STED microscopy uncovers a delicate balance of stability and volatility of stable spines in mouse cortex over 1 month.
Abstract
Excitatory synapses on dendritic spines of pyramidal neurons are considered a central memory locus. To foster both continuous adaption and the storage of long-term information, spines need to be plastic and stable at the same time. Here, we advanced in vivo STED nanoscopy to superresolve distinct features of spines (head size and neck length/width) in mouse neocortex for up to 1 month. While LTP-dependent changes predict highly correlated modifications of spine geometry, we find both, uncorrelated and correlated dynamics, indicating multiple independent drivers of spine remodeling. The magnitude of this remodeling suggests substantial fluctuations in synaptic strength. Despite this high degree of volatility, all spine features exhibit persistent components that are maintained over long periods of time. Furthermore, chronic nanoscopy uncovers structural alterations in the cortex of a mouse model of neurodegeneration. Thus, at the nanoscale, stable dendritic spines exhibit a delicate balance of stability and volatility.
INTRODUCTION
Synapses on spines of principal neurons are a major locus of memory formation and maintenance in cortical circuits (1–4). To serve this function, spine synapses, as nanophysiological information processing devises (5), must be dynamic to change during learning and experience and simultaneously exhibit features of long-term persistence to maintain memory traces. The structural and molecular basis of this dualism at the level of the individual spine, however, remains incompletely understood. Spine dynamics are classically studied using in vivo two-photon imaging, but due to limitations in optical resolution, they are mainly treated as binary entities (2, 5) that are either present or not present. As such, it is, for instance, known that consistent with the synaptic trace theory of memory formation, which posits that activity-induced changes on synapses underlie long-lasting information storage, cortical engagement in learning tasks is often accompanied by a transient peak in spine generation, followed by the selective stabilization of newly formed spines (6, 7). In addition to this plain spine turnover, the size of cortical spines can also vary in response to learning-induced and/or spontaneous processes (3, 8, 9). These alterations can affect all features of postsynaptic organization from receptor complement, postsynaptic scaffold (10) to the actin cytoskeleton, maintaining the spine’s morphology (11–13). Inducing synaptic long-term potentiation (LTP) in vitro, for instance, simultaneously leads to remodeling of the postsynaptic density (PSD) and to changes in spine morphology, including the concerted expansion in head size and neck width and shortening of the neck length (14–16). Distinct from such activity-dependent changes in spine size, there is evidence arguing for “intrinsic” spontaneous morphological volatility, independent of neuronal impulse activity and synaptic transmission, endowing the system with a large level of flexibility (8–10).
The contribution of both processes to structural remodeling under baseline conditions in vivo, however, remains poorly understood to date. If activity-dependent mechanisms underlie the bulk of ongoing in vivo spine remodeling, one would expect that ongoing changes in spine head size, neck length, and neck width are effectively controlled by a single underlying master process and therefore are tightly correlated, such as observed for LTP (16). Such concerted changes would optimally orchestrate the contributions of spine geometry changes to synaptic potentiation because synaptic strength is predicted to substantially increase by shortening and widening of the spine neck (17). Remodeling of the spine as well as of the PSD is dependent on the actin cytoskeleton and is driven by postsynaptic Ca2+ influx (13). As the actin cytoskeleton is composed of several pools of F-actin, all of which undergo continuous assembly and disassembly, not only the spine head but also the entire spine morphology can be expected to exhibit spontaneous intrinsic fluctuations. However, while actin dynamics are well studied in the spine head, little is known about its dynamics in the spine neck. Depending on whether and how alterations of head and neck geometry are coordinated, such changes in total spine geometry may either enhance or suppress fluctuations of synaptic strength. In the past, studies on synaptic plasticity exclusively used the spine total fluorescence as a proxy of head volume and thus synaptic strength, owing to limitations of optical resolution (18–21). The advent of superresolution now enables the assessment of their dynamics and the remodeling of their components at unprecedented resolution (22–24). Imaging in tissue imposes challenges to all superresolution light microscopy techniques, and the first approach to overcome these in the mouse brain was stimulated emission depletion (STED) microscopy (22). So far, however, time periods relevant to long-term memory storage have remained challenging. Thus, the questions of whether activity-driven or spontaneous remodeling is dominant in vivo, of whether there are one or many drivers of spine geometry remodeling, and whether such drivers are independent or controlled by a single master process remain unanswered to date.
To address these questions, we developed an approach for long-term STED imaging in the cortex, which enabled us to monitor individual spines at superresolution for up to 1 month in the cortex of living mice, an order of magnitude longer than previous studies (23). Our data demonstrate that stable spines undergo strong morphological fluctuations in all their geometrical features comparable in magnitude to activity-dependent processes, even under baseline conditions. We show that the distribution of spine head size and neck width is approximately log-normal, indicating multiplicative dynamics, while neck length does not. We found that the dynamics of some spine geometry features, such as neck length and head size, is uncorrelated, while changes in head size and neck width are correlated. All spine features also exhibit persistent components that are maintained over long periods of time. Furthermore, spines in a mouse model of neurodegeneration [amyotrophic lateral sclerosis (ALS)] display a pronounced reduction in density, paralleled by an increase in head size of the remaining stable spines, thereby demonstrating alterations at the level of individual spine morphological features.
RESULTS
Chronic window implant for in vivo STED microscopy
To achieve nanoscale resolution to assess structural correlates of synapses over extended periods of time in vivo, we built a custom-designed STED microscope (Fig. 1A). Our microscope consists of an upright stand to which we attach a blue excitation (one-photon) laser to excite enhanced green fluorescent protein (EGFP) and an orange laser for STED. A vortex phase plate in the STED laser beam creates a doughnut-shaped focal intensity pattern for superresolution in the xy plane. Epifluorescence is detected via a confocal pinhole and single-photon detector. The achieved superresolution during chronic in vivo imaging was around 96 nm (fig. S1), which is 5 to 10 times higher than that of a conventional two-photon microscope. In vivo STED requires a mechanically stable and thermally isolated microscope to avoid thermal drift. To this end, we designed a mounting plate with a large heat sink (25). The cranial window needs to be of highest quality because small optical aberrations can massively deteriorate image quality. The craniotomy needs to be as atraumatic as possible. We thus optimized the cranial window preparation (see Methods) to achieve a minimal distance between the coverslip and brain surface under chronic conditions. This aspect is crucial to minimize optical aberrations on the one hand and to avoid motion artifacts, associated with, e.g., brain vessel pulsation, on the other hand. To affix the mouse’s head underneath the objective, we designed a dedicated head bar (fig. S1A), which can be cemented flush to the skull to allow access to a high–numerical aperture (NA; 1.3), short–working distance objective.
Fig. 1. Repetitive superresolution of the mouse motor cortex using STED microscopy.
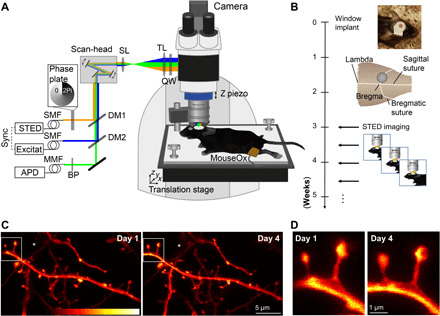
(A) Microscope design: A custom-made STED microscope is attached to a microscope stand. The pulsed 483-nm excitation is temporally synchronized electronically with the 595-nm STED light pulses and merged spatially by dichroic mirrors (DM). After passing two galvanic mirrors in the scan-head, the light is imaged by a scan (SL) and tube lens (TL) before being focused by a glycerol immersion objective (numerical aperture, 1.3) with a correction collar. The mouse is mounted via a head bar on an adjustable heating plate. Vital functions are controlled by a pulse oximeter (MouseOx). (B) After window implantation and a 3-week recovery period, the mouse was imaged twice a week. (C) Representative raw data example of an apical dendrite of a pyramidal neuron in the motor cortex of a Thy1-GFP-M mouse imaged at day 1 (left) and day 4 (right). An axon captured in the same field of view is marked by (*). (D) Magnification of marked region in (C). Images are maximum-intensity projections of six frames. APD, avalanche photodiode detector; BP, band-pass filter; MMF, multimode fiber; QW, quarter wave plate; SMF, single-mode fiber. Colorbar, 0 to 212 photon counts. Photo credit: Waja Wegner, University Medical Center Göttingen.
Longitudinal in vivo superresolution STED microscopy
In vivo STED imaging commenced after a recovery period of 3 to 4 weeks (Fig. 1B). The same fields of view were revisited twice a week. Mice were anesthetized using a combination of midazolam, metedomidin, and fentanyl. Through a tiltable mounting plate, we accurately aligned the cranial window perpendicular to the optical axis of the microscope using a laser-based alignment tool (25). Upon optimization of the window implantation procedure, we achieved a high success rate of imageable windows, allowing for chronic imaging (fig. S1B). STED microscopy in the motor cortex of a Thy1-GFP-M transgenic (tg) mouse (GFP-M line) (26) yielded crisp images depicting dendritic spine morphology in cortical layer 1 at nanoscale resolution (Fig. 1, C and D). STED image resolution critically hinges on the spectroscopic properties of the fluorescent molecule and on the STED focal doughnut. To correct for spherical aberrations, we adapted the correction collar of the objective at each field of view. To determine the resolution most accurately, we measured the full width at half maximum (FWHM) within the in vivo images. The smallest/thinnest structures were axons (such as “*” in Fig. 1C) and spine necks, which could be resolved at a FWHM of 96 nm (fig. S1C), a conservative estimate of the resolution. With these settings, we recorded STED microscopy images over a period of up to 28 days (Fig. 2), which enabled us to monitor fine changes in spine morphology, such as the spine neck [Fig. 2, A (insets) to C] that are typically obscured when using, e.g., two-photon microscopy. In addition, our imaging approach also enabled us to detect individual dendritic spines of all sizes with high precision and to observe morphological phenomena such as clustered spine formation (fig. S2A) or “touching heads” (fig. S2B). To estimate the noise in our measurements contributed by incomplete realignment, photon or detector noise, or manual selection of the area of interest, we repeatedly scanned dendrites in fixed brain sections obtained from the same GFP-M mouse line, also used for in vivo experiments. Between individual scans, we removed the slide and realigned it again before the subsequent scan (fig. S3). The procedure revealed an almost perfect correlation between the obtained data (r = 0.99), and we thus conclude that measurement noise related to the technique and alignment is minimal.
Fig. 2. Chronic STED imaging of dendritic stretches in layer 1 of the motor cortex.
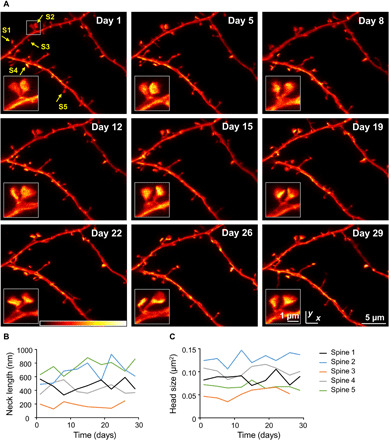
(A) Superresolution reveals changes of spine nanoplasticity of large, mushroom-type, stable spines (inset). Images are maximum-intensity projections of raw data. Colorbar, 0 to 120 photon counts. (B and C) Trajectories of neck length (B) and head size (C) of spines marked in (A).
Distribution and interdependency of spine parameters
All dendritic spines, emanating laterally from the dendrite within the same or a consecutive focal plane to the dendrite, were analyzed across all time points (TPs) (Fig. 3A). Spines present at all imaging TPs were considered “stable” and included in the analysis (see Methods and table S1). The size of the spine head cross section was measured by encircling the head area (Fig. 3A) in its focal plane within a z stack. Neck length was defined as the distance from the base of the spine neck to the beginning of the spine head (Fig. 3A). The neck width represents the thinnest extent of the spine neck (Fig. 3A). We observed a large range of head sizes [median: 0.41 μm2; interquartile range (IR): 0.29 to 0.57 μm2] and neck lengths (median: 812 nm; IR: 503 to1154 nm), spanning in total an order of magnitude (fig. S4, A and B), while the neck width (median: 238 nm; IR: 208 to 271 nm) was less variable (fig. S4C). All three parameters are positively skewed (fig. S4, A to C). By plotting the log10 values of these parameters, we observed a log-normal distribution for head size (Fig. 3B) and neck width (Fig. 3D), but not for neck length (Fig. 3C).
Fig. 3. Spine morphometric parameters are largely uncorrelated, while head size and neck width, but not neck length, exhibit multiplicative dynamics.
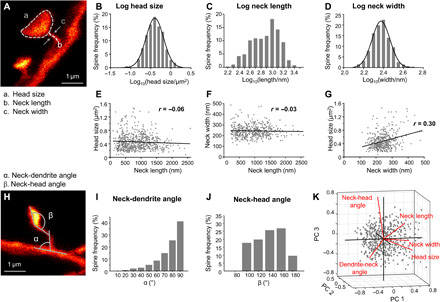
(A) Features assessed of stable spines are head size, spine neck length, and neck width. (B to D) Histogram of the logarithmic spine head sizes (B), spine neck lengths (C), and neck width (D). (B and D) The log10 data are normally distributed indicated by a Gaussian fit (black line). (E to G) Correlation between spine parameters. Spine head size (E) and neck width (F) as a function of neck length, and head size as function of neck width (G); linear regression (black) and Pearson’s correlation coefficient r. (H to J) The angle between the dendrite and the spine neck (α, neck-dendrite angle) (I) and between the neck and the spine head (β, neck-head angle) (J) is measured. (K) Principal components (PC) analysis of five morphological parameters. (B to G and I to K) All data of all time points pooled. Number of analyzed spines are listed in table S1.
To assess whether the morphological parameters were interdependent, we investigated their correlation. The parameter neck length did neither correlate with head size (r = −0.06, P = 0.14; Fig. 3E) nor with neck width (r = −0.03, P = 0.52; Fig. 3F), meaning that, e.g., large spine heads can have either short or long necks and that the width of a spine neck is independent of its length. A weak, but highly significant, positive correlation, however, was observed for the parameters head size and neck width (r = 0.30, P < 0.0001; Fig. 3G). We then measured the angle between the spine neck and dendrite (neck-dendrite angle) as well as the angle between the base of the spine head and the neck (neck-head angle; Fig. 3, H to J). The majority of spines emanated from the dendrite at an angle of ~90°. A principal components analysis (PCA) of the five morphological parameters (order: head size, neck width, neck length, neck-dendrite angle, and neck-head angle; Fig. 3K) of all spines was performed on normalized (z scored) data and revealed an almost even distribution of the contributions of individual components (variance explained reflecting the contribution of each PC to the total variance: PC1 29%, PC2 21.4%, PC3 19.4%, PC4 17.3%, and PC5 13%). The first component is mainly driven by head size and neck width and length (coefficients of PC1: 0.63, 0.61, 0.43, −0.16, and 0.1), while the second component is strongly determined by neck-dendrite and neck-head angles (coefficients of PC2: 0.05, 0.28, −0.34, 0.69, and 0.58).
In addition to spines bearing a defined spine head, we could also superresolve filopodia [long and thin protrusions (min. length, 750 nm) lacking an obvious head], which we analyzed separately (fig. S4, D to H). Filopodia were on average 2301 nm (±762 nm SD) long (fig. S3E), with a neck width of 236 nm (±49 nm SD; fig. S3F) and emanated from the dendrite at a median angle of 75.8° [64.9° to 79°, 95% confidence interval (CI)] (fig. S4G). We did not find a correlation between the width and length of filopodia (fig. S4H). All filopodia in our dataset occurred only once and thus had a lifetime of less than 3 days (likely rather minutes to hours).
Temporal changes of stable spine morphology
We next asked how morphological parameters change over time. To this end, we followed spines being present at all TPs over time and assessed the changes in head size, neck length, and neck width (Fig. 4, A to L). On average, all measures remained constant across all TPs, indicating the absence of phototoxic effects (neck length P = 0.88, head size P = 0.25, neck width P = 0.10; Kruskal-Wallis and Dunn’s multiple comparison test; Fig. 4B). The majority of spines underwent relative changes exceeding ±10% of the initial spine head size, neck length, or neck width over a period of 3 to 4 days (Fig. 4C). More specifically, only 26% of spines displayed a minor head size change within ±10%, while 45% of spines decreased in head size more than 10%, and 29% increased in head size exceeding 10%. Twenty-five percent of spines underwent a neck length variation within ±10%, while 39% of spines decreased in neck length and 36.3% of spines increased in neck length exceeding 10% (Fig. 4C). Spine neck widths were similarly dynamic, with 31% varying within ±10% size change, while 36% of spines underwent a thinning of the neck widths that exceeded 10%, and 32% displayed a thickening of the neck exceeding 10% size change (Fig. 4C). On average, spines grew in head size by 24% (median; IR: 9 to 46%; fig. S5A) and neck length by 22% (median; IR: 9 to 45%; fig. S5B), up to a maximum of >200%. The median shrinkage of head size was −22% (IR: −10 to −35%; fig. S5A), and that of neck length was −23% (IR: −11 to −34%; fig. S5B). Next, we asked whether size fluctuations would hinge on initial size. Therefore, we plotted head size (Fig. 4D), neck length (Fig. 4E), and neck width (fig. S5C) after lag times of 3.5, 7, 10.5, and 14 days over their initial size. The slope of the linear regression was below 1 for all three parameters with a positive y intercept, indicating a tendency for shrinkage of large spines and growth of small spines. This finding is further substantiated by plotting the size changes occurring within 3 to 4 days as a function of initial size, which yields a negative linear regression (fig. S5, D to F). Moreover, the magnitude of change in head size and neck length seems to hinge on initial size, as indicated by the dashed lines (fig. S5, D and E), arguing for multiplicative dynamics. We then computed the Pearson’s correlation coefficient of the spine parameters for the different lag times (Fig. 4, D and E, and fig. S5C) and plotted those r values as a function of lag time for head size (Fig. 4F), neck length (Fig. 4G), and neck width (Fig. 4H). These data illustrate the steep decline in correlation occurring between the first two TPs, and a shallower regression over the subsequent TPs, with a large offset for head size and neck length and low offset for the neck width. The presence of such an offset in our measurement (Fig. 4F) indicates that large heads mainly remain large and small heads remain small over the imaging period of 14 days and similarly so for neck length (Fig. 4G). Therefore, the main fluctuations in size occur at time scales of 3 to 4 days or shorter. Much larger fluctuations were observed for neck widths (Fig. 4H).
Fig. 4. In stable spines, head size and neck length fluctuate independently and are more persistent than fluctuations in neck width, while changes in head size and neck width do correlate.
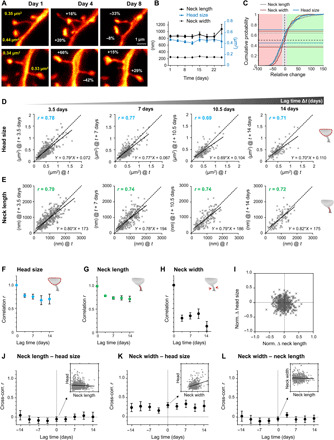
(A) Representative examples of changes in spine head size. (B) The morphological parameters are stable over the observation period of 24 days. Data are means ± SEM. (C) Cumulative distribution of relative changes of neck length (gray), neck width (red), and spine head size (blue) over 3 to 4 days. Fraction of spines that changed spine head size within ±10% (gray area) is indicated by blue horizontal lines, while the same for neck length is indicated by gray and for neck width by red horizontal lines (light red area denotes relative change in size to lower than −10%, while changes exceeding 10% are indicated by the green area). (D and E) Head size (D) and neck length (E) after four different time intervals Δt plotted against their size at time t. Straight line is a linear regression, r is the Pearson’s correlation coefficient, and dashed is the line of unity. (F to H) Pearson’s correlation coefficient plotted over lag time Δt for head size (F), neck length (G), and neck width (H). (I) Normalized changes of head size and neck length are not correlated. (J to L) Pearson’s cross-correlation between different spine parameter for different lag times. Neck length to head size (J) and neck length to neck width (L) are uncorrelated, while neck width to head size (K) shows a significant correlation for up to 14 days. Inset scatter plot and linear regression at lag time Δt = 0 for illustration. (F to H and J to L). Error bars are SD of bootstrapped data. (B to L) All data of the same time interval were pooled. Numbers of analyzed spines are listed in table S1.
We next wondered whether fluctuations in certain parameters occurred in a concerted fashion. We thus plotted the normalized relative changes over 3 to 4 days of head size as a function of neck length alterations, which was uncorrelated (Fig. 4I). Please note that the normalization restricts the changes to a range of +1 and −1; the distribution is therefore symmetric. To assess whether fluctuations in spine geometry parameters correlate between the different TPs, we calculated the Pearson’s correlation coefficient between different pairs of parameters, i.e., the cross-correlation, for lag times of 3.5, 7, 10.5, and 14 days. The cross-correlation was close to zero between head size and neck length (Fig. 4J) as well as between neck length and width (Fig. 4L). However, a significant correlation (min: r = 0.16, max: r = 0.33; Fig. 4K) for time intervals up to 14 days was observed between head size and neck width (Fig. 4K).
Transient spines have smaller heads than stable spines
Next, we asked whether spine morphology and its fluctuations are related to the spine’s fate or its previous history. We analyzed all spines over three consecutive TPs and categorized them into stable, gained, and lost spines based on their lifetime (Fig. 5A). Corroborating earlier publications using two-photon imaging (5, 27–29), stable spines (present on all three imaging TPs) had an almost threefold larger head (head size measured at the first TP) compared to spines just gained (gained) or spines measured at the TP before their loss (lost) [median and 95% CI: head size stable spines, 0.40 (0.35 to 0.48) μm2; gained, 0.16 (0.12 to 0.19) μm2; and lost spines, 0.18 (0.14 to 0.21) μm2 (stable versus gained, P < 0.0001; stable versus lost, P < 0.0001; Kruskal-Wallis test with Dunn’s multiple comparison test; Fig. 5B]. Similarly, stable spines had a longer neck [median and 95% CI: neck length stable spines, 737 (642 to 945) nm; gained, 542 (437 to 573) nm; and lost spines, 509 (437 to 576) nm (stable versus gained, P = 0.005; stable versus lost, P = 0.001; Kruskal-Wallis test with Dunn’s multiple comparisons test; Fig. 5C] and a larger neck width [median and 95% CI: neck width stable spines, 247 (235 to 259) nm; gained, 209 (195 to 232) nm; and lost spines, 226 (200 to 240) nm] compared with gained and lost spines (stable versus gained, P = 0.0005; stable versus lost, P = 0.023; Kruskal-Wallis test with Dunn’s multiple comparison test; Fig. 5D).
Fig. 5. Spine parameters differ between transient and stable spines.

(A) Spine categories based on lifetime (open circle, spine not present; filled circle, spine present). (B to D) Gained and lost spines show significantly smaller head sizes (B), smaller neck length (C), and thinner neck width (D). *P < 0.05; **P < 0.01; ***P < 0.001; ****P < 0.0001. Data are median ± 95% CI. Numbers of analyzed spines are listed in table S1.
Probing ultrastructural morphological abnormalities of dendritic spines in a transgenic mouse model of ALS
Last, we applied chronic STED microscopy to investigate morphological alterations of dendritic spines in a disease model. To this end, we investigated spines on layer V pyramidal neurons in the motor cortex of a tg mouse model of ALS, a fatal motor neuron disease characterized by the degeneration of upper and lower motor neurons in the motor cortex and spinal cord, respectively (30, 31). We used a well-characterized mouse model of the disease that is based on the overexpression of the mutated superoxide dismutase 1 gene [SOD1G93A; hereafter called SOD (32)]. These mice recapitulate key phenotypic features of ALS and die prematurely due to paralysis. Earlier work indicates that upper motor neurons, which reside in cortical layer V, are also affected in the mouse model, but actual insight into ultrastructural abnormalities and the dynamics of those changes in vivo is lacking to date (33–35). We crossed SOD1G93A with GFP-M mice and examined both the SOD1G93A expressing mice as well as their non-SOD tg littermates (called control “CTR” hereafter). First, we assessed the spine density on apical tufts of layer V pyramidal neurons over three consecutive TPs (Fig. 6A) and found a significant decrease in spine density in SOD tg mice [effect of group: F1,56 = 14.34, P = 0.0007; effect of time: F2,56 = 2, P = 0.15; group-by-time interaction effect: F2,56 = 1.39, P = 0.26, two-way repeated-measures analysis of variance (ANOVA); Fig. 6B]. Overall, spine density remained stable within the 8-day imaging period. A detailed morphological assessment of stable spines (those being present at all imaging TPs) revealed that the distribution of the parameters head size, neck length, and neck width in SOD mice was like those found in CTR mice (fig. S5, A to F). We observed a log-normal distribution only for head size and neck width, while the neck length was not log-normally distributed (fig. S6, D to F). On average, spine head size was increased [median and 95% CI: CTR, 0.41 (0.39 to 0.43) μm2; SOD, 0.44 (0.42 to 0.47) μm2; P = 0.035, Mann-Whitney test; Fig. 6, C and F]. The neck length, on the other hand, did not differ significantly between genotypes [median and 95% CI: CTR, 812 (762 to 886) nm; SOD, 822 (766 to 874) nm; P = 0.78, Mann-Whitney test; Fig. 6D]. The neck width showed a small, but significant, decrease in average size for SOD mice [median and 95% CI: CTR, 238 (232 to 245) nm; SOD, 229 (224 to 235) nm; P = 0.037, Mann-Whitney test; Fig. 6E). We then probed the stability of these parameters over time and assessed whether fluctuations would correlate. To this end, we computed the cross-correlation between neck length, head size, and neck width (fig. S6, G to I) and the autocorrelations (fig. S6, J to L) of these parameters for different lag times. For all parameters, we found comparable correlations between CTR and SOD mice. A large offset for the autocorrelation of head size and neck length (fig. S6, J and K) indicates a high degree of stability like in CTR mice. A cross-correlation of ~20 to 40% for neck width and head size supports correlated changes between these parameters, which we also observed for CTR mice. To test whether the increase in head size would at least partially compensate for the loss of spines in SOD mice, we computed the total synaptic area per dendritic unit and observed a partial compensation, such that the difference in synaptic area was not statistically significant between SOD and CTR mice (effect of group: F1,56 = 1.58, P = 0.22; effect of time: F2,56 = 0.77, P = 0.47; group-by-time interaction effect: F2,56 = 0.21, P = 0.81, two-way repeated-measures ANOVA; fig. S6M).
Fig. 6. Nanoscopy in SOD transgenic mice.
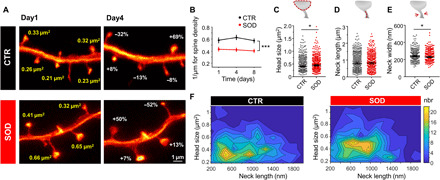
(A) Representative example of a GFP-expressing dendrite in a CTR and a SOD transgenic mouse showing changes in spine head size within 3 days (images are maximum intensity projections). (B) CTR mice show higher spine density than SOD mice. (C to F) Changes in spine parameter. While the head size (C) of stable spines was increased in SOD mice, no difference was observed in neck length (D), but neck width (E) was decreased in SOD mice. (F) Contour plot of head size and neck length shows a shift to larger head sizes for SOD mice (nbr, number). *P < 0.05. Data in (B) are means ± SEM and in (C to E) are median ± 95% CI. Numbers of analyzed spines are listed in table S1. Data were pooled.
DISCUSSION
Establishing chronic in vivo STED nanoscopy to superresolve dendritic spines in mouse neocortex for up to 1 month enabled us to provide the first characterization of fluctuations in spine geometry at nanoscale resolution over unprecedented periods of time in vivo. We found that all assessed geometric features exhibit fluctuations of substantial magnitude even on stable spines, which are believed to represent persistent, mature synapses (5, 27, 36, 37). Changes in spine geometry occur mainly within 3 to 4 days and are of a magnitude similar to alterations caused by LTP induction, indicative of substantial modifications in synaptic strength. Over longer periods up to 14 days, head sizes and neck lengths were very stable, which is consistent with the assumption that geometric features fluctuate around mean values that are spine specific and maintained over periods on the order of months at least (5, 29, 36). The maintained components of spine head size and spine neck width were matched such that larger spine heads are systematically associated with wider spine necks, despite substantial ongoing fluctuations. For neck length and head size, temporal fluctuations around their respective mean values were statistically independent of each other, indicating the existence of multiple independent drivers of geometric remodeling. Together, these findings provide a picture of in vivo spine dynamics exhibiting a delicate balance of stability and volatility at the nanoscale level.
Longitudinal STED nanoscopy to characterize dendritic spine nanostructure
Key to the establishment of chronic in vivo STED imaging is a stable cranial window preparation, with minimal amounts of fluid in between the coverslip and the cortical surface as well as minimal regrowth of adhesive tissue/bone or dural thickening, which we achieved by an atraumatic craniotomy, the right size of the coverslip, resin cement, and the design of a novel head bar, allowing access to a high-NA objective. This optimized protocol enabled us to successfully scan 100% of the mice implanted for this study, chronically over at least 3 weeks, thus recording two to eight TPs.
Our in vivo STED data are well in line with electron microscopy (EM) data obtained in fixed tissue. For instance, the average neck diameter in our data was 238 nm, and the median head size was 0.41 μm2, corresponding to a spine head volume of 0.20 μm3 (assuming a spherical volume), consistent with EM data (38). However, in sharp contrast to EM, our STED approach allows for a longitudinal assessment of synaptic structures in vivo. The log-normal distributions we observe for head size and neck width are well explained by mathematical models (37, 39). The right-skewed but not log-normally distributed neck length, however, does not fit to these models.
Stability and volatility of spine geometry of persistent spines
We here analyzed temporal changes of dendritic spine geometry chronically in cortex of living mice for up to 1 month. In contrast to most longitudinal in vivo two-photon studies, which assess structural plasticity by considering spines as binary entities, our nanoscopy approach enabled us to investigate dedicated spine parameters at nanoscale resolution. It is well accepted that the size of a dendritic spine scales with the strength of the synapse and is predictive of its lifetime (5, 28, 36, 40). However, recent evidence also argues for a critical impact of other parameters, such as neck length or width in determining the function of a synapse (14, 16, 17). As such, earlier studies demonstrated that the spine neck most critically determines dendritic spine compartmentalization (16) and synaptic strength (21) and that the neck length shortens in response to LTP induction (14, 15). Our dataset, acquired in vivo, under baseline conditions (that is without the application of a dedicated learning/memory paradigm), shows that both heads and necks of spines fluctuate substantially within a few days. We particularly focused our investigation on stable spines, which are believed to embody structural correlates of learning and memory (2, 7). The majority of those (~80%) underwent a fluctuation in head size and neck length of more than 10% (~40% even of more than 30%) within 3 to 4 days. While these alterations might seem small at first glance, they could readily affect the strength of the corresponding synapse. For comparison, synaptic potentiation triggered by chemical LTP stimulation or glutamate uncaging has been shown to cause an increase in spine head size between 20 and 40% in a large fraction of spines (41–44), as well as a decrease in neck length by ~20 to 30% (14, 16) and an increase in neck width by 30% (16). The interpretation of those data, however, is complicated by the fact that these studies are conducted in vitro and rely on artificial and probably highly potent stimulation protocols. Our data demonstrate that similar effect sizes occur in vivo, already under baseline conditions, even in the absence of a dedicated stimulation protocol and could well be explained by mathematical models of random cooperative assembly/disassembly, emphasizing the stochastic nature of synaptic protein remodeling and turnover of postsynaptic macromolecular complexes (45, 46).
We observed such large changes within 3 to 4 days but also witnessed a large offset of ~70% in the autocorrelation of head size and spine length over 14 days, indicating overall size stability. This suggests that spine geometry might be volatile within rather short time frames of days but is able to maintain a mean value over larger periods of time. Somewhat surprisingly, we found that the geometric parameters as well as their dynamics were largely independent of each other. A small, yet persistent, positive correlation was only observed between neck width and head size, which could indicate activity-dependent remodeling. The independent nature of these dynamics supports the notion that the F-actin network of dendritic spines (13) is regulated differently between spine heads and necks and thus rejects the predominant idea that the spine cytoskeleton is a passive component downstream to and driven by the primary processes of synaptic plasticity (13).
While most of the time-lapse studies to date address changes in spine volume or spine head size, little is known about spine neck changes in vitro and in vivo. We observed that absolute changes of spine head size and neck length increased with their initial size and that changes were negatively correlated, indicating that small spines tend to increase, whereas large spines tend to shrink. This relationship supports statistic models such as based on a Kesten process (39) or pure multiplicative dynamics (37).
Transient spines differ morphologically from persistent spines
Spine morphology is indicative of synaptic strength and also of its lifetime, but insight into the actual dimensions/magnitude is still lacking. We found a notable difference in head size, with persistent spines having an almost three times larger head compared with transient spines. They also possessed a 35% longer and 25% thicker neck compared with transient spines. These data thus do not only corroborate earlier work conducted with two-photon imaging (5, 28, 29, 40), but now much more accurately determine the actual size difference of multiple spine parameters. We also occasionally observed clustered spine formation, a feature reported in two-photon studies in the past, which has been suggested to cause nonlinear summation of synaptic inputs and to increase computing power (1, 47, 48).
The most extreme case of thin spines are filopodia, which lack an actual spine head. Because of their lean shape, they are difficult to characterize in vivo with two-photon imaging. In our data, these structures are on average 2.3 μm long and have a neck width of 236 nm. All identified filopodia in our dataset only occurred once and thus have a lifetime shorter than 3 to 4 days [see also (36)]. While we cannot fully exclude that newly formed spines initially underwent a stage reminiscent of a filopodium, our current data argue against a major role of filopodia acting as precursors of dendritic spines in adult mice under baseline conditions. Most newly formed spines were much shorter than filopodia and stable, mature spines. Future studies are thus needed to explore the relevance of those immature structures.
Nanoscale alterations of dendritic spines in SOD transgenic mice
We also performed nanoscopy in a transgenic mouse model of ALS, a neurodegenerative disease that is characterized by the loss of upper and lower motor neurons (30, 31). Before frank neuronal degeneration and loss, motor neurons are likely already impaired over a prolonged period of time in this disease. However, little is known to date about how neurodegenerative processes in ALS affect dendritic spines. We observed a pronounced decrease in spine density on apical tufts of layer V cortical neurons in SOD mice (49), which remained stable throughout the imaging period. Our data corroborate earlier studies conducted in a more aggressive mouse model [higher copy number (33, 34)] using Golgi Cox staining. The remaining stable spines in SOD mice had on average larger spine heads and thinner necks compared with CTR mice. The head size increase we observed could well represent homeostatic scaling processes to counteract the loss of synaptic input (50, 51), as a recent study demonstrated an increase in synaptic size and a broader distribution of the same in silenced neuronal networks (52). This notion is corroborated by our finding that the synaptic area per dendrite is not significantly different between SOD and CTR mice. Collectively, these results argue for structural modifications of upper motor neurons, which precede overt neuronal loss and symptom onset and substantiate the notion of ALS being a synaptopathy (53).
Together, we here established long-term in vivo STED microscopy in the cortex of mice. Our data demonstrate that hitherto believed stable dendritic spines undergo pronounced morphological changes. Individual morphological features are largely independent, suggesting diverse drivers of synaptic plasticity.
METHODS
Animals
All mouse experiments were performed according to the guidelines of the national law (Tierschutzgesetz der Bundesrepublik Deutschland, TierSchG) regarding animal protection procedures and approved by the responsible authorities, the Niedersächsisches Landesamt für Verbraucherschutz und Lebensmittelsicherheit (LAVES, AZ 33.19-42502-04-17/2479) and Regierung von Oberbayern (AZ 55.2-1-54-2532-11-2016). Thy1-GFP (M-line, hereafter called CTR) (26) and crosses of SOD1G93A (32) and GFP-M mice (referred to as SOD) were used in groups of up to five mice per cage, with ad libitum access to food and water. Mice were kept at a 12/12-hour light/dark cycle. Mice were implanted at the age of 19 to 23 weeks, and imaging commenced at 23 to 27 weeks of age. In total, 7 males and 4 females, 5 CTR (2 males and 3 females), 6 SOD (5 males and 1 female), were used, out of which 10 of the 11 mice were imaged eight times, and 1 mouse was imaged only five times. However, not all field of views could be imaged during each session due to the appearance of motion artifacts linked to blood vessel pulsation. The maximum number of consecutive imaging TPs per field of view assessed per mouse was as follows: two TP (one mouse), four TP (three mice), six TP (one mouse), seven TP (one mouse), and eight TP (five mice).
Mouse surgical procedure
The mouse was anesthetized by intraperitoneal injection of a mixture of fentanyl (0.05 mg/kg), midazolam (5 mg/kg), and medetomidine (0.5 mg/kg). Once anesthetized, the mouse was placed on a heating plate and shaved on the hind leg, as well as the surgical area on the scalp. During surgery and in vivo imaging, vital functions and depth of anesthesia were controlled: The body temperature was monitored with a rectal temperature probe, and O2 saturation of the blood and heart rate were monitored using a pulse oximeter (MouseOx STARR, STARR Life Science Corp., Oakmont, PA) placed on the shaved thigh. A mixture of 50 volume % N2, 47.5 volume % O2, and 2.5 volume % CO2 was administered over a cone in front of the mouse’s nose to keep the oxygen saturation at ~98%. The mouse was positioned in a stereotaxic frame (Narishige, Tokyo, Japan), and the skin above the skull was removed. After sealing the edges of the skin with the tissue adhesive n-butyl cyanoacrylate (Histoacryl, B. Braun Melsungen AG, Melsungen, Germany), the skull was cleaned using a micro curette (Fine Science Tools GmbH, Heidelberg, Germany, 10082-15) or drill. Next, the head bar was fixed with dental cement (Super-Bond C&B, Sun Medical Co. Ltd., Japan) to the skull. After hardening of the cement, the mouse was moved to an adjustable and heated mounting plate with the head bar screwed to the head holder (Fig. 1A and fig. S1A). A circular craniotomy (4-mm diameter) was then performed (Drill: 216804; RUDOLF FLUME Technik GmbH, Essen, Germany; drilling head: HP 310 104 001 001 007; Hager & Meisinger GmbH, Neuss, Germany) centered over the motor cortex. The dura was gently removed with a fine biology tipped forceps (Dumont #5 biology, Fine Science Tools GmbH, Heidelberg, Germany). Care was taken to not damage the cortical surface and to avoid blood cell deposits at the region of interest. The cover glass of 4-mm diameter (Warner Instruments, CT, USA) was fit in tightly into the opening and affixed to the skull using tissue adhesive. We compared two different sizes of coverslips (4- and 5-mm diameter). We observed less regrowth and thus superior image quality using a 4-mm cover glass. Because of the curvature of the skull, the 4-mm coverslip was easier to fit into the craniotomy directly in contact with the brain surface. Once held in place, the window was firmly fixed using dental cement. We tested a two-component, ultraviolet light curable dental cement (Paladur) and a self-curing adhesive resin cement (SuperBond C&B). In our hands, SuperBond C&B was superior to the Paladur cement and resulted in an improved window quality, with less motion-related artifacts. The surface of the coverslip facing the cortical surface was coated with poly-l-lysine (P4707; Sigma-Aldrich, Taufkirchen, Germany) and a sparse layer of 40-nm fluorescent beads (yellow-green FluoSpheres, F8795; Thermo Fisher Scientific, Waltham, MA) to render it visible for fluorescence widefield, confocal, and STED imaging. Last, a thin layer of silicon polymer (First Contact; Photonic Cleaning Technologies, Platteville, WI) was applied to the outer side of the cover glass to protect the glass surface from dirt and scratches until the actual imaging commenced.
In vivo STED microscope and chronic in vivo imaging
We built a scanning STED microscope attached to an upright microscope stand (Leica Microsystems GmbH, Wetzlar, Germany), as previously described (24, 54). In brief, STED light was delivered by a Ti:Sapphire laser (MaiTai; Spectra-Physics, Santa Clara, CA), followed by an OPO (APE, Berlin, Germany), emitting 80-MHz pulses at 595-nm wavelength. The pulses were stretched to ~300 ps by dispersion in a glass rod and a 120-m-long polarization-preserving fiber (OZ Optics, Ottawa, Canada). A helical phase delay of 0 to 2π was introduced by transmitting the STED beams through a vortex phase plate (RPC Photonics, Rochester, NY). For excitation, a pulsed laser diode operating at 483 nm, emitting pulses of 100-ps duration (PiLas, Advanced Laser Diode Systems, Berlin, Germany), was used. After combining the excitation and STED beam via a dichroic mirror, both beams were passing a Yanus scan head (Till Photonics-FEI, Gräfelfing, Germany), consisting of two galvanometric scanners and relay optics, and then were focused into the 1.3 NA objective lens (PL APO, 63×, glycerol; Leica, Wetzlar, Germany). Temporal overlap was ensured by triggering the excitation laser diode with the STED light pulses. The back-projected fluorescent light was filtered with a 525/50-nm band pass and focused on a multimode fiber for confocal detection, connected to an avalanche photodiode detector (APD; Excelitas, Waltham, MA).
In vivo STED imaging was initiated upon a recovery period of 3 to 4 weeks after window implantation. Mice were anesthetized as stated above. The head of the mouse was screwed to the tiltable head holder on the mounting plate. The window was aligned perpendicular to the optical axis of the microscope with the help of a home-built optical alignment device (25). The mouse was then moved to the microscope and placed on a motorized translation stage (MS-2000, Applied Scientific Instruments, Eugene, OR). For each dendrite, an overview image was recorded and the position of the micrometer stage was noted. Blood vessels on the cortical surface served as landmarks for the realignment of the translation stage between imaging sessions, which were used for calibrating the coordinates of the translation stage for each imaging spot. A confocal xz image, covering the dendrite of interest and the fluorescent beads adhering to the coverslip, allowed for the accurate determination of the depth of the image plane. Typically, imaging was performed at a cortical depth of 15 to 35 μm. The correction collar of the objective (PL APO, 63×, glycerol, 1.3 NA; Leica, Wetzlar, Germany) was adjusted at each depth to compensate for spherical aberrations in the tissue to optimize the STED resolution. The excitation and STED laser power were kept at a minimum to avoid phototoxicity, typically evidenced as blebbing of neurites. After completion of the imaging session, the window was covered again with silicone and anesthesia was antagonized with atipamezole (2.5 mg/kg) and buprenorphine (0.1 mg/kg). In vivo STED imaging was conducted twice a week, i.e., at 3- to 4-day intervals.
Imaging parameters: GFP was excited with 4.5 μW and depleted with an average STED power of 11.3 to 14 mW at the back aperture of the objective. Z stacks were recorded at 600-nm increments and at a pixel size of 30 × 30 nm in x and y and a pixel dwell time of 5 μs. Images with a signal >5 Mc/s were corrected for the actual count rate according to the instructions of the manufacturer of the detector (APD, SPCM-AQRH-13). At the end of the experiment, mice were perfused transcardially with 4% paraformaldehyde in phosphate-buffered saline. Seventy-micrometer-thick sections were prepared using a Vibratome (VT1000S, Leica Microsystems, Wetzlar, Germany).
Image analysis
Spine morphology was analyzed manually using Fiji (55). Only motion artifact-free image stacks were selected for data analysis. Spines, emanating laterally from the parent dendrite and captured within one or two adjacent imaging frames and appearing in at least two consecutive TPs, were analyzed. Dynamics of spine morphological parameters were assessed in stable spines. Spine neck length was measured by drawing a line along the neck from its base at the dendrite to the beginning of the spine head using the freehand line tool (Fig. 3A). The spine neck width was measured as the FWHM of a line profile (average of three lines) of the spine neck at its thinnest position. Profiles of <5 photon counts were rejected, and thus, the number of analyzed neck width is lower than spine head size or neck length. To analyze the spine head size, we smoothed (average over 3 pixels) the image stacks and encircled the spine heads with the freehand selection tool to compute the head area (Fig. 3A). Each spine was analyzed in its focal plane.
Normalized relative changes at TP “tx+1” were calculated to the previous TP “tx” by computing [M(tx+1) − M(tx)]/[M(tx+1) + M(tx)]. Percent changes were computed by [M(tx+1) − M(tx)]/M(tx)*100%. “M” stands for the measured spine parameter spine head size, neck length, or neck width.
The Pearson’s correlation coefficient r was computed by (Eq. 1)
| (1) |
M stands for the measured spine parameter spine head size, neck length, or neck width; N stands for the total number of spines; ∆t is the lag time; and is the average size of all spines.
The autocorrelation was computed with M1 = M2, and the cross-correlation was computed with M1 and M2 denoting different spine parameters. To estimate the error of the correlation coefficient r, we bootstrapped the data. Thus, we resampled the data and repeatedly chose N spines from our original sample, with replacement, and calculated the (auto/cross-)correlation. The bootstrapping was repeated 100 times, and the SD of these dataset was plotted as error bar.
The angle at which a spine emanated from the parent dendrite (neck-dendrite angle) was measured using the angle tool in Fiji (note that this angle is limited to 90° as always the smaller angle is reported; Fig. 3H). The angle formed between the spine neck and the spine head (neck-head angle) was also measured using the angle tool (Fig. 3H). Also, here, the smaller angle is reported, i.e., the maximum value of this angle 180°.
The PCA was performed in MATLAB, using z scored values of the five parameters head size, neck width, neck length, neck-dendrite angle, and neck-head angle for all stable spines at all TPs.
To assess overall spine density, all spines along the captured dendrites were counted over three consecutive TPs.
Statistics
Statistical analyses were performed either in MATLAB or in GraphPad Prism. The statistical test and precision measure are specified in the corresponding Results section together with the P value. Significance was defined by *P < 0.05, **P < 0.01, ***P < 0.001, and ****P < 0.0001. The total number of mice, analyzed spines, and number of dendrites per graph are summarized in table S1.
Acknowledgments
We would like to thank S. Rode for help with Fig. 1A. Funding: This work was supported by the Deutsche Forschungsgemeinschaft (DFG; German Research Foundation) under Germany’s Excellence Strategy within the framework of the Munich Cluster for Systems Neurology (EXC 2145 SyNergy, ID 390857198; to S.Lie.), and the Goettingen Cluster for Multiscale Bioimaging (EXC 2067/1-390729940; to K.I.W.), the DFG Emmy Noether Programme (to S. Lie.), the DFG Research Center and Cluster of Excellence (EXC 171, Area A1) “Nanoscale Microscopy and Molecular Physiology of the Brain” (to K.I.W., H.S., A.C.M., and W.W.), and the Deutsche Gesellschaft für Muskelkranke e.V. (to S.Lie.). Author contributions: Conceptualization: S. Lie., F.W., and K.I.W.; surgery: A.C.M., H.S., V.W.Y.K., and P.Š.; STED microscope: K.I.W.; imaging: A.C.M., W.W., and K.I.W.; formal analysis: S. Li, F.W., and K.I.W.; writing: S.Lie., F.W., and K.I.W.; funding acquisition: S.Lie. and K.I.W.; supervision: K.I.W. Competing interests: The authors declare that they have no competing interests. Data and materials availability: All data needed to evaluate the conclusions in the paper are present in the paper and the Supplementary Materials. No material was generated.
SUPPLEMENTARY MATERIALS
Supplementary material for this article is available at http://advances.sciencemag.org/cgi/content/full/7/24/eabf2806/DC1
REFERENCES AND NOTES
- 1.Fu M., Yu X., Lu J., Zuo Y., Repetitive motor learning induces coordinated formation of clustered dendritic spines in vivo. Nature 483, 92–95 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Hofer S. B., Mrsic-Flogel T. D., Bonhoeffer T., Hübener M., Experience leaves a lasting structural trace in cortical circuits. Nature 457, 313–317 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Holtmaat A., Svoboda K., Experience-dependent structural synaptic plasticity in the mammalian brain. Nat. Rev. Neurosci. 10, 647–658 (2009). [DOI] [PubMed] [Google Scholar]
- 4.Sala C., Segal M., Dendritic spines: The locus of structural and functional plasticity. Physiol. Rev. 94, 141–188 (2014). [DOI] [PubMed] [Google Scholar]
- 5.Holtmaat A., Trachtenberg J. T., Wilbrecht L., Shepherd G. M., Zhang X., Knott G. W., Svoboda K., Transient and persistent dendritic spines in the neocortex in vivo. Neuron 45, 279–291 (2005). [DOI] [PubMed] [Google Scholar]
- 6.Yang G., Pan F., Gan W. B., Stably maintained dendritic spines are associated with lifelong memories. Nature 462, 920–924 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Xu T., Yu X., Perlik A. J., Tobin W. F., Zweig J. A., Tennant K., Jones T., Zuo Y., Rapid formation and selective stabilization of synapses for enduring motor memories. Nature 462, 915–919 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Mongillo G., Rumpel S., Loewenstein Y., Intrinsic volatility of synaptic connections — A challenge to the synaptic trace theory of memory. Curr. Opin. Neurobiol. 46, 7–13 (2017). [DOI] [PubMed] [Google Scholar]
- 9.Chambers A. R., Rumpel S., A stable brain from unstable components: Emerging concepts and implications for neural computation. Neuroscience 357, 172–184 (2017). [DOI] [PubMed] [Google Scholar]
- 10.Ziv N. E., Brenner N., Synaptic tenacity or lack thereof: Spontaneous remodeling of synapses. Trends Neurosci. 41, 89–99 (2018). [DOI] [PubMed] [Google Scholar]
- 11.Urban N. T., Willig K. I., Hell S. W., Nägerl U. V., STED nanoscopy of actin dynamics in synapses deep inside living brain slices. Biophys. J. 101, 1277–1284 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Zito K., Knott G., Shepherd G. M. G., Shenolikar S., Svoboda K., Induction of spine growth and synapse formation by regulation of the spine actin cytoskeleton. Neuron 44, 321–334 (2004). [DOI] [PubMed] [Google Scholar]
- 13.Chazeau A., Giannone G., Organization and dynamics of the actin cytoskeleton during dendritic spine morphological remodeling. Cell. Mol. Life Sci. 73, 3053–3073 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Araya R., Vogels T. P., Yuste R., Activity-dependent dendritic spine neck changes are correlated with synaptic strength. Proc. Natl. Acad. Sci. U.S.A. 111, E2895–E2904 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Tanaka J.-i., Horiike Y., Matsuzaki M., Miyazaki T., Ellis-Davies G. C. R., Kasai H., Protein synthesis and neurotrophin-dependent structural plasticity of single dendritic spines. Science 319, 1683–1687 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Tønnesen J., Katona G., Rózsa B., Nägerl U. V., Spine neck plasticity regulates compartmentalization of synapses. Nat. Neurosci. 17, 678–685 (2014). [DOI] [PubMed] [Google Scholar]
- 17.Cartailler J., Kwon T., Yuste R., Holcman D., Deconvolution of voltage sensor time series and electro-diffusion modeling reveal the role of spine geometry in controlling synaptic strength. Neuron 97, 1126–1136.e10 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Harris K., Stevens J., Dendritic spines of CA 1 pyramidal cells in the rat hippocampus: Serial electron microscopy with reference to their biophysical characteristics. J. Neurosci. 9, 2982–2997 (1989). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Knott G. W., Holtmaat A., Wilbrecht L., Welker E., Svoboda K., Spine growth precedes synapse formation in the adult neocortex in vivo. Nat. Neurosci. 9, 1117–1124 (2006). [DOI] [PubMed] [Google Scholar]
- 20.Matsuzaki M., Ellis-Davies G. C., Nemoto T., Miyashita Y., Iino M., Kasai H., Dendritic spine geometry is critical for AMPA receptor expression in hippocampal CA1 pyramidal neurons. Nat. Neurosci. 4, 1086–1092 (2001). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Nusser Z., Lujan R., Laube G., Roberts J. D. B., Molnar E., Somogyi P., Cell type and pathway dependence of synaptic AMPA receptor number and variability in the hippocampus. Neuron 21, 545–559 (1998). [DOI] [PubMed] [Google Scholar]
- 22.Berning S., Willig K. I., Steffens H., Dibaj P., Hell S. W., Nanoscopy in a living mouse brain. Science 335, 551–551 (2012). [DOI] [PubMed] [Google Scholar]
- 23.Pfeiffer T., Poll S., Bancelin S., Angibaud J., Inavalli V. K., Keppler K., Mittag M., Fuhrmann M., Nägerl U. V., Chronic 2P-STED imaging reveals high turnover of dendritic spines in the hippocampus in vivo. eLife 7, e34700 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Willig K. I., Steffens H., Gregor C., Herholt A., Rossner M. J., Hell S. W., Nanoscopy of filamentous actin in cortical dendrites of a living mouse. Biophys. J. 106, L01–L03 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Steffens H., Wegner W., Willig K. I., In vivo STED microscopy: A roadmap to nanoscale imaging in the living mouse. Methods 174, 42–48 (2020). [DOI] [PubMed] [Google Scholar]
- 26.Feng G., Mellor R. H., Bernstein M., Keller-Peck C., Nguyen Q. T., Wallace M., Nerbonne J. M., Lichtman J. W., Sanes J. R., Imaging neuronal subsets in transgenic mice expressing multiple spectral variants of GFP. Neuron 28, 41–51 (2000). [DOI] [PubMed] [Google Scholar]
- 27.Holtmaat A., Wilbrecht L., Knott G. W., Welker E., Svoboda K., Experience-dependent and cell-type-specific spine growth in the neocortex. Nature 441, 979–983 (2006). [DOI] [PubMed] [Google Scholar]
- 28.Majewska A. K., Newton J. R., Sur M., Remodeling of synaptic structure in sensory cortical areas in vivo. J. Neurosci. 26, 3021–3029 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Loewenstein Y., Yanover U., Rumpel S., Predicting the dynamics of network connectivity in the neocortex. J. Neurosci. 35, 12535–12544 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Hardiman O., Al-Chalabi A., Chio A., Corr E. M., Logroscino G., Robberecht W., Shaw P. J., Simmons Z., van den Berg L. H., Amyotrophic lateral sclerosis. Nat. Rev. Dis. Primers 3, 17071 (2017). [DOI] [PubMed] [Google Scholar]
- 31.Robberecht W., Philips T., The changing scene of amyotrophic lateral sclerosis. Nat. Rev. Neurosci. 14, 248–264 (2013). [DOI] [PubMed] [Google Scholar]
- 32.Gurney M. E., Pu H., Chiu A. Y., Dal Canto M. C., Polchow C. Y., Alexander D. D., Caliendo J., Hentati A., Kwon Y. W., Deng H. X., Chen W., Zhai P., Sufit R. L., Siddique T., Motor neuron degeneration in mice that express a human Cu, Zn superoxide dismutase mutation. Science 264, 1772–1775 (1994). [DOI] [PubMed] [Google Scholar]
- 33.Fogarty M. J., Noakes P. G., Bellingham M. C., Motor cortex layer V pyramidal neurons exhibit dendritic regression, spine loss, and increased synaptic excitation in the presymptomatic hSOD1 G93A mouse model of amyotrophic lateral sclerosis. J. Neurosci. 35, 643–647 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Fogarty M. J., Mu E. W. H., Noakes P. G., Lavidis N. A., Bellingham M. C., Marked changes in dendritic structure and spine density precede significant neuronal death in vulnerable cortical pyramidal neuron populations in the SOD1(G93A) mouse model of amyotrophic lateral sclerosis. Acta Neuropathol. Commun. 4, 77 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Jara J. H., Stanford M. J., Zhu Y., Tu M., Hauswirth W. W., Bohn M. C., Devries S. H., Özdinler P. H., Healthy and diseased corticospinal motor neurons are selectively transduced upon direct AAV2-2 injection into the motor cortex. Gene Ther. 23, 272–282 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Berry K. P., Nedivi E., Spine dynamics: Are they all the same? Neuron 96, 43–55 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Loewenstein Y., Kuras A., Rumpel S., Multiplicative dynamics underlie the emergence of the log-normal distribution of spine sizes in the neocortex in vivo. J. Neurosci. 31, 9481–9488 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Arellano J. I., Benavides-Piccione R., Defelipe J., Yuste R., Ultrastructure of dendritic spines: Correlation between synaptic and spine morphologies. Front. Neurosci. 1, 131–143 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Statman A., Kaufman M., Minerbi A., Ziv N. E., Brenner N., Synaptic size dynamics as an effectively stochastic process. PLOS Comput. Biol. 10, e1003846 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Zuo Y., Lin A., Chang P., Gan W.-B., Development of long-term dendritic spine stability in diverse regions of cerebral cortex. Neuron 46, 181–189 (2005). [DOI] [PubMed] [Google Scholar]
- 41.Kopec C. D., Li B., Wei W., Boehm J., Malinow R., Glutamate receptor exocytosis and spine enlargement during chemically induced long-term potentiation. J. Neurosci. 26, 2000–2009 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Otmakhov N., Tao-Cheng J. H., Carpenter S., Asrican B., Dosemeci A., Reese T. S., Lisman J., Persistent accumulation of calcium/calmodulin-dependent protein kinase II in dendritic spines after induction of NMDA receptor-dependent chemical long-term potentiation. J. Neurosci. 24, 9324–9331 (2004). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Hruska M., Henderson N., Le Marchand S. J., Jafri H., Dalva M. B., Synaptic nanomodules underlie the organization and plasticity of spine synapses. Nat. Neurosci. 21, 671–682 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Henry F. E., Hockeimer W., Chen A., Mysore S. P., Sutton M. A., Mechanistic target of rapamycin is necessary for changes in dendritic spine morphology associated with long-term potentiation. Mol. Brain 10, 50 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Shomar A., Geyrhofer L., Ziv N. E., Brenner N., Cooperative stochastic binding and unbinding explain synaptic size dynamics and statistics. PLOS Comput. Biol. 13, e1005668 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Ranft J., Almeida L. G., Rodriguez P. C., Triller A., Hakim V., An aggregation-removal model for the formation and size determination of post-synaptic scaffold domains. PLOS Comput. Biol. 13, e1005516 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Frank A. C., Huang S., Zhou M., Gdalyahu A., Kastellakis G., Silva T. K., Lu E., Wen X., Poirazi P., Trachtenberg J. T., Silva A. J., Hotspots of dendritic spine turnover facilitate clustered spine addition and learning and memory. Nat. Commun. 9, 422 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Chen J. L., Villa K. L., Cha J. W., So P. T. C., Kubota Y., Nedivi E., Clustered dynamics of inhibitory synapses and dendritic spines in the adult neocortex. Neuron 74, 361–373 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Acevedo-Arozena A., Kalmar B., Essa S., Ricketts T., Joyce P., Kent R., Rowe C., Parker A., Gray A., Hafezparast M., Thorpe J. R., Greensmith L., Fisher E. M. C., A comprehensive assessment of the SOD1G93A low-copy transgenic mouse, which models human amyotrophic lateral sclerosis. Dis. Model. Mech. 4, 686–700 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Keck T., Keller G. B., Jacobsen R. I., Eysel U. T., Bonhoeffer T., Hübener M., Synaptic scaling and homeostatic plasticity in the mouse visual cortex in vivo. Neuron 80, 327–334 (2013). [DOI] [PubMed] [Google Scholar]
- 51.Turrigiano G., Homeostatic synaptic plasticity: Local and global mechanisms for stabilizing neuronal function. Cold Spring Harb. Perspect. Biol. 4, a005736 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Hazan L., Ziv N. E., Activity dependent and independent determinants of synaptic size diversity. J. Neurosci. 40, 2828–2848 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Fogarty M., Amyotrophic lateral sclerosis as a synaptopathy. Neural Regen. Res. 14, 189–192 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Wegner W., Mott A. C., Grant S. G. N., Steffens H., Willig K. I., In vivo STED microscopy visualizes PSD95 sub-structures and morphological changes over several hours in the mouse visual cortex. Sci. Rep. 8, 219 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Schindelin J., Arganda-Carreras I., Frise E., Kaynig V., Longair M., Pietzsch T., Preibisch S., Rueden C., Saalfeld S., Schmid B., Tinevez J.-Y., White D. J., Hartenstein V., Eliceiri K., Tomancak P., Cardona A., Fiji: An open-source platform for biological-image analysis. Nat. Methods 9, 676–682 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary material for this article is available at http://advances.sciencemag.org/cgi/content/full/7/24/eabf2806/DC1


