Abstract
INTRODUCTION:
Mounting evidence demonstrates potential for fecal–oral transmission of severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2). The US Food and Drug Administration now requires SARS-CoV-2 testing of potential feces donors before the use of stool manufactured for fecal microbiota transplantation. We sought to develop and validate a high-sensitivity SARS-CoV-2 reverse transcriptase polymerase chain reaction (RT-PCR) procedure for testing stool specimens.
METHODS:
A modified extraction method was used with an RT-PCR assay adapted from the Centers for Disease Control and Prevention PCR protocol for respiratory specimens. Contrived specimens were created using pre-COVID-19 banked stool specimens and spiking in known concentrations of SARS-CoV-2-specific nucleic acid. The highest transcript concentration at which 2/2 or 1/2 SARS-CoV-2 targets were detected in 9/10 replicates was defined as the dual-target limit and single-target limit of detection, respectively. The clinical performance of the assay was evaluated with stool samples collected from 17 nasopharyngeal swab RT-PCR-positive patients and 14 nasopharyngeal RT-PCR-negative patients.
RESULTS:
The dual-target and single-target limit of detection were 56 copies/μL and 3 copies/μL, respectively. SARS-CoV-2 was detected at concentrations as low as 0.6 copies/μL. Clinical stool samples from known COVID-19-positive patients demonstrated the detection of SARS-CoV-2 in stool up to 29 days from symptom onset with a high agreement with nasopharyngeal swab tests (kappa statistic of 0.95, P value < 0.001).
DISCUSSION:
The described RT-PCR test is a sensitive and flexible approach for the detection of SARS-CoV-2 in stool specimens. We propose an integrated screening approach that incorporates this stool test to support continuation of fecal microbiota transplantation programs.
INTRODUCTION
Coronavirus-2019 (COVID-19) has emerged as a global pandemic. Since the recognition of severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) on December 12, 2019, more than 64 million people have been infected with more than 1.4 million attributed deaths globally as of December 2, 2020 (1).
SARS-CoV-2 has been detected in a plethora of tissue types and clinical specimens, including stool (2). In addition to the detection of SARS-CoV-2 nucleic acid, active replication within enterocytes (3,4), abundant gastrointestinal glandular cell ACE-2 expression (the target receptor for SARS-CoV-2), and recovery of viable viruses from stool have been reported (4,5). This evidence suggests potential for fecal–oral transmission of SARS-CoV-2. In response, the US Food and Drug Administration (FDA) issued a safety alert about the risk of transmission of SARS-CoV-2 through fecal microbiota transplantation (FMT) procedures. The FDA also issued a partial clinical hold for all investigational new drug studies using stool products manufactured after December 1, 2019 (6). To address this risk, the FDA now requires screening potential donors for SARS-CoV-2 symptoms, previous positive tests, and exposures, in addition to testing donors and/or stool for SARS-CoV-2. The FDA also requires discussion of limitations of these testing procedures and risk of SARS-CoV-2 transmission from FMT as part of the informed consent process. Such measures are prudent and timely, given the recent safety concerns from transmission of other pathogens after FMT procedures (7,8). However, many of the currently used COVID-19 diagnostic tests granted Emergency Use Authorization may not apply to testing of stool because it may not be considered an authorized emergent need. Commercially available platforms can have had unpredictable supply chain shortages and may be less flexible for pooling or other screening strategies, demonstrating a need to develop a high-sensitivity stool PCR assay for SARS-CoV-2 (9,10).
Herein, we describe the development and validation of a high-sensitivity reverse transcriptase polymerase chain reaction (RT-PCR) procedure for the detection of SARS-CoV-2 in stool and screening of FMT donors to support adaptation of FMT banking programs to the era after the emergence of COVID-19.
METHODS
This study was conducted at the Emory University Clinical Virology Research Laboratory. The Clinical Virology Research Laboratory is a The Clinical Laboratory Improvement Amendments-certified laboratory that serves as the diagnostic support laboratory for the Emory Microbiome Enrichment Program, which was founded in 2012 and has coordinated administration of over 400 FMTs. Stool samples were obtained from a convenience sample of discarded clinical microbiology laboratory stool specimens from admitted patients with respiratory testing for SARS-CoV-2 (STUDY00001658) and participants in an ongoing clinical trial (institutional review board 00022371). The samples were collected between May 2, 2020, and September 30, 2020, at Emory University Hospital in Atlanta, Georgia.
Laboratory methods
Stool samples were suspended in Qiagen PowerBead lysis buffer and Qiagen CI buffer (Qiagen, Germantown, MD). Using a shaker set at 37 °C, the samples were incubated and underwent bead beating for 15 minutes, followed by centrifugation for 1 min at 1,300 relative centrifugal force. Next, 200 μL of lysed and bead-beaten sample was extracted using the Qiagen EZ1 advanced automated extractor with the Virus DSP Kit plus the Virus Card (Qiagen). Extracted samples underwent RT-PCR for selected gene regions of the SARS-CoV-2 virus nucleocapsid (N1, N2) and human RNase P gene using a protocol adapted from the Centers for Disease Control and Prevention (11,12). Briefly, after RNA extraction, 20 μL reactions were set up, containing 5 μL of sample RNA, 8.5 μL of nuclease free water, 1.5 μL of combined primer/probe mix, and 5 μL of TaqPath 1-Step RT-qPCR Master Mix (Thermo Fisher, Waltham, MA). Thermal cycling was performed at 25 °C for 2 minutes, followed by 50 °C for 15 minutes, followed by an initial denaturation at 95 °C for 2 minutes, followed by 45 cycles of amplification at 95 °C for 3 seconds and 55.0 °C for 30 seconds (Thermo Fisher 7500). A previously quantitated SARS-CoV-2 plasmid material (Integrated DNA Technologies, Coralville, IA) at a concentration of 50 copies/µL was extracted and tested concurrently as a positive control on all runs. Exponential growth curves that crossed the threshold line within 40 cycles (cycle threshold [CT] < 40) were considered positive.
Validation study
Our assay was previously validated on upper and lower respiratory and serum samples (see Supplementary Material, Supplementary Digital Content 1, http://links.lww.com/CTG/A636). We performed a limit of detection (LoD) range-finding experiment in stool samples by testing serial dilutions (140, 111, 56, 28, 6, 3 and 0.6 copies/µL) of quantitated SARS-CoV-2 plasmid material (Integrated DNA Technologies). Specimens were created using pre-COVID-19 banked stool specimens and spiking in plasmid material. A confirmation of the LoD was determined by repeating initial LoD at 10 replicates. The highest dilution of transcript at which both SARS-CoV-2 targets were detected in 9/10 replicates was defined as the dual target LoD of our assay. The highest dilution of transcript at which any SARS-CoV-2 target was detected (motivated by intent to exclude stool samples with any positive SARS-CoV-2 result) was defined as the single target LoD; the acceptance criteria for the performance was prespecified as 90% agreement. We performed interinstrument reproducibility studies using 2 Applied Biosystems Integrated 7500 PCR instruments (Thermo Fisher). To assess specimen stability, we tested contrived positive samples refrigerated (4 °C) at 7 days.
Clinical specimen testing
The clinical performance of the assay was evaluated with stool samples collected from 17 nasopharyngeal swab RT-PCR-positive patients and 14 nasopharyngeal RT-PCR-negative patients. Clinical information was available to the performer/reader of this assay. A subset of these (5 NP RT-PCR-positive and 3 NP RT-PCR-negative patients) were tested in duplicate on a previously described triplex laboratory-developed test (LDT) that targets N2, envelope, and the human RNAase P genes (13). Clinical information was not available to the performers/readers of the triplex test. NP swab testing was performed by the molecular section of Emory Medical Laboratories as part of routine clinical care on the Roche 6,800 cobas assay (Roche, Basel, Switzerland) which tests targets ORF1 a/b nonstructural region that is unique to SARS-CoV-2 and a pan-Sarbecovirus conserved region in the structural protein envelope E-gene. Days from symptom onset were retrieved retrospectively from the electronic medical record through an Emory University institutional review board-approved protocol STUDY00000260.
Statistical analysis
As previously performed specificity testing has shown no cross reactivity with N1, N2 primer set of the assay, a positive result was determined as either a positive N1 or N2 target. CT values from SARS-CoV-2-specific targets, ORF1 a/b from NP specimens, and N2 (or in cases where N2 was negative N1 [n = 1]) from stool specimens where compared. For subjects who had multiple NP test results, the test closest to the date of stool testing was chosen. Cohen kappa coefficient was used to assess the agreement between stool and NP swab RT-PCR results. NP RT-PCR was considered the gold standard test based on previous work by our group showing high negative predictive value (100%) of NP RT-PCR when compared with unbiased metagenomic sequencing (14). Wilcoxon matched-pairs signed-rank 2-tailed test was used to compare the mean N1/N2 and ORF1 a/b CT values. Negative predictive values were calculated using clinical specimens with known respiratory SARS-CoV-2 results. The performance of stool RT-PCR compared with NP RT-PCR was compared using a receiver operating characteristic curve analysis, and the area under the receiver operating characteristic curve was determined. Statistical analysis was performed using R version 4.0.2 (Vienna, Austria) and the RStudio interface version 1.3.1073 (Boston, MA).
RESULT
LoD
We tested serial dilutions of quantified RNA prepared previously described to determine the test LoD. The dual-target LoD of our assay was found to be 56 copies/µL, and the single-target LoD was found to be 3 copies/µL (Table 1). SARS-CoV-2 was detected at concentrations as low as 0.6 copies/µL (Table 1).
Table 1.
Limit of SARS-CoV-2 target sequence detection with high-sensitivity stool RT-PCR (n = 59)
| Target | Percent positivity (%) | Mean CTa (SD) | |||||
| N1 | N2 | RNAseP | Overall | N1 | N2 | RNAseP | |
| RNA conc. (Copies/μL) | |||||||
| 140 (n = 5) | 100 | 100 | 100 | 100 | 29.1 (1.2) | 31.1 (1.6) | 32.5 (1.4) |
| 111 (n = 9) | 100 | 100 | 89 | 100 | 30.3 (1.6) | 32.3 (0.8) | 31.0 (4.2) |
| 56 (n = 10) | 100 | 100 | 90 | 100 | 31.6 (1.3) | 34.5 (0.8) | 33.8 (3.9) |
| 28 (n = 5) | 100 | 100 | 100 | 100 | 32.6 (0.9) | 35.0 (0.9) | 35.7 (2.4) |
| 6 (n = 10) | 80 | 90 | 100 | 100 | 34.4 (1.1) | 36.2 (1.5) | 34.5 (1.2) |
| 3 (n = 10) | 60 | 70 | 90 | 90 | 34.3 (0.5) | 36.2 (1.1) | 34.3 (1.1) |
| 0.6 (n = 10) | 40 | 30 | 90 | 50 | 34.0 (0.7) | 36.2 (1.7) | 34.4 (1.6) |
CT, cycle threshold; RT-PCR, reverse transcriptase polymerase chain reaction; SARS-CoV-2, severe acute respiratory syndrome coronavirus 2.
Calculated for detected samples.
Clinical testing
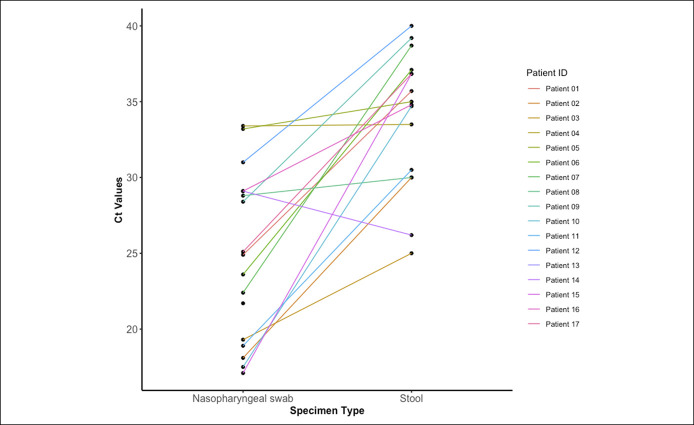
Among NP RT-PCR-positive patients (n = 17), SARS-CoV-2 was detected in 16 (94%) with 87.5% (14/16) testing positive for both targets. All NP RT-PCR-positive patients underwent NP swab and stool RT-PCR testing within a median (range) of 7 (1–43) days and 11 (3–29) days, respectively, from symptom onset (see Supplementary Figure 1, Supplemental Table 1, Supplementary Digital Content 1, http://links.lww.com/CTG/A636). Demographic and clinical characteristics are summarized in Supplementary Digital Content 1, (see Supplemental Table 2, http://links.lww.com/CTG/A636). SARS-CoV-2 was not detected in stool collected from any NP RT-PCR-negative patients (0/14, 0%). This resulted in a high agreement between NP and stool RT-PCR testing (kappa statistic of 0.94, P value < 0.001) with an area under the receiver operating characteristic curve of 0.971, a sensitivity of 1.00 (95% confidence interval [CI]: 0.79–1.00), specificity of 0.93 (95% CI: 0.79–1.00), a positive predictive value of 0.94 (95% CI: 0.71–1.00), and a negative predictive value of 1.00 (95% CI: 0.77–1.00) using respiratory testing as a gold standard (see Supplementary Table 1, Supplementary Digital Content 1, http://links.lww.com/CTG/A636). Median stool CT value was significantly higher than NP swab ORF1 a/b CT value (34.9 [6.5] vs 24.9 [9.8]; P value < 0.001) (Figure 1; Supplementary Figure 2, 3,http://links.lww.com/CTG/A636). Stool CT values did not correlate with NP CT values (R2: −0.04, P value = 0.50) (Table 1; see Supplementary Figure 4, Supplementary Digital Content 1, http://links.lww.com/CTG/A636). Storage at 4 °C and retesting of 2 positive and 1 negative stool specimens yielded similar results as initial testing.
Figure 1.
Paired plot of stool CT and NP swab CT values for NP RT-PCR-positive patients (n = 17). CT, cycle threshold; NP, nasopharyngeal; RT-PCR, reverse transcriptase polymerase chain reaction.
DISCUSSION
In this validation of a high-sensitivity SARS-CoV-2 stool RT-PCR test, we were able to frequently detect SARS-CoV-2 in stool at concentrations as low as 0.6 copies/µL. Furthermore, we were able to detect COVID-19 in the stool of COVID-19 NP RT-PCR-positive patients up to 29 days from symptom onset with a high agreement with NP RT-PCR testing. These results enhance the confidence of SARS-CoV-2 screening measures for FMT programs after the emergence of COVID-19.
As with SARS and middle east respiratory syndrome-CoV, the fecal–oral route of transmission has been suspected with SARS-CoV-2 (15). Not only have recent studies been able to detect SARS-CoV-2 and culture virus in the stool of patients with COVID-19 (16), they have detected SARS-CoV-2 RNA for longer periods than in upper respiratory samples (17). Moreover, a transmission cluster in a high-rise building which was investigated by epidemiologic tracing, environmental testing, and release of tracer gas led the authors to conclude the source of the outbreak to be fecal aerosols generated during toilet flushing after use by the index patients (18).
Given the above data, direct PCR testing for SARS-CoV-2 of stool specimens may add value for public health surveillance of sewage streams (19) and in the clinical scenarios of negative respiratory tract testing with high clinical suspicion of COVID-19. One such scenario that may reflect higher risk from false-negative diagnostic results is in screening of potential organ donors for potential donor-derived infections before procurement. Bypassing lower respiratory tract sampling with stool tests may also have additional value in avoiding exposure risks of aerosol-generating procedures or for pediatric patients for whom respiratory specimen collection may be more challenging (10). Outside of its role in clinical diagnostics, stool testing capacity for SARS-CoV-2 is an important step toward safe resumption of FMT in clinical and research settings.
FMT, the process of transplanting stool from a healthy donor to a diseased recipient, is highly effective for the treatment of recurrent Clostridium difficile infection (RCDI) and generally associated with mild adverse effects. FMT is approximately 90% efficacious for the treatment of RCDI (20) and has been included in major society guidelines for RCDI management (21). Use of FMT has expanded in research settings to multiple disease models such as inflammatory bowel disease, hepatic encephalopathy, and eradication of multidrug-resistant organism colonization and subsequent infection (22). When efficacy is observed, FMT is often followed by shifts in microbial community structure and function from configurations associated with disease toward those that more closely resemble healthy individuals. Although adverse events after FMT are generally mild and infrequent, a 2019 report highlighted the risk for transmission of multidrug-resistant Escherichia coli with a resultant fatal infection and a 2020 report described multiple transmissions of E. coli pathotypes (7,8). These safety events underscored the importance of an iterative approach to feces donor screening practices, which included expanded stool testing for multidrug-resistant organisms and specific pathotypes. Although we are aware of no reported cases of SARS-CoV-2 transmission related to FMT, there is a clear need for a highly sensitive stool test, given evidence supporting the potential for fecal–oral spread.
Despite the clear need and benefit of stool testing for clinical diagnosis and FMT screening, many potential hurdles and unanswered questions remain. Many available commercial tests have been validated on (and thus approved solely for) upper and lower respiratory sample testing. Only recently have such platforms been evaluated on stool specimens (10). However, the utility of these commercial high throughput platforms may be hindered by supply chain limitations. In addition, differences in LoD have been observed for upper respiratory samples between our LDTs and similar commercial platforms (10 vs 250 copies/µL) (23). Moreover, the viral load of SARS-CoV-2 may fluctuate throughout the clinical course and dip below the LoD of a test with poor analytical sensitivity. To improve our LoD, we used a separate stool-specific extraction step, which may aid with PCR inhibitors found in stool such as complex bile salts, urea, and glycolipids (24). With continued sustained SARS-CoV-2 transmission in communities globally, evidence of nondurable immunity, and reinfection, FMT centers must adapt screening practices to minimize potential transmission risks.
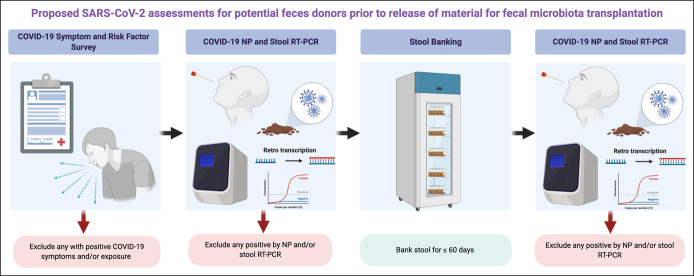
We propose a multidimensional screening protocol that applies the principles for stool donor screening from the pre-COVID-19 era (Figure 2). First, donors with symptoms and epidemiological risk factor should be excluded. Second, bookend respiratory testing should be performed. Finally, SARS-CoV-2 PCR should be performed on each donated stool specimen (even in the absence of positive symptoms or risk factors) before release for administration. At our center, we also include serological screening at the beginning and end of each banking period to contextualize potential donor risk of SARS-CoV-2 infection. Isolated positive SARS-CoV-2 serology test results could represent previous infection or expected response after vaccination and should not necessarily be exclusionary in the absence of contemporaneous respiratory or stool PCR test results but may add qualitative value in characterizing donor risk of infection. At this time, we have not evaluated the timing of further evaluation of donors with positive screening questions, exposures, or positive tests, although, as more of the global population is expected to be exposed with ongoing transmission, this will likely become an increasingly important consideration. We agree with the FDA recommendation to destroy any stool specimens collected during a bookend banking period and 4 weeks before any positive SARS-CoV-2 PCR result, even if a single target is detected. A recently published simulation modeling study suggested that the proportion of positive samples after testing every donation would have the lowest proportion of positive released samples but would be balanced by the release of fewer negative samples because of potential false positives (25). Prospective validation of this approach could support a more streamlined algorithm (e.g., PCR testing of stool alone without respiratory sample testing), but we suggest erring on the side of caution given unpredictable features of SARS-CoV-2 shedding, transmission, and increased attention to safety of FMT, given the recent pathogen transmission events (7,8).
Figure 2.
Proposed SARS-CoV-2 assessments for potential feces donors before the release of material for FMT. Outlining initial screening questionnaire for exposures and symptoms for COVID-19 (a), followed by initial nasopharyngeal and stool testing of SARS-CoV-2 (b), followed by banking and storage of aliquots for SARS-CoV-2 testing of each sample for less than 60 days (c), followed by release of SARS-CoV-2 nasopharyngeal, and stool testing 2–4 weeks after last stool donation. Figure created with Biorender.com. COVID-19, Coronavirus 2019; FMT, fecal microbiota transplant; NP, Nasopharyngeal; RT-PCR, Reverse transcriptase polymerase chain reaction; SARS-CoV-2, severe acute respiratory syndrome coronavirus 2.
Although our sample size is limited, the rigor of our validation approach is similar to other validated diagnostics and we confirm the ability to adapt our current upper respiratory LDT assay with strong performance for excluding SARS-CoV-2-positive stool. Further studies are required to explore the true prevalence of fecal shedding in those with confirmed COVID-19, duration of shedding in stool and, most importantly, the role of virus found (via RT-PCR or culture) as a source of transmission. In the meantime, continued development of stool diagnostics and screening algorithms are required to ensure safe and efficacious delivery of FMT to patients with conditions with limited alternative treatment options.
CONFLICTS OF INTEREST
Guarantor of the article: Michael H. Woodworth, MD, MSc.
Specific author contributions: M.H.W. conceived the study. A.B. and M.H.W. designed the study. A.S.W., K.J.B., and M.W.A. collected the samples. J.M.I. and V.S. performed the experiments and J.J.W. and C.S.K. provided laboratory supervision. A.B. and M.H.W. analyzed the data. A.B., C.S.K., J.M.I., and M.H.W. interpreted the data. A.B. and M.H.W. drafted the manuscript. All authors reviewed and critically revised the manuscript for important intellectual content.
Financial support: This work was supported in part by the Center for AIDS Research (P30 AI050409) and the National Institute of Allergy and Infectious Diseases of the National Institutes of Health under Award Number K23AI144036. The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institutes of Health.
Potential competing interests: None to report.
Study Highlights.
WHAT IS KNOWN
✓ Severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) can be found in stool and may be transmissible by fecal microbiota transplanation.
✓ The US Food and Drug Administration now requires screening potential stool donors for SARS-CoV-2.
WHAT IS NEW HERE
✓ A high-sensitivity polymerase chain reaction assay can detect SARS-CoV-2 at concentrations as low as 0.6 copies/μL.
✓ Stool SARS-CoV-2 test results are highly concordant with nasopharyngeal test results.
✓ SARS-CoV-2 can be detected in stool up to 29 days from symptom onset.
✓ Fecal microbiota transplanation donor screening should incorporate high-sensitivity stool and respiratory testing with COVID-19 symptom screening.
Supplementary Material
Footnotes
SUPPLEMENTARY MATERIAL accompanies this paper at http://links.lww.com/CTG/A636
Contributor Information
Jessica M. Ingersoll, Email: jingers@emory.edu.
Max W. Adelman, Email: max.w.adelman@emory.edu.
Andrew S. Webster, Email: andrew.scott.webster@emory.edu.
Kari J. Broder, Email: kari.jordan.broder@emory.edu.
Victoria Stittleburg, Email: victoria.d.simmons@emory.edu.
Jesse J. Waggoner, Email: jesse.waggoner@emoryhealthcare.org.
Colleen S. Kraft, Email: coleen.kraft@emory.edu.
Michael H. Woodworth, Email: michael.holmes.woodworth@emory.edu.
REFERENCES
- 1.Dong E, Du H, Gardner L. An interactive web-based dashboard to track COVID-19 in real time. Lancet Infect Dis 2020;20:533–4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Wang W, Xu Y, Gao R, et al. Detection of SARS-CoV-2 in different types of clinical specimens. JAMA 2020;323:1843–4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Wölfel R, Corman VM, Guggemos W, et al. Virological assessment of hospitalized patients with COVID-2019. Nature 2020;581:465–9. [DOI] [PubMed] [Google Scholar]
- 4.Xiao F, Tang M, Zheng X, et al. Evidence for gastrointestinal infection of SARS-CoV-2. Gastroenterology 2020;158:1831–33.e3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Jeong HW, Kim SM, Kim HS, et al. Viable SARS-CoV-2 in various specimens from COVID-19 patients. Clin Microbiol Infect 2020;26:1520–24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.US Food and Drug Administration. Safety Alert: Fecal Microbiota for Transplantation: New Safety Information—Regarding Additional Protections for Screening Donors for COVID-19 and Exposure to SARS-CoV-2 and Testing for SARS-CoV-2. Food and Drug Administration: Silver Spring, MD, 2020. [Google Scholar]
- 7.DeFilipp Z, Bloom PP, Torres Soto M, et al. Drug-resistant E. coli bacteremia transmitted by fecal microbiota transplant. N Engl J Med 2019;381:2043–50. [DOI] [PubMed] [Google Scholar]
- 8.US Food and Drug Administration. Safety Alert Regarding Use of Fecal Microbiota for Transplantation and Risk of Serious Adverse Events Likely Due to Transmission of Pathogenic Organisms. Food and Drug Administration: Silver Spring, MD, 2020. [Google Scholar]
- 9.Babiker A, Myers CW, Hill CE, et al. SARS-CoV-2 testing. Am J Clin Pathol 2020;153:706–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Szymczak WA, Goldstein DY, Orner EP, et al. Utility of stool PCR for the diagnosis of COVID-19: Comparison of two commercial platforms. J Clin Microbiol 2020;58:e01369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Lu X, Wang L, Sakthivel SK, et al. US CDC real-time reverse transcription PCR panel for detection of Severe Acute Respiratory Syndrome Coronavirus 2. Emerg Infect Dis 2020;26:1654–65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Centers for Disease Control and Prevention. CDC 2019-Novel Coronavirus (2019-nCoV) Real-Time RT-PCR Diagnostic Panel. CDC/DDID/NCIRD/Division of Viral Diseases. Centers for Diseases Control and Prevention: Atlanta, GA, 2020. [Google Scholar]
- 13.Waggoner JJ, Stittleburg V, Pond R, et al. Triplex real-time RT-PCR for Severe Acute Respiratory Syndrome Coronavirus 2. Emerg Infect Dis 2020;26:1633–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Babiker A, Bradley HL, Stittleburg VD, et al. Metagenomic sequencing to detect respiratory viruses in persons under investigation for COVID-19. J Clin Microbiol 2020;59:e02142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Yeo C, Kaushal S, Yeo D. Enteric involvement of coronaviruses: Is faecal-oral transmission of SARS-CoV-2 possible? Lancet Gastroenterol Hepatol 2020;5:335–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Xiao F, Sun J, Xu Y, et al. Infectious SARS-CoV-2 in feces of patient with severe COVID-19. Emerg Infect Dis 2020;26:1920–2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Wu Y, Guo C, Tang L, et al. Prolonged presence of SARS-CoV-2 viral RNA in faecal samples. Lancet Gastroenterol Hepatol 2020;5:434–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kang M, Wei J, Yuan J, et al. Probable evidence of fecal aerosol transmission of SARS-CoV-2 in a high-rise building. Ann Intern Med 2020;173:974–80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Amin N, Liu P, Foster T, et al. Pathogen flows from on-site sanitation systems in low-income urban neighborhoods, dhaka: A quantitative environmental assessment. Int J Hyg Environ Health 2020;230:113619. [DOI] [PubMed] [Google Scholar]
- 20.Cammarota G, Masucci L, Ianiro G, et al. Randomised clinical trial: Faecal microbiota transplantation by colonoscopy vs. vancomycin for the treatment of recurrent Clostridium difficile infection. Aliment Pharmacol Ther 2015;41:835–43. [DOI] [PubMed] [Google Scholar]
- 21.McDonald LC, Gerding DN, Johnson S, et al. Clinical practice guidelines for Clostridium difficile infection in adults and children: 2017 update by the Infectious Diseases Society of America (IDSA) and Society for Healthcare Epidemiology of America (SHEA). Clin Infect Dis 2018;66:987–94. [DOI] [PubMed] [Google Scholar]
- 22.Woodworth MH, Hayden MK, Young VB, et al. The role of fecal microbiota transplantation in reducing intestinal colonization with antibiotic-resistant organisms: The current landscape and future directions. Open Forum Infect Dis 2019;6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Broder K, Babiker A, Myers C, et al. Test agreement between Roche cobas 6800 and cepheid GeneXpert xpress SARS-CoV-2 assays at high cycle threshold ranges. J Clin Microbiol 2020;58. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Schrader C, Schielke A, Ellerbroek L, et al. PCR inhibitors–occurrence, properties and removal. J Appl Microbiol 2012;113:1014–26. [DOI] [PubMed] [Google Scholar]
- 25.Olesen SW, Zaman A, Osman M, et al. Modeling donor screening strategies to reduce the risk of Severe Acute Respiratory Syndrome Coronavirus 2 transmission via fecal microbiota transplantation. Open Forum Infect Dis 2020;7:ofaa499. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.