Adult muscles and their associated stem cells retain functional positional memory based on developmental origin.
Abstract
Muscle stem cells (satellite cells) are distributed throughout the body and have heterogeneous properties among muscles. However, functional topographical genes in satellite cells of adult muscle remain unidentified. Here, we show that expression of Homeobox-A (Hox-A) cluster genes accompanied with DNA hypermethylation of the Hox-A locus was robustly maintained in both somite-derived muscles and their associated satellite cells in adult mice, which recapitulates their embryonic origin. Somite-derived satellite cells were clearly separated from cells derived from cranial mesoderm in Hoxa10 expression. Hoxa10 inactivation led to genomic instability and mitotic catastrophe in somite-derived satellite cells in mice and human. Satellite cell–specific Hoxa10 ablation in mice resulted in a decline in the regenerative ability of somite-derived muscles, which were unobserved in cranial mesoderm–derived muscles. Thus, our results show that Hox gene expression profiles instill the embryonic history in satellite cells as positional memory, potentially modulating region-specific pathophysiology in adult muscles.
INTRODUCTION
Skeletal muscle is the contractile tissue that occupies approximately 40% of an individual’s body weight and is distributed throughout the body. Skeletal muscle has functionally heterogeneous properties that are specific to muscle type. Limb and trunk muscles function in body posture, locomotion, and respiration; while craniofacial muscles mainly control facial expression, speech, feeding activity, and eye movement. Most skeletal muscles of the trunk and limbs are derived from myogenic precursor cells that migrate from somites during embryonic development in vertebrates, whereas head musculature including masseter (MAS) muscles arises mainly from the cranial mesoderm (1).
Resident muscle stem cells, also called satellite cells, are located between the basal lamina and the plasmalemma of myofibers (2–4). Satellite cells provide myonuclei for postnatal muscle growth and hypertrophy, repair, and regeneration in adult muscle (5). Recent studies revealed that satellite cells are a functional heterogeneous population in different muscles and dependent not only on fiber types but also on embryonic origin (3, 4). We previously reported that satellite cells isolated from the extensor digitorum longus (EDL) muscle, which originate from somite, differ in their fate determination dynamics during proliferation, differentiation, and self-renewal compared with those from MAS muscle that originate from the cranial mesoderm (6). Satellite cells in the nasopharynx muscle of the head constitutively proliferate and contribute to myonuclear turnover without muscle injury (7), whereas these phenomena are rarely observed in limb muscles. The heterogeneity in the satellite cell population among muscles may therefore be related to region-specific pathophysiological phenotypes of muscle diseases (8).
There are distinct genetic networks in pre-myogenic progenitors between head and trunk/limb muscles during embryonic development (1). Paired-box protein Pax3 regulates limb myogenesis, whereas T-box transcription factor Tbx1, bicoid-related homeodomain transcription factor Pitx2, and musculin/Tcf21 play important roles in the specification of head muscle progenitor cells (9–14). Satellite cells also share the ontogeny of the muscle, and topographic genes expressed in satellite cells in the head are distinct from limb (6, 9, 15). Although satellite cell heterogeneity among muscles may be linked to body region–specific pathological phenotypes in muscle diseases (3, 4, 8), functional topographical genes remain unidentified. In addition, the epigenetic regulation of topographical gene expression in muscles and satellite cells is largely unknown. DNA methylation is a stable epigenetic mark that is associated with silencing and up-regulation of gene expression (16).
Here, we performed global DNA methylome and transcriptome analyses on different muscles and their associated satellite cell–derived myoblasts from adult mice to comprehensively uncover region-specific epigenetic and transcriptional properties. Homeobox (Hox) genes are well known as master regulators of the animal body plan that guide morphogenesis during development by controlling the genetic network program for temporal and spatial development of tissues and organs (17). Vertebrates have 13 Hox paralogs grouped in four gene clusters (A to D). We found that adult muscles and their satellite cells expressed Hox-A cluster genes, the expression patterns of which reflected their anatomical location and embryonic history. We also identified the Hoxa10 gene as a topographically functional gene: Postnatal deletion of Hoxa10 in satellite cells led to genomic instability and abnormal mitosis, resulting in a decline in the regionally specific regenerative ability of muscles. Thus, our findings suggest that Hox gene expression profiles are maintained in regionally specific patterns as positional memory in both adult muscles and their satellite cells.
RESULTS
Hox-A cluster locus is hypermethylated in limb muscle and associated satellite cells
To compare regionally specific DNA methylation between the head and limb muscles, we conducted the DNA methylome analysis using MAS muscle tissue from the head and tibialis anterior (TA) muscle tissue from the hindlimb in adult mice (Fig. 1A). We performed the microarray-based integrated analysis of methylation by isoschizomers (MIAMI) (18) and found that 7 probes for Hox-A cluster genes including Hoxa2, Hoxa3, and Hoxa5 were listed as the top 10 probes of DNA hypermethylated genes in the TA muscle compared with the MAS muscle (Fig. 1B). To validate the reliability of our MIAMI data, we performed additional DNA methylome analysis using the post-bisulfite adaptor tagging (PBAT)–mediated targeted methylome sequencing (19, 20) on the samples from MAS and TA muscles and associated satellite cell–derived myoblasts (positive for Pax7 and MyoD) cultured in growth media (GM) for 4.5 days (Fig. 1A). DNA methylation levels of CpG sites (Fig. 1C), principal components analysis (PCA) (Fig. 1D), and unsupervised hierarchical clustering (Fig. 1E) indicated similar methylation patterns among the samples (n = 3 mice) and that the global methylation status in muscle tissues was relatively higher than that in satellite cells. We performed Gene Ontology (GO) enrichment analysis based on hyper– or hypo–DNA methylated genes, which were described with embryonic development–related GO terms, such as anterior/posterior pattern specification, embryonic skeletal system morphogenesis, proximal/distal pattern formation, and embryonic hindlimb morphogenesis (fig. S1, A and B). Vertebrates have 13 Hox paralogs with four gene clusters (A to D), which are located on different chromosomes (Fig. 1F). Corresponding to the MIAMI data, only the Hox-A cluster was remarkably hypermethylated in both muscle tissues and satellite cells from TA muscles compared with satellite cells from MAS muscles (Fig. 1G). DNA methylation profiles of the Hox-A locus demonstrated that Hox-A genes, including Hoxa2, Hoxa3, and Hoxa5, are hypermethylated in the TA muscle and its associated satellite cells (Fig. 1H).
Fig. 1. Hox-A cluster locus is hypermethylated in limb muscles and associated satellite cells.
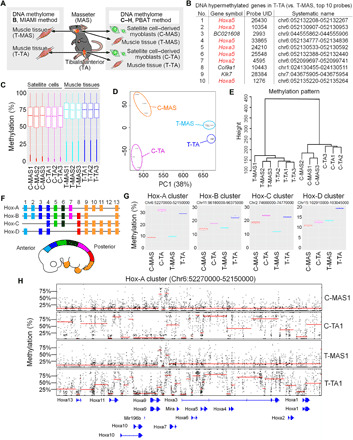
(A) Schematic illustration of DNA methylome analysis in adult mice. Global DNA methylation status in MAS or TA muscle tissues and their associated satellite cell–derived myoblasts was evaluated using the MIAMI method (B) and PBAT method (C to H). (B) The top 10 probes of DNA hypermethylated genes identified from TA muscle (versus MAS muscle). (C) Boxplot of mean CpG methylation level of 1-kb sidling windows. (D) PCA analysis for DNA methylation. (E) Hierarchical clustering using Euclidean distance across the entire sample set. (F) Schematic illustration of Hox cluster genes in the mouse embryo. (G) DNA methylation profiles for each Hox cluster. (H) DNA methylation profiles for Hox-A cluster. Black dots indicate the methylation levels of individual cytosines in the CpG context covered by at least five reads. Red lines indicate the domains calculated with a changepoint detection method, and their mean methylation levels are reflected in their positions in y axis [(B) n = 1; (C to H) n = 3].
Hox gene expression is maintained in adult muscles and their associated satellite cells
Hox-A and Hox-C cluster genes are known to be expressed in satellite cells in adult mice (21–24). Given that the hypermethylation status at Hox-A cluster locus was observed in limb muscles (Fig. 1), we next determined whether the Hox gene expression profile differed in various muscles and their associated satellite cells. We performed a microarray using different muscle tissues in adult mice and found a unique expression pattern of Hox-A and Hox-C cluster genes (Fig. 2A). Recent studies reported that DNA hypermethylation at the HOX-A locus is positively correlated with the up-regulation of HOX-A cluster genes in human cancer cells (25, 26). Consistent with this finding, we show that Hox-A and Hox-C cluster genes were abundantly expressed in both forelimb and hindlimb muscles, which originated from somite in development. However, only a few genes in the Hox-A and Hox-C clusters were detected in MAS, digastric (DIG), and trapezius (TRP) muscles that originated from the cranial mesoderm (Fig. 2A) (6, 9, 15, 27). We used four representative adult muscle tissues and their associated satellite cells, which included MAS and TRP with cranial mesoderm origin, supraspinatus (SS) and TA with somite origin (Fig. 2, B and C), and TRP and SS that are anatomically adjacent but have distinct origins. Genes that modulate the limb skeleton pattern are known to be regulated by the Hox paralogs Hox9 to 13 (17). Quantitative polymerase chain reaction (qPCR) analysis revealed that several Hox-A cluster genes such as Hoxa9 to 13 were detected in SS and TA limb muscle tissues and appear to be expressed in an anterior-posterior–dependent manner (Fig. 2B). TRP muscle abundantly expressed Hoxa5–7 but not Hoxa9–13 genes, whereas Hoxa5–13 genes were barely detected in MAS muscle (Fig. 2B). Muscle tissues and their associated satellite cells similarly expressed Hox-A cluster genes (Fig. 2, B and C).
Fig. 2. Hoxa10 gene is highly expressed in limb muscles and associated satellite cells.
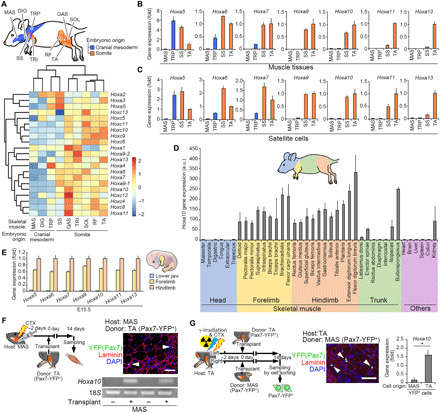
(A) Schematic illustration of cranial mesoderm–derived head muscles and somite-derived limb muscles in adult mice. Heatmap generated from microarray analysis demonstrated expression patterns of Hox-A and Hox-C cluster genes. (B and C) qPCR analysis of Hoxa5–13 genes expressed in muscle tissues (B) and cultured satellite cells (C) in adult mice (n = 4 mice). (D) qPCR analysis of Hoxa10 gene expression in muscle tissues, including the head, forelimb, hindlimb, trunk, and other organs from adult mice (n = more than 3 mice for each). a.u., arbitrary units. (E) qPCR analysis of Hoxa5–13 gene expression in the lower jaw, forelimb, and hindlimb at E15.5 (n = 5 mouse embryos). (F) Schematic illustration of the transplantation assay. TA muscle homogenates (donor: Pax7-YFP mice) were engrafted into MAS muscle (host: WT mice) preinjured with a CTX injection. Representative immunohistochemical image of MAS transverse sections for laminin, YFP (Pax7), and DAPI. Scale bar, 50 μm. A representative image of an agarose gel electrophoresis containing PCR products from Hoxa10 and 18S genes 14 days following transplantation (n = 5 mice). (G) Schematic illustration of the transplantation assay. TA muscle tissues (host: WT mice) were locally exposed to 18 Gy of γ-irradiation and injected with CTX before transplantation of either MAS or TA muscle tissue homogenates (donor: Pax7-YFP mice). Scale bar, 50 μm. Grafted YFP+ satellite cells were re-sorted from TA muscles from host mice 14 days following transplantation. qPCR analysis for Hoxa10 gene expression (n = 4 mice). All error bars show the SEM.
Because Hoxa10 expression clearly separated somite-derived muscles and cranial mesoderm–derived muscles (Fig. 2A), we further determined the expression pattern of Hoxa10 in muscles throughout the body in adult mice (Fig. 2D). qPCR analysis confirmed that all forelimb and hindlimb muscles as well as some trunk muscles expressed the Hoxa10 gene, while it was undetected in head muscles, which is consistent with the Hoxa10 expression pattern at embryonic developmental regions (Fig. 2E). Thus, these results suggest that adult muscles and their satellite cells retain Hox gene expression–based regional identities whose distribution almost recapitulates their embryonic origin, which we call “positional memory.”
To test how robust Hox genes are maintained in satellite cells upon grafting in a heterologous location, we performed a transplantation study using MAS (Hoxa10-negative niche) and TA (Hoxa10-positive niche) muscles from Pax7-YFP knock-in mice (28). Pax7-YFP mouse–derived TA muscle homogenates that contained yellow fluorescent protein (YFP) (Pax7+) satellite cells were transplanted into cardiotoxin (CTX)–injected preinjured MAS muscle (Fig. 2F). Immunohistochemical analysis confirmed that TA muscle–derived YFP (Pax7+) cells repopulated the MAS niche 2 weeks after transplantation (Fig. 2F). Expression of Hoxa10 mRNA was detected only in the MAS muscle tissue transplanted with TA muscle homogenates (Fig. 2F). Next, Pax7-YFP mouse–derived MAS or TA muscle homogenates were transplanted into TA muscle preinjured with CTX immediately after 18 grays (Gy) of γ-irradiation of the hindlimbs (Fig. 2G). Two weeks following transplantation, immunohistochemistry confirmed that MAS or TA muscle homogenate–derived YFP (Pax7+) cells repopulated the TA muscle niche. We analyzed the effect of ectopic transplantation at a 2-week period because engrafted satellite cells were expected to almost return to the resting phase even though the muscles are not completely regenerated. Repopulated YFP (Pax7+) cells were then re-sorted, and qPCR analysis revealed that the higher levels of Hoxa10 gene expression were detected in re-sorted cells from the TA muscle engrafted with TA muscle tissues containing YFP (Pax7+) cells compared with MAS muscle tissues containing YFP (Pax7+) cells (Fig. 2G).
Postnatal Hoxa10 deletion in satellite cells decreased region-specific muscle regeneration
Because Hoxa10 is highly expressed only in limb muscles but not in head muscles, we assessed whether genes conferring positional memory have a role in adult mice. Hoxa10-deficient mice exhibit abnormal morphogenesis of sexual organs, spinal nerves, and femurs during development (29, 30). To investigate the function of Hoxa10 in satellite cells from adult muscle, we constructed a Hoxa10-floxed mouse line (fig. S2, A and B). We then generated satellite cell–specific and tamoxifen (TMX)–inducible Hoxa10 knockout mice (scKO) by crossing Pax7CreERT2/+ mice (31) with Hoxa10-floxed mice. Genetic inactivation of Hoxa10 was induced by repeated intraperitoneal injection of TMX in Pax7CreERT2/+;Hoxa10f/f mice. A muscle injury was then induced in both head and hindlimb muscles using a CTX injection (Fig. 3A). The number of Pax7+ satellite cells was unaltered in intact TA muscles from scKO mice 24 days following the first TMX injection (Fig. 3B). TA muscle mass was significantly reduced in scKO mice compared with those of Hoxa10f/f (CON) mice; however, MAS muscle was unaffected (Fig. 3, C and D), which was consistent with the Hoxa10 gene expression in limb but not head muscles. Cross-sectional area (CSA) of regenerating myofibers in TA but not MAS muscle significantly decreased at both early (5 days; Fig. 3F) and late (14 days; Fig. 3, E and G) phases of muscle regeneration following CTX injection.
Fig. 3. Hoxa10 is required for limb muscle regeneration.
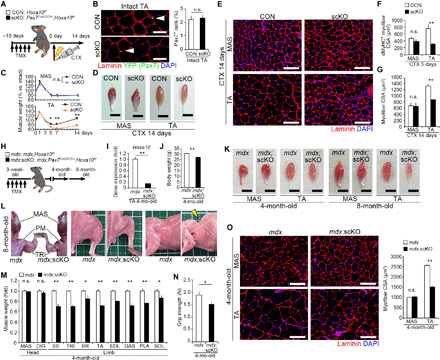
(A to G) Hoxa10 deletion in satellite cells was induced by five serial intraperitoneal injections of TMX in Pax7CreERT2/+;Hoxa10f/f mice before induction of muscle regeneration of MAS and TA muscles by CTX injection. (A) Time course. (B) Representative immunohistochemical images of TA muscle sections 24 days following the first TMX injection. Pax7+ cell number per DAPI+ nucleus (ratio) (n = 3 mice each). Scale bars, 50 μm. (C) Weight of regenerated muscles normalized to intact muscles before and after CTX injection (n = 4 mice each). (D) Representative images of regenerated muscles 14 days after CTX injection. Scale bars, 5 mm. (E) Immunohistochemistry of MAS and TA muscle sections 14 days after CTX injection. CSA of regenerating myofibers was visualized by staining for developmental MyHC (dMyHC) (F) 5 days (TA, n = 5 mice; MAS, n = 4 mice) or for laminin (G) 14 days (n = 5 mice each) after CTX injection. Scale bars, 100 μm. (H to O) To perform TMX-inducible ablation of Hoxa10 in satellite cells in mdx mice, mdx;Pax7CreERT2/+;Hoxa10f/f (mdx;scKO) mice were generated. (H) Time course. (I) qPCR analysis of Hoxa10 expression in TA muscle (n = 5 mice each). (J) Mean body weight. (K) Representative images of muscles. Scale bars, 5 mm. (L) At 8 months, mdx;scKO mice exhibited kyphosis and severe muscle atrophy throughout the body except in head muscles. Grid size (solid line), 1 cm by 1 cm. PM, pectoralis major. (M) Muscle weight of mdx;scKO mice normalized to those of mdx mice (n = 7 mice each). (N) Grip strength (n = 7 mice each). (O) Immunohistochemistry for laminin to measure myofiber CSA (n = 5 mice each). Scale bars, 100 μm. All error bars show the SEM. Photo credit: Kiyoshi Yoshioka, Kumamoto University.
We further examined how satellite cell–specific deletion of the Hoxa10 gene affects muscle regeneration throughout the body in mdx mice, which is a Duchenne muscular dystrophy (DMD) model used to monitor chronic muscle degeneration and regeneration throughout the body (Fig. 3H). Hoxa10 inactivation in satellite cells of mdx (mdx;scKO) mice was induced by a serial injection of TMX at 3 weeks of age and then analyzed at 4 or 8 months of age. mdx;scKO mice displayed reduced grip strength and severe muscle atrophy throughout the body except in head muscles (Fig. 3, I to O). At 8 months, the mdx;scKO mice showed gradual muscle weakness and noticeable kyphosis, which is a hallmark of extensive and severe muscle wasting in the trunk muscles (Fig. 3L). These results indicate that satellite cell–specific deletion of Hoxa10 perturbed efficient regeneration in limb muscles in vivo, demonstrating that a regionally specific gene can control satellite cell function in a site-specific manner.
Hoxa10 deletion triggers mitotic catastrophe
Given that satellite cell–specific inactivation of Hoxa10 caused a severe regeneration defect in limb muscles, even though Hox paralogs are often functionally redundant (17), we tested how satellite cell function is affected by the absence of Hoxa10 ex vivo. To inactivate Hoxa10 gene in satellite cells, Pax7CreERT2/+;Hoxa10f/f mice were treated for 5 days with TMX. Individual myofibers from EDL muscle were freshly isolated 5 days after the last injection (designated as 0 hours) and then cultured floating in a mitogen-rich media for up to 72 hours or plating in GM for 6 days (Fig. 4A). Immunocytochemistry revealed that the number of Pax7+ quiescent satellite cells associated with myofibers was unchanged in the absence of Hoxa10 (Fig. 4B). There was no difference in the total number of Pax7+ and/or MyoD+ satellite cells per myofiber between CON and scKO at 24 hours (Fig. 4B), thereby indicating that Hoxa10 was not required for satellite cell activation. However, after 48 hours, the total number of satellite cells per myofiber significantly decreased compared with cells expressing Hoxa10 (Fig. 4B). Under adherent culture conditions, we further observed minimal migrating and proliferating satellite cells from myofibers isolated from scKO mice compared with those from CON (Fig. 4C) as well as satellite cells in the niche (Fig. 4B). These data suggest that Hoxa10 plays an important role in the expansion of activated satellite cells and that the satellite cell niche does not affect the proliferative ability of Hoxa10-ablated satellite cells.
Fig. 4. Inactivation of Hoxa10 leads to mitotic catastrophe in satellite cells.
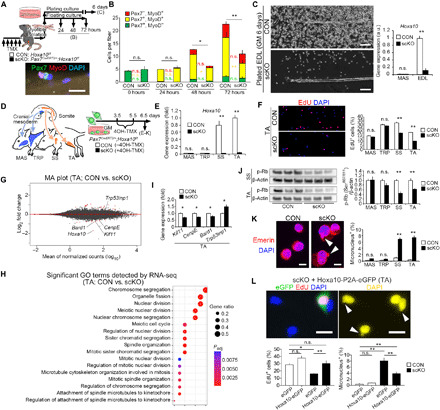
(A) Satellite cells associated with myofibers were isolated from muscles of Pax7CreERT2/+;Hoxa10f/f mice and cultured under floating or adherent conditions. (B) The number of Pax7+ and/or MyoD+ satellite cells per EDL myofiber (n = more than 3 mice, 15 to 20 myofibers per mouse were counted). (C) Isolated myofibers were cultured with GM for the adherent condition. Hoxa10 gene expression in satellite cells that migrated from the myofibers was determined by qPCR (n = 4 mice each). (D to K) Satellite cells were isolated from the cranial mesoderm–derived (MAS and TRP) or somite-derived (SS and TA) muscles of Pax7CreERT2/+;Hoxa10f/f mice and treated with 4OH-TMX under GM culture conditions. (D) Time schedule. (E) qPCR analysis (n = 6 mice). (F) Proliferation ability of cultured satellite cells determined by 3 hours of EdU pulse-chase analysis (MAS and TRP, n = 3 mice; SS and TA, n = 6 mice). (G and H) RNA-seq analysis was performed in satellite cells isolated from TA muscle. Mean MA plots depicting fold change in gene expression (G) and significant GO terms (H) (n = 9 mice). Padj, adjusted P value. (I) qPCR analysis of mitosis and chromosome segregation–related gene expression (n = 6 mice). (J) Immunoblot analysis for the expression of phosphorylated (Ser807/811) Rb in satellite cells from scKO mice normalized to phosphorylated levels from control mice (n = 6 mice). (K) Micronuclei were quantified by staining for Emerin and DAPI (n = 5 mice). Arrowheads show micronuclei. Scale bars, 10 μm. (L) pCMV-Hoxa10-P2A-eGFP vector–mediated exogenous expression of Hoxa10 in TA scKO cells. Arrowheads show micronuclei in untransfected (eGFP–) cells. EdU+ cells or micronucleus+ cells were quantified (n = 4 mice). Scale bars, 20 μm. All error bars show the SEM.
To further evaluate the effect of Hoxa10 inactivation on satellite cell function, we used muscles with different developmental origins, such as TRP and MAS muscles that originate from the cranial mesoderm and SS and TA muscles with a somite origin (Fig. 4D and fig. S3A). These cell types are clearly distinguishable by their Hoxa10 expression levels (Figs. 2C and 4E). A cell cycle assay including EdU (5-ethynyl-2′-deoxyuridine) pulse-chase analysis revealed that proliferation was impaired in both SS- and TA-derived scKO satellite cells, while both TRP- and MAS-derived scKO satellite cells were unaffected (Fig. 4F and fig. S3, B, C, E, and F). A terminal deoxynucleotidyl transferase–mediated deoxyuridine triphosphate nick end labeling (TUNEL) assay showed that apoptosis was not involved in the cell number decline observed in scKO TA satellite cells (fig. S3G).
Given that proliferation in Hoxa10-deficient satellite cells was decreased compared with those expressing Hoxa10, we next examined how population expansion of limb-derived satellite cells was impaired by the loss of Hoxa10. RNA sequencing (RNA-seq) analysis revealed that mitosis-related gene sets for chromosomal segregation, meiotic nuclear division, and spindle organization significantly changed in Hoxa10-deficient satellite cells compared with CON cells (Fig. 4, G and H). Kif11, CenpE, and Bard1 regulate mitotic spindle formation and genomic stability, and Trp53inp1 is up-regulated by cellular stress and induces cell cycle arrest. qPCR analysis indicated notable changes in the expression of these genes in TA-derived scKO satellite cells (Fig. 4I). Dephosphorylation of Rb, which is known to be induced by genomic instability, arrests the cell cycle. Our immunoblot analysis confirmed that Hoxa10 ablation led to a marked decrease in the level of phosphorylated Rb in limb-derived scKO satellite cells (Fig. 4J). Micronuclei and chromosomal bridges are hallmarks of chromosome misalignment and mitotic catastrophe. We found that the ratio of micronuclei and chromosomal bridges increased in only somite-derived scKO satellite cells and not cranial mesoderm–derived scKO satellite cells (Fig. 4K and fig. S3, D, H, and I). Note that genetic ablation of Hoxa10 did not influence myogenic differentiation or self-renewal of TA-derived satellite cells but that differentiating myotubes contained abundant micronuclei (fig. S3, J to N). To determine the effect of exogenous expression of Hoxa10, satellite cells were isolated from TA or MAS muscles of Pax7CreERT2/+;Hoxa10f/f mice, transfected with pCMV-Hoxa10-P2A-eGFP or pCMV-eGFP (control) vectors, treated with 4-hydroxy TMX (4OH-TMX), and analyzed for 6.5 days under GM culture conditions after isolation. The decreased proportion of EdU+ proliferative cells and the increased proportion of micronucleus+ cells in somite-derived scKO cells were rescued by pCMV-Hoxa10-P2A-eGFP vector–mediated exogenous Hoxa10 expression in TA-derived satellite cells (Fig. 4L). We also confirmed that plasmid-mediated expression of other Hox genes (Hoxa5, Hoxa9, and Hoxc10) did not rescue the proliferative defect in limb-derived Hoxa10 scKO satellite cells (fig. S3W). Last, these findings in our genetic ablation models were confirmed by small interfering RNA (siRNA)–mediated knockdown of Hoxa10 in limb-derived satellite cells in wild-type (WT) mice (fig. S3, O to V). Together, our data indicate that postnatal reduction of Hoxa10 in somite-derived satellite cells led to genomic instability and mitotic catastrophe, resulting in a reduction in proliferation ability.
HOXA10 expression and function are conserved in human satellite cells
We evaluated whether the function of HOXA10 characterized in mice was conserved in satellite cells from adult humans. We used semitendinosus (ST) muscle from the lower limb and MAS and buccinator (BUC) muscles from the head of adult humans (Fig. 5A). qPCR analysis confirmed that PAX3 or TCF21 genes were detected in ST limb or MAS head muscles, respectively (Fig. 5B), similar to the results from mouse satellite cells (6). We showed that the HOXA10 gene was detectable in ST muscle and in its associated satellite cells (Fig. 5, B and C). It is likely that HOXA10 gene expression is robustly maintained in ST satellite cells because long-term passaging did not influence its expression level (Fig. 5D).
Fig. 5. Expression and function of HOXA10 are preserved in human satellite cells.
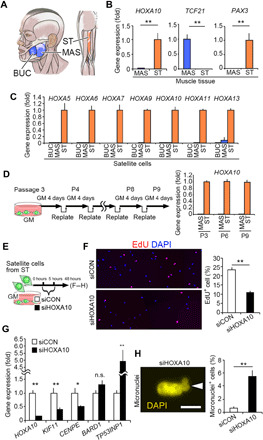
(A to D) Satellite cells were isolated from the lower limb ST muscle and MAS and BUC muscles from the heads of adult humans (A). (B and C) qPCR analysis of gene expression in skeletal muscle tissues (B) (n = 4 individuals for each muscle) and satellite cells (C) (BUC, MAS, and ST: n = 3, n = 4, and n = 3 individuals, respectively). (D) Satellite cells isolated from MAS and ST muscles were repeatedly passaged for up to nine passages in culture. Samples were collected at the third (P3), sixth (P6), and ninth (P9) passages, and HOXA10 gene expression was determined by qPCR (the value of P3 = 1, P6 and P9 relative to P3). (E to H) To examine the effect of HOXA10 knockdown, siRNA targeting HOXA10 was transfected into satellite cells derived from ST muscle in vitro. (E) Time schedule of siRNA transfection. (F) Quantification of EdU+ cells (n = 4 individuals). (G) qPCR analysis of mitosis and chromosome segregation–related gene expression (n = 12 individuals). (H) A representative image of micronucleus+ satellite cells and their quantification (n = 4 individuals). All error bars show the SEM.
To further examine the function of HOXA10 in ST-derived satellite cells, siRNA-mediated knockdown of HOXA10 was performed (Fig. 5E). Consistent with the mouse phenotypes, reducing HOXA10 expression levels resulted in the impairment of proliferation ability, as well as a down-regulation in the expression of KIF11 and CENPE gene in limb satellite cells, while TP53INP1 was up-regulated compared with the control siRNA–transfected cells (Fig. 5, F and G). Notably, we confirmed that the ratio of micronuclei increased in HOXA10 knocked-down satellite cells (Fig. 5H). Thus, we conclude that HOXA10 is expressed in limb satellite cells and functions in population expansion of satellite cells in human limb muscle.
DISCUSSION
Expression of region-specific genes, including Hox genes, has been observed in muscles both during embryonic development and in adult mice (6, 9, 15, 21–23, 32, 33). However, whether regionally specific genes have a function in satellite cells of adult muscle remains to be elucidated. In the present study, our comprehensive analysis demonstrated that Hox gene expression profiles in adult muscles almost mirror embryonic origin. These regionally specific Hox expression patterns were regulated epigenetically, which could act as a molecular signature that reflects the embryonic history in both adult skeletal muscles and resident muscle stem cells. Our ectopic transplantation study shows that MAS-derived satellite cells maintained low Hox status 2 weeks following TA niche exposure. However, Evano et al. (24) reported that satellite cells isolated from the extraocular muscle, which is derived from the cranial mesoderm, express Hox-A cluster genes at very low levels, but these genes are up-regulated 3 to 4 weeks after transplantation into CTX preinjected TA muscle. This discrepancy may be explained by the different time periods after transplantation or different cell types: A longer duration of exposure to different niches may gradually influence Hox status, or extraocular muscle–derived satellite cells may adapt to different niches compared to MAS-derived satellite cells. We also note that for efficient cell engraftment, we depleted intrinsic satellite cells in TA muscle tissue with γ-irradiation before transplantation, while Evano et al. (24) did not use radiation. Thus, these irradiated and nonirradiated transplantation conditions may differentially affect the regenerative niche and plasticity of engrafted satellite cells in the host muscle tissues. Therefore, we need to further investigate the molecular and cellular mechanisms underlying the plasticity of Hox expression in satellite cells in an ectopic transplantation study with a nonirradiated condition and a longer period.
Recent studies reported that DNA hypermethylation on the HOX-A locus is positively correlated with the up-regulation of HOX-A cluster genes in human cancer cells (25, 26). This correlation suggests that DNA hypermethylation permits the up-regulation of HOX-A cluster genes. Our DNA methylome analysis also revealed hypermethylation of the Hox-A loci and high expression levels of Hox-A cluster genes in adult limb muscles and their associated satellite cell–derived myoblasts. Although these loci are not uniformly hypermethylated and the DNA hypermethylation patterns do not simply correlate with the expression levels of Hox-A cluster genes, our findings are further supported by the study depicting the hypermethylation status of Hox-A loci in hindlimb-derived satellite cells in mice (24). In addition to DNA methylation, histone modifications may also be involved in regulating Hox-A gene cluster expression in satellite cells (23, 34). A recent study by Schwörer et al. (23) described the up-regulation of Hoxa9 in limb satellite cells during aging, and up-regulated Hoxa9 perturbed cell cycle entry through the misregulation of genes involved in development, such as Wnt, transforming growth factor–β, and JAK/STAT pathways. However, it remains unknown whether the up-regulation of Hoxa9 in satellite cells with aging is related to DNA hypermethylation status in Hox-A loci (35). Because our data demonstrated that genetic loss of Hoxa10 in satellite cells of young adult mice resulted in a decrease in proliferation with genomic instability, Hoxa9 and Hoxa10 have entirely distinct function in satellite cells, even though both are adjacent in the Hox-A cluster genes. Consistent with the study by Schwörer et al. (23), we also confirmed that the overexpression of Hoxa9 or Hoxc10 suppressed satellite cell population expansion. Currently, it remains unclear how structurally similar Hox proteins exert distinct functions. In support of our findings, functional equivalence among Hox paralog genes does not always seem to be the rule (36): HOXA5 inhibits endothelial cell differentiation and blocks angiogenesis (37), whereas its paralog HOXB5 promotes vascular growth and differentiation of endothelial cells (38). In addition, Hoxa2 and Hoxb2 have opposing roles in hindbrain oligodendrocyte patterning: Hoxa2 suppresses oligodendrogenesis, but Hoxb2 accelerates it during mouse brain development (39). Considering these findings, we speculate that the expression levels of individual Hox genes and combinations of Hox gene subsets determine satellite cell function in a context-dependent manner. It would be valuable to identify other genes that may substitute for Hoxa10 in cranial mesoderm–derived satellite cells. We have sought to uncover such mechanisms in head-derived satellite cells using DNA methylome and RNA-seq data combined with RNA interference–mediated knockdown analysis but have not yet found any interesting genes. Alternatively, we think that there may be no compensating mechanism in MAS-derived satellite cells: These cells can proliferate like fibroblasts or other types of cells independent of Hoxa10 (it is also unclear why, in an evolutionary context, limb satellite cells need Hoxa10 to proliferate, which would be an interesting issue to solve). Furthermore, protein-protein interaction analysis revealed that some Hox proteins associate with DNA repair and DNA replication through the nontranscriptional activities of Hox proteins (36); for example, HOXB7 and HOXC4 proteins regulate DNA repair via a direct interaction of nonhomologous end joining proteins (40, 41); HOXC13 and HOXD13 proteins directly interact with proteins related to DNA replication origins (42, 43). Therefore, further investigations by proteome analysis will be needed to determine the nontranscriptional activities of Hoxa10 in regulating proliferation and genomic stability in satellite cells.
We demonstrated that muscles and their associated satellite cells retain positional memory based on developmental origin and anatomical location, which dictates stem cell function in a regionally specific manner in adults. In our recent study, we showed an example for the connection of position-specific gene expression profiles with position-specific properties of satellite cells using an androgen receptor (AR)–highly expressing peripheral muscle (androgen-sensitive) and an AR–lower expressing TA (androgen-insensitive) muscle (44). Our ectopic transplantation study revealed that perineal satellite cell–derived regenerated TA muscles are more severely affected by an androgen reduction than hindlimb satellite cell–derived muscles in a castrated mouse model (44). These findings suggest that functional positional identities may link gene expression profiles with hormonal sensitivities, although the data do not necessarily account for the robustness of Hox-based positional memory in satellite cells. Muscular dystrophy is characterized by progressive muscle weakness and wasting that vary in severity and in body regions affected (8). Patients with DMD are preferentially inflicted with a severe pathology in limb and trunk muscles, but not in craniofacial muscles, particularly eye muscles. Patients with Emery-Dreifuss muscular dystrophy (EDMD) show progressive muscle weakness, especially in the arms and lower legs. Moreover, particular muscles are known to be affected in patients with oculopharyngeal muscular dystrophy (OPMD), limb girdle muscular dystrophy (LGMD), and facioscapulohumeral muscular dystrophy (FSHD). Ectopic expression of the transcriptional factor DUX4 is observed in muscles of patients with FSHD, and DUX4 expression recapitulates the FSHD pathology (45). Intriguingly, DUX4 contains two adjacent DNA binding homeodomains, where structural similarities to some of Hox proteins are suggested (45). We therefore speculate that region-specific Hox expression profiles influence DUX4 transcriptional activity, which might be associated with the body region–specific pathology of patients with FSHD. The body region–specific muscle pathology is also observed during aging and microgravity. For instance, MAS muscle from the head is resistant to atrophy during aging and spaceflight (46, 47), whereas head muscles regenerate slower than the limb muscles (44, 48). In this study, we identified Hoxa10 as a region-specific gene that regulates genomic stability and expansion of satellite cell populations. On the basis of these data, Hox-based functional positional memory may explain the varying regenerative capacity between different muscle groups and account for the susceptibility of specific regions of muscles to diseases. Further studies will elucidate the link between positional memory and region-specific pathologies in muscle disorders.
MATERIALS AND METHODS
Animals
The Experimental Animal Care and Use Committee of Nagasaki University and Kumamoto University approved the animal experimentation (ref. no. 1203190970 and A30-098). The BRUCE-4 ES cell line (C57/BL6J) was used to generate the Hoxa10-floxed mouse line. A targeting vector was generated to modify the Hoxa10 locus by inserting loxP sequences upstream of exon 2 and downstream of exon 3 in the Hoxa10 gene (fig. S2A). To induce conditional ablation of the Hoxa10 gene, Hoxa10-floxed mice were crossed with Pax7CreERT2/+ mice (31). Pax7-YFP (Pax7YFP/YFP) mice were used to visualize satellite cells expressing Pax7-YFP fusion proteins (28). mdx mice were obtained from CREA Japan Inc. (Tokyo, Japan) and were crossed with scKO mice to generate mdx;scKO mice. In Pax7CreERT2/+;Hoxa10f/f mice, inactivation of Hoxa10 was induced by repeated intraperitoneal injection of TMX (Sigma-Aldrich) dissolved in corn oil (5 μl/g, 20 mg/ml) as previously described (49).
For muscle injury, mice were anesthetized, and the hair on the lower jaw and lower leg areas was removed with a depilatory cream (Epilat, Kracie). The facial nerve was percutaneously identified to avoid needle damage. To induce muscle injury, 10 μM CTX (Sigma-Aldrich) (50 μl for TA and 40 μl for MAS) was injected into the muscles of anesthetized mice. CTX-injected and noninjected (intact) muscles were harvested for transverse sectioning and immunostaining. For cell transplantation into TA muscle, mice were anesthetized, and 18 Gy of γ-radiation was exposed to the hind legs to prevent population expansion of endogenous satellite cells. The rest of the body was protected by a 2-cm-thick lead plate. However, γ-radiation could not be used for transplantation into the MAS muscle because it severely affects cells in the head, including the cells in the central nervous system.
Grip strength
To test muscle strength, forelimb grip strength was measured using a grip strength meter (Columbus Instruments, Columbus, OH) as previously described (50) with minor modification. Peak tension (in newton) was recorded when the mouse released its grip. Three sets of five successive measurements were performed for each mouse, and the peak value was defined as mouse whole-limb grip strength.
Antibodies
Mouse anti-Pax7 (1:1000; Pax7, sc-81648), rabbit anti-MyoD (1:800; M-318, sc-760), and rat anti-laminin a2 (1:2000; 4H8-2, sc-59854) antibodies were purchased from Santa Cruz (Santa Cruz, CA). Mouse anti-MyHC antibody (1:1000; MF20, MAB4470) was purchased from R&D Systems (Minneapolis, MN). Rabbit anti-GFP antibody (1:1000; A-11122) was purchased from Invitrogen (Carlsbad, CA). Rat anti-Ki67 antibody (1:1000; TEC-3, M7249) was purchased from DAKO (Glostrup, Denmark). Mouse anti–phospho-Rb (1:2000; Ser807/811, #9308), rabbit anti-Emerin (1:1000; D3B9G, #30853), and rabbit anti–phospho-Aurora A (Thr288)/Aurora B (Thr232)/Aurora C (Thr198) (1:2000; D13A11, #2914) were purchased from Cell Signaling Technology (Danvers, MA). Mouse anti-tubulin antibody (1:5000; B512, #T5168) was purchased from Sigma-Aldrich (St. Louis, MO). Mouse anti–β-actin (1:5000; 6D1, #M177-3) was purchased from Medical & Biological Laboratories Co. Ltd. (MBL) (Nagoya, Japan).
Methylome analysis
For MIAMI analysis, a genome-wide analysis of DNA methylation was performed on the basis of the protocol (http://epigenome.dept.showa.gunma-u.ac.jp/~hatada/miami/image/MIAMI%20Protocol%20V4.pdf) (18) as descried previously (51). Briefly, genomic DNA was extracted from MAS and TA muscle tissues of mice via the standard proteinase K method. Purified genomic DNA was digested with methylation-sensitive Hpa II and methylation-insensitive Msp I, followed by adaptor ligation and PCR amplification. Amplified DNA from MAS and TA tissues was labeled with Cy3 and Cy5, respectively. Labeled DNA was then co-hybridized using the Oligo aCGH/ChIP-on-chip Hybridization kit (Agilent Technologies, CA), and the difference in the Hpa II/Msp I signal ratio was determined as the methylation value. Values of <0.5 and >2 denoted DNA hypomethylation and hypermethylation, respectively.
For further methylome analysis, the PBAT-mediated whole-genome bisulfite sequencing was performed as previously described (19, 20). For preparation of PBAT libraries, genomic DNAs were extracted from MAS and TA muscle tissues and plated satellite cells of adult mice. Sequencing was performed by Macrogen Japan Corp. (Kyoto, Japan) using the HiSeq X Ten, and the reads were mapped to the mouse mm10 reference genome using BMap (http://itolab.med.kyushu-u.ac.jp/BMap/index.html) with default parameter settings. Methylation levels of CG sites were calculated for only those covered by five or more reads. Methylation levels of 1-kb sliding windows, promoters [<2 kb upstream of transcriptional start site (TSS)], gene bodies, CG islands (CGIs), and TSS-proximal regions were determined by averaging the methylation levels of CG sites in individual features and were analyzed using two unsupervised classification methods, namely, hierarchical clustering and PCA. Differentially methylated regions (DMRs) were identified using the program metilene (52) with default parameters. DMRs were filtered so that they have at least five CpGs with a q value of less than 0.05 and methylation differences larger than 20%. The nearest neighbor gene of a DMR was defined as the gene associated with the DMR. All functional enrichment analysis with GO biological process terms was performed using TopGO (R package version 2). A P value of 0.01 was set as the significance level for gene set enrichment with an additional requirement that at least five genes are involved in the annotated biological function. Raw DNA methylation datasets are available from the Gene Expression Omnibus (GEO) public depository under the accession number GSE154056. In Fig. 1H, red lines indicate the domains calculated with a changepoint detection method (53), and their mean methylation levels are reflected in their positions in y axis.
Transcriptome analysis
For microarray experiments, total RNA was extracted from MAS, DIG, TRP, SS, GAS (gastrocnemius), TRI (triceps brachii), SOL (soleus), RF (rectus femoris), and TA. Microarray analysis was performed using the Affimetrix mouse gene 2.0 ST array (Affymetrix, Santa Clara, CA). The complementary RNA was amplified, labeled, and hybridized to a 60K Agilent 60-mer oligomicroarray according to the manufacturer’s instructions. All hybridized microarray slides were scanned by an Agilent scanner. Relative hybridization intensities and background hybridization values were calculated using Agilent Feature Extraction Software (9.5.1.1).
For RNA-seq analysis, total RNA was obtained from satellite cells grown under the GM culture condition. Library preparation and sequence analysis were performed according to the manufacturer’s protocol (NEBNext Ultra Directional RNA Library Prep Kit for Illumina) (54).
Raw microarray datasets are available from the GEO public depository under the accession number GSE139315. Raw RNA-seq datasets are available from the GEO public depository under the accession number GSE139639.
Mouse satellite cell culture
Satellite cells were isolated from muscles using either the single myofiber method (55) or the preplating method (56) as previously described. The single myofiber method was used only for EDL muscle except the siRNA study (fig. S3, O to R), where satellite cells were purified using the preplating method followed by the single myofiber method. Isolated satellite cells were cultured in GM [Dulbecco’s modified Eagle’s medium (DMEM; C11995500, Thermo Fisher Scientific, Tokyo, Japan) supplemented with 30% fetal bovine serum (Thermo Fisher Scientific), 1% chick embryo extract (C3999, USBiological, MA), basic fibroblast growth factor (10 ng/ml; NIB 47079000, Oriental Yeast, Tokyo, Japan), and 1% penicillin-streptomycin (168-23191, Wako, Osaka, Japan)] at 37°C under 5% CO2 in culture dishes coated with Matrigel (354230, BD Biosciences, NJ). Myogenic differentiation was induced in differentiation medium (DMEM supplemented with 5% horse serum and 1% penicillin-streptomycin) at 37°C under 5% CO2. For induction of Cre recombinase in vitro, 1 μM 4OH-TMX (Sigma-Aldrich) was added to culture medium for 2 days. For floating culture, isolated myofibers from EDL muscle were cultured in floating medium [DMEM supplemented with 10% horse serum (Thermo Fisher Scientific), 0.5% chick embryo extract, and 1% penicillin-streptomycin] at 37°C under 5% CO2.
For transplantation, satellite cell populations were also isolated by fluorescence-activated cell sorting (FACS) using a FACSAria II flow cytometer (BD Immunocytometry Systems) as previously described (28). Debris and dead cells were excluded by forward scatter, side scatter, and propidium iodide (PI) gating.
Human muscle tissue and satellite cell culture
The use of human tissue samples was approved by the Nagasaki University Ethics Committee (ref. 16052306; ref. 0738-8). ST muscles were collected from patients undergoing reconstructive surgery, MAS and BUC muscles were collected from patients undergoing orthognathic surgery, and human satellite cells were isolated from muscle tissues by the preplating techniques and cultured in GM at 37°C under 5% CO2 as previously described (56).
Transfection
Transfection of siRNA was performed as previously described (49). Cells were plated in a six-well plate at 30 to 40% confluency and transfected with siRNA. siRNA duplexes (Stealth siRNA; Thermo Fisher Scientific) were diluted in OptiMEM (Thermo Fisher Scientific) to 10 pmol per well and incubated with RNAiMAX (Thermo Fisher Scientific) diluted in OptiMEM according to the manufacturer’s instructions. The following siRNA sequences were used: mouse Hoxa10 siRNA-1: 5′-AAUUAAAGUUGGCUGUGAGCUCCCG-3′; mouse Hoxa10 siRNA-2: 5′-UAAGGUACAUGUUGAAUAGAAACUC-3′; human HOXA10 siRNA: 5′-GAGUUUCUGUUCAAUAUGUACCUUA-3′. A control siRNA selected by Thermo Fisher Scientific was also used in this experiment. For forced expression of the Hoxa10 gene, cells were transfected with the pCMV-Hoxa10-P2A-eGFP vector using Lipofectamine LTX (Thermo Fisher Scientific) according to the manufacturer’s instructions. The pCMV-eGFP vector was used as a control.
Immunostaining and imaging
Immunocytochemistry of satellite cells and isolated myofibers was performed as previously described (49). Samples were incubated with primary antibodies at 4°C overnight following fixation in 2 to 4% paraformaldehyde and blocked/permeabilized in phosphate-buffered saline (PBS) containing 0.3% Triton X-100 and 5% goat serum for 30 min at room temperature (20° to 25°C).
For immunohistochemistry, frozen cryosections of muscle tissues were fixed in 4% paraformaldehyde, blocked with 5% goat serum or the M.O.M. kit (Vector Laboratories, CA) for 30 min at room temperature, and incubated in primary antibodies at 4°C overnight. All immunostaining samples were visualized using appropriate species-specific Alexa Fluor 488 and/or 546 fluorescence–conjugated secondary antibodies (Thermo Fisher Scientific). Samples were mounted in medium containing 4′,6-diamidino-2-phenylindole (DAPI) for nuclear staining (Nacalai Tesque, Kyoto, Japan). Samples were viewed on an Olympus fluorescence microscope IX83 (Olympus, Tokyo, Japan) or a CellInsight CX5 (Thermo Fisher Scientific). Digital images were acquired and quantified with a DP80 camera using cellSens software (Olympus). Images were optimized globally and assembled into figures using Adobe Photoshop. Immunostaining for laminin or hematoxylin-eosin staining was used to measure the CSA of myofibers, and the CSA was quantified using ImageJ software [National Institutes of Health (NIH), Bethesda, MD].
For the EdU incorporation assay, cells were incubated for 3 hours with 10 μM EdU in GM at 37°C, 5% CO2 and then fixed in 4% paraformaldehyde in PBS. EdU detection was performed according to the manufacturer’s instructions using a Click-iT EdU Imaging Kit (Thermo Fisher Scientific). EdU-positive cell quantification was performed using CellInsight (Thermo Fisher Scientific). For TUNEL staining of apoptotic cells, satellite cells were fixed in 4% paraformaldehyde for 10 min and then subjected to the TUNEL reaction using MEBSTAIN Apoptosis TUNEL Kit Direct (MBL, Nagoya, Japan) following the instructions provided by the manufacturer.
Real-time PCR
Total RNA was isolated with Isogen II (Nippon Gene), and complementary DNA was synthesized with ReverTra Ace kit with genomic DNA remover (Toyobo). Real-time PCR was performed with the Thunderbird SYBR qPCR mix (Toyobo) and CFX96 Touch real-time PCR detection system (Bio-Rad). The expression levels of selected genes were quantified on the basis of the standard curve method, and the values were normalized with TATA box binding protein (TBP), GAPDH, or 18S ribosomal RNA (18S). Primer sequences are listed in table S1.
Immunoblotting
Protein lysates were obtained from muscle tissues or cultured cells with radioimmunoprecipitation assay buffer (Wako). Primary antibodies were diluted in CanGetSignal solution A (Toyobo) and incubated at 4°C overnight. Horseradish peroxidase (HRP)–conjugated secondary antibodies diluted in CanGetSignal solution B (Toyobo) were incubated at room temperature for 1 hour. HRP-conjugated secondary antibodies were visualized by chemiluminescence regents (Atto, Tokyo, Japan) with a digital luminescent image analyzer LAS-4000 (GE Healthcare, Tokyo, Japan). The intensities of protein bands were quantified using ImageJ software (NIH).
Statistical analysis
Statistical analysis was performed in Microsoft Excel or Prism 8 (GraphPad Software, La Jolla, CA). For statistical comparisons of two conditions, Student’s t test was used. For comparisons of more than two groups, the data were analyzed with one-way or two-way analysis of variance (ANOVA), according to the experimental design, followed by Sidak’s multiple comparison test. P values of *P < 0.05 and **P < 0.01 were considered statistically significant. Results are presented as means ± SEM. n.s. indicates results that were not statistically significant.
Acknowledgments
We thank all laboratory members for technical support. We thank D. Wellik, P. Zammit, S. Takeda, and S. Tajbakhsh for fruitful discussions and helpful comments. Funding: This work was supported by the Japan Agency for Medical Research and Development (AMED) under grant numbers 16bm0704010 and 19bm0704036. This work was also supported, in part, by AMED under grant number 18ek0109383, the Grants-in-Aid for Scientific Research KAKENHI (16J09948, 17H03608, 18H03193, 18K19749, 20J01490, 20K19537, and 20H00456) from the Japan Society for the Promotion of Science, the Takeda Science Foundation, Japanese Physical Therapy Association, the International Research Core for Stem Cell-based Developmental Medicine (Kumamoto University), the program of the Joint Usage/Research Center for Developmental Medicine (Institute of Molecular Embryology and Genetics, Kumamoto University), the Cooperative Research Project Program of the Medical Institute of Bioregulation (Kyushu University), and the Platform Project for Supporting Drug Discovery and Life Science Research [Basis for Supporting Innovative Drug Discovery and Life Science Research (BINDS)] from AMED under grant number JP20am0101103 (support number 1963). Author contributions: K.Y. designed and performed the experiments, analyzed the data, and wrote the manuscript. H.N., Y.Ki., D.S., J.N., Y.T., Y.Ka., F.M., H.A., and Y.Oh. performed the experiments and analyzed the data. N.O., A.Y., S.O., Y.S., K.C., K.I., I.A., Y.Og., and T.I. provided analytical tools and gave technical support. Y.On. conceived the project, designed and performed the experiments, analyzed the data, assembled the input data, and wrote the main manuscript. All authors read and approved the final manuscript. Competing interests: The authors declare that they have no competing interests. Data and materials availability: All data needed to evaluate the conclusions in the paper are present in the paper and/or the Supplementary Materials. Raw DNA methylation datasets are available from the GEO public depository under the accession number GSE154056. Raw microarray datasets are available from the GEO public depository under the accession number GSE139315. Raw RNA-seq datasets are available from the GEO public depository under the accession number GSE139639. Additional data related to this paper may be requested from the authors.
SUPPLEMENTARY MATERIALS
Supplementary material for this article is available at http://advances.sciencemag.org/cgi/content/full/7/24/eabd7924/DC1
REFERENCES AND NOTES
- 1.Sambasivan R., Kuratani S., Tajbakhsh S., An eye on the head: The development and evolution of craniofacial muscles. Development 138, 2401–2415 (2011). [DOI] [PubMed] [Google Scholar]
- 2.Mauro A., Satellite cell of skeletal muscle fibers. J. Biophys. Biochem. Cytol. 9, 493–495 (1961). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Morgan J. E., Zammit P. S., Direct effects of the pathogenic mutation on satellite cell function in muscular dystrophy. Exp. Cell Res. 316, 3100–3108 (2010). [DOI] [PubMed] [Google Scholar]
- 4.Randolph M. E., Pavlath G. K., A muscle stem cell for every muscle: Variability of satellite cell biology among different muscle groups. Front. Aging Neurosci. 7, 190 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Yin H., Price F., Rudnicki M. A., Satellite cells and the muscle stem cell niche. Physiol. Rev. 93, 23–67 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Ono Y., Boldrin L., Knopp P., Morgan J. E., Zammit P. S., Muscle satellite cells are a functionally heterogeneous population in both somite-derived and branchiomeric muscles. Dev. Biol. 337, 29–41 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Randolph M. E., Phillips B. L., Choo H.-J., Vest K. E., Vera Y., Pavlath G. K., Pharyngeal satellite cells undergo myogenesis under basal conditions and are required for pharyngeal muscle maintenance. Stem Cells 33, 3581–3595 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Emery A. E., The muscular dystrophies. BMJ 317, 991–995 (1998). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Harel I., Nathan E., Tirosh-Finkel L., Zigdon H., Guimarães-Camboa N., Evans S. M., Tzahor E., Distinct origins and genetic programs of head muscle satellite cells. Dev. Cell 16, 822–832 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Kelly R. G., Jerome-Majewska L. A., Papaioannou V. E., The del22q11.2 candidate gene Tbx1 regulates branchiomeric myogenesis. Hum. Mol. Genet. 13, 2829–2840 (2004). [DOI] [PubMed] [Google Scholar]
- 11.Tajbakhsh S., Rocancourt D., Cossu G., Buckingham M., Redefining the genetic hierarchies controlling skeletal myogenesis: Pax-3 and Myf-5 act upstream of MyoD. Cell 89, 127–138 (1997). [DOI] [PubMed] [Google Scholar]
- 12.Relaix F., Rocancourt D., Mansouri A., Buckingham M., A Pax3/Pax7-dependent population of skeletal muscle progenitor cells. Nature 435, 948–953 (2005). [DOI] [PubMed] [Google Scholar]
- 13.Shih H. P., Gross M. K., Kioussi C., Cranial muscle defects of Pitx2 mutants result from specification defects in the first branchial arch. Proc. Natl. Acad. Sci. U.S.A. 104, 5907–5912 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Lu J. R., Bassel-Duby R., Hawkins A., Chang P., Valdez R., Wu H., Gan L., Shelton J. M., Richardson J. A., Olson E. N., Control of facial muscle development by MyoR and capsulin. Science 298, 2378–2381 (2002). [DOI] [PubMed] [Google Scholar]
- 15.Sambasivan R., Gayraud-Morel B., Dumas G., Cimper C., Paisant S., Kelly R. G., Tajbakhsh S., Distinct regulatory cascades govern extraocular and pharyngeal arch muscle progenitor cell fates. Dev. Cell 16, 810–821 (2009). [DOI] [PubMed] [Google Scholar]
- 16.Ehrlich M., Lacey M., DNA methylation and differentiation: Silencing, upregulation and modulation of gene expression. Epigenomics 5, 553–568 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Rux D. R., Wellik D. M., Hox genes in the adult skeleton: Novel functions beyond embryonic development. Dev. Dyn. 246, 310–317 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Hatada I., Fukasawa M., Kimura M., Morita S., Yamada K., Yoshikawa T., Yamanaka S., Endo C., Sakurada A., Sato M., Kondo T., Horii A., Ushijima T., Sasaki H., Genome-wide profiling of promoter methylation in human. Oncogene 25, 3059–3064 (2006). [DOI] [PubMed] [Google Scholar]
- 19.Miura F., Enomoto Y., Dairiki R., Ito T., Amplification-free whole-genome bisulfite sequencing by post-bisulfite adaptor tagging. Nucleic Acids Res. 40, e136 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Miura F., Shibata Y., Miura M., Sangatsuda Y., Hisano O., Araki H., Ito T., Highly efficient single-stranded DNA ligation technique improves low-input whole-genome bisulfite sequencing by post-bisulfite adaptor tagging. Nucleic Acids Res. 47, e85 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Houghton L., Rosenthal N., Regulation of a muscle-specific transgene by persistent expression of Hox genes in postnatal murine limb muscle. Dev. Dyn. 216, 385–397 (1999). [DOI] [PubMed] [Google Scholar]
- 22.Machado L., Esteves de Lima J., Fabre O., Proux C., Legendre R., Szegedi A., Varet H., Ingerslev L. R., Barrès R., Relaix F., Mourikis P., In situ fixation redefines quiescence and early activation of skeletal muscle stem cells. Cell Rep. 21, 1982–1993 (2017). [DOI] [PubMed] [Google Scholar]
- 23.Schwörer S., Becker F., Feller C., Baig A. H., Köber U., Henze H., Kraus J. M., Xin B., Lechel A., Lipka D. B., Varghese C. S., Schmidt M., Rohs R., Aebersold R., Medina K. L., Kestler H. A., Neri F., von Maltzahn J., Tümpel S., Rudolph K. L., Epigenetic stress responses induce muscle stem-cell ageing by Hoxa9 developmental signals. Nature 540, 428–432 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Evano B., Gill D., Hernando-Herraez I., Comai G., Stubbs T. M., Commere P.-H., Reik W., Tajbakhsh S., Transcriptome and epigenome diversity and plasticity of muscle stem cells following transplantation. PLOS Genet. 16, e1009022 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kurscheid S., Bady P., Sciuscio D., Samarzija I., Shay T., Vassallo I., Criekinge W. V., Daniel R. T., van den Bent M. J., Marosi C., Weller M., Mason W. P., Domany E., Stupp R., Delorenzi M., Hegi M. E., Chromosome 7 gain and DNA hypermethylation at the HOXA10 locus are associated with expression of a stem cell related HOX-signature in glioblastoma. Genome Biol. 16, 16 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Su J., Huang Y.-H., Cui X., Wang X., Zhang X., Lei Y., Xu J., Lin X., Chen K., Lv J., Goodell M. A., Li W., Homeobox oncogene activation by pan-cancer DNA hypermethylation. Genome Biol. 19, 108 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Lescroart F., Hamou W., Francou A., Théveniau-Ruissy M., Kelly R. G., Buckingham M., Clonal analysis reveals a common origin between nonsomite-derived neck muscles and heart myocardium. Proc. Natl. Acad. Sci. U.S.A. 112, 1446–1451 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Kitajima Y., Ono Y., Visualization of PAX7 protein dynamics in muscle satellite cells in a YFP knock-in-mouse line. Skelet. Muscle 8, 26 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Rijli F. M., Matyas R., Pellegrini M., Dierich A., Gruss P., Dolle P., Chambon P., Cryptorchidism and homeotic transformations of spinal nerves and vertebrae in Hoxa-10 mutant mice. Proc. Natl. Acad. Sci. U.S.A. 92, 8185–8189 (1995). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Satokata I., Benson G., Maas R., Sexually dimorphic sterility phenotypes in Hoxa10-deficient mice. Nature 374, 460–463 (1995). [DOI] [PubMed] [Google Scholar]
- 31.Lepper C., Fan C. M., Inducible lineage tracing of Pax7-descendant cells reveals embryonic origin of adult satellite cells. Genesis 48, 424–436 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Ngo-Muller V., Bertrand A., Concordet J. P., Daegelen D., Mouse muscle identity: the position-dependent and fast fiber-specific expression of a transgene in limb muscles is methylation-independent and cell-autonomous. Dev. Dyn. 228, 594–605 (2003). [DOI] [PubMed] [Google Scholar]
- 33.Grieshammer U., Sassoon D., Rosenthal N., A transgene target for positional regulators marks early rostrocaudal specification of myogenic lineages. Cell 69, 79–93 (1992). [DOI] [PubMed] [Google Scholar]
- 34.Liu L., Cheung T. H., Charville G. W., Hurgo B. M. C., Leavitt T., Shih J., Brunet A., Rando T. A., Chromatin modifications as determinants of muscle stem cell quiescence and chronological aging. Cell Rep. 4, 189–204 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Hernando-Herraez I., Evano B., Stubbs T., Commere P.-H., Jan Bonder M., Clark S., Andrews S., Tajbakhsh S., Reik W., Ageing affects DNA methylation drift and transcriptional cell-to-cell variability in mouse muscle stem cells. Nat. Commun. 10, 4361 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Rezsohazy R., Saurin A. J., Maurel-Zaffran C., Graba Y., Cellular and molecular insights into Hox protein action. Development 142, 1212–1227 (2015). [DOI] [PubMed] [Google Scholar]
- 37.Rhoads K., Arderiu G., Charboneau A., Hansen S. L., Hoffman W., Boudreau N., A role for Hox A5 in regulating angiogenesis and vascular patterning. Lymphat. Res. Biol. 3, 240–252 (2005). [DOI] [PubMed] [Google Scholar]
- 38.Winnik S., Klinkert M., Kurz H., Zoeller C., Heinke J., Wu Y., Bode C., Patterson C., Moser M., HoxB5 induces endothelial sprouting in vitro and modifies intussusceptive angiogenesis in vivo involving angiopoietin-2. Cardiovasc. Res. 83, 558–565 (2009). [DOI] [PubMed] [Google Scholar]
- 39.Miguez A., Ducret S., di Meglio T., Parras C., Hmidan H., Haton C., Sekizar S., Mannioui A., Vidal M., Kerever A., Nyabi O., Haigh J., Zalc B., Rijli F. M., Thomas J. L., Opposing roles for Hoxa2 and Hoxb2 in hindbrain oligodendrocyte patterning. J. Neurosci. 32, 17172–17185 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Rubin E., Wu X., Zhu T., Cheung J. C.-Y., Chen H., Lorincz A., Pandita R. K., Sharma G. G., Ha H. C., Gasson J., Hanakahi L. A., Pandita T. K., Sukumar S., A role for the HOXB7 homeodomain protein in DNA repair. Cancer Res. 67, 1527–1535 (2007). [DOI] [PubMed] [Google Scholar]
- 41.Schild-Poulter C., Pope L., Giffin W., Kochan J. C., Ngsee J. K., Traykova-Andonova M., Haché R. J.-G., The binding of Ku antigen to homeodomain proteins promotes their phosphorylation by DNA-dependent protein kinase. J. Biol. Chem. 276, 16848–16856 (2001). [DOI] [PubMed] [Google Scholar]
- 42.Marchetti L., Comelli L., D’Innocenzo B., Puzzi L., Luin S., Arosio D., Calvello M., Mendoza-Maldonado R., Peverali F., Trovato F., Riva S., Biamonti G., Abdurashidova G., Beltram F., Falaschi A., Homeotic proteins participate in the function of human-DNA replication origins. Nucleic Acids Res. 38, 8105–8119 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Salsi V., Ferrari S., Ferraresi R., Cossarizza A., Grande A., Zappavigna V., HOXD13 binds DNA replication origins to promote origin licensing and is inhibited by geminin. Mol. Cell. Biol. 29, 5775–5788 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Yoshioka K., Kitajima Y., Seko D., Tsuchiya Y., Ono Y., The body region specificity in murine models of muscle regeneration and atrophy. Acta Physiol (Oxford) 231, e13553 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Bosnakovski D., Xu Z., Ji Gang E., Galindo C. L., Liu M., Simsek T., Garner H. R., Agha-Mohammadi S., Tassin A., Coppée F., Belayew A., Perlingeiro R. R., Kyba M., An isogenetic myoblast expression screen identifies DUX4-mediated FSHD-associated molecular pathologies. EMBO J. 27, 2766–2779 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Norton M., Verstegeden A., Maxwell L. C., McCarter R. M., Constancy of masseter muscle structure and function with age in F344 rats. Arch. Oral Biol. 46, 139–146 (2001). [DOI] [PubMed] [Google Scholar]
- 47.Philippou A., Minozzo F. C., Spinazzola J. M., Smith L. R., Lei H., Rassier D. E., Barton E. R., Masticatory muscles of mouse do not undergo atrophy in space. FASEB J. 29, 2769–2779 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Pavlath G. K., Thaloor D., Rando T. A., Cheong M., English A. W., Zheng B., Heterogeneity among muscle precursor cells in adult skeletal muscles with differing regenerative capacities. Dev. Dyn. 212, 495–508 (1998). [DOI] [PubMed] [Google Scholar]
- 49.Ono Y., Urata Y., Goto S., Nakagawa S., Humbert P. O., Li T. S., Zammit P. S., Muscle stem cell fate is controlled by the cell-polarity protein Scrib. Cell Rep. 10, 1135–1148 (2015). [DOI] [PubMed] [Google Scholar]
- 50.Seko D., Ogawa S., Li T. S., Taimura A., Ono Y., μ-Crystallin controls muscle function through thyroid hormone action. FASEB J. 30, 1733–1740 (2016). [DOI] [PubMed] [Google Scholar]
- 51.Yuan X., Tsujimoto K., Hashimoto K., Kawahori K., Hanzawa N., Hamaguchi M., Seki T., Nawa M., Ehara T., Kitamura Y., Hatada I., Konishi M., Itoh N., Nakagawa Y., Shimano H., Takai-Igarashi T., Kamei Y., Ogawa Y., Epigenetic modulation of Fgf21 in the perinatal mouse liver ameliorates diet-induced obesity in adulthood. Nat. Commun. 9, 636 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Jühling F., Kretzmer H., Bernhart S. H., Otto C., Stadler P. F., Hoffmann S., metilene: Fast and sensitive calling of differentially methylated regions from bisulfite sequencing data. Genome Res. 26, 256–262 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Yokoyama T., Miura F., Araki H., Okamura K., Ito T., Changepoint detection in base-resolution methylome data reveals a robust signature of methylated domain landscape. BMC Genomics 16, 594 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Harada A., Maehara K., Ono Y., Taguchi H., Yoshioka K., Kitajima Y., Xie Y., Sato Y., Iwasaki T., Nogami J., Okada S., Komatsu T., Semba Y., Takemoto T., Kimura H., Kurumizaka H., Ohkawa Y., Histone H3.3 sub-variant H3mm7 is required for normal skeletal muscle regeneration. Nat. Commun. 9, 1400 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Kitajima Y., Ogawa S., Ono Y., Visualizing the functional heterogeneity of muscle stem cells. Methods Mol. Biol. 1516, 183–193 (2016). [DOI] [PubMed] [Google Scholar]
- 56.Yoshioka K., Kitajima Y., Okazaki N., Chiba K., Yonekura A., Ono Y., A modified pre-plating method for high-yield and high-purity muscle stem cell isolation from human/mouse skeletal muscle tissues. Front. Cell Dev. Biol. 8, 793 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary material for this article is available at http://advances.sciencemag.org/cgi/content/full/7/24/eabd7924/DC1


