Abstract
Automatic segmentation of lung opacification from computed tomography (CT) images shows excellent potential for quickly and accurately quantifying the infection of Coronavirus disease 2019 (COVID-19) and judging the disease development and treatment response. However, some challenges still exist, including the complexity and variability features of the opacity regions, the small difference between the infected and healthy tissues, and the noise of CT images. Due to limited medical resources, it is impractical to obtain a large amount of data in a short time, which further hinders the training of deep learning models. To answer these challenges, we proposed a novel spatial- and channel-wise coarse-to-fine attention network (SCOAT-Net), inspired by the biological vision mechanism, for the segmentation of COVID-19 lung opacification from CT images. With the UNet++ as basic structure, our SCOAT-Net introduces the specially designed spatial-wise and channel-wise attention modules, which serve to collaboratively boost the attention learning of the network and extract the efficient features of the infected opacification regions at the pixel and channel levels. Experiments show that our proposed SCOAT-Net achieves better results compared to several state-of-the-art image segmentation networks and has acceptable generalization ability.
Keywords: COVID-19, Convolutional neural network, Segmentation, Lung opacification, Attention mechanism
1. Introduction
The coronavirus disease 2019 (COVID-19), which is caused by severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2), has become an ongoing pandemic [1], [2]. As of 9 September 2020, there have been 212 countries with outbreaks, a total of 27,486,960 cases diagnosed, and 894,983 deaths, and the number of infected people continues to increase [3]. Clinically, reverse transcription-polymerase chain reaction (RT-PCR) is the gold standard for diagnosing COVID-19 [4], but it also has the disadvantages of a high false-negative rate [5], [6] and the inability to provide information about the patients condition.
COVID-19 has certain typical visible imaging features, such as lung opacification caused by ground-glass opacities (GGO), consolidation, and pulmonary fibrosis, which can be observed in thoracic computed tomography (CT) images [6], [7]. Therefore, CT can be used as an essential tool for clinical diagnosis. CT can also directly reflect changes in lung inflammation during the treatment process and is a crucial indicator for evaluating the treatment effect [4]. However, in the course of treatment, the need for repeated inspections leads to a sharp increase in the workload of radiologists. In addition, the assessment of inflammation requires a comparison of the region of lesions before and after treatment. Quantitative diagnosis by radiologists is inefficient and subjective and is difficult to be widely promoted. Artificial intelligence (AI) technology may gradually come to play an important role in CT evaluation of COVID-19 by enabling the evaluation to be carried out more quickly and accurately. AI can also realize the rapid response by integrating multiple functionalities, such as diagnosis [8], [9], segmentation [10], [11], and quantitative analysis [12], [13], assisting doctors in rapid screening, differential diagnosis, disease course tracking, and efficacy evaluation to improve the ability to handle COVID-19. In this study, we focus on the segmentation of COVID-19 lung opacification from CT images.
Benefiting from the rapid development of deep learning [14], many excellent convolutional neural networks (CNNs) have been applied to medical image analysis tasks and have achieved the most advanced performance [8], [15]. CNNs can be applied in various image segmentation tasks due to their excellent expression ability and data-driven adaptive feature extraction model. However, the success of any CNN is inseparable from the accurate manual labeling of a large number of training images by medical personnel, so CNNs are not suitable for all tasks. COVID-19 lung opacification segmentation based on CT images is an arduous task that has the following problems. First, in the emergency situation of the COVID-19 outbreak, it is difficult to obtain enough data with accurate labels to train deep learning models in a short time due to limited medical resources. Second, the infection areas in a CT slice show various features such as different sizes, positions, and textures, and there is no distinct boundary, which increases the difficulty of segmentation. Third, due to the complexity of the medical images, the lung opacity area is quite similar to other lung tissues and structures, making it challenging to identify. Several works [16], [17], [18] have tried to solve these challenges from the perspectives of reducing manual depiction time, using noisy labels, and implementing semi-supervised learning, and have achieved specific results.
Our approach in this study is derived from the attention learning mechanism, which makes full use of the inherent extraordinary attention ability of CNN to make the network generate attention maps and make the attention vectors in the training process weight the spatial domain feature and channel domain feature. We will show that the spatial and channel domain features activated by the network can characterize the target area more accurately.
The attention mechanism stems from the study of biological vision mechanisms [19], particularly selective attention, a characteristic of human vision. The feature integration theory proposed by Treisman and Gelade [20] uses a spotlight to describe the spatial selectivity of attention metaphorically. This model points out that visual processing is divided into two stages. In the first stage, visual processing quickly and spontaneously performs low-level feature extraction, including orientation, brightness, and color, from the visual input in various dimensions in a parallel manner. In the second stage, visual processing will locate objects based on the features of the previous stage, generate a map of locations, and dynamically assemble the low-level features of each dimension of the activation area into high-level features. Generally speaking, essential areas attract the attention of the visual system more strongly. Wolfe et al. [21] believe that the attention mechanism uses not only the bottom-up information of the image but also top-down information of the high-level visual organization structure, and the high-level information can effectively filter out a large amount of irrelevant information.
In our attention mechanism inspired model, we first use a traditional CNN to extract local image features spontaneously. After that, we generate an attention map based on the low-level features of the previous stage to activate the spatial response of the feature, then calculate the attention vector based on the feature interdependence of the activation area to activate the channel response of the feature, and finally reorganize of the high-level features. The attention map and attention vector contain top-down information fed back to the current local features in the form of gating. It is clear that this coarse-to-fine attention process is a hybrid domain attention mode that includes spatial-wise and channel-wise attention modules.
The attention learning method proposed above is specially designed to tackle the challenges faced by the task of COVID-19 lung opacification segmentation. The lung CT slices of patients with pneumonia contain tissue structures easily confused with inflammation areas such as the trachea, blood vessels, emphysema background, and the existing CNN based methods complete segmentation mainly based on local information, leading inevitably to the overfitting of irrelevant information. In contrast, we designed the spatial-wise module to generate attention maps in feature extraction, suppressing irrelevant information, and enhancing essential information in the spatial domain. Given the large intra-class differences between opacity regions, our channel-wise module is designed to select and reorganize the spatial domain features. On the whole, we use a CNN with strong generalization ability to capture all the salient areas of lung CT images and then gradually enhance relevant and suppress irrelevant spatial and channel domain features. It is like the process of radiologists searching for the target area, i.e., first finding the approximate search range through the relevant tissue structures, and then checking one-by-one whether each salient area belongs to the target [22]. Our method is more in line with such diagnostic experience of the radiologists.
Our experimental results will show that compared with traditional CNNs, our so-called spatial- and channel-wise coarse-to-fine attention network (SCOAT-Net) recognizes the opacity area better when segmenting COVID-19 lung opacification. The contributions of this paper are threefold:
-
•
A novel coarse-to-fine attention network is proposed for segmentation of COVID-19 lung opacification from CT images, which utilizes embedded spatial-wise and channel-wise attention modules and achieves state-of-the-art performance (i.e., an average Dice similarity coefficient, or DSC, of 0.8899).
-
•
We use the attention mechanism so that the neural network can generate attention maps without external region of interest (ROI) supervision. We use these attention maps to understand the training process of the network by observing the areas that the network focuses on in different stages and increasing the interpretability of the neural network.
-
•
The generalization ability and compatibility of the proposed SCOAT-Net are validated on two external datasets, showing that the proposed model has specific data migration capability and can quantitatively assess the pulmonary involvement, a difficult task for radiologists.
2. Related works
2.1. Segmentation networks
Deep neural networks (DNNs) have shown excellent performance for many automatic image segmentation tasks. Zhao et al. [23] proposed the pyramid scene parsing network (PSPNet), which introduces global pyramid pooling into the fully convolutional network (FCN) to make the global and local information act on the prediction target together. DeeplabV3 [24] proposed the ASPP (atrous spatial pyramid pooling) module to make the segmentation model perform better on multi-scale objects. U-Net [10] was introduced by Ronneberger et al. based on the encoder-decoder structure that is widely used in medical image segmentation due to its excellent performance. It uses skip connections to connect the high-level low-resolution semantic feature map and the low-level high-resolution structural feature map of the encoder and decoder so that the network output has a better spatial resolution. Oktay et al. [25] proposed the attention gate model and applied it to the U-Net model, which improved the sensitivity and prediction accuracy of the model without increasing the calculation cost. UNet++ [26] uses a series of nested and dense skip paths to connect the encoder and decoder sub-networks based on the U-NET framework, which further reduces the semantic relationship between the encoder and decoder and achieves better performance in liver segmentation tasks.
2.2. Artificial intelligence for COVID-19 based on CT
The segmentation of lung opacification based on CT images is an integral part of COVID-19 image processing, and there are many related works on this topic. Using the lungs and pulmonary opacities manually segmented by experts as standards, Oulefki et al. [12] developed a CT image prediction model based on CNNs to monitor COVID-19 disease development, and it showed excellent potential for the quantification of lung involvement. Some studies [27], [28], [29] trained segmentation or detection models with CT and segmentation templates of abnormal lung cases, which can extract the areas related to lung diseases, making the learning process of pneumonia type classification easier in the next steps. The deep learning model relies on a large amount of data training, and it is impractical to collect a large amount of data with professional labels in a short time. Some studies [30], [31] use comparative learning as an entry point, which uses self-supervised comparative learning to obtain transformation-invariant representation features on limited-sample, effectively diagnosing COVID-19. Several research groups [16], [17], [18] attempted to solve this challenge from the perspectives of reducing manual delineation time, using noisy labels, and implementing semi-supervised learning. VB-Net [16] has a perfect effect on the segmentation of COVID-19 infection regions. The mean percentage of infection (POI) estimation error for automatic segmentation and manual segmentation on the verification set is only 0.3%. In particular, it adopts a human-in-the-loop strategy to reduce the time of manual delineation significantly. Wang et al. [17] proposed noise-robust Dice loss and applied it in COPLE-Net, which surpasses other anti-noise training methods to learn COVID-19 pneumonia lesion segmentation in noisy labels. Inf-Net [18] uses a parallel partial decoder to aggregate high-level features and generate a global map to enhance the boundary area. It also uses a semi-supervised segmentation framework to achieve excellent performance in lung infection area segmentation.
2.3. Attention mechanism
More and more attempts have been focused on the combination of deep learning and visual attention mechanisms, which can be roughly divided into two categories: exogenous-attention mechanisms and endogenous-attention mechanisms. An exogenous-attention mechanism allows the network to learn to generate an attention map during the training process by conducting ROI supervision externally so that the region activated by the network can accurately diagnose disease changes. Ouyang et al. [32] applied this mechanism to the diagnosis of COVID-19 and glaucoma respectively, and the sensitivity was greatly improved. In contrast, a endogenous-attention mechanism does not rely on exogenous ROI supervision but rather exploits the intrinsic endogenous-attention ability of CNN. Endogenous-attention consists of two parts, among which spatial-wise attention [25], [33], [34] redistributes the networks attention at the pixel level of the feature map to achieve more precise location, and channel-wise attention [35] redistributes the attention at the channel level to instruct the network in selecting practical features. In Lei et al. [36] and Fu et al. [37], spatial and channel dimension attention were combined with parallel mode to jointly guide network training, which captured rich contextual dependencies to address the segmentation task. Zhang et al. [38] proposed an attention learning method with the higher layer feature as the attention mask of the lower layer feature, which can achieve the best performance in skin lesion classification.
Recently, some studies also employed attention mechanisms to solve the segmentation of COVID-19 lesions. For example, Zhou et al. [39] and Zhao et al. [40] integrated the spatial and channel attention mechanisms into U-Net to obtain better feature representation. Unlike these studies, the proposed attention modules in our SCOAT-Net are not parallelly but serially connected. Our design is inspired by the feature integration theory [20], which suggests that the attributes of a certain object are processed in sequence, i.e., the pre-attentive and the focused attention stages. For the segmentation of COVID-19 lung opacification, the spatial pre-attention in our SCOAT-Net helps reduce significantly the irrelevant area features and hence decrease the difficulty of optimizing the channel attention for local feature extraction. Another recent model proposed in Mahmud et al. [41] not only includes spatial- and channel-level attentions but also introduces pixel-level attention to supplement the low-level features, which adds more model parameters. In contrast, to realize the integration of context features of various levels, our SCOAT-Net introduces skip connections to integrate the features of lower level with that of current level, without introducing additional parameters. These integrated features are then used to effectively calculate the interdependence between the channel-wise attention modules and adaptively recalibrate the response.
3. Method
UNet++ is an excellent image segmentation network which has achieved high-grade performance in medical imaging tasks [26]. It contains dense connections that make the contextual information of different scales closely related. However, although this complicated connection method improves the generalization ability of the model, it also causes information redundancy and weak convergence of the loss function on a small data set. Medical images have the characteristics of high complexity and noise, which cause model overfitting when the amount of training data is insufficient. The SCOAT-Net proposed in this work redesigns the connection structure of UNet++ and introduces the more biologically plausible attention learning mechanism. It extracts the spatial and channel features from coarse to fine with only a few added parameters and obtains more accurate segmentation results.
3.1. Structure of the lung opacification segmentation network
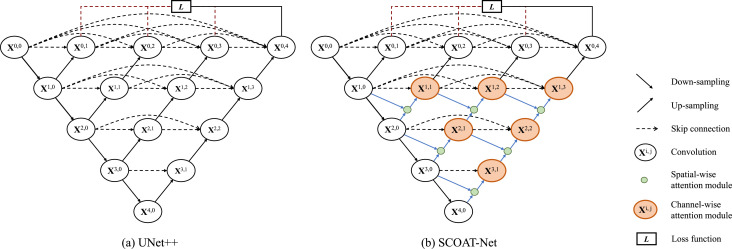
Fig. 1 compares the basic structures of UNet++ and the proposed SCOAT-Net. Inheriting the basic structure of UNet++, SCOAT-Net is composed of an encoder and a decoder connected by skip connections. The encoder extracts the information of the semantic level of the image and provides a relatively coarse location, using a max-pooling layer as a down-sampling module. The decoder reconstructs the segmentation template from the semantic information. It uses U-shaped skip connections to receive the corresponding low-level features of the encoder and calculate the final segmentation result. The upsampling module of the decoder uses the bilinear interpolation layer instead of the deconvolution layer to improve the resolution of the feature map. This method dramatically reduces the number of parameters as well as the calculation cost, and it has good performance on small-scale datasets.
Fig. 1.
Comparison of UNet++ (a) and the proposed SCOAT-Net (b). The main difference in the network structure between the two models is that our SCOAT-Net introduces the new spatial-wise attention module (the light-green nodes in (b)) and extends some convolution units to the channel-wise module (the light-orange nodes in (b)). (For interpretation of the references to colour in this figure legend, the reader is referred to the web version of this article.)
We reconstruct the connection at the top of the network (except for the bottom layer ) and introduce the attention module. This causes the calculation of the attention mechanism to act on the high-level semantic information and keep the bottom layer of the detailed image information as much as possible, resulting in fine, high-resolution segmentation. The proposed attention module consists of two parts: the spatial-wise attention module and the channel-wise attention module.
We use context feature maps with different resolutions as information of different dimensions for the spatial-wise attention module, as shown in the green circle of Fig. 1, which can combine all the multi-dimensional feature maps extracted by all the filters to calculate the attention map of the image and adjust the target area of the network adaptively. The output of the spatial-wise attention module is contacted with the feature maps of the same layer to enter the channel-wise attention module, as shown in the orange circle. The channel-wise attention module calculates the interdependence between the channels and adaptively recalibrates the information response of the channel. Additionally, in each convolution module, we use the residual block to train our network.
3.2. Spatial-wise attention
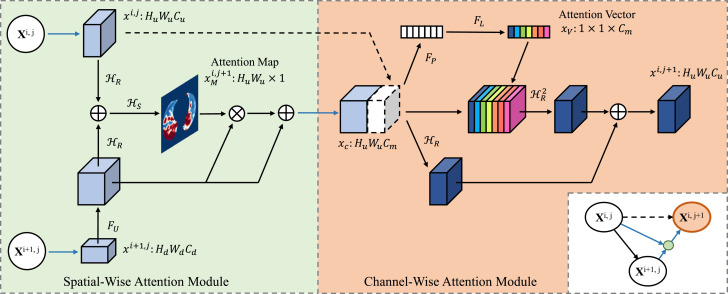
The proposed spatial-wise attention module emphasizes attention at the pixel level, making the network pay attention to the key formation and ignore irrelevant information. Normally, in a CNN, the features extracted by the network change from simple low-level features to complex high-level features with the deepening of the convolutional layers. When calculating the attention map, we can not only use the information of single-layer features but also combine the upper and lower features of different resolutions. The final output of this module is expressed as , which is given by (1) and (2):
| (1) |
| (2) |
where the function stands for the convolution of size followed by a batch normalization and a ReLU, used for feature integration. denotes the convolution of size followed by a batch normalization and a sigmoid activation function, used for feature integration and generation of the attention maps. is the up-sampling operation with a bilinear interpolation function. The input of this module is composed of the upper layer feature and the lower layer feature , where represents the output of each convolution module . is the attention map generated by this module, which uses the saliency information in the spatial position to weigh the input features to complete the redistribution of the feature attention at the pixel level. The attention map generated by the sigmoid function is normalized between 0 and 1, and the output response will be weakened after point multiplication with the current feature map. Nested structure uses of this method will lead to over-fitting or the degradation of model performance caused by the gradient’s disappearance. To improve this phenomenon, inspired by the ResNet, we add the original features after weighting them by , as shown in (2). The final output is sent to the next channel-wise attention module, as shown in Fig. 2 .
Fig. 2.
The detailed structures of the proposed spatial-wise attention module and channel-wise attention module.
3.3. Channel-wise attention
The input of the proposed channel-wise attention module is obtained by concatenating the spatial-wise attention module’s output with the feature map of the same layer, as in (3):
| (3) |
where represents concatenation. is the channel-wise statistical information calculated by through a global average pooling layer, as in (4), which can reflect the response degree on each feature map.
| (4) |
We want the module to adaptively learn the feature channels that require more attention, and we also want it to learn the interdependence between channels. Inspired by the SENet [35], we pass through two fully connected (FC) layers with parameters and to obtain the attention vector of the channel, as in (5):
| (5) |
where refers to the ReLU activation function, and refers to the sigmoid activation function. A structure containing two fully connected layers, which reduces the complexity and improves the generalization ability of the model, is adopted here. The fully connected layer of parameter reduces the feature channels’ dimension with reduction ratio ( in this experiment). In contrast, the fully connected layer of parameter recombines the feature channels to increase its dimension to . The attention vector finally weights the input feature map , and after the convolution operation completes the feature extraction, it is added to itself to obtain the final output , as in (6):
| (6) |
where represents the two-layer convolution for feature extraction.
3.4. Loss function
By combining binary cross-entropy (BCE) loss and Dice coefficient loss [42], we use a hybrid loss function for segmentation as follows:
| (7) |
where denotes the ground truths, denotes the predicted probabilities, indicates the batch size, and corresponds to the sigmoid activation function. This hybrid loss includes pixel-level and batch-level information, which helps the network parameters to be better optimized.
3.5. Evaluation metrics
To evaluate the performance of lung opacification segmentation, we measure the Dice similarity coefficient (DSC), sensitivity (SEN), positive predicted value (PPV), volume accuracy (VA), regional level precision (RLP), regional level recall (RLR), and 95% HD between the segmentation results and the ground truth in 3D space, which are defined as follows.
| (8) |
where and refer to the segmented volumes by the model and the ground truth, respectively.
In addition to the above voxel-level evaluation indicators, we also design the regional-level evaluation indicators RLP and RLR, as in (9):
| (9) |
where denotes the total number of connected regions of the model prediction result, denotes the number of real opacity regions predicted by the model, denotes the total number of real opacitiy regions, and denotes the number of real opacity regions predicted by the model. If the center of the connected area predicted by the model is in a real opacity region, then we accept that the predicted connected area is correct. We calculate the center of the connected area as:
| (10) |
where represents the point set of a single connected area of the prediction result, and represents the point set of its edge.
We use 95% HD (hausdorff distance) to measure the boundary accuracy of the segmentation results. HD is calculated as follows [17]:
| (11) |
| (12) |
where and represent the surface point set of the segmentation result and ground truth VOIs, respectively. For 95% HD, the 95th percentile in (11) is taken.
4. Experiment and results
4.1. Data and implementation
This study and its procedures were approved by the local ethics committees. All methods were performed in accordance with the relevant guidelines and regulations. The entire experiment followed the Helsinki Declaration. Informed consent was not required for this retrospective study (i.e., those discharged or who died). Written informed consent from the involved patients was not required. The data contains 19 lung CT scans of COVID-19 patients obtained using SOMATOM Definition AS. Volumes of interest (VOIs) of the opacity areas were manually delineated at voxel level by a radiologist with 5-year experience in chest CT interpretation using medical image processing and navigation software 3D Slicer (version 4.8.0, Brigham and Womens Hospital), and subsequently confirmed (modified or re-delineated) by another radiologist with 12-year experience in chest CT interpretation. Large vessels and bronchioles were excluded from the VOIs. Because the margin of the infected lesions was ill-defined, we delineated the VOI as precise as possible. The same delineate method has been published in [28].
Additionally, we prepared two external datasets to test the generalization ability of our model. One is an image set containing 8 lung CT scans of two patients scanned at different times using SOMATOM go.Top from Wuhan Red Cross Hospital (named as WUHAN dataset). Another is a public dataset1 containing 9 axial volumetric CT scans with the segmented templates (named as KAGGLE dataset).
The input images are single-layer CT images with the size of pixels, obtained from Dicom format files. For the high dynamic range of CT images, we used a pulmonary window with a width of 1200 and a level of -600 to normalize the input images in the range [0, 1]. In addition, we used the random horizontal flip to augment the data before being sent to the network. The sketch templates of the radiologists served as the ground truth, so they were used to calculate the loss function with the final output of the network. We used the gradient descent algorithm with Adam to optimize the loss function that updates the network parameters. The learning rate was set to 0.01, which was multiplied by 0.1 after every ten epoch decays. When the iterative result converged, we adjusted the learning rate to 0.001 for training again. The learning rate decay strategy remained unchanged, and the iteration was set to 50 times. The final results of training in this warm-up [43] method will be slightly improved. All experiments were conducted on an NVIDIA RTX GPU, and the proposed SCOAT-Net2 was implemented based on a Pytorch framework. We performed five-fold cross-validation to test the results.
4.2. Results on lung opacification segmentation
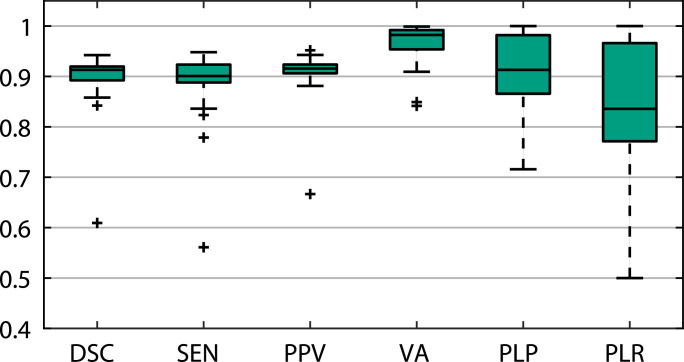
The aim of this experiment was to evaluate the performance of our proposed SCOAT-Net with different loss functions for lung opacification segmentation. We used six different loss functions, namely MSE, IOU [44], BCE, Dice [42], Focal [45], and BCE-Dice, to train the proposed network with the same strategy and hyper-parameters, and the quantitative comparison is listed in Table 1 . It is evident that BCE, Dice, and Focal had excellent segmentation performance, and their DSCs were the highest. Among them, BCE was superior to Dice and Focal in terms of DSC, VA and RLR but slightly inferior in terms of SEN and RLP. It is worth noting that Dice had a more significant performance in terms of SEN and RLR. Dice can predict the entire opacity area better, but it also causes the PPV and RLP performance to decline because it yields more false-positive predictions. The hybrid loss function combining BCE and Dice with parameter ( is empirically set to be 0.5 in the experiments) produced the best results. Except for SEN and RLP, which were slightly lower than Dice, the other indicators were the best. The box plot shown in Fig. 3 demonstrates the performance of our proposed network with the BCE-Dice loss function. In 19 cases, the model we proposed exhibited excellent performance. The medians of DSC, SEN, PPV and PLP were all higher than 0.9, and the medians of VA was higher than 0.95, even though one or two cases did not achieve excellent results.
Table 1.
Quantitative evaluation of SCOAT-Net with different loss functions for lung opacification segmentation.
| Loss functions | Results (%) |
|||||
|---|---|---|---|---|---|---|
| DSC | SEN | PPV | VA | RLP | RLR | |
| MSE | 83.22 | 71.89 | 83.86 | 87.00 | 81.68 | 81.75 |
| IOU [44] | 75.29 | 71.72 | 81.57 | 76.76 | 80.69 | 76.23 |
| BCE | 87.76 | 80.04 | 89.62 | 94.37 | 89.35 | 84.97 |
| Dice [42] | 84.61 | 88.03 | 86.43 | 87.13 | 85.26 | 83.85 |
| Focal [45] | 85.38 | 84.27 | 89.22 | 86.84 | 86.16 | 80.37 |
| BCE-Dice | 88.99 | 87.85 | 90.28 | 96.25 | 90.87 | 84.83 |
Fig. 3.
The segmentation performances of SCOAT-Net with BCE-Dice loss function.
4.3. Comparison of different networks
We compared our proposed SCOAT-Net with other popular segmentation algorithms for lung opacification segmentation. The BCE-Dice loss function was used to train these networks. The quantitative evaluation of these networks was calculated by cross-validation, as shown in Table 2 . PSPNet had excellent PPV and RLP but the lowest SEN. Although most of the predicted regions were correct, the voxel prediction could not cover all the opacity regions. ESPNetv2 had good PPV and RLR, but RLP was extremely low, which shows that the light-weight models could not achieve excellent region-level segmentation results on complex medical image segmentation tasks. DeepLabV3+ achieved an excellent result in Table 2, which perhaps results from the good adaptability of its atrous spatial pyramid pooling module designed for semantic segmentation. U-Net, which has an excellent performance in many medical image segmentation tasks, achieved general results in this work. Compared with U-Net, which has a more complex structure and more connections, UNet++ had slightly improved RLR performance, but it had a significant drop in other indexes. This indicates that its dense connection improved the models generality but did not achieve excellent results on the relatively small dataset used in this work.
Table 2.
Quantitative evaluation of different networks for lung opacification segmentation. The BCE-Dice loss was used for training.
| Methods | DSC (%) | SEN (%) | PPV (%) | VA (%) | RLP (%) | RLR (%) | 95% HD(mm) |
|---|---|---|---|---|---|---|---|
| PSPNet [23] | 80.86 | 75.67 | 88.87 | 84.42 | 89.12 | 76.24 | 59.93 |
| ESPNetv2 [46] | 83.19 | 79.77 | 88.61 | 89.03 | 67.84 | 78.31 | 63.96 |
| DenseASPP [47] | 86.87 | 85.76 | 88.98 | 94.83 | 88.62 | 78.71 | 51.96 |
| DeepLabV3+ [24] | 85.26 | 83.97 | 88.33 | 93.75 | 89.16 | 78.57 | 53.61 |
| U-Net [10] | 83.61 | 82.96 | 85.57 | 92.57 | 86.18 | 76.48 | 73.50 |
| COPLE-Net [17] | 83.70 | 84.27 | 83.42 | 93.45 | 77.46 | 74.60 | 59.21 |
| CE-Net [48] | 85.78 | 84.46 | 87.88 | 94.70 | 82.79 | 79.45 | 55.85 |
| Attention U-Net [25] | 82.66 | 79.95 | 86.58 | 90.43 | 88.20 | 75.22 | 60.97 |
| UNet+ [26] | 81.83 | 80.29 | 84.03 | 91.87 | 80.30 | 76.72 | 74.32 |
| Proposed | 88.99 | 87.85 | 90.28 | 96.25 | 90.87 | 84.83 | 29.16 |
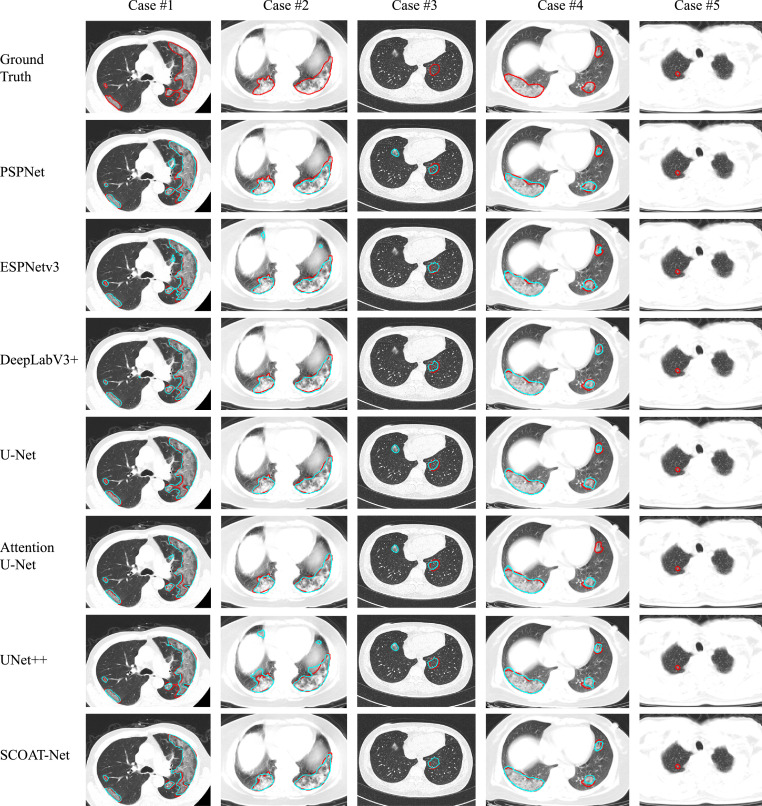
Our proposed SCOAT-Net achieved the best performance among the compared networks. In particular, our model identified and segmented the pulmonary opacities more effectively by using spatial and channel-wise attention modules. Fig. 4 shows a visual comparison of the results of each network. In the case #1 to the case #4, SCOAT-Net had the best segmentation performance, not only effectively hitting the target opacity region but also producing the least difference between the segmentation area and the ground truth. However, SCOAT-Net also returned some unsatisfactory segmentation results, as shown in the case #5 of Fig. 4. All the models, including our model, failed to predict this tiny opacity region.
Fig. 4.
Visual comparison of segmentation performance of different models trained with BCE-Dice loss function. The red curves represent the ground truth, and the cyan curves represent the results of different models. (For interpretation of the references to colour in this figure legend, the reader is referred to the web version of this article.)
4.4. Effectiveness of the attention module
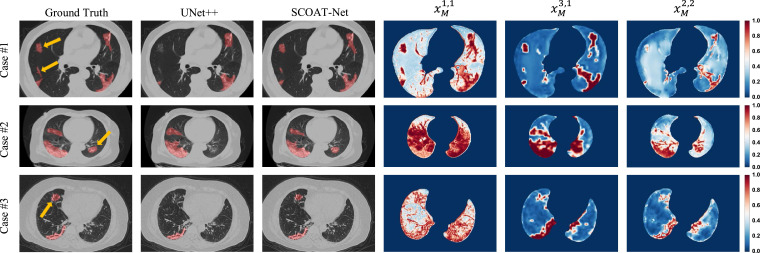
In this experiment, we verified the performance of the attention module on the lung opacification segmentation task. Our SCOAT-Net uses a total of six spatial-wise attention modules, as shown in the green circle in Fig. 1. These modules can adaptively generate attention maps with the focused area information of the network. The early stage of our network is defined as the position that closes to the input and passes fewer convolution layers. The later stage is defined as the position that closes to the output and passes more convolution layers. We selected three different stages of attention maps for display, and the order from the early stage to the late stage is , , and , as shown in Fig. 5 . For better display, we only show the lung area. We can see that our SCOAT-Net had better performance in lung opacification recognition than UNet++. From the attention map, we can see that focuses on all the salient areas of the lungs, basically covering all the structures of the lung. In constract, dramatically reduces the significant area, and the attention of the network is more concentrated on the restricted area at the semantic level. With the compensation of the lower features, further suppresses the attention area of the network in detail. As the training phase progressed, the attention regions of SCOAT-Net gradually became smaller. Additionally, for the opacity region that UNet++ did not recognize (the region indicated by the yellow arrow), SCOAT-Net adequately identified the target area, and on all the attention maps, much attention focused on the target area. The attention module we designed not only effectively weights the feature map but also further helps us understand the training process of the neural network, which improves its interpretability.
Fig. 5.
Visualization of the segmentation results of Unet++ and SCOAT-Net (the left three columns) and the attention maps of our SCOAT-Net (the right three columns) on three COVID-19 cases. The red areas on the images of the left three columns are the lung opacification segmentation of the ground truth and the results of UNet++ and our SCOAT-Net, and the yellow arrows highlight some local differences of the segmentation results. (For interpretation of the references to colour in this figure legend, the reader is referred to the web version of this article.)
Furthermore, we also introduced the attention module from other studies [25], [34] into UNet and UNet++ and compared the results with that of our SCOAT (spatial- and channel-wise coarse-to-fine attention) method, as shown in Table 3 . A1 imitates the connection structure of Attention UNet [25], and A2 uses the pyramid attention module of [34]. Compared with the U-Net, we found that the model with A1 or A2 attention module did not improve the performance. With the SCOAT module, the performance of DSC, Sen and VA is improved, but the overall performance is not significantly improved. Note that the performance of the module is not effectively reflected due to the defect of U-Net structure, which lacks the calculation module between the encoder and the decoder, resulting in the mismatch of low-level features to high-level features. Compared with U-Net, although UNet++ has relatively weaker performance sometimes, it has the potential of providing a more robust generalization performance for having a series of nested connection structures. Compared with the baseline UNet++, all the networks with the attention module obtained improved performance. SCOAT and A1 had an outstanding performance on DSC, SEN, and RLP, and SCOAT, A1 and A2 had significantly improved VA. The results show that the attention module can improve the segmentation performance while only increasing a few parameters, especially for the recognition of the target area.
Table 3.
Quantitative evaluation of different attention module for segmentation. The baseline network is UNet++.
| Methods | Params | Results (%) |
|||||
|---|---|---|---|---|---|---|---|
| DSC | SEN | PPV | VA | RLP | RLR | ||
| U-Net | 30.01M | 83.61 | 82.96 | 85.57 | 92.57 | 86.18 | 76.48 |
| U-Net&A1 | 32.08M | 82.66 | 79.95 | 86.58 | 90.43 | 88.20 | 75.22 |
| U-Net&A2 | 30.03M | 83.58 | 80.83 | 87.42 | 91.37 | 86.85 | 77.77 |
| U-Net&SCOAT | 32.97M | 85.74 | 85.16 | 86.47 | 95.72 | 85.57 | 77.36 |
| UNet+ | 35.05M | 81.83 | 80.29 | 84.03 | 91.87 | 80.30 | 76.72 |
| UNet+&A1 | 37.69M | 86.10 | 84.76 | 87.69 | 95.78 | 88.97 | 79.66 |
| UNet+&A2 | 35.09M | 82.64 | 81.89 | 83.67 | 93.37 | 80.47 | 77.29 |
| UNet+&SCOAT | 39.15M | 88.99 | 87.85 | 90.28 | 96.25 | 90.87 | 84.83 |
4.5. Validation on external datasets
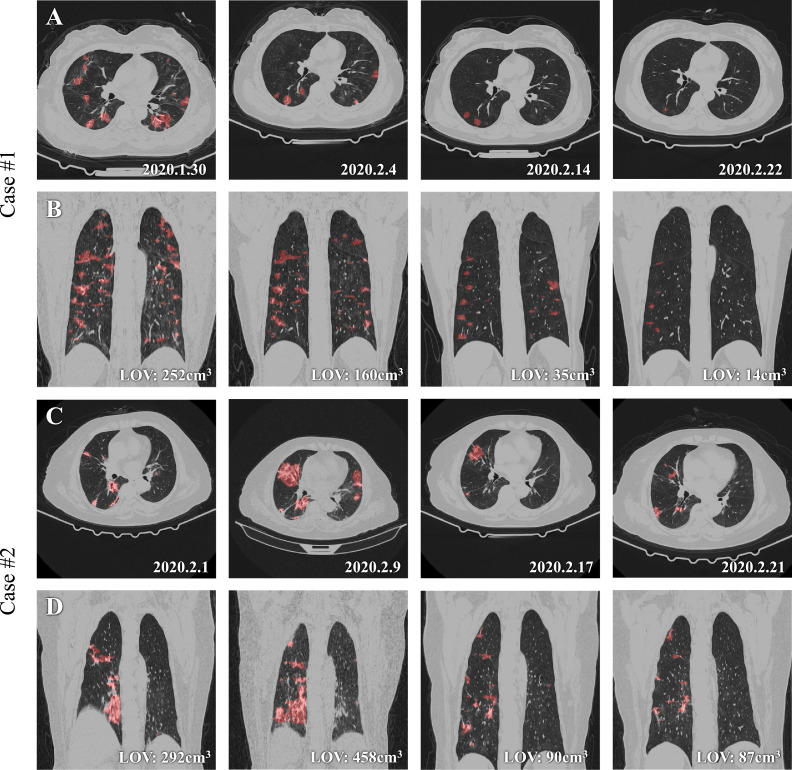
First, we used an external dataset of another center, i.e., the WUHAN dataset introduced above, to test the robustness and compatibility of the proposed SCOAT-Net. The scans in this dataset are different from the scans used for training. Fig. 6 presents the lung CT scans of two cases under treatment. COVID-19 is clinically divided into four stages [49]: early stage, progressive stage, peak stage, and absorption stage. The clinical report of the first case shows that it was in the absorption stage at all four time points. From the result of our model, we can see that on both the axial unenhanced and coronal reconstruction CT images, the opacity regions were significantly reduced, which was further verified by the lung opacification volumes (LOVs) displayed on the lower-right corners of the coronal images. The clinical report of the second case shows that the patient was in the early stage at the first time point, the progressive stage at the second time point, and the absorption stage at the third and fourth time points. Our calculated LOV was highest at the second time point, and there was a significant decrease in the third time point, which matched the diagnosis report of the patient.
Fig. 6.
Qualitative evaluation of the results of SCOAT-Net on two cases from other type of CT scan. A and B show the evolution of one COVID-19 case during the 24-day treatment period. C and D show the evolution of another case during the 21-day treatment period. A and C are axial unenhanced chest CT images at four time points (dates are annotated in the lower-right corner of each panel); B and D are the coronal reconstructions at the same time points. The segmentation of pulmonary opacities derived from SCOAT-Net is displayed in red, and the volumetric assessment of our results (i.e., lung opacification volume (LOV)) is annotated in the lower-right corners of the images of B and C. (For interpretation of the references to colour in this figure legend, the reader is referred to the web version of this article.)
Furthermore, we evaluated our model on the public KAGGLE dataset mentioned earlier, which includes 9 axial volumetric CT scans and is segmented the infected areas by a radiologist. In the experiment, we directly verified the model trained on our own data set, and the results on the 9 cases are shown in Table 4 . Overall, our model achieves the highest performance in terms of average DSCs compared with other models and gives better predictions for the most cases, which further indicates the better generalization ability of our model. Note that for case #8, because it is a tiny lesion area that is very difficult to predict, the results of all the models listed here are unsatisfactory. Although our model gave the best prediction for this case, it is far from complete segmentation.
Table 4.
Validation of different networks for lung infection segmentation on the KAGGLE dataset.
| Methods | DSC (%) |
|||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Case #1 | Case #2 | Case #3 | Case #4 | Case #5 | Case #6 | Case #7 | Case #8 | Case #9 | Average | |
| PSPNet | 68.27 | 67.41 | 78.14 | 64.06 | 78.61 | 48.92 | 48.52 | 0.00 | 76.38 | 58.92 |
| ESPNetv2 | 69.27 | 72.93 | 75.33 | 55.16 | 70.34 | 54.98 | 62.95 | 8.07 | 68.64 | 59.74 |
| DenseASPP | 62.82 | 67.98 | 74.16 | 68.26 | 62.95 | 37.46 | 58.04 | 14.33 | 59.30 | 56.15 |
| DeepLabV3+ | 66.73 | 70.10 | 73.14 | 63.80 | 61.11 | 38.13 | 60.58 | 14.01 | 63.07 | 56.74 |
| U-Net | 65.65 | 78.36 | 54.95 | 68.04 | 76.30 | 62.37 | 54.55 | 11.47 | 78.50 | 61.13 |
| COPLE-Net | 59.72 | 43.36 | 75.90 | 54.48 | 59.92 | 14.71 | 54.30 | 6.62 | 49.18 | 46.47 |
| CE-Net | 68.20 | 79.73 | 72.69 | 58.37 | 78.76 | 52.37 | 62.04 | 0.69 | 72.47 | 60.59 |
| Attention U-Net | 66.95 | 80.39 | 72.67 | 70.36 | 79.44 | 62.28 | 62.83 | 8.89 | 77.97 | 64.64 |
| UNet+ | 67.39 | 76.52 | 73.44 | 63.95 | 78.57 | 65.26 | 60.98 | 8.84 | 70.67 | 62.85 |
| Proposed | 68.74 | 79.12 | 79.98 | 70.88 | 77.63 | 57.91 | 64.89 | 27.72 | 80.43 | 67.48 |
In summary, our proposed SCOAT-Net was validated on two different external datasets, proving that it has the ability to provide an objective assessment of pulmonary involvement and therapy response in COVID-19.
4.6. Attempt at fine-grained segmentation of lung opacification
This study aims to establish a model for segmenting the lung opacification of COVID-19, and the opacity areas has visible imaging manifestation caused by GGO, consolidation, and pulmonary fibrosis. Fine-grained segmentation of the opacity areas will of course provide further help to the clinic. In this experiment, the dataset in Zhang et al. [50] was adopted, containing 750 slices with GGO and consolidation segmentation templates from 150 CT scans. We used 100 samples for training and 50 samples for testing. Compared with the results of KISEG as well as other two methods reported in Liu et al. [51], our model achieved acceptable GGO and consolidation segmentation results, as shown in Table 5 . Note that we selected KISEG for comparison because this method is a state-of-theart specifically designed for finegrained segmentation of lung infection.
Table 5.
Quantitative evaluation of different networks for GGO and consolidation segmentation.
Table 5 indicates that compared with others, the proposed method achieves a certain degree of performance on fine-grained opacity area segmentation, especially the higher IOU for consolidation, but it is not ideal for GGO segmentation. On the one hand, fine-grained segmentation of lung opacification is still a challenging task due to the slight difference in imaging manifestation between GGO and consolidation. On the other hand, our SCOAT-Net does not have a specific design for this task, but for the segmentation of abnormal areas of the lungs. In the future, to obtain a better fine-grained segmentation performance we will attempt to design a customized attention module that uses differences in the shape and density of various types of lung opacification to increase the distance between classes in the feature domain.
5. Discussion and conclusion
CNNs have been widely used in various medical image segmentation tasks due to their excellent performance [10], [25], [26], [44]. Some networks have been improved from the perspective of connection structure (e.g., U-Net [10]), and others have been improved from the perspective of combining multi-scale features (e.g., PSPNet [23]). These improvements have enhanced the expression ability of the models to a certain extent. However, due to the particularity of medical image-related tasks, only a small amount of applicable data can be obtained, making it impossible to converge when training conventional DNNs, which is a common problem. In addition to augmenting the data [53], some studies show that attention mechanisms can be more effective in enhancing the generalization capacity of models.
The main difference between the proposed SCOAT-Net and the traditional segmentation network is our specially designed attention modules, which can continuously suppress irrelevant features and enhance useful features in the image space and channel domain during the training process. The better image segmentation performance than state-of-the-art CNNs, shown in Table 4, indicates that our method has great application potential in complex medical scenarios. Furthermore, we compared the influences of two types of attention modules in other models and the proposed attention modules in our network on this task. The network incorporating the attention modules has improved performance to varying degrees compared to the baseline network. It is worth mentioning that the attention modules we proposed generate a series of attention maps. We can observe the changes of the focused regions at different stages, which contributes to the better interpretability of the neural network.
Furthermore, we compared the influences of two types of attention modules in other models and the proposed attention modules in our network on this task. The network incorporating the attention modules has improved performance to varying degrees compared to the baseline network. Also, we verified the robustness and compatibility of our network on different types of CT equipments and confirmed that it has excellent data migration capability. Our network can accurately segment lung opacity regions in CT images at different time-points during the treatment. It provides a quantitative assessment of pulmonary involvement, which is a difficult task for radiologists but is essential to the clinical follow-up of patient disease development and treatment response.
Despite the superiority mentioned above, our network still has shortcomings, e.g., failure of predicting certain tiny opacity regions, as shown in case #5 of Fig. 4. This suggests that we can continue to enhance our network’s recognition of targets of different scales by using multi-scale feature fusion or cascading convolution in different receptive field sizes.
CRediT authorship contribution statement
Shixuan Zhao: Methodology, Software, Writing - original draft. Zhidan Li: Software, Validation. Yang Chen: Writing - review & editing. Wei Zhao: Data curation, Writing - review & editing. Xingzhi Xie: Data curation, Investigation. Jun Liu: Conceptualization, Supervision, Resources. Di Zhao: Conceptualization, Supervision, Writing - review & editing. Yongjie Li: Conceptualization, Project administration, Writing - review & editing.
Declaration of Competing Interest
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Acknowledgment
This work was supported by Key Area R&D Program of Guangdong province with Grant No. 2018B03033801, National Key Research and Development Project (2018ZX10723203-001-002), Key Emergency Project of Pneumonia Epidemic of novel coronavirus infection (2020SK3006), Emergency Project of Prevention and Control for COVID-19 of Central South University (160260005) and Foundation from Changsha Scientific and Technical bureau, China (kq2001001).
We would also like to thank LetPub for its linguistic assistance during the preparation of this manuscript.
Biographies
Shixuan Zhao received the B.S degree from the University of Electronic Science and Technology of China (UESTC). He is now a Ph.D. student with the MOE Key Laboratory for Neuroinformation, School of Life Science and Technology, UESTC, China. His research interests are medical image analysis and computer vision.
Zhidan Li received the B.S degree from China Medical University. He is now a master student with the MOE Key Laboratory for Neuroinformation, School of Life Science and Technology, University of Electronic Science and Technology of China, China. His research interests are medical image classification and segmentation.
Yang Chen received the B.S. degree from Harbin Medical University, the M.S. degree from Sichuan University, and the Ph.D. degree from the University of Electronic Science and Technology of China. She has worked in the Imaging department of West China Hospital of Sichuan University for more than ten years and is now a postdoctoral fellow at the West China Biomedical Big Data Center. Her research interests artificial intelligence analysis of medical images.
Wei Zhao received the Ph.D. degree in imaging and nuclear medicine from Fudan University, China. He is a radiologist of The Second Xiangya Hospital. His research interests include chest CT imaging, radiomics and deep learning.
Xingzhi Xie received the B.S. degree in clinical medicine from Central South University, China. She is a graduate student in imaging and nuclear medicine at The Second Xiangya Hospital. Her research interests include CT imaging, radiomics and deep learning.
Jun Liu is the director of the radiology department of The Second Xiangya Hospital. He is also the leader of 225 subjects in Hunan Province, a National member of the Neurology Group of the Chinese Society of Radiology, National Committee of the Neurology Group of the Radiological Branch of the Chinese Medical Association. His research interests include brain functional imaging, radiomics and deep learning.
Zhao Di received his Ph.D. degree in computational science from Louisiana Tech University. Zhao Di has been engaged in post-doctoral research at Columbia University and Ohio State University. He is undertaking a number of national, provincial and ministerial research projects. He has good research experience in ”deep learning for medical image analysis”, and has published 25 academic journal papers and academic conference papers. He published one book and one translation. He holds a number of academic positions.
Yongjie Li received his Ph.D. degree in biomedical engineering from the University of Electronic Science and Technology of China (UESTC) in 2004. He is currently a Professor with the MOE Key Laboratory for Neuroinformation, School of Life Science and Technology, UESTC, China. He has published more than 90 peer-reviewed international journals and conference papers including Neuroimage, IEEE TPAMI, IEEE TIP, IEEE TBME, ICCV, CVPR, etc. He is also an active reviewer for more than ten leading journals and conferences. His research interests include visual mechanism modeling, and the applications in image processing for computer vision and medical diagnosis.
Footnotes
References
- 1.Wu J.T., Leung K., Leung G.M. Nowcasting and forecasting the potential domestic and international spread of the 2019-nCoV outbreak originating in Wuhan, China: a modelling study. Lancet. 2020;395(10225):689–697. doi: 10.1016/S0140-6736(20)30260-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Wu Z., McGoogan J.M. Characteristics of and important lessons from the coronavirus disease 2019 (COVID-19) outbreak in China: summary of a report of 72 314 cases from the chinese center for disease control and prevention. JAMA. 2020;323(13):1239–1242. doi: 10.1001/jama.2020.2648. [DOI] [PubMed] [Google Scholar]
- 3.World-Health-Organization, Weekly operational update coronavirus disease 2019 (COVID-19), 2020, ([EB/OL]). https://www.who.int/docs/default-source/coronaviruse/weekly-updates/wou-9-september-2020-cleared-14092020.pdf?sfvrsn=68120013_2.
- 4.Zu Z.Y., Jiang, Xu P.P., Chen W., Ni Q.Q., Lu G., Zhang L.J. Coronavirus disease 2019 (COVID-19): a perspective from China. Radiology. 2020;296(2):e200490. doi: 10.1148/radiol.2020200490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Chan J.F.W., Yuan S., Kok K., To K.K.W., Chu H., Yang J., Xing F., Liu J., Yip C.C., Poon R.W.S., et al. A familial cluster of pneumonia associated with the 2019 novel coronavirus indicating person-to-person transmission: a study of a family cluster. Lancet. 2020;395(10223):514–523. doi: 10.1016/S0140-6736(20)30154-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Ai T., Yang Z., Hou H., Zhan C., Chen C., Lv W., Tao Q., Sun Z., Xia L. Correlation of chest CT and RT-PCR testing in coronavirus disease 2019 (COVID-19) in China: a report of 1014 cases. Radiology. 2020;296(2):e200642. doi: 10.1148/radiol.2020200642. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Chung M., Bernheim A., Mei X., Zhang N., Huang M., Zeng X., Cui J., Xu W., Yang Y., Fayad Z.A., et al. CT imaging features of 2019 novel coronavirus (2019-nCoV) Radiology. 2020;295(1):e200230. doi: 10.1148/radiol.2020200230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Esteva A., Kuprel B., Novoa R.A., Ko J.M., Swetter S.M., Blau H.M., Thrun S. Dermatologist-level classification of skin cancer with deep neural networks. Nature. 2017;542(7639):115–118. doi: 10.1038/nature21056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Minaee S., Kafieh R., Sonka M., Yazdani S., Jamalipour Soufi G. Deep-COVID: predicting COVID-19 from chest x-ray images using deep transfer learning. Med. Image Anal. 2020;65:101794. doi: 10.1016/j.media.2020.101794. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Ronneberger O., Fischer P., Brox T. MICCAI. 2015. U-Net: convolutional networks for biomedical image segmentation; pp. 234–241. [Google Scholar]
- 11.Wang G., Song T., Dong Q., Cui M., Huang N., Zhang S. Automatic ischemic stroke lesion segmentation from computed tomography perfusion images by image synthesis and attention-based deep neural networks. Med. Image Anal. 2020;65:101787. doi: 10.1016/j.media.2020.101787. [DOI] [PubMed] [Google Scholar]
- 12.Oulefki A., Agaian S., Trongtirakul T., Kassah Laouar A. Automatic COVID-19 lung infected region segmentation and measurement using CT-scans images. Pattern Recognit. 2020:107747. doi: 10.1016/j.patcog.2020.107747. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Kickingereder P., Isensee F., Tursunova I., Petersen J., Neuberger U., Bonekamp D., Brugnara G., Schell M., Kessler T., Foltyn M., et al. Automated quantitative tumour response assessment of MRI in neuro-oncology with artificial neural networks: a multicentre, retrospective study. Lancet Oncol. 2019;20(5):728–740. doi: 10.1016/S1470-2045(19)30098-1. [DOI] [PubMed] [Google Scholar]
- 14.Lecun Y., Bengio Y., Hinton G.E. Deep learning. Nature. 2015;521(7553):436–444. doi: 10.1038/nature14539. [DOI] [PubMed] [Google Scholar]
- 15.Xie Y., Xia Y., Zhang J., Song Y., Feng D., Fulham M.J., Cai W. Knowledge-based collaborative deep learning for benign-malignant lung nodule classification on chest CT. IEEE Trans. Med. Imaging. 2019;38(4):991–1004. doi: 10.1109/TMI.2018.2876510. [DOI] [PubMed] [Google Scholar]
- 16.Shan F., Gao Y., Wang J., Shi W., Shi N., Han M., Xue Z., Shi Y. Abnormal lung quantification in chest CT images of COVID-9 patients with deep learning and its application to severity prediction. Med Phys. 2020 doi: 10.1002/mp.14609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Wang G., Liu X., Li C., Xu Z., Ruan J., Zhu H., Meng T., Li K., Huang N., Zhang S. A noise-robust framework for automatic segmentation of COVID-19 pneumonia lesions from CT images. IEEE Trans. Med. Imaging. 2020;39(8):2653–2663. doi: 10.1109/TMI.2020.3000314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Fan D., Zhou T., Ji G., Zhou Y., Chen G., Fu H., Shen J., Shao L. Inf-Net: automatic COVID-19 lung infection segmentation from CT images. IEEE Trans. Med. Imaging. 2020;39(8):2626–2637. doi: 10.1109/TMI.2020.2996645. [DOI] [PubMed] [Google Scholar]
- 19.Itti L., Koch C. A saliency-based search mechanism for overt and covert shifts of visual attention. Vision Res. 2000;40(10):1489–1506. doi: 10.1016/S0042-6989(99)00163-7. [DOI] [PubMed] [Google Scholar]
- 20.Treisman A., Gelade G. A feature-integration theory of attention. Cogn. Psychol. 1980;12(1):97–136. doi: 10.1016/0010-0285(80)90005-5. [DOI] [PubMed] [Google Scholar]
- 21.Wolfe J.M., Võ M.L.-H., Evans K.K., Greene M.R. Visual search in scenes involves selective and nonselective pathways. Trends Cogn. Sci. 2011;15(2):77–84. doi: 10.1016/j.tics.2010.12.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Wu C.-C., Wolfe J.M. Eye movements in medical image perception: a selective review of past, present and future. Vision. 2019;3(2):32. doi: 10.3390/vision3020032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.H. Zhao, J. Shi, X. Qi, X. Wang, J. Jia, Pyramid scene parsing network, arXiv preprint arXiv:1612.01105 (2016).
- 24.Chen L.-C., Zhu Y., Papandreou G., Schroff F., Adam H. ECCV. 2018. Encoder-decoder with atrous separable convolution for semantic image segmentation; pp. 801–818. [Google Scholar]
- 25.O. Oktay, J. Schlemper, L.L. Folgoc, M.C.H. Lee, M.P. Heinrich, K. Misawa, K. Mori, S. Mcdonagh, N. Hammerla, B. Kainz, et al., Attention U-Net: learning where to look for the pancreas, arXiv preprint arXiv:1804.03999 (2018).
- 26.Zhou Z., Siddiquee M.M.R., Tajbakhsh N., Liang J. DLMIA. Springer; 2018. Unet++: a nested U-net architecture for medical image segmentation; pp. 3–11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Gao K., Su J., Jiang Z., Zeng L.-L., Feng Z., Shen H., Rong P., Xu X., Qin J., Yang Y., Wang W., Hu D. Dual-branch combination network (DCN): towards accurate diagnosis and lesion segmentation of COVID-19 using CT images. Med. Image Anal. 2021;67:101836. doi: 10.1016/j.media.2020.101836. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Wang M., Xia C., Huang L., Xu S., Qin C., Liu J., Cao Y., Yu P., Zhu T., Zhu H., Wu C., Zhang R., Chen X., Wang J., Du G., Zhang C., Wang S., Chen K., Liu Z., Xia L., Wang W. Deep learning-based triage and analysis of lesion burden for COVID-19: a retrospective study with external validation. Lancet Digit. Health. 2020;2(10):e506–e515. doi: 10.1016/S2589-7500(20)30199-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.He K., Zhao W., Xie X., Ji W., Liu M., Tang Z., Shi Y., Shi F., Gao Y., Liu J., Zhang J., Shen D. Synergistic learning of lung lobe segmentation and hierarchical multi-instance classification for automated severity assessment of COVID-19 in CT images. Pattern Recognit. 2021;113:107828. doi: 10.1016/j.patcog.2021.107828. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Chen X., Yao L., Zhou T., Dong J., Zhang Y. Momentum contrastive learning for few-shot COVID-19 diagnosis from chest CT images. Pattern Recognit. 2021;113:107826. doi: 10.1016/j.patcog.2021.107826. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Li J., Zhao G., Tao Y., Zhai P., Chen H., He H., Cai T. Multi-task contrastive learning for automatic CT and x-ray diagnosis of COVID-19. Pattern Recognit. 2021:107848. doi: 10.1016/j.patcog.2021.107848. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Ouyang X., Huo J., Xia L., Shan F., Liu J., Mo Z., Yan F., Ding Z., Yang Q., Song B., Shi F., Yuan H., Wei Y., Cao X., Gao Y., Wu D., Wang Q., Shen D. Dual-sampling attention network for diagnosis of COVID-19 from community acquired pneumonia. IEEE Trans. Med. Imaging. 2020;39(8):2595–2605. doi: 10.1109/TMI.2020.2995508. [DOI] [PubMed] [Google Scholar]
- 33.Liu L., Kurgan L., Wu F.-X., Wang J. Attention convolutional neural network for accurate segmentation and quantification of lesions in ischemic stroke disease. Med. Image Anal. 2020;65:101791. doi: 10.1016/j.media.2020.101791. [DOI] [PubMed] [Google Scholar]
- 34.Wang W., Zhao S., Shen J., Hoi S.C., Borji A. CVPR. 2019. Salient object detection with pyramid attention and salient edges; pp. 1448–1457. [Google Scholar]
- 35.Hu J., Shen L., Albanie S., Sun G., Wu E. CVPR. 2018. Squeeze-and-excitation networks; pp. 7132–7141. [Google Scholar]
- 36.Lei B., Huang S., Li H., Li R., Bian C., Chou Y.-H., Qin J., Zhou P., Gong X., Cheng J.-Z. Self-co-attention neural network for anatomy segmentation in whole breast ultrasound. Med. Image Anal. 2020;64:101753. doi: 10.1016/j.media.2020.101753. [DOI] [PubMed] [Google Scholar]
- 37.Fu J., Liu J., Tian H., Li Y., Bao Y., Fang Z., Lu H. CVPR. 2019. Dual attention network for scene segmentation; pp. 3146–3154. [Google Scholar]
- 38.Zhang J., Xie Y., Xia Y., Shen C. Attention residual learning for skin lesion classification. IEEE Trans. Med. Imaging. 2019;38(9):2092–2103. doi: 10.1109/TMI.2019.2893944. [DOI] [PubMed] [Google Scholar]
- 39.Zhou T., Canu S., Ruan S. Automatic COVID-19 CT segmentation using U-Net integrated spatial and channel attention mechanism. Int J Imaging Syst Technol. 2021;31(1):16–27. doi: 10.1002/ima.22527. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.X. Zhao, P. Zhang, F. Song, G. Fan, Y. Sun, Y. Wang, Z. Tian, L. Zhang, G. Zhang, D2a U-Net: automatic segmentation of COVID-19 lesions from CT slices with dilated convolution and dual attention mechanism, arXiv preprint arXiv:2102.05210 (2021). [DOI] [PMC free article] [PubMed]
- 41.Mahmud T., Alam M.J., Chowdhury S., Ali S.N., Rahman M.M., Fattah S.A., Saquib M. Covtanet: a hybrid tri-level attention based network for lesion segmentation, diagnosis, and severity prediction of COVID-19 chest CT scans. IEEE Trans. Ind. Inf. 2020 doi: 10.1109/TII.2020.3048391. [DOI] [PMC free article] [PubMed] [Google Scholar]; 1–1
- 42.Milletari F., Navab N., Ahmadi S.-A. IEEE Int. Conf. 3D Vision. 2016. V-Net: fully convolutional neural networks for volumetric medical image segmentation; pp. 565–571. [Google Scholar]
- 43.A. Gotmare, N.S. Keskar, C. Xiong, R. Socher, A closer look at deep learning heuristics: learning rate restarts, warmup and distillation, arXiv preprint arXiv:1810.13243 (2018).
- 44.Huang H., Lin L., Tong R., Hu H., Zhang Q., Iwamoto Y., Han X., Chen Y.-W., Wu J. ICASSP. 2020. UNet 3+: a full-scale connected UNet for medical image segmentation; pp. 1055–1059. [Google Scholar]
- 45.Lin T.-Y., Goyal P., Girshick R., He K., Dollár P. ICCV. 2017. Focal loss for dense object detection; pp. 2980–2988. [DOI] [PubMed] [Google Scholar]
- 46.Mehta S., Rastegari M., Shapiro L., Hajishirzi H. CVPR. 2019. ESPNetv2: a light-weight, power efficient, and general purpose convolutional neural network; pp. 9190–9200. [Google Scholar]
- 47.Yang M., Yu K., Zhang C., Li Z., Yang K. CVPR. 2018. DenseASPP for semantic segmentation in street scenes; pp. 3684–3692. [Google Scholar]
- 48.Gu Z., Cheng J., Fu H., Zhou K., Hao H., Zhao Y., Zhang T., Gao S., Liu J. CE-Net: context encoder network for 2D medical image segmentation. IEEE Trans. Med. Imaging. 2019;38(10):2281–2292. doi: 10.1109/TMI.2019.2903562. [DOI] [PubMed] [Google Scholar]
- 49.Li H., Liu S., Xu H., Cheng J. Guideline for medical imaging in auxiliary diagnosis of coronavirus disease 2019. Chin. J. Med. Imaging Technol. 2020;36(3):321–331. [Google Scholar]
- 50.Zhang K., Liu X., Shen J., Li Z., Sang Y., Wu X., Zha Y., Liang W., Wang C., Wang K., et al. Clinically applicable AI system for accurate diagnosis, quantitative measurements, and prognosis of COVID-19 pneumonia using computed tomography. Cell. 2020;181(6):1423–1433. doi: 10.1016/j.cell.2020.04.045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Liu X., Wang K., Wang K., Chen T., Zhang K., Wang G. MICCAI. 2020. KISEG: a three-stage segmentation framework for multi-level acceleration of chest CT scans from COVID-19 patients; pp. 25–34. [Google Scholar]
- 52.A. Paszke, A. Chaurasia, S. Kim, E. Culurciello, ENet: a deep neural network architecture for real-time semantic segmentation, arXiv preprint arXiv:1606.02147 (2016).
- 53.Zhao A., Balakrishnan G., Durand F., Guttag J.V., Dalca A.V. CVPR. 2019. Data augmentation using learned transformations for one-shot medical image segmentation; pp. 8535–8545. [Google Scholar]