Abstract
Incidence of melanoma continues to rise in the United States with ~100,000 new cases diagnosed in 2019. While the 5-year survival rate of melanoma is 99% when localized, the rate of survival drops to 22.5% when distant disease is detected. As such, an area of great interest is understanding the mechanisms that promote melanoma metastasis so that better potential therapeutic targets can be discovered. Herein, we demonstrate that activation of NRF2 by FAM129B contributes to increased metastatic potential of BRAF V600E mutant melanoma cells. Specifically, FAM129B induces NRF2 by competing for KEAP1 binding (the negative regulator of NRF2) via an ETGE motif. Furthermore, we show that phosphorylation of FAM129B plays a role in mediating the interaction between FAM129B and KEAP1, as the phosphorylation status of FAM129B dictates its subcellular localization. When phosphorylated, FAM129B is found primarily in the cytosol where it can bind to KEAP1, but upon inhibition of MEK activity, FAM129B is localized to the cell membrane and no longer interacts with KEAP1. In BRAF V600E mutant melanoma, the MAPK pathway leads to hyperphosphorylation of FAM129B, and therefore FAM129B localizes to the cytosol, binds KEAP1, and upregulates NRF2. Importantly, genetic modulation or pharmacological inhibition that results in a decrease in FAM129B protein level or its phosphorylation decreases migration and invasion of mutant melanoma in an NRF2-dependent manner. Overall, these data indicate that phosphorylation of FAM129B plays a significant role in driving the metastatic potential of BRAF V600E melanoma via upregulation of the NRF2 signaling pathway.
Keywords: NRF2, FAM129B, KEAP1, melanoma, metastasis
1. Introduction
In the United States, melanoma is the fifth most diagnosed cancer, resulting in over 7,000 deaths per year. When detected early, localized melanomas can be surgically removed, with patients having a 99% 5-year survival rate1. However, if melanoma metastasizes, the 5-year survival rate decreases to only 22.5% and requires more aggressive therapeutic approaches (i.e., invasive surgery, chemotherapy, immunotherapy, radiotherapy). Therefore, understanding potential mechanisms that promote metastasis and identifying targets could offer valuable insight into novel therapeutic approaches to treat metastatic melanoma and extend patient lifespan. One protein critical for the progression of melanoma is the mitogen-activated protein kinase (MAPK) signaling pathway member: serine/threonine protein kinase B-raf (BRAF)2. In malignant melanoma, BRAF is found to be mutated in ~50% of all cases, 90% of which contain a V600E mutation3–5. Mutation of valine (V) to glutamic acid (E) acts as a phosphomimetic, resulting in hyperactivation of BRAF and its downstream targets, including mitogen-activated protein kinase kinase (MEK) and extracellular signal-regulated kinase (ERK)6. Importantly, constitutive BRAF-driven MAPK signaling has been shown to increase cancer cell growth, migration, and invasion7–9. Vemurafenib is an FDA approved drug for the treatment of late-stage melanoma that specifically targets the ATP binding pocket of V600E mutant BRAF10,11. While the use of vemurafenib has been effective in prolonging patient survival, overall patient outlook could be further improved by targeting malignant tumors early to prevent the onset of metastasis to other organs. Furthermore, even though the role of the MAPK pathway is well identified in driving cell proliferation in melanoma, its specific role in promoting malignant melanoma cell metastasis still needs further elucidation.
Prior studies have linked activation of the MAPK pathway to increased expression of NRF2 (nuclear factor (erythroid-derived 2)-like 2), a transcription factor known to regulate tumor progression and survival, including cancer cell migration and invasion12,13; however, the mechanistic relationship between NRF2, MAPK, and invasion remains to be determined. As a critical mediator of cellular homeostasis, NRF2 regulates a variety of genes involved in mediating the antioxidant response (glutamate-cysteine ligase modifier subunit/glutamate-cysteine ligase catalytic subunit [GCLM/GCLC]), cell detoxification (NAD(P)H quinone dehydrogenase 1 [NQO1]), protein homeostasis (proteasome maturation protein/proteasome subunit alpha 1 [POMP/PSMA1]), heme metabolism (heme oxygenase-1 [HO-1]), and DNA damage repair (RAD51 homolog 1/p53 binding protein 1 [RAD51/53BP1])14–21. Under basal conditions, NRF2 levels are kept low by its negative regulator: Kelch-like ECH-associated protein 1 (KEAP1)22. KEAP1 serves as a substrate adaptor protein for a Cullin 3/Ring-box 1 (CUL3/RBX1) E3 ubiquitin ligase that facilities the ubiquitylation of NRF2 and its p97-mediated recruitment to the 26S proteasome for degradation23–25. NRF2 binds to a KEAP1 dimer through two motifs (ETGE and DLG)26; however, other proteins with similar motifs have been shown to compete with NRF2 for KEAP1 binding (i.e. NRF1, p62)26–29. By outcompeting NRF2 binding to KEAP1, ETGE-containing proteins can prevent NRF2 from being ubiquitylated and degraded, thus allowing newly synthesized NRF2 to accumulate, translocate to the nucleus, and activate transcription of its target genes. Therefore, it is of great interest to identify aberrant expression or localization of ETGE-containing proteins in cancers with elevated NRF2, as these could be mechanisms of increased survival and metastasis of cancer cells.
A recent study demonstrated that BRAF is involved in the phosphorylation of Family With Sequence Similarity 129 Member B (FAM129B), and that FAM129B is primarily localized in the cytosol of BRAF mutant malignant melanoma cells, increasing their ability to invade30. Additionally, FAM129B, a protein thought to be anti-apoptotic and involved in cell-cell junctions, contains an ETGE motif31,32. Therefore, we hypothesized that BRAF-dependent phosphorylation of FAM129B increases its interaction with KEAP1 due to aberrant cytosolic localization. As such, in hyperactive BRAF melanomas, hyperphosphorylation of FAM129B will outcompete NRF2 for KEAP1 binding due to changes in the localization of FAM129B, resulting in enhanced NRF2 signaling and a subsequent increase in metastatic potential. Herein, we demonstrate that in V600E mutant BRAF A375 melanoma cells, FAM129B binds to KEAP1 exclusively via the ETGE motif, resulting in NRF2 upregulation. Specifically, cellular localization of FAM129B, dictated by phosphorylation downstream of BRAF, plays a role in its interaction with KEAP1, resulting in nuclear localization of NRF2. In contrast, cell-cell junction localized FAM129B does not interact with KEAP1, allowing canonical NRF2 regulation. More importantly, we show a mechanism by which BRAF mutant melanomas have increased metastatic potential via cytosolic FAM129B-based binding of KEAP1 and NRF2 induction.
2. Materials and Methods
2.1. Materials, antibodies, and cell culture
Brusatol and tBHQ (Sigma-Aldrich) were dissolved in autoclaved water prior to treatment. U0126 was dissolved in dimethyl sulfoxide (DMSO) (Sigma-Aldrich). HEK293 (human embryonic kidney) and A375 (metastatic human melanoma) cell lines were obtained from the American Type Culture Collection (ATCC) and cultured in DMEM (Corning Cellgro) with L-Glutamine, 4.5 g/L Glucose, and Sodium Pyruvate with 10% FBS (Genesee Scientific), as well as 100 unit/mL Pen Strep (Gibco). Antibodies were purchased from Santa Cruz Biotechnology (NRF2, KEAP1, NQO1, BRAF, GAPDH, p-ERK, ERK, SOX9), Cell Signaling Technology (Tubulin, FAM129B), MyBioSource (p-FAM129B), BD Biosciences (β-catenin), New England Biolabs (CBD), and Trevigen (HA). Alexa Fluor 488 anti-mouse and 594 anti-rabbit secondaries were purchased from Invitrogen. Hoechst 33342 was obtained from ThermoFisher Scientific.
2.2. Immunoblotting
For immunoblotting experiments, cells were collected in 1X Sample Buffer (50 mM Tris-HCl [pH 6.8], 2% sodium dodecyl sulfate [SDS], 10% glycerol, 100 mM dithiothreitol [DTT], 0.1% bromophenol blue), boiled for 10 minutes, and then were sonicated via the Bioruptor (Diagenode) for 20 minutes. Samples were run on a 7.5% SDS-PAGE gel, followed by transfer to a nitrocellulose membrane (Prometheus). Membranes were blocked for 1 hr in 5% milk, prior to incubation with primary antibody overnight at 4° C. Membranes were then washed 4 times for 15 minutes in 1X phosphate-buffered saline (PBS) then incubated with secondary antibody for 1 hour in 5% milk. Again, membranes were washed with PBS (6 times, 10 minutes each), then developed using an enhanced chemiluminescent (ECL) horseradish peroxidase reaction (Thermo Fisher Scientific) and imaged using the Azure c600 (Azure Biosystems).
Immunoprecipitation and pull-down experiments were done by cotransfecting HEK293 cells with 100 ng HA-FAM129B (or muFAM129B) and KEAP1-CBD for 24 hours. Cell lysates were collected and incubated with either HA-beads (Sigma-Aldrich) or CBD-beads (New England Biolabs). After incubating with rotation at 4° C overnight, beads were washed with radioimmunoprecipitation assay (RIPA) buffer (1% NP-40, 1% sodium deoxycholate, 0.1% SDS, 0.15 M NaCl, 0.01 M sodium phosphate [pH 7.2], 2 mM EDTA) three times. 1X Sample Buffer was then added to the beads, followed by boiling and immunoblot analysis for HA and CBD.
2.3. Transfection
HEK293 cells were transfected with 100 ng Vector (control), FAM129B, or muFAM129B, and/or NQO1-ARE-driven firefly luciferase and TK-driven-Renilla luciferase plasmids using Lipofectamine 3000 (Invitrogen) as per the manufacturer’s instructions. Briefly, cells were plated and 24 hours later each plasmid construct plus Lipofectamine 3000 was added to cells. The next day, cells were harvested for immunoblot analysis or luciferase activity was assessed using a luciferase reporter assay (Promega). Transfection of small interfering RNA (siRNA) was done using Hiperfect (Qiagen) as per the manufacturer’s instructions using either non-targeted (Qiagen), FAM129B (Qiagen), BRAF (Santa Cruz Biotechnology), or NRF2 (Qiagen) siRNA for 72 hours.
2.4. Immunofluorescence
For indirect immunofluorescence, A375 cells were grown on glass cover slips (Fisher Scientific) to 70–90% confluence in 35-mm plastic cell culture dishes. After treatment for 4 hours, cells were fixed using ice cold methanol for 20 minutes then washed with PBS thrice, and incubated in primary antibody diluted in 10% FBS in PBS for 1 hour. Next, cover slips were washed again in PBS, then blocked with secondary (Alexa Fluor 488 [mouse] or 594 [rabbit]) diluted in 10% FBS in PBS for 1 hour. Cells were then mounted to glass slides using antifade mounting medium and imaged. All images were taken using the Zeiss Axio Observer.Z1 microscope using Slidebook 4.2.0.11 computer software (Intelligent Imaging Innovations, Inc.).
2.5. Invasion and Migration
For migration experiments, A375 cells were transiently transfected with 10 nM control or FAM129B siRNA for 72 hours, or 10 nM control or NRF2 siRNA for 72 hours then left untreated or treated with 10 μM U0126 for 4 hours. Cells were serum starved overnight prior to scratching. Images were taken every 6 hours for 24 hours via the IncuCyte (Essen Biosciences). Wound percent closure was calculated via ImageJ (NIH) as a measure of the scratch width at each timepoint compared to the initial scratch width. For invasion, A375 cells under the same treatment conditions used for migration experiments were serum starved overnight and seeded into a 2.13 mg/mL Matrigel solution in the top chamber of a ClearView Cell Migration plate (Essen Biosciences). FBS was added to the bottom chamber as a chemoattractant. Invasion is represented as the number of cells that were counted in the bottom chamber at 48 hours compared to the initial number of cells in the top chamber as calculated via IncuCyte ZOOM software (Essen Biosciences).
2.6. Statistical analysis
Mean values were calculated and error was reported as the Standard Error of the Mean (SEM). Significance was determined to be p < 0.05 as calculated by the Student’s t-test (n.s. = no significance).
3. Results
3.1. BRAF mutations correlate with increased NRF2 target gene expression in metastatic melanoma
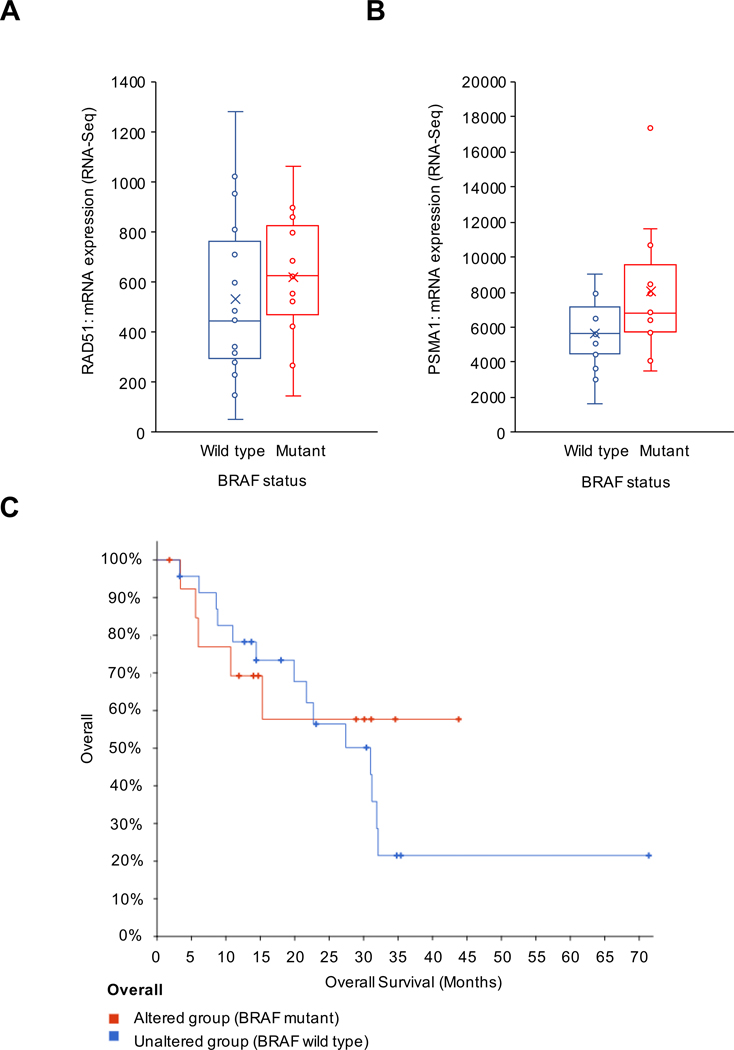
Metastatic melanoma is often associated with mutations in BRAF; however, the exact downstream mechanisms of BRAF that increase metastatic potential remain unclear. Investigation of a previously published study curated by The Cancer Genome Atlas (TCGA) indicated that in patients with metastatic melanomas where a BRAF gain-of-function mutation existed, there was a corresponding increase in mRNA expression of RAD51 and PSMA1 (Fig. 1A–B)33. RAD51 and PSMA1 are well defined NRF2 target genes indicating that NRF2 may be upregulated in BRAF mutant melanomas. Furthermore, BRAF mutations resulted in a decreased trend in overall median survival for patients with metastatic melanoma compared to the wild type group (Fig. 1C). Therefore, the effects of BRAF mutations on NRF2 activity were explored.
Figure 1: BRAF mutations correlate with increased NRF2 target gene expression in metastatic melanoma.
All data were acquired from a 2016 UCLA study of metastatic melanoma derived from TCGA. RAD51 (A) and PSMA1 (B) mRNA expression were detected in wild type versus gain-of-function mutant BRAF metastatic melanoma. (C) Overall survival of patients in BRAF mutated (altered) versus wild type (unaltered) groups was measured over 5 years.
3.2. FAM129B binds to KEAP1 and induces NRF2 activation
Initially, using mass spectrometry to analyze a KEAP1-CBD pulldown, our group identified that FAM129B is a binding partner of KEAP1 (data not shown). Of the KEAP1-interacting proteins identified, FAM129B was of particular interest because of its implications in cancer. Investigation of the protein amino acid sequence for FAM129B revealed the presence of an ETGE motif, which is the same motif that allows NRF2 to bind to KEAP1; this indicated that FAM129B might compete with NRF2 for KEAP1 binding. To confirm the interaction between FAM129B and KEAP1, HEK293 cells were transfected with HA-FAM129B and KEAP1-CBD and co-immunoprecipitation analysis was performed using either an anti-CBD (Fig. 2A) or anti-HA (Fig. 2B) antibody. To validate that binding between the two proteins occurred in an ETGE-dependent manner, the ETGE sequence in FAM129B was mutated (muFAM129B [ETGE to AAAA]), and the mutated FAM129B lost interaction with KEAP1 (Fig. 2C). Therefore, like NRF2, the ETGE motif is necessary for FAM129B to interact with KEAP1. Next, to examine the effect of FAM129B-KEAP1 binding on NRF2 levels, HEK293 cells were transfected with either the wild type HA-FAM129B or HA-muFAM129B plasmid. Overexpression of the wild type, but not mutant, FAM129B led to a clear increase in both NRF2 and NQO1 levels similar to treatment with the NRF2 inducer tBHQ (Fig. 2D); this suggests that FAM129B competitivity binds to KEAP1 resulting in NRF2 accumulation. To verify that FAM129B binding to KEAP1 resulted in activation of the NRF2 pathway, a reporter gene assay was performed in HEK293 cells cotransfected with NQO1-ARE-driven luciferase, TK-driven-Renilla luciferase, and either wild type FAM129B or muFAM129B. Like the tBHQ positive control, wild type FAM129B increased luciferase activity, whereas muFAM129B did not (Fig. 2E). Therefore, FAM129B is an ETGE-containing protein that can bind with KEAP1 and enhance NRF2-mediated transcription.
Figure 2: FAM129B binds to KEAP1 and induces NRF2 activation.
HEK293 cells were co-transfected with HA-FAM129B and KEAP1-CBD and protein-protein interaction was assessed via co-immunoprecipitation analysis. Cell lysates were incubated with either (A) HA-beads or (B) CBD beads and immunoprecipitated complexes were then immunoblotted using either anti-HA or anti-CBD antibodies. (C) HEK293 cells were cotransfected with either wild type or mutant HA-FAM129B in the presence or absence of KEAP1-CBD for 24 hr, and proteins in the KEAP1-containing complexes were pulled down with CBD beads, and then immunoblotted using either anti-HA or anti-CBD antibodies. (D) HEK293 cells were transfected with HA-FAM129B or HA-muFAM129B for 24 hr prior to collection; NRF2, HA-FAM129B, and NQO1 levels were determined via immunoblot analysis. (E) FAM129B or muFAM129B was co-transfected with NQO1-ARE-driven firefly luciferase and TK-driven-Renilla luciferase in HEK293 cells for 24 hr prior to measurement of dual luciferase activity. For both (D) and (E), cells were treated with 50 μM tBHQ for 24 hr as a positive control for NRF2 activation.
3.3. FAM129B knockdown or pharmacologically induced membrane localization decreases NRF2 protein levels
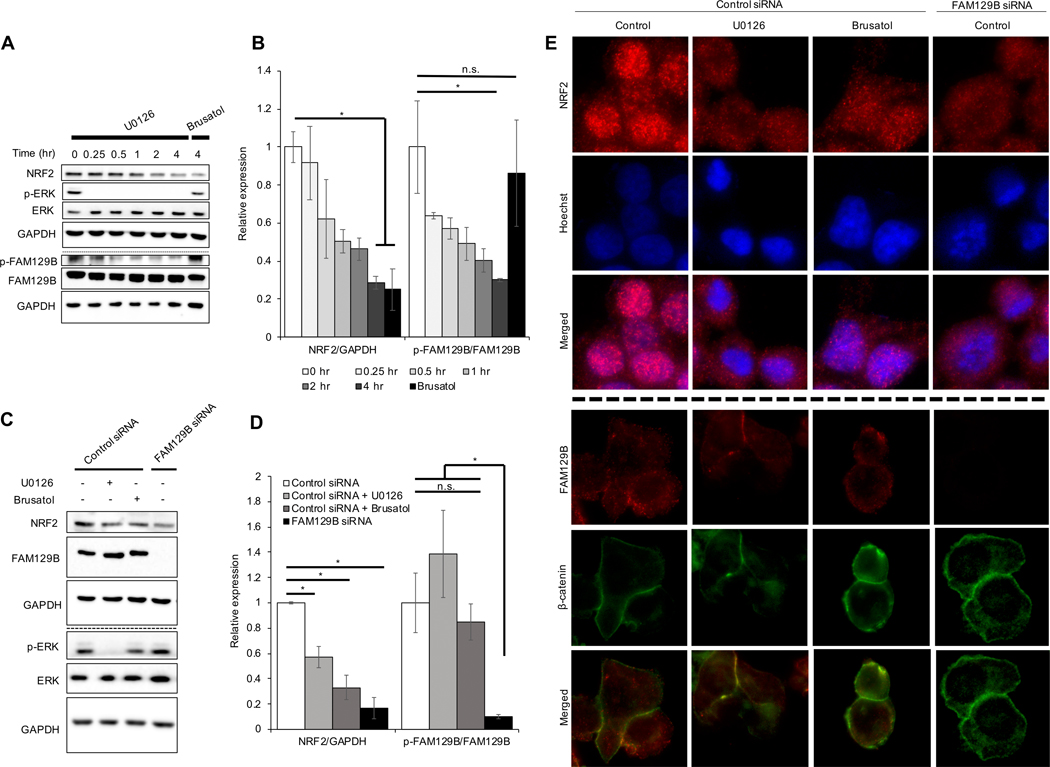
Next, the melanoma cell line A375 was chosen for further analysis because this cell line (1) contains a BRAF V600E mutation and high NRF2 expression34; (2) FAM129B plays a role in carcinogenesis of BRAF mutant melanoma. To determine if modulation of FAM129B localization or expression could reduce NRF2 expression in a mutant BRAF melanoma setting, two approaches were used. First, A375 cells were treated with U0126 (MEK inhibitor) for 4 hours, which resulted in decreased p-ERK and p-FAM129B, and also correlated with decreased NRF2 levels (Fig. 3A–B); phosphorylation of FAM129B was detected at serine residues which were previously shown to be phosphorylated by ERK30. Second, knockdown of FAM129B in A375 cells had a similar effect on NRF2 levels as treatment with U0126 and brusatol (NRF2 inhibitor) (Fig. 3C–D). Previous work indicated that MEK inhibition resulted in decreased FAM129B activity, and while ERK levels remained constant across treatment groups, U0126 treatment inhibited phosphorylation of ERK, indicating that inhibition of MAPK signaling may be inhibiting NRF2. Next, the effect of FAM129B knockdown on nuclear localization of NRF2 was assessed via immunofluorescence. In A375 cells treated with non-targeted siRNA, NRF2 was primarily localized in the nucleus; however, upon knockdown of FAM129B, localization of NRF2 was no longer nuclear similar to U0126 treatment (Fig. 3E). Furthermore, immunofluorescence analysis of FAM129B levels and localization indicated that FAM129B was mainly cytosolic in control A375 cells; however, upon treatment with U0126, FAM129B localized instead to the cell membrane at cell-cell junctions (β-catenin was used as a localization control for cell-cell junctions at membrane). Conversely, brusatol had no effect on FAM129B localization as compared to control (Fig. 3E). These results suggest that the MAPK pathway may mediate the effect of FAM129B on NRF2 protein expression by dictating the cellular localization of FAM129B.
Figure 3: FAM129B knockdown or pharmacologically induced membrane localization decreases NRF2 protein levels.
(A) A375 cells were treated with 10 μM U0126 for 0.25, 0.5, 1, 2, and 4 hr or 40 nM brusatol for 4 hr and then subjected to immunoblot analysis of NRF2, p-ERK, ERK, p-FAM129B, and FAM129B. (B) Protein levels of NRF2/GAPDH and p-FAM129B/FAM129B were quantified from immunoblot analyses in (A) (n=3). (C) At 72 hr post siRNA transfection, cells were treated with 10 μM U0126 or 40 nM brusatol (used as a control for NRF2 inhibition) for 4 hr before NRF2, FAM129B, p-ERK, and ERK protein levels were detected via immunoblot analysis. (D) NRF2, FAM129B, p-ERK, and ERK protein levels were quantified from immunoblot analyses in (C) (n=3). (E) Cells described in (C) were subjected to indirect immunofluorescence analysis of NRF2 (top) and Hoechst (middle); merged (bottom) panel indicates NRF2 nuclear localization. Additionally, cells from (C) underwent indirect immunofluorescent staining for FAM129B (top) and β-catenin (middle); merged (bottom) panel indicates FAM129B/β-catenin colocalization.
3.4. Inhibition of FAM129B phosphorylation affects the cellular localization of NRF2
As A375 cells contain a BRAF V600E gain-of-function mutation, and FAM129B has been shown to be phosphorylated downstream of the BRAF kinase cascade, the role of FAM129B phosphorylation status on its localization and NRF2 levels was assessed. To determine how inhibition of BRAF affects this phenotype, A375 cells were treated with 80 nM BRAF siRNA twice over a 72 hr period prior to collection. Knockdown of BRAF resulted in decreased NRF2 and p-FAM129B levels without decreasing total FAM129B protein expression (Fig. 4A–B). Additionally, FAM129B was cytosolic in control siRNA treated A375 cells compared to when BRAF was knocked down, which resulted in increased β-catenin colocalization of FAM129B. Treatment with brusatol had no effect on FAM129B localization; however, treatment with U0126 resulted in FAM129B localizing to cell-cell junctions in control siRNA cells. As expected, knockdown of BRAF caused FAM129B to localize to the cell membrane regardless of pharmacological intervention (Fig. 4C). In addition, in A375 cells, FAM129B and KEAP1 colocalize in the cytosol; however, after treatment with U0126, FAM129B is mainly localized at cell-cell junctions, while KEAP1 remains cytosolic (Fig. 4D). This supports the argument that only phosphorylated FAM129B that localizes in the cytosol can bind with KEAP1. Consistently, in A375 cells, phosphorylation of FAM129B by mutant BRAF increased the FAM129B-KEAP1 interaction resulting in constitutive upregulation of NRF2, whereas BRAF knockdown suppressed NRF2 nuclear localization by about 60% (Fig. 4E–F). Overall, this indicates that BRAF-dependent phosphorylation of FAM129B controls its cellular localization and thus its ability to bind to KEAP1 to block NRF2 degradation.
Figure 4: Inhibition of FAM129B phosphorylation affects the cellular localization of NRF2.
(A) A375 cells were transfected with 80 nM BRAF siRNA twice over 72 hours prior to immunoblot analysis of BRAF, p-FAM129B, FAM129B, and NRF2. (B) NRF2/GAPDH and BRAF/GAPDH protein levels were quantified from immunoblot analyses in (A) (n=3). (C) A375 cells were transiently transfected with either 80 nM control or BRAF siRNA twice over 72 hours prior to 4 hr treatment with 10 μM U0126 or 40 nM brusatol; cells were then analyzed using indirect immunofluorescence for FAM129B (left) and β-catenin (middle). (D) Indirect immunofluorescence analysis for FAM129B (left) and KEAP1 (middle) at 4 hr post 10 μM U0126 treatment; merged panel (right) indicates colocalization of FAM129B and KEAP1. (E) A375 cells were transfected with BRAF siRNA and then subjected to indirect immunofluorescence analysis of NRF2 and nuclear counterstain Hoechst. (F) NRF2 nuclear staining was quantified as the amount of cells that had clear NRF2 nuclear localization compared to the total amount of cells (n=~50 cells across 5–6 images were quantified).
3.5. FAM129B knockdown decreases metastatic potential of melanoma in an NRF2-dependent manner
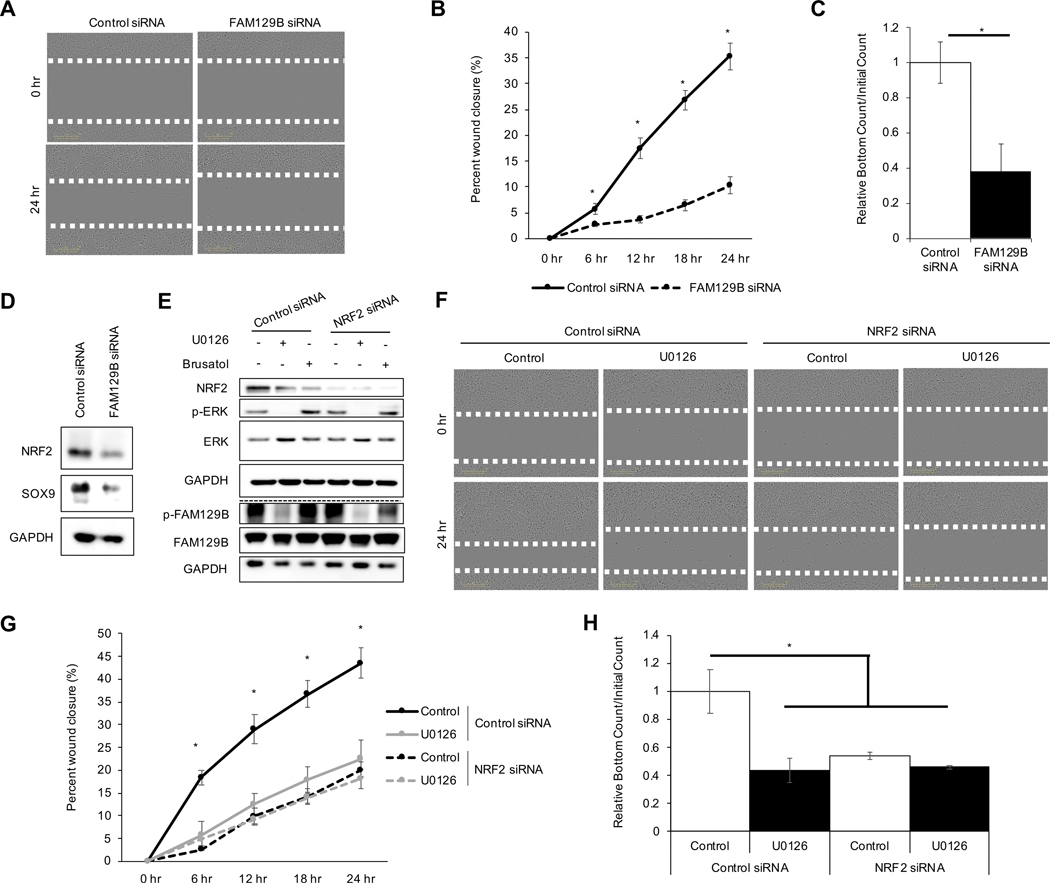
To gain insight into the possible role of FAM129B-mediated NRF2 activation in the metastatic potential of malignant melanoma cells, migration and invasion following manipulation of both FAM129B and NRF2 protein levels was explored. Several proteins linked to metastasis have been correlated to constitutive activation of the NRF2 signaling pathway35,36. Knockdown of FAM129B resulted in decreased migration of A375 cells into the scratch as indicated by an ~25% decrease in percent wound closure at 24 hours (Fig. 5A–B). Additionally, knockdown of FAM129B decreased invasion of A375 cells by ~60% (Fig. 5C). Recently, SRY-box 9 (SOX9), a transcription factor shown to be involved in metastasis, was identified to be regulated by the NRF2 signaling pathway35; herein, loss of FAM129B resulted in a decrease in SOX9 protein expression that correlated with decreased NRF2 levels (Fig. 5D). Finally, the dependence of FAM129B-mediated NRF2 upregulation on metastatic potential of melanoma cells was explored. A375 cells were treated with control or NRF2 siRNA for 72 hr and treated with U0126 or brusatol (used as control for NRF2 inhibition) for 4 hours before cells were subjected to immunoblot analysis for NRF2 protein levels as well as ERK and FAM129B phosphorylation status (Fig. 5E). Treatment of A375 cells with U0126 resulted in ~20% wound closure at 24 hours, like the effect of NRF2 siRNA (Fig. 5F–G). Interestingly, U0126 did not further decrease wound closure in NRF2 siRNA-transfected A375 cells, indicating that the effect of U0126 on A375 cell migration is NRF2-dependent. However, invasion was also significantly decreased (~60%) in A375 cells following treatment with U0126 in A375 control siRNA transfected cells, or in NRF2 siRNA transfected A375 cells (Fig. 5H). Once again, U0126 did not further decrease invasion in NRF2 siRNA transfected A375 cells, indicating the NRF2 dependence of these inhibitors in this context. Overall, these findings indicate a relationship between FAM129B phosphorylation status and elevated NRF2 levels in driving the metastatic potential of malignant melanoma cells.
Figure 5: FAM129B knockdown decreases metastatic potential of melanoma in an NRF2-dependent manner.
For panels (A-D), A375 cells were transiently transfected with 10 nM of either control or FAM129B siRNA for 72 hr prior to experimentation. (A) Cells were scratched to create a wound; images shown are at time of initial scratch (0 h) and 24 hr later. (B) Percent wound closure was calculated every 6 hr up to 24 hr post scratch from the cells/images in (A) (n=3). (C) Cells were subjected to a chemotactic invasion assay; values shown are the relative number of cells that migrated into the lower well at 48 hr post seeding normalized to the initial number of cells in the upper well (n=3). (D) Immunoblot analysis of NRF2 and SOX9 protein levels. For panel (E-H) A375 cells were treated with 10 nM control or NRF2 siRNA for 72 hr, then 10 μM U0126 or 40 nM brusatol for 4 hours prior to collection. (E) Immunoblot analysis of NRF2, p-FAM129B, FAM129B, p-ERK, and ERK protein levels. (F) Cells were scratched and imaged every 6 hr for 24 hr. Images displayed represent initial scratch (0 h) and 24 hr later. (G) Wound percent closure was quantified from cells/images in (F) (n=3). (H) The cells were monitored for invasion via the chemotactic invasion assay. Invasion is represented by the number of cells that migrated to the lower well at 48 hr normalized to the initial number of cells in the upper well (n=3).
4. Discussion
Understanding mechanisms that drive increased NRF2 expression in cancer is critical, as high NRF2 levels are associated with enhanced chemoresistance, metastasis, tumor aggressiveness, and patient mortality. While previous work has identified compounds for targeting NRF2 in cancer, none have progressed to clinical trials. Therefore, identifying other more druggable targets that regulate the NRF2 signaling pathway is of interest. As mentioned above, NRF2 binds to KEAP1 via an ETGE and a DLG motif; however, several proteins that contain ETGE motifs have been identified that can compete with NRF2 for binding with KEAP126–29. Among these, FAM129B was shown to bind to KEAP1 in an ETGE-dependent manner and consequently induce NRF2 protein expression (Fig. 2). As such, like many other ETGE-containing proteins, FAM129B could potentiate prolonged activation of NRF2 in certain cancer contexts and lead to tumor progression and metastasis. Prior work has implicated the importance of the relationship between NRF2 and FAM129B, as increased FAM129B expression activated NRF2 and led to chemoresistance37. In this study, we have identified that the FAM129B-KEAP1 interaction is an important mechanism by which hyperactive BRAF drives metastasis of melanoma: hyperactivated BRAF leads to NRF2 induction via phosphorylation of FAM129B, which competitively binds to KEAP1. As BRAF mutations have been linked with increased NRF2 target gene expression and decreased patient survival, the role of BRAF mutations in driving NRF2 activity was of great interest (Fig. 1). Specifically, we propose that BRAF-dependent activation of NRF2 via FAM129B-KEAP1 binding could be a key determinant of melanoma malignancy.
Previous work has shown that phosphorylation status plays a key role in dictating the cellular localization of FAM129B; however, how phosphorylation of FAM129B mediates NRF2 activity has not been shown30. To understand this relationship, the localization of FAM129B and NRF2 were examined in BRAF V600E mutant melanoma cells (A375), as FAM129B is constitutively phosphorylated in this cell line. Interestingly, under basal conditions FAM129B was primarily localized in the cytosol, while NRF2 localized in the nucleus; however, upon knockdown of FAM129B, NRF2 levels not only decreased, but its localization also shifted from nuclear to cytosolic (Fig. 3). This suggests that in A375 cells, NRF2 levels are basally high due to the presence of FAM129B. Additionally, use of BRAF siRNA or U0126 treatment to inhibit phosphorylation of FAM129B also resulted in a decrease of NRF2 protein levels. Intriguingly, treatment with U0126 caused FAM129B to localize to the cell membrane and no longer colocalize with KEAP1 in the cytosol. Similarly, knockdown of BRAF resulted in a significant shift in localization of FAM129B from the cytosol to cell-cell junctions, regardless of pharmacological intervention (Fig. 4). Together, these data suggest that the phosphorylation status of FAM129B dictates its ability to interact with KEAP1 by changing its cellular localization, thus controlling its ability to upregulate NRF2. Overall, this indicates that in BRAF V600E mutant melanoma, hyperphosphorylation of FAM129B could increase malignancy, as these cancers would have constitutive upregulation of NRF2 and its cytoprotective targets.
Both NRF2 and FAM129B have recently been linked to increased metastasis; however, the mechanisms by which these proteins increase metastatic potential remain elusive38. Previous work showed that increased expression of FAM129B resulted in activation of focal adhesion kinase (FAK) and increased invasion of non-small cell lung cancer and that loss of FAM129B slowed wound healing in vivo39,40. Additionally, studies have shown that phosphorylation of FAM129B promoted invasion of BRAF mutant melanoma but offered limited explanation as to what drove this invasive behavior30. Here, we showed that inhibiting phosphorylation of FAM129B, as well as knocking it down via siRNA, resulted in decreased migration and invasion of A375 melanoma cells by lowering NRF2 protein levels (Fig. 5). Previous work has shown that activation of the NRF2 signaling pathway results in increased metastatic potential, and with our findings that knockdown of FAM129B resulted in decreased migration and invasion, this could suggest that FAM129B increases metastatic potential via constitutive activation of the NRF2 signaling pathway35. As targeting the NRF2 signaling pathway has previously been shown to decrease tumor aggressiveness, yet no NRF2-targeted inhibitors are currently approved for use in humans, this mechanism indicates that the BRAF-FAM129B cascade could be a viable therapeutic target in KEAP1 wild type NRF2-upregulated cancers41. Furthermore, as metastasis of melanoma significantly decreases overall patient survival, targeting this cascade could decrease the metastatic potential of hyperactive BRAF melanoma, and thus increase patient survival.
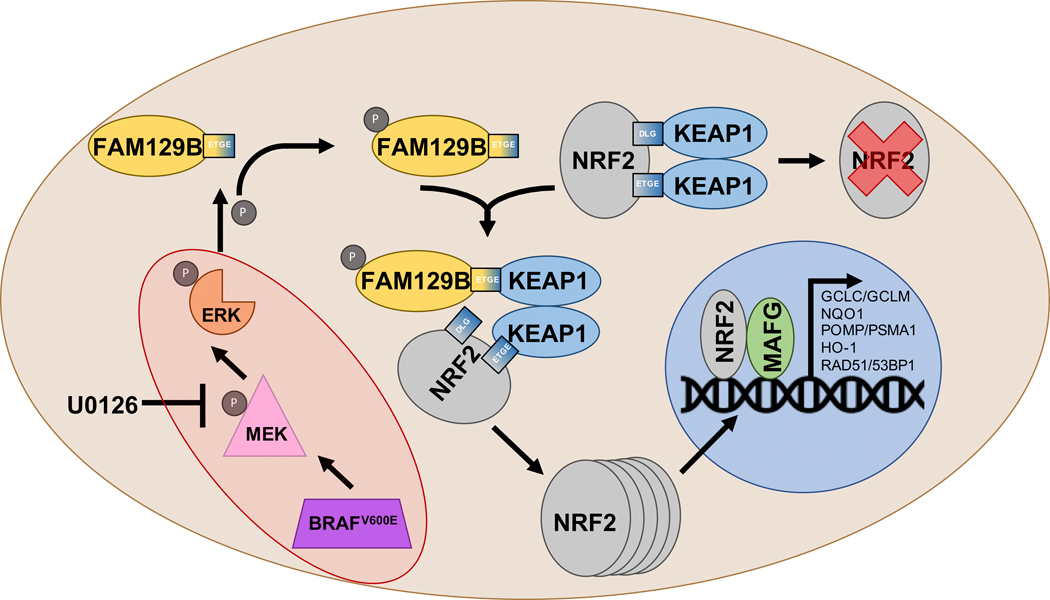
Overall, these findings demonstrate a mechanism of NRF2 induction driven by a BRAF-FAM129B cascade (Fig. 6). This implies that NRF2 could play a significant role in the survival, metastasis, and chemoresistance of tumors with hyperactivation of the MAPK pathway, including BRAF mutant melanoma. While our data demonstrate this phenomenon in melanoma cells with constitutively active BRAF, this mechanism could apply to any tumor with hyperactive MAPK signaling. For example, previous work also implicated Epidermal Growth Factor Receptor (EGFR) in mediating the cytosolic localization of FAM129B via tyrosine phosphorylation; therefore, EGFR mutant tumors could also become NRF2-addicted as a result of constitutive FAM129B phosphorylation42. Also, while no NRF2 targeted therapies are approved by the FDA for use in cancer treatment, there are inhibitors of EGFR (i.e. gefitinib, cetuximab) and BRAF (i.e. vemurafenib) that are currently used in cancer treatment43–45. Therefore, by targeting the upstream effectors of FAM129B, NRF2 protein levels can be modulated, thus altering metastatic potential of tumors, and potentially increasing patient survival.
Figure 6: Schematic of NRF2 induction via FAM129B in mutant BRAF cells.
Constitutive activation of the MAPK signaling pathway via a mutation in BRAF (V600E) increases phosphorylation of FAM129B and mediates its cytosolic localization. In turn, FAM129B binds to KEAP1, preventing association between KEAP1 and NRF2, thus NRF2 protein accumulates, translocates to the nucleus, and activates transcription of its target genes.
Acknowledgments
Metastatic melanoma patient data was acquired via The Cancer Genome Atlas (TCGA).
Funding
This research is support by funding from the National Institutes of Health: T32 ES007091 (C.J.S.), P42 ES004940 (D.D.Z.), and R35 ES031575 (D.D.Z.).
Footnotes
Data Availability Statement
The data to support the current study is available upon request of the corresponding author. TCGA data was accessed via the cBIOPortal and comparison and survival plots were generated therein: [https://www.cbioportal.org/results/oncoprint?genetic_profile_ids_PROFILE_MUTATION_EXTENDED=mel_ucla_2016_mutations&genetic_profile_ids_PROFILE_COPY_NUMBER_ALTERATION=mel_ucla_2016_gistic&genetic_profile_ids_PROFILE_MRNA_EXPRESSION=mel_ucla_2016_rna_seq_mrna_median_Zscores&cancer_study_list=mel_ucla_2016&Z_SCORE_THRESHOLD=2.0&RPPA_SCORE_THRESHOLD=2.0&data_priority=0&profileFilter=0&case_set_id=mel_ucla_2016_3way_complete&gene_list=BRAF%250ARAD51%250APSMA1&geneset_list=%20&tab_index=tab_visualize&Action=Submit].
Disclosure
We have no conflicts of interests to declare.
References
- 1.American Cancer Society. Cancer facts & figures. In. Atlanta, GA: The Society:volumes. [Google Scholar]
- 2.Dong J, Phelps RG, Qiao R, et al. BRAF oncogenic mutations correlate with progression rather than initiation of human melanoma. Cancer Res. 2003;63(14):3883–3885. [PubMed] [Google Scholar]
- 3.Davies H, Bignell GR, Cox C, et al. Mutations of the BRAF gene in human cancer. Nature. 2002;417(6892):949–954. [DOI] [PubMed] [Google Scholar]
- 4.Brose MS, Volpe P, Feldman M, et al. BRAF and RAS mutations in human lung cancer and melanoma. Cancer Res. 2002;62(23):6997–7000. [PubMed] [Google Scholar]
- 5.Ascierto PA, Kirkwood JM, Grob JJ, et al. The role of BRAF V600 mutation in melanoma. J Transl Med. 2012;10:85. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Wan PT, Garnett MJ, Roe SM, et al. Mechanism of activation of the RAF-ERK signaling pathway by oncogenic mutations of B-RAF. Cell. 2004;116(6):855–867. [DOI] [PubMed] [Google Scholar]
- 7.Makrodouli E, Oikonomou E, Koc M, et al. BRAF and RAS oncogenes regulate Rho GTPase pathways to mediate migration and invasion properties in human colon cancer cells: a comparative study. Mol Cancer. 2011;10:118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Arozarena I, Sanchez-Laorden B, Packer L, et al. Oncogenic BRAF induces melanoma cell invasion by downregulating the cGMP-specific phosphodiesterase PDE5A. Cancer Cell. 2011;19(1):45–57. [DOI] [PubMed] [Google Scholar]
- 9.Lu H, Liu S, Zhang G, et al. Oncogenic BRAF-Mediated Melanoma Cell Invasion. Cell Rep. 2016;15(9):2012–2024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.McCain J. The MAPK (ERK) Pathway: Investigational Combinations for the Treatment Of BRAF-Mutated Metastatic Melanoma. P T. 2013;38(2):96–108. [PMC free article] [PubMed] [Google Scholar]
- 11.Garbe C, Eigentler TK. Vemurafenib. Recent Results Cancer Res. 2018;211:77–89. [DOI] [PubMed] [Google Scholar]
- 12.Tao S, Wang S, Moghaddam SJ, et al. Oncogenic KRAS confers chemoresistance by upregulating NRF2. Cancer Res. 2014;74(24):7430–7441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Rojo de la Vega M, Chapman E, Zhang DD. NRF2 and the Hallmarks of Cancer. Cancer Cell. 2018;34(1):21–43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.MacLeod AK, McMahon M, Plummer SM, et al. Characterization of the cancer chemopreventive NRF2-dependent gene battery in human keratinocytes: demonstration that the KEAP1-NRF2 pathway, and not the BACH1-NRF2 pathway, controls cytoprotection against electrophiles as well as redox-cycling compounds. Carcinogenesis. 2009;30(9):1571–1580. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Itoh K, Chiba T, Takahashi S, et al. An Nrf2/small Maf heterodimer mediates the induction of phase II detoxifying enzyme genes through antioxidant response elements. Biochem Biophys Res Commun. 1997;236(2):313–322. [DOI] [PubMed] [Google Scholar]
- 16.Kwak MK, Wakabayashi N, Greenlaw JL, Yamamoto M, Kensler TW. Antioxidants enhance mammalian proteasome expression through the Keap1-Nrf2 signaling pathway. Mol Cell Biol. 2003;23(23):8786–8794. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Jang J, Wang Y, Kim HS, Lalli MA, Kosik KS. Nrf2, a regulator of the proteasome, controls self-renewal and pluripotency in human embryonic stem cells. Stem Cells. 2014;32(10):2616–2625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Jayakumar S, Pal D, Sandur SK. Nrf2 facilitates repair of radiation induced DNA damage through homologous recombination repair pathway in a ROS independent manner in cancer cells. Mutat Res. 2015;779:33–45. [DOI] [PubMed] [Google Scholar]
- 19.Dodson M, de la Vega MR, Cholanians AB, Schmidlin CJ, Chapman E, Zhang DD. Modulating NRF2 in Disease: Timing Is Everything. Annu Rev Pharmacol Toxicol. 2019;59:555–575. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Hayes JD, Dinkova-Kostova AT. The Nrf2 regulatory network provides an interface between redox and intermediary metabolism. Trends Biochem Sci. 2014;39(4):199–218. [DOI] [PubMed] [Google Scholar]
- 21.Alam J, Stewart D, Touchard C, Boinapally S, Choi AM, Cook JL. Nrf2, a Cap’n’Collar transcription factor, regulates induction of the heme oxygenase-1 gene. J Biol Chem. 1999;274(37):26071–26078. [DOI] [PubMed] [Google Scholar]
- 22.Itoh K, Wakabayashi N, Katoh Y, et al. Keap1 represses nuclear activation of antioxidant responsive elements by Nrf2 through binding to the amino-terminal Neh2 domain. Genes Dev. 1999;13(1):76–86. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Zhang DD, Lo SC, Cross JV, Templeton DJ, Hannink M. Keap1 is a redox-regulated substrate adaptor protein for a Cul3-dependent ubiquitin ligase complex. Mol Cell Biol. 2004;24(24):10941–10953. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kobayashi A, Kang MI, Okawa H, et al. Oxidative stress sensor Keap1 functions as an adaptor for Cul3-based E3 ligase to regulate proteasomal degradation of Nrf2. Mol Cell Biol. 2004;24(16):7130–7139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Tao S, Liu P, Luo G, et al. p97 Negatively Regulates NRF2 by Extracting Ubiquitylated NRF2 from the KEAP1-CUL3 E3 Complex. Mol Cell Biol. 2017;37(8). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Tong KI, Katoh Y, Kusunoki H, Itoh K, Tanaka T, Yamamoto M. Keap1 recruits Neh2 through binding to ETGE and DLG motifs: characterization of the two-site molecular recognition model. Mol Cell Biol. 2006;26(8):2887–2900. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Tian W, Rojo de la Vega M, Schmidlin CJ, Ooi A, Zhang DD. Kelch-like ECH-associated protein 1 (KEAP1) differentially regulates nuclear factor erythroid-2-related factors 1 and 2 (NRF1 and NRF2). J Biol Chem. 2018;293(6):2029–2040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Komatsu M, Kurokawa H, Waguri S, et al. The selective autophagy substrate p62 activates the stress responsive transcription factor Nrf2 through inactivation of Keap1. Nat Cell Biol. 2010;12(3):213–223. [DOI] [PubMed] [Google Scholar]
- 29.Lau A, Wang XJ, Zhao F, et al. A noncanonical mechanism of Nrf2 activation by autophagy deficiency: direct interaction between Keap1 and p62. Mol Cell Biol. 2010;30(13):3275–3285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Old WM, Shabb JB, Houel S, et al. Functional proteomics identifies targets of phosphorylation by B-Raf signaling in melanoma. Mol Cell. 2009;34(1):115–131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Chen S, Evans HG, Evans DR. FAM129B/MINERVA, a novel adherens junction-associated protein, suppresses apoptosis in HeLa cells. J Biol Chem. 2011;286(12):10201–10209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Hast BE, Goldfarb D, Mulvaney KM, et al. Proteomic analysis of ubiquitin ligase KEAP1 reveals associated proteins that inhibit NRF2 ubiquitination. Cancer Res. 2013;73(7):2199–2210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Hugo W, Zaretsky JM, Sun L, et al. Genomic and Transcriptomic Features of Response to Anti-PD-1 Therapy in Metastatic Melanoma. Cell. 2016;165(1):35–44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Jessen C, Kress JKC, Baluapuri A, et al. The transcription factor NRF2 enhances melanoma malignancy by blocking differentiation and inducing COX2 expression. Oncogene. 2020;39(44):6841–6855. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Schmidlin CJ, Zeng T, Liu P, et al. Chronic arsenic exposure enhances metastatic potential via NRF2-mediated upregulation of SOX9. Toxicol Appl Pharmacol. 2020;402:115138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Lignitto L, LeBoeuf SE, Homer H, et al. Nrf2 Activation Promotes Lung Cancer Metastasis by Inhibiting the Degradation of Bach1. Cell. 2019;178(2):316–329 e318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Cheng KC, Lin RJ, Cheng JY, et al. FAM129B, an antioxidative protein, reduces chemosensitivity by competing with Nrf2 for Keap1 binding. EBioMedicine. 2019;45:25–38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Wang H, Liu X, Long M, et al. NRF2 activation by antioxidant antidiabetic agents accelerates tumor metastasis. Sci Transl Med. 2016;8(334):334ra351. [DOI] [PubMed] [Google Scholar]
- 39.Zhou X, Yang F, Zhang Q, et al. FAM129B promoted tumor invasion and proliferation via facilitating the phosphorylation of FAK signaling and associated with adverse clinical outcome of non-small cell lung cancer patients. Onco Targets Ther. 2018;11:7493–7501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Oishi H, Itoh S, Matsumoto K, et al. Delayed cutaneous wound healing in Fam129b/Minerva-deficient mice. J Biochem. 2012;152(6):549–555. [DOI] [PubMed] [Google Scholar]
- 41.Ren D, Villeneuve NF, Jiang T, et al. Brusatol enhances the efficacy of chemotherapy by inhibiting the Nrf2-mediated defense mechanism. Proc Natl Acad Sci U S A. 2011;108(4):1433–1438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Ji H, Lee JH, Wang Y, et al. EGFR phosphorylates FAM129B to promote Ras activation. Proc Natl Acad Sci U S A. 2016;113(3):644–649. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Sosman JA, Kim KB, Schuchter L, et al. Survival in BRAF V600-mutant advanced melanoma treated with vemurafenib. N Engl J Med. 2012;366(8):707–714. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Hosomi Y, Morita S, Sugawara S, et al. Gefitinib Alone Versus Gefitinib Plus Chemotherapy for Non-Small-Cell Lung Cancer With Mutated Epidermal Growth Factor Receptor: NEJ009 Study. J Clin Oncol. 2020;38(2):115–123. [DOI] [PubMed] [Google Scholar]
- 45.Cohen R, Sroussi M, Pilati C, Houry S, Laurent-Puig P, Andre T. Unresectable metastatic colorectal cancer patient cured with cetuximab-based chemotherapy: a case report with new molecular insights. J Gastrointest Oncol. 2018;9(4):E23–E27. [DOI] [PMC free article] [PubMed] [Google Scholar]