyses
Genomes of fluorescent recombinant F1 progeny or the fluorescent F2 segregants of isolated recombinant F1 progeny from ICR assays were extracted by single worm lysis. Individual hermaphrodites were picked into single 10μL aliquots of worm lysis buffer (50mM KCl, 10mM TrisHCl pH 8.2, 2.5mM MgCl2, 0.45% IGEPAL, 0.45% Tween20, 0.3μg/μL proteinase K in ddH2O). Each suspended worm was then serially frozen and thawed three times by immersion in a 95% ethanol and dry ice bath followed by a 65°C water bath. Each lysate was incubated at 60°C for one h and then incubated at 95°C for 15 min to heat inactivate proteinase K. Final lysates were diluted with 10μL of ddH2O.
Recombinant loci were PCR amplified using OneTaq 2x Master Mix (New England Biolabs). Specificity of PCR reactions was determined by gel electrophoresis. Desired amplicons were extracted by PCR purification (Zymo PCR Purification Kit) if only one band was observed by electrophoresis, or gel extraction (Thermo Scientific Gel Extraction Kit) if multiple amplicons were observed. Purified amplicons were submitted for Sanger sequencing (Sequetech) with sequencing primers specific to the locus (Key Resources Table). Sequencing files were aligned to reference GFP sequences with Benchling alignment software to detect converted polymorphisms.
KEY RESOURCES TABLE
| REAGENT or RESOURCE | SOURCE | IDENTIFIER |
|---|---|---|
| Bacterial and Virus Strains | ||
| OP50 Escherichia coli | CGC | OP50 |
| TOP10 Escherichia coli chemically competent cells | Invitrogen | C4040-06 |
| Chemicals, Peptides, and Recombinant Proteins | ||
| ≥99.8% pure Tris base (Tris[hydroxymethyl]aminomethane or Trimethamine) | Bio-Rad | Cat#1610716EDU; CAS 77-86-1 |
| Dimethylsulfoxide ≥99.9% (DMSO) | VWR | 97063-136; CAS 67-68-5 |
| GeneRuler 1kb Ladder | ThermoFisher Scientific | Cat#SM0311 |
| Hydrochloric Acid, Certified ACS Plus, 36.5 to 38.0% (HCl) | ThermoFisher Scientific | 40233; CAS 7647-01-0 |
| IGEPAL® CA-630 | Sigma-Aldrich | Cat#I8896; CAS 9002-93-1 |
| Magnesium chloride (MgCl2) | Sigma-Aldritch | Cat#M8266; CAS 7786-30-3 |
| OneTaq Quick-Load 2X Master Mix w/ Standard Buffer | New England Biolabs | Cat#M0486 |
| Potassium Chloride (KCl) | VWR | Cat#MK6858-04; CAS 7447-40-7 |
| Proteinase K, Molecular Biology Grade | New England Biolabs | Cat#P8107S |
| RNAse A | Sigma Aldrich | Cat#R6148 |
| Tween® 20 | Sigma-Aldrich | Cat#P9416; CAS 9005-64-5 |
| Tris-HCl | Sigma | Cat#93363, CAS 1185-53-1 |
| Ammonium Sulfate (NH4)2SO4 | Sigma | Cat#A4915, CAS 7783-20-2 |
| Magnesium chloride (MgCl2) | Sigma | Cat#M9272, CAS 7791-18-6 |
| β-mercaptoethanol | Sigma | Cat#63689, CAS 60-24-2 |
| EDTA | Sigma | Cat#03690, CAS 60-00-4 |
| dNTPs | Sigma | Cat#DNTP100A |
| non-acetylated BSA | Ambion/Life Technologies | Cat#AM2616 |
| Tris base | Fluka | Cat#08656 |
| Cloned Pfu DNA polymerase | Agilent | Cat#600154 |
| Kapa Taq polymerase | Kapa Biosystems | Cat#BK1002 |
| Experimental Models: Organisms/Strains | ||
| C. elegans: Strain AV554 (dpy-13(e184sd) unc-5(ox171::Mos1)/ nT1 (qIs51) IV; KrIs14(Phsp-16.48::MosTransposase; lin-15B; Punc-122::GFP) / nT1 (qIs51) V) | Villeneuve Lab | AV554 |
| C. elegans: Strain CB791 (unc-5(e791) IV) | Caenorhabditis Genetics Center | CB791 |
| C. elegans: Strain DLW14 (unc-5(lib1 [intersister repair assay Pmyo-3::GFP(−) + unc-119(+) + Pmyo-2::GFP(Mos1)]) IV; KrIs14(Phsp-16.48::MosTransposase; lin-15B; Punc-122::GFP) V) | This Study | DLW14 |
| C. elegans DLW75: Strain (xpf-1(tm2842) II; unc-5(lib1 [intersister repair assay Pmyo-3::GFP(−) + unc-119(+) + Pmyo-2::GFP(Mos1)]) IV; KrIs14(Phsp-16.48::MosTransposase; lin-15B; Punc-122::GFP) V) | This Study | DLW75 |
| C. elegans: Strain DLW82 (xpf-1(tm2842) II; unc-5(e791) IV) | This Study | DLW82 |
| C. elegans: Strain EN909 (KrIs14(Phsp-16.48::MosTransposase; lin-15B; Punc-122::GFP) V) | CGC | EN909 |
| C. elegans: Strain N2 (Wild-type) | CGC | N2 |
| C. elegans: Strain TG1660 (xpf-1(tm2842) II) | CGC | TG1660 |
| Oligonucleotides | ||
| DLO546 (5’-AGTTGGTAATGGTAGCGACC-3’) | This Study | DLO546 |
| DLO638 (5’-ACGAAGGAGGGTAGGTGTTG-3’) | This Study | DLO638 |
| DLO640 (5’-TTGAGCCGGCTTCTTCACTA-3’) | This Study | DLO640 |
| DLO641 (5’-TTAGAAGTCAGAGGCACGGG-3’) | This Study | DLO641 |
| DLO695 (5’-TGGCCAAAGGACCCAAAG-3’) | This Study | DLO695 |
| DLO822 (5’-ATTTTAACCCTCGGGGTACG-3’) | This Study | DLO822 |
| DLO823 (5’-TCCATGCCATGTGTAATCCCA-3’) | This Study | DLO823 |
| DLO824 (5’-AGATCCATCTAGAAATGCCGGT-3’) | This Study | DLO824 |
| Recombinant DNA | ||
| pMG1 | This Study | pMG1 |
| pMG3 | This Study | pMG3 |
| pMG13 | This Study | pMG13 |
| pDL23 | GenScript | pDL23 |
| pJW1285 | Jorgensen Lab | pJW1285 |
| pCFJ104 | Jorgensen Lab | pCFJ104 |
| Software and Algorithms | ||
| Benchling Align Sequences Tool | Benchling | https://help.benchling.com/en/ |
| DescTools [v0.99.37] | R package | https://cran.r-project.org/web/packages/DescTools/index.html |
| Illustrator | Adobe | https://www.adobe.com/ |
| reshape2 [v1.4.4] | R package | https://cran.r-project.org/web/packages/reshape2/index.html |
| RStudio | RStudio Team | https://rstudio.com/ |
| Tidyverse | R package | https://www.tidyverse.org/ |
| Other | ||
| QIAquick PCR Purification Kit | Qiagen | 28104 |
| Monarch® PCR & DNA Cleanup Kit | New England Biolabs | T1030S |
| DNeasy Blood and Tissue Kit | Qiagen | 69504 |
| GeneJET Gel Extraction Kit | Thermo Fisher | R1341 |
| Gibson Assembly HiFi 1 Step Master Mix | SGI-DNA | GA1100-03 |
The most efficient and effective primer set for amplifying pmyo-2 (pharynx) GFP+ loci was DLO822 + DLO823. In addition, pmyo-2 (pharynx) GFP+ loci were amplified using DLO640+DLO641. The most efficient and effective primer set for amplifying crossover loci was DLO824+DLO546.
Not all fluorescent progeny lysed were able to be PCR amplified or successfully sequenced. We were able to completely sequence 68/87 wild-type NCO recombinants, 5/14 wild-type CO recombinants, 60/70 xpf-1(tm2842) NCO recombinants, and 10/14 xpf-1(tm2842) CO recombinants.
Interhomolog Assay
The interhomolog assay was replicated following the protocol outlined in Rosu et al.4 In brief, parent (P0) hermaphrodites for interhomolog assays were generated by crossing AV554 males to CB791 hermaphrodites to generate dpy-13(e184sd) unc-5(ox171:: Mos1)/+ unc-5(e791) IV; KrIs14/+ V F1 progeny. Heat shock was performed by placing P0 hermaphrodites in an air incubator (refrigerated Peltier incubator, VWR Model VR16P) at 34° C for one h. Following heat shock, hermaphrodites were incubated at 20°C for 10 h and then were picked to individual NGM plates seeded with OP50. After 12 h each P0 hermaphrodite was transferred to a new NGM plate. P0 hermaphrodites were similarly passaged to new NGM plates every 12 h for a total of 6 transfers. Following transfer, the number of eggs laid by each hermaphrodite was scored. ~48–60 h following transfer, F1 progeny were scored for recombinant phenotypes. For details in determining noncrossover and crossover progeny, see Rosu et al.4
Ionizing radiation treatment and quantification of both brood viability and ovulation rates
L4 stage hermaphrodites were picked 16–18 h before irradiation and incubated overnight at 15°C. Irradiation was performed using a 137Cs source (University of Oregon). Following irradiation, hermaphrodites were singled to individual NGM plates with OP50 lawns and were maintained at 20°C. At 10 h and 46 h following irradiation, the hermaphrodites were transferred to new NGM plates seeded with OP50. The proportion of hatched F1 progeny, dead eggs, and unfertilized oocytes were scored 36-48 h following hermaphrodite removal. Brood viability was calculated as (Hatched Progeny) / (Hatched Progeny + Dead Eggs). Normalized brood viability was calculated by dividing the brood viability of each irradiated hermaphrodite within each scored time point (10-22 h, 22-46 h) by the mean brood viability of unirradiated hermaphrodites. Brood viability experiments were replicated three times for each genotype and irradiation dose, with the broods of n = 5 hermaphrodites scored per replicate. To ensure that ovulation was similar between all genotypes assessed, progeny and unfertilized oocytes laid 0-10 h following irradiation treatment were additionally scored but were not included in brood viability calculations or analyses.
Quantification of ICR assay F2 segregant phenotypes
Fluorescent recombinant ICR assay progeny were identified following the protocols described above using a total of 28 parent hermaphrodites generated through cross scheme #2. However, instead of performing the full time course, parent hermaphrodites were discarded following the 22-34 h time point. n = 11 F1 progeny were identified that expressed GFP both in the body wall and in the pharynx and n = 1 F1 progeny was identified that expressed GFP in the body wall only. Each of these recombinants was placed on an individual NGM plate seeded with OP50 and was incubated at 20°C. Each recombinant was monitored daily to determine if it had laid eggs. If > 30 eggs were visible on the plate or the F1 recombinant was visually egg laying defective, identified by internal egg hatching inside of the F1, the F1 recombinant was lysed for PCR analysis (Figure S4A). F2 segregants were maintained at 20°C for an additional 24 h, and then were scored for fluorescent phenotypes using an Axio Zoom V16 fluorescent dissection microscope (Zeiss).
QUANTIFICATION AND STATISTICAL ANALYSIS
All statistics were calculated in R (v4.0.3). Data wrangling was performed using the Tidyverse package (v1.3.0). Proportions of recombinant intersister/intrachromatid repair assay or interhomolog repair assay progeny and proportions of ‘short’ and ‘long’ conversion tracts (Figures 1B, 2, and 3C) were compared using Fisher’s Exact Test. Brood viability between time points within the same genotype and between genotypes within the same time points were compared using Mann-Whitney U tests (Figure 4). Segregation ratios of F2 progeny from F1 ICR assay recombinants (Figure S4B) were compared to an expected distribution for mendelian segregation of a dominant phenotype arising from a single locus (75% parental phenotype, 25% no GFP expression) by Chi Square Tests of Goodness of Fit. For all tests, statistical significance was determined as a p value equal to or less than 0.05 following correction for multiple comparisons, if applicable. 95% confidence intervals (Figures 1B, 2, 3C, and S4B; Tables S1 and S2) were calculated using the DescTools package (v0.99.30).
Supplementary Material
Highlights.
New assay to monitor DNA break repair with sister chromatid or same DNA molecule
Sister chromatid or same DNA molecule used late in prophase I as repair partner
Sequencing of noncrossover conversion tracts of homolog-independent repair events
XPF-1 promotes late homolog-independent chromatid repair events
ACKNOWLEDGMENTS
We thank the CGC(funded by National Institutes of Health P40 OD010440) for strains. We thank A. Villeneuve, C. Cahoon, and N. Kurhanewicz for comments on the manuscript. We also thank K. Sugioka for his insights into the longrange interactions of the myosin promoters within the ICR assay. We also thank O. Rog and D. Almanzar for sharing their manuscript and data prior to publication. This work was supported by the National Institutes of Health T32GM007413 and Advancing Science in America (ARCS) Foundation Award to E.T.; National Institutes of Health R25HD070817 to C.C., A.H., and A.S.; National Institutes of Health R01HD098129 to F.C., and National Institutes of Health R00HD076165 and R35GM128890 to D.E.L. T.P. was supported by the Cockrell Endowment Fellowship. D.E.L. is also a recipient of a March of Dimes Basil O’Connor Starter Scholar award and Searle Scholar Award.
Footnotes
DECLARATION OF INTERESTS
The authors declare that they have no competing interests.
SUPPLEMENTAL INFORMATION
Supplemental information can be found online at https://doi.org/10.1016/j.cub.2021.03.008.
REFERENCES
- 1.Handel MA, and Schimenti JC (2010). Genetics of mammalian meiosis: regulation, dynamics and impact on fertility. Nat. Rev. Genet. 11,124–136. [DOI] [PubMed] [Google Scholar]
- 2.Gray S, and Cohen PE (2016). Control of meiotic crossovers: from double-strand break formation to designation. Annu. Rev. Genet. 50,175–210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Lao JP, and Hunter N (2010). Trying to avoid your sister. PLoS Biol. 8, e1000519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Rosu S, Libuda DE, and Villeneuve AM (2011). Robust crossover assurance and regulated interhomolog access maintain meiotic crossover number. Science 334, 1286–1289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Hayashi M, Chin GM, and Villeneuve AM (2007). C. elegans germ cells switch between distinct modes of double-strand break repair during meiotic prophase progression. PLoS Genet. 3, e191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Lemmens BBLG, Johnson NM, and Tijsterman M (2013). COM-1 promotes homologous recombination during Caenorhabditis elegans meiosis by antagonizing Ku-mediated non-homologous end joining. PLoS Genet. 9, e1003276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Goldfarb T, and Lichten M (2010). Frequent and efficient use of the sister chromatid for DNA double-strand break repair during budding yeast meiosis. PLoS Biol. 8, e1000520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Robert VJ, Davis MW, Jorgensen EM, and Bessereau J-L (2008). Gene conversion and end-joining-repair double-strand breaks in the Caenorhabditis elegans germline. Genetics 180, 673–679. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Fasullo MT, and Davis RW (1987). Recombinational substrates designed to study recombination between unique and repetitive sequences in vivo. Proc. Natl. Acad. Sci. USA 84, 6215–6219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Kadyk LC, and Hartwell LH (1992). Sister chromatids are preferred over homologs as substrates for recombinational repair in Saccharomyces cerevisiae. Genetics 132, 387–402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Pierce AJ, Johnson RD, Thompson LH, and Jasin M (1999). XRCC3 promotes homology-directed repair of DNA damage in mammalian cells. Genes Dev. 13, 2633–2638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Johnson RD, Liu N, and Jasin M (1999). Mammalian XRCC2 promotes the repair of DNA double-strand breaks by homologous recombination. Nature 401, 397–399. [DOI] [PubMed] [Google Scholar]
- 13.Johnson RD, and Jasin M (2000). Sister chromatid gene conversion is a prominent double-strand break repair pathway in mammalian cells. EMBO J. 19, 3398–3407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Bessereau JL, Wright A, Williams DC, Schuske K, Davis MW, and Jorgensen EM (2001). Mobilization of a Drosophila transposon in the Caenorhabditis elegans germ line. Nature 413, 70–74. [DOI] [PubMed] [Google Scholar]
- 15.Robert V, and Bessereau J-L (2007). Targeted engineering of the Caenorhabditis elegans genome following Mos1-triggered chromosomal breaks. EMBO J. 26, 170–183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Engels WR, Johnson-Schlitz DM, Eggleston WB, and Sved J (1990). High-frequency P element loss in Drosophila is homolog dependent. Cell 62, 515–525. [DOI] [PubMed] [Google Scholar]
- 17.McVey M, Larocque JR, Adams MD, and Sekelsky JJ (2004). Formation of deletions during double-strand break repair in Drosophila DmBlm mutants occurs after strand invasion. Proc. Natl. Acad. Sci. USA 101, 15694–15699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Davis MW, Hammarlund M, Harrach T, Hullett P, Olsen S, and Jorgensen EM (2005). Rapid single nucleotide polymorphism mapping in C. elegans. BMC Genomics 6, 118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Almanzar DE, Gordon SG, and Rog O (2021). Meiotic sister chromatid exchanges are rare in C. elegans. Curr. Biol Published online March 18, 2021. 10.1016/j.cub.2020.11.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Lemmens BBLG, and Tijsterman M (2011). DNA double-strand break repair in Caenorhabditis elegans. Chromosoma 120, 1–21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Macaisne N, Kessler Z, and Yanowitz JL (2018). Meiotic double-strand break proteins influence repair pathway utilization. Genetics 210, 843–856. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Adamo A, Montemauri P, Silva N, Ward JD, Boulton SJ, and La Volpe A (2008). BRC-1 acts in the inter-sister pathway of meiotic double-strand break repair. EMBO Rep. 9, 287–292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Jaramillo-Lambert A, Ellefson M, Villeneuve AM, and Engebrecht J (2007). Differential timing of S phases, X chromosome replication, and meiotic prophase in the C. elegans germ line. Dev. Biol. 308, 206–221. [DOI] [PubMed] [Google Scholar]
- 24.Saito TT, Youds JL, Boulton SJ, and Colaiácovo MP (2009). Caenorhabditis elegans HIM-18/SLX-4 interacts with SLX-1 and XPF-1 and maintains genomic integrity in the germline by processing recombination intermediates. PLoS Genet. 5, e1000735. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.O’Neil NJ, Martin JS, Youds JL, Ward JDR, Petalcorin MI, Rose AM, and Boulton SJ (2013). Joint molecule resolution requires the redundant activities of MUS-81 and XPF-1 during Caenorhabditis elegans meiosis. PLoS Genet. 9, e1003582. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Saito TT, Lui DY, Kim HM, Meyer K, and Colaiácovo MP (2013). Interplay between structure-specific endonucleases for crossover control during Caenorhabditis elegans meiosis. PLoS Genet. 9, e1003586. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Agostinho A, Meier B, Sonneville R, Jagut M, Woglar A, Blow J, Jantsch V, and Gartner A (2013). Combinatorial regulation of meiotic holliday junction resolution in C. elegans by HIM-6 (BLM) helicase, SLX-4, and the SLX-1, MUS-81 and XPF-1 nucleases. PLoS Genet. 9, e1003591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Manandhar M, Boulware KS, and Wood RD (2015). The ERCC1 and ERCC4 (XPF) genes and gene products. Gene 569, 153–161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Bhargava R, Onyango DO, and Stark JM (2016). Regulation of single-strand annealing and its role in genome maintenance. Trends Genet. 32, 566–575. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Bae W, Hong S, Park MS, Jeong HK, Lee MH, and Koo HS (2019). Single-strand annealing mediates the conservative repair of double-strand DNA breaks in homologous recombination-defective germ cells of Caenorhabditis elegans. DNA Repair (Amst.) 75, 18–28. [DOI] [PubMed] [Google Scholar]
- 31.Yin Y, and Smolikove S (2013). Impaired resection of meiotic double-strand breaks channels repair to nonhomologous end joining in Caenorhabditis elegans. Mol. Cell. Biol. 33, 2732–2747. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Hicks WM, Kim M, and Haber JE (2010). Increased mutagenesis and unique mutation signature associated with mitotic gene conversion. Science 329, 82–85. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Fleck O, Lehmann E, Schär P, and Kohli J (1999). Involvement of nucleotide-excision repair in msh2 pms1-independent mismatch repair. Nat. Genet. 21,314–317. [DOI] [PubMed] [Google Scholar]
- 34.Crown KN, McMahan S, and Sekelsky J (2014). Eliminating both canonical and short-patch mismatch repair in Drosophila melanogaster suggests a new meiotic recombination model. PLoS Genet. 10, e1004583. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Marsolier-Kergoat MC, Khan MM, Schott J, Zhu X, and Llorente B (2018). Mechanistic view and genetic control of DNA recombination during meiosis. Mol. Cell 70, 9–20.e6. [DOI] [PubMed] [Google Scholar]
- 36.Peterson SE, Keeney S, and Jasin M (2020). Mechanistic insight into crossing over during mouse meiosis. Mol. Cell 78, 1252–1263.e3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Yin Y, and Petes TD (2014). The role of Exo1p exonuclease in DNA end resection to generate gene conversion tracts in Saccharomyces cerevisiae. Genetics 197, 1097–1109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Ertl HA, Russo DP, Srivastava N, Brooks JT, Dao TN, and LaRocque JR (2017). The role of Blm helicase in homologous recombination, gene conversion tract length, and recombination between diverged sequences in Drosophilamelanogaster. Genetics 207, 923–933. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Carvajal-Garcia J, Crown KN, Ramsden DA, and Sekelsky J (2020). DNA polymerase theta suppresses mitotic crossing over. bioRxiv. 10.1101/2020.11.27.400952. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.van Schendel R, Roerink SF, Portegijs V, van den Heuvel S, and Tijsterman M (2015). Polymerase ϴ is a key driver of genome evolution and of CRISPR/Cas9-mediated mutagenesis. Nat. Commun. 6, 7394. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Volkova NV, Meier B, González-Huici V, Bertolini S, Gonzalez S, Vöhringer H, Abascal F, Martincorena I, Campbell PJ, Gartner A, and Gerstung M (2020). Mutational signatures are jointly shaped by DNA damage and repair. Nat. Commun. 11, 2169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Bickel JS, Chen L, Hayward J,Yeap SL,Alkers AE, and Chan RC (2010). Structural maintenance of chromosomes (SMC) proteins promote homolog-independent recombination repair in meiosis crucial for germ cell genomic stability. PLoS Genet. 6, e1001028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Okkema PG, Harrison SW, Plunger V, Aryana A, and Fire A (1993). Sequence requirements for myosin gene expression and regulation in Caenorhabditis elegans. Genetics 135, 385–404. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The published article includes all ICR assay datasets generated or analyzed in this study. Datasets detailing the per-hermaphrodite brood viability and ovulation counts used to generate Figures 4 and S2 are available upon request.


