Differential recognition of glycan isomers by proteins is key to innate and adaptive immunity.
Abstract
The recognition of oligomannose-type glycans in innate and adaptive immunity is elusive due to multiple closely related isomeric glycan structures. To explore the functions of oligomannoses, we developed a multifaceted approach combining mass spectrometry assignments of oligomannose substructures and the development of a comprehensive oligomannose microarray. This defined microarray encompasses both linear and branched glycans, varying in linkages, branching patterns, and phosphorylation status. With this resource, we identified unique recognition of oligomannose motifs by innate immune receptors, including DC-SIGN, L-SIGN, Dectin-2, and Langerin, broadly neutralizing antibodies against HIV gp120, N-acetylglucosamine-1-phosphotransferase, and the bacterial adhesin FimH. The results demonstrate that each protein exhibits a unique specificity to oligomannose motifs and suggest the potential to rationally design inhibitors to selectively block these protein-glycan interactions.
INTRODUCTION
Oligomannose-type glycans are key components of eukaryotic cells, prokaryotic cells, and many viruses such as HIV and severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) (1–5). In addition to playing a key structural role, oligomannoses have profound biological functions in maintaining cellular homeostasis. They facilitate the trafficking of lysosomal hydrolases through interaction of phosphorylated oligomannoses with mannose-6-phosphate (Man-6-P) receptors (6). Oligomannoses are also key components of protein folding and quality control in the endoplasmic reticulum (e.g., calnexin and calreticulin) and are recognized by a wide variety of endogenous lectins (1, 7).
Oligomannoses are functionally important in mediating immune recognition between the host and microorganisms. Expressed on human urinary epithelial cells, oligomannoses are ligands for FimH, an adhesin located on the tip of the type 1 fimbriae of uropathogenic Escherichia coli, enabling the attachment and the subsequent infection of the bacteria (8–11). Oligomannoses on yeast and microorganisms, on the other hand, are pathogen-associated molecular patterns (PAMPs), which are recognized by a variety of host innate immune receptors including C-type lectin receptors (CLRs), such as DC-SIGN, L-SIGN, Dectin-2, and Langerin (12, 13). Such interaction could promote endocytosis of the pathogens and production of cytokines and chemokines, leading to the activation of adaptive immunity. Oligomannose glycans are also antigenic. A group of broadly neutralizing antibodies (bNAbs) binds the mannose patch of HIV gp120, and some directly target oligomannoses or hybrid N-glycans (14, 15). Despite being a key mediator in these immune recognition events, the precise oligomannose structures important for each interaction have not been fully elucidated.
Comprising a vast number of isomers varying in sequence, linkages, and branching pattern, oligomannoses represent an elusive class of glycans that is particularly challenging to characterize structurally. Negative-ion mode electrospray ionization–mass spectrometry (ESI-MS) with collision-induced dissociation (CID) or higher-energy collisional dissociation (HCD) holds great potential in structural analysis of complex glycans (16–18). It can produce cross-ring fragmentation and other diagnostic ions that are valuable in differentiating isomeric glycans (19–21). Negative-ion ESI-CID/HCD-MS/MS has been successfully applied to structural assignment of reducing glycans (17, 18, 22–26) and reduced glycan alditols (20, 21, 27–29). Fragmentation features of some naturally occurring oligomannose N-glycans have been partly described by Harvey (17), but it has not been systematically investigated whether such a method can discriminate closely related oligomannose isomers that differ only in linkages and arm positions.
Glycan microarrays are a reliable platform to study glycan recognition, but the number and the variety of oligomannose structures available to date have been relatively limited (14, 30–32). To unravel the biological and pathological functions of oligomannoses, it is highly desirable to have a robust oligomannose microarray that includes a larger number of structures representing diversity in the number, linkage, and stereochemistry and modifications of the mannose residues.
Here, we describe a multifaceted approach that combines negative-ion mode ESI-HCD-MS/MS sequencing and glycan microarray to screen and pinpoint the differential recognition of oligomannose isomers. With a comprehensive panel of oligomannose glycans, we developed a robust ESI-HCD-MS/MS method enabling the structural characterization of isomers varying in linkages and branching patterns. These sequence-defined oligomannoses were used for the construction of a comprehensive oligomannose microarray, which also included naturally occurring asparagine (Asn)–linked high mannose-type N-glycans and phosphorylated oligomannoses generated by chemoenzymatic approaches. Experimental studies using the oligomannose microarray developed here provide advanced insights into the binding specificities of mammalian innate immune receptors, bacterial FimH, and bNAbs against HIV. In such studies, we identified a trisaccharide that is a potential inhibitor of FimH. Overall, our results offer several translational opportunities as they can guide the rational design of inhibitors to block these oligomannose interactions.
RESULTS
To explore oligomannose recognition by glycan-binding proteins (GBPs) and their structural differentiation, we prepared a comprehensive oligomannose glycan library (fig. S1). Seventy-nine (#1 to 79) glycans were obtained through chemical synthesis, among which 67 were linear and branched oligomannose variants (#1 to 67) and 12 were high mannose-type N-glycans without the chitobiose core (#68 to 79). These, together with five control glycans (#80 to 84), comprised oligomannose library 1. Furthermore, we expanded this library using a combined enzymatic and chemoenzymatic approach to prepare high mannose-type N-glycans Man4 to Man9 with the chitobiose core linked to Asn (#85 to 90; detailed preparation is in Materials and Methods and fig. S2), the oligomannose phosphodiesters (#91 to 94; described below), and phosphomonoesters (#95 to 100). All glycans (#1 to 100) were included to comprise oligomannose library 2 (fig. S1).
With a large number of closely related oligomannose isomers, this comprehensive library comprised the reference structures for this study (Fig. 1). We explored the MS/MS sequencing strategy to identify diagnostic fragmentation pattern that can unambiguously characterize the oligomannose isomers. We also generated sequence-defined oligomannose microarrays to assign the unique recognitions of these oligomannose structures.
Fig. 1. A systematic approach combining negative-ion ESI-HCD-MS/MS sequencing and a glycan microarray to characterize the differential recognition of oligomannose isomers.
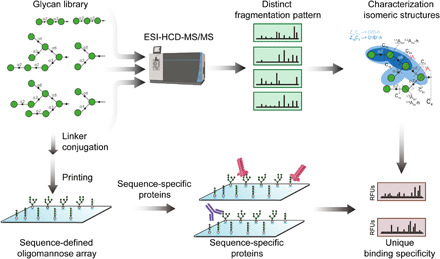
Oligomannose glycans are analyzed by ESI-HCD-MS/MS, which produces diagnostic fragmentations that can be used to characterize isomeric glycan structures. These glycans are also used in fabrication of a sequence-defined oligomannose array subsequent to conjugation with amine-functionalized linkers. Proteins involved in innate and adaptive immunity are interrogated on the microarray, leading to unique binding specificities, reflecting the differential recognition of oligomannose isomers.
Structural assignment of linear and branched oligomannose isomers by negative-ion ESI-HCD-MS/MS
We analyzed glycans within the oligomannose glycan library by negative-ion ESI-HCD-MS/MS, which is known to generate tandem MS spectra with diagnostic A-type cross-ring fragment ions, next to C- and B-type ions derived from glycosidic bond cleavage.
We first investigated whether negative-ion ESI-HCD-MS/MS can facilitate linkage determination in linear oligomannose isomers. Specific fragmentation signatures were observed for the five trisaccharide isomers and the four tetrasaccharide isomers (Fig. 2, A to E, and fig. S3, A to D). Accordingly, the spectra of the tri- and tetrasaccharides contain a set of C-type ions at mass/charge ratio (m/z) 179, 341, and 503. The α1,2-linked Man is accompanied by a series of peaks with a neutral loss of 18, 78, and 120 Da from the C-type ions, corresponding to the B, 0,4A-h, and 1,3A ions, respectively (-h denotes dehydration) (Fig. 2, A to C and F). Previously, 1,3A was correlated with the substitutions on the 3-arm of high mannose-type N-glycans (17), which only contains α1,2-linked Man. The α1,6-linked Man is associated with a series of cross-ring fragmentations, 0,2A, 0,3A, and 0,4A, with neutral losses of 60, 90, and 120 Da from the C-ion, respectively (Fig. 2, C to E). Among them, the 0,2A and 0,4A pair are more abundant than 0,3A. The same pattern has been seen in 6-linked poly-N-acetyllactosamine glycans (26). By comparison, the α1,3-linked Man did not present distinct A- or B-type fragmentations, leaving the C ions the dominant peaks (Fig. 2, B, D, and F). The same rule also applies to tetrasaccharide isomers (fig. S3, A to D). Notably, these diagnostic fragment patterns (Fig. 1F) are similar to glucans of corresponding linkages (24).
Fig. 2. Oligomannose linkages and branching patterns determined by negative-ion ESI-HCD-MS/MS.
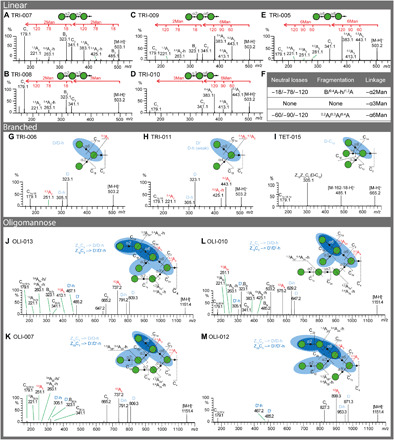
(A to E) ESI-HCD-MS-MS product-ion spectra of five linear trimannosyl isomers TRI-007 (A), TRI-008 (B), TRI-009 (C), TRI-010 (D), and TRI-005 (E). The key fragment ions for linkage determination were labeled on top of each spectrum. (F) Summary of the characteristic ions and their fragmentation origins of 1,2-, 1,3-, and 1,6-linkages. (G to I) ESI-HCD-MS-MS product-ion spectra of two branched trimannosyl isomers TRI-006 (G) and TRI-011 (H) and the tetramannosyl glycan TET-015 (I). (J to M) ESI-HCD-MS-MS product-ion spectra of four Man7 isomers OLI-013 (J), OLI-007 (K), OLI-010 (L), and OLI-012 (M). The fragmentation pattern of each oligomannose is shown on top of the spectrum. The key fragment ions D and D′ ions are highlighted in blue. The label -h denotes dehydration. Other key cross-ring fragmentations are highlighted in red. To have a closer comparison, the low–molecular weight region (m/z 150 to 500) of OLI-013, OLI-007, and OLI-010 was zoomed in and shown in fig. S5, together with MS3 of m/z 827.3 of OLI-012.
We then sought to differentiate the branched oligomannose tri- and tetrasaccharide isomers. The D ion, derived from C/Z double cleavage, and the further loss of H2O (D-h) are prominent for high mannose-type N-glycans (17). The branched oligomannose trimer TRI-006 with an α1,3-/α1,6-doubly substituted core Man presented a dominant pair at m/z 323 and 305 (Fig. 2G). They are accompanied by a peak 72 Da lower to the D ion (m/z 251), in accordance with a 0,3A cleavage of the core. These three ions provide structural information on the 6-arm and are characteristic of α1,3-/α1,6-branched oligomannoses. By contrast, a trimannose isomer with an α1,4-/α1,6-doubly substituted core Man, TRI-011, presented a strong D ion, a less abundant D-h, and a strong cross-ring cleavage 0,4A of the core with a neutral loss of 102 from the D ion (m/z 323, 305, and 221, respectively; Fig. 2H). The same phenomenon is also observed in the α1,4-/α1,6-doubly substituted pentasaccharide PEN-012 (m/z 485, 467, and 383, respectively; fig. S4A). In addition, a pair of peaks at m/z 443/425 and 767/749 is also prominent in the product ion spectrum of TRI-006 and PEN-012 (Fig. 2H and fig. S4A). They are characteristic to the 1,4-linked Man derived from 0,2A and 0,2A-h of the branched Man, respectively (26). With these diagnostic fragmentation patterns, we could successfully discriminate branched oligomannose isomers of tetrasaccharides (fig. S3, E and F), pentasaccharides (fig. S4, A to D), and hexasaccharides (fig. S4, E and F) that differ only in linkages.
We also analyzed the product spectrum of a tetrasaccharide with α1,3-/α1,4-/α1,6-triply substituted Man (Fig. 2I). The spectrum is dominated by the fragment ion at m/z 305, corresponding to D ion-162-18. This is likely derived from a loss of α1,4-linked Man subsequent to the formation of D ion (Z1βZ1γC2 cleavage), similar to that of the bisecting N-acetylglucosamine (GlcNAc)–containing N-glycans, which have a 1,4-linkage but in β configuration (GlcNAcβ1-4Man) (18). Notably, this fragmentation also occurs to the reduced bisecting N-glycans (21). In addition, the spectrum also contains a peak at m/z 485, indicating a loss of hexose from the tetrasaccharide. Future studies are needed to identify where the loss occurs.
Thus, the D-type double cleavage fragmentation, together with the unique combinations of A-type cross-ring cleavage and B/C-type glycosidic bond cleavages (as shown in fig. S3G), facilitates the identification of branching pattern and assignment of linkages in oligomannose glycans. This information can be conveniently applied to discriminate isomeric oligomannose structures.
Differentiation of high mannose-type N-glycan isomers
While naturally occurring high mannose-type N-glycans have been studied by negative-ion ESI-CID-MS/MS (17), not all possible arrangements of mannose residues were previously available for comparison. We therefore investigated a comprehensive set of high mannose isomers ranging from Man5 to Man9 (see data file S1 for details). Although they are structurally complex, high mannose-type N-glycans are organized in a similar fashion, with an α1,3/α1,6 disubstituted Man present at the branching point. Thus, the overall pattern of these spectra looks similar. The four Man7 isomers—OLI-013, OLI-007, OLI-010, and OLI-012—are discussed as examples to illustrate the fragmentation patterns (Fig. 2, J to M).
The four spectra are dominated by a diagnostic D/D-h ion pair, indictive of the composition of the 6-arm. This ion pair divides the four isomers into three categories: 809/791 presented by OLI-013 and OLI-007 (Fig. 2, J and K), 647/629 by OLI-010 (Fig. 2L), and 971/953 by OLI-012 (Fig. 2M), suggesting a tetra-, tri-, and pentasaccharide present on the 6-arm, respectively. In addition, the high abundant 0,3A ions of the α1,3/α1,6 disubstituted Man (m/z 737, 575, and 899; Δ = 72 Da from the corresponding D ions) and the C ions derived from the glycosidic cleavage at the 6-branch (m/z 665, 503, and 827; Δ = 144 Da from the corresponding D ions), further confirming the α1,3/α1,6-branched Man and the composition of the 6-arm. As these high mannose isomers have additional α1,3/α1,6-branched Man on the 6-linked branch, a closer investigation of the low m/z region, in combination with MS3 fragmentation of either D, 0,3A, or Cα ions, reveals more structural information (fig. S5). A second set of D/D-h (designated D′/D′-h) and 0,3A fragment ions (D′-72) are the most abundant peaks in this region. OLI-013 and OLI-012 presented m/z 485/467 and 413, while OLI-007 and OLI-010 showed m/z 325/305 and 251, corresponding to a disaccharide and monosaccharide 6-linked to the α1,3/α1,6-branched core Man on the nonreducing terminal, respectively (fig. S5, A and D, and B and C, respectively). Consequently, the four isomers are differentiated from each other. Additional ions are attributed to the Manα1-2Man- (for OLI-013, OLI-007, and OLI-012) or Manα1-2Manα1-2Man- (OLI-010) linear extensions, which results in a characteristic B, 0,4A-h, and 1,3A ion triplex (Δ18, 78, and 120 from the C-ion precursor; fig. S5). Although relatively weak, these triplex peaks are still identifiable with acceptable signal-to-noise ratio.
To corroborate the applicability of our method, we also purified two chitobiose core-containing high mannose-type N-glycans (designated Man6-1 and Man8-1) from ribonuclease (RNase) B. They are dominant isomers of the composition Man6-GlcNAc2 and Man8-GlcNAc2, respectively. Although the MS/MS spectra of both the singly and doubly charged molecular ions are dominated by fragmentation derived from the chitobiose core, MS3 still yields information useful for structure assignment (figs. S6 and S7). The similarity between the MS3 spectrum of Man6-1 and HEX-002 suggests the Man6-1 carries an additional α1,2-linked Man residue to the lower α1-3 linked arm of Man5 (fig. S6, C and D, and data file S1). Likewise, Man8-1 is assigned as Man8 (D1D3) based on the similarity to OLI-015 (fig. S7, C and D, and data file S1). These results are consistent with previous reports of the dominant isomers of high mannose glycans in RNase B (33, 34).
Collectively, diagnostic fragmentation patterns generated by ESI-HCD-MS/MS reveal linkage information and branching patterns in oligomannose glycans and can be used to discriminate isomeric oligomannose structures.
GNPTAB is specific to characteristic oligomannose isomers
Isomeric oligomannoses are valuable for investigating the sequence selectivity of glycoenzymes such as glycosyltransferases and glycosidases. An important example we sought to characterize is the specificity of N-acetylglucosamine-1-phosphotransferase subunits alpha/beta (GNPTAB), which catalyzes the addition of GlcNAc-1-phosphate (GlcNAc-P) on high mannose N-glycans of lysosomal hydrolases. While it is known that GNPTAB recognizes a specific protein conformation (35), its specificity toward oligomannose glycans is unclear.
We investigated the enzymatic activity of GNPTAB in a high-performance liquid chromatography (HPLC) peak-shift assay enabled by the conjugation to a bifunctional fluorescent tag, 2-amino-N-(2-amino-ethyl)-benzamide (AEAB).. Compared to mock reaction, oligomannose conjugates reactive with GNPTAB show additional peaks (asterisk in Fig. 3A) in their individual HPLC chromatogram. Among all oligomannoses tested, three trisaccharides (TRI-005, TRI-007, and TRI-009), a tetrasaccharide (TET-007), and a hexasaccharide (HEX-002) are good substrates of the enzyme. In addition, the reaction with TET-007 ran close to completion, suggesting that the 6-linked A arm is preferable by GNPTAB. Therefore, the potential phosphorylation sites on a Man9 template are deduced as shown in Fig. 3B (circled in red). Our observations are in accordance with previous reports that GNPTAB requires the γ subunit that is encoded by GNPTG for optimal enzymatic activity (36). Lack of double phosphorylated products, such as previously reported (37, 38), is also likely due to the involvement of the γ subunit in the addition of the second GlcNAc-P (36), which requires further investigation.
Fig. 3. Highly selective glycosylation catalyzed by GNPTAB toward oligomannose isomers.
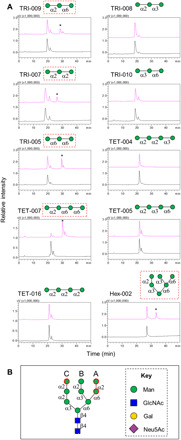
(A) HPLC analysis of the mock reaction (black) and the GNPTAB reaction mixture (pink) with tri-, tetra-, and hexa-mannosides. The product peaks are labeled with an asterisk. (B) Potential glycosylation sites on the high mannose N-glycan Man9-GlcNAc2 deduced from the information obtained from linear isomers included in the assay. Glycan structures reacted with GNPTAB in (A) are highlighted in red square; potential glycosylation on Man9-GlcNAc2 is in red circle.
Together, our results suggest that GNPTAB specifically catalyzes the addition of GlcNAc-P on the A and C arms of oligomannose isomers, and thus, specific features of the oligomannose glycans are required for efficient modification by this enzyme.
Oligomannose microarray construction
To investigate the differential recognition of these isomeric oligomannoses, we conjugated AEAB (#1 to 79 and #91 to 94) and fluorenylmethyloxycarbonate 3-(methoxyamino)propylamine (F-MAPA) (#95 to 100) to glycans to construct an oligomannose microarray (Fig. 1 and fig. S1). Whereas AEAB labeling is through glycan reduction, F-MAPA derivatization retains the integrity of their reducing terminal glycan (39). Probes prepared with oligomannose library 1 (#1 to 84) comprised oligomannose array set 1, whereas probes prepared with glycans in oligomannose library 2 (#1 to 100) comprised oligomannose array set 2.
Various plant lectins displayed specific interactions with mannose-containing glycans (40); thus, the oligomannose microarray is a useful platform for deciphering such interactions. We selected a few lectins as representatives to interrogate on the oligomannose microarray (fig. S8, A to E). The binding preferences of these lectins are identical on oligomannose array sets 1 and 2 (data file S2). As expected, Concanavalin A lectin (ConA) binds almost all glycans, while Ricinus communis I agglutinin (RCA) (note- ‘Ricinus communis’ should be italicized) only binds the two biantennary N-glycans terminating in Gal or 2,6-linked sialic acid, which further confirms the quality of the array. Three mannose-binding lectins—GNA (Galanthus nivalis lectin), NPA (Narcissus pseudonarcissus lectin, Daffodil), and HHL (Hippeastrum hybrid lectins, AL)—display similar binding profiles on the oligomannose microarray (fig. S8, A to C and G). Previous studies reported that α-mannosides were inhibitors of lectin binding, particularly Manα1-6Manα1-6Man and Manα1-6(Manα1-3)Manα-O-Me (41). Many of the glycans with the trimannosyl core Manα1-6(Manα1-3)Man are strongly bound by the three lectins (i.e., #26 and 81). Our data also demonstrate that linear mannobiose glycans with a β-linked core GlcNAc on the reducing terminal are among the top binders (e.g., #10 and 16), demonstrating that the Man residues located close to the core are important for the binding.
Characterization of the binding specificities of innate immune receptors
Many innate immune receptors are known to recognize mannose-containing glycans expressed by hosts or pathogens (42, 43). With the newly assembled oligomannose array, we investigated the binding preferences of DC-SIGN, L-SIGN, Dectin-2, and Langerin (Fig. 4). As closely related CLRs, DC-SIGN and L-SIGN were previously found to share similar binding profiles toward oligomannose glycans with branched trimannosyl core Manα1-6(Manα1-3)Manα1-R (43). Both proteins show strong binding signals on the oligomannose array, and high mannose N-glycans are among the top binders (#70 to 79; Fig. 4, A and B). L-SIGN, in addition, strongly binds some shorter oligomannoses (#38, 46, 62, 54, 61, 47, 66, 65, and 19; Fig. 4B), many of which contain the chitobiose core. On the basis of the common structural motifs bound by individual proteins, we deduced their binding specificities on the template of Man9-GlcNAc2 (highlighted in red; Fig. 4, A and B). While DC-SIGN prefers the B arm (Manα1-2Manα1-3Manα1-), additional α1,6-linked Man on the A arm and α1,2-linked Man on the C arm boost the interactions. L-SIGN, on the other hand, shows better binding with the α1,2-linked Man on the C arm (Manα1-3Manα1-) of the trimannosyl core, and the outmost α1,2-linked Man on the C arm enhances the binding. However, L-SIGN differs from DC-SIGN in its preference to the reducing end penultimate GlcNAc (Fig. 4B), suggesting that it has a stricter specificity toward the N-glycan core region. This is consistent with previous observations that the L-SIGN binding is mainly restricted to the oligomannose-type N-glycans, whereas DC-SIGN has a more relaxed binding preference (43).
Fig. 4. Distinct binding selectivity of the innate immune receptors to oligomannose glycans.
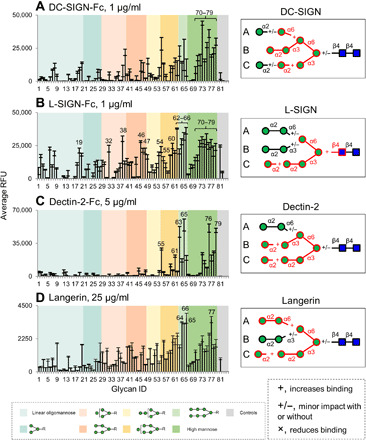
(A to D) Microarray analysis of human IgG Fc-tagged DC-SIGN (A), L-SIGN (B), and Dectin-2 (C) and His-tagged Langerin (D) on oligomannose array. Detection was with Alexa Fluor 488–labeled goat anti-human IgG, Cy3-labeled goat anti-human IgG, or mouse anti-His IgG followed by Alexa Fluor 633–labeled goat anti-mouse IgG (H+L). Results are shown as relative fluorescence units (RFUs) by averaging the background-subtracted fluorescence signals of four replicate spots; error bars represent the SD among the four values. The glycans are arranged by their structural features and color-coded as listed at the bottom. Binding preferences of the corresponding proteins to oligomannose glycans deduced from microarray analyses are shown on the right. The epitopes common to the top binders are highlighted in red, with +, +/−, and × suggesting that the presence of the corresponding structures increases, not changes, or decreases the binding intensities.
Dectin-2 has been reported to bind high mannose-type N-glycans, particularly Man9-GlcNAc2 and Man8-GlcNAc2 (44). Consistent with this, we observed that Dectin-2 binds Man8 with B and C arms, whereas Man9 gave saturated binding (#76 and 79, respectively; Fig. 4C). However, other Man8 isomers (#77 and 78) are only modestly bound. Therefore, Dectin-2 requires the presence of both B and C arm on the trimannosyl core (Fig. 4C). This assignment is also supported by saturated binding given by probes #65 and 63. Binding to other glycans, such as #55 and 61, suggests that the nonreducing terminal α1,2-linked Man on B and C arms is not absolutely essential for Dectin-2 binding but can increase the binding signal (Fig. 4C).
Langerin, in contrast to Dectin-2, binds a broader range of the glycans on the array. Oligomannose glycans with A and C arms are among the top binders (#66, 77, and 64; Fig. 4D). Probes with B and C arms (#65) are also strongly bound. Thus, the binding preference of Langerin resembles L-SIGN, in which the C arm is essential and the α1,2-linked Man extension on the C arm and Manα1-2Manα1- on the A arm can increase its binding intensity (Fig. 4D). Together, these innate immune receptors exhibit clear binding preferences to distinct oligomannose isomers.
Characterization of the binding specificities of bacterial adhesin FimH
The oligomannose array is well suited for the characterization of bacterial adhesins recognizing mannosyl glycans. Crucial to urinary pathogenic E. coli, FimH was suggested to bind high mannose N-glycans and terminal α1,2-, α1,3-, and α1,6-linked Man (9, 10). However, the specific binding epitopes are only partly understood.
We tested three constructs of FimH that were recombinantly expressed in E. coli. These constructs include an isolated lectin domain with a C-terminal His tag (FimHL-His), a full-length FimH with an N-terminal extension of FimG (FimH.DsG), and a full-length FimH with an N-terminal His–glutathione S-transferase (GST) tag (without FimG portion, FimH-His). Our initial test with the two His-tagged proteins showed no detectable binding when relying on the His epitope for detection. Subsequently, arrays were probed with biotinylated FimH.DsG and FimHL-His; these analyses reveal robust binding by only FimH.DsG-Biotin at both 50 and 200 μg/ml (Fig. 5A and fig. S6F). Among the top-ranking glycans are Man3 and Man5 with the chitobiose core or at least one GlcNAc residue (#81, 86, 24, 80, and 25). Those glycans without GlcNAc linkage (#68 to 79) exhibit only low binding signals. These top binders all contain the motif Manα1-3Manβ1-4GlcNAc with no linear extensions on the α1,3-linked Man on the C branch. The results indicate that FimH.DsG prefers to bind Manα1-3Manβ1-4GlcNAcβ1-R. An additional 6-branch to the core Man and GlcNAc to the core GlcNAc increases binding. While the presence of A and B arms is favorable, the α1,2-linked mannose extension on the C arm is unfavorable (Fig. 4A). We also performed a titration series of FimH to investigate its dose response. Differential binding to some representative oligomannoses is shown in fig. S9A. The three with strong and moderate binding curves are HEX-008 (#51), Man5GlcNc2-AEAB (#81), and TRI-015 (#10), all carrying the minimal binding epitope. TRI-016 (#11), a closely related isomer to TRI-015 (#10), exhibits weak binding.
Fig. 5. Restricted binding of the full-length FimH and HIV bNAbs on the oligomannose array.
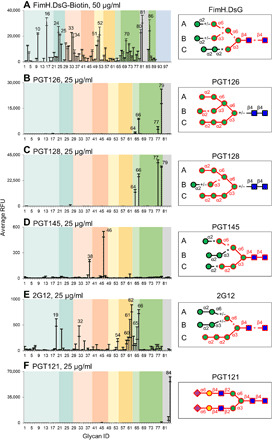
(A to F) Microarray analysis of the biotinylated FimH (A), PGT126 (B), PGT128 (C), PGT145 (D), 2G12 (E), and PGT121 (F) on the oligomannose array. Detection was with Cy5-streptavidin or Cy3-labeled anti-human IgG. Results are shown as RFUs by averaging the background-subtracted fluorescence signals of four replicate spots; error bars represent the SD among the four values. The glycans are arranged by their structural features and color-coded as shown in the legend to Fig. 4. Binding preferences of the corresponding proteins to oligomannose glycans deduced from microarray analyses are shown on the right. The epitopes common to the top binders are highlighted in red, with +, +/−, and × suggesting that the presence of the corresponding structures increases, not changes, or decreases the binding intensities, respectively.
These results identifying the binding epitope of FimH are consistent with the crystal structures (45). While previous results suggested that FimHL binds α-mannosides with a 2000-fold higher affinity compared with full-length FimH.DsG (μM versus nM in Kd) (10), our results indicate that the full-length FimH.DsG can still bind high mannose-type N-glycans. Whether the lectin domain alone has similar binding preference warrants further investigation.
Binding specificities of HIV bNAbs
Because of the unusual high density of high mannose-type N-glycans on HIV envelope glycoprotein gp120, the glycans and the underlying peptides become primary targets for human glycan-dependent bNAbs against HIV (15, 46–49). Although many bNAbs have been studied previously on glycan microarrays, only a limited number of oligomannose glycans were available. Thus, we investigated five of the best characterized bNAbs—PGT121, PGT126, PGT128, PGT145, and 2G12—on the oligomannose array.
Unexpectedly, all bNAbs bind a relatively restricted set of glycans except for 2G12 (Fig. 5, B to F). The binding profiles of PGT126 and PGT128 were almost identical and restricted to #77, 79, 64, and 66 (Fig. 5, B and C). Both antibodies bind Man9 in which all the A, B, and C arms are present (#79). Loss of the terminal α1,2-linked Man substantially reduced the binding of PGT126 but not PGT128 (#77; Fig. 5, B and C, respectively). Complete lack of the B arm diminished the binding of PGT126 and moderately reduced PGT128 (#64), thus suggesting that the specificity of PGT126 is more restricted. These results are consistent with the previous observations that the A and C arms are indispensable for the binding of PGT128 (46). In addition, our results demonstrate that neither antibody requires the absolute presence of the chitobiose core, although the equivalent structures with the chitobiose core did give higher signals (#66 versus #64; Fig. 5, B and C).
Conversely, PGT145 does not bind high mannose-type glycans that lack the chitobiose core (#68 to 79; Fig. 5D). Weak binding with probes #46 and 38 suggests that it recognizes the chitobiose core-containing Man3 with an α1,2-linked Man extension on the C branch and an α1,6-linked Man linked to core (Fig. 5D). Additional extensions on these two branches or the presence of B arm diminish the binding.
2G12 has a broader binding spectrum mainly toward the oligomannose but not toward the high mannose-type N-glycans lacking the chitobiose core (Fig. 5E). Previous studies concluded that the Manα1-2Man epitope on the C arm is crucial to 2G12 binding (49). Top binders of 2G12 (#62, 66, 19, 61, 65, 60, and 54) share the same epitope with the intact C arm (Manα1-2Manα1-2Manα1-3Manβ1-4) (Fig. 5E). Additional α1,6-linked Man and GlcNAc to the core region increased binding intensity. Using a series of titrations of 2G12, two representative glycans exhibiting good concentration-dependent binding curves were OLI-018 (#62) and HEX-013 (#19), both containing the C arm as well as the chitobiose core (fig. S9B). Our data suggest that the chitobiose core is also essential to 2G12 binding, as low or no binding was detected with a pentasaccharide, PEN-007 (#53), or the high mannose glycans lacking the chitobiose core (#68 to 79). Interestingly, and distinct from the above bNAbs, PGT121 did not recognize oligomannose glycans on the array but strongly bound α2,6-disialylated biantennary N-glycan (Fig. 5F).
While these bNAbs target different regions on gp120, our results demonstrate that many display binding preferences toward glycans of unique isomeric structures. Previous reports suggested that some HIV bNAbs prefer to bind hybrid N-glycans (14). Although no such glycans were present on the oligomannose array, the binding preferences of PGT-145 and 2G12 toward the C branch suggest that a hybrid N-glycan could be accommodated and thus warrants further study.
Inhibition of glycan-protein interactions
To shed light on the avidity-based glycan-protein interaction, we tested the inhibitory activities of closely related oligomannose isomers (fig. S10). FimH and 2G12 were selected based on their unique binding specificities described above.
For FimH, although at high concentration (20 mM) both TRI-015 and TRI-016 strongly inhibit its binding, Manα1-3Man containing TRI-015 is a better inhibitor in comparison to its isomeric counterpart at medium and low concentrations (2 and 0.2 mM, respectively; fig. S10A). The strong potency indicates that it is an interesting lead compound for further investigation. Similarly, although neither TET-004 nor TET-007 contains the chitobiose core, which is key to the strong engagement with 2G12, TET-004 (Manα1-2Manα1-2Manα1-3Man) is a stronger inhibitor relative to TET-007 (fig. S10B).
Together, these results corroborate our observations on microarray binding studies and suggest candidate inhibitors that deserve further exploration.
DISCUSSION
The results presented here illustrate the unique glycan-binding specificities of CLRs, bacterial adhesin FimH, and HIV bNAbs toward oligomannose isomers. These new insights were enabled by the development of a comprehensive and well-defined oligomannose library encompassing closely related oligomannose isomers on the basis of a robust ESI-HCD-MS/MS structural characterization method. The oligomannose ligands identified in this study are essential players in innate and adaptive immunity, and in pathogenic events, which shed light on their therapeutic potential.
The fragmentation patterns identified in negative-ion ESI-HCD-MS/MS enabled differentiation of isomeric oligomannose glycans. They are valuable references to the MS2 and MSn spectra of large glycans and glycopeptides and therefore advance their structural assignment. Although observed in HCD, a similar set of diagnostic fragmentations may be generated by conventional CID with adjusted collision energy settings as partly shown previously (17). Our results also provide a comparative dataset for other dissociation methods such as various electron-based dissociation techniques, infrared multiple photon dissociation (IRMPD), and ultraviolet photodissociation (UVPD) (50). De novo structural characterization of glycans often requires a combination of a variety of methodologies and analytical techniques. Our results are of particular value in conjunction with a diverse selections of liquid chromatography methods (34, 51) or ion mobility MS (52–54) that enables isomer separation. Moreover, as demonstrated in the present study, the information provided by ESI-HCD-MS/MS is orthogonal to that obtained by glycan microarrays. In conjunction with exoglycosidase and sequence-defined lectins and antibodies (26, 55), glycan microarrays can be especially effective in predicting the presence of certain glycan epitopes, which greatly facilitates the structural assignments in approaches such as shotgun glycomics.
We observed differences in glycan-binding preference on the oligomannose array. Whereas GNPTAB and bNAbs have relatively restricted binding toward specific oligomannose isomers, innate immune receptors and pathogen adhesin FimH, overall, tend to have a broader binding specificity (10). This likely reflects the difference in their mannose binding pockets. Thus, restricted specificity aids the exquisite regulation of the relevant biological events, avoiding the generation of invalid enzymatic products and cross-reactivity of the antibody responses. The relaxed binding of CLRs and the pathogen adhesin, on the other hand, broadens the targets to which immune cells and pathogens can attach, therefore expanding the breadth of immune responses and perhaps facilitating microbial survival. No two CLRs showed identical specificities, suggesting the uniqueness of these CLRs as part of our immune system. It will be interesting to conduct molecular modeling and co-crystallization studies using these proteins and their different oligomannose ligands.
The natural ligands identified in this study offer insights into potential lead compounds as therapeutic inhibitors. CLRs such as DC-SIGN and L-SIGN can promote the “trans” infection of viruses such as HIV and SARS-CoV (56, 57) and the release of proinflammatory cytokines and chemokines (58). A blocking reagent rationally designed based on the ligands of these CLRs may block the detrimental effects. In this regard, a recent study suggested that mannose can ameliorate autoimmune diabetes and airway inflammation by inducing regulatory T cells in a mouse model (59). As the mechanism is unclear, it would be interesting to see whether certain oligomannose-based drugs have a stronger effect.
Mannose has also been tested for its ability to prevent recurrent and incidental urinary tract infection by blocking FimH from adhesion to urinary epithelial cells, but the doses are very high (11, 60). The better binding trisaccharide identified in our study could facilitate the design of a glycomimetic reagent with reduced dosage amounts. Uromodulin, an abundant protein in human urine, was reported to be a multivalent decoy for FimH, resulting in pathogen clearance (61). It would be interesting to identify whether uromodulin carries such a binding motif and whether it will change with aging and with pathological status. Moreover, many other pathogens such as Klebsiella pneumoniae, Shigella flexneri, and Salmonella typhimurium also express type 1 fimbriae. Whether these pathogens can interact with oligomannoses warrants further exploration.
The oligomannose array developed in this study holds promise in identification and characterization of all types of mannose-binding proteins. Apart from the CLRs, bacterial adhesins, and anti-glycan antibodies exemplified in this study, there is a vast number of glycosyltransferases and glycosidases present in the genome of microbes that are largely unexplored in terms of substrate specificity. Encompassing a comprehensive group of closely related isomers with defined linkages, our oligomannose array provides a suitable platform for the scientific community to study the hitherto understudied mammalian and microbial mannose-binding proteins and glycoenzymes.
MATERIALS AND METHODS
Materials
Glycans numbered #1 to 79 in a quantity of 50 to 500 μg were obtained through SBIR-STTR grant from Omicron Biochemicals Inc. These glycans were produced by chemical synthesis and analyzed by nuclear magnetic resonance, HPLC, and MS by the company (the detailed results are provided in data file S3). Results suggested that the purity of each compound was >95%. Each glycan was reconstituted in H2O to prepare a stock solution. Five nanomoles of each glycan was taken for ESI-MS/MS analysis, and the remaining samples were lyophilized for AEAB or F-MAPA conjugation. The plasmid encoding the soluble domain of the glycosyltransferase GNPTAB following a secretion signal and N-terminal His and green fluorescent protein tags (pGEn2-DEST vector) was provided by K. Moremen (62). Human DC-SIGN (Fc-tag), Langerin (His-tag), DC-SIGNR (Fc-tag), and Dectin-2 (Fc-tag) were purchased from R&D Systems (161-DC-050, 2088-LN-050) and Sino Biological (10559-H01H, 10250-H01H), respectively. Among the three recombinant FimH constructs, the C-terminal His tag (FimHL-His) and the full-length FimH with N-terminal extension of FimG (FimH.DsG) were generated in house as previously reported (63, 64). A full-length FimH with N-terminal His-GST tag (FimH-His) was purchased from Signalway Antibody (#AP71165). Biotinylated plant lectins ConA, GNA, GNA, NPA, and HHL were from Vector Laboratories. The cyanine 5-streptavidin–labeled streptavidin (Cy5-SA) was purchased from Invitrogen.
GNPTAB was generated following protocols published previously (65). In brief, suspension and serum-free adapted human embryonic kidney (HEK) 293 cells (Freestyle 293-F cells, Invitrogen) were transiently transfected with the GNPTAB plasmid using polyethyleneimine. Five to 7 days after transfection, protein was purified from the cultural supernatant with His-Pur Ni-NTA resin (Thermo Fisher Scientific). After elution with imidazole-containing buffer [50 mM sodium phosphate, 300 mM sodium chloride, and 400 mM imidazole (pH 8.0)], GNPTAB was dialyzed against storage buffer [20 mM tris (pH 7.5) with 300 mM sodium chloride], snap-frozen, and stored at −80°C until use.
ESI-MS/MS analysis
Negative-ion ESI-MS analysis and HCD/CID-MS/MS were performed on a Fusion Lumos mass spectrometer from Thermo Fisher Scientific (San Jose, CA). The glycan samples were dissolved in acetonitrile:H2O (1:1, v/v) and injected directly into a mass spectrometer at a flow rate of 3 μl/min. The ion source was set at 1.8-kV spray voltage in negative-ion mode and ion transfer tube temperature at 200°C for efficient desolvation and ionization. MS scans were performed in the orbitrap at a resolution of 120,000. A radio frequency (RF) lens was set at 60%, and automatic gain control (AGC) target was set at 5E4 for the full MS scan. The corresponding precursors were manually selected for MS/MS analysis. HCD collision energy was adjusted between 2 and 18 for optimal fragmentation (detailed settings used for individual samples are listed in table S1). MS/MS and MS3 scans were performed in orbitrap at a resolution of 120,000 or in ion trap, within a mass range of m/z 150—molecular ion. AGC target was set at 5E4. The acquired spectra were summed for presentation. All the spectra were manually annotated and interpreted. All tandem MS spectra of the glycans analyzed in this study are available under the following link: ftp://massive.ucsd.edu/MSV000086808/ (we recommend using Firefox browser to access this collection of large raw data files).
Conjugation of oligomannose glycans
AEAB conjugation and purification were performed following the protocols published previously (66) with some modifications. AEAB and NaBH3CN stock solutions were prepared with acetic acid:DMSO (dimethyl sulfoxide) (3:7, v/v) at 88 and 64 mg/ml, respectively. To lyophilized glycans (30 to 200 nmol), 200 equiv AEAB and 400 equiv NaBH3CN were added. The reaction mixture was vortexed for 1 min and incubated on a shaker at 65°C for 4 hours. To the reaction mixture, 10 volumes of acetonitrile were added, and the mixture was left in −20°C for 2 hours. The supernatant was removed following centrifugation. The pellet was reconstituted in H2O and loaded onto a prewashed Sep-Pak Vac NH2 cartridge (Waters Corp.). The column was washed with acetonitrile and 85% acetonitrile, and the AEAB conjugate was eluted with 100 mM NH4OAc and lyophilized to dry. The glycan-AEAB conjugate was reconstituted in H2O and loaded on a prewashed Sep-Pak C18 cartridge (Waters Corp.) for desalting. The column was washed with H2O, and the purified glycan-AEAB conjugate was eluted with 50% acetonitrile (with 0.1% trifluoroacetic acid) and dried by lyophilization.
F-MAPA conjugation was performed following the protocol (39) with the six phosphorylated oligomannose glycans (#91 to 96). F-MAPA, sodium acetate, and 2-amino-5-methoxybenzoic acid (2-AM) stock solutions were prepared with acetic acid:DMSO (3:7, v/v) at 350 mM, 500 mM and 10 mM, respectively. To each lyophilized glycan, 15, 11, and 0.8 μl of the F-MAPA, NaOAc, and 2-AM stock solutions were added. The reaction mixture was vortexed for 1 min and incubated on a shaker at 65°C for 4 hours. After the reaction, 10 volumes of ethyl acetate were added and the mixture was left at −20°C for 1 hour. The reaction mixture was centrifuged, and the supernatant was removed. The pellet was reconstituted in H2O and loaded onto a Sep-Pak C18 cartridge (Waters Corp.) prewashed with 10 column volumes of acetonitrile and H2O. The column was washed with H2O, and the F-MAPA conjugate was eluted with 30% acetonitrile. The acetonitrile was removed by Speed-Vac, and the sample was dried by lyophilization.
The purified AEAB and F-MAPA glycan conjugates were analyzed by HPLC and matrix-assisted laser desorption/ionization (MALDI)–MS as shown below before further use. The estimated yields of the glycan-AEAB conjugates and the glycan-F-MAPA conjugates were 30 to 60% and 10 to 30%, respectively. Upon passing the quality test, the glycan-AEAB conjugates can be directly used in microarray printing, whereas the glycan–F-MAPA conjugates were further treated with 5% methylpiperidine for 30 min to remove the Fmoc. The reaction mixture was passed through a Sep-Pak C18 cartridge, the H2O elution fractions were pooled and lyophilized, and the glycan-MAPA conjugate was ready for microarray printing.
Enzymatic reactions with GNPTAB
AEAB-conjugated oligomannose glycans (2 nmol) were lyophilized in Eppendorf tube before they were taken up in tris buffer [100 mM (pH 7.5)] with 50 mM MgCl2 and 50 mM MnCl2. To the solution, 1-μl uridine diphosphate (UDP)–GlcNAc (50 mM) and 1-μl GNPTAB stock solutions (1 mg/ml) were added and the mixture was incubated at 37°C. At 12 hours, 1 μl each of the UDP-GlcNAc and enzyme stock was added to continue the reaction for another 12 hours. For preparation purpose, 50 nmol of TRI-005 and TRI-009 and 40 nmol of TRI-007 and TET-007 were reconstituted in tris buffer, in which stocks of UDP-GlcNAc and GNPTAB were added to a final concentration of 10 mM and 0.2 mg/ml, respectively. The final concentration of the glycan was 1 nmol/μl. Incubation was at 37°C for 60 hours. The reaction was stopped by freezing at −80°C, and an aliquot of each reaction mixture was subjected to HPLC analysis. The products of TRI-005, TRI-009, TRI-007, and TET-007 were purified by semipreparative HPLC as shown below.
Generation of the Asn-linked chitobiose-containing Man4-GlcNAc2 to Man9-GlcNAc2
Extensive pronase treatment was performed to generate Asn-linked chitobiose-containing high mannose N-glycans. RNase B is known to contain high mannose-type glycans, and their structures have been extensively studied previously (34, 54). Purified bovine RNase B and RNase A mixture (234.7 mg; Worthington Biochemical Corp.) was dissolved in tris buffer [100 mM (pH 8.0)], to which 8.8 mg of pronase (EMD Millipore) was added. The incubation was at 55°C for 72 hours, and the same amount of the enzyme was added every 12 hours. The product was loaded on Sep-Pak C18 (Waters Corp.) preconditioned with acetonitrile and H2O. The flow-through and H2O wash were pooled and loaded on a Supelclean ENVI-Carb column (Sigma-Aldrich) prewashed with acetonitrile and H2O. The column was washed with H2O and eluted with 10, 20, and 50% acetonitrile. Fractions were monitored by MALDI-MS, and those containing targeted glycans were combined and dried by Speed-Vac and lyophilization.
The mixture was labeled with Fmoc as described previously (65) to facilitate fine separation. Semipreparative HPLC on amine-functionalized solid phase gave rise to >3 μmol Fmoc-labeled Man5-GlcNAc2-Asn to Man9-GlcNAc2-Asn (described below). Moreover, a small amount of Man4-GlcNAc2-Asn-Fmoc was also purified for the first time. All purified components were analyzed by HPLC and MALDI–time-of-flight (TOF) MS and confirmed their purity (fig. S2). These glycans were subject to de-Fmoc reaction as described for F-MAPA to yield the purified Man4-GlcNAc2-Asn to Man9-GlcNAc2-Asn.
High-performance liquid chromatography
Analysis and semipreparation of the AEAB- and F-MAPA–glycan and the reaction product by GNPTAB were performed on a Shimadzu HPLC CBM-20A system, equipped with a UV detector (SPD-20AV) and a fluorescence detector (RF-20A). For analysis, the samples were chromatographed on a Luna 5-μm NH2 column (250 × 4.6 mm, Phenomenex) with a concentration of 16 to 40% H2O and 4 to 50% 250 mM ammonium acetate over 60 min. The GNPTAB products had later elution time in comparison to the substrate. For semipreparation, the separation was on a Luna 5-μm NH2 column (250 mm by 10 mm, Phenomenex) with either the same gradient or a shallower gradient to resolve the Fmoc-labeled Man4-GlcNAc2-Asn to Man9-GlcNAc2-Asn using 16 to 24% H2O and 4 to 21% 250 mM ammonium acetate over 50 min. Detection of the AEAB-glycans was at UV 330 nm, fluorescence at excitation (Ex) = 330 nm, and emission (Em) = 420 nm; detection of F-MAPA-glycans was at UV 254 nm, fluorescence Ex = 254, and Em = 340, respectively. LNnT-AEAB and LNnT-F-MAPA conjugates with known concentrations were used for quantification.
Matrix-assisted laser desorption/ionization–mass spectrometry
MALDI-TOF MS analyses of the glycan derivatives were performed on an UltrafleXtreme MALDI-TOF/TOF system from Bruker Daltonics (Billerica, MA) equipped with a Smartbeam II laser. The analyte was reconstituted in H2O, and an aliquot (~100 pmol) was taken to mix with 2,5-dihydroxybenzoic acid (10 mg/ml in MeOH:H2O = 1:1). The mixture was deposited to a target plate (Bruker Daltonics) and subject to MALDI-TOF MS analysis. Spectrum between m/z 500 and 3000 was acquired under reflectron positive mode. Each MS spectrum presented an accumulation of more than 10,000 laser shots.
Microarray printing and analyses
Microarray printing and analyses were conducted largely as reported previously (65). In brief, the purified glycan conjugates were reconstituted in phosphate buffer [100 mM sodium phosphate (pH 8.5)] at a final concentration of 100 μM (probes 1 to 79, 81, and 83 to 84), 50 μM (probes 80 and 82), or 20 μM (probes 85 to 100). Each conjugate was printed in four replicates, ~330 pl each, on N-hydroxysuccinimide (NHS)–coated glass slides (Schott AG) using a sciFLEXARRAYER S11 instrument (Scienion, Germany). The slides were left for reaction to proceed overnight at 70% relative humidity, followed by blocking with 50 mM ethanolamine in 100 mM borate buffer (pH 8.5) for 1 hour and washing with phosphate-buffered saline (0.05% Tween 20) and H2O. Then, the slides were sealed and stored at −20°C until use.
The microarray slides were rehydrated with TSM buffer (20 mM tris-HCl, 150 mM sodium chloride, 2 mM calcium chloride, and 2 mM magnesium chloride) before incubation with biotinylated lectins, CLRs, bacterial adhesin, and anti-HIV bNAbs. Biotinylated plant lectins GNA, NPA, and HHL were used at 10 μg/ml, while ConA and RCA were used at 1 μg/ml. The recombinant human CLRs were tested at the following concentrations: DC-SIGN-Fc, 1 and 5 μg/ml; L-SIGN-Fc, 1 and 10 μg/ml; Dectin-2-Fc, 1 and 5 μg/ml; and Langerin-His, 1 and 25 μg/ml. All the protein samples above were prepared with 1% bovine serum albumin (BSA) in TSM buffer with additional Ca2+ (TSMBB-Ca, 20 mM tris-HCl, 150 mM sodium chloride, 5 mM calcium chloride, 2 mM magnesium chloride, and 0.05% Tween 20). The recombinant FimH.DsG and FimHL-His were biotinylated with EZ-Link Sulfo-NHS-LC-LC-biotin (Thermo Fisher Scientific) and quantified on NanoDrop before use at 50 and 200 μg/ml for microarray analysis. The anti-HIV broadly neutralizing monoclonal antibodies (mAbs) were tested at 25 μg/ml. The FimH and the mAbs were prepared with 1% BSA in TSM buffer (TSMBB; 20 mM tris-HCl, 150 mM sodium chloride, 2 mM calcium chloride, 2 mM magnesium chloride, and 0.05% Tween 20). The binding of biotinylated plant lectins and the biotinylated FimH.DsG and FimHL were detected with Cy5-SA (Invitrogen) at 0.5 μg/ml. The human immunoglobulin G (IgG) Fc-tagged proteins DC-SIGN and Dectin-2 were detected with Alexa Fluor 488–labeled goat anti-human IgG (H+L) (Invitrogen) at 5 μg/ml. The His-tagged Langerin was detected with mouse anti-His IgG (H8, Invitrogen MA1-21315) at 5 μg/ml followed by Alexa Fluor 633 goat anti-mouse IgG (H+L) (Invitrogen A21052) at 5 μg/ml. The Fc-tagged L-SIGN and HIV mAbs were detected with cyanine 3–goat anti-human IgG (Jackson ImmunoResearch 109-165-008, Fcγ specific) at 5 μg/ml.
The signal intensities were quantified using GenePix Pro 7 that is associated with the microarray scanner and processed by Excel (Microsoft). The results are shown as relative fluorescence units (RFUs) by averaging the background-subtracted fluorescence signals of four replicate spots, with error bars representing the SD among the four values.
Statistical analysis
The microarray data were processed and analyzed by Excel (Microsoft). For correlation analysis between test samples, GLAD toolkit was used (67).
Acknowledgments
A significant portion of oligomannose glycans were obtained from Omicron Biochemicals through the NIH/NCI/NIGMS SBIR/STTR programs to support glycan availability for binding studies (project number 261201300038C). The following reagents were obtained through the NIH AIDS Reagent Program, Division of AIDS, NIAID, NIH: anti–HIV-1 gp120 monoclonal antibodies 2G12 from Polymun Scientific, and PGT121, PGT126, PGT128, and PGT145 from IAVI. Funding: This work was supported by NIH grants P41GM103694 and R24GM137763 to R.D.C. Author contributions: C.G. and R.D.C. conceived the project. C.G. performed the negative-ion ESI-MS/MS analysis and, together with K.S., summarized the data. B. Eck, C.G., and M.W. performed glycan conjugation, and N.J. helped in analyzing the quality of the conjugates. C.G. purified high mannose N-glycans from RNase B and performed the enzymatic reaction with GNPTAB, which was produced by M.S.H. C.G., T.R.M., A.M.M., and K.J.B. performed the glycan microarray experiments and, together with A.Y.M. and J.H.-M., analyzed the data. C.G. and Y.M. performed the dose response and inhibition assays and analyzed the data. D.E. and B. Er generated the recombinant FimH. R.D.C. supervised all experiments. C.G. wrote the first draft of the manuscript, and all authors contributed to the final version. Competing interests: The authors declare that they have no competing interests. Data and materials availability: All data are available in the manuscript or in the Supplementary Materials, and microarray data will be publicly available online through the National Center for Functional Glycomics website. All tandem MS spectra of the glycans analyzed in this study are available under the following link, and we recommend using Firefox browser to access these large files: ftp://massive.ucsd.edu/MSV000086808/.
SUPPLEMENTARY MATERIALS
Supplementary material for this article is available at http://advances.sciencemag.org/cgi/content/full/7/24/eabf6834/DC1
REFERENCES AND NOTES
- 1.P. Stanley, N. Taniguchi, M. Aebi, N-glycans, Chapter 9 in Essentials of Glycobiology, A. Varki, R. D. Cummings, J. D. Esko, P. Stanley, G. W. Hart, M. Aebi, A. G. Darvill, T. Kinoshita, N. H. Packer, J. H. Prestegard, R. L. Schnaar, P. H. Seeberger, Eds. (Cold Spring Harbor NY, Cold Spring Harbor Laboratory Press, ed. 3, 2017). [Google Scholar]
- 2.Rollenske T., Szijarto V., Lukasiewicz J., Guachalla L. M., Stojkovic K., Hartl K., Stulik L., Kocher S., Lasitschka F., al-Saeedi M., Schröder-Braunstein J., von Frankenberg M., Gaebelein G., Hoffmann P., Klein S., Heeg K., Nagy E., Nagy G., Wardemann H., Cross-specificity of protective human antibodies against Klebsiella pneumoniae LPS O-antigen. Nat. Immunol. 19, 617–624 (2018). [DOI] [PubMed] [Google Scholar]
- 3.Gow N. A. R., van de Veerdonk F. L., Brown A. J. P., Netea M. G., Candida albicans morphogenesis and host defence: Discriminating invasion from colonization. Nat. Rev. Microbiol. 10, 112–122 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Doores K. J., Bonomelli C., Harvey D. J., Vasiljevic S., Dwek R. A., Burton D. R., Crispin M., Scanlan C. N., Envelope glycans of immunodeficiency virions are almost entirely oligomannose antigens. Proc. Natl. Acad. Sci. U.S.A. 107, 13800–13805 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.C. Gao, J. Zeng, N. Jia, K. Stavenhagen, Y. Matsumoto, H. Zhang, J. Li, A. J. Hume, E. Mühlberger, I. van Die, J. Kwan, K. Tantisira, A. Emili, R. D. Cummings, SARS-CoV-2 spike protein interacts with multiple innate immune receptors. bioRxiv 2020.07.29.227462 [Preprint]. 30 July 2020. 10.1101/069187. [DOI]
- 6.Dahms N. M., Lobel P., Kornfeld S., Mannose 6-phosphate receptors and lysosomal enzyme targeting. J. Biol. Chem. 264, 12115–12118 (1989). [PubMed] [Google Scholar]
- 7.Tannous A., Pisoni G. B., Hebert D. N., Molinari M., N-linked sugar-regulated protein folding and quality control in the ER. Semin. Cell Dev. Biol. 41, 79–89 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Jones C. H., Pinkner J. S., Roth R., Heuser J., Nicholes A. V., Abraham S. N., Hultgren S. J., FimH adhesin of type 1 pili is assembled into a fibrillar tip structure in the Enterobacteriaceae. Proc. Natl. Acad. Sci. U.S.A. 92, 2081–2085 (1995). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Zhou G., Mo W. J., Sebbel P., Min G., Neubert T. A., Glockshuber R., Wu X. R., Sun T. T., Kong X. P., Uroplakin Ia is the urothelial receptor for uropathogenic Escherichia coli: Evidence from in vitro FimH binding. J. Cell Sci. 114, 4095–4103 (2001). [DOI] [PubMed] [Google Scholar]
- 10.Sauer M. M., Jakob R. P., Luber T., Canonica F., Navarra G., Ernst B., Unverzagt C., Maier T., Glockshuber R., Binding of the bacterial adhesin FimH to its natural, multivalent high-mannose type glycan targets. J. Am. Chem. Soc. 141, 936–944 (2019). [DOI] [PubMed] [Google Scholar]
- 11.Hatton N. E., Baumann C. G., Fascione M. A., Developments in mannose-based treatments for Uropathogenic Escherichia coli induced urinary tract infections. Chembiochem 22, 613–629 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.van Kooyk Y., Rabinovich G. A., Protein-glycan interactions in the control of innate and adaptive immune responses. Nat. Immunol. 9, 593–601 (2008). [DOI] [PubMed] [Google Scholar]
- 13.Brown G. D., Willment J. A., Whitehead L., C-type lectins in immunity and homeostasis. Nat. Rev. Immunol. 18, 374–389 (2018). [DOI] [PubMed] [Google Scholar]
- 14.Shivatare S. S., Chang S. H., Tsai T. I., Tseng S. Y., Shivatare V. S., Lin Y. S., Cheng Y. Y., Ren C. T., Lee C. C. D., Pawar S., Tsai C. S., Shih H. W., Zeng Y. F., Liang C. H., Kwong P. D., Burton D. R., Wu C. Y., Wong C. H., Modular synthesis of N-glycans and arrays for the hetero-ligand binding analysis of HIV antibodies. Nat. Chem. 8, 338–346 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Walker L. M., Huber M., Doores K. J., Falkowska E., Pejchal R., Julien J. P., Wang S. K., Ramos A., Chan-Hui P. Y., Moyle M., Mitcham J. L., Hammond P. W., Olsen O. A., Phung P., Fling S., Wong C. H., Phogat S., Wrin T., Simek M. D., Principal Investigators P. G., Koff W. C., Wilson I. A., Burton D. R., Poignard P., Broad neutralization coverage of HIV by multiple highly potent antibodies. Nature 477, 466–470 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Reinhold V. N., Reinhold B. B., Costello C. E., Carbohydrate molecular weight profiling, sequence, linkage, and branching data: ES-MS and CID. Anal. Chem. 67, 1772–1784 (1995). [DOI] [PubMed] [Google Scholar]
- 17.Harvey D. J., Fragmentation of negative ions from carbohydrates: Part 2. Fragmentation of high-mannose N-linked glycans. J. Am. Soc. Mass Spectrom. 16, 631–646 (2005). [DOI] [PubMed] [Google Scholar]
- 18.Harvey D. J., Fragmentation of negative ions from carbohydrates: Part 3. Fragmentation of hybrid and complex N-linked glycans. J. Am. Soc. Mass Spectrom. 16, 647–659 (2005). [DOI] [PubMed] [Google Scholar]
- 19.Ashwood C., Lin C.-H., Thaysen-Andersen M., Packer N. H., Discrimination of isomers of released N- and O-glycans using diagnostic product ions in negative ion PGC-LC-ESI-MS/MS. J. Am. Soc. Mass Spectrom. 29, 1194–1209 (2018). [DOI] [PubMed] [Google Scholar]
- 20.Everest-Dass A. V., Kolarich D., Campbell M. P., Packer N. H., Tandem mass spectra of glycan substructures enable the multistage mass spectrometric identification of determinants on oligosaccharides. Rapid Commun. Mass Spectrom. 27, 931–939 (2013). [DOI] [PubMed] [Google Scholar]
- 21.Everest-Dass A. V., Abrahams J. L., Kolarich D., Packer N. H., Campbell M. P., Structural feature ions for distinguishing N- and O-linked glycan isomers by LC-ESI-IT MS/MS. J. Am. Soc. Mass Spectrom. 24, 895–906 (2013). [DOI] [PubMed] [Google Scholar]
- 22.Chai W., Piskarev V., Lawson A. M., Negative-ion electrospray mass spectrometry of neutral underivatized oligosaccharides. Anal. Chem. 73, 651–657 (2001). [DOI] [PubMed] [Google Scholar]
- 23.Chai W., Lawson A., Piskarev V., Branching pattern and sequence analysis of underivatized oligosaccharides by combined MS/MS of singly and doubly charged molecular ions in negative-ion electrospray mass spectrometry. J. Am. Soc. Mass Spectrom. 13, 670–679 (2002). [DOI] [PubMed] [Google Scholar]
- 24.Palma A. S., Liu Y., Zhang H., Zhang Y., McCleary B. V., Yu G., Huang Q., Guidolin L. S., Ciocchini A. E., Torosantucci A., Wang D., Carvalho A. L., Fontes C. M. G. A., Mulloy B., Childs R. A., Feizi T., Chai W., Unravelling glucan recognition systems by glycome microarrays using the designer approach and mass spectrometry. Mol. Cell. Proteomics 14, 974–988 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Harvey D. J., Crispin M., Scanlan C., Singer B. B., Lucka L., Chang V. T., Radcliffe C. M., Thobhani S., Yuen C. T., Rudd P. M., Differentiation between isomeric triantennary N-linked glycans by negative ion tandem mass spectrometry and confirmation of glycans containing galactose attached to the bisecting (β1-4-GlcNAc) residue in N-glycans from IgG. Rapid Commun. Mass Spectrom. 22, 1047–1052 (2008). [DOI] [PubMed] [Google Scholar]
- 26.Gao C., Zhang Y., Liu Y., Feizi T., Chai W., Negative-ion electrospray tandem mass spectrometry and microarray analyses of developmentally regulated antigens based on Type 1 and Type 2 backbone sequences. Anal. Chem. 87, 11871–11878 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Möginger U., Grunewald S., Hennig R., Kuo C. W., Schirmeister F., Voth H., Rapp E., Khoo K. H., Seeberger P. H., Simon J. C., Kolarich D., Alterations of the human skin N- and O-glycome in basal cell carcinoma and squamous cell carcinoma. Front. Oncol. 8, 70 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Madunić K., Zhang T., Mayboroda O. A., Holst S., Stavenhagen K., Jin C., Karlsson N. G., Lageveen-Kammeijer G. S. M., Wuhrer M., Colorectal cancer cell lines show striking diversity of their O-glycome reflecting the cellular differentiation phenotype. Cell. Mol. Life Sci. 78, 337–350 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Sethi M. K., Thaysen-Andersen M., Smith J. T., Baker M. S., Packer N. H., Hancock W. S., Fanayan S., Comparative N-glycan profiling of colorectal cancer cell lines reveals unique bisecting GlcNAc and α-2,3-linked sialic acid determinants are associated with membrane proteins of the more metastatic/aggressive cell lines. J. Proteome Res. 13, 277–288 (2014). [DOI] [PubMed] [Google Scholar]
- 30.Zhu Y., Yan M., Lasanajak Y., Smith D. F., Song X., Large scale preparation of high mannose and paucimannose N-glycans from soybean proteins by oxidative release of natural glycans (ORNG). Carbohydr. Res. 464, 19–27 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Lubkowski J., Durbin S. V., Silva M. C. C., Farnsworth D., Gildersleeve J. C., Oliva M. L. V., Wlodawer A., Structural analysis and unique molecular recognition properties of a Bauhinia forficata lectin that inhibits cancer cell growth. FEBS J. 284, 429–450 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Gao C., Wei M., McKitrick T. R., McQuillan A. M., Heimburg-Molinaro J., Cummings R. D., Glycan microarrays as chemical tools for identifying glycan recognition by immune proteins. Front. Chem. 7, 833 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Prien J. M., Ashline D. J., Lapadula A. J., Zhang H., Reinhold V. N., The high mannose glycans from bovine ribonuclease B isomer characterization by ion trap MS. J. Am. Soc. Mass Spectrom. 20, 539–556 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Wei J., Tang Y., Bai Y., Zaia J., Costello C. E., Hong P., Lin C., Toward automatic and comprehensive glycan characterization by online PGC-LC-EED MS/MS. Anal. Chem. 92, 782–791 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.van Meel E., Lee W. S., Liu L., Qian Y., Doray B., Kornfeld S., Multiple domains of GlcNAc-1-phosphotransferase mediate recognition of lysosomal enzymes. J. Biol. Chem. 291, 8295–8307 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Qian Y., Lee I., Lee W. S., Qian M., Kudo M., Canfield W. M., Lobel P., Kornfeld S., Functions of the α, β, and γ subunits of UDP-GlcNAc:Lysosomal enzyme N-acetylglucosamine-1-phosphotransferase. J. Biol. Chem. 285, 3360–3370 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Varki A., Kornfeld S., Structural studies of phosphorylated high mannose-type oligosaccharides. J. Biol. Chem. 255, 10847–10858 (1980). [PubMed] [Google Scholar]
- 38.Song X., Lasanajak Y., Olson L. J., Boonen M., Dahms N. M., Kornfeld S., Cummings R. D., Smith D. F., Glycan microarray analysis of P-type lectins reveals distinct phosphomannose glycan recognition. J. Biol. Chem. 284, 35201–35214 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Wei M., McKitrick T. R., Mehta A. Y., Gao C., Jia N., McQuillan A. M., Heimburg-Molinaro J., Sun L., Cummings R. D., Novel reversible fluorescent glycan linker for functional glycomics. Bioconjug. Chem. 30, 2897–2908 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Barre A., Bourne Y., Van Damme E. J. M., Rougé P., Overview of the structure–function relationships of mannose-specific lectins from plants, algae and fungi. Int. J. Mol. Sci. 20, 254 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Kaku H., Van Damme E. J. M., Peumans W. J., Goldstein I. J., Carbohydrate-binding specificity of the daffodil (Narcissus pseudonarcissus) and amaryllis (Hippeastrum hybr.) bulb lectins. Arch. Biochem. Biophys. 279, 298–304 (1990). [DOI] [PubMed] [Google Scholar]
- 42.Feinberg H., Mitchell D. A., Drickamer K., Weis W. I., Structural basis for selective recognition of oligosaccharides by DC-SIGN and DC-SIGNR. Science 294, 2163–2166 (2001). [DOI] [PubMed] [Google Scholar]
- 43.Guo Y., Feinberg H., Conroy E., Mitchell D. A., Alvarez R., Blixt O., Taylor M. E., Weis W. I., Drickamer K., Structural basis for distinct ligand-binding and targeting properties of the receptors DC-SIGN and DC-SIGNR. Nat. Struct. Mol. Biol. 11, 591–598 (2004). [DOI] [PubMed] [Google Scholar]
- 44.McGreal E. P., Rosas M., Brown G. D., Zamze S., Wong S. Y. C., Gordon S., Martinez-Pomares L., Taylor P. R., The carbohydrate-recognition domain of Dectin-2 is a C-type lectin with specificity for high mannose. Glycobiology 16, 422–430 (2006). [DOI] [PubMed] [Google Scholar]
- 45.Wellens A., Garofalo C., Nguyen H., van Gerven N., Slättegård R., Hernalsteens J. P., Wyns L., Oscarson S., de Greve H., Hultgren S., Bouckaert J., Intervening with urinary tract infections using anti-adhesives based on the crystal structure of the FimH–oligomannose-3 complex. PLOS ONE 3, e2040 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Pejchal R., Doores K. J., Walker L. M., Khayat R., Huang P. S., Wang S. K., Stanfield R. L., Julien J. P., Ramos A., Crispin M., Depetris R., Katpally U., Marozsan A., Cupo A., Maloveste S., Liu Y., McBride R., Ito Y., Sanders R. W., Ogohara C., Paulson J. C., Feizi T., Scanlan C. N., Wong C. H., Moore J. P., Olson W. C., Ward A. B., Poignard P., Schief W. R., Burton D. R., Wilson I. A., A potent and broad neutralizing antibody recognizes and penetrates the HIV glycan shield. Science 334, 1097–1103 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Julien J.-P., Sok D., Khayat R., Lee J. H., Doores K. J., Walker L. M., Ramos A., Diwanji D. C., Pejchal R., Cupo A., Katpally U., Depetris R. S., Stanfield R. L., McBride R., Marozsan A. J., Paulson J. C., Sanders R. W., Moore J. P., Burton D. R., Poignard P., Ward A. B., Wilson I. A., Broadly neutralizing antibody PGT121 allosterically modulates CD4 binding via recognition of the HIV-1 gp120 V3 base and multiple surrounding glycans. PLOS Pathog. 9, e1003342 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Calarese D. A., Scanlan C. N., Zwick M. B., Deechongkit S., Mimura Y., Kunert R., Zhu P., Wormald M. R., Stanfield R. L., Roux K. H., Kelly J. W., Rudd P. M., Dwek R. A., Katinger H., Burton D. R., Wilson I. A., Antibody domain exchange is an immunological solution to carbohydrate cluster recognition. Science 300, 2065–2071 (2003). [DOI] [PubMed] [Google Scholar]
- 49.Dunlop D. C., Bonomelli C., Mansab F., Vasiljevic S., Doores K. J., Wormald M. R., Palma A. S., Feizi T., Harvey D. J., Dwek R. A., Crispin M., Scanlan C. N., Polysaccharide mimicry of the epitope of the broadly neutralizing anti-HIV antibody, 2G12, induces enhanced antibody responses to self oligomannose glycans. Glycobiology 20, 812–823 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Gray C. J., Migas L. G., Barran P. E., Pagel K., Seeberger P. H., Eyers C. E., Boons G. J., Pohl N. L. B., Compagnon I., Widmalm G., Flitsch S. L., Advancing solutions to the carbohydrate sequencing challenge. J. Am. Chem. Soc. 141, 14463–14479 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Veillon L., Huang Y., Peng W., Dong X., Cho B. G., Mechref Y., Characterization of isomeric glycan structures by LC-MS/MS. Electrophoresis 38, 2100–2114 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Pagel K., Harvey D. J., Ion mobility–mass spectrometry of complex carbohydrates: Collision cross sections of sodiated N-linked glycans. Anal. Chem. 85, 5138–5145 (2013). [DOI] [PubMed] [Google Scholar]
- 53.Hofmann J., Hahm H. S., Seeberger P. H., Pagel K., Identification of carbohydrate anomers using ion mobility–mass spectrometry. Nature 526, 241–244 (2015). [DOI] [PubMed] [Google Scholar]
- 54.Harvey D. J., Seabright G. E., Vasiljevic S., Crispin M., Struwe W. B., Isomer information from ion mobility separation of high-mannose glycan fragments. J. Am. Soc. Mass Spectrom. 29, 972–988 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Cummings R. D., Pierce J. M., The challenge and promise of glycomics. Chem. Biol. 21, 1–15 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Geijtenbeek T. B. H., Kwon D. S., Torensma R., van Vliet S. J., van Duijnhoven G. C., Middel J., Cornelissen I. L., Nottet H. S., KewalRamani V. N., Littman D. R., Figdor C. G., van Kooyk Y., DC-SIGN, a dendritic cell–specific HIV-1-binding protein that enhances trans-infection of T cells. Cell 100, 587–597 (2000). [DOI] [PubMed] [Google Scholar]
- 57.Yang Z.-Y., Huang Y., Ganesh L., Leung K., Kong W. P., Schwartz O., Subbarao K., Nabel G. J., pH-dependent entry of severe acute respiratory syndrome coronavirus is mediated by the spike glycoprotein and enhanced by dendritic cell transfer through DC-SIGN. J. Virol. 78, 5642–5650 (2004). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Sato K., Yang X. L., Yudate T., Chung J. S., Wu J., Luby-Phelps K., Kimberly R. P., Underhill D., Cruz P. D. Jr., Ariizumi K., Dectin-2 is a pattern recognition receptor for fungi that couples with the Fc receptor γ chain to induce innate immune responses. J. Biol. Chem. 281, 38854–38866 (2006). [DOI] [PubMed] [Google Scholar]
- 59.Zhang D., Chia C., Jiao X., Jin W., Kasagi S., Wu R., Konkel J. E., Nakatsukasa H., Zanvit P., Goldberg N., Chen Q., Sun L., Chen Z. J., Chen W. J., D-mannose induces regulatory T cells and suppresses immunopathology. Nat. Med. 23, 1036–1045 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Lenger S. M., Bradley M. S., Thomas D. A., Bertolet M. H., Lowder J. L., Sutcliffe S., D-mannose vs other agents for recurrent urinary tract infection prevention in adult women: A systematic review and meta-analysis. Am. J. Obstet. Gynecol. 223, 265.e1–265.e3 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Weiss G. L., Stanisich J. J., Sauer M. M., Lin C. W., Eras J., Zyla D. S., Trück J., Devuyst O., Aebi M., Pilhofer M., Glockshuber R., Architecture and function of human uromodulin filaments in urinary tract infections. Science 369, 1005–1010 (2020). [DOI] [PubMed] [Google Scholar]
- 62.Moremen K. W., Ramiah A., Stuart M., Steel J., Meng L., Forouhar F., Moniz H. A., Gahlay G., Gao Z., Chapla D., Wang S., Yang J.-Y., Prabhakar P. K., Johnson R., dela Rosa M., Geisler C., Nairn A. V., Seetharaman J., Wu S.-C., Tong L., Gilbert H. J., Baer J. L., Jarvis D. L., Expression system for structural and functional studies of human glycosylation enzymes. Nat. Chem. Biol. 14, 156–162 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Rabbani S., Jiang X., Schwardt O., Ernst B., Expression of the carbohydrate recognition domain of FimH and development of a competitive binding assay. Anal. Biochem. 407, 188–195 (2010). [DOI] [PubMed] [Google Scholar]
- 64.Sauer M. M., Jakob R. P., Eras J., Baday S., Eriş D., Navarra G., Bernèche S., Ernst B., Maier T., Glockshuber R., Catch-bond mechanism of the bacterial adhesin FimH. Nat. Commun. 7, 10738 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Gao C., Hanes M. S., Byrd-Leotis L. A., Wei M., Jia N., Kardish R. J., McKitrick T. R., Steinhauer D. A., Cummings R. D., Unique binding specificities of proteins toward isomeric asparagine-linked glycans. Cell Chem. Biol. 26, 535–547.e4 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Song X., Xia B., Stowell S. R., Lasanajak Y., Smith D. F., Cummings R. D., Novel fluorescent glycan microarray strategy reveals ligands for galectins. Chem. Biol. 16, 36–47 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Mehta A. Y., Cummings R. D., GLAD: GLycan array dashboard, a visual analytics tool for glycan microarrays. Bioinformatics 35, 3536–3537 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary material for this article is available at http://advances.sciencemag.org/cgi/content/full/7/24/eabf6834/DC1


