Abstract
Background and aims
The novel SARS-CoV-2 has been rattling the world since its outbreak in December 2019, leading to the COVID-19 pandemic. The learning curve of this new virus has been steep, with a global scientific community desperate to learn how the virus is transmitted, how it replicates, why it causes such a wide spectrum of disease manifestations, resulting in none or few symptoms in some. Others are burdened by an intense immune response that resembles the cytokine storm syndrome (CSS), which leads to severe disease manifestations, often complicated by fatal acute respiratory distress syndrome and death. Research efforts have been focusing on finding effective cures and vaccinations for this virus.
The presence of SARS-CoV-2 in the gastrointestinal (GI) tract, represented by several GI manifestations, has led to its investigation as a target for the virus and as an indicator of disease severity. The response of the microbiome (which is heavily linked to immunity) to the novel SARS-CoV-2 virus, and its role in igniting the exaggerated immune response has therefore become a focus of interest. The objective of our study was to gather the data connecting between the microbiome, the GI tract and COVID-19 and to investigate whether these reported alterations in the gut microbiome bear any resemblance to those seen in lupus, the prototypical autoimmune disease. Confirming such changes may become the steppingstone to potential therapies that may prevent transmission, progression and immune related manifestations of COVID-19, via manipulation of the gut microbiota.
Methods
We performed an extensive literature review, utilizing the Pubmed search engine and Google Scholar for studies evaluating the microbiome in COVID-19 patients and compared results with studies evaluating the microbiome in lupus. We searched for the terms: microbiome, dysbiosis, COVID-19, SARS-CoV-2, gastrointestinal as well as lupus and autoimmune. While there were hundreds of articles which referred to gastrointestinal manifestations in COVID-19, to date only 4 studies investigated the gastrointestinal microbiome in this setting. We compared the similarities between microbiome of COVID-19 patients and lupus patients.
Results
We found that there are several similar processes of immune dysregulation in patients with COVID-19 and in those with lupus, with several other alterations seen in other pathological states. Some of these similarities include loss of microbiota biodiversity, increased representation of pathobionts, which are microbes associated with inflammation and disease (i.e Proteobacteria) and a relative decrease of symbionts, which are protective microbes, associated with anti-inflammatory properties (i.e Lactobacillus). Compromise to the intestinal barrier has also been reported in both.
Conclusions
We conclude that the gastrointestinal tract contributes to the disease manifestations in COVID-19. Whether gastrointestinal dysbiosis is the cause or effect of gastrointestinal manifestations and several severe systemic manifestations, which may be the response to an increased pro-inflammatory environment, is still debatable and warrants further investigation. Given the resemblance of the microbiome in COVID-19 patients to that seen in lupus patients, it becomes clearer why several therapies used in autoimmune conditions are currently under investigation for the treatment of COVID-19 patients. Moreover, these findings should promote further investigating the utility of manipulation of the microbiome, via nutritional supplementation or even fecal transplantations, interventions that may alter the course of the disease, and potentially prevent disease transmission at low cost and low risk.
Keywords: Microbiome, Dysbiosis, SLE, Lupus, Autoimmunity, COVID-19, SARS-CoV-2
1. Introduction
The outbreak of the novel strain of coronavirus in December 2019, originating in Wuhan, China, was the start of a catastrophic pandemic, which has since then been rattling the world. This virus, now known as SARS-CoV-2 or Covid-19 is characterized by a wide range of presentations, from asymptomatic or flu like illness causing fever, dry cough, myalgias and or extreme fatigue, to severe pneumonia with sepsis leading to an acute respiratory distress syndrome (ARDS) in up to 20% of cases, with subsequent respiratory failure requiring mechanical ventilation. Multi-organ involvement has also been reported in severe cases and includes hematological, gastrointestinal, neurological and cardiovascular complications, leading to high death rates, which are thought to be the result of a cytokine storm [[1], [2], [3], [4], [5]].
To enter the human host cells, SARS-CoV-2 utilizes its spike protein (S- protein), which binds to the angiotensin converting enzyme-2 (ACE-2) and is then processed by the transmembrane protease serine-2 (TMPRSS-2), which primes the S-protein [6]. This prompts an immune response from the new host, which leads to activation of the immune system in an effort to detect, recognize and react to the novel antigens, produced by the new virus. The intensity of the immune response is thought to affect disease phenotype, and it is an exaggerated immune response, which is thought to contribute to severe disease manifestations [5,7], often complicated by fatal acute respiratory distress syndrome and death. This exaggerated immune response, described in COVID-19 patients, resembles that of the cytokine storm syndrome (CSS), which has been described in several other diseases including viral infections (i.e Influenza A, cytomegalovirus), autoimmune disorders (i.e lupus, Still's disease, juvenile idiopathic arthritis, macrophage activation syndrome), malignancies and complications in medical interventions such as immunotherapy and stem cell transplant [8,9]. Other serious complications of COVID-19 that are also thought to be the result of a hyperactive immune response include hypercoagulable states as well as several autoimmune phenomena, which are being frequently reported [10,11].
The scientific community has been closely studying the SARS-CoV-2 virus, its infectivity, modes of transmission, mechanisms of action as well as the virus's interaction with its host. As data has been fast accumulating, and conclusions rapidly changing, the immune response, provoked by SARS-CoV-2, is a leading suspect as the culprit of severe disease manifestations. Clinical clues, which support the theory of an exaggerated immune response include elevated inflammatory markers and pro-inflammatory cytokines such as serum ferritin, erythrocyte sedimentation rate (ESR), C-reactive protein (CRP) and interleukin 6 (IL)-6, all of which have been documented consistently in patients with severe disease manifestations [12,13]. Several additional pro-inflammatory cytokines have been noted in association with SARS-CoV-2 including interferon gamma (INF-γ), IL-8, tumor necrosis factor-α (TNF-α), IL-10, IL-2 and others [[14], [15], [16], [17], [18], [19], [20]]. IL-6 and IL-10 have been reliable indicators of disease severity, yet not clinically available, and a study of 102 COVID-19 patients, demonstrated a clear association between high levels of pro-inflammatory cytokines and more severe disease [20]. Additionally, the ARDS picture described in COVID-19 patients, has been thought to be the result of an auto-inflammatory cytokine storm, which has been similarly described as macrophage activation syndrome observed in patients with SARS-CoV and MERS-CoV [7,21,22].
2. COVID-19 and autoimmunity
Viruses have been known to provoke immune responses, which may present similarly to those seen in autoimmune diseases, by disrupting immunological tolerance via several well-known mechanisms such as molecular mimicry, bystander activation, epitope spreading and presentation of cryptic antigens [[23], [24], [25], [26]]. The viral infection, via these mechanisms, may lead to activation of antigen presenting cells that may in turn activate pre-primed auto reactive T cells, thus leading to the production of pro-inflammatory mediators, ultimately causing tissue damage [26]. Some examples of viruses, linked to autoimmune conditions and auto-inflammatory states, include hepatitis C, associated with cryoglobulinemic vasculitis and Sjögren's-like syndrome [27,28], and herpesviruses, Ebstein-Barr virus (EBV), associated with systemic lupus erythematosus (SLE), rheumatoid arthritis (RA) and adult-onset Still's disease (AOSD) [[29], [30], [31]]. In addition, enteric viruses are associated with type I diabetes [32] and influenza viruses are associated with acute disseminated encephalomyelitis [33]. Despite being a novel virus, SARS-CoV-2 has already been linked to a long list of auto-inflammatory and autoimmune conditions, including Guillain-Barré syndrome (GBS) [[34], [35], [36], [37]], autoimmune hemolytic anemia, immune thrombocytopenic purpura [[38], [39], [40], [41]], and Kawasaki disease (KD) [42,43]. An additional autoimmune phenomenon reported in association with COVID-19, which bears a striking resemblance to the inflammatory symptoms in KD, but does not completely overlap, has been termed by the World Health Organization (WHO) as the Multisystem Inflammatory Syndrome (MIS). This syndrome, which had been initially reported in children (MIS-C) [44], but later described in the adult population (MIS-A) [45] is a devastating complication of COVID-19, associated with a significant need for critical care, a high rate of cardiac involvement and a 2% overall mortality rate [44].
The fact that immune cells and mediators have been early targets for disease modification in COVID-19, comes as no surprise given the strong influence the SARS-CoV-2 has on the host immune system. Since early on in the course of the pandemic, there have been strong associations made between COVID-19 and various cells of the immune system, both in quality and quantity, some associated with the disease itself and others suggested as indicators of disease severity and prognosis. The characteristic lymphopenia in patients with COVID-19 (and which is commonly seen in other viral illnesses), may lead to failure in the maintenance of peripheral tolerance, resulting in activation of effector T cells with autoimmune potential. Loss of self-tolerance is a mechanism, which highlights the paradoxical association between lymphopenia and autoimmunity [46,47] and may explain the link between these two features in viral infections [16,48]. Decrease in eosinophils, natural killer cells and antigen presenting cells have also been demonstrated [19,49]. One study, for example, which analyzed broncho-alveolar lavage fluid transcriptome from COVID-19 patients, revealed an increase in dendritic cells (DCs) and activated neutrophils [50], while another revealed higher percentage of inflammatory monocytes in patients with severe lung pathology [15].
3. Interactions between viruses and the human gut
The human microbiome has come to be known as a key player in the modulation of its host's immunity and thought to take part in the pathogenesis of various chronic diseases and autoimmune conditions. The virome however, which includes the gastrointestinal presence of viruses and their interaction with the host have been less explored and only recently data has been emerging regarding intestinal luminal viruses and bacteriophages [51]. The mechanism by which viruses interact with the host is via penetration and integration of their genetic material into the host genome [52]. Viral invasion into the gastrointestinal (GI) lumen may induce diarrhea, compromising the mucosal barrier, thus increasing gut permeability, disturbing the gastrointestinal microbiotal homeostasis and effecting microbiome composition, diversity and function. Ultimately, disruption of the mucosal barrier may lead to immunological alterations that may promote hyper-inflammatory responses and autoimmune phenomena, as described above [10,11,53]. We believe that several manifestations of COVID-19 might be related to interactions between the novel SARS-CoV-2 virus and its host's microbiome, ultimately leading to immune dysregulation.
4. COVID-19, the gut and the lung-gut axis
Like other coronaviruses, SARS-CoV-2 utilizes ACE-2, a cell surface receptor, for entry into host cells [54,55]. The abundance of ACE-2 in the lung tissue can explain the common respiratory manifestation associated with the disease; they are also present in several extrapulmonary tissues including the heart, liver, kidney and intestines [[56], [57], [58]]. In the intestines, where ACE-2 acts as a coreceptor for nutrient uptake and amino acid absorption [58], high levels of the receptor are found on the luminal surface of differentiated epithelial cells in the small intestine, with lower levels found in the crypt cells and in the colon [[58], [59], [60], [61]]. It is the presence of ACE-2 along the gastrointestinal tract, which enables penetration of SARS-CoV-2 into the GI tract, sometimes resulting in the presence of GI manifestations such as diarrhea, nausea, vomiting [1,[62], [63], [64], [65], [66]] anorexia, decreased appetite, abdominal pain [67,68] or some combination of these. COVID-19 patients who present with GI manifestations, have been found to be more likely to excrete viral RNA in stool [[69], [70], [71], [72]], and sometimes have prolonged time to viral clearance [73], even long after respiratory viral RNA is cleared [74,75]. Since the initial cases reported in Wuhan China, which described low rates of GI symptoms in COVID-19 patients [1,76], the presence of marked digestive symptoms has been described at much higher rates (10% to over 50%) in other cohorts, some out of Wuhan itself [6,77,78]. One study investigating the clinical characteristics of COVID-19 in 204 patients, reported vomiting, diarrhea and abdominal pain in 50.5% of cases [68]. In some reports, SARS-CoV-2 associated diarrhea occurred prior to the onset of respiratory symptoms [65,79]. In others, GI symptoms were reported concurrently with respiratory symptoms, most commonly cough and surprisingly, some studies reported that up to 25% of patients presented with GI symptoms in the absence of significant respiratory symptoms, with or without other constitutional symptoms (such as fever, reported in one study in >80% of patients with GI symptoms) [62]. A positive correlation between GI manifestations and overall disease severity was also reported by some [[80], [81], [82]]. Given the variable presentation of those with GI symptoms, it is not surprising that delayed diagnosis has been reported in this subgroup of patients who presented with predominant GI symptoms in the absence of more classic disease manifestations [[83], [84], [85]].
Despite our understanding of how SARS-CoV-2 may penetrate the GI system as a possible cause of gastrointestinal manifestations [6,76,86], the question of whether or not these manifestations are the result of direct viral infectivity remains unanswered. In fact, several other mechanisms are plausible, such as provocation of an inflammatory response by the viral particles or medication associated GI manifestations. The understanding of these mechanisms is important because it may lead to a potential target for prevention. Understanding which factors impact the abundance of the ACE-2 cell receptors is important as well, given their role in virus penetration into the cells. Interestingly, alterations to the microbiome may contribute to increase in intestinal ACE-2 in COVID-19 patients [58,87]. One example is the increased presence of Coprobacillus, a microbe that has been shown to upregulate colonic ACE-2 in murine models [88]. ACE-2 itself has been found to have a role in amino acid transport, a role in modulating the microbiotal composition, and furthermore animal models with ACE-2 mutants were found to exhibit decreased expression of antimicrobial peptides and showed altered gut microbial composition [58,87,88].
Aside from the shared presence of ACE-2 in both the intestines and the lungs, there are additional links between these two systems, known as the gut-lung-axis, which may contribute to some of the phenomena seen in patients with COVID-19. This axis is also thought to be modulated by the microbiome [[89], [90], [91]] as demonstrated in a murine model, which showed that germ free mice that lack intestinal microbes had decreased pathogenic clearance in the lungs [92]. Several mechanisms involving the interactions of the gut microbiome with other organs, including the lungs, and their bidirectional influence on one another, have been proposed [91]. This axis is thought to be immune regulated, through the gut microbiota and its metabolome, which are phagocytosed by antigen presenting cells, ultimately activating the adaptive immune system. An additional mechanism may involve translocation of bacterial/viral antigens into the lymphatic system or distant organs, via the blood stream, as a result of increased gut permeability, leading to an immunological response that ultimately leads to tissue damage in the target organ [87]. Like the gut microbiota, the lung is host of a distinct microbiota [89,93], dominated by similar phyla, including Bacteroidetes, Firmicutes and Proteobacteria [94] and their metabolites, might also affect the composition, integrity and function of the gut microbiome, in a similar mechanism. The gut-lung-axis has already been described in several pulmonary diseases. Several studies have suggested that gut dysbiosis plays a key role in the pathogenesis of sepsis and acute respiratory distress syndrome (ARDS) [95], by loss of microbial diversity, leading to dysbiosis that in turn may lead to immune dysregulation, causing ARDS, similar to that seen in severe COVID-19 infection [96]. One study found that microbial action on dietary fiber is known to increase short chain fatty acids (SCFA) in blood, thereby protecting against allergic inflammation in the lungs [97], while another demonstrated that various prebiotics were associated with increased butyrate levels, thereby reducing inflammation and improving conditions in asthma and cystic fibrosis [98]. One murine model even demonstrated that depletion of certain gut microbes by antibiotics leads to increased susceptibility to influenza pneumonia [99]. These make it reasonable to consider [100] that COVID-19 pneumonia and ARDS associated with severe COVID-19 [101], may be caused by abnormal interactions in the lung-gut-axis as a result of dysbiosis. Only a couple of studies have explored the changes that occur in the microbiome of the lung in COVID-19 patients, and a few others explored the changes in the gut microbiome in this cohort of patients. One study, which analyzed post-mortem biopsies from 20 deceased COVID-19 patients, showed that the most common bacterial genera in the lung microbiome were Acinetobacter, Chryseobacterium, Bukholderia, Brevundimonas, Sphingobium and Enterobacteriaceae. Another study demonstrated similarities in the lung microbiome of COVID-19 patients and in those with community-acquired bacterial pneumonia, both of which were enriched with pathobionts, defined as microbes, which inhabit the gastrointestinal tract and are associated with chronic inflammation [102]. The presumed bidirectional cross-talk between the gut microbiota and the lungs [103], which occurs via microbial metabolites, endotoxins and cytokines, allowing one system to impact the other systemically [104], is the proposed mechanism by which SARS-CoV-2 impacts the gut microbiota, likely contributing not only to the development of GI manifestations, but also to the many autoimmune phenomena, associated with COVID-19.
5. The microbiotal link between COVID-19 and autoimmunity
The strong association between SARS-CoV-2 and autoimmunity, has crowned it “the virus of autoimmunity” [10,11,105], and it is postulated that intestinal dysbiosis may be the cause of the exaggerated immune response which leads to these phenomena. The Human Functional Genomics Project (HFGP) has, in fact demonstrated that differences in composition and function of the microbiome contribute to inter-individual variation in cytokine responses to pathogen exposure. This is likely the result of microbial-derived mediators, which are products metabolic pathways, facilitated by the gut microbiota [106,107]. In some cases, the inflammatory response can be overaggressive, causing a ‘cytokine storm’, which results in widespread tissue damage, septic shock and multi organ failure [[108], [109], [110]]. Another proposed player, to contribute to dysbiosis and severe systemic manifestations is the dysfunctional mitochondria. It is proposed, that like in other hyper-inflammatory conditions, which induce hyperferritinemia, as a result of iron dysregulation, COVID-19 too is a hyperferritinemic state [13,21], which promotes oxidative stress. The dysfunctional mitochondria may directly cause several disease manifestations such as hypercoagulability, may contribute to microbiota dysbiosis, which in turn may directly cause severe disease manifestations seen in COVID-19. Additionally, the microbiota can further lead to a hyper-inflammatory environment, thereby exacerbating oxidative stress and mitochondrial dysfunction, causing a devastating vicious cycle [111]. It turns out that there are several similar alterations in the microbiome of COVID-19 patients and in that of patients with autoimmune conditions, which may be the shared culprit to the dysregulated immune system seen in both conditions, ultimately leading to increased disease severity and to autoimmune manifestations [105]. As mentioned above, like other respiratory infections, which have been associated with dysbiosis [112], several changes have been demonstrated in the microbiome of COVID-19 patients. Some of these changes include decreased richness, which is a decrease in the total number of microbial species present in the microbiome of COVID-19 patients, as well as decreased diversity, which refers to the relative amount of each microbial species. These changes, however, may have been the result of confounding factors, such as medication use, age and comorbid conditions. As results have been variable and in fact, one study that controlled for use of antibiotics, did not demonstrate significant differences in richness and diversity between COVID-19 patients and healthy controls [110]. In fact, some of the most widely used medications during the COVID-19 pandemic independently affect diversity and richness of the microbiome, most notorious is azithromycin, which rapidly reduces bacterial richness and diversity [113].
Other compositional changes that have been noted in the gut microbiome of COVID-19 group patients included the domination of Streptococcus, Rothia, Veillonella, Erysipelatoclostridium, and Actinomyces, whereas the microbiome of healthy controls was dominated by the genera Romboutsia, Faecalibacterium, Fusicatenibacter, and Eubacterium hallii group. Ultimately, the five species that were selected to distinguish between COVID-19 patients and healthy controls included in one study, Fusicatenibacter, Romboutsia, Intestinibacter, Actinomyces, Erysipelatoclostridium. These microbiotal changes were demonstrated not only in those with prolonged GI symptoms, but also those with severe respiratory disease [114]. The association between the microbiome and autoimmunity has been well studied and therefore we were not surprised to find that several features of dysbiosis, which were demonstrated in COVID-19 patients, are similar to those seen in lupus, which the prototypical autoimmune disease in humans. Diversity and richness are affected, one case-control study found that compared to healthy controls, COVID-19 patients had significantly reduced bacterial diversity, a significantly higher relative abundance of opportunistic pathogens (i.e Streptococcus, Actinomyces) and a lower relative abundance of beneficial symbionts, all findings which have been demonstrated in lupus and other autoimmune conditions [105,115]. Another small study from China demonstrated dysbiosis in COVID-19 patients characterized by decreased abundance of Lactobacillus and Bifidobacterium [116]. Both microbes are usually associated with anti-inflammation and both have been found decreased in a large number of lupus studies, in both murine and human models [105,117]. An additional study from Wuhan, China, described a link between the composition of the gut microbiome and the predisposition of healthy individuals to COVID-19 [80]. Increased levels of Lactobacillus species correlated to higher levels of the anti-inflammatory interlukin IL-10 and improved disease prognosis, while increased levels of proinflammatory microbes, such as Klebsiella, Streptococcus, and Ruminococcus gnavus, correlated with elevated levels of proinflammatory cytokines and increased disease severity. These species have all been described in association with several autoimmune conditions and Ruminococcus gnavus specifically, has been associated with active lupus [[118], [119], [120]]. Additionally, increased abundance of Coprobacillus, Clostridium ramosum and Clostridium hathewayi, has been found to positively correlate with overall disease severity in COVID-19 patients [[121], [122], [123], [124], [125]], the latter being reported in association with kidney disease in lupus [122]. On the other hand, Faecalibacterium prausnitzii was found to be inversely correlated with disease severity in both lupus patients and COVID-19 patients [105,121,[126], [127], [128], [129], [130]]. Alistipes onderdonkii is another beneficial bacterial species, which showed a negative correlation with COVID-19 severity and has been reported to be decreased in lupus patients as well [131,132]. The effect of the fecal SARS-CoV-2 load on the composition and function of the microbiome was also studied and it seems that the microbiome of those with a high fecal SARS CoV-2 load also had abundance of opportunistic pathogens such as Collinsella aerofaciens, Collinsella tanakaei, Streptococcus infantis, Morganella morganii.
It is well known that an altered Firmicutes/Bacteroidetes (F/B) ratio effects the mucosal barrier, which leads to intestinal permeability by promoting functional changes on the molecular level in the gut, ultimately promoting inflammation [132,133], as is found in lupus and several other auto-inflammatory conditions. An interesting finding revealed that an abundance of bacteria from the Firmicutes phyla positively correlated with disease severity, including the genus Coprobacillus, the species Clostridium ramosum and Clostridium hathewayi, data that might be explained by the diverse role of the Firmicutes bacteria in regulation of ACE-2 expression in the murine gut [88,134]. The abundance of Bacteroidetes species, A. onderdonkii and B. ovatus and 4 species from the genus Bacteroides: B. thetaiotaomicron, B. massiliensis, B. ovatus and B. dorei, negatively correlated with COVID-19 severity. These microbes, which are reported to take part in host immune regulation [135,136] and reported to suppress colonic ACE-2 expression [88], showed inverse correlation with fecal viral load of SARS-CoV-2. In lupus patients, on the other hand, most studies have found that the abundance of Firmicutes (F) was decreased, whereas the abundance of Bacteroidetes (B) increases, and most commonly note that the ratio F/B is decreased [137], a ratio that has not been specifically discussed in regards to COVID-19 patients. One study did demonstrate that at the phylum level, members of the Bacteroidetes were more relatively abundant in patients with COVID-19 when compared to healthy controls, whereas Actinobacteria were more relatively abundant in healthy controls [110]. Some studies investigating the microbiome in this population, reported a lower abundance of Bacteroides species than healthy individuals [[138], [139], [140], [141]]. It is worth noting that an altered composition of the gut microbiome has also been shown to be a potential risk factor for COVID-19 infection in healthy individuals, thus stressing the potential benefit of treatments that will modulate the composition of the microbiome as a preventive measure for disease contraction [142].
Some functional similarities are described in the microbiome of COVID-19 patients and in those with lupus. Compromise to the mucosal barrier is thought to be one of the key features promoting immunological alterations in COVID-19, a theory that is well-established in several autoimmune conditions, including lupus as well as in several chronic medical condition [105,117,143]. Additionally, one study found that COVID-19 patients with a high SARS-CoV-2 fecal viral load was associated with higher functional capacity for nucleotide de novo biosynthesis, amino acid biosynthesis and glycolysis, whereas decreased or absence of virus in the feces was associated with more short-chain fatty acid producing bacteria, such as Parabacteroides merdae, Bacteroides stercoris, Alistipes onderdonkii and Lachnospiraceae bacterium [132]. These metabolic pathways have a significant effect on the integrity of the mucosal barrier in the gut, affect the dynamics between enteric pathogens and the microbiome and influence immune responses. (see Table 1 )
Table 1.
Microbiotal similarities between COVID-19 and lupus.
| Microbiotal changes | COVID-19 | LUPUS |
|---|---|---|
| Dysbiosis | + | + |
| Microbial diversity | ↓ | ↓ |
| Altered F/B ratio | + | + |
| Gut Premeability | ↑ | ↑ |
| Opportunistic pathogens | ↑ | ↑ |
| Ruminoccocus ganvus | ↑ | ↑ |
| Clostridium ssp | ↑ | ↑ |
| Symbionts | ↓ | ↓ |
| Lactobacillus | ↓ | ↑↓ |
| Bifidobacterium | ↓ | ↓ |
| Faecalibacterium prausnitzii | ↓ | ↓ |
| Alistipes onderdonkii | ↓ | ↓ |
6. The microbial link between COVID-19 and aging
Whether the altered microbiome is a cause or consequence in disease pathogenesis or severity of COVID-19, altogether, data suggests a potential role for the microbiome in determining response to SARS-CoV-2 infection and disease intensity [133,144]. These interesting findings also make sense of the fact that those individuals who are at highest risk for severe disease, complications and death from COVID-19 are the elderly and frail [145], a risk that is multifactorial, but likely impacted by the altered microbiome. In fact, also this population shares several features of dysbiosis with lupus patients (Fig. 1 ) [117]. The vulnerability of the elderly and frail, is thought to result from immunosenescence; a condition which refers to the decline in immune regulation brought on by aging, affecting both the innate and adaptive immune system. Immunosenescence leads to decreased capacity of the host to respond to infection, resulting in an increased risk for severe disease manifestations and a decreased tolerance to auto- antigens, ultimately leading to autoimmunity. One of the proposed mechanisms leading to immune aging is thought to be related to the microbiotal changes, which include loss of diversity, and decreased prevalence of beneficial microbes, that have anti-inflammatory properties, all changes that are described in lupus and COVID-19 patients as well, as described above [117,146].
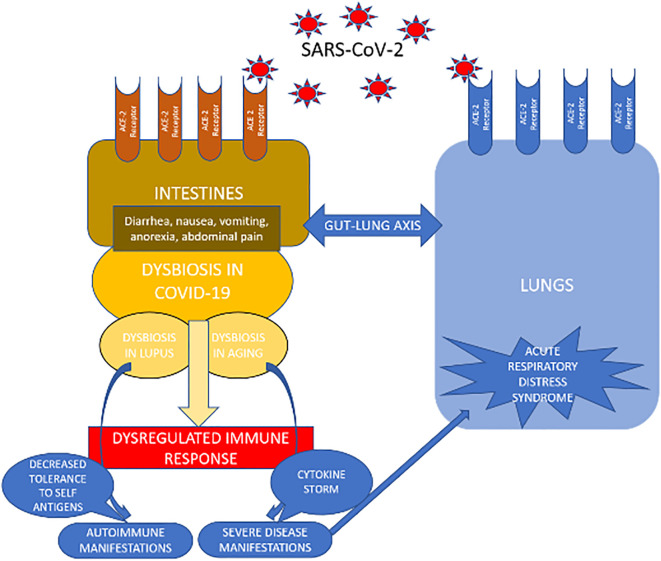
Fig. 1.
Illustrates the effect of SARS-CoV-2 on the lungs and the gastrointestinal (GI) tract and demonstrates the dynamic interplay between the lungs and the GI tract in response to SARS-CoV-2.
7. The microbiome and COVID-19 therapy
While prevention remains the cornerstone of treatment for the COVID-19 pandemic, several therapeutic measures have been investigated, some targeting the virus itself, while others attempt to modulate the inflammatory response associated with severe disease [147]. Remdesivir - a nucleoside analogue that targets the virus, inhibits SARS-CoV-2 RNA dependent RNA polymerase activity and viral replication, has some benefit in hospitalized patients [148]. Other early therapies included intravenous immunoglobulin (IVIG), and more commonly convalescent plasma (specific IVIG), extracted from patients who recovered from COVID-19, which have been used for severely ill patients. The rationale for the former is that it contains an array of anti-viral antibodies while the latter contains specific COVID-19 antibodies, both of which may benefit patients with severe COVID-19 [149]. In addition, hyperimmune globulin, which is extraction of the anti-COVID-19 antibodies from convalescent plasma, hence specific and concentrated IVIG may also be beneficial for COVID-19 disease. Interestingly, IVIG has also been shown to attenuate immune responses in several autoimmune conditions and COVID-19 patients, and therefore, could contribute to halting an unwelcomed, immune hyperactivity in this setting [[150], [151], [152], [153], [154], [155]].
Additional therapies targeted at the exaggerated immune response, which commonly accompanies SARS-CoV-2 in severely ill patients include dexamethasone and targeted immune inhibitors, like sarilumab, an anti-interleukin 6 (IL-6) receptor antibody. Type 1 interferons (especially interferon-alpha), which have broad antiviral activity have been successful treatments in in-vitro studies against of SARS-CoV-2 [156] and have since then been evaluated in clinical trials to treat SARS-CoV-2 [14,157].
A therapeutic approach targeting the microbiome is worth investigating, as it may prove effective on all three fronts: prevention, decreasing viral penetration and progression through the gastrointestinal tract as well as blunting an exaggerated immune response. Modulating the microbiome towards a more anti-inflammatory environment, may allow an appropriate immune response towards the virus, both within the gut and systemically without the consequences of immune dysregulation and autoimmunity. Although the microbiotal changes discussed above can only be interpreted associatively at this time [115], in theory, adjuvant therapy, which targets the reshaping of the gut microbiome may prove beneficial for patients suffering from severe COVID-19 symptoms, whether manifested inside the GI tract, the respiratory tract or systemically in the form of autoimmunity [83]. Several nutrients have already been associated with disease severity. Vitamin D levels have been strongly linked to the immune response in SARS-CoV-2, affecting severity of COVID-19 infection and mortality [[158], [159], [160], [161], [162]]. These data, however, are not without limitations and the role of vitamin D deficiency in COVID-19 remains controversial [163]. Vitamin C has also been a nutrient of interest in patients with critical illness and has been suggested to be protective in patients with severe COVID-19, although data is insufficient [164]. The essential trace mineral Zinc (Zn), which is known for its antioxidant, anti-inflammatory, immunomodulatory, and antiviral activities has been contemplated as an adjunct treatment for COVID-19 patients [165]. Special interest in this trace mineral has risen because increasing intracellular levels of Zn, have been shown to inhibit the replication of several RNA viruses including that of SARS-Co-V, in vitro, suggesting a similar effect on SARS-CoV-2 [166]. Selenium too has similar contribution to the immune system and several observational studies have found an association between selenium deficiency and patients with COVID-19 [167,168]. The microbiome may be the common pathway leading to the contribution of these nutrients, as all alter the gut microbiome towards a reduced inflammatory environment. As an example, in chicken models, Zn deficiency has been associated with significant alterations in the gut microbiome and decreased overall species richness and diversity, leading to dysbiosis, similar to that seen in several diseases [169]. In addition, Zn supplementation has been associated with a reduction of pathobionts and a rise of symbionts [170]. Safe use of any such supplementation still requires further investigation. In fact, a protective effect may even be dose dependent, for example excess in dietary Zn has been shown to negatively alter the gut microbiome by decreasing microbial diversity and promoting dysbiosis, which was associated with increase in susceptibility to C. difficile infection [171]. These nutrients along with several other nutritional dietary components with known anti-inflammatory and antioxidant properties have been proposed as supplementations for protection against COVID-19 infection and against severe disease manifestations [172]. The mechanism by which they offer this protection is thought to be via interaction with the gut microbiome [173]. Other proposed interventions include prebiotics and probiotics, both of which may shift the balance of the microbiotal composition towards protective microbes, which harbor anti-inflammatory properties [174,175]. Some prebiotics have demonstrated ability to regulate cytokine levels [100], such as whole grains, which demonstrated ability to decrease IL-6 levels [176] and butylated high amylose maize starch, which has shown ability to increase levels of IL-10 in human cohorts [177]. Even fecal transplantation is contemplated from healthy donors to critically ill COVID-19 patients, as a way to promote a healthier immune response in these patients, although the presence of SARS-CoV-2 in the feces must first be ruled out [178]. It is worth noting that several of the above proposed treatments have also been discussed as potential treatments in many autoimmune diseases, lupus included [105,128].
Nutritional strategies or other interventions targeting the microbiome may be beneficial to regulate the balance of intestinal microbiota and reduce immune dysregulation, altering the course of COVID-19 in high risk patients, and may even prove effective in preventing the disease. However, it is important to note that while attempting to modulate the microbiome towards a more anti-inflammatory environment is very appealing, treatment options, have not been extensively studied in COVID-19 and this warrants further investigation [179].
8. Conclusion
Our study emphasized the significance of the interactions between SARS-CoV-2 and the gastrointestinal tract. Presence of the SARS-CoV-2 virus not only leads to GI manifestations in some patients but may also alter the body's immune response to COVID-19. Additionally, given the resemblance of the microbiome in COVID-19 patients to that seen in lupus patients, it becomes clearer why several therapies used in autoimmune conditions are also currently under investigation for the treatment of COVID-19 patients. Manipulation of the microbiome for prevention and disease alteration is yet to be established.
References
- 1.Guan W.J., Ni Z.Y., Hu Y., et al. Clinical characteristics of coronavirus disease 2019 in China. NEJM. 2020;382:1708–1720. doi: 10.1056/NEJMoa2002032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Wang L., Wang Y., Ye D., Liu Q. Review of the 2019 novel coronavirus (SARS-CoV-2) based on current evidence. Int J Antimicrob Agents. 2020;55:105948. doi: 10.1016/j.ijantimicag.2020.105948. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Hojyo S., Uchida M., Tanaka K., Hasebe R., Tanaka Y., Murakami M., et al. How COVID-19 induces cytokine storm with high mortality. Inflamm Regen. 2020;40:37. doi: 10.1186/s41232-020-00146-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Shoenfeld Y. Corona (COVID-19) time musings: our involvement in COVID-19 pathogenesis, diagnosis, treatment and vaccine planning. Autoimmun Rev. 2020;19:102538. doi: 10.1016/j.autrev.2020.102538. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Vaninov N. In the eye of the COVID-19 cytokine storm. Nat Rev Immunol. 2020;20:277. doi: 10.1038/s41577-020-0305-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.D’Amico F., Baumgart D.C., Danese S., Peyrin-Biroulet L. Diarrhea during covid-19 infection: pathogenesis, epidemiology, prevention, and management. Clin Gastroenterol Hepatol. 2020;18:1663–1672. doi: 10.1016/j.cgh.2020.04.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Moore J.B., June C.H. Cytokine release syndrome in severe COVID19. Lessons from arthritis and cell therapy in cancer patients point to therapy for severe disease. Science. 2020;368:473–474. doi: 10.1126/science.abb8925. [DOI] [PubMed] [Google Scholar]
- 8.Behrens E.M., Koretzky G.A. Review: cytokine storm syndrome: looking toward the precision medicine era. Arthritis Rheum. 2017;69:1135–1143. doi: 10.1002/art.40071. [DOI] [PubMed] [Google Scholar]
- 9.Fajgenbaum D.C., June C.H. Cytokine Storm. N Engl J Med. 2020;383:2255–2273. doi: 10.1056/NEJMra2026131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Halpert G., Shoenfeld Y. SARS-CoV-2, the autoimmune virus. Autoimmun Rev. 2020;19:102695. doi: 10.1016/j.autrev.2020.102695. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Ehrenfeld M., Tincani A., Andreoli L., et al. Covid-19 and autoimmunity. Autoimmun Rev. 2020;19:102597. doi: 10.1016/j.autrev.2020.102597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Zeng F., Huang Y., Guo Y., et al. Association of inflammatory markers with the severity of COVID-19: a meta-analysis. Int J Infect Dis. 2020;96:467–474. doi: 10.1016/j.ijid.2020.05.055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Ruscitti P., Berardicurti O., Di Benedetto P., Cipriani P., Iagnocco A., Shoenfeld Y., et al. Severe covid-19, another piece in the puzzle of the hyperferritinemic syndrome. An immunomodulatory perspective to alleviate the storm. Front Immunol. 2020;11:1130. doi: 10.3389/fimmu.2020.01130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Sun X., Wang T., Cai D., et al. Cytokine storm intervention in the early stages of COVID-19 pneumonia. Cytokine Growth Factor Rev. 2020;53:38–42. doi: 10.1016/j.cytogfr.2020.04.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Zhou Y., Fu B., Zheng X., et al. Pathogenic T-cells and inflammatory monocytes incite inflammatory storm in severe COVID-19 patients. Natl Sci Rev. 2020;7(6) doi: 10.1093/nsr/nwaa041. nwaa041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Mazzoni A., Salvati L., Maggi L., et al. Impaired immune cell cytotoxicity in severe COVID-19 is IL-6 dependent. J Clin Invest. 2020;130:4694–4703. doi: 10.1172/JCI138554. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Blanco-Melo D., Nilsson-Payant B.E., Liu W.C., et al. Imbalanced host response to sars-cov-2 drives development of covid-19. Cell. 2020;181 doi: 10.1016/j.cell.2020.04.026. 1036–1045.e9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Liu Y., Zhang C., Huang F., et al. Elevated plasma levels of selective cytokines in COVID-19 patients reflect viral load and lung injury. Natl Sci Rev. 2020;7:1003–1011. doi: 10.1093/nsr/nwaa037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Rodríguez Y., Novelli L., Rojas M., et al. Autoinflammatory and autoimmune conditions at the crossroad of COVID-19. J Autoimmun. 2020;114:102506. doi: 10.1016/j.jaut.2020.102506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Han H., Ma Q., Li C., et al. Profiling serum cytokines in COVID-19 patients reveals IL-6 and IL-10 are disease severity predictors. Emerg Microbes Infect. 2020;9:1123–1130. doi: 10.1080/22221751.2020.1770129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Colafrancesco S., Alessandri C., Conti F., Priori R. COVID-19 gone bad: a new character in the spectrum of the hyperferritinemic syndrome? Autoimmun Rev. 2020;19:102573. doi: 10.1016/j.autrev.2020.102573. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Henderson L.A., Canna S.W., Schulert G.S., et al. On the alert for cytokine storm: immunopathology in Covid-19. Arthritis Rheum. 2020;72:1059–1063. doi: 10.1002/art.41285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Rojas M., Restrepo-Jiménez P., Monsalve D.M., Pacheco Y., Acosta-Ampudia Y., Ramírez-Santana C., et al. Molecular mimicry and autoimmunity. J Autoimmun. 2018;95:100–123. doi: 10.1016/j.jaut.2018.10.012. [DOI] [PubMed] [Google Scholar]
- 24.Smatti M.K., Cyprian F.S., Nasrallah G.K., Al Thani A.A., Almishal R.O., Yassine H.M. Viruses and autoimmunity: a review on the potential interaction and molecular mechanisms. Viruses. 2019;11:762. doi: 10.3390/v11080762. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Pacheco Y., Acosta-Ampudia Y., Monsalve D.M., Chang C., Gershwin M.E., Anaya J.M. Bystander activation and autoimmunity. J Autoimmun. 2019;103:102301. doi: 10.1016/j.jaut.2019.06.012. [DOI] [PubMed] [Google Scholar]
- 26.Fujinami R.S., von Herrath M.G., Christen U., Whitton J.L. Molecular mimicry, bystander activation, or viral persistence: infections and autoimmune disease. Clin Microbiol Rev. 2006;19:80–94. doi: 10.1128/CMR.19.1.80-94.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Ramos-Casals M., Loustaud-Ratti V., De Vita S., Zeher M., Bosch J.A., Toussirot E., et al. Sjögren syndrome associated with hepatitis C virus: a multicenter analysis of 137 cases. Medicine (Baltimore) 2005;84:81–89. doi: 10.1097/01.md.0000157397.30055.c9. [DOI] [PubMed] [Google Scholar]
- 28.Chen J., Zhang H., Chen P., Lin Q., Zhu X., Zhang L., et al. Correlation between systemic lupus erythematosus and cytomegalovirus infection detected by different methods. Clin Rheumatol. 2015;34:691–698. doi: 10.1007/s10067-015-2868-3. [DOI] [PubMed] [Google Scholar]
- 29.Dostál C., Newkirk M.M., Duffy K.N.W., Palečková A., Bošák V., Černá M., et al. Herpes viruses in multicase families with rheumatoid arthritis and systemic lupus erythematosus. Ann N Y Acad Sci. 1997;815:334–337. doi: 10.1111/j.1749-6632.1997.tb52078.x. [DOI] [PubMed] [Google Scholar]
- 30.Wouters J.M.G.W., van der Veen J., van de Putte L.B., de Rooij D.J. Adult onset Still’s disease and viral infections. Ann Rheum Dis. 1988;47:764–767. doi: 10.1136/ard.47.9.764. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Filippi C.M., von Herrath M.G. Viral trigger for type 1 diabetes: pros and cons. Diabetes. 2008;57:2863–2871. doi: 10.2337/db07-1023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Sellers S.A., Hagan R.S., Hayden F.G., Fischer W.A., 2nd. The hidden burden of influenza: a review of the extra-pulmonary complications of influenza infection. Influenza Other Respir Viruses. 2017;11:372–393. doi: 10.1111/irv.12470. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Alberti P., Beretta S., Piatti M., et al. Guillain-Barré syndrome related to COVID-19 infection. Neurol Neuroimmunol Neuroinflamm. 2020;7 doi: 10.1212/NXI.0000000000000741. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Toscano G., Palmerini F., Ravaglia S., et al. Guillain-Barré syndrome associated with SARS-CoV-2. N Engl J Med. 2020;382:2574–2576. doi: 10.1056/NEJMc2009191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.De Sanctis P., Doneddu P.E., Viganò L., Selmi C., Nobile-Orazio E. Guillain-Barré syndrome associated with SARS-CoV-2 infection. A systematic review. Eur J Neurol. 2020;27:2361–2370. doi: 10.1111/ene.14462. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Arnaud S., Budowski C. Ng wing tin S, Degos B. post SARS-CoV-2 Guillain-Barré syndrome. Clin. Neurophysiol. 2020;131:1652–1654. doi: 10.1016/j.clinph.2020.05.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Zulfiqar A.A., Lorenzo-Villalba N., Hassler P., Andrès E. Immune thrombocytopenic purpura in a patient with Covid-19. N Engl J Med. 2020;382 doi: 10.1056/NEJMc2010472. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Lopez C., Kim J., Pandey A., Huang T., DeLoughery T.G. Simultaneous onset of COVID-19 and autoimmune haemolytic anaemia. Br J Haematol. 2020;190:31–32. doi: 10.1111/bjh.16786. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Bhattacharjee S., Banerjee M. Immune thrombocytopenia secondary to COVID-19: a systematic review. SN Compr Clin Med. 2020:1–11. doi: 10.1007/s42399-020-00521-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Bomhof G., Mutsaers P.G.N.J., Leebeek F.W.G., et al. COVID-19-associated immune thrombocytopenia. Br J Haematol. 2020;190:e61–e64. doi: 10.1111/bjh.16850. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Jones V.G., Mills M., et al. COVID-19 and Kawasaki disease: novel virus and novel case. Hosp Pediatr. 2020;10:537–540. doi: 10.1542/hpeds.2020-0123. [DOI] [PubMed] [Google Scholar]
- 42.Verdoni L., Mazza A., Gervasoni A., et al. An outbreak of severe Kawasaki-like disease at the Italian epicentre of the SARS-CoV-2 epidemic: an observational cohort study. Lancet. 2020;395:1771–1778. doi: 10.1016/S0140-6736(20)31103-X. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Feldstein L.R., Rose E.B., Horwitz S.M., et al. Multisystem inflammatory syndrome in U.S. children and adolescents. N Engl J Med. 2020;383:334–346. doi: 10.1056/NEJMoa2021680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Khoruts A., Fraser J.M. A causal link between lymphopenia and autoimmunity. Immunol Lett. 2005;98:23–31. doi: 10.1016/j.imlet.2004.10.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Azkur A.K., Akdis M., Azkur D., et al. Immune response to SARS-CoV-2 and mechanisms of immunopathological changes in COVID-19. Allergy. 2020;75:1564–1581. doi: 10.1111/all.14364. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Theofilopoulos A.N., Dummer W., Kono D.H. T cell homeostasis and systemic autoimmunity. J Clin Invest. 2001;108:335–340. doi: 10.1172/JCI12173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Zhang J.J., Dong X., Cao Y.Y., Yuan Y.D., Yang Y.B., Yan Y.Q., et al. Clinical characteristics of 140 patients infected with SARS-CoV-2 in Wuhan, China. Allergy. 2020;75:1730–1741. doi: 10.1111/all.14238. [DOI] [PubMed] [Google Scholar]
- 48.Zhou Z., Ren L., Zhang L., et al. Heightened innate immune responses in the respiratory tract of Covid-19 patients. Cell Host Microbe. 2020;27 doi: 10.1016/j.chom.2020.04.017. 883–890.e2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Mills S., Shanahan F., Stanton C., Hill C., Coffey A., Ross R.P. Movers and shakers: influence of bacteriophages in shaping the mammalian gut microbiota. Gut Microbes. 2013;4:4–16. doi: 10.4161/gmic.22371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Manrique P., Dills M., Young M.J. The human gut phage community and its implications for health and disease. Viruses. 2017;9:141. doi: 10.3390/v9060141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Lerner A., Ramesh A., Matthias T. David and Goliath war revival in the enteric viruses and microbiota struggle. Potential implication for celiac disease. Microorganisms. 2019;7:173. doi: 10.3390/microorganisms7060173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Ge X.Y., et al. Isolation and characterization of a bat SARS-like coronavirus that uses the ACE2 receptor. Nature. 2013;503:535–538. doi: 10.1038/nature12711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Zhou P., Yang X.L., Wang X.G., et al. A pneumonia outbreak associated with a new coronavirus of probable bat origin. Nature. 2020;579:270–273. doi: 10.1038/s41586-020-2012-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Crackower M.A., Sarao R., Oudit G.Y., et al. Angiotensin-converting enzyme 2 is an essential regulator of heart function. Nature. 2002;417:822–828. doi: 10.1038/nature00786. [DOI] [PubMed] [Google Scholar]
- 55.Hamming I., Timens W., Bulthuis M.L., Lely A.T., Navis G., van Goor H. Tissue distribution of ACE2 protein, the functional receptor for SARS coronavirus. A first step in understanding SARS pathogenesis. J Pathol. 2004;203:631–637. doi: 10.1002/path.1570. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Hashimoto T., Perlot T., Rehman A., et al. ACE2 links amino acid malnutrition to microbial ecology and intestinal inflammation. Nature. 2012;487:477–481. doi: 10.1038/nature11228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Liang W., Feng Z., Rao S., Xiao C., Xue X., Lin Z., et al. Diarrhoea may be underestimated: a missing link in 2019 novel coronavirus. Gut. 2020;69:1141–1143. doi: 10.1136/gutjnl-2020-320832. [DOI] [PubMed] [Google Scholar]
- 58.Harmer D., Gilbert M., Borman R., Clark K.L. Quantitative mRNA expression profiling of ACE 2, a novel homologue of angiotensin converting enzyme. FEBS Lett. 2002;532:107–110. doi: 10.1016/s0014-5793(02)03640-2. [DOI] [PubMed] [Google Scholar]
- 59.Leung W.K., To KF, Chan P.K., et al. Enteric involvement of severe acute respiratory syndrome-associated coronavirus infection. Gastroenterology. 2003;125:1011–1017. doi: 10.1016/j.gastro.2003.08.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Jin X., Lian J.S., Hu J.H., et al. Epidemiological, clinical and virological characteristics of 74 cases of coronavirus-infected disease 2019 (COVID-19) with gastrointestinal symptoms. Gut. 2020;69:1002–1009. doi: 10.1136/gutjnl-2020-320926. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Cholankeril G., Podboy A., Aivaliotis V.I., et al. High prevalence of concurrent gastrointestinal manifestations in patients with severe acute respiratory syndrome coronavirus 2: early experience from California. Gastroenterology. 2020;159:775–777. doi: 10.1053/j.gastro.2020.04.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Goyal P., Choi J.J., Pinheiro L.C., et al. Clinical characteristics of Covid-19 in New York City. N Engl J Med. 2020;382:2372–2374. doi: 10.1056/NEJMc2010419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Lin L., Jiang X., Zhang Z., Huang S., Zhang Z., Fang Z., et al. Gastrointestinal symptoms of 95 cases with SARS-CoV-2 infection. Gut. 2020;69:997–1001. doi: 10.1136/gutjnl-2020-321013. [DOI] [PubMed] [Google Scholar]
- 64.Fu B., Qian K., Fu X. SARS-CoV-2-induced vomiting as onset symptom in a patient with COVID-19. Dig Dis Sci. 2020;65:1568–1570. doi: 10.1007/s10620-020-06285-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Patel K.P., Patel P.A., Vunnam R.R., et al. Gastrointestinal, hepatobiliary, and pancreatic manifestations of COVID-19. J Clin Virol. 2020;128:104386. doi: 10.1016/j.jcv.2020.104386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Pan L., Mu M., Yang P., Sun Y., Wang R., Yan J., et al. Clinical characteristics of COVID-19 patients with digestive symptoms in Hubei, China: a descriptive, cross-sectional, multicenter study. Am J Gastroenterol. 2020;115:766–773. doi: 10.14309/ajg.0000000000000620. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Chan J.F., Kok K.H., Zhu Z., Chu H., To KK, Yuan S., et al. Genomic characterization of the 2019 novel human-pathogenic coronavirus isolated from a patient with atypical pneumonia after visiting Wuhan. Emerg Microbes Infect. 2020;9:221–236. doi: 10.1080/22221751.2020.1719902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Suárez-Fariñas M., Tokuyama M., Wei G., et al. Intestinal inflammation modulates the expression of ACE2 and TMPRSS2 and potentially overlaps with the pathogenesis of SARS-CoV-2-related disease. Gastroenterology. 2021;160 doi: 10.1053/j.gastro.2020.09.029. 287–301.e20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Zhou J., Li C., Zhao G., et al. Human intestinal tract serves as an alternative infection route for Middle East respiratory syndrome coronavirus. Sci Adv. 2017;3 doi: 10.1126/sciadv.aao4966. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Xiao F., Tang M., Zheng X., Liu Y., Li X., Shan H. Evidence for gastrointestinal infection of SARS-CoV-2. Gastroenterology. 2020;158:1831–1833. doi: 10.1053/j.gastro.2020.02.055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Cheung K.S., Hung I.F.N., Chan P.P.Y., et al. Gastrointestinal manifestations of sars-cov-2 infection and virus load in fecal samples from a Hong Kong cohort: systematic review and meta-analysis. Gastroenterology. 2020;159:81–95. doi: 10.1053/j.gastro.2020.03.065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Chan K.H., Poon L.L., Cheng V.C., et al. Detection of SARS coronavirus in patients with suspected SARS. Emerg Infect Dis. 2004;10:294–299. doi: 10.3201/eid1002.030610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Wu Y., Guo C., Tang L., Hong Z., Zhou J., Dong X., et al. Prolonged presence of SARS-CoV-2 viral RNA in faecal samples. Lancet Gastroenterol Hepatol. 2020;5:434–435. doi: 10.1016/S2468-1253(20)30083-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Chen N., Zhou M., Dong X., et al. Epidemiological and clinical characteristics of 99 cases of 2019 novel coronavirus pneumonia in Wuhan, China: a descriptive study. Lancet. 2020;395:507–513. doi: 10.1016/S0140-6736(20)30211-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Wang D., Hu B., Hu C., et al. Clinical characteristics of 138 hospitalized patients with 2019 novel coronavirus-infected pneumonia in Wuhan, China. JAMA. 2020;323:1061–1069. doi: 10.1001/jama.2020.1585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Fang D., Ma J., Guan J., Wang M., Song Y., Tian D., et al. Manifestations of digestive system in hospitalized patients with novel coronavirus pneumonia in Wuhan, China: a single-center, descriptive study. Chin J Digest. 2020;E005 [Google Scholar]
- 77.Song Y., Liu P., Shi X.L., Chu Y.L., Zhang J., Xia J., et al. SARS-CoV-2 induced diarrhoea as onset symptom in patient with COVID-19. Gut. 2020;69:1143–1144. doi: 10.1136/gutjnl-2020-320891. [DOI] [PubMed] [Google Scholar]
- 78.Zhang Y., Ma P., Zhang X., Pei Z., Wang H., Dou X. Association of digestive symptoms with severity and mortality of COVID-19. Medicine. 2020;99 doi: 10.1097/MD.0000000000022736. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Wan Y., Li J., Shen L., Zou Y., Hou L., Zhu L., et al. Enteric involvement in hospitalized patients with COVID-19 outside Wuhan. Lancet Gastroenterol Hepatol. 2020;5:534–535. doi: 10.1016/S2468-1253(20)30118-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Liu J., Cui M., Yang T., Yao P. Correlation between gastrointestinal symptoms and disease severity in patients with COVID-19: a systematic review and meta-analysis. BMJ Open Gastroenterol. 2020;7 doi: 10.1136/bmjgast-2020-000437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Han C., Duan C., Zhang S., Spiegel B., Shi H., Wang W., et al. Digestive symptoms in COVID-19 patients with mild disease severity: clinical presentation, stool viral RNA testing, and outcomes. Am J Gastroenterol. 2020;115:916–923. doi: 10.14309/ajg.0000000000000664. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Gu J., Han B., Wang J. COVID-19: gastrointestinal manifestations and potential fecal-oral transmission. Gastroenterology. 2020;158:1518–1519. doi: 10.1053/j.gastro.2020.02.054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Zhang X., Tang C., Tian D., Hou X., Yang Y. Management of digestive disorders and procedures associated with COVID-19. Am J Gastroenterol. 2020;115:1153–1155. doi: 10.14309/ajg.0000000000000728. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Ding S., Liang T.J. Is SARS-CoV-2 also an enteric pathogen with potential fecal-oral transmission? A COVID-19 virological and clinical review. Gastroenterology. 2020;159:53–61. doi: 10.1053/j.gastro.2020.04.052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Yang T., Chakraborty S., Saha P., et al. Gnotobiotic rats reveal that gut microbiota regulates colonic mRNA of Ace2, the receptor for SARS-CoV-2 infectivity. Hypertension. 2020;7(6):e1–e3. doi: 10.1161/HYPERTENSIONAHA.120.15360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Geva-Zatorsky N., Sefik E., Kua L., et al. Mining the human gut microbiota for immunomodulatory organisms. Cell. 2017;168:928–943. doi: 10.1016/j.cell.2017.01.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Dang A.T., Marsland B.J. Microbes, metabolites, and the gut-lung axis. Mucosal Immunol. 2019;12:843–850. doi: 10.1038/s41385-019-0160-6. [DOI] [PubMed] [Google Scholar]
- 88.Enaud R., Prevel R., Ciarlo E., Beaufils F., Wieërs G., Guery B., et al. The gut-lung axis in health and respiratory diseases: a place for inter-organ and inter-kingdom crosstalks. Front Cell Infect Microbiol. 2020;10:9. doi: 10.3389/fcimb.2020.00009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Budden K.F., Gellatly S.L., Wood D.L., Cooper M.A., Morrison M., Hugenholtz P., et al. Emerging pathogenic links between microbiota and the gut-lung axis. Nat Rev Microbiol. 2017;15:55–63. doi: 10.1038/nrmicro.2016.142. [DOI] [PubMed] [Google Scholar]
- 90.Fagundes C.T., Amaral F.A., Vieira A.T., Soares A.C., Pinho V., Nicoli J.R., et al. Transient TLR activation restores inflammatory response and ability to control pulmonary bacterial infection in germfree mice. J Immunol. 2012;188:1411–1420. doi: 10.4049/jimmunol.1101682. [DOI] [PubMed] [Google Scholar]
- 91.Bingula R., Filaire M., Radosevic-Robin N., et al. Desired turbulence? Gut-lung axis, immunity, and lung cancer. J Oncol. 2017;2017:5035371. doi: 10.1155/2017/5035371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Zhang D., Li S., Wang N., Tan H.Y., Zhang Z., Feng Y. The cross-talk between gut microbiota and lungs in common lung diseases. Front Microbiol. 2020;11:301. doi: 10.3389/fmicb.2020.00301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Dickson R.P. The microbiome and critical illness. Lancet Respir Med. 2016;4:59–72. doi: 10.1016/S2213-2600(15)00427-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Mosca A., Leclerc M., Hugot J.P. Gut microbiota diversity and human diseases: should we reintroduce key predators in our ecosystem? Front Microbiol. 2016;7:455. doi: 10.3389/fmicb.2016.00455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Trompette A., Gollwitzer E.S., Yadava K., Sichelstiel A.K., Sprenger N., Ngom-Bru C., et al. Gut microbiota metabolism of dietary fiber influences allergic airway disease and hematopoiesis. Nat Med. 2014;20:159–166. doi: 10.1038/nm.3444. [DOI] [PubMed] [Google Scholar]
- 96.Anand S., Mande S.S. Diet, microbiota and gut-lung connection. Front Microbiol. 2018;9:2147. doi: 10.3389/fmicb.2018.02147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Looft T., Allen H.K. Collateral effects of antibiotics on mammalian gut microbiomes. Gut Microbes. 2012;3:463–467. doi: 10.4161/gmic.21288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Dhar D., Mohanty A. Gut microbiota and Covid-19- possible link and implications. Virus Res. 2020;285:198018. doi: 10.1016/j.virusres.2020.198018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Lake M.A. What we know so far: COVID-19 current clinical knowledge and research. Clin Med (Lond) 2020;20:124–127. doi: 10.7861/clinmed.2019-coron. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Shen Z., Xiao Y., Kang L., et al. Genomic diversity of severe acute respiratory syndrome-coronavirus 2 in patients with coronavirus disease 2019. Clin Infect Dis. 2020;71:713–720. doi: 10.1093/cid/ciaa203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Keely S., Talley N.J., Hansbro P.M. Pulmonary-intestinal cross-talk in mucosal inflammatory disease. Mucosal Immunol. 2012;5:7–18. doi: 10.1038/mi.2011.55. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Dumas A., Bernard L., Poquet Y., Lugo-Villarino G., Neyrolles O. The role of the lung microbiota and the gut-lung axis in respiratory infectious diseases. Cell Microbiol. 2018;20 doi: 10.1111/cmi.12966. [DOI] [PubMed] [Google Scholar]
- 103.Katz-Agranov N., Zandman-Goddard G. The microbiome and systemic lupus erythematosus. Immunol Res. 2017;65:432–437. doi: 10.1007/s12026-017-8906-2. [DOI] [PubMed] [Google Scholar]
- 104.Schirmer M., Smeekens S.P., Vlamakis H., et al. Linking the human gut microbiome to inflammatory cytokine production. Cell. 2016;167 doi: 10.1016/j.cell.2016.10.020. 1125–1136.e8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Mendes V., Galvão I., Vieira A.T. Mechanisms by which the gut microbiota influences cytokine production and modulates host inflammatory responses. J Interf Cytokine Res. 2019;39:393–409. doi: 10.1089/jir.2019.0011. [DOI] [PubMed] [Google Scholar]
- 106.Kalantar-Zadeh K., Ward S.A., Kalantar-Zadeh K., El-Omar E.M. Considering the effects of microbiome and diet on sars-cov-2 infection: nanotechnology. ACS Nano. 2020;14:5179–5182. doi: 10.1021/acsnano.0c03402. [DOI] [PubMed] [Google Scholar]
- 107.Tay M.Z., Poh C.M., Rénia L., et al. The trinity of COVID-19: immunity, inflammation and intervention. Nat Rev Immunol. 2020;20:363–374. doi: 10.1038/s41577-020-0311-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Yeoh Y.K., Zuo T., Lui G.C., et al. Gut microbiota composition reflects disease severity and dysfunctional immune responses in patients with COVID-19. Gut. 2021;70:698–706. doi: 10.1136/gutjnl-2020-323020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Saleh J., Peyssonnaux C., Singh K.K., Edeas M. Mitochondria and microbiota dysfunction in COVID-19 pathogenesis. Mitochondrion. 2020;54:1–7. doi: 10.1016/j.mito.2020.06.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Groves H.T., Higham S.L., Moffatt M.F., Cox M.J., Tregoning J.S. Respiratory viral infection alters the gut microbiota by inducing inappetence. mBio. 2020;11 doi: 10.1128/mBio.03236-19. e03236–19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Donati Zeppa S., Agostini D., Piccoli G., Stocchi V., Sestili P. Gut microbiota status in covid-19: an unrecognized player? Front Cell Infect Microbiol. 2020;10:576551. doi: 10.3389/fcimb.2020.576551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Segal J.P., Mak J.W.Y., Mullish B.H., Alexander J.L., Ng S.C., Marchesi J.R. The gut microbiome: an under-recognised contributor to the COVID-19 pandemic? Ther Adv Gastroenterol. 2020;13 doi: 10.1177/1756284820974914. 1756284820974914. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Gu S., Chen Y., Wu Z., et al. Alterations of the gut microbiota in patients with coronavirus disease 2019 or H1N1 influenza. Clin Infect Dis. 2020;71:2669–2678. doi: 10.1093/cid/ciaa709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Xu K., Cai H., Shen Y., Ni Q., Chen Y., Hu S., et al. Management of corona virus disease-19 (COVID-19): the Zhejiang experience. Zhejiang Da Xue Xue Bao Yi Xue Ban. 2020;49:147–157. doi: 10.3785/j.issn.1008-9292.2020.02.02. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Katz-Agranov N., Zandman-Goddard G. The microbiome links between aging and lupus. Autoimmun Rev. 2021 Mar;20:102765. doi: 10.1016/j.autrev.2021.102765. [DOI] [PubMed] [Google Scholar]
- 116.Azzouz D., Omarbekova A., Heguy A., Schwudke D., Gisch N., Rovin B.H., et al. Lupus nephritis is linked to disease-activity associated expansions and immunity to a gut commensal. Ann Rheum Dis. 2019;78:947–956. doi: 10.1136/annrheumdis-2018-214856. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Niers L.E., Timmerman H.M., et al. Identification of strong interleukin-10 inducing lactic acid bacteria which down-regulate T helper type 2 cytokines. Clin Exp Allergy. 2005;35:1481–1489. doi: 10.1111/j.1365-2222.2005.02375.x. [DOI] [PubMed] [Google Scholar]
- 118.Chow J., Tang H., Mazmanian S.K. Pathobionts of the gastrointestinal microbiota and inflammatory disease. Curr Opin Immunol. 2011;23:473–480. doi: 10.1016/j.coi.2011.07.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Zuo T., Zhang F., et al. Alterations in gut microbiota of patients with COVID-19 during time of hospitalization. Gastroenterology. 2020;159 doi: 10.1053/j.gastro.2020.05.048. 944–55.e8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Lau W.L., Chang Y., Vaziri N.D. The consequences of altered microbiota in immune-related chronic kidney disease. Nephrol Dial Transplant. 2020 May 21 doi: 10.1093/ndt/gfaa087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Manzoor S.E., McNulty C.A.M., Nakiboneka-Ssenabulya D., Lecky D.M., Hardy K.J., Hawkey P.M. Investigation of community carriage rates of Clostridium difficile and Hungatella hathewayi in healthy volunteers from four regions of England. J Hosp Infect. 2017;97:153–155. doi: 10.1016/j.jhin.2017.05.014. [DOI] [PubMed] [Google Scholar]
- 122.Du Z., Hudcovic T., Mrazek J., Kozakova H., Srutkova D., Schwarzer M., et al. Development of gut inflammation in mice colonized with mucosa-associated bacteria from patients with ulcerative colitis. Gut Pathog. 2015;7:32. doi: 10.1186/s13099-015-0080-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Nishino K., Imaeda H., Sakai S., Ohno M., Nishida A., Andoh A. The abundance of Clostridium hathewayi, a potent inducer of t helper 17 (Th17) cells, is associated with the disease severity of Crohn’s disease. Gastroenterology. 2017;152:S993. [Google Scholar]
- 124.He L.H., Ren L.F., Li J.F., Wu Y.N., Li X., Zhang L. Intestinal flora as a potential strategy to fight SARS-CoV-2 infection. Front Microbiol. 2020;11:1388. doi: 10.3389/fmicb.2020.01388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Miyake S., Kim S., Suda W., et al. Dysbiosis in the gut microbiota of patients with multiple sclerosis, with a striking depletion of species belonging to clostridia Xiva and iv clusters. PLoS One. 2015;10 doi: 10.1371/journal.pone.0137429. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.De Luca F., Shoenfeld Y. The microbiome in autoimmune diseases. Clin Exp Immunol. 2019;195:74–85. doi: 10.1111/cei.13158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127.Li Y., Wang H.F., Li X., Li H.X., Zhang Q., Zhou H.W., et al. Disordered intestinal microbes are associated with the activity of Systemic Lupus Erythematosus. Clin Sci (Lond) 2019;133:821–838. doi: 10.1042/CS20180841. [DOI] [PubMed] [Google Scholar]
- 128.Sokol H., Pigneur B., Watterlot L., et al. Faecalibacterium prausnitzii is an anti-inflammatory commensal bacterium identified by gut microbiota analysis of Crohn disease patients. Proc Natl Acad Sci. 2008;105:16731–16736. doi: 10.1073/pnas.0804812105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129.Luo X.M., Edwards M.R., Mu Q., et al. Gut microbiota in human systemic lupus erythematosus and a mouse model of lupus. Appl Environ Microbiol. 2018;84 doi: 10.1128/AEM.02288-17. e02288–17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130.Zuo T., Liu Q., Zhang F., et al. Depicting SARS-CoV-2 faecal viral activity in association with gut microbiota composition in patients with COVID-19. Gut. 2021;70:276–284. doi: 10.1136/gutjnl-2020-322294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131.Terruzzi I., Senesi P. Does intestinal dysbiosis contribute to an aberrant inflammatory response to severe acute respiratory syndrome coronavirus 2 in frail patients? Nutrition. 2020;79–80 doi: 10.1016/j.nut.2020.110996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132.Rajput S., Paliwal D., Naithani M., Kothari A., Meena K., Rana S. COVID-19 and gut microbiota: a potential connection. Indian J Clin Biochem. 2021:1–12. doi: 10.1007/s12291-020-00948-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133.Vatanen T., Kostic A.D., d’Hennezel E. Variation in microbiome LPS immunogenicity contributes to autoimmunity in humans. Cell. 2016;165:842–853. doi: 10.1016/j.cell.2016.04.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 134.Yoshida N., Emoto T., Yamashita T. Bacteroides vulgatus and Bacteroides dorei reduce gut microbial lipopolysaccharide production and inhibit atherosclerosis. Circulation. 2018;138:2486–2498. doi: 10.1161/CIRCULATIONAHA.118.033714. [DOI] [PubMed] [Google Scholar]
- 135.Feng Y., Huang Y., Wang Y., Wang P., Song H., Wang F. Antibiotics induced intestinal tight junction barrier dysfunction is associated with microbiota dysbiosis, activated NLRP3 inflammasome and autophagy. PLoS One. 2019;14 doi: 10.1371/journal.pone.0218384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 136.Turnbaugh P.J., Ley R.E., Mahowald M.A. An obesity-associated gut microbiome with increased capacity for energy harvest. Nature. 2006;444:1027. doi: 10.1038/nature05414. [DOI] [PubMed] [Google Scholar]
- 137.Emoto T., Yamashita T., Sasaki N. Analysis of gut microbiota in coronary artery disease patients: a possible link between gut microbiota and coronary artery disease. J Atheroscler Thromb. 2016;23:908–921. doi: 10.5551/jat.32672. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 138.Yang T., Santisteban M.M., Rodriguez V. Gut dysbiosis is linked to hypertension. Hypertension. 2015;65:1331–1340. doi: 10.1161/HYPERTENSIONAHA.115.05315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 139.Ley R.E., Turnbaugh P.J., Klein S. Human gut microbes associated with obesity. Nature. 2006;444:1022–1023. doi: 10.1038/4441022a. [DOI] [PubMed] [Google Scholar]
- 140.Gou W., Fu Y., Yue L., Chen G.D., Cai X., Shuai M., et al. Gut microbiota may underlie the predisposition of healthy individuals to COVID-19. medRxiv. 2020 [Google Scholar]
- 141.Giron L.B., Dweep H., Yin X., et al. Severe COVID-19 is fueled by disrupted gut barrier integrity. medRxiv. 2020 11.13.20231209. [Google Scholar]
- 142.Villapol S. Gastrointestinal symptoms associated with COVID-19: impact on the gut microbiome. Transl Res. 2020;226:57–69. doi: 10.1016/j.trsl.2020.08.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 143.Jordan R.E., Adab P., Cheng K.K. COVID-19: risk factors for severe disease and death. BMJ. 2020;368:m1198. doi: 10.1136/bmj.m1198. [DOI] [PubMed] [Google Scholar]
- 144.Nagpal R., Mainali R., Ahmadi S., et al. Gut microbiome and aging: physiological and mechanistic insights. Nutr Healthy Aging. 2018;4:267–285. doi: 10.3233/NHA-170030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 145.Mehta P., McAuley D.F., Brown M., et al. COVID-19: consider cytokine storm syndromes and immunosuppression. Lancet. 2020;395:1033–1034. doi: 10.1016/S0140-6736(20)30628-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 146.Beigel J.H., Tomashek K.M., Dodd L.E., et al. Remdesivir for the treatment of Covid-19 - final report. N Engl J Med. 2020;383:1813–1826. doi: 10.1056/NEJMoa2007764. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 147.Krause I., Wu R., Sherer Y., Patanik M., Peter J., Shoenfeld Y. In vitro antiviral and antibacterial activity of commercial intravenous immunoglobulin preparations - a potential role for adjuvant intravenous immunoglobulin therapy in infectious diseases. Transfus Med. 2002;12:133–139. doi: 10.1046/j.1365-3148.2002.00360.x. [DOI] [PubMed] [Google Scholar]
- 148.Zandman-Goddard G., Krauthammer A., Levy Y., et al. Long-term therapy with intravenous immunoglobulin is beneficial in patients with autoimmune diseases. Clinic Rev Allerg Immunol. 2012;42:247–255. doi: 10.1007/s12016-011-8278-7. [DOI] [PubMed] [Google Scholar]
- 149.Perricone C., Triggianese P., Bursi R., et al. Intravenous immunoglobulins at the crossroad of autoimmunity and viral infections. Microorganisms. 2021;9:121. doi: 10.3390/microorganisms9010121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 150.Esen F., Özcan P.E., Orhun G., Polat Ö., Anaklı İ., Alay G., et al. Effects of adjunct treatment with intravenous immunoglobulins on the course of severe COVID-19: results from a retrospective cohort study. Curr Med Res Opin. 2021;37:543–548. doi: 10.1080/03007995.2020.1856058. [DOI] [PubMed] [Google Scholar]
- 151.Maor Y., Cohen D., Paran N., et al. Compassionate use of convalescent plasma for treatment of moderate and severe pneumonia in COVID-19 patients and association with IgG antibody levels in donated plasma. EClinicalMedicine. 2020 Sep;26:100525. doi: 10.1016/j.eclinm.2020.100525. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 152.Joyner M.J., Carter R.E., Senefeld J.W., et al. Convalescent plasma antibody levels and the risk of death from Covid-19. N Engl J Med. 2021;384:1015–1027. doi: 10.1056/NEJMoa2031893. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 153.Perotti C., Baldanti F., Bruno R., et al. Mortality reduction in 46 severe Covid-19 patients treated with hyperimmune plasma. A proof of concept single arm multicenter trial. Haematologica. 2020;105:2834–2840. doi: 10.3324/haematol.2020.261784. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 154.Mantlo E., Bukreyeva N., Maruyama J., Paessler S., Huang C. Antiviral activities of type I interferons to SARS-CoV-2 infection. Antivir Res. 2020;179:104811. doi: 10.1016/j.antiviral.2020.104811. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 155.Martinez M.A. Compounds with therapeutic potential against novel respiratory 2019 coronavirus. Antimicrob Agents Chemother. 2020 Apr 21;64 doi: 10.1128/AAC.00399-20. e00399–20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 156.Ribeiro H., Santana K.V.S., Oliver S.L., et al. Does vitamin D play a role in the management of Covid-19 in Brazil? Rev Saude Publica. 2020;54:53. doi: 10.11606/s1518-8787.2020054002545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 157.Aygun H. Vitamin D can prevent COVID-19 infection-induced multiple organ damage. Naunyn Schmiedeberg’s Arch Pharmacol. 2020;393:1157–1160. doi: 10.1007/s00210-020-01911-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 158.Mitchell F. Vitamin-D and COVID-19: do deficient risk a poorer outcome? Lancet Diabetes Endocrinol. 2020;8:570. doi: 10.1016/S2213-8587(20)30183-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 159.Hastie C.E., Mackay D.F., Ho F., et al. Vitamin D concentrations and COVID-19 infection in UK biobank. Diabetes Metab Syndr. 2020;14:561–565. doi: 10.1016/j.dsx.2020.04.050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 160.Laird E., Rhodes J., Kenny R.A. Vitamin D and inflammation: potential implications for severity of Covid-19. Ir Med J. 2020;113:81. [PubMed] [Google Scholar]
- 161.Rubin R. Sorting out whether vitamin D deficiency raises COVID-19 risk. JAMA. 2021;325:329–330. doi: 10.1001/jama.2020.24127. [DOI] [PubMed] [Google Scholar]
- 162.Holford P., Carr A.C., Jovic T.H., et al. Vitamin C-an adjunctive therapy for respiratory infection, Sepsis and COVID-19. Nutrients. 2020;12:3760. doi: 10.3390/nu12123760. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 163.Pal A., Squitti R., Picozza M., et al. Zinc and COVID-19: basis of current clinical trials. Biol Trace Elem Res. 2020:1–11. doi: 10.1007/s12011-020-02437-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 164.te Velthuis A.J., van den Worm S.H., Sims A.C., Baric R.S., Snijder E.J., van Hemert M.J. Zn(2+) inhibits coronavirus and arterivirus RNA polymerase activity in vitro and zinc ionophores block the replication of these viruses in cell culture. PLoS Pathog. 2010;6 doi: 10.1371/journal.ppat.1001176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 165.Majeed M., Nagabhushanam K., Gowda S., Mundkur L. An exploratory study of selenium status in healthy individuals and in patients with COVID-19 in a south Indian population: the case for adequate selenium status. Nutrition. 2021;82:111053. doi: 10.1016/j.nut.2020.111053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 166.Moghaddam A., Heller R.A., Sun Q., Seelig J., Cherkezov A., Seibert L. Selenium deficiency is associated with mortality risk from COVID-19. Nutrients. 2020;12:2098. doi: 10.3390/nu12072098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 167.Reed S., Neuman H., Moscovich S., Glahn R.P., Koren O., Tako E. Chronic zinc deficiency alters chick gut microbiota composition and function. Nutrients. 2015:9768–9784. doi: 10.3390/nu7125497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 168.Yang Q., Liang Q., Balakrishnan B., Belobrajdic D.P., Feng Q.J., Zhang W. Role of dietary nutrients in the modulation of gut microbiota: a narrative review. Nutrients. 2020;12:381. doi: 10.3390/nu12020381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 169.Zackular J.P., Moore J.L., Jordan A.T., et al. Dietary zinc alters the microbiota and decreases resistance to Clostridium difficile infection [published correction appears in Nat Med. 2016;22:1502] Nat Med. 2016;22:1330–1334. doi: 10.1038/nm.4174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 170.Ferreira C., Viana S.D., Reis F. Gut microbiota dysbiosis-immune hyperresponse-inflammation triad in coronavirus disease 2019 (COVID-19): impact of pharmacological and Nutraceutical approaches. Microorganisms. 2020;8:1514. doi: 10.3390/microorganisms8101514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 171.Yamamoto E.A., Jørgensen T.N. Relationships between vitamin D, gut microbiome, and systemic autoimmunity. Front Immunol. 2020;10:3141. doi: 10.3389/fimmu.2019.03141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 172.Olaimat A.N., Aolymat I., Al-Holy M., et al. The potential application of probiotics and prebiotics for the prevention and treatment of COVID-19. NPJ Sci Food. 2020;4:17. doi: 10.1038/s41538-020-00078-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 173.Baud D., Dimopoulou Agri V., Gibson G.R., Reid G., Giannoni E. Using probiotics to flatten the curve of coronavirus disease COVID-2019 pandemic. Front Public Health. 2020;8:186. doi: 10.3389/fpubh.2020.00186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 174.Collado M.C., Engen P.A., Bandin C., Cabrera-Rubio R., Voigt R.M., Green S.J., et al. Timing of food intake impacts daily rhythms of human salivary microbiota: a randomized, crossover study. FASEB J. 2018;32:2060–2072. doi: 10.1096/fj.201700697RR. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 175.West N.P., Christophersen C.T., Pyne D.B., Cripps A.W., Conlon M.A., Topping D.L., et al. Butyrylated starch increases colonic butyrate concentration but has limited effects on immunity in healthy physically active individuals. Exerc Immunol Rev. 2013;19:102–119. [PubMed] [Google Scholar]
- 176.Liu F., Ye S., Zhu X., et al. Gastrointestinal disturbance and effect of fecal microbiota transplantation in discharged COVID-19 patients. J Med Case Rep. 2021;15:60. doi: 10.1186/s13256-020-02583-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 177.Goloviznin M., Gurevich V., Churilov L.P., Shoenfeld Y. Novelties in the field of autoimmunity–1st Saint Petersburg congress of autoimmunity, the bridge between east and west. Autoimmun Rev. 2017;16:1175–1184. doi: 10.1016/j.autrev.2017.10.001. [DOI] [PubMed] [Google Scholar]
- 178.Reed S., Neuman H., Moscovich S., Glahn R.P., Koren O., Tako E. Chronic zinc deficiency alters Chick gut microbiota composition and function. Nutrients. 2015;7:9768–9784. doi: 10.3390/nu7125497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 179.Din A.U., Mazhar M., Wasim M., et al. SARS-CoV-2 microbiome dysbiosis linked disorders and possible probiotics role. Biomed Pharmacother. 2020;133:110947. doi: 10.1016/j.biopha.2020.110947. [DOI] [PMC free article] [PubMed] [Google Scholar]