Mesenchymal vesicular GHR supports alveolar epithelial progenitor cell renewal and limits lung fibrosis.
Abstract
Recent studies have identified impaired type 2 alveolar epithelial cell (ATII) renewal in idiopathic pulmonary fibrosis (IPF) human organoids and severe fibrosis when ATII is defective in mice. ATIIs function as progenitor cells and require supportive signals from the surrounding mesenchymal cells. The mechanisms by which mesenchymal cells promote ATII progenitor functions in lung fibrosis are incompletely understood. We identified growth hormone receptor (GHR) is mainly expressed in mesenchymal cells, and its expression is substantially decreased in IPF lungs. Higher levels of GHR expression correlated with better lung function in patients with IPF. Profibrotic mesenchymal cells retarded ATII growth and were associated with suppressed vesicular GHR expression. Vesicles enriched with Ghr promote ATII proliferation and diminished pulmonary fibrosis in mesenchymal Ghr-deficient mice. Our findings demonstrate a previously unidentified mesenchymal paracrine signaling coordinated by GHR that is capable of supporting ATII progenitor cell renewal and limiting the severity of lung fibrosis.
INTRODUCTION
The direct and indirect interactions between mesenchymal cells and epithelial progenitor cells in the developmental and adult lung are an evolutionarily conserved phenomenon (1–4). Normal mesenchymal cells can positively contribute to the maintenance and renewal of epithelial progenitor cells in development and homeostasis (5). In contrast, fibrotic mesenchymal cells are less supportive of epithelial progenitor cell renewal (4). Mesenchymal cells can secrete cytokines and growth factors that influence epithelial progenitor cell functions (6, 7). The nature of epithelial-mesenchymal interactions that either promote or deter progenitor cell renewal is incompletely understood. Understanding the mesenchymal contribution to the regenerative capacity of alveolar progenitor cells under homeostatic and disease states could provide important new clues into the renewal of epithelial progenitors and pathogenesis of fibrosis.
Recent studies using elegant lineage tracing and/or single-cell analysis revealed that diverse mesenchymal cells influence the epithelial progenitor cell niche under homeostatic conditions. Parabronchial smooth muscle progenitors lineaged by Myh11 in the distal mesenchyme have been shown to secrete Fgf10 to promote Scgb1a1 cell proliferation after naphthalene injury (8). Pdgfra+ cells play an important role in lung development and alveolarization (9). Pdgfra+ cells facilitate the type 2 alveolar epithelial cell (ATII) formation of sphere-like colonies in vitro (10). Lgr5-expressing mesenchymal cells promote alveolar differentiation, and Lgr6-expressing cells direct airway differentiation of Scgb1a1+ progenitors (3). Axin2+/Pdgfra+ cells support alveolar growth and regeneration through functional signaling pathways, including interleukin-6 (IL-6)/Stat3, Bmp, and Fgf (4, 6). Fibroblasts provide short-range Wnt signals to neighboring ATII stem cells and maintain their stemness (7). Ectopic hedgehog activation in the Pdgfra+ fibroblasts led to loss of distal alveoli and airspace enlargement (11). Transforming growth factor–β (TGF-β), a central mediator of fibrogenesis, impairs fibroblast ability to support adult lung epithelial progenitor cell organoid formation in vitro (12). These reports raise the possibility that the impairment of ATII renewal in idiopathic pulmonary fibrosis (IPF) lung could be due to the loss of supportive functions in profibrotic fibroblasts. However, it remains unclear what changes occur in secreted supportive factors from mesenchymal cells during fibrosis and what the subsequent impact on the epithelial niche is.
Growth hormone receptor (GHR) belongs to the class I cytokine receptor family and has been shown to signal, at least in part, through the Janus-family tyrosine kinase–signal transducer and activator of transcription (STAT) pathway (13). The only ligand of GHR is growth hormone (GH). GH is secreted by the pituitary gland and released into circulation at different concentrations over 24-hour periods (14, 15). A clinical case report showed that a male with mutation in GHR exon 5(p.D137N) developed severe lymphocytic interstitial pneumonitis, parenchymal fibrosis, bronchiectasis, and emphysema, and treatment with GH had no clear improvement (16). Ghr signaling can promote the regeneration of hepatocytes in adult liver (17). GHR deficiency exacerbates fibrosis in a mouse model of inflammatory cholestasis and liver fibrosis (18–20). Fibroblasts express GHR and through secretion of insulin-like growth factor 1 (IGF-1) can modulate mammary epithelial growth in breast tissue, especially in puberty (18). Although Ghr is expressed in the adult lung, it is unknown whether there is a role in regeneration under homeostatic and disease conditions.
Juxtaposing cells communicate by using extracellular vesicles (EVs) (21, 22), which are heterogeneous complex cargoes containing proteins, lipids, and nucleic acids. EVs can deliver their content into target cells, thereby influencing the behavior of the recipient cells (23). Exosomes are one type of EV, with size ranges from 30 to 150 nm. EVs derived from different cell types have different functions, and EVs have been found to play a role in the pathogenesis of lung fibrosis (21, 24). Evidence that stromal-derived exosomes support hematopoietic stem cells in vitro has been reported recently (25). Cancer stromal cell RNA can transfer to breast tumor cells through exosomes (26). A recent report showed that the lung spheroid cell secretome and exosomes could attenuate and resolve bleomycin- and silica-induced fibrosis by reestablishing normal alveolar structure and decreasing both collagen accumulation and myofibroblast proliferation (27). However, it is unclear whether the niche mesenchymal cells can execute functions through exosomes in solid organ regeneration and repair.
In the current study, we identified a previously unknown role for mesenchymal Ghr in supporting the epithelial progenitor cell niche in the adult lung in homeostasis and after lung injury. We have investigated both human and mouse lungs with single-cell RNA sequencing (RNA-seq) and found that GHR is mainly expressed in the mesenchymal cells in the lung and expression was decreased in fibrotic lungs. We then analyzed a clinical database of a cohort of patients with interstitial lung disease (ILD) and IPF and found that GHR is significantly reduced in pulmonary fibrosis and positively correlated with lung functions in patients with IPF. Profibrotic mouse and human mesenchymal cells suppress epithelial progenitor cell renewal due to the diminished cellular and exosomal expression of Ghr. We found that Ghr-enriched vesicles promoted ATII regeneration and attenuated pulmonary fibrosis in mesenchymal Ghr-deficient mice. Our study establishes a new role for mesenchymal Ghr in the regeneration of epithelial progenitor cell function and regulation of pulmonary fibrosis in mice. Restoring Ghr by vesicles that abrogate pulmonary fibrosis could provide a new potential therapeutic approach to fibrosing lung diseases.
RESULTS
GHR is down-regulated in severe fibrotic human lung
We mined publicly available global gene expression databases attained from lung tissues of 254 individuals with ILD and 108 controls, who went for surgery for the investigation of a nodule and have no chronic lung disease by computed tomography or pathology [Lung Genomics Research Consortium (LGRC) and Gene Expression Omnibus (GEO) number GSE47460]. These data indicated that patients with ILD exhibited significantly lower GHR mRNA levels in whole lung tissue (Fig. 1A). Next, we analyzed GHR expression in human lung tissue collected from seven IPF and seven matched controls by immunohistochemistry. Table S1 shows the demographic and clinical characteristics of participants in the immunofluorescence and RNAscope experiments. Consistent with the LGRC data, immunofluorescence revealed decreased GHR protein expression levels in severe fibrotic areas of human IPF lungs (Fig. 1, B and C). Furthermore, we performed RNAscope, a method that can detect mRNA expression in tissue sections. The GHR mRNA levels were significantly reduced in severe fibrotic areas of human IPF lungs compared with normal lungs (Fig. 1, D and E). The protein levels of GHR in IPF and healthy lung lysates did not show significant differences, but a trend toward reduced GHR in IPF lung lysates was observed (fig. S1A). In IPF lungs, where patchy interstitial fibrosis alternates with nonfibrotic regions, we found that GHR expression was significantly decreased in the fibrotic areas compared to the nonfibrotic regions (Fig. 1, F and G). Together, these findings suggest that GHR down-regulation is a feature of fibrotic lungs.
Fig. 1. Decreased expression of GHR in fibrotic human lungs.
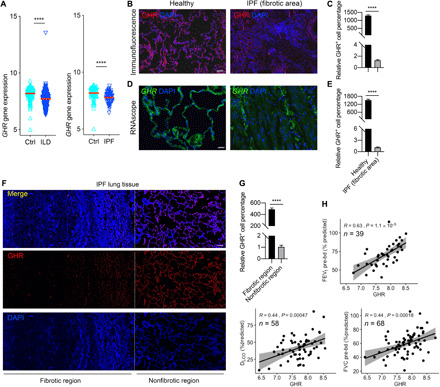
(A) GHR gene expression in lung tissues of healthy and individuals with ILD or IPF (Ctrl, n = 136; ILD, n = 255; IPF, n = 68) in LGRC cohort. (B and C) Representative images of immunofluorescence (B) and quantitative analysis (C) for the protein levels of GHR in lung tissues of healthy and IPF individuals. (D and E) RNAscope analysis (D) and quantitative analysis (E) for the RNA levels of GHR in lung tissues of healthy and IPF individuals. (F) Representative confocal microscopy GHR immunostaining for IPF lung tissues shows decreased GHR protein expression in severe fibrotic area of IPF lung tissues. (G) Quantification of Ghr protein expression with DNA stain [4′,6-diamidino-2-phenylindole (DAPI), blue] from staining in (F). (H) Pearson correlation coefficients of FEV1 and FVC before bronchodilator (bd) and DLCO as a function of GHR gene expression in the lungs of patients with IPF. R and P values, as well as the numbers of patients analyzed, are indicated in the figures. Scale bars, 100 μm (D), 200 μm (B), and 500 μm (F). ****P < 0.0001 by unpaired two-tailed Student’s t test, Bars represent means ± SEM.
To determine whether the GHR gene expression levels are associated with pulmonary function in individuals with IPF, we analyzed lung functions including forced expiratory volume in one second (FEV1) and forced vital capacity (FVC) before bronchodilator usage, as well as the diffusing capacity of the lung for carbon monoxide (DLCO) from an IPF patient cohort derived from the LGRC dataset. Using Pearson correlation analysis, we found that decreased GHR gene expression was positively correlated with disease severity, as reflected by FEV1, FVC, and DLCO (Fig. 1H). These data suggest there may be clinically relevant lung function differences in patients with IPF based on the gene expression levels of GHR.
GHR is mainly expressed in mesenchymal cells and is significantly decreased in fibrotic mesenchymal cells
We next sought to determine the cellular source of GHR. To determine the cell-specific expression of GHR in IPF and normal human lung tissue, we performed dual immunofluorescence staining of GHR with the mesenchymal cell marker COL1A1, ATII marker HTII-280, or endothelial cell marker CD31. GHR was ubiquitously expressed in the cell membrane and had good colocalization with the COL1A1+ mesenchymal cells in normal human lungs (Fig. 2A) but was markedly decreased in the COL1A1+ mesenchymal cells in the IPF lungs (Fig. 2B). Very few HTII-280+ ATIIs expressed GHR in normal human lungs and was even more rarely found in IPF fibrotic lungs (Fig. 2, C and D). Few CD31+ endothelial cells expressed GHR in the normal human lungs, and even fewer CD31+GHR+ endothelial cells were detected in IPF fibrotic lungs (Fig. 2, E and F). These data suggest that GHR-expressing cells are mainly mesenchymal cells in the human lungs, and GHR expression is down-regulated in mesenchymal cells, ATIIs, and endothelial cells of severe fibrotic lungs.
Fig. 2. Most of the mesenchymal cells, and very few ATIIs, are GHR positive in normal lung but not in IPF fibrotic area.
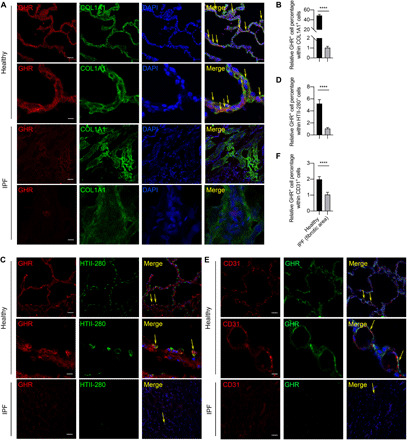
(A) Representative confocal microscopy images of GHR and COL1A1 coimmunostaining for healthy and IPF human lung tissues. (B) Quantification of GHR and COL1A1 coexpression by colocalization calculated from staining in (A). (C) GHR and HTII-280 coimmunostaining reveals that a small portion of HTII-280+ATII cells are GHR positive in normal human lungs and no GHR+HTII-280+ cells in fibrotic human lungs. (D) Quantification of GHR and HTII-280 coexpression by flow cytometry or by colocalization calculated from staining in (C). (E) Rare CD31+GHR+ endothelial cells were found in normal lungs but not IPF lungs. (F) Quantification of GHR and CD31 coexpression by colocalization calculated from staining in (E). Arrows show the coexpressing cells. Scale bars, 100 μm (A to C, lower power images) and 10 μm (A to C, higher power images). Bars represent means ± SEM, n = 7 biological replicates. ****P < 0.0001 by unpaired two-tailed Student’s t test.
Next, we evaluated Ghr expression in murine lungs. We analyzed our single-cell RNA-seq datasets (GSE104154 and GSE134948) and found that within the total lung single cells, Ghr is mainly expressed in the Col1a1-expressing mesenchymal cell cluster (Fig. 3A). We then performed subset analysis and reclustered the mesenchymal cells defined by Col1a1 and Acta2 (CAMC), and these CAMC cells clustered into at least three subpopulations (Fig. 3B). Ghr-expressing cells were scattered in all these clusters, and most of them (~88.35%, within 2756 Ghr+ cells, 2435 of them are Col1a1+) coexpress Col1a1 (Fig. 3B). These single-cell RNA-seq data suggested that similar to human lungs, Ghr expression is mainly in the mesenchymal cells of the mouse lungs. We further examined the relationship between Ghr and previously defined niche-supporting mesenchymal cell markers, including Pdgfra, Tcf21, Axin2, Lgr5, and Lgr6. We found that Pdgfra, Tcf21, and Axin2 are mainly expressed in Col1a1-expressing mesenchymal clusters (CMC), whereas Lgr5 and Lgr6 are mainly expressed in Acta2-expressing mesenchymal clusters (AMC). Ghr+Pdgfra+ cells (1743 cells) represent 76.81% of the total Ghr+ cells (2269 cells) in CMC (fig. S2A). Ghr and Tcf21 coexpression data showed that around 60% (1354 cells) of the total Ghr+ cells are Tcf21+ in the CMC (fig. S2B). There are 706 Axin2+ cells in the CMC of our dataset, and ~45% of them are Ghr+ (fig. S2C). In the AMC, there are 180 Lgr5+ cells and 200 Lgr6+ cells (fig. S2D). A total of 56% of the Lgr5+ cells and 62% of the Lgr6+ cells are Ghr+ (fig. S2E). These data suggest that most of the Ghr+ mesenchymal cells are niche-supporting mesenchymal cells.
Fig. 3. Mouse lung mesenchymal expression of Ghr.
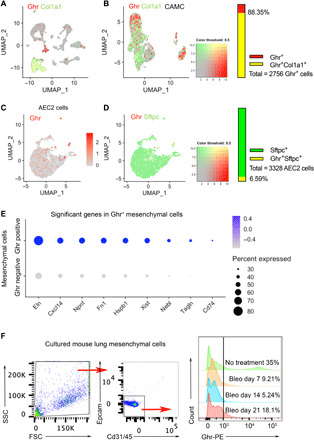
(A) Single-cell transcriptomic sequencing of wild-type (WT) mouse lung total single cells. Ghr gene expression is mainly in Col1a1+ cell cluster. (B) CAMC (mesenchymal cells defined by Col1a1 and Acta2) were reclustered and plotted for Ghr and Col1a1 coexpression. Around 88.35% of the total Ghr+ cells are Col1a1+. (C) Ghr expression in ATII cells by single-cell RNA-seq analysis. About 6.59% Sftpc+ ATII cells are Ghr+. (D) ATII expression of Ghr and Sftpc. (E) Top up-regulated genes for Ghr-positive mesenchymal cells compared with Ghr-negative mesenchymal cells. (F) Fluorescence-activated cell sorting (FACS) analysis of Ghr on cell surface of cultured passage 3 primary lung mesenchymal cells (Epcam+Cd31/45+ cells were excluded) from normal and bleomycin-treated mice (7, 14, and 21 days after bleomycin administration).
To determine whether Ghr is expressed in mouse ATII cells, we reclustered ATII cells as defined by Sftpc+ expression and found a small portion (~6.59%, of the 3122 Sftpc+ cells, 206 of them are Ghr+) of ATII cells are expressing Ghr (Fig. 3, C and D). These data suggest that Ghr is mainly expressed in mesenchymal cells but rarely expressed in ATII cells in mouse lungs, which is consistent with the Ghr cell type expression patterns we found in human lungs. Ghr+ mesenchymal cells were enriched with genes Eln, Cxcl14, Npnt, Fn1, Hspb1, Xist, Nebl, Tagln, and Cd74 (Fig. 3E). Of these, Eln, Cxcl14, Npnt, and Nebl are genes enriched in a previously defined cohort of Col13a1-expressing matrix fibroblasts (28). Npnt is a gene enriched in a recently defined distal mesenchymal cell subset with a proposed role in alveolar stem cell regeneration (11).
We then evaluated Ghr expression in mesenchymal cells of mouse lungs following bleomycin-induced injury. We harvested mesenchymal cells from mouse lungs 7, 14, and 21 days after injury and cultured them in vitro. Ghr-positive cells represented 35% of the total normal mesenchymal cells by fluorescence-activated cell sorting (FACS) analysis, which decreased to 9.21% of the total mesenchymal cells from bleomycin-treated mouse lung on day 7, 5.24% of the day 14 mesenchymal cells, and 18.1% within the day 21 mesenchymal cells (Fig. 3F). These data demonstrated that Ghr expression is diminished in profibrotic mesenchymal cells from mouse lungs in vitro following lung injury.
Mesenchymal Ghr loss of function inhibited epithelial cell renewal and is associated with reduced chemokine expression
We then sought to determine whether mesenchymal Ghr provides a supportive signal in the epithelial progenitor cell niche. To determine whether mesenchymal Ghr can influence epithelial colony formation ex vivo, we performed colony formation assays by coculturing Ghr-deficient (Ghr−/−) and littermate control mesenchymal cells with Scgb1a1+ Club cells and Sftpc+ ATII cells (Fig. 4A). The in vitro colony formation experiments showed that Ghr deficiency in mesenchymal cells led to the reduced colony formation, in terms of both colony number and size, in both Scgb1a1+ Club cells and Sftpc+ ATII cells (Fig. 4B), suggesting that lack of mesenchymal Ghr retards epithelial progenitor cell colony formation. To determine whether epithelial progenitor cell differentiation can be affected by mesenchymal Ghr deficiency, reverse transcription polymerase chain reaction (RT-PCR) was performed to evaluate the gene expression of different types of differentiated epithelial cells. Coculturing with Ghr−/− mesenchymal cell resulted in decreased expression of Scgb1a1 (Club cell marker), Sftpc (ATII cell marker), Pdpn (AEC1 cell marker), and Aqp5 (AEC1 cell marker) in Scgb1a1+ Club cell colonies (Fig. 4C). In addition, Ghr−/− mesenchymal cells reduced Sftpc and Aqp5 expression but increased expression of the cell cycle checkpoint molecule Cdkn1a in Sftpc+ ATII cell colonies (Fig. 4D). Moreover, we used pegvisomant (PEG), a GHR antagonist that can bind to GHR and interfere with functional GHR signaling, to treat mesenchymal cells before and during the coculture of ATII progenitors. The number of ATII colonies in the presence of Ghr+/+ mesenchymal cells significantly decreased with the treatment of PEG in a dose-dependent manner (Fig. 4E). Lung mesenchymal cells isolated from Ghr−/− mice do not express surface Ghr (Fig. 4F), which is consistent with a previous report (29). Ghr−/− mesenchymal cells showed significantly reduced Ghr, Ghr transcript variant (TV) 1 and 2, and Tbx4 expression but increased cell migration–inducing and hyaluronan-binding protein (Cemip) and Tgfbr3 mRNA expression (Fig. 4G). Ghr−/− mesenchymal cells also expressed reduced Has2 gene expression, with no change in Has1 and Has3 mRNA expression (Fig. 4H). Ghr has been reported to signal through STATs, and STATs can activate downstream chemokines (30). We found that Ghr−/− mesenchymal cells demonstrated diminished gene expression of Cxcl1, Cxcl4, Cxcl5, Cxcl12, Cxcl14, and Cxcl16 (Fig. 4I). IL-6 has been reported to promote ATII renewal, and its early intervention in bleomycin-induced lung injury reduced lung fibrosis (31). We found that Ghr deficiency suppressed Il-6 gene expression in mouse lung mesenchymal cells (Fig. 4J). Together, our observations indicate that mesenchymal Ghr is required for supporting epithelial progenitor cell proliferation and differentiation and is associated with regulation of Cxc chemokines and cytokines.
Fig. 4. Ghr-deficient mesenchymal cells are less supportive for epithelial progenitor cell colony formation.
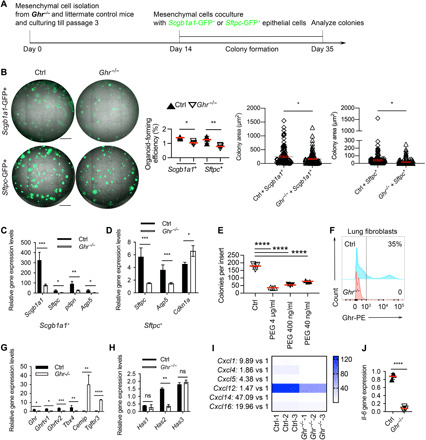
(A) Schematic of in vitro coculture colony formation assay for experiments in (B) to (D). (B) Scgb1a1-GFP+ (green fluorescent protein–positive) Club cell and Sftpc-GFP+ ATII organoids cocultured with mesenchymal cells isolated from Ghr−/− or control mouse lungs. The number and size of Scgb1a1-GFP+ Club cell and Sftpc-GFP+ ATII organoids were decreased when cocultured with Ghr−/− mesenchymal cells. (C) Expression of Scgb1a1, Sftpc, Pdpn, and Aqp5 in Scgb1a1+ Club cell organoids cocultured with lung mesenchymal cells isolated from Ghr−/− and control mice. (D) Expression of Sftpc, Pdpn, and Cdkn1a in Sftpc+ ATII cell organoids cocultured with lung mesenchymal cells isolated from Ghr−/− and control mice. (E) Pegvisomant (PEG) treatment for mesenchymal cells cocultured with ATII colonies. (F) FACS confirmation of Ghr expression on lung mesenchymal cells from Ghr−/− and control mice. (G) Ghr, Ghr TV1, Ghr TV2, Tbx4, Cemip, and Tgfbr3 expression in lung mesenchymal cells from Ghr−/− and control mice. (H) Has1, Has2, and Has3 expression in lung mesenchymal cells from Ghr−/− and control mice. (I) Chemokine gene (Cxcl1, Cxcl4, Cxcl5, Cxcl12, Cxcl14, and Cxcl16) expression in Ctrl and Ghr−/− mouse lung mesenchymal cells. (J) Il-6 gene expression in lung mesenchymal cells from Ghr−/− and control mice. Scale bars, 1 mm (B). n = 3, *P < 0.05, **P < 0.005, ***P < 0.001, and ****P < 0.0001 by unpaired two-tailed Student’s t test [B (panels 3 and 4) and J], one-way analysis of variance (ANOVA) (E), and two-way ANOVA [B (panel 2), C, D, G, and H], means ± SEM.
GHR gain of function promotes ATII colony formation and increases CXC chemokine expression
To demonstrate the translatability of GHR to human lung and investigate gain-of-function assays, we interrogated the functions of GHR by using human lung fibroblasts in the epithelial colony formation assay. The endogenous GHR function in human mesenchymal cells was tested using a GHR overexpressing (GHRTg) human normal lung fibroblast (hLF) cell line generated by lentivirus transfection (Fig. 5A). GHRTg hLFs significantly increased ATII colony formation. We found that GHRTg hLF augmented the numbers of both GhrLo and GhrHi ATII colonies (Fig. 5B), indicating that GHR overexpression in mesenchymal cells is beneficial for ATII cell growth and renewal. We further examined whether GHR overexpression alters CXC chemokine expression. We found that GHRTg hLFs demonstrated increased expression of chemokine CXCL1, CXCL2, CXCL3, CXCL5, CXCL6, CXCL8, and CXCL12 mRNAs (Fig. 5C). Wnt5a from juxtaposing mesenchymal cells can maintain ATII stemness (7). We found that WNT5A gene expression were increased in GHRTg hLFs (Fig. 5D). Furthermore, consistent with the CXC chemokine mRNA expression, GHR overexpression augmented CXCL1, CXCL2, CXCL8, CXCL10, and CXCL12 chemokine protein secretion in the culture medium of GHRTg hLFs (Fig. 5E). GHR is reported to be upstream of IGF-1 (30), and GHR deficiency is associated with reduced serum levels of IGF-1 (32). We detected increased release of IGF-1 from GHRTg hLFs compared to control lung mesenchymal cells in response to tumor necrosis factor–α (TNF-α) and IL-6 stimulation (Fig. 5F). In addition, we found that IL-6 secretion was elevated in GHRTg hLF supernatants (Fig. 5G). Moreover, we found that gene expression levels of CXCL1, CXCL2, CXCL3, CXCL8, CXCL16, and IL-6 were decreased in lung tissues from the LGRC ILD patient cohort (Fig. 5H). Together with GHR reduction in ILD lung, these data suggest that the distinct molecular profiles correlated with clinical features. Collectively, Ghr mesenchymal deficiency and overexpression regulated epithelial colony formation and were associated with chemokine expression in human mesenchymal cells, suggesting that there could be a role for the Ghr axis in regulating pulmonary fibrosis.
Fig. 5. GHR-enriched human lung mesenchymal cells promote ATII organoids associated with elevated CXC chemokine and cytokine expression.
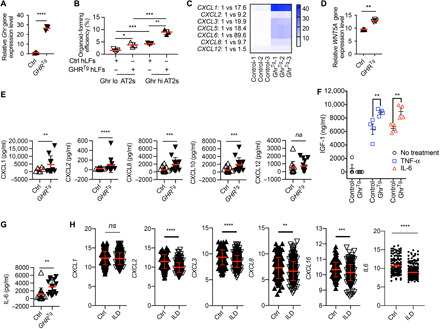
(A) Expression of GHR gene in GHR overexpressing (GHRTg) and control (Ctrl) human lung mesenchymal cells (n = 3 in each group). (B) Organoid culture of GhrLo and GhrHi ATIIs with Ctrl and GHRTg hLF (human lung fibroblasts). (C) CXC chemokine gene (CXCL1, CXCL2, CXCL3, CXCL4, CXCL6, CXCL8, and CXCL12) expression in GHRTg and Ctrl human lung mesenchymal cells (n = 3 in each group). (D) WNT5A gene expression in GHRTg and Ctrl human lung mesenchymal cells. (E) Bio-Plex Pro Human Chemokine analysis of secreted CXCL1, CXCL2, CXCL8, CXCL10, and CXCL12 levels in supernatants of cultured GHRTg and Ctrl human lung mesenchymal cells (Ctrl, n = 15; GHRTg, n = 13). (F) Enzyme-linked immunosorbent assay analysis of IGF-1 protein secretion in the supernatants of GHRTg and Ctrl human lung mesenchymal cells upon TNF-α and IL-6 stimulation (n = 4 in each group). (G) IL-6 cytokine secretion in supernatants of cultured GHRTg and Ctrl human lung mesenchymal cells (Ctrl, n = 15; GHRTg, n = 13). (H) CXCL1, CXCL2, CXCL3, CXCL8, CXCL16, and IL-6 gene expression in lung tissues of ILD (n = 255) and Ctrl (n = 136) individuals in the LGRC cohort. *P < 0.05, **P < 0.005, ***P < 0.001, and ****P < 0.0001 by unpaired two-tailed Student’s t test (A, D, E, G, and H) and two-way ANOVA (B and F), means ± SEM. ns, not significant.
GHR negatively mediates the activation and invasive capacity of lung fibroblasts
We next evaluated the role of GHR in mediating fibroblast activation and invasion of extracellular matrix (ECM). We have previously demonstrated that a subset of fibrogenic fibroblasts will invade ECM in vitro and promote fibrosis in vivo (33, 34). We found that smooth muscle actin (α-SMA) accumulation was similar in both GHRTg human lung fibroblasts and control fibroblasts at baseline. Upon TGF-β treatment, the expression of α-SMA was significantly increased in control fibroblasts, while GHRTg human lung fibroblasts demonstrated a marked reduction in α-SMA expression compared to control fibroblasts (fig. S3, A and B).
To gain additional insights into the potential mechanisms by which GHR regulates fibroblast behavior, we examined the effect of GHR on mouse lung fibroblast invasion. We isolated primary lung fibroblasts from Ghr−/− mice and littermate control and analyzed their invasive capacity. We found that invasive capacity was significantly enhanced in Ghr-null mouse lung fibroblasts (fig. S3, C and D). Collectively, these data suggest that up-regulation of GHR can prevent the activation of fibroblasts, and loss of Ghr can promote an invasive fibroblast phenotype and severe fibrosis.
Loss of Ghr in lung subepithelial mesenchyme increases mouse susceptibility to pulmonary fibrosis
We next sought to determine whether the loss of the niche-supporting factor Ghr from mesenchymal cells can affect mouse models of pulmonary fibrosis. We generated GhrΔBMC (Tbx4LME-CreER;Ghrfl/fl) mice to delete Ghr in lung mesenchymal cells and examined the impact in the bleomycin-induced fibrosis model (Fig. 6A). We have previously demonstrated that Tbx4-expressing mesenchymcal cells expand after lung injury and promote the invasive fibroblast phenotype (35). We initially deleted mesenchymal Ghr at baseline before injury to determine the basal impact on lung injury and fibrosis. This strategy resulted in 80% knockdown of Ghr in cultured lung mesenchymal cells of GhrΔBMC compared to littermate control mice (Fig. 6B). GhrΔBMC mice were more susceptible to bleomycin-induced fibrosis at day 21 (Fig. 6C), suggesting that mesenchymal Ghr regulates fibrogenesis. To gain additional insights into potential pathogenic mechanisms, we examined the impact of mesenchymal Ghr deficiency on Cxc chemokines in vivo. We collected bronchoalveolar lavage fluid (BALF) from mouse lungs and examined Cxc chemokine protein concentrations. Secretion of Cxcl1, Cxcl2, Cxcl5, Cxcl10, Cxcl11, and Cxcl12 proteins was decreased in the BALF collected from GhrΔBMC mice 21 days after lung injury (Fig. 6D), which is consistent with our in vitro data that Ghr−/− mesenchymal cells show diminished Cxc chemokine expression. We observed greater structural distortion and collagen deposition in GhrΔBMC mice (Fig. 6, E and F), suggesting that mesenchymal Ghr deficiency may retard ATII renewal leading to enhanced fibrosis. These data suggest that mesenchymal Ghr is a significant regulator in vivo of fibrogenesis.
Fig. 6. Mesenchymal Ghr-deficient mice are more susceptible to pulmonary fibrosis.
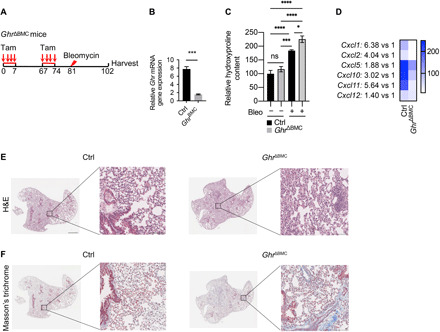
(A) A schematic diagram depicting the administration of bleomycin into the lungs of GhrΔBMC and littermate control mice. Mouse lungs were harvested on day 21. (B) Quantitative RT-PCR analysis of Ghr mRNA expression (means ± SEM) in the lungs of GhrΔBMC and littermate control mice. (C) Hydroxyproline content in the lungs of GhrΔBMC and littermate control mice harvested before and 21 days after belomycin injection (Ctrl, n = 3; GhrΔBMC, n = 3; Ctrl + Bleo, n = 19; GhrΔBMC + Bleo, n = 15). (D) Cxcl1, Cxcl2, Cxcl5, Cxcl10, Cxcl11, and Cxcl12 protein secretion in BALF of GhrΔBMC and littermate control mice harvested on day 21 after bleomycin injury. (E and F) Representative images of hematoxylin and eosin (H&E) staining (E) and Masson’s trichrome staining (F) in GhrΔBMC and littermate control mouse lungs 21 days after bleomycin injury (Ctrl, n = 19; GhrΔBMC, n = 15). Ctrl, control. Scale bars, 10 μm (E and F, higher power images) and 100 μm (E and F, lower power images). *P < 0.05, ***P < 0.001, and ****P < 0.0001 by unpaired two-tailed Student’s t test (B) and two-way ANOVA (C), means ± SEM.
Fibrotic mesenchymal cells demonstrate diminished secreted vesicular GHR levels
To determine whether profibrotic mesenchymal cells have the same supportive ability as normal mesenchymal cells for epithelial progenitor cell colony formation, human primary mesenchymal cell lines from normal and individuals with IPF were isolated and cocultured with ATIIs for colony formation assays (table S1). ATII cells cultured with IPF mesenchymal cells yielded significantly reduced colony number and size compared to healthy mesenchymal cells (Fig. 7, A to C). We sought to determine whether GHR mRNA could be secreted in vesicles from mesenchymal cells as a potential means of cell communication. We found that both IPF and normal lung fibroblasts secreted vesicles and GHR mRNA was selectively reduced in IPF mesenchymally derived vesicles (Fig. 7D). These data suggest that human fibrotic mesenchymal cells retard epithelial progenitor cell renewal and proliferation through a Ghr-mediated mechanism.
Fig. 7. Fibrotic lung mesenchymal cells retard epithelial regeneration and express less GHR.
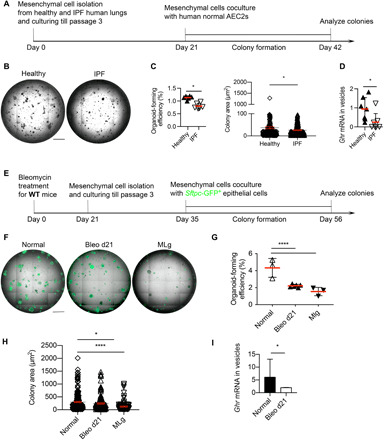
(A) Schematic of in vitro coculture human ATII colony formation assay for experiments in (B) and (C). (B) Representative images of ATII organoids cocultured with lung mesenchymal cells from healthy and IPF individuals. (C) Bar graph depicting the organoid-forming efficiency and colony area in (B). (D) GHR mRNA expression in secreted vesicles from cultured lung mesenchymal cells of healthy and IPF individuals (n = 7 in each group). (E) Schematic of in vitro cocultured mouse ATII colony formation assay for experiments in (F) to (H). (F) Representative images of ATII organoids cocultured with mesenchymal cells isolated from normal and bleomycin-treated mice (21 days after treatment), as well as the MLg mouse fibroblast cell line. (G) Organoid-forming efficiency of ATII cocultured with normal, bleo d21, and MLg mesenchymal cells. (H) Colony size of ATII cocultured with normal, bleo d21, and MLg mesenchymal cells. (I) Vesicular expression of Ghr from normal and bleomycin-treated day 21 mouse lung mesenchymal cells cultured in vitro. Scale bars, 1 mm (B and F). *P < 0.05 and ****P < 0.0001 by unpaired two-tailed Student’s t test (C, D, and I) and one-way ANOVA (G and H), means ± SEM.
To explore whether these effects could extend to mouse primary mesenchymal cell lines, colony formation assays for epithelial progenitor cells cocultured with normal and bleomycin-treated day 21 mouse lung mesenchymal cells were performed (Fig. 7E). Fibrotic mesenchymal cells showed similar levels of colony supportive ability for Sftpc+ ATII cell colony growth when compared to the MLg cells (mouse lung fibroblast cell line) but reduced colony formation compared to unchallenged mesenchymal cells (Fig. 7, F to H). To determine whether vesicles from profibrotic mesenchymal cells have different Ghr gene expression levels compared with normal mesenchymal cells, we isolated vesicles from supernatants of profibrotic mesenchymal cells and unchallenged mesenchymal cells cultured in vitro. Secreted vesicles from bleomycin day 21 mesenchymal cells demonstrated reduced Ghr mRNA expression compared with normal mesenchymal cells, consistent with the human data (Fig. 7I). These data suggest that diminished vesicular Ghr from fibrotic mouse mesenchymal cells could have a functional impact on the renewal and proliferation of the epithelial progenitor cells.
Ghr-enriched vesicles facilitate ATII colony formation in vitro
We have shown that endogenous mesenchymal Ghr plays a supportive role in the epithelial progenitor cell niche. We now sought to determine whether vesicles can transport functional Ghr to recipient ATII cells and whether exogenous Ghr-enriched vesicles can promote epithelial colony forming in vitro. To address these questions, we overexpressed the Ghr zipcode plasmid in human embryonic kidney (HEK) 293 cells and harvested vesicles secreted in the culture medium. We found that the zipcode plasmid significantly increased Ghr expression in vesicles compared to the control plasmid (fig. S4A). The morphology of vesicles was analyzed by transmission electron microscopy (TEM). The vesicles showed a round morphology and a characteristic central depression in the vesicles (fig. S4B). Nanosight analysis delineated the number and the size of the mouse lung mesenchymal cells (fig. S4C). It has been reported that Ghr only exists as mRNA in mouse vesicles (36). Ghr protein expression was detected in mesenchymal cell lysate but not in vesicles from mouse lung mesenchymal cells (fig. S4D). The Sftpc+ ATII colony formation was significantly augmented following Ghr vesicle treatment compared with control vesicles and controls without treatment (fig. S4E). Expression of Ghr at both RNA (fig. S4F) and protein levels (fig. S4G) was significantly elevated in Sftpc+ ATII colonies treated with Ghr vesicles. Furthermore, we found that the cell surface Ghr expression on epithelial colonies was increased upon Ghr vesicle treatment (fig. S4H), suggesting that vesicle Ghr can be expressed on the epithelial cell surface following transfer. We further sought to determine whether fibroblasts respond to Ghr vesicle treatment. Expression of CXCL1 and IL-6 was significantly increased in human normal lung mesenchymal cells upon Ghr vesicle treatment (fig. S4I). These data suggested that Ghr-enriched vesicles can enhance Ghr expression on epithelial cells following transfer and promote ATII proliferation in vitro and may be a relevant contributor to cell-cell communication via the Ghr axis.
Ghr-enriched vesicles ameliorate pulmonary fibrosis in mesenchymal Ghr deficiency
We have shown that administration of exogenous Ghr vesicles play a supportive role in the epithelial progenitor cell niche in vitro. We sought to determine whether Ghr-enriched vesicles could attenuate pulmonary fibrosis in mesenchymal Ghr-deficient mice. We first determined whether epithelial progenitor cells can take up vesicles following intratracheal administration. Confocal images of mouse lungs treated with mCherry-tagged Ghr vesicles showed the uptake of vesicles by Scgb1a1+ and Sftpc+ epithelial cells (Fig. 8A). To further examine whether Ghr vesicles carry Ghr mRNA to ATIIs, we sorted ATIIs from mice treated with mCherry-tagged Ghr vesicles or control vesicles. A real-time quantitative RT-PCR was used to evaluate the Ghr mRNA expression on ATIIs treated with Ghr or control vesicles. Ghr vesicles markedly increased Ghr mRNA expression on recipient ATIIs (Fig. 8B). The total Ghr protein was also elevated in ATIIs receiving Ghr vesicles compared to control vesicles (Fig. 8C). These results suggest that vesicles can carry functional Ghr to epithelial progenitor cells in vivo. We further examined the therapeutic potential for Ghr-enriched vesicles by intratracheally administrating Ghr-enriched vesicles to GhrΔBMC mice after bleomycin injury. These mice were pretreated with two courses of tamoxifen to knockdown Ghr in Tbx4+ mesenchymal cells before the bleomycin challenge (Fig. 8D). The GhrΔBMC mice treated with Ghr vesicles showed decreased hydroxyproline content compared to control vesicle–treated GhrΔBMC mice (Fig. 8E). The BALF collected from GhrΔBMC mice with Ghr vesicle treatments showed elevated concentrations of Cxcl1, Cxcl2, Cxcl5, Cxcl10, Cxcl11, Cxcl12, and Cxcl16 in relative to the control vesicle–treated group (Fig. 8F). GhrΔBMC mice with Ghr vesicle treatment also exhibited less damaged lung morphology compared to the control vesicle group (Fig. 8, G and H). These data support the concept that Ghr vesicles can ameliorate pulmonary fibrosis in mice with mesenchymal Ghr deficiency (Fig. 8I).
Fig. 8. Ghr vesicles reduce pulmonary fibrosis in mesenchymal Ghr-deficient mice.
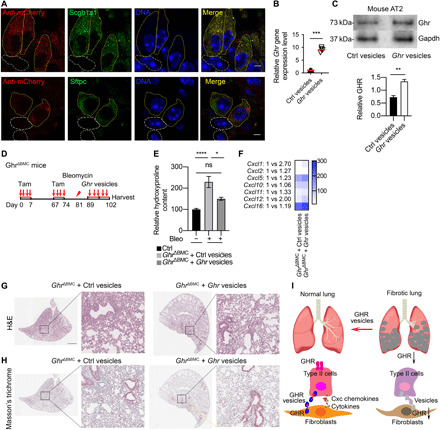
(A) Immunostaining of red fluorescent protein, Scgb1a1, Sftpc, and DAPI in mouse lungs administrated with mCherry sequence containing Ghr vesicles. Yellow solid line, mCherry and Scgb1a1 or Sftpc colocalized cells; white dotted line, Scgb1a1- or Sftpc-negative cells. (B) Ghr mRNA expression in ATII cells sorted from mouse lungs treated with Ghr or control vesicles. (C) Ghr protein expression in ATII cells sorted from mouse lungs treated with Ghr or control vesicles. (D) A schematic diagram depicting the administration of Ghr or control vesicles into the lungs of GhrΔBMC mice after bleomycin treatment. Mouse lungs were harvested on day 21. (E) Significantly decreased hydroxyproline levels were detected in Ghr vesicles and control vesicle–administered GhrΔBMC mouse lungs (Ctrl, n = 18; GhrΔBMC + Ctrl vesicle, n = 16; GhrΔBMC + Ghr vesicle, n = 18). (F) Cxcl1, Cxcl2, Cxcl5, Cxcl10, Cxcl11, Cxcl12, and Cxcl16 protein secretion in BALF of GhrΔBMC mice with Ghr or control vesicle treatments. (G and H) Representative images of H&E staining (G) and trichrome staining (H) in Ghr or control vesicle–treated GhrΔBMC mouse lung 21 days after bleomycin injury. (I) A diagram illustrating how subepithelial mesenchymal cells support ATII cells through Ghr vesicle and Ghr-coordinated CXC chemokines in normal and fibrotic conditions in the lung. Scale bars, 10 μm (A), 100 μm (G and H, higher power), and 1 mm (G and H, lower power). *P < 0.05, **P < 0.005, ***P < 0.001, and ****P < 0.0001 by unpaired two-tailed Student’s t test (B and C) and one-way ANOVA (E), means ± SEM.
DISCUSSION
One of the hurdles to developing novel therapeutics for IPF is the insufficient understanding of the molecular mechanisms that promote epithelial progenitor cell renewal. While U.S. Food and Drug Administration–approved therapies have been shown to reduce the loss of lung function, there is no evidence that fibrosis improves (37–39). This may be because both pirfenidone and nintedanib target fibroblast production of ECM but do not affect the underlying cause of IPF that may be ATII failure (40, 41). Emerging data have shown that mesenchymal cells can provide niche supportive signals to promote epithelial progenitors in the normal lung (3, 4, 7). To date, no data have delineated whether epithelial progenitors can receive supportive signals from mesenchymal cells in fibrotic lungs or whether key supportive signals are missing. In the current study, we demonstrate that mesenchymal GHR contributes to ATII renewal in vitro, is lost in fibrotic mesenchyme, and, when restored by exogenous vesicle delivery, ameliorates pulmonary fibrosis in vivo.
GHR is significantly reduced in profibrotic lung tissue, and its expression is correlated with lung function in a cohort of patients with IPF. We further found that GHR is mainly expressed in mesenchymal cells and the gain and loss of GHR function in mesenchymal cells promotes or retards epithelial progenitor function, respectively, suggesting that mesenchymal cell GHR is supportive of the epithelial niche under homeostatic conditions. In bleomycin-induced lung fibrosis, the vesicular delivery of Ghr transfers mRNA to epithelial progenitor cells and restores diminished Ghr expression in fibrotic mesenchymal cells. These data suggest that GHR supports the mesenchymal epithelial niche, and deficiency in mesenchymal cells suppresses epithelial renewal in fibrotic lung.
Our results support a model in which Ghr-expressing mesenchymal cells provide paracrine signals through vesicular Ghr transfer to epithelial progenitor cells. Epithelial progenitor cells can receive and transcribe vesicular-conserved Ghr mRNA that then function in promoting progenitor cell renewal and regeneration. GHR is expressed in very few ATII cells in the lungs, and mesenchymal GHR may serve as the reservoir for supplying epithelial progenitors GHR. The observation that GHR mRNA levels in AT2 are low but present and could mean that most of the ATIIs themselves are not producing GHR mRNA, only the ATIIs that are more receptive to receiving mesenchymal GHR mRNA and are expressing GHR protein by translating GHR mRNAs from the transporting vesicles. It is also possible that these GHR-positive ATII cells may be ATII progenitors. It is suspected that the progenitors in the healthy adult lung only represent around 1% of the total population, and slow renewal is observed in homeostasis (10). The demand for high GHR in these progenitors may be low in homeostasis. Last, it could mean rapid translation of mRNA in this cell population. GHR expression on ATIIs can presumably interact with circulating GH to stimulate cell growth and regeneration. Profibrotic mesenchymal cells are unable to provide the nutrient GHR to promote epithelial progenitor cell function. These data support mesenchymal Ghr as an important niche-supporting signal in health and disease.
Mesenchymal cells can provide a microenvironment to support, maintain, and regulate the functions and fate of epithelial progenitors (3, 4, 7, 10). It has been shown that the functions of vesicles reflect, at least in part, those of their originating cells (42). Vesicles/exosomes play an important role in intracellular communication and are capable of modifying the activity of target cells through horizontal transfer of genetic information (43). Vesicles are involved in cellular processes such as cell proliferation and immune regulation (44). Vesicles/exosomes are therefore particularly attractive for their therapeutic potential. Our current data indicate that Ghr mRNA conserved in vesicles can change the behavior of epithelial progenitor cells. Vesicular Ghr can act as a paracrine signal capable of modulating the epithelial progenitor cell microenvironment, such as promoting CXC chemokine release, and potentially orchestrating epithelial progenitor cell renewal.
In addition to the direct effect that mesenchymal GHR provides to support epithelial progenitor renewal through vesicular transport of GHR mRNA to AT2 cells, there could be indirect effects through secreted factors, such as Wnts and cytokines/chemokines. Our data show that the lung mesenchymal cells from Ghr−/− mice expressed reduced Cxcl1, Cxcl4, Cxcl5, Cxcl12, Cxcl14, and Cxcl16 and were less supportive of epithelial progenitor cell colony formation. GHR-enriched human lung mesenchymal cells expressed elevated levels of CXCL1, CXCL2, CXCL3, CXCL5, CXCL6, CXCL8, CXCL12, WNT5A, IL-6, and IGF-1 and were more supportive of epithelial progenitor cell colony formation. CXCL1, CXCL2, CXCL3, CXCL8, and CXCL16 gene expression was significantly reduced in the lung tissues of patients with ILD compared with control individuals in the LGRC cohort. Mesenchymal Ghr-deficient mice had reduced Cxcl1, Cxcl2, Cxcl5, Cxcl10, Cxcl11, and Cxcl12 secretion in BALF and increased pulmonary fibrosis 21 days after bleomycin treatment. When administrating Ghr-enriched EVs, these mice showed restored levels of Cxcl1, Cxcl2, Cxcl5, Cxcl10, Cxcl11, Cxcl12, and Cxcl16 and mitigated pulmonary fibrosis. Ghr-enriched EVs induced CXCL1 and IL-6 expression in human lung fibroblasts and promoted epithelial progenitor cell colony formation. These results indicate that mesenchymal Ghr is required for optimal epithelial progenitor cell niche function, through the direct effect of EV Ghr mRNA, and potentially an indirect effect by providing Cxc chemokines. Ghr deficiency in mesenchymal cells decreased BALF Cxc chemokines and promoted pulmonary fibrosis, while Ghr-enriched EVs restored the Cxc chemokines and alleviated pulmonary fibrosis.
Niche mesenchymal cells are not mesenchymal stem cells. Lung mesenchymal cells are local mesenchymal cells that contribute to the epithelial progenitor niche. Mesenchymal stem cells are recruited hematopoietic cells to sites of inflammation during injury and repair. The EVs derived from mesenchymal stem cell have shown to be beneficial in acute respiratory distress syndrome (ARDS), asthma, pulmonary arterial hypertension, pneumonia, and experimental models of pulmonary fibrosis (45–51). We found that mesenchymal GHR is beneficial for the epithelial progenitor niche. Vesicles that are enriched with Ghr are supportive of the epithelial progenitor niche, and Ghr vesicle delivery can reduce pulmonary fibrosis in bleomycin-induced mouse models.
The 25-nucleotide zipcode can shuttle specific RNA cargo into secreted vesicles (52). We used this method to enrich Ghr in vesicles. The Ghr expression in zipcode-transfected vesicles was around 100-fold higher than nonzipcode-transfected vesicles. The Ghr-enriched vesicles were able to induce ATII colony formation, increase the cellular and cell surface expression of Ghr in recipient ATII cells, and stimulate the expression of CXCL1 and IL-6 in human lung mesenchymal cells. Multiple intratracheal instillations of Ghr-enriched vesicles reduced bleomycin-induced pulmonary fibrosis in long-term mesenchymal Ghr-deficient mice and increased Cxc chemokine secretion in BALF. These results suggest that Ghr-enriched vesicles have the ability to ameliorate pulmonary fibrosis presumably through supporting the epithelial progenitor cell niche. In wild-type (WT) mice, we observed a trend toward reduced fibrosis with the administration of Ghr-enriched vesicles but not to the same magnitude as in the mesenchymal Ghr-deficient mice (fig. S5A). This may be related to the degree of Ghr deficiency needed to observe a treatment effect. Collectively, these data suggest that there is a relative Ghr deficiency in IPF and fibrotic mouse models, and Ghr-enriched vesicles facilitate epithelial progenitor cell niche functions in vitro through direct contact with epithelial progenitor cells and reduce the fibrotic response in vivo.
Our data suggest that the IPF lung demonstrates mesenchymal loss of GHR. The role of GH secretion lung fibrosis is unknown. GH is secreted from the pituitary gland in the brain (53). It has been reported that GH can regulate lung size and hypoxemia. Deficient pituitary secretion of GH led to a decrease in lung size and restrictive ventilatory impairment in adult patients with hypopituitarism (54). A clinical case report showed that a patient lost GH secretion after the surgical removal of craniopharyngioma-developed hypoxemia, and GH replacement therapy significantly increased the serum GH level and improved hypoxemia (55). IPF is an aging-related lung disease, most of the patients are more than 65 years of age (56). Normal aging is accompanied by a gradual decline in GH levels (57). Old rodents showed a lower average peak of GH pulses than young rodents (58). Further studies will be needed to determine whether GH levels are impaired in patients with IPF.
In conclusion, these data suggest a novel Ghr-mediated axis that regulates epithelial repair and importantly offers a new potential therapeutic approach to fibrosing lung diseases. As approaches to deliver vesicles to human diseases are further developed, restoring Ghr to the IPF lung could alter the natural history of this unremitting disease.
MATERIALS AND METHODS
Ethical compliance
All mouse maintenance and procedures were done under the guidance of the Cedars-Sinai Medical Center Institutional Animal Care and Use Committee in accordance with the institutional and regulatory guidelines. All human lung experiments were approved by the Cedars-Sinai Medical Center Institutional Review Board (IRB) and were in accordance with the guidelines outlined by the Board. Informed consent was obtained from each subject.
Mice
The Sftpc–green fluorescent protein (GFP) mice were described previously (31). The genotyping and generation of Scgb1a1CreER mice have previously been described (59). RosaGFP were purchased from the Jackson laboratory. Scgb1a1CreER mice were crossed with RosaGFP mice to generate Scgb1a1CreER-GFP mice. The Ghr−/− mouse line was a gift from V. Chesnokova and has been previously described (29). Ghrfl/fl were purchased and rederived from KOMP KO-6206 Ghrtm1c(KOMP)Wtsi line (The Jackson laboratory). Tbx4LME-CreER has been described previously (35). Tbx4LME-CreER were crossed with Ghrfl/fl to generate Ghr△BMC (Tbx4LME-CreER;Ghrfl/fl) mice. All mice were on a C57BL/6 background.
All the mice were housed in micro-isolator cages, up to five per cage, in Cedars-Sinai Medical Center (CSMC) pathogen-free facilities. All the mice received standard chow and autoclaved sterile drinking water.
Age-matched adult (8 to 16 weeks old) Ghr△BMC mice (and littermate controls) were given two courses of intragastric tamoxifen (0.2 mg/g body weight per dose, two courses of four doses within 2-month period; T5648, Sigma-Aldrich, dissolved in corn oil) to induce Cre-mediated recombination (60) before any injury.
Animal injury experiments
Mice were given one dose of intratracheal bleomycin (1.25 U/kg body weight; Hospira, dissolved in saline) to induce noninfectious murine lung alveolar injury.
Histology, immunofluorescence, and confocal imaging
At the time of tissue collection, mice were euthanized by intraperitoneal injection of a mixture of ketamine (100 mg/kg; Fort Dodge) and xylazine (10 mg/kg; Shenandoah). The chest cavity was exposed, and the lungs were cleared of blood by perfusion with cold phosphate-buffered saline (PBS) via the right ventricle. Lungs were inflated with 10% formaldehyde under constant pressure and allowed to fix for 16 hours. Tissues were then prepared for optimal cutting temperature (O.C.T.) compound embedding or paraffin embedding. Cryosections were used for immunofluorescent detection and staining. Antibodies used in immunofluorescent staining were Sftpc (Goat and Rabbit; 1:200; B. Stripp laboratory), COL1A1 (NB600-408; 1:200; Novus Biologicals), HTII-280 (1:200; a gift of the L. G. Dobbs laboratory, University of California, San Francisco, San Francisco, CA), CD31 (1:200; mouse, 303101, BioLegend), GHR (Goat, AF1210; 1:200; R&D Systems), red fluorescent protein (Rabbit; 1:200; 600-401-379, Rockland), and correspondent secondary antibodies conjugated to Alexa 546 or 488 (1:500; A-21085, A-11056, and A-11008, Thermo Fisher Scientific). Paraffin sections were used for hematoxylin and eosin (H&E) and Masson’s trichrome staining.
RNAscope
GHR mRNA on human lung tissue was detected by an RNAscope Fluorescent Multiplex kit (320850, Advanced Cell Diagnostics). Probe region 1136-2136 of NM_000163.4 was used to design probes for targeting human GHR (444211, Advanced Cell Diagnostics). Frozen sections were used for GHR mRNA detection, according to the RNAscope assay protocol.
Hydroxyproline assay
Collagen content in mouse lungs was measured with a conventional hydroxyproline method (35). Lung tissues were vacuum-dried and hydrolyzed with 6N hydrochloric acid at 120°C for overnight. Hydroxyproline concentration was expressed as micrograms per milliliter per milligram dry lung weight, unless specified otherwise. The ability of the assay to completely hydrolyze and recover hydroxyproline from collagen was confirmed by using standards containing known amounts of purified collagen.
Mesenchymal cell isolation and culture
Mouse lung mesenchymal cells were isolated as described (33). The lungs from either unchallenged mice or mice 7, 14, and 21 days after bleomycin treatment were minced, digested for 30 min at 37°C in digestion buffer, which contains Dulbecco’s modified Eagle’s medium (DMEM) with deoxyribonuclease (DNase) I (100 μg/ml), type IV collagenase (1 mg/ml), and bovine serum albumin (1 mg/ml), passed through a 100-μm filter, centrifuged at 1500 rpm for 10 min, and plated in DMEM supplemented with 10% fetal bovine serum (FBS), penicillin (100 U/ml), and streptomycin (100 μg/ml). Experiments were performed using mesenchymal cells at passages 3 to 7. Ghr-deficient mesenchymal cells were isolated from Ghr−/− mice, and control cells were from littermate control mice.
Human lung mesenchymal cells were isolated from surgical lung biopsies or lung transplant explants obtained from patients with IPF (35). The diagnosis of IPF was according to the American Thoracic Society recommendations (61). The specimens were obtained under the auspices of IRB-approved protocols. The tissues were minced and cultured in DMEM supplemented with 15% FBS, penicillin (100 U/ml), streptomycin (100 μg/ml), gentamicin (5 μg/ml), and amphotericin B (0.25 μg/ml). The cells of passages 5 to 8 were used for colony formation assays and exosome/secreted vesicle isolation. All experiments were approved by the Cedars-Sinai Medical Center IRB and in accordance with the guidelines outlined by the board.
Human lung mesenchymal cell line overexpressing GHR
Lentivirus plasmid encoding GHR TV1 (NM_000163) open reading frame with mCherry fused on the N-terminal was constructed. Lentivirus particles were produced by cotransfection of GHR lentivirus plasmid and packaging plasmids to HEK293 cells by Lipofectamine 3000 (L3000015, Thermo Fisher Scientific). Human normal lung mesenchymal CCD-19Lu cell line (American Type Culture Collection) was purchased and infected with GHR lentivirus particles to generate GHRTg human lung mesenchymal cells.
Flow cytometry analysis
HTII-280 and GHR expression on single human lung cells, Ghr expression on lung mesenchymal cells from bleomycin-treated and control mice, Ghr−/− and littermate control mice, and Ghr expression on ATII colonies treated with Ghr or control vesicles (cultured for 14 to 21 days) were analyzed with Fortessa (BD Biosciences).
Flow cytometry sorting for mouse epithelial cells
Single-cell suspensions from mouse lungs were isolated as previously described (31). Mouse lungs were perfused with 10 ml of PBS and digested with elastase (4 U/ml; Worthington Biochemical Corporation) and DNase I (100 U/ml; Sigma-Aldrich) to obtain single-cell suspensions. Antibodies against CD31 (1:40; 102404, BioLegend), CD34 (1:20; 119304, BioLegend), CD45 (1:200; 103104, BioLegend), Sca-1 (1:200; 122511, BioLegend), and CD24 (1:50; 103104, BioLegend) and secondary antibody anti-streptavidin (1:150) were all from BioLegend (San Diego, CA). Mouse anti-EpCAM (1:200; 324221, BioLegend) and annexin V (1:100; BDB556419, BD Biosciences) were purchased. Mouse anti-GHR antibody (B-10) phycoerythrin (PE) (sc-137185 PE; 1:200) was from Santa Cruz Biotechnology. 7-aminoactinomycin D (7-AAD) (BDB559925; 1:20) was from BD Biosciences (San Diego, CA) and was used to discriminate dead cells. Flow cytometry staining methods were performed according to previous reports (35). Briefly, primary antibodies, including CD31-Biotin, CD34-Biotin, CD45-Biotin, EpCAM-PE-Cy7, and Ghr-PE, were added to incubate cells. Biotin-conjugated antibodies were detected following incubation with streptavidin-APC-Cy7 (47-4317-82, eBioscience). Dead cells were discriminated by 7-AAD staining. Flow cytometry was performed using a Fortessa flow cytometer and FACSAria III sorter (BD Immunocytometry Systems, San Jose, CA) and analyzed using FlowJo 10.5.3 software (Tree Star, Ashland, OR).
Ex vivo epithelial colony culture and measurement
Flow-sorted mouse 7AAD−Epcam+CD31/34/45−, Sftpc-GFP ATIIs, and Scgb1a1-GFP Club cells were cultured in Matrigel (354230, BD Biosciences) and medium (1:1) mixture in the presence of MLg lung mesenchymal cells (31, 62). Matrigel medium mix (100 μl) containing 3 × 103 epithelial progenitor cells and 2 × 105 mesenchymal cells were plated into each 0.4-μm Transwell insert of a 24-well plate (82050-022, Greiner Bio-One), and 400 μl of medium was added in the lower chambers. For PEG experiments, we treated the mesenchymal cells with PEG at different concentrations 30 min before and during the coculture of ATII progenitors. Colonies were cultured in a humidified 37°C and 5% CO2 incubator. A Zeiss AXIO inverted fluorescent microscope (Carl Zeiss AG, Oberkochen, Germany) was used to visualize the colonies. Colonies derived from Matrigel culture were pictured, and numbers were counted manually at days 10 to 14 after plating. Colonies with a diameter of ≥50 μm were counted, and the colony-forming efficiency was calculated by the percentage of colonies derived from input ATIIs at day 12 after plating. If all colonies in a well could not be measured, then several random nonoverlapping pictures were acquired from each well. ImageJ software was used to measure the surface area of the pictured organoids. Organoid perimeters for area measurements were defined manually. Automated determination of the defined area was calculated by the Analyze Particle function of ImageJ software, with investigator verification of the automated determinations, as automated measurements allowed for unbiased analyses of increased numbers of organoids. For automated size measurements, images were set as monochrome for organoid threshold identification. The largest and smallest organoid sizes were measured manually, and their areas were used as the reference values for setting the minimal and maximal particle sizes. Organoids touching the edge of the images were excluded from counting.
Quantitative real-time PCR
Lung tissues or ex vivo–cultured lung colonies and vesicles were collected and immediately snap-frozen and stored at −80°C. The RNeasy Mini Kit (74106, QIAGEN) was used for both mouse lung tissue and cultured colonies RNA isolation. Extracted RNA was stored at −80°C. RT was performed with a high-capacity cDNA RT kit with ribonuclease inhibitor by using 0.1 μg of total RNA (4374966, Thermo Fisher Scientific). Quantitative real-time PCR analysis was performed by using the SYBR Green PCR Master Mix per its manual (4364346, Thermo Fisher Scientific). Specific primers were used to detect gene expression levels (table S2).
Single-cell RNA-seq
10x Genomics single-cell RNA-seq was performed according to the manufacturer’s protocol as described previously (28). Lung single cells were sorted to eliminate dead cells and subsequently loaded onto a GemCode single-cell instrument (10x Genomics) to generate single-cell gel beads in emulsion (GEMs). GEM-RT were performed in a Veriti 96-well thermal cycler (Thermo Fisher Scientific). After RT, GEMs were harvested, and the cDNAs were amplified and cleaned with the SPRIselect Reagent Kit (Beckman Coulter). Indexed sequencing libraries were constructed using the Chromium Single-Cell 30 Library Kit (10x Genomics) for enzymatic fragmentation, end-repair, A-tailing, adaptor ligation, ligation cleanup, sample index PCR, and PCR cleanup. The barcoded sequencing libraries were quantified by quantitative PCR using the KAPA Library Quantification Kit (KAPA Biosystems). Sequencing libraries were loaded on a NextSeq500 (Illumina) with a custom sequencing setting (26 bp for read 1 and 98 bp for read 2). The GEO accession numbers for the raw data files of the single-cell RNA-seq analyses reported in this paper are GSE134948 and GSE104154.
Bio-Plex Pro human and mouse chemokine assay
Human chemokines including CXCL1, CXCL2, CXCL8, CXCL10, and CXCL12, as well as human IL-6, were analyzed by customized Bio-Plex Pro Human Chemokine Assay (Bio-Rad). Nondiluted 50 μl of serum-free medium collected from 48-hour cultured GHRTg and control cells were used. Mouse chemokines including Cxcl1, Cxcl2, Cxcl5, Cxcl10, Cxcl11, Cxcl12, and Cxcl16 were analyzed by Bio-Plex Pro Mouse Chemokine Assay (Bio-Rad). BALF collected from mice 21 days after bleomycin injury was used. The assay procedures were carried out according to the manual and were automatically analyzed by the Bio-Plex 200 system (Bio-Plex Multiplex Immunoassay System, Bio-Rad).
Enzyme-linked immunosorbent assay
The level of IGF-1 was analyzed for the 48-hour cultured serum-free medium collected from GHRTg and control cells by a human IGF-1 Duoset enzyme-linked immunosorbent assay kit (DY291-05, R&D).
LGRC dataset analysis
GHR, CXCL1, CXCL2, CXCL3, CXCL8, and CXCL16 gene expression in lungs from patients with ILD and healthy controls were analyzed by using the LGRC dataset (63) (GEO: GSE47460).
Matrigel invasion assay
The invasive behavior of fibroblasts isolated from Ghr−/− and littermate control mouse lungs was performed essentially as described previously (33, 35). Equal numbers (5 × 104) of fibroblasts were plated onto the BioCoat Matrigel Invasion Chamber (BD Biosciences), and the cell invasion was performed in the presence of 10% FBS complete medium. After 24 hours of incubation in a CO2 incubator, media were removed, and the polycarbonate filters with the invaded cells were washed once with PBS followed by fixing and staining with the Protocol Hema 3 stain set. Matrigel matrix and noninvading cells on the upper surface of the filter were removed by wiping with a cotton swab, and the filters were removed from the insert by a scalpel blade and were mounted onto glass slides. The slides were imaged by using the ECL system (Bio-Rad), the Red Green Blue (RGB) images were then separated by www.dcode.fr/rgb-channels, and red channel images were analyzed by ImageJ for density value.
Exosome/vesicle isolation and electron microscopy
The cell culture supernatant was centrifuged at 300g for 10 min at 4°C, followed by 2000g for 10 min at 4°C, and then 10,000g for 30 min at 4°C. The supernatant was then centrifuged at 100,000g for 70 min at 4°C, washed in PBS, and then centrifuged at 100,000g for 70 min at 4°C. Exosomes were resuspended in PBS. Sample aliquots of 6 μl were pipetted onto a 200-mesh copper grid (catalog no. 1GC200, Ted Pella Inc.) with carbon-coated formvar film and incubated for 1 hour. Excess liquid was removed by blotting. The exosome sample was fixed in 2% glutaraldehyde (v/v; G7526, Sigma-Aldrich, Saint Louis, MO) for 1 min. The grid was washed twice by brief contact with 100 μl of MilliQ water, each time for 1 min. Next, the grid was placed on 30 μl of 1.5% uranyl acetate (w/v) for 12 s, followed by using filter paper to remove excess liquid and air dry. JEOL 100CX TEM was used to acquire the images of the morphology of the exosome. The method was used for experiments in fig. S4B.
OptiPrep buoyant density gradient isolation of exosomes/vesicles
To obtain discontinuous density gradients, Optiprep (D1556, Sigma-Aldrich) solution was diluted with PBS to obtain 40, 30, 25, 15, 10, and 5% iodixanol solutions. Exosomes resuspended in 200 μl of PBS were mixed with 1 ml of 50% iodixanol, the aliquots of 2 ml with increasing density were loaded layered atop sequentially in 13.2-ml ultracentrifuge tubes, and centrifuged at 100,000g at 4°C for 16 hours. Six 2-ml fractions were collected from the top and centrifuged at 100,000g at 4°C for 1 hour. The pellet was collected for experiments in fig. S4 (B and C).
Western blot analysis of exosomes/vesicles
Vesicle samples were lysed in 10× cell lysis buffer (Cell Signaling Technology, catalog no. 9803). Western blot was performed as described previously (62). To detect protein expression of Ghr, exosome cargo protein TSG101 (involved in multivesicular biogenesis), CD63, and CD81 (tetraspanins), 50 μg of exosome lysate was run through NuPAGE 4 to 12% bis-tris gel (Thermo Fisher Scientific, Waltham, MA) and blotted onto a polyvinylidene difluoride membrane (Thermo Fisher Scientific, Waltham, MA). Polyclonal antibody to Ghr (Boster Biological Technology, PA1726; R&D, AF1210), monoclonal antibody to Ghr (sc-137185, Santa Cruz Biotechnology; ab134078, Abcam), TSG101 (Ab125011, Abcam), CD63 (Ab68418, Abcam), and CD81 (catalog no. 10630D, Thermo Fisher Scientific, Waltham, MA), as well as associated horseradish peroxidase–conjugated secondary antibodies (Cell Signaling Technology), were used as per the manufacturer’s instructions. The bands were imaged and analyzed using the ECL system (Bio-Rad, Hercules, CA).
Zipcode plasmid, transfection, and Ghr-enriched vesicle isolation
To insert GHR mRNA with an N-terminal mCherry tag into the secreted vesicles. “Zipcode” technology was used; two tandem copies of the zipcode sequence (5′- ACCCTGCCGCCTGGACTCCGCCTGT-3′) were inserted at the 3′ untranslated region of the GHR gene under the control of the cytomegalovirus promoter (64). This plasmid was used for the transient transfection of HEK293 cells. Cell medium was changed to serum-free medium 2 days after transfection. The cell culture supernatant was collected from the 2-day cell culture, sequential centrifugation steps at 500g for 10 min and 2000g for 10 min. The resulting supernatant was then filtered using 0.2-μm filters, Last, exosomes were collected by spinning at 100,000g for 70 min twice. Alternatively, serum-free medium was collected for vesicle isolation using ExoQuick-TC (EXOTC50A-1, SBI System Biosciences). The serum-free medium was collected and centrifuged at 3000g for 15 min to remove cells and cell debris. Then, medium and ExoQuick-TC were combined at the ratio of 5:1, incubated overnight at 4°C, and centrifuged at 1500g for 30 min to isolate vesicles. The pellet was collected or lysed in lithium dodecyl sulfate (LDS) loading buffer for Western blotting or in lysis buffer from the RNeasy Mini Kit (74104, QIAGEN) for isolating RNA for experiments in Figs. 7 (D and I) and 8 (A to H) and figs. S4 (A and D to I) and S5.
Nanosight exosome analysis
Purified exosomes/vesicles were diluted 1:100 before Nanosight NS300 (Malvern Panalytical) for analysis. One-milliliter diluted sample was introduced to the viewing unit with a disposable syringe. The remnant media from EV isolation were collected and tested for particle existence to ensure the EV enrichment/purity.
Secreted vesicle treatment
For vesicle treatment, vesicles were freshly used or with one- to three-time thaw and freeze cycle. GHR vesicles or control vesicles 4 × 108/ml were added to the colony culture medium, and the medium was changed every other day. Five doses of GHR and Ctrl vesicles (4 × 1010/ml, 1 μl/g mouse weight per dose) were administrated to Ghr△BMC and littermate control mice through nonsurgical intratracheally instillation start from day 7 after bleomycin injury every other day. Five doses of GHR and Ctrl vesicles (4 × 108/ml, 1 μl/g mouse weight per dose) were intratracheally administrated to WT mice through nonsurgical instillation start from day 7 after bleomycin injury every other day.
Statistics and reproducibility
Data are expressed as the means ± SEM. Differences in measured variables between experimental and control groups were assessed by unpaired two-tailed student’s t tests or Wilcoxon rank-sum test with nonparametric data. One-way or two-way analysis of variance (ANOVA) followed by Sidak’s multiple comparison test with Bonferroni test was used for multiple comparisons. The survival curves were compared using the log-rank test. The results were considered statistically significant at P ≤ 0.05. GraphPad Prism software was used for statistical analysis.
Acknowledgments
We acknowledge the assistance of the CSMC Comparative Medicine. We also thank S. Melmed, V. Chesnokova, C. Zhou, E. Marbán, T. Antes, A. L. Coelho, A. Chin, and D. Vizio of Cedars-Sinai Medical Center for sharing protocols, reagents, and instruments. We would like to acknowledge N. Hassanzadeh-Kiabi, A. Lopez, and J. Suda at the Flow Cytometry Core of Cedars-Sinai Medical Center in Los Angeles for assistance with flow cytometry cell sorting. We are also grateful for the assistance provided by C. Santiskulvong at Genomics Core of Cedars-Sinai Medical Center for single-cell RNA-seq. We thank C. Zhu and J. Abad at UCLA electron microscopy core facility for help with the electron microscopy imaging. We are grateful to J. Gornbein in the Statistics Core at UCLA and C. Bresee in the Biostatistics and Bioinformatics Core of Cedars-Sinai Medical Center for assistance with the biostatistics analyses. Funding: This study was supported by NIH grants P01 HL108793, R01 HL060539, R01AI052201 (to P.W.N.), HL122068 (to D.J.), AHA no. 19CDA34660211 (to T.X.), and Marie Skłodowska-Curie grant agreement no. 797209 (to S.R.). Author contributions: The conceptualization, methodology, and supervision were carried out by P.W.N., D.J., and T.X.; the investigation was carried out by T.X., V.K., N.L., N.D., Y.W., C.Y., G.H., X.L., S.R., F.T., J.L., P.C., C.H., and B.S.; writing of the original draft was handled by T.X.; and the draft was reviewed and edited by all the authors. Competing interests: The authors declare that they have no competing interests. Data and materials availability: The accession number for the raw data files of the RNA-seq and microarray analyses reported in this paper is in the GEO database with the accession numbers GSE104154 and GSE131800 (RNA-seq) and GSE47460 (microarray). All other data needed to evaluate the conclusions in the paper are present in the paper and/or the Supplementary Materials. All R code generated during and/or used in the current study is available in the Supplementary Materials. Additional data related to this paper may be requested from the authors.
SUPPLEMENTARY MATERIALS
Supplementary material for this article is available at http://advances.sciencemag.org/cgi/content/full/7/24/eabg6005/DC1
REFERENCES AND NOTES
- 1.Cao Z., Ye T., Sun Y., Ji G., Shido K., Chen Y., Luo L., Na F., Li X., Huang Z., Ko J. L., Mittal V., Qiao L., Chen C., Martinez F. J., Rafii S., Ding B. S., Targeting the vascular and perivascular niches as a regenerative therapy for lung and liver fibrosis. Sci. Transl. Med. 9, eaai8710 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Lechner A. J., Driver I. H., Lee J., Conroy C. M., Nagle A., Locksley R. M., Rock J. R., Recruited monocytes and type 2 immunity promote lung regeneration following pneumonectomy. Cell Stem Cell 21, 120–134.e7 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Lee J.-H., Tammela T., Hofree M., Choi J., Marjanovic N. D., Han S., Canner D., Wu K., Paschini M., Bhang D. H., Jacks T., Regev A., Kim C. F., Anatomically and functionally distinct lung mesenchymal populations marked by Lgr5 and Lgr6. Cell 170, 1149–1163.e12 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Zepp J. A., Zacharias W. J., Frank D. B., Cavanaugh C. A., Zhou S., Morley M. P., Morrisey E. E., Distinct mesenchymal lineages and niches promote epithelial self-renewal and myofibrogenesis in the lung. Cell 170, 1134–1148.e10 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Leeman K. T., Fillmore C. M., Kim C. F., Lung stem and progenitor cells in tissue homeostasis and disease. Curr. Top. Dev. Biol. 107, 207–233 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Chung M. I., Bujnis M., Barkauskas C. E., Kobayashi Y., Hogan B. L. M., Niche-mediated BMP/SMAD signaling regulates lung alveolar stem cell proliferation and differentiation. Development 145, dev163014 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Nabhan A. N., Brownfield D. G., Harbury P. B., Krasnow M. A., Desai T. J., Single-cell Wnt signaling niches maintain stemness of alveolar type 2 cells. Science 359, 1118–1123 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Volckaert T., Dill E., Campbell A., Tiozzo C., Majka S., Bellusci S., De Langhe S. P., Parabronchial smooth muscle constitutes an airway epithelial stem cell niche in the mouse lung after injury. J. Clin. Invest. 121, 4409–4419 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Endale M., Ahlfeld S., Bao E., Chen X., Green J., Bess Z., Weirauch M. T., Xu Y., Perl A. K., Temporal, spatial, and phenotypical changes of PDGFRα expressing fibroblasts during late lung development. Dev. Biol. 425, 161–175 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Barkauskas C. E., Cronce M. J., Rackley C. R., Bowie E. J., Keene D. R., Stripp B. R., Randell S. H., Noble P. W., Hogan B. L. M., Type 2 alveolar cells are stem cells in adult lung. J. Clin. Invest. 123, 3025–3036 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Wang C., de Mochel N. S. R., Christenson S. A., Cassandras M., Moon R., Brumwell A. N., Byrnes L. E., Li A., Yokosaki Y., Shan P., Sneddon J. B., Jablons D., Lee P. J., Matthay M. A., Chapman H. A., Peng T., Expansion of hedgehog disrupts mesenchymal identity and induces emphysema phenotype. J. Clin. Invest. 128, 4343–4358 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Ng-Blichfeldt J. P., de Jong T., Kortekaas R. K., Wu X., Lindner M., Guryev V., Hiemstra P. S., Stolk J., Konigshoff M., Gosens R., TGF-β activation impairs fibroblast ability to support adult lung epithelial progenitor cell organoid formation. Am. J. Phys. Lung Cell. Mol. Phys. 317, L14–L28 (2019). [DOI] [PubMed] [Google Scholar]
- 13.Rosenfeld R. G., Belgorosky A., Camacho-Hubner C., Savage M. O., Wit J. M., Hwa V., Defects in growth hormone receptor signaling. Trends Endocrinol. Metab. 18, 134–141 (2007). [DOI] [PubMed] [Google Scholar]
- 14.Dehkhoda F., Lee C. M. M., Medina J., Brooks A. J., The growth hormone receptor: Mechanism of receptor activation, cell signaling, and physiological aspects. Front. Endocrinol. 9, 35 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.N. C. Olarescu, K. Gunawardane, T. K. Hansen, N. Moller, J. O. L. Jorgensen, in Endotext, K. R. Feingold, B. Anawalt, A. Boyce, G. Chrousos, W. W. de Herder, K. Dhatariya, K. Dungan, A. Grossman, J. M. Hershman, J. Hofland, S. Kalra, G. Kaltsas, C. Koch, P. Kopp, M. Korbonits, C. S. Kovacs, W. Kuohung, B. Laferrère, E. A. McGee, R. M. Lachlan, J. E. Morley, M. New, J. Purnell, R. Sahay, F. Singer, C. A. Stratakis, D. L. Trence, D. P. Wilson, Eds. (South Dartmouth, MA, 2000). [Google Scholar]
- 16.Pugliese-Pires P. N., Tonelli C. A., Dora J. M., Silva P. C., Czepielewski M., Simoni G., Arnhold I. J., Jorge A. A., A novel STAT5B mutation causing GH insensitivity syndrome associated with hyperprolactinemia and immune dysfunction in two male siblings. Eur. J. Endocrinol. 163, 349–355 (2010). [DOI] [PubMed] [Google Scholar]
- 17.Zerrad-Saadi A., Lambert-Blot M., Mitchell C., Bretes H., Collin de l’Hortet A., Baud V., Chereau F., Sotiropoulos A., Kopchick J. J., Liao L., Xu J., Gilgenkrantz H., Guidotti J. E., GH receptor plays a major role in liver regeneration through the control of EGFR and ERK1/2 activation. Endocrinology 152, 2731–2741 (2011). [DOI] [PubMed] [Google Scholar]
- 18.Macias H., Hinck L., Mammary gland development. Wiley Interdiscip. Rev. Dev. Biol. 1, 533–557 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.L. B. Patricia Stiedl, V. Stanek, J. Svinka, R. M. Mahon, G. Zollner, T. Claudel, M. Mueller, W. Mikulits, H. Esterbauer, R. Eferl, J. Haybaeck, M. Trauner, E. Casanova, in Proceedings: AACR Annual Meeting 2014 (2014), vol. 74. [Google Scholar]
- 20.Stiedl P., McMahon R., Blaas L., Stanek V., Svinka J., Grabner B., Zollner G., Kessler S. M., Claudel T., Muller M., Mikulits W., Bilban M., Esterbauer H., Eferl R., Haybaeck J., Trauner M., Casanova E., Growth hormone resistance exacerbates cholestasis-induced murine liver fibrosis. Hepatology 61, 613–626 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Martin-Medina A., Lehmann M., Burgy O., Hermann S., Baarsma H. A., Wagner D. E., De Santis M. M., Ciolek F., Hofer T. P., Frankenberger M., Aichler M., Lindner M., Gesierich W., Guenther A., Walch A., Coughlan C., Wolters P., Lee J. S., Behr J., Konigshoff M., Increased extracellular vesicles mediate wnt5a signaling in idiopathic pulmonary fibrosis. Am. J. Respir. Crit. Care Med. 198, 1527–1538 (2018). [DOI] [PubMed] [Google Scholar]
- 22.Bourdonnay E., Zaslona Z., Penke L. R., Speth J. M., Schneider D. J., Przybranowski S., Swanson J. A., Mancuso P., Freeman C. M., Curtis J. L., Peters-Golden M., Transcellular delivery of vesicular SOCS proteins from macrophages to epithelial cells blunts inflammatory signaling. J. Exp. Med. 212, 729–742 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Maas S. L. N., Breakefield X. O., Weaver A. M., Extracellular vesicles: Unique intercellular delivery vehicles. Trends Cell Biol. 27, 172–188 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Borges F. T., Melo S. A., Ozdemir B. C., Kato N., Revuelta I., Miller C. A., Gattone V. H. II, LeBleu V. S., Kalluri R., TGF-β1–Containing exosomes from injured epithelial cells activate fibroblasts to initiate tissue regenerative responses and fibrosis. J. Am. Soc. Nephrol. 24, 385–392 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Stik G., Crequit S., Petit L., Durant J., Charbord P., Jaffredo T., Durand C., Extracellular vesicles of stromal origin target and support hematopoietic stem and progenitor cells. J. Cell Biol. 216, 2217–2230 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Nabet B. Y., Qiu Y., Shabason J. E., Wu T. J., Yoon T., Kim B. C., Benci J. L., DeMichele A. M., Tchou J., Marcotrigiano J., Minn A. J., Exosome RNA unshielding couples stromal activation to pattern recognition receptor signaling in cancer. Cell 170, 352–366.e13 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Dinh P.-U. C., Paudel D., Brochu H., Popowski K. D., Gracieux M. C., Cores J., Huang K., Hensley M. T., Harrell E., Vandergriff A. C., George A. K., Barrio R. T., Hu S., Allen T. A., Blackburn K., Caranasos T. G., Peng X., Schnabel L. V., Adler K. B., Lobo L. J., Goshe M. B., Cheng K., Inhalation of lung spheroid cell secretome and exosomes promotes lung repair in pulmonary fibrosis. Nat. Commun. 11, 1064 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Xie T., Wang Y., Deng N., Huang G., Taghavifar F., Geng Y., Liu N., Kulur V., Yao C., Chen P., Liu Z., Stripp B., Tang J., Liang J., Noble P. W., Jiang D., Single-cell deconvolution of fibroblast heterogeneity in mouse pulmonary fibrosis. Cell Rep. 22, 3625–3640 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Chesnokova V., Zonis S., Zhou C., Recouvreux M. V., Ben-Shlomo A., Araki T., Barrett R., Workman M., Wawrowsky K., Ljubimov V. A., Uhart M., Melmed S., Growth hormone is permissive for neoplastic colon growth. Proc. Natl. Acad. Sci. U.S.A. 113, E3250–E3259 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Ranke M. B., Wit J. M., Growth hormone - past, present and future. Nat. Rev. Endocrinol. 14, 285–300 (2018). [DOI] [PubMed] [Google Scholar]
- 31.Liang J., Zhang Y., Xie T., Liu N., Chen H., Geng Y., Kurkciyan A., Mena J. M., Stripp B. R., Jiang D., Noble P. W., Hyaluronan and TLR4 promote surfactant-protein-C-positive alveolar progenitor cell renewal and prevent severe pulmonary fibrosis in mice. Nat. Med. 22, 1285–1293 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Hinrichs A., Kessler B., Kurome M., Blutke A., Kemter E., Bernau M., Scholz A. M., Rathkolb B., Renner S., Bultmann S., Leonhardt H., de Angelis M. H., Nagashima H., Hoeflich A., Blum W. F., Bidlingmaier M., Wanke R., Dahlhoff M., Wolf E., Growth hormone receptor-deficient pigs resemble the pathophysiology of human Laron syndrome and reveal altered activation of signaling cascades in the liver. Mol. Metab. 11, 113–128 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Li Y., Jiang D., Liang J., Meltzer E. B., Gray A., Miura R., Wogensen L., Yamaguchi Y., Noble P. W., Severe lung fibrosis requires an invasive fibroblast phenotype regulated by hyaluronan and CD44. J. Exp. Med. 208, 1459–1471 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Geng Y., Liu X., Liang J., Habiel D. M., Kulur V., Coelho A. L., Deng N., Xie T., Wang Y., Liu N., Huang G., Kurkciyan A., Liu Z., Tang J., Hogaboam C. M., Jiang D., Noble P. W., PD-L1 on invasive fibroblasts drives fibrosis in a humanized model of idiopathic pulmonary fibrosis. JCI Insight 4, e125326 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Xie T., Liang J., Liu N., Huan C., Zhang Y., Liu W., Kumar M., Xiao R., D’Armiento J., Metzger D., Chambon P., Papaioannou V. E., Stripp B. R., Jiang D., Noble P. W., Transcription factor TBX4 regulates myofibroblast accumulation and lung fibrosis. J. Clin. Invest. 126, 3063–3079 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Keerthikumar S., Chisanga D., Ariyaratne D., Al Saffar H., Anand S., Zhao K., Samuel M., Pathan M., Jois M., Chilamkurti N., Gangoda L., Mathivanan S., ExoCarta: A web-based compendium of exosomal cargo. J. Mol. Biol. 428, 688–692 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Richeldi L., du Bois R. M., Raghu G., Azuma A., Brown K. K., Costabel U., Cottin V., Flaherty K. R., Hansell D. M., Inoue Y., Kim D. S., Kolb M., Nicholson A. G., Noble P. W., Selman M., Taniguchi H., Brun M., Le Maulf F., Girard M., Stowasser S., Schlenker-Herceg R., Disse B., Collard H. R.; INPULSIS Trial Investigators , Efficacy and safety of nintedanib in idiopathic pulmonary fibrosis. N. Engl. J. Med. 370, 2071–2082 (2014). [DOI] [PubMed] [Google Scholar]
- 38.Nathan S. D., Costabel U., Albera C., Behr J., Wuyts W. A., Kirchgaessler K. U., Stauffer J. L., Morgenthien E., Chou W., Limb S. L., Noble P. W., Pirfenidone in patients with idiopathic pulmonary fibrosis and more advanced lung function impairment. Respir. Med. 153, 44–51 (2019). [DOI] [PubMed] [Google Scholar]
- 39.Costabel U., Albera C., Lancaster L. H., Lin C. Y., Hormel P., Hulter H. N., Noble P. W., An open-label study of the long-term safety of pirfenidone in patients with idiopathic pulmonary fibrosis (RECAP). Respiration 94, 408–415 (2017). [DOI] [PubMed] [Google Scholar]
- 40.King T. E. Jr., Bradford W. Z., Castro-Bernardini S., Fagan E. A., Glaspole I., Glassberg M. K., Gorina E., Hopkins P. M., Kardatzke D., Lancaster L., Lederer D. J., Nathan S. D., Pereira C. A., Sahn S. A., Sussman R., Swigris J. J., Noble P. W.; ASCEND Study Group , A phase 3 trial of pirfenidone in patients with idiopathic pulmonary fibrosis. N. Engl. J. Med. 370, 2083–2092 (2014). [DOI] [PubMed] [Google Scholar]
- 41.Wolters P. J., Blackwell T. S., Eickelberg O., Loyd J. E., Kaminski N., Jenkins G., Maher T. M., Molina-Molina M., Noble P. W., Raghu G., Richeldi L., Schwarz M. I., Selman M., Wuyts W. A., Schwartz D. A., Time for a change: Is idiopathic pulmonary fibrosis still idiopathic and only fibrotic? Lancet Respir. Med. 6, 154–160 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Whiteside T. L., Extracellular vesicles isolation and their biomarker potential: Are we ready for testing? Ann. Transl. Med. 5, 54 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Tetta C., Bruno S., Fonsato V., Deregibus M. C., Camussi G., The role of microvesicles in tissue repair. Organogenesis 7, 105–115 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Akyurekli C., Le Y., Richardson R. B., Fergusson D., Tay J., Allan D. S., A systematic review of preclinical studies on the therapeutic potential of mesenchymal stromal cell-derived microvesicles. Stem Cell Rev. Rep. 11, 150–160 (2015). [DOI] [PubMed] [Google Scholar]
- 45.Lee C., Mitsialis S. A., Aslam M., Vitali S. H., Vergadi E., Konstantinou G., Sdrimas K., Fernandez-Gonzalez A., Kourembanas S., Exosomes mediate the cytoprotective action of mesenchymal stromal cells on hypoxia-induced pulmonary hypertension. Circulation 126, 2601–2611 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Lee J. W., Fang X., Krasnodembskaya A., Howard J. P., Matthay M. A., Concise review: Mesenchymal stem cells for acute lung injury: Role of paracrine soluble factors. Stem Cells 29, 913–919 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Mansouri N., Willis G. R., Fernandez-Gonzalez A., Reis M., Nassiri S., Mitsialis S. A., Kourembanas S., Mesenchymal stromal cell exosomes prevent and revert experimental pulmonary fibrosis through modulation of monocyte phenotypes. JCI Insight 4, e128060 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Matthay M. A., Goolaerts A., Howard J. P., Lee J. W., Mesenchymal stem cells for acute lung injury: Preclinical evidence. Crit. Care Med. 38, S569–S573 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Monsel A., Zhu Y. G., Gennai S., Hao Q., Hu S., Rouby J. J., Rosenzwajg M., Matthay M. A., Lee J. W., Therapeutic effects of human mesenchymal stem cell-derived microvesicles in severe pneumonia in mice. Am. J. Respir. Crit. Care Med. 192, 324–336 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Prado N., Marazuela E. G., Segura E., Fernandez-Garcia H., Villalba M., Thery C., Rodriguez R., Batanero E., Exosomes from bronchoalveolar fluid of tolerized mice prevent allergic reaction. J. Immunol. 181, 1519–1525 (2008). [DOI] [PubMed] [Google Scholar]
- 51.Zhu Y. G., Feng X. M., Abbott J., Fang X. H., Hao Q., Monsel A., Qu J. M., Matthay M. A., Lee J. W., Human mesenchymal stem cell microvesicles for treatment of Escherichia coli endotoxin-induced acute lung injury in mice. Stem Cells 32, 116–125 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Bolukbasi M. F., Mizrak A., Ozdener G. B., Madlener S., Strobel T., Erkan E. P., Fan J. B., Breakefield X. O., Saydam O., miR-1289 and "Zipcode"-like sequence enrich mRNAs in microvesicles. Mol. Ther. Nucleic Acids 1, e10 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Gossard F., Dihl F., Pelletier G., Dubois P. M., Morel G., In situ hybridization to rat brain and pituitary gland of growth hormone cDNA. Neurosci. Lett. 79, 251–256 (1987). [DOI] [PubMed] [Google Scholar]
- 54.De Troyer A., Desir D., Copinschi G., Regression of lung size in adults with growth hormone deficiency. Q. J. Med. 49, 329–340 (1980). [PubMed] [Google Scholar]
- 55.Qi W., Gu F., Wu C., Growth hormone replacement therapy improves hypopituitarism-associated hypoxemia in a patient after craniopharyngioma surgery: A case report. Medicine (Baltimore) 98, e14101 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Pardo A., Selman M., Lung fibroblasts, aging, and idiopathic pulmonary fibrosis. Ann. Am. Thorac. Soc. 13 (Suppl. 5), S417–S421 (2016). [DOI] [PubMed] [Google Scholar]
- 57.Hennessey J. V., Espaillat R., Diagnosis and management of subclinical hypothyroidism in elderly adults: A review of the literature. J. Am. Geriatr. Soc. 63, 1663–1673 (2015). [DOI] [PubMed] [Google Scholar]
- 58.Takahashi S., Gottschall P. E., Quigley K. L., Goya R. G., Meites J., Growth hormone secretory patterns in young, middle-aged and old female rats. Neuroendocrinology 46, 137–142 (1987). [DOI] [PubMed] [Google Scholar]
- 59.Chen H., Matsumoto K., Brockway B. L., Rackley C. R., Liang J., Lee J. H., Jiang D., Noble P. W., Randell S. H., Kim C. F., Stripp B. R., Airway epithelial progenitors are region specific and show differential responses to bleomycin-induced lung injury. Stem Cells 30, 1948–1960 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Gao X., Bali A. S., Randell S. H., Hogan B. L., GRHL2 coordinates regeneration of a polarized mucociliary epithelium from basal stem cells. J. Cell Biol. 211, 669–682 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Raghu G., Collard H. R., Egan J. J., Martinez F. J., Behr J., Brown K. K., Colby T. V., Cordier J. F., Flaherty K. R., Lasky J. A., Lynch D. A., Ryu J. H., Swigris J. J., Wells A. U., Ancochea J., Bouros D., Carvalho C., Costabel U., Ebina M., Hansell D. M., Johkoh T., Kim D. S., King T. E. Jr., Kondoh Y., Myers J., Muller N. L., Nicholson A. G., Richeldi L., Selman M., Dudden R. F., Griss B. S., Protzko S. L., Schunemann H. J.; ATS/ERS/JRS/ALAT Committee on Idiopathic Pulmonary Fibrosis , An official ATS/ERS/JRS/ALAT statement: Idiopathic pulmonary fibrosis: Evidence-based guidelines for diagnosis and management. Am. J. Respir. Crit. Care Med. 183, 788–824 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Xie T., Liang J., Geng Y., Liu N., Kurkciyan A., Kulur V., Leng D., Deng N., Liu Z., Song J., Chen P., Noble P. W., Jiang D., MicroRNA-29c prevents pulmonary fibrosis by regulating epithelial cell renewal and apoptosis. Am. J. Respir. Cell Mol. Biol. 57, 721–732 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Bauer Y., Tedrow J., de Bernard S., Birker-Robaczewska M., Gibson K. F., Guardela B. J., Hess P., Klenk A., Lindell K. O., Poirey S., Renault B., Rey M., Weber E., Nayler O., Kaminski N., A novel genomic signature with translational significance for human idiopathic pulmonary fibrosis. Am. J. Respir. Cell Mol. Biol. 52, 217–231 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Wang J.-H., Forterre A. V., Zhao J., Frimannsson D. O., Delcayre A., Antes T. J., Efron B., Jeffrey S. S., Pegram M. D., Matin A. C., Anti-HER2 scFv-directed extracellular vesicle-mediated mRNA-based gene delivery inhibits growth of HER2-positive human breast tumor xenografts by prodrug activation. Mol. Cancer Ther. 17, 1133–1142 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary material for this article is available at http://advances.sciencemag.org/cgi/content/full/7/24/eabg6005/DC1


