Testis-specific lncRNA regulates expression of mouse Y chromosome genes and plays a critical role in offspring sex ratio.
Abstract
Heat shock factor 2 (HSF2) regulates the transcription of the male-specific region of the mouse Y chromosome long arm (MSYq) multicopy genes only in testes, but the molecular mechanism underlying this tissue specificity remains largely unknown. Here, we report that the testicular germ cell–specific long noncoding RNA (lncRNA), NR_038002, displays a characteristic spatiotemporal expression pattern in the nuclei of round and elongating spermatids. NR_038002-knockout male mice produced sperm with abnormal head morphology and exhibited reduced fertility accompanied by a female-biased sex ratio in offspring. Molecular analyses revealed that NR_038002 interacts with HSF2 and thereby activates expression of the MSYq genes. We designate NR_038002 as testicular germ cell–specific HSF2-interacting lncRNA (Teshl). Together, our study is the first to demonstrate that the testis specificity of HSF2 activity is regulated by the lncRNA Teshl and establishes a Teshl-HSF2-MSYq molecular axis for normal Y-bearing sperm qualities and consequent balanced offspring sex ratio.
INTRODUCTION
Long noncoding RNAs (lncRNAs) are a class of transcripts longer than 200 nucleotides (nt) with limited protein-coding potential (1). They have been implicated in diverse biological processes, including differentiation and development (2), and regulation of gene expression through cross-talk with DNA, RNA, and proteins (3). Compared with mRNAs, lncRNAs display less sequence conservation (4) and greater tissue-specific expression patterns (5). Although numerous lncRNAs have been reported to be specifically expressed in testes (6), only a few have been characterized (7–15).
Mammalian spermatogenesis is a tightly regulated series of developmental processes that occur in successive mitotic, meiotic, and postmeiotic phases (spermiogenesis) (16). During the pachytene stage of meiosis, autosomes are abundantly transcribed, whereas sex chromosomes are transcriptionally silenced owing to meiotic sex chromatin inactivation (MSCI)—a surveillance mechanism for the largely nonhomologous regions of X and Y chromosomes (17–19). The repressed state of sex chromosomes partially persists throughout postmeiotic phases (20). Nevertheless, some X- and Y-linked genes, specifically those with indispensable functions in sperm differentiation, are transcribed during the postmeiotic phase. For example, loss of the male-specific region of the mouse Y chromosome long arm (MSYq), which contains the multicopy spermatid-expressed genes, Ssty1 and Ssty2 (spermiogenesis-specific transcript on the Y 1 and Y 2, respectively) and Sly (Sycp3-like Y-linked), is associated with sperm head abnormalities and male infertility (21).
During spermatogenesis, heat shock factor 2 (HSF2), a transcription factor involved in corticogenesis and spermatogenesis (22), is expressed in a stage-specific manner in the nuclei of early pachytene spermatocytes and postmeiotically in round spermatids (23). Hsf2–knockout (KO) mice exhibit abnormal sperm chromatin compaction, sperm head abnormalities, and subfertility (24). HSF2 binds the promoters of a number of multicopy gene targets on sex chromosomes and regulates their transcription in the postmeiotic phase (24). However, how the ubiquitously expressed HSF2 regulates the expression of multicopy genes only in testes is largely unknown.
On the basis of these observations, we tested whether NR_038002, testicular germ cell–specific lncRNA, plays a critical role in regulating the expression of HSF2-dependent sex chromosome genes during postmeiotic phases. To explore the functional importance of this testicular germ cell–specific lncRNA, we used CRISPR-Cas9 genome editing to disrupt the genomic locus of NR_038002. Our investigation of transcriptome profiles and results of various biological analyses demonstrate that NR_038002 interacts with HSF2 to activate Y chromosomal multicopy genes during the postmeiotic phase, a function that is critical for regulation of offspring sex ratio.
RESULTS
NR_038002 is a testicular germ cell–specific lncRNA conserved between mice and humans
We previously reported that NR_038002, transcribed from the 1700027A15Rik gene on mouse chromosome 1, is a testicular germ cell–specific lncRNA predicted to have the highest expression levels among identified specific lncRNAs (6). For convenience, we use the term “NR_038002” for both the gene and transcript in this study. To further examine NR_038002, we performed Northern blot analyses on various mouse tissues (Fig. 1A). NR_038002 exhibited a testis-specific expression pattern, with a distinct ~600-nt band. Rapid amplification of complementary DNA ends (RACE), used to identify potential isoforms and 5′ and 3′ ends of NR_038002 (fig. S1A), showed the presence of a few transcripts with slight differences in the 5′ sequence. This is likely due to random variation because only a single TATA box was found. We found two isoforms for NR_038002, differing in the length of poly A tail (fig. S1B). We found that NR_038002 transcription was terminated at position +416, followed by polyadenylation. In silico programs were used to investigate the coding potential of NR_038002. CPAT (25), but not CPC2 (26), predicted that translation of NR_038002 into a protein was highly unlikely (fig. S1C). Sucrose gradient–based polysome fractionation of mouse testes followed by Northern blotting further revealed the presence of NR_038002 transcripts only in low–molecular weight, polysome-free fractions (Fig. 1B), suggesting that NR_038002 does not interact with the translational machinery.
Fig. 1. Identification and characterization of the mouse testis–specific lncRNA, NR_038002.
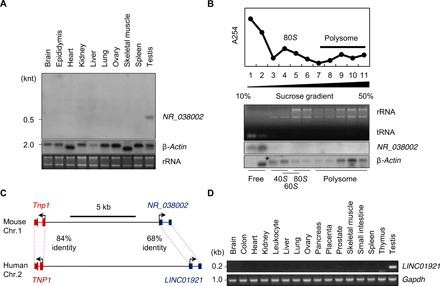
(A) Tissue distribution of NR_038002 in various mouse tissues, as determined by Northern blotting. Middle and bottom panels represent a Northern blot of mouse β-actin mRNA and a denaturing agarose gel image of mouse 28S and 18S ribosomal RNA (rRNA), respectively (loading controls). (B) Polysome fractionation of NR_038002 lncRNA and β-actin mRNA in mouse testes followed by Northern blotting. Top graph: Absorbance at 254 nm for each fraction. Middle: Denaturing agarose gel electrophoresis of RNA extracted from each fraction. tRNA, transfer RNA. (C) Schematic representation of the genomic locus of NR_038002 and LINC01921 adjacent to Tnp1 in humans and mice, respectively. (D) Tissue distribution of LINC01921 in various human tissues, determined by reverse transcription polymerase chain reaction (RT-PCR).
We next examined the sequence conservation of NR_038002 based on PhastCons and PhyloP scores (27) and found that exonic regions of the NR_038002 gene are conserved among diverse mammalian species (fig. S1D). In particular, we identified a human locus, termed LINC01921, that transcribes an RNA with a nucleotide sequence very similar to that of NR_038002 (68% identity in exonic sequence) (Fig. 1C). Notably, such a level of conservation is observed in lncRNAs that are known to be well conserved between mice and humans, including Malat (79%), H19 (71%), Hotair (69%), and Xist (67%). We also found syntenic localization of NR_038002 and LINC01921 genes in mouse and human genomes, respectively, with both being located close to the genomic locus of the Tnp1 (transition protein 1) gene (28) (Fig. 1C). A structural analysis using RNAfold (29) predicted that both transcripts form stem-loops and have an overall similar structure (fig. S1E). Last, a tissue distribution analysis showed that LINC01921 is also specifically expressed in testes (Fig. 1D), indicating additional conservation at the expression level.
NR_038002 is located in nuclei of round and elongating spermatids
We previously reported that NR_038002 is expressed in a germ cell–specific manner, and its expression is limited to the postmeiotic phase (6). To determine exactly when and where NR_038002 is expressed in germ cell developmental stages, we performed in situ hybridization. Signals obtained using an antisense probe were distinctly detected in germ cells of seminiferous tubules, but not in testicular somatic cells, such as Sertoli and Leydig cells (Fig. 2A). In spermatogenic cells, NR_038002 was expressed at the I–III and VII–XII stages of spermatogenesis (7 to 15 steps of spermiogenesis), corresponding to late-stage round spermatids and entire-stage elongating spermatids (Fig. 2B). NR_038002 was located only in nuclei of the spermatids, indicating that NR_038002 is a developmentally regulated nuclear lncRNA expressed in round and elongating spermatids (Fig. 2C).
Fig. 2. Spatiotemporal expression patterns of NR_038002 in mouse testes.
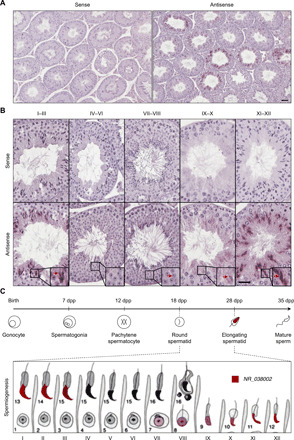
(A) In situ hybridization analysis of NR_038002 in mouse testes using sense and antisense probes. Scale bar, 20 μm. (B) Higher magnification of in situ hybridization in serial sections for determination of expression stages and localization of NR_038002 in seminiferous tubules. NR_038002 expression was detected in nuclei of late round spermatids (weak signal) and elongating spermatids (strong signal). Note that the NR_038002 nuclear signal is different from hematoxylin and eosin (H&E) stain in color and shape (blurred), as indicated with red arrow in VII to VIII. Scale bar, 20 μm. (C) Diagram of 12-stage spermatogenesis represented by Roman numerals and 16-step spermiogenesis indicated by Arabic numbers in mice (53) showing the stages in which NR_038002 (red) is expressed.
To investigate the transcriptional regulatory function of NR_038002 in mouse germ cells, we overexpressed NR_038002 transcripts in GC-2 cells and then performed a microarray analysis of the cells. However, this analysis showed no significant changes in the expression of genes related to spermatogenesis (table S1).
Male mice lacking NR_038002 are subfertile and produce sperm with an abnormal head morphology
To investigate the in vivo function of NR_038002, we generated NR_038002-mutant mice using CRISPR-Cas9 genome editing (Fig. 3A). We ultimately obtained four founder mice, all of which were confirmed through cloning and sequencing of genomic DNAs (fig. S2, A and B). For phenotypic analyses, we selected a single mouse line in which the 894–bp (base pair) genomic region of NR_038002 was deleted and inbred these founder mice to obtain homozygous NR_038002-KO mice. The lack of NR_038002 transcripts was confirmed by Northern blotting and quantitative reverse transcription polymerase chain reaction (qRT-PCR) analyses (Fig. 3, B to D). NR_038002-KO mice were viable and developed normally. There was no significant difference in testis size or testis-to-body weight ratio between wild-type (WT) and KO mice (Fig. 3, E and F), and histological analyses revealed no disruptions in seminiferous tubules in NR_038002-KO mice (Fig. 3G). To further investigate the phenotype of NR_038002-KO mice, we evaluated mature sperm collected from the cauda epididymis and vas deferens of WT and KO mice. Although the number of sperm was not different between WT and KO mice (Fig. 3H), a microscopic analysis revealed defects in the head morphology of sperm from NR_038002-KO mice (Fig. 3, I and J). The most commonly observed abnormalities were small, flattened, and misshapen heads (Fig. 3J), implying the involvement of NR_038002 in sperm head formation. Quantitative analyses showed that sperm with abnormal heads accounted for ~46.7% of the total sperm population in NR_038002-KO mice compared with ~3.7% in WT mice. To determine whether the loss of NR_038002 affects male fertility, we performed a fertility test, as detailed in fig. S2C. NR_038002-KO male mice were subfertile compared with WT mice (8.6 pups versus 5.9 pups) (Fig. 3K), and the degree of the sperm head abnormality was correlated with the decline in fertility in each individual KO mouse (Fig. 3L). These results demonstrate that an NR_038002 deficiency leads to sperm head malformation and a consequent reduction in male fertility.
Fig. 3. Phenotypic analysis of NR_038002-KO male mice.
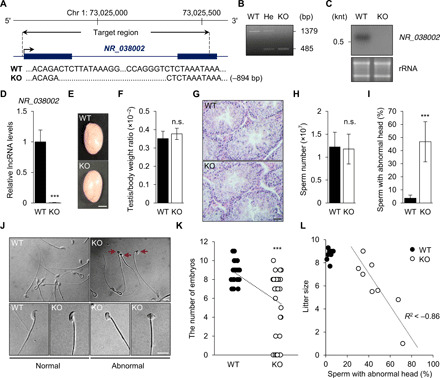
(A) Schematic diagram of the genomic locus of NR_038002 and the region deleted using CRISPR-Cas9 genome editing. The deleted region (−894 bp) of NR_038002 is indicated in the sequencing results. (B) Genotyping of NR_038002+/+ [wild-type (WT)], NR_038002+/− (He, heterozygote), and NR_038002−/− (KO) mice. (C) Northern blot of NR_038002 from WT and NR_038002-KO testes. (D) Quantitative RT-PCR (qRT-PCR) analyses of NR_038002 in WT and NR_038002-KO testes. Gapdh mRNA was used as a control. Data are presented a means ± SD (n = 4, ***P < 0.001, two-tailed Student’s t test). (E) Macroscopic appearance of adult testes from 8-week-old WT and NR_038002-KO mice. Scale bar, 0.2 mm. (F) Testis–to–body weight ratio in 8-week-old WT and NR_038002-KO mice (n = 6, n.s. P > 0.05, two-tailed Student’s t test). n.s., not significant. (G) H&E staining of seminiferous tubules of WT and NR_038002-KO mice. Scale bar, 40 μm. (H) Number of mature sperm from WT and NR_038002-KO mice (n = 6, n.s. P > 0.05, two-tailed Student’s t test). (I) Percentage of sperm with abnormal head morphology in WT and NR_038002-KO mice (n = 8, ***P < 0.001, two-tailed Student’s t test). (J) Representative spermatozoa from WT and NR_038002-KO mice. Red arrows indicate sperm with abnormal heads. (K) Number of embryos after mating with WT (n = 10) and NR_038002-KO (n = 8) male mice. Each dot represents an individual female mouse used for mating with WT (n = 28) or NR_038002-KO (n = 33) mice. Vaginal plugs were observed in all mated females. Data are presented a means ± SD (***P < 0.001, two-tailed Student’s t test). (L) Correlation between sperm with abnormal heads and litter size (n = 8). The dotted line shows the correlation between sperm head abnormality and litter size.
The abnormal head morphology of the sperm could be related to a defect in chromatin condensation, reflecting stepwise replacement of histones during spermiogenesis, first with transition proteins and lastly with protamines (30). To test this, we analyzed changes in the expression of transition proteins and protamines in NR_038002-KO sperm. We found that expression levels of transition proteins 1/2 (Tnp1/2) and protamine 1/2 (Prm1/2) were unchanged (fig. S3A). Consistently, PRM1 and PRM2 protein expression levels and processing were also normal in NR_038002-KO sperm (fig. S3B). We tested whether NR_038002-KO sperm are sensitive to chemical disruption, releasing genomic DNA fragments upon treatment with SDS under nonreducing conditions. We found that NR_038002-KO sperm did not differ from WT sperm with respect to this chromatin property (fig. S3C). Thus, altered sperm head formation in the absence of NR_038002 is unlikely due to incomplete chromatin assembly.
Loss of NR_038002 causes up-regulation of X chromosomal gene expression
To comprehensively investigate the function of NR_038002 during male germ cell development, we performed RNA sequencing (RNA-seq) analyses of WT and KO testes. Deletion of NR_038002 was accompanied by significant changes in the expression (fold change >1.5 and P < 0.05) of a total of 82 genes in testes (Fig. 4A and table S2), 65 of which were up-regulated and 17 of which were down-regulated. Notably, 69 of 82 differentially expressed genes (DEGs) were located on X or Y chromosome. All 54 X-linked genes were up-regulated in NR_038002-KO testes, whereas expression of the 15 Y-linked genes was reduced in KO testes (Fig. 4B). Because some sex-linked genes were previously known as multicopy genes, we analyzed the DEGs whether they are multicopy genes. All of the 15 Y-linked DEGs belonged to multicopy gene families, such as Rbm31y, Sly, and Ssty, whereas 28 of 54 (51.9%) X-linked genes were the members of multicopy gene families, including Btbd35 (BTB domain containing 35), Gm6812, H2al1 (H2A histone family member L1), Mageb (melanoma antigen, family b), Samt1 (spermatogenesis-associated multipass transmembrane protein 1), Slx/Slxl1 (Scyp3-liked, X-linked/Slx-like 1), and Tgif2lx (TGFB-induced factor homeobox 2–like, X-linked) (Fig. 4C and table S2). Gene Ontology analyses of these DEGs showed that the corresponding encoded proteins are localized to the nucleus, nuclear chromatin, and nucleosomes and are functionally involved in DNA binding, chromatin silencing, spermatid development, and gamete generation (Fig. 4D and table S3). Using qRT-PCR, we validated changes in the expression of some X-linked DEGs with fold change >2 (Fig. 4E). The 54 X-linked DEGs included H2al1 variants and H2al3, both of which are known to be highly expressed in elongating spermatids (31). A recent study demonstrated that H2al2, which is closely related to H2al1, participates in chromatin organization during spermiogenesis (32). These data suggest that NR_038002 plays a role in transcriptional repression of X chromosomal genes in mouse testes.
Fig. 4. Loss of NR_038002 leads to aberrant up-regulation of X chromosomal genes.
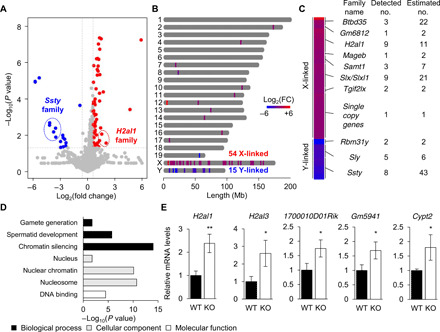
(A) Volcano plots showing differentially expressed genes (DEGs) (fold change >1.5 and *P < 0.05) in WT and NR_038002-KO testes. (B) Distribution and heatmap of DEGs along the mouse chromosome, determined using ChromoMap. (C) Family names and detected and estimated numbers of multicopy sex-linked DEGs. (D) Gene Ontology enrichment analysis of DEGs. (E) qRT-PCR analysis of up-regulated X chromosomal genes. Gapdh mRNA was used as a control. Data are presented a means ± SD (n = 3, P > 0.05, *P < 0.05, and **P < 0.01, two-tailed Student’s t test).
Loss of NR_038002 down-regulates Y chromosomal multicopy gene expression in the MSYq and in turn activates X chromosomal gene expression
In the context of repression of X chromosomal gene expression, it should be noted that the protein encoded by the Y chromosome–located multicopy gene, Sly, has been found to regulate the expression of X and Y chromosome–encoded genes in mice (33). In addition to Sly, MSYq contains several multicopy genes, including Srsy (serine-rich secreted, Y-linked) as well as Ssty1 and Ssty2. These genes exist as hundreds of copies of an ampliconic unit (Fig. 5A) (34) and, with the exception of Srsy, are expressed predominantly in mouse testes (35). Similarly, multicopy genes encoding SLX and SLXL1 are located on the mouse X chromosome and regulate X- and Y-linked genes, but their products antagonize the regulatory influence of SLY (36). SLY acts a transcription corepressor, interacting with TBL1XR1, a member of the SMRT/N-CoR repressor complex (37), and represses the expression of a number of X and Y chromosomal genes (33), including histone variants, similar to our findings in NR_038002-KO mice. Sly–knockdown (KD) mice are subfertile and produce sperm with abnormal head morphology (33).
Fig. 5. Loss of NR_038002 leads to aberrant down-regulation of Y chromosomal multicopy genes and a female-biased sex ratio in offspring.
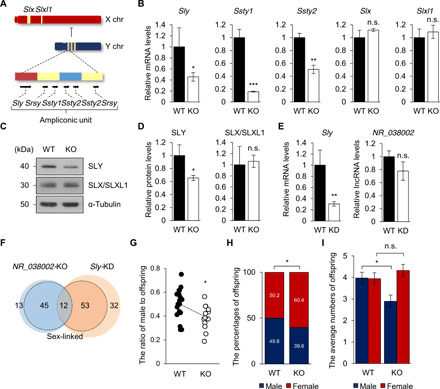
(A) Gene content and structure of mouse X and Y chromosomes. Representation of the ampliconic unit in the mouse Y chromosome, consisting of red, yellow, and blue core blocks. (B) qRT-PCR analysis of X and Y chromosomal multicopy genes in WT and NR_038002-KO mouse testes. Gapdh mRNA was used as a control. Data are presented as means ± SD (n = 3, n.s. P > 0.05, *P < 0.05, **P < 0.01, and ***P < 0.001, two-tailed Student’s t test). n.s., not significant. (C) Western blot of SLY and SLX/SLXL1 in WT and NR_038002-KO testes. (D) Densitometric analysis of SLY and SLX/SLXL1 in Western blots, normalized to α-tubulin. Data are presented as means ± SD (n = 3, n.s. P > 0.05, *P < 0.05, two-tailed Student’s t test). (E) qRT-PCR analysis of Sly and NR_038002 in WT and Sly–knockdown (KD) testes (n = 3, n.s. P > 0.05, **P < 0.01, two-tailed Student’s t test). (F) Venn diagram indicating overlapping DEGs in NR_038002-KO and Sly-KD testes. DEGs (fold change >1.5 and *P < 0.05) in sex chromosomes are indicated by the bold line. (G) Each dot represents the male pup ratio after mating with male WT (n = 18) and NR_038002-KO (n = 13) mice (*P < 0.05, two-tailed Student’s t test). (H) Bar graph showing the percentage of offspring of each sex, obtained by summation of the total number of pups (*P < 0.05, two-tailed Fisher’s exact t test). (I) Bar graph showing the average numbers of offspring of each sex, analyzed in (G) and (H). Data are presented as the means ± SEM (n.s. P > 0.05, *P < 0.05, two-tailed Student’s t test).
Our RNA-seq data demonstrated the down-regulation of Sly and Ssty in NR_038002-KO testes. Using qRT-PCR to obtain a more accurate measurement of expression levels, we reinvestigated the expression of multicopy sex chromosomal genes in NR_038002-KO testes. Both Sly mRNA and protein were slightly, but significantly, down-regulated in NR_038002-KO testes (Fig. 5, B to D). Ssty1 and Ssty2 transcript levels were also markedly reduced in NR_038002-KO testes (Fig. 5B). Unlike the case for Sly, Ssty1, and Ssty2, expression of Slx/Slxl1 was normal in the absence of NR_038002 (Fig. 5, B to D). To analyze the relationship of NR_038002 with Sly, we examined the expression of NR_038002 in Sly-KD testes. NR_038002 transcript levels were not changed in Sly-KD testes (Fig. 5E), suggesting that the regulation of X chromosomal genes by SLY occurs downstream of NR_038002 activity. Thus, it is likely that the up-regulation of X chromosomal genes in NR_038002-KO testes is directly attributable to Sly down-regulation. A profile analysis of DEGs (fold change >1.5 and P < 0.05) revealed that 12 sex-linked genes were misregulated in both NR_038002-KO and Sly-KD testes (Fig. 5F and table S4). Not all X and Y chromosomal genes were similarly regulated in NR_038002-KO and Sly-KD testes, likely owing to the modest reduction in the amount of Sly mRNA and protein in NR_038002-KO mice (Fig. 5, B to D). Moreover, expression levels of Ssty1 and Ssty2, which are up-regulated in the absence of Sly (33), were decreased in NR_038002-KO testes (Fig. 5B). Thus, it is possible that NR_038002 regulates the expression of Ssty1 and Ssty2 as well as Sly, but the regulation of Ssty1 and Ssty2 by NR_038002 is independent of Sly. SSTY1 and SSTY2 appear to be functionally important, with properties similar to the H3K4me3 reader function of spindlin (38). Moreover, SLY and SLX/SLXL1 were recently reported to compete for interaction with SSTY1 at the promoter of thousands of spermatid-specific multicopy genes, exerting an opposite regulatory effect on their expression (38). Our results, together with previous Sly studies, suggest that NR_038002 activates the expression of the Y chromosomal multicopy genes, Sly and Ssty1/2, which in turn repress X chromosomal gene expression.
Lack of NR_038002 results in a female-biased offspring sex ratio
Sly and Slx/Slxl1 are transmission distorters, involved in an intragenomic conflict that favors their own transmission (38). Because the expression levels of Sly and Slx/Slxl1 were unbalanced in NR_038002-KO mice owing to down-regulation of SLY, we rigorously examined offspring for a distortion in their sex ratio. A count of the number of offspring obtained from mating with female mice showed that the proportions of male and female pups for KO males were 0.39 and 0.61, respectively (n = 13), compared with 0.5 and 0.5 for WT males (n = 18) (Fig. 5G). We then calculated the sex ratio distortion based on the total number of offspring. This analysis showed that KO male mice sired more female offspring (60.4%; 198 of 328) compared with WT male mice (50.2% female; 139 of 277), a difference that approached statistical significance in a Fisher’s exact test (P < 0.05) (Fig. 5H). A female-biased sex ratio was also observed in Sly-KD mice, 2/3MSYq− mice, and YRIIIqdel mice, findings at odds with the male-biased ratio observed in Slxl1-KO mice (33, 39, 40). Our results suggest that deregulation of multicopy genes on the X or Y chromosome in NR_038002-KO male mice causes an X-versus-Y sperm fitness difference, thereby resulting in distortion of the sex ratio toward female offspring. In this regard, it should be noted that the sex ratio distortion produced by the absence of NR_038002 appears to reflect a decrease in the number of male offspring (4.0 and 2.9 male offspring from WT and KO males, respectively), rather than a change in female progeny number (3.9 and 4.3 female offspring from WT and KO males, respectively) (Fig. 5I and table S5). A decrease in male progeny number is congruent with the degree of reduced fertility in male NR_038002-KO mice (Fig. 3K), implying impaired fertility of Y-bearing sperm.
NR_038002 interacts with HSF2 to activate Y chromosomal genes during spermiogenesis
Previous studies have shown that transcription of the sex chromosomal multicopy genes, Slx, Sly, Ssty1, and Ssty2, is regulated by HSF1 and HSF2 (24, 41). In Hsf2-KO testes, both Ssty2 and Sly mRNA levels are reduced, but Slx mRNA levels are increased (24). To investigate how NR_038002 regulates the expression level of genes on sex chromosomes, we first examined whether NR_038002 regulates mRNA and protein expression levels of Hsf1 and Hsf2 in mouse testes, but found no significant change in expression of either at the mRNA or protein level in NR_038002-KO testes (Fig. 6, A to C). Next, we performed a biotin-labeled RNA pull-down assay to test whether NR_038002 interacts with HSF1 and HSF2 in mouse testes (Fig. 6D). Notably, we found that biotinylated sense NR_038002 specifically bound to HSF2, but not HSF1, in testes (Fig. 6E). To identify the HSF2-binding region of NR_038002, we constructed three truncated fragments of NR_038002 and tested them in RNA pull-down assay (Fig. 6F). These analyses revealed that the 5′ fragment (1 to 100 nt) clearly interacted with HSF2, whereas the other fragments (101 to 230 nt and 231 to 416 nt) did not (Fig. 6F). To validate the interaction between NR_038002 and HSF2, we performed RNA immunoprecipitation (RIP) in mouse testes using an anti-HSF2 antibody. We found that NR_038002 was highly enriched in anti-HSF2 antibody immunoprecipitates compared with control immunoglobulin G (IgG) immunoprecipitates (Fig. 6, G and H). A previous chromatin immunoprecipitation (ChIP)-chip analysis showed that promoters located upstream of Sly, Ssty1, and Ssty2 are occupied by HSF2 only in testes, and not in the brain, muscle, or kidney, suggesting that the target promoter sequences alone are not sufficient to specify HSF2 binding (24). A comparison of HSF2 binding to the promoter sequences of Sly, Ssty1, and Ssty2 in NR_038002 WT and KO testes showed greater occupancy of these promoters by HSF2 in WT testes than in KO testes (Fig. 6I), indicating the importance of NR_038002 for HSF2 binding to Sly, Ssty1, and Ssty2 promoters. Further investigation is needed to determine whether NR_038002 and HSF2 together bind to the promoters or NR_038002 only stabilizes the binding of HSF2 to the promoters. Collectively, our results suggest that the interaction of NR_038002 with HSF2 is critical for the expression of Y chromosomal multicopy genes, whose postmeiotic expression, in turn, is crucial for normal sperm maturation and fertility (Fig. 6J). On the basis of these findings, we newly designate NR_038002 as testicular germ cell–specific HSF2-interacting lncRNA (Teshl).
Fig. 6. NR_038002 interacts with HSF2 to activate Y chromosomal multicopy genes.
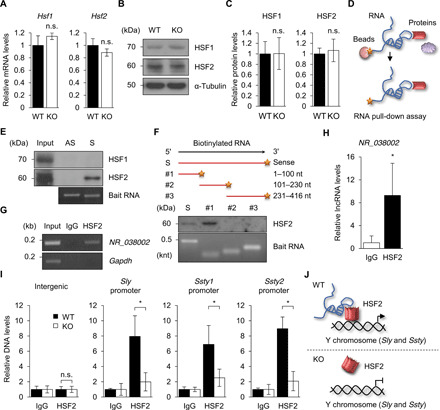
(A) qRT-PCR analysis of Hsf1 and Hsf2 in WT and NR_038002-KO testes. Gapdh mRNA was used as a control. Data are presented as means ± SD (n = 3, n.s. P > 0.05, two-tailed Student’s t test). n.s., not significant. (B) Western blot of HSF1 and HSF2 in WT and NR_038002-KO testes. (C) Densitometric analysis of HSF1 and HSF2 in Western blots, normalized to α-tubulin. Data are presented as means ± SD (n = 3, n.s. P > 0.05, two-tailed Student’s t test). (D) Schematic diagram of RNA pull-down assay. (E) Western blot of eluates obtained by RNA pull-down using specific anti-HSF1 and anti-HSF2 antibodies. Bottom panel shows biotinylated full-length antisense and sense strands of NR_038002. (F) Western blot of eluates obtained by RNA pull-down using the anti-HSF2 antibody. Bottom panel shows sense strands of biotinylated full-length and fragmented [1 to 100 nt, 101 to 30 nt, and 231 to 416 nt] NR_038002. (G) RNA immunoprecipitation (RIP) PCR analysis of NR_038002, which binds to HSF2 in mouse testes. Gapdh mRNA, which does not bind to HSF2, served as the negative control. IgG, immunoglobulin G. (H) RIP-qPCR analysis of NR_038002. Data are presented as means ± SD (n = 3, *P < 0.05, two-tailed Student’s t test). (I) Chromatin immunoprecipitation qPCR analysis of Sly, Ssty1, and Ssty2 promoter regions, which bind HSF2, in WT and NR_038002-KO mouse testes. The intergenic region, which does not bind HSF2, served as a negative control. Data are presented as means ± SD (n = 3, n.s. P > 0.05, *P < 0.05, two-tailed Student’s t test). (J) Model of NR_038002 function, showing that NR_038002 binding to HSF2 activates multicopy Y chromosomal Sly and Ssty genes.
DISCUSSION
In the present study, we report the novel finding that the conserved testicular germ cell–specific lncRNA NR_038002, hereafter Teshl, activates the Y chromosomal multicopy genes, Sly and Ssty, through interaction with the transcription factor HSF2 in mouse testes, and showed that this process is required for production of normal Y-bearing sperm. Deletion of Teshl in mouse testes caused impaired sperm head formation and resulted in sex ratio distortion toward females in offspring (Fig. 7). Disruption of Hsf2 exhibited similar phenotypes, such as altered expression of multicopy genes, sperm head abnormalities, and subfertility (24, 42).
Fig. 7. Schematic diagram showing production of normal Y-bearing sperm by NR_038002 (Teshl)–dependent HSF2 action on the male-specific Y chromosome long arm.
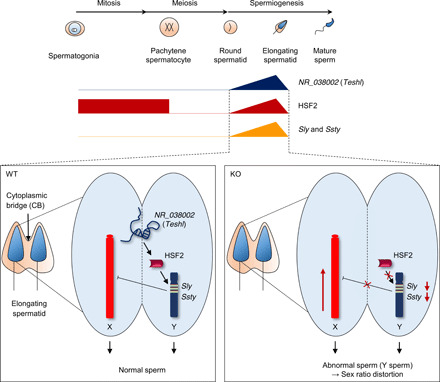
NR_038002 (Teshl) exhibits spatiotemporally expression patterns in the nuclei of elongating spermatids in WT mouse testes. HSF2 is expressed during early spermatogenesis, inactivated during meiosis, and reactivated beginning in round spermatids. Teshl plays an important role in activating the Y chromosomal multicopy genes Sly and Ssty by directing the binding of HSF2 to target regions, which results in down-regulation of X chromosome genes. In Teshl-KO mouse testes, HSF2 cannot bind to the promoter regions of Sly and Ssty, resulting in a significant decrease in their expression levels and consequent up-regulation of spermatid-expressed genes located on the X chromosome. Therefore, Y-bearing sperm with abnormal head morphology are produced, and offspring exhibit a female-biased sex ratio.
The deletion of Teshl led to deregulation of distinct sets of sex chromosomal multicopy genes, as evidenced by the significant up-regulation of X-linked genes and down-regulation of Y-linked genes. Sex chromosomes are transcriptionally silenced during meiosis by MSCI and then partially reactivated at genes (17) with important roles during sperm maturation. Although sex chromosomal multicopy genes have been proposed to reactivate sex chromosomes during this postmeiotic period, the detailed molecular mechanism is largely unknown. Our study provides the first evidence for operation of a Teshl-HSF2-MSYq axis in this process, showing that Teshl-dependent HSF2 activity promotes expression of Y chromosomal multicopy genes in MSYq during the postmeiotic phase. Both HSF1 and HSF2 are transcription factors that regulate the transcription of multicopy genes located in the MSYq and share ~15% of targets (24, 41, 43). Our data showed that Teshl interacts with HSF2 but not HSF1 (Fig. 6E), indicating that Teshl-dependent HSF2 transcriptional regulation is independent of HSF1. HSF1 activation by heat shock is mediated by a ribonucleoprotein complex containing the translation elongation factor eEF1A and the lncRNA, HSR1 (44), which may serve as a thermosensor with its constitutive expression pattern. However, HSF2 is transiently activated by Teshl to regulate postmeiotic target genes during spermiogenesis. Teshl may act as a developmental cue with its spatiotemporal expression pattern.
The temporal pattern of Teshl expression is very similar to that of its nearest gene, Tnp1 (elongating spermatids), suggesting the possibility that both genes are transcribed by a coregulatory mechanism. If so, the histone-to-protamine exchange process necessary for chromatin condensation and the transient expression of sex chromosomal multicopy genes involved in spermiogenesis may be temporally coordinated.
The current model for equal transmission of X- and Y-bearing sperm to offspring posits that a balanced copy number of Slx/Slxl1 and Sly/Ssty genes is necessary for normal X- and Y-bearing sperm differentiation (38). For example, Slx/Slxl1-KO male mice exhibit a male-biased sex ratio in offspring, whereas males carrying Slx/Slxl1 duplications display a distortion toward female offspring (40), indicating the importance of Slx/Slxl1 copy number. Male mice with a deletion of the MSYq exhibit severely impaired sperm differentiation and a female-skewed offspring sex ratio (39). Sly-KD male mice also show a female-biased sex ratio in offspring (33). Sex ratio distortion toward females in MSYq-deleted mice is likely attributable to the absence of Y chromosomal multicopy genes rather than the partial loss of the Y chromosome. Likewise, our Teshl-KO mice exhibited a female-biased distortion in sex ratio among offspring owing to substantial inactivation of the Y chromosomal multicopy genes, Sly and Ssty. Misregulation of most of the MSYq multicopy genes in Teshl KO mice suggests chromosome-wide–level regulation of gene expression by Teshl-HSF2. Cytoplasmic bridges connecting spermatids are known to facilitate the sharing of haploid gene products (see also Fig. 7) (45). The question is, how does down-regulation of Y chromosomal multicopy genes cause a distortion in the sex ratio in offspring? A recent analysis of sex chromosomal transcripts at the single-cell level after meiosis showed that thousands of transcripts, including Slx, Sly, and Ssty, are not equally shared despite the presence of cytoplasmic bridges (46). This suggests that spermatids do not display absolute phenotypic equivalence with regard to gene products of sex chromosomes. The sex ratio distortion observed in Teshl-KO mice is not attributable to an imbalance in the proportion of X- or Y-bearing sperm because the total number of sperm was not changed in Teshl-KO mice (Fig. 3H). Furthermore, the sex ratio changes in offspring of male Teshl-KO mice compared with WT male mice was attributable to a decrease in the number of male progeny, not an increase in the number of female progeny (table S5). X- and Y-bearing sperm from male mice with a deletion of the MSYq have altered morphology and motility (47). Similarly, the distortion in the sex ratio toward females in offspring of Teshl-KO mice may be related to the abnormal head morphology and/or motility defects of Y-bearing sperm. Therefore, our study strongly suggests that, by activating Y chromosomal multicopy genes whose products contribute to normal spermatogenesis, Teshl plays a crucial role in conferring the normal fertility and integrity of Y-bearing sperm (Fig. 7).
One of the most common genetic causes of spermatogenesis failure in humans is deletion of the MSYq, which leads to copy number variations of Y chromosomal multicopy genes and, ultimately, male infertility (48). In this study, we identified LINC01921 as a human ortholog of Teshl (Fig. 1D). It was recently reported that LINC01921 is significantly decreased or differentially expressed in patients with nonobstructive azoospermia (49). Although further investigation is required to determine whether a decrease in the expression levels of LINC01921 is directly involved in nonobstructive azoospermia, this human lncRNA Teshl ortholog could be considered a suitable biomarker for diagnosis of human infertility.
In conclusion, we identified a new testicular germ cell–specific lncRNA and defined its biological and molecular nature, demonstrating the importance of the Teshl-Hsf2-MSYq axis in postmeiotic gene expression mechanisms on the Y chromosome. Our study further contributes to our understanding of sex ratio variations and suggests the potential for the development of future fertility therapeutics
MATERIALS AND METHODS
Animals
All animal experiments were performed in accordance with Korean Food and Drug Administration guidelines. Protocols were reviewed and approved by the Institutional Animal Care and Use Committee of Gwangju Institute of Science and Technology (GIST) (permit number: GIST-2020-034). Each tissue sample was snap frozen in liquid nitrogen for later extraction.
Northern blotting
Total RNA was isolated from each mouse tissue using TRIzol reagent (Ambion). RNA was heated at 65°C for 5 min and resolved on a denaturing agarose gel. After equilibrating the gel for 15 min in distilled water and then twice for 10 min each in 10× saline sodium citrate (SSC) buffer, total RNA was transferred to Hybond-XL membranes (GE Healthcare). After ultraviolet cross-linking, the blot was prehybridized for 1 hour at 65°C in Rapid-hyb Buffer (Amersham), followed by hybridization for 2 hours at 65°C in the presence of a cDNA probe. The probe, derived from products amplified using gene-specific primers (table S6), was purified using ProbeQuant G-50 Micro Columns (GE Healthcare) and labeled with [α-32P]dCTP (Perkin Elmer) using a Prime-It II Random Primer Labeling Kit (Agilent) according to the manufacturer’s instructions. Blots were washed two to four times in 2× SSC and 0.05% SDS at 37°C for 10 min and twice in 0.1× SSC and 0.1% SDS at 65°C for 10 min, after which they were exposed to FUJI Medical X-ray Film (Fujifilm) at −70°C.
Rapid amplification of cDNA ends
Deoxyribonuclease I (DNase I)–treated total RNA from a mouse testis was used for first-strand cDNA synthesis. The SMARTer RACE cDNA Amplification Kit (Clontech) was used to perform 5′- and 3′-RACE according to the manufacturer’s instructions. The gene-specific primers used for RACE are listed in table S6. RACE products were cloned into the linearized pRACE vector using the In-Fusion HD cloning kit and then sequenced.
Sucrose gradient–based polysome fractionation
Beckman ultracentrifuge tubes were loaded with 2.2 ml of a 10 to 50% sucrose (Sigma-Aldrich) gradient in salt buffer [2 mM tris (pH 7.5), 10 mM NaCl, and 0.5 mM MgCl2] and incubated horizontally at 4°C overnight. Two pieces of mouse testes were homogenized in 1 ml of polysome extraction buffer [20 mM tris (pH 7.5), 100 mM KCl, 5 mM MgCl2, 0.5% (v/v) NP-40, protease inhibitor cocktail, RNasin (Promega), and cycloheximide (100 μg/ml)]. After a 10-min centrifugation, clarified lysates were layered onto ultracentrifuge tubes containing 10 to 50% sucrose gradients and centrifuged at 39,000 rpm for 90 min in an SW41 rotor (Beckman Coulter) at 4°C. Gradients were collected in 12 × 1–ml fractions using a Pasteur pipet, and absorbance was measured at 254 nm. RNA was isolated by extraction using TRIzol reagent. RNA quality and amount were determined using an ND-1000 spectrophotometer (Thermo Fisher Scientific).
Prediction of RNA secondary structure
Secondary structure analyses of lncRNAs were performed using RNAfold (http://rna.tbi.univie.ac.at/cgi-bin/RNAWebSuite/RNAfold.cgi) with default parameters.
Conventional RT-PCR
DNase I–treated tissue RNA was reverse transcribed using an Omniscript RT kit (Qiagen) according to the manufacturer’s instructions. RT-PCR was carried out using primers listed in table S6, and results were normalized to mouse Gapdh mRNA. Analyses of a human cDNA panel used 5 μl of cDNA purchased from Clontech (Human MTC panels I and II cDNA panel). A human GAPDH primer pair in the kit was used as a control.
Quantitative RT-PCR
Testicular samples from Sly-KD mice were prepared as previously described (33). Total testis RNA was reverse transcribed using a QuantiNova RT kit (Qiagen) according to the manufacturer’s instructions. qRT-PCR was carried out using TOPreal qPCR 2X premix (Enzynomics) with primers listed in table S6. Amplifications were performed in at least triplicate for each sample. Relative gene expression levels were evaluated using the 2−ΔΔCt method (50) and were normalized to the level of Gapdh mRNA. Data are presented as means ± SD. Statistical analyses were performed using two-tailed Student’s t tests, and P ≤0.05 was taken as indicating a significant difference.
In situ hybridization
Sense and antisense NR_038002 probes were amplified from mouse testis cDNA using the primer pair indicated in table S6. After cloning into the pGEM T Easy Vector (Promega), the plasmid was linearized by digesting with Sac II or Pst I, generating sense and antisense probes, respectively. Digoxigenin (DIG)–labeled RNA probes were synthesized using a DIG RNA Labeling Kit (Roche) with SP6 RNA polymerase as a sense probe and T7 RNA polymerase as an antisense probe. Adult mouse testes were fixed in Bouin’s solution (Sigma-Aldrich) overnight at room temperature (RT), then dehydrated in an ethanol series, embedded in paraffin, cut into 5-μm-thick sections, and loaded onto slide glasses. Sections were deparaffinized, rehydrated, and stained with hematoxylin and eosin Y (H&E; Sigma-Aldrich). For in situ hybridization, sections were deparaffinized and air dried for 10 min, and then fixed with 4% paraformaldehyde (PFA) for 10 min and treated with proteinase K (1 μg/ml) (GeneAll) at 37°C for 5 min. After washing in 1× phosphate-buffered saline (PBS), sections were postfixed by incubating with 4% PFA for 5 min. Thereafter, sections were acetylated by incubating in acetylation buffer [1.35% (v/v) triethanolamine (Sigma-Aldrich), 1.75% (v/v) concentrated hydrochloric acid (Sigma-Aldrich), and 0.25% (v/v) acetic anhydride (Sigma-Aldrich)] for 10 min, and then washed with 1× PBS. The sections were prehybridized with hybridization buffer [deionized formamide (500 μl/ml) (Sigma-Aldrich), Denhardt’s solution (20 μl/ml) (Roche), baker’s yeast RNA (0.25 μg/ml) (Sigma-Aldrich), salmon sperm DNA (0.5 μg/ml) (Roche), and 1× SSC] for 1 hour at RT. After denaturing DIG-labeled probes (1 μg) at 80°C for 5 min, sections were hybridized with DIG-labeled probes in hybridization buffer by incubating overnight at 68°C. After hybridization, the sections were washed once in 5× SSC for 10 min at 65°C, twice in 0.2× SSC for 1 hour at 65°C, and once in 0.2× SSC for 5 min at RT. After washing in B1 buffer [0.1 M tris (pH 7.4) and 1.67 M NaCl] for 5 min at RT, sections were blocked by incubating with 10% (v/v) heat-inactivated normal goat serum (HINGS) in B1 buffer for 1 hour, and then incubated overnight at 4°C in B1 buffer containing 1% HINGS and anti-DIG antibody (Roche), diluted 1:5000. Sections were then washed three times in B1 buffer and equilibrated in B2 buffer [0.1 M tris (pH 9.5) and 0.1 M NaCl]. Last, sections were incubated with NBT/BCIP (Promega), diluted 1:50, in B2 buffer for several hours until the signal was detectable and then observed under an Aperio ScanScope (Leica).
Cell culture and transfection
Mouse GC-2 cells were obtained from the American Type Culture Collection (ATCC) and cultured with medium and serum as recommended by the ATCC. For transfection, full-length NR_038002 was amplified from mouse testis cDNA using primers incorporating Kpn I and Age I restriction sites (table S6), and cloned into the pcDNA3.1A vector. Transfection was performed using Lipofectamine 2000 (Invitrogen) according to the manufacturer’s instructions.
Microarray
RNA purity and integrity were evaluated using an ND-1000 spectrophotometer (Thermo Fisher Scientific) and Agilent 2100 Bioanalyzer (Agilent). The Affymetrix whole transcript expression array process was executed according to the manufacturer’s protocol (Affymetrix). cDNA was synthesized using the GeneChip WT (Whole Transcript) Amplification kit as described by the manufacturer. The sense cDNA was then fragmented and biotin labeled with TdT (terminal deoxynucleotidyl transferase) using a GeneChip WT Terminal Labeling Kit. Approximately 5.5 μg of labeled DNA target was hybridized to the Affymetrix GeneChip Mouse 2.0 ST Array at 45°C for 16 hours. Hybridized arrays were washed and stained on a GeneChip Fluidics Station 450 and scanned on a GCS3000 Scanner (Affymetrix). Signal values were computed using Affymetrix GeneChip Command Console software. Raw data were extracted automatically with the Affymetrix data extraction protocol using the provided Affymetrix GeneChip Command Console Software. After importing CEL files, the data were summarized and normalized using the robust multiaverage method implemented in Affymetrix Expression Console Software. Comparative analyses of test and control samples were carried out using fold change >2 and a P <0.05 as criteria. Statistical analyses and visualization of DEGs were implemented in R software and Galaxy (https://usegalaxy.org/).
Generation of NR_038002-KO mice
Cas9 protein tagged with a nuclear localization signal and guide RNAs (gRNAs) targeting exon 1 and exon 2 of the NR_038002 gene were purchased from Macrogen. The activity of gRNAs (exon 1_gRNA: CGT GTC ACA GAC TCT TAT AAA GG; exon 2_gRNA: GAG GTT TTA TTT AGA GAC CCT GG) was validated by assessing deletion of NR_038002 using in vitro cleavage reactions. Briefly, amplified NR_038002, used as a template, was incubated for 90 min at 37°C with Cas9 protein (20 nM) and single gRNA (sgRNA) (40 nM) in 1× NEB 3.1 buffer. Reactions were stopped with 6× stop solution containing 30% glycerol, 1.2% SDS, and 100 mM EDTA. Cleavage activity was confirmed by electrophoresis of reaction mixtures. NR_038002-KO mice, generated by Macrogen, were interbred and maintained at Macrogen facilities under pathogen-free conditions. For breeding, C57BL/6 female mice were treated with pregnant mare serum gonadotropin and human chorionic gonadotropin. After 48 hours, treated female mice were mated with C57BL/6 male mice. The next day, female mice were checked for vaginal plugs, after which mice were euthanized and fertilized embryos were harvested. One-cell embryos were then microinjected with a mixture of sgRNA and Cas9 protein and incubated at 37°C for 1 to 2 hours. Fourteen to 16 injected one-cell-stage embryo were transplanted into oviducts of pseudopregnant recipient mice (ICR). After founder offspring were born, they were genotyped by PCR analysis of tail-clip samples using the primers listed in table S6, followed by sequencing.
Phenotypic analyses of NR_038002-KO mice
Testis integrity was analyzed by fixing testes from WT and NR_038002-KO male mice by immersion in Bouin’s solution overnight. The tissue was then dehydrated, embedded in paraffin, and sectioned (5 μm thick). Testis sections from male WT and NR_038002-KO mice were H&E stained and scanned using an Aperio ScanScope CS2 System (Leica Biosystems). For sperm counts, sperm from the cauda epididymis and vas deferens of male WT and NR_038002-KO mice were collected and counted in a hemocytometer under a light microscope. For analysis of sperm head morphology, mature sperm were isolated from the cauda epididymis. Sperm from male WT (n = 8) and NR_038002-KO (n = 8) mice were spread onto microscope slides and allowed to air dry. About 200 sperm on each slide were analyzed under a light microscope. For fertility tests, adult C57BL/6 WT females (8 weeks old) were mated with sexually mature male WT (n = 10) and NR_038002 KO (n = 8) mice for 14 days and checked daily for vaginal plugs. The fertility rate was calculated from the number of fetuses produced by pregnant females, counted 12 to 16 days after mating.
Analysis of DNA fragmentation and sperm nuclear basic proteins
Genomic DNA (gDNA) was obtained by incubating isolated sperm in lysis buffer containing 2% SDS and proteinase K (0.5 mg/ml) for 2 hours at 55°C. gDNAs in remaining sperm nuclei were released by incubating nuclei in lysis buffer containing 20 mM dithiothreitol for 6 hours. Released gDNA was subsequently purified on a DNeasy column (Qiagen). Chromatin was isolated from sperm from the cauda epididymis and vas deferens by treatment with cetyltrimethylammonium bromide, and basic proteins were extracted with NaCl, urea, and guanidine hydrochloride. Proteins were precipitated with trichloroacetic acid, separated on a 15% acid-urea polyacrylamide gel, and stained with Coomassie blue.
RNA sequencing
Libraries were prepared from total RNA using the NEBNext Ultra II Directional RNA-Seq Kit (NEB). mRNA used for cDNA synthesis and shearing was isolated using a Poly(A) RNA Selection Kit (LEXOGEN) following the manufacturer’s instructions. Indexing was performed using Illumina indexes 1 to 12. After performing a PCR-based enrichment step, libraries were evaluated for mean fragment size using the Agilent 2100 Bioanalyzer (DNA High Sensitivity Kit), and the library was quantified using a StepOne Real-Time PCR System (Life Technologies). High-throughput paired-end sequencing (100-bp reads) was performed using HiSeq 2500 (Illumina). Alignment files were obtained by mapping mRNA-Seq reads using the HISAT2 software tool. Alignment files, in turn, were used as input for the Cufflinks program for assembling transcripts, estimating their abundances, and detecting DEGs and isoforms. Expression levels of gene regions were determined using the FPKM (fragments per kilobase of exon per million fragments) method, and the resulting FPKM data were normalized by trimmed mean of M values (TMM) normalization using the EdgeR module in R software. Raw and processed RNA-seq data have been deposited in the Gene Expression Omnibus (GEO) database at the National Center for Biotechnology Information (NCBI) and can be accessed using the GEO accession number, GSE162652. To predict the gene families of the DEGs, we first obtained the sequence information of each gene and subsequently investigated a copy number with an identity greater than 95%, using BLASTN program with reference selected from NCBI Reference Sequence Database (Refseq). Isoforms, pseudogenes, and noncoding genes were excluded. If more than one gene was detected through BLASTN search, we referred to such a gene as a multicopy gene (gene family).
Western blotting
Testis lysates were prepared by homogenizing testes in radioimmunoprecipitation assay (RIPA) lysis buffer (Pierce) containing a protease inhibitor cocktail, and protein concentration was determined using a bicinchoninic acid (BCA) assay (Pierce). Equivalent amounts of protein in each sample were resolved by SDS–polyacrylamide gel electrophoresis (PAGE) and transferred to polyvinylidene difluoride membranes (Millipore). Membranes were blocked in 5% nonfat milk and incubated at 4°C overnight with primary antibodies against the following proteins, as indicated in the text: SLY (diluted 1:2000) (51), SLX/SLXL1 (diluted 1:1000) (52), α-tubulin (diluted 1:10,000; Millipore, #05-829), HSF1 (diluted 1:100; Santa Cruz Biotechnology, #sc-17756), and HSF2 (diluted 1:200; R&D Systems, #AF5227). Membranes were subsequently incubated with horseradish peroxidase–conjugated anti-rabbit IgG (diluted 1:10,000; Promega, #W401B), anti-mouse IgG (diluted 1:1000; Santa Cruz Biotechnology, #sc-516102), or anti-goat IgG (diluted 1:10,000; Santa Cruz Biotechnology, #sc-2354) secondary antibody, as appropriate. Scans of full-size immunoblots are presented in Source Data.
RNA pull-down assay
RNA pull-down assays were performed using a Pierce Magnetic RNA-Protein Pull-Down Kit (Thermo Fisher Scientific) according to the manufacturer’s instructions. Briefly, RNA was transcribed in vitro using a HiScribe T7 High Yield RNA Synthesis Kit (NEB), followed by DNase I (NEB) digestion and LiCl (Sigma-Aldrich) purification. RNA was biotinylated using a Pierce RNA 3′-End Desthiobiotinylation Kit (Thermo Fisher Scientific) according to the manufacturer’s instructions and purified using a NucAway spin column (Invitrogen). Testis extracts were prepared using the Pierce T-PER Tissue protein extraction reagent (Thermo Fisher Scientific). Fifty picomol of biotinylated RNA was captured with streptavidin magnetic beads and then incubated with 1 mg of testis lysates at 4°C for 3 hours. The beads were then washed three times with wash buffer and boiled in 2× Laemmli loading buffer. The retrieved proteins were resolved by SDS-PAGE and Western blotting.
RNA immunoprecipitation
RIP was performed using an EZ-Magna RIP Kit (Millipore) according to the manufacturer’s instructions. Briefly, RNAs were immunoprecipitated using 10 μg of goat anti-HSF2 antibody (R&D Systems, #AF5227); normal goat IgG (R&D Systems, #AB-108-C) was used as a control. After proteinase K digestion, immunoprecipitated RNA was extracted, purified, and used as a template for preparation of cDNA using a QuantiNova Reverse Transcription Kit (Qiagen). Enrichment of immunoprecipitated RNA was assessed by qRT-PCR using Gapdh mRNA as a negative control. The primer pairs used are shown in table S6.
Chromatin immunoprecipitation
ChIP assays were performed using a Magna ChIP A/G Kit (Millipore) according to the manufacturer’s instructions. In brief, adult testes from WT and NR_038002-deficient mice were collected and cross-linked with 1% formaldehyde, followed by quenching with glycine. Chromatin was sheared on ice to an average length of 200 to 1000 bp with a Vibra-Cell VCX-600 ultrasonic processor (20% amplitude, 10-s pulse on/30-s off, 20 cycles), and centrifuged at 10,000g at 4°C for 10 min. Immunoprecipitation with 5 μg of normal goat IgG (R&D Systems, #AB-108-C) and goat anti-HSF2 antibody (R&D Systems, #AF5227), washing, and elution were carried out according to the protocol. Regarding ribonuclease (RNase) treatment, no RNase was treated before or during ChIP. Primers for the ChIP assay were designed to amplify the promoter regions of the target genes in table S6. Fold enrichment was calculated by PCR products with anti-Hsf2 antibody to those with the normal goat IgG.
Statistical analysis
All experiments were performed at least in triplicate for each sample. Data are presented as the means ± SD. After analysis of variance with F-test, statistical analysis was performed using Student’s t test, and P values were indicated in the figure legends.
Acknowledgments
Funding: This work was supported by a National Research Foundation of Korea (NRF) grant funded by the Korea Ministry of Science and ICT (2019R1A1084354), a GIST Research Institute (GRI) IIBR grant funded by GIST, Korea in 2020, and Agence Nationale de la Recherche (ANR-17-CE12-0004-01 to J.C.). Author contributions: C.C. conceived the study. S.H.H. designed and carried out the experiments. G.H., S.J.L., and J.C. provided experimental materials. S.H.H. and C.C. wrote the manuscript. Competing interests: The authors declare that they have no competing interests. Data and materials availability: All data needed to evaluate the conclusions in the paper are present in the paper and/or the Supplementary Materials. The anti-SLY antibody, anti-SLX/SLXL1 antibody, and Sly-KD mice can be provided by J.C., pending a scientific review and a completed material transfer agreement. Requests for these materials should be submitted to julie.cocquet@inserm.fr.
SUPPLEMENTARY MATERIALS
Supplementary material for this article is available at http://advances.sciencemag.org/cgi/content/full/7/24/eabg5177/DC1
REFERENCES AND NOTES
- 1.Kapranov P., Cheng J., Dike S., Nix D. A., Duttagupta R., Willingham A. T., Stadler P. F., Hertel J., Hackermuller J., Hofacker I. L., Bell I., Cheung E., Drenkow J., Dumais E., Patel S., Helt G., Ganesh M., Ghosh S., Piccolboni A., Sementchenko V., Tammana H., Gingeras T. R., RNA maps reveal new RNA classes and a possible function for pervasive transcription. Science 316, 1484–1488 (2007). [DOI] [PubMed] [Google Scholar]
- 2.Fatica A., Bozzoni I., Long non-coding RNAs: New players in cell differentiation and development. Nat. Rev. Genet. 15, 7–21 (2014). [DOI] [PubMed] [Google Scholar]
- 3.Long Y., Wang X., Youmans D. T., Cech T. R., How do lncRNAs regulate transcription? Sci. Adv. 3, eaao2110 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Kutter C., Watt S., Stefflova K., Wilson M. D., Goncalves A., Ponting C. P., Odom D. T., Marques A. C., Rapid turnover of long noncoding RNAs and the evolution of gene expression. PLOS Genet. 8, e1002841 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Necsulea A., Soumillon M., Warnefors M., Liechti A., Daish T., Zeller U., Baker J. C., Grutzner F., Kaessmann H., The evolution of lncRNA repertoires and expression patterns in tetrapods. Nature 505, 635–640 (2014). [DOI] [PubMed] [Google Scholar]
- 6.Hong S. H., Kwon J. T., Kim J., Jeong J., Kim J., Lee S., Cho C., Profiling of testis-specific long noncoding RNAs in mice. BMC Genomics 19, 539 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Anguera M. C., Ma W., Clift D., Namekawa S., Kelleher R. J. 3rd, Lee J. T., Tsx produces a long noncoding RNA and has general functions in the germline, stem cells, and brain. PLOS Genet. 7, e1002248 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Li L., Wang M., Wang M., Wu X., Geng L., Xue Y., Wei X., Jia Y., Wu X., A long non-coding RNA interacts with Gfra1 and maintains survival of mouse spermatogonial stem cells. Cell Death Dis. 7, e2140 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Hosono Y., Niknafs Y. S., Prensner J. R., Iyer M. K., Dhanasekaran S. M., Mehra R., Pitchiaya S., Tien J., Escara-Wilke J., Poliakov A., Chu S. C., Saleh S., Sankar K., Su F., Guo S., Qiao Y., Freier S. M., Bui H. H., Cao X., Malik R., Johnson T. M., Beer D. G., Feng F. Y., Zhou W., Chinnaiyan A. M., Oncogenic role of THOR, a conserved cancer/testis long non-coding RNA. Cell 171, 1559–1572.e20 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Kataruka S., Akhade V. S., Kayyar B., Rao M. R. S., Mrhl long noncoding RNA mediates meiotic commitment of mouse spermatogonial cells by regulating Sox8 expression. Mol. Cell. Biol. 37, e00632-16 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Kurihara M., Otsuka K., Matsubara S., Shiraishi A., Satake H., Kimura A. P., A testis-specific long non-coding RNA, lncRNA-Tcam1, regulates immune-related genes in mouse male germ cells. Front. Endocrinol. 8, 299 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Hu K., Zhang J., Liang M., LncRNA AK015322 promotes proliferation of spermatogonial stem cell C18-4 by acting as a decoy for microRNA-19b-3p. In Vitro Cell. Dev. Biol. Anim. 53, 277–284 (2017). [DOI] [PubMed] [Google Scholar]
- 13.Satoh Y., Takei N., Kawamura S., Takahashi N., Kotani T., Kimura A. P., A novel testis-specific long noncoding RNA, Tesra, activates the Prss42/Tessp-2 gene during mouse spermatogenesis. Biol. Reprod. 100, 833–848 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Lewandowski J. P., Dumbovic G., Watson A. R., Hwang T., Jacobs-Palmer E., Chang N., Much C., Turner K. M., Kirby C., Rubinstein N. D., Groff A. F., Liapis S. C., Gerhardinger C., Bester A., Pandolfi P. P., Clohessy J. G., Hoekstra H. E., Sauvageau M., Rinn J. L., The Tug1 lncRNA locus is essential for male fertility. Genome Biol. 21, 237 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Liang M., Wang H., He C., Zhang K., Hu K., LncRNA-Gm2044 is transcriptionally activated by A-MYB and regulates Sycp1 expression as a miR-335-3p sponge in mouse spermatocyte-derived GC-2spd(ts) cells. Differentiation 114, 49–57 (2020). [DOI] [PubMed] [Google Scholar]
- 16.Eddy E. M., Male germ cell gene expression. Recent Prog. Horm. Res. 57, 103–128 (2002). [DOI] [PubMed] [Google Scholar]
- 17.Turner J. M. A., Meiotic sex chromosome inactivation. Development 134, 1823–1831 (2007). [DOI] [PubMed] [Google Scholar]
- 18.Burgoyne P. S., Mahadevaiah S. K., Turner J. M. A., The consequences of asynapsis for mammalian meiosis. Nat. Rev. Genet. 10, 207–216 (2009). [DOI] [PubMed] [Google Scholar]
- 19.Turner J. M., Meiotic silencing in mammals. Annu. Rev. Genet. 49, 395–412 (2015). [DOI] [PubMed] [Google Scholar]
- 20.Turner J. M., Mahadevaiah S. K., Ellis P. J., Mitchell M. J., Burgoyne P. S., Pachytene asynapsis drives meiotic sex chromosome inactivation and leads to substantial postmeiotic repression in spermatids. Dev. Cell 10, 521–529 (2006). [DOI] [PubMed] [Google Scholar]
- 21.Toure A., Clemente E. J., Ellis P., Mahadevaiah S. K., Ojarikre O. A., Ball P. A., Reynard L., Loveland K. L., Burgoyne P. S., Affara N. A., Identification of novel Y chromosome encoded transcripts by testis transcriptome analysis of mice with deletions of the Y chromosome long arm. Genome Biol. 6, R102 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Akerfelt M., Morimoto R. I., Sistonen L., Heat shock factors: Integrators of cell stress, development and lifespan. Nat. Rev. Mol. Cell Biol. 11, 545–555 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Sarge K. D., Park-Sarge O. K., Kirby J. D., Mayo K. E., Morimoto R. I., Expression of heat shock factor 2 in mouse testis: Potential role as a regulator of heat-shock protein gene expression during spermatogenesis. Biol. Reprod. 50, 1334–1343 (1994). [DOI] [PubMed] [Google Scholar]
- 24.Akerfelt M., Henriksson E., Laiho A., Vihervaara A., Rautoma K., Kotaja N., Sistonen L., Promoter ChIP-chip analysis in mouse testis reveals Y chromosome occupancy by HSF2. Proc. Natl. Acad. Sci. U.S.A. 105, 11224–11229 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Wang L., Park H. J., Dasari S., Wang S., Kocher J. P., Li W., CPAT: Coding-potential assessment tool using an alignment-free logistic regression model. Nucleic Acids Res. 41, e74 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Kang Y. J., Yang D. C., Kong L., Hou M., Meng Y. Q., Wei L., Gao G., CPC2: A fast and accurate coding potential calculator based on sequence intrinsic features. Nucleic Acids Res. 45, W12–W16 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Karolchik D., Barber G. P., Casper J., Clawson H., Cline M. S., Diekhans M., Dreszer T. R., Fujita P. A., Guruvadoo L., Haeussler M., Harte R. A., Heitner S., Hinrichs A. S., Learned K., Lee B. T., Li C. H., Raney B. J., Rhead B., Rosenbloom K. R., Sloan C. A., Speir M. L., Zweig A. S., Haussler D., Kuhn R. M., Kent W. J., The UCSC Genome Browser database: 2014 update. Nucleic Acids Res. 42, D764–D770 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Yu Y. E., Zhang Y., Unni E., Shirley C. R., Deng J. M., Russell L. D., Weil M. M., Behringer R. R., Meistrich M. L., Abnormal spermatogenesis and reduced fertility in transition nuclear protein 1-deficient mice. Proc. Natl. Acad. Sci. U.S.A. 97, 4683–4688 (2000). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Lorenz R., Bernhart S. H., Siederdissen C. H. Z., Tafer H., Flamm C., Stadler P. F., Hofacker I. L., ViennaRNA Package 2.0. Algorithms Mol. Biol. 6, 26 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Cho C., Willis W. D., Goulding E. H., Jung-Ha H., Choi Y. C., Hecht N. B., Eddy E. M., Haploinsufficiency of protamine-1 or −2 causes infertility in mice. Nat. Genet. 28, 82–86 (2001). [DOI] [PubMed] [Google Scholar]
- 31.Govin J., Escoffier E., Rousseaux S., Kuhn L., Ferro M., Thevenon J., Catena R., Davidson I., Garin J., Khochbin S., Caron C., Pericentric heterochromatin reprogramming by new histone variants during mouse spermiogenesis. J. Cell Biol. 176, 283–294 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Barral S., Morozumi Y., Tanaka H., Montellier E., Govin J., de Dieuleveult M., Charbonnier G., Coute Y., Puthier D., Buchou T., Boussouar F., Urahama T., Fenaille F., Curtet S., Hery P., Fernandez-Nunez N., Shiota H., Gerard M., Rousseaux S., Kurumizaka H., Khochbin S., Histone variant H2A.L.2 guides transition protein-dependent protamine assembly in male germ cells. Mol. Cell 66, 89–101.e8 (2017). [DOI] [PubMed] [Google Scholar]
- 33.Cocquet J., Ellis P. J., Yamauchi Y., Mahadevaiah S. K., Affara N. A., Ward M. A., Burgoyne P. S., The multicopy gene Sly represses the sex chromosomes in the male mouse germline after meiosis. PLoS Biol. 7, e1000244 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Soh Y. Q., Alfoldi J., Pyntikova T., Brown L. G., Graves T., Minx P. J., Fulton R. S., Kremitzki C., Koutseva N., Mueller J. L., Rozen S., Hughes J. F., Owens E., Womack J. E., Murphy W. J., Cao Q., de Jong P., Warren W. C., Wilson R. K., Skaletsky H., Page D. C., Sequencing the mouse Y chromosome reveals convergent gene acquisition and amplification on both sex chromosomes. Cell 159, 800–813 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Ellis P. J. I., Bacon J., Affara N. A., Association of Sly with sex-linked gene amplification during mouse evolution: A side effect of genomic conflict in spermatids? Hum. Mol. Genet. 20, 3010–3021 (2011). [DOI] [PubMed] [Google Scholar]
- 36.Cocquet J., Ellis P. J., Yamauchi Y., Riel J. M., Karacs T. P., Rattigan A., Ojarikre O. A., Affara N. A., Ward M. A., Burgoyne P. S., Deficiency in the multicopy Sycp3-like X-linked genes Slx and Slxl1 causes major defects in spermatid differentiation. Mol. Biol. Cell 21, 3497–3505 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Moretti C., Serrentino M. E., Ialy-Radio C., Delessard M., Soboleva T. A., Tores F., Leduc M., Nitschke P., Drevet J. R., Tremethick D. J., Vaiman D., Kocer A., Cocquet J., SLY regulates genes involved in chromatin remodeling and interacts with TBL1XR1 during sperm differentiation. Cell Death Differ. 24, 1029–1044 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Moretti C., Blanco M., Ialy-Radio C., Serrentino M. E., Gobe C., Friedman R., Battail C., Leduc M., Ward M. A., Vaiman D., Tores F., Cocquet J., Battle of the sex chromosomes: Competition between X- and Y-chromosome encoded proteins for partner interaction and chromatin occupancy drives multi-copy gene expression and evolution in muroid rodents. Mol. Biol. Evol. 37, 3453–3468 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Ellis P. J., Clemente E. J., Ball P., Toure A., Ferguson L., Turner J. M., Loveland K. L., Affara N. A., Burgoyne P. S., Deletions on mouse Yq lead to upregulation of multiple X- and Y-linked transcripts in spermatids. Hum. Mol. Genet. 14, 2705–2715 (2005). [DOI] [PubMed] [Google Scholar]
- 40.Kruger A. N., Brogley M. A., Huizinga J. L., Kidd J. M., de Rooij D. G., Hu Y. C., Mueller J. L., A neofunctionalized X-linked ampliconic gene family is essential for male fertility and equal sex ratio in mice. Curr. Biol. 29, 3699–3706.e5 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Akerfelt M., Vihervaara A., Laiho A., Conter A., Christians E. S., Sistonen L., Henriksson E., Heat shock transcription factor 1 localizes to sex chromatin during meiotic repression. J. Biol. Chem. 285, 34469–34476 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Wang G., Zhang J., Moskophidis D., Mivechi N. F., Targeted disruption of the heat shock transcription factor (hsf)-2 gene results in increased embryonic lethality, neuronal defects, and reduced spermatogenesis. Genesis 36, 48–61 (2003). [DOI] [PubMed] [Google Scholar]
- 43.Bjork J. K., Sandqvist A., Elsing A. N., Kotaja N., Sistonen L., miR-18, a member of Oncomir-1, targets heat shock transcription factor 2 in spermatogenesis. Development 137, 3177–3184 (2010). [DOI] [PubMed] [Google Scholar]
- 44.Shamovsky I., Ivannikov M., Kandel E. S., Gershon D., Nudler E., RNA-mediated response to heat shock in mammalian cells. Nature 440, 556–560 (2006). [DOI] [PubMed] [Google Scholar]
- 45.Ventela S., Toppari J., Parvinen M., Intercellular organelle traffic through cytoplasmic bridges in early spermatids of the rat: Mechanisms of haploid gene product sharing. Mol. Biol. Cell 14, 2768–2780 (2003). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Bhutani K., Stansifer K., Ticau S., Bojic L., Villani A. C., Slisz J., Cremers C. M., Roy C., Donovan J., Fiske B., Friedman R. C., Widespread haploid-biased gene expression enables sperm-level natural selection. Science 371, eabb1723 (2021). [DOI] [PubMed] [Google Scholar]
- 47.Rathje C. C., Johnson E. E. P., Drage D., Patinioti C., Silvestri G., Affara N. A., Ialy-Radio C., Cocquet J., Skinner B. M., Ellis P. J. I., Differential sperm motility mediates the sex ratio drive shaping mouse sex chromosome evolution. Curr. Biol. 29, 3692–3698.e4 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Krausz C., Y chromosome and male infertility. Andrologia 37, 219–223 (2005). [DOI] [PubMed] [Google Scholar]
- 49.Xie Y., Yao J., Zhang X., Chen J., Gao Y., Zhang C., Chen H., Wang Z., Zhao Z., Chen W., Lv L., Li Y., Gao F., Xie M., Zhang J., Zhao L., Wang Z., Liang X., Sun X., Zou X., Deng C., Liu G., A panel of extracellular vesicle long noncoding RNAs in seminal plasma for predicting testicular spermatozoa in nonobstructive azoospermia patients. Hum. Reprod. 35, 2413–2427 (2020). [DOI] [PubMed] [Google Scholar]
- 50.Livak K. J., Schmittgen T. D., Analysis of relative gene expression data using real-time quantitative PCR and the 2−ΔΔCT method. Methods 25, 402–408 (2001). [DOI] [PubMed] [Google Scholar]
- 51.Reynard L. N., Cocquet J., Burgoyne P. S., The multi-copy mouse gene Sycp3-like Y-linked (Sly) encodes an abundant spermatid protein that interacts with a histone acetyltransferase and an acrosomal protein. Biol. Reprod. 81, 250–257 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Reynard L. N., Turner J. M., Cocquet J., Mahadevaiah S. K., Toure A., Hoog C., Burgoyne P. S., Expression analysis of the mouse multi-copy X-linked gene Xlr-related, meiosis-regulated (Xmr), reveals that Xmr encodes a spermatid-expressed cytoplasmic protein, SLX/XMR1. Biol. Reprod. 77, 329–335 (2007). [DOI] [PubMed] [Google Scholar]
- 53.Meistrich M. L., Hess R. A., Assessment of spermatogenesis through staging of seminiferous tubules. Methods Mol. Biol. 927, 299–307 (2013). [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary material for this article is available at http://advances.sciencemag.org/cgi/content/full/7/24/eabg5177/DC1


