Abstract
Summary
We present bedtk, a new toolkit for manipulating genomic intervals in the BED format. It supports sorting, merging, intersection, subtraction and the calculation of the breadth of coverage. Bedtk uses implicit interval tree, a data structure for fast interval overlap queries. It is several to tens of times faster than existing tools and tends to use less memory.
Availability and implementation
The source code is available at https://github.com/lh3/bedtk.
1 Introduction
Processing genomic intervals is a routine task and has many practical applications in bioinformatics. Some common uses of genomic intervals include computing coverage and depth of specific genomic features and finding intersections of new sequencing results with known genomic features such as exons or genes. BEDTools (Quinlan and Hall, 2010) is a popular toolkit to manipulate intervals in the BED format. However, it can be slow given large datasets. BEDOPS (Neph et al., 2012) addresses this issue by streaming sorted BED files. While this strategy improves performance, it is less convenient to use and is limited to a subset of interval operations. These observations motivated us to develop bedtk that achieves high performance without requiring sorting.
2 Materials and methods
Efficiently finding overlapping intervals is a core functionality behind all interval processing tools. Bedtk uses implicit interval tree, a data structure extending the concept of array layouts of searching tree (Brodal et al., 2002; Khuong and Morin, 2017), to perform interval overlap queries against a static list of intervals.
2.1 Implicit binary search trees
A binary search tree (BST) is a binary tree where each node is associated with a key and this key is no less than all keys in the left subtree and no greater than all keys in the right subtree. Here, we show that a BST can be implicitly represented by a sorted array. In this implicit BST, each node is an element in the array and the tree topology is inferred from the index of each element.
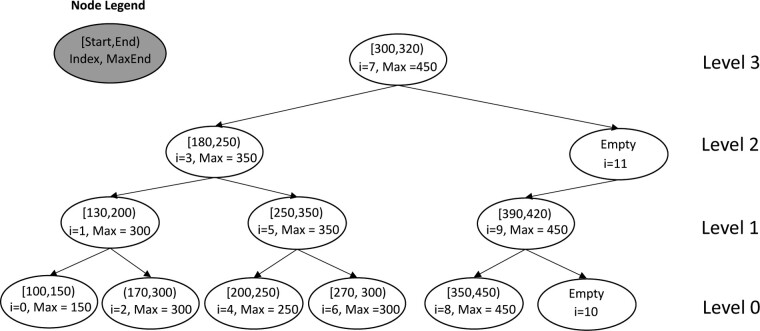
More specifically, consider a sorted array consisting of elements. This array can implicitly represent a binary tree with levels, with leaves put at level 0 and the root at level (Fig. 1). This implicit BST has the following properties:
Fig. 1.
Example of implicit interval tree
At level , the first node is indexed by . As a special case, the root of the tree is indexed by .
For a node indexed by at level , its left child is indexed by , and its right child indexed by .
For a node indexed by at level , it is a left child if its th bit is 0 (i.e. is an even number), and its parent node is . Node is a right child if its th bit is 1 (i.e. is an odd number), and its parent node is .
For a node indexed by at level , there are nodes descending from (including ). The left-most leaf in this subtree is .
With these properties, we can identify the parent and the children of a node in time. Figure 1 shows an example of implicit BST for the start positions of a list of intervals which is sorted by the start positions. When there are fewer than elements in the array, we still assume a full binary tree with some nodes set to ‘empty’. Notably in this case, an empty node may have a non-empty child (e.g. in Fig. 1, empty node 11 has a non-empty child node 9).
2.2 Implicit interval trees
An augmented interval tree is a data structure for interval overlap queries. It is an extension to BST where a node corresponds to an interval and each node additionally keeps a MaxEnd field which is the largest end position in the subtree descending from the node. Because a BST can be implicitly represented with an array, an augmented interval tree can be represented with an array as well (Fig. 1).
Sorting input intervals takes time. Computing MaxEnd requires a bottom-up tree traversal in time. The time complexity of searching is per query, where is the number of intervals in the tree and is the number of overlapping intervals.
3 Results
We implemented implicit interval tree in bedtk along with few other common operations such as sorting and interval merging. We compared bedtk to BEDTools v2.29.2 and BEDOPS v2.4.39 on two BED files: (i) 1 194 285 exons from GenCode annotations and (ii) 9 944 559 alignment intervals of long RNA-seq reads. Both are available at https://github.com/lh3/cgranges/releases.
As is shown in Table 1, bedtk is consistently faster than BEDTools and BEDOPS for all the evaluated operations, even when we discount sorting time for the BEDOPS ‘intersection’ operation. The performance gap between bedtk and BEDTools is even larger for unsorted input (‘intersect’ and ‘coverage’). This exemplifies the advantage of the implicit interval tree. BEDOPS takes the least memory for sorting potentially because it uses advanced encoding; it uses even less memory for ‘intersect’ as it streams the input instead of loading one or both input files into memory. However, requiring sorted input complicates data processing pipelines and wastes working disk space. And even with sorted input, BEDOPS is slower than bedtk.
Table 1.
Performance of interval operations
| Operation | Bedtk | BEDTools | BEDOPS |
|---|---|---|---|
| Sort | 5.35 s/393 MB | 21.14 s/2731 MB | 11.25 s/205 MB |
| Intersect | 6.42 s/19 MB | 54.87 s/397 MB | 8.78 s/3 MB |
| Coverage | 10.91 s/19 MB | 257.11 s/678 MB |
Note: Each cell gives the CPU time in sections and peak memory in megabases (MB) measured on a Linux server with two AMD EPYC 7301 CPUs at 2.2 GHz. The ‘sort’ operation sorts file 2. ‘intersect’ reports intervals in file 2 that overlaps intervals in file 1. For each interval in file 2, ‘coverage’ computes the number of bases covered by intervals in file 1. BEDOPS does not support the ‘coverage’ operation. It also requires sorted input for ‘intersect’; sorting time is excluded. All operations are CPU bounded, not I/O bounded.
4 Discussions
An implicit interval tree can be implemented in <100 lines of C++ code. It is a simple yet efficient data structure for fast interval queries. Daniel Jones alters the memory layout of implicit interval tree to van Emde Boas for improved cache locality (https://github.com/dcjones/coitrees). It speeds up query at the cost of more memory and unsorted query output. Meanwhile, Mike Lin gives up the standard top-down interval tree query and instead allows to start a query from any node in the tree (https://github.com/mlin/iitii). This improves the performance on practical data. In additional to interval trees, nested containment list (Alekseyenko and Lee, 2007) and augmented interval list (Feng et al., 2019) are alternative data structures for fast interval overlap queries. However, no standalone user-oriented tools have implemented these advanced algorithms. Bedtk is the first toolkit that is designed for end users and outperforms popular tools for common interval operations.
Acknowledgements
The authors thank Daniel C. Jones and Mike Lin for further improving the performance of implicit interval trees.
Funding
This work was supported by National Institutes of Health [R01HG010040].
Conflict of Interest: none declared.
Contributor Information
Heng Li, Department of Data Science, Dana-Faber Cancer Institute, Boston, MA 02215, USA; Department of Biomedical Informatics, Harvard Medical School, Boston, MA 02215, USA.
Jiazhen Rong, Department of Biomedical Informatics, Harvard Medical School, Boston, MA 02215, USA.
References
- Alekseyenko A.V., Lee C.J. (2007) Nested Containment List (NCList): a new algorithm for accelerating interval query of genome alignment and interval databases. Bioinformatics, 23, 1386–1393. [DOI] [PubMed] [Google Scholar]
- Brodal G.S. et al. (2002) Cache oblivious search trees via binary trees of small height. In SODA '02: Proceedings of the Thirteenth Annual ACM-SIAM Symposium on Discrete Algorithms, January. pp. 39–48, San Francisco, California.
- Feng J. et al. (2019) Augmented Interval List: a novel data structure for efficient genomic interval search. Bioinformatics, 35, 4907–4911. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khuong P.V., Morin P. (2017) Array layouts for comparison-based searching. ACM Journal of Experimental Algorithmic, 22, 1.3:1-39. [Google Scholar]
- Neph S. et al. (2012) BEDOPS: high-performance genomic feature operations. Bioinformatics, 28, 1919–1920. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Quinlan A.R., Hall I.M. (2010) BEDTools: a flexible suite of utilities for comparing genomic features. Bioinformatics, 26, 841–842. [DOI] [PMC free article] [PubMed] [Google Scholar]