Abstract
PURPOSE
Randomized trials established the superiority of ibrutinib-based therapy over chemoimmunotherapy in chronic lymphocytic leukemia. Durability of progression-free survival (PFS) with ibrutinib can vary by patient subgroup. Clinical tools for prognostication and risk-stratification are needed.
PATIENTS AND METHODS
Patients treated with ibrutinib in phase II and III trials provided the discovery data set and were subdivided into discovery and internal validation cohorts. An external validation cohort included 84 patients enrolled in our investigator-initiated phase II trial. Univariable analysis of 18 pretreatment parameters was performed using PFS and overall survival (OS) end-points. Multivariable analysis and machine-learning algorithms identified four factors for a prognostic model that was validated in internal and external cohorts.
RESULTS
Factors independently associated with inferior PFS and OS were as follows: TP53 aberration, prior treatment, β-2 microglobulin ≥ 5 mg/L, and lactate dehydrogenase > 250 U/L. Each of these four factors contributed one point to a prognostic model that stratified patients into three risk groups: three to four points, high risk; two points, intermediate risk; zero to one point, low risk. The 3-year PFS rates for all 804 patients combined were 47%, 74%, and 87% for the high-, the intermediate-, and the low-risk group, respectively (P < .0001). The 3-year OS rates were 63%, 83%, and 93%, respectively (P < .0001). The model remained significant when applied to treatment-naïve and relapsed/refractory cohorts individually. For 84 patients in the external cohort, BTK and PLCG2 mutations were tested cross-sectionally and at progression. The cumulative incidences of mutations were strongly correlated with the model. In the external cohort, Richter’s transformation occurred in 17% of the high-risk group, and in no patient in the low-risk group.
CONCLUSION
Patients at increased risk of ibrutinib failure can be identified at treatment initiation and considered for clinical trials.
INTRODUCTION
Chronic lymphocytic leukemia (CLL) is a clonal expansion of mature B cells driven by constitutive activation of B-cell receptor (BCR) signaling1,2 and overexpression of the prosurvival BCL-2 protein.3 Randomized trials demonstrated that selective inhibition of BCR signaling or BCL-2 can substantially improve the outcome of patients with CLL when compared with chemoimmunotherapy in the first-line as well as in relapsed disease settings.4-8 To date, the US Food and Drug Administration has approved five targeted agents for the treatment of CLL: ibrutinib, acalabrutinib, idelalisib, duvelisib, and venetoclax. Of these, ibrutinib, a Bruton’s tyrosine kinase inhibitor, was the first to be approved and has the longest and broadest clinical data available.9-12
CONTEXT
Key Objective
We sought to develop a prognostic scoring system of survival for patients with chronic lymphocytic leukemia (CLL) treated with ibrutinib.
Knowledge Generated
TP53 aberration, relapsed/refractory CLL, β-2 microglobulin ≥ 5 mg/L, and lactate dehydrogenase > 250 U/L were independently associated with inferior progression-free and overall survival and were used to build a four-factor scoring system that stratified patients into high-, intermediate-, and low-risk groups. At 3 years, 53% of the patients in the high-risk group had progressed or died, in contrast to 13% in the low-risk group. BTK or PLCG2 mutations and Richter transformation were found most frequently in the high-risk group.
Relevance
In clinical practice, the score can help set expectations, guide monitoring frequency, and direct patients to risk-adapted treatment approaches on the basis of the predicted risk of early progression. In clinical trials, stratifying patients by risk score could more rapidly and clearly answer whether novel treatments improve outcomes for patients in greatest need.
Although ibrutinib is highly active in CLL, single-agent ibrutinib is not curative, and drug resistance can emerge. The clinical course of ibrutinib-resistant CLL can be aggressive,13 and the durability of response to alternative treatments is limited; most patients relapse within 2 years after starting venetoclax14 or CD19 chimeric antigen receptor-modified T cells.15 The most commonly identified mechanism for the emergence of ibrutinib-resistant CLL is an expansion of mutant subpopulations carrying BTK and/or PLCG2 mutations that reconstitute BCR signaling despite the presence of ibrutinib.16-18 The application of high-sensitivity testing can detect these mutations as early as 15 months before clinical progression.19 Moreover, ibrutinib-resistant CLL is clonally heterogeneous, which supports clinical observations related to a limited duration of response to alternative therapy administered after ibrutinib.
Risk stratification can help identify high-risk disease up front and enable informed therapeutic decision making. Equally important is the identification of low-risk patients who are likely to achieve durable responses to ibrutinib monotherapy. Although several biologic factors have been associated with the risk of disease progression,20-22 a robust, validated clinical tool that could be used for risk-adapted treatment approaches is lacking. Notably, the CLL International Prognostic Index (CLL-IPI) was validated in patients with treatment-naïve CLL (TN-CLL) in the chemoimmunotherapy era.23 Another prognostic tool estimated the survival of patients with relapsed/refractory CLL (RR-CLL) treated with heterogeneous regimens of both chemotherapy and targeted agents.24 None of the existing prognostic models have systematically explored biomarkers of ibrutinib resistance in the context of proposed clinical models. Here, we report a four-factor prognostic model predictive of progression-free survival (PFS) and overall survival (OS) and its validation in 804 patients with CLL treated uniformly with ibrutinib 420 mg per day.
PATIENTS AND METHODS
Study Design and Participants
The discovery data set was composed of 720 patients with CLL treated with ibrutinib in clinical trials: the phase Ib/II (ClinicalTrials.gov identifier: NCT01105247), RESONATE (ClinicalTrials.gov identifier: NCT01578707), RESONATE-2 (ClinicalTrials.gov identifier: NCT01722487), RESONATE-17 (ClinicalTrials.gov identifier: NCT01744691), and iLLUMINATE (ClinicalTrials.gov identifier: NCT02264574). Efficacy and safety outcomes have been published.6,10-12,25 For external validation, we used data from 84 patients treated with single-agent ibrutinib in a phase II trial at the National Institutes of Health (NIH; ClinicalTrials.gov identifier: NCT01500733).22,26 The clinical trials were approved by the responsible institutional review boards, and participants provided written informed consent. These trials tested ibrutinib-based treatment of CLL at 420 mg once per day and had a median follow-up of more than 30 months.
Statistical Analysis
We randomly divided the discovery data set into training (75%) and internal validation (25%) cohorts. In the training cohort, we performed traditional univariable and multivariable analyses for PFS and OS using stepwise selection. In parallel, we applied two supervised machine-learning methods using the lasso-based regularized Cox model27 and the random survival forests algorithm.28 Traditional and machine-learning methods consistently identified four independent variables that were used to construct a prognostic index. The model was tested in the internal and external validation cohorts, as well as in the combined cohort including all 804 patients. Detailed statistical methods are provided in the Data Supplement (online only).
Sequencing of BTK and PLCG2 Mutations
Samples collected in the NIH cohort were subjected to high-sensitivity sequencing of BTK and PLCG2 as described previously.19,29 The estimated sensitivity of the assay was 0.1%. Detailed testing methods are provided in the Data Supplement.
RESULTS
Patients, Treatment, and Predictors of Outcome
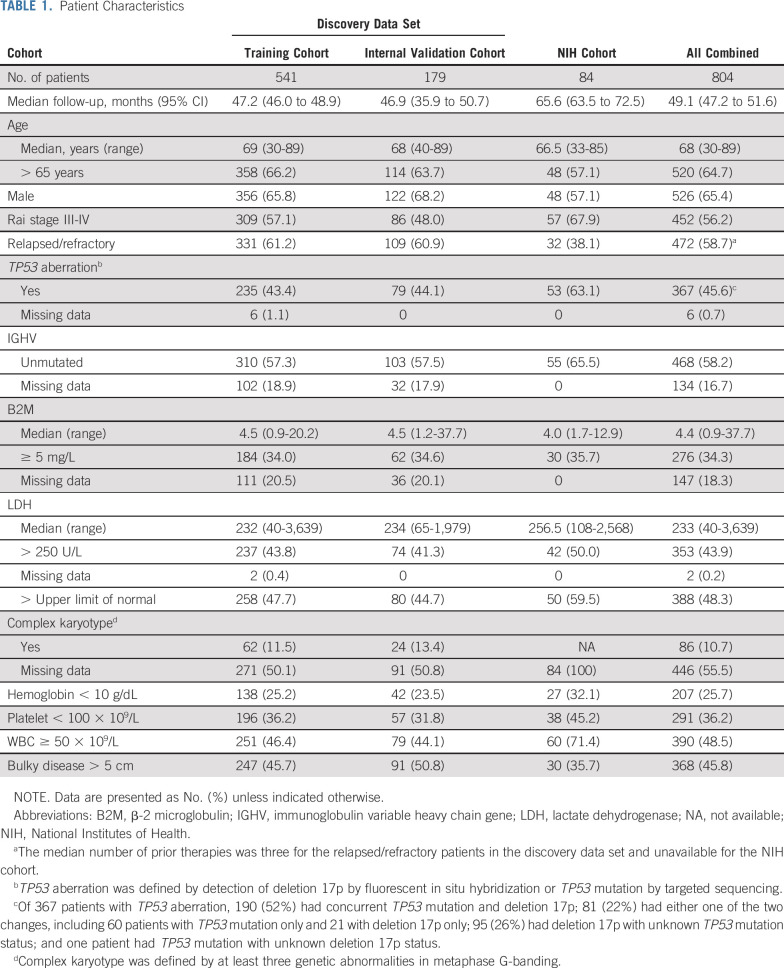
Table 1 lists the baseline characteristics of the 804 patients. Data for 720 patients in the discovery data set were aggregated from five industry-sponsored trials (Data Supplement). A majority of these patients had RR-CLL (61%) or an advanced Rai stage (55%). TP53 aberration was detected in 44%. Concurrent TP53 mutation and deletion 17p were detected in 71% of the patients with TP53 aberration who were tested for both. Eighty-four percent received single-agent ibrutinib, and 16%, ibrutinib with obinutuzumab. We also included 84 patients treated with single-agent ibrutinib in an investigator-initiated trial at the NIH. The NIH cohort was enriched with patients with TN-CLL (62%) and TP53 aberration (63%) per inclusion criteria. The median follow-up was 49 months, and the median OS was not reached for the entire data set.
TABLE 1.
Patient Characteristics
Univariable analysis of 18 baseline factors in the discovery cohort revealed 11 factors associated with inferior OS and PFS (Data Supplement). Rai stage was associated with inferior OS but not PFS. Conversely, younger age, unmutated immunoglobulin variable heavy-chain gene (IGHV), and lower absolute lymphocyte count were associated with inferior PFS but not OS. Complex karyotype was associated with both inferior PFS and inferior OS; no centralized testing was conducted, and data were missing in one half of the patients, precluding additional analyses.
For multivariable analysis, the discovery cohort was randomly divided 3:1 into training and internal validation cohorts. The four factors independently associated with both inferior PFS and inferior OS in the discovery cohort were as follows: RR-CLL, TP53 aberration, β-2 microglobulin (B2M) ≥ 5 mg/L, and lactate dehydrogenase (LDH) > 250 U/L. With each additional adverse factor present at baseline, PFS and OS probability decreased (P < .0001; Fig 1). All four factors demonstrated consistent prognostic effects in training and validation cohorts (Data Supplement).
FIG 1.
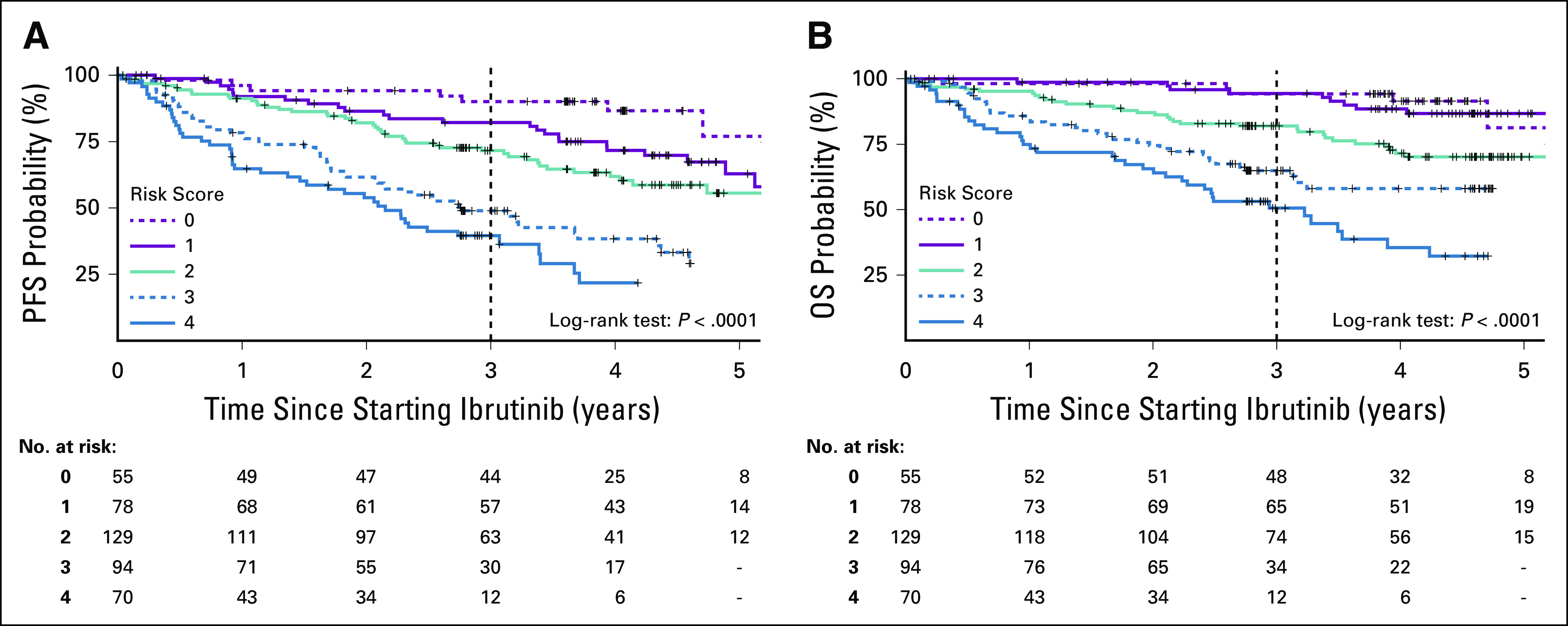
Survival by the number of risk factors present at the start of ibrutinib. Survival analysis was limited to patients treated with ibrutinib monotherapy. (A) Kaplan-Meier estimates of progression-free survival (PFS) in the combined cohort. (B) Kaplan-Meier estimates of overall survival (OS) in the combined cohort.
Machine-Learning Methods
We used two supervised machine-learning methods to confirm the validity of the factors identified from the traditional Cox regression analysis within the discovery data set. To determine the optimal number of factors included in a model, we used lasso Cox regression, which provides constraints to model complexity (Data Supplement). The four-factor model was the simplest model, with regression coefficients and hazard ratios (HRs) for PFS and OS within one standard deviation of those with the minimum cross-validation error. The random survival forests method, which used recursive binary splitting and 10-fold cross-validation, reaffirmed the top four variables for both PFS and OS as TP53 aberration, RR-CLL, high LDH, and high B2M (Data Supplement). Using continuous values for LDH and B2M revealed the same four factors. There was no interaction between B2M and LDH, confirming them as independent prognostic factors (Data Supplement).
Different B2M cutoffs have been adopted in existing prognostic tools. We tested 10 B2M cutoffs from 1 to 10 mg/L in the combined cohort using lasso Cox regression. HRs were similar among B2M cutoffs of 5, 6, and 7 mg/L and lower for ≤ 4 mg/L and ≥ 8 mg/L. We chose a B2M cutoff of 5 mg/L for the final model, given that the value was close to the median. Similarly, LDH > 250 U/L was chosen, given that 250 U/L was the lowest value to be selected in the model based on lasso Cox regression and was close to the median of the data set (233 U/L).
A Prognostic Index for Patients Treated With Ibrutinib
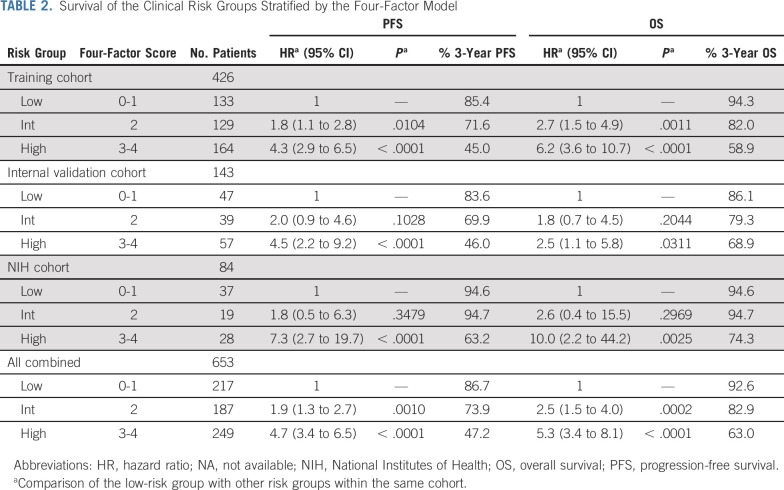
We used the four individually validated factors, which were TP53 aberration, RR-CLL, high LDH, and high B2M, to build a prognostic index (online calculator for the CLL 4-factor model is available at https://dir.nhlbi.nih.gov/lab/LLM/CLL4fxmodel/). On the basis of the separation of outcomes in the training cohort by the cumulative number of risk factors (Fig 1), we distinguished three prognostic risk groups: low risk, with zero to one factor present at the start of ibrutinib; intermediate risk, with two factors; and high risk, with three to four factors. In the training cohort, the intermediate- and high-risk groups had significantly inferior PFS and OS when compared with the low-risk group (Table 2; Fig 2; Data Supplement). In both internal and external validation cohorts, the high-risk group had significantly inferior PFS and OS when compared with the low-risk group. Likewise, HRs for PFS and OS were increased (range, 1.8-2.6) in the intermediate-risk group compared with the low-risk group; however, owing to the smaller cohort sizes, these differences did not consistently reach statistical significance. When combining all cohorts, the 3-year PFS rates for low-, intermediate-, and high-risk groups were 87%, 74%, and 47%, and the 3-year OS rates were 93%, 83%, and 63%, respectively (P < .0001).
TABLE 2.
Survival of the Clinical Risk Groups Stratified by the Four-Factor Model
FIG 2.
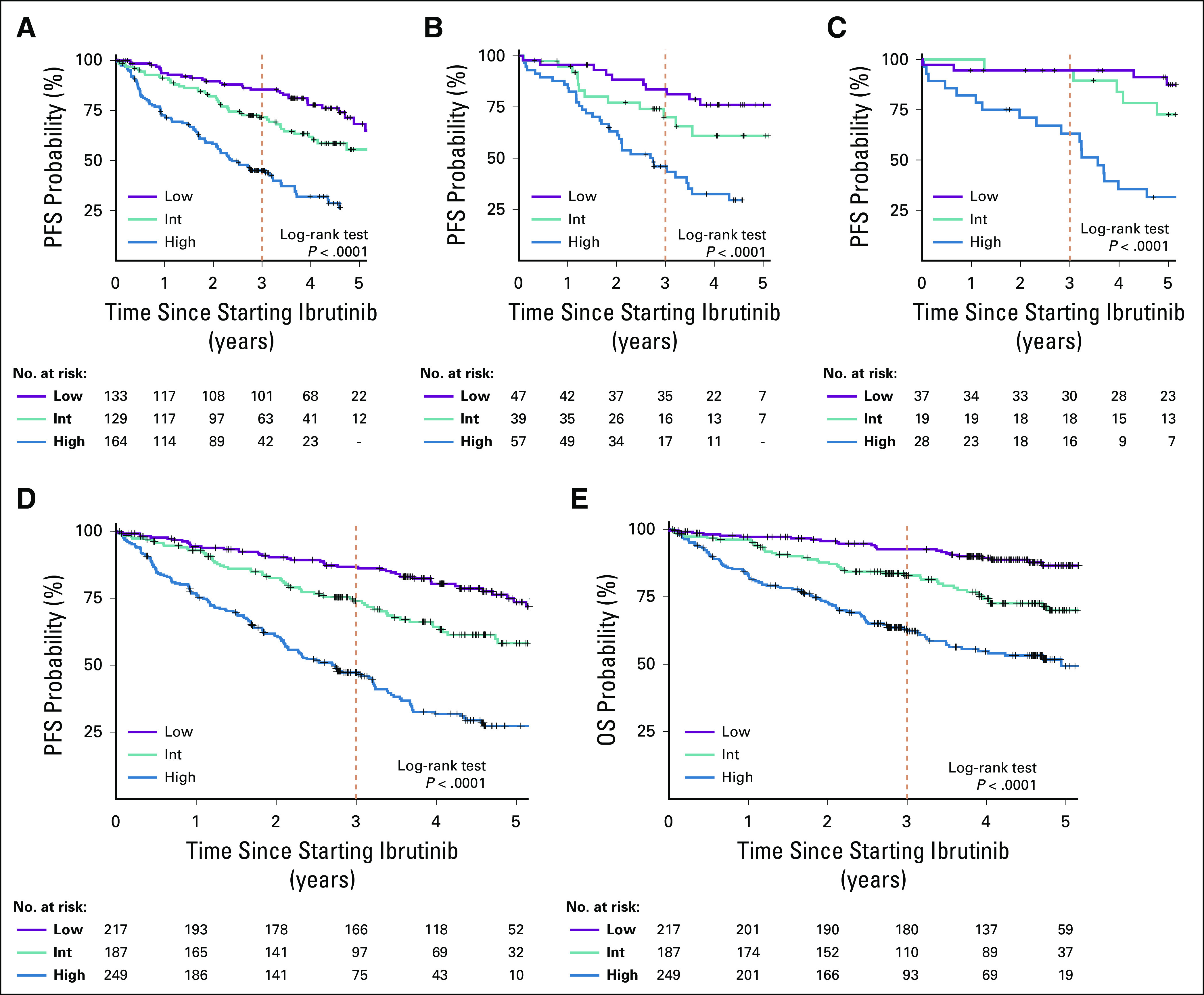
Validation of the prognostic index of survival in patients treated with ibrutinib. Survival analysis was limited to patients treated with ibrutinib monotherapy. Kaplan-Meier estimates of progression-free survival (PFS) in (A) the training cohort, (B) the internal validation cohort, and (C) the external validation cohort. (D) PFS and (E) overall survival (OS) in the combined cohort. High, high-risk group; Int, intermediate-risk group; Low, low-risk group.
Although RR-CLL contributes one factor to the prognostic index, the model retained statistical significance for PFS and OS in patients treated with ibrutinib in first-line therapy (Data Supplement). As expected, PFS and OS of the TN-CLL subset were more favorable across the risk groups compared with those of the RR-CLL subset.
To account for variation in reference ranges, we tested LDH > upper limit of normal (ULN) as an alternative to > 250 U/L. The modified model using LDH > ULN performed equally as well as the original four-factor model in predicting PFS and OS (Data Supplement). Thus, LDH cutoff > ULN can be used in settings in which the ULN does not approximate 250 U/L.
Comparison of the CLL-IPI With the Four-Factor Model
Two factors in this four-factor model are also used in the CLL-IPI, a weighted scoring system that considers age, Rai stage, B2M, IGHV, and TP53 aberration.23 We compared the performance of the CLL-IPI and the four-factor model in the 531 patients who had all the data points needed for risk categorization by both prognostic tools. The CLL-IPI assigned most patients to the high- (39%) or very high-risk groups (49%); 12% to the intermediate-risk group, and only three patients (0.6%) to the low-risk group (Data Supplement). More than a third (35%) of the patients in the very high-risk group of the CLL-IPI fell into the low- or intermediate-risk category of the four-factor model, demonstrating the limits of the CLL-IPI for patients treated with ibrutinib. The concordance test confirmed the lack of substantial agreement between the two prognostic tools (weighted κ = 0.45). The patients in the highest risk group by CLL-IPI had better PFS and OS than did those in the high-risk group by the four-factor model. For instance, 3-year PFS rates were 59% and 49% for the highest-risk group of the CLL-IPI and the four-factor model, respectively. Similarly, HRs observed in the four-factor model were consistently higher than those in the CLL-IPI, leading to improved discrimination of PFS (C-statistic = 0.69 for four-factor, 0.63 for CLL-IPI).
Incidence of BTK and/or PLCG2 Mutations in the Prognostic Subgroups
BTK and/or PLCG2 mutations are found in approximately 80% of patients who develop ibrutinib-resistant CLL and are often detectable many months before clinical progression.19,30 We cross-sectionally tested BTK and PLCG2 mutations using archived samples from the NIH cohort; 63% of this cohort had a TP53 aberration, and 62% had TN-CLL at baseline. With a cumulative observation time of 387 person-years, BTK/PLCG2 sequencing was performed 146 times, including 77 tests using cell-free DNA. Overall, 69 (82%) of 84 patients in the NIH cohort were tested at least once, and 40 patients (48%), at least twice. The frequency of testing was similar across the prognostic risk groups (P = .71). Each variant was manually reviewed and annotated for predicted pathogenicity (Data Supplement). The cumulative incidence of mutations was 50%, 40%, and 17% for the high-, the intermediate-, and the low-risk group, respectively (Fig 3A; Data Supplement). In the high-risk group, an additional 20% of patients had progression with no detectable mutations, often manifesting as Richter’s transformation. The cumulative incidence of Richter’s transformation was 17% in the high-, 5% in the intermediate-, and 0% in the low-risk group. Time to initial detection of mutations and time to clinical progression were highly correlated (ρ = 0.93, P < .0001) with up to 15 months from detection until clinical progression (Figs 3B-3C). The median time to initial detection of mutations was 36 months (range, 6-71.6 months). Across all risk groups, BTK/PLCG2 mutations were nearly three times more common in patients with the TP53 aberration (38%) than in those without the aberration (13%; Fig 3D).
FIG 3.
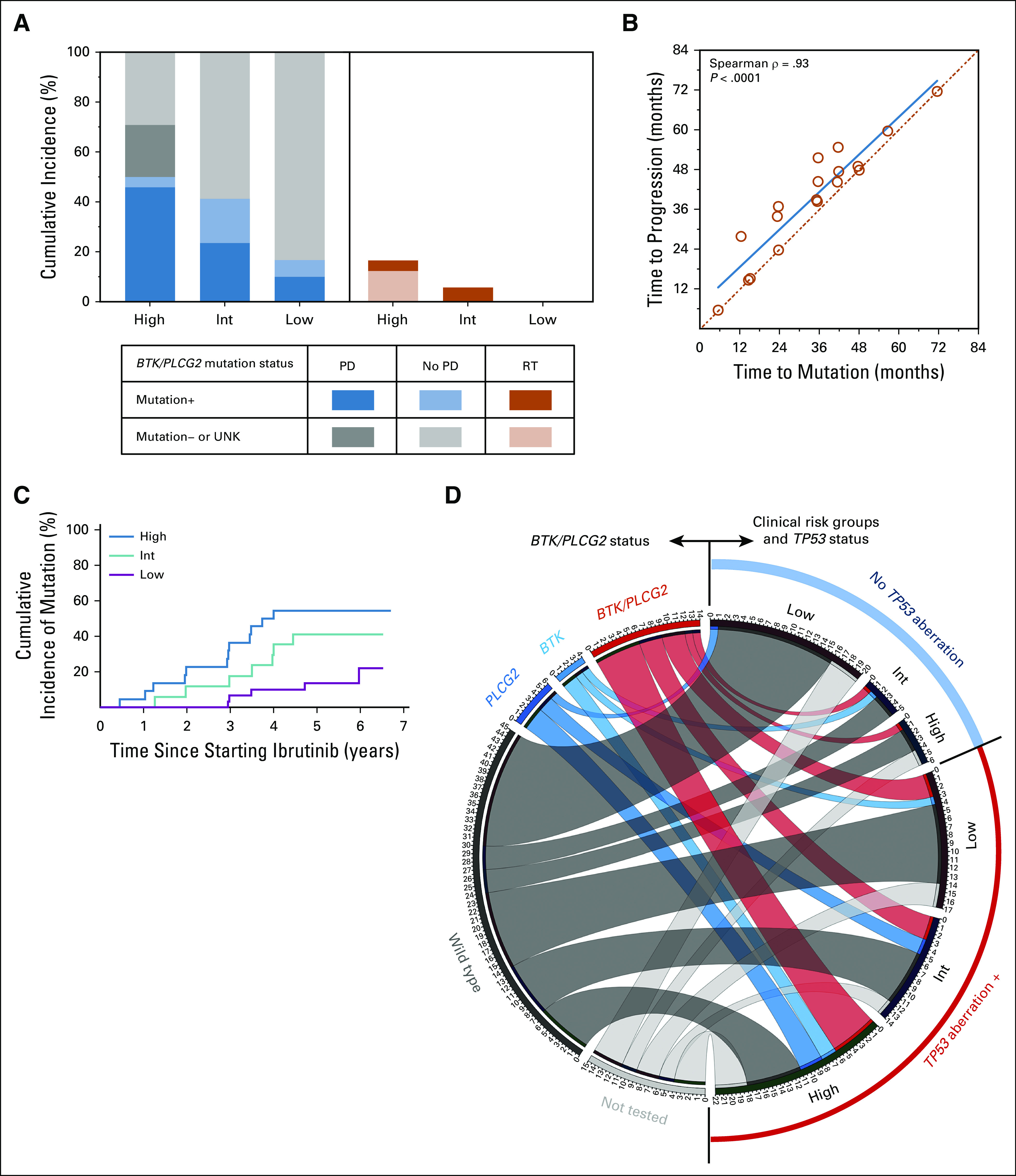
BTK and PLCG2 mutations in the National Institutes of Health cohort. (A) Cumulative incidence of BTK and/or PLCG2 mutations (left panel) and Richter’s transformation (RT; right panel) by prognostic risk groups. (B) Correlation of time to the first detection of BTK and/or PLCG2 mutations (time to mutation) and clinical progression (time to progression). (C) Cumulative incidence of BTK and/or PLCG2 mutations in the prognostic risk groups. (D) Circos plot showing types of mutations for patients in each clinical risk group subsetted by the status of TP53 aberration (TP53 mutation or deletion 17p). High, high-risk group; Int, intermediate-risk group; Low, low-risk group; PD, progressive disease with chronic lymphocytic leukemia or RT; UNK, unknown mutational status because of sample unavailability.
DISCUSSION
We developed a prognostic scoring system highly predictive of survival in patients with CLL treated with ibrutinib. TP53 aberration, RR-CLL, B2M ≥ 5 mg/L, and LDH > 250 U/L were independently associated with inferior PFS and OS. The four-factor scoring system stratified the prognosis of more than 800 patients into three tiers: high-, intermediate-, and low-risk groups. At 3 years, one half (53%) of the patients in the high-risk group had progressed or died, in contrast to the 13% observed in the low-risk group.
Several strengths distinguish this proposed prognostic model from others reported previously in CLL. First, the four-factor model is practical and easy to implement in general practice because it is built on parameters recommended for universal testing in CLL.31 Second, the patients in this study were uniformly treated with ibrutinib, either as monotherapy or in combination with obinutuzumab, and because they received long-term ibrutinib for more than 5 years, they provide a unique insight into long-term prognostic factors for ibrutinib treatment. Third, to our knowledge, this is the first study to apply machine-learning methods to optimize a prognostic model in CLL. Application of lasso-based regularized Cox and random survival forests allowed objective selection of variables and complemented results from the traditional Cox regression analysis. Finally, this study systematically explored biomarkers of ibrutinib resistance. Other studies reporting BTK and PLCG2 mutations in CLL retrospectively tested patients who developed clinical relapse, rather than taking an unbiased, cross-sectional approach such as that adopted in this study.18,30
The prognostic model presented herein redefines high-risk CLL for patients receiving ibrutinib therapy. In our analysis, several classic prognostic factors, such as age, IGHV, bulky disease, and Rai stage, did not have consistent or independent prognostic value. Applying the CLL-IPI to our data led to inflated risk predictions in many patients because CLL-IPI includes unmutated IGHV and age over 65 years as adverse factors. As a result, 88% of the patients fell into either the high- or the very high-risk CLL-IPI category, limiting effective risk stratification. Our four-factor model provided more meaningful risk stratification, distributing patients more evenly across risk groups, and allowing the separation of patients with markedly different survival outcomes and distinct patterns of disease progression. For instance, median PFS was 33 months for the high-risk group and was not reached for the low-risk group. BTK/PLCG2 mutations and Richter’s transformation were found most frequently in the high-risk group (50% and 17%, respectively), and least frequently in the low-risk group (17% and 0%, respectively). Although 40% of the intermediate-risk group had BTK/PLCG2 mutations, a more durable response to ibrutinib was observed, with a median PFS not reached in this group.
BTK and/or PLCG2 mutations are found in most patients with CLL progressing on ibrutinib.19,30 Monitoring for these mutations could identify patients who will eventually progress. However, routine monitoring has limitations because of its cost and for biologic and clinical reasons, and it is currently unknown whether treatment interventions before progressive disease have clinical benefits. BTK and PLCG2 mutations are not detectable at baseline, making them poor biomarkers for upfront risk stratification.18 There is high interpatient variability in the time from treatment initiation to when such mutations become detectable (Fig 3B). Moreover, CLL progression in the absence of the mutations has been reported in up to 20% of patients treated with ibrutinib. Taken together, the usefulness of BTK/PLCG2 mutations is currently limited to correlative biomarkers, rather than as disease-defining markers.19,30 Our study tested patients in the NIH cohort for these mutations at progression and also cross-sectionally during ongoing ibrutinib therapy. We observed a wide variation in the time between starting ibrutinib and the first detection of a mutation (range, 6 months to 6 years) and in the time between first mutation and clinical progression (range, 0-15 months). Notably, BTK and/or PLCG2 mutations were detected more frequently and earlier in the high-risk group than in the low-risk group, suggesting that the four-factor model can capture the subsequent risk of clonal evolution.32
Our study has limitations inherent to the trial population and the extent of data available for analyses. It is possible that the four-factor model could be improved by the inclusion of additional parameters, such as complex karyotype20,33 and CD49d expression,21 which were unavailable in the majority of patients in our data set. However, karyotype and CD49d are not tested consistently, nor are they recommended as a part of the baseline evaluation by International Workshop on CLL guidelines.31 Furthermore, a consensus definition of complex karyotype has not been established, and there are conflicting data on the predictive value of complex karyotype in patients treated with ibrutinib.20,33,34 Another limitation of the four-factor model is its inclusion of prior treatment history as an adverse risk factor, and thus, the model predictions of survival differ in TN- and RR-CLL. In addition, nearly 30% of the patients with TN-CLL were excluded from the subset analysis, most often because of missing B2M. TP53 aberration was relatively uncommon among patients with TN-CLL in the discovery cohort (12%) and highly enriched in the NIH cohort (40%). Because of its study design and the completeness of the B2M data collection, the NIH cohort is overrepresented in a small group of high-risk patients with TN-CLL (78% of the high-risk group [seven of nine patients] and 3% of the TN-CLL subset [seven of 208 patients]). Despite these limitations, the high-risk group continues to separate itself from other groups within the TN-CLL subset. Longer follow-up times and larger numbers of patients will be needed to better estimate differences in outcome in TN-CLL.
The proposed four-factor model enables informed clinical decision making and patient counseling and can provide a platform for the investigation of risk-adapted treatment approaches. In clinical practice, the score can help set expectations, guide the frequency of monitoring, and direct patients to risk-adapted treatment approaches on the basis of the predicted risk of early progression. In clinical trials, stratifying patients by risk score could more rapidly and clearly answer whether novel treatments improve outcomes for those patients in greatest need. High-risk patients who progress while receiving ibrutinib are also at increased risk of progressing with alternative targeted agents.14,35,36 These patients could benefit from participation in clinical trials investigating novel regimens or can be considered for adoptive cellular therapy once targeted agents have failed.15,37,38 Although treatment intensification for high-risk patients may be beneficial, it is equally important to identify low-risk patients who are well served by ibrutinib monotherapy. Multidrug combinations tend to increase toxicity and potentially limit dose adherence. Randomized trials comparing doublet or triplet regimens that are based on targeted agents will be informative in assessing the efficacy, safety, and quality of life attained by combination approaches (ClinicalTrials.gov identifiers: NCT03701282 and NCT03737981).
ACKNOWLEDGMENT
We thank the patients and their families who participated in the clinical trials. We thank the investigators who collected data from the trials and made them available for analyses. We thank Suneel Kudaravalli, who was at Pharmacyclics LLC, an AbbVie Company, at the time of this study, for supporting the National Institutes of Health trial and for analyses of the industry database.
Presented in part at the 58th Annual Meeting of American Society of Hematology, San Diego, CA, December 3-6, 2016; and the 60th Annual Meeting of American Society of Hematology, San Diego, CA, December 1-4, 2018.
SUPPORT
Supported by the Intramural Research Program of the National Institutes of Health; Pharmacyclics LLC, an AbbVie Company (A.W.); the American Society of Hematology Scholar Award (I.E.A.); and Acerta LLC, a member of the AstraZeneca Group (A.W.).
See accompanying editorial on page 551
AUTHOR CONTRIBUTIONS
Conception and design: Inhye E. Ahn, David Ipe, Mei Cheng, James P. Dean, Adrian Wiestner
Provision of study material or patients: Maher Albitar, Erika M. Gaglione
Collection and assembly of data: Inhye E. Ahn, David Ipe, Mei Cheng, Maher Albitar, Wanlong Ma, Sarah E. M. Herman, Erika M. Gaglione, Susan Soto, James P. Dean
Data analysis and interpretation: Inhye E. Ahn, Xin Tian, David Ipe, Mei Cheng, L. Claire Tsao, Lei Zhang, James P. Dean, Adrian Wiestner
Manuscript writing: All authors
Final approval of manuscript: All authors
Accountable for all aspects of the work: All authors
AUTHORS' DISCLOSURES OF POTENTIAL CONFLICTS OF INTEREST
Prediction of Outcome in Patients With Chronic Lymphocytic Leukemia Treated With Ibrutinib: Development and Validation of a Four-Factor Prognostic Model
The following represents disclosure information provided by authors of this manuscript. All relationships are considered compensated unless otherwise noted. Relationships are self-held unless noted. I = Immediate Family Member, Inst = My Institution. Relationships may not relate to the subject matter of this manuscript. For more information about ASCO's conflict of interest policy, please refer to www.asco.org/rwc or ascopubs.org/jco/authors/author-center.
Open Payments is a public database containing information reported by companies about payments made to US-licensed physicians (Open Payments).
David Ipe
Employment: AbbVie
Stock and Other Ownership Interests: AbbVie
Mei Cheng
Employment: Pharmacyclics
Stock and Other Ownership Interests: AbbVie stock through Pharmacyclics
Maher Albitar
Employment: Genomic Testing Cooperative
Leadership: Genomic Testing Cooperative
Stock and Other Ownership Interests: Genomic Testing Cooperative
Patents, Royalties, Other Intellectual Property: Multiple patents on methods in diagnostics
Travel, Accommodations, Expenses: Genomic Testing Cooperative
L. Claire Tsao
Employment: AbbVie/Pharmacyclics
Stock and Other Ownership Interests: AbbVie/Pharmacyclics
Travel, Accommodations, Expenses: AbbVie/Pharmacyclics
Lei Zhang
Employment: AbbVie
Stock and Other Ownership Interests: AbbVie/Pharmacyclics
James P. Dean
Employment: Pharmacyclics
Stock and Other Ownership Interests: AbbVie
Adrian Wiestner
Research Funding: Pharmacyclics (Inst), Acerta Pharma (Inst), Genmab (Inst), Verastem (Inst), Merck (Inst), Nurix (Inst)
No other potential conflicts of interest were reported.
REFERENCES
- 1.Herishanu Y, Pérez-Galán P, Liu D, et al. The lymph node microenvironment promotes B-cell receptor signaling, NF-kappaB activation, and tumor proliferation in chronic lymphocytic leukemia. Blood. 2011;117:563–574. doi: 10.1182/blood-2010-05-284984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Dühren-von Minden M, Übelhart R, Schneider D, et al. Chronic lymphocytic leukaemia is driven by antigen-independent cell-autonomous signalling. Nature. 2012;489:309–312. doi: 10.1038/nature11309. [DOI] [PubMed] [Google Scholar]
- 3.Souers AJ, Leverson JD, Boghaert ER, et al. ABT-199, a potent and selective BCL-2 inhibitor, achieves antitumor activity while sparing platelets. Nat Med. 2013;19:202–208. doi: 10.1038/nm.3048. [DOI] [PubMed] [Google Scholar]
- 4.Woyach JA, Ruppert AS, Heerema NA, et al. Ibrutinib regimens versus chemoimmunotherapy in older patients with untreated CLL. N Engl J Med. 2018;379:2517–2528. doi: 10.1056/NEJMoa1812836. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Shanafelt TD, Wang XV, Kay NE, et al. Ibrutinib-rituximab or chemoimmunotherapy for chronic lymphocytic leukemia. N Engl J Med. 2019;381:432–443. doi: 10.1056/NEJMoa1817073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Moreno C, Greil R, Demirkan F, et al. Ibrutinib plus obinutuzumab versus chlorambucil plus obinutuzumab in first-line treatment of chronic lymphocytic leukaemia (iLLUMINATE): A multicentre, randomised, open-label, phase 3 trial. Lancet Oncol. 2019;20:43–56. doi: 10.1016/S1470-2045(18)30788-5. [DOI] [PubMed] [Google Scholar]
- 7.Fischer K, Al-Sawaf O, Bahlo J, et al. Venetoclax and obinutuzumab in patients with CLL and coexisting conditions. N Engl J Med. 2019;380:2225–2236. doi: 10.1056/NEJMoa1815281. [DOI] [PubMed] [Google Scholar]
- 8.Sharman JP, Egyed M, Jurczak W, et al. Acalabrutinib with or without obinutuzumab versus chlorambucil and obinutuzumab for treatment-naive chronic lymphocytic leukaemia (ELEVATE TN): a randomised, controlled, phase 3 trial. Lancet. 2020;395:1278–1291. doi: 10.1016/S0140-6736(20)30262-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Advani RH, Buggy JJ, Sharman JP, et al. Bruton tyrosine kinase inhibitor ibrutinib (PCI-32765) has significant activity in patients with relapsed/refractory B-cell malignancies. J Clin Oncol. 2013;31:88–94. doi: 10.1200/JCO.2012.42.7906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Byrd JC, Furman RR, Coutre SE, et al. Targeting BTK with ibrutinib in relapsed chronic lymphocytic leukemia N Engl J Med 36932–42.2013[Erratum: N Engl J Med 370:786, 2014] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Byrd JC, Brown JR, O’Brien S, et al. Ibrutinib versus ofatumumab in previously treated chronic lymphoid leukemia. N Engl J Med. 2014;371:213–223. doi: 10.1056/NEJMoa1400376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Burger JA, Tedeschi A, Barr PM, et al. Ibrutinib as initial therapy for patients with chronic lymphocytic leukemia. N Engl J Med. 2015;373:2425–2437. doi: 10.1056/NEJMoa1509388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Maddocks KJ, Ruppert AS, Lozanski G, et al. Etiology of ibrutinib therapy discontinuation and outcomes in patients with chronic lymphocytic leukemia. JAMA Oncol. 2015;1:80–87. doi: 10.1001/jamaoncol.2014.218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Jones JA, Mato AR, Wierda WG, et al. Venetoclax for chronic lymphocytic leukaemia progressing after ibrutinib: An interim analysis of a multicentre, open-label, phase 2 trial. Lancet Oncol. 2018;19:65–75. doi: 10.1016/S1470-2045(17)30909-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Turtle CJ, Hay KA, Hanafi LA, et al. Durable molecular remissions in chronic lymphocytic leukemia treated with CD19-specific chimeric antigen receptor-modified T cells after failure of ibrutinib. J Clin Oncol. 2017;35:3010–3020. doi: 10.1200/JCO.2017.72.8519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Furman RR, Cheng S, Lu P, et al. Ibrutinib resistance in chronic lymphocytic leukemia N Engl J Med 3702352–2354.2014[Erratum: N Engl J Med 370:2547, 2014] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Woyach JA, Furman RR, Liu TM, et al. Resistance mechanisms for the Bruton’s tyrosine kinase inhibitor ibrutinib. N Engl J Med. 2014;370:2286–2294. doi: 10.1056/NEJMoa1400029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Burger JA, Landau DA, Taylor-Weiner A, et al. Clonal evolution in patients with chronic lymphocytic leukaemia developing resistance to BTK inhibition. Nat Commun. 2016;7:11589. doi: 10.1038/ncomms11589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Ahn IE, Underbayev C, Albitar A, et al. Clonal evolution leading to ibrutinib resistance in chronic lymphocytic leukemia. Blood. 2017;129:1469–1479. doi: 10.1182/blood-2016-06-719294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Thompson PA, O’Brien SM, Wierda WG, et al. Complex karyotype is a stronger predictor than del(17p) for an inferior outcome in relapsed or refractory chronic lymphocytic leukemia patients treated with ibrutinib-based regimens. Cancer. 2015;121:3612–3621. doi: 10.1002/cncr.29566. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Tissino E, Benedetti D, Herman SEM, et al. Functional and clinical relevance of VLA-4 (CD49d/CD29) in ibrutinib-treated chronic lymphocytic leukemia. J Exp Med. 2018;215:681–697. doi: 10.1084/jem.20171288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Ahn IE, Farooqui MZH, Tian X, et al. Depth and durability of response to ibrutinib in CLL: 5-Year follow-up of a phase 2 study. Blood. 2018;131:2357–2366. doi: 10.1182/blood-2017-12-820910. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.International CLL-IPI Working Group An international prognostic index for patients with chronic lymphocytic leukaemia (CLL-IPI): A meta-analysis of individual patient data. Lancet Oncol. 2016;17:779–790. doi: 10.1016/S1470-2045(16)30029-8. [DOI] [PubMed] [Google Scholar]
- 24.Soumerai JD, Ni A, Darif M, et al. Prognostic risk score for patients with relapsed or refractory chronic lymphocytic leukaemia treated with targeted therapies or chemoimmunotherapy: A retrospective, pooled cohort study with external validations. Lancet Haematol. 2019;6:e366–e374. doi: 10.1016/S2352-3026(19)30085-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.O’Brien S, Jones JA, Coutre SE, et al. Ibrutinib for patients with relapsed or refractory chronic lymphocytic leukaemia with 17p deletion (RESONATE-17): A phase 2, open-label, multicentre study. Lancet Oncol. 2016;17:1409–1418. doi: 10.1016/S1470-2045(16)30212-1. [DOI] [PubMed] [Google Scholar]
- 26.Farooqui MZ, Valdez J, Martyr S, et al. Ibrutinib for previously untreated and relapsed or refractory chronic lymphocytic leukaemia with TP53 aberrations: A phase 2, single-arm trial. Lancet Oncol. 2015;16:169–176. doi: 10.1016/S1470-2045(14)71182-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Tibshirani R. The lasso method for variable selection in the Cox model. Stat Med. 1997;16:385–395. doi: 10.1002/(sici)1097-0258(19970228)16:4<385::aid-sim380>3.0.co;2-3. [DOI] [PubMed] [Google Scholar]
- 28.Ishwaran H, Kogalur UB, Blackstone EH, et al. Random survival forests. Ann Appl Stat. 2008;2:841–860. [Google Scholar]
- 29.Albitar A, Ma W, DeDios I, et al. Using high-sensitivity sequencing for the detection of mutations in BTK and PLCγ2 genes in cellular and cell-free DNA and correlation with progression in patients treated with BTK inhibitors. Oncotarget. 2017;8:17936–17944. doi: 10.18632/oncotarget.15316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Woyach JA, Ruppert AS, Guinn D, et al. BTKC481S-mediated resistance to ibrutinib in chronic lymphocytic leukemia. J Clin Oncol. 2017;35:1437–1443. doi: 10.1200/JCO.2016.70.2282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Hallek M, Cheson BD, Catovsky D, et al. iwCLL guidelines for diagnosis, indications for treatment, response assessment, and supportive management of CLL. Blood. 2018;131:2745–2760. doi: 10.1182/blood-2017-09-806398. [DOI] [PubMed] [Google Scholar]
- 32.Landau DA, Sun C, Rosebrock D, et al. The evolutionary landscape of chronic lymphocytic leukemia treated with ibrutinib targeted therapy. Nat Commun. 2017;8:2185. doi: 10.1038/s41467-017-02329-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Baliakas P, Jeromin S, Iskas M, et al. Cytogenetic complexity in chronic lymphocytic leukemia: Definitions, associations, and clinical impact. Blood. 2019;133:1205–1216. doi: 10.1182/blood-2018-09-873083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.O’Brien S, Furman RR, Coutre S, et al. Single-agent ibrutinib in treatment-naïve and relapsed/refractory chronic lymphocytic leukemia: A 5-year experience. Blood. 2018;131:1910–1919. doi: 10.1182/blood-2017-10-810044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Stilgenbauer S, Eichhorst B, Schetelig J, et al. Venetoclax for patients with chronic lymphocytic leukemia with 17p deletion: Results from the full population of a phase II pivotal trial J Clin Oncol 361973–1980.2018[Erratum: J Clin Oncol 37:2299, 2019] [DOI] [PubMed] [Google Scholar]
- 36.Roberts AW, Ma S, Kipps TJ, et al. Efficacy of venetoclax in relapsed chronic lymphocytic leukemia is influenced by disease and response variables. Blood. 2019;134:111–122. doi: 10.1182/blood.2018882555. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Dreger P, Ghia P, Schetelig J, et al. High-risk chronic lymphocytic leukemia in the era of pathway inhibitors: Integrating molecular and cellular therapies. Blood. 2018;132:892–902. doi: 10.1182/blood-2018-01-826008. [DOI] [PubMed] [Google Scholar]
- 38.Mato AR, Hill BT, Lamanna N, et al. Optimal sequencing of ibrutinib, idelalisib, and venetoclax in chronic lymphocytic leukemia: Results from a multicenter study of 683 patients. Ann Oncol. 2017;28:1050–1056. doi: 10.1093/annonc/mdx031. [DOI] [PubMed] [Google Scholar]